Abstract
Interleukin-enhancer binding factor 3 (ILF3) is a double-stranded RNA-binding protein that has been reported to contribute to the occurrence and progression of various malignant tumors. The aim of the present study was to evaluate the prognostic value of ILF3 and to apply this knowledge to avoid excessive medical treatment in patients with lung adenocarcinoma (LUAD). ILF3 expression in a discovery set consisting of tumor and peri-tumor tissue microarrays was analyzed using immunohistochemical methods. The mRNA level of ILF3 was subsequently analyzed in a validation set downloaded from The Cancer Genome Atlas. The Kaplan-Meier method, univariate and multivariate Cox analyses, decision curve analysis and nomogram models were used to evaluate the prognostic value of ILF3. ILF3 expression was upregulated in tumor tissues compared with peri-tumor tissues and was negatively associated with the overall survival time of patients with LUAD in the discovery and validation sets. Moreover, ILF3 expression was used for risk stratification in patients with tumor-node-metastasis stages II–IV and poor-to-moderate tumor differentiation. ILF3 expression was identified as an independent predictor of adverse prognosis for patients with LUAD in the discovery and validation sets. Finally, nomogram models for the 3- and 5 year survival time of patients with LUAD revealed that ILF3 expression may be used to improve the predictive accuracy of the prognosis and to avoid excessive medical treatment for certain patients with the disease. Overall, the data obtained in the current study revealed that high ILF3 expression was associated with poor prognosis, and demonstrated that ILF3, as a potential independent risk factor, may improve the hierarchical postoperative management of patients with LUAD.
Keywords: interleukin-enhancer binding factor 3, lung adenocarcinoma, overall survival, risk factor, prognosis
Introduction
Lung cancer is the most commonly diagnosed malignant tumor and one of the leading causes of cancer-associated mortality worldwide and in China (1,2). Non-small-cell lung cancer (NSCLC) represents ~85–90% of the total number of lung cancer cases (3) and may be divided into three pathological subtypes: Adenocarcinoma, squamous cell carcinoma and large cell carcinoma (4). Among these, lung adenocarcinoma (LUAD) is the most common pathological subtype and accounts for ~40% of all lung cancer cases (5). Despite progress in diagnosis and treatment, the 5 year survival rate of patients with NSCLC remains low (~15%), which is mainly attributed to the low rate of diagnosis in the early stages of the disease and a high rate of cancer recurrence and metastasis (3,6). Therefore, the identification and validation of novel biomarkers may improve the prognosis of patients with LUAD.
Interleukin-enhancer binding factor 3 (ILF3) plays an important role in modulating numerous aspects of RNA metabolism, primarily due to its double-stranded RNA-binding motifs (7). Consequently, ILF3 participates in various cellular biological processes, including cell cycle regulation, DNA metabolism, transcription, translation, mRNA stability, microRNA expression and circular RNA (circRNA) regulation (8–13). Moreover, ILF3 has been linked to the occurrence and progression of various malignant tumors (14–18). For example, ILF3 has been shown to contribute to the occurrence of breast cancer by maintaining the expression of the urokinase-type plasminogen activator (14). Importantly, ILF3 was found to be highly expressed in advanced breast cancer tissues, and the upregulation of ILF3 was negatively associated with the distant metastasis-free survival of patients with breast cancer (15). In human epithelial ovarian cancer, the expression level of ILF3 was increased in tumor tissues compared with peri-tumor tissues and was significantly higher in serous carcinomas compared with mucinous, endometrial and clear cell carcinomas (16). Furthermore, compared with early-stage or well-differentiated ovarian cancer, the expression level of ILF3 was increased in late-stage or poorly-differentiated ovarian cancer (17). It has also been reported that ILF3 sustains the epidermal growth factor receptor-mediated signaling pathway in NSCLC, indicating that ILF3 may serve an important role in the occurrence of cancer (18).
To the best of our knowledge, the prognostic value of ILF3 and its potential predictive significance for guiding clinical practice have not yet been investigated. Therefore, the aim of the present study was to assess the prognostic value of ILF3 and to apply this knowledge to avoid excessive medical treatment of patients with LUAD. The results revealed the prognostic value of ILF3 and established a new prognostic model for overall survival (OS) time in patients with LUAD.
Materials and methods
Study cohorts
The current present involved two independent sets of patients with LUAD. The discovery set consisted of 143 patients, with pathologically confirmed LUAD, who did not receive chemotherapy or radiotherapy prior to surgical resection of the tumor. A total of 143 LUAD and 40 adjacent non-cancerous tissues (2 cm from the tumor margin) were collected following surgery at the Affiliated Hospital of Nantong University (Jiangsu, China) between January 2009 and December 2011. All samples were pathologically confirmed by two pathologists from the Department of Pathology, Affiliated Hospital of Nantong University (Nantong, China). Of the 143 patients, 73 were men while 70 were women, and the mean age of patients at the time of surgery was 61 years (range, 39–83 years). The patient clinical data, including gender, smoking status, age, tumor differentiation and tumor-node-metastasis (TNM) stage, were retrieved from the hospital records. All patients were staged according to the 8th edition of the TNM staging system for lung cancer (19). The OS time was defined as the time between surgery and mortality from any cause. The present retrospective study was approved by the Clinical Research Ethics Committee of The Affiliated Hospital of Nantong University (Jiangsu, China; approval no. 2017-K025) and all patients provided written informed consent.
The validation set was downloaded from The Cancer Genome Atlas (TCGA) database (20), accessed on November 1st, 2017. The patient inclusion criteria for the present study were as follows: Availability of clinical data (including TNM stage, survival status, follow-up time, smoking status, gender, age and ethnicity), a diagnosis of pathologically confirmed LUAD and the availability of mRNA-seq data for ILF3. Patients who died on the day of surgery were excluded from the study. A total of 501 tumor and 59 peri-tumor unpaired samples were selected for subsequent study. A total of 228 patients were men while 273 were women, and the mean age of patients at the time of surgery was 65.40 years (range, 33–88 years).
Tissue microarrays (TMAs) and immunohistochemistry (IHC)
Surgical tissue samples was fixed with 10% formalin for 24 h at room temperature. Formalin-fixed paraffin-embedded surgical tissue samples were used for TMAs and IHC. To construct the TMAs, tissue cylinders (2 mm in diameter) were removed from each sample and selected tissue cylinders were grouped into a single array block using an Unitma Quick-Ray tissue microarrayer (cat. no. UT06; Unitma Co., Ltd). Each TMA specimen was subsequently cut into 4 µm tissue sections, which were mounted on microscope slides. IHC staining was performed as previously described (21), except an anti-ILF3 antibody (ab92355; 1:400; Abcam) was used as the primary antibody. A total of two pathologists blindly evaluated the percentage of ILF3-positive samples using the NDP.view2 software (version 2.6.13; Japan SLC, Inc.), as well as the intensity of ILF3 IHC staining. A semi-quantitative immunoreactivity scoring system was used to evaluate the staining (22). The semi-quantitative H-score (0–300) was calculated as the product of the intensity (0, negative; 1, weak; 2, moderate; 3, strong) and the percentage of ILF-3-positive samples (0–100).
Statistical analysis
X-tile (version 3.6.1; Yale University), GraphPad Prism (version 7.00; GraphPad Software), SPSS (version 20.0; IBM Corp.) and R (version 3.5.2; R Foundation for Statistical Computing) software were used for the statistical analyses. Patients in the discovery and validation sets were stratified into high and low expression groups according to cut-off points (80 and 1,1628.1 for the discovery and validation sets, respectively) that were computed using X-tile as previously described (23). Differential expression of ILF3 between tumor and peri-tumor samples was analyzed using the Mann-Whitney-Wilcoxon test. Survival curves and forest plots were plotted using GraphPad Prism, and the difference in survival between the groups was evaluated using the log-rank test. The χ2 or Fisher's exact tests were used to analyze the association between ILF3 expression and clinicopathological parameters. The Cox proportional hazards regression model was used for univariate and multivariate analyses of factors affecting patient prognosis. The factors that were associated with prognosis in the univariate analysis were subsequently included in a multivariate analysis. Nomograms and calibration curves were plotted using R software with the regression modeling strategies package (version 5.0.0; http://www.r-project.org). Harrell's concordance index (C-index) and the Akaike information criterion (AIC) were calculated to assess and compare the accuracy of the prognostic models (24). All data are expressed as the mean ± standard error mean. P<0.05 was considered to indicate a statistically significant difference.
Results
Association between ILF3 expression, clinicopathological parameters and OS time in patients with LUAD
ILF3-positive staining was predominantly observed in the cytoplasm and nucleus of the LUAD tissue samples (Fig. 1A). The mean expression level of ILF3 in the 143 tumor samples was significantly increased compared with the 40 peri-tumor samples (P<0.001; Fig. 1B). Furthermore, the mRNA level of ILF3 was analyzed in 501 tumor and 59 peri-tumor tissue samples in TCGA database. In line with the IHC staining data, the mRNA level of ILF3 was increased in the tumor samples compared with the peri-tumor samples (P<0.001; Fig. 1C).
Figure 1.
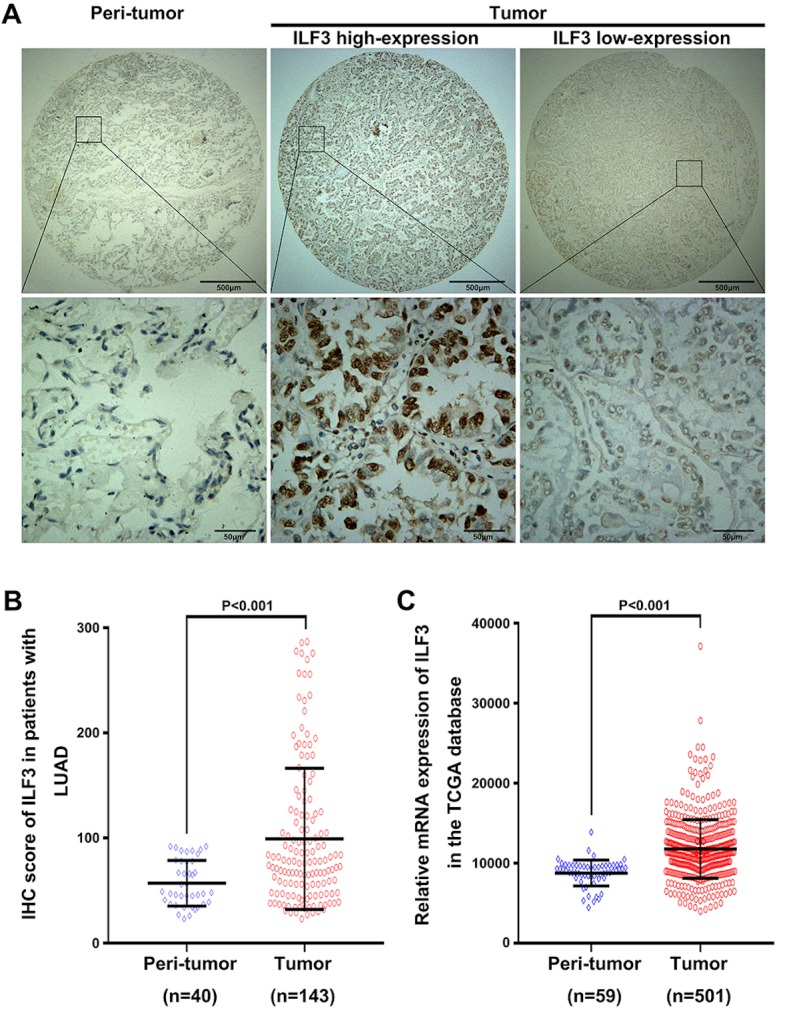
Expression level of ILF3 in LUAD and normal tissues. (A) Representative images of ILF3 staining captured at ×40 (top) and ×400 (bottom) magnification. Scale bars, 500 µm (top) and 50 µm (bottom). (B) ILF3 staining intensity in tumor and peri-tumor tissues in the discovery set. (C) ILF3 mRNA levels in tumor and peri-tumor tissues in the TCGA validation set. ILF3, interleukin-enhancer binding factor 3; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; IHC, immunohistochemistry.
The association between ILF3 expression and clinicopathological parameters in patients with LUAD is presented in Table I. In the discovery set, a high ILF3 expression level was associated with the TNM stage (P=0.041). However, no significant association was found between ILF3 expression and the other clinical parameters examined (tumor differentiation, gender, age and smoking status). Furthermore, no significant association between ILF3 expression and the clinical parameters examined in the TCGA validation set was observed.
Table I.
Association between ILF3 expression and clinical characteristics in patients with lung adenocarcinoma.
| A, Discovery set | |||
|---|---|---|---|
| Characteristics | Low ILF3 expression, n (%) | High ILF3 expression, n (%) | P-value |
| Sex | 0.661 | ||
| Female | 39 (27.3) | 31 (21.7) | |
| Male | 38 (26.6) | 35 (24.5) | |
| Age (years) | 0.080 | ||
| <60 | 38 (26.6) | 23 (16.1) | |
| ≥60 | 39 (27.3) | 43 (30.1) | |
| Smoking | |||
| Yes | 14 (9.8) | 18 (12.6) | 0.193 |
| No/unknown | 63 (44.1) | 48 (33.6) | |
| TNM stage | 0.041a | ||
| I | 49 (34.3) | 28 (19.6) | |
| II | 19 (13.3) | 22 (15.4) | |
| III | 9 (6.3) | 14 (9.8) | |
| IV | 0 (0.0) | 2 (1.4) | |
| Differentiation | 0.711 | ||
| Well | 17 (11.9) | 11 (7.7) | |
| Moderately | 44 (30.8) | 41 (28.7) | |
| Poorly | 16 (11.2) | 14 (9.8) | |
| B, Validation set (TCGA) | |||
| Characteristics | Low ILF3 expression, n (%) | High ILF3 expression, n (%) | P-value |
| Gender | 0.445 | ||
| Female | 153 (30.5) | 120 (24.0) | |
| Male | 120 (24.0) | 108 (21.6) | |
| Age (years) | 0.329 | ||
| <60 | 65 (13.0) | 63 (12.6) | |
| ≥60 | 208 (41.5) | 165 (32.9) | |
| Smoking status | 0.591 | ||
| Yes | 99 (19.8) | 88 (17.6) | |
| No/unknown | 174 (34.7) | 140 (27.9) | |
| TNM stage | 0.590 | ||
| I | 144 (28.7) | 128 (25.6) | |
| II | 72 (14.4) | 49 (9.8) | |
| III | 45 (9.0) | 38 (7.6) | |
| IV | 12 (2.4) | 13 (2.6) | |
| Ethnicity | 0.665 | ||
| Caucasian | 68 (13.6) | 53 (10.6) | |
| Non-caucasian | 205 (40.9) | 175 (34.9) | |
ILF3, interleukin-enhancer binding factor 3. P-values were calculated using χ2 test.
P<0.05 was considered to indicate a statistically significant difference.
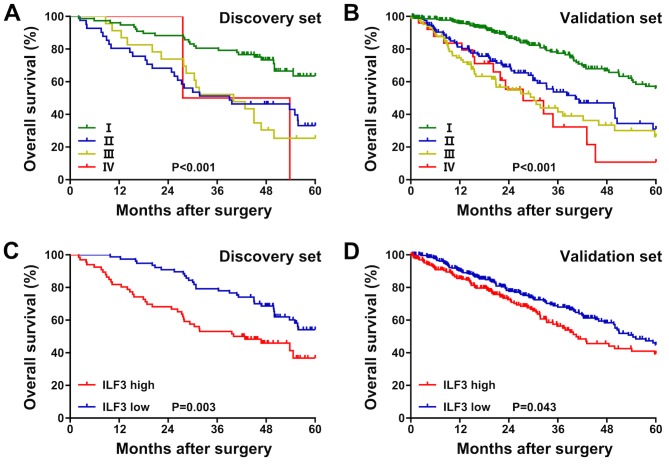
Moreover, in the discovery set, the OS time of patients with TNM stage I was longer than that of patients with TNM stages II–III (P<0.001; Fig. 2A). In the validation set, the OS time of patients with TNM stage I was longer than that of patients with TNM stages II–IV (P<0.001; Fig. 2B). Patients with a low ILF3 expression level exhibited an increased OS time compared with patients with high expression, both in the discovery (P=0.003; Fig. 2C) and validation (P=0.043; Fig. 2D) sets.
Figure 2.
Kaplan-Meier survival plots in patients with lung adenocarcinoma. Kaplan-Meier survival plots for the (A) discovery and (B) validation sets stratified by the tumor-node-metastasis stage. Kaplan-Meier survival plots for the (C) discovery and (D) validation sets stratified by ILF3 expression. ILF3, interleukin-enhancer binding factor 3.
Subgroup analysis of the association between ILF3 expression and the OS time in patients with different TNM stages
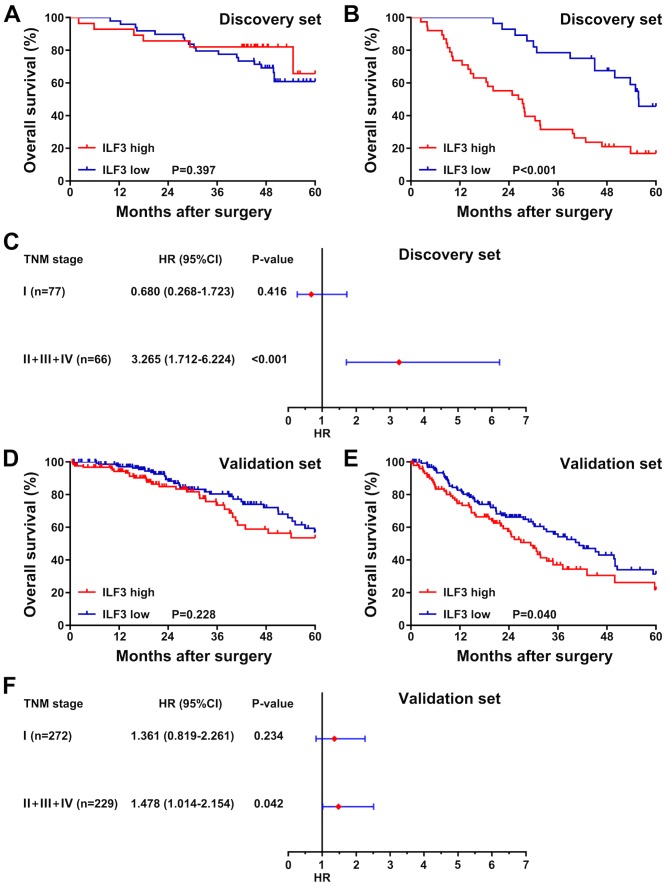
To determine whether the OS time of patients with LUAD with different TNM stages was associated with the ILF3 expression level, the patients in the discovery and validation sets were divided into two subgroups based on TNM stage. The first subgroup consisted of patients with TNM stage I, and the second subgroup consisted of patients with TNM stages II–IV. Survival analysis was subsequently performed for each subgroup, and the association between ILF3 expression and the OS time of patients was presented as a forest plot based on the univariate analysis. In the discovery set, no significant difference in survival time was observed between patients with high or low ILF3 expression in the TNM stage I subgroup (P=0.397; Fig. 3A). However, in the TNM stages II–IV subgroup, the OS time of patients with high ILF3 expression was shorter than that of patients with low ILF3 expression (P<0.001; Fig. 3B). Furthermore, the ILF3 expression level was significantly associated with the OS time of patients in the TNM stages II–IV subgroup (P<0.001; Fig. 3C), but not in the TNM stage I subgroup (P=0.416).
Figure 3.
Subgroup analysis of the association between ILF3 expression and the overall survival time of patients with different TNM stages. Representation of the survival in the TNM (A) stage I and (B) stages II–IV subgroups in the discovery set stratified by ILF3 expression (high and low). (C) Forest plot based on ILF3 expression in the discovery set. Representation of the survival time in the TNM (D) stage I and (E) stages II–IV patient subgroups in the validation set stratified by ILF3 expression (high and low). (F) Forest plot based on ILF3 expression in the validation set. ILF3, interleukin-enhancer binding factor 3; TNM, tumor-node-metastasis; HR, hazard ratio; CI, confidence interval.
In the validation set, as in the discovery set, ILF3 expression was not associated with the OS time of patients in the TNM stage I subgroup (P=0.228; Fig. 3D). By contrast, in the TNM stages II–IV subgroup, patients with low ILF3 expression had an improved OS time than those with high ILF3 expression (P=0.040; Fig. 3E). Moreover, in the validation set, the ILF3 expression level was significantly associated with the OS time of patients with TNM stages II–IV (P=0.042; Fig. 3F), but not TNM stage I (P=0.234).
Subgroup analysis of the association between ILF3 expression and the OS time in patients with different tumor differentiation
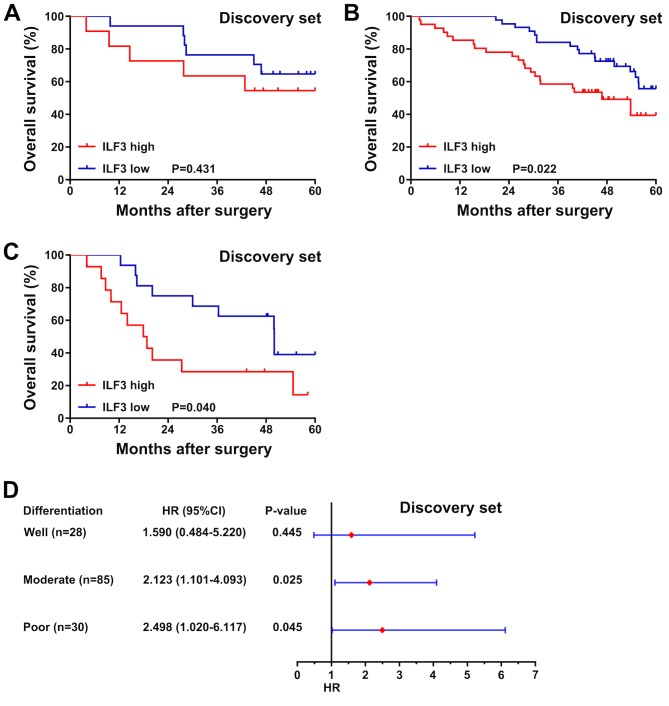
In the discovery set, no significant association between ILF3 expression and the OS time was observed in patients with well-differentiated tumors (P=0.431; Fig. 4A). Nevertheless, among patients with moderately or poorly differentiated tumors, those with a low ILF3 expression level had an increased OS time compared with patients with high expression (P=0.022 and P=0.040, respectively; Fig. 4B and C). Moreover, a forest plot based on a univariate analysis revealed that ILF3 expression was associated with the OS time of patients with moderate or poor tumor differentiation (P=0.025 and P=0.045, respectively; Fig. 4D), but not with well-differentiated tumors (P=0.445).
Figure 4.
Subgroup analysis of the association between ILF3 expression and the overall survival time of patients with different tumor differentiation. Representation of the survival time of patients in the discovery set with (A) well, (B) moderate and (C) poor tumor differentiation stratified by ILF3 expression (high and low). (D) Forest plot based on ILF3 expression in the discovery set. ILF3, interleukin-enhancer binding factor 3; HR, hazard ratio; CI, confidence interval.
ILF3 expression is an independent risk factor for patients with LUAD
The results of univariate and multivariate analyses are presented in Table II. In the univariate analysis, ILF3 expression (P=0.003 and P=0.044, for the discovery and validation sets, respectively) and the TNM stage (P<0.001, for both discovery and validation sets) were significant risk factors for OS time. Moreover, tumor differentiation (P=0.017) and smoking status (P=0.033) in the discovery set and ethnicity (P=0.013) in the validation set were identified as significant risk factors for OS time.
Table II.
Univariate and multivariate analyses of overall survival time in patients with lung adenocarcinoma.
| A, Univariate analysis | ||||
|---|---|---|---|---|
| Discovery set | Validation set (TCGA) | |||
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, years | ||||
| ≥60 vs. <60 | 0.868 (0.541–1.394) | 0.558 | 0.882 (0.627–1.240) | 0.470 |
| Sex | ||||
| Male vs. female | 1.437 (0.890–2.320) | 0.138 | 1.062 (0.785–1.436) | 0.697 |
| Smoking | ||||
| Yes vs. no/unknown | 1.765 (1.048–2.972) | 0.033a | 1.209 (0.884–1.653) | 0.236 |
| Ethnicity | ||||
| Caucasian vs. non-caucasian | 1.610 (1.103–2.350) | 0.013a | ||
| Differentiation | ||||
| Poor and moderate vs. well | 1.599 (1.087–2.350) | 0.017a | ||
| TNM stage | ||||
| II, III and IV vs. I | 1.682 (1.300–2.175) | <0.001a | 1.648 (1.428–1.903) | <0.001a |
| ILF3 expression | ||||
| High vs. low | 2.050 (1.270–3.311) | 0.003a | 1.363 (1.008–1.844) | 0.044a |
| B, Multivariate analysis | ||||
| Discovery set | Validation set (TCGA) | |||
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Smoking status | ||||
| Yes vs. no | 1.512 (0.893–2.561) | 0.124 | ||
| Ethnicity | 1.594 (1.093–2.326) | 0.016a | ||
| Caucasian vs. Non-caucasian | ||||
| Differentiation | ||||
| Poor and moderate vs. well | 1.542 (1.042–2.283) | 0.030a | ||
| TNM stage | ||||
| II, III and IV vs. I | 1.544 (1.184–2.015) | 0.001a | 1.634 (1.417–1.883) | <0.001a |
| ILF3 expression | ||||
| High vs. low | 1.761 (1.075–2.885) | 0.025a | 1.376 (1.017–1.862) | 0.038a |
HR, hazard ratio; CI, confidence interval; TCGA, The Cancer Genome Atlas; TNM, tumor-node-metastasis; ILF3, interleukin-enhancer binding factor 3.
P<0.05 was considered to indicate a statistically significant difference.
Subsequently, all the significant risk factors for OS time identified in the univariate analysis were used for multivariate analysis, and the results were presented as a forest plot showing risk factors for prognosis in patients with LUAD. This analysis revealed that ILF3 expression was an independent risk factor for OS time in the discovery (P=0.025) and validation (P=0.038) sets. Furthermore, the TNM stage (P=0.001 and P<0.001, for the discovery and validation sets, respectively), tumor differentiation (P=0.030, for the discovery set) and ethnicity (P=0.016, for the validation set) were identified as underlying independent risk factors for OS time in patients with LUAD (Fig. 5).
Figure 5.
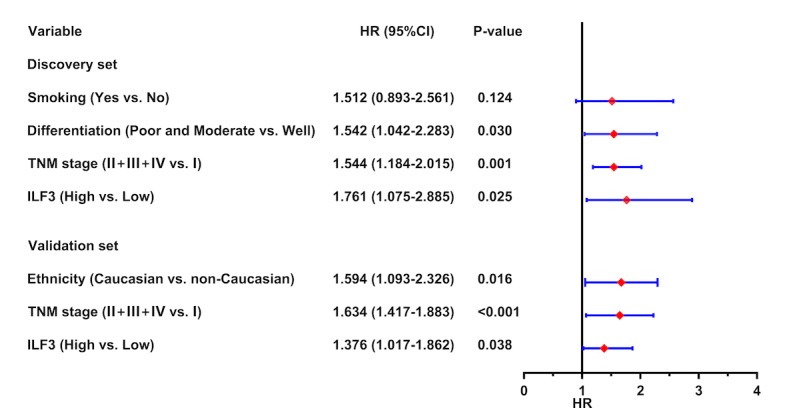
Forest plot based on ILF3 expression and other risk factors. In the discovery set, ILF3 expression (based on immunohistochemistry), tumor differentiation and TNM stage were identified as independent risk factors for OS time in patients with LUAD. In the validation set, ILF3 expression (based on the mRNA level), TNM stage and ethnicity were identified as independent risk factors for OS time in patients with LUAD. ILF3, interleukin-enhancer binding factor 3; TNM, tumor-node-metastasis; OS, overall survival; LUAD, lung adenocarcinoma; HR, hazard ratio; CI, confidence interval.
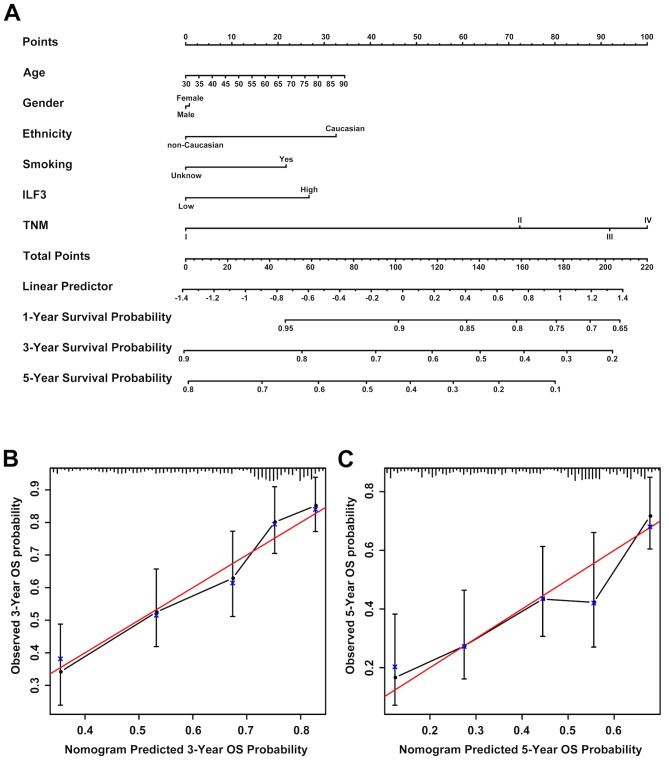
Prognostic nomograms for OS time in patients with LUAD
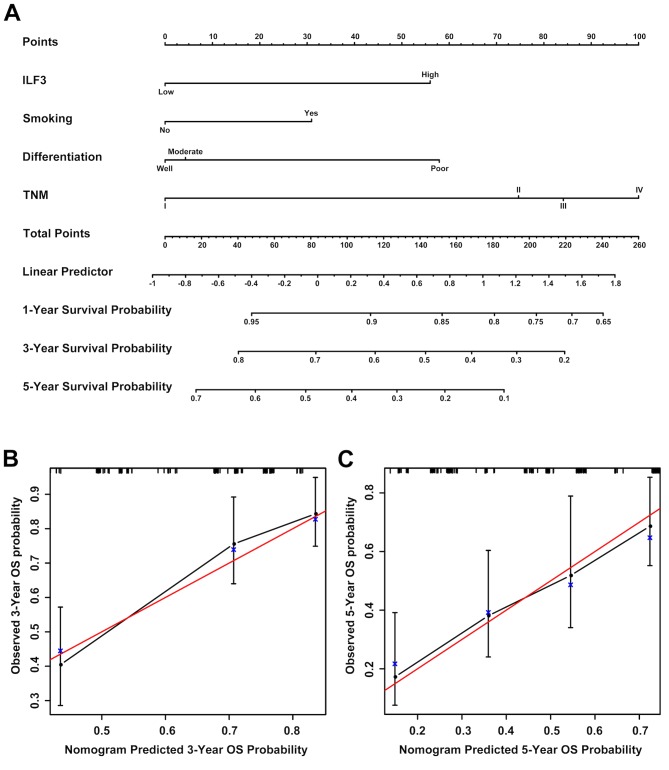
Prognostic nomograms estimating the OS time of patients with LUAD were generated for the discovery (Fig. 6A) and validation (Fig. 7A) sets based on the results obtained in the univariate and multivariate analyses. The nomogram based on the discovery set data integrated ILF3 expression, TNM stage, tumor differentiation and smoking status, whereas the nomogram based on the validation set data integrated all the risk factors, including ILF3 expression, TNM stage, age, gender, ethnicity and smoking status. Each patient received points based on each risk factor, and the total number of points (i.e., the sum of the points received for each risk factor) was used to predict the OS time. A high number of total points were associated with advanced age, smoking, high ILF3 expression, poor differentiation, females and the Caucasian race. It was also associated with a high risk and poor prognosis. The calibration curve for the probability of survival time at 3 or 5 years revealed a strong association between the predicted outcome from the nomogram and the actual observed outcome for the discovery (Fig. 6B and C) and validation (Fig. 7B and C) sets.
Figure 6.
Prognostic nomogram for patients with LUAD based on the discovery set data. (A) A prognostic nomogram integrating ILF3 expression (based on immunohistochemistry) and other risk factors (including smoking status, tumor differentiation and TNM stage) was generated to predict the clinical outcome of patients with LUAD. Calibration curves of the prognostic nomogram for the (B) 3- and (C) 5 year probability of survival time are presented. The red line represents an ideal model, and the vertical bars represent the 95% confidence interval. An ideal model suggests that the predicted outcome from the nomogram is completely identical to that of the actual observed outcome and it characterized by sensitivity 100%, specificity 100% and veracity 100%. LUAD, lung adenocarcinoma; ILF3, interleukin-enhancer binding factor 3; TNM, tumor-node-metastasis.
Figure 7.
Prognostic nomogram for patients with LUAD based on the validation set data. (A) A prognostic nomogram integrating ILF3 expression (based on the mRNA level) and other risk factors (including smoking status, ethnicity, age, gender and TNM stage) was generated to predict the clinical outcome of patients with LUAD. Calibration curves of the prognostic nomogram for the (B) 3- and (C) 5 year probability of survival time are presented. The lines on the top of (B) and (C) represent the distribution of patients. The red line represents an ideal model, and vertical bars represent the 95% confidence interval. LUAD, lung adenocarcinoma; ILF3, interleukin-enhancer binding factor 3; TNM, tumor-node-metastasis.
The predictive accuracy of the prognostic nomograms based on ILF3 expression was evaluated using the C-index and AIC. The results of the comparison between the prognostic model based on ILF3 expression and the conventional prognostic model based on the TNM stage are presented in Table III. For the discovery set, the C-index for the probability of survival at 3 and 5 years was increased to 0.7045 and 0.6823, respectively, for the model based on ILF3 expression, while the AIC was decreased to 433.0039 and 624.1688, respectively. Similarly, for the validation set, the C-index for the probability of survival time at 3 and 5 years was increased to 0.6970 and 0.6909, respectively, for the model based on ILF3 expression, while the AIC was decreased to 1506.3810 and 1827.3640, respectively. Therefore, the nomogram based on ILF3 expression provided an improved predictive accuracy compared with the prognostic model based on the TNM stage in both the discovery and validation sets.
Table III.
Comparison of the accuracy of the prognostic models.
| A, Discovery set | ||||
|---|---|---|---|---|
| 3-year OS time | 5-year OS time | |||
| Model | C-index | AIC | C-index | AIC |
| TNM | 0.6263 | 445.7592 | 0.6269 | 619.0112 |
| ILF3-based model | 0.7045 | 443.0039 | 0.6823 | 624.1688 |
| B, Validation set (TCGA) | ||||
| 3-year OS time | 5-year OS time | |||
| Model | C-index | AIC | C-index | AIC |
| TNM | 0.6679 | 1506.5460 | 0.6613 | 1831.0630 |
| ILF3-based model | 0.6970 | 1506.3810 | 0.6909 | 1827.3640 |
OS, overall survival; C-index, Harrell's concordance index; AIC, Akaike information criterion; TNM, tumor-node-metastasis; ILF3, interleukin-enhancer binding factor 3; TCGA, The Cancer Genome Atlas.
As presented in Fig. S1, a decision curve analysis for the prediction of OS time was performed. Compared with the model based on the TNM stage, a model combining the analysis of the TNM stage and ILF3 expression benefited patients with a survival probability of <49 or >74%. For example, if a survival probability of 80% was used as a threshold, the net benefit of the combined model was ~0.05, which was greater than the net benefit of the model based on the TNM stage (0.01).
Discussion
The ILF3 family consists of four members (NF90a, NF90b, NF110a and NF110b), which result from the mutually exclusive alternative splicing of the ILF3 gene transcript (25–27). NF90 and NF110 serve important roles in the regulation of circRNA biogenesis and the antiviral immune response (13). Furthermore, ILF3 interacts with Nanog homeobox mRNA to regulate pluripotency in embryonic stem cells and has potential roles in sustaining embryonic stem cell self-renewal and cell fate determination (28). While a previous study indicated that ILF3 expression may be a novel risk factor for venous thromboembolism, stroke and coronary artery disease (29), ILF3 autoantibodies have been identified as potential diagnostic biomarkers for human autoimmune disease (30). Furthermore, ILF3 interacts with interleukin-2 in T cells to upregulate synoviolin in rheumatoid synovial cells and is therefore a potential therapeutic target for rheumatoid arthritis (31). To the best of our knowledge, the current study is the first to report that ILF3 was an independent risk factor for OS time in patients with LUAD.
Increasing evidence suggests that ILF3 may contribute to the aggressiveness and progression of certain malignant tumors, including hepatocellular carcinoma, NSCLC and breast and ovarian cancer (14,16,32,33). In the current study, ILF3 expression was significantly associated with the TNM stage and OS time of patients with LUAD. Furthermore, subgroup analyses revealed that patients with TNM stages II–IV and poor or moderate tumor differentiation may be stratified according to ILF3 expression. Collectively, these results suggested that ILF3 expression may significantly affect the prognosis of patients with LUAD.
Moreover, when the conventional prognostic model based on the TNM stage was applied to patients with LUAD, the C-index of the discovery set was low compared with that of the validation set. This phenomenon may be associated with differences in the levels of economic development and medical services provided in different regions. Nevertheless, compared with the conventional prognostic model based on the TNM stage, the nomogram model based on ILF3 expression exhibited improved predictive accuracy for the OS time of patients with LUAD, in both the discovery and validation sets.
LincIN, a novel NF90-binding long non-coding RNA, is upregulated in advanced breast tumors and is involved in metastasis (34). NF90 is a member of ILF3 family, and LincIN is a novel long non-coding RNA which binding NF90. So LincIN is associated with the current study. Upregulated expression of ILF2 in NSCLC is associated with tumor cell proliferation and poor prognosis (35). The present study suggested that ILF3 is a potential independent adverse prognostic factor for post-operative survival time in patients with LUAD and may be beneficial for the postoperative hierarchical management of patients with the disease.
Further work is required to strengthen the results obtained in the present study. First, due to a lack of information on disease recurrence or progression, the present study only reported the analysis of the association between ILF3 expression and OS time. Additional work is required to establish further associations with disease recurrence and progression. Secondly, the prognostic value of ILF3 expression for patients with LUAD requires further validation in more extensive prospective multi-center clinical trials, which may improve the reliability of the prognostic nomogram based on ILF3 expression. Thirdly, the ILF3 expression level was not investigated in patients with suspected LUAD and this requires further investigation. Furthermore, the present study lacked differentiation data of patients in the validation set, which may have affected the predictive accuracy of the nomogram, based on ILF3. Finally, the role of ILF3 in the development and progression of LUAD remains unclear, and further studies are required to elucidate the underlying mechanisms.
Overall, the present study identified ILF3 as a predictor of adverse prognosis in patients with LUAD. Furthermore, determining the expression level of ILF3 may improve the hierarchical post-operative management of patients with LUAD based on the TNM stage or tumor differentiation. Moreover, the prognostic nomogram based on ILF3 expression and other risk factors presented a significant improvement in the predictive accuracy of the survival time of patients with LUAD compared with the model based on the TNM stage and may avoid excessive medical treatment for patients with the disease.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ILF3
interleukin-enhancer binding factor 3
- LUAD
lung adenocarcinoma
- TCGA
The Cancer Genome Atlas
- OS
overall survival
- TNM
tumor-node-metastasis
- NSCLC
non-small-cell lung cancer
- circRNA
circular RNA
- TMA
tissue microarray
- IHC
immunohistochemistry
- C-index
the Harrell's concordance index
- AIC
Akaike information criterion
Funding
The present study was funded by the National Natural Science Foundation of China (grant no. 81770266) and the ‘Six-One’ Project for High-Level Health Talents of the Jiangsu Province (grant no. LGY2016037).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available in TCGA database (https://www.cancer.gov).
Authors' contributions
JS and HZ designed the present study. YL, JZ and ZW acquired the data. XL, ZX, YL, JZ, ZW and HH analyzed and interpreted the data. CJ and XJL performed statistical analysis. ZX drafted the initial manuscript. HZ and XL revised the initial manuscript for important intellectual content. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The current study was approved by the Clinical Research Ethics Committee of The Affiliated Hospital of Nantong University (Jiangsu, China) and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-Small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao S, Zhong L, Liu J, Feng J, Bian T, Zhang Q, Chen J, Lv X, Chen J, Liu Y. Prognostic value of decreased GRK6 expression in lung adenocarcinoma. J Cancer Res Clin Oncol. 2016;142:2541–2549. doi: 10.1007/s00432-016-2244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang JT, Lee YM, Huang RS. The impact of the cancer genome atlas on lung cancer. Transl Res. 2015;166:568–585. doi: 10.1016/j.trsl.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minguet J, Smith KH, Bramlage P. Targeted therapies for treatment of non-small cell lung cancer-recent advances and future perspectives. Int J Cancer. 2016;138:2549–2561. doi: 10.1002/ijc.29915. [DOI] [PubMed] [Google Scholar]
- 7.Castella S, Bernard R, Corno M, Fradin A, Larcher JC. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip Rev RNA. 2015;6:243–256. doi: 10.1002/wrna.1270. [DOI] [PubMed] [Google Scholar]
- 8.Xu YH, Leonova T, Grabowski GA. Cell cycle dependent intracellular distribution of two spliced isoforms of TCP/ILF3 proteins. Mol Genet Metab. 2003;80:426–436. doi: 10.1016/j.ymgme.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Zhuang J, Shen L, Yang L, Huang X, Lu Q, Cui Y, Zheng X, Zhao X, Zhang D, Huang R, et al. TGFβ1 promotes gemcitabine resistance through regulating the LncRNA-LET/NF90/miR-145 signaling axis in bladder cancer. Theranostics. 2017;7:3053–3067. doi: 10.7150/thno.19542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larcher JC, Gasmi L, Viranaicken W, Edde B, Bernard R, Ginzburg I, Denoulet P. Ilf3 and NF90 associate with the axonal targeting element of tau mRNA. FASEB J. 2004;18:1761–1763. doi: 10.1096/fj.04-1763fje. [DOI] [PubMed] [Google Scholar]
- 11.Pei Y, Zhu P, Dang Y, Wu J, Yang X, Wan B, Liu JO, Yi Q, Yu L. Nuclear export of NF90 to stabilize IL-2 mRNA is mediated by AKT-dependent phosphorylation at Ser647 in response to CD28 costimulation. J Immunol. 2008;180:222–229. doi: 10.4049/jimmunol.180.1.222. [DOI] [PubMed] [Google Scholar]
- 12.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, Chen LL. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–227. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 14.Hu Q, Lu YY, Noh H, Hong S, Dong Z, Ding HF, Su SB, Huang S. Interleukin enhancer-binding factor 3 promotes breast tumor progression by regulating sustained urokinase-type plasminogen activator expression. Oncogene. 2013;32:3933–3943. doi: 10.1038/onc.2012.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Yang C, Zhang M, Liu H, Gong C, Zhang J, Xu S, Zou J, Kai Y, Li Y. Interleukin enhancer-binding factor 3 and HOXC8 co-activate cadherin 11 transcription to promote breast cancer cells proliferation and migration. Oncotarget. 2017;8:107477–107491. doi: 10.18632/oncotarget.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, Fu P, Zhu H, Reed E, Remick SC, Petros W, Mueller MD, Yu JJ. Correlations among ERCC1, XPB, UBE2I, EGF, TAL2 and ILF3 revealed by gene signatures of histological subtypes of patients with epithelial ovarian cancer. Oncol Rep. 2012;27:286–292. doi: 10.3892/or.2011.1483. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Yu JJ. Gene expression patterns in the histopathological classification of epithelial ovarian cancer. Exp Ther Med. 2010;1:187–192. doi: 10.3892/etm_00000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng CC, Chou KF, Wu CW, Su NW, Peng CL, Su YW, Chang J, Ho AS, Lin HC, Chen CG, et al. EGFR-mediated interleukin enhancer-binding factor 3 contributes to formation and survival of cancer stem-like tumorspheres as a therapeutic target against EGFR-positive non-small cell lung cancer. Lung Cancer. 2018;116:80–89. doi: 10.1016/j.lungcan.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Detterbeck FC, Chansky K, Groome P, Bolejack V, Crowley J, Shemanski L, Kennedy C, Krasnik M, Peake M, Rami-Porta R, et al. The IASLC lung cancer staging project: Methodology and validation used in the development of proposals for revision of the stage classification of NSCLC in the forthcoming (Eighth) edition of the TNM classification of lung cancer. J Thorac Oncol. 2016;11:1433–1446. doi: 10.1016/j.jtho.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 20.Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu K, Wang S, Liu Y, Gu J, Gu S, Xu Z, Zhang R, Wang Z, Ma H, Chen Y, Ji L. Overexpression of MYCN promotes proliferation of non-small cell lung cancer. Tumour Biol. 2016;37:12855–12866. doi: 10.1007/s13277-016-5236-2. [DOI] [PubMed] [Google Scholar]
- 22.Ji L, Li H, Gao P, Shang G, Zhang DD, Zhang N, Jiang T. Nrf2 pathway regulates multidrug-resistance-associated protein 1 in small cell lung cancer. PLoS One. 2013;8:e63404. doi: 10.1371/journal.pone.0063404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 24.Weiss A, Chavez-MacGregor M, Lichtensztajn DY, Yi M, Tadros A, Hortobagyi GN, Giordano SH, Hunt KK, Mittendorf EA. Validation study of the American joint committee on cancer eighth edition prognostic stage compared with the anatomic stage in breast cancer. JAMA Oncol. 2018;4:203–209. doi: 10.1001/jamaoncol.2017.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duchange N, Pidoux J, Camus E, Sauvaget D. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene. 2000;261:345–353. doi: 10.1016/S0378-1119(00)00495-9. [DOI] [PubMed] [Google Scholar]
- 26.Reichman TW, Parrott AM, Fierro-Monti I, Caron DJ, Kao PN, Lee CG, Li H, Mathews MB. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J Mol Biol. 2003;332:85–98. doi: 10.1016/S0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- 27.Chaumet A, Castella S, Gasmi L, Fradin A, Clodic G, Bolbach G, Poulhe R, Denoulet P, Larcher JC. Proteomic analysis of interleukin enhancer binding factor 3 (Ilf3) and nuclear factor 90 (NF90) interactome. Biochimie. 2013;95:1146–1157. doi: 10.1016/j.biochi.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Guo C, Xue Y, Yang G, Yin S, Shi W, Cheng Y, Yan X, Fan S, Zhang H, Zeng F. Nanog RNA-binding proteins YBX1 and ILF3 affect pluripotency of embryonic stem cells. Cell Biol Int. 2016;40:847–860. doi: 10.1002/cbin.10539. [DOI] [PubMed] [Google Scholar]
- 29.Hinds DA, Buil A, Ziemek D, Martinez-Perez A, Malik R, Folkersen L, Germain M, Malarstig A, Brown A, Soria JM, et al. Genome-Wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum Mol Genet. 2016;25:1867–1874. doi: 10.1093/hmg/ddw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bremer HD, Landegren N, Sjoberg R, Hallgren A, Renneker S, Lattwein E, Leonard D, Eloranta ML, Ronnblom L, Nordmark G, et al. ILF2 and ILF3 are autoantigens in canine systemic autoimmune disease. Sci Rep. 2018;8:4852. doi: 10.1038/s41598-018-23034-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izumi T, Fujii R, Izumi T, Nakazawa M, Yagishita N, Tsuchimochi K, Yamano Y, Sato T, Fujita H, Aratani S, et al. Activation of synoviolin promoter in rheumatoid synovial cells by a novel transcription complex of interleukin enhancer binding factor 3 and GA binding protein alpha. Arthritis Rheum. 2009;60:63–72. doi: 10.1002/art.24178. [DOI] [PubMed] [Google Scholar]
- 32.Jiang W, Huang H, Ding L, Zhu P, Saiyin H, Ji G, Zuo J, Han D, Pan Y, Ding D, et al. Regulation of cell cycle of hepatocellular carcinoma by NF90 through modulation of cyclin E1 mRNA stability. Oncogene. 2015;34:4460–4470. doi: 10.1038/onc.2014.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo NL, Wan YW, Tosun K, Lin H, Msiska Z, Flynn DC, Remick SC, Vallyathan V, Dowlati A, Shi X, et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14:8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Z, Slater CM, Zhou Y, Devarajan K, Ruth KJ, Li Y, Cai KQ, Daly M, Chen X. LincIN, a novel NF90-binding long non-coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res. 2017;19:62. doi: 10.1186/s13058-017-0853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni T, Mao G, Xue Q, Liu Y, Chen B, Cui X, Lv L, Jia L, Wang Y, Ji L. Upregulated expression of ILF2 in non-small cell lung cancer is associated with tumor cell proliferation and poor prognosis. J Mol Histol. 2015;46:325–335. doi: 10.1007/s10735-015-9624-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available in TCGA database (https://www.cancer.gov).