Narendra et al. demonstrate that APEX can be used as the first genetically encoded reporter for isotope ratio imaging mass spectrometry. Bulk protein turnover is measured in individual lysosomes demonstrating unanticipated heterogeneity among lysosomes within a single cell.
Abstract
Quantification of stable isotope tracers after metabolic labeling provides a snapshot of the dynamic state of living cells and tissue. A form of imaging mass spectrometry quantifies isotope ratios with a lateral resolution <50 nm, using a methodology that we refer to as multi-isotope imaging mass spectrometry (MIMS). Despite lateral resolution exceeding diffraction-limited light microscopy, lack of contrast has largely limited use of MIMS to large or specialized subcellular structures, such as the nucleus and stereocilia. In this study, we repurpose the engineered peroxidase APEX2 as the first genetically encoded marker for MIMS. Coupling APEX2 labeling of lysosomes and metabolic labeling of protein, we identify that individual lysosomes exhibit substantial heterogeneity in protein age, which is lost in iPSC-derived neurons lacking the lysosomal protein progranulin. This study expands the practical use of MIMS for cell biology by enabling measurements of metabolic function from stable isotope labeling within individual organelles in situ.
Introduction
Stable isotope quantification after metabolic labeling allows assessment of the dynamic state of living cells and tissues without the use of radioactivity. The advent of the NanoSIMS instrument has enabled a new form of imaging mass spectrometry (MS) with a lateral resolution <50 nm, using a method we call multi-isotope imaging mass spectrometry (MIMS; Slodzian et al., 1992; Lechene et al., 2006). This form of secondary ion MS is capable of measuring seven secondary ions simultaneously, allowing precise assessment of up to three isotope ratios, which can be expanded to four isotope ratios with peak switching methods (Guillermier et al., 2014, 2017).
The NanoSIMS instrument measures ions from a submicron spot of a sample surface by first probing the sample with a focused beam of positively charged cesium ions. The primary beam sputters and ionizes molecular fragments and atoms (secondary ions), representative of the uppermost atomic layers. The negatively charged secondary ions are extracted and separated according to their masses in a magnetic sector. For imaging, the primary beam is rastered across the sample, and the image contrast results from variation in the secondary ion signal at each probed location.
This method has been used to measure nitrogen fixation within individual bacteria, dopamine distribution within individual vesicles, protein turnover in hair-cell stereocilia, stem cell division, lipid movement across capillaries, lipid metabolism within lipid droplets, nuclear pore turnover, and metabolism of atherosclerotic plaques, among other applications (Lechene et al., 2007; Steinhauser et al., 2012; Zhang et al., 2012; Kim et al., 2014; Bailey et al., 2015; Schreiber et al., 2016; Lovrić et al., 2017; He et al., 2018; Guillermier et al., 2019; Gyngard and Steinhauser, 2019; Toyama et al., 2019). Despite submicron lateral resolution, however, lack of contrast has made identification of common subcellular structures, such as lysosomes, ER, and mitochondria, cumbersome, limiting the practical use of MIMS for cell biology. As localized alterations in bulk protein turnover are a hallmark of neurodegenerative disease (e.g., resulting from dysfunction of lysosomes, accumulation of protein aggregates, and/or decreased turnover of mitochondrial protein by mitophagy), methods that allow assessment of protein turnover at the level of individual organelles would represent a substantial advance.
The genetically encoded peroxidase APEX2 was recently developed as a tag for EM and proximity labeling (Martell et al., 2012, 2017; Lam et al., 2015; Hung et al., 2014; Han et al., 2017). APEX2 can catalyze the H2O2-dependent polymerization of DAB, which in turn can precipitate electron-opaque transitional metals such as OsO4, thereby providing contrast for transmission electron microscopy (TEM; Palade, 1952).
In this study, we demonstrate that APEX2 could similarly serve as a genetically encoded marker for MIMS. We demonstrate its utility with measurements of protein turnover in individual organelles in situ, revealing unanticipated heterogeneity in protein turnover among individual lysosomes within a single cell.
Results
APEX2 localization by direct measurement of osmium by MIMS
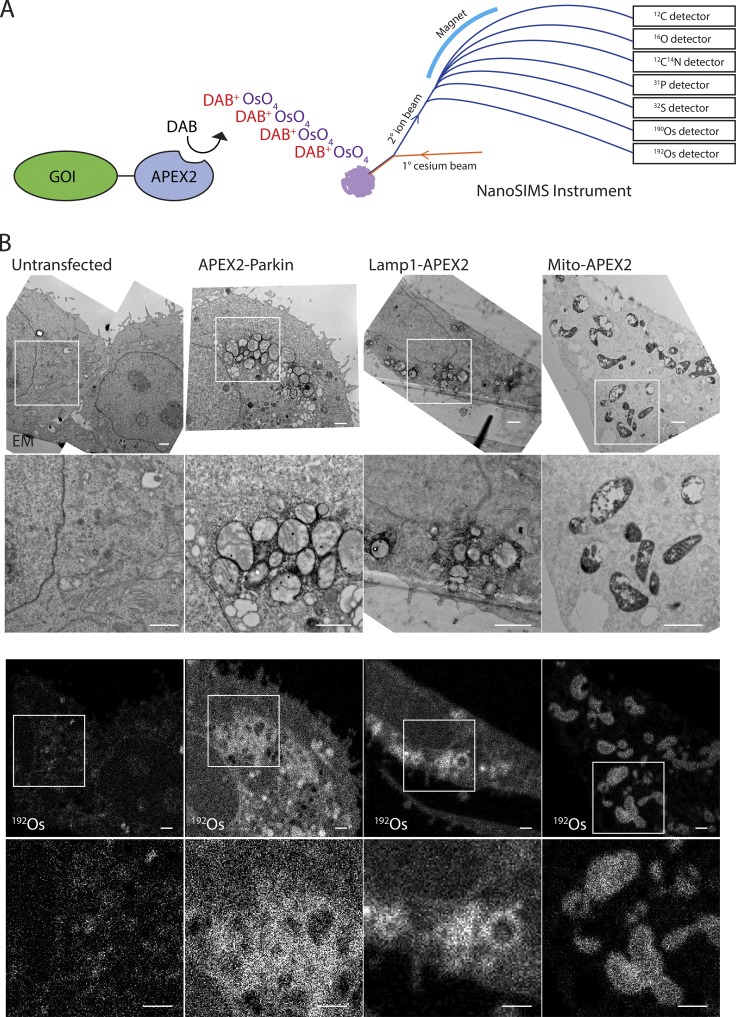
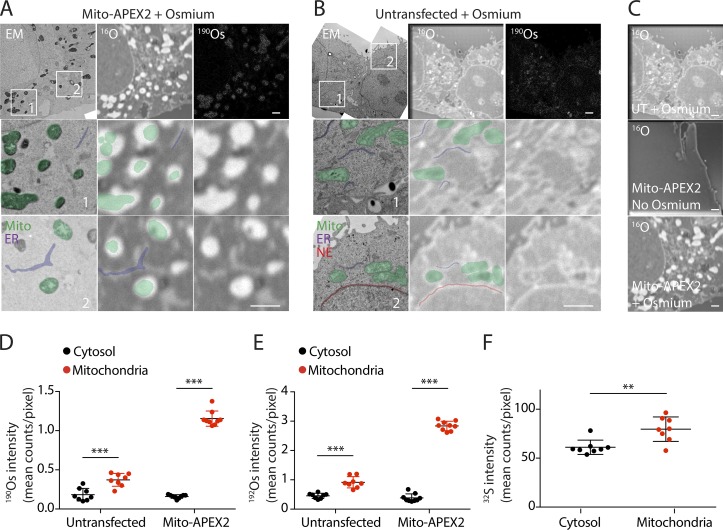
APEX2 produces TEM contrast through deposition of osmium, and since osmium is detectable by NanoSIMS (Meibom and Frei, 2002), we hypothesized that tuning one of the detectors to the major osmium isotopes (i.e., 190Os or 192Os) would allow detection of APEX2 fusion proteins (Fig. 1 A). To test this hypothesis, we expressed three APEX2-fused proteins in HeLa cells: APEX2 fused to the Cox8a mitochondria targeting sequence, directing APEX2 to the mitochondrial matrix; Lamp1-APEX2, targeting APEX2 to the lysosomal membrane; and APEX2-Parkin, which is in the cytosol under basal conditions but is recruited to the outer mitochondrial membrane following collapse of the mitochondrial membrane potential (Narendra et al., 2008). Sequential thin sections were prepared for correlated TEM and MIMS. TEM confirmed targeting of each of the three fusion proteins to the expected structure (Fig. 1 B). In each case, osmium enhancement of the intended structure was apparent, with the greatest osmium staining observed in the mito-APEX2 sample by TEM.
Figure 1.
Detection of APEX2 fusion proteins by MIMS. (A) Schematic depicting detection of APEX2 by the NanoSIMS instrument in parallel with ions abundant in biological samples. APEX2 fused to the gene of interest (GOI) catalyzes the polymerization of DAB, which in turn precipitates OsO4. The sample is probed with a primary beam of positively charged cesium ions. Ionized atoms and polyatomic fragments from the sample surface are focused into a secondary ion beam, which is separated in a magnetic field. Seven parallel detectors are tuned to detect specific ions of interest. (B) Adjacent ultrathin sections of HeLa cells untransfected or transfected with APEX2-Parkin, Lamp1-APEX2, or Mito-APEX2. Samples were treated with DAB and OsO4 and analyzed by TEM or MIMS for osmium ions (190Os or 192Os) or 12C14N. APEX2-Parkin and untransfected cells were treated with the mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP) 10 µM for 5 h to induce translocation of Parkin to the outer mitochondrial membrane. Representative images from a single cell are shown for each condition. These are representative of 14 untransfected HeLa cells (in two technical/biological replicates), 2 APEX2-Parkin transfected HeLa cells (in one technical/biological replicate), 11 Mito-APEX2 transfected HeLa cells (in three technical/two biological replicates), 6 Lamp1-Apex2 HeLa cells (in two technical replicates/one biological replicate). Scale bar in all images = 1 µm.
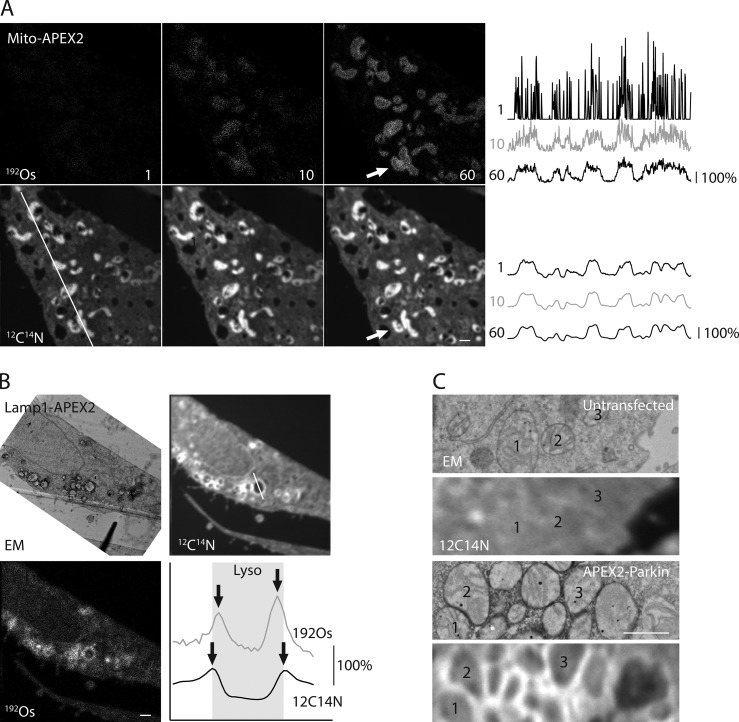
In the mito-APEX2 sample, osmium was detectable in a faint mitochondrial pattern in the first image, which correlated well with the mitochondrial pattern observed in the adjacent ultrathin section analyzed by TEM (Figs. 1 B and 2 A). Each imaging plane is generated from analysis of just the surface atomic layers of the sample; thus, this pattern became more distinct upon summing 60 sequential images. The quality of the 190Os and 192Os images were similar, with signal-to-noise ratios of 7.22 and 7.38, respectively (Fig. S1, A–C). 192Os counts were 236% higher than 190Os counts, consistent with the approximately twofold greater abundance of 192Os compared with 190Os. Generally, the margins of mitochondria could be differentiated in the summed 192Os image, but in some cases, the border between closely positioned mitochondria was indiscernible (arrow in Fig. 2 A). No mitochondrial pattern was observed in an untransfected sample or in the Lamp1-APEX2 transected samples, demonstrating that the osmium pattern in the mito-APEX2 transfected sample is attributable to the localization of APEX2 to this compartment. Similarly, 190Os and/or 192Os was detectable in the expected outer mitochondrial membrane or in a lysosomal pattern following transfection with APEX2-Parkin or Lamp1-APEX2, respectively, and matched that observed by TEM in the adjacent ultrathin sections (Fig. 1 B).
Figure 2.
APEX2 is detected on single organelles by local enhancement of 12C14N. (A) 192Os (top, left) and 12C14N (bottom, left) MIMS images of HeLa cells expressing Mito-APEX2. 1, 10, or 60 consecutive images were summed, and their intensity was auto scaled. Rightmost panels are reproduced from Fig. 1 B. Arrow indicates a cluster of mitochondria with clear boundaries in the 12C14N image but not the 192Os image. Line scans of the region depicted in the leftmost image are shown for 192Os (top, right) and 12C14N (bottom, right). Intensity is relative to the average intensity within each scanned region. Image is representative of 11 Mito-APEX2 transfected HeLa cells (in three technical/two biological replicates). (B) EM, 12C14N, and 192Os MIMS images of HeLa cells expressing Lamp1-Apex2. Left panels are reproduced from Fig. 1 B. Line scan through a single lysosome is shown in the lower right. Arrows depict maximal local intensity of 192Os and 12C14N, indicating Lamp1-positive membranes of lysosomes, and shaded region depicts the limits of the lysosome. Image is representative of six Lamp1-APEX2 HeLa cells (in two technical replicates/one biological replicate). (C) HeLa cells untransfected (top) or transfected with APEX2-Parkin (bottom) and treated with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to induce translocation of Parkin to the outer mitochondrial membrane. Individual mitochondria are identified in the correlated EM (top) and MIMS (bottom) images. Image is representative of two APEX2-Parkin transfected HeLa cells (in one technical/biological replicate). Scale bar in all images = 1 µm.
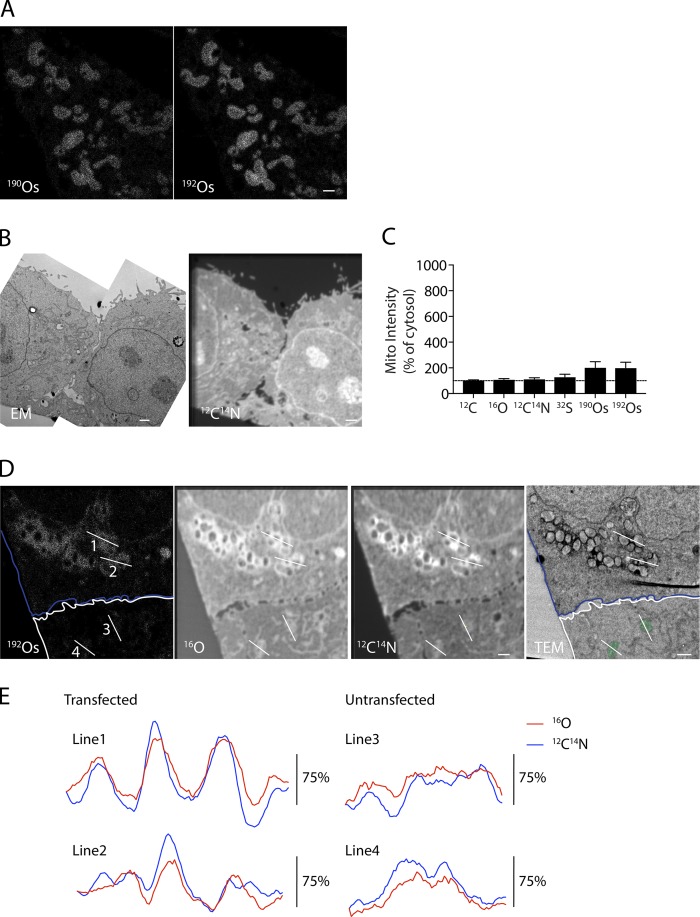
Figure S1.
Detection of APEX2 by MIMS. (A) 190Os (left) and 192Os (right) MIMS images of HeLa cells expressing Mito-APEX2. 192Os panel, which is the same as in Fig. 1 B, is reproduced for clarity. (B and C) Correlated TEM and 12C14N MIMS images of an untransfected HeLa cell. Panels are the same as in Fig. 1 B, reproduced for clarity. (C) Quantification of indicated ion intensities for individual mitochondria relative to cytosol in an untransfected HeLa cell. For this analysis, eight mitochondria from a representative untransfected cell were measured. Error bars represent SD. (D) Field with APEX2-Parkin transfected cell (blue outline) and untransfected cell (white outline) treated with the mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to recruit APEX2-Parkin to the outer mitochondrial membrane. Lines in D indicate position of line scans in E through APEX2-Parkin–labeled mitochondria (lines 1 and 2) and mitochondria identifiable in the 16O image in the absence of a protein marker. Scale bars = 1 µm.
APEX2 localization by transition metal–dependent enhancement of other ions
Unexpectedly, we found enhanced intensity of other ions (e.g., 12C14N) in areas of DAB/OsO4 deposition (Fig. 2 A). As the secondary ion counts of 12C14N were ∼100-fold greater than 192Os, what appeared as a cluster of mitochondria with indistinct boundaries in the summed 192Os image could be seen as three separate mitochondria in the initial 12C14N image. Similar 12C14N enhancement was observed in Lamp1-APEX2– and APEX2-Parkin–transfected but not untransfected samples (Fig. 2, B and C; and Fig. S1, B and C). In each case, individual mitochondria or lysosomes could be identified from 12C14N signal often more distinctly than from the direct 190Os or 192Os signal at identical acquisition times. In images with APEX2 on the cytosolic side of lysosomal or the outer mitochondrial membrane, the membranes could be differentiated from their lumens based on the 12C14N signal (Fig. 2, B and C). Of note, there was greater background in the enhanced 12C14N images compared with the 192Os images; thus, the 12C14N images and 192Os images are complementary, with the former providing superior definition of the organelle boundary and the latter exhibiting decreased background in the summed images.
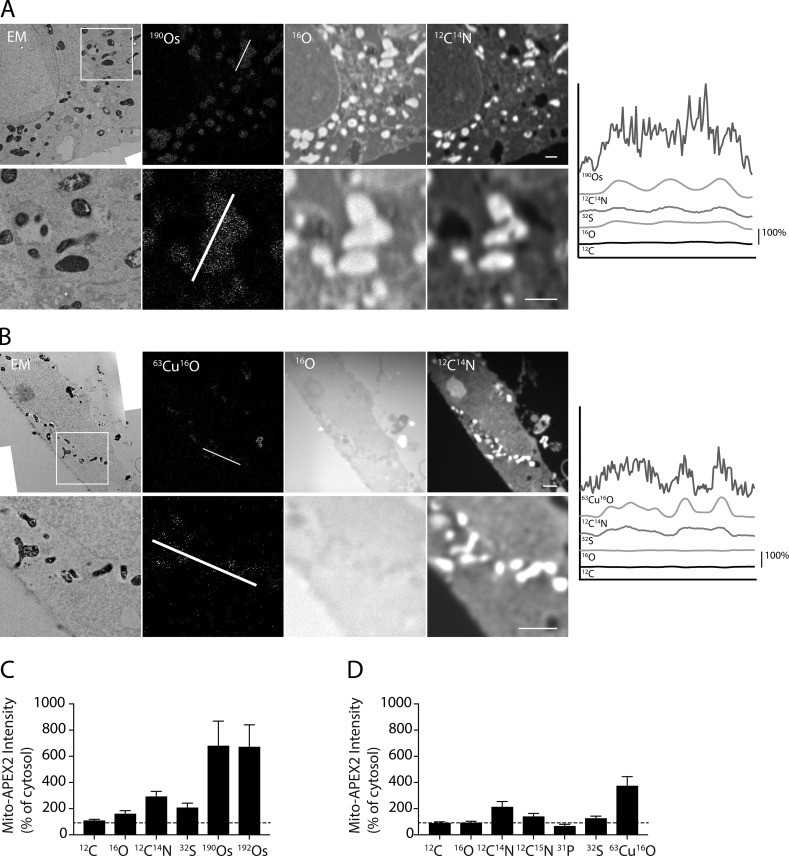
In addition to OsO4, DAB polymers can precipitate other metal-containing compounds, such as CuSO4. To test whether CuSO4 could also be used for the detection of APEX2 by MIMS, DAB-treated cells expressing Mito-APEX2 were either fixed with osmium or stained with CuSO4. As expected, membranes were no longer visible by TEM in the absence of the lipid fixative OsO4, but a mitochondrial pattern was visible in the Mito-APEX2 sample due to the electron-dense copper enhancement of DAB (Fig. 3, A and B; Sabatini et al., 1963). 63Cu16O was directly detectable in a mitochondrial pattern, suggesting that CuSO4 can also be used for APEX2 detection by MIMS (Fig. 3 B). Notably, an enhancement of 12C14N was observed in the region of CuSO4 deposition, similar to what was observed in the OsO4 stained sample. The 12C14N signal allowed for clearer identification of the APEX2-labeled structure than did direct detection of 63Cu16O at the acquisition times used in this study.
Figure 3.
CuSO4 and OsO4 enhance ionization in their vicinity by a matrix effect. (A) HeLa cell transfected with Mito-APEX2, fixed with OsO4, and imaged in adjacent ultrathin sections by EM and MIMS. Location of line scan is indicated in second image. Image is representative of 11 Mito-APEX2 transfected HeLa cells (in three technical/two biological replicates). (B) HeLa cell transfected with Mito-APEX2, stained with CuSO4 (in the absence of OsO4), and imaged in adjacent ultrathin sections by EM and MIMS. Location of line scan indicated in second image. Image is representative of three Mito-APEX2 transfected HeLa cells (in one technical/biological replicate). (C and D) Average intensity of ions from mitochondrial ROIs compared with random ROIs from the cytosol for samples fixed with OsO4 (C) or stained with CuSO4 in the absence of OsO4 (D). 14 mitochondria from one representative cell in A and B were analyzed in each condition. Dotted lines indicate 100% of cytosol intensity. Error bars represent SD. Scale bar in all images = 1 µm.
To further explore the DAB/OsO4 and DAB/CuSO4-related enhancement, we examined the effect of OsO4 and CuSO4 on ions that are relatively abundant in biological samples (12C, 16O, 12C14N, 12C15N [in a sample labeled with 15N-leucine], 31P, and 32S; Fig. 3, C and D). Enhancement of 12C14N, 12C15N, and 32S signals was observed in both DAB/OsO4 and DAB/CuSO4, samples with a mitochondrial pattern matching the TEM image in both MIMS images. Interestingly, 16O was enhanced in the DAB/OsO4 but not the DAB/CuSO4 samples. Enhancement to a lesser extent was also observed for 12C in the DAB/OsO4 sample but not in the DAB/CuSO4 sample.
Notably, 32S is not an element in DAB/OsO4, suggesting that its enhancement is not due to detection of 32S from one of the staining components. This suggests that OsO4 and CuSO4 deposition may have a matrix effect leading to enhanced detection of several elements. In addition to the matrix effect, increased 12C14N may be due in part to direct detection of these elements from DAB. Consistent with this view, the 12C14N (which can be derived from either DAB or protein) exhibited a stronger increase in the vicinity of CuSO4 than did 12C15N (which should have a greater contribution from 15N-leucine labeled protein).
Label-free identification of mitochondria and ER by MIMS
Examination of areas of Mito-APEX2 expression in correlated TEM and 16O MIMS images revealed that the ER membranes were also visible in the 16O MIMS image (Fig. 4 A). This led us to reexamine the TEM and 16O MIMS images from untransfected cells (Fig. 4 B). In the 16O MIMS images that lacked the APEX2 label, ER and mitochondrial membranes were readily identified. The ER appeared as linear or serpentine 16O-intense structures, whereas mitochondria appeared as closed loops of membrane. The nuclear envelope was likewise identifiable as a large curved structure in the center of the cell. These membranes were not visible in the CuSO4-treated samples that lacked OsO4 treatment (with or without APEX2 expression), suggesting that OsO4 fixation is necessary for detection of 16O-intense membranes (Fig. 4 C). Although organelles such as mitochondria were identifiable label-free in the 16O ion images, APEX2 directed to the outer mitochondrial membrane provided greater contrast for identifying mitochondrial boundaries (Fig. S1, D and E).
Figure 4.
ER and mitochondrial membranes are 16O intense in OsO4 fixed samples. (A and B) ER, mitochondria (mito), and nuclear envelop (NE) are identified in correlated EM and MIMS images from HeLa cells transfected with Mito-APEX2 (A) or untransfected (B). Images in A are reproduced from Fig. 3 A, and images in B are reproduced from Fig. 1 B. Images are representative of 5 HeLa cells transfected with Mito-APEX2 (in one technical/biological replicate) and 14 untransfected HeLa cells (in two technical/biological replicates). (C) 16O MIMS images of HeLa cells transfected with Mito-APEX2 (middle and bottom) or untransfected (top) and fixed with OsO4 (top and bottom) or unfixed (middle). Images are reproduced from Figs. 1 B, 4 A, and 4 B. UT, untransfected. Images are representative of 14 untransfected HeLa cells stained with osmium (in two biological/technical replicates) and 1 untransfected HeLa cell stained with CuSO4 in the absence of osmium (in one biological/technical replicate). (D–F) Counts of 190Os (D), 192Os (E), and 32S (F) from mitochondrial and cytosol ROIs of HeLa cells transfected with Mito-APEX2 or untransfected. **, P < 0.01; ***, P < 0.001. For this analysis, seven mitochondria from a representative untransfected cell and nine mitochondria from a representative Mito-APEX2 transfected cell were measured. Scale bar in all images = 1 µm.
Definition of mitochondrial boundaries in the 16O MIMS image should allow measurement of other elements detected in parallel from individual mitochondria. To test this, we selected regions of interest (ROIs) within individual mitochondria from the 16O MIMS images from both untransfected and mito-APEX2–transfected samples and measured 190Os and 192Os abundance (Fig. 4, D and E). We found that 190Os and 192Os abundance in individual mitochondria, which possess OsO4 stained membranes, was approximately twofold greater than that of cytosol. 190Os and 192Os were also more abundant in mitochondria expressing Mito-APEX2 than in individual mitochondria from the untransfected sample (approximately sevenfold of cytosol compared with approximately twofold of cytosol). Thus, the APEX2-induced OsO4 deposition can also be measured by MIMS. As mitochondria are known to contain iron-sulfur clusters, we also hypothesized that the 32S may be more abundant in mitochondria compared with the cytosol (Johnson et al., 2005). Consistent with this hypothesis, we found that the mean 32S concentration was 30% higher in mitochondria than in the cytosol (Fig. 4 F). Together, these findings demonstrated that elemental abundance can be measured in individual mitochondria by MIMS.
Subset of sulfur-rich lysosomes are identifiable label-free by MIMS
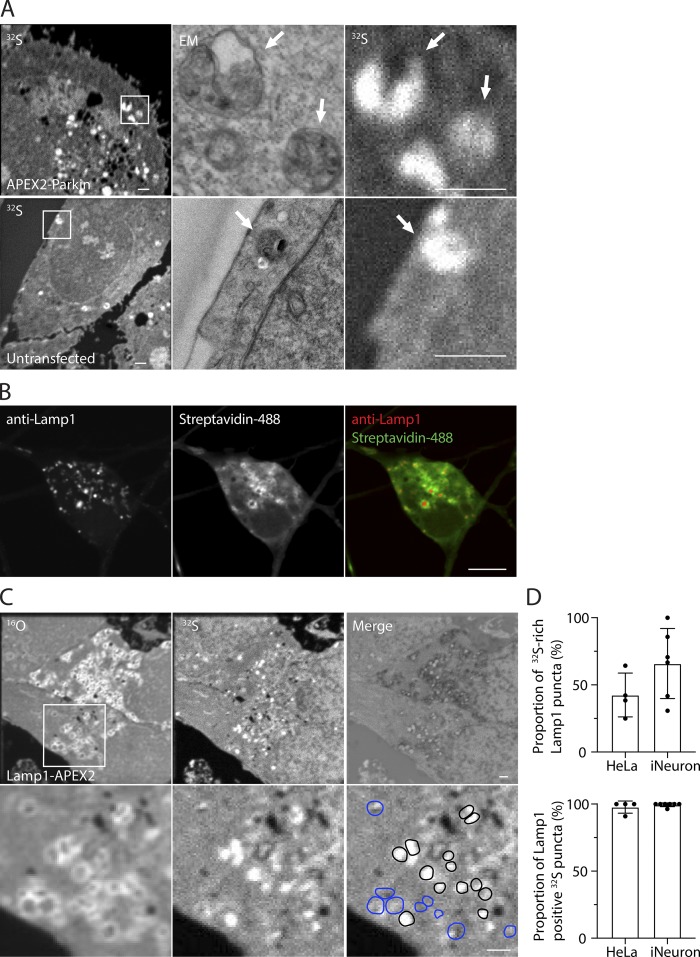
Further examination of samples revealed additional structures that are identifiable in the MIMS images of biologically abundant elements. In HeLa cells from multiple samples, 32S-rich puncta were apparent in the cytosol (Fig. 5 A). Several of the 32S-rich puncta that could be aligned with TEM of adjacent thin sections were consistently identifiable as lysosomes. Sulfur ion counts in 32S-rich lysosomes were significantly greater than in mitochondria from the same cells (213 vs. 130% of cytosol, P = 7.8 × 10−12; n = 20 mitochondria and 11 lysosomes from three HeLa cells). We used transient expression of Lamp1-APEX2 in HeLa cells and induced pluripotent stem cell (iPSC)-derived neurons (iNeurons) stably expressing Lamp1-APEX2 to assess the frequency of putative 32S-rich lysosomes (Fig. 5, B–D). A fluorescent APEX2-dependent substrate (Streptavidin-488) colocalized with Lamp1 immunostaining in iNeurons by confocal microscopy (Fig. 5 B), and Lamp1-APEX2 was apparent as ring-shaped structures by MIMS (Fig. 5 C). A subset of Lamp1-puncta was 32S rich in all cells analyzed, ranging from 30.8 to 100% in iNeurons and 25.0 to 64.3% in HeLa cells (Fig. 5, C and D, top graph). In both HeLa cells and iNeurons expressing Lamp1-Apex2, the majority of these 32S-rich puncta were Lamp1 positive (96.7 and 98.1%, respectively; Fig. 5, C and D, bottom graph).
Figure 5.
Lysosomes are sulfur rich. (A) HeLa cells transfected with APEX2-Parkin (left) or untransfected (right) and treated with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) to induce Parkin translocation to the outer mitochondrial membrane. Arrows identify individual mitochondria in correlated EM and indicated MIMS images. Scale bar in images = 1 µm. Images are representative of 2 APEX2-Parkin transfected cells (in one technical/biological replicate) and 14 untransfected cells (in two technical/biological replicates). (B) Staining of Lamp1-APEX biotinylated prey proteins in iPSC-derived neurons. Representative images from a single cell are shown. Scale bar in image = 10 µm. (C) Neurons derived from WT iPSC cells stably expressing Lamp1-APEX2 imaged by MIMS. Enhancement of 12C14N (top) shows membrane of lysosomes, many of which have relatively abundant 32S in their lumen (middle). Merged image of the inverse of the 12C14N image with 32S image (bottom). Scale bar in images = 1 µm. (D) Top: Proportion of Lamp1-positive lysosomes with sulfur counts >2SD above the mean detected in the cytosol. Bottom: Proportion of sulfur-rich puncta that were Lamp1 positive. 83 Lamp1-positive puncta and 30 sulfur-rich puncta were analyzed from four HeLa cells (WT or ATG5 KO), and 61 Lamp1-positive lysosomes and 53 sulfur-rich puncta were analyzed from six iNeuron cells (WT or GRN−/−). Error bars represent SD. HeLa and iNeuron experiments were performed separately, each in one technical/biological replicate.
Protein turnover assessed in single lysosomes reveals heterogeneity in protein age
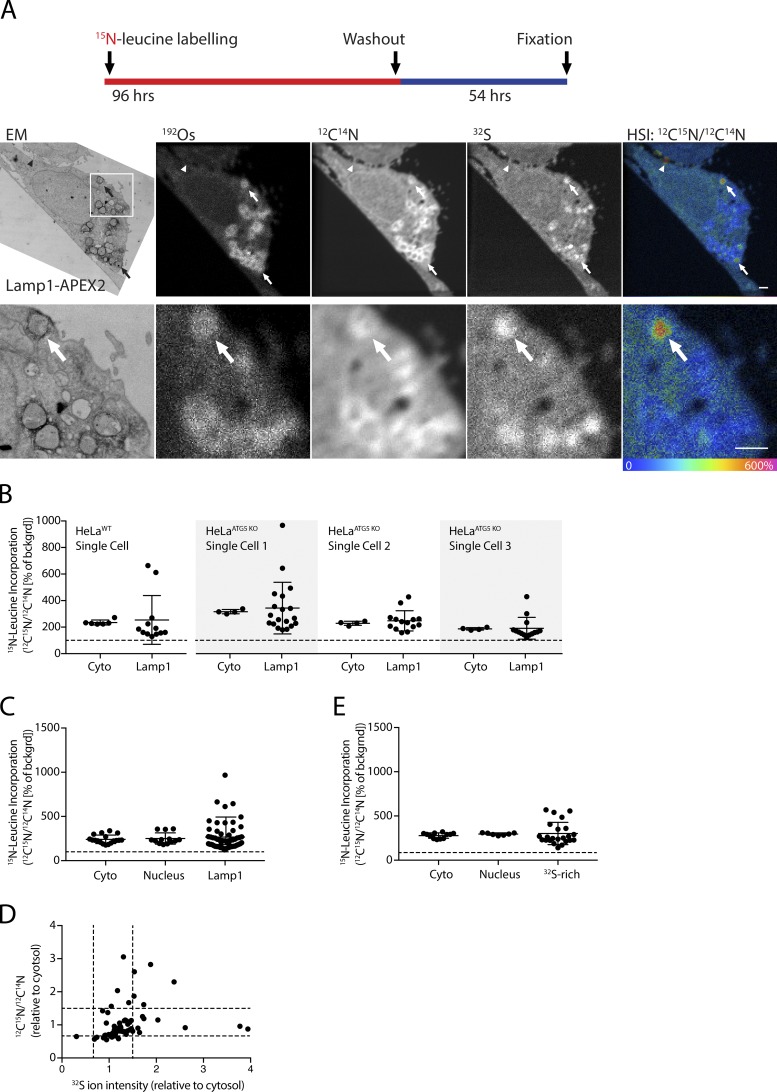
To assess whether APEX2 could be used in conjunction with metabolic labeling of protein, we incubated WT and/or ATG5 KO HeLa cells transfected with Lamp1-Apex2 or Mito-APEX2 in medium containing 15N-leucine as the only source of this essential amino acid in a pulse-chase experiment. Whereas no difference in the pattern of 15N-leucine incorporation was noted between the WT and ATG5 KO cells, in both cell lines there was relative accumulation of 15N-leucine within specific puncta in the cell. When these puncta were adjacent to the plasma membrane, they appeared to localize with a subset of filipodia (Fig. 6 A, arrowhead). In contrast, cytoplasmic 15N-leucine–intense puncta were typically surrounded by a Lamp1-positive membrane, and a lysosome could be identified in the adjacent section by TEM (Fig. 6 A, arrows). This pattern was noted in both Lamp1-APEX2– and Mito-APEX2–expressing HeLa cells and untransfected cells, suggesting that it was not an artifact of Lamp1-APEX2 expression.
Figure 6.
15N-leucine incorporation is heterogeneous among lysosomes in single HeLa cells. (A) Images of HeLa cells expressing Lamp1-APEX2 metabolically labeled with 15N-leucine for 4 d before washout for 54 h and fixation. Arrows indicate lysosomes with high incorporation of 15N-leucine. Arrowhead indicates a filipodia, which also exhibits high 15N-leucine incorporation. HSI image represents the 12C15N/12C14N ratio as a hue and ion counts as an intensity for each pixel. Representative images from a single cell are shown. (B) Quantification of 15N-leucine incorporation into areas of cytosol or individual Lamp1-APEX2–positive lysosomes for single cells treated as described in A. (C) Quantification of 15N-leucine incorporation into areas of cytosol, nucleus, or individual lysosomes (identified by Lamp1-APEX2 labeling) for experiment described in A. Dotted line indicates natural abundance of 12C15N relative to 12C14N. In B and C, four cells (one HeLaWT and three HeLa ATG5 KO) were analyzed (in two technical and one biological replicates). (D) Incorporation of 15N-leucine and 32S abundance of individual lysosomes in experiment described in A. Four cells (one HeLaWT and three HeLaATG5 KO) were analyzed (in two technical and one biological replicates). (E) HeLa cells expressing mito-APEX2 were labeled as in A and 15N-leucine incorporation into areas of cytosol, nucleus, or individual lysosomes (identified by 32S intensity) was measured. Dotted line indicates natural abundance of 12C15N relative to 12C14N. A total of three HeLaWT cells were analyzed (in one technical/biological replicate). Error bars in all graphs represent SD. Scale bar in all images = 1 µm.
To investigate this phenomenon further, 15N-leucine incorporation was assessed in Lamp1-positive lysosomes in HeLa cells. As no difference was observed between WT and ATG5 KO HeLa cells, data from these samples were pooled. This analysis revealed that 15N incorporation into lysosomes was heterogeneous (Fig. 6, A–C). Lysosomes within individual cells exhibited either substantially increased 15N-leucine incorporation or substantially decreased 15N-leucine incorporation relative to the cytosol (Fig. 6, B and C). There was a poor correlation between the sulfur content of Lamp1-positive lysosomes and their incorporation of 15N-leucine (Pearson correlation coefficient [r] = 0.049; Fig. 6 D). Lysosomes identified as 32S-rich structures in unlabeled samples were similarly heterogeneous with respect to their 15N-leucine incorporation, suggesting that the heterogeneity is not an artifact of Lamp1-APEX2 overexpression (Fig. 6 E). Together, these findings suggested that 15N-leucine incorporation into individual lysosomes is heterogeneous within a single cell.
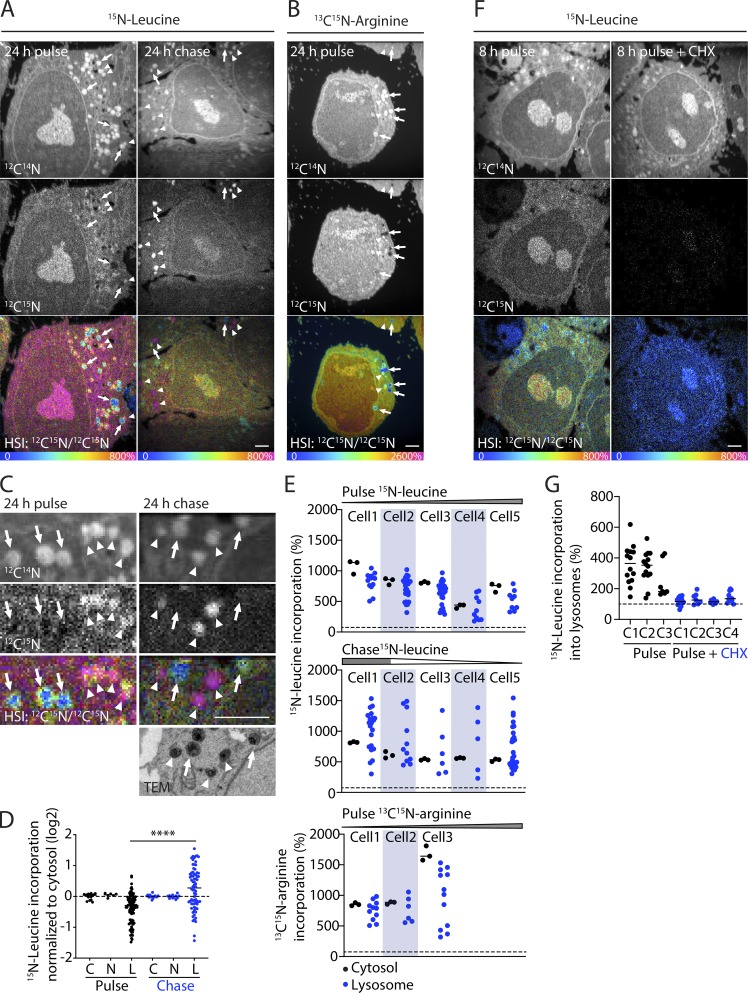
To further assess the robustness of the observed lysosomal heterogeneity, we tested alternative labeling protocols in HeLa cells expressing Lamp1-APEX2 (Fig. 7). We reasoned that if lysosomal heterogeneity in 15N-leucine incorporation reflects differential 15N-leucine metabolism within individual lysosomes, the distributions of 15N-leucine lysosomal incorporation in 24-h pulse and 24-h chase experiments should mirror one another. As expected, with both pulse and chase protocols, individual lysosomes within a given cell often exhibited greater or lesser 15N-leucine incorporation compared with the cytosol (Fig. 7, A–D). This was evident both in the 12C15N/12C14N hue-saturated intensity (HSI) ratio images as well as the 12C15N image alone. In the pulse experiment in particular, puncta with relatively low 12C15N counts (arrows in Fig. 7 B, left) could be observed next to puncta with relatively high 12C15N counts (arrow heads in Fig. 7 B, left), both of which colocalized with Lamp1-APEX2–enhanced puncta in the 12C14N image. Notably, even though heterogeneity was observed with either the pulse or chase protocols, the distributions mirrored each other, with significantly higher mean 15N-leucine incorporation in the chase protocol compared with pulse protocol (75.8 vs. 119% of cytosol, P < 0.001; Fig. 7 D). This indicated that, on average, leucine turnover was slower in lysosomes compared with the cytosol, and that the finding of heterogeneity in 15N-leucine metabolism among lysosomes was durable under different labeling schemes.
Figure 7.
15N-amino acid labeling heterogeneity reflects bulk protein turnover by individual lysosomes. (A–E) HeLa cells transiently expressing Lamp1-APEX2 were metabolically labeled with 15N-leucine (A, left) or 13C15N-arginine (B) before being fixed (24-h pulse) or chased for 24 h following labeling with 15N-leucine for >7 d and fixed (24 h chase; A, right). (A–C) Representative MIMS images are shown for indicated pulse and chase experiments. Arrows indicate Lamp1 puncta with relatively decreased 15N incorporation, and arrowheads indicate Lamp1 puncta with relatively increased 15N incorporation. (C) Magnified areas of representative 15N-leucine pulse and chase MIMS and correlated TEM images. (D) Quantification of 15N-leucine incorporation into the cytosol (C), nucleus (N), and lysosomes (L) normalized to cytosol in each cell and depicted on a log2 scale. Data from five cells from each condition were included in the analysis (from one technical/biological replicate). ****, P < 0.0001. Dotted line indicates natural abundance of 12C15N relative to 12C14N. (E) Quantification of 15N-leucine incorporation into cytoplasm ROIs and lysosomes in single cells. Data from five cells from each condition were included in the 15N-leucine analysis and data from three cells were included for 13C15N-arginine analysis (each from one technical/biological replicate). (F and G) Representative MIMS images (F) and quantification (G) of HeLa cells transiently expressing Lamp1-APEX2 and metabolically labeled with 15N-leucine for 8 h in the presence or absence of 100 µM cycloheximide (CHX). Four untreated HeLa cells and three CHX-treated HeLa cells, each from one technical/biological replicate, were analyzed. Dotted line indicates natural abundance of 12C15N relative to 12C14N. Scale bars in all images = 2.5 µm.
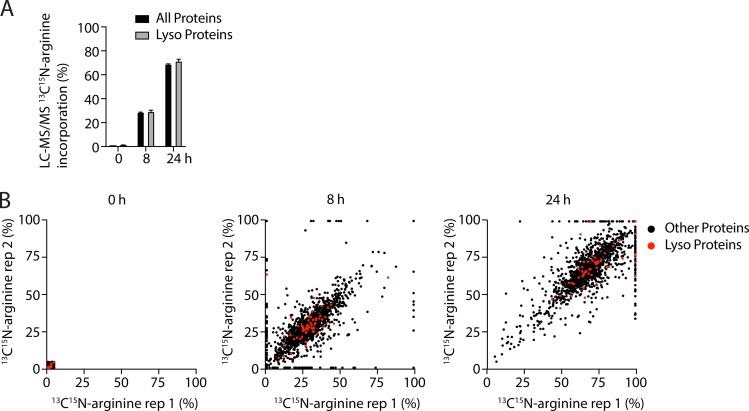
The observed 12C15N signal in lysosomes may be derived from 15N-leucine incorporated into bulk protein in lysosomes; alternatively, it may be derived from free 15N-leucine or one of its metabolites (e.g., acetyl-CoA; Schepartz and Turczyn, 1963). To distinguish between these possibilities, we pulsed HeLa cells with an amino acid, 13C15N-arginine, that is differentially metabolized within lysosomes (and notably, is not converted to acetyl-CoA). 13C15N-arginine exhibited a similar pattern of heterogeneous amino acid incorporation into lysosomes within individual cells, consistent with the 12C15N/12C14N ratio reflecting predominantly bulk protein turnover (Fig. 7, B and E). We next assessed 13C15N-arginine incorporation into tryptic peptides monitored by liquid chromatography (LC)-MS/MS (Fig. S2, A and B; and Table S1). 13C15N-arginine was used in this experiment instead of 15N-leucine, because the majority of tryptic peptides contain at most one arginine residue, and 13C15N-arginine produces a mass shift of 10 D (compared with 1 D for each 15N-leucine residue), both of which simplify proteomic analysis. As expected, we observed 13C15N-arginine progressively incorporated into proteins during the pulse, labeling on average 71.3% of arginine residues in lysosomal proteins and 68.8% in all other proteins at 24 h (P = 0.213). These findings demonstrated that after a 24-h pulse, the majority of HeLa cell proteins have incorporated and are not saturated by 15N-labeled amino acids. Taken together, these data are consistent with the 12C15N signal reflecting 15N-amino acid incorporation into protein.
Figure S2.
13C15N-arginine incorporation into proteins detected by LC-MS/MS. (A and B) Whole-cell lysates from HeLa cells pulsed with 13C15N-arginine for 0, 8, or 24 h were digested to tryptic peptides and measured by LC-MS/MS. Experiment was performed in two technical replicates/one biological replicate. (A) Graph represents average 13C15N-arginine incorporation into the 1,890 quantified proteins that passed quality control, and 55 proteins resident to the lysosome (based on inclusion in the Human Lysosome Gene Database; Brozzi et al., 2013). Technical replicates were averaged. Error bars represent SD. (B) Scatterplots represent percent incorporation into individual lysosomal (red) or other (black) proteins in each of the two replicates at the three time points.
To further verify that the 12C15N/12C14N ratio reflected protein turnover using an orthogonal approach, HeLa cells were pulsed with 15N-leucine for 8 h in the presence or absence of protein translation inhibitor cycloheximide. We reasoned that if 12C15N/12C14N ratios in lysosomes reflect bulk protein turnover, this ratio should depend on protein translation. Consistently, the average 12C15N/12C14N ratio within Lamp1-positive structures exceeded natural abundance after the 8-h pulse in the absence but not the presence of cycloheximide (329 vs. 125% of natural abundance, P < 0.0001; for n = 35 lysosomes from three cells and n = 49 lysosomes from four cells, respectively; Fig. 7, F and G). This demonstrated that the 12C15N/12C14N signal predominantly reflects 15N-leucine incorporated into bulk protein during the pulse. Considered together, these findings established that protein turnover is heterogeneous among individual lysosomes within the same cell.
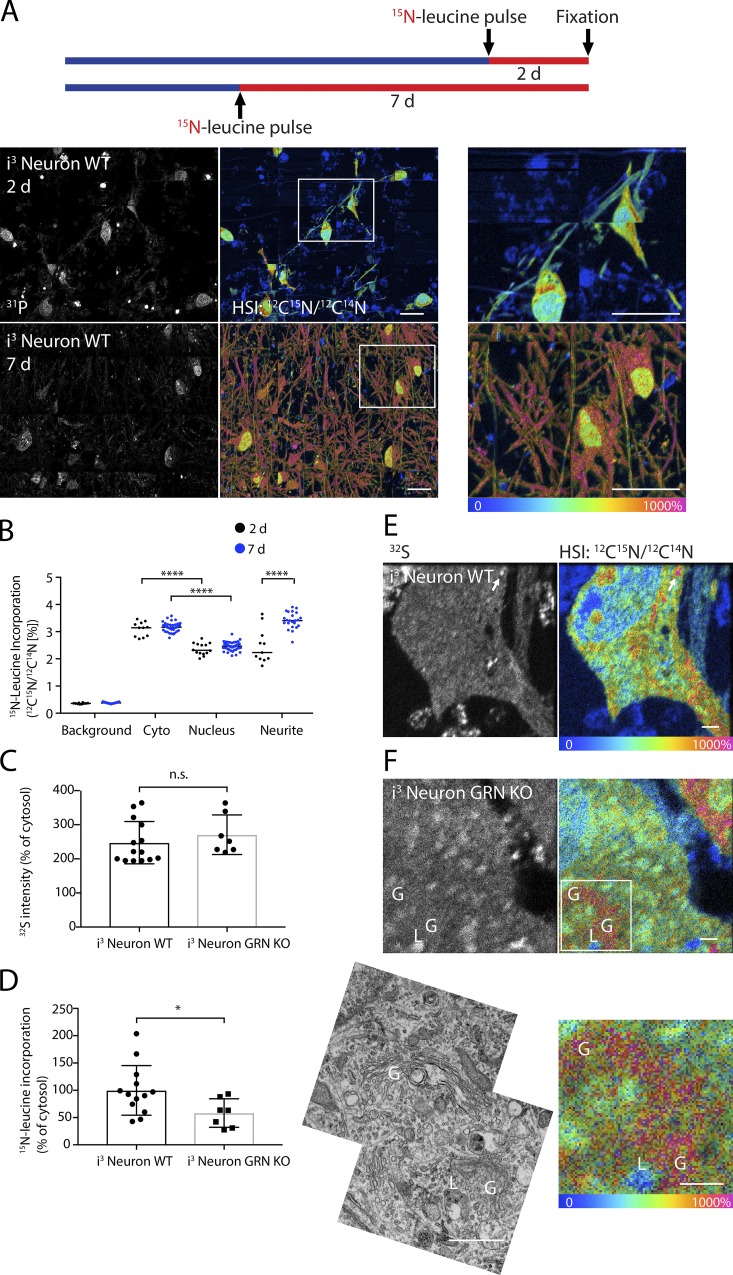
To assess whether lysosomal heterogeneity is a general phenomenon, we next examined 15N-leucine incorporation in a postmitotic cell type: neurons induced from iPSC cells by expression of the transcription factor, neurogenin-2 (Zhang et al., 2013). Unlike the rapidly dividing HeLa cells pulsed for 24 h, WT neurons exhibited substantially less 15N-leucine incorporation in the nucleus compared with the cytosol after a 2-d pulse (Fig. 7, A and D; and Fig. 8, A and B). This likely reflects relative stability of protein within the nucleus of these postmitotic cells, in contrast to high turnover of nuclear protein in the rapidly dividing cancer cell line. After 7 d of labeling, there was no additional incorporation of 15N-leucine in the nucleus or the cytosol of the cell soma compared with that at 2 d, suggesting that protein incorporation was maximal in these regions by 2 d. This was in contrast to the surrounding neurites, which had decreased incorporation of 15N-leucine at 2 d but reached levels similar to the cell soma cytoplasm by 7 d (Fig. 8, A and B). This delay in the incorporation of 15N-leucine into neurites compared with the cell soma likely reflects a lag in the arrival of newly synthesized proteins from the soma to the surrounding neurites.
Figure 8.
Young protein accumulates in a subset of lysosomes in cultured neurons. (A) Mosaic MIMS images of WT iPSC-derived neurons metabolically labeled with 15N-leucine for 2 or 7 d before fixation. Cell nuclei, which have abundant nucleic acid, are readily identified in 31P images. HSI image represents the 12C15N/12C14N ratio as a hue and ion counts as an intensity for each pixel. Scale bar = 20 µm. (B) Quantification of 15N-leucine incorporation into the cytosol, nucleus, and neurites at 2 and 7 d. 14 neurons and 50 neurons, respectively, were analyzed for 2- and 7-d 15N-leucine pulse groups, each in one technical/biological replicate. ****, P < 0.0001. (C and D) 32S content (C) and 15N-leucine incorporation (D) of lysosomes (identified as 32S-enriched puncta) from GRN+/+ and GRN−/− neurons. n.s., not significant; *, P < 0.05. Lysosomes from four cells of each genotype were measured, each in one technical/biological replicate. Error bars in all graphs depict SD. (E and F) MIMS images of GRN+/+ (E) and GRN−/− neurons (F). Arrow indicates lysosome in E. Golgi apparatus (G) and a lysosome (L) are identified in F. Scale bar = 1 µm.
As was observed in HeLa cells, 15N-leucine incorporation into lysosomes within individual WT neurons was more heterogeneous than was observed in the cytosol, with one or more 15N-leucine–intense/32S-rich puncta identified in most WT neurons (Fig. 8, C–E). In neurons, we also occasionally encountered a crescent-shaped structure with very avid incorporation of 15N-leucine (Fig. 8 F). TEM of the adjacent sections showed these structures to be Golgi apparatus, suggesting that the concentration of 15N-leucine in these structures likely reflects newly synthesized proteins destined for the plasma membrane or secretion.
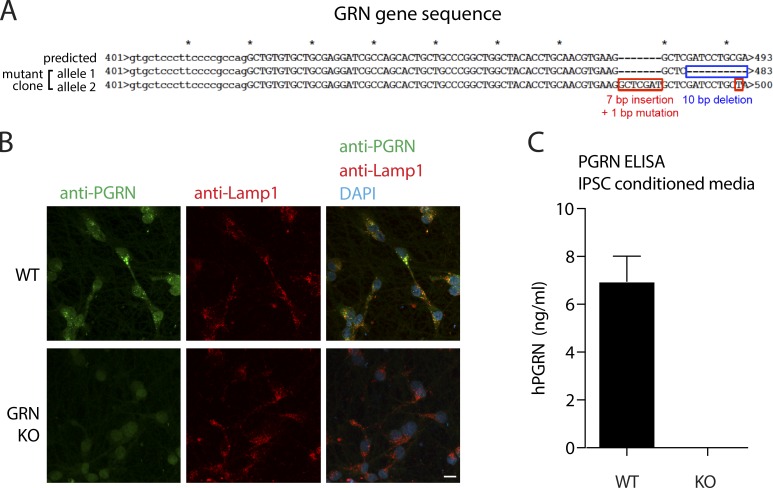
We next assessed whether protein turnover is altered in neurons lacking the lysosomal protein progranulin, a protein implicated in familial lysosomal storage diseases (Smith et al., 2012; Ward et al., 2017; Valdez et al., 2017). Disruption of the granulin gene GRN was verified by Sanger sequencing (Fig. S3 A); loss of progranulin protein was confirmed by immunocytochemistry and absence of secreted progranulin protein in conditioned medium (Fig. S3, B and C). In contrast to the WT neurons, 32S-positive lysosomes in neurons lacking the lysosomal protein progranulin exhibited decreased 15N-leucine incorporation and decreased lysosomal heterogeneity, likely as a consequence of the known lysosomal dysfunction phenotype in this model (Fig. 8, C and F; Smith et al., 2012; Ward et al., 2017; Valdez et al., 2017). This was in contrast to the 32S content, which was elevated to a similar degree in WT and progranulin-null neurons (Fig. 8 C). Together, these findings demonstrate that metabolic information can be measured from individual organelles in situ, identified by APEX2-labeling or unique ion signatures in standard TEM-prepared samples.
Figure S3.
Characterization of GRN−/− iPSCs. (A) GRN genotyping. Sanger sequencing of PCR-amplified GRN alleles was performed for an iPSC clone after transfection with Cas9 nickase and GRN-targeted sgRNAs. The sequences of each GRN allele from a single iPSC clone are shown compared with the predicted GRN sequence. (B) Progranulin immunostaining in WT and GRN-KO iPSC-derived neurons. Neurons were differentiated from WTC11 and GRN−/− iPSCs, followed by anti-progranulin and anti-Lamp1 antibodies. Intralysosomal progranulin (insets) was detected in WTC11 iPSC-derived neurons but not GRN−/− iPSC-derived neurons. (C) Progranulin ELISA of conditioned medium harvested from WT and GRN-KO iPSCs. Error bar represents SEM. n = 6. Scale bars = 10 µm.
Discussion
MIMS is unique in its ability to track and quantify nonradioactive, stable isotope labels by measurement of isotope ratios at submicron resolution. However, the generalizability of this technique in cell biology has been limited by the lack of contrast for distinguishing common organelles, such as mitochondria, ER, and lysosomes, from which metabolic information is to be extracted. Here, we introduce APEX2 as a genetically encoded reporter for MIMS, greatly expanding the MIMS toolkit for identification of subcellular structures within biological samples. When coupled with quantification of stable isotope tracers, parallel interrogation of metabolic function became readily feasible.
In this study, we demonstrated that APEX2 was detectable either by direct measurement of a DAB-precipitated transition metal or by discrete enhancement of 12C14N in the vicinity of the transition metal. Mitochondria and lysosomes were readily identified using APEX2 tagging of classic marker proteins, as well as Parkin, a cytosolic protein that is recruited to mitochondria under conditions of mitochondrial stress. We also observed that OsO4 fixation alone identified mitochondria, ER, and lysosomes, presumably due to the known interaction between OsO4 and lipid-containing membranes (Sabatini et al., 1963). Alternative strategies to identify subcellular structures with NanoSIMS include detection of metal-conjugated antibodies and correlative imaging approaches, each with its complementary advantages and drawbacks (Lechene et al., 2007; Angelo et al., 2014; Thiery-Lavenant et al., 2014). Identification of antigens with lanthanide- or nanogold-conjugated antibodies allows identification of structures without genetic modification. However, the challenge of immunostaining resin-embedded sections limits the range of detectable antigens; sparse staining limits definition of membrane boundaries; and there is frequently a tradeoff between antigen accessibility and preservation of membrane structure. Correlative imaging requires cumbersome alignment and can be particularly challenging at submicron resolution. The APEX-MIMS strategy has the advantage of generalizability to proteins tolerating the APEX protein tag, while providing contrast that is colocalized with functional metabolic measurements. Potential drawbacks of the APEX-MIMS technique include the need to fuse the APEX2 tag to a target protein, which may alter its behavior, and the need to genetically modify the cell or organism. The expanded toolkit, nonetheless, makes it more likely that a solution can be found for a given biological question benefiting from MIMS analysis.
We were surprised to find that APEX2 could be identified not only through direct detection of osmium but also through enhancement of abundant ions in its vicinity, such as 12C14N. We considered the possibility that this enhancement was attributable to increased local abundance of elements contained in the DAB/OsO4 complexes. Indeed, the particularly strong 12C14N signal relative to the 12C15N signal in samples enriched with 15N-leucine is consistent with some contribution from 14N-rich DAB. If true, then the diluting effect of DAB may result in less reliable 12C15N/12C14N ratio measurements in the direct area of enhancement. Because the enhancement of 12C14N is highly localized to the area of DAB deposition, however, any such dilution effect should also be highly localized. As a practical matter, this suggests that a membrane-targeted APEX2-fusion protein may be superior to a luminal marker for accurate measurement of isotope ratios in the organelle lumen. It also raises the intriguing possibility that direct detection of stable isotope–labeled DAB may provide an alternative means of identifying APEX2-labeled structures.
An alternative explanation for enhancement of ions in the immediate vicinity of transition metal–containing compounds is a matrix effect, as has been observed previously in applications of secondary ion MS (Wu and Odom, 1996). The lack of enhancement of carbon, which is an elemental component of DAB, together with the observed enhancement of ions not contained in DAB/OsO4 complexes (e.g., sulfur), suggests that the matrix effect is indeed a contributing factor. Irrespective of the underlying mechanism, however, the enhancement of 12C14N provides several advantages. First, it greatly increases analytical throughput. Physical positioning of detectors to measure 12C15N and 12C14N is not possible when one detector is tuned to masses in the range of the osmium isotopes. This necessitates more time-intensive sequential analysis of the isotope ratio followed by osmium. While this concern is obviated when APEX2 is identified by CuSO4 (because the lighter copper isotopes can be captured in parallel with 12C15N and 12C14N), even greater gains in efficiency are achievable by using enhancement of CN, which is much more abundant than the osmium or copper signals. Second, shorter acquisition translates into improved depth resolution, because fewer cumulative planes are required to achieve a sufficient signal. This may prove critical for analyses of small structures that are limited in the z-plane, for which acquisition of sufficient osmium counts might not be feasible.
We unexpectedly observed that organelles such as mitochondria and ER can be identified label-free in the 16O ion image in OsO4-fixed samples. This may prove useful for identifying organelles in the presence or absence of APEX2-labeled structures. The label-free identification can be regarded as complementary to the APEX2 labeling of organelles, which provides greater contrast and additionally can label molecularly distinct structures (e.g., depolarized mitochondria that have recruited Parkin) that are not identifiable by morphology alone.
With an enhanced capacity to identify subcellular structures, we examined the intracellular distribution of 15N-leucine incorporation. The contrast for lysosomes provided by Lamp1-APEX2 coupled with the submicron spatial resolution of the NanoSIMS instrument for the first time enabled measurement of 15N-leucine incorporation by individual lysosomes, revealing marked heterogeneity of lysosomal 15N-labeling within individual cells. As protein translation was required to observe 15N-leucine incorporation into lysosomes, and similar labeling was observed with 13C15N-arginine, we interpret the heterogeneous incorporation of 15N-leucine into lysosomes to reflect differential protein turnover among lysosomes within a single cell. Mean 15N-leucine incorporation into lysosomes was greater during a 15N-leucine chase compared with a 15N-leucine pulse of the same length, indicating that protein turnover in lysosomes, while heterogeneous, was on average decreased compared with the cytosol. Notably, there was poor correlation between lysosomal 32S content and 15N-leucine incorporation, suggesting heterogeneity among individual lysosomes within single cells along at least two dimensions. Collectively, these data are consistent with previously unappreciated heterogeneity of lysosomes within individual cells, inclusive of marked heterogeneity of lysosomal protein age.
Among the population of heterogeneously labeled lysosomes, a subpopulation had younger protein than the cytosol, incorporated either as part of lysosomal biogenesis or alternatively arising from the delivery of young protein to the lysosome for turnover. Although the proteasome is traditionally thought to regulate the turnover of short-lived proteins, recent studies suggest that the lysosome may also be used as a quality control pathway for misfolded protein in the secretory pathway (Satpute-Krishnan et al., 2014; Omari et al., 2018). The subpopulation of lysosomes with higher protein turnover appeared to represent a functional population, as protein turnover was decreased in cells lacking the lysosomal protein progranulin. Heterozygous loss-of-function progranulin mutations cause frontotemporal dementia, an adult-onset neurodegenerative disease, and homozygous mutations cause neuronal ceroid lipofuscinosis, a pediatric lysosomal storage disorder. These observations have strongly suggested that progranulin acts to regulate key aspects of lysosomal biology, and that loss-of-function progranulin mutations cause neurodegeneration primarily through lysosomal dysfunction (Baker et al., 2006; Smith et al., 2012; Ward et al., 2017; Valdez et al., 2017). Notably, the majority of lysosomes in progranulin-null neurons contained protein that was older than the surrounding cytoplasm, perhaps reflecting a general decrease in protein turnover. This finding is consistent with prior observations that progranulin-deficient cells accumulate undigested protein aggregates within lysosomes (Ward et al., 2017; Valdez et al., 2017).
In summary, we repurpose APEX2 as the first genetically encoded marker for MIMS and demonstrate its use with stable isotope labeling to measure protein metabolism within single organelles in situ. This strategy should be generalizable to any subcellular structure that can be identified by a fusion protein and any molecule that can be labeled with a stable isotope and detected by MIMS. The method should also be generalizable to the detection of other reactions involving DAB, such as HRP-conjugated secondary antibodies and in situ detection of enzyme activities, such as that of the electron-transport chain complex, cytochrome c oxidase (Seligman et al., 1968). As APEX2 labeling has recently been demonstrated ex vivo in tissues from animal models, such as fruitflies and mice, this APEX2-MIMS method should also enable measurement of in vivo metabolic fluxes at the level of individual organelles (Chen et al., 2015; Daigle et al., 2018). We anticipate this method will greatly expand the MIMS toolkit, enabling measurement of metabolism within single organelles in situ.
Materials and methods
Cell culture
Matched HeLa cells ATG5 knockout (KO) and WT were generously donated by R.J. Youle (National Institutes of Health, Bethesda, MD) and were described previously (Nezich et al., 2015). HeLa cells were cultured in DMEM with high glucose, l-glutamine, and pyruvate (Gibco) completed with 10% FBS and penicillin/streptomycin (Gibco; complete DMEM). Complete DMEM lacking leucine (Sigma-Aldrich) was supplemented with 15N-l-leucine (Cambridge Isotopes) for metabolic labeling experiments or 13C6,15N4-l-arginine (Cambridge Isotopes) in SILAC (stable isotope labeling with amino acids in cell culture) DMEM media lacking l-lysine and l-arginine (Thermo Fisher Scientific). The WTC11 iPSC line, original derived from a healthy 30-yr-old Japanese man, was used for all iPSC-related experiments (Miyaoka et al., 2014). For routine culture, iPSCs were plated on Matrigel-coated dishes (Corning), dissociated with Accutase when needed, and cultured in E8 medium (Life Technologies) with Y-27632 rock-inhibitor (Millipore). The GRN gene was deleted from the parental WTC11 line using CRISPR-Cas9 technology. Briefly, cells were transfected via nucleofection with dual Cas9 Nickase (plasmid pX335) and plasmids expressing two sgRNAs targeting the GRN gene (5′-GAAGGCTCGATCCTGCGAGAAGG-3′, 5′-CGTTGCAGGTGTAGCCAGCCGGG-3′) from a U6 promoter. Individual clones were picked and screened for indels in the GRN gene via PCR. A single clone, in which a 7-bp insertion in one GRN allele and 10-bp deletion in the other GRN allele occurred and were predicted to cause loss of GRN expression, was chosen for further characterization. Loss of GRN expression in this clone was confirmed by progranulin ELISA and progranulin immunostaining. Next, we integrated a doxycycline-inducible mouse NGN2 transgene into the AAVS1 safe harbor locus (Fernandopulle et al., 2018) and a doxycycline-inducible Lamp1-APEX2 transgene into the CLYBL safe harbor locus using site-specific TALENs (transcription activator-like effector nucleases) in the GRN mutation iPSC line and parental control WTC11 iPSC line. iPSC clones were picked, expanded, and validated for doxycycline-induced neuronal differentiation and Lamp1-APEX localization and activity via anti-Lamp1 and streptavidin-488 immunostaining. For metabolic labeling experiments, iPSC-derived neurons were incubated in leucine-free DMEM/F-12 (Sigma-Aldrich) supplemented with 15N-l-leucine (Cambridge Isotopes).
Cloning/transfection
APEX2-Parkin, Lamp1-APEX2, and Mito-APEX2 were generated by restriction digestion cloning from YFP-Parkin (Narendra et al., 2008), Lamp1-GFP (Addgene, plasmid 16290, gift from R. Vale [University of California San Francisco, San Francisco, CA]), and Cox8a-EYFP vectors (Clontech), into the HindIII and BamHI sites of Vimentin-APEX2 (Addgene, plasmid 66170) or APEX2-Tubulin (Addgene, plasmid 66171), which were gifts from A. Ting (Stanford University, Palo Alto, CA). The Lamp1-APEX2 targeting vector for the CLBYL safe harbor site was generated by scarless cloning of the PCR product containing Lamp1-APEX2 into a CLYBL targeting vector modified from pC113N-iCAG.copGFP (Addgene, plasmid 66577, gift from J. Zhou [National Heart, Lung, and Blood Institute Intramural Program, National Institutes of Health, Bethesda, MD]), using NEBuilder HiFi DNA Assembly (NEB). For transient transfection of HeLa cells, cells were seeded onto uncoated 6- or 12-well plates and transfected with the indicated APEX2-fusion protein plasmids using Fugene HD (Promega), Lipofectamine 2000 (Invitrogen), or XtremeGENE9 (Roche), following the manufacturer’s instruction. In the case of iPSC neurons expressing Lamp1-APEX2, a stable cell line was created by integration into the CLYBL safe-harbor site, using pZT-C13-L1 and pZT-C13-R1 (Addgene plasmids 62196 and 62197, gifts of J. Zhou), encoding TALENs targeting the CLYBL site, and the Lamp1-APEX2 donor vector. Cells were seeded onto 6- or 12-well plates that were coated with poly-ornithine.
Processing for TEM/MIMS
For labeling with DAB, cells were fixed at the indicated time points with 2% glutaraldehyde (Electron Microscopy Services) in EM buffer (100 mM sodium cacodylate and 2 mM CaCl2). Cells were washed three times in EM buffer and stained with ImPACT DAB (Vector) for 2.5–10 min while the reaction was monitored by bright-field microscopy. The reaction was stopped by washing three times with EM buffer. Cells were subsequently refixed for ≥24 h in 2% glutaraldehyde in EM buffer. For experiments with CuSO4 counterstaining, samples were treated with the CuSO4-based DAB Enhancer BOND (Leica Biosystems) for 10 min after DAB polymerization. Except where specified, cells were osmicated with reduced 1% OsO4/1.5% KCN for 30 min, followed by several washes with dH2O. Samples were dehydrated with a graded cold EtOH series up to 100%. For epoxy embedding, cells were lifted from the culture dishes with propylene oxide and pelleted in an Eppendorf tube before addition of TAAB-812 epoxy resin mixture. Alternatively, cells were epoxy resin embedded directly in the tissue culture plate. Alternating thin (80-nm) and thick (200-nm) sections were cut from the sample with an ultrasectioning diamond knife and mounted onto an EM grid and silicon chip, respectively, such that the top surfaces of the two sections were adjacent. For TEM, thin sections were imaged with a JEOL 1200 EXII Transmission Electron Microscope.
NanoSIMS
Analyses were performed at the Brigham and Women’s Hospital Center for NanoImaging with the NanoSIMS 50L (Cameca). The standard analysis utilizes a 16-keV cesium ion beam for the analysis of negatively charged secondary ions, similar to previously published analytical methods (Steinhauser et al., 2012; Kim et al., 2014). The detectors were configured in various combinations to measure 12C, 16O, 12C14N, 12C15N, 31P, 32S, and/or one of two transition metals: Cu or Os. Os was quantified as either the 190Os or 192Os isotopic variant. Similar to our prior work with Zn (Kumar et al., 2016; Lu et al., 2018), Cu was captured as an oxide (63Cu16O). Images were visualized and analyzed using the freely available custom plugin to FIJI/ImageJ (National Institutes of Health) called OpenMIMS, version 3.0 (https://github.com/BWHCNI/OpenMIMS/wiki/Installation). For each field analyzed, images within the z-stack were first aligned using the “Autotrack” function in the “Stack Editing” tab. Data in successive images were then summed using the “Compress” function, also in the “Stack Editing” tab. HSI images representing the 12C15N/12C14N ratios were generated using the “Display HSI” function in the “Process” tab. The lower end of the ratio range was set to 37, corresponding to the natural abundance for 12C15N of ∼0.037%, and the upper end was adjusted to a multiple of 37 that placed average cytosol enrichment approximately within the middle of the range. ROIs were drawn in the MIMS ROI Manager, and the corresponding counts of the desired ions were extracted using the “table” function in the “Tomography” tab.
LC-MS/MS
Whole-cell lysates from HeLa cells were electrophoresed into the top of an SDS-PAGE gel but not further separated. A single gel slice from each sample was digested with trypsin. Extracted peptides were desalted and used for LC-MS/MS data acquisition on an Orbitrap Luminos mass spectrometer (Thermo Fisher Scientific) coupled with a 3000 Ultimate HPLC instrument (Thermo Fisher Scientific). Peptides were separated on an ES803 column (Thermo Fisher Scientific) with mobile phase B increasing from 2 to 27% over 200 min. The LC-MS/MS data were acquired in data-dependent mode. The resolution of the survey scan (375–1,600 m/z) was set at 120,000 at m/z 400, with a target value of 10 × 106 ions. Collision-induced dissociation was performed on as many precursor ions as allowed in 3 s. The isolation window is 1.6 D. Database search and heavy/light ratio calculation were performed using Proteome Discoverer 2.3 (Thermo Fisher Scientific) against Sprot Human database. Oxidation (M) and label:13C(6)15N(4) (R) were included in the database search. Proteins that were not detected in all six samples (three time points in duplicate) and were measured to have >5% 13C15N-arginine incorporation at 0 h or <5% 13C15N-arginine incorporation at 24 h were excluded from further analysis. Proteins were determined to be lysosomal if they were found in the Human Lysosome Gene Database (Brozzi et al., 2013).
Human progranulin ELISA
We coated high-binding 96-well enzyme immunoassay/radioimmunoassay plates (Corning) overnight with a monoclonal antibody raised against the C-terminus of human progranulin (PGRN; 1.5 µg/ml; a gift of Laura Mitic, Bluefield Project, San Francisco, CA) and blocked with 1% BSA for 1 h at 37°C. We then incubated plates with conditioned medium collected for 24 h from GRN+/+ and GRN−/− iPSCs or recombinant human PGRN (0–32 ng/ml, R&D Systems) for 1 h at 37°C. We then incubated plates with an N-terminal monoclonal PGRN antibody (1.5 µg/ml; a gift of Laura Mitic), anti-mouse biotinylated IgG (1:5,000, Vector Laboratories), and streptavidin-HRP conjugate (1:10,000, Thermo Fisher Scientific). We developed the reactions at room temperature using 3,3′,5,5′-tetramethylbenzidine substrate (Thermo Fisher Scientific), quenched reactions with 1 N HCl, and performed analysis at 450 nm on a SpectraMax M5 spectrophotometer (Molecular Devices).
Progranulin immunostaining
WTC11 and isogenic GRN−/− iPSCs were differentiated by addition of doxycycline in neuronal induction medium, as described by Fernandopulle et al. (2018). 3 d after induction, neurons were dissociated with Accutase and replated on poly-ornithine–coated coverslips in neuronal media. Neurons were grown for an additional 7 d, followed by incubation with phenol-biotin, 2 s of H2O2, fixation, Lamp1/streptavidin immunostaining to detect lysosomes, and APEX-labeled. In brief, after fixation for 30 min with freshly prepared 4% PFA in PBS, coverslips were washed three times in PBS, then blocked/permeabilized in PBS + 0.1% saponin + 3% BSA, then incubated overnight at 4°C with anti-PGRN antibody (1:300, goat polyclonal; R&D Systems) and anti-Lamp1 (1:1,000, mouse monoclonal H4A3). Coverslips were washed three times in PBS, stained with anti-goat Alexa Fluor 488, anti-mouse Alexa Fluor 555, and DAPI in blocking solution for 1 h at room temperature, washed three times with PBS, mounted, and imaged on an inverted Nikon spinning-disc confocal microscope.
Streptavidin staining
WTC11 iPSCs expressing doxycycline-inducible NGN2 and Lamp1-APEX were differentiated into neurons as described above in the presence of doxycycline for the duration of culture to maintain transgene expression. Then, neurons were incubated with phenol biotin (CAS 41994-02-9, Adipogen) for 30 min at 37°C, followed by brief treatment with 1 mM H2O2 for 2 s and immediate quenching/fixation in PBS + 10 mM sodium azide + 4% PFA. Although similar results were obtained with 1-min treatment, the shorter pulse was found to be sufficient for signal and minimized background. Cells were fixed for 10 min, followed by Lamp1 immunostaining and streptavidin Alexa Fluor 488 staining of biotinylated APEX prey proteins. Staining was done as described in the progranulin immunostaining method, except that anti-PGRN was omitted and Streptavidin Alexa Fluor 488 (Life Technologies) was incubated during the secondary antibody incubation step at 1:300. Imaging was performed using a Nikon Eclipse Ti2 spinning disk equipped with a 100× oil objective lens at room temperature (NA 1.4).
Statistical analysis
For all statistical analyses, two-sample t tests (two-tailed) were performed in Excel (Microsoft) or Prism (GraphPad).
Online supplemental material
Fig. S1 provides further evidence for detection of APEX2 by MIMS. Fig. S2 demonstrates incorporation of 13C15N-arginine into proteins quantified by LC-MS/MS. Fig. S3 depicts characterization of GRN−/− iPSC cells. Table S1 provides details on incorporation of 13C15N-arginine into individual proteins.
Supplementary Material
Acknowledgments
We thank Virginia Crocker and the National Institute of Neurological Disorders and Stroke (NINDS) EM Facility and Louise Trakimas and the Harvard Medical School EM Facility for technical assistance with sample preparation for MIMS and TEM. We thank Yan Li at the NINDS Protein/Peptide Sequencing Facility for the SILAC-based proteomics analysis. We thank Ichi Miyaoka and Bruce Conklin (Gladstone Institute, University of California, San Francisco, San Francisco, CA) for providing the WTC11 and isogenic WTC11 GRN−/− iPSC lines.
This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Neurological Disorders and Stroke; extramural National Institutes of Health (grant DP2-CA216362), the Brigham and Women’s Hospital Department of Medicine Evergreen Innovation Award, and by a kind gift from the Bluefield Project to Cure FTD.
The authors declare no competing financial interests.
Author contributions: D.P. Narendra, C. Guillermier, and M.L. Steinhauser conceptualized the study; D.P. Narendra and M.E. Ward performed experiments and analyzed and interpreted data; C. Guillermier, F. Gyngard, M.E. Ward, and M.L. Steinhauser analyzed and interpreted data; X. Huang performed experiments; D.P. Narendra and M.L. Steinhauser wrote the paper; C. Guillermier and M.E. Ward contributed to review and editing of the paper; D.P. Narendra and M.L. Steinhauser supervised, provided resources, and acquired funding for the paper; and all authors approved the manuscript.
References
- Angelo M., Bendall S.C., Finck R., Hale M.B., Hitzman C., Borowsky A.D., Levenson R.M., Lowe J.B., Liu S.D., Zhao S., et al. . 2014. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20:436–442. 10.1038/nm.3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A.P., Koster G., Guillermier C., Hirst E.M.A., MacRae J.I., Lechene C.P., Postle A.D., and Gould A.P.. 2015. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell. 163:340–353. 10.1016/j.cell.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. . 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 442:916–919. 10.1038/nature05016 [DOI] [PubMed] [Google Scholar]
- Brozzi A., Urbanelli L., Germain P.L., Magini A., and Emiliani C.. 2013. hLGDB: a database of human lysosomal genes and their regulation. Database (Oxford). 2013:bat024 10.1093/database/bat024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-L., Hu Y., Udeshi N.D., Lau T.Y., Wirtz-Peitz F., He L., Ting A.Y., Carr S.A., and Perrimon N.. 2015. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. USA. 112:12093–12098. 10.1073/pnas.1515623112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle T.L., Madisen L., Hage T.A., Valley M.T., Knoblich U., Larsen R.S., Takeno M.M., Huang L., Gu H., Larsen R., et al. . 2018. A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell. 174:465–480.e22. 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandopulle M.S., Prestil R., Grunseich C., Wang C., Gan L., and Ward M.E.. 2018. Transcription Factor-Mediated Differentiation of Human iPSCs into Neurons. Curr. Protoc. Cell Biol. 79:e51 10.1002/cpcb.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermier C., Doherty S.P., Whitney A.G., Babaev V.R., Linton M.F., Steinhauser M.L., and Brown J.D.. 2019. Imaging mass spectrometry reveals heterogeneity of proliferation and metabolism in atherosclerosis. JCI Insight. 4:e128528 10.1172/jci.insight.128528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermier C., Poczatek J.C., Taylor W.R., and Steinhauser M.L.. 2017. Quantitative imaging of deuterated metabolic tracers in biological tissues with nanoscale secondary ion mass spectrometry. Int. J. Mass Spectrom. 422:42–50. 10.1016/j.ijms.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermier C., Steinhauser M.L., and Lechene C.P.. 2014. Quasi-simultaneous acquisition of nine secondary ions with seven detectors on NanoSIMS50L: application to biological samples. Surf. Interface Anal. 46(Suppl 1):150–153. 10.1002/sia.5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyngard F., and Steinhauser M.L.. 2019. Biological explorations with nanoscale secondary ion mass spectrometry. J. Anal. At. Spectrom. 34:1534–1545. 10.1039/C9JA00171A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Udeshi N.D., Deerinck T.J., Svinkina T., Ellisman M.H., Carr S.A., and Ting A.Y.. 2017. Proximity Biotinylation as a Method for Mapping Proteins Associated with mtDNA in Living Cells. Cell Chem. Biol. 24:404–414. 10.1016/j.chembiol.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Weston T.A., Jung R.S., Heizer P., Larsson M., Hu X., Allan C.M., Tontonoz P., Reue K., Beigneux A.P., et al. . 2018. NanoSIMS Analysis of Intravascular Lipolysis and Lipid Movement across Capillaries and into Cardiomyocytes. Cell Metab. 27:1055–1066.e3. 10.1016/j.cmet.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Zou P., Rhee H.-W., Udeshi N.D., Cracan V., Svinkina T., Carr S.A., Mootha V.K., and Ting A.Y.. 2014. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell. 55:332–341. 10.1016/j.molcel.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.C., Dean D.R., Smith A.D., and Johnson M.K.. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247–281. 10.1146/annurev.biochem.74.082803.133518 [DOI] [PubMed] [Google Scholar]
- Kim S.M., Lun M., Wang M., Senyo S.E., Guillermier C., Patwari P., and Steinhauser M.L.. 2014. Loss of white adipose hyperplastic potential is associated with enhanced susceptibility to insulin resistance. Cell Metab. 20:1049–1058. 10.1016/j.cmet.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Rajagopalan S., Sarkar P., Dorward D.W., Peterson M.E., Liao H.-S., Guillermier C., Steinhauser M.L., Vogel S.S., and Long E.O.. 2016. Zinc-Induced Polymerization of Killer-Cell Ig-like Receptor into Filaments Promotes Its Inhibitory Function at Cytotoxic Immunological Synapses. Mol. Cell. 62:21–33. 10.1016/j.molcel.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., and Ting A.Y.. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 12:51–54. 10.1038/nmeth.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechene C., Hillion F., McMahon G., Benson D., Kleinfeld A.M., Kampf J.P., Distel D., Luyten Y., Bonventre J., Hentschel D., et al. . 2006. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J. Biol. 5:20 10.1186/jbiol42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechene C.P., Luyten Y., McMahon G., and Distel D.L.. 2007. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science. 317:1563–1566. 10.1126/science.1145557 [DOI] [PubMed] [Google Scholar]
- Lovrić J., Dunevall J., Larsson A., Ren L., Andersson S., Meibom A., Malmberg P., Kurczy M.E., and Ewing A.G.. 2017. Nano Secondary Ion Mass Spectrometry Imaging of Dopamine Distribution Across Nanometer Vesicles. ACS Nano. 11:3446–3455. 10.1021/acsnano.6b07233 [DOI] [PubMed] [Google Scholar]
- Lu J., Baccei A., Lummertz da Rocha E., Guillermier C., McManus S., Finney L.A., Zhang C., Steinhauser M.L., Li H., and Lerou P.H.. 2018. Single-cell RNA sequencing reveals metallothionein heterogeneity during hESC differentiation to definitive endoderm. Stem Cell Res. (Amst.). 28:48–55. 10.1016/j.scr.2018.01.015 [DOI] [PubMed] [Google Scholar]
- Martell J.D., Deerinck T.J., Lam S.S., Ellisman M.H., and Ting A.Y.. 2017. Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat. Protoc. 12:1792–1816. 10.1038/nprot.2017.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., Ellisman M.H., and Ting A.Y.. 2012. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30:1143–1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom A., and Frei R.. 2002. Evidence for an ancient osmium isotopic reservoir in Earth. Science. 296:516–518. 10.1126/science.1069119 [DOI] [PubMed] [Google Scholar]
- Miyaoka Y., Chan A.H., Judge L.M., Yoo J., Huang M., Nguyen T.D., Lizarraga P.P., So P.-L., and Conklin B.R.. 2014. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods. 11:291–293. 10.1038/nmeth.2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.-F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich C.L., Wang C., Fogel A.I., and Youle R.J.. 2015. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 210:435–450. 10.1083/jcb.201501002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omari S., Makareeva E., Roberts-Pilgrim A., Mirigian L., Jarnik M., Ott C., Lippincott-Schwartz J., and Leikin S.. 2018. Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc. Natl. Acad. Sci. USA. 115:E10099–E10108. 10.1073/pnas.1814552115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G.E. 1952. A study of fixation for electron microscopy. J. Exp. Med. 95:285–298. 10.1084/jem.95.3.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D.D., Bensch K., and Barrnett R.J.. 1963. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J. Cell Biol. 17:19–58. 10.1083/jcb.17.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Ajinkya M., Bhat S., Itakura E., Hegde R.S., and Lippincott-Schwartz J.. 2014. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell. 158:522–533. 10.1016/j.cell.2014.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepartz B., and Turczyn M.. 1963. Oxidation of L-Amino Acids and Incorporation into Protein in Homogenates of Brain at Two Stages of Development. J. Neurochem. 10:825–829. 10.1111/j.1471-4159.1963.tb11907.x [DOI] [PubMed] [Google Scholar]
- Schreiber F., Littmann S., Lavik G., Escrig S., Meibom A., Kuypers M.M.M., and Ackermann M.. 2016. Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat. Microbiol. 1:16055 10.1038/nmicrobiol.2016.55 [DOI] [PubMed] [Google Scholar]
- Seligman A.M., Karnovsky M.J., Wasserkrug H.L., and Hanker J.S.. 1968. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J. Cell Biol. 38:1–14. 10.1083/jcb.38.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slodzian G., Daigne B., Girard F., Boust F., and Hillion F.. 1992. Scanning secondary ion analytical microscopy with parallel detection. Biol. Cell. 74:43–50. 10.1016/0248-4900(92)90007-N [DOI] [PubMed] [Google Scholar]
- Smith K.R., Damiano J., Franceschetti S., Carpenter S., Canafoglia L., Morbin M., Rossi G., Pareyson D., Mole S.E., Staropoli J.F., et al. . 2012. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 90:1102–1107. 10.1016/j.ajhg.2012.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser M.L., Bailey A.P., Senyo S.E., Guillermier C., Perlstein T.S., Gould A.P., Lee R.T., and Lechene C.P.. 2012. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 481:516–519. 10.1038/nature10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery-Lavenant G., Guillermier C., Wang M., and Lechene C.. 2014. Detection of immunolabels with multi-isotope imaging mass spectrometry (MIMS). Surf. Interface Anal. 46(Suppl 1):147–149. 10.1002/sia.5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama B.H., Arroyo e Drigo R., Lev-Ram V., Ramachandra R., Deerinck T.J., Lechene C., Ellisman M.H., and Hetzer M.W.. 2019. Visualization of long-lived proteins reveals age mosaicism within nuclei of postmitotic cells. J. Cell Biol. 218:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez C., Wong Y.C., Schwake M., Bu G., Wszolek Z.K., and Krainc D.. 2017. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum. Mol. Genet. 26:4861–4872. 10.1093/hmg/ddx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M.E., Chen R., Huang H.-Y., Ludwig C., Telpoukhovskaia M., Taubes A., Boudin H., Minami S.S., Reichert M., Albrecht P., et al. . 2017. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci. Transl. Med. 9:eaah5642 10.1126/scitranslmed.aah5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.J., and Odom R.W.. 1996. Matrix-enhanced secondary ion mass spectrometry: a method for molecular analysis of solid surfaces. Anal. Chem. 68:873–882. 10.1021/ac950717i [DOI] [PubMed] [Google Scholar]
- Zhang D.-S., Piazza V., Perrin B.J., Rzadzinska A.K., Poczatek J.C., Wang M., Prosser H.M., Ervasti J.M., Corey D.P., and Lechene C.P.. 2012. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature. 481:520–524. 10.1038/nature10745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. . 2013. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 78:785–798. 10.1016/j.neuron.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.