Abstract
Triple-negative breast cancer (TNBC) is a subtype of breast cancer that is characterized by aggressive and metastatic clinical characteristics and generally leads to earlier distant recurrence and poorer prognosis than other molecular subtypes. Accumulating evidence has demonstrated that long non-coding RNAs (lncRNAs) serve a crucial role in a wide variety of biological processes by interacting with microRNAs (miRNAs) as competing endogenous RNAs (ceRNAs) and, thus, affect the expression of target genes in multiple types of cancer. Seven datasets from the Gene Expression Omnibus (GEO) database, including 444 tumor and 88 healthy tissue samples, were utilized to investigate the underlying mechanisms of TNBC and identify prognostic biomarkers. Differentially expressed genes (DEGs) were further validated in The Cancer Genome Atlas database and the associations between their expression levels and clinical information were analyzed to identify prognostic values. A potential lncRNA-miRNA-mRNA ceRNA network was also constructed. Finally, 69 mRNAs from the integrated Gene Expression Omnibus datasets were identified as DEGs using the robust rank aggregation method with |log2FC|>1 and adjusted P<0.01 set as the significance cut-off levels. In addition, 29 lncRNAs, 21 miRNAs and 27 mRNAs were included in the construction of the ceRNA network. The present study elucidated the mechanisms underlying the progression of TNBC and identified novel prognostic biomarkers for TNBC.
Keywords: triple-negative breast cancer, long non-coding RNA, microRNA, competing endogenous RNA
Introduction
Triple-negative breast cancer (TNBC) is a typical molecular subtype of breast cancer that lacks the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) and accounts for 10–20% of all types of breast cancer (1,2). TNBC is also well known for its aggressive and metastatic clinical characteristics and generally leads to early distant recurrence and poor prognosis (3,4). Currently, no specific targeted therapy is available for TNBC (5). Therefore, it is crucial to identify potential biomarkers and novel therapeutic targets for the development of a more efficient treatment.
Emerging evidence has indicated that long non-coding RNAs (lncRNAs) play a vital role in a large variety of biological processes, including genetic transcription, chromosome modification, cell cycle, cell differentiation and migration (6–8). Various studies have indicated that specific miRNAs may participate in tumor progression and function as oncogenes or tumor suppressor genes (9–11). Moreover, the competing endogenous RNA (ceRNA) hypothesis, which suggests that non-coding RNAs and pseudogene transcripts are able to compete for the same miRNA response elements in order to regulate each other and communicate with mRNAs, has gained increasing interest (12). Thereafter, this hypothesis was validated experimentally by further studies (13,14). However, published studies with large sample sizes and specific biomarkers for TNBC are limited. Therefore, the ceRNA network of TNBC has not yet been fully investigated and requires further exploration.
In the present study, published microarray and sequencing data were searched in the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases and gene expression profiling data were collected from a large sample size of patients with TNBC, in order to identify candidate RNA signatures in TNBC. A predictable ceRNA network was also constructed based on the ceRNA hypothesis, in order to identify the TNBC-specific RNAs involved in the ceRNA crosstalk. These integrated analyses aimed to detect novel lncRNA/miRNA/mRNA biomarkers of TNBC and reveal the underlying molecular regulatory mechanisms of TNBC pathogenesis and progression.
Materials and methods
Data mining
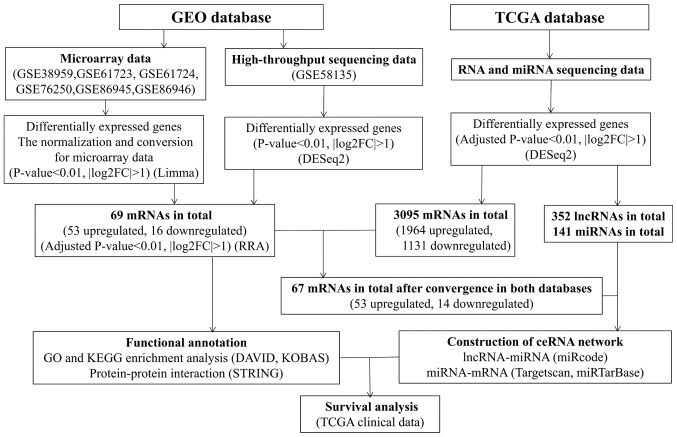
Microarray datasets, including GSE38959 (15), GSE61723 (16), GSE61724 (16), GSE76250 (17), GSE86945 (18) and GSE86946 (18), and one dataset obtained by expression profiling via high-throughput sequencing, namely GSE58135 (19), were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). In order to be included, all datasets had to consist of at least 20 samples and employ tissue samples collected from patients with TNBC and corresponding adjacent or healthy tissues. Large sample sizes are considered to provide more reliable results in the screening of differentially expressed genes (DEGs). It has been reported that small sample sizes are one of the major challenges in microarray analysis and, thus, recent integrated bioinformatics studies tend to use datasets with relatively large sample sizes (20). Therefore, GEO datasets containing at least 20 samples were chosen for further analysis in the present study. RNA-sequencing (RNA-seq) and microRNA-sequencing (miRNA-seq) data of patients with TNBC, comprising 116 patients with TNBC with complete expression profiles and clinical information, were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). Finally, 126 RNA-seq samples, including 115 tumor tissues and 11 healthy tissues, and 122 miRNA-seq samples, containing 113 tumor tissues and 9 healthy tissues, were obtained. The present study was performed in accordance with the publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). A flowchart of the data collection process and method implementation is presented in Fig. 1.
Figure 1.
Flowchart of data collection and method implementation in this study. GEO, Gene Expression Omnibus; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate; TCGA, The Cancer Genome Atlas; DAVID, Database for Annotation, Visualization and Integrated Discovery; STRING, Search Tool for the Retrieval of Interacting Genes; KOBAS, KEGG Orthology Based Annotation System; FC, fold change; miRNAs, microRNAs; lncRNAs, long non-coding RNAs; ceRNA, competing endogenous RNA.
Analysis of DEGs
The Limma package of R software (version 3.5.2) (21) was used for the normalization and log base 2 transformation of microarray data from the GEO datasets and screening of DEGs between tumor and healthy tissues. Three datasets, specifically GSE76250, GSE86945 and GSE86946, were merged into one group for normalization since they were all profiled on the Affymetrix Human Transcriptome Array 2.0 platform according to the GPL17586 platform (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL17586). A boxplot of the normalization and log base 2 transformation corresponding to this group is presented in Fig. S1. The GSE58135 dataset was employed for differential expression analysis between tumor and healthy tissues, which was performed using the DESeq2 package v.1.26.0 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) of R software (version 3.5.2). Gene integration analysis of the DEGs identified from the seven GEO datasets was conducted using the RobustRankAggreg package version 1.1 (22) based on a robust rank aggregation (RRA) method. Genes were then regarded as DEGs following RRA analysis based on the following significance cut-off levels: Adjusted P<0.01 and |log2FC|>1. The RRA method that was applied screens genes that are ranked consistently better than expected based on the null hypothesis of uncorrelated inputs (22). Thus, the gene expression values of samples from different datasets were not integrated into this analysis. In accordance with a number of published papers based on the RobustRankAggreg package (23,24), batch effect correction was not conducted in the present study.
The TCGA datasets were used for differential expression analyses of mRNAs, lncRNAs and miRNAs, which were compared between tumor and healthy tissues using the DESeq2 package (version 1.26.0) of R software (version 3.5.2). An adjusted P<0.01 and |log2FC|>1 were set as the cut-off criteria.
Functional enrichment analysis
Enrichment analysis of DEGs that were screened from the GEO database based on the RRA method was conducted using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. The GO enrichment analysis and functional annotation of the DEGs, which included the terms molecular function (MF), biological process (BP) and cellular component (CC), were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; version 6.8, (http://david.abcc.ncifcrf.gov/) (25). The KEGG Orthology Based Annotation System (KOBAS; version 3.0; http://kobas.cbi.pku.edu.cn/) was used to evaluate the statistical enrichment of DEGs in KEGG pathways. P<0.01 was considered to indicate significantly enriched DEGs.
Protein-protein interaction (PPI) network analysis
The Search Tool for the Retrieval of Interacting Genes (STRING version 11.0) (https://string-db.org/) database provides information regarding the predicted and experimental interactions of proteins (26), thus it was used for the identification of the protein-protein interactions among the identified DEGs. In the present study, the DEGs were mapped into PPIs and a combined score of ≥0.4 was considered as the cut-off level. Moreover, Cytoscape software (version 3.7.1) was used to construct the PPI networks (27).
Construction of the ceRNA network
The intersection of DEmRNAs between GEO and TCGA databases obtained by using the VennDiagram package (version 1.6.20; http://CRAN.R-project.org/package=VennDiagram), DElnc RNAs and DEmiRNAs from the TCGA database were used to construct the ceRNA network. The miRcode database version 11.0; (http://www.mircode.org/) was used to collect predicted and experimentally validated lncRNA-targeted miRNAs. MiRTarBase version 6.0 (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and TargetScan version 7.2 (http://www.targetscan.org/vert_72/) were cooperatively utilized to determine miRNA-targeted mRNAs. Based on the above lncRNA-miRNA and miRNA-mRNA interactions, visualization of the lncRNA-miRNA-mRNA network was performed using Cytoscape.
Survival analysis
Gene expression data and survival information obtained from the TCGA database were assessed by Kaplan-Meier survival analysis and a log-rank test was performed using the survival package version 2.44–1.1 (https://www.rdocumentation.org/packages/survival/versions/2.44–1.1) in R software. All the samples were categorized into high and low expression groups based on the median expression level of each DEG. Regarding the log-rank test results, P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs
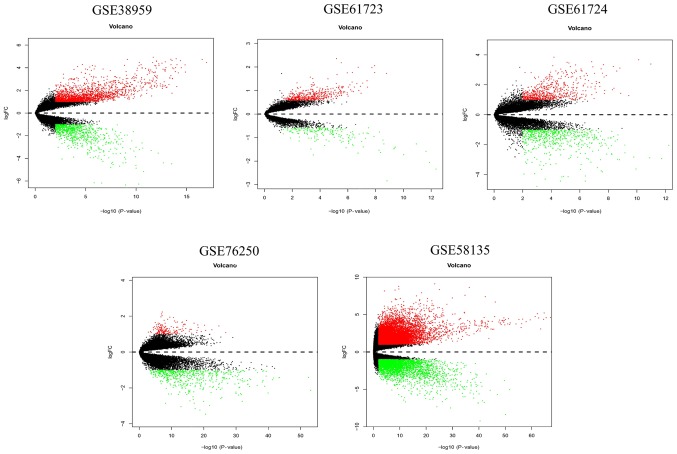
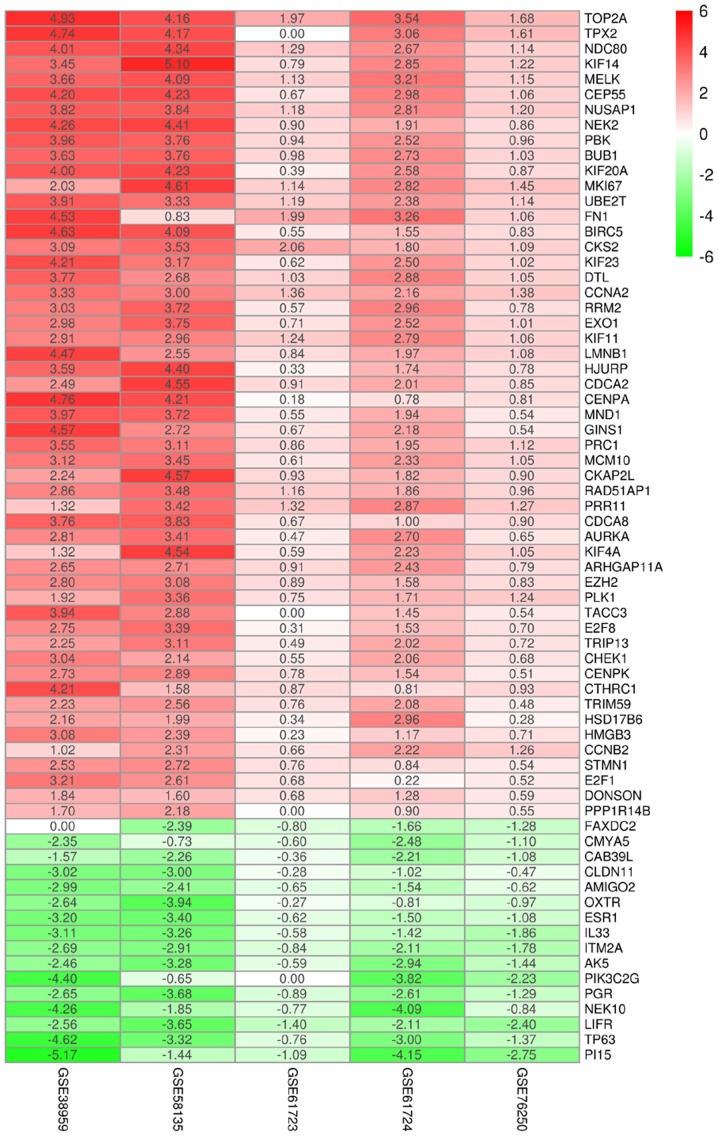
Information of the seven datasets that were downloaded from the GEO database and included in the current study is shown in Table I. Volcano plots revealed the number of differentially expressed genes identified from each dataset according to the set cut-off levels (P<0.01 and |log2FC|>1) (Fig. 2). Based on different cut-off levels (adjusted P<0.01 and |log2FC|>1), a total of 69 differentially expressed mRNAs (DEmRNAs), consisting of 16 downregulated and 53 upregulated genes, were identified following integrated analysis of these GEO datasets using the RobustRankAggreg package and further analyzed (Fig. 3).
Table I.
Details of datasets from the Gene Expression Omnibus database.
| Author, year | PMID | Record | Tissue | Platform | Healthy | Cancer | (Refs.) |
|---|---|---|---|---|---|---|---|
| Komatsu et al, 2013 | 23254957 | GSE38959 | Triple-negative | GPL4133 Agilent-014850 Whole Human Genome | 13 | 30 | (15) |
| breast cancer | Microarray 4×44K G4112F (Feature Number version) | ||||||
| Mathe et al, 2015 | 26537449 | GSE61723 | Triple-negative | GPL16686 [HuGene-2_0-st] Affymetrix Human Gene 2.0 | 17 | 33 | (16) |
| breast cancer | ST Array [transcript (gene) version] | ||||||
| Mathe et al, 2015 | 26537449 | GSE61724 | Triple-negative | GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 | 4 | 16 | (16) |
| breast cancer | ST Array [transcript (gene) version] | ||||||
| Liu et al, 2016 | 26813360 | GSE76250 | Triple-negative | GPL17586 [HTA-2_0] Affymetrix Human Transcriptome | 33 | 165 | (17) |
| breast cancer | Array 2.0 [transcript (gene) version] | ||||||
| Romero-Cordoba et al, 2018 | 30115973 | GSE86945 | Triple-negative | GPL17586 [HTA-2_0] Affymetrix Human Transcriptome | 0 | 100 | (18) |
| breast cancer | Array 2.0 [transcript (gene) version] | ||||||
| Romero-Cordoba et al, 2018 | 30115973 | GSE86946 | Triple-negative | GPL17586 [HTA-2_0] Affymetrix Human Transcriptome | 0 | 58 | (18) |
| breast cancer | Array 2.0 [transcript (gene) version] | ||||||
| Varley et al, 2014 | 24929677 | GSE58135 | Triple-negative | GPL11154 Illumina HiSeq 2000 (Homo sapiens) | 21 | 42 | (19) |
| breast cancer |
Figure 2.
Volcano plots of differentially expressed genes in each Gene Expression Omnibus dataset.
Figure 3.
Log2FC heatmap of the image data of each expression microarray. The abscissa corresponds to the GEO ID, while the ordinate corresponds to the gene name. Red represents log2FC>0, green represents log2FC<0, and the values represent the log2FC values in each GEO dataset. FC, fold change; GEO, Gene Expression Omnibus.
In the TCGA database, a total of 1,964 upregulated and 1,131 downregulated mRNAs, 352 differentially expressed lncRNAs (DElncRNAs) and 141 differentially expressed miRNAs (DEmiRNAs) were identified based on the aforementioned cut-off levels. Heatmaps of the top 200 DEmRNAs and DElncRNAs on the basis of adjusted P-value and overall 141 DEmiRNAs are demonstrated in Figs. S2–S4, respectively.
Functional enrichment analysis
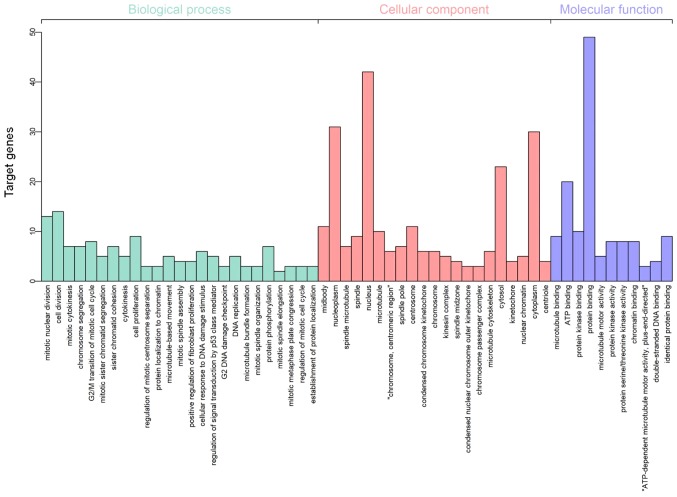
Following gene integration analysis, GO term functional enrichment analysis of the 69 DEGs was conducted using DAVID with P<0.01 set as the significance cut-off level. The following categories or ontologies were included in the analysis: i) Biological processes; ii) cellular component; and iii) molecular function. In terms of the biological processes, the DEGs were significantly enriched in ‘mitotic nuclear division’ and ‘cell division’. In the cellular component analysis, the DEGs were markedly enriched in the ‘nucleoplasm’ and ‘nucleus’. According to the molecular function, the DEGs were significantly enriched in ATP binding and protein binding (Fig. 4). With respect to KEGG pathway enrichment analysis, performed using KOBAS with P<0.01 set as the cut-off level, the 69 genes were found to be mainly enriched in ‘cell cycle’, ‘progesterone-mediated oocyte maturation’, ‘oocyte meiosis’ and ‘microRNAs in cancer’ terms (Table II).
Figure 4.
Gene Ontology terms of the differentially expressed genes identified from the GEO database, including biological process, cellular component and molecular function. GEO, Gene Expression Omnibus.
Table II.
Enriched Kyoto Encyclopedia of Genes and Genomes pathways of the differentially expressed genes.
| Term | Count | P-value | FDR | Genes |
|---|---|---|---|---|
| hsa04110: Cell cycle | 6 | 1.55×10−6 | 1.01×10−4 | PLK1, CCNB2, BUB1, CCNA2, CHEK1, E2F1 |
| hsa04914: Progesterone-mediated oocyte maturation | 5 | 9.06×10−6 | 2.94×10−4 | CCNB2, BUB1, PLK1, PGR, CCNA2 |
| hsa04114: Oocyte meiosis | 5 | 2.43×10−5 | 5.26×10−4 | CCNB2, BUB1, PLK1, AURKA, PGR |
| hsa05206: MicroRNAs in cancer | 5 | 1.00×10−3 | 1.33×10−2 | STMN1, TP63, EZH2, E2F1, KIF23 |
| hsa04115: P53 signaling pathway | 3 | 1.03×10−3 | 1.33×10−2 | CCNB2, RRM2, CHEK1 |
| hsa05222: Small cell lung cancer | 3 | 2.02×10−3 | 2.18×10−2 | FN1, E2F1, CKS2 |
| hsa05161: Hepatitis B | 3 | 8.31×10−3 | 7.71×10−2 | E2F1, BIRC5, CCNA2 |
Term, enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway; count, the number of differentially expressed genes of each term; P-value, P-value of enrichment analysis; FDR, false discovery rate.
PPI network analysis
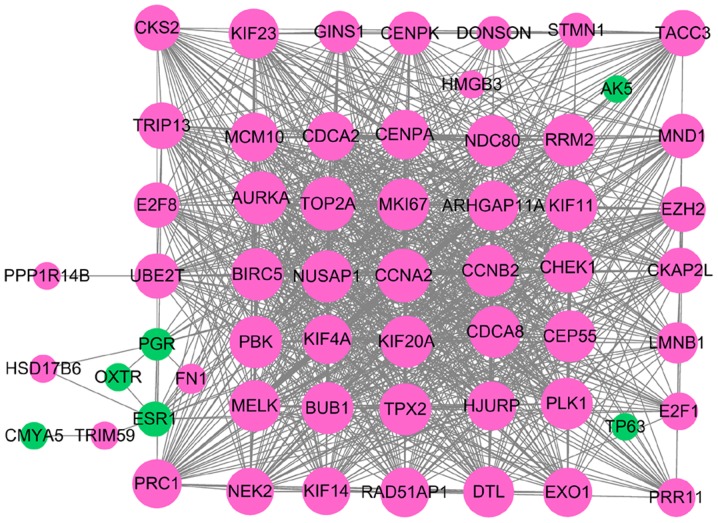
The DEGs in TNBC were examined using the STRING database to construct PPI networks. A total of 69 DEGs were identified from the GEO database using the RRA method, including 53 upregulated and 16 downregulated DEGs. Following removal of the isolated and partially connected nodes, a complex PPI network was formulated consisting of 58 nodes and 926 interactions, which were obtained with a combined score >0.4 (Fig. 5).
Figure 5.
Protein-protein interaction network of the differentially expressed genes identified from the Gene Expression Omnibus database. Pink nodes represent upregulated genes and green nodes represent downregulated genes. Node size is positively associated with degree, which is the number of DEGs the node/genes can interact with.
Construction of the ceRNA network
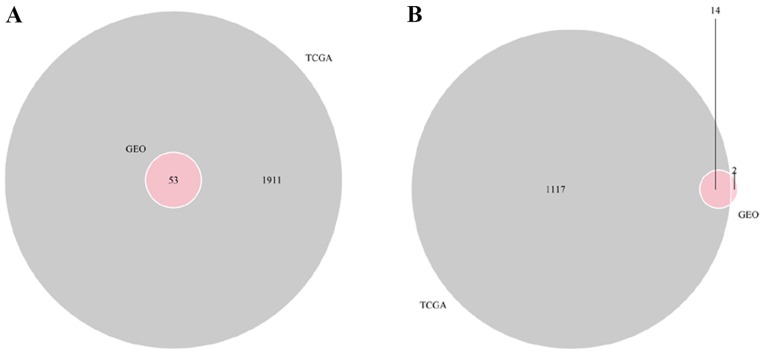
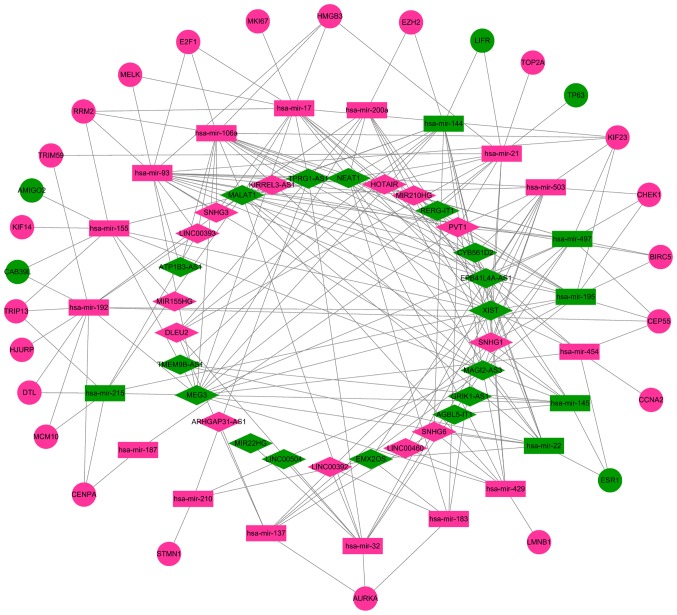
A total of 67 DEmRNAs from the intersections of the GEO and TCGA databases and 352 DElncRNAs and 141 DEmiRNAs from the TCGA database were selected in order to construct a ceRNA network. A Venn diagram demonstrated the intersections of DEmRNAs between the GEO and TCGA databases (Fig. 6). To further understand the functions of the DElncRNAs, DEmiRNAs and DEmRNAs, a ceRNA network was constructed. In Table III, it is shown that 31 DElncRNAs interacted with 29 DEmiRNAs retrieved from the miRcode database. Subsequently, DEmRNAs were searched in the miRTarBase and TargetScan databases based on the 29 DEmiRNAs. According to results from both databases, a total of 27 DEmRNAs capable of interacting with 21 of the 29 DEmiRNAs were chosen (Table IV). Following removal of the remaining eight DEmiRNAs and the corresponding lncRNAs, 29 DElncRNAs, 21 DEmiRNAs and 27 DEmRNAs were used to establish a ceRNA network (Fig. 7).
Figure 6.
Venn diagram of the intersections of differentially expressed mRNAs between the Gene Expression Omnibus and The Cancer Genome Atlas databases. (A) Intersection of upregulated mRNAs; (B) Intersection of downregulated mRNAs. GEO, Gene Expression Omnibus.
Table III.
miRNAs that may be targeted by specific lncRNAs in triple-negative breast cancer.
| lncRNA | miRNA |
|---|---|
| XIST | miR-503, miR-301b, miR-454, miR-93, miR-106a, miR-96, miR-137, miR-140, miR-141, miR-200a, miR-144, miR-155, miR-195, miR-497, miR-17, miR-192, miR-215, miR-429, miR-204, miR-21, miR-22, miR-32, miR-338 |
| MEG3 | miR-551a, miR-301b, miR-454, miR-93, miR-106a, miR-96, miR-140, miR-141, miR-200a, miR-143, miR-144, miR-145, miR-155, miR-195, miR-497, miR-17, miR-192, miR-215, miR-429, miR-204, miR-21, miR-22, miR-338 |
| MALAT1 | miR-503, miR-93, miR-106a, miR-96, miR-140, miR-141, miR-200a, miR-143, miR-144, miR-145, miR-155, miR-195, miR-497, miR-17, miR-192, miR-215, miR-429, miR-204, miR-21, miR-22, miR-32, miR-338 |
| NEAT1 | miR-503, miR-301b, miR-454, miR-93, miR-106a, miR-96, miR-140, miR-141, miR-200a, miR-143, miR-144, miR-195, miR-497, miR-17, miR-183, miR-429, miR-204, miR-22, miR-338 |
| MAGI2-AS3 | miR-503, miR-93, miR-106a, miR-137, miR-141, miR-200a, miR-143, miR-144, miR-145, miR-155, miR-195, miR-497, miR-429, miR-204, miR-210, miR-22, miR-32 |
| PVT1 | miR-503, miR-551a, miR-93, miR-106a, miR-140, miR-143, miR-145, miR-195, miR-497, miR-17, miR-183, miR-187, miR-21 |
| SNHG1 | miR-503, miR-137, miR-140, miR-141, miR-200a, miR-143, miR-144, miR-145, miR-195, miR-497, miR-204, miR-21, miR-32 |
| EPB41L4A-AS1 | miR-503, miR-93, miR-106a, miR-141, miR-200a, miR-195, miR-497, miR-17, miR-183, miR-429, miR-22, miR-338 |
| DLEU2 | miR-551a, miR-96, miR-137, miR-141, miR-200a, miR-143, miR-144, miR-21, miR-32 |
| CYB561D2 | miR-503, miR-93, miR-106a, miR-140, miR-144, miR-22, miR-338 |
| HOTAIR | miR-301b, miR-454, miR-143, miR-17, miR-93, miR-204, miR-21 |
| SNHG | miR-93, miR-106a, miR-141, miR-200a, miR-17, miR-338 |
| MIR210HG | miR-551a, miR-93, miR-106a, miR-145, miR-195, miR-497 |
| TPRG1-AS1 | miR-93, miR-106a, miR-17, miR-210, miR-32, miR-338 |
| SNHG6 | miR-137, miR-144, miR-429, miR-204, miR-22 |
| EMX2OS | miR-503, miR-143, miR-183, miR-210, miR-22 |
| ATP1B3-AS1 | miR-93, miR-106a, miR-96, miR-204 |
| LINC00393 | miR-93, miR-106a, miR-192, miR-215 |
| LINC00460 | miR-503, miR-143, miR-429, miR-338 |
| GRIK1-AS1 | miR-145, miR-204, miR-338 |
| LINC00504 | miR-140, miR-32, miR-338 |
| MIR155HG | miR-155, miR-204, miR-338 |
| TMEM9B-AS1 | miR-144, miR-145, miR-22 |
| AGBL5-IT1 | miR-145, miR-204 |
| C6orf99 | miR-140, miR-338 |
| KIRREL3-AS1 | miR-144, miR-338 |
| LINC00392 | miR-183, miR-32 |
| UCA1 | miR-96, miR-143 |
| ARHGAP31-AS1 | miR-137 |
| MIR22HG | miR-32 |
| RERG-IT1 | miR-21 |
lncRNA, long non-coding RNA; miRNA/miR, microRNA.
Table IV.
mRNAs that may be targeted by specific miRNAs in triple-negative breast cancer.
| miRNA | mRNA |
|---|---|
| miR-192 | CAB39L, CEP55, MCM10, TRIM59, CENPA, HJURP, TRIP13, DTL |
| miR-17 | KIF23, HMGB3, RRM2, E2F1, MKI67, MELK |
| miR-93 | RRM2, E2F1, KIF23, HMGB3, BIRC5, MELK |
| miR-21 | TOP2A, TRIM59, LIFR, TP63, HMGB3 |
| miR-155 | TRIP13, RRM2, CAB39L, AMIGO2, KIF14 |
| miR-215 | TRIP13, CENPA, MCM10, DTL |
| miR-195 | KIF23, CHEK1, CEP55, BIRC5 |
| miR-497 | CHEK1, KIF23, CEP55, BIRC5 |
| miR-106a | RRM2, HMGB3, E2F1, KIF23 |
| miR-454 | ESR1, CEP55, CCNA2 |
| miR-503 | KIF23, CHEK1 |
| miR-144 | LIFR, EZH2 |
| miR-32 | AURKA |
| miR-145 | ESR1 |
| miR-183 | AURKA |
| miR-200a | EZH2 |
| miR-187 | CENPA |
| miR-429 | LMNB1 |
| miR-137 | AURKA |
| miR-210 | STMN1 |
| miR-22 | ESR1 |
miRNA/miR, microRNA; mRNA, messenger RNA.
Figure 7.
The lncRNA-miRNA-mRNA competing endogenous RNA network for triple-negative breast cancer. Diamonds represent lncRNAs; rectangles represent miRNAs; ellipses represent mRNAs; red indicates upregulated genes and green represents downregulated genes. lncRNA, long non-coding RNA; miRNA, microRNA.
Survival analysis
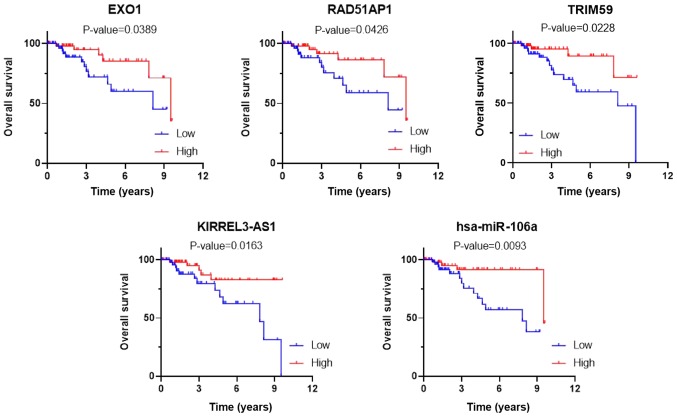
To determine whether the expression levels of the 69 DEmRNAs identified from the GEO database and the DEGs included in the ceRNA network were related to the overall survival of patients with TNBC, Kaplan-Meier curves were generated based on the gene expression data and survival information retrieved from the TCGA database. As a result, three DEmRNAs, tripartite motif containing 59 (TRIM59), exonuclease 1 (EXO1) and RAD51-associated protein 1 (RAD51AP1), one DElncRNA, KIRREL3-antisense RNA 1 (KIRREL3-AS1), and one DEmiRNA, hsa-mir-106a, were found to be significantly associated with the prognosis of patients with TNBC (P<0.05; Fig. 8). Moreover, TRIM59, KIRREL3-AS1 and hsa-mir-106a were found to be involved in the ceRNA network. All five survival-related genes were upregulated and the high expression levels of all five genes were related to better prognosis.
Figure 8.
Kaplan-Meier curves of three DEmRNAs, TRIM59, EXO1 and RAD51AP1, one DElncRNA, KIRREL3-AS1, and one DEmiRNA, hsa-mir-106a, associated with overall survival. DE, differentially expressed; lncRNAs, long non-coding RNAs; TRIM59, tripartite motif containing 59; EXO1, exonuclease 1; RAD51AP1, RAD51-associated protein 1; KIRREL3-AS1, KIRREL3-antisense RNA 1.
Discussion
Five distinct intrinsic breast cancer subtypes have been identified based on gene expression: i) Luminal A; ii) luminal B; iii) HER2 overexpressing; iv) basal-like; and v) healthy breast tissue-like (28–30). Prat et al (31) have also identified a new breast cancer intrinsic subtype known as claudin-low or mesenchymal-like. These six subtypes of breast cancer defined by immunohistochemistry (IHC) demonstrate distinct differences in terms of breast cancer survival (32). TNBC is regarded as an aggressive subtype of breast cancer, since it is characterized by higher rates of progression, metastasis and recurrence than the other molecular subtypes. This subtype is more commonly diagnosed in young women (33). The main therapeutic strategies for the treatment of patients with TNBC are surgery and chemotherapy. One of the primary factors that contribute to poor prognosis of patients with TNBC is that few effective therapeutic targets are available. Therefore, studies on the underlying mechanisms of the pathogenesis and progression of TNBC are required for the development of more effective therapeutic strategies.
Compared with the original long-term fundamental experimental studies, integrated mining of microarray data and high-throughput sequencing from public databases appears to be more comprehensive and highly informative. Furthermore, since a number of studies have indicated the involvement of ceRNA crosstalk in diverse biological processes, including tumorigenesis, progression and metastasis, the comprehensive analysis of lncRNA-miRNA-mRNA ceRNA networks has become more widely used in the prediction of candidate RNA signatures in various cancer types (34–36), including TNBC (37–39). Nonetheless, non-coding RNA-associated ceRNA networks based on whole-genome gene expression profiling with large-scale microarray and sequencing data of patients with TNBC have not been described.
In the current study, seven datasets from studies on TNBC were downloaded from the publicly available GEO database. The published original studies, from which the data were obtained, are discussed in the following text. Komatsu et al (15) identified abnormal spindle microtubule assembly and centromere protein K as novel molecular targets for TNBC therapy, since the absence of these genes was found to cause an arrest in the G2/M and G0/G1 phases of the cell cycle, respectively, and subsequently induced cell death in TNBC cells. Mathe et al (16) compared the genes that were found to be TNBC-specific from their cohort which consists of patients with TNBC and the The Cancer Genome Atlas (TCGA) cohort which is an external validation cohort including patients with TNBC from the TCGA database, and it was found that four of the genes, namely ankyrin repeat domain 30A, acidic nuclear phosphoprotein 32 family member E, desmocollin 2 and interleukin 6 signal transducer (IL6ST), were common to both TNBC cohorts. The survival curves revealed that high expression of IL6ST was significantly associated with improved survival outcomes. Liu et al (17) successfully developed an mRNA and an integrated mRNA-lncRNA signature based on eight mRNAs and two lncRNAs. The data from those two signatures suggested that the lncRNAs HIST2H2BC and SNRPEP4 promoted cell proliferation and invasion, thus contributing to paclitaxel resistance in TNBC cells. Romero-Cordoba et al (18) identified a set of altered miRNAs and experimentally confirmed that a specific miRNA, hsa-mir-342-3p, was downregulated in TNBC compared with other phenotypes. In addition, loss of function of miR-342-3p resulted in monocarboxylate transporter 1 overexpression and contributed to oncogenic metabolic reprogramming in TNBC. Varley et al (19) analyzed sequencing data in order to identify breast cancer-associated read-through fusion transcripts. This analysis led to the identification of SCNN1A-TNFRSF1A and CTSDIFITM10, two recurrent read-through fusion transcripts found to involve membrane proteins, which raised the possibility of them being breast cancer-specific cell surface markers. In order to fully mine the information of these datasets, multistep processing and integrated bioinformatics were applied to reveal DEGs in TNBC. More importantly, a robust rank aggregation (RRA) method was used to identify stable DEGs among different studies. This method analyzes prioritized gene lists and finds commonly overlapping genes, which are ranked consistently better than expected by chance (40).
Finally, 69 dysregulated mRNAs were identified from the GEO datasets, including 53 upregulated and 16 downregulated genes, which are demonstrated in Fig. 3. Subsequently, GO and KEGG functional enrichment analyses were performed and a PPI network of these DEGs was constructed in order to demonstrate their characteristics and specific biological significance. DEGs were further validated in the TCGA database. Eventually, 29 lncRNAs, 21 miRNAs and 27 mRNAs were selected to construct a potential lncRNA-miRNA-mRNA ceRNA network by biological prediction in order to elucidate the interactions and regulatory mechanisms of DEGs. Finally, Kaplan-Meier curves were generated using the gene expression data and survival information provided by the TCGA database to detect prognostic indicators for patients with TNBC. As a result, three DEmRNAs, namely TRIM59, EXO1 and RAD51AP1, one DElncRNA, KIRREL3-AS1 and one DEmiRNA, hsa-mir-106a, were considered to be closely associated with the prognosis of patients with TNBC. Moreover, TRIM59, KIRREL3-AS1 and hsa-mir-106a were involved in the ceRNA network. The expression levels of all five genes were found to be upregulated and related to longer survival times. Nevertheless, the determination of the nature of protective factors or risk factors depends on gene expression at both the transcription and translation levels. Certain DEGs identified in the present study have been reported to be associated with the prognosis of breast cancer. As observed by gene expression profiling, IHC analysis has revealed that the expression of certain proliferation markers, including Ki67 and Aurora A kinase, is associated with poor prognosis in ER+ disease (41,42). In addition, as assessed by IHC, BCL2 expression is a powerful predictor of favorable prognosis in breast cancer across different molecular subtypes (43). Despite these findings, the routine assessment of IHC markers in addition to ER, PR and HER2 has yet to be implemented in standard clinical treatment guidelines. Therefore, the remaining survival-related genes identified in the current study should be further explored in future studies.
In the present study, miRcode was used to collect predicted and experimentally validated miRNAs that are targeted by lncRNAs. In addition, TargetScan and miRTarBase were cooperatively utilized to determine mRNAs that are targeted by miRNAs. According to accumulating biological data, numerous computational models for potential miRNA-disease association inference have been developed, which are highly useful in research of the underlying molecular mechanism of human diseases and the development of new drugs for disease treatment (44–46). In particular, Chen et al (47) proposed the model of the Ensemble of Decision Tree-based MiRNA-Disease Association prediction (EDTMDA) for the identification of miRNA-disease associations by inputting features that were extracted from integrated miRNA similarity, disease similarity and known miRNA-disease associations. It is believed that EDTMDA is able to make reliable predictions and guide experiments to reveal further miRNA-disease associations. However, limited studies on computational models for the prediction of potential lncRNA-disease associations are available. The involvement of lncRNAs in numerous biological processes and their important roles in a variety of complex human diseases have been demonstrated. Therefore, the development of more effective computational models to identify potential relationships between lncRNAs and diseases may aid further understanding of disease mechanisms at the lncRNA molecular level. Furthermore, the development of a computational model capable of directly determining mRNAs that are targeted by lncRNAs would notably reduce the time required for analysis. In conclusion, research on computational models for the prediction of potential diseases at the level of gene expression regulation, post-translational protein modification and cellular environment may promote the diagnosis, treatment, prognosis and prevention of human diseases, including TNBC.
Of note, gene expression profiling was conducted using large-scale microarray and sequencing data of patients with TNBC. A robust rank aggregation (RRA) method was applied to comprehensively screen stable DEGs among the different studies. Furthermore, a network of predicted ceRNA interactions derived from integrative bioinformatics analysis was successfully constructed. The reliability of the results was secured by the use of reasonable screening criteria, sufficient sample size and appropriate visualization of the results. There are also limitations to the current study. First, it would be more appropriate to include paired samples in order to eliminate error among different patients since each gene expression level varied substantially in different patients. Secondly, the complex ceRNA network was constructed by biological prediction based on the hypothesis that lncRNAs may serve as ceRNAs and, therefore, affect the expression of target genes by merging miRNAs. Therefore, further research is required for the verification of the current study results and further understanding of the molecular mechanisms of the identified RNA signatures in TNBC.
In conclusion, the present study identified a large number of cancer-specific and several survival-related DEGs by integrated analysis of large-scale gene expression profiles from the GEO and TCGA databases. The predicted lncRNA-miRNA-mRNA ceRNA network may provide guidance for further studies on the molecular pathogenesis and progression mechanisms underlying TNBC.
Supplementary Material
Acknowledgements
The authors would like to thank Mr. Jiwei Li (Lifegenes Biotechnology, Shanghai, China) for his advice and technical assistance.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PY and LFT wrote the manuscript and were responsible for data analysis. GT, LL and PY conceived and designed the study. GT and LL revised and drafted the manuscript. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4–13. doi: 10.3121/cmr.2008.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California cancer registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 3.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 4.Rakha EA, Elrehim DA, Paish C, Green AR, Lee AH, Robertson JF, Blamey RW, Macmillan D, Ellis IO. Basal phenotype identifes a poor prognostic subgroup of breast cancer of clinical importance. Eur J Cancer. 2006;42:3149–3156. doi: 10.1016/j.ejca.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Zaky SS, Lund MJ, May KA, Godette KD, Beitler JJ, Holmes LR, O'Regan RM, Yu ES, Yu DS, Landry JC. The negative effect of triple-negative breast cancer on outcome after breast-conserving therapy. Ann Surg Oncol. 2011;18:2858–2865. doi: 10.1245/s10434-011-1669-4. [DOI] [PubMed] [Google Scholar]
- 6.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 7.Deva Magendhra Rao AK, Patel K, Korivi Jyothiraj S, Meenakumari B, Sundersingh S, Sridevi V, Rajkumar T, Pandey A, Chatterjee A, Gowda H, Mani S. Identification of lncRNAs associated with early stage breast cancer and their prognostic implications. Mol Oncol. 2019;13:1342–1355. doi: 10.1002/1878-0261.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Yan CC, Zhao X, You ZH. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief Bioinform. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haakensen VD, Nygaard V, Greger L, Aure MR, Fromm B, Bukholm IR, Luders T, Chin SF, Git A, Caldas C, et al. Subtype-specific micro-RNA expression signatures in breast cancer progression. Int J Cancer. 2016;139:1117–1128. doi: 10.1002/ijc.30142. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Xie D, Zhao Q, You ZH. Micrornas and complex diseases: From experimental results to computational models. Brief Bioinform. 2019;20:515–539. doi: 10.1093/bib/bbx130. [DOI] [PubMed] [Google Scholar]
- 12.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Zhou J, Wang Z, Wang P, Gao X, Wang Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed Pharmacother. 2018;104:451–457. doi: 10.1016/j.jpba.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu M, Yoshimaru T, Matsuo T, Kiyotani K, Miyoshi Y, Tanahashi T, Rokutan K, Yamaguchi R, Saito A, Imoto S, et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int J Oncol. 2013;42:478–506. doi: 10.3892/ijo.2012.1744. [DOI] [PubMed] [Google Scholar]
- 16.Mathe A, Wong-Brown M, Morten B, Forbes JF, Braye SG, Avery-Kiejda KA, Scott RJ. Novel genes associated with lymph node metastasis in triple negative breast cancer. Sci Rep. 2015;5:15832. doi: 10.1038/srep15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YR, Jiang YZ, Xu XE, Hu X, Yu KD, Shao ZM. Comprehensive transcriptome profiling reveals multigene signatures in triple-negative breast cancer. Clin Cancer Res. 2016;22:1653–1662. doi: 10.1158/1078-0432.CCR-15-1555. [DOI] [PubMed] [Google Scholar]
- 18.Romero-Cordoba SL, Rodriguez-Cuevas S, Bautista-Pina V, Maffuz-Aziz A, D'Ippolito E, Cosentino G, Baroni S, Iorio MV, Hidalgo-Miranda A. Loss of function of miR-342-3p results in MCT1 over-expression and contributes to oncogenic metabolic reprogramming in triple negative breast cancer. Sci Rep. 2018;8:12252. doi: 10.1038/s41598-018-29708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varley KE, Gertz J, Roberts BS, Davis NS, Bowling KM, Kirby MK, Nesmith AS, Oliver PG, Grizzle WE, Forero A, et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat. 2014;146:287–297. doi: 10.1007/s10549-014-3019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moradifard S, Hoseinbeyki M, Ganji SM, Minuchehr Z. Analysis of microRNA and gene expression profiles in Alzheimer's disease: A meta analysis approach. Sci Rep. 2018;8:4767. doi: 10.1038/s41598-018-20959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi KQ, Lin Z, Chen XJ, Song M, Wang YQ, Cai YJ, Yang NB, Zheng MH, Dong JZ, Zhang L, Chen YP. Hepatocellular carcinoma associated microRNA expression signature: Integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget. 2015;6:25093–25108. doi: 10.18632/oncotarget.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Han S, Huang W, Chen T, Liu Y, Pan S, Li S. A meta-analysis of microRNA expression in liver cancer. PLoS One. 2014;9:e114533. doi: 10.1371/journal.pone.0114533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 29.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 34.Long J, Bai Y, Yang X, Lin J, Yang X, Wang D, He L, Zheng Y, Zhao H. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019;19:90. doi: 10.1186/s12935-019-0817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Zhang W, Jiang Y, Liu K, Ran L, Song F. Identification of functional lncRNAs in gastric cancer by integrative analysis of GEO and TCGA data. J Cell Biochem. 2019;120:17898–17911. doi: 10.1002/jcb.29058. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Ding Y, Da B, Fei Y, Feng G. Identification of potential prognostic long non-coding RNA signatures based on a competing endogenous RNA network in lung adenocarcinoma. Oncol Rep. 2018;40:3199–3212. doi: 10.3892/or.2018.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H, Dai M, Chen X, Chen X, Qin S, Dai S. Integrated analysis of the potential roles of miRNA-mRNA networks in triple negative breast cancer. Mol Med Rep. 2017;16:1139–1146. doi: 10.3892/mmr.2017.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang R, Xing L, Wang M, Chi H, Zhang L, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs/mRNAs and miRNAs with associated cerna networks in triple-negative breast cancer. Cell Physiol Biochem. 2018;50:473–488. doi: 10.1159/000494162. [DOI] [PubMed] [Google Scholar]
- 39.Yuan N, Zhang G, Bie F, Ma M, Ma Y, Jiang X, Wang Y, Hao X. Integrative analysis of lncRNAs and miRNAs with coding RNAs associated with ceRNA crosstalk network in triple negative breast cancer. Onco Targets Ther. 2017;10:5883–5897. doi: 10.2147/OTT.S149308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Võsa U, Kolde R, Vilo J, Metspalu A, Annilo T. Comprehensive meta-analysis of microrna expression using a robust rank aggregation approach. Methods Mol Biol. 2014;1182:361–373. doi: 10.1007/978-1-4939-1062-5_28. [DOI] [PubMed] [Google Scholar]
- 41.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali HR, Dawson SJ, Blows FM, Provenzano E, Pharoah PD, Caldas C. Aurora kinase A outperforms Ki67 as a prognostic marker in ER-positive breast cancer. Br J Cancer. 2012;106:1798–1806. doi: 10.1038/bjc.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, Le Quesne J, Baglietto L, Severi G, Giles GG, McLean CA, et al. BCL2 in breast cancer: A favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer. 2010;103:668–675. doi: 10.1038/sj.bjc.6605921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Huang L. LRSSLMDA: Laplacian regularized sparse subspace learning for miRNA-disease association prediction. PLoS Comput Biol. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Yin J, Qu J, Huang L. MDHGI: Matrix decomposition and heterogeneous graph inference for miRNA-disease association prediction. PLoS Comput Biol. 2018;14:e1006418. doi: 10.1371/journal.pcbi.1006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Wang L, Qu J, Guan NN, Li JQ. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Zhu CC, Yin J. Ensemble of decision tree reveals potential miRNA-disease associations. PLoS Comput Biol. 2019;15:e1007209. doi: 10.1371/journal.pcbi.1007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.