Abstract
The loci encoding B and T cell Ag receptors are generally distinct in commonly studied mammals, with each receptor’s gene segments limited to intralocus, cis chromosomal rearrangements. The nurse shark (Ginglymostoma cirratum) represents the oldest vertebrate class, the cartilaginous fish, with adaptive immunity provided via Ig and TCR lineages, and is one species among a growing number of taxa employing Ig-TCRδ rearrangements that blend these distinct lineages. Analysis of the nurse shark Ig-TCRδ repertoire found that these rearrangements possess CDR3 characteristics highly similar to canonical TCRδ rearrangements. Furthermore, the Ig-TCRδ rearrangements are expressed with TCRγ, canonically found in the TCRδ heterodimer. We also quantified BCR and TCR transcripts in the thymus for BCR (IgHV-IgHC), chimeric (IgHV-TCRδC), and canonical (TCRδV-TCRδC) transcripts, finding equivalent expression levels in both thymus and spleen. We also characterized the nurse shark TCRαδ locus with a targeted bacterial artifical chromosome sequencing approach and found that the TCRδ locus houses a complex of V segments from multiple lineages. An IgH-like V segment, nestled within the nurse shark TCRδ translocus, grouped with IgHV-like rearrangements we found expressed with TCRδ (but not IgH) rearrangements in our phylogenetic analysis. This distinct lineage of TCRδ-associated IgH-like V segments was termed “TAILVs.” Our data illustrate a dynamic TCRδ repertoire employing TCRδVs, NARTCRVs, bona fide trans-rearrangements from shark IgH clusters, and a novel lineage in the TCRδ-associated Ig-like V segments.
The nurse shark, Ginglymostoma cirratum, is a representative of the oldest vertebrate class, the chondrichthyans, with an adaptive immune system based on Ig superfamily (IgSF) Ag receptor somatic gene rearrangement in lymphocytes. The Ag receptors of B and T cells have significant similarities through all jawed vertebrate lineages, including the cartilaginous fishes (1–3). Shark BCR, or Ig, and TCR genes employ RAG-mediated V(D)J recombination with segmental junctional diversified by TdT activity. In sharks, both Ig and TCR (4) genes are modified by activation-induced cytidine deaminase–catalyzed somatic hypermutation (SHM) in response to Ag and repertoire generation, respectively (5–11). Shark IgH gene loci exist in many clusters, such as 15 in nurse shark (12) but possibly >100 in other species (13). The segmental ordering of an IgH cluster is VH-D1-D2-JH followed by a single set of C region exons, which delineate each cluster as either IgM (Cμ), IgW [Cω, the ancestor of IgD (14, 15)], or IgNAR (CNAR). This deviates from the typical IgH translocon topography (V1-n-D1-n-J1-n. ….Cμ-Cδ-Cγ-Cε-Cα) found in Euteleostomi lineages (16). Although nurse shark IgH rearrangements are generally intracluster, occurring within a single VDDJ cluster, class switch recombination was discovered to alter the C region class of VDJ rearrangements in the nurse shark (17).
The nurse shark has been used as a “primitive” model of the adaptive immune system for decades (18). In this species, polygenic and polymorphic MHC (19, 20); four TCR chains, including a doubly rearranging NARTCRδ (21); and multiple IgH and IgL isotypes have been well characterized (3, 6, 22, 23). Previously, we described unusual cDNA rearrangements of IgHV to TCRδDJC and extremely rare cases of IgHV to TCRδD to TCRαJC rearrangements (24). These could be attributed to either trans-rearrangements, occurring across vast genomic distances between distinct TCR and Ig receptor loci, or rearrangements originating from an IgH variable segment or cluster nested in the TCRδ locus.
Rare (1 out of >200,000 PBLs) interchromosomal, interarm, or distal intrachromosomal trans-rearrangements occur between different Ag receptor loci in mammals, despite regulatory mechanisms in place to prevent them (25). An inversion of human chromosome 7 can bring TCRβ and γ loci, located at distinct telomeres, into proximity, facilitating trans-rearrangements (26, 27). Likewise, an inversion on human chromosome 14 juxtaposes the IgH and TCRαδ loci, enabling similar chimeric rearrangements (28). Such trans-rearrangements are associated with ataxia telangiectasia and childhood acute B lymphoblastic leukemia (29). The rates of trans-rearrangement can increase 50- to 100-fold when double-strand breaks occur during chemotherapy or radiation treatment of lymphoma and leukemia patients (reviewed in Ref. 30).
There is some logic to IgH-TCRδ rearrangements, as both Ag receptor chains have been documented to interact with nonpeptide Ags and display similar CDR3 length distributions (31). The presence of VHδ segments (V segments more similar to IgHV than TCRδV located in the TCRδ loci of a growing number of vertebrate lineages) furthers the narrative of TCRδ rearrangements tolerating an Ig V domain. In the frog Xenopus, “VHδ” genes abound in the TCRα/δ locus and are commonly found in TCRδ transcripts (32). Such VHδ segments have also been detected in the TCRαδ locus of the platypus, passeriform birds, and the coelacanth (33–35). In galliform birds, VHδ segments are encoded within a distinct locus with a separate TCRδ C region. The membrane-distal domain of shark NARTCRδ employs a V segment most similar to those of the cartilaginous fish–exclusive IgH class, IgNAR (21, 22). Formulation of a functional NARTCRδ requires two RAG-mediated VDJ recombination events. The membrane-distal V domain is formed by the rearrangement of VDJ segments of the NARTCR lineage, which exist in VDJ clusters (21). This NARTCR domain exon is then spliced into a V domain exon formed by rearrangement between a leaderless, NARTCR-supporting TCRδV, encoding an additional Cys residue, and canonical TCRδD and TCRδJ segments. The rearranged NARTCR-supporting V domain exon is then spliced to the TCRδ C region, yielding a final receptor encoded by NARTCRV-NARTCRD-NARTCRJ–supporting TCRδV-TCRδD-TCRδJ-TCRδC (21). Nonplacental mammals (marsupials and monotremes) have a TCR, TCRμ, with a quaternary structure similar to that proposed for NARTCR (36, 37): TCRμ also has two V domains, but both V domains of TCRμ are Ig-like with the membrane-proximal V encoded by a germline-fused VDJ gene. The TCRμ C region is clearly most similar to TCRδ but exists in a locus distinct from the TCRαδ locus. These Ig-TCRδ hybrid receptors corroborate earlier suggestions, based on CDR length, that γδTCR binding is structurally more akin to that of Ig receptors than MHC-restricted αβTCR (38).
In previous studies, we documented trans-rearrangements involving two IgMV and two IgWV rearrangements to TCRδDJ yielding Ig-TCRδ chimeric receptor rearrangements in the nurse shark. In the current study, we aimed to further characterize the repertoire breadth and prevalence of Ig-TCRδ receptors, in addition to elucidating the genomic organization facilitating Ig-TCRδ rearrangements with a draft of the nurse shark TCRδ locus. To this end, we tested multiple hypotheses to conclude that Ig-TCRδ rearrangements were virtually indistinguishable from their canonical TCRδ counterparts in terms of CDR3 length and diversity and expression in both primary and secondary lymphoid tissue. Additionally, some, but not all, Ig-TCRδ rearrangements originate from within the TCRδ locus in nurse sharks. This study outlines Ig-TCRδ rearrangements that substantially contribute to the shark’s TCRδ repertoire diversity, with prevalent expression levels suggesting their importance in an ancient adaptive immune system.
Materials and Methods
Animals
Nurse sharks (G. cirratum) were captured off the coast of the Florida Keys and maintained in artificial seawater at ~28°C in large indoor tanks at the Institute of Marine and Environmental Technology in Baltimore, MD, as per animal protocol (University of Maryland Institutional Animal Care and Use Committee no. 1012003, Texas A&M Institutional Animal Care and Use Committee no. 2013–0001). Animals were euthanized and bled out after an overdose of MS222. Tissues were harvested, and cells, DNA, RNA, and frozen histology blocks were prepared, as have been previously described (39–42).
PCR, cloning, and sequencing of IgHV-TCRδDJC transcripts
cDNA primed with reverse TCRδC (primer FLAJ710, Supplemental Table I) was made with SuperScript III Reverse Transcriptase (Life Technologies, Carlsbad, CA) from 5 μg of total RNA from thymus, spleen, and spiral valve (intestine) of a mature (10-y-old) female nurse shark (“Joanie”). These cDNAs were used as a template for standard PCR amplification using GoTaq DNA polymerase (Promega, Madison, WI), with forward primers for IgM (FLAJ1701) or IgW (FLAJ1699 and MFC185) variable genes and reverse primers for the TCRδ C region gene (FLAJ767). After an initial 2-min 95°C denaturation, samples were cycled 35 times through a 30-s 95°C denaturation, 30-s annealing, and 1 min at 72°C, followed by a final 7-min extension at 72°C. Annealing temperatures used were 47°C for FLAJ1701, 51°C for FLAJ1699, and 58°C for MFC185. Bands were purified with the GeneCatcher (Gel Company, San Francisco, CA) isolation system and cloned into pCRII-TOPO (Life Technologies) or pBluescript II KS(+) (Stratagene, La Jolla), using One Shot TOP10 competent cells (Life Technologies). ZR Plasmid Minipreps (Zymo Research, Irvine, CA) of clones of appropriate size were sequenced using BigDye Terminator v1.1 Cycle (Life Technologies) through the Texas A&M University DNA Technologies Core Laboratory. Questionable base calls were corrected based upon other aligned sequences if they clearly occurred within V, J, or constant regions, but the sequence was excluded if ambiguous bases were in N/P regions or the D segment of CDR3. Sequence data were managed with Bioedit (www.mbio.ncsu.edu/BioEdit/bioedit.html) or Geneious Version 9.1 (Biomatters, Auckland, New Zealand) and submitted to GenBank (https://www.ncbi.nlm.nih.gov/popset?DbFrom=nuccore&Cmd=Link&LinkName=nuccore_popset&IdsFromResult=374080000, accession numbers JF507709.1–JF507661.1). The CDR3 length was calculated using the “CDR3 length = exclusive number of amino acids from C (of V segment YxC motif) to F (of J segment FGxG motif)” ImMunoGeneTics equation (43). For example, the first clone (T0006M2J09) in Fig. 1A would have a CDR3 length of 21 aa.
FIGURE 1.
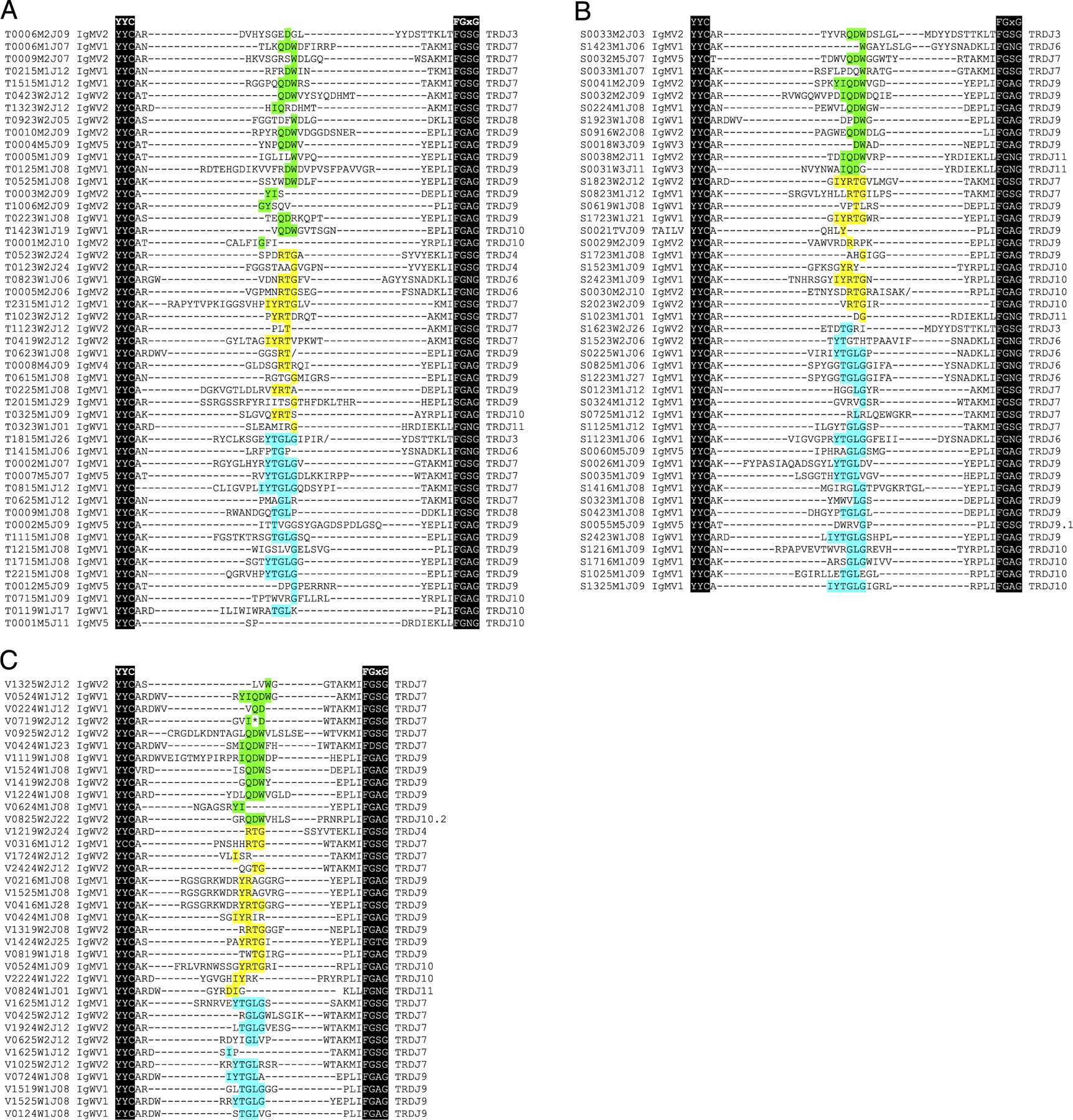
Diverse, in-frame, IgHV-TCRDJC rearrangements are expressed in thymus and periphery of nurse shark. Amino acid translation showing CDR3 regions amplified from IgHV forward and TCRδ reverse primers from (A) thymus, (B) spleen, and (C) spiral valve. Clone name is to the left of alignment, followed by the IgH V segment origin, with the TCRδJ used to the right of the alignment. Sequences are aligned to the conserved cysteine of the YxC motif of the V domain and the FGxG motif of the J. Nucleotides within CDR3 are aligned in the center, and conserved residues of the genomic D segment are highlighted by frame (first frame, cyan; second frame, green; third frame, yellow) and flanked by nontemplate and palindromic residues. Asterisks (*) denote stop codons, and slashes (/) denote frameshifts.
Bacterial artificial chromosome sequencing of the nurse shark TCRαδ locus
The G. cirratum bacterial artificial chromosome (BAC) library was probed using cloned segments of TCRαV, TCRαC, TCRδV, TCRδC, and NARTCR probes from splenic transcripts amplified with 32P, as described under high-stringency conditions (42, 44). Selected BAC clones, positive for TCRα, TCRδ, and/or NARTCR components, were isolated from 500-ml cultures using the Qiagen Large Construct Kit (Qiagen, Hilden, Germany). Purified BAC DNA was sent to the Duke Center for Genomic and Computational Biology for PacBio RSII large insert (15–20 kb) library sequencing. Assemblies were performed with the PacBio Corrected Reads pipeline, Celera Assembler version 8.3, using both raw PacBio reads, corrected in the PacBio Corrected Reads pipeline, and PacBio circular consensus sequences with minimum a read length of 500 bp were used as input files for the assembly (45, 46). Options to merge haplotypes to a single consensus were used to account for the BAC library containing both parental chromosomes. Reads were deposited into the Sequence Read Archive under Bioproject PRJNA530194. Annotation was performed using the Geneious software suite version 9.1. We employed a combination of BLAST, using a custom database of expressed nurse shark Ig and TCR sequences, and manual searches of assembled contigs for recombination signal sequences unveiled V, D, and J segments not yet in public databases.
Quantitative real-time PCR
Quantitative PCR (qPCR) was performed with 50 ng of random hexamer–primed cDNA generated with SuperScript III from thymic and spleen RNA samples of sharks ranging in age from 9 mo to 10 y. We used the SYBR Green PCR Master Mix (Bio-Rad, Hercules, CA), following the manufacturer’s recommendations. Triplicate wells were assayed in a Bio-Rad CFX96 thermocycler for 40 cycles, annealing at 55°C. For the absolute quantification of each sample, a standard curve was created using serial dilutions of certain concentrations of a single-copy gene cloned into a vector. Additional interexonic, real-time PCR primers can also be found in Supplemental Table I. The resulting quantities, given in copy number per 50 ng of cDNA, were split into three groups (canonical BCR, canonical TCR, and chimeric Ig-TCR) for statistical analyses. Significance was determined via the median-based Kruskal-Wallis test, with post hoc Dunn test to determine specific differences between the groups.
Generation of anti-IgHV polyclonal antisera
Polyclonal nurse shark IgMV1, IgWV1, and IgWV2 group (47) Abs were generated in rabbits by Cocalico Biologicals (Reamstown, PA) by immunizing them with IgHV–maltose binding protein (MBP) fusion proteins. The IgHV sequences were amplified from shark (Joanie) spleen cDNA with 35 cycles of PCR, annealing at 59°C (IgM) or 63°C (IgW). Restriction endonuclease site–engineered primers used were MFC180 and MFC181 for IgMVI, MFC182 and MFC183 for IgWVI, and MFC184 and MFC183 for IgWVII (Supplemental Table I). These products were cloned into the pMAL-c2x (New England Biolabs, Beverly, MA) expression vector using SHuffle Express competent cells (New England Biolabs). Recombinant protein was produced in bacteria and cleared supernatant passed through amylose resin columns twice. Fusion proteins were eluted with maltose and precipitated with saturated ammonium sulfate, resuspended in PBS, and dialyzed in Slide-A-Lyzer cassettes (Pierce, Rockford, IL). Sizes were verified by 12% SDS-PAGE using Coomassie and silver staining. The immune serum of recombinant protein-immunized rabbits was passed through an MBP affinity column to remove MBP-specific Abs. Affinity purification of Abs to shark IgHV in the cleared sera was performed with the same immunizing Ag immobilized in an agarose bead column (AminoLink, Pierce) and verified by SDS-PAGE and Western blotting.
Flow cytometry
Thymocytes and splenocytes (5 × 105 cells per treatment) were stained with either biotinylated mouse mAb LK14 (48) (against G. cirratum IgL) or unlabeled anti-IgHV rabbit polyclonal (against G. cirratum IgMV1, IgWV1, or IgWV2) at 1:100 in staining buffer (1% BSA in shark PBS) for 1 h at 4°C. Cells were then washed three times with staining buffer before staining with streptavidin-allophycocyanin (eBioscience, San Diego, CA) at 1:1500 and anti-rabbit Alexa Fluor 488 at 1:500 (Southern Biotech, Birmingham, AL), respectively, for 30 min at 4°C. All samples were washed and resuspended in 300 μl of staining buffer containing 0.1% sodium azide and examined by flow cytometry on a BD LSR II instrument (BD Biosciences, San Jose, CA). Fifty thousand events were collected, gated for live cells, and analyzed using the FlowJo software (Tree Star, Ashland, OR). Identical signal thresholds could be applied to all samples except the 120-mo shark, which had lower fluorescent intensity across all experiments.
Phylogenetic analyses
V segment alignments for both cartilaginous fish and vertebrate lineage trees were performed in Geneious using ClustalW. Amino acid alignments containing the entire V segment, from FR1 and to the conserved Cys of FR3, were used in the cartilaginous fish alignments. The multispecies alignments used nucleotide sequences of only the framework regions as CDR length and composition vary greatly across multiple vertebrate lineages (49, 50). The resulting nucleotide alignment was manually adjusted to fit the b strand ImMunoGeneTics protein display for V domains, as previously described (51).
Maximum likelihood trees were constructed for all alignments in MEGAX using 1000 bootstrap replicates (52). For the cartilaginous fish phylogeny, we used a Poisson correction model with bootstrap values displayed for bifurcations with >50% consensus tree support. The multispecies tree used a general time reversal model, and the substitution rate was γ distributed with invariant sites with six discrete γ categories.
Stellaris RNA FISH
Frozen tissue sections were prepared from OCT embedded thymus tissue of a 10-y-old nurse shark, as previously described (24). Three tissue sections of the thymus, cut to a thickness of 10 μm, were probed using the Stellaris (LGC Biosearch Technologies, Middlesex, U.K.) RNA FISH system. Custom probe sets were designed using the Stellaris custom probe designer for the IgWV1 segment, TCRδ C region, and TCRγ C region. The fluorophores used for each probe set were CAL Fluor Red 610 (IgWV1), Quasar 670 (TCRδC), and CAL Fluor Orange 560 (TCRγC). Nuclei were stained with DAPI (Sigma-Aldrich, St. Louis, MO). The probed slides were imaged on a Zeiss Stallion Digital Imaging Workstation, as previously described (4). Images were processed using ImageJ software version1.7 (53).
Results
Repertoire sequencing reveals additional IgHV usage
Transcripts in which an IgHV segment is rearranged to TCRδ gene segments have been previously documented in the nurse shark (24). To determine the breadth of this chimeric receptor repertoire, 5′ RACE PCR was performed using a TCRδ C region primer. Nested forward primers were designed to selectively amplify the Ig-TCRδ hybrid receptors (Supplemental Table I). The sequenced amplicons totaled 137 unique IgV-TCRδDJ rearrangements (chimeric, or trans-rearranged, clones are abbreviated Ig-TCRδ) from spiral valve (n = 38), spleen (n = 51), and thymus (n = 50) transcripts, displaying a diverse repertoire of Ig-TCRδ in both primary and secondary lymphoid tissues (Fig. 1A–C, Supplemental Fig. 1). Most of the recovered transcripts (95.6%) were in frame, indicative of selection mechanisms prohibiting nonfunctional rearrangements or mutations. We identified nine IgM/W V segments rearranged with TCRδ; all but one segment, S0021TVJ09, could be found in BCR rearrangements bearing an IgM (or IgW) C region. This sequence encoded an IgH-like V segment and was most similar (68% nucleotide identity) to the nurse shark IgH group 7 pseudogene cluster (EU312153). This genomic V segment was not found expressed in any rearrangement and was not determined to be the source of this peculiar V segment used in transcript S0021TVJ09. Accession numbers for IgH V segments found in both IgH and TCRδ rearrangements were AY609260 (IgMV1), DQ857389 (IgMV2.1), AY609247.1 (IgMV2.2), AY609259.1 (IgMV4), AY609249.1 (IgMV5), KF192877.1 (IgWV1), KF192883.1 (IgWV2), and KF192884.1 (IgWV3).
Nurse shark Ig-TCRδ and canonical TCRδ repertoires possess equivalent CDR3 metrics
We compared the Ig-TCRδ CDR3 transcript metrics to that of the canonical TCRδ repertoire to ascertain whether chimeric Ig-TCRδ rearrangements bore similar CDR3 metrics to their canonical counterparts (Fig. 2, Table I). Two aberrant transcripts with extraordinarily long CDR3 lengths (S2223W1J08 and V0924W1J20 with CDR3 lengths of 41 and 49 aa) were removed from statistical analyses. The distribution of CDR3 length for functional TCRδ (n = 55) and Ig-TCRδ (n = 135) was nearly identical and clearly distinguishable from TCRβ (n = 41) and IgH (n = 173) (Fig. 2A). Mean CDR3 length of Ig-TCRδ rearrangements was 17.44 ± 0.42 aa, statistically indistinguishable from that of canonical TCRδ (17.40 ± 0.67 aa), with each TCRδ subset encoding repertoire CDR3 content significantly longer than that of both TCRβ(11.68 ± 1.01 aa) and IgH (12.55 ± 0.25) determined via ANOVA (Fig. 2B, Table I). The ranges of CDR3 lengths of receptors free from MHC restriction (TCRγδ and Ig and especially IgHV and TCRδ) are typically much longer than that of TCRαβ (38). Indeed, the range of the Ig-TCRδ subset was comparable to that of canonical TCRδ receptors (Fig. 2C, Table I), with both TCRδ subsets encompassing a larger range than either TCRβ or IgH (Fig. 2C). Finally, we analyzed the isoelectric point (iP) of CDR3 residues and found that TCRδ (iP = 8.35) and Ig-TCRδ (iP = 8.38) both averaged slightly higher iP than either IgH (iP = 6.86) or TCRβ (iP = 5.98) (Fig. 2D). These data show that chimeric Ig-TCRδ and canonical TCRδ receptors are comparable not only in CDR3 length but possess similar charge signatures integral to epitope–paratope interactions as well. These results were unsurprising as both Ig-TCRδ and canonical TCRδ receptors draw from the same TCR D and J segment pool, contributing heavily to the CDR3 similarities outlined above (Fig. 1, Supplemental Fig. 1). Additionally, we elucidated that the IgWV1 gene segment, found expressed in Ig-TCRδ rearrangements, was found in individual cells also expressing TCRγ using RNA FISH (Supplemental Fig.2). The highly similar CDR3 metrics between Ig-TCRδ and canonical TCRδ is indicative of an IgHV segment, with divergent CDR1 and CDR2 loops, which use provides substantial expansion of potential TCRδ paratopes.
FIGURE 2.
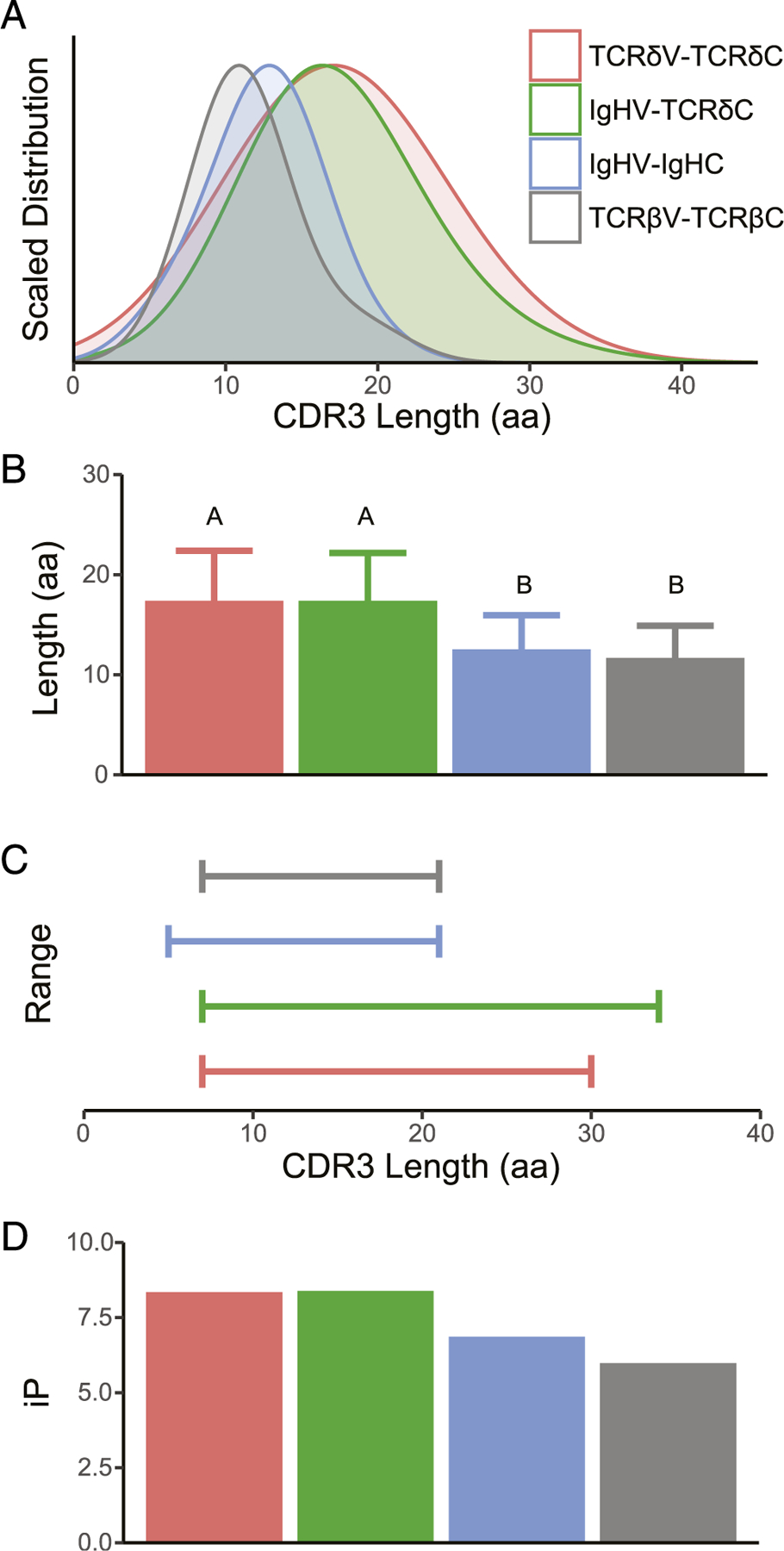
Chimeric Ig-TCRδ and canonical TCRδ encode comparable CDR3 metrics. Analysis of CDR3 for rearranged transcripts of TCRδV-TCRδC (red), IgHV-TCRδC (green), IgHV-IgHC (blue), and TCRβV-TCRβC (gray) showcases the near-identical distribution (A), average length (B), range (C), and iP (D) of chimeric Ig-TCRδ and canonical TCRδ. Significance of CDR3 length indicated by lettering (B) was determined via ANOVA with post hoc Tukey honestly significant difference test, with each TCRδ subset being significantly longer than IgH and TCR2β.
Table I.
CDR3 lengths of Ig-TCRδ chimeric receptor chains
| Total Ig-TCRδ | Thymus Ig-TCRδ | Spleen Ig-TCRδ | Spiral Valve Ig-TCRδ | Canonical TCRδa | |
|---|---|---|---|---|---|
| Median | 17 | 17 | 18 | 15 | 18 |
| Mean | 17.4 | 18.0 | 17.8 | 16.3 | 17.4 |
| Variance | 22.8 | 26.4 | 23.6 | 19.6 | 25.0 |
| Maximum | 34 | 34 | 28 | 29 | 30 |
| Minimum | 7 | 8 | 7 | 11 | 7 |
| Range | 27 | 26 | 21 | 18 | 23 |
From Criscitiello et al. (24).
Absolute quantification of TCRδ by real-time PCR shows equivalent levels for canonical and chimeric transcripts
Although the number and diversity of chimeric clones suggested that nurse shark Ig-TCRδ rearrangements were of significance for the shark immune system, we performed absolute qPCR to determine their prevalence in comparison with other TCRδ rearranged transcripts. Specific primers for each V segment (TCRδ, IgM, and IgW) were used with either a TCRδ or IgH C region primer (Supplemental Table I). Interestingly, no significant differences were found between expression levels of canonical TCRδ and chimeric Ig-TCRδ transcripts in the thymus (Fig. 3A). Although expression of BCR was decidedly lower than either TCR group in the thymus, we found no significant differences between the three groups in the thymus of young sharks aged 10 and 16 mo (Fig. 3A). Expression of transmembrane IgH message in the thymus of developing sharks has been documented, but consistent with our new quantitative data, such expression diminishes to background levels as the animals mature (6, 23). Expression of BCR in the spleen compared with thymus was markedly higher than both TCRδ groups, yet there remained no significant difference between canonical TCRδ and chimeric Ig-TCRδ expression in the periphery (Fig. 3B). Furthermore, flow cytometry performed on thymocytes, using anti-IgHV polyclonal and the LK14 mAb specific for κ L chains, provided preliminary evidence that chimeric receptors are present as a surface receptor (Supplemental Fig. 3). Although IgW has been shown to prefer an IgL other than κ (54), qPCR confirmed that expression of either IgWV segment was negligible in the thymus of these sharks (Fig. 3A). Importantly, these data show Ig-TCRδ and canonical TCRδ mRNA expression to be equivalent in both primary and secondary lymphoid tissues and suggest that they are used as a surface Ag receptor.
FIGURE 3.
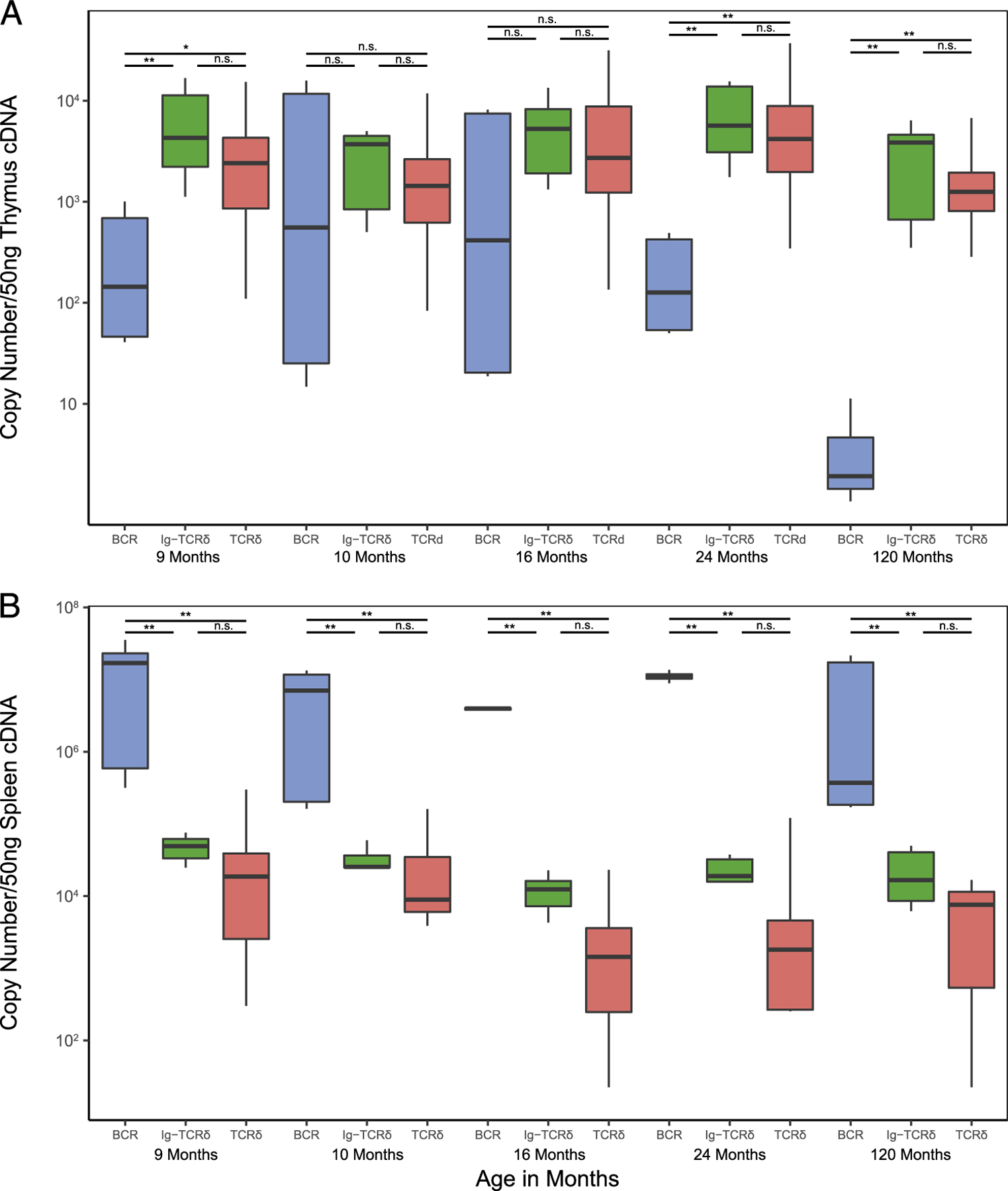
Absolute qPCR reveals chimeric Ig-TCRδ expression to be comparable to canonical TCRδ in both primary (thymus) and secondary (spleen) lymphoid tissue. Absolute transcript values (displayed on the y-axis, calculated using known concentrations of an amplicon cloned into plasmid) are compared within each shark (displayed on the x-axis indicated by age in months) for (A) thymus and (B) spleen samples. To compare, rearrangements were classified as canonical B cell (BCR, blue), chimeric (Ig-TCR, green), and canonical TCRδ transcripts (TCRδ, red). The boxplot extends from the first to the third quartiles (25th and 75th percentiles, respectively), with centerline indicating median value. The whisker of each boxplot extends to the value no. >1.5 times the interquartile range (difference between the first and third quartiles), values beyond these whiskers are potential outliers, indicating a significant skew or preference for a given rearrangement. Because of the relatively high proportion of outliers present, the nonparametric Kruskall-Wallace test with post hoc Dunn test was used to determine significant differences in groups (BCR, chimeric, TCR) within a given shark. Significance is indicated by asterisks (*). **p < 0.01, *p < 0.05, n.s. (p > 0.05).
Draft assembly of the G. cirratum TCRδ locus reveals a novel V segment lineage
Because IgH V segments have been found to be associated with TCRδ loci in a growing number of vertebrate lineages, we also performed a draft assembly of the nurse shark TCRδ locus with targeted BAC sequencing. To this end, the G. cirratum BAC library was probed, and selected BAC clones, positive for TCRδ, TCRα, and NARTCR components, were selected for long-read sequencing. The resulting assembly yielded a draft TCRδ translocus assembly totaling 169 kb with an average coverage depth of 61, with TCRα J segments lying downstream. Although the TCRα C region was not assembled within the contig housing the TCRδ translocus (Fig. 4, contig 10), the presence of TCRα J segments downstream of the TCRδ translocus indicate a linked TCRαδ locus in this ancient model of IgSF receptor loci. This was corroborated by the BAC library screening, which identified multiple BACs probing positive for TCRδ, TCRα, and NARTCR gene segments (data not shown) and the linkage of TCRα and NARTCR in contigs that could not be linked to the grander TCRδ translocus (Fig. 4, contigs 36, 65, and 112). Although the targeted BAC sequencing approach allows confident assembly of IgSF loci, the technique is hindered in instances in which probed BACs contain no overlapping regions. Furthermore, without a nurse shark karyotype to probe chromosomes, selection of positive BACs with overlap is a random method that does not account for vast genomic regions without TCR gene segments.
FIGURE 4.
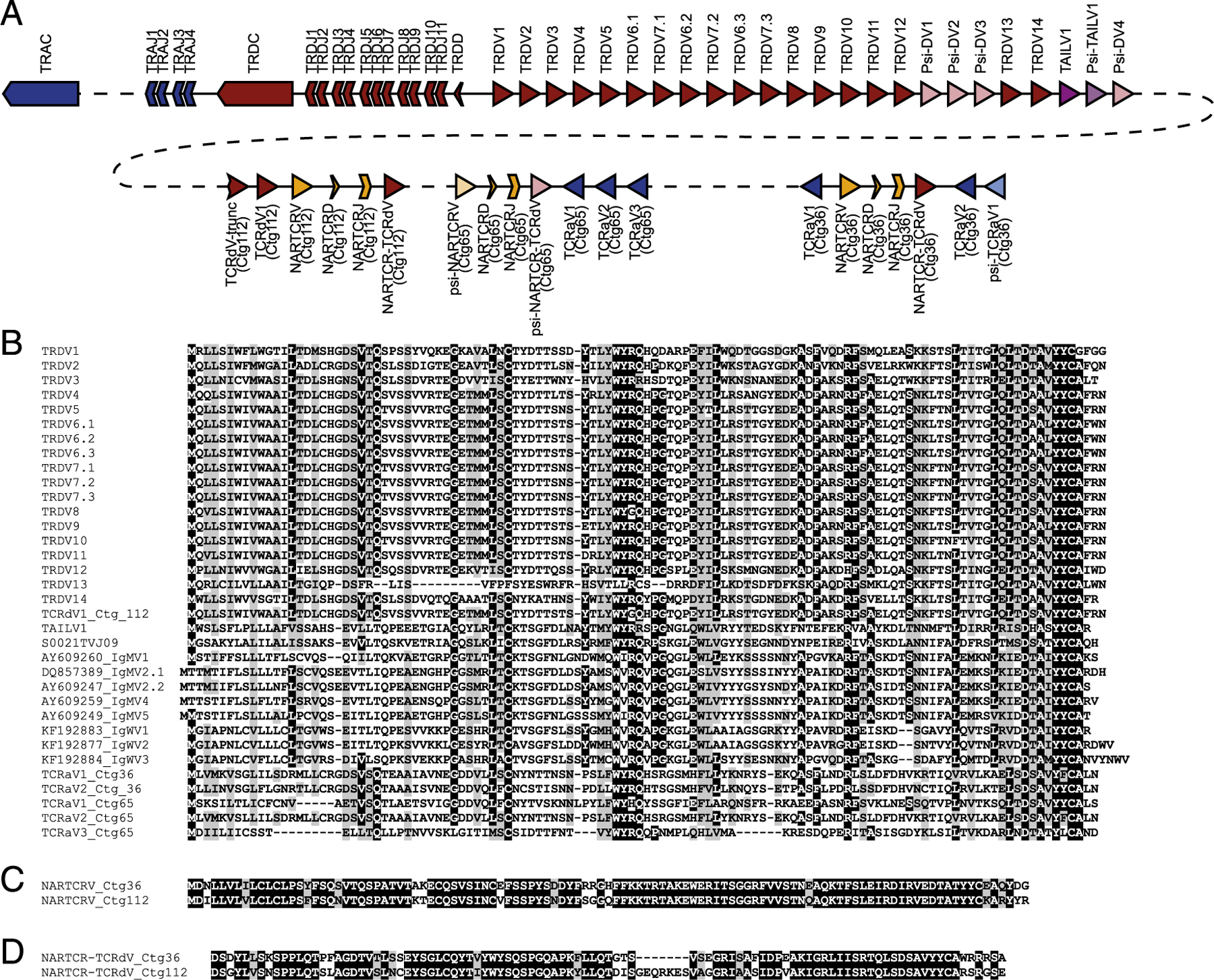
Assembly and mapping of TCRδ loci reveals IgH-like V segment bearing similarity to those found rearranged with TCRδ but not IgH. (A) Segments located within the assembled nurse shark TCRδ locus are color coded according to lineage-differentiating TCRδ (red), TCRα (blue), and IgH/TAILV segments (purple). Pseudogene segments are indicated by lighter shading. The contigs in which the segments are housed are boxed and labeled beneath the map. The prospective location of the TCRα C region is also indicated by the light blue box downstream of the TCRαJs on contig 10. (B and C) Alignment of V segments identified in assemblies, including the TAILV and IgHVs found in both TCRδ and IgH rearrangements (B), as well as NARTCRV (C), and NARTCR supporting TCRV (D).
The draft assembly of the nurse shark included four contigs with a total of 29 functional V segments, 4 D segments, 18 J segments, and 1 TCRδ C region. The largest contig (contig 10) revealed a translocon including 24 V segments (19 functional and five pseudo-V segments), a single TCRδ D segment, 11 TCRδ J segments, 4 TCRα J segments, and the TCRδ C region (Fig. 4A). We also assembled fragments with segments shown to be involved in formulating the TCRδ repertoire in three additional, nonoverlap-ping BACs. These BACs included three NARTCR VDJ blocks, with two functional NARTCRVs (contigs 36 and 112) and one pseudo-NARTCRV (contig 65). Each NARTCR block was found upstream of a supporting TCRδ V segment (functional on contigs 36 and 112, pseudogenized on contig 65). Two additional TCRδV segments were identified upstream of the NARTCR block in contig 112. The first TCRδV on contig 112 was truncated by the assembly, and the 164 bp assembled contained no frameshifts or stop codons; however, the second TCRδV is functional. Presumptively, these indicate an additional TCRδV translocon stretch unidentified thus far. Finally, we identified 6 TCRα V segments all oriented in reverse orientation in relation to the NARTCR blocks. Three functional TCRαVs were located downstream of the NARTCR block on contig 65. The remaining three were located on contig 36. One functional TCRαV was found upstream of the NARTCR block, whereas one functional TCRαV and a single pseudo-TCRαV were located downstream.
Additionally, we identified one functional IgHV-like segment and one pseudo-IgHV–like segment nested in the translocon stretch of TCRδVs (Fig. 4A). Although the IgHV-like segment is putatively functional, it could not be confidently ascribed to any Ig-TCRδ rearrangement in our expression dataset; however, it was similar to the novel IgHV-like segment we found only in TCRδ rearrangements (Fig. 4B). In phylogenetic analyses of cartilaginous fish IgH and TCRδ V segments, these Ig-like V segments (not found in rearrangements bearing IgH C regions) interestingly grouped together in a clade nestled between the IgM and IgW segments (Fig. 5A). We dubbed the segments belonging to this clade TCRδ-associated Ig-like V segments (TAILVs) to distinguish them from the V segments found on both TCRδ and IgH receptors. We also performed a phylogenetic analysis that included TCRα, TCRδ, VHδ, IgH, and IgL genomic V segments from all vertebrate lineages. The nurse TAILV representative sequence was found in a branch that included nurse and whale shark IgMVs, recapitulating the findings in our cartilaginous fish tree (Fig. 5B, 5C). This multispecies phylogeny also unveiled a clade of V segments, previously labeled TCRδV, sharing a common ancestry with Ig V segments rather than TCRV. The lack of a clade solely composed of Ig V segments found in TCRδ loci indicates that they have arisen multiple times through the course of vertebrate evolution.
FIGURE 5.
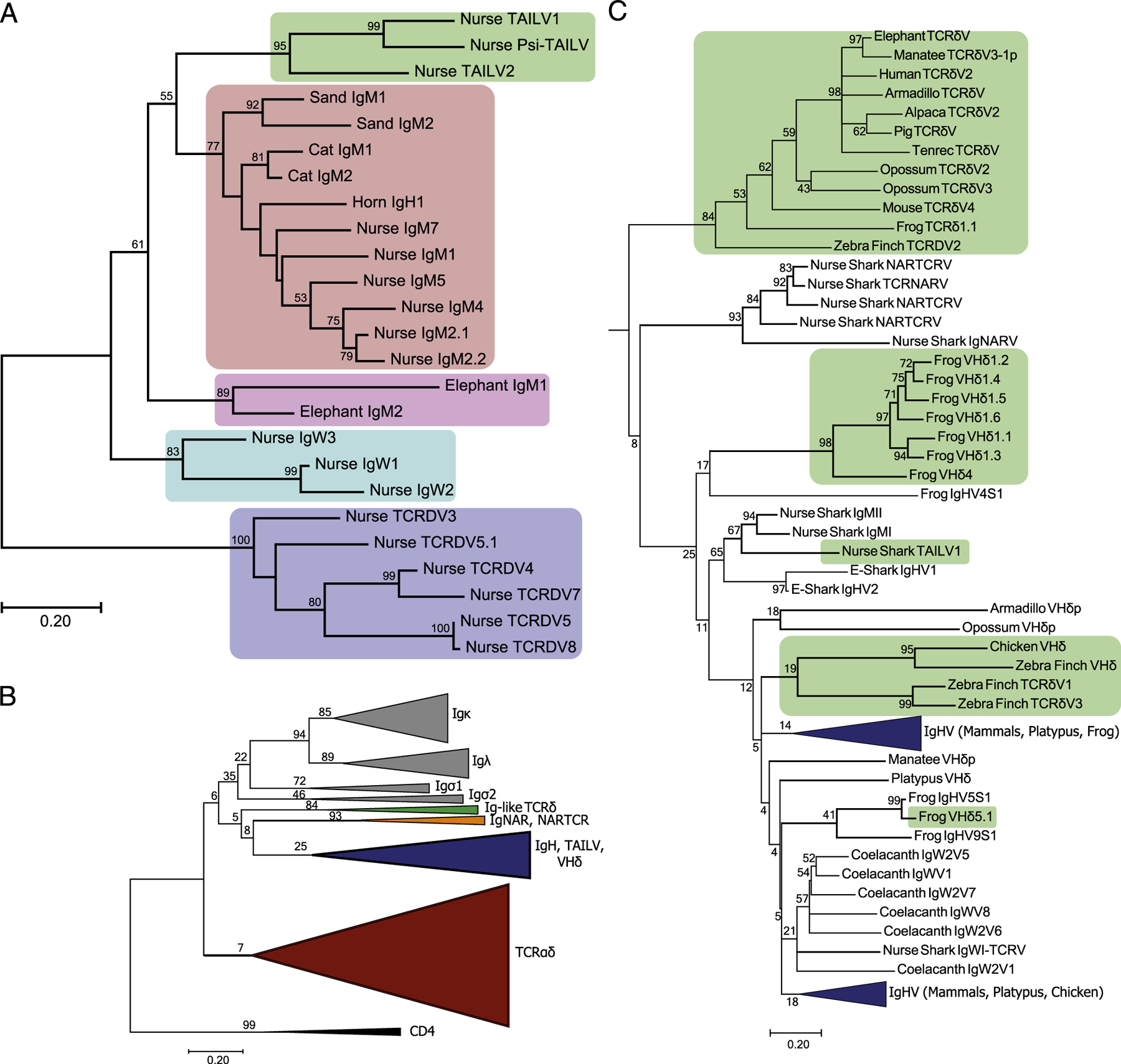
Phylogenetic analysis of Ag receptor V segments unveils a novel lineage of TCRδ-associated IgH-like V segments (TAILV) in cartilaginous fish. (A) An amino acid alignment of chondrichthyes V segments, trimmed to the start of FR1 and ended at the conserved Cys of FR3, was used to construct a maximum likelihood tree. Species included in the alignment were Ginglymostoma cirratum (nurse), Scyliorhinus canicula (cat), Heterodontus francisci (horn), Carcharhinus plumbeus (sand), and Callorhinchus milii (elephant). The percentage of trees supporting branches after 1000 Bootstrap replications is displayed for bifurcations with >50%. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The shaded regions indicate different V segment lineages found in Chondrichthyes, including elasmobranch TAILVs (green), elasmobranch IgMV (red), holocephali IgMV (pink), elasmobranch IgW (teal), and elasmobranch TCRδV (blue). (B) Topology of the jawed vertebrate Ag receptor tree. V segment lineage indicated by color for IgL (gray), Ig-like TCRδV (green), IgNARV/NARTCRV (orange), IgH (blue, includes TAILV and VHδ), and TCRαδV (red). The tree includes representative sequences from human, mouse, pig, tenrec, elephant, alpaca, manatee, opossum, platypus, zebra finch, chicken, frog, coelacanth, nurse shark, and elephant shark. (C) Subtree of the IgH and Ig-like V segments. Green shading indicates lineages of Ig-like V segments that are exclusively found on TCR rearrangements, highlighting multiple exchanges of Ig V segments into TCR loci throughout vertebrate evolution.
Comparative analysis of cartilaginous fish TCRδ loci
A number of cartilaginous fish, including the whale shark, Rhincodon typus (55), bamboo shark, Chiloscyllium plagiosum, and cloudy cat shark, Scyliorhinus torazame (56), have joined the first cartilaginous fish genome of the elephant shark, Callorhinchus milii (57). Of the recently sequenced genomes, only the bamboo shark has a scaffold (BEZZ01002038.1) in which multiple TCRδVs could be located; however, no TCRδ J segments or C region was found. This left only the elephant shark assembly with a TCRδ on the 694-kb scaffold NW_006890273 that is flanked by two IgMV segments found in typical cluster arrangement: one cluster possessing a full complement of IgM CH exons, the other with only a single CH exon. Also present in the elephant shark genome are NARTCRV and supporting TCRδV blocks, allowing syntenic comparisons to those found on smaller contigs in the nurse shark (Fig. 6A) (58). The absence of a TCRδ J segment and C region in either assembly made it impossible to confirm whether the TCRδC proximal V translocon was inverted in other cartilaginous fish species, as well. Nonetheless, the elephant shark scaffold was still useful to formulate a predictive placement for drafted nurse shark NARTCR contigs, allowing us to posit the topography of a complete nurse shark TCRδ locus (Fig. 6A). Furthermore, the proximity of the IgM clusters to the TCRδ locus in the elephant shark allowed us to formulate a potential model for the locale of (at least some) IgH clusters used in Ig-TCRδ rearrangements (Fig. 6B). Finally, we formulated a model of all nurse shark Ig-TCRδ rearrangements elucidated thus far: one intralocus TAILV and bona fide translocus rearrangements between IgWV, from three clusters, and IgMV, from five clusters, which contribute to the TCRδ repertoire (Fig. 6B).
FIGURE 6.
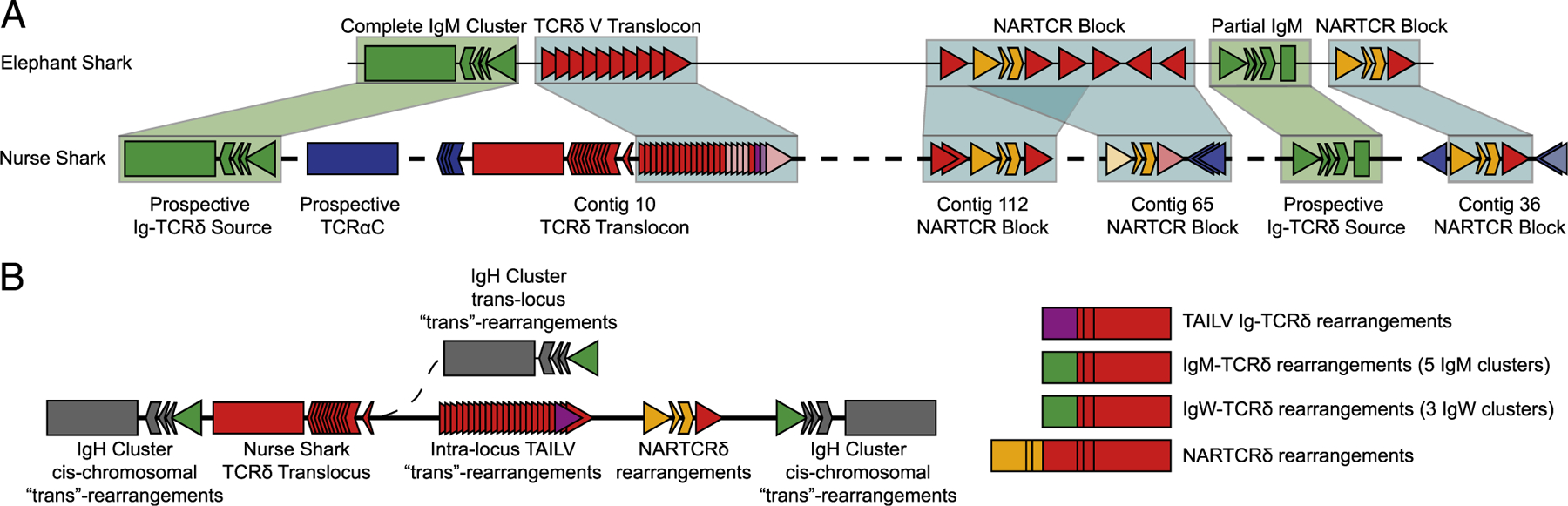
Comparative synteny between holocephali and elasmobranchii TCRδ loci and model of nurse shark noncanonical TCRδ rearrangement origins. (A) Sequence comparison of the TCRδ locus structure using elephant shark scaffold NW_006890273.1 as a map for assembled nurse shark contigs. Light blue shading indicates syntenic nurse shark contigs from the draft assembly, with associated contigs labeled beneath the box. Green shading indicates prospective positions of nurse shark IgH V segments used in Ig-TCR rearrangements based on position within the elephant shark scaffold. (B) Model of the multiple sources of Ig-TCRδ rearrangements, including TAILV rearrangements from within the TCRδ locus, bona fide trans-rearrangements between the eight IgH clusters whose location in relation to the TCRδ locus is unknown, and the doubly rearranging NARTCRδ.
Discussion
From what is known of cartilaginous fish IgH isotypes and the Ag recognition modes of vertebrate γδ T cells, there is a clear logic in IgMV and IgWV rearranging to TCRδ. Both share the same intrinsic structure of the IgSF V domain and the capacity to interact with free Ag. Drawing from the same pool of D and J segments as canonical TCRδ rearrangements, the Ig-TCRδ repertoire was found to be just as diverse as their canonical counterparts, characterized by large nontemplate TdT additions at the V-D and D-J junctions (Fig. 1, Supplemental Fig. 1). The large nontemplate additions found in Ig-TCRδ rearrangements were constrained to encode CDR3 lengths consistent with that of canonical TCRδ rearrangements in the nurse shark (Fig. 2, Table I). We were surprised to find that the predicted CDR3 iP in the two TCRδ rearrangement classes were also roughly equivalent, despite the bulk of CDR3 residues stemming from nontemplate nucleotides (Fig. 2). Similarities observed in CDR3s of Ig-TCRδ and canonical TCRδ suggest the two groups make use of the same dimeric partner chain. The heterodimeric TCR complex, complete with signal transduction machinery, is required of all T cells during thymic selection (59–61). We have determined that Ig-TCRδ rearrangements also pair with TCRγ; this is likely due to conservation of the Cys residue in TCRδ C regions, facilitating the interchain disulfide bond linking γδTCR complexes (Supplemental Fig. 2)(62). Any Ig-TCRδ chain rearrangements bearing incompatibilities with a partner chain, which prohibited paratope formation, would be selectively eliminated from the repertoire. Differences in the IgV domain are seemingly well tolerated by the partner chain, as the mRNA expression levels of Ig-TCRδ and canonical TCRδ were indistinguishable (Fig. 3). Thymic expression of aberrant receptors could be attributed to remnants of nonfunctional rearrangements (Fig. 3A). Equivalent expression levels for Ig-TCRδ and canonical TCRδ in the spleen of all five sharks (Fig. 3B) indicate that the Ig-TCRδ rearrangements regularly pass selection. Furthermore, the presence of Ig-TCRδ receptor rearrangements for at least three IgHV segments was identified on sorted thymocytes even in sharks that lacked support for BCR expression in the thymus (Supplemental Fig. 3).
Altogether, Ig-TCRδ rearrangements in the nurse shark are neither aberrant nor rare. With CDR3 metrics and expression levels so closely aligned to canonical TCRδ, it appears that Ig-TCRδ rearrangements, via their Ig-derived CDR1 and CDR2, are well tolerated by the partner chain and would diversify the recognition potential of shark TCRδ receptors. Such diversity would likely be disallowed in MHC-restricted αβ T cells but is tolerated in γδ T cells unbound from these restrictions.
Recombination signal sequences are the regulatory elements governing rearrangements in sharks as well as in mammals (62–65). If an IgHV were in proximity to the TCRδ locus, one could expect Ig-TCRδ rearrangements to occur between IgH and TCRδ loci. Indeed, this was the case, as several V segments identified in our Ig-TCRδ dataset originate from canonical IgH clusters. We found that bona fide trans-rearrangements between IgH and TCRδ loci are regular occurrences in the nurse shark. However, these were not the only type of chimeric Ig-TCRδ rearrangements we documented, as several V segments were not found in any IgH cluster nor expressed in the nurse shark Ab repertoire. Our draft nurse shark TCRδ locus assembly identified a single IgHV-like segment integrated in a translocon stretch of TCRδVs, with no downstream Ig C region, that likewise was not found expressed as an IgH (Fig. 4). Phylogenetic analysis of shark TCRδ and IgH V segments determined that these peculiar segments both clustered in a distinct clade in the phylogenetic analysis (Fig. 5). As both are clearly more Ig-like than TCR, we decided to call these V segments TAILVs. Presence of TAILVs in the TCRδ locus in one of the earliest iterations of IgSF-based adaptive immunity adds to the narrative that Ig and TCRδ V segments are largely interchangeable. This is also consistent with the emerging paradigm of IgHV segments associated with TCRδ loci of a growing number of species. Indeed, we identified a clade of canonically labeled TCRδVs (located in the TCRδ locus of tetrapod, avian, and mammalian lineages) with a shared Ig ancestry (Fig. 5B, 5C). This evidence highlights implementation of Ig-like V segments on TCRδ receptors in all jawed vertebrate lineages from cartilaginous fish to eutherian mammals. Phylogenetic evidence did not support TAILV as the progenitor of VHδ nor the aforementioned Ig-like TCRδV clade (Fig. 5C) but illustrates how IgH and TCRδ have freely exchanged gene segments over the course of vertebrate evolution. In fact, the location of the holocephalan (the most ancient chondrichthyes lineage) elephant shark IgMVs, which flank a TCRδ locus with no evidence of TAILVs, in the cartilaginous fish phylogeny suggest these proximal IgH clusters could be the ancient precursors to TAILVs (Fig. 6). Confirmation of this hypothesis via identification of syntenic blocks will require a more-complete elephant shark assembly and completion of the nurse shark TCRδ locus. Although we cannot claim that these IgMVs gave rise to the nurse shark TAILVs, we outlined hypothetical origins of TAILV as well as potential sources of IgHV segments found in both IgH clusters and Ig-TCRδ rearrangements in Fig. 6. This hypothetical synteny also provided evidence that the NARTCR blocks are likely localized to the TCRδ locus in all chondrichthyan species with confirmed NARTCR expression.
The TCRα and TCRγ loci of sharks have been shown to employ SHM (4, 66, 67), a process normally associated with IgH/L loci. Although IgHV sequences in our dataset contained minor differences within identical CDR3 regions, we found no conclusive evidence (via mutations in the IgHV of transcripts with identical CDR3 content) suggesting that SHM acts to expand diversity within the IgHV segments used in chimeric rearrangements. This was surprising, as one would assume the Ig V segments would be primed for diversifying mutations, suggesting that the anatomic and developmental windows of activation-induced cytidine deaminase expression are tightly regulated in the shark as well as endothermic vertebrates.
In summary, although the evolutionary history of Ig-TCRδ remains complex in vertebrates, we have confirmed that Ig-TCRδ rearrangements contribute to the peripheral T cell repertoire. The cellular contribution of Ig-TCRδ within the nurse shark immune system was highlighted in sequence diversity and expression in an assortment of tissues. Indeed, at all ages included in this study, Ig-TCRδ receptors are as prevalent as canonical TCRδ rearrangements. Additionally, the Ig-TCRδ repertoire makes use of Ig V segments both from IgH clusters and from a separate lineage of IgHV-like segments, termed TAILVs because of their exclusive association with TCRδ (Fig. 6B). Comparative TCRδ locus analysis of cartilaginous fish, both holocephali and elasmobranchii that split ~420 million years ago (68), exemplifies the versatility of TCRδ: a TCR employing receptors bearing the V domain of canonical TCRδ, IgHV (through chimeric Ig-TCRδ rearrangements originating both within the TCRδ locus [TAILV] and from IgH clusters of unknown proximity), and the doubly rearranging NARTCR (Fig. 6B).
Supplementary Material
Acknowledgments
We acknowledge Becky Lohr for initial cloning of an IgWV-TCRδC transcript in the nurse shark and Shehnaz Lokhandwala for assistance in annotation and submission of sequences to GenBank.
This work was supported by grants from the National Institutes of Health to M.F.C. (AI56963) and M.F.F. (AI027877) and the National Science Foundation to M.F.C. (IOS-1257829 and IOS-1656870).
Abbreviations used in this article:
- BAC
bacterial artificial chromosome
- IgSF
Ig superfamily
- iP
isoelectric point
- MBP
maltose binding protein
- qPCR
quantitative PCR
- SHM
somatic hypermutation
- TAILV
TCRδ-associated Ig-like V segment
Footnotes
The sequences presented in this article have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers JF507709.1–JF507661.1 and MN061599.1–MN061634.1 and to the Sequence Read Archive under Bioproject accession number PRJNA530194.
Disclosures
The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Litman GW, Hinds K, Berger L, Murphy K, and Litman R. 1985. Structure and organization of immunoglobulin VH genes in Heterodontus, a phylogenetically primitive shark. Dev. Comp. Immunol 9: 749–758. [DOI] [PubMed] [Google Scholar]
- 2.Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, and Litman GW. 1997. alpha, beta, gamma, and delta T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity 6: 1–11. [DOI] [PubMed] [Google Scholar]
- 3.Criscitiello MF, and Flajnik MF. 2007. Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur. J. Immunol 37: 2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott JA, Castro CD, Deiss TC, Ohta Y, Flajnik MF, and Criscitiello MF. 2018. Somatic hypermutation of T cell receptor a chain contributes to selection in nurse shark thymus. eLife 7: e28477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein RM, Schluter SF, Bernstein H, and Marchalonis JJ. 1996. Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc. Natl. Acad. Sci. USA 93: 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, and Flajnik MF. 2001. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc. Natl. Acad. Sci. USA 98: 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz M, Greenberg AS, and Flajnik MF. 1998. Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc. Natl. Acad. Sci. USA 95: 14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster DM, Gookin JL, Poore MF, Stebbins ME, and Levy MG. 2004. Outcome of cats with diarrhea and Tritrichomonas foetus infection. J. Am. Vet. Med. Assoc 225: 888–892. [DOI] [PubMed] [Google Scholar]
- 9.Foster BJ, Shults J, Zemel BS, and Leonard MB. 2004. Interactions between growth and body composition in children treated with high-dose chronic glucocorticoids. Am. J. Clin. Nutr 80: 1334–1341. [DOI] [PubMed] [Google Scholar]
- 10.Zhu C, and Hsu E. 2010. Error-prone DNA repair activity during somatic hypermutation in shark B lymphocytes. J. Immunol 185: 5336–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz M, Velez J, Singh M, Cerny J, and Flajnik MF. 1999. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int. Immunol 11: 825–833. [DOI] [PubMed] [Google Scholar]
- 12.Lee V, Huang JL, Lui MF, Malecek K, Ohta Y, Mooers A, and Hsu E. 2008. The evolution of multiple isotypic IgM heavy chain genes in the shark.J. Immunol 180: 7461–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litman GW, Anderson MK, and Rast JP. 1999. Evolution of antigen binding receptors. Annu. Rev. Immunol 17: 109–147. [DOI] [PubMed] [Google Scholar]
- 14.Berstein RM, Schluter SF, Shen S, and Marchalonis JJ. 1996. A new high molecular weight immunoglobulin class from the carcharhine shark: implications for the properties of the primordial immunoglobulin. Proc. Natl. Acad. Sci. USA 93: 3289–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta Y, and Flajnik M. 2006. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc. Natl. Acad. Sci. USA 103: 10723–10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinds KR, and Litman GW. 1986. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature 320: 546–549. [DOI] [PubMed] [Google Scholar]
- 17.Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, and Hsu E. 2012. Origin of immunoglobulin isotype switching. Curr. Biol 22: 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clem IW, De Boutaud F, and Sigel MM. 1967. Phylogeny of immunoglobulin structure and function. II. Immunoglobulins of the nurse shark.J. Immunol 99: 1226–1235. [PubMed] [Google Scholar]
- 19.Kasahara M, Vazquez M, Sato K, McKinney EC, and Flajnik MF. 1992. Evolution of the major histocompatibility complex: isolation of class II A cDNA clones from the cartilaginous fish. Proc. Natl. Acad. Sci. USA 89: 6688–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartl S, and Weissman IL. 1994. Isolation and characterization of major histocompatibility complex class IIB genes from the nurse shark. Proc. Natl. Acad. Sci. USA 91: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Criscitiello MF, Saltis M, and Flajnik MF. 2006. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc. Natl. Acad. Sci. USA 103: 5036–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, and Flajnik MF. 1995. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374: 168–173. [DOI] [PubMed] [Google Scholar]
- 23.Rumfelt LL, Diaz M, Lohr RL, Mochon E, and Flajnik MF. 2004. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol 173: 1129–1139. [DOI] [PubMed] [Google Scholar]
- 24.Criscitiello MF, Ohta Y, Saltis M, McKinney EC, and Flajnik MF. 2010. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J. Immunol 184: 6950–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster DB, Huang R, Hatch V, Craig R, Graceffa P, Lehman W, and Wang CL. 2004. Modes of caldesmon binding to actin: sites of caldesmon contact and modulation of interactions by phosphorylation. J. Biol. Chem 279: 53387–53394. [DOI] [PubMed] [Google Scholar]
- 26.Foster PS, and Harrison DW. 2004. Cerebral correlates of varying ages of emotional memories. Cogn. Behav. Neurol 17: 85–92. [DOI] [PubMed] [Google Scholar]
- 27.Foster PH 2004. Health disparities affecting African American. J. Health Care Poor Underserved 15: vii–viii. [PubMed] [Google Scholar]
- 28.Denny CT, Yoshikai Y, Mak TW, Smith SD, Hollis GF, and Kirsch IR. 1986. A chromosome 14 inversion in a T-cell lymphoma is caused by site-specific recombination between immunoglobulin and T-cell receptor loci. Nature 320: 549–551. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi Y, Tycko B, Soreng AL, and Sklar J. 1991. Transrearrangements between antigen receptor genes in normal human lymphoid tissues and in ataxia telangiectasia. J. Immunol 147: 3201–3209. [PubMed] [Google Scholar]
- 30.Allam A, and Kabelitz D. 2006. TCR trans-rearrangements: biological significance in antigen recognition vs the role as lymphoma biomarker. J. Immunol 176: 5707–5712. [DOI] [PubMed] [Google Scholar]
- 31.Chien Y-H, Jores R, and Crowley MP, 1996. Recognition by γ/δ T cells. Annu. Rev. Immunol 14: 511–532. [DOI] [PubMed] [Google Scholar]
- 32.Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF, and Miller RD. 2010. The dynamic TCRδ: TCRδ chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur. J. Immunol 40: 2319–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parra ZE, Lillie M, and Miller RD. 2012. A model for the evolution of the mammalian t-cell receptor α/δ and μ loci based on evidence from the duckbill Platypus. Mol. Biol. Evol 29: 3205–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parra ZE, Mitchell K, Dalloul RA, and Miller RD. 2012. A second TCRδ locus in Galliformes uses antibody-like V domains: insight into the evolution of TCRδ and TCRμ genes in tetrapods. J. Immunol 188: 3912–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saha NR, Ota T, Litman GW, Hansen J, Parra Z, Hsu E, Buonocore F,Canapa A, Cheng JF, and Amemiya CT. 2014. Genome complexity in the coelacanth is reflected in its adaptive immune system. J. Exp. Zoolog. B Mol. Dev. Evol 322: 438–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parra ZE, Baker ML, Schwarz RS, Deakin JE, Lindblad-Toh K, and Miller RD. 2007. A unique T cell receptor discovered in marsupials. Proc. Natl. Acad. Sci. USA 104: 9776–9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Parra ZE, and Miller RD. 2011. Platypus TCRμ provides insight into the origins and evolution of a uniquely mammalian TCR locus. J. Immunol 187: 5246–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rock EP, Sibbald PR, Davis MM, and Chien YH. 1994. CDR3 length in antigen-specific immune receptors. J. Exp. Med 179: 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rumfelt LL, McKinney EC, Taylor E, and Flajnik MF. 2002. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand. J. Immunol 56: 130–148. [DOI] [PubMed] [Google Scholar]
- 40.Dooley H, Buckingham EB, Criscitiello MF, and Flajnik MF. 2010. Emergence of the acute-phase protein hemopexin in jawed vertebrates. Mol. Immunol 48: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Criscitiello MF, Ohta Y, Graham MD, Eubanks JO, Chen PL, and Flajnik MF. 2012. Shark class II invariant chain reveals ancient conserved relationships with cathepsins and MHC class II. Dev. Comp. Immunol 36: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo M, Kim H, Kudrna D, Sisneros NB, Lee SJ, Mueller C, Collura K,Zuccolo A, Buckingham EB, Grim SM, et al. 2006. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics 7: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lefranc MP, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L,Thouvenin-Contet V, and Lefranc G. 2003. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol 27: 55–77. [DOI] [PubMed] [Google Scholar]
- 44.Mertz LM, and Rashtchian A. 1994. Nucleotide imbalance and polymerase chain reaction: effects on DNA amplification and synthesis of high specific activity radiolabeled DNA probes. Anal. Biochem 221: 160–165. [DOI] [PubMed] [Google Scholar]
- 45.Miller JR, Delcher AL, Koren S, Venter E, Walenz BP, Brownley A,Johnson J, Li K, Mobarry C, and Sutton G. 2008. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24: 2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, and Phillippy AM. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol 30: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rumfelt LL, Lohr RL, Dooley H, and Flajnik MF. 2004. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenberg A 1994. Evolution of the Antigen Receptor Family. University of Miami, Coral Gables, Florida. [Google Scholar]
- 49.Lefranc M-P, Giudicelli V, Ginestoux C, Bodmer J, Müller W, Bontrop R, Lemaitre M, Malik A, Barbié V, and Chaume D. 1999. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 27: 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefranc M-P, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, et al. 2015. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 43(Database issue): D413–D422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mashoof S, Pohlenz C, Chen PL, Deiss TC, Gatlin D III, Buentello A, and Criscitiello MF. 2014. Expressed IgH μ and τ transcripts share diversity segment in ranched Thunnus orientalis. Dev. Comp. Immunol 43: 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, and Tamura K, 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider CA, Rasband WS, and Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenberg AS, Hughes AL, Guo J, Avila D, McKinney EC, and Flajnik MF. 1996. A novel “chimeric” antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur. J. Immunol 26: 1123–1129. [DOI] [PubMed] [Google Scholar]
- 55.Read TD, Petit RA III, Joseph SJ, Alam MT, Weil MR, Ahmad M, Bhimani R, Vuong JS, Haase CP, Webb DH, et al. 2017. Draft sequencing and assembly of the genome of the world’s largest fish, the whale shark: Rhincodon typus Smith 1828. [Published erratum appears in 2017 BMC Genomics 18: 755.] BMC Genomics 18: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hara Y, Yamaguchi K, Onimaru K, Kadota M, Koyanagi M, Keeley SD,Tatsumi K, Tanaka K, Motone F, Kageyama Y, et al. 2018. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol 2: 1761–1771. [DOI] [PubMed] [Google Scholar]
- 57.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB,Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, et al. 2014. Elephant shark genome provides unique insights into gnathostome evolution. [Published erratum appears in 2014 Nature 513: 574.] Nature 505: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, et al. 2007. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 5: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayes SM, Shores EW, and Love PE. 2003. An architectural perspective on signaling by the pre-, alphabeta and gammadelta T cell receptors. Immunol. Rev 191: 28–37. [DOI] [PubMed] [Google Scholar]
- 60.Smith-Garvin JE, Koretzky GA, and Jordan MS. 2009. T cell activation. Annu. Rev. Immunol 27: 591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ribeiro ST, Ribot JC, and Silva-Santos B. 2015. Five layers of receptor signaling in γδ T-cell differentiation and activation. Front. Immunol 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flajnik MF 2018. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol 18: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flajnik MF, and Kasahara M. 2010. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet 11: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsu E 2016. Assembly and expression of shark Ig genes. J. Immunol 196: 3517–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu E 2009. V(D)J recombination: of mice and sharks. Adv. Exp. Med. Biol 650: 166–179. [DOI] [PubMed] [Google Scholar]
- 66.Chen H, Bernstein H, Ranganathan P, and Schluter SF. 2012. Somatic hypermutation of TCR γ V genes in the sandbar shark. Dev. Comp. Immunol 37: 176–183. [DOI] [PubMed] [Google Scholar]
- 67.Chen H, Kshirsagar S, Jensen I, Lau K, Covarrubias R, Schluter SF, and Marchalonis JJ. 2009. Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proc. Natl. Acad. Sci. USA 106: 8591–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Renz AJ, Meyer A, and Kuraku S. 2013. Revealing less derived nature of cartilaginous fish genomes with their evolutionary time scale inferred with nuclear genes. PLoS One 8: e66400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


