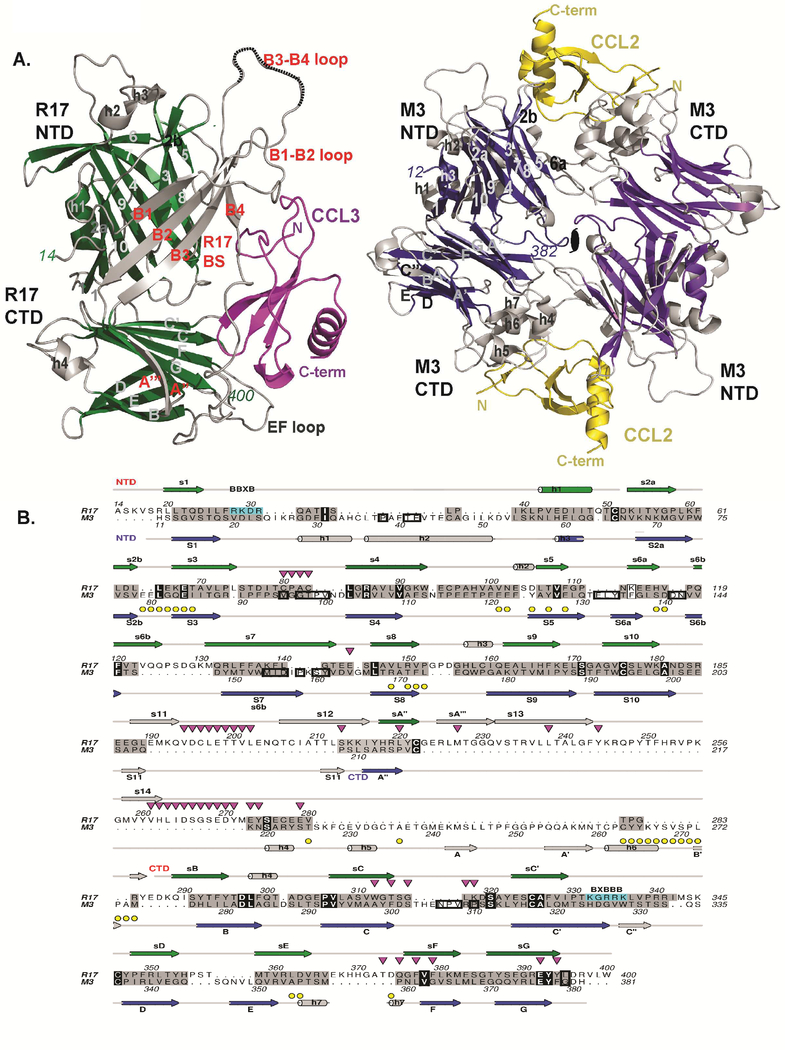
Figure 2: Structural comparison of R17 with M3.
(A) Comparison of CCL3 (magenta) bound R17 with CCL2 (yellow) bound M3 where shared core secondary structure elements are depicted in green for R17 and dark blue for M3. Divergent structural elements are depicted in light gray in both R17 and M3. (B) Structure-based sequence alignment of R17 with M3. Secondary structure elements of R17 are on top while secondary structure elements of M3 are on the bottom. Both are colored as in Figure 1A. Structurally similar residues are colored grey, while identical residues including conserved three cysteines are in black. Yellow circles denote M3 chemokine binding interface residues, while down-pointing magenta triangles denote R17 chemokine binding interface residues. Residues buried in the M3 dimer are boxed. BBXB motifs on R17 are boxed in cyan. See also Figure S2.