SUMMARY
Fragment crystallizable (Fc) region of immunoglobulin G (IgG) antibody binds to specific Fc receptors (FcγRs) to control antibody effector functions. Currently, engineered specific Fc-FcγR interactions are validated with a static conformation derived from the crystal structure. However, computational evidence suggests that the conformational variability of Fcs plays an important role in receptor recognition. Here we elucidate Fc flexibility of IgG1, IgG2, and IgG1 Fc with mutations (M255Y/S257T/T259E) in solution by small-angle X-ray scattering (SAXS). Measured SAXS profiles and experimental parameters show variations in flexibility between Fc isotypes. We develop and apply a modeling tool for accurate conformational sampling of Fcs followed by SAXS fitting. Revealed conformational variability of the CH2 domain as low as 10 Å in displacement, illustrates the power of the atomistic modeling combined with SAXS. This inexpensive SAXS-based approach offers to improve the engineering of antibodies for tailoring Fc receptor interactions through altering and measuring Fc flexibility.
Graphical Abstract

In Brief
Fragment crystallizable (Fc) region of immunoglobulin G controls antibody effector function through specific Fc receptors interaction. Remesh et al. developed and applied an all-atom solution scattering-based modeling tool to reveal biologically relevant conformational plasticity of the Fc isotypes. This inexpensive SAXS-based approach can also aid typical antibody engineering efforts.
INTRODUCTION
The fragment crystallizable (Fc) region of immunoglobulin G (IgG) serves to trigger immunological events following interaction with Fc receptors (FcγR) (Jiang et al., 2011; Schwab and Nimmerjahn, 2013) and there is in silico evidence suggesting that differential conformational variability of Fcs, particularly CH2 domain flexibility (Frank et al., 2014), may influence FcγR binding which results in different IgG-mediated immune activation. Although, several crystal structures of the Fc region are available (Krapp et al., 2003; Matsumiya et al., 2007; Oganesyan et al., 2008; Teplyakov et al., 2013), missing hinge regions or large disorders in the CH2 domain make interpretations about the role of these regions using crystal structures alone quite challenging. Fc is a homodimer of N-linked glycopeptide chains comprised of two immunoglobulin domains (CH2 and CH3) that dimerize via inter-heavy chain disulfide bridges at the N-terminal hinge region and non-covalent interactions between the C-terminal CH3 domains. The overall shape of the Fc is similar to that of a “horseshoe,” with the majority of the internal space filled by the glycan chains only attached to the conserved asparagine 297 residues. The flexibility of Fc domains does not appear to involve large-scale motions like those of Fab domains in the full-length IgG. The relatively small conformational flexibility of CH2 domains was previously reported by molecular dynamics (MD) simulations (Frank et al., 2014), and is evident by the fact that the crystal structures of the various Fcs show variations in the tilt and twist of the CH2 domains (Kiyoshi et al., 2015; Krapp et al., 2003; Matsumiya et al., 2007; Oganesyan et al., 2008; Teplyakov et al., 2013). However, it is difficult to separate true conformational differences among different isotypes or engineered variants from those of the crystal-packing effects. Experimental validation of the Fc flexibility in solution remains unexplored, primarily due to the lack of an accurate experimental technique that can track small protein-domain motions in solution.
Single-particle (electron microscopy and tomography) techniques suffer from low contrast and low signal-to-noise ratio, bottlenecks that prevent accurate characterization of small domain motions. Effects of glycan modification on the rotational correlation times and chemical shift overlap have limited the utility of nuclear magnetic resonance (NMR) in studying flexible glycosylated systems as well (Cahour et al., 1984; Homans, 1990). Current improvements in small-angle X-ray scattering (SAXS) have allowed a study of macromolecule flexibility including multi-domain protein and RNA or DNA-protein complexes (Hammel, 2012; Pelikan et al., 2009; Rambo and Tainer, 2011, 2013). Using structures of individual domains from X-ray crystallography or NMR together with advanced computational approaches, domain displacement with less than 10 Å movement were visualized by SAXS (Chen et al., 2010; Duda et al., 2008; Hammel et al., 2002, 2016).
Here we show that SAXS is capable of detecting small motions of individual Fc domains. By combining modeling of CH2 domain conformational flexibility with the experimental SAXS data, we have derived Fc ensemble atomistic models in solution. We quantified the conformational flexibility of the CH2 domain for two IgG isotypes (Fc1 and Fc2) and the Fc1-YTE mutant with enhanced binding to the neonatal Fc receptor (Dall’Acqua et al., 2006; Majumdar et al., 2015; Oganesyan et al., 2008). In summary, our atomistic modeling, in combination with the SAXS data, provides experimental validation of Fc flexibility in solution, which is linked to its receptor selection.
RESULTS
SAXS Data Indicate Conformational Differences between Fc Isotypes
We collected SAXS data of two Fc isotypes (Fc1 and Fc2) and Fc1-YTE (M255Y/S257T/T259E), a mutant with enhanced binding to the neonatal Fc receptor (Dall’Acqua et al., 2006). Data were collected in the q range between 0.01 and 0.5 Å−1 for the protein concentrations within the range of 1 to 14 mg/mL. To assess the quality of the data for further accurate structural interpretations, we checked variation in radius of gyration (Rg) with increased protein concentration. Very small Rg variation (within 0.5 Å range) indicates a negligible influence of structure factors at high protein concentrations (Figure S1A). Nevertheless, we derived interference-free SAXS profiles for each Fc by merging curves of high and low protein concentrations (Figure S1B). The fact that molecular weights determined from the experimental scattering profiles are identical with the theoretical values (Table S1), and that Guinier plots are linear (Figure S1C), confirm an intact Fc homodimer and aggregation-free state of the samples (Table S1). Thus, the differences in the experimental Rg values (Table S1) and the SAXS profiles (Figure S1B) are indicative of conformational differences between the Fc isotypes rather than the different state of the samples.
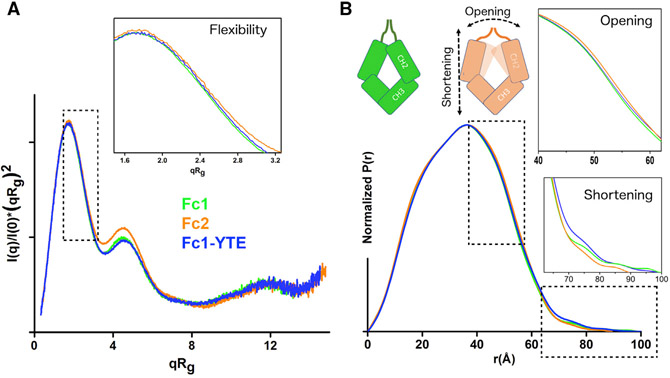
Next, we compared normalized Kratky plots for all Fcs. Identical shapes of the Kratky plots show the similar overall architecture of all Fc isotypes. However, the subtle peak shift, particularly in case of Fc2, indicates different flexibility between the isotypes (Durand et al., 2010; Receveur-Bréchot and Durand, 2012) (Figure 1A). Furthermore, broadening of the Fc2 pair distance distribution function (P(r)) shows a more open conformation (Figure 1B). The tail of the P(r) functions between 75 and 90 Å represents the extended conformations of the N-terminal hinge region. This protein region is not visible in the known crystal structures of Fcs (Krapp et al., 2003; Matsumiya et al., 2007; Oganesyan et al., 2008; Teplyakov et al., 2013) and is most likely unstructured and flexible in the solution (Figure 1B). Interestingly, the P(r) tail is significantly smaller in Fc2, which leads to a smaller maximal dimension of the entire glycoprotein by ~10 Å. The presence of two disulfide bonds and a number of residues in the hinges are similar for all three measured Fcs (Figure S2). Thus, the observed shortening of the P(r) tail is linked to the opening of the Fc2 molecule (Figure 1B).
Figure 1. SAXS Data Analysis to Assess Conformational Differences between Fc Isotypes.
(A) Normalized Kratky plots of Fc1 (green), Fc-YTE (blue), and Fc2 (orange) showing peak shifts for F1-YTE and Fc2, and indicating an increase in protein flexibility. The inset shows a zoomed view of normalized Kratky plots. In the normalized Kratky plot, the data were scaled using reciprocal space I(0) and Rg.
(B) Pair distribution function calculated for the SAXS profiles from (A) indicates differences in the inter-domain distances and maximal dimensions as described in the cartoon. The inset shows a zoomed view of the P(r) broadening at the distances r ~ 40–60 Å, indicating the opening of Fc2 or Fc1-YTE. The second inset shows a zoomed view of the P(r) tail at the r ~ 60–100 Å, indicating shortening of Fc2.
Modeling of Fc1 Flexibility in Solution
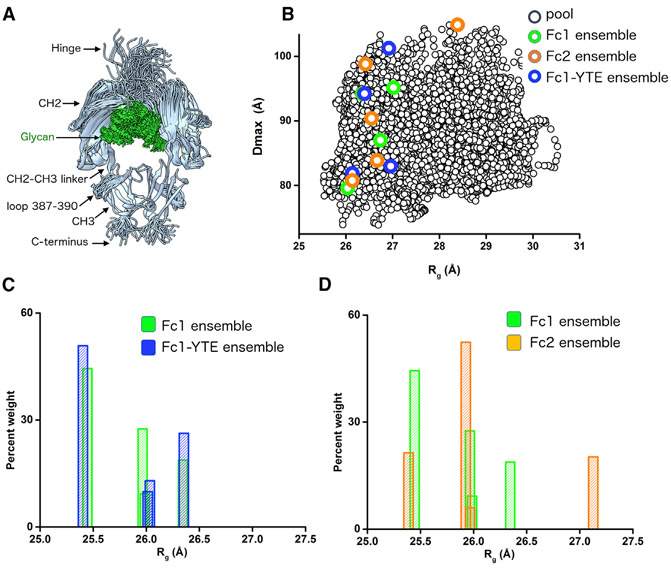
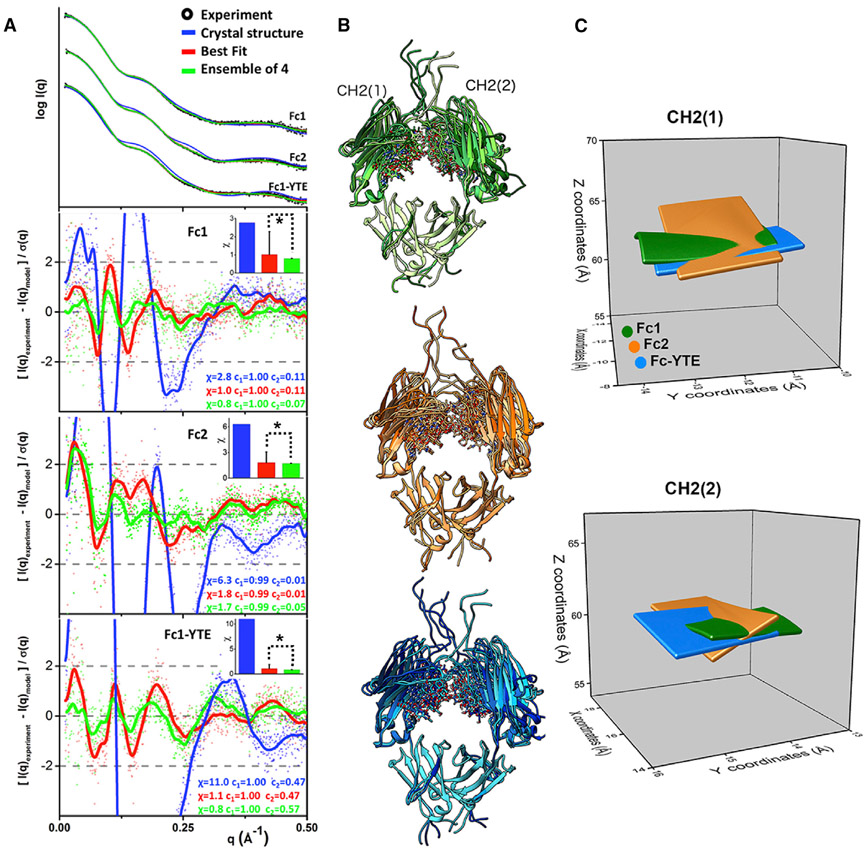
We developed and applied an all-atom modeling to determine Fc flexibility in solution. A SAXS modeling approach for glycosylated proteins was previously developed to study the flexibility of glycan chains (Guttman et al., 2013). However, problems with handling protein domain motions together with glycan flexibility directed our efforts for development of the conformational sampling approach, BILBOMD (Pelikan et al., 2009). The BILBOMD approach is based on the minimal MD simulation (Brooks et al., 2009). In the conformational sampling we used Fc1 crystal structure (PDB: 3AVE) (Matsumiya et al., 2011) with an added hinge region containing two disulfide bridges and three C-terminal residues as an initial model. In the first simulation, we performed conformational sampling of the three-residue hinge between CH2-CH3, the N-terminal hinge region with the disulfide bridges, three C-terminal residues, and a short flexible loop in the CH3 domain (Figures 2A and S2). We generated 3,000 conformers with the conformational sampling restrained by Rg limits of 25–28 Å (Figure 2B). Calculation of the SAXS profiles and multi-conformational analysis (Schneidman-Duhovny et al., 2016) were performed directly after the conformational sampling. Poor fit of the initial model (χ = 2.8) shows that the crystal structure does not represent the solution state of the molecule (Figure 3A, blue). A best-fit conformer selected from the entire pool of the models fits the experimental SAXS profile well, with χ = 1.0 (Figure 3A, red). Next, we selected an ensemble of four models to mimic CH2 flexibility in the solution (Figure 3B). A residual plot (Trewhella et al., 2017) was used to compare the quality of the fit with either best-fit or an ensemble model (Figure 3A, red versus green). The significant smoothening of the fit residuals at q < 0.25 Å−1 (Figure 3A, red versus green) shows the importance of matching the SAXS data with the multi-conformational model. The small improvement in χ values (χ = 0.8) was statistically significant when the χ values of top 10 best-fit models and top 10 ensemble models were compared (Figure 3A, inset). The ensemble of 4 conformers with significant variation in tilt and twist of the CH2 domain relative to the CH3 domain accurately represents the solution state of Fc1 (Figure 3B). To confirm that the results obtained are robust and reproducible the conformational sampling and SAXS fitting were repeated with the crystal structure of Fc1 PDB: 4W4N as a starting model. The derived best-fit and ensemble model fits experimental SAXS very similarly (Figure S3A). Percent weighted Rg distribution of four conformers from the ensemble model compared well with the distribution obtained from analysis performed with 3AVE model (Figure S3B).
Figure 2. Conformational Sampling and Percent Weighted Distribution of Rg of Fc Isotypes.
(A) Representative 100 conformers of Fc1 derived by BILBOMD sampling.
(B) Maximal dimension (Dmax) values of the 3,000 conformers in the entire pool (black) with respect to Rg. Rg and Dmax values of selected conformers in the ensemble matching SAXS profiles for Fc1 (green), Fc2 (orange), and Fc1-YTE, (blue) are indicated.
(C and D) Comparison of Fc1 (green) and Fc1-YTE (blue), or Fc2 (orange) weighted distribution of four models from the selected ensemble with respect to the Rg values of the model.
Figure 3. Multi-conformational Model of Fc isotypes from Error-Weighted Residual Fits to Experimental SAXS Profiles.
(A) Fits and the error-weighted residuals of experimental (black dots) and theoretical SAXS profile for the initial (blue), the best-fit (red), and ensemble model (green) of Fc1(3AVE), Fc2(4HAG),and Fc1-YTE(3FJT). The residuals are also shown after the FFT smoothing (lines). Insets: bar charts show the comparison of the χ value for the initial model (blue), with the mean for the χ value of the top-ten best-fit or top-ten ensemble models with SD. * indicates that, at p < 0.05, the population of the χ values between best-fit and ensemble models are significantly different.
(B) Four conformers from the selected multistate ensemble of Fc1 (shades of green), Fc1-YTE (shades of blue), and Fc2 (shades of orange) superimposed on the static CH3 domain indicate the motion of CH2 in solution.
(C) The plot of the 3D matrix derived from X, Y, and Z coordinates of the center of mass (COM) of CH2 domains in the selected multistate ensemble model obtained with four models of Fc1 (green), Fc-YTE (blue), and Fc2 (orange). Top and bottom plots show the matrix for left and right CH2 as indicated in (B).
To illustrate the sensitivity of SAXS to determine Fc flexibility, we measured the displacement of the center of mass (COM) of the CH2 domain in the multi-conformational model. The fact that the maximal displacement of the CH2 domain is less than 10 Å relative to its position in initial Fc1 model demonstrates the sensitivity of the all-atom modeling to determine CH2 domain movement (Figure 3C and S4A).
Restricted Motion of Glycans
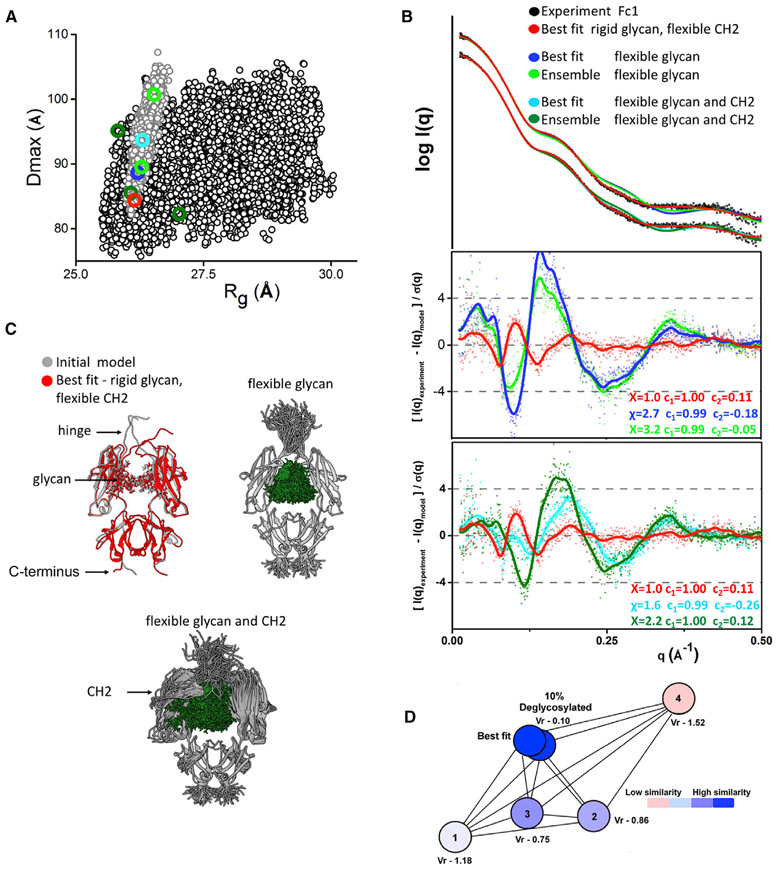
To test whether the glycan moiety of Fc1 is flexible in the solution we performed conformational sampling of the two Fc glycan chains, followed by SAXS fitting as described above. First, we performed sampling in the context of rigid CH2-CH3 domains using the initial arrangement as determined in the crystal structure. We generated 2,000 conformers mimicking the flexibility of the glycan chain through the flexible connection between asparagine 297 and individual monosaccharides bound. In this sampling, we also allowed flexibility of N-terminal hinge regions (Figures 4A and 4C). Neither single nor ensemble model fit SAXS data well (Figure 4B). Comparison with the best-fit obtained from rigid glycans sampling shows that the flexibility of glycans and N-terminal hinge is not sufficient to explain the discrepancy between the crystal and solution states (Figure 4B). Next, we extended the conformational sampling of glycans by including the flexibility of the CH2-CH3 linker (Figures 4A and 4C) and modeled an additional 6,000 conformers mimicking the flexibility of both CH2 domains and glycan chains. To our surprise, neither single nor ensemble model fit SAXS data better than the best-fit model with rigid glycans (Figure 4B). Finally, we combined all conformational samplings to perform a selection of the best-fit and ensemble model from the pool of 11,000 conformers (Figure 4A). None of the models with glycans displaced from the internal space between CH2 domains were selected to fit the SAXS data. Selected models from the combined pool were identical with the selection from the pool with the rigid glycans as described above (Figure 3B). This result indicates that the glycans do not undergo large movement in solution and are located between CH2 domains as seen in the crystal structure. Although previous NMR studies (Barb and Prestegard, 2011; Yamaguchi et al., 1998) report the dynamic behavior of N-glycan moiety, SAXS fitting presented here can only detect large displacement of the glycan. Thus, we assume that multiple hydrophobic and hydrophilic interactions (Raju, 2008) between an amino acid and sugar residues are responsible for occluding glycans in the Fc cavity, but still allowing potential dynamics of glycans, particularly of the more flexible α1-3MAN branch (Barb and Prestegard, 2011).
Figure 4. Evaluating the Origin of Flexibility in Fcs.
(A) Dmax and Rg values for the 3,000 conformers derived by the sampling of flexible glycan chain and hinge region (gray dot) in comparison with the 6,000 conformers derived by the sampling of flexible CH2-CH3 linker, glycan, and hinge region (black). Rg and Dmax values of selected best-fit and multistate ensemble models are indicated and colored as in (B).
(B) Poor fits and the ratios of the experimental (black dots) and theoretical SAXS profiles for the best-fit (blue and cyan), and multistate ensemble models (green and dark green), selected from two samplings with flexible glycan chains. Fit for the best-fit model selected from sampling with rigid glycan chains is shown for the comparison (red).
(C) Comparison of initial (gray) and best-fit model (red) selected from sampling with rigid glycan chains. The type of the flexibility that was explored in the sampling of flexible glycan chains or combined glycan and CH2-CH3 flexibility is shown by the representative models.
(D) The force plot of the Vr values for the SAXS profiles mimicking 100% and 90% of glycosylation in the comparison with the conformers from the Fc1 multistate ensemble model (models 1, 2, 3, and 4) indicates how similar or not are the theoretical SAXS profiles. Vr values were assigned a gradient color with blue (high similarity) and red (low similarity).
Comparison of CH2 Flexibility in Fc Isotypes
After validation of the methodology for Fc1 and establishing the flexibility of the CH2 domain as a primary source of flexibility we further investigated the solution state of Fc2 and Fc1-YTE. The analysis of experimental SAXS curves and P(r) functions already indicates different conformational states of the Fc2 isotype and YTE mutant (Figure 1). However, differences observed in SAXS curves may also reflect the heterogeneity in the glycosylation. Therefore, we tested if a small percentage of de-glycosylated species in the Fc1-YTE (3.6%) and Fc2 (3.7%) determined by mass spectroscopy (Table S2) can lead to the differences in the SAXS curves. We applied the volatility ratios (Vr) (Hura et al., 2013) to validate the effect of de-glycosylation on the dissimilarity between the SAXS profiles. We calculated SAXS profiles for Fc with 100% and 90% glycosylation and compared them with the theoretical SAXS of four Fc1 models from the multi-conformational model. The force plot of Vr (Figure 4D) shows that the profiles mimicking 100% and 90% of glycosylation are closely identical (Vr = 0.1). On other hand, SAXS profiles of the four models significantly differ from each other with 10-fold larger Vr values (Vr = 0.75–1.52). Thus, the small percentage of de-glycosylation in Fc1-YTE and Fc2 have a negligible effect on SAXS profiles, and the observed differences between the Fc isotypes reflect variations in the CH2 flexibility rather than small heterogeneity in the glycosylation.
The sequences of the Fc isotypes are closely identical with only few variations, mostly in the N-terminal hinge region (Teplyakov et al., 2013) (Figure S2). The CH2 or CH3 domains of the Fcs adopt an identical fold (Krapp et al., 2003; Matsumiya et al., 2007; Oganesyan et al., 2008). The fact that SAXS is sensitive to changes in the overall domain arrangement, rather than the small perturbation in the fold, allowed us to apply the pool of generated Fc1 conformers to the Fc2 and Fc1-YTE modeling. We used the pool of the Fc1 conformers from the conformational sampling of the flexible CH2-CH3 hinge (Figures 2A and 2B) to fit Fc2 or Fc1-YTE SAXS curves. Initially, we analyzed differences between Fc isotypes by comparing theoretical Rg values of the four selected conformers. It must be emphasized that the conformations themselves are not of significance, but the observation that an “ensemble” of conformations is a key factor in describing the solution state of the Fc isotypes. The Rg distribution of Fc1 conformers is in the range 25.4–26.4 Å (Figure 2C), whereas the Rg distribution of Fc1-YTE conformers is shifted toward higher Rg values. This shift correlates with slightly higher experimental Rg of Fc1-YTE (Table S1). The Rg distribution of Fc2 conformers is also skewed toward higher values (Figure 2D) with a significantly broader range (25.4–27.2 Å). Also, the presence of conformers with larger Rg = 27.2 Å agreed with the observed shift in the Kratky plot and broadening of the P(r) function (Figure 1).
The atomic coordinates of the CH2 domain in the selected ensembles (Figure 3B) were further simplified to describe domain movement in 3D. We compared the positions of the COM of the CH2 domain with respect to the stationary CH3 region (Figures 3C and S4). By plotting the COM within the surface matrix, we were able to compare relative differences of CH2 displacement for each Fc. We found that, while the CH2 domain of Fc1 and Fc1-YTE appear to move more or less along a single plane, the CH2 domain of Fc2 moves in multiple perpendicular planes and may have certain tilt and twist angles that are unique features of Fc2 flexibility. Further, the presence of more open CH2 conformations in Fc1-YTE may arise from M255Y/ S257T/T259E mutation located in the CH2-CH3 cleft (Figure S2), and indicates the role of the CH2-CH3 interface in the controlling of the Fc flexibility.
DISCUSSION
The flexibility of Fc isotypes reported here allows for improved understanding FcγR-mediated recognition of IgG and will benefit the efforts to design therapeutic antibodies. Our studies show previously unseen conformational flexibility of Fcs in solution. Access to high-quality SAXS data, Fc crystal structures, and our all-atom modeling method allowed us to visualize CH2 domain motion in solution.
By matching the multi-Fc conformations to the experimental SAXS we show that CH2 domains undergo enhanced motion with variations in tilt and twist angles relative to the CH3 domain. Motions observed here strongly correlate with previously suggested Fc conformational variability observed by MD simulation (Frank et al., 2014). The spatial parameters of a CH2 domain, as seen through X-ray crystallography (Krapp et al., 2003; Matsumiya et al., 2007; Oganesyan et al., 2008; Teplyakov et al., 2013), are the product of crystal-packing restraint and represent just one snapshot within the intrinsic range of their conformations.
Altering of CH2 domain opening has become a target in the engineering of therapeutic antibodies (Higel et al., 2016; Irani et al., 2015; Le et al., 2016; Mizushima et al., 2011). As suggested by X-ray crystallography, the perturbation of CH2 opening by mutations in protein or glycan moiety (Borrok et al., 2012; Crispin et al., 2009; Krapp et al., 2003; Nagae and Yamaguchi, 2012; Raju, 2008) directly affects the interaction with the FcγR receptor. The Fc flexibility characterized here indicates that it is not static perturbation that alters FcγR receptor interaction, but rather the differential conformational space adopted by the CH2 domains. The receptor needs to screen a range of Fc structures in solution to select complementary conformers, and altering of CH2 flexibility appears to tune desirable receptor affinities.
The YTE mutation has a direct impact on the interaction of Fc1-YTE with neonatal Fc receptor (FcRn) (Dall’Acqua et al., 2006). FcRn interacts with Fc1-YTE primarily through two sites, one in the CH2 domain close to the YTE mutation, and one in the CH3 domain located at His433 (Martin et al., 2001). We speculate that the shift of conformational space toward more open CH2 conformers results in adjustable distances between the two binding sites. In general, the mutation of the CH2-CH3 cleft that alters CH2 flexibility can also influence FcyR interaction located near the N-terminal hinge. On the other hand, the sequence difference of the Fc2 N-terminal hinge region seems to lead to the different motion of the CH2 domains (Figure 3C). The N-terminal hinge restricts the range of CH2 motions by acting as a tether. We speculate that the unique P-X-X-G-P motif of the Fc2 N-terminal hinge is responsible for mediating high-amplitude bending in Fc2 (Jacob et al., 1999; Krieger et al., 2005). The bending of the hinge region may allow opening of the CH2 domain, as seen through the broadening of the P(r) function (Figure 1B), and lead to unique Fc2 flexibility (Figure 3C). These differences, which are genetically encoded, can contribute to the Fc receptor selection. The role of the N-terminal hinge region sequence as an important regulator of CH2 motion can provide novel strategies in the engineering of therapeutics Fcs. However, the SAXS-based approach presented here is an inexpensive technique to validate domain motions and can be used in the screening of desirable Fc flexibility and elsewhere. Efficient screening of designed Fcs will lead to improvements in therapeutic antibody efficacy, pharmacokinetics, production, and to a deeper understanding of the role of Fc flexibility in the immune system.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contacts - Michal Hammel (mhammel@lbl.gov).
METHOD DETAILS
Fc Protein Expression and Purification
The recombinant human IgG2 Fc, IgG1 Fc and IgG1 Fc-YTE fragments were purchased from Sino Biological Inc. (Beijing China) using their proprietary expression system and purification protocols. Briefly, the DNA encoding the mature Fc fragments were cloned into a CMV promoter driven expression vector and the proteins were expressed in HEK293 cells by transient transfection, then purified on a protein A column (Sino). The Fc proteins were dialyzed into a final buffer of 20 mM Tris pH 7.5, 50 mM NaCl
SAXS Data Collection and Evaluation
SAXS data were collected at the ALS beamline 12.3.1 LBNL Berkeley, California (Classen et al., 2013). X-ray wavelength was λ=1.03 Å and the sample-to-detector distance was set to 1.5 m resulting in scattering vectors, q, ranging from 0.01 Å−1 to 0.5 Å−1. The scattering vector is defined as q = 4π sinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20°C and data were processed as described (Hura et al., 2009) . The SAXS profiles for Fcs in the concentration range 1 - 14 mg/ml were investigated by comparing radius of gyrations Rg derived by the Guinier approximation I(q) = I(0) exp(−q2Rg2/3) with the limits qRg<1.6 (Figure S1C). Rg variation with less than 0.5Å indicates the negligible influence of structure factor or aggregations at high protein concentration (Figure S1). Nevertheless, we derived interference free SAXS profile by merging profiles obtained for high and low concentration (Figure S1B) using program SCÅTTER. The program GNOM (Svergun, 1992) was used to compute the P(r) function.
Solution Structure Modeling
The crystal structure of Fc1 PDB ID: 3AVE (Matsumiya et al., 2007, 2011) and PDB ID: 4W4N were used to build an atomistic model for solution structure modeling by filling missing residues in the hinge and loop region by MODELLER (Sali and Blundell, 1993). In our updated rigid body modeling strategy BILBOMD (Pelikan et al., 2009), simplified molecular dynamics simulations were used to explore conformational space adopted by CH2 domains and glycan chains. Multiple conformational samplings with altered flexible regions (Figure S2) were explored to validate flexibility of CH2 domain, N-terminal hinge and glycan chain. FoXS (Schneidman-Duhovny et al., 2013) was used to pre-calculate SAXS profile for all recorded conformers. MultiFoXS (Schneidman-Duhovny et al., 2016) was used to identify the multistate model required to best fit the experimental data. Structures were visualized in CHIMERA (Pettersen et al., 2004).
Mass Spectroscopy
Approximately 100 ng of intact mAb was injected onto a Waters Acquity I-Class UPLC Protein BEH C4 1 x 50 mm, 1.7 mm, part no: 186005589) using Acquity I-Class UPLC (Waters Corp.) at a flow rate of 0.250 mL/min. The column was kept at 70°C. Mass spectrometry grade HPLC solvents (0.1% Formic acid and B: 100% ACN in 0.1% Formic acid) were purchased from VWR (part no: LC 452-1, LC 441-1). The column effluent was introduced into a Waters Vion Qtof mass spectrometer via electrospray ionization using a spray voltage of 2.0 kV and cone voltage of 120 V. The resulting chromatographic peaks were integrated to generate summed mass spectra, which were deconvoluted by maximum entropy method (MaxEnt1) using Unifi software (Waters) to yield observed masses. The mass spectrometer was calibrated with NaCsI (Waters, part no: 700002646-8) within 1 ppm mass accuracy. As a secondary control, molecular weight of a commercial monoclonal antibody (Waters, part no: 186006552) was confirmed (within <30 ppm) prior to the experiment. The molecular mass data were interpreted using the theoretical mass of the mAb sequence considering modifications such as disulfides, N-terminal pyroglutamate and various glyco-forms. The glycol-forms were quantified using the relative ion currents.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical test performed for this study was the Student’s t-test to measure the level of significant different between the best fit and ensemble model. OriginLab was used for this purpose Origin (OriginLab, Northampton, MA).
DATA AND SOFTWARE AVAILABILITY
SAXS data were deposited in the Small Angle Scattering Biological Data Bank. The accession codes are - SASDDT4 (IgG1 Fc), SASDDU4 (IgG2 Fc2) and SASDDV4 (IgG1 Fc- M135Y/S137T/T139E mutant). All software used for the modeling studies are open-source and can be downloaded from websites as indicated in the Key Resource Table.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human IgG1 Fc | Sino Biological, Inc. | IgG1 Fc |
| Human IgG2 Fc | Sino Biological, Inc. | IgG2 Fc |
| Human IgG1 Fc-YTE | Sino Biological, Inc. | IgG1 Fc-YTE |
| Deposited Data | ||
| IgG1 Fc | Small Angle Scattering Biological Data Bank | SASDDT4 |
| IgG2 Fc | Small Angle Scattering Biological Data Bank | SASDDU4 |
| IgG1 Fc-YTE | Small Angle Scattering Biological Data Bank | SASDDV4 |
| Software and Algorithms | ||
| Blu-Ice | Classen et al., 2013 | https://www.ncbi.nlm.nih.gov/pubmed/23396808 |
| SCÅTTER | Developed by Robert Rambo at the Diamond Light Source (Didcot, UK) | http://www.bioisis.net/tutorial/9 |
| GNOM | Svergun, 1992 | https://www.embl-hamburg.de/biosaxs/gnom.html |
| MODELLER | Sali and Blundell, 1993 | https://salilab.org/modeller/ |
| BILBOMD | Pelikan et al., 2009 | http://bl1231.als.lbl.gov/bilbomd |
| FoXS | Schneidman-Duhovny et al., 2013 | https://modbase.compbio.ucsf.edu/foxs/ |
| Multi-FoXS | Schneidman-Duhovny et al., 2016 | https://modbase.compbio.ucsf.edu/multifoxs/ |
| CHIMERA | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| OriginLab | Origin (OriginLab, Northampton, MA) | https://www.originlab.com/ |
| Unifi Software | Waters Corp. | http://www.waters.com/ |
| Other | ||
| Waters Vion Qtof mass spectrometer | Waters Corp. | http://www.waters.com/ |
Supplementary Material
Highlights.
Experimental visualization of Fc flexibility in solution
Flexibility variation observed between Fc isotypes in solution
All-atom modeling provides accurate conformational sampling of glycoproteins
SAXS utilized to model <10 Å domain displacement
ACKNOWLEDGMENTS
This work was supported in part by Janssen R&D, the DOE BER Integrated Diffraction Analysis Technologies (IDAT) program, and NIGMS grant P30 GM124169-01, ALS-ENABLE. SAXS at the Advanced Light Source SIBYLS beamline is supported by NIH grant CA92584. We thank Dr. Gregory Hura and Ms. Katherine Brunett for aiding data collection, and members of the SIBYLS beamline for comments and suggestions for preparation of this paper.
Footnotes
DECLARATION OF INTERESTS
None declared.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.str.2018.03.017.
REFERENCES
- Barb AW, and Prestegard JH (2011). NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat. Chem. Biol 7, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrok MJ, Jung ST, Kang TH, Monzingo AF, and Georgiou G (2012). Revisiting the role of glycosylation in the structure of human IgG Fc. ACS Chem. Biol 7, 1596–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. (2009). CHARMM: the biomolecular simulation program. J. Comput. Chem 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahour A, Debeire P, Hartmann L, Montreuil J, van Halbeek H, and Vliegenthart JF (1984). Primary structure of the major glycans of the N-acetyllactosamine type derived from the human immunoglobulins M from two patients with Waldenström’s macroglobulinemia. FEBS Lett. 170, 343–349. [DOI] [PubMed] [Google Scholar]
- Chen H, Ricklin D, Hammel M, Garcia BL, McWhorter WJ, Sfyroera G, Wu YQ, Tzekou A, Li S, Geisbrecht BV, et al. (2010). Allosteric inhibition of complement function by a staphylococcal immune evasion protein. Proc. Natl. Acad. Sci. USA 107, 17621–17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, Dyer K, Hammel M, Meigs G, Frankel KA, et al. (2013). Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. J. Appl. Crystallogr 46, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispin M, Bowden TA, Coles CH, Harlos K, Aricescu AR, Harvey DJ, Stuart DI, and Jones EY (2009). Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. J. Mol. Biol 387, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Dall’Acqua WF, Kiener PA, and Wu H (2006). Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J. Biol. Chem 281, 23514–23524. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, and Schulman BA (2008). Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D, Vives C, Cannella D, Perez J, Pebay-Peyroula E, Vachette P, and Fieschi F (2010). NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol 169, 45–53. [DOI] [PubMed] [Google Scholar]
- Frank M, Walker RC, Lanzilotta WN, Prestegard JH, and Barb AW (2014). Immunoglobulin G1 Fc domain motions: implications for Fc engineering. J. Mol. Biol 426, 1799–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Weinkam P, Sali A, and Lee KK (2013). All-atom ensemble modeling to analyze small-angle X-ray scattering of glycosylated proteins. Structure 21, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M (2012). Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS). Eur. Biophys. J 41, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Amlanjyoti D, Reyes FE, Chen JH, Parpana R, Tang HY, Larabell CA, Tainer JA, and Adhya S (2016). HU multimerization shift controls nucleoid compaction. Sci. Adv 2, e1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Kriechbaum M, Gries A, Kostner GM, Laggner P, and Prassl R (2002). Solution structure of human and bovine beta(2)-glycoprotein I revealed by small-angle X-ray scattering. J. Mol. Biol 321, 85–97. [DOI] [PubMed] [Google Scholar]
- Higel F, Seidl A, Sorgel F, and Friess W (2016). N-Glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins. Eur. J. Pharm. Biopharm 100, 94–100. [DOI] [PubMed] [Google Scholar]
- Homans SW (1990). Oligosaccharide conformations: application of NMR and energy calculations. Prog. Nucl. Magn. Reson. Spectrosc 22, 55–81. [Google Scholar]
- Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, and Tainer JA (2013). Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat. Methods 10, 453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL 2nd, Tsutakawa SE, Jenney FE Jr., Classen S, Frankel KA, Hopkins RC, et al. (2009). Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani V, Guy AJ, Andrew D, Beeson JG, Ramsland PA, and Richards JS (2015). Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol. Immunol 67, 171–182. [DOI] [PubMed] [Google Scholar]
- Jacob J, Duclohier H, and Cafiso DS (1999). The role of proline and Glycine in determining the backbone flexibility of a channel-forming peptide. Biophys. J 76, 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.-r., Song A, Bergelson S, Arroll T, Parekh B, May K, Chung S, Strouse R, and Mire-sluis A (2011). Advances in the assessment and control of the effector functions of therapeutic antibodies. Nat. Rev. Drug Discov 10, 101–111. [DOI] [PubMed] [Google Scholar]
- Kiyoshi M, Caaveiro JM, Kawai T, Tashiro S, Ide T, Asaoka Y, Hatayama K, and Tsumoto K (2015). Structural basis for binding of human IgG1 to its high-affinity human receptor FcγRI. Nat. Commun 6, 6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp S, Mimura Y, Jefferis R, Huber R, and Sondermann P (2003). Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol 325, 979–989. [DOI] [PubMed] [Google Scholar]
- Krieger F, Möglich A, and Kiefhaber T (2005). Effect of proline and Glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc 127, 3346–3352. [DOI] [PubMed] [Google Scholar]
- Le NP, Bowden TA, Struwe WB, and Crispin M (2016). Immune recruitment or suppression by glycan engineering of endogenous and therapeutic antibodies. Biochim. Biophys. Acta 1860, 1655–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R, Esfandiary R, Bishop SM, Samra HS, Middaugh CR, Volkin DB, and Weis DD (2015). Correlations between changes in conformational dynamics and physical stability in a mutant IgG1 mAb engineered for extended serum half-life. MAbs 7, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WL, West AP, Gan L, and Bjorkman PJ (2001). Crystal structure at 2. 8 Å of an FcRn/heterodimeric Fc complex: mechanism of pH dependent binding. Mol. Cell 7, 866–877. [DOI] [PubMed] [Google Scholar]
- Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, and Kato K (2007). Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J. Mol. Biol 368, 767–779. [DOI] [PubMed] [Google Scholar]
- Matsumiya S, Yamaguchi Y, Saito J.-i., Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, and Kato K (2011). Corrigendum to “structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1”. J. Mol. Biol 386, 767–779. [DOI] [PubMed] [Google Scholar]
- Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, and Kato K (2011). Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells 16, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, and Yamaguchi Y (2012). Function and 3D structure of the N-glycans on glycoproteins. Int. J. Mol. Sci 13, 8398–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan V, Gao C, Shirinian L, Wu H, and Dall’Acqua WF (2008). Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Crystallogr. D. Biol. Crystallogr 64, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, and Hammel M (2009). Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys 28, 174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Raju TS (2008). Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr. Opin. Immunol 20, 471–478. [DOI] [PubMed] [Google Scholar]
- Rambo RP, and Tainer JA (2011). Characterizing flexible and instrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, and Tainer JA (2013). Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu. Rev. Biophys 42, 415–441. [DOI] [PubMed] [Google Scholar]
- Receveur-Bréchot V, and Durand D (2012). How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci 13, 55–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, and Blundell TL (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A (2013). Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J 105, 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, and Sali A (2016). FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab I, and Nimmerjahn F (2013). Intravenous immunoglobulin therapy: how does IgG modulate the immune system? Nat. Rev. Immunol 13, 176–189. [DOI] [PubMed] [Google Scholar]
- Svergun D (1992). Determination of the regularization Parameter in Indirect-Transform Methods using perceptual criteria. J. Appl. Cryst 25, 495–503. [Google Scholar]
- Teplyakov A, Zhao Y, Malia TJ, Obmolova G, and Gilliland GL (2013). IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface. Mol. Immunol 56, 131–139. [DOI] [PubMed] [Google Scholar]
- Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, et al. (2017). 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update. Acta Crystallogr. D. Struct. Biol 73, 710–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Kato K, Shindo M, Aoki S, and Furusho K (1998). Dynamics of the carbohydrate chains attached to the Fc portion of immunoglobulin G as studied by NMR spectroscopy assisted by selective C labeling of the glycans. J. Biomol. NMR 12, 385–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SAXS data were deposited in the Small Angle Scattering Biological Data Bank. The accession codes are - SASDDT4 (IgG1 Fc), SASDDU4 (IgG2 Fc2) and SASDDV4 (IgG1 Fc- M135Y/S137T/T139E mutant). All software used for the modeling studies are open-source and can be downloaded from websites as indicated in the Key Resource Table.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human IgG1 Fc | Sino Biological, Inc. | IgG1 Fc |
| Human IgG2 Fc | Sino Biological, Inc. | IgG2 Fc |
| Human IgG1 Fc-YTE | Sino Biological, Inc. | IgG1 Fc-YTE |
| Deposited Data | ||
| IgG1 Fc | Small Angle Scattering Biological Data Bank | SASDDT4 |
| IgG2 Fc | Small Angle Scattering Biological Data Bank | SASDDU4 |
| IgG1 Fc-YTE | Small Angle Scattering Biological Data Bank | SASDDV4 |
| Software and Algorithms | ||
| Blu-Ice | Classen et al., 2013 | https://www.ncbi.nlm.nih.gov/pubmed/23396808 |
| SCÅTTER | Developed by Robert Rambo at the Diamond Light Source (Didcot, UK) | http://www.bioisis.net/tutorial/9 |
| GNOM | Svergun, 1992 | https://www.embl-hamburg.de/biosaxs/gnom.html |
| MODELLER | Sali and Blundell, 1993 | https://salilab.org/modeller/ |
| BILBOMD | Pelikan et al., 2009 | http://bl1231.als.lbl.gov/bilbomd |
| FoXS | Schneidman-Duhovny et al., 2013 | https://modbase.compbio.ucsf.edu/foxs/ |
| Multi-FoXS | Schneidman-Duhovny et al., 2016 | https://modbase.compbio.ucsf.edu/multifoxs/ |
| CHIMERA | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| OriginLab | Origin (OriginLab, Northampton, MA) | https://www.originlab.com/ |
| Unifi Software | Waters Corp. | http://www.waters.com/ |
| Other | ||
| Waters Vion Qtof mass spectrometer | Waters Corp. | http://www.waters.com/ |