Abstract
F-box and WD repeat domain-containing protein 7 (FBW7), also known as FBXW7, AGO or hCDC4, is an F-box protein with seven tandem WD40 repeats. FBW7 is a key substrate recognition subunit of the Skp1-Cul1-F-box-protein E3 ubiquitin ligase. FBW7 targets for ubiquitination and destruction of numerous crucial transcription factors and protooncogenes, including cyclin E, c-Myc, c-Jun, Notch and MCL-1. FBW7 is a well-characterized tumor suppressor, and its gene is frequently mutated or deleted in various types of human cancer, including colorectal cancer, gastric cancer, ovarian cancer and different types of leukemia. Accumulating evidence indicates that the aberrant expression of FBW7 is involved in the development of hematological tumors, including T cell acute lymphoblastic leukemia, adult T cell leukemia/lymphoma, chronic lymphocytic leukemia and multiple myeloma. The present review will describe the latest findings on the role of FBW7 in hematological tumors, in order to identify a novel target for future therapies.
Keywords: F-box and WD repeat domain-containing protein 7, ubiquitin-proteasome system, hematological tumor, T cell acute lymphoblastic leukemia, adult T cell leukemia/lymphoma, chronic lymphocytic leukemia, multiple myeloma
1. Introduction
The ubiquitin-proteasome system (UPS) is the major regulatory pathway of protein degradation in eukaryotic cells (1). Defects in UPS function can result in several diseases, including cancer (2). F-box and WD repeat domain-containing protein 7 (FBW7), also known as FBW7, AGO or hCDC4, is an essential component of the Skp1-Cul1-F-box (SCF)-type ubiquitin ligase (E3) complex, which comprises four subunits: Skp1, cullin1, Rbx1 and an F-box protein that determines substrate specificity (3). FBW7 is a member of the F-box protein family (4). Previous studies demonstrated that dysregulation of FBW7 serves a crucial role in the development of hematological tumors (5–7) (Fig. 1).
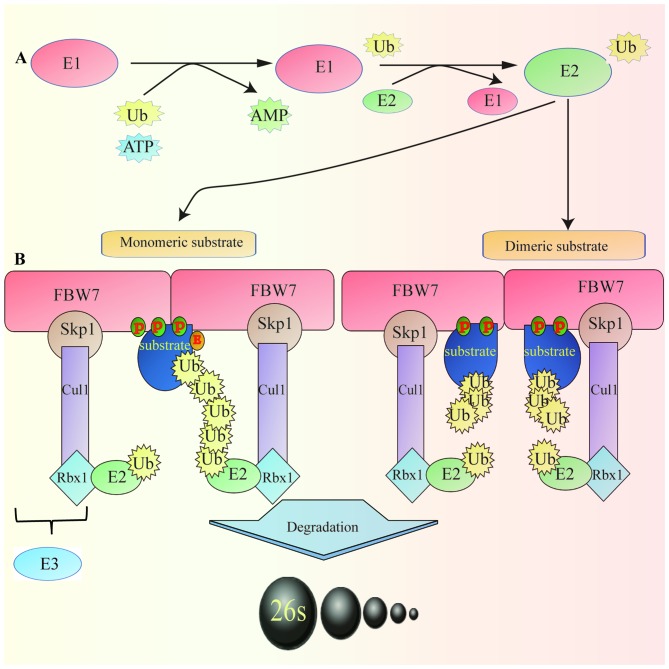
Figure 1.
Ubiquitin-proteasome system and FBW7 (monomeric and dimeric) degrading specific substrate. (A) E1 activates Ub protein that is transferred to the cysteine residue of E2 to form the E2-Ub complex. Subsequently, in a step catalyzed by E3, Ub covalently binds to the target protein, forming a polyubiquitin chain. The ubiquitinated protein is recognized by the 26S proteasome and degraded in an ATP-dependent manner. (B) FBW7 is present as a monomer and a dimer. Dimerization of the FBW7 protein enhances the activity and the binding of FBW7 to substrates, particularly to substrates with weak CDC4 phosphodegron, including cyclin-dependent kinase inhibitor Sicl. However, certain protein substrates, including cyclin E and c-Myc, can be efficiently degraded by monomeric FBW7. Cul1, cullin1; FBW7, F-box and WD repeat domain-containing protein 7; P, phosphorus; Rbx1, ring-box 1; Skp1, S-phase kinase associated protein 1; Ub, ubiquitin; E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin ligase
2. Structure and activity of FBW7
The human FBW7 gene is located on chromosome 4 (4q31.3) and encodes three transcripts named FBW7α, FBW7β and FBW7γ (8). The corresponding three proteins possess identical catalytic function but distinct subcellular localizations. FBW7α is mostly localized in the nucleus, whereas FBW7β is found in the cytoplasm and FBW7γ in the nucleolus (9). These three subtypes have different amino acid sequences in the N-terminal region but contain conserved interaction regions (F-box and WD40 repeats) in the C-terminal region (9–11). All isoforms share three essential domains that have distinct functions: i) D domain; ii) F-box domain and iii) seven tandem WD40 (tryptophan-aspartic acid 40) repeats (9) (Fig. 2). The D domain promotes FBW7 dimerization and facilitates its binding to substrates (9). The F-box domain is necessary for direct binding of FBW7 to Skp1, enabling the recruitment of other components of SCF (9). The WD40 repeats are required for substrate recognition (12).
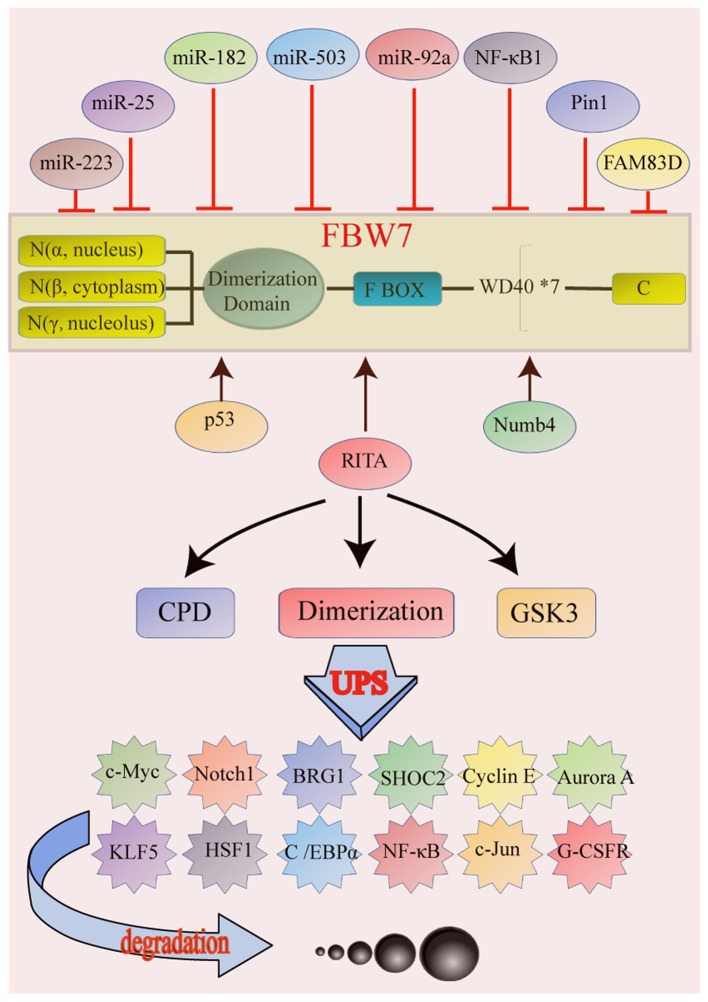
Figure 2.
Structure and function of FBW7, and genes affected by FBW7. (A) FBW7 exists as three protein isoforms, α, β and γ, which have different amino acid sequences in the N-terminal region but contain conserved interaction regions (F-box and WD40 repeats) in the C-terminal region. The isoforms share three essential domains with distinct functions: i) D domain, ii) F-box domain and iii) 7 tandem WD40 (tryptophan-aspartic acid 40) repeats. (B) p53, RITA and Numb4 are activators of FBW7, whereas miR-223, miR-25, miR-182, miR-503, miR-92a, NF-κB 1, Pin1 and FAM83D inhibit FBW7 activation. FBW7 is a direct transcriptional target of p53. RITA increased protein expression of FBW7 by inhibiting Notch signaling. Numb4 promotes assembly and activation of FBW7. miRNAs bind directly to the 3′UTR of FBW7 and decrease its expression. NF-κB 1 decreases FBW7 mRNA level by inhibiting transcription factor E2F1 (E2F1)-mediated FBW7 promoter transcriptional activation. Pin1 binds to FBW7 in a phoshorylation-dependent manner and promotes FBW7 self-ubiquitination and protein degradation by inhibiting FBW7 dimerization. FAM83D physically interacts with and downregulates FBW7 protein levels. (C) CDPs binding, FBW7 dimerization and GSK3-β catalyzation are all important mechanisms in FBW7 ubiquitination and degradation. BRG1, brahma-related gene-1; C/EBPα, CCAAT/enhancer-binding protein α; FAM83D, family with sequence similarity 83 member D; FBW7, FBW7, F-box and WD repeat domain-containing protein 7; G-CSFR, granulocyte colony-stimulating factor receptor; GSK3, glycogen synthase 3; HSF1, heat shock transcription factor 1; KLF5, Kruppel like factor 5; miR, micro RNA; RITA, RBP-J-interaction and tubulin-associated protein; SHOC2, SHOC2 leucine rich repeat scaffold protein.
The target proteins degraded by FBW7 contain a conserved sequence of phosphorylated amino acids named CDC4 phosphodegron (CDP). CDPs bind to the WD40 repeats, allowing the recognition of the substrate by ubiquitin ligase for its subsequent degradation (10). Furthermore, dimerization of FBW7 increases the binding of FBW7 to substrates, particularly to those with weak phosphodegrons (13). In addition, glycogen synthase 3β (GSK3β) serves a crucial role in the degradation of substrate by FBW7. GSK3β catalyzes the phosphorylation of threonine in the substrate CDPs, promoting FBW7 binding and accelerating the degradation of the substrate (14,15) (Fig. 2).
3. Substrates of FBW7 in cancer
FBW7 is an important substrate adaptor responsible for recognition and binding of the substrate proteins in the ubiquitin-proteasome degradation pathway. Many of its substrates are oncogenes. For example, c-Myc is an important substrate for ubiquitination by FBW7 (16,17).
In general, decreased expression of FBW7 results in a significant increase in the intracellular content of c-Myc protein, whereas overexpression of FBW7 promotes c-Myc ubiquitination and degradation, decreasing its cellular level (14). In the Burkitt's lymphoma cell line, c-Myc is most commonly mutated at T58, causing the failure of FBW7 to regulate c-Myc protein degradation, ultimately leading to c-Myc protein accumulation and tumorigenesis (14). Cyclin E is another classical substrate of FBW7. By binding to cyclin E-dependent kinase 2 (CDK2), cyclin E forms a complex that promotes the G/S phase transition, leading to uncontrolled cell proliferation (18). FBW7 can also ubiquitinate various other target proteins, including Notch1 (19), NF-κB (20), c-Jun (21), granulocyte colony stimulating factor receptor (22), SHOC2 leucine rich repeat scaffold protein (23), brahma-related gene-1 (24), Aurora A (25), Kruppel like factor 5 (26), heat shock transcription factor 1 (27) and CCAAT/enhancer-binding protein α (28) (Table I).
Table I.
Factors regulated by FBW7.
| Author, year | Factors | Function | Reference |
|---|---|---|---|
| Yada et al, 2004 Welcker et al, 2004 | c-Myc | Transcription factor that regulates the expression of many genes | (16,17) |
| Koepp et al, 2001 | Cyclin E | Regulates cell-cycle entry and progression | (18) |
| Weng et al, 2004 | Notch1 | Transcriptional regulators of cell fate and differentiation | (19) |
| Fukushima et al, 2012 | NF-κB | Important factor in many fundamental cellular processes | (20) |
| Wei et al, 2005 | c-Jun | Transcription factor which regulates the cell cycle | (21) |
| Lochab et al, 2013 | G-CSFR | Controls the production, differentiation, and function of granulocytes | (22) |
| Xie et al, 2019 | SHOC2 | Involved autophagy induction and cell survival | (23) |
| Huang et al, 2018 | BRG1 | Involved in governing chromatin structure and gene transcription | (24) |
| Finkin et al, 2008 | Aurora-A | Involved in the cell mitosis and cell cycle regulatory process | (25) |
| Liu N et al, 2010 | KLF5 | Promotes differentiation and self-renewal of fat cells | (26) |
| Kourtis et al, 2015 | HSF1 | Regulates the heat-shock response and supports malignancy | (27) |
| Bengoechea-Alonso et al, 2010 | C/EBPα | Involved in lipid metabolism | (28) |
BRG1, brahma-related gene-1; C/EBPα, CCAAT/enhancer-binding protein α; HSF1, heat shock transcription factor 1; KFL5, Kruppel like factor 5; SHOC2, SHOC2 leucine rich repeat scaffold protein.
4. Regulation of FBW7
The main mechanisms of FBW7 disruption in cancer are deletion, mutation and promoter methylation (29–31) and the gene mutation is the most common. A previous study demonstrated that FBW7 is mutated in ~6% of human tumors, and in 31% of acute T-cell lymphocytic leukemia cases (32). It has been demonstrated that conditional FBW7 knockout in the T-cell lineage of mice leads initially to thymic hyperplasia and subsequently to the development of thymic lymphoma, indicating that the loss of FBW7 function is an important factor responsible for tumorigenesis in the blood system (33). In addition, FBW7 expression and activity can be regulated by numerous genes, including p53 (34). FBW7 is a p53-dependent tumor suppressor, and p53 activation can enhance the ubiquitination-mediated degradation of oncoproteins (34). Furthermore, previous studies have identified numerous micro (mi)RNAs that can regulate FBW7 expression, including miR-223 (35), miR-25 (36), miR-182 (37), miR-503 (37) and miR-92a (38). In addition, in T-cell acute lymphoblastic leukemia (T-ALL), miR-223 acts as an oncogene by inhibiting the FBW7 expression. The expression of FBW7 is also controlled by the RBP-J-interaction and tubulin-associated (RITA) protein. Overexpression of RITA results in increased expression of FBW7 and decreased expression of cyclin E, cyclin D1, CDK2, Hes-1 and NF-kBp65 (39). In addition, NF-κB1 (40), Pin1 (41), family with sequence similarity 83 member D (FAM83D) (42) and Numb4 (43) are also regulators of FBW7 expression (Table II).
Table II.
Factors regulating FBW7.
| Author, year | Factors | Activator/Inhibitor | Reference |
|---|---|---|---|
| Kimura et al, 2003 | p53 | Activator | (34) |
| Mansour et al, 2013 | miRNAs | Inhibitor | (35–38) |
| Xiang et al, 2015 | |||
| Li et al, 2014 | |||
| Zhou et al, 2015 | |||
| Wang et al, 2014 | RITA | Inhibitor | (39) |
| Huang et al, 2014 | NF-κB1 | Inhibitor | (40) |
| Min et al, 2012 | Pin1 | Inhibitor | (41) |
| Wang et al, 2013 | FAM83D | Inhibitor | (42) |
| Jiang et al, 2012 | Numb4 | Activator | (43) |
FAM83D, family with sequence similarity 83 member D; miRNA, microRNA; RITA, RBP-J-interaction and tubulin-associated protein.
5. FBW7 mRNA expression in different types of human cancer
In order to determine whether FBW7 is differently expressed in tumor tissues compared with normal tissues, the Oncomine database (https://www.oncomine.org) was used to analyze FBW7 mRNA levels in different tumor and normal tissues. The results indicated that FBW7 expression is higher in leukemia (Fig. 3A), suggesting that FBW7 may be associated with the initiation and progression of certain hematological tumors. The Timer database (https://cistrome.shinyapps.io/timer/) was then used to analyze FBW7 mRNA levels in diffuse large B cell lymphoma (DLBCL). The result demonstrated no significant difference in FBW7 expression level in DLBCL. This may be due to different expression levels of FBW7 in various hematological tumors (Fig. 3B).
Figure 3.
FBW7 expression levels in different types of human cancer. (A) FBW7 expression in different types of cancer compared with normal tissues obtained from Oncomine dataset. (B) Human FBW7 expression levels in different types of tumor obtained from The Cancer Genome Atlas database were determined by TIMER (https://cistrome.shinyapps.io/timer/). *P<0.05, **P<0.01 and ***P<0.001. FBW7, FBW7, F-box and WD repeat domain-containing protein 7.
6. Role of the FBW7 gene in common hematological tumors
T-ALL
T-ALL is a highly proliferative hematologic malignancy caused by the malignant transformation of T-cell progenitors (44). Patients with T-ALL typically present with aggressive clinical features correlated with poor prognosis, including inhibition of normal hematopoietic function, high white blood cell counts, pleural effusions and central nervous system involvement (45). T-ALL accounts for 15 and 25% of the total number of childhood and adult cases of acute lymphoblastic leukemia, respectively (46). The rate of T-ALL complete remission can reach 94%, and the long-term survival rate can be as high as 85% (47); however, 20% pediatric patients and 40% adult patients are prone to recurrence and ultimately develop refractory leukemia (48). Understanding the underlying mechanisms of T-ALL development is therefore critical. T-ALL is the only known malignant cancer that can be induced by FBW7 deletion without the requirement of other tumor-promoting factors (49,50). Furthermore, T-ALL development is accelerated by the simultaneous loss of p53 or Phosphatase and TENsin homolog or by concurrent activation of Notch (49,51,52). However, it was demonstrated that FBW7 hotspot mutation knock-in mice (FBW7mut/+) do not develop spontaneous leukemia, suggesting that the FBW7 missense mutation and FBW7 homologous deletion present subtle differences (53). FBW7 deletion/mutation increases the protein level of Notch and c-Myc (49,50,53,54). However, Notch1 alone is not sufficient to induce or maintain T-ALL in the absence of c-Myc function; however, c-Myc deletion in established T-ALL specifically ablates leukemia-initiating cells (LIC), and the inhibition of c-Myc induction by small molecule inhibitors of bromodomain and extra-terminal motif/bromodomains can suppress the proliferation of mouse and human T-ALL cells, suggesting that c-Myc could be considered as a key oncogene driving T-ALL (53). It was reported that >60% of T-ALL cases present with abnormal activation of the Notch1 signaling pathway, indicating that Notch1 might be the most common oncogene in T-ALL (55). Mutations of Notch1 occur mainly in the heterodimeric domain region and the proline, glutamine, serine, and threonine domain (56,57). Activating mutations of Notch1 gene result in sustained expression of genes regulating T cell differentiation, including Hes family bHLH transcription factor 1 and CD25, changing the expression balance of c-Myc and p27 (58–60), upregulating the NF-κB signaling pathway (5), enhancing the expression of the anti-apoptotic X-linked inhibitor of apoptosis protein associated with ubiquitination and degradation, and activating the PKB/Akt/mTOR signaling pathway-mediated inhibition of p53 (61). All the aforementioned signaling pathways inhibit apoptosis and cause abnormal proliferation of non-functional T cells, thereby directly leading to the development of T-ALL. In addition, Notch and NF-κB bind to the promoter of miR-223, activating its expression (62). Notch-mediated upregulation of miR-223 subsequently inhibits FBW7 gene expression in T-ALL, and miR-223 and FBW7 expression are negatively correlated in T-ALL patient-derived xenografts (62). In addition, it has been demonstrated that FBW7 loss of function leads to an upregulation of the glucocorticoid receptor in primary T-ALL cells, thereby enhancing their sensitivity to glucocorticoids and improving the prognosis of patients with T-ALL (63). MCL-1 is an anti-apoptotic protein of the BCL-2 family that promotes cancer by inhibiting apoptosis (64). Deletion of FBW7 in T-ALL cell line results in an increased expression level of MCL-1. MCL-1 upregulation is sensitive to various kinase inhibitors, including sorafenib, but resistant to the BCL2 inhibitor ABT-737. However, when FBW7 function is restored or MCL-1 is deleted, cell sensitivity to ABT-737 is restored (65). MCL-1 also serves a role in the chemotherapy efficacy of anti-tubulin drugs. Treatment with paclitaxel and similarly acting compounds induces MCL-1 phosphorylation, which is then recognized by FBW7 and degraded by ubiquitination. Subsequently, the intracellular MCL-1 protein content is significantly decreased and promotes therefore apoptosis. In addition, when FBW7 is inactivated or downregulated in the tumor, the stability of MCL-1 protein is enhanced, thereby increasing resistance of the tumor to microtubule-targeted drugs and decreasing the effectiveness of chemotherapy (66). The cellular content of MCL-1 protein is also affected by the deubiquitinating enzyme named ubiquitin specific peptidase 9 X-Linked (USP9X) (67). Since USP9X decreases the ubiquitination of MCL-1 and increases its stability, USP9X downregulation and decreased MCL-1 protein expression could increase tumor cell sensitivity to ABT-737 treatment.
Adult T cell leukemia/lymphoma (ATL)
ATL is a malignant T cell monoclonal proliferative disease caused by the human T-cell leukemia virus type 1 (HTLV-1). ATL represents a rare type of lymphocytic leukemia/lymphoid tumor affecting T cells (68). FBW7 acts as a tumor suppressor in ATL cells; however mutations can transform FBW7 into an oncogenic protein (69). Mutations in the WD40 domain of FBW7 were identified in 25% (8/32) of acute ATL cases (69). Furthermore, the FBW7 D510E and D527G mutants are capable of ubiquitinating proteolysis of endogenous cyclin E, MCL-1, and c-Myc. However, these mutants are ineffective in degrading the Notch intracellular domain (NICD) in ATL cells, resulting in the activation of Notch1 signaling. The increased Notch1 signaling can subsequently promote ATL cell proliferation and tumorigenesis (69). The same mutants present significant carcinogenic activity when co-expressed with HTLV-1 Tax, mutated p53 R276H or c-Myc38C (69). In addition, previous studies reported the downregulation of FBW7 expression in ATL, which leads to c-Myc accumulation and initiation of ATL cell proliferation (70). The c-Myc-FBW7 axis pathway could therefore represent a potential target for the treatment of ATL.
Chronic lymphocytic leukemia (CLL)
CLL is the most common type of leukemia in Western countries (71). CLL is primarily characterized by the accumulation of monoclonal CD5+ mature B cells in lymphoid tissues and peripheral blood (72). Despite significant advances in its treatment, CLL remains incurable (73). In CLL, the mutation rate of FBW7 is 2–6% (74–76), and these mutations are associated with chromosome 12 trisomy aberrations of mature CD19+ B cells (75). A previous study identified FBW7 mutations in 36 out of 905 untreated patients with CLL. All these mutations are heterozygous, missense in 78% of cases and primarily affect the substrate-binding domain of WD40 (77). Furthermore, 10% of the mutations were located on the first exon of the α-transcript. In order to identify the target protein of FBW7 in CLL, a WD40 domain-truncated CLL cell line (HG-3) was generated using the CRISPR/Case9 method (77). Homozygous truncation of FBW7 results in increased levels of activated Notch1-NICD and c-Myc proteins, and in enhanced activity of HIF1-α (77). In primary cells derived from patients with CLL carrying the FBW7 mutation, the level of activated Notch1-NICD was elevated and remained stable after inhibition of translation (77). In addition, FBW7 mutation was combined with enhanced expression of Notch1 target genes (77). FBW7 mutations in CLL cells may therefore explain the dysregulation of Notch1 signaling in certain patients with CLL, indicating that FBW7 could be a cancer-driving gene in CLL.
Multiple myeloma (MM)
MM, which is an incurable malignant plasma cell disease, is one of the most common hematological malignancies in adults, accounting for 15% of malignant hematological tumors (78). MM is primarily characterized by clonal proliferation and abnormal accumulation of plasma cells in the bone marrow, combined with the secretion of a large number of monoclonal immunoglobulins (79). The etiology of MM is complex. NF-κB may serve a crucial role in supporting MM cell survival. The NF-κB signaling pathway is activated in the majority of patients with MM, whereas NF-κB signaling pathway blockage using some inhibitors of NF-κB kinase subunit β (IKK2) can inhibit MM cell proliferation (80,81). In non-canonical NF-κB pathway, IKKα is activated by NIK and directly phosphorylates p100. Phosphorylated p100 is therefore degraded in a 26S proteasome-dependent manner (82). It has been demonstrated that p100 degradation in the nucleus is crucial to activate the non-canonical NF-κB pathway (83). During this reaction, FBW7 recognizes the CPD sequence of p100 and promotes p100 degradation in a GSK-3 phosphorylation-dependent manner (15). Furthermore, FBW7 overexpression enhances the activity of NF-κB, whereas FBW7 downregulation can upregulate p100 expression (15). In MM, FBW7 silencing leads to an increased expression of p100 and partly activates MM cell apoptosis (15,84). FBW7 may therefore be an oncogene in MM. In addition, fibroblasts from patients with MM are crucial in the progression of the disease and in drug resistance (85,86). For example, it was demonstrated that miR-27b-3p is significantly upregulated in the fibroblasts of bone marrow from patients with MM where it controls FBW7 activity, which in turn regulates MCL-1 that can promote fibroblast proliferation (87).
7. Conclusions and perspectives
FBW7 is an important tumor suppressor that regulates multiple oncogenes, including cyclin E, c-Myc, Notch, c-Jun and mTOR. In addition, FBW7 is regulated by p53, certain miRNAs, including miR-223, miR-25, miR-182, miR-503 and miR-92a, RITA, FAM83D and Numb4. Mutation, deletion and hypermethylation are the main mechanisms of FBW7 disruption in cancer, leading to tumor progression. Mutation of FBW7 may promote tumorigenesis and increase the tumor resistance to chemotherapy. Furthermore, the carcinogenic effects of FBW7 have also been identified in certain types of hematological tumor, including ATL and MM. These findings have important implications for the understanding of hematopoietic mechanisms, development of diagnostic reagents, and design and optimization of therapeutic drugs. The detection of FBW7 mutations is clinically relevant, and FBW7 could serve as a potential target for the treatment of hematological tumors. Further investigation into FBW7 mutations will have a positive impact on the prevention of hematological tumors and the development of personalized treatments.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Science and Technology Major Project for New Drug (grant no. 2017ZX301033), the National Natural Science Foundation of China (grant nos. 81874049, 81602179 and 81570198), the Co-construction of Provincial and Department Project (grant no. WKJ-ZJ-1919) and the Zhejiang Provincial Natural Science Foundation of China (grant no. LY19H160036).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
QZ, LH, XT and QX conceived and designed the review. YG and ZX were involved in designing the review. QZ, LH, YG, ZX, XT and QX were involved in the collection of references. QZ and LH collected and assembled the data presented in Tables I and II. QZ and LH designed the figures. QZ and LH wrote the manuscript. YG collected and assembled the data present in Tables I and II. ZX was involved in designing the figures and analysing the data of figure 3. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing interests.
References
- 1.Hershko A, Ciechanover A, Varshavsky A. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: The role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu K, Nihira NT, Inuzuka H, Wei W. Physiological functions of FBW7 in cancer and metabolism. Cell Signal. 2018;46:15–22. doi: 10.1016/j.cellsig.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uddin S, Bhat AA, Krishnankutty R, Mir F, Kulinski M, Mohammad RM. Involvement of F-BOX proteins in progression and development of human malignancies. Semin Cancer Biol. 2016;36:18–32. doi: 10.1016/j.semcancer.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welcker M, Clurman BE. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 7.Tan Y, Sangfelt O, Spruck C. The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett. 2008;271:1–12. doi: 10.1016/j.canlet.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 8.Spruck CH, Strohmaier H, Sangfelt O, Müller HM, Hubalek M, Müller-Holzner E, Marth C, Widschwendter M, Reed SI. hCDC4 gene mutations in endometrial cancer. Cancer Res. 2002;62:4535–4539. [PubMed] [Google Scholar]
- 9.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Welcker M, Larimore EA, Swanger J, Bengoechea-Alonso MT, Grim JE, Ericsson J, Zheng N, Clurman BE. Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 2013;27:2531–2536. doi: 10.1101/gad.229195.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crusio K M, King B, Reavie L B, Aifantis I. The ubiquitous nature of cancer: the role of the SCF Fbw7 complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 14.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O'Connor O, Hoffmann A, Elenitoba-Johnson KS, Pagano M. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol. 2012;14:375–385. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 18.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 19.Weng AP, Ferrando AA, Lee W, Morris JP, IV, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima H, Matsumoto A, Inuzuka H, Zhai B, Lau AW, Wan L, Gao D, Shaik S, Yuan M, Gygi SP, et al. SCF(Fbw7) modulates the NFkB signaling pathway by targeting NFkB2 for ubiquitination and destruction. Cell Rep. 2012;1:434–443. doi: 10.1016/j.celrep.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Lochab S, Pal P, Kapoor I, Kanaujiya JK, Sanyal S, Behre G, Trivedi AK. E3 ubiquitin ligase Fbw7 negatively regulates granulocytic differentiation by targeting G-CSFR for degradation. Biochim Biophys Acta. 2013;1833:2639–2652. doi: 10.1016/j.bbamcr.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Xie CM, Sun Y. The MTORC1-mediated autophagy is regulated by the FBXW7-SHOC2-RPTOR axis. Autophagy. 2019;15:1470–1472. doi: 10.1080/15548627.2019.1609864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L-Y, Zhao J, Chen H, Wan L, Inuzuka H, Guo J, Fu X, Zhai Y, Lu Z, Wang X, et al. SCFFBW7-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat Commun. 2018;9:3569. doi: 10.1038/s41467-018-06038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkin S, Aylon Y, Anzi S, Oren M, Shaulian E. Fbw7 regulates the activity of endoreduplication mediators and the p53 pathway to prevent drug-induced polyploidy. Oncogene. 2008;27:4411–4421. doi: 10.1038/onc.2008.77. [DOI] [PubMed] [Google Scholar]
- 26.Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, Wang Y, Wang W, Wang R, Wang Q, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kourtis N, Moubarak RS, Aranda-Orgilles B, Lui K, Aydin IT, Trimarchi T, Darvishian F, Salvaggio C, Zhong J, Bhatt K, et al. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat Cell Biol. 2015;17:322–332. doi: 10.1038/ncb3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengoechea-Alonso MT, Ericsson J. The ubiquitin ligase Fbxw7 controls adipocyte differentiation by targeting C/EBPalpha for degradation. Proc Natl Acad Sci USA. 2010;107:11817–11822. doi: 10.1073/pnas.0913367107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akhoondi S, Lindström L, Widschwendter M, Corcoran M, Bergh J, Spruck C, Grandér D, Sangfelt O. Inactivation of FBW7/hCDC4-beta expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 2010;12:R105. doi: 10.1186/bcr2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 33.Perry JM, Li L. Self-renewal versus transformation: Fbxw7 deletion leads to stem cell activation and leukemogenesis. Genes Dev. 2008;22:1107–1109. doi: 10.1101/gad.1670708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura T, Gotoh M, Nakamura Y, Arakawa H. hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 2003;94:431–436. doi: 10.1111/j.1349-7006.2003.tb01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansour MR, Sanda T, Lawton LN, Li X, Kreslavsky T, Novina CD, Brand M, Gutierrez A, Kelliher MA, Jamieson CH, et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med. 2013;210:1545–1557. doi: 10.1084/jem.20122516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang J, Hang JB, Che JM, Li HC. MiR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int J Clin Exp Pathol. 2015;8:9147–9153. [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Sarver AL, Khatri R, Hajeri PB, Kamenev I, French AJ, Thibodeau SN, Steer CJ, Subramanian S. Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J Pathol. 2014;234:488–501. doi: 10.1002/path.4407. [DOI] [PubMed] [Google Scholar]
- 38.Zhou C, Shen L, Mao L, Wang B, Li Y, Yu H. miR-92a is upregulated in cervical cancer and promotes cell proliferation and invasion by targeting FBXW7. Biochem Biophys Res Commun. 2015;458:63–69. doi: 10.1016/j.bbrc.2015.01.066. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Yang Z, Liu C, Huang S, Wang H, Chen Y, Chen G. RBP-J-interacting and tubulin-associated protein induces apoptosis and cell cycle arrest in human hepatocellular carcinoma by activating the p53-FBW7 pathway. Biochem Biophys Res Commun. 2014;454:71–77. doi: 10.1016/j.bbrc.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 40.Huang H, Ma L, Li J, Yu Y, Zhang D, Wei J, Jin H, Xu D, Gao J, Huang C. NF-κB1 inhibits c-Myc protein degradation through suppression of FBW7 expression. Oncotarget. 2014;5:493–505. doi: 10.18632/oncotarget.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, Shaik S, Lee DY, Finn G, Balastik M, et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell. 2012;46:771–783. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Liu Y, Zhang P, Zhang W, Wang W, Curr K, Wei G, Mao JH. FAM83D promotes cell proliferation and motility by downregulating tumor suppressor gene FBXW7. Oncotarget. 2013;4:2476–2486. doi: 10.18632/oncotarget.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang X, Xing H, Kim TM, Jung Y, Huang W, Yang HW, Song S, Park PJ, Carroll RS, Johnson MD. Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem Cells. 2012;30:1313–1326. doi: 10.1002/stem.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peirs S, Van der Meulen J, Van de Walle I, Taghon T, Speleman F, Poppe B, Van Vlierberghe P. Epigenetics in T-cell acute lymphoblastic leukemia. Immunol Rev. 2015;263:50–67. doi: 10.1111/imr.12237. [DOI] [PubMed] [Google Scholar]
- 45.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 46.Vadillo E, Dorantes-Acosta E, Pelayo R, Schnoor M. T cell acute lymphoblastic leukemia (T-ALL): New insights into the cellular origins and infiltration mechanisms common and unique among hematologic malignancies. Blood Rev. 2018;32:36–51. doi: 10.1016/j.blre.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Allen A, Sireci A, Colovai A, Pinkney K, Sulis M, Bhagat G, Alobeid B. Early T-cell precursor leukemia/lymphoma in adults and children. Leuk Res. 2013;37:1027–1034. doi: 10.1016/j.leukres.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 49.Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, Mashimo Y, Oguro H, Nitta E, Ito K, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22:986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alborán IM, Nakayama K, Nakayama KI. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Jr, Liu Y, Huang Y, Kang HC, DelRosario R, Jen KY, et al. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834–844. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C. Regulation of leukemia-initiating cell activity by the ubiquitin ligase FBXW7. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tosello V, Ferrando AA. The NOTCH signaling pathway: Role in the pathogenesis of T-cell acute lymphoblastic leukemia and implication for therapy. Ther Adv Hematol. 2013;4:199–210. doi: 10.1177/2040620712471368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arruga F, Gizdic B, Bologna C, et al. Mutations in NOTCH1 PEST domain orchestrate CCL19-driven homing of chronic lymphocytic leukemia cells by modulating the tumor suppressor gene DUSP22. 2017;31:1882. doi: 10.1038/leu.2016.383. [DOI] [PubMed] [Google Scholar]
- 57.Chiang MY, Radojcic V, Maillard I. Oncogenic Notch signaling in T-cell and B-cell lymphoproliferative disorders. Curr Opin Hematol. 2016;23:362–370. doi: 10.1097/MOH.0000000000000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 59.Suresh S, Irvine AE. The NOTCH signaling pathway in normal and malignant blood cell production. J Cell Commun Signal. 2015;9:5–13. doi: 10.1007/s12079-015-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar V, Palermo R, Talora C, Campese AF, Checquolo S, Bellavia D, Tottone L, Testa G, Miele E, Indraccolo S, et al. Notch and NF-kB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:2324–2335. doi: 10.1038/leu.2014.133. [DOI] [PubMed] [Google Scholar]
- 63.Malyukova A, Brown S, Papa R, O'Brien R, Giles J, Trahair TN, Dalla Pozza L, Sutton R, Liu T, Haber M, et al. FBXW7 regulates glucocorticoid response in T-cell acute lymphoblastic leukaemia by targeting the glucocorticoid receptor for degradation. Leukemia. 2013;27:1053–1062. doi: 10.1038/leu.2012.361. [DOI] [PubMed] [Google Scholar]
- 64.Yang-Yen HF. Mcl-1: A highly regulated cell death and survival controller. J Biomed Sci. 2006;13:201–204. doi: 10.1007/s11373-005-9064-4. [DOI] [PubMed] [Google Scholar]
- 65.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 67.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 68.Katsuya H, Ishitsuka K, Utsunomiya A, Hanada S, Eto T, Moriuchi Y, Saburi Y, Miyahara M, Sueoka E, Uike N, et al. ATL-Prognostic Index Project Treatment and survival among 1594 patients with ATL. Blood. 2015;126:2570–2577. doi: 10.1182/blood-2015-03-632489. [DOI] [PubMed] [Google Scholar]
- 69.Yeh C-H, Bellon M, Pancewicz-Wojtkiewicz J, Nicot C. Oncogenic mutations in the FBXW7 gene of adult T-cell leukemia patients. Proc Natl Acad Sci USA. 2016;113:6731–6736. doi: 10.1073/pnas.1601537113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mihashi Y, Mizoguchi M, Takamatsu Y, Ishitsuka K, Iwasaki H, Koga M, Urabe K, Momosaki S, Sakata T, Kiyomi F, et al. C-MYC and its main ubiquitin ligase, FBXW7, influence cell proliferation and prognosis in adult T-cell leukemia/lymphoma. Am J Surg Pathol. 2017;41:1139–1149. doi: 10.1097/PAS.0000000000000871. [DOI] [PubMed] [Google Scholar]
- 71.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 72.Kipps TJ, Stevenson FK, Wu CJ, Croce CM, Packham G, Wierda WG, O'Brien S, Gribben J, Rai K. Chronic lymphocytic leukaemia. Nat Rev Dis Primers. 2017;3:16096. doi: 10.1038/nrdp.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bosch F, Dalla-Favera R. Chronic lymphocytic leukaemia: From genetics to treatment. Nat Rev Clin Oncol. 2019;16:684–701. doi: 10.1038/s41571-019-0239-8. [DOI] [PubMed] [Google Scholar]
- 74.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Böttcher S, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, Alpermann T, Roller A, Kohlmann A, Haferlach T, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28:108–117. doi: 10.1038/leu.2013.263. [DOI] [PubMed] [Google Scholar]
- 76.Quijada-Álamo M, Hernández-Sánchez M, Robledo C, Hernández-Sánchez JM, Benito R, Montaño A, Rodríguez-Vicente AE, Quwaider D, Martín AÁ, García-Álvarez M, et al. Next-generation sequencing and FISH studies reveal the appearance of gene mutations and chromosomal abnormalities in hematopoietic progenitors in chronic lymphocytic leukemia. J Hematol Oncol. 2017;10:83. doi: 10.1186/s13045-017-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Close V, Close W, Kugler SJ, Reichenzeller M, Yosifov DY, Bloehdorn J, Pan L, Tausch E, Westhoff MA, Döhner H, et al. FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood. 2019;133:830–839. doi: 10.1182/blood-2018-09-874529. [DOI] [PubMed] [Google Scholar]
- 78.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 79.Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC, Chng WJ, Moreau P, Attal M, Kyle RA, et al. Treatment of multiple myeloma with high-risk cytogenetics: A consensus of the International Myeloma Working Group. Blood. 2016;127:2955–2962. doi: 10.1182/blood-2016-01-631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jourdan M, Moreaux J, De Vos J, Hose D, Mahtouk K, Abouladze M, Robert N, Baudard M, Rème T, Romanelli A, et al. Targeting NF-kappaB pathway with an IKK2 inhibitor induces inhibition of multiple myeloma Cell Proliferation. Br J Haematol. 2007;138:160–168. doi: 10.1111/j.1365-2141.2007.06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun SC. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Senftleben U, Cao Y, Xiao G, Greten FR, Krähn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 84.Busino L, Millman SE, Pagano M. SCF-mediated degradation of p100 (NF-κB2): Mechanisms and relevance in multiple myeloma. Sci Signal. 2012;5:pt14. doi: 10.1126/scisignal.2003408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frassanito MA, Rao L, Moschetta M, Ria R, Di Marzo L, De Luisi A, Racanelli V, Catacchio I, Berardi S, Basile A, et al. Bone marrow fibroblasts parallel multiple myeloma progression in patients and mice: In vitro and in vivo studies. Leukemia. 2014;28:904–916. doi: 10.1038/leu.2013.254. [DOI] [PubMed] [Google Scholar]
- 86.Frassanito MA, De Veirman K, Desantis V, Di Marzo L, Vergara D, Ruggieri S, Annese T, Nico B, Menu E, Catacchio I, et al. Halting pro-survival autophagy by TGFβ inhibition in bone marrow fibroblasts overcomes bortezomib resistance in multiple myeloma patients. Leukemia. 2016;30:640–648. doi: 10.1038/leu.2015.289. [DOI] [PubMed] [Google Scholar]
- 87.Frassanito MA, Desantis V, Di Marzo L, Craparotta I, Beltrame L, Marchini S, Annese T, Visino F, Arciuli M, Saltarella I, et al. Bone marrow fibroblasts overexpress miR-27b and miR-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J Pathol. 2019;247:241–253. doi: 10.1002/path.5187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.