Abstract
The present study examined SMAD family member 4 (Smad4), SMAD family member 2 (Smad2) and phosphorylated (p-)Smad2 expression in biopsy specimens from patients with invasive breast ductal carcinoma, in order to assess their abilities as prognostic markers. A total of 126 tissue samples were selected, and the expression of Smad2, p-Smad2 and Smad4 in carcinoma tissues was detected by immunostaining, and the association between protein expression and clinicopathological variables was analyzed. Smad4 expression was negatively correlated with human epidermal growth factor receptor 2 in carcinoma tissues, and Smad4 expression was consistent with that of p-Smad2. Although multivariate analysis revealed that Smad2, p-Smad2 and Smad4 were not independent predictors, Kaplan-Meier curves demonstrated that Smad4 positivity was correlated with a longer overall survival (OS) and progression-free survival (PFS) time. However, upon analysis of combined markers, there was a significant difference between the p-Smad2/Smad4 co-positive and co-negative patients; the latter tended to exhibit a shorter OS and PFS time, and multivariate analysis revealed that the combined expression of p-Smad2 and Smad4 may be used as an independent prognostic factor. These results suggested that the assessment of p-Smad2 and Smad4 protein expression in breast ductal carcinoma biopsy specimens may provide additional prognostic information.
Keywords: breast ductal carcinoma, phosphorylated-SMAD family member 2, SMAD family member 4, immunochemistry assay
Introduction
Breast carcinoma is the most common malignant tumor and leading cause of cancer mortality in women worldwide (1). Breast tissue biopsies remain the best way to diagnose breast carcinoma. When malignant breast lumps are localized to the breast tissue, the relative cure rate of radical mastectomy or radiotherapy and chemotherapy is high. However, if breast carcinoma is detected at an advanced stage, and the carcinoma cells have spread outside the breast tissue, the prognosis for survival is substantially decreased (2). Although the precise molecular mechanism of breast carcinoma progression remains unclear, numerous studies have revealed that transforming growth factor-β (TGF-β)/SMAD family member (Smad) signaling pathways that regulate cell growth, differentiation, proliferation and apoptosis serve an important function in the progression of breast carcinoma (3–5).
The TGF-β signaling pathway is activated when TGF-β directly binds to transmembrane TGF-β type II receptors (TβRIIs); subsequently, TβRII recruits and activates TβRI. In turn, Smad2 or Smad3 transiently bind to TβRI and become activated by TβRI-induced phosphorylation in the cytoplasm. Phosphorylated (p-)Smad2 or p-Smad3 form a heterologous complex with a co-Smad (Smad4), which is translocated from the cytoplasm into the nucleus and binds to specific DNA sequences to regulate particular gene transcription (6).
Activated Smad2 or Smad3 exert different effects on the biological function of carcinoma cells (7). In gastric carcinoma, Smad2 is considered to protect the gastric mucosal epithelium from malignant transformation, whereas Smad3 is not directly associated with the initiation of gastric carcinoma, but is associated with the epithelial-mesenchymal transition (EMT) in gastric epithelial cells (8). In MDA-MB-231 breast carcinoma cells, Smad2 and Smad3 have diametrically opposite effects, with Smad3 knockdown resulting in a delayed bone metastasis of carcinoma cells, and Smad2 knockdown resulting in an enhanced invasive ability of MDA-MB-231 cells (9).
Smad4 has been identified as a tumor suppressor gene, and its mutation inactivation or decreased expression is often observed in tumor tissues, including colorectal and pancreatic carcinomas (10,11). So far, information regarding the function of Smad4 in breast carcinoma is very limited. A previous study revealed that the expression of Smad4 in breast carcinoma tissue was lower compared with that of surrounding normal adjacent breast epithelial tissue, but the survival time of patients who were Smad4-negative was longer (12). In addition, certain scholars believe that Smad4 may inhibit the growth of breast carcinoma cells by inducing apoptosis (13). However, subsequent to studying the MCF10 cell series, corresponding to different stages of breast cancer progression, it was revealed that the expression level of the Smad4 protein increased from non-malignant to highly malignant in highly invasive cells (14). Above all else, the aforementioned studies indicate that the function of Smad4 protein in the progression of breast carcinoma is very complex.
In the present study, the ductal carcinoma subtype with the highest incidence in breast cancer was selected as the subject of the study (15). Immunohistochemistry was used to examine the expression of Smad2, p-Smad2 and Smad4 in 126 invasive breast ductal carcinoma tissues, in order to investigate the correlation between the expression of these proteins and various clinicopathological parameters, in addition to the consistency of expression among them, analyze combined markers and identify prognostic factors for patients with breast ductal carcinoma.
Materials and methods
Patients and tissue specimens
A total of 126 breast ductal carcinoma specimens were collected from the Department of General Surgery of Beihua University Affiliated Hospital (Jilin, China) between January 2009 and December 2010. All specimens were confirmed by hematoxylin-eosin staining for invasive breast ductal carcinoma. No preoperative radiotherapy, chemotherapy or other antitumor treatments were available. All patients were female, aged 28–84 years (median age, 48 years). Out of all cases, 17 were well-differentiated, 79 moderately differentiated and 30 poorly differentiated. All patients were followed up until December 2014. The overall survival (OS) time was defined as the time between initial surgery and mortality or last follow-up. The OS time was 8.5–69.5 months, and the median survival time 49 months. The progression-free survival (PFS) time was defined as the time between initial surgery and deterioration (relapse or metastasis) or mortality. The PFS time was 5–69.5 months, and the median progression time 34.5 months.
A portion of the samples were fixed in 10% buffered formalin at 37°C for 3 h and embedded in paraffin. Specimens were stained with hematoxylin for 5 min and eosin for 20–30 sec at room temperature and examined histopathologically. Clinicopathological parameters, including age, tumor size, lymph node metastasis, distant metastasis, histological grade, estrogen (ER), progesterone (PR) and human epidermal growth factor receptor 2 (HER2) were available for all patients.
Immunohistochemical assay
Immunohistochemical assays were performed using the conventional streptavidin-peroxidase method. In brief, 5 µm thick paraffin-embedded tissues were dewaxed using xylene and dehydrated using ethanol gradient (100, 95 and 80%). Antigen retrieval buffer (citric acid and sodium citrate preparation) was used for incubating tissue sections. Sections were boiled (95°C) for 20 min and then cooled until they reached room temperature. Endogenous peroxidase was blocked using 3% hydrogen peroxide (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) for 10 min at 37°C. To eliminate non-specific staining, the sections were incubated with goat serum (Fuzhou Maixin Biotech Co., Ltd.) for 20 min at room temperature. They were then incubated with rabbit polyclonal anti-Smad2 (cat. no. ab63576; Abcam, Cambridge, UK), rabbit polyclonal anti-Smad2 (phospho S467; cat. no. ab53100; Abcam) and rabbit monoclonal anti-Smad4 (cat. no. ab40759; Abcam) antibodies at 4°C overnight. These antibodies were diluted at a ratio of 1:100. Following 3 washes in PBS, biotinylated anti-rabbit immunoglobulin (cat. no. KIT-9707; Fuzhou Maixin Biotech Co., Ltd.) were applied for 10 min at room temperature. Processing was then conducted in a humidified chamber at room temperature by the addition of streptavidin-peroxidase (cat. no. KIT-9707; Fuzhou Maixin Biotech Co., Ltd.) for 10 min. Next, the specimens were washed in PBS and color-developed with 3,3′-diaminobenzidine (Fuzhou Maixin Biotech Co., Ltd.) and hydrogen peroxide for 5 min. Subsequently, they were counterstained with Mayer's hematoxylin (Fuzhou Maixin Biotech Co., Ltd.) for 30 sec-1 min at room temperature. Negative controls were obtained by omission of the primary antibody and substitution of the primary antibody with normal serum. The positive control comprised a section of a breast ductal carcinoma block previously demonstrated to be positive for the marker, which was incorporated in each run. Stained specimens were imaged under a light microscope (magnifications ×200 and ×400).
Immunohistochemistry evaluation
All immunohistochemistry-stained sections were scored by at least 3 of the 4 independent experienced pathologists involved in the present study (Dr Nannan Liu, Dr Chunyan Yu, Dr Dongxue Qi and Dr Jihong Zhang) who had no prior knowledge of the clinicopathologic parameters and clinical outcomes of the patients. The distribution and intensity of Smad2, p-Smad2 and Smad4 staining were observed using light microscopy (CX31; Olympus Corporation, Tokyo, Japan), and at least 5 fields (×400) were analyzed for each tissue section. Staining was evaluated using the Taubert scoring system as previously described (16), according to the proportion of stained cells and staining intensity. The number of immunopositive cells was semi-quantitatively estimated as follows: i) The percentage of positive cells was scored as 1 (1–10%), 2 (11–50%), 3 (51–75%) and 4 (>75%); ii) Staining intensity was scored as 0 (absent), 1 (weak), 2 (moderate) and 3 (intense). The immunoreactive scores (IRS) were calculated by multiplying the scores of i) and ii) (17), and were as follows: 0, no staining; 1–4, weak staining; 5–8, moderate staining; and 9–12, strong staining. An IRS of <1 was considered to indicate a negative staining score for p-Smad2 and Smad4. The median score (6.3) was selected as the cutoff point for the separation of ‘high Smad2 expression’ (score > median) from ‘low Smad2 expression’ (score ≤ median) tissue sections. Results were confirmed by a repeat of the staining experiment on sequential sections from the same block.
Statistical analysis
Statistical analysis was conducted using SPSS software version 25 for Windows (IBM Corp., Armonk, NY, USA). The data were presented as the means of IRS values calculated for each section. The Spearman's rank correlations coefficient was calculated for analyzing the correlation between the expression of Smad2, p-Smad2 and Smad4 and various clinicopathological parameters. Cohen's κ coefficient was used to evaluate inter-observer consistency in the quantification. The Kaplan-Meier method (log-rank test) was used for OS and PFS curves. Cox regression was used for univariate and multivariate analysis for OS and PFS. P<0.05 was considered to indicate a statistically significant difference.
Results
Association between Smad2, p-Smad2 and Smad4 expression and clinicopathological parameters
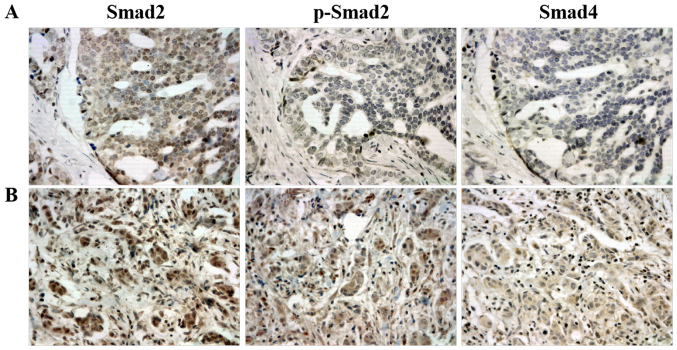
Immunohistochemistry was performed to determine the expression of Smad2, p-Smad2 and Smad4 in 126 primary breast ductal carcinoma tissue specimens. As expected, immunohistochemistry staining exhibited a predominantly cytoplasmic pattern of the Smad4 protein expression, while Smad2 and p-Smad2 were revealed in the cytoplasm and nuclear compartments (Fig. 1). According to the IRS criteria, no cases of Smad2-negative expression were observed among the evaluated specimens. The median score of all cases was 5.7, with 60 (47.6%) carcinoma tissues being Smad2-low (IRS<5.7) and 66 (52.4%) being Smad2-high (IRS≥5.7). However, p-Smad2 and Smad4 were negatively expressed in a proportion of cells. A total of 25 (19.8%) carcinoma tissues were p-Smad2-negative (IRS<1), while 101 (80.2%) stained positive (1≤IRS≤12). In addition, 21 (16.7%) carcinoma tissues were Smad4-negative (IRS<1) and 105 (83.3%) stained positive (1≤IRS≤12).
Figure 1.
Representative immunohistochemical staining of Smad2, p-Smad2 and Smad4 in (A) and (B) two different breast ductal carcinoma specimens. The expression of Smad2, p-Smad2 and Smad4 were strong, negative and negative in (A) specimens, respectively. The expression of Smad2, p-Smad2 and Smad4 were strong, moderate and weak in (B) specimens, respectively. Smad2, SMAD family member 2; Smad4, SMAD family member 4; p-, phosphorylated.
The association between the expression of Smad2, p-Smad2 and Smad4 and clinicopathological parameters was further analyzed. Significant correlations were identified between Smad4 and HER2 expression (r=−0.179, P=0.044; Table I), but there was no statistically significant correlation between Smad4 expression and any other clinicopathological parameters, including age, tumor size, lymph node metastasis, distant metastasis, histological grade, ER and PR (P>0.05; Table I). Furthermore, no statistically significant correlation was discovered between the expression of p-Smad2 and Smad2 and any of the various clinicopathological parameters (P>0.05; Table I).
Table I.
Association of clinicopathological parameters with Smad2, p-Smad2 and Smad4 expression in 126 patients with breast ductal carcinoma.
| Smad2 | p-Smad2 | Smad4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Cases | Low | High | r-value | P-value | Negative | Positive | r-value | P-value | Negative | Positive | r-value | P-value |
| Age (years) | 60 | 66 | 25 | 101 | 21 | 105 | |||||||
| <48 | 62 | 31 | 31 | 0.047 | 0.602 | 11 | 51 | −0.052 | 0.564 | 10 | 52 | −0.014 | 0.875 |
| ≥48 | 64 | 29 | 35 | 14 | 50 | 11 | 53 | ||||||
| Histological grade | |||||||||||||
| I | 17 | 8 | 9 | −0.022 | 0.806 | 3 | 14 | −0.048 | 0.595 | 2 | 15 | −0.027 | 0.762 |
| II | 79 | 37 | 42 | 15 | 64 | 14 | 65 | ||||||
| III | 30 | 15 | 15 | 7 | 23 | 5 | 25 | ||||||
| Tumor size | |||||||||||||
| T2 | 99 | 51 | 48 | 0.149 | 0.095 | 19 | 80 | −0.031 | 0.729 | 16 | 83 | −0.026 | 0.773 |
| T3 | 27 | 9 | 18 | 6 | 21 | 5 | 22 | ||||||
| Lymph node metastasis | |||||||||||||
| N0 | 42 | 21 | 21 | 0.023 | 0.801 | 12 | 30 | 0.130 | 0.146 | 10 | 32 | 0.104 | 0.247 |
| N1 | 37 | 17 | 20 | 6 | 31 | 5 | 32 | ||||||
| N2 | 24 | 11 | 13 | 3 | 21 | 2 | 22 | ||||||
| N3 | 23 | 11 | 12 | 4 | 19 | 4 | 19 | ||||||
| Distant metastasis | |||||||||||||
| M0 | 118 | 57 | 61 | 0.053 | 0.557 | 24 | 94 | 0.048 | 0.594 | 20 | 98 | 0.029 | 0.746 |
| M1 | 8 | 3 | 5 | 1 | 7 | 1 | 7 | ||||||
| ER | |||||||||||||
| Negative | 92 | 43 | 49 | −0.029 | 0.747 | 20 | 72 | 0.078 | 0.384 | 18 | 72 | 0.141 | 0.114 |
| Positive | 34 | 17 | 17 | 5 | 29 | 3 | 33 | ||||||
| PR | |||||||||||||
| Negative | 89 | 44 | 45 | 0.056 | 0.530 | 16 | 73 | −0.072 | 0.420 | 14 | 75 | −0.039 | 0.665 |
| Positive | 37 | 16 | 21 | 9 | 28 | 7 | 30 | ||||||
| HER2 | |||||||||||||
| Negative | 94 | 46 | 48 | 0.045 | 0.615 | 15 | 79 | −0.167 | 0.062 | 12 | 82 | −0.179 | 0.044a |
| Positive | 32 | 14 | 18 | 10 | 22 | 9 | 23 | ||||||
P<0.05. Smad, SMAD family member; p-, phosphorylated; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Consistent association among Smad4, p-Smad2 and Smad2 expression
A p-Smad2/Smad4 co-positive expression was observed in 100/126 cases. A p-Smad2/Smad4 co-negative expression was observed in 20/126 cases. In addition, 5/126 cases had a p-Smad2(−)/Smad4(+) and 1 had a p-Smad2(+)/Smad4(−) expression. Of note, according to the κ consistency test results, a significant agreement in the classification for Smad4 and p-Smad2 was revealed in all tissue specimens (κ=0.841, P<0.001; Table II). However, the consistent association was not presented in classification for between Smad2 and p-Smad2 (κ=0.024, P=0.394; data not shown) or Smad2 and Smad4 (κ=−0.013, P=0.632; data not shown).
Table II.
Agreement between Smad4 and p-Smad2 expression.
| p-Smad2 | |||||
|---|---|---|---|---|---|
| Smad4 | Cases | Negative | Positive | κ | P-value |
| Negative | 21 | 20 | 1 | 0.841 | 0.001a |
| Positive | 105 | 5 | 100 | ||
| Total | 126 | 25 | 101 | ||
P<0.05. Smad, SMAD family member; p-, phosphorylated.
Association between Smad2, p-Smad2 and Smad4 expression and the survival of patients with breast ductal carcinoma
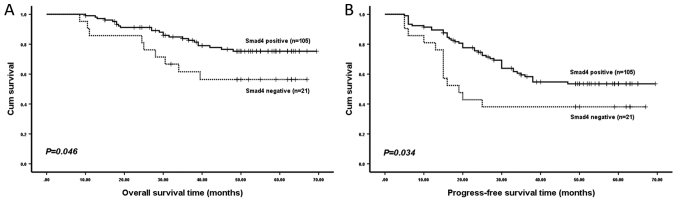
Kaplan-Meier analysis revealed that patients with an Smad4-negative expression were likely to have a significantly shorter OS time (P=0.046; Fig. 2) and PFS time (P=0.034; Fig. 2) in all 126 specimens compared with Smad4-positive patients. However, no significant difference in OS time (P=0.181; data not shown) or PFS time (P=0.063; data not shown) was detected between patients with a positive and negative p-Smad2 expression. In addition, no significant difference was identified in OS time (P=0.617; data not shown) or PFS time (P=0.552; data not shown) between patients with a positive and negative Smad2 expression.
Figure 2.
(A) Overall and (B) progression-free survival time of patients stratified according to Smad4 expression. Smad4, SMAD family member 4.
Univariate and multivariate analysis of prognostic variables in 126 patients with breast ductal carcinoma
Univariate analysis revealed that distant metastasis significantly predicted an increased risk of breast carcinoma progression (P=0.017; Table III) and poor OS time (P=0.010; Table III). However, the risk of breast carcinoma progression was significantly decreased in patients with Smad4-positive expression (P=0.040; Table III), an effect that was not observed for OS time (P=0.051; Table III). Other clinicopathological parameters did not exhibit any statistically significant difference in prognostic risk assessment (P>0.05; Table III). Multivariate Cox analysis revealed that Smad4 expression did not remain a statistically significant marker of deterioration [hazard ratio (HR), 0.539; 95% confidence interval (CI), 0.290–1.002; P=0.051; data not shown] once distant metastasis had been taken into account, whereas distant metastasis remained a significant independent predictor (HR, 2.945; 95% CI, 1.257–6.902; P=0.013; data not shown).
Table III.
Univariate analysis of clinicopathological parameters for overall and progression-free survival in 126 breast ductal carcinoma patients.
| Overall survival time (months) | Progress-free survival time (months) | ||||
|---|---|---|---|---|---|
| Variable | Cases | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (year) | 126 | 0.502 | 0.982 | ||
| <48 | 62 | 1 | 1 | ||
| ≥48 | 64 | 1.270 (0.632–2.554) | 0.994 (0.594–1.664) | ||
| Histological grade | 0.093 | 0.130 | |||
| I | 17 | 1 | 1 | ||
| II | 79 | 2.211 (0.515–9.492) | 1.075 (0.475–2.430) | ||
| III | 30 | 4.164 (0.922–18.809) | 1.881 (0.785–4.509) | ||
| Tumor size | 0.158 | 0.535 | |||
| T2 | 99 | 1 | 1 | ||
| T3 | 27 | 1.742 (0.806–3.768) | 1.216 (0.656–2.255) | ||
| Lymph node metastasis | 0.964 | 0.702 | |||
| N0 | 42 | 1 | 1 | ||
| N1 | 37 | 0.842 (0.339–2.093) | 1.344 (0.710–2.543) | ||
| N2 | 24 | 1.018 (0.394–2.626) | 0.891 (0.411–1.932) | ||
| N3 | 23 | 1.099 (0.406–2.971) | 1.148 (0.530–2.489) | ||
| Distant metastasis | 0.017a | 0.010a | |||
| M0 | 118 | 1 | 1 | ||
| M1 | 8 | 3.618 (1.262–10.369) | 3.072 (1.313–7.185) | ||
| ER | 0.819 | 0.970 | |||
| Negative | 92 | 1 | 1 | ||
| Positive | 34 | 1.094 (0.506–2.365) | 0.989 (0.549–1.780) | ||
| PR | 0.384 | 0.766 | |||
| Negative | 89 | 1 | 1 | ||
| Positive | 37 | 0.701 (0.315–1.560) | 1.087 (0.628–1.882) | ||
| HER2 | 0.098 | 0.563 | |||
| Negative | 94 | 1 | 1 | ||
| Positive | 32 | 1.832 (0.895–3.747) | 1.185 (0.666–2.109) | ||
| Smad2 | 0.618 | 0.558 | |||
| Low | 60 | 1 | 1 | ||
| High | 66 | 0.838 (0.419–1.617) | 1.168 (0.695–1.965) | ||
| p-Smad2 | 0.187 | 0.070 | |||
| Negative | 25 | 1 | 1 | ||
| Positive | 101 | 0.595 (0.275–1.286) | 0.581 (0.322–1.046) | ||
| Smad4 | 0.051 | 0.040a | |||
| Negative | 21 | 1 | 1 | ||
| Positive | 105 | 0.465 (0.215–1.005) | 0.523 (0.282–0.971) | ||
P<0.05. Smad, SMAD family member; p-, phosphorylated; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; CI, confidence interval.
Association between co-negative and co-positive p-Smad2/Smad4 expression and the survival of patients with breast ductal carcinoma
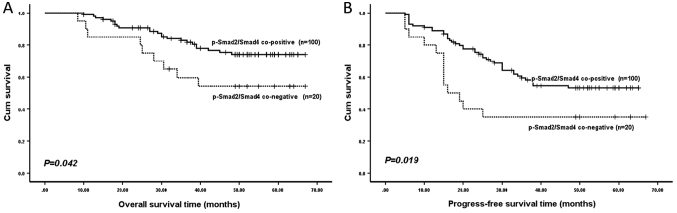
Since Smad4 expression was significantly consistent with p-Smad2 expression, as confirmed by Cohen's κ coefficient, patients were divided into two subgroups, according to their co-negative or co-positive p-Smad2/Smad4 expression. As revealed by Kaplan-Meier analysis, patients in the p-Smad2/Smad4 co-negative expression subgroups had a significantly shorter OS time (P=0.042; Fig. 3) and PFS time (P=0.019; Fig. 3) compared with those in the co-positive expression subgroups.
Figure 3.
(A) Overall and (B) progression-free survival time of patients stratified according to p-Smad2/Smad4 co-positive and co-negative expression. Smad4, SMAD family member 4; p-Smad2, phosphorylated SMAD family member 2.
Univariate and multivariate analysis of prognostic variables in patients with breast ductal carcinoma with a co-negative and co-positive p-Smad2/Smad4 expression
Univariate analysis results indicated that distant metastasis and p-Smad2/Smad4 co-expression were significantly associated with OS time (P=0.022 and P=0.047, respectively; Table IV) and PFS time (P=0.012 and P=0.023, respectively; Table IV). Nevertheless, the other clinicopathological parameters evaluated, including age, tumor size, lymph node metastasis, histological type, ER, PR and HER2 expression did not significantly affect OS and PFS time (Table IV). Furthermore, multivariate Cox regression analysis indicated that distant metastasis and co-negative or co-positive p-Smad2/Smad4 expression were also independent predictors for OS time (P=0.024 and P=0.049, respectively; Table V) and PFS time (P=0.017 and P=0.030, respectively; Table V) of patients with breast ductal carcinoma.
Table IV.
Univariate analysis of clinicopathological parameters for overall and progression-free survival in patients with breast ductal carcinoma with a co-negative and co-positive p-Smad2/Smad4 expression.
| Overall survival time (months) | Progress-free survival time (months) | ||||
|---|---|---|---|---|---|
| Variable | Cases | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (year) | 120 | 0.516 | 0.999 | ||
| <48 | 59 | 1 | 1 | ||
| ≥48 | 61 | 0.793 (0.395–1.596) | 1 (0.592–1.689) | ||
| Histological grade | 0.084 | 0.284 | |||
| I | 16 | 1 | 1 | ||
| II | 76 | 2.163 (0.504–9.287) | 1.047 (0.463–2.367) | ||
| III | 28 | 4.186 (0.927–18.904) | 1.660 (0.682–4.038) | ||
| Tumor size | 0.158 | 0.447 | |||
| T2 | 94 | 1 | 1 | ||
| T3 | 26 | 1.744 (0.806–3.772) | 1.272 (0.684–2.367) | ||
| Lymph node metastasis | 0.917 | 0.683 | |||
| N0 | 40 | 1 | 1 | ||
| N1 | 36 | 0.806 (0.324–2.005) | 1.215 (0.637–2.316) | ||
| N2 | 23 | 0.998 (0.387–2.575) | 0.781 (0.351–1.740) | ||
| N3 | 21 | 1.176 (0.435–3.181) | 1.243 (0.573–2.694) | ||
| Distant metastasis | 0.022a | 0.012a | |||
| M0 | 112 | 1 | 1 | ||
| M1 | 8 | 3.412 (1.190–9.777) | 2.986 (1.274–6.998) | ||
| ER | 0.770 | 0.919 | |||
| Negative | 89 | 1 | 1 | ||
| Positive | 31 | 1.122 (0.519–2.425) | 0.969 (0.529–1.774) | ||
| PR | 0.463 | 0.675 | |||
| Negative | 85 | 1 | 1 | ||
| Positive | 35 | 0.741 (0.333–1.650) | 1.127 (0.643–1.976) | ||
| HER2 | 0.125 | 0.796 | |||
| Negative | 89 | 1 | 1 | ||
| Positive | 31 | 1.751 (0.856–3.583) | 1.081 (0.598–1.954) | ||
| Smad2 | 0.750 | 0.543 | |||
| Low | 59 | 1 | 1 | ||
| High | 61 | 0.893 (0.446–1.788) | 1.178 (0.695–1.995) | ||
| p-Smad2 and Smad4 | 0.047a | 0.023a | |||
| p-Smad2/Smad4 co-negative | 20 | 1 | 1 | ||
| p-Smad2/Smad4 co-positive | 100 | 0.458 (0.212–0.991) | 0.486 (0.261–0.905) | ||
P<0.05. Smad, SMAD family member; p-, phosphorylated; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; CI, confidence interval.
Table V.
Multivariate analysis of clinicopathological parameters for overall and progression-free survival in patients with breast ductal carcinoma with a co-negative and co-positive p-Smad2/Smad4 expression.
| Variable | Comparison | HR | 95% CI | P-value |
|---|---|---|---|---|
| Overall survival time (months) | ||||
| Distant metastasis | M0 vs. M1 | 3.383 | 1.178–9.713 | 0.024a |
| p-Smad2 and Smad4 | p-Smad2/Smad4 co-negative vs. p-Smad2/Smad4 co-positive | 0.461 | 0.213–0.997 | 0.049a |
| Progression-free survival time (months) | ||||
| Distant metastasis | M0 vs. M1 | 2.840 | 1.209–6.668 | 0.017a |
| p-Smad2 and Smad4 | p-Smad2/Smad4 co-negative vs. p-Smad2/Smad4 co-positive | 0.502 | 0.269–0.937 | 0.030a |
P<0.05. Smad, SMAD family member; p-, phosphorylated; HR, hazard ratio; CI, confidence interval.
Discussion
The results of the present study revealed that the Smad4 expression was significantly negatively correlated with the expression of HER2 in breast carcinoma tissues; as the positive rate of Smad4 expression gradually decreased, the positive expression rate of HER2 increased correspondingly, suggesting that the expression characteristics of Smad4(−)/HER2(+) were present in a portion of breast ductal carcinomas specimens. Although no direct correlation has been reported between Smad4 and HER2, it has been reported that the TGF-β/Smad and the HER2/Ras/extracellular signal-regulated kinase (Erk) mitogen-activated protein kinase (MAPK) pathway often directly interact and mutually regulate the activities or expression of each other (18). One function of elevated HER2 activity in breast carcinoma is to impede the growth inhibitory function of TGF-β via the phosphorylation of R-Smad proteins by Erk MAPK (19). It means that HER2-mediated signal transformation may involve the shutdown of the TGF-β/Smad tumor suppressive pathway. Subsequently, the inactivation of Smad4 may further interfere with TGF-β-induced growth inhibition. Similarly, in human pancreatic ductal adenocarcinoma, the activation of HER2 and inactivation of Smad4 were also the most frequent gene alterations, which enabled human pancreatic ductal epithelial cells to acquire sustaining proliferative signaling, evade growth suppressors and finally result in tumorigenic transformation (20). This may not explain why patients with an Smad4-negative expression were more likely to be HER2-positive in breast ductal carcinoma tissues, but it may be important for understanding the expression characteristics of Smad4(−)/HER2(+) in certain breast carcinoma specimens. In addition, the study revealed that patients with breast carcinoma with a high HER2 expression were generally associated with a poor prognosis and high carcinoma cell proliferation (21), suggesting that HER2 has a positive regulatory effect on the proliferation of cancer cells. By contrast, Smad4 functions as a tumor-suppressor gene, which results in a negative regulatory effect on the proliferation of cancer cells (22). Therefore, there is expected to be a negative correlation between the expression of Smad4 and HER2 in breast carcinoma tissues, but the detailed underlying molecular mechanism and co-regulation signal loop need to be further studied.
Although Smad2 is not so much regarded as a tumor-suppressor gene, certain studies have demonstrated that the functional inactivation of Smad2 is sufficient to inhibit the physiological function of TGF-β (23,24). Furthermore, one previous study tested Smad2-targeted knockout mouse keratinocytes and revealed that Smad2−/− mice did not naturally develop into skin tumor types, but that Smad2 may accelerate tumor formation and malignant transformation in chemical carcinogenesis experiments, and that Smad2−/− skin cancer was poorly differentiated and exhibited an increase in EMT, indicating that an Smad2-deficient epithelium was more likely to form a tumor and malignant transformation (25). Accordingly, Smad2-deficiency in mice did not cause intestinal tumor types, but it may accelerate the malignant progression of late-stage invasive tumor types (26). In the present study, Smad2 was not negatively expressed in any of the 126 breast ductal carcinoma tissues, but 25 tissues were p-Smad2-negative. It was clear that these 25 specimens were due to a lack of phosphorylation, not due to a loss of Smad2 expression. More importantly, of the p-Smad2-negative cases, 20 exhibited a p-Smad2/Smad4 co-negative expression. Previous studies have revealed that the frequency of Smad4 gene mutant inactivation was high in solid tumor types, including breast carcinoma (27–29), which may partially explain the loss of Smad4 expression in the present study. Similar results have been discovered in the study of prostate carcinoma cell lines with different invasive and metastatic capacities: Smad2 was highly expressed in these cells, but the expression levels of p-Smad2 and Smad4 were different from each other, while a loss of expression was observed for each of them (30). Based on this, it was speculated that the progression of the malignant tumor types may be due to the interruption of the Smad activation process in the TGF-β signaling pathway.
In the present study, although neither p-Smad2 nor Smad4 were independent predictors, a substantial agreement in classification was observed between p-Smad2 and Smad4 expression. In addition, a Kaplan-Meier curve and Cox proportional hazards model revealed that p-Smad2/Smad4 co-positive patients had a longer OS and PFS time, in addition to a better prognosis, compared with p-Smad2/Smad4 co-negative patients. Based on the aforementioned results, it was speculated that the progression of these p-Smad2/Smad4 co-negative patients may have been due to the inability of Smad2 to phosphorylate, and at the same time the loss of Smad4 expression, which made Smad2 unable to form a heterologous complex with Smad4, and thus unable to form a transcription complex for the regulation of target gene transcription; as a result, the TGF-β/Smad pathway was disrupted, resulting in cell growth inhibition prevention and cell proliferation initiation (31). Another study revealed that the expression of the interstitial marker proteins vimentin and N-cadherin in breast carcinoma cells were increased following the inhibition of Smad2 expression, while the expression of the epithelial marker protein E-cadherin was decreased, suggesting the occurrence of EMT. It was through EMT that epithelial cells acquired an invasive ability, which promoted malignant tumor cell metastasis (32). In turn, the co-activation of p-Smad2 and Smad4 may serve a synergistic function in tumor suppression by transmitting TGF-β signaling in the 100 p-Smad2/Smad4 co-positive expression breast carcinoma cases, resulting in a longer survival time and better prognosis. In a similar study, although the effect of p-Smad2 and Smad4 co-expression or co-inactivation on prognosis was not observed, the OS time of patients with breast carcinoma which was p-Smad2-positive was significantly longer compared with that of p-Smad2-negative patients (33), which may have been due to the distinct specimen groups, the cut-offs used for the assessment or the antibodies used. Furthermore, the close correlation between p-Smad2 and Smad4 expression was also discovered in other types of cancer, including osteosarcoma; Smad4 expression was significantly associated with p-Smad2 expression, and they each co-regulated the expression of the cell cycle inhibitor p21/waf1 to inhibit tumor cell growth (34).
In conclusion, the results of the present study indicated that Smad4-positive patients had a better prognosis compared with Smad4-negative patients, although Smad4 expression could not be used as an independent predictor. Of note, at least in a part of breast ductal carcinomas, patients with a p-Smad2/Smad4 co-negative expression had a poor prognosis, as compared with p-Smad2/Smad4 co-positive patients. In addition, according to the results of the present study, p-Smad2 and Smad4 co-expression or co-inactivation may be seen as an independent predictor of prognosis in patients with invasive breast ductal carcinoma. Since, in most cases, the primary diagnosis of breast ductal carcinomas is based on histopathological sections, such breast cancer samples are therefore critical in guiding the clinical management of the disease, potentially helping to avoid improper or excessive treatment. Of course, it is also limited to detecting the expression of proteins in paraffin samples only by immunohistochemistry, and future research will attempt to confirm the results of the present study by analyzing Smad2, p-Smad2 and Smad4 mRNA and protein levels in fresh breast ductal carcinoma samples.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Science and Technology projects in Jilin Province Department of Education (grant no. JJKH20180356KJ), the Department of Science and Technology of Jilin Province (grant no. 20180623018TC) and the Jilin Province Health and Family Planning Commission (grant no. 2017J084).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
NL and CY contributed to the study design. DQ, CY, JZ and NL contributed to the data analysis. DQ and JJ contributed to the collection of the tissue samples and patient data. NL and CY wrote the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was ethically approved by the Ethics Committee of the Affiliated Hospital of Beihua University (Jilin, China), and written informed consent was obtained from each patient once the purpose and nature of the study had been fully explained.
Patient consent for publication
Informed consent was obtained from all patients regarding the publication of the data and associated images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res. 2017;50:33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leone BA, Vallejo CT, Romero AO, Machiavelli MR, Pérez JE, Leone J, Leone JP. Prognostic impact of metastatic pattern in stage IV breast cancer at initial diagnosis. Breast Cancer Res Treat. 2017;161:537–548. doi: 10.1007/s10549-016-4066-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen HS, Bai MH, Zhang T, Li GD, Liu M. Ellagic acid induces cell cycle arrest and apoptosis through TGF-β/Smad3 signaling pathway in human breast cancer MCF-7 cells. Int J Oncol. 2015;46:1730–1738. doi: 10.3892/ijo.2015.2870. [DOI] [PubMed] [Google Scholar]
- 4.Lo PK, Zhang Y, Yao Y, Wolfson B, Yu J, Han SY, Duru N, Zhou Q. Tumor-associated myoepithelial cells promote the invasive progression of ductal carcinoma in situ through activation of TGF β signaling. J Biol Chem. 2017;292:11466–11484. doi: 10.1074/jbc.M117.775080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang KM, Kim W, Bae E, Gim J, Weist BM, Jung Y, Hyun JS, Hernandez JB, Leem SH, Park T, et al. DRAK2 participates in a negative feedback loop to control TGF-β/Smads signaling by binding to type I TGF-β receptor. Cell Rep. 2012;2:1286–1299. doi: 10.1016/j.celrep.2012.09.028. [DOI] [PubMed] [Google Scholar]
- 6.Massagué J. TGF β signaling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ungefroren H, Groth S, Sebens S, Lehnert H, Gieseler F, Fändrich F. Differential roles of Smad2 and Smad3 in the regulation of TGF-β1-mediated growth inhibition and cell migration in pancreatic ductal adenocarcinoma cells: Control by Rac1. Mol Cancer. 2011;10:67. doi: 10.1186/1476-4598-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Li Q, Zhou X, Yu J, Mu Y, Munker S, Xu C, Shen Z, Müllenbach R, Liu Y, et al. Decreased levels of active SMAD2 correlate with poor prognosis in gastric cancer. PLoS One. 2012;7:e35684. doi: 10.1371/journal.pone.0035684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen M, Pardali E, van der Horst G, Cheung H, van den Hoogen C, van der Pluijm G, Ten Dijke P. Smad2 and Smad3 have opposing roles in breast cancer bone metastasis by differentially affecting tumor angiogenesis. Oncogene. 2010;29:1351–1361. doi: 10.1038/onc.2009.426. [DOI] [PubMed] [Google Scholar]
- 10.Yan P, Klingbiel D, Saridaki Z, Ceppa P, Curto M, McKee TA, Roth A, Tejpar S, Delorenzi M, Bosman FT, Fiocca R. Reduced expression of SMAD4 is associated with poor survival in colon cancer. Clin Cancer Res. 2016;22:3037–3047. doi: 10.1158/1078-0432.CCR-15-0939. [DOI] [PubMed] [Google Scholar]
- 11.Yamada S, Fujii T, Shimoyama Y, Kanda M, Nakayama G, Sugimoto H, Koike M, Nomoto S, Fujiwara M, Nakao A, Kodera Y. SMAD4 expression predicts local spread and treatment failure in resected pancreatic cancer. Pancreas. 2015;44:660–664. doi: 10.1097/MPA.0000000000000315. [DOI] [PubMed] [Google Scholar]
- 12.Stuelten CH, Buck MB, Dippon J, Roberts AB, Fritz P, Knabbe C. Smad4-Expression is decreased in breast cancer tissues: A retrospective study. BMC Cancer. 2006;6:25. doi: 10.1186/1471-2407-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Wu L, Oelschlager DK, Wan M, Stockard CR, Grizzle WE, Wang N, Chen H, Sun Y, Cao X. Smad4 inhibits tumor growth by inducing apoptosis in estrogen receptor-alpha-positive breast cancer cells. J Biol Chem. 2005;280:27022–27028. doi: 10.1074/jbc.M505071200. [DOI] [PubMed] [Google Scholar]
- 14.So JY, Lee HJ, Kramata P, Minden A, Suh N. Differential expression of key signaling proteins in MCF10 cell lines, a human breast cancer progression model. Mol Cell Pharmacol. 2012;4:31–40. [PMC free article] [PubMed] [Google Scholar]
- 15.Barroso-Sousa R, Metzger-Filho O. Differences between invasive lobular and invasive ductal carcinoma of the breast: Results and therapeutic implications. Ther Adv Med Oncol. 2016;8:261–266. doi: 10.1177/1758834016644156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taubert H, Heidenreich C, Holzhausen HJ, Schulz A, Bache M, Kappler M, Eckert AW, Würl P, Melcher I, Hauptmann K, et al. Expression of survivin detected by immunohistochemistry in the cytoplasm and in the nucleus is associated with prognosis of leiomyosarcoma and synovial sarcoma patients. BMC Cancer. 2010;10:65. doi: 10.1186/1471-2407-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 18.Luo K. Signaling cross talk between TGF-β/Smad and other signaling pathways. Cold Spring Harb Perspect Biol. 2017;9:a022137. doi: 10.1101/cshperspect.a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kretzschmar M. Transforming growth factor-beta and breast cancer: Transforming growth factor-beta/SMAD signaling defects and cancer. Breast Cancer Res. 2000;2:107–115. doi: 10.1186/bcr42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Z, Li Z, Wang X, Kang Y, Yuan Y, Niu J, Wang H, Chatterjee D, Fleming JB, Li M, et al. Deciphering the mechanisms of tumorigenesis in human pancreatic ductal epithelial cells. Clin Cancer Res. 2013;19:549–559. doi: 10.1158/1078-0432.CCR-12-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldarella A, Crocetti E, Bianchi S, Vezzosi V, Urso C, Biancalani M, Zappa M. Female breast cancer status according to ER, PR and HER2 expression: A population based analysis. Pathol Oncol Res. 2011;17:753–758. doi: 10.1007/s12253-011-9381-z. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Zhong J, Liu JH, Liao DF, Shen YY, Zhong XL, Xiao X, Ding WJ, Peng XD, Xiong W, Zu XY. Pokemon inhibits transforming growth factor β-Smad4-related cell proliferation arrest in breast cancer through specificity protein 1. J Breast Cancer. 2019;22:15–28. doi: 10.4048/jbc.2019.22.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying Z, Tian H, Li Y, Lian R, Li W, Wu S, Zhang HZ, Wu J, Liu L, Song J, et al. CCT6A suppresses SMAD2 and promotes prometastatic TGF-β signaling. J Clin Invest. 2017;127:1725–1740. doi: 10.1172/JCI90439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Wahdan-Alaswad R, Danielpour D. Critical role of Smad2 in tumor suppression and transforming growth factor-beta-induced apoptosis of prostate epithelial cells. Cancer Res. 2009;69:2185–2190. doi: 10.1158/0008-5472.CAN-08-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoot KE, Lighthall J, Han G, Lu SL, Li A, Ju W, Kulesz-Martin M, Bottinger E, Wang XJ. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118:2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamamoto T, Beppu H, Okada H, Kawabata M, Kitamura T, Miyazono K, Kato M. Compound disruption of smad2 accelerates malignant progression of intestinal tumors in apc knockout mice. Cancer Res. 2002;62:5955–5961. [PubMed] [Google Scholar]
- 27.Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14:111–123. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voorneveld PW, Kodach LL, Jacobs RJ, Liv N, Zonnevylle AC, Hoogenboom JP, Biemond I, Verspaget HW, Hommes DW, de Rooij K, et al. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology. 2014;147:196–208. doi: 10.1053/j.gastro.2014.03.052. [DOI] [PubMed] [Google Scholar]
- 29.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. SMAD4-Dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu KJ, Zhu GF, Zhang D, Zeng J, Wang XY, Xue Y, Zhang LL, He DL. Identification of TGF-beta/Smads pathway in human prostate cancer cell lines and its significance. Zhonghua Nan Ke Xue. 2009;15:920–924. (In Chinese) [PubMed] [Google Scholar]
- 31.Mhawech-Fauceglia P, Kesterson J, Wang D, Akers S, DuPont NC, Clark K, Lele S, Liu S. Expression and clinical significance of the transforming growth factor-β signalling pathway in endometrial cancer. Histopathology. 2011;59:63–72. doi: 10.1111/j.1365-2559.2011.03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv ZD, Kong B, Li JG, Qu HL, Wang XG, Cao WH, Liu XY, Wang Y, Yang ZC, Xu HM, Wang HB. Transforming growth factor-β 1 enhances the invasiveness of breast cancer cells by inducing a Smad2-dependent epithelial-to-mesenchymal transition. Oncol Rep. 2013;29:219–225. doi: 10.3892/or.2012.2111. [DOI] [PubMed] [Google Scholar]
- 33.Xie W, Mertens JC, Reiss DJ, Rimm DL, Camp RL, Haffty BG, Reiss M. Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: A tissue microarray study. Cancer Res. 2002;62:497–505. [PubMed] [Google Scholar]
- 34.Won KY, Kim YW, Park YK. Expression of smad and its signalling cascade in osteosarcoma. Pathology. 2010;42:242–247. doi: 10.3109/00313021003631288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.