Abstract
Purpose
To investigate the occurrence and genetic characteristics of the blaIMP-26-positive plasmid from a multidrug-resistant clinical isolate, Enterobacter hormaechei L51.
Methods
Species identification was determined by MALDI-TOF MS and Sanger sequencing. Antimicrobial susceptibility testing was performed by the agar dilution and broth microdilution. Whole-genome sequencing was conducted using Illumina HiSeq 4000-PE150 and PacBio Sequel platforms, and the genome was annotated by the RAST annotation server. The ANI analysis of genomes was performed using OAT. Phylogenetic reconstruction and analyses were performed using the Harvest suite based on the core-genome SNPs of 61 publicly available E. hormaechei genomes.
Results
The E. hormaechei L51 genome consists of a 5,018,729 bp circular chromosome and a 343,918 bp conjugative IncHI2/2A plasmid pEHZJ1 encoding blaIMP-26 which surrounding genetic context was intI1-blaIMP-26-ltrA-qacE∆1-sul1. A new sequence type (ST1103) was assigned for the isolate L51 which was resistant to cephalosporins, carbapenems, but sensitive to piperacillin-tazobactam, amikacin, tigecycline, trimethoprim-sulfamethoxazole and colistin. Phylogenetic analysis demonstrated that E. hormaechei L51 belonged to the same subspecies as the reference strain E. hormaechei SCEH020042, however 18,248 divergent SNP were identified. Resistance genes in pEHZJ1 including aac(3)-IIc, aac(6ʹ)-IIc, blaSHV-178, blaDHA-1, blaTEM-1, blaIMP-26, ereA2, catII, fosA5, qnrB4, tet(D), sul1 and dfrA19.
Conclusion
In our study, we identified a conjugative IncHI2/2A plasmid carrying blaIMP-26 and blaSHV-178 in E. hormaechei ST1103, a novel multidrug-resistant strain isolated from China, and describe the underlying resistance mechanisms of the strain and detailed genetic context of mega plasmid pEHZJ1.
Keywords: Enterobacter hormaechei, multidrug-resistant, blaIMP-26, IncHI2/2A, genomics
Introduction
Enterobacter hormaechei is a species of gram-negative bacterium, belonging to the Enterobacter cloacae complex (ECC) and intestinal flora 75,1 which can exist in the intestinal tract of humans and animals, and is an opportunistic infectious pathogen. ECC was first described as nosocomial pathogens in the 1970s.2 Then, due to the dissemination of ESBLs and carbapenemases, ECC has become the third crucial drug-resistant pathogen in Enterobacteriaceae.3 In addition, it is one of the most common species that produces IMPs in Enterobacteriaceae.4 As the predominant species and the most commonly isolated nosocomial pathogen of ECC,5–7 the infection rate of E. hormaechei is increasing year by year.8 So far, ESBLs and carbapenemases were detected in E. hormaechei, including Klebsiella pneumoniae carbapenemases-2 (KPC-2),9 KPC-4,7 New Delhi metallo-β-lactamase-1 (NDM-1),10 NDM-7,11 German-Imipenemase-1(GIM-1),12 extended spectrum beta-lactamases CTX-M-(15, 9, 2),9 SHV-12, TEM-1B,7 and so on. However, detection of Metallo-β-lactamase IMP-26 has not been described.
IMP-26 was firstly reported in Pseudomonas aeruginosa isolated from Singapore in 2008, which was similar to IMP-4 and this sequence was previously stored in the GenBank database as IMP-4, from an Acinetobacter calcoaceticus strain (No. ABC24668.1).13 Since then, this metallo-β-lactamase type has appeared in China,14,15 Australia, Philippines,16 Malaysia17 and Vietnam18 in succession. However, these reports indicated that IMP-26 is located on the chromosome or without reference to its location, and only two reports ever described a copy of blaIMP-26 on the plasmid.19,20
Here, we identified a clinical isolate of E. hormaechei L51 producing both blaIMP-26 and blaSHV-178, and described the complete sequence of a conjugative IncHI2/2A plasmid. In addition, we performed phylogenetic analyses, S1-PFGE and southern blotting and drug resistance analysis to reveal the homology of isolate and the potential transmission mechanisms of blaIMP-26.
Materials and Methods
Bacterial Isolate and Susceptibility Testing
The isolate L51 was discovered in a routine screening of the intestinal colonization by bacteria resistant to carbapenems in the First Affiliated Hospital of Zhejiang University (FAHZU) in Hangzhou since 2016. The species identity of the bacteria strain was determined using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker, Bremen, Germany) and Sanger sequencing. The MIC values were determined using the agar dilution method and broth microdilution method. Results were interpreted following the Clinical and Laboratory Standards Institute (CLSI) standards,21 except tigecycline and colistin which were interpreted according to the 2018 EUCAST clinical breakpoints (http://www.eucast.org/clinical_breakpoints/).
MLST, Conjugation Experiments, and S1-PFGE-Southern Blot Hybridization
Multilocus sequence typing (MLST) was conducted as described previously.22 New alleles and sequence type were submitted to the MLST database and approved by PubMLST (http://pubmlst.org/ecloacae). Conjugation experiments were carried out by co-incubation of E. hormaechei L51 and recipient E. coli EC600 at a 1:10 ratio in the LB broth at a temperature of 25°C. The transconjugants were coated on Mueller-Hinton medium with rifampicin (100 mg/L) and meropenem (2 mg/L). Colonies were selected from the selective medium and verified as E. coli EC600, then screened for blaIMP-26 by PCR and sequencing. The size and number of plasmids of E. hormaechei L51 were identified by S1-PFGE, as described previously,23,24 in addition, the location of blaIMP-26 was demonstrated according to southern blot and hybridization experiments using DIG-labeled blaIMP-probe.
Whole-Genome Sequencing and Bioinformatics Analysis
Genomic DNA was extracted using the Bacterial DNA Kit (Omega Bio-Tek, USA). Library construction was performed using a 350-bp small fragment genomic DNA library25 and a 10-kb fragment library. The sequencing was conducted using the Illumina HiSeq 4000-PE150 and PacBio Sequel platforms (Beijing Novogene Bioinformatics Technology Company, China), to obtain short-read data and long-read data, respectively. Hybrid assembly was performed using Unicycler v0.4.2 producing an Illumina assembly graph by short reads and building bridges by long reads.26 Genomic sequences were annotated using RAST 2.0 (http://rast.nmpdr.org/). The Average Nucleotide Identity (ANI) values were calculated using OAT (Orthologous Average Nucleotide Identity Tool) and the proposed cut-off for species demarcation was 95~96%. The resistome analysis was carried out using CARD (https://card.mcmaster.ca/). Replicon type of plasmid was determined by Plasmid Finder 2.0 (https://cge.cbs.dtu.dk/services/PlasmidFinder/), insertion sequence (IS) elements and transposons were identified using ISfinder (http://www-is.biotoul.fr/), prophages were predicted by PHAST (http://phast.wishartlab.com/) and CRISPR arrays were identified using CRISPR Finder (http://crispr.i2bc.paris-saclay.fr/). Plasmid sequence alignment to the GenBank database was performed using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). A circular map of the plasmids pEHZJ1 and pIMP26 was plotted using the BLAST Ring Image Generator (BRIG).27
Phylogenetic Analysis
A phylogenic tree was constructed using the Harvest suite, including Parsnp, a fast core-genome multi-aligner, and Gingr, a dynamic visual platform based on core-genome SNPs approach28 that incorporates 61 publicly available E. hormaechei genomes (Table S1). The genome of E. hormaechei SCEH020042 (NZ_CP028538.1) was used as the reference isolate.
GenBank Accession Numbers
The complete genome sequence of E. hormaechei L51 has been deposited to GenBank under the accession CP033102-CP033103.
Results
Isolation and Identification of E. hormaechei L51 Harboring blaIMP-26 and blaSHV-178
A female patient, 16-year-old, was admitted to the department of gastroenterology of FAHZU in March 2016 and was diagnosed with Crohn disease. The patient had abdominal pain for more than 2 months. After admission, the amount of excrement of stool was very small, and it was mainly mushy. Stool was collected at the time of admission examination, then a strain of Enterobacteriaceae was isolated from the selection medium with meropenem (2mg/L) and designated as L51. The isolate was confirmed as E. hormaechei subsp. hoffmannii harboring blaIMP-26 and blaSHV-178 by MALDI-TOF MS and ANI analysis.
Antimicrobial Susceptibility Profiles
AST revealed that the strain was resistant to amoxicillin-clavulanate, cefotaxime, ceftazidime, cefepime, meropenem, imipenem, ertapenem, aztreonam, gentamicin, tobramycin, ciprofloxacin, tetracycline, chloramphenicol and nitrofurantoin, and was intermediate to levofloxacin and fosfomycin. Among them, resistance to cefotaxime, ceftazidime, tetracycline and chloramphenicol was greater than 128 mg/L. In addition, the strain was sensitive to piperacillin-tazobactam, amikacin, tigecycline, trimethoprim-sulfamethoxazole and colistin (Table 1).
Table 1.
Antimicrobial Susceptibility Profile of Enterobacter hormaechei L51
| Drug Class | Antimicrobial Drug | MIC (mg/L) | R/I/S |
|---|---|---|---|
| Penicillins | Amoxicillin-clavulanate | 128/64 | R |
| Piperacillin-tazobactama | 16/4 | S | |
| Cephalosporins | Cefotaxime | ≥128 | R |
| Ceftazidime | ≥128 | R | |
| Cefepime | 16 | R | |
| Cefpirome | 32 | R | |
| Carbapenems | Meropenem | 8 | R |
| Imipenem | 8 | R | |
| Ertapenem | 8 | R | |
| Monobactams | Aztreonam | 128 | R |
| Aminoglycosides | Gentamicin | 128 | R |
| Amikacin | 2 | S | |
| Tobramycin | 16 | R | |
| Fluoroquinolones | Ciprofloxacin | 1 | R |
| Levofloxacin | 0.5 | I | |
| Tetracyclines | Tetracycline | ≥128 | R |
| Tigecycline | 1 | S | |
| Others | Trimethoprim-sulfamethoxazole | ≥8 | S |
| Chloramphenicol | ≥128 | R | |
| Nitrofurantoin | 32 | R | |
| Fosfomycin | 128 | I | |
| Colistin | 0.5 | S |
Note: aTazobactam at a fixed concentration of 4mg/L.
Abbreviations: R, resistant; I, intermediary; S, susceptible.
Molecular Characteristics
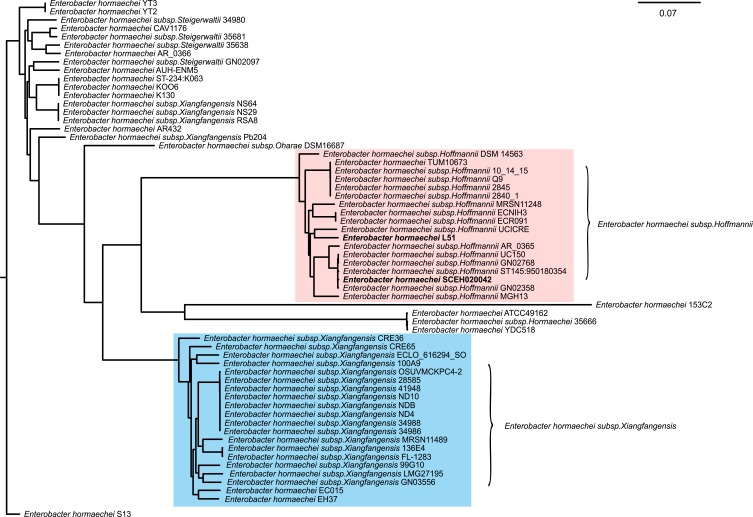
PCR and sequencing further demonstrated the presence of blaIMP-26 in E. hormaechei L51. Based on sequence analysis of seven housekeeping genes, L51 was assigned to a new sequence type ST1103 (59-47-new338-new379-70-new146-6). The genome of L51 consisted of a circular chromosome of 5,018,729 bp and a plasmid of 343,918 bp. The GC content of chromosomes and plasmids was 54.8% and 48.4%, respectively. Four phages were found on the chromosome and two were found on the plasmid. In addition, two CRISPR arrays were identified on the chromosome (Table S2). A large number of drug-resistant genes were found in E. hormaechei L51, of these, 15 were encoded in the plasmid (Table 2). The phylogenetic analysis of 61 E. hormaechei isolates (Figure 1) showed that various subspecies of E. hormaechei were separately aggregated, and E. hormaechei L51 and the reference isolate E. hormaechei SCEH020042 belonged to E. hormaechei subsp. hoffmannii. However, some E. hormaechei subsp. steigerwaltii isolates and some E. hormaechei subsp. xiangfangensis isolates were classified in the same cluster. In addition, 18,248 SNPs were identified between E. hormaechei L51 and the reference strain E. hormaechei SCEH020042.
Table 2.
Antibiotic Resistance Genes in Plasmid of Enterobacter hormaechei L51
| Drug Class | Coding Genes | Gene Family, Description | Position |
|---|---|---|---|
| Aminoglycoside | aac(3)-IIc | AAC(3) | 297,252–298,061 |
| aac(6ʹ)-IIc | AAC(6ʹ) | 300,146–300,727 | |
| Beta-lactamase | blaSHV-178 | SHV beta-lactamase | 270,538–271,398 |
| blaDHA-1 | DHA beta-lactamase | 278,802–279,941 | |
| blaTEM-1 | TEM beta-lactamase | 117,616–118,476 | |
| blaIMP-26 | IMP beta-lactamase | 130,800–131,540 | |
| Macrolide | ereA2 | Macrolide esterase | 299,713–299,940 |
| Streptogramin | mrx | Macrolide phosphotransferase (MPH) | 90,384–91,622 |
| mph(A) | 91,619–92,524 | ||
| Phenicol | catII | Chloramphenicol acetyltransferase (CAT) | 261,382–262,023 |
| Fosfomycin | fosA5 | Fosfomycin thiol transferase | 142,659–143,078 |
| Fluoroquinolone | qnrB4 | Quinolone resistance protein (qnr) | 284,062–284,709 |
| Tetracycline | tet(D) | Major facilitator superfamily (MFS) antibiotic efflux pump | 258,586–259,770 |
| Sulphonamide | sul1 | Sulfonamide resistant sul | 293,427–294,266 |
| Trimethoprim | dfrA19 | Trimethoprim resistant dihydrofolate reductase dfr | 128,462–129,031 |
Figure 1.
The phylogenetic tree of 61 completed E. hormaechei genome generated by the Harvest suite, where Enterobacter hormaechei SCEH020042 is used as the standard strain. Enterobacter hormaechei subsp. hoffmannii strains and E. hormaechei subsp. xiangfangensis strains are marked in pink and blue, respectively.
Characteristics of IncHI2/2A Plasmid pEHZJ1
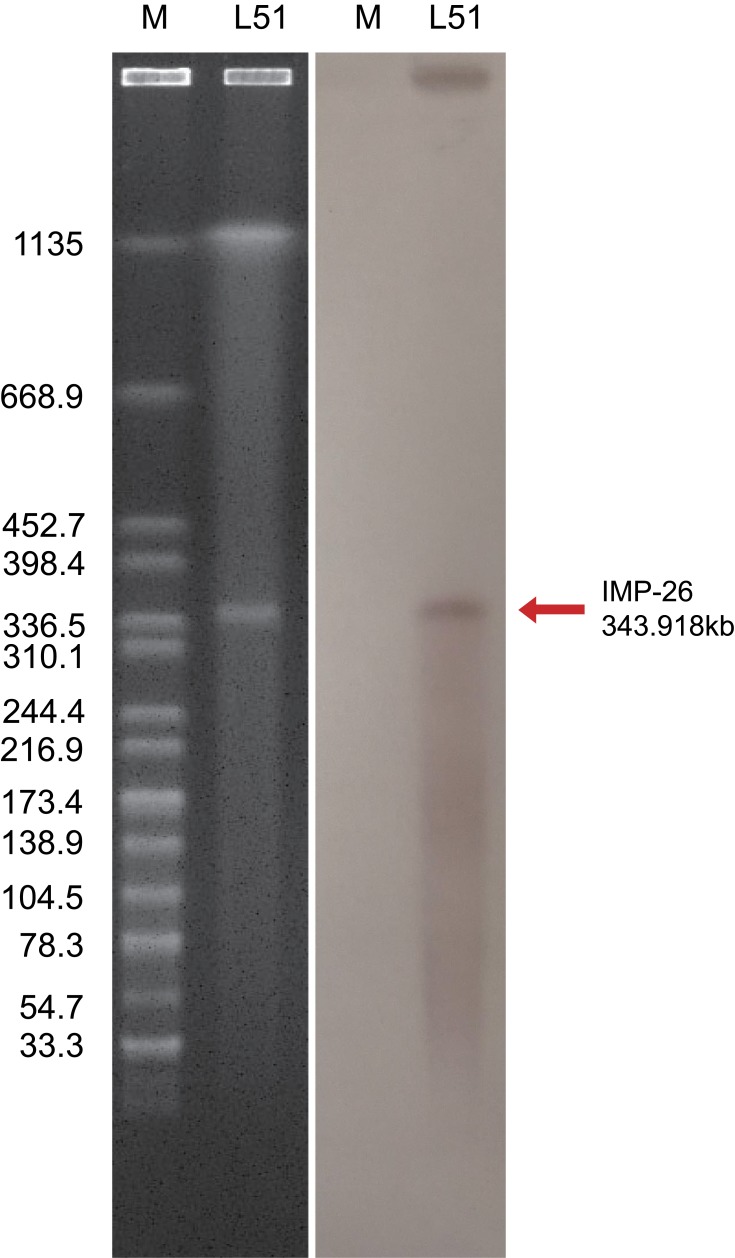
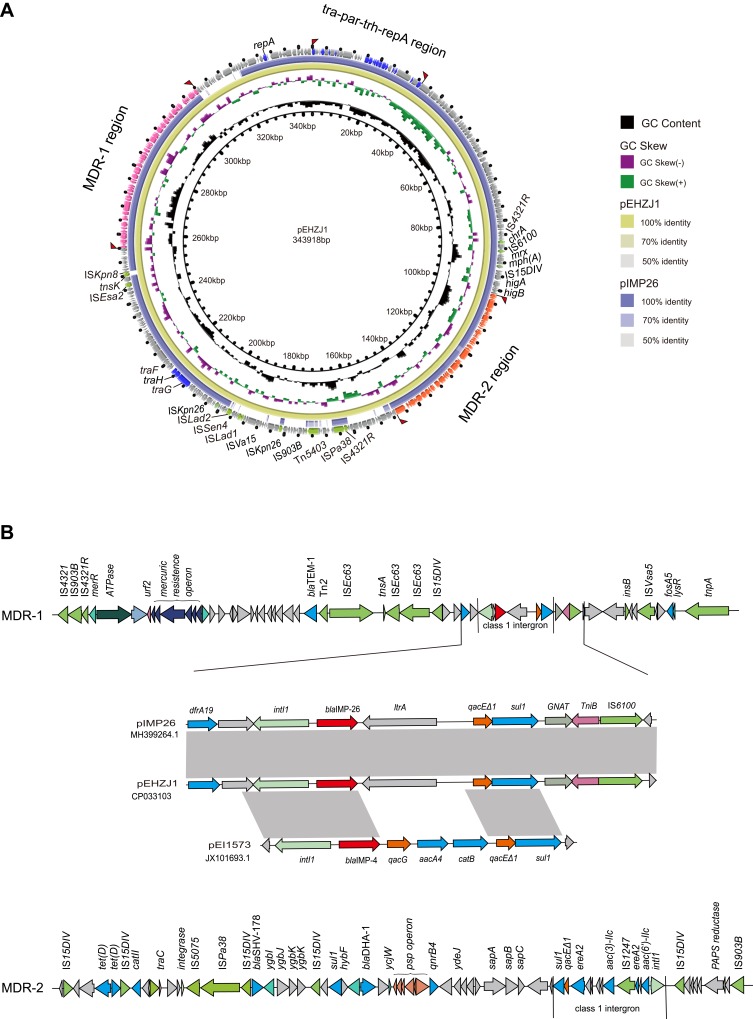
S1-PFGE and southern blot hybridization confirmed that the mega plasmid, pEHZJ1, contained blaIMP-26 (Figure 2). pEHZJ1 was 343,918 bp in length, with 318 CDSs and an average GC content of 48.4%. Replicon typing indicated that pEHZJ1 belonged to the IncHI2/2A plasmid. Conjugation experiments revealed that the transmission of blaIMP-26 from E. hormaechei L51 into E. Coli 600 was successful. pEHZJ1 carried a collection of replication initiation and conjugative transfer assembly proteins (tra-par-trh-rep region) and transcriptional regulators (padR, tetR, merR, lysR, ygbI, ycjW) and a large number of insertion sequences (ISs), which together constituted the structural backbone of the plasmid. In addition, the plasmid contained tellurium (~15 kb), mercury (~4 kb), arsenic (~4.8 kb) operon that confers resistance to tellurium, mercury and arsenic, and the copies of higA and higB which encode toxin-antitoxin system proteins. pEHZJ1 also contained two-large MDR regions (MDR-1 and MDR-2) of ~46 kb and ~52 kb, respectively. In MDR-1 and MDR-2 regions, there were two copies of class 1 integron and numerous antibiotic resistance genes including to beta-lactam (blaSHV-178, blaDHA-1, blaTEM-1, blaIMP-26), to aminoglycoside (aac(3)-IIc, aac(6ʹ)-IIc), to tetracycline (tet(D)), to chloramphenicol (catII), to fosfomycin (fosA5), to fluoroquinolone (qnrB4), to sulphonamide (sul1) and to trimethoprim (dfrA19).
Figure 2.
The identification of plasmid size using S1-PFGE (left) and southern blot and hybridization (right). The pEHZJ1 plasmid was between 336.5kbp and 398.4kbp, and was positive for a probe against blaIMP-26.
Class 1 Integron Carrying blaIMP-26 Located in MDR-1 Region of pEHZJ1
In MDR-1 region of pEHZJ1, a class 1 integron carrying gene cassette contained blaIMP-26, and a group II intron reverse transcriptase. And it was bracketed by ISs (IS110, IS5, IS6 and IS4) and contained transposons belonging to the Tn3 family (Figure 3). Moreover, the 5′-conserved segment (5′-CS) of the integron was adjacent to a resistance gene (dfr19), and the 3′-CS contained two coding sequences-qacE∆1 and sul1 (orf5 was absent) and was adjacent to fosA5. The MDR-2 region contained another class 1 integron carrying gene cassette aac(6ʹ)-IIc-ereA2-aac(3)-IIc-ereA2, which was inserted into an IS1247 backbone. For the integron in MDR-2, its 5′-CS was adjoining to IS15DIV and its 3′-CS region contained qacE∆1 and sul1. The genes surrounding blaIMP-26 in pEHZJ1 were identical to pIMP26 (accession: MH399264), but different from blaIMP-4 in pEI1573 (accession: JX101693). In this study, blaIMP-26 or blaIMP-4 in pEHZJ1, pIMP26 or pEI1573 were closely followed by intI1, but the gene cassette qacG, aacA4 and catB carried by the integron in pEI1573 was absent in pEHZJ1 (Figure 3B).
Figure 3.
The genetic features of the pEHZJ1 plasmid.
Notes: (A) Comparison of the pEHZJ1 plasmid sequence with pIMP26 using BRIG. The MDR-1 region, MDR-2 region and tra-par-trh-rep region are indicated in orange, pink and dark blue, respectively. Insert sequence and transposons are indicated in green. (B) Genetic features of MDR regions (MDR-1 and MDR-2) and a comparison of genes surrounding blaIMP-26 on pEHZJ1, pIMP26 (GenBank: MH399264.1) and pEI1573 (GenBank: JX101693.1). Gray regions denote regions that are homologous with >95% nucleotide similarity. blaIMP-26 is indicated in red and class 1 integrons are marked by straight lines.
Discussion
ECC is a complex group of bacterial species and is one of the leading causes of clinical nosocomial infections worldwide.29 Among Enterobacter spp. ECC is the most important complex group in human diseases, accounting for 65–75% of infections.2 As one of the most frequently isolated bacteria of ECC, E. hormaechei has become an important pathogen causing human clinical infection.30,31 Some reports exhibited that an E. hormaechei outbreak strain ever occurred nationwide in the Netherlands,32 in addition, this bacterium also appeared in several outbreaks of sepsis, most notably in the United States and Brazil.33 At present, it is recognized as capable of persisting in the hospital environment and developing resistance to multiple antibiotics.14,34 These facts imply that it will be a big potential threat to human health and may serve as a reservoir for drug resistance transmission in nosocomial infections.
Carbapenems were widely used as effective drugs in the treatment of multidrug-resistant bacterial infections. However, the presence and dissemination of carbapenemases in Enterobacteriaceae have emerged as a major challenge to modern medicine.12,35 In this study, we obtained a multidrug-resistant strain of E. hormaechei L51, harboring blaIMP-26 carbapenem resistance gene and blaSHV-178 ESBL gene. AST results showed this strain was high-level resistance to a variety of antimicrobial agents and were consistent with the genomic genes of the acquired resistance to antibiotics, suggesting that the drug resistance genes in chromosomes and pEHZJ1 were the major reason for multi-drug resistance of isolate L51 and our attention should be paid to it.
The phylogenetic analysis of 61 E. hormaechei isolates showed that some E. hormaechei subsp. steigerwaltii isolates and some E. hormaechei subsp. xiangfangensis isolates were classified in the same cluster. This may have been caused by the complex species composition of ECC making it difficult to identify the subspecies of a strain. In addition, 18,248 SNPs were identified between E. hormaechei L51 and the reference strain E. hormaechei SCEH020042, despite both strains belonged to the same subspecies. This suggested that strains within the subspecies of E. homaechei may be more divergent than expected. More complete genome sequences of Enterobacter isolates are required to improve the accuracy of identifying E. hormaechei.
Previous report has documented that SHV-type ESBL has strong genetic plasticity,36 in addition, pEHZJ1 IncHI2/2A plasmid contains both blaSHV-178 ESBL gene and blaIMP-26 carbapenem resistance gene, which was consistent with the hypothesis that IncHI2 plasmids carrying ESBL genes are more likely to acquire carbapenem resistance genes through horizontal transfer from integrons.3 Interestingly, it has ever demonstrated that IncHI2-type plasmids were involved in the propagation of blaIMP-4 among Enterobacteriaceae in Australia.37 Previous studies have shown that most of IncHI2 plasmids are transferable3 and frequently harbor carbapenemase genes,38,39 which idea was verified again in our study. In addition, transcriptional regulators, tra-par-trh-rep region, transposon and insertion sequences, and tellurium, mercury, arsenic operons were found in pEHZJ1, which backbone was similar to pIMP26,20 with 82% query coverage and 99% nucleotide identity. The presence of tellurium, mercury, and arsenic operons in pEHZJ1 was consistent with the conclusion that IncHI2 plasmids usually harbor toxin-antitoxin systems and heavy metal resistance from, Aoki et al.40 These metal-resistant operons may make the strains carrying IncHI2-type plasmid to be more viable in diverse environments.
Thus far, two reports have studied the genetic features surrounding blaIMP-26 in P. aeruginosa,17,18 and one in Enterobacter cloacae.20 Among them, the genetic context surrounding blaIMP-26 in P. aeruginosa was intI1-blaIMP-26-qacG-aacA4-aac(6ʹ)-orf-catB3 and intI1-blaIMP-26-qacG-aac(6ʹ)-Ib-aac(6ʹ)-orf3-orf4-catB3-dfrA1-tnpA-istB-orf5, respectively. In our study, the genetic environment of blaIMP-26 was intI1-blaIMP-26-ltrA-qacE∆1-sul1, which was the same as in E. cloacae.20 Integron was first proposed by Strokes and Hall in 1989, which was a mobile DNA molecule with unique structure to capture and integrate exogenous genes.41–43 There was one literature ever report that bacteria positive for integrons have higher incidences of resistance against antibacterial drugs beta-lactamase, aminoglycosides, monobactams and fluoroquinolones compared to bacteria without integrons.44 In recent years, the transfer of integrons harboring multiple drug-resistant gene cassettes or the joint transfer of two or more integrons into one isolate appears to be the basis for the rapid growth of MDR isolates.45 Our conjugative plasmid pEHZJ1 possesses two class 1 integrons and an array of drug resistance genes, which may well pose a threat for the treatment of nosocomial infection. Previous reports showed that blaIMP-26 has high genetic similarity to blaIMP-4, with only one position substitution at 145 (G-to-T change),13 moreover, blaIMP-26 and blaIMP-4 were generally carried by a class 1 integron.17,18,46 So, we selected pIMP26, pEI1573 and our plasmid pEHZJ1 for comparative analysis of genes surrounding blaIMP-4 and blaIMP-26. The results revealed that pIMP26 had the same genetic context surrounding blaIMP-26 as pEHZJ1, however genetic environment of blaIMP-4 in pEI1573 was intI1-blaIMP-4-qacG-aacA4-catB-qacE∆1-sul1. Although molecular characteristics of blaIMP-4 and blaIMP-26 are very similar, their genetic context also could be different. Future research is required to understand whether the genes surrounding blaIMP affect the resistance of different strains.
Conclusion
In summary, we describe the complete sequence of a mega conjugative IncHI2/2A plasmid carrying both blaIMP-26 and blaSHV-178 of E. hormaechei L51. Phylogenetic analysis based on core-genome SNPs revealed the huge differences between the subspecies of E. hormaechei and provided a reference view that more whole-genome sequencing data are needed for the more accurate identification of E. hormaechei subspecies. The spread of the IncHI2-type plasmid carrying IMP resistance genes in ECC is worth worrying about, although only one case was reported in this study. Our results also suggest that focusing on E. hormaechei will be important in future studies.
Funding Statement
This research was funded by the National Natural Science Foundation of China, grant number 81741098; Henan Science and Technology Department, grant number 162102310509; Henan medical science and technology project, grant number 201701006; Key scientific research project plan of institutions of higher learning of Henan province, grant number 19A320058; the National Key Research and Development Program of China, grant number 2016YFD0501105; the Zhejiang Provincial Natural Science Foundation of China, grant number LY17H190003.
Ethical Approval
The ethical protocol was approved by the Ethics Committee of First Affiliated Hospital of Zhejiang University (no. 2018-1031).
Written Informed Consent
Written informed consent was obtained from the patient described in the study for the publication of the case details.
Data Sharing Statement
The datasets generated for this study can be found in NCBI, CP033102-CP033103.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.O’Hara CM, Steigerwalt AG, Hill BC, et al. Enterobacter hormaechei, a new species of the family Enterobacteriaceae formerly known as enteric group 75. J Clin Microbiol. 1989;27:2046–2049. doi: 10.1128/JCM.27.9.2046-2049.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders WE, Sanders CC. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev. 1997;10:220–241. doi: 10.1128/CMR.10.2.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou K, Yu W, Cao X, et al. Characterization of the population structure, drug resistance mechanisms and plasmids of the community-associated Enterobacter cloacae complex in China. J Antimicrob Chemother. 2018;73:66–76. doi: 10.1093/jac/dkx361 [DOI] [PubMed] [Google Scholar]
- 4.Matsumura Y, Peirano G, Motyl MR, et al. Global molecular epidemiology of IMP-producing enterobacteriaceae. Antimicrob Agents Chemother. 2017;61:e02729–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmas J, Breysse F, Devulder G, et al. Rapid identification of Enterobacteriaceae by sequencing DNA gyrase subunit B encoding gene. Diagn Microbiol Infect Dis. 2006;55:263–268. doi: 10.1016/j.diagmicrobio.2006.02.003 [DOI] [PubMed] [Google Scholar]
- 6.Paauw A, Caspers MP, Schuren FH, et al. Genomic diversity within the Enterobacter cloacae complex. PLoS One. 2008;3:e3018. doi: 10.1371/journal.pone.0003018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavda KD, Chen L, Fouts DE, et al. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio. 2016;7. doi: 10.1128/mBio.02093-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohad S, Block C, Kravitz V, et al. Rapid identification of Enterobacter hormaechei and Enterobacter cloacae genetic cluster III. J Appl Microbiol. 2014;116:1315–1321. doi: 10.1111/jam.12439 [DOI] [PubMed] [Google Scholar]
- 9.Andrade LN, Siqueira TES, Martinez R, et al. Multidrug-resistant CTX-M-(15, 9, 2)- and KPC-2-producing enterobacter hormaechei and enterobacter asburiae isolates possessed a set of acquired heavy metal tolerance genes including a chromosomal sil operon (For acquired silver resistance). Front Microbiol. 2018;9:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B, Feng Y, McNally A, et al. Occurrence of Enterobacter hormaechei carrying blaNDM-1 and blaKPC-2 in China. Diagn Microbiol Infect Dis. 2018;90:139–142. doi: 10.1016/j.diagmicrobio.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 11.Villa J, Carretero O, Viedma E, et al. Emergence of NDM-7-producing multi-drug-resistant Enterobacter hormaechei sequence type ST-78 in Spain: a high-risk international clone. Int J Antimicrob Agents. 2019;53:533–534. doi: 10.1016/j.ijantimicag.2018.11.009 [DOI] [PubMed] [Google Scholar]
- 12.Wendel AF, Meyer S, Deenen R, et al. Long-term, low-frequency cluster of a german-imipenemase-1-producing Enterobacter hormaechei ssp. steigerwaltii ST89 in a Tertiary Care Hospital in Germany. Microb Drug Resist. 2018;24:1305–1315. doi: 10.1089/mdr.2017.0433 [DOI] [PubMed] [Google Scholar]
- 13.Koh TH, Khoo CT, Tan TT, et al. Multilocus sequence types of carbapenem-resistant Pseudomonas aeruginosa in Singapore carrying metallo-beta-lactamase genes, including the novel bla(IMP-26) gene. J Clin Microbiol. 2010;48:2563–2564. doi: 10.1128/JCM.01905-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyrouthy R, Barets M, Marion E, et al. Novel Enterobacter Lineage as leading cause of nosocomial outbreak involving carbapenemase-producing strains. Emerg Infect Dis. 2018;24:1505–1515. doi: 10.3201/eid2408.180151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia Y, Liang Z, Su X, et al. Characterization of carbapenemase genes in Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a university hospital in Chongqing, China. Ann Lab Med. 2012;32:270–275. doi: 10.3343/alm.2012.32.4.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lascols C, Peirano G, Hackel M, et al. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–136. doi: 10.1128/AAC.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MJ, Bae IK, Jeong SH, et al. Dissemination of metallo-beta-lactamase-producing Pseudomonas aeruginosa of sequence type 235 in Asian countries. J Antimicrob Chemother. 2013;68:2820–2824. doi: 10.1093/jac/dkt269 [DOI] [PubMed] [Google Scholar]
- 18.Tada T, Nhung PH, Miyoshi-Akiyama T, et al. Multidrug-resistant sequence Type 235 pseudomonas aeruginosa clinical isolates producing IMP-26 with increased carbapenem-hydrolyzing activities in Vietnam. Antimicrob Agents Chemother. 2016;60:6853–6858. doi: 10.1128/AAC.01177-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumura Y, Peirano G, Bradford PA, et al. Genomic characterization of IMP and VIM carbapenemase-encoding transferable plasmids of Enterobacteriaceae. J Antimicrob Chemother. 2018;73:3034–3038. doi: 10.1093/jac/dky303 [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Zhou K, Xiao S, et al. A multidrug resistance plasmid pIMP26, carrying blaIMP-26, fosA5, blaDHA-1, and qnrB4 in Enterobacter cloacae. Sci Rep. 2019;9:10212. doi: 10.1038/s41598-019-46777-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CaLS I. Performance standards for antimicrobial susceptibility testing—Twenty-eighth informational supplement M100-S28. CLSI 2018. 2018.
- 22.Woo PCY, Miyoshi-Akiyama T, Hayakawa K, et al. Multilocus Sequence Typing (MLST) for characterization of Enterobacter cloacae. PLoS One. 2013;8:e66358. doi: 10.1371/journal.pone.0066358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng B, Zhang J, Ji J, et al. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother. 2015;59:7086–7089. doi: 10.1128/AAC.01363-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Zhang W, Liang H, et al. Whole-genome sequence of a multidrug-resistant clinical isolate of Acinetobacter lwoffii. J Bacteriol. 2011;193:5549–5550. doi: 10.1128/JB.05617-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillippy AM, Wick RR, Judd LM, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alikhan NF, Petty NK, Ben Zakour NL, et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Treangen TJ, Ondov BD, Koren S, et al. The Harvest suite for rapid core-genome and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gajdacs M, Urban E. Resistance Trends and Epidemiology of Citrobacter-Enterobacter-Serratia in Urinary Tract Infections of Inpatients and Outpatients (RECESUTI): a 10-year survey. Medicina (Kaunas). 2019;55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7:887–902. doi: 10.2217/fmb.12.61 [DOI] [PubMed] [Google Scholar]
- 31.Monahan LG, DeMaere MZ, Cummins ML, et al. High contiguity genome sequence of a multidrug-resistant hospital isolate of Enterobacter hormaechei. Gut Pathog. 2019;11:3. doi: 10.1186/s13099-019-0288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leverstein-van Hall MA, Blok HE, Paauw A, et al. Extensive hospital-wide spread of a multidrug-resistant enterobacter cloacae clone, with late detection due to a variable antibiogram and frequent patient transfer. J Clin Microbiol. 2006;44:518–524. doi: 10.1128/JCM.44.2.518-524.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morand PC, Billoet A, Rottman M, et al. Specific distribution within the Enterobacter cloacae complex of strains isolated from infected orthopedic implants. J Clin Microbiol. 2009;47:2489–2495. doi: 10.1128/JCM.00290-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davin-Regli A, Pages JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol. 2015;6:392. doi: 10.3389/fmicb.2015.00392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng B, Lv T, Xu H, et al. Discovery and characterisation of an escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int J Antimicrob Agents. 2018;51:273–275. doi: 10.1016/j.ijantimicag.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 36.Bourouis A, Ben Moussa M, Belhadj O. Multidrug-resistant phenotype and isolation of a novel SHV- beta-Lactamase variant in a clinical isolate of Enterobacter cloacae. J Biomed Sci. 2015;22:27. doi: 10.1186/s12929-015-0131-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sidjabat HE, Heney C, George NM, et al. Interspecies transfer of blaIMP-4 in a patient with prolonged colonization by IMP-4-producing Enterobacteriaceae. J Clin Microbiol. 2014;52:3816–3818. doi: 10.1128/JCM.01491-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miro E, Segura C, Navarro F, et al. Spread of plasmids containing the bla(VIM-1) and bla(CTX-M) genes and the qnr determinant in Enterobacter cloacae, Klebsiella pneumoniae and Klebsiella oxytoca isolates. J Antimicrob Chemother. 2010;65:661–665. doi: 10.1093/jac/dkp504 [DOI] [PubMed] [Google Scholar]
- 39.Fischer J, Rodríguez I, Schmoger S, et al. Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J Antimicrob Chemother. 2013;68:478–480. doi: 10.1093/jac/dks393 [DOI] [PubMed] [Google Scholar]
- 40.Aoki K, Harada S, Yahara K, et al. Molecular characterization of IMP-1-producing Enterobacter cloacae complex isolates in Tokyo. Antimicrob Agents Chemother. 2018;62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/mmi.1989.3.issue-12 [DOI] [PubMed] [Google Scholar]
- 42.Recchia GD, Hall RM. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015 [DOI] [PubMed] [Google Scholar]
- 43.Li J, Liu C, Li B, et al. Identification and molecular characterization of a novel DyP-type peroxidase from Pseudomonas aeruginosa PKE117. Appl Biochem Biotechnol. 2012;166:774–785. doi: 10.1007/s12010-011-9466-x [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Freijo P, Fluit AC, Schmitz FJ, et al. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998;42:689–696. doi: 10.1093/jac/42.6.689 [DOI] [PubMed] [Google Scholar]
- 45.Nógrády N, Gadó I, Pászti J, et al. Analysis of gene cassettes of streptomycin-spectinomycin resistance of Hungarian Salmonella enterica serotype Typhimurium strains. Acta Vet Hung. 2003;51:137–151. doi: 10.1556/AVet.51.2003.2.2 [DOI] [PubMed] [Google Scholar]
- 46.Partridge SR, Ginn AN, Paulsen IT, et al. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother. 2012;56:6029–6032. doi: 10.1128/AAC.01189-12 [DOI] [PMC free article] [PubMed] [Google Scholar]