Abstract
The Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway is involved in cell immunity, division and death, as well as in tumor formation. The expression of key genes in the JAK-STAT signaling pathway in different types of cancer serves different roles. However, few reports are available on the prognostic value of the genes of the JAK-STAT signaling pathway in skin cutaneous melanoma (SKCM). The potential prognostic value of gene expression in the JAK-STAT signaling pathway in patients with SKCM was analyzed in the present study using data obtained from The Cancer Genome Atlas. To predict the potential functions and mechanisms of these genes in SKCM, gene set enrichment analysis (GSEA) and bioinformatics analysis were performed. A nomogram model including gene expression level and high risk factors was used to predict the risk level of prognostic. High expression levels of STAT1, STAT3, STAT4 and STAT5B, and low expression levels of STAT6 were associated with favorable prognosis [adjusted P<0.001; hazard ratio (HR), 0.595; 95% confidence interval (CI), 0.455–0.778; adjusted P=0.018; HR, 0.725; 95% CI, 0.555–0.947; adjusted P<0.001; HR, 0.590; 95% CI, 0.450–0.773; adjusted P=0.007; HR, 0.690; 95% CI, 0.526–0.940; and adjusted P=0.026; HR, 0.737, 95% CI, 0.563–0.964, respectively]. GSEA results demonstrated that these genes were involved in cell differentiation, invasion, adhesion, migration, cycle, colony formation and mitogen-activated protein kinase signaling. The combination of genes with favorable prognosis had a better effect on the overall survival (univariate survival analysis, P<0.05). The results of the present study suggest that STAT1, STAT3, STAT4, STAT5B and STAT6 gene expression may be used as a potential prognostic biomarker of SKCM, and the combined outcomes may exhibit a stronger interaction and higher survival time for SKCM.
Keywords: melanoma, Janus kinase-signal transducer and activator of transcription signaling pathway, prognosis
Introduction
Skin cutaneous melanoma (SKCM) is a highly aggressive skin cancer, which arises from the malignant transformation of melanocytes in the basal layer of the epidermis (1,2) and has a poor prognosis, with a 5-year overall survival (OS) of 91.8% worldwide (3). The number of new cases of SKCM that will emerge in 2019 was estimated to be 96,480, with the mortality estimated to be 7,230 (4,5). Therefore, there is an urgent need to identify novel biomarkers and prognostic predictive indicators for the detection and management of SKCM.
The JAK-STAT signaling pathway serves a crucial role in cell immunity, division and death, and in tumor formation (6). The two key components involved in this pathway are Janus kinases (JAKs) and signal transducer and activator of transcription proteins (STATs), which are encoded by the genes JAK (JAK1, JAK2, JAK3 and TYK2) and STAT (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6), respectively (6).
A previous study has demonstrated that the key genes involved in the JAK-STAT signaling pathway are associated with several types of cancer, including breast (7,8), ovarian, lung, brain (9) and colorectal (10) cancer, and that their differential expression may result in different prognosis outcomes in different types of cancer. However, few reports about the association between these genes and SKCM are available, and further investigation analyzing the prognostic value of gene expression in the JAK-STAT signaling pathway in SKCM is needed. The aim of the present study was to identify the prognostic values of the expression of genes involved in the JAK-STAT signaling pathway in patients with SKCM based on data derived from public databases and bioinformatics analysis, and to explore the underlying mechanism that may affect the outcome in SKCM prognosis.
Materials and methods
Data preparation
The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov; accessed on September 10th 2018 was used to obtain the gene expression and clinical data of patients with SKCM, including sex, age and tumor stage. A total of 458 SKCM cases were selected after removing the cases with missing mRNA expression or clinical data and 0-day survival time.
Functional analysis of key genes in the JAK-STAT signaling pathway
The Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway map was generated using the KEGG website (https://www.kegg.jp; accessed on September 10th 2018 (11–13). Gene ontology (GO) term analysis, including biological function (BP), molecular function (MF) and cellular component (CC), as well as KEGG enrichment analysis for JAK and STAT gene families, were performed using the Database for Annotation, Visualization and Integrated Discovery version 6.8 (DAVID; https://david.ncifcrf.gov/tools.jsp; accessed on September 13th 2018. The official gene symbol was used as the identifier; the species was Homo sapiens (14,15).
Gene interaction and association analysis
Gene-gene interaction analysis was performed using gene multiple association network integration algorithm (GeneMANIA; http://genemania.org; accessed on September 15th 2018 using the default parameters (16,17). The correlation between JAK and STAT pathway gene expression in SKCM was evaluated using the Pearson's correlation coefficient; P<0.01 was considered to indicate a significant correlation. Protein-protein interaction analysis was performed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org; accessed on September 25th 2018. The minimum required interaction score was 0.400 (18,19).
Patient grouping based on gene expression level
Patients with SKCM were divided into high or low expression groups using the median value, and the vertical scatter plots were generated.
Survival analysis
Kaplan-Meier survival plots with log-rank test were used to evaluate the OS for the high and low-expression groups of each gene and clinicopathological characteristics. In addition, the Cox proportional hazard regression model was used for univariate and multivariate survival analyses, and 95% confidence intervals (CIs) and hazard ratios (HRs) were calculated. Joint effects analysis was performed for the combination of genes identified as significant by the survival analysis.
Nomogram
A nomogram was used to evaluate the association between JAK and STAT gene expression and clinical information in SKCM. In addition, the risk rank for each gene and clinicopathological characteristic, including age, sex and tumor stage, was evaluated by the risk points and total points. Survival rates (1-, 3- and 5-year) were also scored. A high score was associated with a low survival rate.
Gene set enrichment analysis (GSEA)
To investigate the potential underlying mechanism of the differential expression of STAT1, STAT3, STAT4, STAT5B and STAT6 in SKCM, GSEA version 3.0 software (http://software.broadinstitute.org/gsea/msigdb/index.jsp; accessed on September 20th 2018 (20) was used, and the difference in the expression levels in the low and high-expression groups for each gene was analyzed with reference gene sets, which were based on the Molecular Signatures Database sets c2 (KEGG gene sets, c2.all.v6.2.symbols.gmt), c5 [Gene Ontology (GO) gene sets, c5.all.v6.2.symbols.gmt] and c6 (oncogenic signatures gene sets, c6.all.v6.2.symbols.gmt) (21). The permutation number was set to 1,000. Those enrichment gene sets revealed by GSEA as exhibiting a nominal P<0.05 and a false discovery rate (FDR) <0.25 were considered to indicate a statistically significant difference. The default parameters were used in GSEA software.
Statistical analysis
Statistical analysis was performed using SPSS v.25.0 software (IBM Corp.) and R 3.3.5 (https://www.r-project.org/). Kaplan-Meier survival analysis and the log-rank test were used to calculate the OS and P-values for all associations. The Cox proportional hazards regression model was used for univariate and multivariate survival analyses. HRs and 95% CIs were calculated using the Cox proportional hazards regression model with adjustment for influential clinical characteristics such as race, sex, age, Tumor-Node-Metastasis stage (22) and body mass index. FDRs in the GSEA were adjusted for multiple testing with the Benjamini-Hochberg procedure to control the FDR (23,24). P<0.05 was considered to indicate a statistically significant difference. Vertical scatter plots and survival curves were generated in GraphPad Prism v.7.0 (GraphPad Software, Inc.).
Results
Clinicopathological characteristics of patients with SKCM
The clinicopathological data of the 458 patients included in the present study are presented in Table I. Race, age and tumor stage were significantly associated with median survival time (P=0.003, P<0.001 and P<0.001, respectively; Table I). After normalization, only JAK1, STAT1, STAT3, STAT4, STAT5B and STAT6 genes mRNA expression were observed.
Table I.
Survival analysis based on clinical information.
| Characteristic | Patients (n=458) | No. of events (%) | MST (days) | HR (95% CI) | Crude P-value |
|---|---|---|---|---|---|
| Race | 0.003a | ||||
| Caucasian | 435 | 210 (48.3) | 2,454 | 0.337 (0.166–0.687) Ref. | |
| Other | 13 | 8 (61.5) | 636 | ||
| Unknown | 10 | ||||
| Sex | 0.345 | ||||
| Male | 284 | 146 (51.4) | 2,421 | Ref. 0.872 (0.657–1.158) | |
| Female | 174 | 73 (42.0) | 2,367 | ||
| Age (years) | <0.001a | ||||
| ≤60 | 239 | 115 (48.1) | 3,196 | 0.587 (0.445–0.773) Ref. | |
| >60 | 219 | 104 (47.5) | 1,864 | ||
| Tumor stageb | <0.001a | ||||
| Early | 231 | 108 (46.8) | 3,195 | 0.547 (0.411–0.728) Ref. | |
| Advanced | 191 | 96 (50.3) | 1,927 | ||
| Unknown | 36 |
P<0.05.
Due to the small sample size of certain tumor stages, two groups were used to reduce the probability of statistical errors. MST, median survival time; HR, hazard ratio; CI, confidence interval; Ref., reference group.
Bioinformatics analysis of JAK and STAT genes
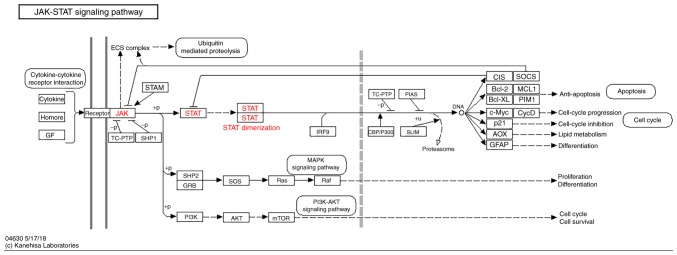
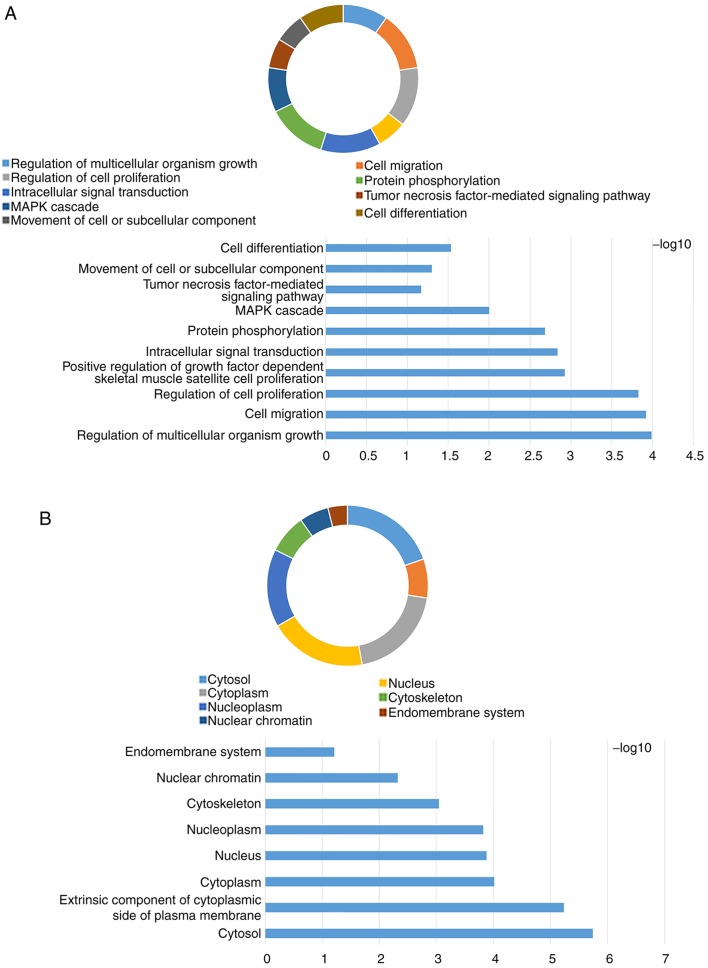
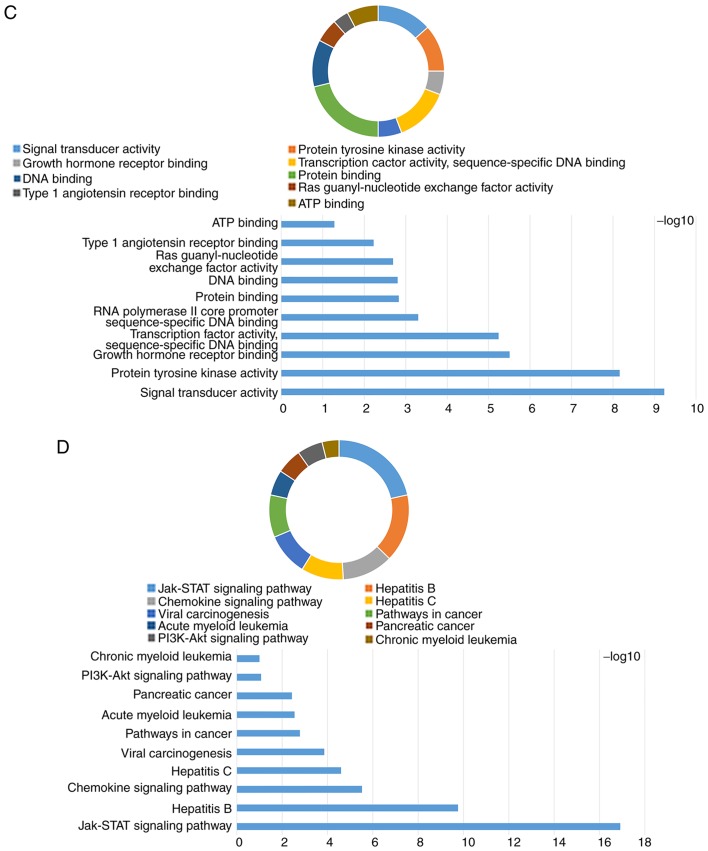
The KEGG pathway map of the JAK-STAT signaling pathway is represented in Fig. 1 (KEGG map no. 04630; http://www.genome.jp/dbget-bin/www_bget?pathway:map04630). The GO term and KEGG enrichment analysis for JAK and STAT genes included BP (Figs. S1A and S2A), CC (Figs. S1B and S2B), MF (Figs. S1C and S2C) and KEGG (Figs. S1D and S2D). The DAVID result for the combination of JAK and STAT gene families suggested that the JAK and STAT signaling pathways were associated with cell migration, the mitogen-activated protein kinase (MAPK) cascade, cell differentiation (Fig. 2A), cytosol, cytoplasm (Fig. 2B), DNA binding, ATP binding, signal transducer activity (Fig. 2C), the PI3K-AKT signaling pathway and the pancreatic cancer signaling pathway (Fig. 2D).
Figure 1.
KEGG pathway map of the JAK-STAT signaling pathway obtained from the KEGG website. KEGG, Kyoto Encyclopedia of Genes and Genomes; JAK, Janus kinase; STAT, signal transducer and activator of transcription.
Figure 2.
Analysis of enriched GO terms and KEGG pathways for JAK and STAT genes obtained using Database for Annotation, Visualization and Integrated Discovery. (A) Biological process results of GO functional enrichment analysis. (B) Cellular component results of GO functional enrichment analysis. (C) Molecular function results of GO functional enrichment analysis. (D) KEGG pathway analysis results. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; JAK, Janus kinase; STAT, signal transducer and activator of transcription; MAPK, mitogen-activated protein kinase.
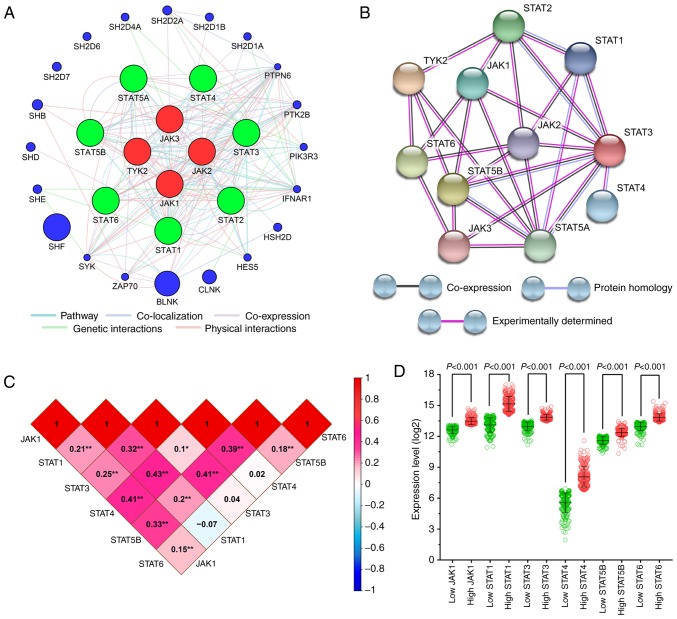
Gene interaction and association analysis
The gene-gene interaction network was analyzed separately in the JAK family (Fig. S3) and in the STAT family (Fig. S4), as well as in their combination (Fig. 3A). The protein-protein interaction network is presented in Fig. 3B.
Figure 3.
Correlation and association analysis for JAK and STAT genes in SKCM (A) Gene interaction network of JAK and STAT genes generated by GeneMANIA. (B) Protein-protein interaction network generated using the STRING database. (C) Pearson's correlation coefficients for JAK1, STAT1, STAT3, STAT4, STAT5B and STAT6 gene expression levels. (D) Scatter plots of JAK1, STAT1, STAT3, STAT4, STAT5B and STAT6 gene expression level in The Cancer Genome Atlas database. GeneMANIA, gene multiple association network integration algorithm; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; STAT, signal transducer and activator of transcription; SKCM, skin cutaneous melanoma; error bars in the scatter plots, mean ± SD.
Pearson's correlation coefficient was used to analyze the correlation between JAK and STAT genes in SKCM tissues based on the TCGA dataset (Fig. 3C). The results suggested that, with the exception of STAT6, the expression of STAT genes (STAT1, STAT3, STAT4 and STAT5B) in SKCM strongly correlated with each other.
Scatter plots of the expression of all JAK and STAT genes in TCGA dataset (using the median as the cut-off value) are presented in Fig. 3D. The difference between the high and low-expression groups was statistically significant (P<0.001).
Survival analysis
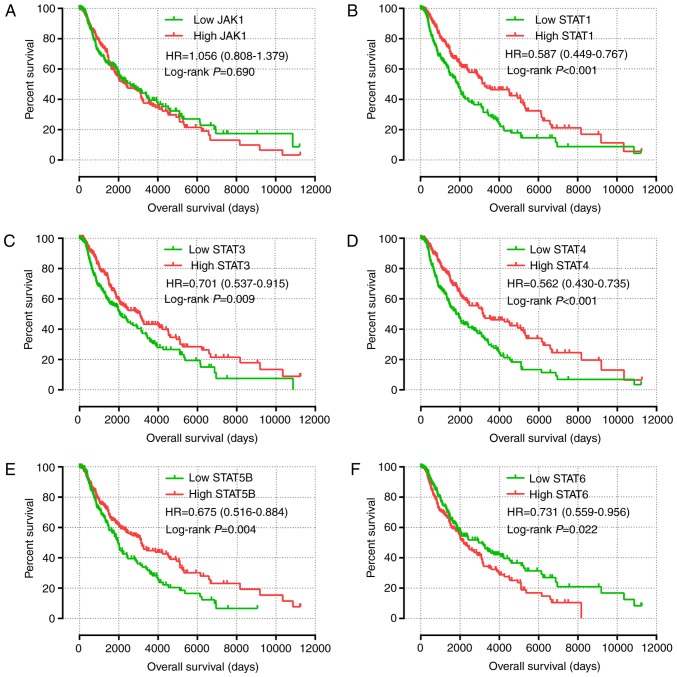
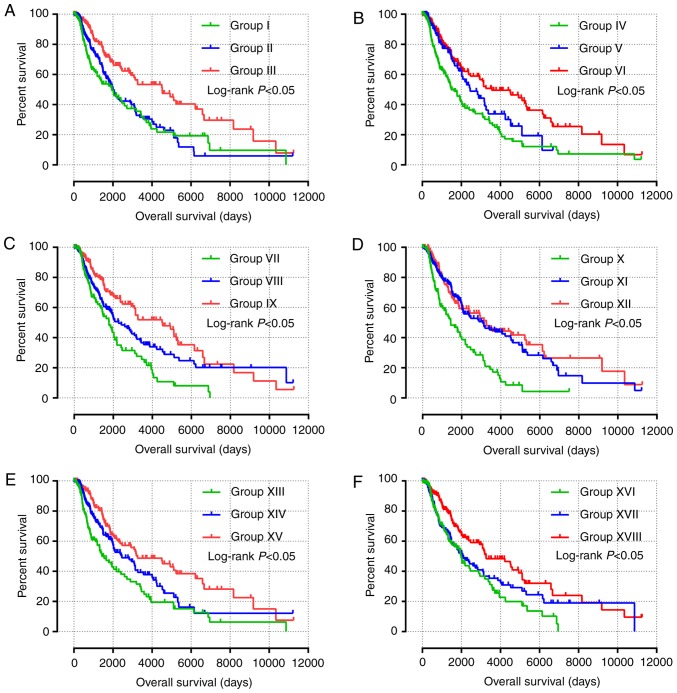
The results of univariate survival analysis of JAK and STAT genes are presented in Fig. 4 and Table II. High expression levels of STAT1, STAT3, STAT4 and STAT5B, and low expression levels of STAT6 were associated with a favorable prognosis (P<0.05). Multivariate Cox proportional hazards regression analysis identified that sex, race, age and tumor stage were associated with the prognosis of patients with SKCM. Multivariate survival analysis, in agreement with univariate survival analysis, demonstrated that high expression of STAT1, STAT3, STAT4 and STAT5B, and low expression of STAT6 was associated with a favorable prognosis (adjusted P<0.001; HR, 0.595; 95% CI, 0.455–0.778; adjusted P=0.018; HR, 0.725; 95% CI, 0.555–0.947; adjusted P<0.001; HR, 0.590; 95% CI, 0.450–0.773; adjusted P=0.007; HR, 0.690; 95% CI, 0.526–0.940; and adjusted P=0.026; HR, 0.737; 95% CI, 0.563–0.964, respectively; Table II).
Figure 4.
Prognostic value of the expression of key genes of the JAK-STAT signaling pathway for overall survival. (A-F) Kaplan-Meier survival plots for all patients with skin cutaneous melanoma according to (A) JAK1, (B) STAT1, (C) STAT3, (D) STAT4, (E) STAT5B and (F) STAT6 expression. JAK, Janus kinase; STAT, signal transducer and activator of transcription.
Table II.
Prognostic survival analysis based on high or low expression of JAK and STAT family genes
| Gene | Patients (n=458) | No. of events (%) | MST (days) | Crude HR (95% CI) | Crude P-value | Adjusted HRb (95% CI) | Adjusted P-valueb |
|---|---|---|---|---|---|---|---|
| JAK1 | |||||||
| Low | 229 | 99 (43.2) | 2,588 | Ref. 1.056 (0.808–1.379) | 0.690 | Ref. 0.950 (0.720–1.253) | 0.716 |
| High | 229 | 120 (52.4) | 2,365 | ||||
| STAT1 | |||||||
| Low | 229 | 119 (52.0) | 1,910 | Ref 0.587(0.449–0.767) | <0.001a | Ref. 0.595 (0.455–0.778) | <0.001a |
| High | 229 | 100 (47.3) | 3,259 | ||||
| STAT3 | |||||||
| High | 229 | 113 (49.3) | 2,030 | Ref. 0.701 (0.537–0.915) | 0.009a | Ref. 0.725 (0.555–0.947) | 0.018a |
| Low | 229 | 106 (46.3) | 3,080 | ||||
| STAT4 | |||||||
| Low | 229 | 121 (52.8) | 1,785 | Ref. 0.562 (0.430–0.735) | <0.001a | Ref. 0.590 (0.450–0.773) | <0.001a |
| High | 229 | 98 (42.8) | 3,176 | ||||
| STAT5B | |||||||
| Low | 229 | 117 (48.1) | 2,022 | Ref. 0.675 (0.516–0.884) | 0.004a | Ref. 0.690 (0.526–0.940) | 0.007a |
| High | 229 | 102 (47.4) | 3,139 | ||||
| STAT6 | |||||||
| High | 229 | 115 (50.4) | 2,184 | Ref. 0.731 (0.559–0.956) | 0.022a | Ref. 0.737 (0.563–0.964) | 0.026a |
| Low | 229 | 104 (47.2) | 3,139 | ||||
P<0.05.
Adjusted for age and tumor stage. JAK1, Janus kinase 1; STAT, signal transducer and activator of transcription; MST, median survival time; HR, hazard ratio; CI, confidence interval; Ref., reference.
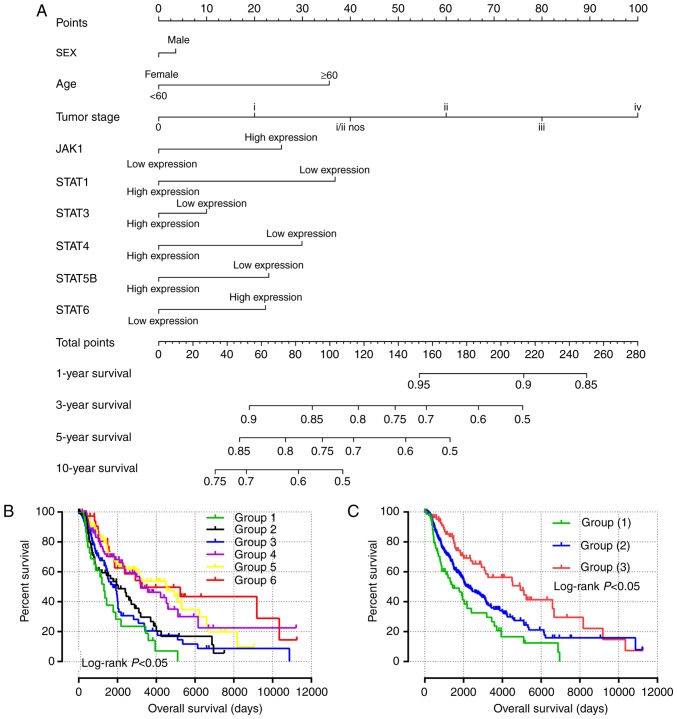
Predictive nomogram and joint effects analysis
Independent factors, including age, sex, tumor stage and mRNA expression, were integrated into the prognostic nomogram to predict the clinical outcomes of patients with SKCM (Fig. 5A). Age, tumor stage, and JAK1, STAT1, STAT4, STAT5B and STAT6 expression levels exhibited major contributions as prognostic signatures in the risk scores (range, 0–100).
Figure 5.
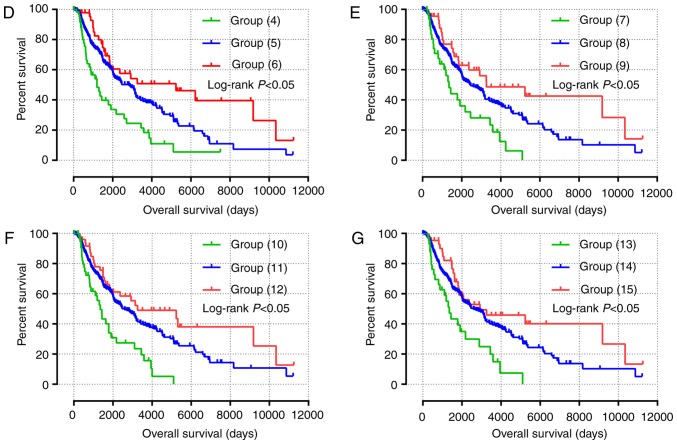
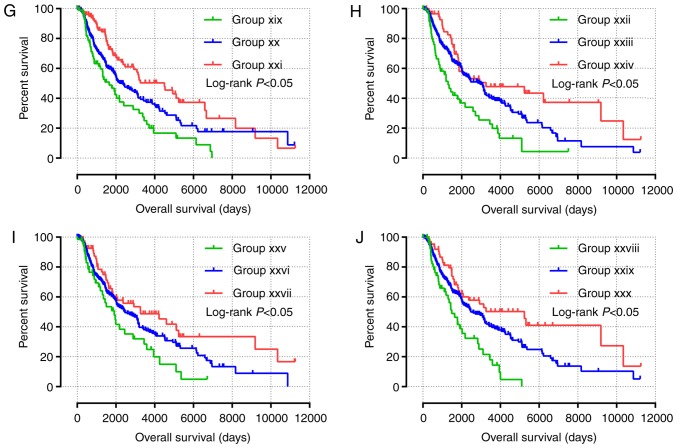
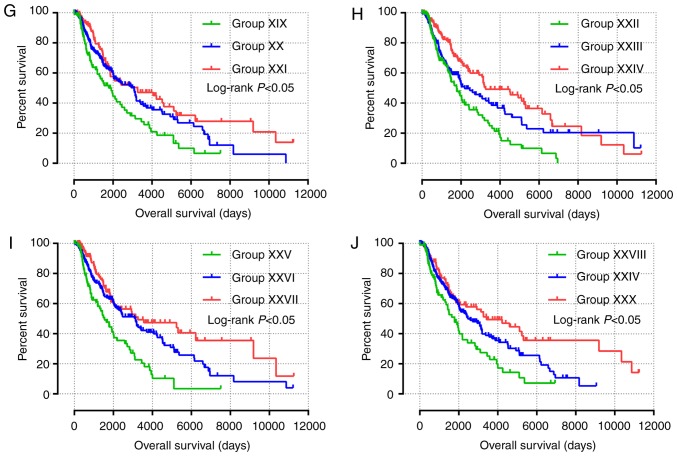
Nomogram and joint-effects survival analysis of JAK and STAT genes. (A) Nomogram for predicting the 1-, 3- and 5-year event (death) based on risk scores and clinical information. (B) Joint effects analysis of the influence of combined gene expression on overall survival in patients with skin cutaneous carcinoma. (C-G) Joint effects analysis of the influence of combined expression of four selected genes on overall survival in patients with skin cutaneous melanoma according to (C) group (1)-(3), (D) group (4)-(6), (E) group (7)-(9), (F) group (10)-(12) and (G) group (13)-(15). JAK1, Janus kinase 1; STAT, signal transducer and activator of transcription. NOS, non-specific.
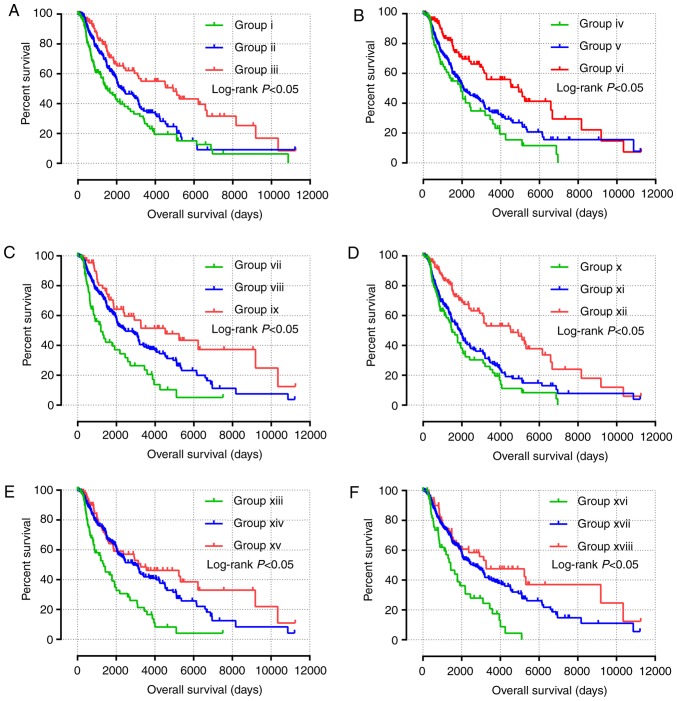
All survival related genes, including STAT1, STAT3, STAT4, STAT5B and STAT6, were selected and grouped to perform joint effects analysis. The grouping information of joint effects analysis is presented in Tables III–VI. The group number with simple Arabic numerals represented all gene selected groups (Table III); Arab numerals with brackets are 4 selected genes (Table IV); Lowercase Roman numerals are 3 selected genes (Table V); Capitalized Roman numerals are 2 selected genes (Table VI). The combination of genes with favorable prognosis had a better effect on the OS (univariate survival analysis; P<0.05; Table VII and Figs. 5B-G, 6A-J and 7A-J).
Table III.
Grouping information for joint effects analysis of all STAT genes.
| Composition | |||||
|---|---|---|---|---|---|
| Group | STAT1 | STAT3 | STAT4 | STAT5B | STAT6 |
| 1 | Low | Low | Low | Low | High |
| 2 | High | Low | Low | Low | High |
| Low | High | Low | Low | High | |
| Low | Low | High | Low | High | |
| Low | Low | Low | High | High | |
| Low | Low | Low | Low | Low | |
| 3 | High | High | Low | Low | High |
| High | Low | High | Low | High | |
| High | Low | Low | High | High | |
| High | Low | Low | Low | Low | |
| Low | High | High | Low | High | |
| Low | High | Low | High | High | |
| Low | High | Low | Low | Low | |
| Low | Low | High | High | High | |
| Low | Low | High | Low | Low | |
| Low | Low | Low | High | Low | |
| 4 | High | High | High | Low | High |
| High | High | Low | High | High | |
| High | Low | High | High | High | |
| Low | High | High | High | High | |
| High | High | Low | Low | Low | |
| High | Low | High | Low | Low | |
| Low | High | High | Low | Low | |
| High | Low | Low | High | Low | |
| Low | High | Low | High | Low | |
| Low | Low | High | High | Low | |
| 5 | High | High | High | High | High |
| High | High | High | Low | Low | |
| High | High | Low | High | Low | |
| High | Low | High | High | Low | |
| Low | High | High | High | Low | |
| 6 | High | High | High | High | Low |
Bold, favorable prognosis groups in univariate survival analysis. STAT, signal transducer and activator of transcription.
Table VI.
Grouping information for joint effects analysis of two selected STAT genes
| Composition | |||||
|---|---|---|---|---|---|
| Group | STAT1 | STAT3 | STAT4 | STAT5B | STAT6 |
| I | Low | Low | – | – | – |
| II | High | Low | – | – | – |
| Low | High | – | – | – | |
| III | High | High | – | – | – |
| IV | Low | – | Low | – | – |
| V | High | – | Low | – | – |
| Low | – | High | – | – | |
| VI | High | – | High | – | – |
| VII | Low | – | – | Low | – |
| VIII | Low | – | – | High | – |
| High | – | – | Low | – | |
| IX | High | – | – | High | – |
| X | Low | – | – | – | High |
| XI | Low | – | – | – | Low |
| High | – | – | – | High | |
| XII | High | – | – | – | Low |
| XIII | – | Low | Low | – | – |
| XIV | – | High | Low | – | – |
| – | Low | High | – | – | |
| XV | – | High | High | – | – |
| XVI | – | Low | – | Low | – |
| XVII | – | Low | – | High | – |
| – | High | – | Low | – | |
| XVIII | – | High | – | High | – |
| XIX | – | Low | – | – | High |
| XX | – | Low | – | – | Low |
| – | High | – | – | High | |
| XXI | – | High | – | – | Low |
| XXII | – | – | Low | Low | – |
| XXIII | – | – | Low | High | – |
| – | – | High | Low | – | |
| XXIV | – | – | High | High | – |
| XXV | – | – | Low | – | High |
| XXVI | – | – | Low | – | Low |
| – | – | High | – | High | |
| XXVII | – | – | High | – | Low |
| XXVIII | – | – | – | Low | High |
| XXIV | – | – | – | Low | Low |
| – | – | – | High | High | |
| XXX | – | – | – | High | Low |
‘−’, gene not selected. Bold, favorable prognosis groups in univariate survival analysis. STAT, signal transducer and activator of transcription.
Table IV.
Grouping information for joint effects analysis of four selected STAT genes.
| Composition | |||||
|---|---|---|---|---|---|
| Group | STAT1 | STAT3 | STAT4 | STAT5B | STAT6 |
| (1) | Low | Low | Low | Low | – |
| (2) | High | Low | Low | Low | – |
| Low | High | Low | Low | – | |
| Low | Low | High | Low | – | |
| Low | Low | Low | High | – | |
| High | High | Low | Low | – | |
| High | Low | High | Low | – | |
| High | Low | Low | High | – | |
| Low | High | High | Low | – | |
| Low | High | Low | High | – | |
| Low | Low | High | High | – | |
| High | High | High | Low | – | |
| High | High | Low | High | – | |
| High | Low | High | High | – | |
| Low | High | High | High | – | |
| (3) | High | High | High | High | – |
| (4) | Low | Low | Low | – | High |
| (5) | High | Low | Low | – | High |
| Low | High | Low | – | High | |
| Low | Low | High | – | High | |
| Low | Low | Low | – | Low | |
| High | High | Low | – | High | |
| High | Low | High | – | High | |
| High | Low | Low | – | Low | |
| Low | High | High | – | High | |
| Low | High | Low | – | Low | |
| Low | Low | High | – | Low | |
| High | High | High | – | High | |
| High | High | Low | – | Low | |
| High | Low | High | – | Low | |
| Low | High | High | – | Low | |
| (6) | High | High | High | – | Low |
| (7) | Low | Low | – | Low | High |
| (8) | High | Low | – | Low | High |
| Low | High | – | Low | High | |
| Low | Low | – | High | High | |
| Low | Low | – | Low | Low | |
| High | High | – | Low | High | |
| High | Low | – | High | High | |
| High | Low | – | Low | Low | |
| Low | High | – | High | High | |
| Low | High | – | Low | Low | |
| Low | Low | – | High | Low | |
| High | High | – | High | High | |
| High | High | – | Low | Low | |
| High | Low | – | High | Low | |
| Low | High | – | High | Low | |
| (9) | High | High | – | High | Low |
| (10) | Low | – | Low | Low | High |
| (11) | High | – | Low | Low | High |
| Low | – | High | Low | High | |
| Low | – | Low | High | High | |
| Low | – | Low | Low | Low | |
| High | – | High | Low | High | |
| High | – | Low | High | High | |
| High | – | Low | Low | Low | |
| Low | – | High | High | High | |
| Low | – | High | Low | Low | |
| Low | – | Low | High | Low | |
| High | – | High | High | High | |
| High | – | High | Low | Low | |
| High | – | Low | High | Low | |
| Low | – | High | High | Low | |
| (12) | High | – | High | High | Low |
| (13) | – | Low | Low | Low | High |
| (14) | – | High | Low | Low | High |
| – | Low | High | Low | High | |
| – | Low | Low | High | High | |
| – | Low | Low | Low | Low | |
| – | High | High | Low | High | |
| – | High | Low | High | High | |
| – | High | Low | Low | Low | |
| – | Low | High | High | High | |
| – | Low | High | Low | Low | |
| – | Low | Low | High | Low | |
| – | High | High | High | High | |
| – | High | High | Low | Low | |
| – | High | Low | High | Low | |
| – | Low | High | High | Low | |
| (15) | – | High | High | High | Low |
‘−’, gene not selected. Bold, favorable prognosis groups in univariate survival analysis. STAT, signal transducer and activator of transcription.
Table V.
Grouping information for joint effects analysis of three selected STAT genes.
| Composition | |||||
|---|---|---|---|---|---|
| Group | STAT1 | STAT3 | STAT4 | STAT5B | STAT6 |
| i | Low | Low | Low | – | – |
| ii | High | Low | Low | – | – |
| Low | High | Low | – | – | |
| Low | Low | High | – | – | |
| High | High | Low | – | – | |
| High | Low | High | – | – | |
| Low | High | High | – | – | |
| iii | High | High | High | – | – |
| iv | Low | Low | – | Low | – |
| v | Low | Low | – | Low | – |
| High | High | – | Low | – | |
| High | High | – | High | – | |
| Low | High | – | Low | – | |
| Low | Low | – | High | – | |
| High | High | – | High | – | |
| vi | High | High | – | High | – |
| vii | Low | Low | – | – | High |
| viii | High | Low | – | – | High |
| Low | High | – | – | High | |
| Low | Low | – | – | Low | |
| High | High | – | – | High | |
| High | Low | – | – | Low | |
| Low | High | – | – | Low | |
| ix | High | High | – | – | Low |
| x | Low | – | Low | Low | – |
| xi | High | – | Low | Low | – |
| Low | – | High | Low | – | |
| Low | – | Low | High | – | |
| High | – | High | Low | – | |
| High | – | Low | High | – | |
| Low | – | High | High | – | |
| xii | High | – | High | High | – |
| xiii | Low | – | Low | – | High |
| xiv | High | – | Low | – | High |
| Low | – | High | – | High | |
| Low | – | Low | – | Low | |
| High | – | High | – | High | |
| High | – | Low | – | Low | |
| Low | – | High | – | Low | |
| xv | High | – | High | – | Low |
| xvi | Low | – | – | Low | High |
| xvii | High | – | – | Low | High |
| Low | – | – | High | High | |
| Low | – | – | Low | Low | |
| High | – | – | High | High | |
| High | – | – | Low | Low | |
| Low | – | – | High | Low | |
| xviii | High | – | – | High | Low |
| xiv | – | Low | Low | Low | – |
| xx | – | High | Low | Low | – |
| – | Low | High | High | – | |
| – | Low | Low | Low | – | |
| – | High | High | High | – | |
| – | High | Low | Low | – | |
| – | Low | High | High | – | |
| xxi | – | High | High | High | – |
| xxii | – | Low | Low | – | High |
| xxiii | – | High | Low | – | High |
| – | Low | High | – | High | |
| – | Low | Low | – | Low | |
| – | High | High | – | High | |
| – | High | Low | – | Low | |
| – | Low | High | – | Low | |
| xxiv | – | High | High | – | Low |
| xxv | – | Low | – | Low | High |
| xxvi | – | High | – | High | High |
| – | Low | – | Low | High | |
| – | Low | – | Low | Low | |
| – | High | – | High | High | |
| – | High | – | High | Low | |
| – | Low | – | Low | Low | |
| xxvii | – | High | – | High | Low |
| xxviii | – | – | Low | Low | High |
| xxix | – | – | High | High | High |
| – | – | Low | Low | High | |
| – | – | Low | Low | Low | |
| – | – | High | High | High | |
| – | – | High | High | Low | |
| – | – | Low | Low | Low | |
| xxx | – | – | High | High | Low |
‘−’, gene not selected. Bold, favorable prognosis groups in univariate survival analysis. STAT, signal transducer and activator of transcription.
Table VII.
Joint effects analysis of the prognostic value of combinations of gene expression in skin cutaneous melanoma.
| Group | No. of genes | Patients (n=458) | No. of events (%) | MST (days) | Log-rank P-value | HR (95% CI) |
|---|---|---|---|---|---|---|
| 1 | 5 | 38 | 24 (63.1) | 1,301 | Ref. | Ref. |
| 2 | 5 | 87 | 46 (52.9) | 2,030 | 0.073 | 0.636 (0.487–1.043) |
| 3 | 5 | 106 | 53 (50.0) | 1,766 | 0.094 | 0.661 (0.407–1.073) |
| 4 | 5 | 98 | 37 (37.8) | 3,196 | <0.001a | 0.352 (0.210–0.590) |
| 5 | 5 | 93 | 42 (45.2) | 4,533 | <0.001a | 0.322 (0.194–0.535) |
| 6 | 5 | 36 | 17 (47.2) | 3,259 | <0.001a | 0.280 (0.149–0.528) |
| (1) | 4 | 71 | 42 (59.2) | 1,441 | Ref. | Ref. |
| (2) | 4 | 301 | 140 (46.4) | 2,192 | 0.014a | 0.648 (0.458–0.915) |
| (3) | 4 | 86 | 37 (43.0) | 4,930 | <0.001a | 0.376 (0.241–0588) |
| (4) | 4 | 61 | 37 (60.7) | 1,197 | Ref. | Ref. |
| (5) | 4 | 351 | 160 (45.6) | 2,829 | <0.001a | 0.515 (0.360–0.738) |
| (6) | 4 | 46 | 22 (47.8) | 5,237 | <0.001a | 0.312 (0.182–0.535) |
| (7) | 4 | 47 | 27 (57.4) | 1,354 | Ref. | Ref. |
| (8) | 4 | 365 | 171 (46.8) | 2,470 | 0.002a | 0.530 (0.352–0.798) |
| (9) | 4 | 46 | 21 (45.7) | 3,266 | <0.001a | 0.370 (0.189–0.601) |
| (10) | 4 | 56 | 34 (60.7) | 1,354 | Ref. | Ref. |
| (11) | 4 | 351 | 160 (45.6) | 2,588 | <0.001a | 0.484 (0.333–0.704) |
| (12) | 4 | 51 | 25 (49.0) | 3,259 | <0.001a | 0.338 (0.200–0.573) |
| (13) | 4 | 42 | 26 (61.9) | 1,354 | Ref. | Ref. |
| (14) | 4 | 372 | 171 (46.0) | 2,588 | 0.001a | 0.498 (0.328–0.755) |
| (15) | 4 | 44 | 22 (50.0) | 2,927 | <0.001a | 0.344 (0.193–0.612) |
| i | 3 | 130 | 75 (57.7) | 1,441 | Ref. | Ref. |
| ii | 3 | 250 | 112 (44.8) | 2,273 | 0.022a | 0.710 (0.529–0.952) |
| iii | 3 | 103 | 46 (44.7) | 4,930 | <0.001a | 0.418 (0.289–0.605) |
| iv | 3 | 91 | 47 (51.6) | 1,949 | Ref. | Ref. |
| v | 3 | 267 | 131 (49.1) | 2,071 | 0.086 | 0.746 (0.534–1.042) |
| vi | 3 | 125 | 51 (40.8) | 4,930 | <0.001a | 0.415 (0.278–0.620) |
| vii | 3 | 78 | 44 (56.4) | 1,197 | Ref. | Ref. |
| viii | 3 | 312 | 146 (46.8) | 2,470 | <0.001a | 0.530 (0.377–0.744) |
| ix | 3 | 93 | 39 (41.9) | 4,526 | <0.001a | 0.328 (0.212–0.508) |
| x | 3 | 103 | 61 (59.2) | 1,487 | Ref. | Ref. |
| xi | 3 | 245 | 127 (51.8) | 1,864 | 0.127 | 0.788 (0.580–1.070) |
| xii | 3 | 135 | 60 (44.4) | 4,533 | <0.001a | 0.388 (0.270–0.557) |
| xiii | 3 | 95 | 55 (57.9) | 1,354 | Ref. | Ref. |
| xiv | 3 | 276 | 152 (551) | 2,889 | <0.001a | 0.480 (0.348–0.661) |
| xv | 3 | 112 | 50 (44.6) | 3,259 | <0.001a | 0.367 (0.249–0.542) |
| xvi | 3 | 69 | 40 (58.0) | 1,429 | Ref. | Ref. |
| xvii | 3 | 324 | 148 (45.7) | 2,588 | <0.001a | 0.502 (0.352–0.715) |
| xviii | 3 | 90 | 40 (44.4) | 3,266 | <0.001a | 0.358 (0.229–0.559) |
| xix | 3 | 87 | 51 (58.6) | 1,655 | Ref. | Ref. |
| xx | 3 | 268 | 121 (45.1) | 2,367 | 0.012a | 0.658 (0.474–0.913) |
| xxi | 3 | 128 | 57 (44.5) | 4,507 | <0.001a | 0.419 (0.286–0.613) |
| xxii | 3 | 70 | 43 (61.4) | 1,301 | Ref. | Ref. |
| xxiii | 3 | 328 | 148 (45.1) | 2,948 | <0.001a | 0.530 (0.377–0.745) |
| xxiv | 3 | 85 | 38 (44.7) | 3,259 | <0.001a | 0.348 (0.223–0.542) |
| xxv | 3 | 66 | 35 (53.0) | 1,910 | Ref. | Ref. |
| xxvi | 3 | 323 | 150 (46.4) | 2,470 | 0.043a | 0.683 (0.472–0.988) |
| xxvii | 3 | 94 | 44 (46.8) | 3,266 | 0.001a | 0.462 (0.294–0.724) |
| xxviii | 3 | 70 | 40 (57.1) | 1,486 | Ref. | Ref. |
| xxix | 3 | 323 | 149 (46.1) | 2,470 | 0.001a | 0.536 (0.376–0.764) |
| xxx | 3 | 90 | 40 (44.4) | 5,237 | <0.001a | 0.349 (0.223–0.545) |
| I | 2 | 139 | 70 (50.4) | 1,949 | Ref. | Ref. |
| II | 2 | 180 | 92 (51.1) | 2,005 | 0.519 | 0.902 (0.660–1.233) |
| III | 2 | 164 | 67 (40.9) | 4,526 | <0.001a | 0.485 (0.346–0.678) |
| IV | 2 | 168 | 96 (57.1) | 1,487 | Ref. | Ref. |
| V | 2 | 122 | 48 (39.3) | 2,454 | 0.017a | 0.655 (0.462–0.928) |
| VI | 2 | 193 | 85 (44.0) | 3,564 | <0.001a | 0.477 (0.356–0.641) |
| VII | 2 | 133 | 70 (52.6) | 1,780 | Ref. | Ref. |
| VIII | 2 | 192 | 90 (46.9) | 2,273 | 0.008a | 0.654 (0.477–0.897) |
| IX | 2 | 133 | 59 (44.4) | 4,507 | <0.001a | 0.449 (0.316–0.638) |
| X | 2 | 125 | 70 (56.0) | 1,438 | Ref. | Ref. |
| XI | 2 | 208 | 94 (45.2) | 3,080 | <0.001a | 0.483 (0.353–0.662) |
| XII | 2 | 125 | 55 (44.0) | 3,259 | <0.001a | 0.435 (0.304–0.623) |
| XIII | 2 | 130 | 75 (57.7) | 1,441 | Ref. | Ref. |
| XIV | 2 | 198 | 84 (42.4) | 2,545 | 0.011a | 0.677 (0.488–0.912) |
| XV | 2 | 130 | 60 (46.2) | 3,259 | <0.001a | 0.466 (0.332–0.656) |
| XVI | 2 | 148 | 72 (48.6) | 1,992 | Ref. | Ref. |
| XVII | 2 | 162 | 80 (49.4) | 2,071 | 0.297 | 0.843 (0.612–1.162) |
| XVIII | 2 | 148 | 67 (45.3) | 3,259 | 0.001a | 0.564 (0.403–0.791) |
| XIX | 2 | 111 | 61 (54.9) | 1,910 | Ref. | Ref. |
| XX | 2 | 236 | 106 (44.9) | 3,080 | 0.008a | 0.652 (0.476–0.894) |
| XXI | 2 | 111 | 52 (46.8) | 3,259 | <0.001a | 0.508 (0.349–0.739) |
| XXII | 2 | 141 | 77 (54.6) | 1,780 | Ref. | Ref. |
| XXIII | 2 | 176 | 78 (44.3) | 2,192 | 0.015a | 0.674 (0.490–0.926) |
| XXIV | 2 | 141 | 64 (45.4) | 3,259 | <0.001a | 0.466 (0.333–0.653) |
| XXV | 2 | 118 | 65 (55.1) | 1,544 | Ref. | Ref. |
| XXVI | 2 | 222 | 106 (47.4) | 3,136 | <0.001a | 0.548 (0.400–0.750) |
| XXVII | 2 | 118 | 48 (40.7) | 3,259 | <0.001a | 0.399 (0.273–0.583) |
| XXVIII | 2 | 105 | 56 (53.3) | 1,780 | Ref. | Ref. |
| XXIX | 2 | 248 | 114 (46.0) | 2,470 | 0.013a | 0.667 (0.484–0.920) |
| XXX | 2 | 105 | 49 (46.7) | 3,424 | <0.001a | 0.466 (0.314–0.692) |
P<0.05. MST, median survival time; HR, hazard ratio; CI, confidence interval; Ref., reference.
Figure 6.
(A-J) Joint effects analysis of the effects of combined expression of three selected genes on overall survival in patients with skin cutaneous melanoma according to (A) group i-iii, (B) group iv-vi, (C) group vi-ix, (D) group x-xii, (E) group xiii-xv, (F) group xvi-xvii, (G) group xix-xxi, (H) group xxii-xxiv, (I) group xxv-xxvii and (J) group xxviii-xxx.
Figure 7.
(A-J) Joint effects analysis of the influence of combined expression of two selected genes on overall survival in patients with skin cutaneous melanoma according to (A) group I–III, (B) group IV–VI, (C) group VI–IX, (D) group X–XII, (E) group XIII–XV, (F) group XVI–XVII, (G) group XIX–XXI, (H) group XXII–XXIV, (I) group XXV–XXVII and (J) group XXVIII–XXX.
GSEA
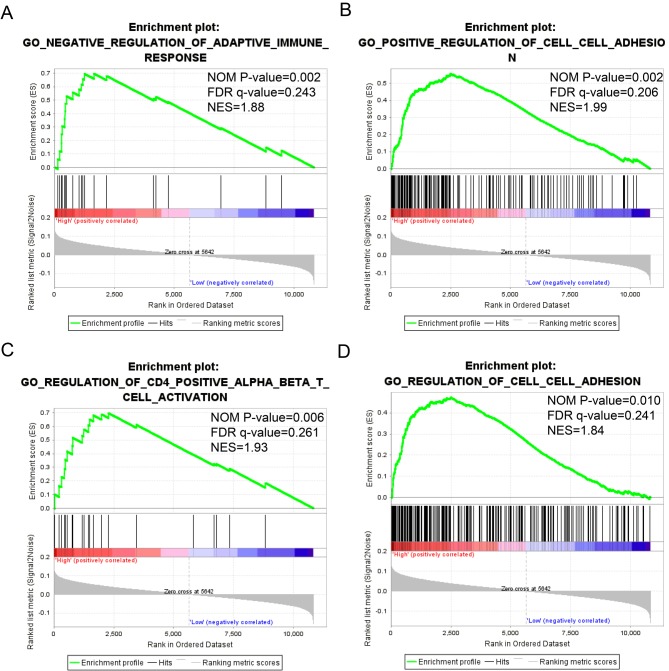
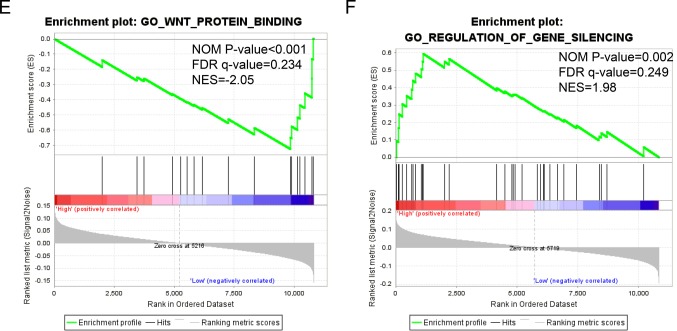
The detailed GSEA results, including KEGG, GO and oncogenic signatures, are shown in Tables SI, SII and SIII, respectively, and in Fig. 8. High expression of STAT1 was significantly associated with immune response (normalized P=0.002; FDR=0.243; Fig. 8A), cell adhesion (normalized P=0.002; FDR=0.206; Fig. 8B; normalized P=0.010; FDR=0.241; Fig. 8D) and WNT protein binging (normalized P=0.006; FDR=0.261; Fig. 8C). By contrast, low expression of STAT4 was associated with WNT protein binding (normalized P=0.001; FDR=−0.234; Fig. 8E), whereas high expression of STAT5B was associated with gene silencing (normalized P=0.002; FDR=0.249; Fig. 8F).
Figure 8.
GSEA of genes expressed in patients with skin cutaneous melanoma. (A-D) GSEA of the c5 reference gene sets for the high STAT1 expression group. (A) c5 item negative regulation of adaptive immune response, (B) c5 item positive regulation of cell-cell adhesion, (C) c5 item regulation of CD4 positive alpha beta T cell activation and (D) c5 item regulation of cell-cell adhesion. (E) GSEA of the c5 reference gene sets for the low STAT4 expression group. (F) GSEA of the c5 reference gene sets for the high STAT5B expression group. STAT, signal transducer and activator of transcription; GSEA, gene set enrichment analysis; NES, normalized enrichment score; FDR, false discovery rate; NOM, nominal.
Discussion
In the Surveillance, Epidemiology, and End Results Program database, the 5-year relative survival is 98% for prostate cancer, 89.9% for breast cancer and 19.4% for lung cancer (3,4). For melanoma, a 91.8% 5-year relative survival rate appears satisfactory; however, the 5-year relative survival for patients with tumor stage IV is only 3% (22,25).
The JAK-STAT signaling pathway serves a crucial role in functions such as cell proliferation, differentiation, migration and apoptosis, cell survival in hematopoiesis, immune cell development, stem cell maintenance and organismal growth processes (26–28). Dysfunction in the JAK-STAT signaling pathway is associated with diseases such as cancer and immune disorders (6,27). In the JAK-STAT signaling pathway, the JAK family comprises JAK1, JAK2, JAK3 and TYK2, and the STAT family comprises STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6 (6,29). A number of diseases are associated with the expression levels of genes of the JAK-STAT signaling pathway. A previous study has demonstrated that increased expression of STAT3 or STAT1, but not of both, was present in melanoma compared with that in benign nevi or skin melanocytes (30). In addition, active STAT3 was present in melanoma (30). By contrast, overexpression of microRNA (miR)-214 in a lung cancer cell line reduced the expression of JAK1, which inhibited cell proliferation and colony formation, and suppressed cell migration and invasion (31). In papillary thyroid cancer, downregulation of JAK1 by miR-520a-3p inactivated the JAK-STAT signaling pathway (32).
However, studies that explored the association between the prognosis of melanoma and the JAK-STAT signaling pathway are limited. In the present study, the expression of JAK and STAT family genes in melanoma was investigated based on TCGA data. The observation that JAK-STAT gene expression is associated with the MAPK signaling pathway is in agreement with a previous study, which demonstrated that the JAK-STAT signaling pathway is integrated with the MAPK signaling pathway (33–35) and is associated with melanoma (36). Expression of JAK2, JAK3, TYK2, STAT2 and STAT5B was not observed after normalizing mRNA expression in the present study, suggesting that these genes were expressed at a low level. High expression of STAT1, STAT3, STAT4 and STAT5B, as well as low expression of STAT6, were associated with a favorable prognosis of patients with SKCM. These results are consistent with other studies, in which high expression of STAT1 was associated with favorable prognosis in high-grade serous ovarian cancer (HGSC) (37), colorectal cancer (38) and esophageal squamous cell carcinoma (39). In addition, high expression of STAT1 in HGSC was significantly associated with the recruitment of intraepithelial CD8+ T cells, which enhanced the prognostic and predictive value of intratumoral CD8+ T cells in HGSC (40), potentially due to tumors with high STAT1 mRNA expression exhibiting elevated expression of genes specific for tumor-associated macrophages and immunosuppressive T lymphocytes (7). The results of the enrichment analysis in the present study also revealed that STAT1 was associated with the immune response, which suggested that STAT1 may accelerate the immune cell response to cancer (41). However, high STAT1 expression was associated with poor prognosis in glioblastoma (42), and breast (7,8), ovarian, lung, blood and brain (9) cancer.
In contrast to that of STAT1, STAT3 expression is downregulated in malignant pleural mesothelioma (43); however, the prognostic value of this association has not been reported. The majority of studies on STAT3 and cancer prognosis have demonstrated that phosphorylated STAT3 is associated with a poor outcome in colorectal cancer (10), multiple myeloma (44) and urothelial carcinoma (45). In addition, STAT4 has been detected in several types of cancer, including prostate (46), breast (47), gastric (48) and ovarian (49) cancer. The upregulation of STAT4 in hepatocellular carcinoma is associated with favorable prognosis, possibly due to the expression of STAT4 in the immune cells; however, the function and mechanism of STAT4 in non-immune cells remains unknown (50). High expression of STAT5B in astrocytoma is associated with poor prognosis (51), whereas high expression of STAT6 is associated with poor prognosis in colon cancer (52).
The potential effects of these associations require further exploration. For instance, colorectal carcinoma cell lines exhibiting low STAT1 and high STAT3 expression levels are associated with enhanced tumor growth in xenografts; by contrast, xenograft cell lines with high STAT1 and low STAT3 levels grew slowly (53). Thus, gene interactions may influence the cancer outcome. The different expression level of these two genes in different types of cancer may serve different roles. Joint effects analysis in the present study suggested that any combination of the tested markers may have a higher prognostic value compared with that of any individual biomarker.
The STAT gene family affects cell differentiation (54), invasion (55–57), adhesion (58) and migration (59), as well as cell cycle (60–62) and colony formation (39,63) through the JAK-STAT signaling pathway; these processes are associated with the occurrence, development and outcome of cancer. The potential mechanism of how these genes affect prognosis should be further studied.
The present study investigated the prognostic value of the key genes in the JAK-STAT signaling pathway; only STAT genes were demonstrated to affect the prognosis of SKCM. In melanoma research, no golden standard exists for diagnosis or prognosis that would serve a similar role as the hairy-related 2 gene in breast cancer or prostate specific antigen in prostate cancer. Certain genes may act as biomarkers to predict the prognosis and mechanism of SKCM, including RAF (Raf proto-oncogene), MEK (MAP/ERK kinase), MAPK, RAS, myelocytomatosis oncogene and S100 calcium binding protein (64–69). However, further research is required to identify a golden standard for predicting melanoma.
There were certain limitations in the present study. Firstly, only JAK expression data were reported following normalization; additional mRNA expression data are needed to further confirm these observations. Secondly, this is an association study. Further research is needed to explore the function and mechanism of the genes of the JAK-STAT signaling pathway identified in the present study in patients with SKCM. Another limitation of the current study is the limited sample size. Improved design and larger sample size studies are necessary to validate these results. Finally, there were no expression standards to measure whether the gene expression was high or low.
To the best of our knowledge, the present study is the first to evaluate the association between the expression of genes of the JAK-STAT signaling pathway and OS in patients with SKCM, and to identify the joint effects of prognostic values among the five identified genes. Overall, the results of the present study provided a novel insight into the function of these genes in SKCM clinical outcomes, and may be further utilized in the clinic for predicting the prognosis of SKCM.
In conclusion, the combination of the highly expressed STAT1, STAT3, STAT4 and STAT5B genes, and the lowly expressed STAT6 gene is associated with a favorable prognosis in patients with SKCM, and may be used as a novel biomarker for predicting the prognosis of patients with SKCM. The expression of the genes in the STAT family may affect the prognosis of SKCM by accelerating the immune response and immune cell activity, and by involving the MAPK signaling pathway. Further studies are required to validate these findings.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Nature Science Foundation of China (grant no. 81760344).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the TCGA repository, https://portal.gdc.cancer.gov/.
Authors' contributions
XZ and FP conceived and designed the study. QW, HM and SL processed the data and performed the statistical analysis. LZ, RH, XW and XL wrote and revised the manuscript and helped to perform the analysis and interpretation of data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study did not involve human or animal subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Goldstein BG, Goldstein AO. Diagnosis and management of malignant melanoma. Am Fam Physician. 2001;63:1359–1368. [PubMed] [Google Scholar]
- 2.Situm M, Buljan M, Kolić M, Vučić M. Melanoma-clinical, dermatoscopical, and histopathological morphological characteristics. Acta Dermatovenerol Croat. 2014;22:1–12. [PubMed] [Google Scholar]
- 3.Grimaldi AM, Simeone E, Ascierto PA. The role of MEK inhibitors in the treatment of metastatic melanoma. Curr Opin Oncol. 2014;26:196–203. doi: 10.1097/CCO.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 7.Tymoszuk P, Charoentong P, Hackl H, Spilka R, Müller-Holzner E, Trajanoski Z, Obrist P, Revillion F, Peyrat JP, Fiegl H, Doppler W. High STAT1 mRNA levels but not its tyrosine phosphorylation are associated with macrophage infiltration and bad prognosis in breast cancer. BMC Cancer. 2014;14:257. doi: 10.1186/1471-2407-14-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Huang J, Li W, Chen Y, Liu X, Wang J. Meta-analysis of STAT3 and phospho-STAT3 expression and survival of patients with breast cancer. Oncotarget. 2018;9:13060–13067. doi: 10.18632/oncotarget.23962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui X, Jing X, Yi Q, Long C, Tan B, Li X, Chen X, Huang Y, Xiang Z, Tian J, Zhu J. Systematic analysis of gene expression alterations and clinical outcomes of STAT3 in cancer. Oncotarget. 2018;9:3198–3213. doi: 10.18632/oncotarget.23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han C, Sun B, Zhao X, Zhang Y, Gu Q, Liu F, Zhao N, Wu L. Phosphorylation of STAT3 promotes vasculogenic mimicry by inducing epithelial-to-mesenchymal transition in colorectal cancer. Technol Cancer Res Treat. 2017;16:1209–1219. doi: 10.1177/1533034617742312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47((D1)):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45((D1)):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 24.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 25.Gerami P, Cook RW, Wilkinson J, Russell MC, Dhillon N, Amaria RN, Gonzalez R, Lyle S, Johnson CE, Oelschlager KM, et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res. 2015;21:175–183. doi: 10.1158/1078-0432.CCR-13-3316. [DOI] [PubMed] [Google Scholar]
- 26.Ghoreschi K, Laurence A, O'Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–287. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison DA. The Jak/STAT pathway. Cold Spring Harb Perspect Biol. 2012;4:a011205. doi: 10.1101/cshperspect.a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zoranovic T, Grmai L, Bach EA. Regulation of proliferation, cell competition, and cellular growth by the drosophila JAK-STAT pathway. JAKSTAT. 2013;2:e25408. doi: 10.4161/jkst.25408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rane SG, Reddy EP. Janus kinases: Components of multiple signaling pathways. Oncogene. 2000;19:5662–5679. doi: 10.1038/sj.onc.1203925. [DOI] [PubMed] [Google Scholar]
- 30.Messina JL, Yu H, Riker AI, Munster PN, Jove RL, Daud AI. Activated stat-3 in melanoma. Cancer Control. 2008;15:196–201. doi: 10.1177/107327480801500302. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Du J, Jiang R, Li L. MicroRNA-214 inhibits the proliferation and invasion of lung carcinoma cells by targeting JAK1. Am J Transl Res. 2018;10:1164–1171. [PMC free article] [PubMed] [Google Scholar]
- 32.Bi CL, Zhang YQ, Li B, Guo M, Fu YL. MicroRNA-520a-3p suppresses epithelial-mesenchymal transition, invasion, and migration of papillary thyroid carcinoma cells via the JAK1-mediated JAK/STAT signaling pathway. J Cell Physiol. 2018;234:4054–467. doi: 10.1002/jcp.27199. [DOI] [PubMed] [Google Scholar]
- 33.Jain N, Zhang T, Fong SL, Lim CP, Cao X. Repression of Stat3 activity by activation of mitogen-activated protein kinase (MAPK) Oncogene. 1998;17:3157–3167. doi: 10.1038/sj.onc.1202238. [DOI] [PubMed] [Google Scholar]
- 34.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 35.Nissan MH, Pratilas CA, Jones AM, Ramirez R, Won H, Liu C, Tiwari S, Kong L, Hanrahan AJ, Yao Z, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies MA, Samuels Y. Analysis of the genome to personalize therapy for melanoma. Oncogene. 2010;29:5545–5555. doi: 10.1038/onc.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koti M, Siu A, Clément I, Bidarimath M, Turashvili G, Edwards A, Rahimi K, Mes-Masson AM, Squire JA. A distinct pre-existing inflammatory tumour microenvironment is associated with chemotherapy resistance in high-grade serous epithelial ovarian cancer. Br J Cancer. 2015;112:1215–1222. doi: 10.1038/bjc.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson JA, Al-Attar A, Watson NF, Scholefield JH, Ilyas M, Durrant LG. Intratumoral T cell infiltration, MHC class I and STAT1 as biomarkers of good prognosis in colorectal cancer. Gut. 2010;59:926–933. doi: 10.1136/gut.2009.194472. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Molavi O, Su M, Lai R. The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma. BMC Cancer. 2014;14:791. doi: 10.1186/1471-2407-14-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Au KK, Le Page C, Ren R, Meunier L, Clément I, Tyrishkin K, Peterson N, Kendall-Dupont J, Childs T, Francis JA, et al. STAT1-associated intratumoural TH1 immunity predicts chemotherapy resistance in high-grade serous ovarian cancer. J Pathol Clin Res. 2016;2:259–270. doi: 10.1002/cjp2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan SR, Rickert CG, Vermi W, Sheehan KC, Arthur C, Allen JA, White JM, Archambault J, Lonardi S, McDevitt TM, et al. Dysregulated STAT1-SOCS1 control of JAK2 promotes mammary luminal progenitor cell survival and drives ERalpha(+) tumorigenesis. Cell Death Differ. 2014;21:234–246. doi: 10.1038/cdd.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thota B, Arimappamagan A, Kandavel T, Shastry AH, Pandey P, Chandramouli BA, Hegde AS, Kondaiah P, Santosh V. STAT-1 expression is regulated by IGFBP-3 in malignant glioma cells and is a strong predictor of poor survival in patients with glioblastoma. J Neurosurg. 2014;121:374–383. doi: 10.3171/2014.4.JNS131198. [DOI] [PubMed] [Google Scholar]
- 43.Arzt L, Kothmaier H, Halbwedl I, Quehenberger F, Popper HH. Signal transducer and activator of transcription 1 (STAT1) acts like an oncogene in malignant pleural mesothelioma. Virchows Arch. 2014;465:79–88. doi: 10.1007/s00428-014-1584-8. [DOI] [PubMed] [Google Scholar]
- 44.Jung SH, Ahn SY, Choi HW, Shin MG, Lee SS, Yang DH, Ahn JS, Kim YK, Kim HJ, Lee JJ. STAT3 expression is associated with poor survival in non-elderly adult patients with newly diagnosed multiple myeloma. Blood Res. 2017;52:293–299. doi: 10.5045/br.2017.52.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li WM, Huang CN, Lee YC, Chen SH, Lin HH, Wu WJ, Li CC, Yeh HC, Chang LL, Hsu WC, Ke HL. Over-expression of activated signal transducer and activator of transcription 3 predicts poor prognosis in upper tract urothelial carcinoma. Int J Med Sci. 2017;14:1360–1367. doi: 10.7150/ijms.17367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni Z, Lou W, Lee SO, Dhir R, DeMiguel F, Grandis JR, Gao AC. Selective activation of members of the signal transducers and activators of transcription family in prostate carcinoma. J Urol. 2002;167:1859–1862. doi: 10.1097/00005392-200204000-00095. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Li L, Zhang Y, Zhang Y, Zhao Y, You X, Lin Z, Zhang X, Ye L. The oncoprotein HBXIP uses two pathways to up-regulate S100A4 in promotion of growth and migration of breast cancer cells. J Biol Chem. 2012;287:30228–30239. doi: 10.1074/jbc.M112.343947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou X, Xia Y, Su J, Zhang G. Down-regulation of miR-141 induced by helicobacter pylori promotes the invasion of gastric cancer by targeting STAT4. Cell Physiol Biochem. 2014;33:1003–1012. doi: 10.1159/000358671. [DOI] [PubMed] [Google Scholar]
- 49.Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: Localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- 50.Wang G, Chen JH, Qiang Y, Wang DZ, Chen Z. Decreased STAT4 indicates poor prognosis and enhanced cell proliferation in hepatocellular carcinoma. World J Gastroenterol. 2015;21:3983–3993. doi: 10.3748/wjg.v21.i13.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo YH, Chen YT, Tsai HP, Chai CY, Kwan AL. Nucleophosmin overexpression is associated with poor survival in astrocytoma. APMIS. 2015;123:515–522. doi: 10.1111/apm.12381. [DOI] [PubMed] [Google Scholar]
- 52.Wang CG, Ye YJ, Yuan J, Liu FF, Zhang H, Wang S. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J Gastroenterol. 2010;16:2421–2427. doi: 10.3748/wjg.v16.i19.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nivarthi H, Gordziel C, Themanns M, Kramer N, Eberl M, Rabe B, Schlederer M, Rose-John S, Knösel T, Kenner L, et al. The ratio of STAT1 to STAT3 expression is a determinant of colorectal cancer growth. Oncotarget. 2016;7:51096–51106. doi: 10.18632/oncotarget.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y, Han Y, Wang X, Wang W, Wang X, Wen M, Xia J, Xing H, Li X, Zhang Z. Correlation of EGFR Del 19 with Fn14/JAK/STAT signaling molecules in non-small cell lung cancer. Oncol Rep. 2016;36:1030–1040. doi: 10.3892/or.2016.4905. [DOI] [PubMed] [Google Scholar]
- 55.Jeon M, You D, Bae SY, Kim SW, Nam SJ, Kim HH, Kim S, Lee JE. Dimerization of EGFR and HER2 induces breast cancer cell motility through STAT1-dependent ACTA2 induction. Oncotarget. 2016;8:50570–50581. doi: 10.18632/oncotarget.10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen G, Wang Y, Wu P, Zhou Y, Yu F, Zhu C, Li Z, Hang Y, Wang K, Li J, et al. Reversibly stabilized polycation nanoparticles for combination treatment of early- and late-stage metastatic breast cancer. ACS Nano. 2018;12:6620–6636. doi: 10.1021/acsnano.8b01482. [DOI] [PubMed] [Google Scholar]
- 57.Merk BC, Owens JL, Lopes MB, Silva CM, Hussaini IM. STAT6 expression in glioblastoma promotes invasive growth. BMC Cancer. 2011;11:184. doi: 10.1186/1471-2407-11-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sumiyoshi H, Matsushita A, Nakamura Y, Matsuda Y, Ishiwata T, Naito Z, Uchida E. Suppression of STAT5b in pancreatic cancer cells leads to attenuated gemcitabine chemoresistance, adhesion and invasion. Oncol Rep. 2016;35:3216–3226. doi: 10.3892/or.2016.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian X, Guan W, Zhang L, Sun W, Zhou D, Lin Q, Ren W, Nadeem L, Xu G. Physical interaction of STAT1 isoforms with TGF-β receptors leads to functional crosstalk between two signaling pathways in epithelial ovarian cancer. J Exp Clin Cancer Res. 2018;37:103. doi: 10.1186/s13046-018-0773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei M, Liu B, Gu Q, Su L, Yu Y, Zhu Z. Stat6 cooperates with Sp1 in controlling breast cancer cell proliferation by modulating the expression of p21(Cip1/WAF1) and p27 (Kip1) Cell Oncol (Dordr) 2013;36:79–93. doi: 10.1007/s13402-012-0115-3. [DOI] [PubMed] [Google Scholar]
- 61.Morgan EL, Wasson CW, Hanson L, Kealy D, Pentland I, McGuire V, Scarpini C, Coleman N, Arthur JSC, Parish JL, et al. STAT3 activation by E6 is essential for the differentiation-dependent HPV18 life cycle. PLoS Pathog. 2018;14:e1006975. doi: 10.1371/journal.ppat.1006975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hosui A, Klover P, Tatsumi T, Uemura A, Nagano H, Doki Y, Mori M, Hiramatsu N, Kanto T, Hennighausen L, et al. Suppression of signal transducers and activators of transcription 1 in hepatocellular carcinoma is associated with tumor progression. Int J Cancer. 2012;131:2774–2784. doi: 10.1002/ijc.27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Cheng X, Liang H, Jin Z. Long non-coding RNA HOTAIR and STAT3 synergistically regulate the cervical cancer cell migration and invasion. Chem Biol Interact. 2018;286:106–110. doi: 10.1016/j.cbi.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 64.Khan MA, El-Gamal MI, Oh CH. A progressive review of V600E-B-RAF-dependent melanoma and drugs inhibiting it. Mini Rev Med Chem. 2017;17:351–365. doi: 10.2174/1389557516666160622213142. [DOI] [PubMed] [Google Scholar]
- 65.Liu LJ, Wang W, Huang SY, Hong Y, Li G, Lin S, Tian J, Cai Z, Wang HD, Ma DL, Leung CH. Inhibition of the Ras/Raf interaction and repression of renal cancer xenografts in vivo by an enantiomeric iridium(iii) metal-based compound. Chem Sci. 2017;8:4756–4763. doi: 10.1039/C7SC00311K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bosserhoff AK. Novel biomarkers in malignant melanoma. Clin Chim Acta. 2006;367:28–35. doi: 10.1016/j.cca.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 67.Kugel CH, III, Aplin AE. Adaptive resistance to RAF inhibitors in melanoma. Pigment Cell Melanoma Res. 2014;27:1032–1038. doi: 10.1111/pcmr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harpio R, Einarsson R. S100 proteins as cancer biomarkers with focus on S100B in malignant melanoma. Clin Biochem. 2004;37:512–518. doi: 10.1016/j.clinbiochem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 69.Xiong TF, Pan FQ, Li D. Expression and clinical significance of S100 family genes in patients with melanoma. Melanoma Res. 2019;29:23–29. doi: 10.1097/CMR.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the TCGA repository, https://portal.gdc.cancer.gov/.