Abstract
Dysregulated expression of long non-coding RNAs has been determined to be important in cancer development; however, their role in tongue squamous cell carcinoma (TSCC) progression and carcinogenesis, to the best of our knowledge, is yet to be elucidated. The present study revealed that long intergenic non-coding RNA 00152 (LINC00152) expression was significantly increased in human TSCC tissues compared with in tissues from matched controls using RT-qPCR. In TSCC cell lines, CAL-27 and SCC-9, LINC00152 was revealed to promote TSCC cell proliferation, enhance cell cycle progression and inhibit cell apoptosis. Additionally, migration and invasion of TSCC cell lines was increased in response to LINC00152 overexpression. Mechanistically, LINC00152 was determined to be localized in the cytoplasm and acted as a microRNA (miR)-193b-3p sponge, and LINC00152 knockdown or miR-193b-3p mimics both inhibited PI3K signaling pathway activation and downstream AKT phosphorylation; therefore, promoting TSCC progression in vitro. Overall, the results of the present study suggested that increased LINC00152 expression in TSCC tissues may act as a sponge of miR-193b-3p to promote cancer progression in vitro.
Keywords: long non-coding RNA, LINC00152, tongue squamous cell carcinoma, microRNA-193b-3p, PI3K
Introduction
Oral cancer is a class of malignant tumors that usually occurs in the oral cavity, and includes tongue, gingival, oropharyngeal and lip cancer. Squamous cell carcinoma is the most common histological type of oral cancer (1–4). Tongue squamous cell carcinoma (TSCC) accounts for 25–40% of all types of oral cancer. Due to its high risk of forming secondary or recurrent tumors in the surrounding areas, the 5-year survival rate of TSCC was 42% in young patients between 2001 and 2011 in Asia (5–7). Although advanced diagnostic strategies and therapies, including surgery, chemotherapy and radiotherapy, are widely used, the possibility of complete recovery is remote (8,9). Therefore, a complete recognition of the molecular mechanisms involved in the multi-step processes of TSCC carcinogenesis and progression could provide more effective therapeutic targets for TSCC treatment (10,11).
Long non-coding RNAs (lncRNAs) are defined as a series of non-coding RNAs >200 nucleotides long (12). Numerous studies have demonstrated that lncRNAs act as biological controllers to regulate a spectrum of cellular activities, including cell proliferation, migration, invasion and apoptosis (13,14). The biological functions and molecular mechanisms of lncRNAs are complex and varied. lncRNAs can act as competing endogenous RNAs (ceRNAs) to inhibit the function of microRNAs (miRNAs/miRs), are involved in chromatin remodeling and histone protein modification, and bind proteins to regulate their function (15–17). Additionally, lncRNA dysregulation has been suggested to be closely associated with several human diseases, including several types of cancer (18). For example, lncRNA metastasis-associated lung adenocarcinoma transcript 1 has been reported to facilitate cancer development by regulating the alternative splicing process of ‘metastatic signature’ genes, such as ZEB1 and ZEB2 (19). Additionally, lncRNA nuclear paraspeckle assembly transcript 1 is highly expressed in hepatocellular carcinoma tissues and closely associated with poor prognosis (20). Downregulation of some cancer-inhibiting lncRNAs and upregulation of some cancer-promoting lncRNAs have also been reported to be important factors in accelerating cancer occurrence and development (4). However, identification of these dysregulated lncRNAs and their role in cancer development is an ongoing process.
Notably, long intergenic non-coding RNA 00152 (LINC00152) has been reported to be increased in numerous types of cancer, including glioblastoma, ovarian cancer, urothelial bladder carcinoma and breast cancer (21–24). Additionally, a previous study revealed that LINC00152 expression is significantly upregulated in TSCC tissues, which is significantly associated with poor prognosis in patients, thereby suggesting the potential importance of LINC00152 in TSCC carcinogenesis and progression (25). Dysregulated LINC00152 expression in TSCC tissues also suggests that it is a potential therapeutic target. However, the biological functions and the underlying molecular mechanisms of LINC00152 in TSCC development remain unclear.
The present study focused on the expression and roles of LINC00152 in TSCC tissues and cell lines, in order to elucidate its function in the cancer biology of TSCC. LINC00152 expression was detected and analyzed in TSCC tissues and its roles in cancer progression were determined in TSCC cell lines. Additionally, the underlying molecular mechanisms of LINC00152 in promoting TSCC were analyzed in mechanistic studies. The present study suggested a potential oncogenic role of LINC00152 in TSCC development.
Materials and methods
Tissue samples
A total of 15 TSCC tissues and paired adjacent non-tumor tongue tissues (1 cm away from TSCC tissues) were obtained from patients who underwent curative surgery at the Shanghai Changzheng Hospital between July 2014 and October 2016. The cohort consisted of 10 male and 5 female patients aged between 42–67, with median age 51. All specimens were diagnosed with TSCC according to histopathological evaluation. None of the patients underwent chemotherapy or radiotherapy prior to surgery. All tissues were stored at −80°C. All patients provided written informed consent for the use of their tissues. The present study was approved by the Ethics Committee of Xinhua Hospital, Shanghai Jiao Tong University School of Medicine.
Cell culture
TSCC cell lines, SCC-9 and CAL-27, were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. SCC-9 cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences) and 10% FBS (Gibco; Thermo Fishes Scientific, Inc.), whereas CAL-27 cells were grown in DMEM (HyClone; GE Healthcare Life Sciences) and 10% FBS. Cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
Subcellular fractionation
The nuclear and cytoplasmic fractions of CAL-27 and SCC-9 TSCC cells cultured under standard conditions (105 cells at 50% confluence) were separated using the RNA Subcellular Isolation kit (Active Motif, Inc.), according to the manufacturer's protocol.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from TSCC tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For lncRNA and mRNA, 300 ng total RNA was reverse transcribed to cDNA via random primers using the PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocols. qPCR analyses were conducted with specific forward and reverse primers using SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.). The thermocycling conditions were: 95°C for 5 sec; followed by 45 cycles of 55°C for 30 sec and 72°C for 30 sec as described previously (26–28). Human GAPDH was used as an internal control. For miRNAs, 300 ng total RNA was reverse transcribed to cDNA via specific reverse primers using the PrimeScript RT Reagent kit. qPCR analyses were performed using a unified reverse primer and specific forward primers using SYBR® Premix Ex Taq™, according to the manufacturer's protocols. U6 small nuclear (sn)RNA was used as an internal control. All assays were performed in triplicate. Relative RNA expression calculated using the 2−ΔΔCq method (28). The primer sequences were as follows: LINC00152, forward, 5′-GAAAATCACGACTCAGCCCC-3′, reverse 5′-AGACCAGCCCATGACCAAAA-3′; GAPDH, forward 5′-GCACCGTCAAGGCTGAGAAC-3′, reverse 5′-GGATCTCGCTCCTGGAAGATG-3′; miR-193a-3p, forward 5′-ACACTCCAGCTGGGAACTGGCCTACAAAGT-3′, reverse 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACTGGGAC-3′; miR-193b-3p, forward 5′-ACACTCCAGCTGGGAACTGGCCCTCAAAGT-3′, reverse 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGCGGGAC-3′; miR-376c-3p, forward 5′-ACACTCCAGCTGGGAACATAGAGGAAATT-3′, reverse 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACGTGGAA-3′; unified reverse primer, 5′-TGGTGTCGTGGAGTCG-3′; and U6 snRNA, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Plasmid construction and transfection
The cDNA, reverse transcribed from total RNA, encoding LINC00152 was PCR-amplified and subcloned into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.). Small interfering RNAs (siRNAs) specifically targeting LINC00152 and the negative control (NC) were constructed by Shanghai GenePharma Co., Ltd, and the sequences were: si-linc00152-1 sense, 5′-GAAUAACUGGGAGAUGAAATT-3′ and antisense, 5′-UUUCAUCUCCCAGUUAUUCTT-3′; si-linc00152-2 sense, 5′-GGUGGUCUGCCUGUGAUAUTT-3′ and antisense, 5′-AUAUCACAGGCAGACCACCTT-3′; si-linc00152-3 sense, 5′-GUCUUAAUCCCUUGUCCUUTT-3′ and antisense 5′-AAGGACAAGGGAUUAAGACTT-3′. hsa-miR-193b-3p and NC mimics were also purchased from Shanghai GenePharma Co., Ltd. In addition, a mutation at the supposed miR-193b-3p response element of LINC00152 was introduced using a QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) according to the manufacturer's protocol as follows: The target sequence 5′-GGCCAGT-3′ was mutated to 5′-GTCAGTC-3′. A total of 1 µg plasmid was used for transfections using a jetPEI kit (Polyplus-transfection SA), according to the manufacturer's protocol. siRNA or miRNA mimics and NCs were transfected into cells using the INTERFERin® kit (Polyplus-transfection SA) at a concentration of 50 nM. Cells were harvested or collected for other assays 48–72 h post-transfection.
Luciferase assay
A total of 1×104 293T cells per-well were plated in a 96-well plate. pGL3-promoter-LINC00152-wild type (WT) or pGL3-promoter-LINC00152-mutant (MUT) (empty vector obtained from Promega Corporation) were transfected into 293T cells under the aforementioned culture conditions along with miR-193b-3p mimics or NC using a jetPEI kit. After 48 h, Renilla luciferase activity was used as an internal control to normalize relative firefly luciferase activity. A dual-luciferase reporter gene assay system (Promega Corporation) was used in this experiment.
Cell proliferation assays
Cell proliferation was detected using a Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay. A total of 1×104 SCC-9 and CAL-27 cells transfected with siRNA (si-)LINC00152/NC or pcDNA3.1-LINC00152/pcDNA3.1 were seeded into 96-well plates. Cell viability was assessed at 24, 48 and 72 h post-transfection, according to the manufacturer's protocol. Absorbance was detected at 450 nm.
Flow cytometric analysis
SCC-9 and CAL-27 cells transfected with si-LINC00152/si-NC or pcDNA3.1-LINC00152/pcDNA3.1 were collected 48 h post-transfection. A total of 5×104 cells were analyzed via flow cytometry (FACScan; BD Biosciences) after staining with the Apoptosis Detection kit (BD Biosciences), according to the manufacturer's protocol. The percentage of apoptosis was calculated based on the number of Annexin-V positive cells. Cell cycle distribution was also analyzed using flow cytometry after staining with propidium iodide using the Cycletest™ Plus DNA Reagent kit (BD Biosciences), according to the manufacturer's protocol. The number of cells in G0/G1, S or G2/M phase were counted using FlowJo software (version 7.6; FlowJo LLC).
Cell migration and invasion assays
Cell migration assays were conducted using a 24-well Transwell chamber (pore size, 8 µm; Corning, Inc.) 24 h post transfection. Cells (~2×105) were suspended in 200 µl serum-free medium and seeded into the upper chamber per well. For the invasion assay, the Transwell chamber was precoated with Matrigel solution (BD Biosciences), and ~4×105 cells were seeded into the upper chamber. Next, 500 µl medium containing 10% FBS was added to the lower chamber. After incubation for 48 h ate 37°C, the cells that remained in the upper chamber were removed and those below the membrane were fixed using formalin for 5 min and stained with 0.1% crystal violet for 5 min both at room temperature. Images of stained cells were captured using a light microscope at ×40 magnification in five randomly chosen fields, as described previously (29).
Bioinformatics analysis
Bioinformatics analysis was conducted using starBase version 3.0 (starbase.sysu.edu.cn). Interactions between LINC00152 and miRNAs were predicted.
Western blot analysis and antibodies
Total protein was isolated using RIPA lysis buffer supplemented with protease inhibitors (Beyotime Institute of Biotechnology). Protein concentration was detected using a bicinchoninic acid kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Total protein (20 µg) was separated by 10% SDS-PAGE and electrophoretically transferred to a PVDF membrane (EMD Millipore). After blocking in PBS containing 0.1% Tween-20 (Beyotime Institute of Biotechnology) and 5% non-fat dry milk for 2 h at room temperature, the membranes were incubated overnight at 4°C with primary antibodies against phosphorylated (p-)p85 (cat. no. 4228), p85 (cat. no. 4257), p-AKT (cat. no. 4060), AKT (cat. no. 4685), cleaved caspase 3 (cat. no. 9661) and GAPDH (cat. no. 5174) (all at a 1:1,000 dilution; Cell Signaling Technology, Inc.). The membranes were then incubated with DyLight™ 800-Labeled Antibody to Mouse/Rabbit IgG (H+L) (1:1,000 dilution; cat. nos. 0412 and 0416, respectively; KPL, Inc.) for 2 h at room temperature. Finally, immunoblots were visualized using ECL solution (Pierce; Thermo Fisher Scientific, Inc.), and images were captured using the FluorChem Imaging system (AlphaView software version 1.0.2; ProteinSimple; Bio-techne).
Statistical analysis
Statistical analyses were conducted using GraphPad Prism software (version 6.0; GraphPad Software, Inc.). Paired-sample or independent-sample t-tests were performed to examine differences. A one-way ANOVA was used to analyze the differences among more than two groups. All data are presented as the mean ± standard deviation of at least three independent repeats. P<0.05 was considered to indicate a statistically significant difference.
Results
LINC00152 expression is significantly upregulated in TSCC tissues
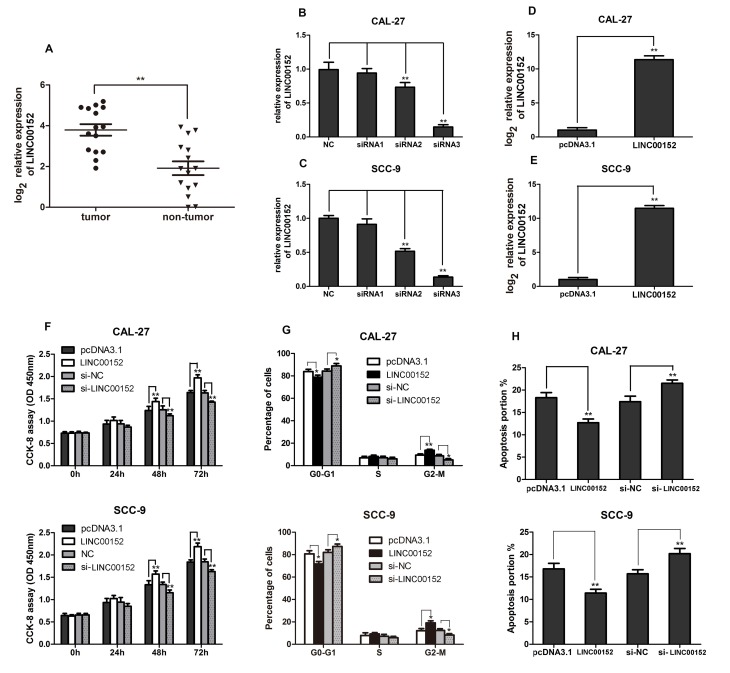
Since the present study primarily focused on the roles of LINC00152 in TSCC development, LINC00152 expression was examined in clinical human TSCC tissues. LINC00152 expression in 15 TSCC tissues and paired adjacent non-tumor tissues was detected using RT-qPCR. As shown in Fig. 1A, LINC00152 expression levels were significantly higher in TSCC tissues compared with in the paired adjacent non-tumor tissues (P<0.01). These findings suggested that LINC00152 expression was significantly upregulated in TSCC.
Figure 1.
Increased LINC00152 levels in TSCC promote cell proliferation, cell cycle progression and apoptosis inhibition. (A) RT-qPCR analysis was performed to detect LINC00152 expression in 15 TSCC and paired non-tumor tissues. Relative LINC00152 expression in (B) CAL-27 and (C) SCC-9 cell lines detected using RT-qPCR 48 h post-transfection with the indicated siRNAs (n=3). Relative LINC00152 expression in (D) CAL-27 and (E) SCC-9 cells detected using RT-qPCR 48 h post-transfection with pcDNA3.1-LINC00152/pcDNA3.1 (n=3). (F) CCK-8 assays were performed to measure cell growth 0, 24, 48 and 72 h post-transfection with si-LINC00152/si-NC or pcDNA3.1-LINC00152/pcDNA3.1 in CAL-27 and SCC-9 cells as indicated (n=6). (G) Flow cytometry was performed to analyze cell cycle progression in CAL-27 and SCC-9 cells (n=3). (H) Annexin V-FITC/propidium iodide staining was performed to measure cell apoptosis in CAL-27 and SCC-9 cells as indicated (n=3). Data are presented as the mean ± standard deviation. *P<0.05; **P<0.01. CCK-8, Cell Counting kit-8; LINC00152, long intergenic non-coding RNA 00152; NC, negative control; OD, optical density; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; TSCC, tongue squamous cell carcinoma.
LINC00152 promotes TSCC cell proliferation and cell cycle progression, and inhibits apoptosis
In order to investigate the biological function of LINC00152 in TSCC pathogenesis, the TSCC cell lines, CAL-27 and SCC-9, were transfected with si-LINC00152 or si-NC, and pcDNA3.1-LINC00152 or empty pcDNA3.1 vector. LINC00152 expression was then confirmed via RT-qPCR. As shown in Fig. 1B and C, LINC00152 expression was significantly decreased in siRNA-3-transfected samples compared with in the NC group. Furthermore, LINC00152 expression in cells transfected with pcDNA3.1-LINC00152 was significantly increased compared with the empty pcDNA3.1-transfected cells (Fig. 1D and E). Using a CCK-8 assay to analyze cell proliferation, it was shown that LINC00152 overexpression significantly promoted the proliferation of the TSCC cell lines, 48 and 72 h post transfection; however, their proliferation was significantly inhibited by LINC00152 knockdown (Fig. 1F). Cell cycle analysis revealed a reduction in the number of cells in G0/G1 phase and an increase in cell numbers in G2/M phase following LINC00152 overexpression, whereas LINC00152 knockdown increased the number of cells in G0/G1 phase and decreased the number of cells in G2/M phase (Fig. 1G). Furthermore, the percentage of apoptotic cells was decreased by LINC00152 overexpression and increased by LINC00152 knockdown in the TSCC cell lines, CAL-27 and SCC-9 (Fig. 1H). Overall, increased LINC00152 expression promoted cell proliferation and cell cycle progression, and inhibited apoptosis in TSCC cells.
LINC00152 promotes migration and invasion of TSCC cells
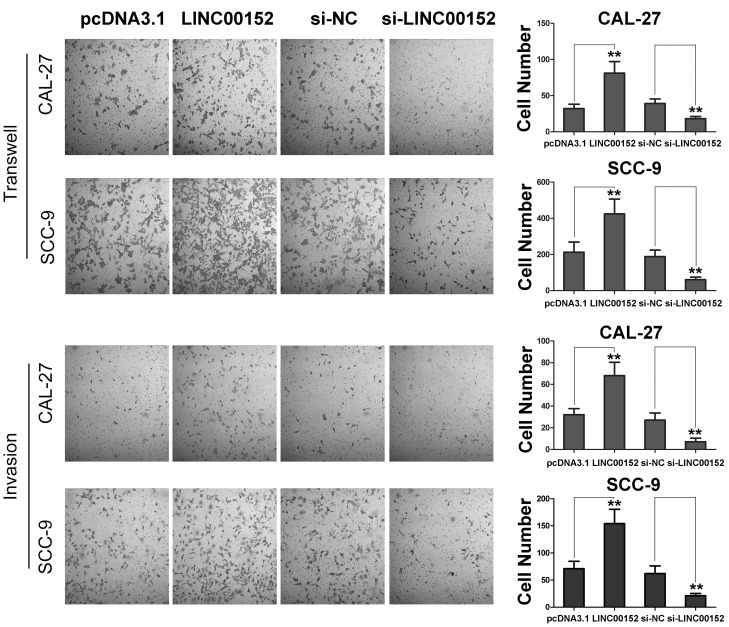
Subsequently, the present study examined whether LINC00152 regulated migration and invasion of TSCC cells. As shown in Fig. 2, LINC00152 overexpression significantly promoted the migration and invasion of the TSCC cell lines, SCC-9 and CAL-27, compared with control-transfected cells. Correspondingly, LINC00152 knockdown inhibited the migration and invasion of TSCC cells. Therefore, these results revealed that LINC00152 promoted the migration and invasion of TSCC cells and that increased LINC00152 expression promoted TSCC progression in vitro.
Figure 2.
LINC00152 promotes migration and invasion of TSCC cells. Transwell chambers with or without Matrigel were used to detect the migration and invasion of TSCC CAL-27 and SCC-9 cells as indicated (n=3). Magnification, ×40. Data are presented as the mean ± standard deviation. **P<0.01. LINC00152, long intergenic non-coding RNA 00152; NC, negative control; si, small interfering RNA; TSCC, tongue squamous cell carcinoma.
LINC00152 acts as a miR-193b-3p sponge to activate the PI3K/AKT signaling pathway in TSCC
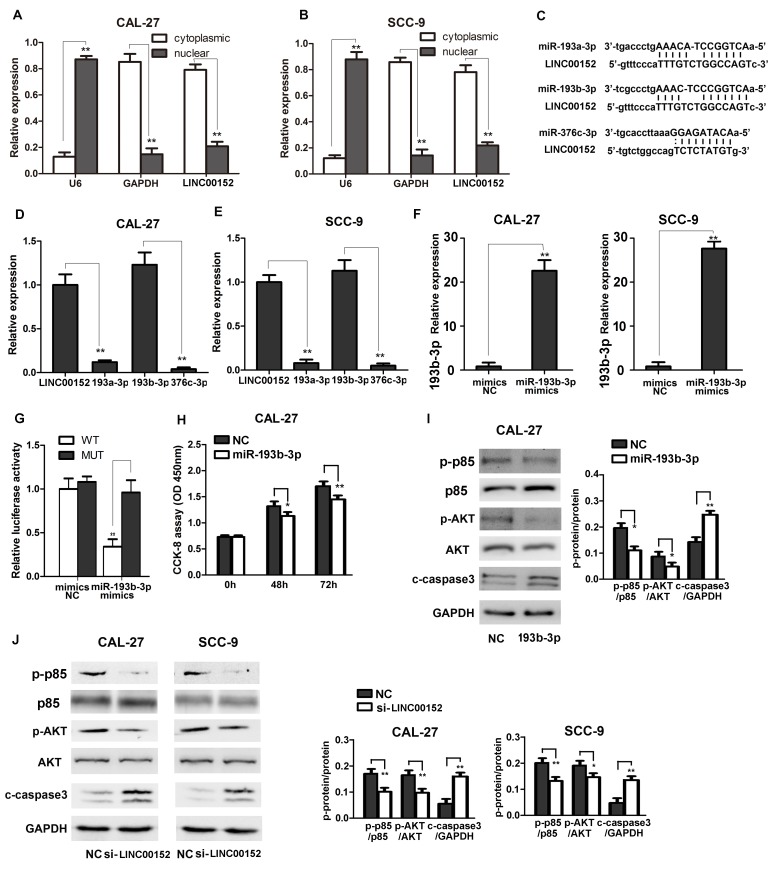
The present study further investigated the mechanisms underlying LINC00152-induced TSCC progression. As shown in Fig. 3A and B, the subcellular localization of LINC00152 was investigated. This confirmed that LINC00152 was mainly found in the cytoplasm of TSCC cells, which suggested its possible interaction with cytoplasmic miRNAs to exert its biological functions. Next, the potential interactions between miRNAs and LINC00152 were analyzed using starBase. Potential interactions between LINC00152 and miR-193a-3p, miR-193b-3p and miR-376c-3p were predicted (Fig. 3C). To validate the interaction between LINC00152 and these miRNAs, relative expression levels of miR-193a-3p, miR-193b-3p and miR-376c-3p were examined in TSCC CAL-27 and SCC-9 cells. As shown in Fig. 3D and E, miR-193b-3p exhibited the highest expression levels in TSCC cells, and the expression level was similar to that of LINC00152. Notably, miRNA absorption (sponge activity) is one of the most important biological functions of lncRNAs and, more importantly, they absorb miRNAs without inhibiting their expression levels (12). Additionally, miRNAs inhibit the expression of their target genes when the expression of a particular miRNA is relatively high (27). Based on this, it may be hypothesized that LINC00152 exerted its function by sponging miR-193b-3p. Subsequently, a luciferase reporter plasmid containing the LINC00152 sequence was constructed and it was revealed that luciferase activity was significantly inhibited in cells transfected with miR-193b-3p mimics; whereas, the luciferase reporter containing the mutated target site was not suppressed by miR-193b-3p mimics, thus demonstrating the interaction between LINC00152 and miR-193b-3p (Fig. 3F and G). Overall, it is possible that LINC00152 acts as a miR-193b-3p sponge in TSCC cells.
Figure 3.
LINC00152 acts as a miR-193b-3p sponge to activate the PI3K/AKT signaling pathway in TSCC cells. Nuclear and cytoplasmic RNAs were separated and relative LINC00152 expression was detected via RT-qPCR in (A) CAL-27 and (B) SCC-9 cells (n=3). (C) Potential LINC00152 miRNAs capable of interaction were predicted using bioinformatics analysis. Relative miR-193a-3p, miR-193b-3p, miR-376c-3p and LINC00152 expression in (D) CAL-27 and (E) SCC-9 cells was detected using RT-qPCR (n=3). (F) Relative miR-193b-3p expression transfected with miR-193b-3p or NC mimics in CAL-27 and SCC-9 cells was detected using RT-qPCR. (G) Relative luciferase activity in 293T cells transfected with LINC00152 WT or LINC00152 MUT and miR-193b-3p or NC mimics was analyzed as indicated using a dual luciferase activity assay (n=3). (H) CCK-8 assays were performed to measure cell growth at the indicated time points after transfection with miR-193b-3p/NC in CAL-27 cells (n=6). (I and J) p-p85 and AKT and c-caspase 3 were detected using western blotting as indicated. Data are presented as the mean ± standard deviation. *P<0.05; **P<0.01. c-, cleaved; CCK-8, Cell Counting kit 8; LINC00152, long intergenic non-coding RNA 00152; miR, microRNA; MUT, mutant type; NC, negative control; OD, optical density; p-, phosphorylated; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; TSCC, tongue squamous cell carcinoma; WT, wild-type.
Since miR-193b-3p has previously been suggested to inhibit cancer progression by activating the PI3K/AKT signaling pathway (30–32) and was confirmed to inhibit TSCC progression. The effect of miR-193b-3p was determined in the TSCC cell. CAL-27 cells transfected with miR-193b-3p exhibited reduced proliferation and PI3K/AKT activation (Fig. 3H and I). Additionally, transfection of miR-139b-3p decreased p85 and AKT phosphorylation (Fig. 3I), LINC00152-regulated activation of the PI3K signaling pathway in TSCC cells was further examined. As shown in Fig. 3J, LINC00152 knockdown significantly inhibited the phosphorylation and activation of p85 and downstream AKT, and activated cleaved caspase 3, thus suggesting that LINC00152 may enhance activation of the PI3K/AKT signaling pathway to promote TSCC cancer progression. Overall, these results revealed that LINC00152 acted as a miR-193b-3p sponge to enhance activation of the PI3K signaling pathway and inactivation of caspase 3.
Discussion
Numerous studies have reported that lncRNAs serve a key role in the development of human cancer (13–20). LINC00152 has been reported to be upregulated in several human tumor tissues, including gastric cancer, hepatocellular carcinoma, colon cancer and gallbladder cancer, and to be associated with poor prognosis (33–36). Functional studies have revealed that LINC00152 can promote cancer progression in a number of these cancer types (33–36). The present study revealed that increased LINC00152 expression in TSCC promoted cell growth, cell cycle progression, and migration and invasion, while reducing cell apoptosis. Therefore, LINC00152 may have considerable potential as a therapeutic target in TSCC progression.
LINC00152 expression was determined to be upregulated in TSCC tissues in the present study. However, the mechanisms underlying increased LINC00152 expression remain unknown. Previous studies have suggested that several gene regulation mechanisms involving cell signal activation, transcription factors, epigenetic factors and gene sequences are dysregulated in cancer cells (19,20). Future studies will continue to focus on the mechanisms that increase LINC00152 expression in TSCC cells, particularly the epigenetic mechanisms, thereby attempting to elucidate TSCC development in more detail.
Growing evidence has also revealed that lncRNAs can promote cancer development by acting as ceRNAs in order to inhibit tumor suppressors (33–36). The present study demonstrated that LINC00152 was mainly located in the cytoplasm and functioned as a ceRNA in TSCC cells (Fig. 3A–3G). LINC00152 was further determined to act as a sponge of miR-193b-3p, which has been reported to be a tumor suppressor (37,38). Currently, the possibility that LINC00152 may also absorb other miRNAs or interact with proteins in TSCC cells to promote cancer progression cannot be excluded. Future studies will focus on this issue as well, since LINC00152 has been reported to regulate gene expression via other mechanisms, including epigenetic modulation and post-transcriptional regulation (39,40). Additionally, LINC00152 was demonstrated to promote TSCC migration and invasion in the present study; however, the roles of miR-193b-3p in the migration and invasion of TSCC cells remain elusive. Hence, the mechanisms underlying LINC00152-mediated migration and invasion need to be further investigated. Furthermore, a previous study reported that miR-193b could enhance tumor proliferation and invasion by targeting neurofibromin 1 in head and neck squamous cell carcinoma (41), which is not in line with the tumor suppressor roles of miR-193b-3p (39,40). This suggests that one miRNA may exert different functions in different types of cancer, and the distinct roles of LINC00152 and miR-193b-3p in various cancer types may form the basis of interesting future investigations elucidating their detailed roles in cancer biology.
In conclusion, the present study demonstrated that the expression levels of the lncRNA, LINC00152, were significantly increased in TSCC tissues. Additionally, increased LINC00152 expression promoted cell growth, cell cycle progression, cell invasion and migration, and inhibited apoptosis, thus enhancing TSCC progression. Furthermore, mechanistic experiments revealed that LINC00152 acted as a miR-193b-3p sponge to promote the phosphorylation and activation of the PI3K signaling pathway and downstream AKT to contribute to TSCC development.
Acknowledgements
The authors would like to thank Ms. Wei Huang (Second Military Medical University, Shanghai, China) for her assistance with the techniques.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81970938) and the Shanghai Natural Science Foundation (grant no. 15ZR1413000).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HL designed and supervised the study. XL, BR, YC and XG performed the experiments. HL and XL analyzed the data and wrote the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Xinhua Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China). All patients provided informed consent for the use of their tissues.
Patient consent for publication
All patients provided written informed consent for the use of their tissues and publication of the research data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.D'souza S, Addepalli V. Preventive measures in oral cancer: An overview. Biomed Pharmacother. 2018;107:72–80. doi: 10.1016/j.biopha.2018.07.114. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Wu H, Ling T. Suppressive effect of microRNA-126 on oral squamous cell carcinoma in vitro. Mol Med Rep. 2014;10:125–130. doi: 10.3892/mmr.2014.2171. [DOI] [PubMed] [Google Scholar]
- 3.O'Callaghan K, Palagano E, Butini S, Campiani G, Williams DC, Zisterer DM, O'Sullivan J. Induction of apoptosis in oral squamous carcinoma cells by pyrrolo-1,5-benzoxazepines. Mol Med Rep. 2015;12:3748–3754. doi: 10.3892/mmr.2015.3832. [DOI] [PubMed] [Google Scholar]
- 4.Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. 2013;7:761–766. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- 5.Ye X, Wang X, Lu R, Zhang J, Chen X, Zhou G. CD47 as a potential prognostic marker for oral leukoplakia and oral squamous cell carcinoma. Oncol Lett. 2018;15:9075–9080. doi: 10.3892/ol.2018.8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeon JH, Kim MG, Park JY, Lee JH, Kim MJ, Myoung H, Choi SW. Analysis of the outcome of young age tongue squamous cell carcinoma. Maxillofac Plast Reconstr Surg. 2017;39:41. doi: 10.1186/s40902-017-0139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu X, Chen S, Chen W, Yang Z, Song M, Li H, Zhang H, Yao F, Su X, Liu T, Yang AK. Clinical analysis of second primary gingival squamous cell carcinoma after radiotherapy. Oral Oncol. 2018;84:20–24. doi: 10.1016/j.oraloncology.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Bachar G, Hod R, Goldstein DP, Irish JC, Gullane PJ, Brown D, Gilbert RW, Hadar T, Feinmesser R, Shpitzer T. Outcome of oral tongue squamous cell carcinoma in patients with and without known risk factors. Oral Oncol. 2011;47:45–50. doi: 10.1016/j.oraloncology.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Vered M, Dayan D, Dobriyan A, Yahalom R, Shalmon B, Barshack I, Bedrin L, Talmi YP, Taicher S. Oral tongue squamous cell carcinoma: Recurrent disease is associated with histopathologic risk score and young age. J Cancer Res Clin Oncol. 2010;136:1039–1048. doi: 10.1007/s00432-009-0749-3. [DOI] [PubMed] [Google Scholar]
- 10.Farquhar DR, Tanner AM, Masood MM, Patel SR, Hackman TG, Olshan AF, Mazul AL, Zevallos JP. Oral tongue carcinoma among young patients: An analysis of risk factors and survival. Oral Oncol. 2018;84:7–11. doi: 10.1016/j.oraloncology.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Sgaramella N, Gu X, Boldrup L, Coates PJ, Fahraeus R, Califano L, Tartaro G, Colella G, Spaak LN, Strom A, et al. Searching for new Targets and Treatments in the battle against squamous cell carcinoma of the head and neck, with specific focus on tumours of the tongue. Curr Top Med Chem. 2018;18:214–218. doi: 10.2174/1568026618666180116121624. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Wu Z, Fu X, Han W. lncRNAs: Insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Li Z. Long non-coding RNA HOTAIR: A novel oncogene (Review) Mol Med Rep. 2015;12:5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Zhang R, Ying K. Long non-coding RNAs: Novel links in respiratory diseases (Review) Mol Med Rep. 2015;11:4025–4031. doi: 10.3892/mmr.2015.3290. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez-Dominguez JR, Hu W, Gromatzky AA, Lodish HF. Long noncoding RNAs during normal and malignant hematopoiesis. Int J Hematol. 2014;99:531–541. doi: 10.1007/s12185-014-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhan A, Mandal SS. Long noncoding RNAs: Emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932–1956. doi: 10.1002/cmdc.201300534. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Peng WX, Mo YY, Luo D. MALAT1-mediated tumorigenesis. Front Biosci (Landmark Ed) 2017;22:66–80. doi: 10.2741/4472. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017:50. doi: 10.1111/cpr.12329. doi: 10.1111/cpr.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu XL, Wang J, He W, Zhao P, Wu WQ. Down-regulation of lncRNA Linc00152 suppressed cell viability, invasion, migration, and epithelial to mesenchymal transition, and reversed chemo-resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:3074–3084. doi: 10.26355/eurrev_201805_15067. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Li D, Gao Y, Tang W, Iw L, Cao Y, Hao B. Long intergenic noncoding RNA 00152 promotes glioma cell proliferation and invasion by interacting with miR-16. Cell Physiol Biochem. 2018;46:1055–1064. doi: 10.1159/000488836. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Yin M, Huang J, Lv Z, Liang S, Miao X, Huang F, Zhao Y. Long noncoding RNA LINC00152 as a novel predictor of lymph node metastasis and survival in human cancer: A systematic review and meta-analysis. Clin Chim Acta. 2018;483:25–32. doi: 10.1016/j.cca.2018.03.034. [DOI] [PubMed] [Google Scholar]
- 24.Haque SU, Niu L, Kuhnell D, Hendershot J, Biesiada J, Niu W, Hagan MC, Kelsey KT, Casper KA, Wise-Draper TM, et al. Differential expression and prognostic value of long non-coding RNA in HPV-negative head and neck squamous cell carcinoma. Head Neck. 2018;40:1555–1564. doi: 10.1002/hed.25136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang Y, Yang L, He Y, Lian Y, Li X, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8:523–530. doi: 10.7150/jca.17510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng IO, Sun H, Qin L, Qiu S, Lee JM, et al. Hepatic RIG-I predicts survival and interferon-α therapeutic response in hepatocellular carcinoma. Cancer Cell. 2014;25:49–63. doi: 10.1016/j.ccr.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Han Y, Liu Q, Hou J, Gu Y, Zhang Y, Chen Z, Fan J, Zhou W, Qiu S, Zhang Y, et al. Tumor-induced generation of splenic erythroblast-like Ter-cells promotes tumor progression. Cell. 2018;173:634–648.e12. doi: 10.1016/j.cell.2018.02.061. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, Lu J, Peng X, Wang J, Liu X, Chen X, Jiang Y, Li X, Zhang B. Integrated analysis of microRNA regulatory network in nasopharyngeal carcinoma with deep sequencing. J Exp Clin Cancer Res. 2016;35:17. doi: 10.1186/s13046-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan ST, Qin Y, Zhou ZW, He ZX, Zhang X, Yang T, Yang YX, Wang D, Qiu JX, Zhou SF. Plumbagin induces G2/M arrest, apoptosis, and autophagy via p38 MAPK- and PI3K/Akt/mTOR-mediated pathways in human tongue squamous cell carcinoma cells. Drug Des Devel Ther. 2015;9:1601–1626. doi: 10.2147/DDDT.S76057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhayadia R, Krowiorz K, Haetscher N, Jammal R, Emmrich S, Obulkasim A, Fiedler J, Schwarzer A, Rouhi A, Heuser M, et al. Endogenous tumor suppressor microRNA-193b: Therapeutic and prognostic value in acute myeloid leukemia. J Clin Oncol. 2018;36:1007–1016. doi: 10.1200/JCO.2017.75.2204. [DOI] [PubMed] [Google Scholar]
- 33.Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, Sun B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–42824. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther. 2016;24:2064–2077. doi: 10.1038/mt.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Qin J, Su Y. miR-193b-3p possesses anti-tumor activity in ovarian carcinoma cells by targeting p21-activated kinase 3. Biomed Pharmacother. 2017;96:1275–1282. doi: 10.1016/j.biopha.2017.11.086. [DOI] [PubMed] [Google Scholar]
- 38.Mets E, Van der Meulen J, Van Peer G, Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De Moerloose B, Benoit Y, et al. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:798–806. doi: 10.1038/leu.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen QN, Chen X, Chen ZY, Nie FQ, Wei CC, Ma HW, Wan L, Yan S, Ren SN, Wang ZX. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16:17. doi: 10.1186/s12943-017-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Liu J, Bai H, Dang Y, Lv P, Wu S. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am J Cancer Res. 2017;7:312–322. [PMC free article] [PubMed] [Google Scholar]
- 41.Lenarduzzi M, Hui AB, Alajez NM, Shi W, Williams J, Yue S, O'Sullivan B, Liu FF. MicroRNA-193b enhances tumor progression via down regulation of neurofibromin 1. PLoS One. 2013;8:e53765. doi: 10.1371/journal.pone.0053765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.