Bajaj et al. review how cancers originate, how their heterogeneity is linked to cancer stem cells, and the signals fundamental to driving these processes.
Abstract
While standard therapies can lead to an initial remission of aggressive cancers, they are often only a transient solution. The resistance and relapse that follows is driven by tumor heterogeneity and therapy-resistant populations that can reinitiate growth and promote disease progression. There is thus a significant need to understand the cell types and signaling pathways that not only contribute to cancer initiation, but also those that confer resistance and drive recurrence. Here, we discuss work showing that stem cells and progenitors may preferentially serve as a cell of origin for cancers, and that cancer stem cells can be key in driving the continued growth and functional heterogeneity of established cancers. We also describe emerging evidence for the role of developmental signals in cancer initiation, propagation, and therapy resistance and discuss how targeting these pathways may be of therapeutic value.
Introduction
Cancers arise from a series of mutations or genomic alterations that provide a cell with an extensive ability to evade pro-apoptotic and growth-inhibitory signals and to be self-sufficient in growth signals that enable them to divide endlessly (Nowell, 1974). Other genetic alterations in these cells aid angiogenesis, tissue invasion, and metastasis (Fearon and Vogelstein, 1990; Hanahan and Weinberg, 2000, 2011). The rarity of cancers and the time needed for them to develop reflect the low probability of any one cell acquiring the correct set and sequence of mutations. In addition, cancer-initiating mutations are likely to arise in primitive tissue stem cells as these naturally self-renew and persist long-term, allowing accumulation of the necessary mutations. Alternatively, transforming events could occur in early progenitors if the mutations confer these cells with self-renewal capacity (Tan et al., 2006). Consistent with this, several groups have experimentally verified that both the resident tissue stem cells and progenitors can serve as cells of origin in hematological cancers as well as in solid tumors.
After establishment and initiation, how a tumor continues to propagate itself is a key question with implications for therapy. The conventional view of tumor propagation has been that most cancer cells have the capability to proliferate extensively and form new tumor cells. This model, however, could not explain why large numbers of cancer cells were needed to initiate cancer in vivo (Bruce and Van Der Gaag, 1963) and the low frequency of colonies seen when cancer cells were plated in vitro. The fact that tumors are heterogeneous, and have a limited subset of cells with the potential to drive cancer growth, was first demonstrated in acute myeloid leukemia (AML; Bonnet and Dick, 1997; Lapidot et al., 1994). The identification of malignant stem cells in leukemia initiated a search for similar populations in solid tumors, and about a decade later, a small population of cells with tumor-initiating properties were identified in mammary cancers (Al-Hajj et al., 2003) and in brain cancers that preferentially gave rise to tumors in immunodeficient mice (Singh et al., 2003, 2004).
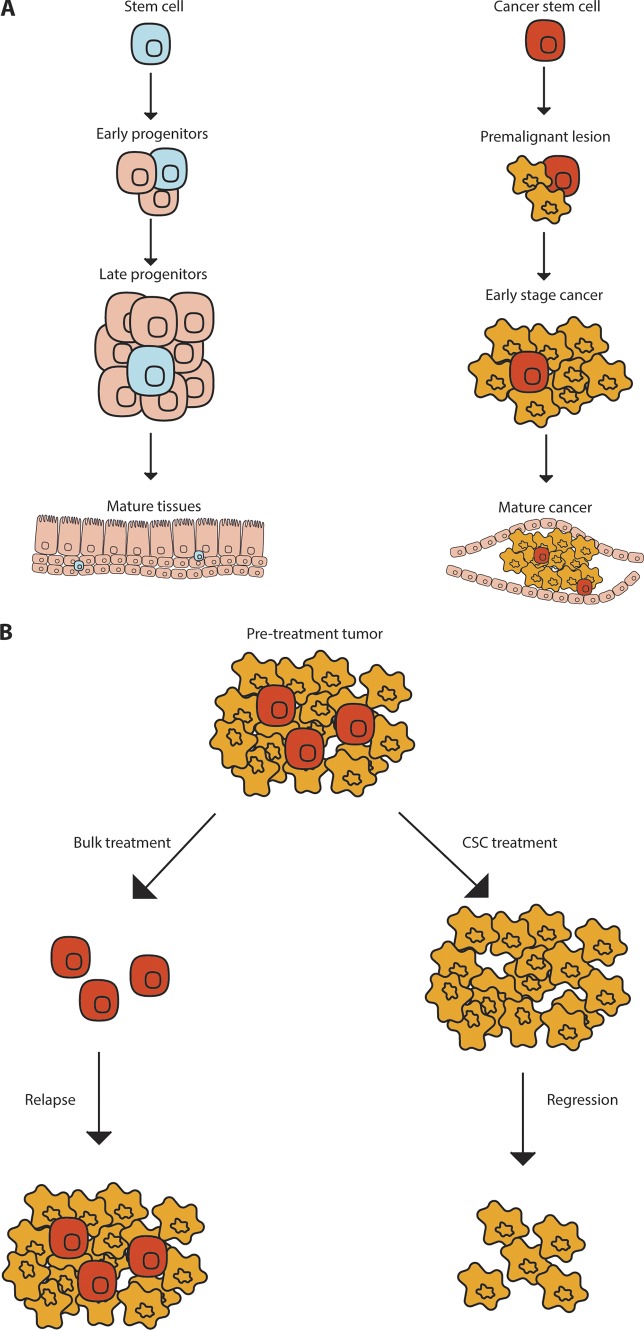
Similar to stem cells, cancer stem cells (CSCs) have been thought of as cells at the top of a hierarchy of more differentiated cell populations (Fig. 1 A). CSCs have also emerged as being particularly drug resistant (Fig. 1 B; Adhikari et al., 2010; Dick, 2008; Hambardzumyan et al., 2006; Liu et al., 2006a; Lytle et al., 2018; Reya et al., 2001), another property enriched in stem cells. Beyond the structural similarities between normal stem cells and CSCs in terms of hierarchical organization, another shared hallmark is the utilization of developmental signaling pathways both during initiation and propagation. Shared gene expression patterns of leukemia (Gentles et al., 2010) and brain tumor stem cells with their normal counterparts (Yan et al., 2011) suggests that they use and depend on developmental and stem cell programs. Since cancers co-opt normal stem cell signals to promote malignant growth, there is increased interest in targeting these pathways to control disease progression. In this review, we discuss the origin of cancer, highlight the functional characterization of cancer initiating cells/CSCs in established tumors, and describe strategies targeting intrinsic stem cell signals, as well as supportive signals from the niche, in an effort to improve therapeutic outcomes.
Figure 1.
Normal and CSC hierarchy. Normal stem cells and CSCs can self-renew and differentiate into more mature cells. (A) Normal stem cells generate the progenitors and mature tissues of the body while CSCs generate more cancer cells. (B) Tumors treated with chemotherapy can leave residual chemoresistant CSCs that can regrow a tumor, whereas CSC targeted therapy may lead to tumor regression and eventual resolution.
Cell of origin
Hematological malignancies
Chronic myeloid leukemia (CML) is the classic example of a disease where the initiating oncogenic event (the BCR-ABL translocation) occurs in the hematopoietic stem cell (HSC). This concept was proposed in the early 1950s because patients were found to have BCR-ABL mutations in multiple hematopoietic lineages (Dameshek, 1951; Tough et al., 1963; Whang et al., 1963). Later experiments confirmed that only BCR-ABL–expressing HSCs and not progenitor cell populations are capable of recapitulating the characteristics of disease in immunodeficient mice (Huntly et al., 2004; Neering et al., 2007). Work using mouse models has indicated that BCR-ABL translocations in the HSCs give rise to CML, while those occurring in more committed progenitors lead to the induction of acute lymphoid leukemia (ALL; Li et al., 1999; Zhao et al., 2007), indicating that the differentiation state of the cell of origin can have a significant impact on disease trajectory and signaling dependencies, independent of the driver mutation.
Like CML, the AML founding clone is generally an HSC or an early progenitor (Bonnet and Dick, 1997), and paired diagnosis and relapse sequencing show that following therapy, tumor cells with either acquired or preexisting mutations that confer resistance survive selectively (Ding et al., 2012). Work in mouse models of MLL-AF9–driven AML indicate that while both the granulocyte macrophage progenitor and the HSC serve as cells of origin, the HSC-derived cancer more closely resembles the human disease (Krivtsov et al., 2006, 2013). Consistent with this observation, HSCs have also been shown to be the cell of origin in human AML in a study where a large cohort of primary human AML patients were sequenced for commonly occurring mutations (Shlush et al., 2014). This identified point mutations in the DNA methyltransferase DNMT3A in ∼25% of the samples, of which ∼90% also had a mutation in the NPM1 gene. While the DNMT3A mutation could also be detected in T cells of some patients, NPM1 could not, indicating that DNMT3A changes probably occur in an ancestral cell like the HSC that can give rise to both myeloid and lymphoid lineages. The fact that only the DNMT3A mutation could be detected in the normal HSCs of patients who had received treatment indicated that these were most likely to be the AML cells of origin (Shlush et al., 2014). It is of interest to note that DNMT3A (Lin et al., 2018; Nangalia et al., 2015) and NPM1 (Bains et al., 2011) mutations alone do not generally give rise to leukemia and are associated with more benign diseases such as myeloproliferative neoplasms and myelodysplastic syndrome. Thus, these data indicate that sequential acquisition of mutations leads to the progression of myeloproliferative neoplasms and/or myelodysplastic syndrome to more aggressive diseases such as AML, as has been shown in the context of colon cancers, where mutations in Apc precede those in the Ras allele (Fearon and Vogelstein, 1990; Vogelstein et al., 1988).
Solid cancers
Consistent with the leukemia paradigm, stem/progenitor populations can often serve as the primary cells of origin in solid cancers. For instance, mutations in Wnt signaling, such as loss of APC, can initiate colon cancer, which then progresses with additional mutations such as activation of Ras and phosphoinositide 3-kinase signaling. The low frequency of these mutations and the time taken for disease progression suggest that the initiating mutation is likely to be in the stem cells (Tomasetti et al., 2017; Vogelstein et al., 1988). Experimental verification of this hypothesis has come from mouse models, where APC loss in the intestinal stem cell compartments results in rapid induction of colon adenomas, indicating that these cells can be the cells of origin of cancer (Barker et al., 2009; Powell et al., 2012). Single tumor gland and single-cell sequencing experiments in human colon cancer have since confirmed that most tumors arise from a single clone, which acquires mutations early during cancer initiation (Dalerba et al., 2011; Sottoriva et al., 2015).
Like colon adenocarcinomas, studies with mouse models have implicated stem cells as cells of origin for squamous cell carcinomas (SCC). Thus, mutations in Kras and p53 (Raimondi et al., 2009), or loss of Pten and Tgfb signaling (Bian et al., 2012), can drive head and neck SCC only from undifferentiated basal cells of the epithelium. Similarly, overexpression of Stat3 and Sox2 oncogenic signals in undifferentiated basal cells results in esophageal SCC formation, while their expression in more differentiated lineages has no impact (Liu et al., 2013). Consistent with this, overexpression of Kras along with loss of p53 can promote cutaneous SCC formation only if the mutations occur in either the inter-follicular epithelium or the bulge stem cells (Lapouge et al., 2011; White et al., 2011). Since the presence of these mutations in epithelial progenitors or transit-amplifying cells has no impact on SCC initiation, these studies collectively indicate that the undifferentiated basal stem cells are the cells of origin for several types of SCCs.
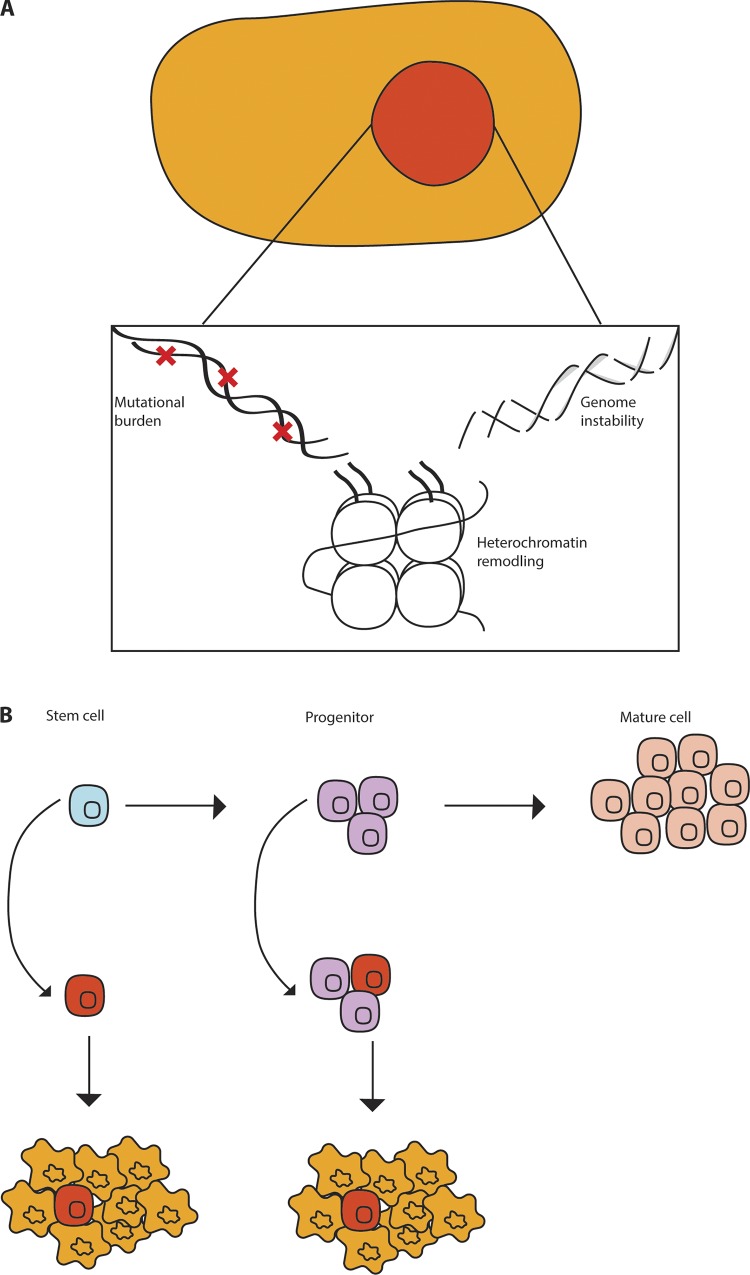
While mutations in either stem or progenitor cells can give rise to tumors, the resulting tumors can differ. For instance, in mouse models of breast cancer, loss of BRCA1 and p53 in luminal progenitors results in adenocarcinomas resembling human disease, while the same mutations in basal stem cells gives rise to malignant adenomyoepitheliomas, a tumor type rarely seen in BRCA patients (Molyneux et al., 2010). In other systems, mutations in either progenitors or stem cells result in distinct tumor types; thus, loss of p53, Nf1, and Pten in neural stem cells, neural progenitors, or oligodendrocyte progenitors gives rise to distinct subtypes of glioblastoma (Alcantara Llaguno et al., 2009; Alcantara Llaguno et al., 2015). These data collectively indicate that the cell of origin has a significant impact on disease type. Interestingly, in some instances, the driver mutations override the cellular context to initiate similar tumor types; thus, activation of hedgehog signaling in either the neuronal stem or precursor cells results in formation of molecularly similar aggressive medulloblastomas (Yang et al., 2008). In addition to stem cells and progenitors, in certain contexts, such as breast cancer and melanoma cell lines, more differentiated cells appear to be able acquire features of stem cells either spontaneously (Chaffer et al., 2011), or upon expression of epigenetic regulators (Roesch et al., 2010). This may indicate that differentiated cells harbor latent potential to acquire properties of stem cells in specific contexts. Collectively, the studies using mouse models of disease or primary human patient samples indicate that while tumors generally arise from normal tissue stem/progenitor cells, the cell of origin as well as the driver mutations can be critical for determining tumor identity (Fig. 2).
Figure 2.
The origin of cancers. (A) Cancer can be driven by a variety of changes such as genetic mutations, genome instability, and heterochromatin changes. (B) These changes often arise in populations such as stem cells or more mature progenitor cells.
Cancer stem cells
The initial identification and characterization of CSCs relied on prior knowledge of normal tissue stem cells since the markers used to isolate the CSCs were often similar to their normal counterparts, e.g., CD34 in leukemic stem cells (LSC; Gentles et al., 2010) and CD133 in brain tumors (Yan et al., 2011). However, it is important to note that surface markers can be divergent between normal and malignant stem cells, and the defining characteristic of a CSC is the functional ability of these cells to give rise to other cell types within the tumor and propagate disease in vivo.
Hematological malignancies
The initial experiments defining leukemia stem cells identified a small population (<1%) within the primary cancer with the ability to engraft disease in immunocompromised mice. This population shared the phenotypic characteristics of normal HSCs (CD34+ CD38–) and could generate more differentiated CD38+ cells, placing it at the top of a hierarchy, much like its normal counterpart (Lapidot et al., 1994). More recently, it has become clear that these cells can also drive relapse after therapy (Shlush et al., 2017). Interestingly, while CML is driven by a population that closely resembles HSCs, the leukemia stem cells in blast crisis CML appear to share characteristics of a common myeloid progenitor cell (Jamieson et al., 2004), indicating that more committed progenitors may acquire the ability to self-renew and propagate disease (Fig. 2 B).
The observations above could not account for the genetic diversity observed in several tumors and the idea that cancers evolve genetically with disease progression. To determine the evolution of genetic diversity in the context of functional heterogeneity, an analysis of the Philadelphia Chromosome copy number variations in primary human BCR-ABL+ ALL samples was performed to define the ancestry of functional stem cell populations (Notta et al., 2011). These experiments showed that some patients may contain several different clonal clusters of LSC populations that evolve independently and contribute toward genetic diversity. Moreover, they may grow at differential rates in xenograft assays and in the patient and have unique responses to therapy. This clonal diversity within the CSC pool presumably allows the tumor to adapt to different environments and highlights the importance of identifying therapeutic approaches that eliminate all stem cell clones (Notta et al., 2011). One way to achieve this goal would be to understand the signals that sustain these cells and identify common signatures, if any, between these pools. Work in AML has indicated that enhanced stem cell gene expression programs are highly predictive of poor therapeutic response and relapse in patients (Eppert et al., 2011; Gentles et al., 2010; Shlush et al., 2017), and it would be of interest to determine if this also holds true for other hematological diseases.
Solid cancers
Work from hematological malignancies drove interest in the identification of CSCs in solid cancers. Much like the LSCs, glioblastoma CSC populations were first identified using markers of normal neuronal stem cells and were shown to be at the top of a hierarchy with the ability to differentiate unidirectionally to more mature glial cells (Singh et al., 2003, 2004). This paradigm is supported by work in several other primary human cancers such as medulloblastoma (Yang et al., 2008), breast (Al-Hajj et al., 2003), ovarian (Zhang et al., 2008), colon (O’Brien et al., 2007; Ricci-Vitiani et al., 2007), prostate (Collins et al., 2005; Patrawala et al., 2007), and pancreatic cancer (Hermann et al., 2007), as well as SCCs of skin (Driessens et al., 2012) and cervix (Bajaj et al., 2011). While the frequency of cancer-propagating cells in several cancers has been reported to be small, the CSC population may be larger in some cancers, such as melanomas (Boiko et al., 2010; Quintana et al., 2008), and may become enriched after therapy and in relapse settings. Differences in the projected extent of the CSC fraction may in part be due to the transplant models used and whether the recipients are immunocompromised or immunocompetent.
In some tumors, the CSCs have been found to be heterogenous with several different sub-clones of CSCs marked by different cell surface markers. For instance, in addition to CD133, markers such as stage-specific embryonic antigen 1 and α6-integrin can also identify populations with the ability to initiate tumors in xenograft models (Lathia et al., 2010; Son et al., 2009). Distinct CSC populations have been observed in colon cancers marked by CD133, CD44, CD26, or any combination of the three (Dalerba et al., 2007; O’Brien et al., 2007; Pang et al., 2010; Ricci-Vitiani et al., 2007). These differences in cell marker profiles indicate that there may be more than one type of CSC population within a given tumor or that CSCs may evolve and acquire different phenotypic markers with disease progression.
The experiments discussed above were largely dependent on transplantation to prove the importance of the CSCs in tumor propagation. To counter criticisms that these may be a transplantation-specific phenomenon, three key studies focused on targeting these subpopulations within autochthonous models to determine whether this led to inhibition of tumor growth. In a mouse model of SCC, the in vivo eradication of the Sox2+ CSC population via diphtheria toxin led to complete regression of the tumor (Boumahdi et al., 2014). Similarly, deleting Nestin+ stem cells arrested the growth of glioblastomas and sensitized them to chemotherapy (Chen et al., 2012). Interestingly, continuous ablation of Lgr5+ colon CSCs not only arrested the growth of primary tumors but also dramatically reduced metastasis (de Sousa e Melo et al., 2017). Collectively, these studies indicate that CSCs are essential for maintaining tumors and driving metastasis and therapy resistance, and support the idea that targeting these cells may significantly improve therapeutic outcomes (Fig. 1 B).
Cancer stem cells in metastasis
Stem cells and stem cell signals have been implicated in metastatic progression of the tumor from the primary site to a distal location (Charafe-Jauffret et al., 2009; Hermann et al., 2007; Pang et al., 2010; Bajaj et al., 2011; Jang et al., 2015; Mani et al., 2008). As cells leave the primary tumor, they undergo an epithelial to mesenchymal transition (EMT) to acquire the characteristics of more mobile mesenchymal cells. Key studies using in vitro breast cancer cells have shown that the acquisition of EMT is accompanied by a transcriptional program indicative of the stem cell state (Mani et al., 2008). These differential transcriptional profiles may be due to inherent changes in the epigenetic landscape of the cell of origin. For instance, the chromatin of genes associated with EMT is more open and accessible in stem cells of SCCs arising from the hair follicle stem cells and not in the less metastatic, more differentiated SCCs that arise from epidermal stem cells (Latil et al., 2017). However, it is also possible that these epigenetic states are more dynamic since SCC cells can express signatures of both the hair follicle and epidermal stem cells at any given point of time, indicating that epigenetic changes can lead to lineage plasticity (Ge et al., 2017).
Consistent with the possibility that metastatic cells harbor stem cell properties, circulating tumor cells with the ability to recreate tumors at a secondary site can also express markers of primary tumor stem cells (Baccelli et al., 2013). Since cells isolated from the primary tumor using markers such as ALDH have an enhanced capacity to metastasize (Charafe-Jauffret et al., 2009), it is likely that the CSCs and metastatic cells have overlapping phenotypic and signaling profiles. This is supported by the observation that in primary pancreatic cancer, cells that express CXCR4 and have the ability to metastasize are a subset of tumor-propagating chemoresistant CD133+ CSCs (Hermann et al., 2007).
Recent studies interrogating populations undergoing EMT at a single-cell level have confirmed the existence of a continuum of cellular states reflective of a fully epithelial, mesenchymal, or several intermediate EMT states (McFaline-Figueroa et al., 2019; Pastushenko et al., 2018). Importantly, the progression of EMT could be halted by disrupting oncogenic developmental signals, such as Ras (McFaline-Figueroa et al., 2019), suggesting that the signals sustaining these distinct metastatic populations may all rely on a core network of stem cell programs. A better understanding of the links between CSCs and metastasis could be a new avenue to define strategies to target metastatic progression.
Stem cell signals in cancer
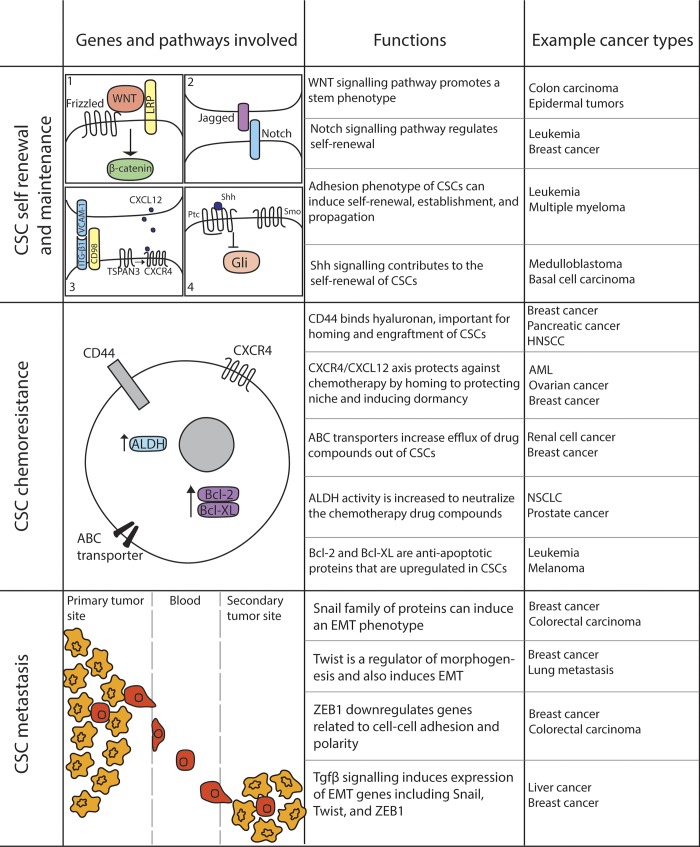
Aberrant activation of signals required for normal development can promote oncogenic transformation. This includes Ptch1 mutations in basal cell carcinoma (Gallatin et al., 1983), APC mutations in colon cancer (Kinzler et al., 1991; Nishisho et al., 1991), and Notch mutations in T-ALL (Ellisen et al., 1991), all developmental pathways that have also been shown to play a critical role in maintaining normal tissue stem cells. Like their normal counterparts, stem cells from many cancers also rely on these developmental signals to maintain self-renewal and evade chemotherapy (Fig. 3).
Figure 3.
Signals sustaining CSCs. Cancer cells are sustained by developmental signaling pathways and interactions with the niche. ABC, ATP binding cassette; HNSCC, head and neck squamous cell carcinoma; LRP, lipoprotein receptor–related protein; NSCLC, non-small-cell lung carcinoma.
Hedgehog signaling
Much of the initial work on Hedgehog signaling in cancer was done in brain tumors, where aberrant activation of this pathway is common. An extensive study on gliomas has revealed that Hedgehog-Gli signaling regulates the expansion of CD133+ glioma stem cells (Clement et al., 2007; Zbinden et al., 2010). In addition, this pathway is also elevated in the more primitive, highly proliferative regions of medulloblastomas, and inhibiting Hedgehog signaling can promote apoptosis of medulloblastoma cells (Bar et al., 2007). Thus, there are clinical trials under way to test the impact of Hedgehog inhibitors in medulloblastomas. A similar requirement for active Hedgehog signaling has been reported in pancreatic cancer (Thayer et al., 2003) and in multiple myeloma, where blocking this pathway results in the loss of clonal expansion and concomitant differentiation (Peacock et al., 2007). In the mammary system, inhibiting Hedgehog signaling reduces not only the ability of mammary CSCs to form primary spheres, but also their ability to self-renew and form secondary spheres (Liu et al., 2006b). Hedgehog signaling has been shown to be critical for the maintenance of stem cells in myeloid leukemia (Zhao et al., 2009); this and other observations formed the basis of trials testing Hedgehog antagonists in myeloid leukemia and ultimately led to the recent approval for Glasdegib in AML (Norsworthy et al., 2019). Since activated Hedgehog signaling can also up-regulate drug transporters on the surface of CSCs and thus promote chemoresistance (Sims-Mourtada et al., 2007), this pathway may regulate several aspects of the stem cell state and broadly promote cancer progression. It is thus not surprising that inhibitors of this pathway are the most advanced in the clinic.
Wnt signaling
The Wnt signaling pathway was originally identified in breast cancer before its role in development was described. After the original observation that Wnt signaling was activated in breast cancer, it has been linked to multiple other cancers, most notably colon cancer, where 95% of cases harbor mutations in APC. The ability of Wnt signaling to influence stem cells in leukemia was shown in the context of CML, where β-catenin–null mice showed profound defects in the ability to develop BCR-ABL–induced CML (Zhao et al., 2007). The few CMLs that did arise were not serially transplantable, highlighting a critical requirement for this pathway for CML stem cell proliferation and self-renewal. Inhibitors of the Wnt pathway targeting the β-catenin/CBP interactions (PRI-724) are in trials for aggressive myeloid diseases (NCT01606579). Consistent with the critical role for Wnt pathway mutations in colon cancer initiation and progression, colon CSCs have also been shown to have high Wnt reporter activity, and exogenously activated Wnt can promote the proliferation of these cells (Vermeulen et al., 2010). In addition, Wnt signaling is critical for sustaining aggressive triple-negative breast CSCs (Jang et al., 2015), and several trials such as those with vantictumab (NCT01345201), (LGK974)189 (NCT01351103), and Ipafricept (Jimeno et al., 2017) have been initiated to target the Wnt pathway in breast cancers.
Notch signaling
Notch signaling has been found to be a critical signal for the development and propagation of multiple cancers including T-ALL, breast cancer, and pancreatic cancer. The possibility that the Notch pathway may have a role in maintaining CSCs was first established in medulloblastomas, where inhibition of Notch cleavage and activation led to a striking reduction in the CD133+ CSCs, and a significant loss in their ability grow in vivo (Hallahan et al., 2004). Consistent with this, the constitutive activation of Notch2 increased the CSC population, and its inhibition completely eradicated stem cells, while leaving the more differentiated populations unaffected (Fan et al., 2006). In addition to medulloblastomas, Notch signaling is essential for the growth and expansion of stem cells from primary glioblastomas both in vitro and in vivo. Inhibition of Notch in murine brain implants of human primary gliomas not only led to the depletion of CSCs but also affected endothelial cells, and thus disrupted tumor angiogenesis, leading to a dramatic reduction in tumor growth (Fan et al., 2010). Importantly, Notch signaling can confer radioresistance to the glioblastoma CD133+ CSC population, highlighting the critical role of this pathway in brain tumor stem cells (Wang et al., 2010). In pancreatic cancer, Mir-34 microRNA–mediated down-regulation of Notch1/2 results in a significant reduction in both tumor growth and the CD44+/CD133+ CSC population (Ji et al., 2009; Mullendore et al., 2009). Consistent with this, Musashi, a regulator of the Numb/Notch signaling axis, marks the chemoresistant CSC populations in pancreatic cancer (Fox et al., 2016) and myeloid leukemias (Ito et al., 2010). Activated Notch signaling is also essential for maintaining the CSC populations in cervical cancers (Bajaj et al., 2011) and ovarian cancers (Hopfer et al., 2005), and inhibitors of the Notch ligand Dll4 are currently in trials for aggressive colon and ovarian carcinomas (NCT03035253). Thus, Notch signaling plays a critical role in promoting therapy-resistant CSC populations across malignancies.
Microenvironmental dependencies
While the role of the microenvironment in sustaining solid cancer growth is well established, its contribution in maintaining the CSC populations is only beginning to be explored. CD44-mediated interactions (Jin et al., 2006) of leukemic progenitors with the osteoblast niche can promote chemoresistance (Lane et al., 2011), and osteopontin–CD44 interactions can support glioma stem cells (Pietras et al., 2014). Adhesive interactions with the endothelial cells are particularly critical for maintaining brain tumor stem cells (Calabrese et al., 2007), and the CD98/VLA-4/VCAM1 (Bajaj et al., 2016) signaling axis between leukemia stem cells and endothelial cells has been shown to be essential for myeloid leukemia propagation in vivo. Since blocking VLA-4–mediated interactions with fibronectin can promote chemosensitivity in AML (Matsunaga et al., 2003), these adhesive interactions are emerging as novel therapeutic targets for aggressive leukemias. New pathways such as Tspan3 are emerging as key points of control for leukemia propagation; these may in part function by mediating interactions of leukemia stem cells with an SDF-1–enriched niche and protecting the undifferentiated state (Kwon et al., 2015). Consistent with this, blocking SDF-1/CXCR4 signaling can influence leukemia differentiation (Tavor et al., 2004). Clinical trials aimed at defining the impact of blocking CD98 (Bixby et al., 2015), CD44 (Vey et al., 2016), and CXCR4 (Martínez-Cuadrón et al., 2018; Uy et al., 2012) signaling have shown some promise and indicate that better targeting agents and/or combinatorial therapy could be beneficial in controlling disease progression.
Future directions
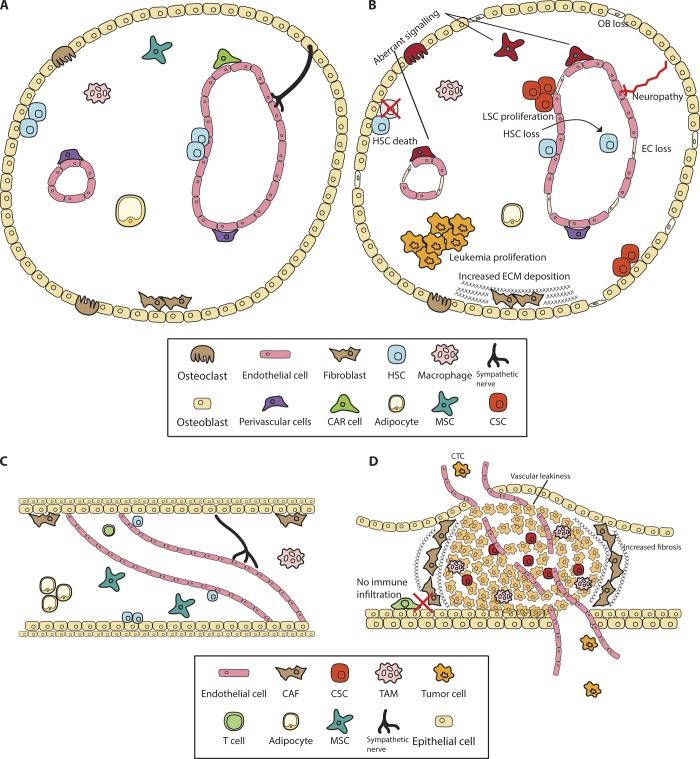
While much has been learned about intrinsic dependencies of CSCs, how they interact with the microenvironment remains largely unknown. Emerging studies on stromal cell populations, such as endothelial cells in leukemias (Bajaj et al., 2016) and brain tumors (Calabrese et al., 2007), stromal myofibroblasts in colon cancer (Vermeulen et al., 2010), cancer-associated fibroblasts in lung and breast cancers (Su et al., 2018), and mesenchymal stromal cells in leukemias (Jacamo et al., 2014), suggest that stromal cells can support CSCs just as they support normal stem cells (Fig. 4). New technologies such as single-cell sequencing aimed toward characterizing the stromal cell niche of cancers in its entirety may be critical to providing a framework for experiments aimed at identifying the specific niche of the CSCs.
Figure 4.
The normal and malignant stem cell niche. Stem cell interactions with the microenvironment lead to changes in the niche that promote cancer progression. (A and C) Normal stem cells reside in niches guided by complex signaling to control their activation and proliferation. (B) LSCs induce cell death and aberrant signaling within the bone marrow microenvironment. (D) Solid tumor stem cells reside in dense stroma and transformed cells that protect them from chemotherapy. Boxes represent niche cells in A and B or C and D. OB, osteoblast; EC, endothelial cells; CAR, CXCL12-abundant reticular cells; CAF, cancer-associated fibroblasts; MSC, mesenchymal stromal cells; TAM, tumor-associated macrophages.
In this context, some recent studies have performed extensive single-cell sequencing on both the normal and preleukemic bone marrow stromal cell populations to determine both the niche of normal HSCs and how it is altered with the initiation of cancer (Baryawno et al., 2019; Wolock et al., 2019). These show that during initiation, leukemias disrupt and alter the niche (Baryawno et al., 2019), consistent with earlier studies showing that CML stem cells can disrupt osteoblast proliferation, leading to increased fibrosis and a supportive growth environment for LSCs at the expense of HSCs (Schepers et al., 2013). Similarly, solid cancers can alter the metastatic niche even before colonization (Kaplan et al., 2005; Costa-Silva et al., 2015; Olmeda et al., 2017). Single-cell sequencing of primary human breast, lung, and pancreatic cancers has revealed heterogeneity not only in the cancer populations but also in immune and other cells of the microenvironment (Bartoschek et al., 2018; Bernard et al., 2019; Elyada et al., 2019; Lawson et al., 2018; Ma et al., 2019; Wagner et al., 2019). Defining the spatial localization of CSCs with respect to specific niche cells, and understanding how these interactions contribute to cancer progression and response to therapy, will be exciting future avenues of work.
While there is some debate on the origin, markers, and frequency of CSCs, a large body of literature now shows that there is significant heterogeneity within multiple cancer types and that subpopulations harbor preferential ability to drive tumor initiation, metastasis, and therapy resistance (Gawad et al., 2014; Saadatpour et al., 2014; Bakker et al., 2016; Tirosh et al., 2016; Darmanis et al., 2017). The work discussed here indicates that such cells not only promote cancer progression but also may be able to adapt and grow in different environments. Understanding both the intrinsic signals that maintain these aggressive stem cell populations and the microenvironmental interactions they depend on will be critical in designing multifaceted therapeutic approaches to target aggressive cancers.
Acknowledgments
J. Bajaj is a recipient of a Scholar Award from the American Society of Hematology and a postdoctoral fellowship from the National Cancer Center, and E. Diaz received support from National Institutes of Health grant T32 GM007752. This work was supported by National Institutes of Health grant R35 CA197699 awarded to T. Reya.
The authors declare no competing financial interests.
References
- Adhikari A.S., Agarwal N., Wood B.M., Porretta C., Ruiz B., Pochampally R.R., and Iwakuma T.. 2010. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 70:4602–4612. 10.1158/0008-5472.CAN-09-3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., and Clarke M.F.. 2003. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 100:3983–3988. 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno S., Chen J., Kwon C.H., Jackson E.L., Li Y., Burns D.K., Alvarez-Buylla A., and Parada L.F.. 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 15:45–56. 10.1016/j.ccr.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno S.R., Wang Z., Sun D., Chen J., Xu J., Kim E., Hatanpaa K.J., Raisanen J.M., Burns D.K., Johnson J.E., and Parada L.F.. 2015. Adult Lineage-Restricted CNS Progenitors Specify Distinct Glioblastoma Subtypes. Cancer Cell. 28:429–440. 10.1016/j.ccell.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccelli I., Schneeweiss A., Riethdorf S., Stenzinger A., Schillert A., Vogel V., Klein C., Saini M., Bäuerle T., Wallwiener M., et al. 2013. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31:539–544. 10.1038/nbt.2576 [DOI] [PubMed] [Google Scholar]
- Bains A., Luthra R., Medeiros L.J., and Zuo Z.. 2011. FLT3 and NPM1 mutations in myelodysplastic syndromes: Frequency and potential value for predicting progression to acute myeloid leukemia. Am. J. Clin. Pathol. 135:62–69. 10.1309/AJCPEI9XU8PYBCIO [DOI] [PubMed] [Google Scholar]
- Bajaj J., Maliekal T.T., Vivien E., Pattabiraman C., Srivastava S., Krishnamurthy H., Giri V., Subramanyam D., and Krishna S.. 2011. Notch signaling in CD66+ cells drives the progression of human cervical cancers. Cancer Res. 71:4888–4897. 10.1158/0008-5472.CAN-11-0543 [DOI] [PubMed] [Google Scholar]
- Bajaj J., Konuma T., Lytle N.K., Kwon H.Y., Ablack J.N., Cantor J.M., Rizzieri D., Chuah C., Oehler V.G., Broome E.H., et al. 2016. CD98-Mediated Adhesive Signaling Enables the Establishment and Propagation of Acute Myelogenous Leukemia. Cancer Cell. 30:792–805. 10.1016/j.ccell.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B., Taudt A., Belderbos M.E., Porubsky D., Spierings D.C., de Jong T.V., Halsema N., Kazemier H.G., Hoekstra-Wakker K., Bradley A., et al. 2016. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 17:115 10.1186/s13059-016-0971-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar E.E., Chaudhry A., Farah M.H., and Eberhart C.G.. 2007. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am. J. Pathol. 170:347–355. 10.2353/ajpath.2007.060066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., and Clevers H.. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 457:608–611. 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Bartoschek M., Oskolkov N., Bocci M., Lövrot J., Larsson C., Sommarin M., Madsen C.D., Lindgren D., Pekar G., Karlsson G., et al. 2018. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 9:5150 10.1038/s41467-018-07582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno N., Przybylski D., Kowalczyk M.S., Kfoury Y., Severe N., Gustafsson K., Kokkaliaris K.D., Mercier F., Tabaka M., Hofree M., et al. 2019. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell. 177:1915–1932.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard V., Semaan A., Huang J., San Lucas F.A., Mulu F.C., Stephens B.M., Guerrero P.A., Huang Y., Zhao J., Kamyabi N., et al. 2019. Single-Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. Clin. Cancer Res. 25:2194–2205. 10.1158/1078-0432.CCR-18-1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y., Hall B., Sun Z.J., Molinolo A., Chen W., Gutkind J.S., Waes C.V., and Kulkarni A.B.. 2012. Loss of TGF-β signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 31:3322–3332. 10.1038/onc.2011.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixby D., Wieduwilt M.J., Akard L.P., Khoury H.J., Becker P.S., van der Horst E.H., Ho W., and Cortes J.E.. 2015. A Phase I Study of IGN523, a Novel Anti-CD98 Monoclonal Antibody in Patients with Relapsed or Refractory Acute Myeloid Leukemia (AML). Blood. 126:3809 10.1182/blood.V126.23.3809.3809 [DOI] [Google Scholar]
- Boiko A.D., Razorenova O.V., van de Rijn M., Swetter S.M., Johnson D.L., Ly D.P., Butler P.D., Yang G.P., Joshua B., Kaplan M.J., et al. 2010. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 466:133–137. 10.1038/nature09161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., and Dick J.E.. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3:730–737. 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., Delatte B., Caauwe A., Lenglez S., Nkusi E., et al. 2014. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 511:246–250. 10.1038/nature13305 [DOI] [PubMed] [Google Scholar]
- Bruce W.R., and Van Der Gaag H.. 1963. A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature. 199:79–80. 10.1038/199079a0 [DOI] [PubMed] [Google Scholar]
- Calabrese C., Poppleton H., Kocak M., Hogg T.L., Fuller C., Hamner B., Oh E.Y., Gaber M.W., Finklestein D., Allen M., et al. 2007. A perivascular niche for brain tumor stem cells. Cancer Cell. 11:69–82. 10.1016/j.ccr.2006.11.020 [DOI] [PubMed] [Google Scholar]
- Chaffer C.L., Brueckmann I., Scheel C., Kaestli A.J., Wiggins P.A., Rodrigues L.O., Brooks M., Reinhardt F., Su Y., Polyak K., et al. 2011. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. USA. 108:7950–7955. 10.1073/pnas.1102454108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charafe-Jauffret E., Ginestier C., Iovino F., Wicinski J., Cervera N., Finetti P., Hur M.H., Diebel M.E., Monville F., Dutcher J., et al. 2009. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 69:1302–1313. 10.1158/0008-5472.CAN-08-2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li Y., Yu T.S., McKay R.M., Burns D.K., Kernie S.G., and Parada L.F.. 2012. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 488:522–526. 10.1038/nature11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V., Sanchez P., de Tribolet N., Radovanovic I., and Ruiz i Altaba A.. 2007. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 17:165–172. 10.1016/j.cub.2006.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.T., Berry P.A., Hyde C., Stower M.J., and Maitland N.J.. 2005. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 65:10946–10951. 10.1158/0008-5472.CAN-05-2018 [DOI] [PubMed] [Google Scholar]
- Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. 2015. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17:816–826. 10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Dylla S.J., Park I.K., Liu R., Wang X., Cho R.W., Hoey T., Gurney A., Huang E.H., Simeone D.M., et al. 2007. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA. 104:10158–10163. 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Kalisky T., Sahoo D., Rajendran P.S., Rothenberg M.E., Leyrat A.A., Sim S., Okamoto J., Johnston D.M., Qian D., et al. 2011. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 29:1120–1127. 10.1038/nbt.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dameshek W. 1951. Some speculations on the myeloproliferative syndromes. Blood. 6:372–375. 10.1182/blood.V6.4.372.372 [DOI] [PubMed] [Google Scholar]
- Darmanis S., Sloan S.A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., et al. 2017. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Reports. 21:1399–1410. 10.1016/j.celrep.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa e Melo F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J., Anderson J.E., Storm E.E., Modrusan Z., Koeppen H., et al. 2017. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 543:676–680. 10.1038/nature21713 [DOI] [PubMed] [Google Scholar]
- Dick J.E. 2008. Stem cell concepts renew cancer research. Blood. 112:4793–4807. 10.1182/blood-2008-08-077941 [DOI] [PubMed] [Google Scholar]
- Ding L., Ley T.J., Larson D.E., Miller C.A., Koboldt D.C., Welch J.S., Ritchey J.K., Young M.A., Lamprecht T., McLellan M.D., et al. 2012. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 481:506–510. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G., Beck B., Caauwe A., Simons B.D., and Blanpain C.. 2012. Defining the mode of tumour growth by clonal analysis. Nature. 488:527–530. 10.1038/nature11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen L.W., Bird J., West D.C., Soreng A.L., Reynolds T.C., Smith S.D., and Sklar J.. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 66:649–661. 10.1016/0092-8674(91)90111-B [DOI] [PubMed] [Google Scholar]
- Elyada E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., Teinor J.A., Belleau P., Biffi G., Lucito M.S., et al. 2019. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 9:1102–1123. 10.1158/2159-8290.CD-19-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. 2011. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17:1086–1093. 10.1038/nm.2415 [DOI] [PubMed] [Google Scholar]
- Fan X., Matsui W., Khaki L., Stearns D., Chun J., Li Y.M., and Eberhart C.G.. 2006. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 66:7445–7452. 10.1158/0008-5472.CAN-06-0858 [DOI] [PubMed] [Google Scholar]
- Fan X., Khaki L., Zhu T.S., Soules M.E., Talsma C.E., Gul N., Koh C., Zhang J., Li Y.M., Maciaczyk J., et al. 2010. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 28:5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E.R., and Vogelstein B.. 1990. A genetic model for colorectal tumorigenesis. Cell. 61:759–767. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Fox R.G., Lytle N.K., Jaquish D.V., Park F.D., Ito T., Bajaj J., Koechlein C.S., Zimdahl B., Yano M., Kopp J., et al. 2016. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature. 534:407–411. 10.1038/nature17988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallatin W.M., Weissman I.L., and Butcher E.C.. 1983. A cell-surface molecule involved in organ-specific homing of lymphocytes. Nature. 304:30–34. 10.1038/304030a0 [DOI] [PubMed] [Google Scholar]
- Gawad C., Koh W., and Quake S.R.. 2014. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc. Natl. Acad. Sci. USA. 111:17947–17952. 10.1073/pnas.1420822111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Gomez N.C., Adam R.C., Nikolova M., Yang H., Verma A., Lu C.P., Polak L., Yuan S., Elemento O., and Fuchs E.. 2017. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell. 169:636–650.e14. 10.1016/j.cell.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Plevritis S.K., Majeti R., and Alizadeh A.A.. 2010. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 304:2706–2715. 10.1001/jama.2010.1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallahan A.R., Pritchard J.I., Hansen S., Benson M., Stoeck J., Hatton B.A., Russell T.L., Ellenbogen R.G., Bernstein I.D., Beachy P.A., and Olson J.M.. 2004. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 64:7794–7800. 10.1158/0008-5472.CAN-04-1813 [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D., Squatrito M., and Holland E.C.. 2006. Radiation resistance and stem-like cells in brain tumors. Cancer Cell. 10:454–456. 10.1016/j.ccr.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2000. The hallmarks of cancer. Cell. 100:57–70. 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hermann P.C., Huber S.L., Herrler T., Aicher A., Ellwart J.W., Guba M., Bruns C.J., and Heeschen C.. 2007. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 1:313–323. 10.1016/j.stem.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Hopfer O., Zwahlen D., Fey M.F., and Aebi S.. 2005. The Notch pathway in ovarian carcinomas and adenomas. Br. J. Cancer. 93:709–718. 10.1038/sj.bjc.6602719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly B.J., Shigematsu H., Deguchi K., Lee B.H., Mizuno S., Duclos N., Rowan R., Amaral S., Curley D., Williams I.R., et al. 2004. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 6:587–596. 10.1016/j.ccr.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Ito T., Kwon H.Y., Zimdahl B., Congdon K.L., Blum J., Lento W.E., Zhao C., Lagoo A., Gerrard G., Foroni L., et al. 2010. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 466:765–768. 10.1038/nature09171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacamo R., Chen Y., Wang Z., Ma W., Zhang M., Spaeth E.L., Wang Y., Battula V.L., Mak P.Y., Schallmoser K., et al. 2014. Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-κB mediates chemoresistance. Blood. 123:2691–2702. 10.1182/blood-2013-06-511527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylla S.J., Muijtjens M., Jones C., Zehnder J.L., Gotlib J., Li K., Manz M.G., Keating A., et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657–667. 10.1056/NEJMoa040258 [DOI] [PubMed] [Google Scholar]
- Jang G.B., Kim J.Y., Cho S.D., Park K.S., Jung J.Y., Lee H.Y., Hong I.S., and Nam J.S.. 2015. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci. Rep. 5:12465 10.1038/srep12465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q., Hao X., Zhang M., Tang W., Yang M., Li L., Xiang D., Desano J.T., Bommer G.T., Fan D., et al. 2009. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 4:e6816 10.1371/journal.pone.0006816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno A., Gordon M., Chugh R., Messersmith W., Mendelson D., Dupont J., Stagg R., Kapoun A.M., Xu L., Uttamsingh S., et al. 2017. A First-in-Human Phase I Study of the Anticancer Stem Cell Agent Ipafricept (OMP-54F28), a Decoy Receptor for Wnt Ligands, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 23:7490–7497. 10.1158/1078-0432.CCR-17-2157 [DOI] [PubMed] [Google Scholar]
- Jin L., Hope K.J., Zhai Q., Smadja-Joffe F., and Dick J.E.. 2006. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 12:1167–1174. 10.1038/nm1483 [DOI] [PubMed] [Google Scholar]
- Kaplan R.N., Riba R.D., Zacharoulis S., Bramley A.H., Vincent L., Costa C., MacDonald D.D., Jin D.K., Shido K., Kerns S.A., et al. 2005. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 438:820–827. 10.1038/nature04186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler K.W., Nilbert M.C., Su L.K., Vogelstein B., Bryan T.M., Levy D.B., Smith K.J., Preisinger A.C., Hedge P., McKechnie D., et al. 1991. Identification of FAP locus genes from chromosome 5q21. Science. 253:661–665. 10.1126/science.1651562 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Twomey D., Feng Z., Stubbs M.C., Wang Y., Faber J., Levine J.E., Wang J., Hahn W.C., Gilliland D.G., et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818–822. 10.1038/nature04980 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Figueroa M.E., Sinha A.U., Stubbs M.C., Feng Z., Valk P.J., Delwel R., Döhner K., Bullinger L., Kung A.L., et al. 2013. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 27:852–860. 10.1038/leu.2012.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.Y., Bajaj J., Ito T., Blevins A., Konuma T., Weeks J., Lytle N.K., Koechlein C.S., Rizzieri D., Chuah C., et al. 2015. Tetraspanin 3 Is Required for the Development and Propagation of Acute Myelogenous Leukemia. Cell Stem Cell. 17:152–164. 10.1016/j.stem.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S.W., Wang Y.J., Lo Celso C., Ragu C., Bullinger L., Sykes S.M., Ferraro F., Shterental S., Lin C.P., Gilliland D.G., et al. 2011. Differential niche and Wnt requirements during acute myeloid leukemia progression. Blood. 118:2849–2856. 10.1182/blood-2011-03-345165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., and Dick J.E.. 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367:645–648. 10.1038/367645a0 [DOI] [PubMed] [Google Scholar]
- Lapouge G., Youssef K.K., Vokaer B., Achouri Y., Michaux C., Sotiropoulou P.A., and Blanpain C.. 2011. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. USA. 108:7431–7436. 10.1073/pnas.1012720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia J.D., Gallagher J., Heddleston J.M., Wang J., Eyler C.E., Macswords J., Wu Q., Vasanji A., McLendon R.E., Hjelmeland A.B., and Rich J.N.. 2010. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 6:421–432. 10.1016/j.stem.2010.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil M., Nassar D., Beck B., Boumahdi S., Wang L., Brisebarre A., Dubois C., Nkusi E., Lenglez S., Checinska A., et al. 2017. Cell-Type-Specific Chromatin States Differentially Prime Squamous Cell Carcinoma Tumor-Initiating Cells for Epithelial to Mesenchymal Transition. Cell Stem Cell. 20:191–204.e5. 10.1016/j.stem.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D.A., Kessenbrock K., Davis R.T., Pervolarakis N., and Werb Z.. 2018. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20:1349–1360. 10.1038/s41556-018-0236-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ilaria R.L. Jr., Million R.P., Daley G.Q., and Van Etten R.A.. 1999. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 189:1399–1412. 10.1084/jem.189.9.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.E., Hou H.A., Tsai C.H., Wu S.J., Kuo Y.Y., Tseng M.H., Liu M.C., Liu C.W., Chou W.C., Chen C.Y., et al. 2018. Dynamics of DNMT3A mutation and prognostic relevance in patients with primary myelodysplastic syndrome. Clin. Epigenetics. 10:42 10.1186/s13148-018-0476-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yuan X., Zeng Z., Tunici P., Ng H., Abdulkadir I.R., Lu L., Irvin D., Black K.L., and Yu J.S.. 2006a Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer. 5:67 10.1186/1476-4598-5-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Dontu G., Mantle I.D., Patel S., Ahn N.S., Jackson K.W., Suri P., and Wicha M.S.. 2006b Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 66:6063–6071. 10.1158/0008-5472.CAN-06-0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Jiang M., Lu Y., Chen H., Sun J., Wu S., Ku W.Y., Nakagawa H., Kita Y., Natsugoe S., et al. 2013. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 12:304–315. 10.1016/j.stem.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle N.K., Barber A.G., and Reya T.. 2018. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer. 18:669–680. 10.1038/s41568-018-0056-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K.Y., Schonnesen A.A., Brock A., Van Den Berg C., Eckhardt S.G., Liu Z., and Jiang N.. 2019. Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response-related genes. JCI Insight. 4:e121387 10.1172/jci.insight.121387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cuadrón D., Boluda B., Martínez P., Bergua J., Rodríguez-Veiga R., Esteve J., Vives S., Serrano J., Vidriales B., Salamero O., et al. 2018. A phase I-II study of plerixafor in combination with fludarabine, idarubicin, cytarabine, and G-CSF (PLERIFLAG regimen) for the treatment of patients with the first early-relapsed or refractory acute myeloid leukemia. Ann. Hematol. 97:763–772. 10.1007/s00277-018-3229-5 [DOI] [PubMed] [Google Scholar]
- Matsunaga T., Takemoto N., Sato T., Takimoto R., Tanaka I., Fujimi A., Akiyama T., Kuroda H., Kawano Y., Kobune M., et al. 2003. Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat. Med. 9:1158–1165. 10.1038/nm909 [DOI] [PubMed] [Google Scholar]
- McFaline-Figueroa J.L., Hill A.J., Qiu X., Jackson D., Shendure J., and Trapnell C.. 2019. A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat. Genet. 51:1389–1398. 10.1038/s41588-019-0489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G., Geyer F.C., Magnay F.A., McCarthy A., Kendrick H., Natrajan R., Mackay A., Grigoriadis A., Tutt A., Ashworth A., et al. 2010. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 7:403–417. 10.1016/j.stem.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Mullendore M.E., Koorstra J.B., Li Y.M., Offerhaus G.J., Fan X., Henderson C.M., Matsui W., Eberhart C.G., Maitra A., and Feldmann G.. 2009. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin. Cancer Res. 15:2291–2301. 10.1158/1078-0432.CCR-08-2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangalia J., Nice F.L., Wedge D.C., Godfrey A.L., Grinfeld J., Thakker C., Massie C.E., Baxter J., Sewell D., Silber Y., et al. 2015. DNMT3A mutations occur early or late in patients with myeloproliferative neoplasms and mutation order influences phenotype. Haematologica. 100:e438–e442. 10.3324/haematol.2015.129510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neering S.J., Bushnell T., Sozer S., Ashton J., Rossi R.M., Wang P.Y., Bell D.R., Heinrich D., Bottaro A., and Jordan C.T.. 2007. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 110:2578–2585. 10.1182/blood-2007-02-073031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishisho I., Nakamura Y., Miyoshi Y., Miki Y., Ando H., Horii A., Koyama K., Utsunomiya J., Baba S., and Hedge P.. 1991. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 253:665–669. 10.1126/science.1651563 [DOI] [PubMed] [Google Scholar]
- Norsworthy K.J., By K., Subramaniam S., Zhuang L., Del Valle P.L., Przepiorka D., Shen Y.L., Sheth C.M., Liu C., Leong R., et al. 2019. FDA Approval Summary: Glasdegib for Newly Diagnosed Acute Myeloid Leukemia. Clin. Cancer Res. 25:6021–6025. 10.1158/1078-0432.CCR-19-0365 [DOI] [PubMed] [Google Scholar]
- Notta F., Mullighan C.G., Wang J.C., Poeppl A., Doulatov S., Phillips L.A., Ma J., Minden M.D., Downing J.R., and Dick J.E.. 2011. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 469:362–367. 10.1038/nature09733 [DOI] [PubMed] [Google Scholar]
- Nowell P.C. 1974. Diagnostic and prognostic value of chromosome studies in cancer. Ann. Clin. Lab. Sci. 4:234–240. [PubMed] [Google Scholar]
- O’Brien C.A., Pollett A., Gallinger S., and Dick J.E.. 2007. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 445:106–110. 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- Olmeda D., Cerezo-Wallis D., Riveiro-Falkenbach E., Pennacchi P.C., Contreras-Alcalde M., Ibarz N., Cifdaloz M., Catena X., Calvo T.G., Cañón E., et al. 2017. Whole-body imaging of lymphovascular niches identifies pre-metastatic roles of midkine. Nature. 546:676–680. 10.1038/nature22977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang R., Law W.L., Chu A.C., Poon J.T., Lam C.S., Chow A.K., Ng L., Cheung L.W., Lan X.R., Lan H.Y., et al. 2010. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 6:603–615. 10.1016/j.stem.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., et al. 2018. Identification of the tumour transition states occurring during EMT. Nature. 556:463–468. 10.1038/s41586-018-0040-3 [DOI] [PubMed] [Google Scholar]
- Patrawala L., Calhoun-Davis T., Schneider-Broussard R., and Tang D.G.. 2007. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 67:6796–6805. 10.1158/0008-5472.CAN-07-0490 [DOI] [PubMed] [Google Scholar]
- Peacock C.D., Wang Q., Gesell G.S., Corcoran-Schwartz I.M., Jones E., Kim J., Devereux W.L., Rhodes J.T., Huff C.A., Beachy P.A., et al. 2007. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc. Natl. Acad. Sci. USA. 104:4048–4053. 10.1073/pnas.0611682104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras A., Katz A.M., Ekström E.J., Wee B., Halliday J.J., Pitter K.L., Werbeck J.L., Amankulor N.M., Huse J.T., and Holland E.C.. 2014. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 14:357–369. 10.1016/j.stem.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., et al. 2012. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 149:146–158. 10.1016/j.cell.2012.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E., Shackleton M., Sabel M.S., Fullen D.R., Johnson T.M., and Morrison S.J.. 2008. Efficient tumour formation by single human melanoma cells. Nature. 456:593–598. 10.1038/nature07567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi A.R., Molinolo A., and Gutkind J.S.. 2009. Rapamycin prevents early onset of tumorigenesis in an oral-specific K-ras and p53 two-hit carcinogenesis model. Cancer Res. 69:4159–4166. 10.1158/0008-5472.CAN-08-4645 [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison S.J., Clarke M.F., and Weissman I.L.. 2001. Stem cells, cancer, and cancer stem cells. Nature. 414:105–111. 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L., Lombardi D.G., Pilozzi E., Biffoni M., Todaro M., Peschle C., and De Maria R.. 2007. Identification and expansion of human colon-cancer-initiating cells. Nature. 445:111–115. 10.1038/nature05384 [DOI] [PubMed] [Google Scholar]
- Roesch A., Fukunaga-Kalabis M., Schmidt E.C., Zabierowski S.E., Brafford P.A., Vultur A., Basu D., Gimotty P., Vogt T., and Herlyn M.. 2010. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 141:583–594. 10.1016/j.cell.2010.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadatpour A., Guo G., Orkin S.H., and Yuan G.C.. 2014. Characterizing heterogeneity in leukemic cells using single-cell gene expression analysis. Genome Biol. 15:525 10.1186/s13059-014-0525-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K., Pietras E.M., Reynaud D., Flach J., Binnewies M., Garg T., Wagers A.J., Hsiao E.C., and Passegué E.. 2013. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 13:285–299. 10.1016/j.stem.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W., et al. 2014. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 506:328–333. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush L.I., Mitchell A., Heisler L., Abelson S., Ng S.W.K., Trotman-Grant A., Medeiros J.J.F., Rao-Bhatia A., Jaciw-Zurakowsky I., Marke R., et al. 2017. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 547:104–108. 10.1038/nature22993 [DOI] [PubMed] [Google Scholar]
- Sims-Mourtada J., Izzo J.G., Ajani J., and Chao K.S.. 2007. Sonic Hedgehog promotes multiple drug resistance by regulation of drug transport. Oncogene. 26:5674–5679. 10.1038/sj.onc.1210356 [DOI] [PubMed] [Google Scholar]
- Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., and Dirks P.B.. 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63:5821–5828. [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., and Dirks P.B.. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Son M.J., Woolard K., Nam D.H., Lee J., and Fine H.A.. 2009. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 4:440–452. 10.1016/j.stem.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A., Kang H., Ma Z., Graham T.A., Salomon M.P., Zhao J., Marjoram P., Siegmund K., Press M.F., Shibata D., and Curtis C.. 2015. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47:209–216. 10.1038/ng.3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Chen J., Yao H., Liu J., Yu S., Lao L., Wang M., Luo M., Xing Y., Chen F., et al. 2018. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 172:841–856.e16. 10.1016/j.cell.2018.01.009 [DOI] [PubMed] [Google Scholar]
- Tan B.T., Park C.Y., Ailles L.E., and Weissman I.L.. 2006. The cancer stem cell hypothesis: a work in progress. Lab. Invest. 86:1203–1207. 10.1038/labinvest.3700488 [DOI] [PubMed] [Google Scholar]
- Tavor S., Petit I., Porozov S., Avigdor A., Dar A., Leider-Trejo L., Shemtov N., Deutsch V., Naparstek E., Nagler A., and Lapidot T.. 2004. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer Res. 64:2817–2824. 10.1158/0008-5472.CAN-03-3693 [DOI] [PubMed] [Google Scholar]
- Thayer S.P., di Magliano M.P., Heiser P.W., Nielsen C.M., Roberts D.J., Lauwers G.Y., Qi Y.P., Gysin S., Fernández-del Castillo C., Yajnik V., et al. 2003. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 425:851–856. 10.1038/nature02009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Venteicher A.S., Hebert C., Escalante L.E., Patel A.P., Yizhak K., Fisher J.M., Rodman C., Mount C., Filbin M.G., et al. 2016. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 539:309–313. 10.1038/nature20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C., Li L., and Vogelstein B.. 2017. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 355:1330–1334. 10.1126/science.aaf9011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tough I.M., Jacobs P.A., Court Brown W.M., Baikie A.G., and Williamson E.R.. 1963. Cytogenetic studies on bone-marrow in chronic myeloid leukaemia. Lancet. 1:844–846. 10.1016/S0140-6736(63)91620-9 [DOI] [PubMed] [Google Scholar]
- Uy G.L., Rettig M.P., Motabi I.H., McFarland K., Trinkaus K.M., Hladnik L.M., Kulkarni S., Abboud C.N., Cashen A.F., Stockerl-Goldstein K.E., et al. 2012. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 119:3917–3924. 10.1182/blood-2011-10-383406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., De Sousa E Melo F., van der Heijden M., Cameron K., de Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H., et al. 2010. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12:468–476. 10.1038/ncb2048 [DOI] [PubMed] [Google Scholar]
- Vey N., Delaunay J., Martinelli G., Fiedler W., Raffoux E., Prebet T., Gomez-Roca C., Papayannidis C., Kebenko M., Paschka P., et al. 2016. Phase I clinical study of RG7356, an anti-CD44 humanized antibody, in patients with acute myeloid leukemia. Oncotarget. 7:32532–32542. 10.18632/oncotarget.8687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M., Nakamura Y., White R., Smits A.M., and Bos J.L.. 1988. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319:525–532. 10.1056/NEJM198809013190901 [DOI] [PubMed] [Google Scholar]
- Wagner J., Rapsomaniki M.A., Chevrier S., Anzeneder T., Langwieder C., Dykgers A., Rees M., Ramaswamy A., Muenst S., Soysal S.D., et al. 2019. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell. 177:1330–1345.e18. 10.1016/j.cell.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wakeman T.P., Lathia J.D., Hjelmeland A.B., Wang X.F., White R.R., Rich J.N., and Sullenger B.A.. 2010. Notch promotes radioresistance of glioma stem cells. Stem Cells. 28:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang J., Frei E. III, Tjio J.H., Carbone P.P., and Brecher G.. 1963. The distribution of the Philadelphia chromosome in patients with chronic myelogenous leukemia. Blood. 22:664–673. 10.1182/blood.V22.6.664.664 [DOI] [PubMed] [Google Scholar]
- White A.C., Tran K., Khuu J., Dang C., Cui Y., Binder S.W., and Lowry W.E.. 2011. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 108:7425–7430. 10.1073/pnas.1012670108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock S.L., Krishnan I., Tenen D.E., Matkins V., Camacho V., Patel S., Agarwal P., Bhatia R., Tenen D.G., Klein A.M., and Welner R.S.. 2019. Mapping Distinct Bone Marrow Niche Populations and Their Differentiation Paths. Cell Reports. 28:302–311.e5. 10.1016/j.celrep.2019.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Ma L., Yi D., Yoon J.G., Diercks A., Foltz G., Price N.D., Hood L.E., and Tian Q.. 2011. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc. Natl. Acad. Sci. USA. 108:1591–1596. 10.1073/pnas.1018696108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.J., Ellis T., Markant S.L., Read T.A., Kessler J.D., Bourboulas M., Schüller U., Machold R., Fishell G., Rowitch D.H., et al. 2008. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 14:135–145. 10.1016/j.ccr.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden M., Duquet A., Lorente-Trigos A., Ngwabyt S.N., Borges I., and Ruiz i Altaba A.. 2010. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 29:2659–2674. 10.1038/emboj.2010.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Balch C., Chan M.W., Lai H.C., Matei D., Schilder J.M., Yan P.S., Huang T.H., and Nephew K.P.. 2008. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 68:4311–4320. 10.1158/0008-5472.CAN-08-0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Blum J., Chen A., Kwon H.Y., Jung S.H., Cook J.M., Lagoo A., and Reya T.. 2007. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 12:528–541. 10.1016/j.ccr.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Chen A., Jamieson C.H., Fereshteh M., Abrahamsson A., Blum J., Kwon H.Y., Kim J., Chute J.P., Rizzieri D., et al. 2009. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 458:776–779. 10.1038/nature07737 [DOI] [PMC free article] [PubMed] [Google Scholar]