Boyer et al. describe reversible nondegradative ubiquitination of the actin polymerase VASP, mediated by a pair of E3 ubiquitin ligases TRIM9 and TRIM67. VASP regulation is necessary for responses of filopodia, growth cones, and axons to the guidance cue netrin-1.
Abstract
Appropriate axon guidance is necessary to form accurate neuronal connections. Axon guidance cues that stimulate cytoskeletal reorganization within the growth cone direct axon navigation. Filopodia at the growth cone periphery have long been considered sensors for axon guidance cues, yet how they respond to extracellular cues remains ill defined. Our previous work found that the filopodial actin polymerase VASP and consequently filopodial stability are negatively regulated via nondegradative TRIM9-dependent ubiquitination. Appropriate VASP ubiquitination and deubiquitination are required for axon turning in response to the guidance cue netrin-1. Here we show that the TRIM9-related protein TRIM67 outcompetes TRIM9 for interacting with VASP and antagonizes TRIM9-dependent VASP ubiquitination. The surprising antagonistic roles of two closely related E3 ubiquitin ligases are required for netrin-1–dependent filopodial responses, axon turning and branching, and fiber tract formation. We suggest a novel model in which coordinated regulation of VASP ubiquitination by a pair of interfering ligases is a critical element of VASP dynamics, filopodial stability, and axon guidance.
Introduction
Axon guidance toward appropriate synaptic partners is critical for the formation of intricate and accurate neuronal networks. The extracellular guidance cues that direct axon navigation are sensed by transmembrane receptors and interpreted via downstream signaling pathways (Kolodkin and Tessier-lavigne, 2011). Guidance receptors often localize to the tips of actin-rich filopodial protrusions in the axonal growth cone, a dynamic, cytoskeleton-rich structure at the distal end of extending axons (Shekarabi and Kennedy, 2002). Directionally biased remodeling and movement of growth cones, coupled with progressive elongation and condensation of axons, produces the turning behavior in axon guidance (Plachez and Richards, 2005). Turning of the growth cone requires tight spatial and temporal regulation of effectors, such as cytoskeletal remodeling proteins (Dent et al., 2011). However, the mechanisms allowing for highly localized regulation in the growth cone are not understood, especially those that allow for rapid alteration of protein function and cytoskeletal dynamics in response to extracellular cues.
The guidance cue netrin-1 and its receptor DCC are required for midline-crossing behavior of many central nervous system axons, including those in the corpus callosum of the placentalian brain (Kennedy et al., 1994; Serafini et al., 1996; Fazeli et al., 1997; Fothergill et al., 2014; Bin et al., 2015). A large body of work demonstrates the function of signaling and cytoskeletal proteins activated during responses to netrin-1 (Boyer and Gupton, 2018). For example, Ena/vasodilator stimulated phosphoprotein (VASP) actin polymerases are critical to filopodia formation and maintenance in neurons (Dent et al., 2007; Kwiatkowski et al., 2007), particularly downstream of DCC and netrin-1 (Lebrand et al., 2004). Recent work demonstrated that negative regulation of downstream effectors primes the neuron for appropriate netrin response (Menon et al., 2015; Plooster et al., 2017). For example, E3 ubiquitin ligase tripartite motif protein 9 (TRIM9) is required for ubiquitination and inhibition of the actin polymerase VASP (Menon et al., 2015). VASP ubiquitination negatively impacts filopodia stability (Menon et al., 2015), a metric of growth cone response to extracellular cues such as netrin-1 (Dent et al., 2004; Gupton and Gertler, 2007; Lebrand et al., 2004). Loss of VASP ubiquitination is necessary for growth cone filopodial response to netrin-1 (Menon et al., 2015); however, the factors that inhibit VASP ubiquitination or promote VASP deubiquitination in the presence of netrin are unknown; furthermore, regulators of TRIM9 have not been identified.
TRIM67 is a class 1 TRIM protein along with TRIM9, sharing identical domain organization and 63.3% sequence identity (Short and Cox, 2006). Our recent work described a line of mice lacking Trim67 and showed that TRIM67 is required in vivo for the appropriate development of several axon tracts including the netrin-sensitive corpus callosum (Boyer et al., 2018). Additionally, we found that TRIM67 interacts with both TRIM9 and the netrin-1 receptor DCC. Little is known about the cellular function of TRIM67, although a previous study in neuroblastoma cells reported TRIM67-dependent ubiquitination of 80K-H, a negative regulator of a Ras protein (Yaguchi et al., 2012). No role has been described for TRIM67 in the regulation of axon guidance.
Here we describe a surprising, antagonistic role for TRIM67 in the ubiquitination of VASP in murine embryonic cortical neurons and demonstrate that appropriate regulation of VASP ubiquitination is required for filopodial and axonal responses to netrin-1. We demonstrate that TRIM67 interacts with the actin polymerase VASP and negatively regulates its TRIM9-dependent ubiquitination. We provide evidence that this antagonism requires TRIM67 ligase function and involves competitive inhibition of the interaction between TRIM9 and VASP. We show that genetic deletion of Trim67 results in increased VASP ubiquitination, basal defects in filopodia dynamics, and loss of acute filopodial and growth cone responses to netrin. Additionally, netrin-dependent axon turning and branching are impaired by Trim67 deletion. We extend these in vitro findings in vivo, where we find a perinatal delay in the developmental completion of the netrin-sensitive corpus callosum. Together, these findings suggest that a pair of closely related TRIMs generate a “yin and yang”-like modulation of VASP function in growth cones, allowing filopodia to effectively search their environment and providing fidelity in netrin-1–dependent axon guidance.
Results
TRIM67 is involved in netrin-dependent axon guidance
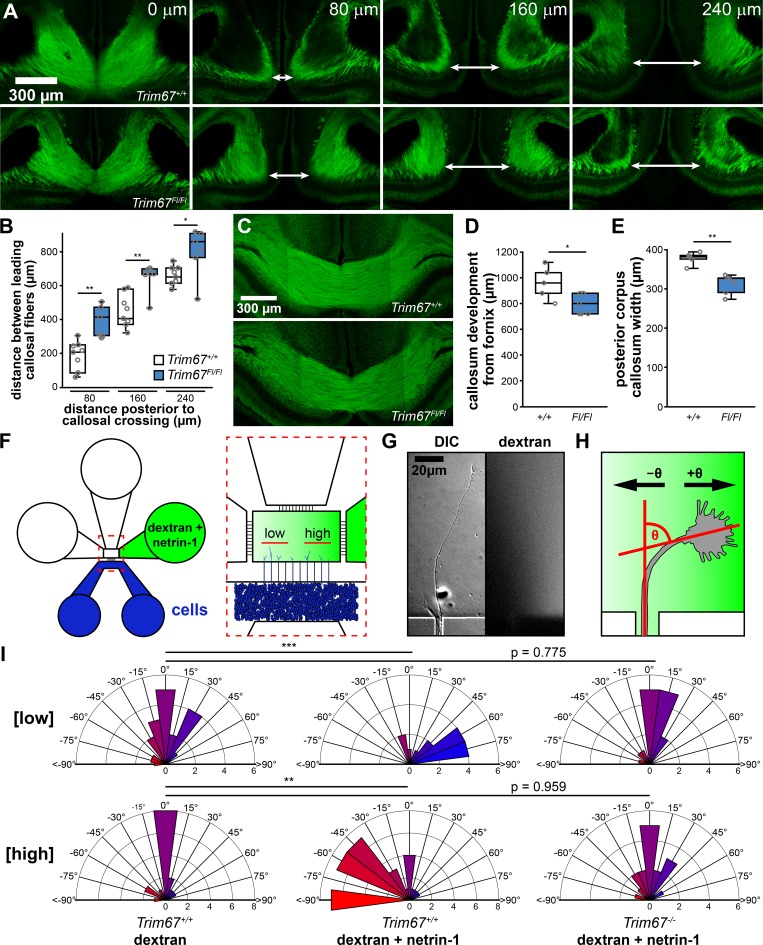
We recently generated mice carrying a Trim67 allele with exon 1 flanked by loxP sites (Trim67Fl/Fl). Germline deletion of Trim67 resulted in a thinner and smaller corpus callosum in the adult (Boyer et al., 2018). In light of this phenotype, we examined the perinatal development of the callosum in newborn (postnatal day 0 [P0]) and P4 Trim67+/+:Nex-Cre:Tau-Lox-STOP-Lox-GFP and Trim67Fl/Fl:Nex-Cre:Tau-Lox-STOP-Lox-GFP littermates. The Nex promoter drives Cre expression, recombination, and GFP expression in excitatory, postmitotic cortical and hippocampal neurons starting at embryonic day 11.5 (E11.5; Goebbels et al., 2006). We analyzed corpus callosum development in serial coronal sections (Fig. 1 A) at P0. Deletion of Trim67 widened the interhemispheric distance between leading callosal fibers (Fig. 1 A, arrows) 80, 160, and 240 µm posterior to the last midline-crossing section of the corpus callosum (Fig. 1 B, P = 0.0055, P = 0.008, and P = 0.045, respectively), suggesting there was a delay in axon extension toward the midline in Trim67Fl/Fl brains. Concordantly, corpus callosum growth was delayed toward the posterior of the brain at P0 (length of corpus callosum from fornix; Trim67+/+ 453 ± 22 µm, Trim67Fl/Fl 360 ± 25 µm; P = 0.034). We performed similar sectioning and staining at P4 (Fig. 1 C), when the callosum completed midline crossing (Wahlsten, 1984). The callosum extended a shorter distance caudally in Trim67Fl/Fl littermates (Fig. 1 D, P = 0.040), and the posterior portion of the callosum was thinner (Fig. 1 E, P = 0.008). These data suggest that TRIM67 is required for the midline-directed outgrowth and/or guidance of cortical axons that form the corpus callosum. A table summarizing population size, mean, and deviance and statistical tests for all panels in this figure and subsequent figures is included (Table S1).
Figure 1.
Trim67 is required for axonal development and guidance in vivo and in vitro. (A) Confocal micrographs of serial coronal sections of GFP in the corpus callosum of murine brains fixed at P0 from Trim67+/+:Nex-Cre:Tau-Lox-STOP-Lox-GFP and Trim67Fl/Fl:Nex-Cre:Tau-Lox-STOP-Lox-GFP littermates. Arrows demarcate leading fibers of the corpus callosum in sections 80, 160, and 240 µm posterior to the final connection of the callosum. (B) Individual data points and box plots of the distance between leading fibers of the corpus callosum at P0. n (animals) = 8 Trim67+/+:Nex-Cre:Tau-Lox-STOP-Lox-GFP, 6 Trim67Fl/Fl:Nex-Cre:Tau-Lox-STOP-Lox-GFP. (C) Confocal micrographs of coronal sections of the corpus callosum at P4, 160 µm anterior to the final connection of the callosum. (D and E) Quantification of the extent of callosal development expressed as individual data points and box plots of the distance from the fornix to the separation of the callosal leading fibers (D) and callosal width at the midline eight sections posterior to the fornix at P4 (E). n (animals) = 5 +/+, 6 Fl/Fl. (F) Schematic of microfluidic axon guidance chambers. (G) Example axon extending from a microgroove into the axon guidance chamber, and fluorescent dextran used to visualize the gradient. (H) Diagram depicting axon turning angle measured between a line bisecting the axonal growth cone and a line overlapping and parallel to the axon as it exits microgroove. (I) Rose plots of embryonic cortical neuron axon turning angles in a gradient of fluorescent dextran or dextran + netrin-1. Low concentration denotes the four microgrooves furthest from the gradient source, while the four microgrooves closest to the source are denoted high concentration (Taylor et al., 2015). Positive turning angles represent axon turning toward the netrin-1 source; negative angles represent axon turning away from the source. Three experiments per genotype/treatment condition. n (axons) = 20 +/+ low [dextran], 17 +/+ high [dextran], 23 +/+ low [netrin], 43 +/+ high [netrin], 15 −/− low [netrin], 17 −/− high [netrin]. *, P < 0.05; **, P < 0.01. Box plots are minimum, Q1, Q2, Q3, maximum.
Since loss of murine Dcc or the gene encoding netrin-1 (Ntn1) results in the failure of formation or agenesis of the corpus callosum (Serafini et al., 1996; Fazeli et al., 1997; Bin et al., 2015; Marsh et al., 2017), and our previous work revealed that TRIM67 interacts with DCC (Boyer et al., 2018), we hypothesized that the TRIM67-dependent defects in the corpus callosum could arise from axon guidance failures. To investigate the role of TRIM67 in netrin-dependent axon turning, we employed microfluidic axon guidance chambers to establish a stable gradient of netrin-1 (Taylor et al., 2015) and measured axon turning angles of cortical neurons cultured from Trim67+/+ and Trim67−/− embryos (Fig. 1, F–I). As previously reported, in Trim67+/+ cells, we observed positive turning angles, indicative of attractive turning, in a low concentration range of the netrin-1 gradient (∼40–220 ng/ml), but not in a dextran-only gradient (Fig. 1 I). At the higher end of the gradient (∼400–600 ng/ml), Trim67+/+ axons exhibited negative turning angles, indicative of repulsion from netrin-1. However, axons of Trim67−/− neurons did not show a turning response to netrin-1 in either range (Fig. 1 I). These results demonstrate a requirement for TRIM67 in netrin-1–dependent axon turning in vitro, consistent with in vivo defects in the formation of the netrin-sensitive corpus callosum.
Netrin increases TRIM67 localization to filopodia tips
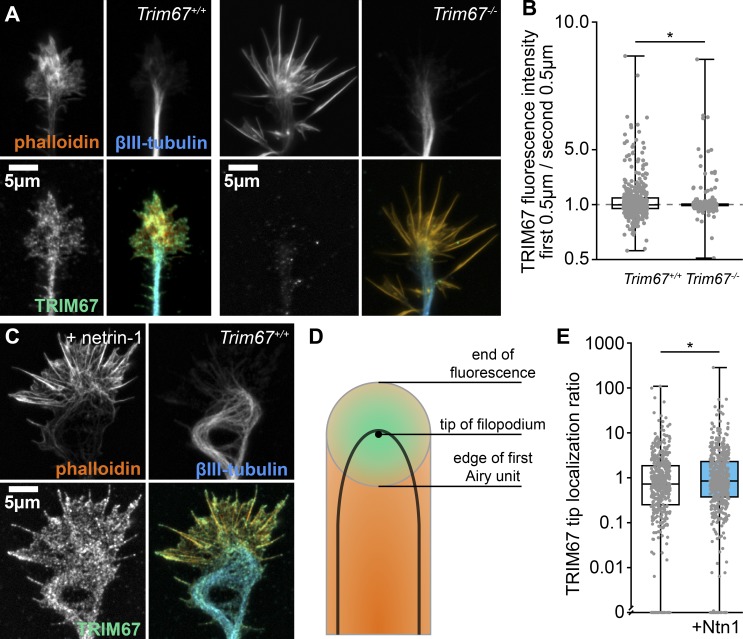
TRIM67 is enriched in the developing cortex and present in axonal growth cones (Boyer et al., 2018). We next characterized the netrin sensitivity of TRIM67 localization in the growth cone. TRIM67 exhibited a punctate pattern of localization in axonal growth cones, specifically in the core region, the edge of the lamellipodium, and the distal 0.5 µm of filopodia tips (Fig. 2, A and B). A similar tip localization is apparent for myc-tagged TRIM67 expressed in embryonic cortical neurons and imaged by structured illumination microscopy (SIM; Fig. S1 A). This is reminiscent of the localization of filopodial tip complex proteins such as TRIM9 (Menon et al., 2015), VASP (Lebrand et al., 2004), and DCC (Shekarabi and Kennedy, 2002), although TRIM67 is not present in all filopodia. Intriguingly, acute netrin-1 treatment enhanced the filopodial tip localization of TRIM67 (Fig. 2, C–E, P = 0.035), suggesting that TRIM67 is located closer to filopodia tips following netrin treatment. This suggests that TRIM67 is poised to modulate netrin-1–dependent axonal responses.
Figure 2.
TRIM67 localization to filopodia tips is enhanced by netrin-1. (A) Immunocytochemistry (ICC) of filamentous actin (phalloidin), β-III-tubulin, and TRIM67 in axonal growth cones of primary neurons isolated from Trim67+/+ and Trim67−/− embryonic cortices. (B) Individual data points and box plots of TRIM67 fluorescence intensity in the first 0.5 µm from the tip of the filopodium to the next 0.5 µm. Three experiments per genotype. n (filopodia) = 463 +/+, 440 −/−. (C) ICC of an axonal growth cone from a Trim67+/+ cortex treated with netrin-1. (D) Diagram showing the Airy disk of a fluorescent protein at the tip of a filopodium (green) and fluorescence of a protein along the filopodium (orange). (E) Individual data points and box-and-whisker plots of tip proximity of TRIM67 in filopodia, quantified as the fluorescence ratio of the center to the edge of the first Airy unit. Three experiments per genotype. n (filopodia) = 444 media, 535 netrin. *, P < 0.05. Data in E are presented on a logarithmic scale due to the presence of large values that obscure the population center, with an axis break to allow display of measures of 0. Box plots are minimum, Q1, Q2, Q3, maximum.
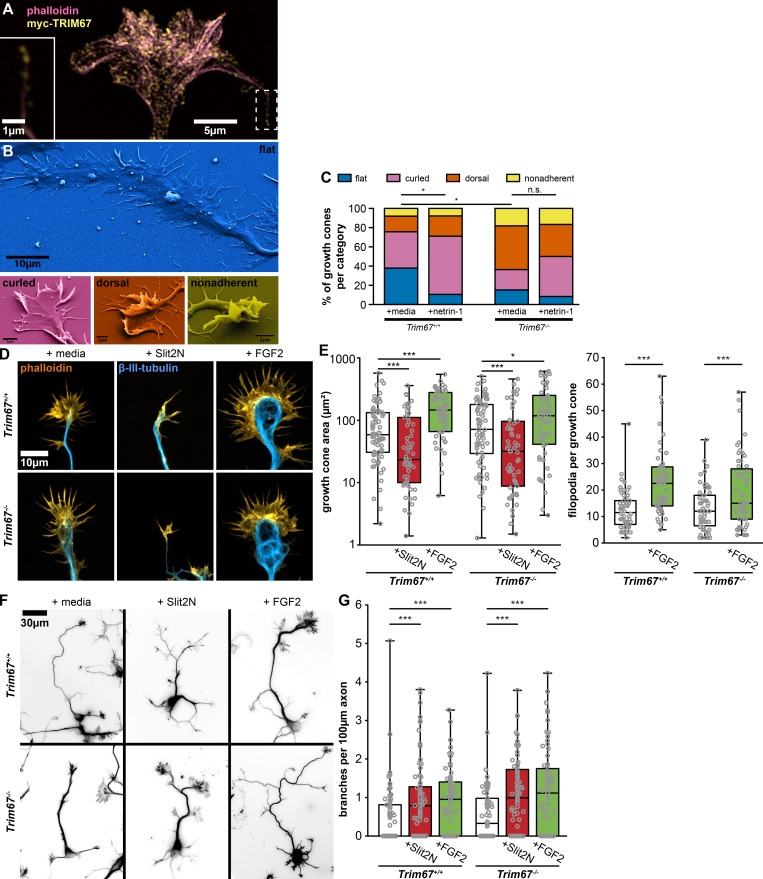
Figure S1.
TRIM67 growth cone response to morphogens. (A) SIM image of myc-TRIM67 and filamentous actin in an axonal growth cone. TRIM67 localizes to the tip of a filopodium (inset, same area as dashed box). (B) Examples of growth cone morphological categories as identified by scanning electron microscopy. (C) Quantification of growth cone morphology distributions of Trim67+/+ and Trim67−/− cortical neurons treated with media or netrin-1. Distributions are compared by Fisher’s exact test. n (cultures) = 3. n (cells) = 37 +/+ media, 38 +/+ netrin, 33 −/− media, 36 −/− netrin. (D) Examples of cortical embryonic axonal growth cones treated with media, Slit2N, or FGF2. (E) Individual data points and box-and-whisker plots showing the data spread in the interquartile range (box) and minimum and maximum (whiskers) of growth cone area and filopodia per growth cone following 40 min of treatment with the indicated guidance cues. n (cultures) = 3. n (cells) = 76 +/+ media, 56 +/+ Slit2N, 56 +/+ FGF2, 76 −/− media, 57 −/− Slit2N, 57 −/− FGF2. (F) Inverted images of neurons combining staining of both filamentous actin (phalloidin) and β-III-tubulin, treated for 24 h with media, Slit2N, or FGF2. (G) Individual data points and box-and-whisker plots showing the data spread in the interquartile range (box) and minimum and maximum (whiskers) of axon branching in response to the indicated guidance cues. n (cultures) = 3. n (cells) = 78 +/+ media, 79 +/+ Slit2N, 71 +/+ FGF2, 73 −/− media, 76 −/− Slit2N, 76 −/− FGF2. *, P < 0.05; ***, P < 0.005; n.s., not significant. Box plots are minimum, Q1, Q2, Q3, maximum.
Axonal netrin-1 responses require TRIM67
We exploited scanning EM of cultured neurons to acquire a 3D gross overview of growth cone responses to netrin and potential defects in Trim67−/− neurons (Fig. 3 A). We hierarchically categorized the 3D morphology of the growth cones as flat (fully apposed to coverslip), curled (peripheral structures away from coverslip), dorsal (possessing lamellipodial ruffles or filopodia on the dorsal surface), and finally, nonadherent (growth cone fully separated from coverslip; Fig. S1 B). Netrin-1 treatment promoted nonflat growth cone morphologies in Trim67+/+ neurons (Fig. S1 C, P = 0.040 by Fisher’s exact test). The growth cones of Trim67−/− axons were shifted toward nonflat morphologies (P = 0.011), and this distribution was not affected by netrin-1 (P = 0.310). These data suggest that addition of netrin-1 alters growth cone adhesion and/or protrusion in a TRIM67-dependent fashion, and further that deletion of Trim67 alters growth cone adhesion and protrusion basally.
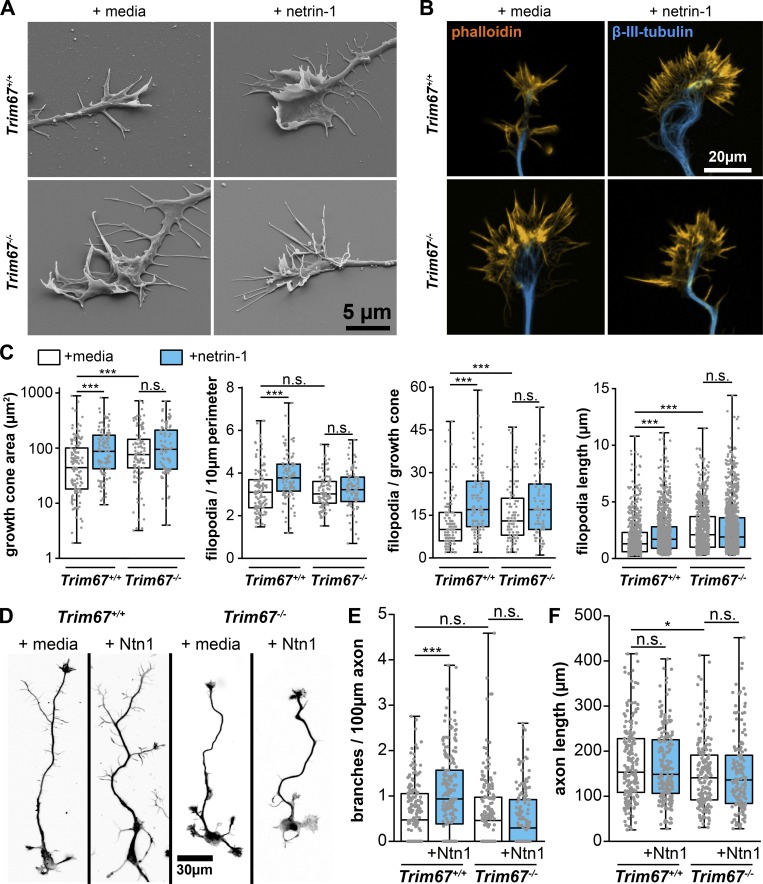
Figure 3.
TRIM67 is required for axon and growth cone responses to netrin-1. (A) Scanning electron micrographs of axonal growth cones from embryonic Trim67+/+ and Trim67−/− cortices treated with sham media or media containing netrin-1 for 40 min after 2 d in vitro. (B) Fluorescent micrographs of growth cones from primary neuronal cultures stained for filamentous actin (phalloidin) and β-III-tubulin. (C) Individual data points and box plots of growth cone responses after acute netrin-1 treatment (40 min), including increase in growth cone area, filopodial density, filopodia number, and filopodia length. Three experiments per genotype/treatment. n (cells) = 127 +/+ media, 103 +/+ netrin, 98 −/− media, 83 −/− netrin. For filopodia length, n (filopodia) = 729 +/+ media, 956 +/+ netrin, 786 −/− media, 1,076 −/− netrin. (D) Fluorescent micrographs of neurons cultured for 3 d in vitro including a final 24 h with addition of media or netrin-1, shown as the combined fluorescence of staining for both filamentous actin (phalloidin) and β-III-tubulin. (E and F) Individual data points and box plots of axon branching per 100-µm axon length (E) and of total axon length (F). Six experiments per genotype/treatment. n (cells) = 188 +/+ media, 186 +/+ netrin, 178 −/− media, 156 −/− netrin. *, P < 0.05; ***, P < 0.005; n.s., P > 0.05. Box plots are minimum, Q1, Q2, Q3, maximum.
The nonresponsiveness of Trim67−/− growth cone morphology to netrin-1 prompted a thorough assessment of growth cone responses to netrin, including size, filopodial number, and filopodial length (Fig. 3, B and C). As reported (Lebrand et al., 2004; Menon et al., 2015), netrin-1 treatment increased growth cone area (P = 5.34 × 10−4), filopodia density (P = 7.08 × 10−7), filopodia number (P = 1.64 × 10−7), and filopodial length (P = 7.97 × 10−12; Fig. 3 C). However, netrin-1 treatment had no effect on these parameters in Trim67−/− growth cones (area, P = 0.896; filopodial density, P = 0.434; filopodial number, P = 0.218; filopodial length, P = 0.808). This indicates that TRIM67 is required for growth cone responses to netrin-1. Basally, filopodia of Trim67−/− growth cones were longer (P = 4.27 × 10−23) and growth cones were larger (P = 0.007) than Trim67+/+ counterparts. We investigated whether TRIM67 was similarly required for responses to other guidance cues (Fig. S1 D). As reported (Szebenyi et al., 2001), treatment with fibroblast growth factor 2 (FGF2) increased growth cone area and number of filopodia (Fig. S1 E; area, P = 8.13 × 10−5; filopodia, P = 2.38 × 10−7). These effects were also observed in Trim67−/− neurons (Fig. S1 E; area, P = 0.034; filopodia, P = 0.007). Slit2N treatment caused a collapse of growth cones, as indicated by a decrease in the growth cone area in both Trim67+/+ (P = 0.003) and Trim67−/− (P = 0.001) neurons (Fig. S1 E).
Netrin also promotes axon branching in cortical neurons (Dent et al., 2004; Winkle et al., 2014). We assessed whether TRIM67 was required for axon branching after a 24-h addition of netrin (Fig. 3, D and E). Netrin-1 increased the density of branches along Trim67+/+ axons (P = 9.43 × 10−7). In Trim67−/− axons, this branching response was absent (P = 0.543), and there was no change in the branch density of untreated axons (P = 0.423). Basally, Trim67−/− axons were shorter than Trim67+/+ counterparts (P = 0.0133), although there was no effect on axon length from netrin-1 treatment in either genotype (Fig. 3 F, Trim67+/+, P = 0.711; Trim67−/−, P = 0.858). To determine whether the regulation of axon branching by TRIM67 was specific to netrin-1, we treated neuron cultures with FGF2 or Slit2N (Fig. S1 F). As reported (Szebenyi et al., 2001; Wang et al., 1999), both morphogens increased axon branching in Trim67+/+ cortical neurons (Fig. S1 G; FGF2, P = 2.78 × 10−4; Slit2N, P = 0.00175). Axons of Trim67−/− neurons were also more branched following treatment with FGF2 (P = 7.43 × 10−4) or Slit2N (P = 0.00173). Along with the results of growth cone analysis, these data suggest that TRIM67 is not required for axonal responses to all guidance cues, but may be specific to netrin-1.
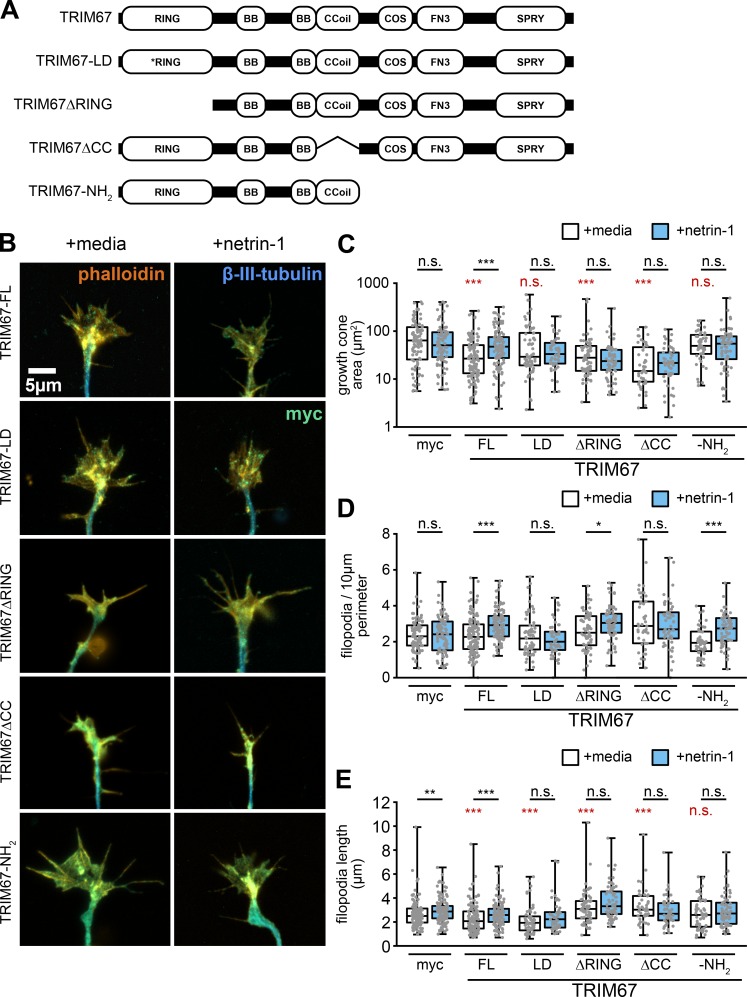
Functional analysis of TRIM67 protein domains
After establishing the necessity for TRIM67 in axonal responses to netrin-1, we performed rescue experiments using full-length TRIM67 or domain mutants of TRIM67 (Fig. S2 A). The localization of most constructs was qualitatively similar to the full-length protein (Fig. S2 B); however, qualitatively the TRIM67-ΔCC and TRIM67-NH2 constructs appeared more diffuse. Only full-length TRIM67 rescued the increase in growth cone area in response to netrin-1 treatment (Fig. S2 C, P = 2.40 × 10−5). Full-length TRIM67 also rescued the increase in filopodial density following netrin-1 treatment (Fig. S2 D; P = 5.36 × 10−7). Intriguingly, the TRIM67-NH2 construct also rescued netrin sensitivity of filopodia density (P = 2.65 × 10−5), whereas the ΔCC did not, suggesting that the coiled-coil domain may be critical to the filipodial regulatory functions of TRIM67. Most constructs did reduce the basal filopodial length, but did not necessarily rescue the netrin-dependent increase in length (Fig. S2 E). These data suggest that many domains of TRIM67, as well as ligase activity, are necessary to fully rescue growth cone responses to netrin-1.
Figure S2.
Multiple domains of TRIM67 are required to fully rescue growth cone response to netrin-1. (A) TRIM67 constructs used in structure–function assays. The RING domain of TRIM proteins contain zinc binding pockets necessary for E3 ubiquitin ligation activity and can mediate oligomerization of TRIM proteins (Freemont, 1993; Koliopoulos et al., 2016; Meroni and Diez-Roux, 2005); we therefore made both a RING-deletion construct (TRIM67ΔRING) and one containing mutations at cysteines 7 and 10 to abolish zinc binding in the RING domain and thus any ligase activity (TRIM67-LD). The coiled-coil (CC) domains of TRIM proteins mediate homo- and heterodimerization with other members of the same TRIM class (Sanchez et al., 2014; Short et al., 2002). In our previous investigation of TRIM9, the CC domain also interacted with the filopodial tip-localized actin polymerase VASP (Menon et al., 2015); therefore, we generated a construct of TRIM67 lacking the coiled-coil domain (TRIM67ΔCC). Finally, we generated a construct possessing only the three N-terminal tripartite motif domains (TRIM67-N). All constructs possessed an N-terminal myc tag. (B) ICC of Trim67−/− growth cones stained for actin, β-III-tubulin, and the indicated myc-TRIM67 construct, showing expression and distribution of each construct. (C–E) Individual data points and box-and-whisker plots showing the data spread in the interquartile range (box) and minimum and maximum (whiskers) of growth cone area (C), filopodial density (n [cells] = 96 myc media, 88 myc netrin, 121 TRIM67 media, 114 TRIM67 netrin, 65 LD media, 52 LD netrin, 68 ΔRING media, 68 ΔRING netrin, 45 ΔCC media, 52 ΔCC netrin, 52 NH2 media, 65 NH2 netrin; D), and filopodia length in neurons expressing each rescue construct (E). All statistical comparisons in red are to myc-expressing, untreated growth cones. n (cells) = 128 myc media, 118 myc netrin, 121 TRIM67 media, 119 TRIM67 netrin, 64 LD media, 50 LD netrin, 68 ΔRING media, 68 ΔRING netrin, 45 ΔCC media, 50 ΔCC netrin, 52 NH2 media, 64 NH2 netrin. n (cultures) = 3. *, P < 0.05; ***, P < 0.005. Box plots are minimum, Q1, Q2, Q3, maximum.
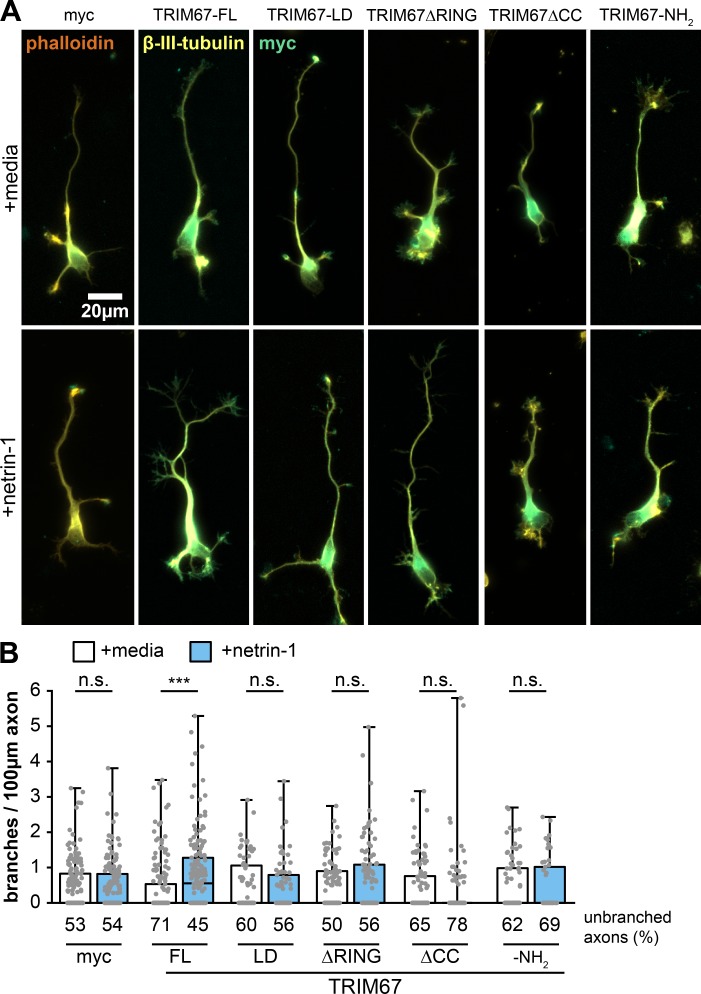
We next explored the function of these TRIM67 domain mutants in netrin-1–dependent axon branching (Fig. S3 A). Axon branching in response to netrin-1 was rescued only by full-length TRIM67 (Fig. S3 B; P = 9.93 × 10−7). Thus, the axonal defects in response to netrin are caused by loss of Trim67. These results suggest that, like growth cone regulation, all domains of TRIM67 are necessary for proper netrin-dependent axon branching. We conclude that TRIM67-dependent regulation of netrin responses is likely complex, as it requires ligase activity and the coiled-coil domain of TRIM67, and thus potentially distinct functions of the protein.
Figure S3.
All domains of TRIM67 are required to rescue netrin-dependent axon branching. (A) ICC of Trim67−/− neurons expressing indicated myc-tagged TRIM67 constructs, stained for myc (green), filamentous actin (phalloidin, red) and β-III-tubulin (yellow). (B) Individual data points and box-and-whisker plots showing the data spread in the interquartile range (box) and minimum and maximum (whiskers) of axon branches per 100-µm axon length. Percentage of neurons with unbranched axons is shown below the x-axis. Note that the percentage of unbranched axons causes many conditions to have a median branching value at 0. All statistical comparisons in red are to myc-expressing, untreated neurons. n (cultures) = 3. n (cells) = 169 myc media, 177 myc netrin, 173 TRIM67 media, 166 TRIM67 netrin, 80 LD media, 77 LD netrin, 108 ΔRING media, 111 ΔRING netrin, 113 ΔCC media, 116 ΔCC netrin, 76 NH2 media, 51 NH2 netrin. ***, P < 0.005. Box plots are minimum, Q1, Q2, Q3, maximum.
Filopodia dynamics are regulated by TRIM67
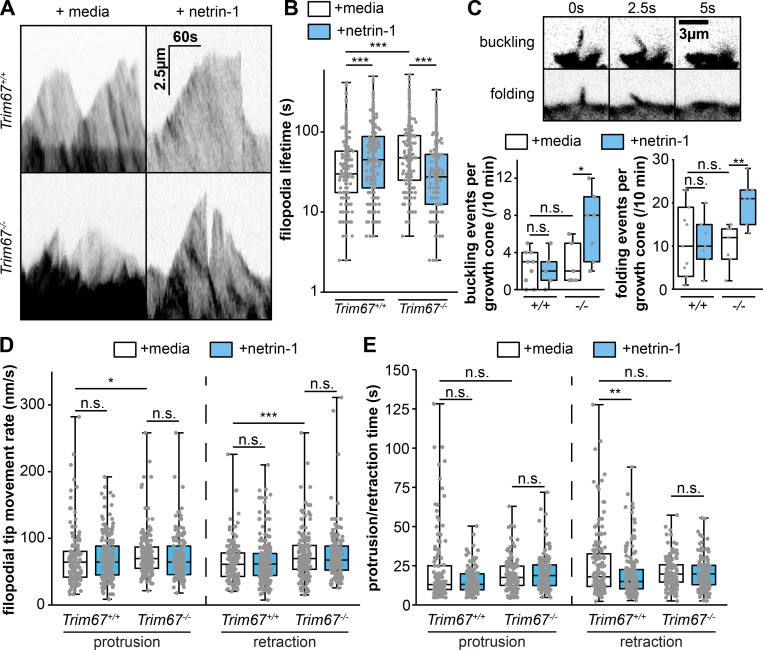
We next assessed the dynamics of axonal growth cone filopodia using time-lapse microscopy (Fig. 4 A and Video 1). In agreement with a previous results (Menon et al., 2015), we found that 40-min treatment with netrin-1 increased the lifetime of filopodia in Trim67+/+ neurons (Fig. 4 B; P = 0.0003). Filopodia lifetime was basally longer in Trim67−/− growth cones (P = 3.63 × 10−6) and reduced by addition of netrin-1 (P = 4.26 × 10−9). This decrease in lifetime could be attributed in part to increased lateral buckling or folding of filopodia following netrin-1 treatment in Trim67−/− neurons (Fig. 4 C; buckling, P = 0.0462; folding, P = 0.0255). Both the protrusion and retraction speed of filopodia tips were higher in Trim67−/− growth cones than in Trim67+/+ growth cones (protrusion, P = 0.00854; retraction, P = 0.00292), but there was no effect on these rates with netrin-1 in either genotype (Fig. 4 D, Trim67+/+ protrusion, P = 0.318; Trim67+/+ retraction, P = 0.882; Trim67−/− protrusion, P = 0.0707; Trim67−/− retraction, P = 0.776). However, the duration of individual filopodial retraction events was shorter in Trim67+/+ growth cones following netrin-1 treatment (P = 0.0148), and this effect was absent in Trim67−/− filopodia (Fig. 4 E; P = 0.884). Together, these data suggest that TRIM67 is required for filopodial dynamics to respond properly to netrin-1.
Figure 4.
TRIM67 regulates filopodial growth and dynamics. (A) Kymographs of filopodia from cultured primary embryonic cortical neurons expressing mCherry. (B) Individual data points and box plots of filopodial lifetime. n (filopodia) = 293 +/+ media, 278 +/+ netrin, 257 −/− media, 226 −/− netrin. (C) Fluorescent micrographs and quantification of filopodial buckling and folding events during the course of 10-min time-lapse of axonal growth cones. (D and E) Individual data points and box-and-whisker plots of rate of filopodial tip protrusions and retractions (D) and duration of individual filopodial protrusion and retraction periods (E) following media sham or netrin treatment. n (events) = protrusion: 142 +/+ media, 193 +/+ netrin, 171 −/− media, 136 −/− netrin; retraction: 151 +/+ media, 226 +/+ netrin, 174 −/− media, 151 −/− netrin. Three experiments per genotype/treatment for all panels. *, P < 0.05; **, P < 0.01; ***, P < 0.005; n.s., P > 0.05. Box plots are minimum, Q1, Q2, Q3, maximum.
Video 1.
Live-cell imaging of filopodial dynamics of axonal growth cones. Cultured embryonic cortical neurons expressing cytoplasmic mCherry, imaged every 2.5 s before or after 40 min of netrin-1 treatment. See Fig. 4.
TRIM67 interacts and localizes with the filopodial actin polymerase VASP
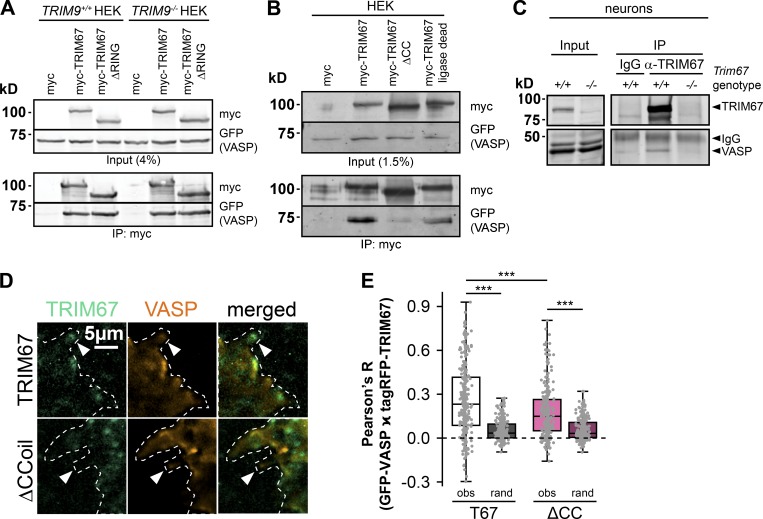
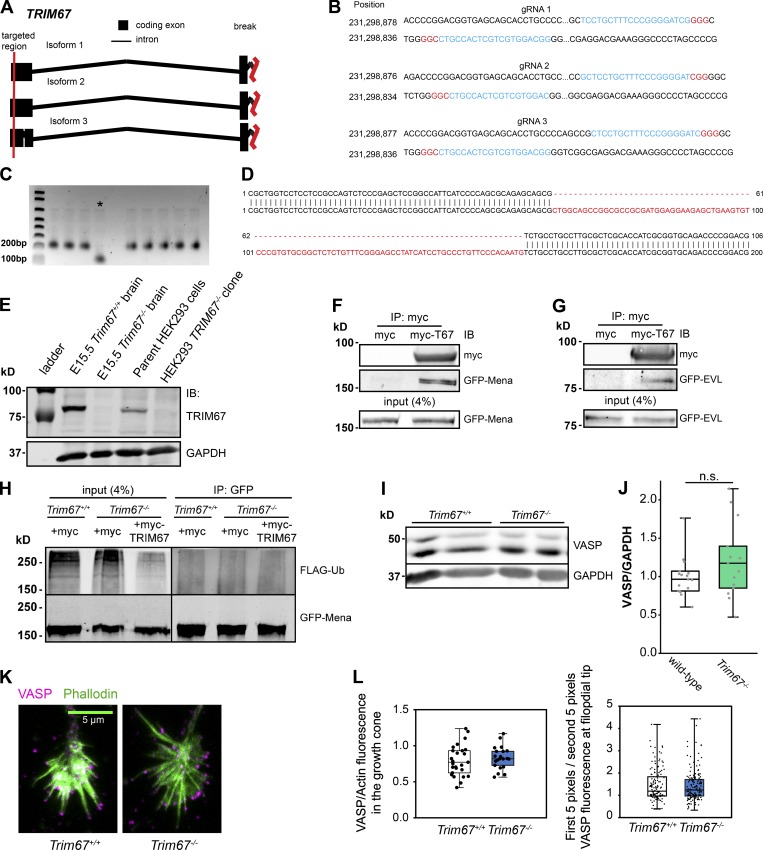
The Ena/VASP family of actin polymerases are required for filopodial responses to netrin-1 (Lebrand et al., 2004) and VASP is specifically regulated by TRIM9-dependent ubiquitination to mediate netrin-1 filopodial response (Menon et al., 2015). Since TRIM9 and TRIM67 are highly similar and interact (Boyer et al., 2018), we hypothesized that TRIM67 may also interact with VASP. Indeed, immunoprecipitation of myc-TRIM67 or a mutant lacking the RING domain (MycTRIM67ΔRING) coprecipitated GFP-VASP from HEK293 cells (Fig. 5 A). The TRIM67:VASP complex was maintained in TRIM9−/− HEK293 cells, indicating that the TRIM67:VASP interaction occurs independently of TRIM9 (Fig. 5 A). To map the domains of TRIM67 necessary for VASP interaction, we generated a HEK293 cell line in which TRIM67 was deleted via CRISPR/Cas9 genome editing (TRIM67−/− HEK293, Fig. S4, A–E) and performed coimmunoprecipitation assays using domain-deletion constructs of TRIM67. The coiled-coil domain of TRIM67 was required for coimmunoprecipitation of VASP, whereas ligase function was not (Fig. 5 B). We confirmed the TRIM67:VASP interaction by immunoprecipitating endogenous TRIM67 from cultured embryonic cortical neurons, which coprecipitated endogenous VASP (Fig. 5 C). We investigated whether tagRFP-tagged TRIM67 and GFP-VASP colocalized in neurons by live total internal reflection fluorescence (TIRF) microscopy (Fig. 5 D). Colocalization was quantified at growth cone filopodia tips and was higher than Fay-randomized controls (Fig. 5 E; RFP-TRIM67, P = 5.57 × 10−30; RFP-TRIM67ΔCC, P = 3.55 × 10−19), indicating that TRIM67 and VASP colocalize at filopodia tips. Consistent with coimmunoprecipitation results suggesting that the coiled-coil domain of TRIM67 is required for interaction with VASP; tagRFP-TRIM67ΔCC showed decreased colocalization with GFP-VASP at filopodial tips (P = 2.17 × 10−5). By coimmunoprecipitation, we also found an interaction between TRIM67 and Ena/VASP family members Mena (Fig. S4 F) and EVL (Fig. S4 G).
Figure 5.
TRIM67 interacts and localizes with VASP. (A) Coimmunoprecipitation assays from TRIM9+/+ or TRIM9−/− HEK293 cells transfected with GFP-VASP and myc or myc-tagged TRIM67 constructs demonstrate an interaction between TRIM67 and VASP that is independent of TRIM9. (B) Coimmunoprecipitation assays from TRIM67−/− HEK293 cells transfected with the indicated TRIM67 and VASP constructs, showing requirement for the TRIM67 coiled-coil domain for the TRIM67:VASP interaction. (C) Immunoprecipitation of endogenous TRIM67 from cultured embryonic cortical neurons, showing coprecipitation of endogenous VASP. (D) Colocalization between GFP-VASP and tagRFP-tagged constructs of TRIM67 in murine embryonic cortical neurons. (E) Individual data points and box plots of the colocalization between VASP and TRIM67 (obs) in filopodia compared with Fay-randomized controls (rand). Three experiments per TRIM67 construct. n (filopodia) = 281 T67, 270 ΔCC. ***, P < 0.005. Box plots are minimum, Q1, Q2, Q3, maximum.
Figure S4.
TRIM67 interacts with all members of the Ena/VASP family and does not regulate VASP protein levels. (A–E) Generation of TRIM67−/− HEK293 cell lines. (A) CRISPR gRNA were designed to target the first exon in all known isoforms of TRIM67. Red vertical lines indicate the regions targeted by the gRNAs. (B) A set of three designed gRNAs (blue) targeting exon 1 of TRIM67. Two complementary gRNAs per set were used with nickase-Cas9 to reduce off target effect potential. The NGG sequence is highlighted in red. (C) PCR of genomic DNA from WT cells (lane 1) and multiple clones indicates a deletion of ∼100 bp in CRISPR clone 3, which is denoted by the asterisk. (D) Sequencing of HEK293 CRISPR clone 3 (top strand) shows a deletion of 94 base pairs in exon 1 of TRIM67. (E) Immunoblot (IB) for TRIM67 and GAPDH in E15.5 murine brain lysate, Trim67−/− murine brain lysate, HEK293 lysates, and CRISPR-Cas9 clone 3 TRIM67−/− HEK293 lysates. (F) Coimmunoprecipitation assay from TRIM67−/− HEK293T cells transfected with GFP-Mena and myc or myc-TRIM67. (G) Similar coimmunoprecipitation with GFP-EVL. (H) Immunoprecipitation of GFP-tagged Mena from denatured TRIM67−/− HEK293T cell lysate coexpressing FLAG-Ub and indicated myc or myc-TRIM67 reveals no heavy molecular weight Mena+ bands or change in ubiquitin levels among conditions. (I) Representative Western blot of VASP in embryonic cortical lysate from two animals of each indicated genotype. (J) Individual data points and box-and-whisker plots showing the data spread in the interquartile range (box) and min and max (whiskers) of VASP protein levels measured by Western blotting of embryonic cortical lysates, normalized to GAPDH. n (embryos) = 16 +/+, 14 −/−. (K) Maximum-projection immunofluorescence images demonstrating VASP abundance and localization in Trim67+/+ and Trim67−/− cortical neurons cultured 2 d in vitro. (L) Individual data points and box-and-whisker plots showing the data spread in the interquartile range (box) and minimum and maximum (whiskers) of total VASP fluorescence from three individual experiments, normalized to phalloidin fluorescence in the growth cone (left; n [cells] = 27 +/+, 21 −/−) and VASP localization to the filopodial tip (right; n [filopodia] = 140 +/+, 169 −/−). Box plots are minimum, Q1, Q2, Q3, maximum.
TRIM67 antagonizes VASP ubiquitination
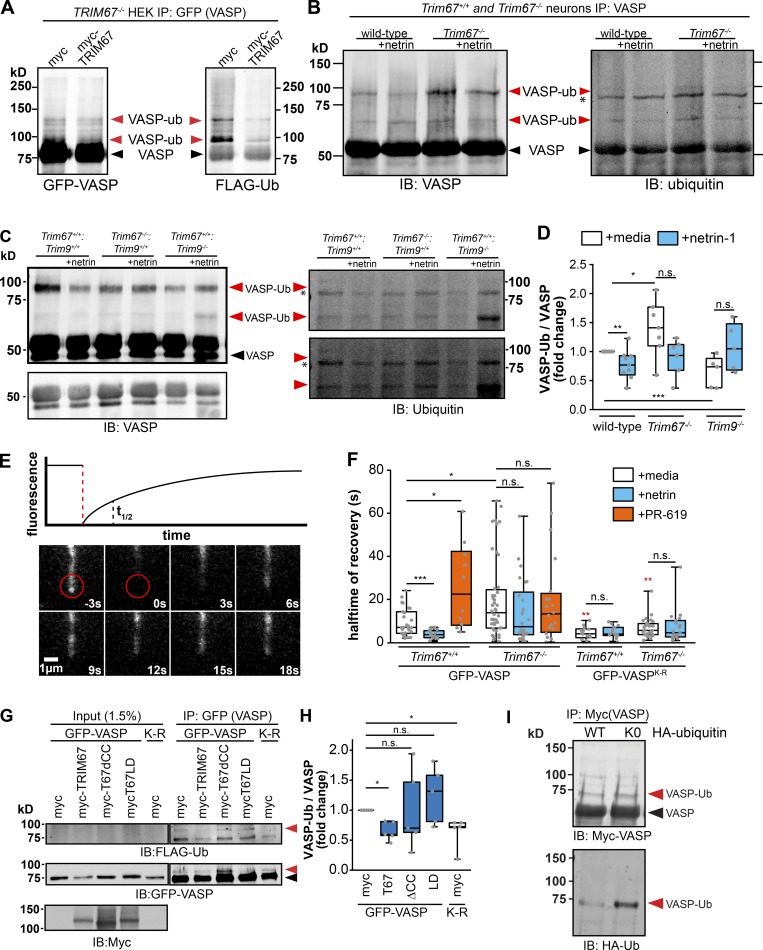
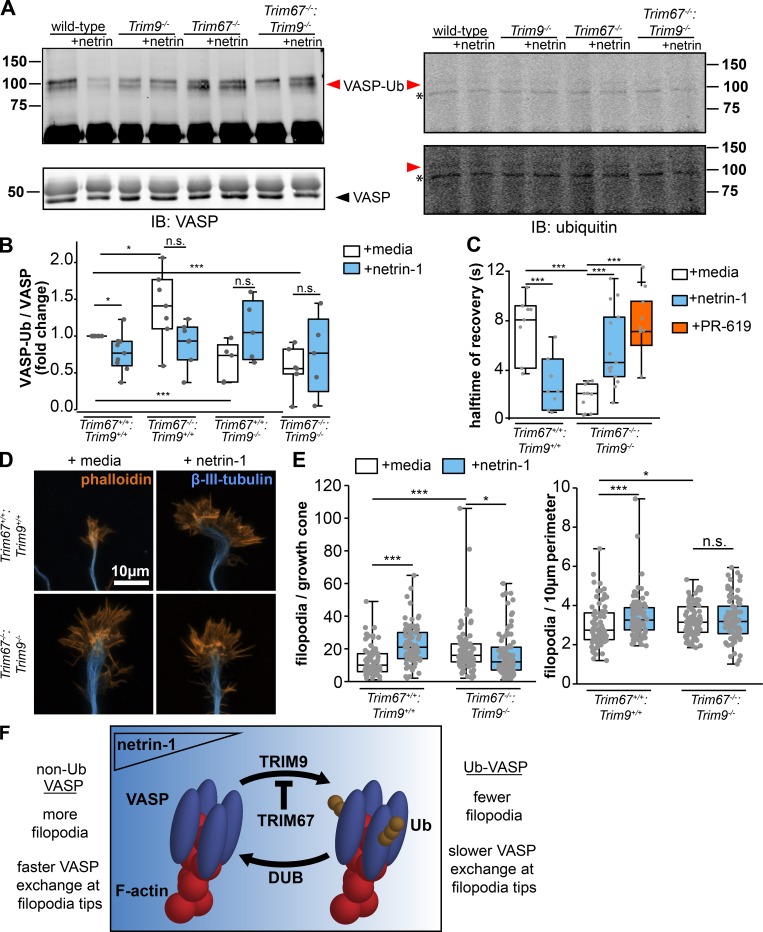
We previously reported that TRIM9-dependent nondegradative ubiquitination of VASP negatively impacted VASP dynamics and filopodial stability, all of which were reversed by netrin (Menon et al., 2015). Ubiquitination of other Ena/VASP family members Mena and EVL was not detected, and similar results were found here with Mena (Fig. S4 H). Due to the structural similarities between TRIM9 and TRIM67 and their conserved coiled-coil mediated interaction with VASP, we investigated how TRIM67 modulated VASP ubiquitination. After a denaturing immunoprecipitation of GFP-VASP from TRIM67−/− HEK293 cells expressing either Myc or MycTRIM67, we observed GFP-VASP at the expected molecular weight (∼75 kD; Fig. 6 A, black arrowheads) and higher molecular weight GFP+ bands that comigrated with Flag-ubiquitin (Fig. 6 A, red arrowheads). These comigrating, high molecular weight bands were interpreted as ubiquitinated VASP (VASP-Ub). This suggested the surprising result that the presence of TRIM67 reduced VASP-Ub. To confirm that endogenous VASP-Ub in neurons demonstrated similar sensitivity to TRIM67, we performed analogous experiments: denaturing immunoprecipitation of VASP from cultured cortical neurons (Fig. 6, B and C). Although detection of ubiquitination of endogenous VASP is more difficult than of VASP expressed in HEK293 cells, consistent with previous work, we found a basal level of VASP-Ub in Trim67+/+ neurons, which decreased following netrin-1 treatment (Fig. 6 D; P = 0.008). Similar to HEK293 cells, in Trim67−/− neurons there was increased VASP-Ub (P = 0.018), but VASP-Ub did not change following netrin-1 treatment. Further, we confirmed previous results that VASP-Ub was TRIM9 dependent, as it decreased in the absence of Trim9 (P = 0.002).
Figure 6.
TRIM67 inhibits the ubiquitination and dynamics of the actin polymerase VASP. (A) Western blot of GFP-VASP immunoprecipitated from denatured TRIM67−/− HEK293 cells expressing myc or MycTRIM67, showing GFP+ bands that comigrate and colabel with ubiquitin (VASP-Ub, red arrowheads), which are heavier than unmodified GFP-VASP (black arrowheads). (B and C) Western blot of VASP immunoprecipitated from denatured cultured embryonic cortical lysate, showing bands that comigrate and colabel with ubiquitin (VASP-Ub, red arrowheads), which migrate at an apparent heavier molecular weight than unmodified VASP (black arrowhead). Asterisk (*) by ubiquitin immunoblot (IB) marks a nonspecific ubiquitin band in all lanes. We note that the endogenous ubiquitinated species are more difficult to detect than ubiquitination of overexpressed and tagged proteins, potentially due to lower protein expression and antibody quality. As recognized in the field, endogenous ubiquitination of proteins is notoriously difficult to detect, particularly when the substrate is not modified by multiple ubiquitins (poly-ubiquitinated). Further, acquiring sufficient material from timed pregnant litters of multiple genotypes is limiting. We thus provide two examples of endogenous ubiquitination assays and multiple intensity exposures to better highlight the bands of interest. (D) Individual data points and box plots of VASP-Ub relative to total VASP levels, normalized to untreated WT of each experiment. Bars are averages of five to seven experiments. n (cultures) = 9 +/+, 5 Trim9−/−, 7 Trim67−/−. (E) Diagram of a fluorescence recovery curve following photobleaching of GFP-VASP at the tip of a filopodium with a representation of the halftime of recovery (t1/2), alongside an image montage of the FRAP of GFP-VASP in a transfected embryonic cortical neuron. (F) Quantification of the FRAP t1/2 of GFP-VASP or GFP-VASPK-R in embryonic cortical neurons treated with netrin-1 or the deubiquitinase inhibitor PR-619. Statistical comparisons in red are with respect to the GFP-VASP FRAP t1/2 in untreated cells of the same genotype. Three to five experiments per genotype/treatment. n (cells) = GFP-VASP: 27 +/+ media, 16 +/+ netrin, 10 +/+ PR-619, 28 −/− media, 26 −/− netrin, 20 −/− PR-619; GFP-VASP(K-R): 14 +/+ media, 15 +/+ netrin, 28 −/− media, 19 −/− netrin. (G) Ubiquitination-precipitation assays of GFP-VASP expressed in HEK293T cells lacking TRIM67 expressing indicated myc-TRIM67 constructs, along with FLAG-ubiquitin. A VASP band that comigrates with FLAG-Ub (red arrowhead) appears heavier than unmodified VASP (black arrowhead). (H) Individual data points and box plots of VASP-Ub (FLAG signal relative to total GFP-VASP) normalized to the myc control condition. n (cultures) = 5 for all conditions. (I) Western blot of Myc-VASP immunoprecipitated from denatured HEK293 cells expressing HA-ubiquitin (WT) or HA-ubiquitin knockout (K0), showing a single predominant Myc+ species that comigrates and colabels with HA-ubiquitin (VASP-Ub, red arrowheads), which is heavier than unmodified Myc-VASP (black arrowhead). *, P < 0.05; **, P < 0.01; ***, P < 0.005; n.s., P > 0.05. Box plots are minimum, Q1, Q2, Q3, maximum.
Experiments in both neurons and HEK cells consistently demonstrated that VASP-Ub ran at a heavier apparent molecular weight than unmodified VASP, yet high molecular weight smears of ubiquitin consistent with polyubiquitination were not observed. Changes in levels of VASP protein were not detected in Trim67−/− or Trim9−/− brain lysates (Fig. S4, I and J), consistent with no degradation of VASP. However, the small proportion of total VASP that was ubiquitinated might impair detection of loss of protein. To examine if loss of Trim67 and the associated increase in VASP-Ub caused local changes to VASP protein levels in the growth cone, we analyzed endogenous VASP localization by immunocytochemistry. This demonstrated no change in VASP protein levels of Trim67+/+ and Trim67−/− growth cones or filopodia (Fig. S4, K and L), potentially consistent with ubiquitination not leading to degradation of this local pool of VASP. We performed ubiquitination assays with Myc-VASP in HEK293 cells expressing either hemagglutinin (HA)-ubiquitin or an HA-ubiquitin mutant that precludes ubiquitin chain formation (HA-ubiquitinK0). A single predominant species of VASP-Ub was apparent in both conditions (Fig. 6 I), further supporting that VASP is not polyubiquitinated, but rather is mono- or multi-monoubiqutinated. In light of the well-documented, unexpected apparent molecular weight shift of VASP caused by phosphorylation (∼4 kD; Reinhard et al., 1992; Butt et al., 1994), we hesitate to suggest the number of ubiquitin moieties ligated to VASP-Ub based on apparent molecular weight shifts.
VASP ubiquitination slows VASP dynamics at filopodia tips
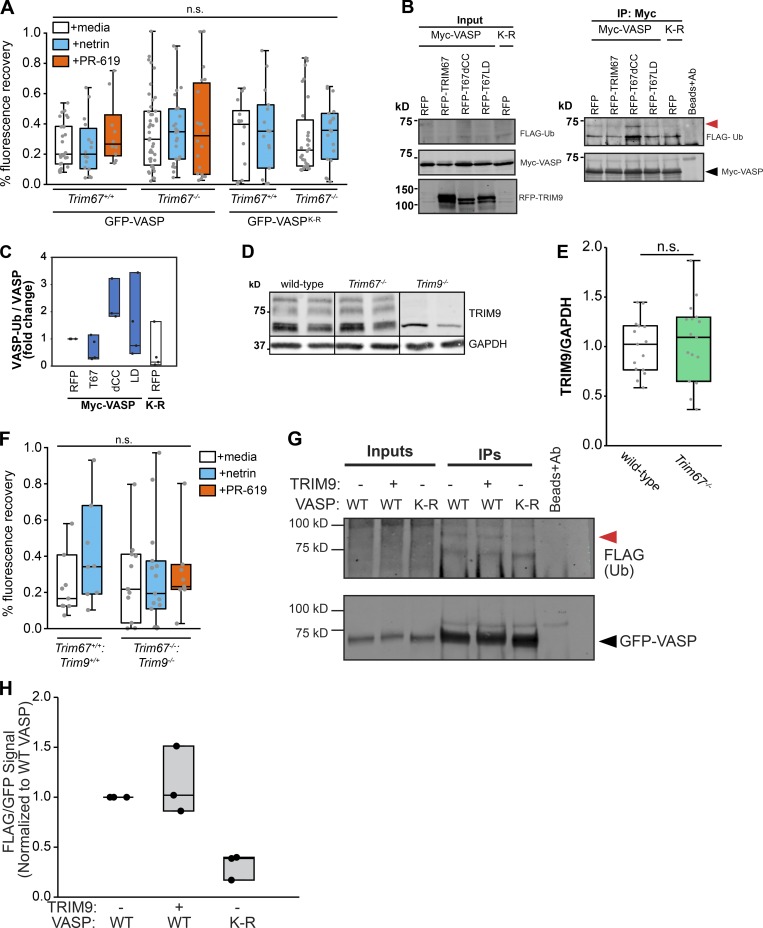
Fluorescence recovery after photobleaching (FRAP) experiments demonstrated that the fluorescence recovery t1/2 of GFP-VASP correlated with the ubiquitination state of VASP (Menon et al., 2015). We performed FRAP assays in embryonic cortical neurons transfected with GFP-VASP (Fig. 6 E) to determine if loss of Trim67 also altered VASP dynamics at filopodia tips. Consistent with previous work, treatment with netrin-1 caused a reduction in the t1/2 of filopodial GFP-VASP (Fig. 6 F; P = 0.007), indicating more rapid dynamics of VASP at filopodial tips when VASP-Ub was reduced (Menon et al., 2015). In Trim67−/− neurons there was a basal increase in FRAP t1/2 (Fig. 6 F; P = 0.014) consistent with increased VASP-Ub, but no change in t1/2 with netrin treatment (P = 0.594). To test whether this effect on FRAP t1/2 was due to ubiquitination, we exploited a construct of GFP-VASP harboring nine lysine residues mutated to arginine (VASPK-R). These mutations decrease VASP-Ub levels (Menon et al., 2015). In both Trim67+/+ and Trim67−/− neurons, the FRAP t1/2 of GFP-VASPK-R was lower than that of GFP-VASP (P = 0.036 and P = 0.026), and there was no effect of netrin on the t1/2 in either genotype (P = 0.95 and P = 0.62). To assay effects of increased VASP-Ub on VASP dynamics, we treated neurons with PR-619, a broad inhibitor of deubiquitinating enzymes (Seiberlich et al., 2012), which increases VASP-Ub (Menon et al., 2015). In Trim67+/+ neurons, PR-619 increased GFP-VASP FRAP t1/2 (Fig. 6 F; P = 0.024), consistent with our published findings (Menon et al., 2015). However, the VASP t1/2 was not affected in Trim67−/− neurons treated with PR-619 (P = 0.925), suggesting that deubiquitinase inhibition did not impact the dynamics of VASP when VASP-Ub was already elevated. We found no difference in the percentage of VASP that recovered after photobleaching between genotypes or treatment conditions (Fig. S5 A). Together, these data suggest that VASP ubiquitination slows VASP dynamics at filopodia tips, and that TRIM67 is required for the proper netrin-1–dependent increase in VASP dynamics at filopodia tips.
Figure S5.
TRIM67 regulates ubiquitination of VASP. (A) Percentage recovery of fluorescence (mobile fraction) in FRAP assays reported in Fig. 6. Three to five experiments per genotype/treatment. n (cells) = GFP-VASP: 27 +/+ media, 16 +/+ netrin, 10 +/+ PR-619, 28 −/− media, 26 −/− netrin, 20 −/− PR-619; GFP-VASP(K-R): 14 +/+ media, 15 +/+ netrin, 28 −/− media, 19 −/− netrin. (B) Ubiquitination-precipitation assays of Myc-VASP expressed in HEK293T cells lacking TRIM67 expressing indicated tagRFP-TRIM67 constructs, along with FLAG-ubiquitin. A FLAG-Ub migrates at a heavier molecular weight (red arrowhead) than unmodified VASP (black arrowhead). (C) Individual data points and box-and-whisker plots showing the data spread in the interquartile range (box) and minimum and maximum (whiskers) of VASP-Ub, quantified from FLAG signal relative to total Myc-VASP, normalized to the RFP control condition. Four experiments/cultures per construct. (D) Representative Western blot of TRIM9 in embryonic cortical lysate from two animals of each indicated genotype. Bottom band in Trim9−/− lysate is a nonspecific band. (E) TRIM9 expression from embryonic cortical lysates. n (embryos) = 15 +/+, 16 −/−. (F) Percentage recovery of fluorescence in FRAP assays reported in Fig. 8. Three experiments per genotype/treatment. n (cells) = 8 +/+ media, 8 +/+ netrin, 12 −/− media, 15 −/− netrin, 8 −/− PR-619. (G) Ubiquitination-precipitation assays of GFP-VASP expressed in HEK293T cells lacking TRIM67 expressing MycTRIM9 where indicated, along with FLAG-ubiquitin. A FLAG-Ub migrates at a heavier molecular weight (red arrowhead) than unmodified VASP (black arrowhead). (H) Quantification of VASP-Ub, quantified from FLAG signal relative to total GFP-VASP, normalized to the WT control condition. Three experiments/cultures per condition. Box plots are minimum, Q1, Q2, Q3, maximum.
Investigation of VASP ubiquitination
We returned to ubiquitination assays of GFP-VASP in TRIM67−/− HEK293 cells to define domains of TRIM67 required for antagonizing VASP ubiquitination. High levels of VASP-Ub occurred in the absence of TRIM67 (Fig. 6, G and H). Introduction of myc-tagged TRIM67 decreased VASP-Ub (P = 0.0300), consistent with results in neurons. This TRIM67-dependent inhibition of VASP ubiquitination required the coiled-coil domain of TRIM67 as well as ligase function, as neither mutant decreased VASP-Ub compared with myc-transfected TRIM67−/− HEK cells (Fig. 6, G and H; ΔCC, P = 0.874; LD, P = 0.656). This suggests that the interaction of TRIM67 with VASP or another protein, such as TRIM9, and the ligase function of TRIM67 are necessary for inhibiting VASP-Ub. In contrast to GFP-VASP, GFP-VASPK-R was not ubiquitinated in the absence of TRIM67 (Fig. 6, G and H; P = 0.0150). GFP contains 20 lysine residues, and ubiquitin-dependent degradation of GFP is documented (Dantuma et al., 2000). Therefore, we repeated ubiquitination assays with myc-VASP and tagRFP-TRIM67 constructs (Fig. S5, B and C). Ubiquitination of myc-VASP was similarly reduced by expression of TRIM67 and increased by expression of TRIM67ΔCC, indicating that ubiquitination of GFP-VASP is a reliable readout.
TRIM67 antagonizes TRIM9-mediated regulation of VASP
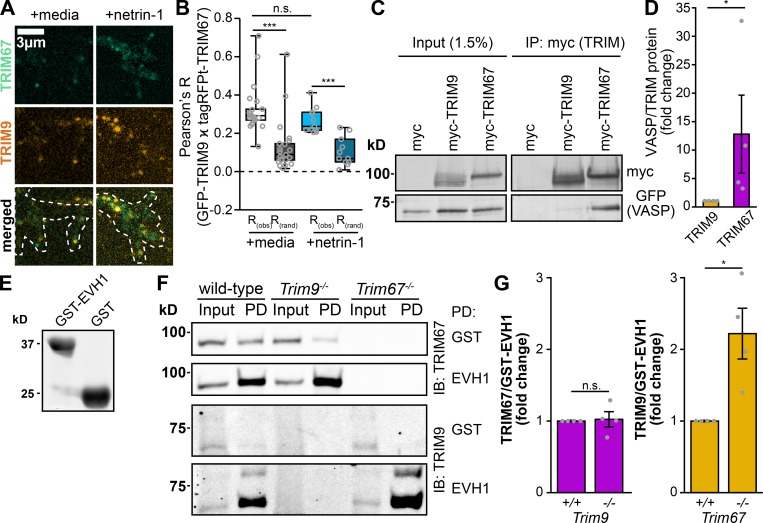
Our data are consistent with the hypothesis that TRIM67 antagonizes the TRIM9-dependent ubiquitination of VASP, and previous work showed that TRIM67 and TRIM9 interact (Boyer et al., 2018). Therefore, we investigated whether TRIM67 and TRIM9 colocalize in neurons. In Trim9−/−:Trim67−/− embryonic cortical neurons, GFP-TRIM9 and tagRFP-TRIM67 colocalized significantly compared with Fay-randomized controls (Fig. 7, A and B). Netrin treatment did not detectably alter the colocalization of these two proteins. Thus, TRIM67 is poised to inhibit TRIM9 function in the growth cone.
Figure 7.
TRIM67 colocalizes with TRIM9 and inhibits the TRIM9:VASP interaction. (A) Colocalization of GFP-TRIM9 and tagRFP-tagged TRIM67 in embryonic cortical neurons. (B) Quantification of colocalization between TRIM9 and TRIM67 (obs) compared with Fay-randomized controls (rand). Three experiments per treatment. n (cells) = 22 media, 11 netrin. (C) Coimmunoprecipitation of myc-tagged TRIM9 or TRIM67 from HEK293 cells showing coprecipitated GFP-VASP. (D) Quantification and individual data points of coprecipitated GFP-VASP relative to myc-tagged TRIM protein normalized to the VASP/TRIM67 ratio of each experiment. n (cultures) = 4 per genotype. Error bars denote SEM. (E) Coomassie-stained gel of recombinant GST or GST-EVH1 domain of VASP, used for pulldowns in F. (F) Pulldowns (PD) from embryonic mouse cortical lysate using either GST or GST-EVH1 domain of VASP, probed for endogenous TRIM67 and TRIM9. IB, immunoblot. (G) Quantification and individual data points of TRIM proteins precipitated by GST-EVH1 from lysates of indicated genotypes, normalized to WT levels. n (cultures) = 4 per condition. Error bars denote SEM. Data are presented as bar charts in D and G, as these datasets contain fewer than the five samples required to produce a box plot without interpolation. *, P < 0.05; ***, P < 0.005; n.s., P > 0.05. Box plots are minimum, Q1, Q2, Q3, maximum.
We hypothesized that TRIM67 might compete with TRIM9 for an interaction with VASP, as deletion of Trim67 did not detectably affect TRIM9 protein levels (Fig. S5, D and E). Coimmunoprecipitation assays suggested a stronger interaction between TRIM67 and VASP than between TRIM9 and VASP (Fig. 7, C and D; P = 0.0211). The Ena/VASP homology domain 1 (EVH1) of VASP interacts with TRIM9 (Menon et al., 2015). Recombinant EVH1 tagged with glutathione S-transferase (GST, Fig. 7 E) precipitated both TRIM67 and TRIM9 from cortical lysate (Fig. 7 F), indicating that both proteins interact with the same domain of VASP. GST-EVH1 enriched indistinguishable amounts of endogenous TRIM67 from WT and Trim9−/− cortical lysate (Fig. 7, F and G; P = 0.878), indicating that TRIM9 did not impair the TRIM67:EVH1 interaction. However, GST-EVH1 precipitated approximately twofold more TRIM9 in the absence of Trim67 (Fig. 7, F and G; P = 0.0211), further suggesting that TRIM67 competes with the interaction between TRIM9 and VASP.
If TRIM67 regulates VASP dynamics and filopodia by antagonizing TRIM9-mediated ubiquitination of VASP, we predicted that in the absence of both TRIM proteins, the phenotype of Trim9−/− neurons (Menon et al., 2015) would dominate. Indeed in Trim9−/−:Trim67−/− neurons, we observed a decrease in VASP-Ub (Fig. 8, A and B; P = 0.002), similar to Trim9 −/− neurons. The FRAP t1/2 of GFP-VASP expressed in Trim9−/−:Trim67−/− embryonic cortical neurons was lower than in untreated WT neurons (P = 0.018) and displayed an increase following addition of PR-619 (Fig. 8 C; P = 0.002), consistent with the hypothesis that VASP has faster dynamics at filopodia tips than VASP-Ub. As with previous FRAP assays, we saw no differences in percentage of recovery with any condition (Fig. S5 F). Analysis of Trim9−/−:Trim67−/− axonal growth cones (Fig. 8 D) showed that similar to those of Trim9−/− neurons, basal filopodial number and filopodial density increased in the absence of both TRIM proteins and did not increase in response to netrin-1 treatment (Fig. 8 E). These data suggest that TRIM67-mediated regulation of VASP and the growth cone involves antagonizing the function of TRIM9. Consistent with TRIM67 regulating VASP via TRIM9, overexpression of TRIM9 in absence of TRIM67 did not further increase VASP-Ub (Fig. S5, G and H).
Figure 8.
TRIM67 antagonizes TRIM9 in the regulation of VASP and filopodia. (A) Western blot of endogenous VASP immunoprecipitated from denatured cultured embryonic cortical lysate of indicated genotypes, showing VASP+ bands that comigrate and colabel with ubiquitin (VASP-Ub, red arrowheads) and run at a heavier apparent molecular weight than unmodified VASP (black arrowhead). Asterisk (*) by ubiquitin immunoblot (IB) marks a nonspecific ubiquitin band in all lanes. A low-intensity (top) and higher-intensity (bottom) scan of anti-ubiquitin blots are included to better reveal bands of interest. (B) Individual data points and box plots of VASP ubiquitination; n (cultures) = 9 +/+ media, 9 +/+ netrin, 7 Trim67−/− media, 7 Trim67−/− netrin, 5 Trim9−/− media, 5 Trim9−/− netrin, 7 Trim67−/−:Trim9−/− media, 5 Trim67−/−:Trim9−/− netrin. (C) Individual data points and box plots of FRAP halftime (t1/2) of GFP-VASP at filopodia tips in embryonic cortical neurons of indicated genotypes treated with netrin-1 or PR-619. Three experiments per condition. n (cells) = 8 +/+ media, 8 +/+ netrin, 10 −/− media, 14 −/− netrin, 8 −/− PR-619. (D) Fluorescent micrographs of growth cones of cortical neurons cultured from Trim67+/+:Trim9+/+ or Trim67−/−:Trim9−/− embryos, treated with either netrin-1 or a media sham. (E) Individual data points and box plots of filopodia number per growth cone and numerical density in embryonic cortical neurons isolated from brains of indicated genotypes. Three experiments per genotype/treatment. n (cells) = 82 +/+ media, 88 +/+ netrin, 93 −/− media, 92 −/− netrin. (F) Working model: VASP is ubiquitinated by TRIM9, resulting in decreased stability of filopodia on axonal growth cones. This ubiquitination is antagonized by TRIM67. We hypothesize that VASP ubiquitination and the resultant short-lived filopodia allow for efficient filopodial exploration of the extracellular environment. Consequently, when a filopodia encounters netrin, TRIM67 is recruited to filopodia tips, where it antagonizes VASP ubiquitination and increases filopodial lifetime, before axon turning. *, P < 0.05; ***, P < 0.005; n.s., P > 0.05. Box plots are minimum, Q1, Q2, Q3, maximum.
Discussion
In this study we demonstrate that TRIM67 antagonizes the TRIM9-dependent nondegradative ubiquitination of VASP crucial for axonal responses to netrin-1. Our working model shown in Fig. 8 F proposes that TRIM9-dependent ubiquitination of VASP slows VASP dynamics and decreases stability of filopodia. This ubiquitination is antagonized by TRIM67, such that in the absence of TRIM67, increased TRIM9-dependent VASP ubiquitination impairs filopodia stability and axon turning. Although the mechanism by which TRIM67 antagonizes VASP ubiquitination is not known, several possibilities are consistent with our data. First, we find that TRIM67 outcompetes TRIM9 for interaction with VASP. Second, the requirement of TRIM67 ligase activity for inhibition of VASP ubiquitination suggests additional levels of regulation. This could occur potentially via TRIM67-mediated ubiquitination of TRIM9, leading to either TRIM9 degradation or altered ligase activity. Although we observed no change in total TRIM9 protein following deletion of Trim67, whether degradation of the local pool of TRIM9 in the growth cone occurs is not known, due to insufficient quality of TRIM9 antibodies for ICC (Winkle et al., 2014). Further, whether an unidentified substrate of TRIM67 regulates TRIM9-mediated ubiquitination of VASP is unknown. We found that ubiquitination and dynamics of VASP, the filopodial morphology, and netrin responses in a Trim67-/:Trim9−/− neurons resemble that of a Trim9−/− neuron. This supports the hypothesis that TRIM67 regulates VASP via antagonizing TRIM9 function (Fig. 8 F). We hypothesize that VASP-Ub and the resultant short-lived filopodia allow for efficient filopodial exploration of the extracellular environment. Consequently, when a filopodia encounters netrin, TRIM67 is recruited to filopodia tips, where it antagonizes VASP ubiquitination and increases filopodial lifetime, before axon turning.
TRIM67 regulates brain development
These experiments support the hypothesis that TRIM67 is critical for aspects of brain development, particularly appropriate midline crossing of callosal axons. In agreement with previous work showing that genetic deletion of Trim67 results in thinning of the adult corpus callosum, we found a reduction in the midline-directed outgrowth of callosal fibers during development. This may be due to diminished sensitivity of Trim67−/− axons to netrin-1 at the midline, as we find that Trim67−/− axons are insensitive to netrin-1 in vitro. The reduced axon length observed in Trim67−/− cultured neurons may also contribute to this midline-crossing deficiency. Similar deficits in axon outgrowth and guidance in response to netrin-1 could contribute to other neuroanatomic defects seen in the brains of Trim67−/− mice (Boyer et al., 2018). Additionally, our observation that TRIM67 is required for netrin-dependent axon branching in vitro may predict a reduction in axon arbor elaboration and subsequently innervation by netrin-1–sensitive neurons. These possibilities are intriguing, as several of the hypotrophies seen in Trim67−/− brains are in regions with evidence of netrin-1 secretion (Boyer et al., 2018; Fazeli et al., 1997; Serafini et al., 1996; Xu et al., 2010; Yung et al., 2015).
TRIM proteins as models of paralog interference
Although TRIM67 may act as an E3 ubiquitin ligase (Yaguchi et al., 2012), our observations suggest it also functions as a competitive inhibitor to a closely related E3 ligase. Based on phylogenetic analysis (Boyer et al., 2018), the TRIM9 and TRIM67 paralog pair likely arose from a gene duplication event, producing two similar proteins in vertebrates. Duplication of gene products that form homodimers, as many TRIM proteins do (Koliopoulos et al., 2016; Sanchez et al., 2014), can lead to a phenomenon known as paralog interference, in which one member of the pair diverges and acts as an inhibitor for the other protein (Baker et al., 2013; Kaltenegger and Ober, 2015). Whereas this behavior is usually seen in relation to transcription factors, the ability of TRIM67 to antagonize the interaction between TRIM9 and VASP and TRIM9-dependent VASP ubiquitination suggests that TRIM9 and TRIM67 may represent an example of paralog interference. This is supported by the observation that the ability of TRIM67 to antagonize TRIM9-dependent VASP ubiquitination in HEK293T cells required the domain of TRIM67 that mediates TRIM dimerization. Class 1 TRIM proteins in mammals are all paralog pairs (Short and Cox, 2006) and, as such, the interaction shown here for TRIM9 and TRIM67 may be relevant for other class 1 TRIM pairs: TRIM1 and TRIM18, TRIM36 and TRIM46. Given the frequency of whole-genome duplication events throughout animal evolution (Glasauer and Neuhauss, 2014; Van de Peer et al., 2009; Robertson et al., 2017; Song et al., 2016) and the consequently conserved dimerization domains of RING domain–containing E3 ligases (Choo and Hagen, 2012; Liew et al., 2010; Metzger et al., 2014), competitive inhibition by paralogs may be a common regulatory mechanism. Future studies will be necessary to explore whether other TRIM proteins display similar inhibition in paralog pairs.
TRIM67 is a protein with diverse functions
The results of our structure–function VASP ubiquitination assay in HEK293 cells suggest that TRIM67 has other functions in addition to competitively inhibiting TRIM9-dependent VASP ubiquitination. The ligase-dead TRIM67 mutant was unable to reduce levels of VASP ubiquitination, suggesting that either the ligase function of TRIM67 is necessary to inhibit TRIM9 or another unknown pathway that regulates VASP-Ub. Our structure–function assays indicate that TRIM67 ligase function is also necessary for filopodial responses to netrin-1, as neither the ligase-dead nor the RING domain–deleted mutants were able to rescue the filopodial phenotype of Trim67−/− neurons. Determining the function and substrates of TRIM67 ligase activity will require further studies.
The puzzle of VASP ubiquitination
A curious apparent molecular weight shift of VASP occurs upon phosphorylation (∼4 kD; Reinhard et al., 1992; Butt et al., 1994). This observation suggests that interpreting the molecular weight shifts associated with VASP-Ub into the number of ubiquitin moieties would be presumptuous. Yet a smear of high molecular weight VASP-Ub that would be predicted for polyubiquitination was not observed. Rather, in HEK293 cells, we saw a single predominant species of Myc-VASP-Ub with either HA-ubiquitin or HA-ubiquitin-KO, consistent with mono- or multi-monoubiquitination of VASP. Our findings here thus bolster the hypothesis that nondegradative ubiquitination of VASP alters or inhibits the functionality of the polymerase in growth cones, and that changes in the degree of ubiquitination of VASP are associated with netrin-1–dependent axonal responses. Previously we found that a cycle of VASP ubiquitination and deubiquitination was necessary for filopodial responses to netrin-1 (Menon et al., 2015). Building on this, our data suggest that the TRIM9-dependent ubiquitination of VASP is inhibited by TRIM67. Therefore, we propose a coordination of VASP ubiquitination by the yin-and-yang–like pair of TRIM9 and TRIM67. This establishes new avenues of inquiry: How does ubiquitination modify VASP function? Are other signaling pathways and cytoskeletal associated proteins regulated in this manner? The requirement of ubiquitination of a relatively small amount of VASP for appropriate netrin responses suggests that ubiquitination potently regulates VASP function. As VASP functions as a tetramer at actin barbed ends, this could be the result of one modified VASP monomer altering the function of the entire tetramer. Further studies need to investigate the number and arrangement of ubiquitin moieties on VASP and how ubiquitination of VASP alters its function on the molecular level or changes the state of the tetramer. Such work could have intriguing implications for the regulation of other actin-associated proteins that function as multimers, such as formins, which recent work has suggested are also regulated by nondegradative ubiquitination (Juanes and Piatti, 2016).
Materials and methods
Experimental model and subject details
Animals
All mouse lines were on a C57BL/6J background and bred at the University of North Carolina (UNC) with approval from the Institutional Animal Care and Use Committee. Timed pregnant females were obtained by placing male and female mice together overnight; the following day was designated as E0.5 if the female had a vaginal plug. Sexes of early postnatal mice used for corpus callosum development assays were not recorded, due to the inaccuracy of standard sexing procedures before P8 (Greenham and Greenham, 1977). Trim9−/−, Trim67−/−, Trim67fl/fl, and Nex-Cre/TauloxP-stop-loxPGFP mice have been described previously (Boyer et al., 2018; Goebbels et al., 2006; Menon et al., 2015; Winkle et al., 2014). Briefly, exon 1 is deleted in both Trim9−/− and Trim67−/− mice, and exon 1 is flanked by loxP sites in respective conditional animals. Nex-Cre/TauloxP-stop-loxPGFP mice express Cre under the Nex promotor, in excitatory neurons of the forebrain. This induces recombination and deletion of the stop codon and expression of GFP in these neurons. Trim9−/−:Trim67−/− mice were generated by crossing Trim9−/− and Trim67−/− mice and crossing resultant heterozygotes. Trim67−/−and Trim67+/+ littermates from Trim67+/− crosses were used for neonatal corpus callosum measurements (Fig. 1). For all culture-based experiments, embryos from separate, time-matched homozygous WT, homozygous single Trim−/−, or homozygous double Trim−/− breeding pairs were used. This was for two reasons. First, for ubiquitination assays, each condition requires at least four embryos, with netrin treatment, which increases the number to eight embryos/genotype per experiment. Thus there would never be a litter large enough to have sufficient homozygous embryos from a heterozygous breeding pair. Second, for imaging/transfection studies, using embryos from heterozygous breeding pairs requires transfection and plating before identifying genotype, which is prohibitively expensive.
Cortical neuron culture
E15.5 mice were removed from pregnant dams by postmortem cesarean section, and dissociated cortical neuron cultures were prepared as described previously (Kwiatkowski et al., 2007). Briefly, cortices were microdissected, and neurons were dissociated with 0.25% trypsin for 15 min at 37°C, followed by quenching of trypsin with neurobasal medium supplemented with 10% FBS and 0.5 mM glutamine. After quenching, cortices were gently triturated 15× in neurobasal medium supplemented with 10% FBS and 0.5 mM glutamine, counted by hemocytometer, and spun in a tabletop centrifuge at 0.1 relative centrifugal force for 7 min at RT. Supernatant was aspirated, and pelleted cells were gently resuspended in neurobasal medium supplemented with B27 (1:50 of manufacturer stock; Invitrogen) and plated on coverglass or tissue culture plastic coated with 1 mg/ml poly-d-lysine (Sigma-Aldrich). To assay growth cones and filopodia, 600 ng/ml recombinant netrin-1, 24 ng/ml recombinant FGF2 (MBL International), or 400 ng/ml Slit2N (PeproTech) was bath applied after 48 h in vitro for 40 min, followed by fixation with 4% PFA in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgSO4, and 0.12 M sucrose). To assay axon length and branching, 250 ng/ml netrin-1, 10 ng/ml FGF2 or 100 ng/ml Slit2N was bath applied for 24 h after 48 h in vitro, followed by fixation with 4% PFA in PHEM buffer. Cells were permeabilized for 10 min in 0.1% Triton X-100, blocked for 30 min in 10% BSA or 10% donkey serum, and stained with indicated primary antibodies for 1 h at RT. Following three washes, species-appropriate fluorescent secondary antibodies were added and allowed to incubate for 1 h at RT. Following three washes, cells were mounted in a TRIS/glycerol/n-propyl-gallate-based mounting medium for imaging. Wide-field epifluorescence images or confocal z-sections of pyramidal-shaped neurons were analyzed. Growth cone perimeter and area were measured using ImageJ (National Institutes of Health). Filopodium length was measured from the filopodium tip to lamellipodial veil. Number of filopodia was counted per growth cone, and density is reported per 10 µm of growth cone perimeter. To assay filopodia dynamics, dynamic colocalization, and FRAP, time-lapse imaging was performed with a stage-top incubator that maintained humidity at 37°C and 5% CO2 (TokaiHit).
HEK293 cell culture
HEK293 cells (female) were obtained from Simon Rothenfußer (Klinikum der Universität München, München, Germany), as were HEK293 cells lacking TRIM9, which were generated from their parental line, as described previously (Menon et al., 2015). HEK293 cells were maintained at 5%CO2/37°C in DMEM with glutamine (Invitrogen), supplemented with 10% FBS (Hyclone).
Generation of TRIM67−/− HEK cells
The generation of HEK293 TRIM67−/− cells was performed using CRISPR/Cas9 gene editing with sgRNA targeting three sets of sequences in exon 1 of the TRIM67 gene in HEK293 cells acquired from the Rothenfußer laboratory: set 1: 5′-TCCTGCTTTCCCGGGGATCG-3′, 5′-GGCAGGTGCTGCTCACCGTC-3′; set 2: 5′-GCTCCTGCTTTCCCGGGGAT-3′, 5′-CAGGTGCTGCTCACCGTC-3′; and set 3: 5′-CTCCTGCTTTCCCGGGGATC-3′, 5′-GGCAGGTGCTGCTCACCGTC-3′.
1.3 × 105 HEK293 cells were seeded and transfected in a 24-well plate with 250 ng of an expression plasmid coding for a CMV promoter-driven nickase-variant Cas9-D10A-puromycin cassette and 250 ng of the mixed U6-driven sgRNAs using Lipofectamine 2000 (Invitrogen) as per manufacturer protocol. After 24 h, transfected cells were selected for by treatment with 1 µg/ml puromycin and incubated for 72 h. Remaining cells were diluted to 0.5 cells/100 µl, and single cells were seeded in 96-well plates. Amplifying clones were expanded. Genomic DNA was extracted, and the target region of interest was amplified by PCR to detect size differences. Positive clones were screened by PCR (primers: 5′-CCTTCTCTCGCCCCTCAATC-3′ and 5′-GTGCTCCAGAGAGGCAGC-3′) and sequenced. A knockout cell clone was identified harboring frameshift mutations in the exon 1 region and verified by Western blot protein analysis.
Plasmids, antibodies, and reagents
Plasmids encoding human TRIM9 cDNA, mouse TRIM67, and GFP-human VASP K-R mutant were described previously (Boyer et al., 2018; Menon et al., 2015; Winkle et al., 2014). The following plasmids were acquired: mCherry (Clontech), FLAG-Ub (Ben Philpot, University of North Carolina, Chapel Hill, Chapel Hill, NC), pmscv-eGFP-Mena, pmscv-eGFP-EVL, pQE-EVH1, pGEX-6P-1-Pro (Frank Gertler, Massachusetts Institute of Technology, Cambridge, MA), and peGFP-N1-VASP (Richard Cheney, University of North Carolina, Chapel Hill, Chapel Hill, NC). pRK5-HA-Ubiquitin-WT and pRK5-HA-Ubiquitin-KO (Lim et al., 2005) were gifts from Ted Dawson (Johns Hopkins University, Baltimore, MD; Addgene plasmid 17608, RRID:Addgene_17608; Addgene plasmid 17603, RRID:Addgene_17603). Myc-VASP was created by partial PCR of GFP-VASP into the pcs2 Myc vector. TRIM67 CC domain sequence (aa 332–369 of murine TRIM67) was cloned into the pGEX-6P-1 plasmid. Myc- and TagRFP-tagged TRIM67 domain deletion constructs were generated by partial PCR of myc-TRIM67, TRIM67ΔRING (aa 159–768), TRIM67ΔCC (aa 1–335, lysine, glutamate, aa 369–768), and TRIM67 N-terminus (aa 1–369). TRIM67 ligase dead construct was generated by QuikChange site-directed mutagenesis of cysteines 7 and 10 to alanine (TGC → GCC).
Antibodies included rabbit polyclonal against TRIM67 (Boyer et al., 2018); rabbit polyclonal antibody against VASP (Frank Gertler); rabbit polyclonal against VASP (sc-13975, SCBT); mouse monoclonal against TRIM9 (H00114088-M01, Abnova); mouse monoclonal against c-Myc (9E10); mouse monoclonal against human β-III-tubulin (TujI SCBT); rabbit polyclonal against FLAG tag (F7425, Sigma-Aldrich); rabbit polyclonal against GFP (A11122, Invitrogen); mouse monoclonal against GFP (75-131, University of California, Davis, Neuromab); chicken polyclonal against GFP (GFP-1010, Aves Labs); rabbit polyclonal against HA (71-5500, Thermo Fisher Scientific); ubiquitin (sc-8017, SCBT); GAPDH (sc-166545, SCBT); fluorescent secondary antibodies and fluorescent phalloidin labeled with Alexa Fluor 488, 561, and 647; PR-619 (2,6-diaminopyridine-3,5-bis(thiocyanate), Abcam), MG132 (81-5-15, American Peptide Company), and Rhodamine B isothiocyanate–dextran (R9379, Sigma-Aldrich).
Transfection procedures
For transfection of plasmids, neurons were resuspended after dissociation in Lonza Nucleofector solution (VPG-1001) and electroporated with an Amaxa Nucleofector according to manufacturer protocol. HEK cells were transfected using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s protocol.
Immunoblotting and precipitation assays
For the ubiquitination assay, MG132- and netrin-1–treated cells were lysed in immunoprecipitation (IP) buffer (20 mM Tris-Cl, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA, 0.5% NP-40, 1% SDS, 2 mM DTT, 5 mM N-ethylmaleimide, 3 mM iodoacetamide, and protease and phosphatase inhibitors, pH 7.3–7.4). MG132 pretreatment for 4 h was used to maintain consistency with previously published ubiquitination assays (Menon et al., 2015). For 5–6 million cells, 270 µl of ubiquitin IP buffer was added and incubated on ice for 10 min. Cells were scraped from the dish and transferred into tubes. 30 µl of 1× PBS was added and gently vortexed. Samples were boiled immediately for 20 min, until clear, then centrifuged at 14,000 rpm for 10 min. The boiled samples were diluted using IP buffer without SDS to reduce the SDS concentration to 0.1%. Approximately 500–1,000 µg of protein was used in each assay. The antibody of interest was added to each sample and incubated overnight at 4°C with rotation. In the morning, Protein A beads (SCBT) were added and incubated for 2 h at 4°C with rotation. The samples were washed two times with lysis buffer for 10 min each.
For binding assays, all recombinant GST-tagged proteins were purified on sepharose-immobilized glutathione beads (Thermo Fisher Scientific). For binding to endogenous TRIM67 or Ena/VASP, E15.5 mouse cortices were lysed in 0.5% NP-40 lysis buffer (50 mM Tris, pH 7.5, 200 mM NaCl, 0.5% NP-40, and phosphatase and protease inhibitors). Lysates were precleared with GST-glutathione-sepharose beads for 1 h at 4°C with agitation and incubated with 5–10 µg of GST fusion protein or GST immobilized onto glutathione-sepharose beads at 4°C overnight. For binding of Myc-tagged TRIM67 variants, HEK293 cells were transfected and, 24 h later, lysed in 1% NP-40 lysis buffer and incubated overnight at 4°C with anti-myc antibody. For all binding assays, precipitated beads were washed three times with lysis buffer or PBS buffer, and bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting.
For coimmunoprecipitation assays, IgG-conjugated A/G beads (SCBT) were used to preclear lysates for 1.5 h at 4°C with agitation. Myc antibody-conjugated A/G beads (SCBT) or protein A/G beads (SCBT) coupled with a mouse anti-GFP antibody (Neuromab) or rabbit anti-VASP antibody (SCBT) were agitated within precleared lysates overnight at 4°C to precipitate target proteins. Beads were washed three times with lysis buffer, and bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting.
Microscope descriptions
All live-cell images and immunofluorescence images were collected on an Olympus IX81-ZDC2 inverted microscope equipped with the following objective lenses: a UPLFLN 40×/1.39-NA differential interference contrast (DIC) objective (Olympus), UAPON 100×/1.49-NA DIC TIRF objective (Olympus), a 20×/0.85-NA UPlanSApo DIC objective lens (Olympus), and a 4×/0.13-NA Plan Apochromat objective (Nikon), an automated XYZ stage (Prior), and an Andor iXon electron multiplying charge-coupled device. Fixed cells were imaged at RT in a glycerol- and n-propyl-gallate–based mounting medium. Cells for live imaging were maintained in culture medium and at 37°C and 5% CO2 with a TokaiHit on-stage cell incubator. Images were procured using Metamorph acquisition software. Neuroanatomical images in Fig. 1 and growth cone images in Fig. S5 were acquired on an inverted laser scanning confocal microscope (Zeiss LSM 780) equipped with a 10×/0.4-NA Plan Apochromat and 63×/1.4-NA Plan Apochromat objective lens and 488- and 561-nm argon lasers, using Zeiss Blue software. These samples were in the glycerol-based mounting medium described above. SIM images were obtained using a DeltaVision OMX SR imaging system (GE Healthcare) with manufacture software and a 60×/1.42-NA oil-immersion objective in 2D-SIM superresolution mode, and were also in the mounting medium described above. Scanning EM images were acquired using a Zeiss Supra 25 field emission scanning EM using a backscatter detector.
Filopodia dynamics measurements
For filopodial dynamics measurements, wide-field epifluorescence images of mCherry were acquired every 2.5 s for 5–10 min. 600 ng/ml netrin-1 was added 40 min before imaging netrin-1–treated neurons. Filopodial protrusion and retraction rates and phase durations were measured from kymographs as the slope and duration of individual events, respectively. Filopodia lifetime was measured as the time from initial filopodial protrusion until retraction into the lamellipodial veil. Filopodial buckling and folding events were both counted manually; buckling events occurred when a filopodium collapsed following a breakage along the length of the filopodium. Folding events occurred when a filopodium collapsed into the veil sideways along its length as opposed to retracting perpendicularly.
FRAP imaging
For FRAP assays, neurons expressing GFP-VASP were imaged after 48 h in vitro using a 491-nm laser in TIRF mode every 0.5 s for 15 s, followed by a 1-s exposure with a solid-state 405-nm laser in FRAP mode (bleach spot ∼1.25 µm in diameter), followed by imaging every 0.5 s with the 491-nm laser in TIRF mode for 60 s. Netrin-1–treated filopodia were imaged within 5 min of addition of 400 ng/ml netrin-1. Deubiquitinase-inhibited filopodia were imaged between 20 and 30 min of addition of 9 µM PR-619.
Neuroanatomic imaging
All mice used for neuroanatomic studies were anesthetized on ice before decapitation, and heads were drop-fixed in 4% PFA for 72 h. Heads were rinsed with 1× PBS twice for 24 h before removal of and vibratome sectioning of the brains. For projection analysis in Nex-Cre/TauloxP-stop-loxPGFP mice, 80-µm coronal sections were cut, and every section was permeabilized in detergent solution (1× PBS, 0.1% Triton X-100, and 0.2% Tween-20) for 1 h on a shaker at RT. Sections were blocked in 10% BSA in 1× PBS for 5 h, then placed in primary antibody solution (anti-GFP chicken [Aves, ab1020, 1:2,000], in 1% BSA in PBS) for 24 h on a shaker at 4°C. Primary antibodies were removed, and sections were rinsed in 1× PBS for 1 h before the addition of secondary antibody solution (Alexa Fluor 488 chicken + 1% BSA in 1× PBS) for 24 h on a covered shaker at 4°C. After postsecondary rinsing with 1× PBS, sections were mounted in Tris/glycerol/n-propyl-gallate and imaged with the 10× objective on the laser scanning confocal microscope described above.
Axon turning assay
Micropass gradient devices were used to measure axon turning. Device preparation and experimental protocol were as previously described (Taylor et al., 2015). Sylgard 184 (Sigma-Aldrich) polymer was prepared as per manufacturer protocol and added to etched silicon wafers. Microfluidic devices were cut to the size of coverslips; fluid chambers were cut out; and devices were cleaned with 3M tape, sprayed with 70% ethanol, and allowed to dry for 45 min. Coverslips were applied to the face of devices with microfluidic grooves, and neuronal culture medium was added to all chambers. After ≥1 h, medium was aspirated, and Trim67+/+ or Trim67−/− E15.5 cortical neurons were plated in devices. After axons entered the axon viewing area (2–4 d), a control gradient of dextran (starting at 1 µM) or gradient of netrin + dextran (600 ng/ml) was established by adding solutions to one side chamber and removing medium from the upper “sink” chamber. DIC (axons) and epifluorescence (dextran) images were acquired every 5 min for 8–18 h at 20× magnification.
Quantification and statistical analysis
All image analysis was performed in ImageJ. No image processing was performed.
Axon turning angle measurement
The angle of axon turning was determined as the angle between (a) a line bisecting the growth cone after 18 h of growth and (b) a line along the initial trajectory of the axon before initiation of the gradient, with positive angles indicating turning toward the source of the gradient (Taylor et al., 2015).
Colocalization and growth cone analysis
We quantified filopodial enrichment of endogenous TRIM67 on the distal 0.5 µm of the filopodia relative to the penultimate 0.5 µm of the filopodia. We measured the proximity of TRIM67 to the filopodial tip after background subtraction. The tip of the filopodia was defined by phalloidin staining, and then TRIM67 intensity was measured at the center and edge of a hypothetical Airy disc localized at the tip (tip localization ratio). This ratio should decrease as the distance of a fluorescent protein to the filopodial tip increases. Pearson’s correlation of colocalization between TRIM67 and Ena/VASP proteins was performed using regions of interest drawn in filopodia and a Colocalization Test plugin for ImageJ with Fay randomization using images acquired with the 100× objective described above. Growth cone area, filopodia number, filopodia density, and filopodia length were measured in ImageJ. Filopodia lengths were measured from the edge of lamellipodial veils to filopodia tips. For filopodia rescue assays, images of neurons expressing comparable levels of Myc (TRIM67 variants) were acquired with the 100× objective, and the number of growth cone filopodia was recorded. TIRF imaging of TRIM67 and Ena/VASP proteins was performed after 48 h in vitro, with the 100× objective and solid-state 491-, 561-nm laser illumination at 100-nm penetration depth. Images were acquired every 0.5 s for 5 min.
Growth cones imaged by scanning EM were divided into categories based on apparent 3D morphology. Categories were hierarchical, starting with “flat” growth cones that possessed no dorsal protrusions. This category was superseded by “curled” growth cones with edges separated from the coverslip surface, and then “dorsal” growth cones with filopodial or lamellipodial protrusions from the dorsal surface. The apical category was “nonadherent” growth cones, which were fully separated from the coverslip surface. The growth cone of the longest axon of each cell was assigned to a single category.
We determined the number of growth cones needed for the FGF2/Slit2N experiments based on previously published experiments in the literature (Szebenyi et al., 2001), in which sample sizes were 50–52 growth cones per group. As the FGF2 and Slit2N effects on growth cones are both quite robust, the estimated number of samples required is considerably less than the sample size predicted to be necessary to confidently detect a statistically significant difference between untreated Trim67+/+ and Trim67−/− growth cone areas. This is due to the small effect size of TRIM67 knockout on growth cone area coupled with the inherently large deviation of these measurements. The sample sizes were larger in the experiment testing netrin-1 effect on Trim67−/− growth cones, and as such we were able to detect this less-apparent increase in growth cone area.
FRAP quantification
FRAP data were analyzed as described previously (Applewhite et al., 2007; Menon et al., 2015). For analyzing FRAP imaging data, background signal was subtracted from all data, and photobleaching was corrected by fitting an exponential decay to the fluorescence of an unbleached control region (F = F0 × e−kt, where F is fluorescence, F0 is initial fluorescence, k is the decay time constant, and t is time). Fluorescence recovery t1/2 and percentage were calculated from an inverse exponential decay (F = A × (1 − e−τt), where F is fluorescence, A is recovery plateau fluorescence, τ is the recovery time constant, and t is time). The data were fitted to a single exponential function as opposed to a higher-order exponential (e.g., F = A × (1 − e−τt − e−τ′t)), as the R2 value for a double exponential (R2 = 0.845) was the same as for a single exponential (R2 = 0.845) fitted to the average of all control data. The percentage recovery was calculated as the plateau fluorescence divided by the average prebleach fluorescence, and t1/2 is equal to ln(2)/τ. Both photobleaching and FRAP curves were fitted to data using the Solver add-in of Microsoft Excel 2013.
Western blot analysis
Western blots were quantified using ImageJ, with the total fluorescence of labeled bands representing relative protein, which was then normalized to the control condition of each experiment. In the case of multiple controls in the same experiment (e.g., VASP protein level quantification), results are normalized to the average of the controls on the same blot to indicate fold change from control. Ubiquitinated VASP levels were measured as the ratio of total fluorescence of high molecular weight VASP bands to the total fluorescence of low molecular weight VASP, with care taken to exclude the antibody heavy chain band.
Statistics
The data shown graphically in all figures is summarized in Table S1, with all numbers of samples, population mean and deviance, statistical tests used, and P values listed. Individual data points and box-and-whisker plots show the data spread in the interquartile range (box) and minimum and maximum (whiskers). At least three independent experiments were performed for each assay. Data distribution normality was determined using the Shapiro–Wilk test. Normally distributed data were compared by unpaired t test for two independent samples, or ANOVA with Tukey post hoc correction, for more than two comparisons. For nonnormal data, the Mann–Whitney U test was used, or Kruskal–Wallis nonparametric ANOVA with Benjamini–Hochberg correction for more than two comparisons. Fisher’s exact test was used to compare growth cone morphology distributions. All data are presented as means ± SEM or as box plots (minimum, Q1, Q2, Q3, maximum) accompanied by all data points. Statistical significance was determined by an α of 0.05, and is represented as not significant (n.s.) at P > 0.05; *, P < 0.05; **, P < 0.01; and ***, P < 0.005.
Online supplemental material
Fig. S1 contains a demonstration of general growth cone response in Trim67+/+ and Trim67−/− cells to morphogens, including netrin-1, Slit2N, and FGF2. Figs. S2 and S3 include structure–function experiments investigating critical domains of TRIM67 for netrin-1 growth cone (Fig. S2) and axon branching (Fig. S3) responses. Fig. S4 shows the generation of TRIM67−/− HEK293 cells by CRISPR/Cas9-mediated gene editing and control experiments with Mena and EVL; Fig. S4 also demonstrates that VASP protein levels and immunocytochemistry are unchanged in Trim67−/− neurons. Fig. S5 includes FRAP recovery percentage data, quantification of TRIM9 protein levels in Trim67−/− neurons, and controls for HEK cell VASP ubiquitination assays. Video 1 demonstrates an example of live-cell imaging for the measurement of filopodia dynamics in growth cone. Table S1 details the quantification and statistical analysis of all experiments included in the article.
Supplementary Material
Acknowledgments
The authors thank Anthony Mangan for assistance with immunoblotting and Caroline Monkiewicz, Vong Thoong, and Chris Hardie for mouse breeding, husbandry, and genotyping.
Funding from the National Institutes of Health supported this research, including grants R01GM108970 (S.L. Gupton), F31NS096823 (N.P. Boyer), F31NS113381 (L.E. McCormick), and F31NS103586 (F.L. Urbina) and funding for the University of North Carolina Neuroscience microscopy core facility, P30 NS045892 and U54 HD079124.
The authors declare no competing financial interests.
Author contributions: Conceptualization, N.P. Boyer, L.E. McCormick, and S.L. Gupton; Methodology, N.P. Boyer, L.E. McCormick, and S.L. Gupton; Investigation, N.P. Boyer, L.E. McCormick, S. Menon, F.L. Urbina, and S.L. Gupton; Writing – Original Draft, N.P. Boyer, F.L. Urbina, and S.L. Gupton; Writing – Review & Editing, N.P. Boyer, L.E. McCormick, S. Menon, and S.L. Gupton; Supervision, S.L. Gupton.
References
- Applewhite D.A., Barzik M., Kojima S., Svitkina T.M., Gertler F.B., and Borisy G.G.. 2007. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell. 18:2579–2591. 10.1091/mbc.e06-11-0990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.R., Hanson-Smith V., and Johnson A.D.. 2013. Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science. 342:104–108. 10.1126/science.1240810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin J.M., Han D., Lai Wing Sun K., Croteau L.-P., Dumontier E., Cloutier J.-F., Kania A., and Kennedy T.E.. 2015. Complete Loss of Netrin-1 Results in Embryonic Lethality and Severe Axon Guidance Defects without Increased Neural Cell Death. Cell Reports. 12:1099–1106. 10.1016/j.celrep.2015.07.028 [DOI] [PubMed] [Google Scholar]
- Boyer N.P., and Gupton S.L.. 2018. Revisiting Netrin-1: One Who Guides (Axons). Front. Cell. Neurosci. 12:221 10.3389/fncel.2018.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer N.P., Monkiewicz C., Menon S., Moy S.S., and Gupton S.L.. 2018. Mammalian TRIM67 functions in brain development and behavior. eNeuro. 5:ENEURO.0186-18.2018. 10.1523/ENEURO.0186-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt E., Abel K., Krieger M., Palm D., Hoppe V., Hoppe J., and Walter U.. 1994. cAMP- and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J. Biol. Chem. 269:14509–14517. [PubMed] [Google Scholar]
- Choo Y.Y., and Hagen T.. 2012. Mechanism of cullin3 E3 ubiquitin ligase dimerization. PLoS One. 7:e41350 10.1371/journal.pone.0041350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma N.P., Lindsten K., Glas R., Jellne M., and Masucci M.G.. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18:538–543. 10.1038/75406 [DOI] [PubMed] [Google Scholar]
- Dent E.W., Barnes A.M., Tang F., and Kalil K.. 2004. Netrin-1 and semaphorin 3A promote or inhibit cortical axon branching, respectively, by reorganization of the cytoskeleton. J. Neurosci. 24:3002–3012. 10.1523/JNEUROSCI.4963-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E.W., Gupton S.L., and Gertler F.B.. 2011. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb. Perspect. Biol. 3:a001800 10.1101/cshperspect.a001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E.W., Kwiatkowski A.V., Mebane L.M., Philippar U., Barzik M., Rubinson D.A., Gupton S., Van Veen J.E., Furman C., Zhang J., et al. 2007. Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 9:1347–1359. 10.1038/ncb1654 [DOI] [PubMed] [Google Scholar]
- Fazeli A., Dickinson S.L., Hermiston M.L., Tighe R.V., Steen R.G., Small C.G., Stoeckli E.T., Keino-Masu K., Masu M., Rayburn H., et al. 1997. Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature. 386:796–804. 10.1038/386796a0 [DOI] [PubMed] [Google Scholar]
- Fothergill T., Donahoo A.L.S., Douglass A., Zalucki O., Yuan J., Shu T., Goodhill G.J., and Richards L.J.. 2014. Netrin-DCC signaling regulates corpus callosum formation through attraction of pioneering axons and by modulating Slit2-mediated repulsion. Cereb. Cortex. 24:1138–1151. 10.1093/cercor/bhs395 [DOI] [PubMed] [Google Scholar]
- Glasauer S.M.K., and Neuhauss S.C.F.. 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics. 289:1045–1060. 10.1007/s00438-014-0889-2 [DOI] [PubMed] [Google Scholar]
- Goebbels S., Bormuth I., Bode U., Hermanson O., Schwab M.H., and Nave K.-A.. 2006. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 44:611–621. 10.1002/dvg.20256 [DOI] [PubMed] [Google Scholar]
- Greenham L.W., and Greenham V.. 1977. Sexing mouse pups. Lab. Anim. 11:181–184. 10.1258/002367777780936620 [DOI] [PubMed] [Google Scholar]
- Gupton S.L., and Gertler F.B.. 2007. Filopodia: the fingers that do the walking. Sci. STKE. 2007:re5 10.1126/stke.4002007re5 [DOI] [PubMed] [Google Scholar]
- Juanes M.A., and Piatti S.. 2016. Control of formin distribution and actin cable assembly by the E3 ubiquitin ligases Dma1 and Dma2. Genetics. 204:205–220. 10.1534/genetics.116.189258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenegger E., and Ober D.. 2015. Paralogue Interference Affects the Dynamics after Gene Duplication. Trends Plant Sci. 20:814–821. 10.1016/j.tplants.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Kennedy T.E., Serafini T., de la Torre J.R., and Tessier-Lavigne M.. 1994. Netrins are diffusible chemotropic factors for commissural axons in the embryonic spinal cord. Cell. 78:425–435. 10.1016/0092-8674(94)90421-9 [DOI] [PubMed] [Google Scholar]
- Koliopoulos M.G., Esposito D., Christodoulou E., Taylor I.A., and Rittinger K.. 2016. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 35:1204–1218. 10.15252/embj.201593741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin A.L., and Tessier-lavigne M.. 2011. Mechanisms and Molecules of Neuronal Wiring: A Primer. Cold Spring Harb. Perspect. Biol. 3:a001727 10.1101/cshperspect.a001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski A.V., Rubinson D.A., Dent E.W., Edward van Veen J., Leslie J.D., Zhang J., Mebane L.M., Philippar U., Pinheiro E.M., Burds A.A., et al. 2007. Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron. 56:441–455. 10.1016/j.neuron.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Lebrand C., Dent E.W., Strasser G.A., Lanier L.M., Krause M., Svitkina T.M., Borisy G.G., and Gertler F.B.. 2004. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 42:37–49. 10.1016/S0896-6273(04)00108-4 [DOI] [PubMed] [Google Scholar]
- Liew C.W., Sun H., Hunter T., and Day C.L.. 2010. RING domain dimerization is essential for RNF4 function. Biochem. J. 431:23–29. 10.1042/BJ20100957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.L., Chew K.C., Tan J.M., Wang C., Chung K.K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C.A., et al. 2005. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25:2002–2009. 10.1523/JNEUROSCI.4474-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh A.P.L., Heron D., Edwards T.J., Quartier A., Galea C., Nava C., Rastetter A., Moutard M.L., Anderson V., Bitoun P., et al. 2017. Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nat. Genet. 49:511–514. 10.1038/ng.3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S., Boyer N.P., Winkle C.C., McClain L.M., Hanlin C.C., Pandey D., Rothenfußer S., Taylor A.M., and Gupton S.L.. 2015. The E3 ubiquitin ligase TRIM9 is a filopodia off switch required for netrin-dependent axon guidance. Dev. Cell. 35:698–712. 10.1016/j.devcel.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M.B., Pruneda J.N., Klevit R.E., and Weissman A.M.. 2014. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta. 1843:47–60. 10.1016/j.bbamcr.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachez C., and Richards L.J.. 2005. Mechanisms of axon guidance in the developing nervous system. Curr. Top. Dev. Biol. 69:267–346. 10.1016/S0070-2153(05)69010-2 [DOI] [PubMed] [Google Scholar]
- Plooster M., Menon S., Winkle C.C., Urbina F.L., Monkiewicz C., Phend K.D., Weinberg R.J., and Gupton S.L.. 2017. TRIM9-dependent ubiquitination of DCC constrains kinase signaling, exocytosis, and axon branching. Mol. Biol. Cell. 28:2374–2385. 10.1091/mbc.e16-08-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M., Halbrügge M., Scheer U., Wiegand C., Jockusch B.M., and Walter U.. 1992. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 11:2063–2070. 10.1002/j.1460-2075.1992.tb05264.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson F.M., Gundappa M.K., Grammes F., Hvidsten T.R., Redmond A.K., Lien S., Martin S.A.M., Holland P.W.H., Sandve S.R., and Macqueen D.J.. 2017. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 18:111 10.1186/s13059-017-1241-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J.G., Okreglicka K., Chandrasekaran V., Welker J.M., Sundquist W.I., and Pornillos O.. 2014. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl. Acad. Sci. USA. 111:2494–2499. 10.1073/pnas.1318962111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiberlich V., Goldbaum O., Zhukareva V., and Richter-Landsberg C.. 2012. The small molecule inhibitor PR-619 of deubiquitinating enzymes affects the microtubule network and causes protein aggregate formation in neural cells: implications for neurodegenerative diseases. Biochim. Biophys. Acta. 1823:2057–2068. 10.1016/j.bbamcr.2012.04.011 [DOI] [PubMed] [Google Scholar]
- Serafini T., Colamarino S.A., Leonardo E.D., Wang H., Beddington R., Skarnes W.C., and Tessier-Lavigne M.. 1996. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 87:1001–1014. 10.1016/S0092-8674(00)81795-X [DOI] [PubMed] [Google Scholar]
- Shekarabi M., and Kennedy T.E.. 2002. The netrin-1 receptor DCC promotes filopodia formation and cell spreading by activating Cdc42 and Rac1. Mol. Cell. Neurosci. 19:1–17. 10.1006/mcne.2001.1075 [DOI] [PubMed] [Google Scholar]
- Short K.M., and Cox T.C.. 2006. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 281:8970–8980. 10.1074/jbc.M512755200 [DOI] [PubMed] [Google Scholar]
- Song X., Tang Y., and Wang Y.. 2016. Genesis of the vertebrate FoxP subfamily member genes occurred during two ancestral whole genome duplication events. Gene. 588:156–162. 10.1016/j.gene.2016.05.019 [DOI] [PubMed] [Google Scholar]
- Szebenyi G., Dent E.W., Callaway J.L., Seys C., Lueth H., and Kalil K.. 2001. Fibroblast growth factor-2 promotes axon branching of cortical neurons by influencing morphology and behavior of the primary growth cone. J. Neurosci. 21:3932–3941. 10.1523/JNEUROSCI.21-11-03932.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Menon S., and Gupton S.L.. 2015. Passive microfluidic chamber for long-term imaging of axon guidance in response to soluble gradients. Lab Chip. 15:2781–2789. 10.1039/C5LC00503E [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Maere S., and Meyer A.. 2009. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10:725–732. 10.1038/nrg2600 [DOI] [PubMed] [Google Scholar]
- Wahlsten D. 1984. Growth of the mouse corpus callosum. Brain Res. 15:59–67. 10.1016/0165-3806(84)90140-8 [DOI] [PubMed] [Google Scholar]
- Wang K.H., Brose K., Arnott D., Kidd T., Goodman C.S., Henzel W., and Tessier-Lavigne M.. 1999. Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell. 96:771–784. 10.1016/S0092-8674(00)80588-7 [DOI] [PubMed] [Google Scholar]
- Winkle C.C., McClain L.M., Valtschanoff J.G., Park C.S., Maglione C., and Gupton S.L.. 2014. A novel Netrin-1-sensitive mechanism promotes local SNARE-mediated exocytosis during axon branching. J. Cell Biol. 205:217–232. 10.1083/jcb.201311003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Goldman J.S., Rymar V.V., Forget C., Lo P.S., Bull S.J., Vereker E., Barker P.A., Trudeau L.E., Sadikot A.F., and Kennedy T.E.. 2010. Critical roles for the netrin receptor deleted in colorectal cancer in dopaminergic neuronal precursor migration, axon guidance, and axon arborization. Neuroscience. 169:932–949. 10.1016/j.neuroscience.2010.05.025 [DOI] [PubMed] [Google Scholar]
- Yaguchi H., Okumura F., Takahashi H., Kano T., Kameda H., Uchigashima M., Tanaka S., Watanabe M., Sasaki H., and Hatakeyama S.. 2012. TRIM67 protein negatively regulates Ras activity through degradation of 80K-H and induces neuritogenesis. J. Biol. Chem. 287:12050–12059. 10.1074/jbc.M111.307678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung A.R., Nishitani A.M., and Goodrich L.V.. 2015. Phenotypic analysis of mice completely lacking netrin 1. Development. 142:3686–3691. 10.1242/dev.128942 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.