Park et al. identify a role for a subset of mRNA splicing factors, including WBP11, in centriole biogenesis. WBP11 is required for the splicing of short, weak introns, including those found in TUBGCP6 pre-mRNA. Splicing of TUBGCP6, as well as other WBP11 targets, allows for faithful mitotic progression and centriole duplication.
Abstract
Centriole duplication occurs once in each cell cycle to maintain centrosome number. A previous genome-wide screen revealed that depletion of 14 RNA splicing factors leads to a specific defect in centriole duplication, but the cause of this deficit remains unknown. Here, we identified an additional pre-mRNA splicing factor, WBP11, as a novel protein required for centriole duplication. Loss of WBP11 results in the retention of ∼200 introns, including multiple introns in TUBGCP6, a central component of the γ-TuRC. WBP11 depletion causes centriole duplication defects, in part by causing a rapid decline in the level of TUBGCP6. Several additional splicing factors that are required for centriole duplication interact with WBP11 and are required for TUBGCP6 expression. These findings provide insight into how the loss of a subset of splicing factors leads to a failure of centriole duplication. This may have clinical implications because mutations in some spliceosome proteins cause microcephaly and/or growth retardation, phenotypes that are strongly linked to centriole defects.
Introduction
Centrioles are the core structural components of the centrosome, the cell’s major microtubule-organizing center that arranges the interphase microtubule cytoskeleton in many cell types and forms the spindle poles during mitosis (Gönczy, 2012). Centrioles are also required to nucleate the axoneme of cilia that play critical roles in cell signaling, fluid movement, and locomotion. In cycling cells, centriole biogenesis is tightly coordinated with cell cycle progression (Fırat-Karalar and Stearns, 2014; Nigg and Holland, 2018). At the start of the cell cycle, each cell contains a pair of parent centrioles. Centriole duplication begins at the G1–S transition, when one new procentriole forms at a single site perpendicular to each existing centriole. The procentriole remains engaged to the parent centriole in an orthogonal configuration and elongates during S and G2 phases. In mitosis, the procentriole disengages from the parent centriole so that each new daughter cell inherits a pair of centrioles that are competent for reduplication in the next cell cycle. Defects in centriole duplication can lead to the generation of excessive numbers of centrioles, promoting mitotic chromosome segregation errors and tumorigenesis in mice (Coelho et al., 2015; Levine et al., 2017; Serçin et al., 2016).
Polo-like kinase 4 (PLK4) is the earliest marker for the site of procentriole assembly and is initially recruited to form a ring around the proximal end of parent centrioles (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Sonnen et al., 2012). At the beginning of S phase, PLK4 transitions from a ring to a single “spot” that marks the site of procentriole assembly (Dzhindzhev et al., 2017; Kim et al., 2013; Ohta et al., 2014; Sonnen et al., 2012). This transformation is a symmetry-breaking reaction that requires the binding of PLK4 to its activator STIL (SCL/TAL1 interrupting locus; Arquint et al., 2015; Leda et al., 2018; Lopes et al., 2015; Moyer et al., 2015). Active PLK4 then phosphorylates STIL in two different regions to promote SAS6 and CPAP binding to initiate the assembly of the procentriole (Dzhindzhev et al., 2014; Kratz et al., 2015; Moyer et al., 2015; Moyer and Holland, 2019; Ohta et al., 2014). Phosphatases must counteract PLK4 activity, but the identity of the phosphatases responsible for controlling the initiation of centriole assembly remains unclear.
Control of centriole biogenesis depends on finely tuning the abundance of centriole duplication proteins. Overexpression of STIL or SAS6 leads to centriole overduplication and, accordingly, the levels of STIL and SAS6 are controlled by cell cycle–regulated protein degradation (Arquint and Nigg, 2014; Arquint et al., 2012; Strnad et al., 2007; Tang et al., 2011; Vulprecht et al., 2012). Centriole duplication is also extremely sensitive to the levels of PLK4; therefore, the abundance of the active kinase is tightly controlled by a negative feedback loop in which the dimeric kinase phosphorylates itself in trans to promote ubiquitination and degradation (Cunha-Ferreira et al., 2013; Guderian et al., 2010; Holland et al., 2010; Klebba et al., 2013). Along with posttranslational control of protein abundance, increasing evidence has linked posttranscriptional control of gene expression to the regulation of centriole biogenesis. Expression of the centriole proteins PLK4 and CP110 is downregulated by the binding of miRNAs to the 3′ UTR of the mRNA (Bao et al., 2018; Cao et al., 2012; Song et al., 2014). Moreover, two alternative isoforms of CEP135 have been shown to play opposing roles in centriole biogenesis, and dysregulation of isoform expression promotes centrosome amplification in breast cancer cells (Dahl et al., 2015; Ganapathi Sankaran et al., 2019). A genome-wide RNAi screen revealed a strong connection between splicing factors and centriole biology, with 14 of 38 genes required for centriole biogenesis playing established roles in mRNA splicing (Balestra et al., 2013). Cells depleted of these splicing factors failed centriole duplication and progressed through the cell cycle to dilute out their centrioles through subsequent cell divisions. Recent work revealed that some splicing factors play direct roles in chromosome segregation in mitosis (Pellacani et al., 2018). Therefore, it remains unclear whether the splicing factors required for centriole assembly are needed for the correct splicing of a subset of pre-mRNAs necessary for centriole biogenesis or have roles independent of their function in splicing.
Here, we identify the pre-mRNA splicing factor WW domain-binding protein 11 (WBP11) as a novel protein required for centriole duplication. We show that WBP11 promotes the splicing of ∼200 short introns with weak 5′ splice sites. Our data indicate that one cause of the centriole biogenesis defects in WBP11-depleted cells is a rapid reduction in the levels of TUBGCP6, a core component of the γ-tubulin ring complex (γ-TuRC) that is required for the nucleation of centriolar microtubules (Bahtz et al., 2012). Importantly, this role in controlling the levels of TUBGCP6 is shared with multiple additional splicing factors that have previously been found to be required for centriole duplication (Balestra et al., 2013). Our study provides a molecular explanation for the essential role of a subset of splicing factors in centriole biogenesis.
Results
WBP11 is required for centriole duplication
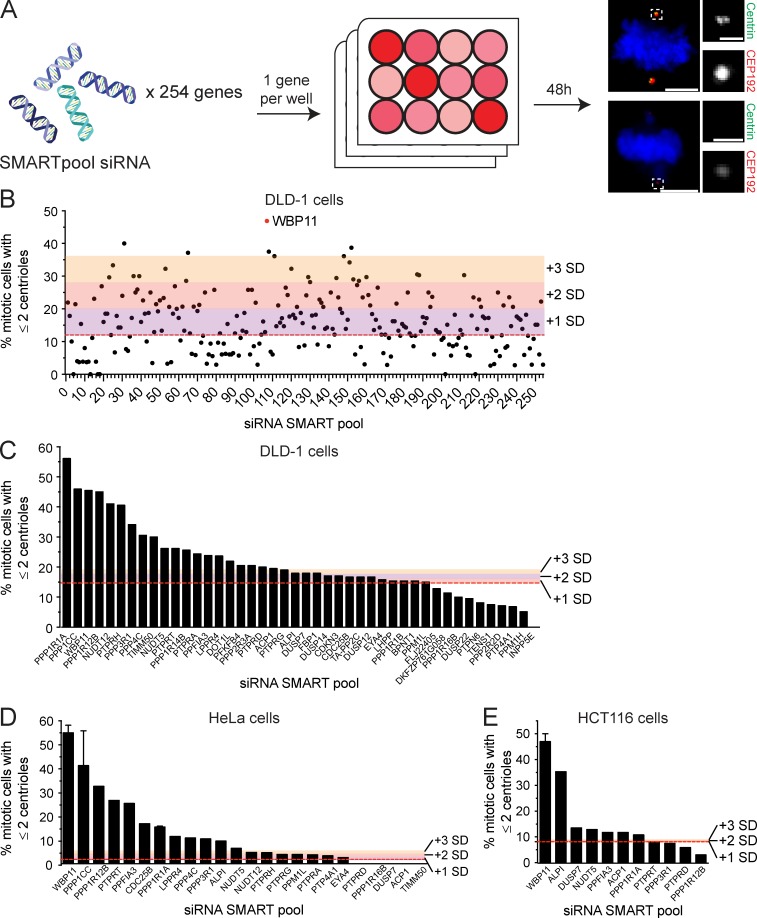
It stands to reason that phosphatases act to antagonize PLK4 kinase activity, but the role of phosphatases in the control of centriole duplication remains poorly defined. To address this question, we performed an siRNA screen in human cells to identify protein phosphatases that function in centriole biogenesis. We used the pseudodiploid human colon cancer DLD-1 cell line and screened 254 genes encoding protein phosphatases or phosphatase interacting proteins using a SMARTpool siRNA library (Table S1). Centriole number was assessed in mitotic cells 2 d after siRNA transfection (Fig. S1 A). To identify hits, we applied a threshold for values that were more than two times the SD from the mean centriole number in control cells. This resulted in 57 genes whose depletion led to the acquisition of five or more centrioles (centriole amplification) and 24 genes whose knockdown reduced centriole number to two or less (centriole loss; Fig. S1 B and Table S1). Increased activity of PLK4 leads to the production of a single parent centriole surrounded by multiple procentrioles (Habedanck et al., 2005). By contrast, cytokinesis failure leads to cells entering into mitosis with four centrosomes, each containing a pair of centrioles. For the genes whose depletion caused centriole amplification, we never observed cells with a single parent centriole surrounded by multiple procentrioles (data not shown). Instead, in cases where extra centrioles were observed, the configuration was consistent with what would be expected following cytokinesis failure. Since the centriole amplification phenotypes we observed are likely to have arisen from cytokinesis failure, we focused our attention on genes whose knockdown led to centriole loss.
Figure S1.
siRNA screen to identify novel phosphatase or phosphatase-interacting proteins required for centriole biogenesis. (A) Schematic representation of the screen design. A SMARTpool of four siRNAs targeting each gene was transfected into DLD-1 cells. 48 h after siRNA transfection, centriole number was analyzed in mitotic cells. A total of 254 genes were screened. Scale bars represent 5 µm; 1 µm in inset. (B) Graph shows the fraction of cells with two or fewer centrioles in mitosis. Each point represents a single gene. Raw data are displayed in Table S1. Colors indicate one, two, or three SDs above the level of centriole underduplication observed in untransfected DLD-1 cells (red line). Hits were considered as genes that are more than two SDs above the control. More than 25 mitotic cells per siRNA were analyzed. (C) Secondary validation of the top hits from the initial screen in DLD-1 cells. Colors indicate one, two, or three SDs above the level of centriole underduplication observed in untransfected DLD-1 cells (red line). n = 1, ≥25 mitotic cells per siRNA were analyzed. (D) Validation of the top hits from the initial screen in HeLa cells. Colors indicate one, two, or three SDs above the level of centriole underduplication observed in untransfected DLD-1 cells (red line). n ≥ 1, ≥25 mitotic cells per siRNA were analyzed. Error bars represent SD. (E) Validation of the top hits from the initial screen in HCT116 cells. Colors indicate one, two, or three SDs above the level of centriole underduplication observed in untransfected DLD-1 cells (red line). n ≥ 1, ≥25 mitotic cells per siRNA were analyzed. Error bars represent SD.
The top centriole loss hit to emerge from the primary screen was the protein phosphatase 1 (PP1) binding protein WBP11. We performed a limited secondary screen in DLD-1, HeLa, and HCT116 cells, and depletion of WBP11 consistently ranked among the top hits that caused centriole duplication failure (Fig. S1, C–E; and Table S1). To our knowledge, WBP11 has not been previously implicated in centriole biogenesis and was therefore selected for further analysis.
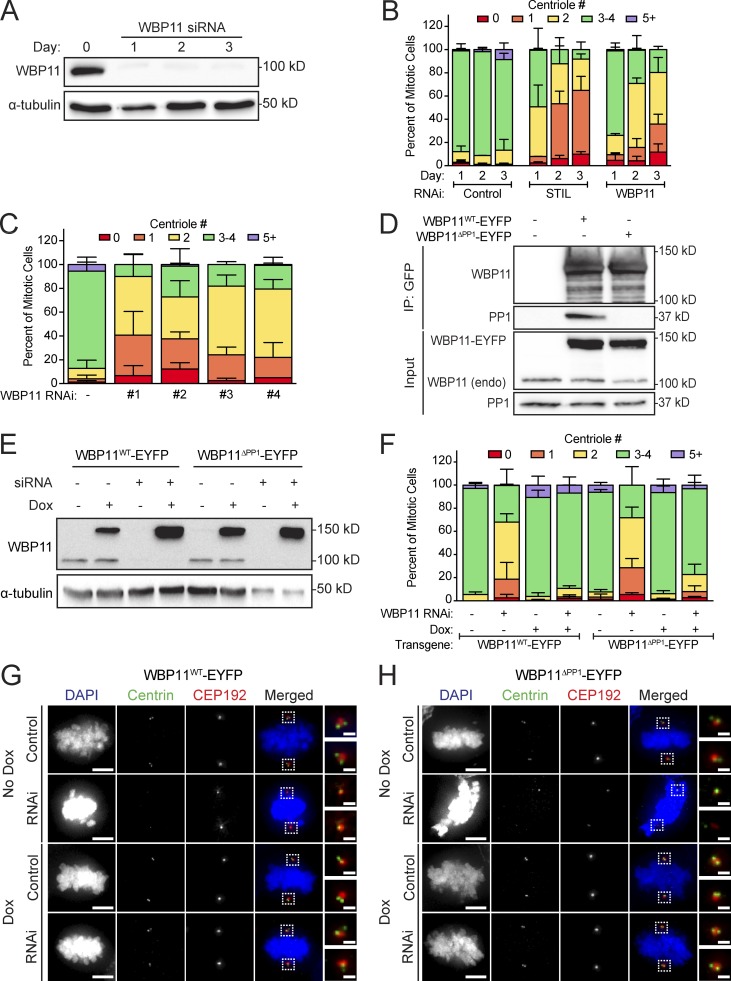
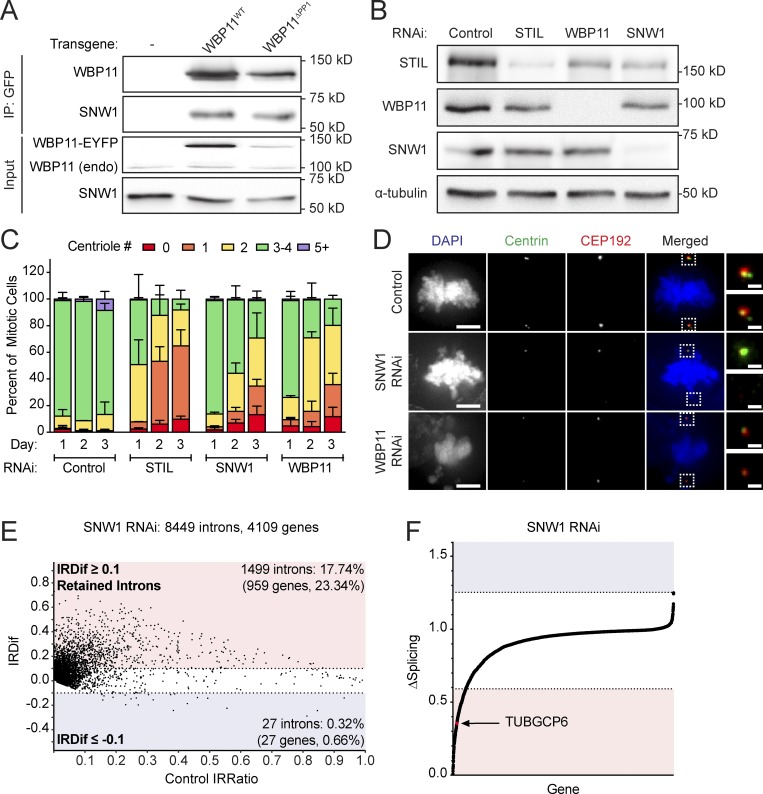
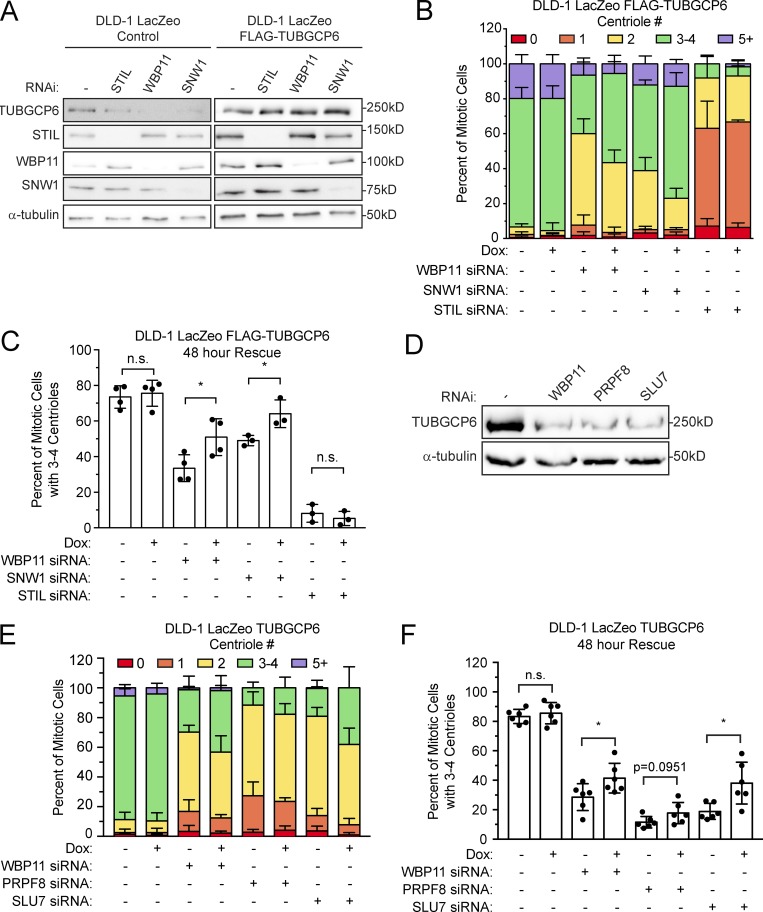
Depletion of WBP11 in DLD-1 cells resulted in 80% of mitotic cells containing two or fewer centrioles by 72 h after siRNA transfection (Fig. 1, A and B). This phenotype was specific for WBP11 depletion, as it was observed with four independent WBP11 siRNAs (Fig. 1 C) and was almost fully rescued by expression of an siRNA-resistant WBP11-EYFP transgene (Fig. 1, E and F). Depletion of WBP11 in RPE-1 cells also caused a failure of centriole duplication, leading to 48% of mitotic cells with two or fewer centrioles by 72 h after siRNA transfection (Fig. S2, A and B). Together, these data show that WBP11 is required for centriole duplication and/or stability.
Figure 1.
WBP11 is required for centriole duplication. (A) Immunoblot showing a time course of siRNA-mediated depletion of WBP11. (B) Quantification of centriole number in mitotic cells 72 h after siRNA-mediated depletion of either STIL or WBP11. n = 3, ≥49 cells per experiment. Error bars represent SD. (C) Quantification of centriole number in mitotic cells 72 h after depletion of WBP11 with one of four independent siRNAs. n = 3, ≥47 cells per experiment. Error bars represent SD. (D) Immunoblot showing coimmunoprecipitation (IP) of endogenous PP1 with WBP11WT-EYFP, but not WBP11ΔPP1-EYFP. (E) Immunoblot showing expression levels of WBP11-EYFP transgenes 72 h after transfection with a WBP11 siRNA. Cells were induced to express the WBP11-EYFP transgenes with doxycycline. (F) Quantification of centriole number in mitotic cells 72 h after siRNA-mediated knockdown of WBP11. Cells were induced to express an RNAi-resistant WBP11 transgene with doxycycline. n = 4, ≥47 cells per experiment. Error bars represent SD. (G) Representative images of cells from F expressing an RNAi-resistant WBP11WT-EYFP transgene. Scale bars represent 5 µm; 1 µm in zoomed-in region. (H) Representative images of cells from F expressing an RNAi-resistant, WBP11ΔPP1-EYFP transgene. Scale bars represent 5 µm; 1 µm in zoomed-in region.
Figure S2.
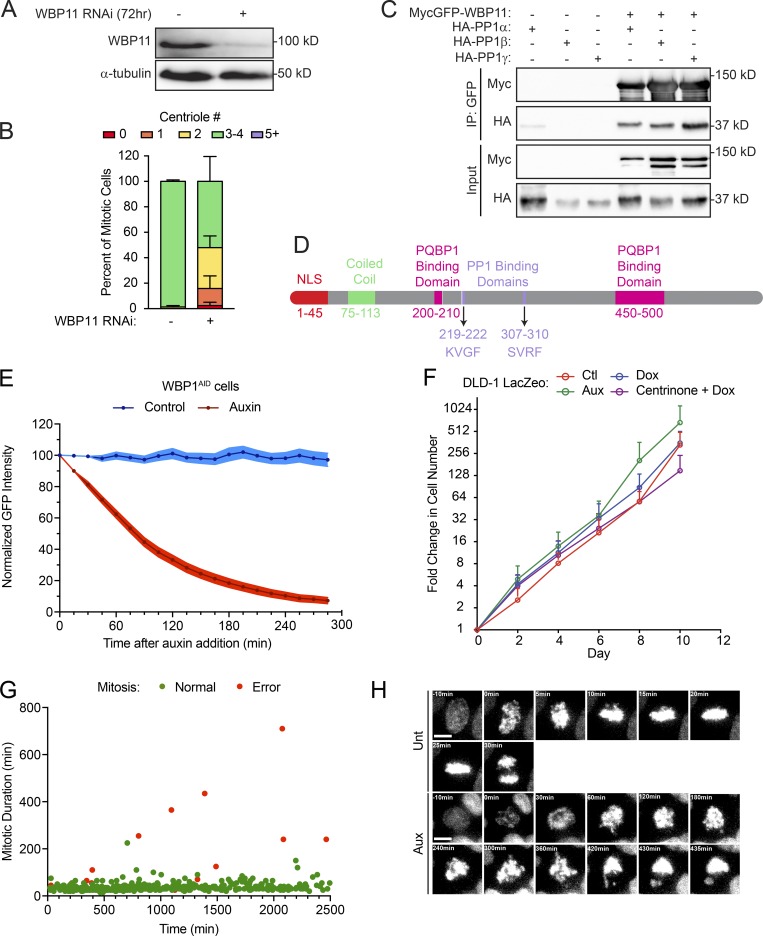
Cells lacking WBP11 show major growth defects. (A) Immunoblot showing expression levels of WBP11 72 h after siRNA transfection in RPE-1 cells. (B) Quantification of centriole number in mitotic RPE-1 cells 72 h after depletion of WBP11 with SMARTpool siRNA. n = 3, ≥50 cells per experiment. Error bars represent SD. (C) Immunoblot showing coimmunoprecipitation (IP) of HA-PP1α, β, and γ with MycGFP-WBP11. (D) Schematic of WBP11 showing its functional domains and the two PP1 binding sites. (E) Quantification of the intensity of the WBP11-mAID-EGFP transgene measured from time-lapse videos of WBP11AID cells after auxin addition. n = 3, 20 cells analyzed per point per replicate. Error bars represent SEM. (F) Growth assay showing the fold increase in cell number of DLD-1 LacZeo cells treated with tetracycline, auxin, or centrinone. Data are means ± SEM, n = 3 (untreated n = 2), performed in triplicate. (G) Quantification of mitotic duration from time-lapse videos of untreated WBP11AID cells expressing H2B-iRFP. The x axis shows how long after the beginning of filming WBP11AID cells entered into mitosis. Green dots mark cells that completed mitosis normally and red dots mark cells that underwent mitotic errors. n = 3, ≥100 cells per experiment. (H) Representative frames from videos of WBP11AID cells stably expressing H2B-iRFP. Cells were either untreated or treated with auxin to induce WBP11AID destruction. Scale bars represent 10 µm.
PP1 binding to WBP11 is not required for centriole biogenesis
Consistent with previous work, we found that WBP11 interacts with all three isoforms of the PP1 catalytic subunit (PP1α, PP1β, and PP1γ; Llorian et al., 2004; Fig. S2 C). WBP11 contains two putative RVxF PP1 binding motifs (Fig. S2 D). To test the requirement of these PP1 binding sites in centriole duplication, we created a WBP11ΔPP1-EYFP construct in which the two residues critical for PP1 binding (valine and phenylalanine) were mutated to alanine at both PP1 binding sites. As expected, the WBP11ΔPP1-EYFP mutant failed to coimmunoprecipitate with PP1 from cells (Fig. 1 D). To evaluate the effect of PP1 binding on centriole biogenesis, siRNA-resistant WT or PP1 binding–defective WBP11-EYFP transgenes were integrated at a predefined genomic locus in a DLD-1 host cell line. Surprisingly, both the WT and PP1 binding–defective WBP11-EYFP transgenes rescued centriole duplication to the same extent in cells depleted of endogenous WBP11 (Fig. 1, E–H). We conclude that PP1 binding to WBP11 is not required for centriole duplication.
Acute depletion of WBP11 leads to centriole duplication failure and mitotic abnormalities
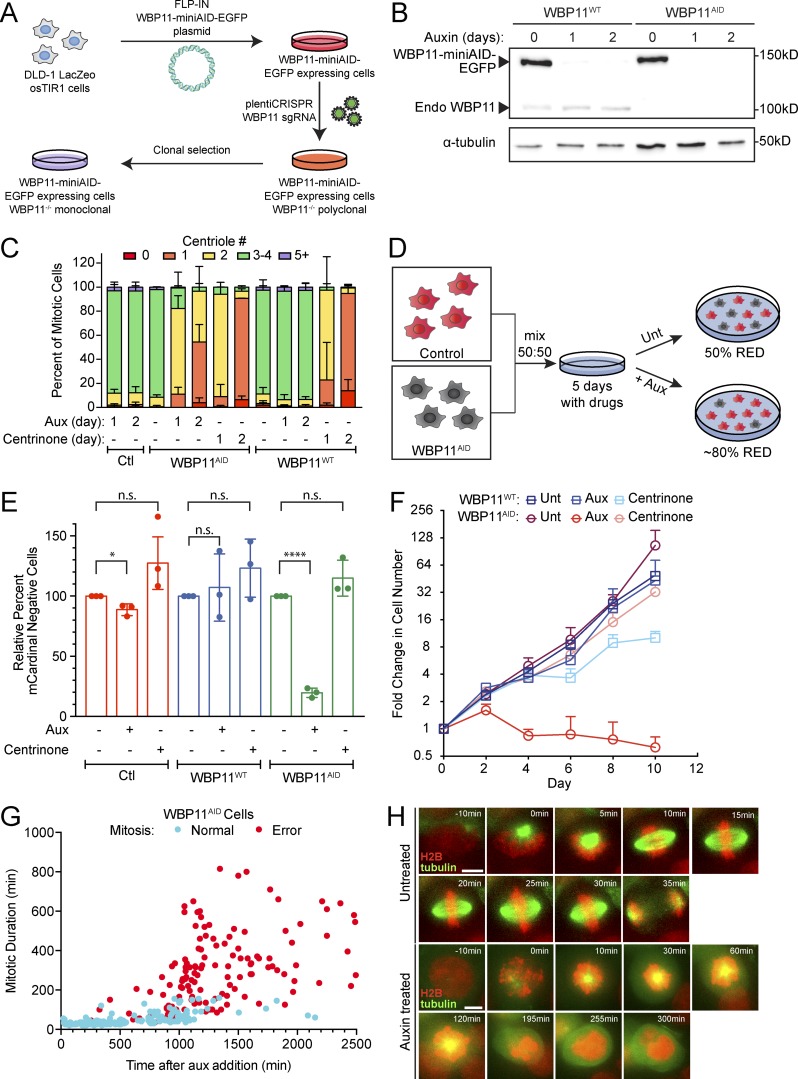
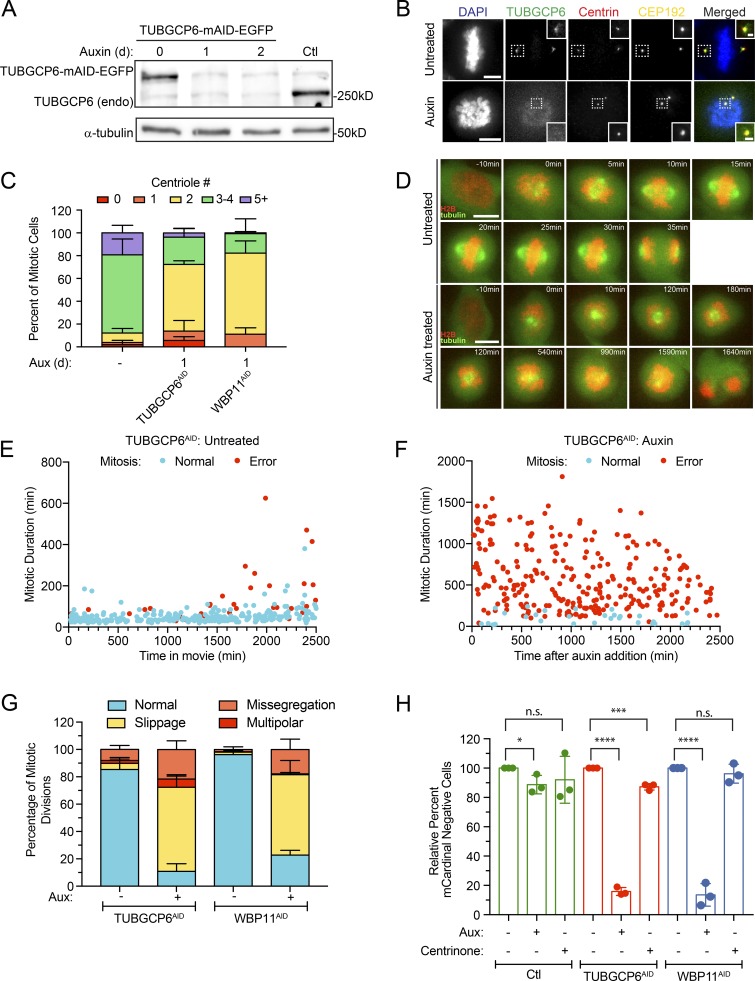
To monitor the acute effect of WBP11 loss on centriole number, we exploited the ability of the auxin-inducible degradation system to achieve rapid protein destruction in mammalian cells (Holland et al., 2012; Nishimura et al., 2009). A WBP11-mAID-EGFP transgene (mAID, mini–auxin-inducible degron) was integrated at a predefined genomic locus in DLD-1 cells, and endogenous WBP11 was then knocked out using CRISPR/Cas9-mediated gene targeting (Fig. 2 A). The resulting WBP11−/−; WBP11-mAID-EGFP DLD-1 cells are hereafter referred to as WBP11AID. As a control, we used a clone of WBP11-mAID-EGFP–expressing cells that went through the same genome-editing procedure but failed to knock out the endogenous WBP11 gene, hereafter referred to as WBP11WT. Addition of auxin led to the degradation of WBP11-mAID-EGFP, with >90% of the protein depleted in 4.5 h (Fig. S2 E). Degradation of WBP11-mAID-EGFP resulted in penetrant centriole duplication failure in WBP11AID, but not WBP11WT, cells: at 24 h after auxin addition, 82% of WBP11AID mitotic cells contained two or fewer centrioles (Fig. 2, B and C).
Figure 2.
WBP11 is required for cell proliferation and timely progression through mitosis. (A) Schematic of WBP11AID cell line design. A WBP11-mAID-EGFP transgene was mutated to be resistant to Cas9 cleavage and integrated into DLD-1 cells at a predefined genetic locus. Cells were then transduced with a lentivirus encoding Cas9 and an sgRNA targeting endogenous WBP11. Monoclonal cell lines were screened for loss of endogenous WBP11. (B) Immunoblot showing expression of the WBP11-mAID-EGFP transgene and its degradation after auxin addition. WBP11WT cells express endogenous WBP11 and the WBP11-mAID-EGFP transgene, while WBP11AID cells express only the WBP11-mAID-EGFP transgene. (C) Quantification of centriole number in mitotic cells 24 or 48 h after degradation of WBP11 with auxin addition or inhibition of PLK4 with centrinone. n = 3, ≥49 cells per experiment. Error bars represent SD. (D) Schematic of a competition growth assay. mCardinal-expressing cells were mixed in a 1:1 ratio with WBP11WT or WBP11AID cells and grown in auxin or centrinone for 5 d. Cells were then analyzed by flow cytometry for mCardinal expression and the ratio of mCardinal-positive to mCardinal-negative cells was determined by flow cytometry. Unt, untreated. (E) Quantification of relative growth of cells using the competition assay following 5 d of auxin treatment. Relative growth was determined by evaluating the fraction of mCardinal-positive cells in the treated compared with untreated populations. n = 3. Unpaired parametric t test. *, P = 0.0160; ****, P < 0.0001. Error bars represent SD. (F) Growth assay showing the fold increase in cell number after auxin or centrinone addition. Data are means ± SEM, n = 3, performed in triplicate. Error bars represent SEM. (G) Quantification of mitotic duration from time-lapse videos of WBP11AID cells expressing H2B-iRFP. The x axis shows how long after addition of auxin WBP11AID cells entered into mitosis. Blue dots mark cells that completed mitosis normally and red dots mark cells that underwent mitotic errors. n = 3, ≥77 cells per experiment. (H) Representative frames from videos of WBP11AID cells stably expressing H2B-iRFP (red) and RFP-tubulin (green). Cells were either untreated or treated with auxin to induce WBP11AID destruction. Scale bars represent 15 µm.
To determine the effect of WBP11 loss on cell proliferation, we examined cell growth using a competition-based growth assay (Fig. 2 D). Centriole loss induced by centrinone treatment had little impact on the short-term growth of DLD-1 cells in this assay (Fig. 2 E). By contrast, the degradation of WBP11-mAID-EGFP reduced the proliferation of WBP11AID cells by 80% but did not affect the growth of WBP11WT cells (Fig. 2 E). Furthermore, although DLD-1 cells continued to proliferate in the absence of centrioles, degradation of WBP11-mAID-EGFP in WBP11AID cells caused a proliferation arrest within 2 d of auxin addition (Fig. 2 F and Fig. S2 F). These data suggest that loss of WBP11 leads to proliferation defects that cannot be explained by centriole duplication failure alone.
To determine why the loss of WBP11 prevents cell proliferation, we filmed WBP11AID cells by time-lapse microscopy following the addition of auxin. Whereas 97% of untreated cells divided normally (Fig. S2, G and H; and Video 1), auxin-treated cells exhibited a dramatic increase in mitotic duration and mitotic errors starting at ∼600 min after auxin addition (Fig. 2, G and H; Fig. S2 H; and Video 2). In contrast to untreated cells (Fig. 2 H and Video 3), cells lacking WBP11 showed reduced centrosome-driven microtubule nucleation and were frequently arrested in mitosis with monopolar spindles (Fig. 2 H and Video 4). The vast majority of these cells underwent mitotic slippage. These data show that the loss of proliferation following degradation of WBP11 occurs primarily as a result of severe cell division defects and not centriole duplication failure.
Video 1.
Untreated WBP11AID cell expressing H2B-iRFP. Still images are represented in Fig. S2 H. One frame captured every 5 min; displayed at 3 frames/s.
Video 2.
Auxin-treated WBP11AID cell expressing H2B-iRFP. Still images are represented in Fig. S2 H. One frame captured every 5 min; displayed at 3 frames/s.
Video 3.
Untreated WBP11AID cell co-expressing H2B-iRFP (red) and RFP-tubulin (green). Still images are represented in Fig. 2 H. One frame captured every 5 min; displayed at 3 frames/s.
Video 4.
Auxin-treated WBP11AID cell co-expressing H2B-iRFP (red) and RFP-tubulin (green). Still images are represented in Fig. 2 H. One frame captured every 5 min; displayed at 3 frames/s.
The WBP11 proximity interactome highlights a core role in pre-mRNA splicing
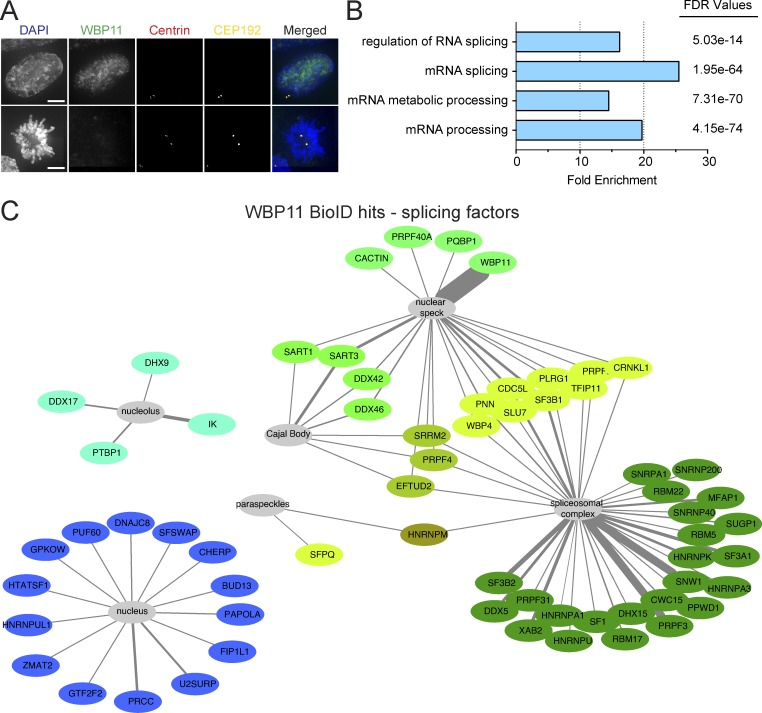
WBP11 is reported to be a nuclear protein, and, accordingly, we did not observe the WBP11-mAID-EGFP transgene localizing to the centriole or centrosome at any stage of the cell cycle (Fig. 3 A; Llorian et al., 2005). To better understand the role of WBP11 in centriole duplication, we conducted biotinylation-dependent proximity mapping (BioID) experiments using DLD-1 cells stably expressing a WBP11-BirA* transgene (Roux et al., 2012). We identified a set of 178 high-confidence proximity interaction partners for WBP11 (Table S2). Previous studies have implicated WBP11 in pre-mRNA splicing and, in accord with this function (Craggs et al., 2001; Llorian et al., 2004, 2005), the top enriched gene ontology (GO) terms for the high-confidence WBP11 proximity interactome were “mRNA processing” (false discovery rate [FDR] = 4.15 × 10−74) and “RNA splicing” (FDR = 4.54 × 10−71; Fig. 3 B). Moreover, 35% (63 of 178) of the high-confidence proximity interactors we identified have putative or known roles in mRNA splicing (Fig. 3 C).
Figure 3.
The proximity interactome of WBP11 contains many splicing factors. (A) Representative image showing WBP11-mAID-EGFP localization in interphase (top panels) and mitosis (bottom panels). Scale bars represent 10 µm. (B) A subset of the top enriched GO terms upon analysis of the WBP11 proximity interactome. PANTHER GO biological process complete analysis was used to calculate the fold enrichments and FDR values. (C) Schematic of known splicing proteins that were present in the WBP11-BirA proximity interactome plotted using Cytoscape. Only high-confidence hits are shown. Proteins with similar interaction profiles are colored the same. Line thickness represents the ratio calculated from the enrichment of WBP11-BirA versus BirA alone. n = 4. Raw data are shown in Table S2.
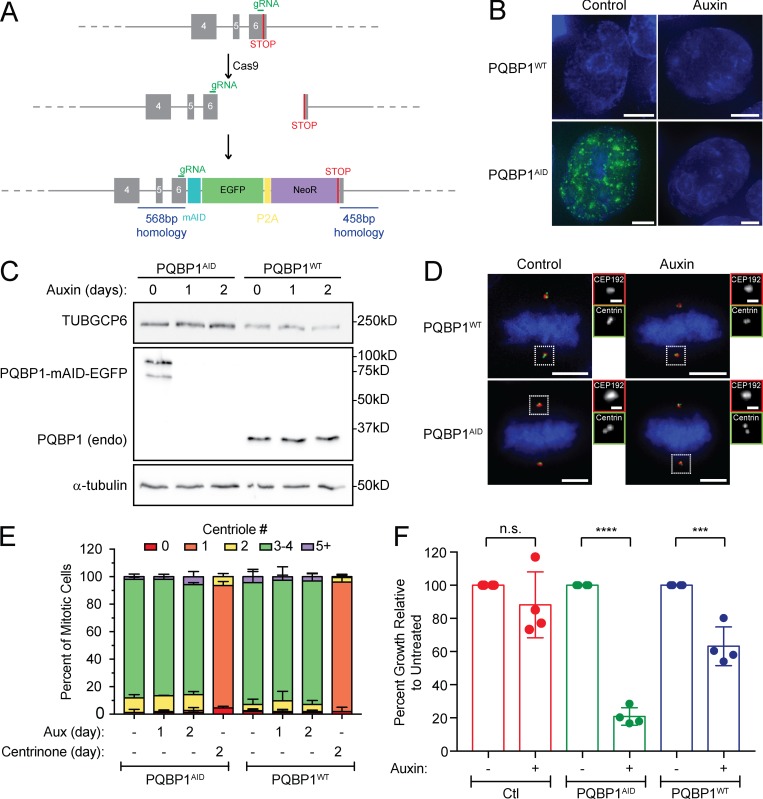
One of the most abundant proteins in the WBP11 proximity interactome was PQBP1, a known binding partner of WBP11 (Komuro et al., 1999). We used CRISPR/Cas9 to tag the C-terminus of both endogenous PQBP1 alleles with mAID-EGFP in DLD-1 cells (Fig. S3, A and B). Degradation of PQBP1 with auxin reduced the long-term growth of cells but did not affect centriole duplication (Fig. S3, C–F). We conclude that WBP11 binding to PQBP1 is not required for centriole duplication to occur.
Figure S3.
PQBP1 is not required for centriole biogenesis. (A) Schematic depicting the strategy for endogenous tagging of the PQBP1 gene. Cells were cotransfected with a plasmid encoding the repair template and a plasmid that expresses Cas9 and an sgRNA. Homozygous PQBP1-AID-EGFP clones were identified by immunoblot. (B) Representative images of a control cell line in which PQBP1 was not tagged (PQBP1WT) and a cell line in which both PQBP1 alleles were endogenously tagged with mAID-EGFP (PQBP1AID). Scale bars represent 5 µm. (C) Immunoblot showing expression of endogenously tagged PQBP1-mAID-EGFP in PQBP1AID cells and its degradation after auxin addition. PQBP1WT cells are shown as a control. (D) Representative images of control cells (PQBP1WT) and cells with endogenously tagged PQBP1-mAID-EGFP (PQBP1AID). Cells were either untreated or treated with auxin to induce PQBP1AID destruction. Scale bars represent 5 µm; 1 µm in zoomed-in regions. (E) Quantification of centriole number in mitotic cells after 48 h of PQBP1-mAID-EGFP degradation by auxin or PLK4 inhibition by centrinone. n = 3, ≥50 cells per experiment. Error bars represent SD. (F) Clonogenic growth assay of the PQBP1AID cell line compared with a nontransduced cell line (Ctl) and to a transduced but not tagged cell line (PQBP1WT). n = 4. Unpaired parametric t test: ***, P = 0.0007; ****, P < 0.0001. Error bars represent SD.
Transcriptome-wide identification of introns whose splicing depends on WBP11
From our proximity interactome, we identified multiple pre-mRNA splicing factors, including SNW1 (Sundaramoorthy et al., 2014; van der Lelij et al., 2014), SON (Ahn et al., 2011), PRPF8 (Grainger and Beggs, 2005), and SLU7 (Frank and Guthrie, 1992), that have previously been shown to be required for centriole duplication (Balestra et al., 2013). Given that the GO analysis of our proximity interactome yielded a high fraction of RNA processing and splicing factors, we hypothesized that WBP11 indirectly regulates centriole duplication through the splicing of pre-mRNAs that encode centriolar proteins. To address this question, we performed mRNA-seq on polyA-containing RNA isolated from DLD-1 cells 72 h after mock or WBP11 siRNA transfection. To exclude changes that may be caused by the siRNA transfection procedure or by off-targets of the WBP11 siRNA, we also isolated polyA-containing RNA from DLD-1 cells and WBP11AID cells that were treated with auxin for 48 h. First, we identified genes whose expression increased or decreased by more than two SDs from the average in both the WBP11 RNAi and WBP11AID samples (Table S3). This analysis identified 29 genes with decreased expression and 49 genes with increased expression. However, none of these genes had known roles in centriole biogenesis (Table S3).
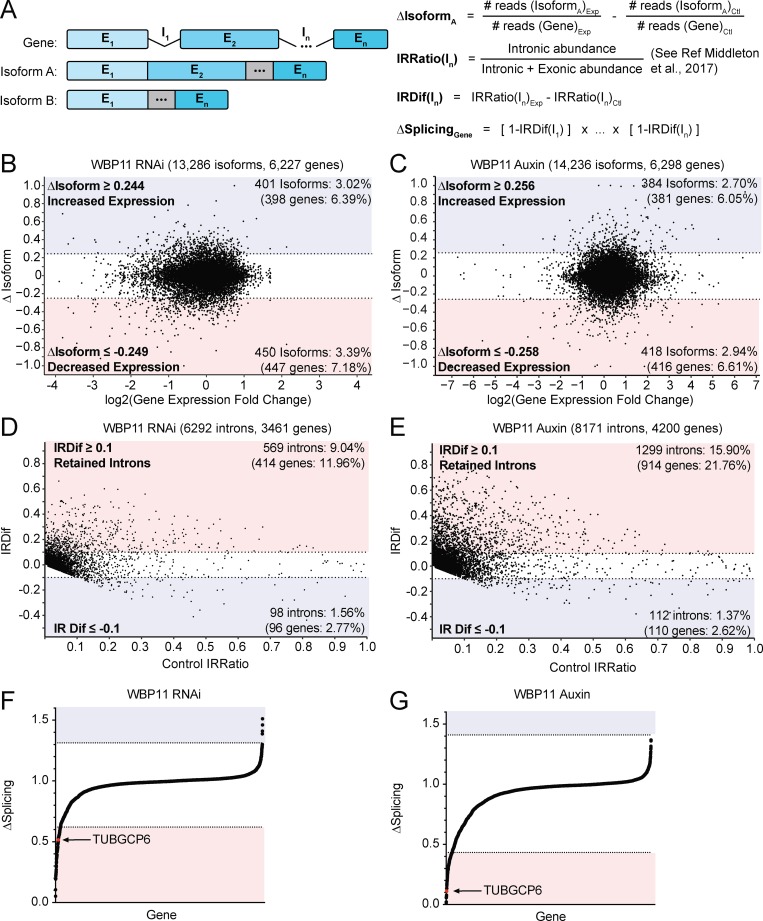
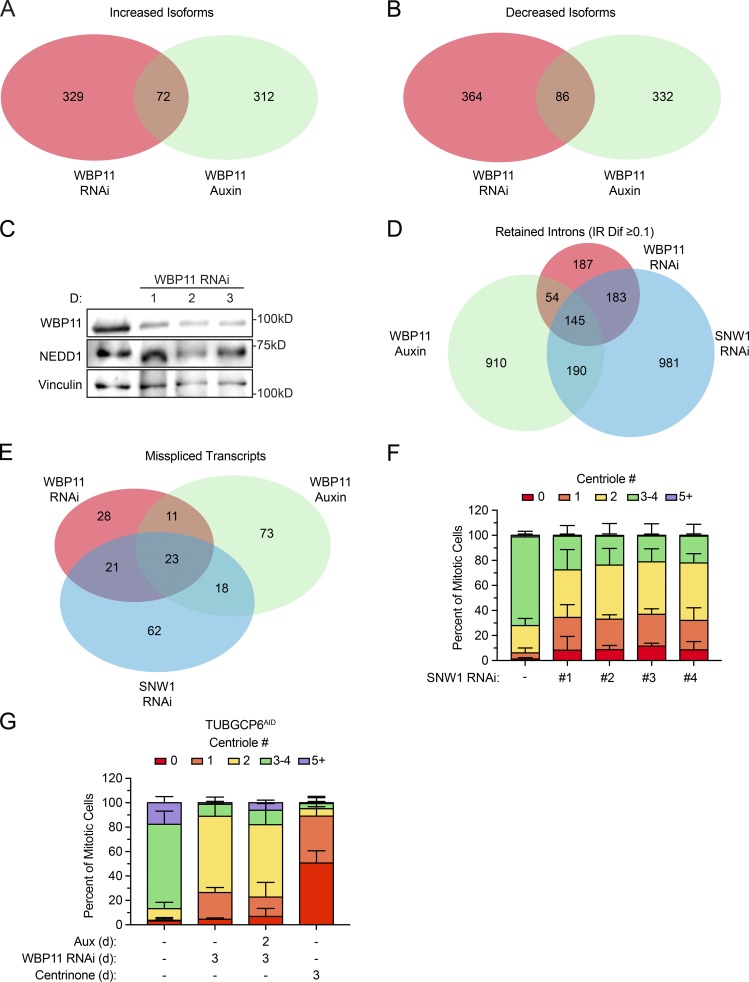
Next, we examined differentially expressed isoforms in both the siRNA and AID datasets. We calculated the fraction of reads for a gene that maps back to each isoform in both the control and experimental sample. We then subtracted the value for each isoform obtained in control cells from the value obtained in cells depleted of WBP11. This produced a ΔIsoform score ranging from −1 to +1 that represents the change in splicing of an isoform after WBP11 depletion: a positive ΔIsoform score represents an isoform with increased expression, and a negative ΔIsoform score corresponds to an isoform with decreased expression (Fig. 4 A). We considered a hit to be any isoform with a ΔIsoform score more than two SDs above or below the average change in isoform expression. This analysis identified 72 isoforms with increased expression and 86 isoforms with decreased expression in both the WBP11 RNAi and auxin samples (Fig. 4, B and C; Fig. S4, A and B; and Table S4). The only centrosomal gene that was present in these hits was NEDD1. However, the NEDD1 protein levels were not altered by WBP11 depletion (Fig. S4 C). Therefore, the differential expression of NEDD1 isoforms cannot explain the centriole duplication failure caused by depletion of WBP11.
Figure 4.
WBP11 is required for proper splicing of a subset of introns. (A) Schematic describing the RNA-seq analysis performed. For a given gene, the change in isoform expression (ΔIsoform) was calculated by subtracting the fraction of reads for a gene that map back to an isoform in the control sample from the corresponding value in the experimental sample. IRratio was calculated using the IRFinder program and measures the frequency with which an intron is retained in an mRNA as a value from 0 to 1 (Middleton et al., 2017). IRDif was calculated by subtracting the IRratio of each intron in the control from the experimental condition. ΔSplicing was calculated by taking the IRDif for each intron and subtracting it from 1 to determine the fraction of mRNAs that are correctly spliced for that intron. We then multiplied this value for each intron in an mRNA to determine the percentage of the mRNAs for each gene that are correctly spliced. (B) Classification of isoforms that were differentially expressed 72 h after WBP11 depletion by RNAi. The ΔIsoform value for each isoform was plotted against fold change in gene expression. Isoforms with a more than two SD change in expression were considered as hits. Colors indicate isoforms with increased (blue) or decreased (red) expression following WBP11 depletion. Raw data are shown in Tables S3 and S4. (C) Classification of isoforms that were differentially expressed 48 h after WBP11 degradation by addition of auxin. The ΔIsoform value for each isoform was plotted against fold change in gene expression. Isoforms with a more than two SD change in expression were considered hits. Colors indicate isoforms with increased (blue) or decreased (red) expression following WBP11 depletion. Raw data are shown in Tables S3 and S4. (D) Classification of introns detected by mRNA-seq 72 h after WBP11 depletion by RNAi. The IRDif score of each intron was plotted against the IRratio of that same intron in the control sample. Colors indicate introns that are excised more efficiently (blue), or retained more often (red), following WBP11 depletion. Raw data are shown in Table S5. (E) Classification of introns detected by mRNA-seq 48 h after WBP11 degradation by addition of auxin. The IRDif score of each intron was plotted against the IRratio of that same intron in the control sample. Colors indicate introns that are excised more efficiently (blue), or retained more often (red), following WBP11 depletion. Raw data are shown in Table S5. (F) Plot showing the change in the fraction of completely spliced mRNA for each gene 72 h after WBP11 depletion by RNAi. Colors indicate three SDs above (blue) or below (red) the average level of mRNA splicing observed. Raw data are shown in Table S6. (G) Plot showing the change in the fraction of completely spliced mRNA for each gene 48 h after WBP11 degradation with auxin. Colors indicate three SDs above (blue) or below (red) the average level of mRNA splicing observed. Raw data are shown in Table S6.
Figure S4.
WBP11 and SNW1 are required for the correct splicing of a common set of pre-mRNAs. (A) Overlapping hits for isoforms with expression increased more than two SDs following WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 72 isoforms that depend on both WBP11 and SNW1 for appropriate expression were identified. (B) Overlapping hits for isoforms with expression decreased more than two SDs following WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 86 isoforms that depend on both WBP11 and SNW1 for appropriate expression were identified. (C) Immunoblot showing NEDD1 protein levels after siRNA-mediated depletion of WBP11. (D) Overlapping hits for retained introns with an IRDif score of ≥0.1 following WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 145 introns that depend on WBP11 and SNW1 for efficient splicing were identified. (E) Overlapping hits for the genes that experienced a greater than three SD decrease of correctly spliced mRNA after WBP11 or SNW1 RNAi or WBP11 degradation with auxin. A set of 23 genes that depend on WBP11 and SNW1 for efficient splicing were identified. (F) Quantification of centriole number in mitotic cells 72 h after depletion of SNW1 with one of four independent siRNAs. n = 3, ≥50 cells per experiment. Error bars represent SD. (G) Quantification of centriole number in mitotic cells 72 h after depletion of WBP11 by siRNA in TUBGCP6AID cells. TUBGCP6AID was degraded by auxin where indicated. n = 3, ≥50 cells per experiment. Error bars represent SD.
Since changes in differential gene and isoform expression could not readily explain the centriole duplication defects observed following the loss of WBP11, we analyzed the effect of WBP11 loss on the splicing of individual introns. We used IRFinder to calculate an intron retention (IR) ratio that scores the frequency with which an intron is retained in an mRNA as a value from 0 to 1 (Middleton et al., 2017). We then subtracted the IRratio for each intron in control cells from the IRratio of the same intron in cells depleted of WBP11. This produced an IR difference (IRDif) score from −1 to +1 that represents the change in IRratio after WBP11 depletion: a positive IRDif score corresponds to an intron that was retained more frequently, and a negative IRDif score corresponds to an intron was spliced more efficiently (Fig. 4 A). To be considered a hit, an intron had to have an IRDif score of ≥0.1 or ≤−0.1. This analysis revealed 569 introns (9% of total introns detected, 85% of all hits) and 1,299 introns (16% of total introns detected, 92% of all hits) with an IRDif score of ≥0.1 after WBP11 depletion by siRNA or auxin, respectively (Fig. 4, D and E; and Table S5). Importantly, we detected only a small number of intronic transcripts whose abundance was decreased following depletion of WBP11, showing that WBP11 is primarily required to promote the efficient splicing of a subset of introns.
35% of the introns retained in the WBP11 RNAi sample were also retained in the auxin-treated WBP11AID samples, providing a high-confidence list of 199 introns in 164 genes that show increased retention of one or more introns following WBP11 depletion (Fig. S4 D). While 87% of WBP11 target genes possess a single intron that relies on WBP11 for efficient splicing, 13% of genes possess multiple introns that are significantly altered following WBP11 knockdown. Genes with greater numbers of introns have a higher probability of being affected by WBP11 knockdown because they are more likely to have at least one intron that requires WBP11 for correct splicing, and because the impact of having multiple partially retained introns increases with the number of introns a gene contains. To determine the effect of WBP11 depletion on each gene, we calculated ΔSplicing as the change in the fraction of completely spliced mRNA for each gene following WBP11 depletion (see Materials and methods; Fig. 4, A, F, and G; and Table S6). We identified 34 high-confidence genes with IR that showed a greater than three SD decrease in the amount of completely spliced mRNA following WBP11 depletion by either siRNA or auxin (Fig. S4 E and Table S6). Only 1 of these 34 genes, TUBGCP6, was a known centrosome protein (see below; Bahtz et al., 2012). These data show that WBP11 loss has a large impact on the splicing of a small number of pre-mRNAs.
WBP11 and SNW1 are required for the splicing of a common set of pre-mRNAs
SNW1 was identified as an abundant proximity-interacting partner of WBP11 (Fig. 3 C), and we confirmed that the WBP11WT-EYFP transgene coimmunoprecipitated with endogenous SNW1 in cells (Fig. 5 A). SNW1 is a pre-mRNA splicing factor that has previously been shown to be required for centriole duplication (Balestra et al., 2013; van der Lelij et al., 2014). After 72 h of SNW1 depletion by SMARTpool siRNA, 71% of cells contained two or fewer centrioles (Fig. 5, B–D). This phenotype was specific for SNW1 depletion as it was observed with four independent SNW1 siRNAs (Fig. S4 F). Since SNW1 and WBP11 interact in cells and produce similar phenotypes after siRNA depletion, we explored the possibility that these two proteins may control the splicing of a common set of pre-mRNAs. To evaluate this, we performed mRNA-seq on cells depleted of SNW1 for 72 h and analyzed IR (Table S5). We found a larger subset of introns retained in SNW1-depleted cells compared with cells depleted of WBP11 (1,499 intron hits in 959 genes; Fig. 5 E). Interestingly, of the introns that increased in abundance in the WBP11 RNAi sample, 57.6% were identical to introns that increased in abundance in the SNW1-depleted cells (Fig. S4 D). Moreover, 44 genes showed a more than three SD decrease in the amount of completely spliced mRNA after siRNA-mediated depletion of WBP11 or SNW1 (Fig. 5 F, Fig. S4 E, and Table S6). These results show that WBP11 and SNW1 promote the removal of a specific subset of introns.
Figure 5.
SNW1 interacts with WBP11 and is required for centriole biogenesis. (A) Immunoblot showing coimmunoprecipitation (IP) of endogenous SNW1 with WBP11WT-EYFP and WBP11ΔPP1-EYFP. (B) Immunoblot showing expression levels of STIL, SNW1, and WBP11 72 h after siRNA transfection. (C) Quantification of centriole number in mitotic cells 72 h after siRNA-mediated depletion of SNW1. Data are shown alongside the data for WBP11 and STIL from Fig. 1 B. n = 3, ≥49 cells per experiment. Error bars represent SD. (D) Representative images of cells from C transfected with a WBP11 or SNW1 siRNA. Scale bars represent 5 µm; 1 µm in zoomed-in regions. (E) Classification of introns detected by mRNA-seq 72 h after SNW1 depletion by RNAi. The IRDif score of each intron was plotted against the IRratio of that same intron in the control sample. Colors indicate introns that are excised more efficiently (blue), or retained more often (red), following SNW1 depletion. Raw data are shown in Table S5. (F) Plot showing the change in the fraction of completely spliced mRNA for each gene 72 h after SNW1 depletion with siRNA. Colors indicate three SDs above (blue) or below (red) the average level of mRNA splicing observed. Raw data are shown in Table S6.
WBP11 promotes the splicing of TUBGCP6
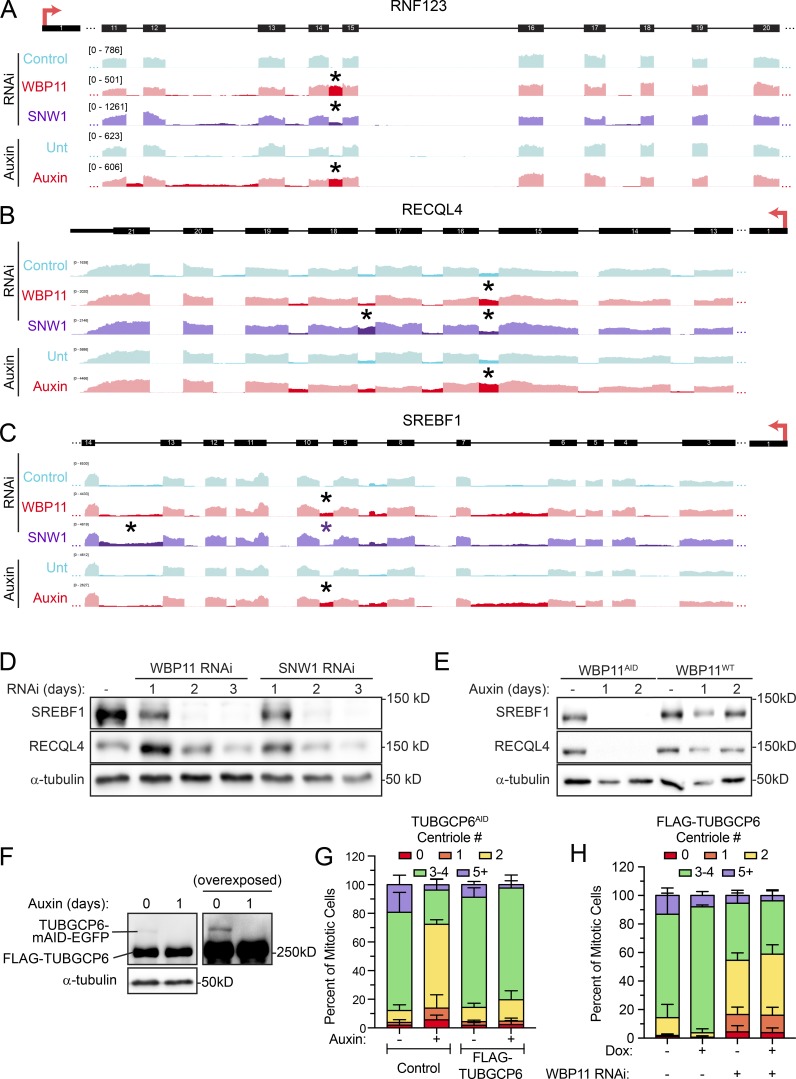
Altogether, we identified 145 introns in 122 genes that were preferentially retained in all three experimental populations (WBP11 siRNA, SNW1 siRNA, and WBP11 auxin; Fig. S4 D and Table S5). Some of these genes, such as RNF123, have a single intron that is highly retained following WBP11 or SNW1 depletion, while other genes, such as RECQL4 and SREBF1, show multiple retained introns (Fig. S5, A–C). Immunoblotting confirmed that defects in splicing led to the expected reduction in SREBF1 and RECQL4 protein levels (Fig. S5, D and E). Although most retained introns require both WBP11 and SNW1 for efficient splicing, there were some introns whose splicing depends on SNW1, but not WBP11 (Fig. S5, B and C).
Figure S5.
WBP11 and SNW1 are required for the correct splicing of a subset of introns. (A) A region of the RNF123 gene is aligned to IGV reads to highlight retained introns (*) observed after loss of WBP11 or SNW1. RNF123 requires WBP11 and, to a lesser extent, SNW1 for efficient splicing of one of its introns (*). (B) A region of the RECQL4 gene is aligned to IGV reads to highlight retained introns (*) observed after loss of WBP11 or SNW1. RECQL4 requires WBP11 and SNW1 for efficient splicing of some introns (*). Note that some introns require SNW1, but not WBP11, for efficient splicing. (C) A region of the SREBF1 gene is aligned to IGV reads to highlight retained introns (*) observed after loss of WBP11 or SNW1. SREBF1 requires WBP11 and SNW1 for efficient splicing of some introns (*). Note that some introns require SNW1, but not WBP11, for efficient splicing. (D) Immunoblot showing expression levels of RECQL4 and SREBF1 after 72 h of SNW1 or WBP11 siRNA knockdown. (E) Immunoblot showing expression levels of RECQL4 and SREBF1 in WBP11WT and WBP11AID cells at 48 h after auxin addition. (F) Immunoblot showing TUBGCP6 expression in TUBGCP6AID cells expressing a FLAG-TUBGCP6 transgene. (G) Quantification of centriole number in mitotic TUBGCP6AID cells 24 h after auxin addition. TUBGCP6AID cells with or without expression of a FLAG-TUBGCP6 transgene were used. Data are shown alongside the TUBGCP6AID data from Fig. 7 C for comparison. n = 3, 50 cells per experiment. Error bars represent SD. (H) Graph showing the percentage of mitotic cells with three to four centrioles after 72 h of WBP11 depletion by siRNA. Expression of the FLAG-TUBGCP6 was induced by addition of doxycycline where indicated. n = 3, 50 cells/experiment. Error bars represent SD.
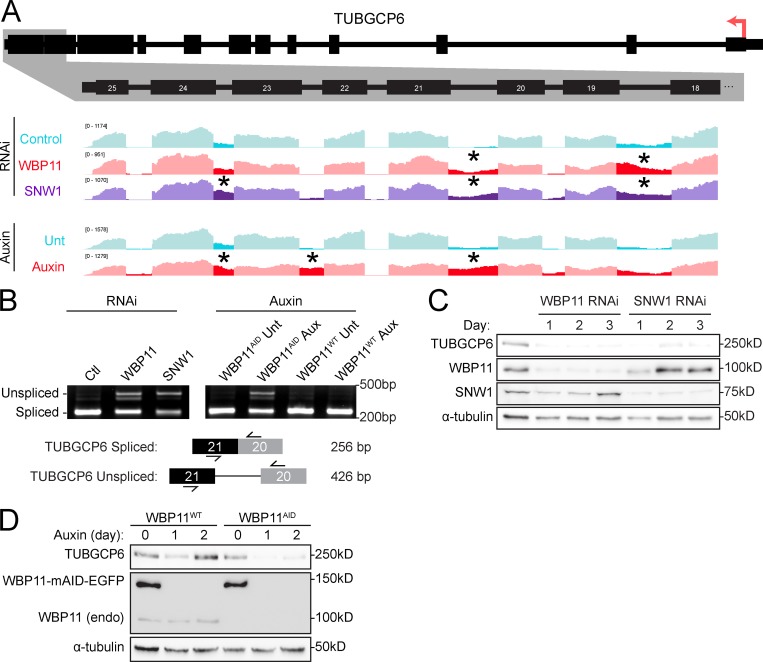
To obtain insight into how WBP11 and SNW1 control centriole duplication, we compared the list of genes that are strongly dependent on both WBP11 and SNW1 for correct splicing with a list of known centrosome proteins (Alves-Cruzeiro et al., 2014). 23 genes across all three experimental conditions (WBP11 siRNA, WBP11 auxin degradation, and SNW1 siRNA) showed a more than three SD decrease in the amount of completely spliced mRNA (Fig. S4 E and Table S6). TUBGCP6 was the only gene within this list known to localize to the centrosome. Correct splicing of the TUBGCP6 pre-mRNA was decreased by 48% following WBP11 siRNA, 89% after WBP11 auxin degradation, and 65% following SNW1 depletion (Figs. 4, F and G; and 5 F). Moreover, the abundance of the TUBGCP6 mRNA was also modestly depleted by 0.2–0.5-fold following loss of WBP11 or SNW1 (Table S3). Manual inspection of the mRNA-seq data aligned to the Integrated Genome Viewer revealed that cells depleted of WBP11 or SNW1 showed the retention of multiple introns toward the 3′ end of the TUBGCP6 mRNA, an effect that was confirmed by RT-PCR analysis (Fig. 6, A and B). TUBGCP6 is an essential component of the γ-TuRC and has a direct role in promoting microtubule nucleation at the centrosome (Bahtz et al., 2012; Farache et al., 2018). Retention of introns at the 3′ end of the TUBGCP6 mRNA is expected to produce a truncated protein of ∼170 kD that would be unable to bind to γ-tubulin. We tested whether the protein levels of TUBGCP6 changed after depletion of SNW1 or WBP11 and found that the abundance of TUBGCP6 was dramatically reduced by 24 h after siRNA-mediated depletion of SNW1 or WBP11 (Fig. 6 C). A similar effect was observed in WBP11AID cells after degradation of the WBP11-mAID-EGFP transgene (Fig. 6 D).
Figure 6.
WBP11 and SNW1 are required for proper splicing of the TUBGCP6 pre-mRNA. (A) Schematic of the TUBGCP6 gene. The 3′ end of the gene is expanded and aligned to Integrative Genomics Viewer (IGV) reads to highlight retained introns (*) observed after loss of WBP11 or SNW1. (B) RT-PCR analysis of splicing of TUBGCP6 intron 21 following depletion of WBP11 or SNW1. Schematic shows the position of the primers used for PCR and the expected product size. (C) Immunoblot showing TUBGCP6 expression levels over a 3-d period following WBP11 or SNW1 siRNA knockdown. (D) Immunoblot showing TUBGCP6 expression levels in WBP11WT or WBP11AID cells after 1 or 2 d of auxin addition.
Depletion of TUBGCP6 leads to similar phenotypes as the loss of WBP11
To determine if depletion of TUBGCP6 leads to a failure of centriole duplication in DLD-1 cells, we used CRISPR/Cas9 to tag the C-terminus of both endogenous TUBGCP6 alleles with a mAID-EGFP degron (Bahtz et al., 2012). Degradation of TUBGCP6 led to a pronounced failure of centriole duplication that was similar to that observed in WBP11AID cells (Fig. 7, A–C). Cells depleted of TUBGCP6 also exhibited mitotic defects similar to those observed following loss of WBP11, including reduced centrosome-driven microtubule nucleation and mitotic arrest with a monopolar spindle (Fig. 2, G and H; Fig. 7, E–G; and Videos 5 and 6). However, while WBP11AID cells divided normally for ∼600 min (10 h) after auxin addition before undergoing abnormal mitotic divisions, TUBGCP6AID cells displayed mitotic abnormalities almost immediately following treatment with auxin (Figs. 2 G and 7 F). This is consistent with the loss of WBP11 indirectly affecting mitotic progression due to defects in the splicing of a limited set of pre-mRNAs. Analysis of mitotic divisions taking place at least 1,000 min after auxin addition showed that both WBP11- and TUBGCP6-depleted cells predominantly undergo mitotic slippage (Fig. 7 G). Similar to WBP11AID cells, the mitotic deficits dramatically reduced the proliferation of cells depleted of TUBGCP6 (Fig. 7 H). Auxin-induced degradation of TUBGCP6 did not exacerbate the failure of centriole duplication observed in cells depleted of WBP11, suggesting these genes function in the same pathway (Fig. S4 G). We, therefore, considered TUBGCP6 to be an attractive candidate to explain how WBP11 loss leads to defects in centriole biogenesis.
Figure 7.
TUBGCP6 depletion produces phenotypes similar to WBP11 loss. (A) Immunoblot showing expression of endogenously tagged TUBGCP6-mAID-EGFP and its degradation after auxin addition. In TUBGCP6AID cells, both endogenous TUBGCP6 alleles were tagged with mAID-EGFP. TUBGCP6WT cells are shown as a control. (B) Representative images of endogenously tagged TUBGCP6-mAID-EGFP. Cells were either untreated or treated with auxin to induce TUBGCP6AID destruction. Scale bars represent 5 µm; 1 µm in zoomed-in regions. (C) Quantification of centriole number in mitotic cells 24 h after TUBGCP6-mAID-EGFP degradation with auxin. Data are shown alongside the 24-h auxin degradation of WBP11AID from Fig. 2 C. n = 3, ≥50 cells per experiment. Error bars represent SD. (D) Representative frames from videos of TUBGCP6AID cells stably expressing H2B-iRFP (red) and EGFP-tubulin (green). Cells were either untreated or treated with auxin to induce TUBGCP6AID destruction. Scale bars represent 10 µm. (E) Quantification of mitotic duration from time-lapse videos of TUBGCP6AID cells expressing H2B-iRFP. The x axis shows the time in the video at which the untreated TUBGCP6AID cells entered into mitosis. Blue dots mark cells that completed mitosis normally and red dots mark cells that underwent mitotic errors. n = 3, ≥100 cells per experiment. (F) Quantification of mitotic duration from time-lapse videos of TUBGCP6AID cells expressing H2B-iRFP. The x axis shows the time after auxin addition that TUBGCP6AID cells entered into mitosis. Blue dots mark cells that completed mitosis normally and red dots mark cells that underwent mitotic errors. n = 3, ≥100 cells per experiment. (G) Quantification of the types of mitotic errors observed in cells depleted of TUBGCP6 or WBP11 for all mitotic divisions after 1,000 min of auxin treatment (images and mitotic time for WBP11 cells shown in Fig. 2). Error bars represent SD. (H) Quantification of relative growth of cells using the competition assay following 5 d of auxin treatment. Relative growth was determined by evaluating the fraction of mCardinal-positive cells in the treated compared with untreated populations. n = 3. Unpaired parametric t test. *, P = 0.0366; ***, P = 0.0006; ****, P < 0.0001. Error bars represent SD.
Video 5.
Untreated TUBGCP6AID cell co-expressing H2B-iRFP (red) and RFP-tubulin (green). Still images are represented in Fig. 7 D. One frame captured every 5 min; displayed at 3 frames/s.
Video 6.
Auxin-treated TUBGCP6AID cell co-expressing H2B-iRFP (red) and RFP-tubulin (green). Still images are represented in Fig. 7 D. One frame captured every 5 min; displayed at 3 frames/s.
Expression of intronless TUBGCP6 partially restores centriole duplication in cells lacking WBP11
To test the hypothesis that the failure of TUBGCP6 pre-mRNA processing is causally linked to the failure of centriole duplication in cells depleted of WBP11, we generated a DLD-1 cell line stably expressing an intronless version of TUBGCP6. We first examined the functionality of the TUBGCP6 transgene by testing its ability to rescue centriole duplication following auxin-induced degradation of endogenous TUBGCP6. Expression of a TUBGCP6 transgene fully rescued centriole duplication in auxin-treated TUBGCP6AID cells, demonstrating that this transgene was able to functionally compensate for the loss of the endogenous protein (Fig. S5, F and G). Importantly, expression of this TUBGCP6 transgene was also able to partially restore centriole duplication 48 h after RNAi-mediated depletion of WBP11 or SNW1 (Fig. 8, A–C). By contrast, expression of a TUBGCP6 transgene had no effect on centriole biogenesis in cells depleted of the centriole protein STIL, indicating that a TUBGCP6 transgene cannot compensate for the loss of core centriole proteins. Expression of TUBGCP6 was unable to rescue centriole biogenesis after 72 h of WBP11 depletion by siRNA (Fig. S5 H). Given that re-expression of intronless TUBGCP6 failed to rescue centriole duplication completely, it is likely that loss of WBP11 affects additional factors required for centriole biogenesis. These results indicate that the defective synthesis of mature TUBGCP6 protein is a contributor to the failure of centriole biogenesis in cells lacking WBP11 and SNW1.
Figure 8.
Expression of intronless TUBGCP6 can partially rescue centriole duplication failure after WBP11 depletion. (A) Immunoblot showing TUBGCP6 levels after 48 h of siRNA-mediated depletion of SNW1 or WBP11. Cells on the right expressed the FLAG-TUBGCP6 transgene. (B) Quantification of centriole number in mitotic cells after siRNA-mediated knockdown of SNW1 or WBP11 for 48 h. FLAG-TUBGCP6 transgene was induced with doxycycline where indicated. n = 3 (SNW1 and STIL), n = 4 (control and WBP11), ≥50 cells per experiment. Error bars represent SD. (C) Graph showing the percentage of mitotic cells with three to four centrioles, as shown in Fig. 8 B. Unpaired parametric t test: *, P = 0.0343 (WBP11) and 0.0353 (SNW1). Error bars represent SD. (D) Immunoblot showing the levels of TUBGCP6 48 h after siRNA-mediated depletion of WBP11, PRPF8, or SLU7. (E) Quantification of centriole number in mitotic cells after siRNA-mediated knockdown of WBP11, PRPF8, or SLU7 for 48 h. FLAG-TUBGCP6 transgene was induced with doxycycline where indicated. n = 6, ≥50 cells per experiment. Error bars represent SD. (F) Graph showing the percentage of mitotic cells with three to four centrioles, as shown in Fig. 8 E. Unpaired parametric t test: *, P = 0.0429 (WBP11) and 0.0115 (SLU7). Error bars represent SD.
The high-confidence WBP11 proximity interactome contained four pre-mRNA splicing factors that have previously been shown to be required for centriole duplication: SNW1, PRPF8, SLU7, and SON (Balestra et al., 2013; Table S2). Given that WBP11 and SNW1 are required for TUBGCP6 pre-mRNA processing, we set out to test whether reduced levels of TUBGCP6 protein may accompany centriole duplication failure in cells depleted of SON, PRPF8, or SLU7. Importantly, depletion of PRPF8 or SLU7 by siRNA significantly reduced TUBGCP6 protein levels (Fig. 8 D). Knockdown of SON led to rapid cell death and was not further analyzed. Importantly, expression of an intronless TUBGCP6 transgene increased the fraction of cells with a normal centriole content 48 h after RNAi-mediated depletion of SLU7 or PRPF8, although for PRPF8 this increase did not reach statistical significance (Fig. 8, E and F). We conclude that TUBGCP6 expression level is sensitive to alterations in the levels of some RNA splicing factors, partly explaining the requirement of these proteins in centriole biogenesis (Balestra et al., 2013).
WBP11-dependent introns possess weak splice sites
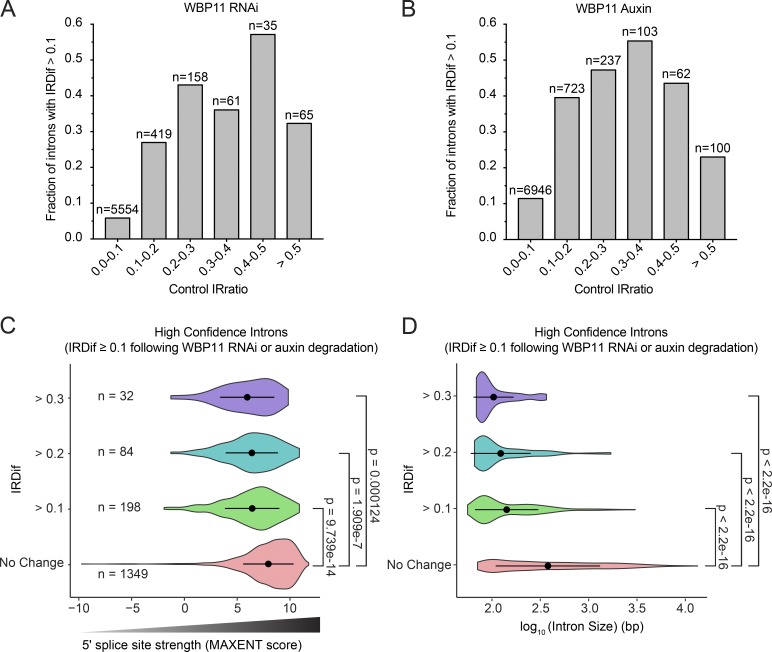
To elucidate the specificity of WBP11 regulation on mRNA splicing, we considered the possibility that WBP11 may be required for the splicing of suboptimal splice sites. We divided all introns from the WBP11 mRNA-seq experiment into six bins based on their IRratio in the control sample. For each of these six categories, we established the proportion of introns that had an IRDif score ≥0.1 and therefore required WBP11 for efficient splicing. If no bias was present, an increasing control IRratio would tend to reduce the IRDif score and, therefore, reduce the proportion of introns with an IRDif score ≥0.1. However, we observed that the probability an intron would be retained generally increased as the control IRratio increased from 0 to 0.5, and a similar trend was observed following auxin-induced degradation of WBP11 (Fig. 9, A and B). This suggests that introns that are poorly spliced in control cells are more likely to be significantly retained following WBP11 depletion.
Figure 9.
WBP11 is required for splicing short introns with weak 5′ splice sites. (A and B) Introns from the WBP11 RNAi RNA-seq experiment (A) or the WBP11 auxin RNA-seq experiment (B) were divided into six bins based on their IRratio in the control sample. For each bin, we calculated the fraction of introns that required WBP11 for efficient splicing (IRDif score ≥0.1). The probability an intron would be retained generally increased as the control IRratio increased from 0 to 0.5. Therefore, introns that are weakly spliced in the control sample are more likely to be retained following WBP11 depletion. (C) Violin plots of MAXENT splice site scores of introns that are significantly retained following WBP11 RNAi and degradation with auxin. Introns were divided into groups based on their IRDif scores. No change = IRDif ≥−0.1 and ≤0.1. Introns that are more highly retained (larger IRDif values) following depletion of WBP11 have weaker splice site scores (lower MAXENT values). Points represent the mean and bars display the SD. P values were calculated using an unpaired parametric t test. (D) Violin plots showing the length of the introns that are significantly retained following WBP11 RNAi and degradation with auxin. Introns were divided into groups based on their IRDif scores. No change = IRDif ≥−0.1 and ≤0.1. Introns that are more highly retained (larger IRDif values) following depletion of WBP11 are smaller in size. Points represent the mean and bars display the SD. P values were calculated using an unpaired parametric t test.
Previous work has shown that 5′ splice sites are critical elements in determining the efficiency of intron-splicing reactions (Roca et al., 2013; Wickramasinghe et al., 2015). To elucidate the specificity of WBP11-dependent RNA splicing, we examined the difference in 5′ splice site strength between a high-confidence list of introns that are retained after WBP11 depletion (IRDif ≥0.1 following WBP11 depletion) and those that are correctly spliced (IRDif ≥−0.1 and ≤0.1 following WBP11 depletion). 5′ splice site strength was estimated for each intron by calculating maximum entropy (MAXENT) scores that predict the likelihood a given splice site will be used (Yeo and Burge, 2004). Consistent with prior work suggesting that IR correlates with lower splice site strength, introns with IRDif values of ≥0.1 had weaker 5′ splice sites (Fig. 9 C; Sakabe and de Souza, 2007). WBP11-dependent introns were also significantly shorter in length (Fig. 9 D). 5′ splice site strength and intron size both decreased as the degree of IR increased (Fig. 9, C and D). Taken together, this suggests that short introns with weak 5′ splice sites show an increased dependence on WBP11 for efficient recognition by the splicing machinery.
Discussion
The vast majority of protein-coding transcripts in human cells must undergo splicing before they can serve as functional templates for protein synthesis. Splicing is therefore essential for all cellular processes, and global defects in spliceosome function are expected to lead to rapid cell-autonomous lethality. Surprisingly, however, a recent genome-wide RNAi screen identified 14 splicing factors that are required for centriole duplication in human cells. It is unclear why mutations in broadly expressed genes encoding spliceosome components result in the specific phenotype of centriole duplication failure.
In this article, we demonstrate a requirement for the pre-mRNA splicing factor WBP11 in centriole biogenesis. WBP11 was required to facilitate efficient splicing of 199 introns. These introns were present in 164 genes and were not enriched in genes of a particular biological pathway or function. Importantly, many of the pre-mRNAs that depend on WBP11 for efficient splicing also require SNW1, a pre-mRNA splicing factor that interacts with WBP11 and was previously shown to be important for centriole duplication. This suggests that SNW1 and WBP11 are required for splicing a subset of introns residing in the pre-mRNA of one or more genes required for centriole function.
Our experiments indicate that reduction in WBP11 or SNW1 leads to defects in centriole duplication in part because of loss of TUBGCP6 protein, thereby providing a molecular explanation for the role of these splicing factors in centriole biogenesis. Several lines of evidence support this conclusion. First, loss of TUBGCP6, WBP11, and SNW1 share phenotypic similarities, including the formation of monopolar spindles, a prolonged mitosis, and centriole duplication failure. Second, we detected aberrant processing of the TUBGCP6 pre-mRNA and severely reduced levels of TUBGCP6 protein in WBP11- and SNW1-depleted cells. Finally, expression of an intronless TUBGCP6 transgene partially rescued the centriole duplication failure phenotype in WBP11- or SNW1-depleted cells. Given that depletion of at least 13 additional splicing factors also leads to defects in centriole biogenesis (Balestra et al., 2013), it is tempting to speculate that compromised processing of the TUBGCP6 pre-mRNA may be a common defect contributing to centriole duplication failure in cells depleted of select splicing factors. Consistent with this hypothesis, depletion of SLU7 and PRPF8, two pre-mRNA splicing factors that are required for centriole duplication and are present in the WBP11 proximity interactome, also decreased the protein levels of TUBGCP6. In addition, the failure of centriole duplication in cells depleted of SLU7 and PRPF8 was partially rescued by expression of a cDNA encoding TUBGCP6.
Perhaps unsurprisingly, expression of an intronless TUBGCP6 cDNA was unable to fully rescue the phenotypes incurred by depletion of WBP11 or SNW1. This is not due to deficiencies in the functionality of the TUBGCP6 transgene, as this transgene could fully rescue loss of the endogenous TUBGCP6 protein. Instead, our data suggest that the defective splicing of other pre-mRNAs contributes to the effects observed following depletion of WBP11 or SNW1. One of the pre-mRNAs most affected by depletion of WBP11 and SNW1 encodes CPSF1, a component required for cleavage and polyadenylation at the 3′ end of pre-mRNAs (Murthy and Manley, 1995). Depletion of CPSF1 is expected to negatively impact the expression of many proteins, highlighting how defective splicing of a relatively small number of pre-mRNA targets can indirectly result in larger changes in gene expression. This may explain why the ability to rescue centriole duplication with the intronless TUBGCP6 transgene is reduced if cells lacking WBP11 or SNW1 are analyzed at later time points after siRNA transfection.
We show that reducing the levels of WBP11 preferentially alters the splicing of only a subset of short introns that contain weak 5′ splice sites. Weak and strong splice sites compete for binding to the spliceosome complex (Munding et al., 2013). It is possible that WBP11 functions to facilitate the binding of the spliceosome to weak 5′ splice sites. An alternative and non–mutually exclusive model is that depletion of WBP11 reduces the concentration/activity of the core spliceosome, leading to alterations in the kinetic equilibrium of spliceosome assembly at strong and weak splice sites, the ultimate consequence of which is an enhanced selection of strong splice sites. We note that some transcripts with retained introns may be rapidly degraded by the nonsense-mediated mRNA decay pathway and would not be detected in our analysis.
IR was traditionally considered to be the consequence of mRNA missplicing. However, recent work has shown IR to be a physiological mechanism of controlling gene expression in diverse processes such as neurogenesis (Braunschweig et al., 2014; Buckley et al., 2011) and hematopoiesis (Wong et al., 2013). In these cases, downregulation of splicing factors leads to the retention of specific introns and reduced expression of a subset of genes. A recent study showed that the temporal control of IR could also provide a mechanism for establishing the timing of gene expression during the cell cycle (Dominguez et al., 2016). One transcript identified to be alternatively spliced in this study was TUBGCP6, which shows preferential retention of an intron in a cell cycle–controlled manner. It is thus plausible that IR in TUBGCP6 mRNA is a regulated process that functions to control the activity of the γ-TuRC and tune the microtubule nucleating capacity of the cell.
The phenotypic link we have described between defects in the splicing machinery and centriole duplication may have clinical significance. The minor spliceosome is responsible for catalyzing the removal of atypical U12-type introns, and mutations in components of this complex have been linked to developmental disorders characterized by microcephaly and dwarfism (Farach et al., 2018; He et al., 2011; Merico et al., 2015). Given that mutations in centriole and centrosome proteins cause developmental disorders that include very similar phenotypes (Chavali et al., 2014), it is tempting to speculate that defects in centriole biogenesis may contribute to the disease pathology in syndromes caused by mutations in splicing factors. In the future, it will be interesting to further explore this relationship by examining centriole number in clinical samples of patients with splicing factor mutations.
Materials and methods
Cell culture and maintenance
All cells lines were grown at 37°C with 5% CO2 and maintained in DMEM (Corning) containing 10% fetal bovine essence (Seradigm), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM L-glutamine. Flp-In TRex–DLD-1 cells were used in all experiments, with the exception of Fig. S2, A and B, where RPE1 cells were used, and cotransfection experiments, where HEK293FT cells were used. Stable, isogeneic Flp-In TRex–DLD-1 cell lines expressing transgenes from a cytomegalovirus promoter were generated using XtremeGene HP and selected for transgene incorporation using Hygromycin (Corning) at a concentration of 400 µg/ml. Transgene expression was induced using 1 µg/ml of tetracycline (Sigma-Aldrich). Flp-In TRex–DLD-1 cells and RPE1 cells lines were authenticated using short tandem repeat genotyping.
For RNAi, cells were seeded at 105 cells per milliliter of medium. Duplexed siRNAs were introduced at a final concentration of 100 nM using RNAiMAX (Life Technologies). siRNA directed against STIL (5′-GCUCCAAACAGUUUCUGCUGGAAU-3′) was purchased from GE Healthcare, a SMARTpool set of four siRNAs directed against WBP11 was purchased from Dharmacon and siRNA #1 (5′-CGUGAAACCUUUGAACGUA-3′) was used for further experiments, and a SMARTpool set of four siRNAs against SNW1 was purchased from Dharmacon and siRNA #1 (5′-GGAAAUGCGUGCCCAAGUA-3′) was used for further experiments. Immediately after transfection, tetracycline was added to induce expression of WBP11-EYFP. 72 h after transfection, cells were harvested for Western blot or fixed for immunofluorescence analysis unless otherwise indicated.
Phosphatase screen
Cells were transfected with Dharmacon ON-TARGETplus SMARTpool siRNA Library–Human Phosphatase against a total of 254 genes following the protocol outlined above. Sequences of the siRNA duplexes used can be found in Table S1. 48 h after transfection, cells were fixed in MeOH at −20°C and incubated for 1 h with centrin and CEP192 antibodies. Cells were imaged using a DeltaVision microscope at 60× magnification, and centrioles were quantified in mitotic cells. Genes that resulted in significant centriole underduplication (greater than two times the SD than in control cells) were validated in DLD-1, HeLa, and HCT-116 cells at 48 h after transfection following the same transfection protocol. The top hit, WBP11, was further quantified at up to 72 h following transfection.
Lentiviral production and transduction
To create lentivirus, the lentiGuide-Puromycin (52963; Addgene) plasmid was cotransfected into HEK293FT cells with the lentiviral packaging plasmids psPAX2 and pMD2.G (12260 and 12259; Addgene). Briefly, 3 × 106 HEK293FT cells were seeded into a poly-L-lysine–coated 10-cm culture dish 24 h before transfection. For each 10-cm dish, 4.5 µg of lentiviral vector, 6 µg of psPAX2, and 1.5 µg of pMD2.G were diluted into 0.6 ml OptiMEM (Thermo Fisher Scientific). Separately, 35 µl of 1 µg/µl 25-kD polyethylenimine (Sigma-Aldrich) was diluted into 0.6 ml OptiMEM, mixed, and incubated at room temperature for 5 min. The DNA and polyethylenimine mixtures were then combined, mixed, and incubated at room temperature for 20 min. During this incubation, the culture medium was replaced with 9 ml prewarmed DMEM + 1% FBS (Sigma-Aldrich). The transfection mixture was added dropwise to the 10-cm dish. Viral particles were harvested 48 h after the medium change and filtered through a 0.45-µm polyvinylidene fluoride syringe filter. The filtered supernatant was either used directly to infect cells or was snap frozen and stored at −80°C. For transduction, 1 ml of virus was added to cells grown in 9 ml of DMEM containing 10 µg/ml polybrene (Sigma-Aldrich).
WBP11-mAID cell line production
A WBP11-mAID-EGFP transgene with mutations at the protospacer-adjacent motif site that render it immune to Cas9 cleavage was integrated at a stable locus in Flp-In TRex–DLD-1 cells that express osTIR1-9xMyc. Stable Flp-In cell lines expressing WBP11-mAID-EGFP were generated as described above. WBP11-mAID-EGFP cells were grown continuously in the presence of tetracycline, and Cas9 along with a single gRNA (sgRNA) targeting endogenous WBP11 (5′-ACCTGCCATACGAAGCATCA-3′) was introduced into the cells using lentiviral transduction. Cells were selected using puromycin (2 µg/ml), passaged into limiting dilution, and clones were screened via Western blot to confirm knockout of WBP11. Degradation of the AID transgene was induced with 500 µM auxin (Sigma-Aldrich). For centrinone controls, cells were incubated with 125 nM centrinone B (Tocris).
PQBP1-mAID and TUBGCP6-mAID cell line production
PQBP1 gene targeting was performed in DLD-1 cells using CRISPR/Cas9. An sgRNA targeting PQBP1 immediately upstream of its stop codon (5′-CCCGAACCAAGCAGCAGGAT-3′) was cloned into the px459 expression vector (48138; Addgene). A donor pUC19 vector was constructed with a mAID-EGFP-P2A-Neo cassette flanked on the 5′ end with a 568-bp homology arm and on the 3′ end with a 458-bp homology arm. DLD-1 cells were transfected with the px459 and pUC19 donor vector and selected in G418 at 600 µg/ml for 10 d. Expression of PQBP1-mAID-EGFP was confirmed by immunofluorescence, and homozygous tagging was confirmed by immunoblot.
TUBGCP6 gene targeting was performed in DLD-1 cells using CRISPR/Cas9. An sgRNA targeting TUBGCP6 immediately upstream of its stop codon (5′-ACAACTACTACCAGGACGCC-3′) was cloned into the px459 expression vector (48138; Addgene). A donor pUC19 vector was constructed with a mAID-EGFP-P2A-Neo cassette flanked on the 5′ end with a 350-bp homology arm and on the 3′ end with a 520-bp homology arm. DLD-1 cells were transfected with the px459 and pUC19 donor vector and selected in G418 at 600 µg/ml for 10 d. Expression of TUBGCP6-mAID-EGFP was confirmed by immunofluorescence and homozygous tagging was confirmed by immunoblot.
BioID and mass spectrometry
Stable Flp-In cell lines expressing FLAG-BirA-WBP11 were generated as described above. Expression of FLAG-BirA-WBP11 was induced using 1 µg/ml of tetracycline (Sigma-Aldrich) when the cells were ∼40% confluent. After 24 h of tetracycline treatment, cells were given fresh medium with 50 µM biotin for 14–18 h. Cells were lysed in buffer containing 50 mM Tris, pH 7.4, 500 mM NaCl, 0.4% SDS, 5 mM EDTA, 1 mM DTT, 2% Triton X-100, 1 mM PMSF, and one Roche Protease Inhibitor tablet. Cells were sonicated twice on ice with 30 pulses at 30% duty cycle. An equal volume of 7.4 mM Tris, pH 7.4, was added, and cells were sonicated again on ice with 30 pulses at 30% duty cycle. Cells were centrifuged, and the remaining supernatant was passed over streptavidin agarose beads and rotated at 4°C overnight. Biotinylated proteins were eluted from streptavidin agarose by resuspension in a solution of 2% SDS, 10% glycerol, and 50 mM Tris, pH 8.1, containing 4 mM biotin with heating at 90°C for 20 min. The protein eluates were precipitated using trichloroacetic acid, washed, dried, digested with trypsin to peptides, desalted, and analyzed by high-performance mass spectrometry exactly as described previously (Lyons et al., 2018). Peptide tandem mass spectra were data-searched against a UniProt (https://www.uniprot.org) human proteome database using the Comet search algorithm, filtered to a 1% FDR using the target-decoy strategy, and used to infer protein identifications exactly as described previously (Kettenbach et al., 2018).
Flow cytometry
Cells were incubated for 5 d in the following conditions: tetracycline (1 µg/ml), auxin (500 µM), or tetracycline (1 µg/ml) and centrinone (250 nM). Cells were then prepared for flow cytometry by trypsinizing, centrifuging for 5 min at 400 g−1, and resuspending cell pellets in 1× PBS. Cell suspensions were analyzed on a Guava EasyCyte 6-2L Benchtop Flow Cytometer. The ratio of mCardinal-negative cells to mCardinal-positive cells was calculated for each coculture condition. Statistical analysis was determined using an unpaired t test.
Antibody usage and production
For Western blot analysis, proteins were separated via SDS-PAGE, transferred onto nitrocellulose membrane with a Trans-Blot Turbo Transfer System (Bio-Rad), and incubated with the following antibodies diluted into 5% milk: YL1/2 (rat anti–α-tubulin, 1:1,000; Pierce Antibodies), rabbit anti-WBP11 (1:1,000; Thermo Fisher Scientific), mouse anti-SNW1 (1:500; Santa Cruz), rabbit anti-STIL (Holland Lab, described in Moyer et al., 2015), mouse anti-PP1 (E9, 1:200; Santa Cruz), rabbit anti-TUBGCP6 (1:1,000; a gift from J. Luders, Institute for Research in Biomedicine Barcelona, Barcelona, Spain), rabbit anti-SREBF1 (1:1,000; Protein Tech), rabbit anti-RECQL4 (1:1,000; Protein Tech), mouse anti–Vinculin H-10 (1:500; Santa Cruz), and mouse anti-NEDD1 (1:500; Santa Cruz).
For immunofluorescence, 18-mm coverslips were fixed in −20°C 100% methanol for 10 min and blocked at room temperature for 1 h. The block solution consists of 2.5% FBS, 200 mM glycine, and 0.1% Triton X-100 in PBS. Coverslips were incubated with the following antibodies diluted in block solution for 1 h at room temperature in the dark: mouse anti-centrin (EMD Millipore, 1:1,000), centrin–Alexa Fluor 555 (directly labeled rabbit, 1:1,000), CEP192-DyL488 (directly labeled goat, raised against CEP192 aa 1–211, 1:1,000), and γ-tubulin–Alexa Fluor 647 (directly labeled goat, raised against γ-tubulin aa 432–451, 1:1,000). Coverslips were washed in triplicate with PBS containing 0.1% Triton-X (PBST) and then, if required, incubated in secondary antibody diluted in block solution for 1 h at room temperature (Invitrogen). Coverslips were washed in triplicate with PBST then incubated in DAPI diluted in PBST for 3 min and mounted onto slides using ProLong Gold Antifade (Invitrogen).
mRNA-seq data collection and analysis
mRNA was purified from cells using Qiagen RNeasy kit with DNase treatment. Purified mRNA was sent to the Genetic Resources Core Facility at Johns Hopkins School of Medicine, and sequencing was done using paired-end Illumina NextSeq with a read length of 75 bp and 60–80 million reads per sample. Reads were aligned and mapped to hg38 using RSEM. The RNA-seq data from this publication was submitted to the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and assigned the accession no. GSE129231.
Differential expression analysis for both genes and isoforms was performed using EBSeq (Leng et al., 2013). For differential gene analysis, hits were considered to be any genes with an expression fold change in the experimental condition that is more than two SDs away from the average expression fold change observed comparing the experimental condition to the control condition. For differential isoform analysis, any genes with only one expressed or detected isoform were excluded from our analysis. We excluded any isoforms for which the FPKM (fragments per kilobase of transcript per million mapped reads) in both conditions (control versus experimental) was <10% of the average FPKM for any given isoform to account for noise. All reads for every detected isoform of a gene were added together to get the total reads. For genes in which there were no reads in one of the two conditions, the gene was excluded from this differential isoform analysis since the gene as a whole is differentially expressed. The proportion of reads for a given gene represented by each isoform was then calculated for both experimental and control conditions. The control condition proportion was subtracted from the experimental condition proportion to get proportion change. Hits were any isoforms where the proportion change was more than two SDs away from the average proportion change.
IRFinder was used to calculate an IRratio that scores the frequency with which an intron is retained in a given mRNA (Middleton et al., 2017). IRratios for introns that failed to meet the IRFinder quality control filter were removed from the analysis. An IRDif score was calculated for each intron by subtracting the IRratio for each intron in the control cells from the IRratio of the same intron in the experimental population. A positive IRDif score corresponds to an intron that was retained more frequently, and a negative IRDif score corresponds to an intron that was spliced more efficiently. Introns with IRDif scores of ≥0.1 or ≤−0.1 were considered hits in our analysis.
To calculate the change in the fraction of spliced mRNA for each gene (ΔSplicing) we used the following formula:
To validate retained introns, RNA was reverse transcribed using SuperScript IV kit (Invitrogen), and the cDNA was PCR-amplified using the following primers: TUBGCP6, 5′-GCTGTCGACTACTTCTTCGTGG-3′ and 5′-GCAGGTACTTGAGAGCGAGG-3′; RNF123, 5′-GATGTCTCTGCTTCGGCTGT-3′ and 5′-GCTAAGCAGAAACTTGCGGG-3′; and RECQL4, 5′-GGGCACTCCCAATACAGCTT-3′ and 5′-GCTCCAGGTAGCACAGCAAA-3′.
MAXENT splice site scores
5′ splice site strength was predicted using MAXENT scores (Yeo and Burge, 2004). The MAXENT framework was generated using large datasets of known human splice sites and takes into account adjacent and nonadjacent dependences. For 5′ splice sites, three exonic base pairs and six intronic base pairs flanking the 5′ splice site are used, as those positions are generally well conserved in the context of splicing. The splice site model assigns a MAXENT score from −20 to +20, with a higher score meaning a higher probability the sequence is a true splice site.
Online supplemental material
Fig. S1 shows the design of the siRNA screen and the hits obtained from the screen. Fig. S2 shows data supporting WBP11’s role in cell proliferation. Fig. S3 shows that PQBP1 is not required for centriole biogenesis. Fig. S4 contains analysis of the RNA-seq data following WBP11 or SNW1 depletion and some follow-up data from this analysis. Fig. S5 contains analysis of some retained introns identified by RNA-seq analysis and follow-up of these hits, particularly of TUBGCP6. Table S1 contains details of the siRNA screen and correlates to Fig. S1. Table S2 contains details of the mass spectrometry analysis and correlates to Fig. 3. Table S3 contains differential gene expression analysis. Table S4 contains differential isoform expression analysis and correlates to Fig. 4, B and C, and Fig. S4, A and B. Table S5 contains IR analysis and correlates to Fig. 4, D and E, and Fig. S4 D. Table S6 contains transcript-wide splicing analysis and correlates to Fig. 4, F and G, and Fig. S4 E. Video 1 shows an untreated WBP11AID cell going through mitosis with H2B-iRFP expressed. Video 2 shows an auxin-treated WBP11AID cell going through mitosis with H2B-iRFP expressed. Video 3 shows an untreated WBP11AID cell going through mitosis with both H2B-iRFP (red) and RFP-tubulin (green) expressed. Video 4 shows an auxin-treated WBP11AID cell going through mitosis with both H2B-iRFP (red) and RFP-tubulin (green) expressed. Video 5 shows a control TUBGCP6AID cell going through mitosis with both H2B-iRFP (red) and RFP-tubulin (green) expressed. Video 6 shows an auxin-treated TUBGCP6AID cell going through mitosis with both H2B-iRFP (red) and RFP-tubulin (green) expressed. Data file 1 contains the source data for the figures. Data file 2 contains information about the statistical tests that were run and their results.
Supplementary Material
Acknowledgments
We are extremely grateful to Jeremy Nathans, Antony Rosen, and the Johns Hopkins Institute for Basic Biomedical Sciences for providing research support.
This work was supported by the National Institutes of Health (R01GM114119 and R01GM133897) and an American Cancer Society Scholar Grant (RSG-16-156-01-CCG) to A.J. Holland and a National Institutes of Health grant (R01 GM122846) to S.A. Gerber. E.M. Park was funded by a National Institutes of Health training grant (T32GM007445) and a National Science Foundation Graduate Research Fellowship.
The authors declare no competing financial interests.
Author contributions: E.M. Park, P.M. Scott, and K. Clutario performed the majority of the experiments. K.B. Cassidy and S.A. Gerber performed the BioID experiments. K. Zhan assisted with data collection and analysis. E.M. Park, P.M. Scott, K.B. Cassidy, S.A. Gerber, and A.J. Holland designed experiments and analyzed the data. E.M. Park and A.J. Holland wrote the manuscript, and all authors edited it.
References
- Ahn E.Y., DeKelver R.C., Lo M.C., Nguyen T.A., Matsuura S., Boyapati A., Pandit S., Fu X.D., and Zhang D.E.. 2011. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol. Cell. 42:185–198. 10.1016/j.molcel.2011.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Cruzeiro J.M., Nogales-Cadenas R., and Pascual-Montano A.D.. 2014. CentrosomeDB: a new generation of the centrosomal proteins database for Human and Drosophila melanogaster. Nucleic Acids Res. 42(Database issue):D430–D436. 10.1093/nar/gkt1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C., and Nigg E.A.. 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 24:351–360. 10.1016/j.cub.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Arquint C., Gabryjonczyk A.M., Imseng S., Böhm R., Sauer E., Hiller S., Nigg E.A., and Maier T.. 2015. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife. 4:e07888 10.7554/eLife.07888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C., Sonnen K.F., Stierhof Y.D., and Nigg E.A.. 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125:1342–1352. 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- Bahtz R., Seidler J., Arnold M., Haselmann-Weiss U., Antony C., Lehmann W.D., and Hoffmann I.. 2012. GCP6 is a substrate of Plk4 and required for centriole duplication. J. Cell Sci. 125:486–496. 10.1242/jcs.093930 [DOI] [PubMed] [Google Scholar]
- Balestra F.R., Strnad P., Flückiger I., and Gönczy P.. 2013. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell. 25:555–571. 10.1016/j.devcel.2013.05.016 [DOI] [PubMed] [Google Scholar]
- Bao J., Yu Y., Chen J., He Y., Chen X., Ren Z., Xue C., Liu L., Hu Q., Li J., et al. 2018. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 9:1045 10.1038/s41419-018-1020-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., and Glover D.M.. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Braunschweig U., Barbosa-Morais N.L., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., and Blencowe B.J.. 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24:1774–1786. 10.1101/gr.177790.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley P.T., Lee M.T., Sul J.Y., Miyashiro K.Y., Bell T.J., Fisher S.A., Kim J., and Eberwine J.. 2011. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 69:877–884. 10.1016/j.neuron.2011.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Shen Y., Zhu L., Xu Y., Zhou Y., Wu Z., Li Y., Yan X., and Zhu X.. 2012. miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat. Cell Biol. 14:697–706. 10.1038/ncb2512 [DOI] [PubMed] [Google Scholar]
- Chavali P.L., Pütz M., and Gergely F.. 2014. Small organelle, big responsibility: the role of centrosomes in development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130468 10.1098/rstb.2013.0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho P.A., Bury L., Shahbazi M.N., Liakath-Ali K., Tate P.H., Wormald S., Hindley C.J., Huch M., Archer J., Skarnes W.C., et al. 2015. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 5:150209 10.1098/rsob.150209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craggs G., Finan P.M., Lawson D., Wingfield J., Perera T., Gadher S., Totty N.F., and Kellie S.. 2001. A nuclear SH3 domain-binding protein that colocalizes with mRNA splicing factors and intermediate filament-containing perinuclear networks. J. Biol. Chem. 276:30552–30560. 10.1074/jbc.M103142200 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Bento I., Pimenta-Marques A., Jana S.C., Lince-Faria M., Duarte P., Borrego-Pinto J., Gilberto S., Amado T., Brito D., et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23:2245–2254. 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- Dahl K.D., Sankaran D.G., Bayless B.A., Pinter M.E., Galati D.F., Heasley L.R., Giddings T.H. Jr., and Pearson C.G.. 2015. A Short CEP135 Splice Isoform Controls Centriole Duplication. Curr. Biol. 25:2591–2596. 10.1016/j.cub.2015.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D., Tsai Y.H., Weatheritt R., Wang Y., Blencowe B.J., and Wang Z.. 2016. An extensive program of periodic alternative splicing linked to cell cycle progression. eLife. 5:e10288 10.7554/eLife.10288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., and Glover D.M.. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24:2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Abdelaziz M., Debski J., Dadlez M., and Glover D.M.. 2017. Two-step phosphorylation of Ana2 by Plk4 is required for the sequential loading of Ana2 and Sas6 to initiate procentriole formation. Open Biol. 7:170247 10.1098/rsob.170247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farach L.S., Little M.E., Duker A.L., Logan C.V., Jackson A., Hecht J.T., and Bober M.. 2018. The expanding phenotype of RNU4ATAC pathogenic variants to Lowry Wood syndrome. Am. J. Med. Genet. A. 176:465–469. 10.1002/ajmg.a.38581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farache D., Emorine L., Haren L., and Merdes A.. 2018. Assembly and regulation of γ-tubulin complexes. Open Biol. 8:170266 10.1098/rsob.170266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fırat-Karalar E.N., and Stearns T.. 2014. The centriole duplication cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130460 10.1098/rstb.2013.0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., and Guthrie C.. 1992. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 6:2112–2124. 10.1101/gad.6.11.2112 [DOI] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Grainger R.J., and Beggs J.D.. 2005. Prp8 protein: at the heart of the spliceosome. RNA. 11:533–557. 10.1261/rna.2220705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G., Westendorf J., Uldschmid A., and Nigg E.A.. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 123:2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., and Nigg E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- He H., Liyanarachchi S., Akagi K., Nagy R., Li J., Dietrich R.C., Li W., Sebastian N., Wen B., Xin B., et al. 2011. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 332:238–240. 10.1126/science.1200587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Fachinetti D., Han J.S., and Cleveland D.W.. 2012. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 109:E3350–E3357. 10.1073/pnas.1216880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., and Cleveland D.W.. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach A.N., Schlosser K.A., Lyons S.P., Nasa I., Gui J., Adamo M.E., and Gerber S.A.. 2018. Global assessment of its network dynamics reveals that the kinase Plk1 inhibits the phosphatase PP6 to promote Aurora A activity. Sci. Signal. 11:eaaq1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Buster D.W., Nguyen A.L., Swatkoski S., Gucek M., Rusan N.M., and Rogers G.C.. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 23:2255–2261. 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A., Saeki M., and Kato S.. 1999. Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J. Biol. Chem. 274:36513–36519. 10.1074/jbc.274.51.36513 [DOI] [PubMed] [Google Scholar]
- Kratz A.S., Bärenz F., Richter K.T., and Hoffmann I.. 2015. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open. 4:370–377. 10.1242/bio.201411023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leda M., Holland A.J., and Goryachev A.B.. 2018. Autoamplification and Competition Drive Symmetry Breaking: Initiation of Centriole Duplication by the PLK4-STIL Network. iScience. 8:222–235. 10.1016/j.isci.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng N., Dawson J.A., Thomson J.A., Ruotti V., Rissman A.I., Smits B.M., Haag J.D., Gould M.N., Stewart R.M., and Kendziorski C.. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 29:1035–1043. 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M.S., Bakker B., Boeckx B., Moyett J., Lu J., Vitre B., Spierings D.C., Lansdorp P.M., Cleveland D.W., Lambrechts D., et al. 2017. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev. Cell. 40:313–322.e5. 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M., Beullens M., Andrés I., Ortiz J.M., and Bollen M.. 2004. SIPP1, a novel pre-mRNA splicing factor and interactor of protein phosphatase-1. Biochem. J. 378:229–238. 10.1042/bj20030950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M., Beullens M., Lesage B., Nicolaescu E., Beke L., Landuyt W., Ortiz J.M., and Bollen M.. 2005. Nucleocytoplasmic shuttling of the splicing factor SIPP1. J. Biol. Chem. 280:38862–38869. 10.1074/jbc.M509185200 [DOI] [PubMed] [Google Scholar]
- Lopes C.A., Jana S.C., Cunha-Ferreira I., Zitouni S., Bento I., Duarte P., Gilberto S., Freixo F., Guerrero A., Francia M., et al. 2015. PLK4 trans-Autoactivation Controls Centriole Biogenesis in Space. Dev. Cell. 35:222–235. 10.1016/j.devcel.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Lyons S.P., Jenkins N.P., Nasa I., Choy M.S., Adamo M.E., Page R., Peti W., Moorhead G.B., and Kettenbach A.N.. 2018. A quantitative chemical proteomic strategy for profiling phosphoprotein phosphatases from yeast to humans. Mol. Cell. Proteomics. 17:2448–2461. 10.1074/mcp.RA118.000822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D., Roifman M., Braunschweig U., Yuen R.K., Alexandrova R., Bates A., Reid B., Nalpathamkalam T., Wang Z., Thiruvahindrapuram B., et al. 2015. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat. Commun. 6:8718 10.1038/ncomms9718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R., Gao D., Thomas A., Singh B., Au A., Wong J.J., Bomane A., Cosson B., Eyras E., Rasko J.E., and Ritchie W.. 2017. IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol. 18:51 10.1186/s13059-017-1184-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer T.C., and Holland A.J.. 2019. PLK4 promotes centriole duplication by phosphorylating STIL to link the procentriole cartwheel to the microtubule wall. eLife. 8:e46054 10.7554/eLife.46054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munding E.M., Shiue L., Katzman S., Donohue J.P., and Ares M. Jr. 2013. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell. 51:338–348. 10.1016/j.molcel.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K.G., and Manley J.L.. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9:2672–2683. 10.1101/gad.9.21.2672 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Holland A.J.. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19:297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellacani C., Bucciarelli E., Renda F., Hayward D., Palena A., Chen J., Bonaccorsi S., Wakefield J.G., Gatti M., and Somma M.P.. 2018. Splicing factors Sf3A2 and Prp31 have direct roles in mitotic chromosome segregation. eLife. 7:e40325 10.7554/eLife.40325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X., Krainer A.R., and Eperon I.C.. 2013. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev. 27:129–144. 10.1101/gad.209759.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., and Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196:801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe N.J., and de Souza S.J.. 2007. Sequence features responsible for intron retention in human. BMC Genomics. 8:59 10.1186/1471-2164-8-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi Sankaran D., Stemm-Wolf A.J., and Pearson C.G.. 2019. CEP135 isoform dysregulation promotes centrosome amplification in breast cancer cells. Mol. Biol. Cell. 30:1230–1244. 10.1091/mbc.E18-10-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serçin Ö., Larsimont J.C., Karambelas A.E., Marthiens V., Moers V., Boeckx B., Le Mercier M., Lambrechts D., Basto R., and Blanpain C.. 2016. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat. Cell Biol. 18:100–110. 10.1038/ncb3270 [DOI] [PubMed] [Google Scholar]
- Song R., Walentek P., Sponer N., Klimke A., Lee J.S., Dixon G., Harland R., Wan Y., Lishko P., Lize M., et al. 2014. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 510:115–120. 10.1038/nature13413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., and Gönczy P.. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213. 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy S., Vázquez-Novelle M.D., Lekomtsev S., Howell M., and Petronczki M.. 2014. Functional genomics identifies a requirement of pre-mRNA splicing factors for sister chromatid cohesion. EMBO J. 33:2623–2642. 10.15252/embj.201488244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.J.C., Lin S.Y., Hsu W.B., Lin Y.N., Wu C.T., Lin Y.C., Chang C.W., Wu K.S., and Tang T.K.. 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30:4790–4804. 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelij P., Stocsits R.R., Ladurner R., Petzold G., Kreidl E., Koch B., Schmitz J., Neumann B., Ellenberg J., and Peters J.M.. 2014. SNW1 enables sister chromatid cohesion by mediating the splicing of sororin and APC2 pre-mRNAs. EMBO J. 33:2643–2658. 10.15252/embj.201488202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., and Krämer A.. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362. 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wickramasinghe V.O., Gonzàlez-Porta M., Perera D., Bartolozzi A.R., Sibley C.R., Hallegger M., Ule J., Marioni J.C., and Venkitaraman A.R.. 2015. Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5′ splice site strength. Genome Biol. 16:201 10.1186/s13059-015-0749-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K., et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 154:583–595. 10.1016/j.cell.2013.06.052 [DOI] [PubMed] [Google Scholar]
- Yeo G., and Burge C.B.. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11:377–394. 10.1089/1066527041410418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.