Many active genes position near nuclear speckles. Kim et al. show that Hsp70 speckle association correlates with increased nascent transcripts. Live-cell imaging shows that increases in nascent Hsp70 transcripts strictly follow speckle association (∼0–2 min), suggesting that speckle association amplifies gene expression.
Abstract
Many active genes reproducibly position near nuclear speckles, but the functional significance of this positioning is unknown. Here we show that HSPA1B BAC transgenes and endogenous Hsp70 genes turn on 2–4 min after heat shock (HS), irrespective of their distance to speckles. However, both total HSPA1B mRNA counts and nascent transcript levels measured adjacent to the transgene are approximately twofold higher for speckle-associated alleles 15 min after HS. Nascent transcript level fold-increases for speckle-associated alleles are 12–56-fold and 3–7-fold higher 1–2 h after HS for HSPA1B transgenes and endogenous genes, respectively. Severalfold higher nascent transcript levels for several Hsp70 flanking genes also correlate with speckle association at 37°C. Live-cell imaging reveals that HSPA1B nascent transcript levels increase/decrease with speckle association/disassociation. Initial investigation reveals that increased nascent transcript levels accompanying speckle association correlate with reduced exosome RNA degradation and larger Ser2p CTD-modified RNA polymerase II foci. Our results demonstrate stochastic gene expression dependent on positioning relative to a liquid-droplet nuclear compartment through “gene expression amplification.”
Introduction
Striking variations in transcriptional activity have been correlated with nuclear compartmentalization. Across multiple species and cell types, lamin-associated domains, as revealed by DamID (DNA adenine methyltransferase identification), show low gene densities and transcriptional activity (Kind et al., 2013). Similarly, across multiple species and cell types, the radial positioning of gene loci within a cell population stochastically closer to the nucleus center is associated with higher transcriptional activity (Kölbl et al., 2012; Takizawa et al., 2008). This stochastic correlation between gene expression and radial positioning may mask a more deterministic relationship between gene expression and gene positioning relative to a specific nuclear body which itself is radially distributed. Nuclear speckles, a RNP-containing, liquid droplet–like nuclear body (Kim et al., 2019) enriched in both RNA processing and transcription-related factors (Lamond and Spector, 2003; Spector and Lamond, 2011), are a prime candidate for such a nuclear body. Nuclear speckles indeed show a radial distribution, with decreased numbers near the nuclear periphery and increased concentration toward the nuclear interior. By electron microscopy, they appear as “Interchromatin Granule Clusters”—clusters of ∼20-nm-diameter RNPs lying between chromatin regions.
Nuclear speckles were suggested to act as a gene expression hub for a subset of genes based on the observation of ∼10/20 highly active genes localizing near the nuclear speckle periphery (Brown et al., 2008; Hall et al., 2006; Shopland et al., 2003). Support for this expression hub model was significantly boosted recently by a new genomic mapping method, TSA (Tyramide Signal Amplification) sequencing (Chen et al., 2018), which demonstrated that chromosome regions localizing most closely with nuclear speckles correspond largely to the A1 Hi-C subcompartment, one of two major transcriptionally active chromosomal subcompartments as mapped by Hi-C (Rao et al., 2014). These nuclear speckle-associated chromosome regions were enriched in the most highly expressed genes, housekeeping genes, and genes with low transcriptional pausing. Another new genomic mapping method, “Split-Pool Recognition of Interactions by Tag Extension” (SPRITE; Quinodoz et al., 2018), also showed that a large fraction of the genome with high levels of active RNA polymerase II (polII) transcription preferentially positioned near nuclear speckles.
This positioning of a subset of genes near nuclear speckles, however, is only a correlation. Despite this genome-wide demonstration of a subset of active genes positioning deterministically near nuclear speckles, there is no evidence that alleles of endogenous genes actually show different expression levels as a function of speckle proximity. Indeed, the prevailing view has been that nuclear speckles instead act primarily as a storage site for RNA processing factors (Lamond and Spector, 2003).
Previously, we demonstrated increased speckle association of bacterial artificial chromosome (BAC) transgenes containing the Hsp70 gene locus, including the genes heat shock protein A1A (HSPA1A), HSPA1B, and HSPA1L, after heat shock (HS; Hu et al., 2009). This increased speckle association was also observed for large, multicopy insertions of plasmid transgenes containing just the HSPA1A gene and shown to depend on the HSPA1A promoter and proximal promoter sequences rather than the actual transcribed sequences (Hu et al., 2010). Live-cell imaging revealed that the increased speckle association after HS for a large ∼700-copy HSPA1A plasmid transgene array occurred either through nucleation of a new nuclear speckle adjacent to the transgene array or, more interestingly, through the actin-dependent, long-range directed movement of the transgene array to a preexisting nuclear speckle (Khanna et al., 2014). Strikingly, a significant increase in the MS2-tagged HSPA1A transcript occurred only after (but within several minutes after) first contact with a nuclear speckle (Khanna et al., 2014).
However, the physiological relevance of the increased nascent transcript signal after speckle association of this large plasmid transgene array remained unclear with regard to the actual behavior of the endogenous Hsp70 locus. Cytologically, like other large, heterochromatic plasmid transgene arrays, this transgene array showed an unusually condensed chromatin mass during interphase, which was also preferentially positioned near the nuclear periphery. Moreover, in contrast to the synchronous induction of transcriptional activation 2–5 min after HS of the endogenous Hsp70 locus, this plasmid transgene array showed a highly asynchronous transcriptional activation over 10–30 min after HS (Hu, 2010). To determine the influence of speckle proximity on transcriptional activation in a more physiological context, we investigated Hsp70 gene activation as a function of speckle association after HS at both the endogenous and BAC transgene loci.
Results and discussion
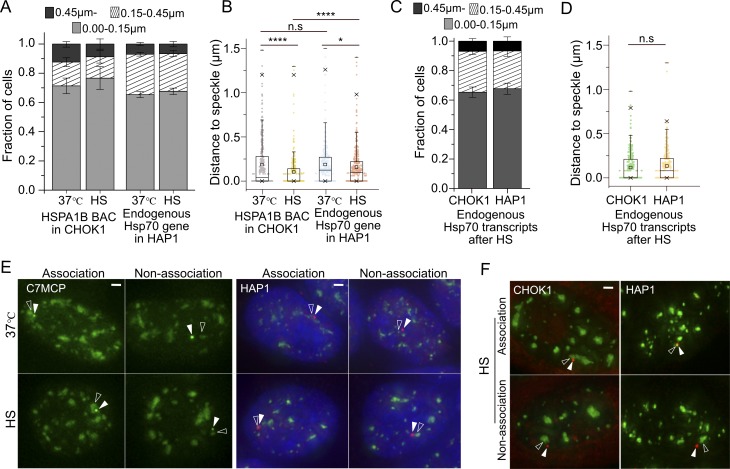
First, using DNA and RNA FISH, we identified human haploid HAP1 and Chinese hamster CHOK1 cell lines in which the endogenous Hsp70 locus showed significant populations of both speckle-associated (∼90%) and non–speckle-associated (∼10%) alleles (≥0.45 µm from the nearest speckle; Fig. S1, A–D). In contrast, human K562, Tig3, WI-38, and HCT116 cells showed near 100% prepositioning of this locus adjacent to nuclear speckles (not depicted; Tasan et al., 2018).
Figure S1.
Positioning of HSPA1B BAC transgene or endogenous Hsp70 gene relative to a nuclear speckle before (37°C) or after HS. (A) Histograms showing fraction of BAC HSPA1B transgenes (lacO) in CHOK1 cell clone C7MCP or endogenous Hsp70 alleles in HAP1 cells (DNA FISH) at varying distances from the nuclear speckle before and after 30-min HS (mean ± SEM, three biological replicates, n = 100–170 per replicate). (B) Boxplots showing distribution of varying distances from speckle shown in histogram A. Mean (square inside box), median (line inside box), box (interquartile range), ends of error bars (upper and lower limit), × (top: 99%, bottom: 1%), – (top: maximum, bottom: minimum). *, P < 0.05; ****, P < 0.00001; n.s, not significant; paired Wilcoxon signed-rank test. (C) Histograms showing fractions of RNA FISH signals from the endogenous Hsp70 locus in CHOK1 or HAP1 cells at varying distances from nuclear speckles after 30-min HS (mean ± SEM, three biological replicates, n = 100–120 per replicate). (D) Boxplots showing distribution of varying distances from speckle shown in histogram C. Box format is same as B. n.s, not significant; paired Wilcoxon signed-rank test. (E) Position of BAC transgene (green, white arrowhead) and nuclear speckle (green, empty arrowhead; left) or endogenous gene DNA FISH signal (red, white arrowhead) and nuclear speckle (green, empty arrowhead; right, DAPI staining, blue) at 37°C (top) or after HS (bottom). Scale bars = 1 µm. (F) Position of RNA FISH of endogenous Hsp70 locus transcripts (red, white arrowheads) and nuclear speckle (green, empty arrowheads) after 30-min HS in CHOK1 cells (left) or HAP1 cells (right). Scale bar = 1 µm.
We next established that integrated human Hsp70 or HSPA1B BAC transgenes in several independently derived CHOK1 cell clones showed similar gene positioning relative to nuclear speckles before and after HS (Hu et al., 2010; Khanna et al., 2014), as seen for the endogenous HSP70 gene loci in both HAP1 and CHOK1 cells (Fig. S1), suggesting a mechanism that targets both the endogenous Hsp70 locus and BAC Hsp70 transgenes to nuclear speckles. Gene positioning was measured by lac operator (lacO)/repressor staining for BAC transgenes and either DNA or RNA immuno-FISH for endogenous genes.
The Hsp70 BAC contains a 171.5-kb human DNA insert encompassing three Hsp70 genes (HSPA1A, HSPA1L, and HSPA1B) and a 256mer lacO repeat inserted 25.1/37.4 kb 5′ of the HSPA1A/HSPA1B gene (Hu et al., 2009). A 8-kb deletion removed the HSPA1A and HSPA1L genes (placing the lacO repeat 29.4 kb upstream of HSPA1B), producing the HSPA1B BAC (Hu et al., 2010), which was further modified by inserting a 24mer MS2 repeat into the HSPA1B 3′ UTR (Khanna et al., 2014). The C7-MS2 coat protein (C7MCP) CHOK1 cell clone was selected for further analysis; C7MCP cells contain one to three HSPA1B BAC copies (quantitative PCR estimate) integrated at a single chromosomal locus and express EGFP-lac repressor to stain the lacO repeat, EGFP-SON to stain nuclear speckles, and mCherry-MCP to stain HSPA1B transcripts.
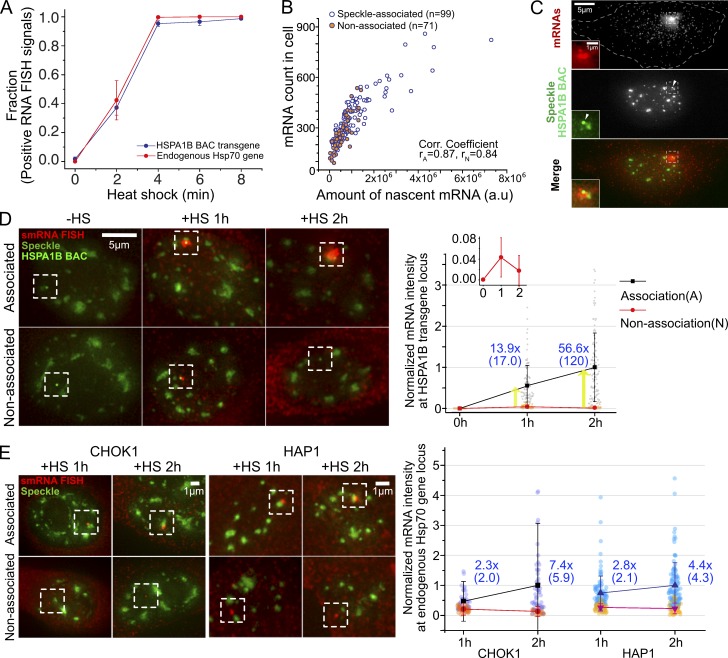
The HSPA1B BAC transgene in CHOK1 cells showed near-identical transcriptional induction dynamics after HS as the endogenous human Hsp70 locus in HAP1 cells (Fig. 1 A), as measured by single-molecule RNA (smRNA) FISH. HSPA1A and HSPA1B genes have no introns. We used a human-specific FISH probe set that did not detect endogenous CHO Hsp70 genes but had the sensitivity to detect single, cytoplasmic Hsp70 RNAs (HSPA1A and HSPA1B) in human cells or CHOK1 cells containing human Hsp70 or HSPA1B BACs. The fraction of nuclei showing a positive RNA FISH Hsp70 nascent transcript signal increased from near 0 to 1 between 0 to 4 min after HS in both haploid HAP1 and CHOK1 C7MCP cells containing the HSPA1B BAC transgene (Fig. 1 A). Similar Hsp70 induction kinetics were observed in CHO DG44 cell clones (Hu, 2010) containing either integrated Hsp70 BAC transgenes or HSPA1B BAC transgenes without MS2 repeats inserted (Fig. S2 A); therefore, the MS2 repeat with bound MCP did not affect these kinetics. Similar kinetics also were observed in C7MCP cells using oligonucleotide probes against the 3′ MS2 repeats (Fig. S2 A); therefore, full-length transcripts are induced with similar kinetics.
Figure 1.
Both HSPA1B BAC transgenes and endogenous genes induce synchronously 2–4 min after HS but show higher transcript levels when associated with nuclear speckles. (A) Transcriptional induction of HSPA1B BAC transgene in CHOK1 cells and endogenous Hsp70 genes in HAP1 cells occurs 0–4 min after HS (SEM, three replicates). n = 110–210 for HAP1, n = 95–150 for CHOK1 cells, each replicate time point. (B) Scatterplot between levels of nascent RNA signals (RNA FISH intensity at HSPA1B locus) versus numbers of mature mRNAs after 15-min HS. n = 99 and 71 for A and N. P value (A versus N) = 8.5 × 10−7 for transcript signal, 1 × 10−6 for mRNA count; paired Wilcoxon signed rank test. rA, Pearson correlation coefficient for speckle-associated locus; rN, for nonassociated locus. (C) smRNA FISH image after 15-min HS. Nascent pre-mRNAs (top) at HSPA1B BAC transgene (arrowhead, middle). White dashes outline cell border. The boxed region around BAC is enlarged on left. (D) Higher pre-mRNA levels (boxed regions) for speckle-associated (top) versus nonassociated (bottom) BAC transgenes. (Left) Representative images of smRNA FISH versus nuclear speckles and BAC transgene at 0, 1, and 2 h after HS. (Right) Normalized pre-mRNA intensities at speckle-associated (black, A) or nonassociated (red, N) BAC transgenes 0, 1, and 2 h after HS, with fold differences (blue) of mean (median) for A versus N. Nascent RNA intensities were normalized by mean intensity of A at 2 h. Cell numbers (A/N) = 101/101, 91/40, and 107/63 at 0, 1, and 2 h. P = 4.5 × 10−11 at 1-h HS and 7.8 × 10−12 at 2-h HS for A versus N; 9.7 × 10−4 for A at 1 versus 2 h; and 8.4 × 10−4 for N at 1 versus 2 h; paired Wilcoxon signed-rank test. (E) Left: Same as in D but for endogenous Hsp70 locus in CHOK1 cells (left two panels) versus haploid human HAP1 cells (right two panels). Nonlinear intensity scaling used for red channel. Right: Same as in D, right, for endogenous Hsp70 locus in CHOK1 (left) versus HAP1 cells (right). Nascent RNA intensities were normalized by mean intensity of A at 2 h in CHOK1 cells and HAP1 cells, respectively. CHOK1: cell numbers (A/N) = 46/33 and 50/28 at 1 and 2 h, P = 1.8 × 10−5 at 1-h HS and 7.5 × 10−8 at 2-h HS for A versus N, 0.0038 for A at 1 versus 2 h, 0.0015 for N at 1 versus 2 h; HAP1: cell numbers (A/N) =102/44 and 112/33 at 1 and 2 h, P = 2.0 × 10−9 at 1-h HS and 6.5 × 10−9 at 2-h HS for A versus N, 4.6 × 10−4 for A at 1 versus 2 h, 0.12 for N at 1 versus 2 h; paired Wilcoxon signed-rank test.
Figure S2.
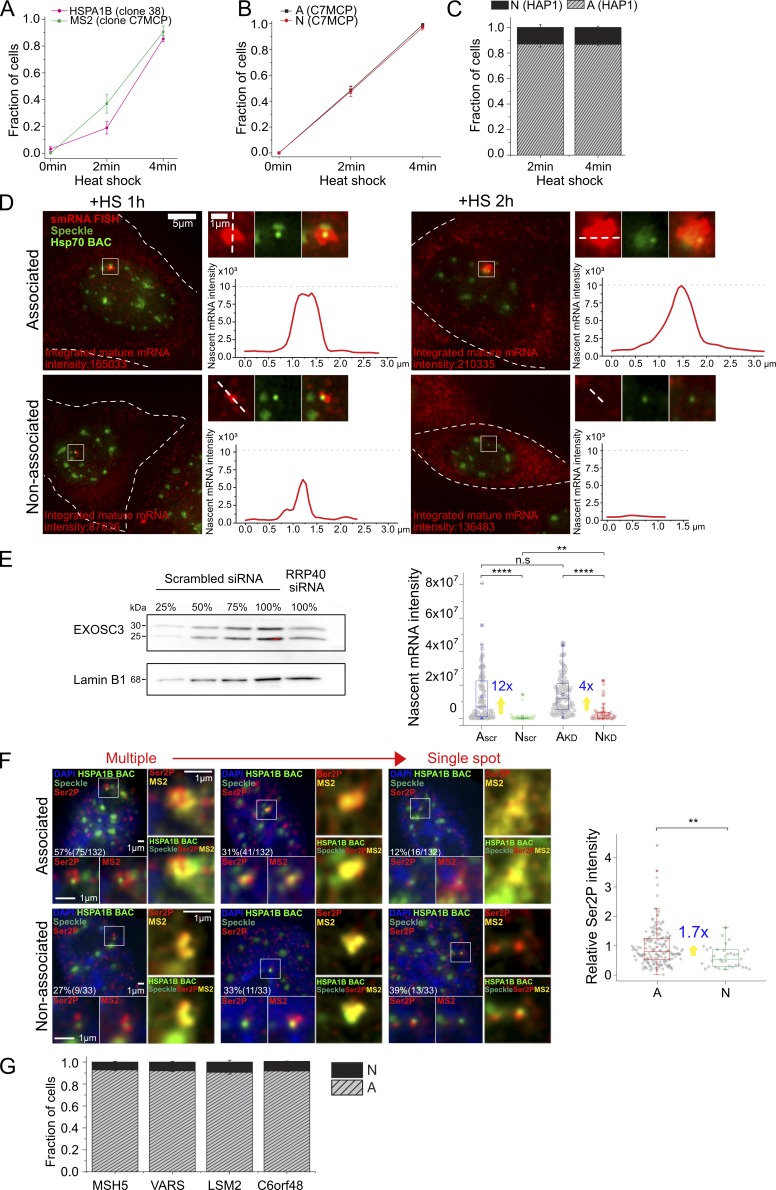
Various supporting data. (A) Transcriptional induction of HSPA1B BAC transgene without MS2 repeats in CHO DG44 cells detected by smRNA FISH against HSPA1B (clone 38, percentage ± SD estimate, n = 42 to 46 each time point, magenta) and of HSPA1B BAC with MS2 repeats in CHOK1 cells detected by smRNA FISH against MS2 (clone C7MCP, percentage ± SD estimate, n = 74 to 96 each time point, green). Full induction occurs 2–4 min after HS in both. (B) Transcriptional induction of speckle-associated (A) and non–speckle-associated (N) HSPA1B BAC transgene in CHOK1 cells (clone C7MCP). Both A and N are induced within 0–4 min after HS (percentage ± SD estimate, n = 360, 211 observed for A; n = 147, 69 for N). (C) Histogram showing fraction of endogenous Hsp70 alleles in HAP1 cells upon speckle association or non–speckle association after 2- and 4-min HS (mean ± SEM, three replicates). At 2-min HS, n = ∼100, 38% activated cells on average among observed 201 to 360 cells, and at 4-min HS, n = 70 to 111, 96.8% activated cells on average among observed 70 to 120 cells for each replicate. (D) Whole-cell images of nucleus shown in Fig. 1 D (left) and intensity profile of nascent mRNA transcripts at HSPA1B BAC transgene locus (right). Integrated mature mRNA signal is proportional to nascent mRNA signal as in Fig. 1 B. Left: White dashes outline cell border. Right, top: Image of nascent transcripts in red, HSPA1B BAC in light green and nuclear speckle in green, and merge. Right, bottom: Intensity profile of nascent transcript measured along the white dash line in top image. Intensities scaled nonlinearly in images to reduce red channel intensity dynamic range (left) but linearly for line scans. (E) Nascent mRNA level change after EXOSC3 knockdown at 1-h HS. Left: Immunoblotting with EXOSC3 antibody to estimate the percentage of knockdown. The EXOSC3 protein in the knockdown samples (100%, RRP40 siRNA treated) was estimated against different percentages (25, 50, 75, and 100%) of scrambled siRNA-treated cell lysates. Lamin B1 was used as the loading control. Right: Boxplot of nascent mRNA intensity (smRNA FISH) at A and N locus for scrambled siRNA or RRP40 siRNA–treated cells with fold differences (blue) of mean for A versus N. Mean (square inside box), median (line inside box), box (interquartile range), ends of error bars (upper and lower limit), × (top: 99%, bottom: 1%), – (top: maximum, bottom: minimum). **, P < 0.01; ****, P < 0.0001; n.s, not significant; paired Wilcoxon signed-rank test. n = 107 and 39 observed for A and N in scrambled; 106 and 43 for RRP40 siRNA–treated cells. (F) RNA polII phosphorylated on serine 2 (Ser2P) and MS2MCP-mCherry signal at 1-h HS. Left: Representative images of Ser2P polII (red, bottom left and right) signals and MS2 signals (red, bottom right, and yellow, right) for BAC transgene (light green) associated (top) versus not associated (bottom) with nuclear speckles (green). Right: Boxplot of relative Ser2P normalized by mean Ser2P intensity of A with fold differences (blue) of mean for A versus N. **, P < 0.01; paired Wilcoxon signed-rank test. n = 165 and 33 observed for A and N. (G) Histograms showing fraction of endogenous Hsp70 flanking gene alleles in HAP1 cells at varying distances from the nuclear speckle (mean ± SEM, three experimental replicates, n = 128 to 289 activated loci observed in each experiment. Scale bars = 1 µm (D, enlarged panels, and F) or 5 µm (D) .
Induction kinetics also were similar for speckle-associated and nonassociated alleles (Fig. S2, A and B). We defined BAC transgenes and endogenous genes here as speckle associated (A) if the transgene and/or nascent transcripts positioned within 0.15 µm from the nuclear speckle edge and non–speckle associated (N) if the transgene located further than 0.45 µm from the nuclear speckle edge or, for endogenous genes, if the nascent transcripts located further than 0.45 µm from the nuclear speckle edge. HSPA1B BAC transgenes in CHOK1 cells showed 49 ± 2.6 A and 48 ± 4.1% N induced alleles at 2 min HS and 99 ± 0.68 A and 97 ± 2.1% N at 4-min HS (percentage of induced alleles ± SEM estimate). In HAP1 cells, without tagged endogenous Hsp70 loci, we cannot visualize transcriptionally inactive alleles by RNA FISH. However, a similar fraction of “on” A and N alleles at 2 min after HS (when only ∼40% of the alleles are on) as at 4 min after HS (when ∼100% of alleles are on; Figs. 1 A and S2 C) also suggests no difference in induction kinetics.
We next compared the magnitude of the integrated RNA FISH nascent transcript signal at both BAC transgenes and endogenous human and CHO loci when they were speckle associated versus non–speckle associated. For the endogenous CHO locus, we used a smRNA FISH probe set specific for Chinese hamster and performed experiments in WT CHOK1 cells.
The integrated nascent transcript signal should be proportional to the transcription rate in a simplified model in which the nascent transcript signal is produced by a steady state between the transcription rate and the transcript release rate from the transcription site, assumed to be constant over time. Indeed, in C7MCP cells, the measured number of HSPA1B mRNAs per cell correlates linearly with the nascent RNA transcript FISH signal (Fig. 1, B and C). Measurements were at 15 min after HS, when the number of individual, dispersed HSPA1B mRNAs are still low enough to count.
However, both the HSPA1B integrated nascent transcript signal and total cellular mRNA count show higher values at 15 min HS for speckle-associated versus nonassociated transgene alleles (Fig. 1 B; 2.1-fold and 1.5-fold higher for transcript signal and mRNA count, respectively, P = 8.4 × 10−7 and 1 × 10−6; paired Wilcoxon signed-rank test). This speckle-associated versus nonassociated nascent transcript level ratio increases to 14-fold at 1 h and 57-fold at 2 h after HS (Fig. 1, D and E; and Fig. S2 D). The ratio increased from 1 to 2 h due to an approximately twofold increase in nascent transcripts for speckle-associated transgenes combined with an approximately twofold decrease in nascent transcripts for nonassociated transgenes (Fig. 1 D, right, inset).
For the endogenous Hsp70 genes in both CHOK1 and HAP1 cells, we observed an approximately threefold (1 h) to approximately sevenfold (2 h) increased level of nascent transcripts from speckle-associated versus nonassociated alleles after HS (Fig. 1 E). Both the endogenous and BAC speckle-associated Hsp70 genes showed roughly a doubling of nascent transcript levels between 1 and 2 h after HS, but only the BAC transgenes not associated with speckles showed a decrease in transcript levels between 1 and 2 h.
Within the assumptions of our simple model, these results would suggest increased transcription with speckle association. Constant transcription but decreased RNA transport rates with speckle association would also increase nascent transcript signals, but would not increase the mRNA count. However, both increased nascent transcript signals and gene expression could occur through a decrease in a nascent transcript degradation rate, rather than through an increased transcription rate. Indeed, nuclear speckles are enriched for nuclear export factors, which may compete with RNA degradation pathways (Fan et al., 2018; Silla et al., 2018); perhaps nuclear speckle-associated nascent transcripts are more stable than nonassociated transcripts.
We used previously validated siRNAs (Fan et al., 2018) to reduce the protein levels of the exosome RRP40 subunit ∼50% (Fig. S2 E, left). Interestingly, after knockdown, we observed a reduction in the relative difference in mean nascent transcript accumulation, measured by smRNA FISH, for speckle-associated versus nonassociated BAC transgenes from ∼12-fold (35-fold, median) for the scrambled siRNA control to ∼4-fold (7-fold, median) at 1 h after HS (Fig. S2 E, right). Nascent transcript levels increased in both cases, but the increase was proportionally larger for N (3.5 mean, 8.3 median) versus A (1.1 mean, 1.7 median) transgenes. Thus, differential RNA degradation rates may account for at least part of the difference in observed nascent transcript and expression levels.
At the same time, we also observed an increased number and intensity of RNA polII Ser2p foci adjacent to and overlapping the lacO spot (29 kb upstream of HSPA1B) and/or MS2 nascent transcript signal for A versus N transgenes at 1 h after HS (Fig. S2 F). A larger fraction of A versus N transgenes show multiple, clustered versus single RNA polII Ser2p foci overlapping the lacO spot and/or MS2 nascent transcript signal and separate from the transgene’s associated nuclear speckle, and a lower fraction of A versus N transgenes show single RNA polII Ser2p spots (Fig. S2 F, left). Moreover, A transgenes show a nearly twofold higher mean integrated intensity of these RNA polII Ser2p foci. To the extent that these RNA polII Ser2p foci are either a consequence of or even causally related to higher transcription rates, these data suggest that the higher nascent transcript levels for speckle-associated transgenes may also be due to higher transcription rates with speckle association. Given the small size of the HSPA1A and HSPA1B transcription units, direct determination of transcription rates and the relative contribution of differential RNA degradation versus transcription rates for speckle-associated versus nonassociated genes is experimentally challenging and awaits future investigations.
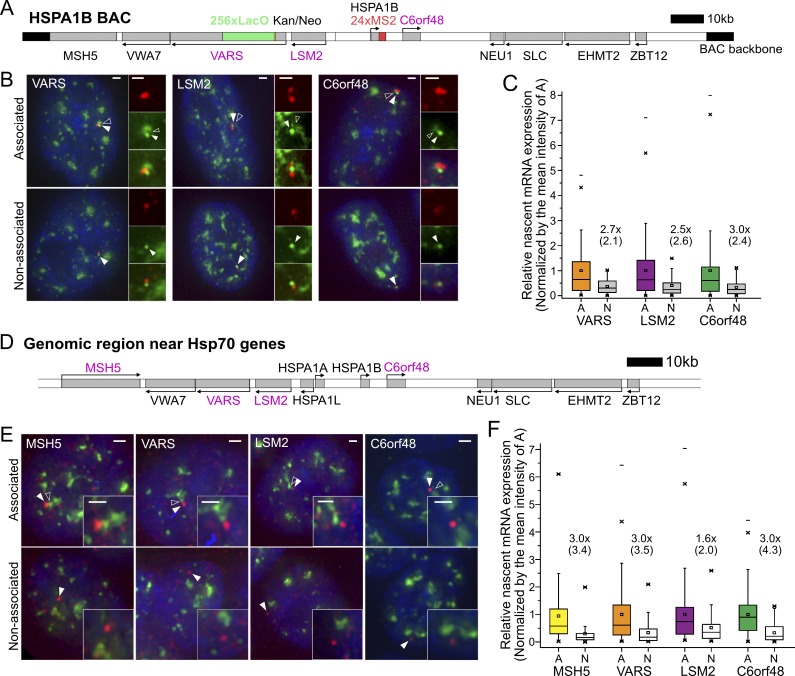
The increased Hsp70 nascent transcripts with speckle association raised the question of whether genes flanking the Hsp70 locus would show similar behavior. We measured nascent transcripts from three active human genes, VARS, LSM2, and C6orf48, flanking HSPA1B on the HSPA1B BAC (Fig. 2, A–C) stably integrated in CHOK1 C7MCP cells. By smRNA FISH, all three genes showed nearly ∼100% of alleles with nascent transcripts and a 2.5–3-fold increase in nascent transcript levels with speckle association (Fig. 2, B and C).
Figure 2.
Transcription amplification of speckle-associated genes flanking Hsp70 gene locus at 37°C. (A and D) Probed gene locations (gray boxes, magenta gene names) relative to Hsp70 genes in BAC construct (A) in CHOK1 cells and at endogenous locus (D) in human HAP1 cells. (B and E) Representative images of smRNA FISH (red) signals for specific BAC transgene (B) or endogenous gene (E) showing nascent transcripts associated (top) versus nonassociated (bottom) with nuclear speckles (intensities scaled differently for top and bottom). White arrowheads point to BAC (B) or nascent transcripts (E); empty arrowheads point to nuclear speckle. Scale bars = 1 µm. (C and F) Boxplots showing nascent transcript levels for three (C) or four (F) genes flanking BAC HSPA1B transgene (C) or endogenous Hsp70 locus (F) as function of speckle association (A) or nonassociation (N). Mean (square inside box), median (line inside box), box (interquartile range), ends of error bars (upper and lower limit), × (top: 99%, bottom: 1%), – (top: maximum, bottom: minimum). Intensities are normalized by the mean intensity at speckle-associated loci: fold differences (×) of the mean (median) between A versus N (black). In C, number of cells (A/N) = 112/44, 143/37, and 185/39; P = 0.0024, 0.013, and 0.016 (paired Wilcoxon signed-rank test) for VARS, LSM2, and C6orf48 BAC transgenes. in D, number of cells (A/N) = 168/64, 144/47, 123/52, and 203/86; P = 2.0 × 10−6, 1.4 × 10−5, 0.23, and 1.5 × 10−11 (paired Wilcoxon signed-rank test) for endogenous MSH5, VARS, LSM2, and C6orf48 genes.
Similarly, for the endogenous human Hsp70 locus in HAP1 cells, we measured 3.0-fold increases with speckle association for each of the VARS, LSM2, and C6orf48 gene nascent transcript levels and 1.7-fold for the MSH5 gene nascent transcript (Fig. 2, D and F). In contrast to C6orf48, which showed constant transcriptional activity, MSH5, LSM2, and VARS are bursting genes that show nascent transcripts in only 90 ± 2%, 46 ± 3%, and 70 ± 3% of these haploid cells (mean ± SEM from three independent experiments). However, for each of these three bursting genes, the fraction of cells with visible nascent transcripts associated with nuclear speckles versus visible nascent transcripts not associated with nuclear speckles (Fig. S2 G) was comparable to the fraction of gene loci mapped by DNA FISH as adjacent to nuclear speckles versus not adjacent to nuclear speckles (Fig. S1 A). Thus, speckle association did not increase the frequency of bursting for these genes, but did enhance their levels of transcription.
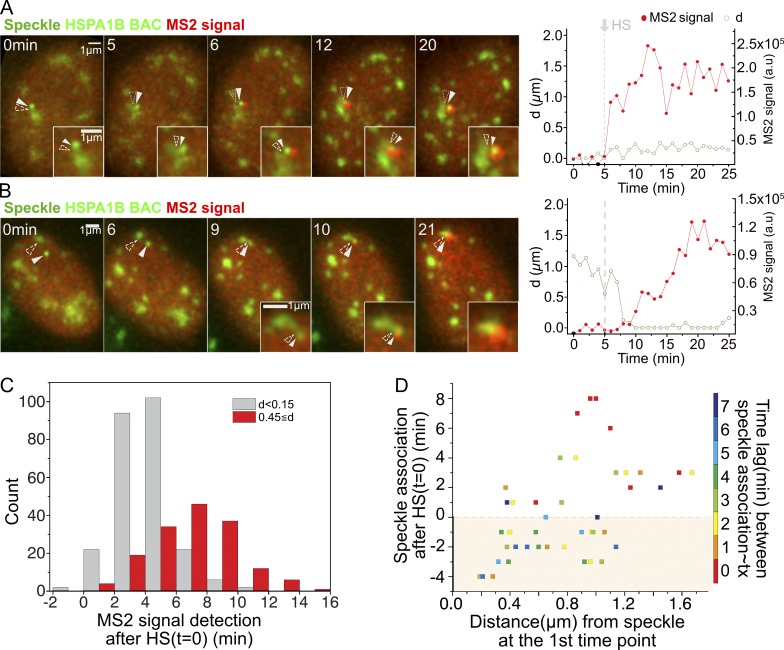
To determine the temporal relationship between speckle association and transcription, we used live-cell imaging of the HSPA1B BAC transgenes in C7MCP cells. From 1,080 cell videos, we obtained 438 in which the BAC transgene, nuclear speckles, and MS2-tagged transcripts could all be tracked over the entire 20- to ∼25-min observation period. The observed dynamics from each of these 438 cells were then sorted into different categories. The three simplest general categories corresponded to cells in which the transgene was always associated with a speckle (Fig. 3 A), cells in which the transgene started distant from a speckle but then moved to and remained associated with a speckle (Fig. 3 B), and cells in which the transgene became associated with a speckle and showed a visible transcription signal, but then moved away from the speckle (Fig. 4 A).
Figure 3.
Strict temporal correlation between speckle association and HSPA1B transcriptional amplification. (A and B) Left: Transgene location (solid arrowheads), nuclear speckles (open arrowheads), and MS2 signal versus time (minutes) after start of observation (heating on at 1 min, stable HS temperature [HS-T] reached at 5 min). Right: Distance (d) of Hsp70 transgene/nascent transcript from closest speckle (green) and nascent RNA level (red). (A) Transgene associated with speckle throughout HS (category 1, 146/438 cells). MS2 signal appears 1 min after reaching HS-T at 5 min (Video 1). (B) Transgene initially unassociated with speckle (category 2, 41/438 cells). MS2 signal appears at 10 min, ∼5 min after reaching HS-T and ∼1 min after moving to and contacting speckle (Video 2). (C) Histogram showing time of MS2 signal appearance after reaching HS-T for transgenes initially speckle associated (gray, n = 251) versus non–speckle associated (red, n = 159). MS2 signal delayed ∼3 min when not associated (mean = 6.5 min) versus associated (mean = 3.6 min). P = 2.2 × 10−16; paired t test. (D) Scatterplot showing timing of speckle–transgene association after reaching HS-T versus initial transgene distance to speckle versus time lag (color) between speckle–gene association and appearance of MS2 signal (n = 42 displayed). Time lags: mean = 3.8 min, typically 0–3 min after reaching HS-T.
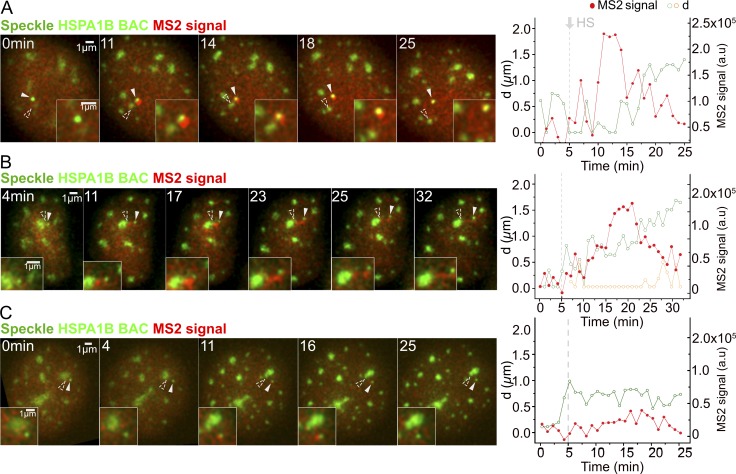
Figure 4.
Temporal correlation between speckle dissociation and decrease in HSPA1B transcription. Display and labeling is the same as in Fig. 3 (A and B). (A) HSPA1B transgene disassociating from speckle (category 3, 28/438 cells). Nascent transcripts decrease and then disappear after transgene separates from speckle (Video 3). (B) HSPA1B transgene disassociating from speckle. d, distance of HSPA1B transgene (green) or nascent transcript (orange) from closest speckle (2/28 cells falling in category 3). Nascent transcripts accumulate in elongated connection between speckle and transgene after their separation (Video 4). (C) No speckle association of HSPA1B transgene after HS (5/438 cells). MS2 signal sometimes transiently rises slightly above background but does not persist.
In the first category (146/438 cells), the BAC transgene remained localized within 0.15 µm of the nuclear speckle during the entire observation period (Fig. 3 A and Video 1). Nascent transcripts became visible above the diffuse background of the MCP typically 2–4 min after the temperature reached 42°C for HS (Fig. 3 C, gray bars), and increased gradually afterward in a typical example (Fig. 3 A and Video 1). In the second category (41/438 cells), the transgene–speckle distance at some point exceeded 0.45 µm, but then the transgene became stably associated with a nuclear speckle, and subsequently, a MS2 signal appeared (Fig. 3 B and Video 2). On average, these cells showed an ∼3-min delay in the appearance of a visible MS2 signal relative to cells in the first category (Fig. 3 C, red bars), due largely to the extra time required for the transgene to move to the speckle. If speckle contact occurred after the temperature had already reached 42°C, a visible MS2 signal typically appeared above background 0–2 min after contact. The time lag in transcription was longer when speckle association occurred before the temperature reached 42°C (Fig. 3 D).
Video 1.
Maximum-intensity 2D projections of 3D image stacks calculated for each 1-min time point. Time given in minutes. Scale bar = 1 µm; 12 fps. Temperature increase begins at 1 min, reaching 42°C at ∼5 min. Transgene always speckle-associated: MS2 signal appears without delay (∼1 min after reaching HS temperature at 5 min, see also Fig. 3 A). Transgenes (green), nuclear speckles (lighter green), and MS2-tagged nascent transcripts (red).
Video 2.
Maximum-intensity 2D projections of 3D image stacks calculated for each 1-min time point. Time given in minutes. Scale bar = 1 µm; 12 fps. Temperature increase begins at 1 min, reaching 42°C at ∼5 min. Appearance of MS2-tagged nascent transcripts (red) is delayed after HS, with MS2 signal appearing after nonassociated transgene (green) moves to and makes contact with nuclear speckle (see also Fig. 3 B). Lighter green, nuclear speckles.
In the third category of speckle movements (28/438 cells), the transgene associated with a nuclear speckle and produced a visible MS2 nascent transcript signal but then moved away from the speckle (Fig. 4 A). Significantly, a decrease or disappearance of the MS2 signal followed this transgene–speckle separation in a typical example (Video 3). Once transgenes moved farther than 0.5–1 µm from a speckle, transcripts typically decayed within 1–2 min. However, when transgenes moved smaller distances away from the speckle, a low level of transcription was maintained longer rather than a rapid and complete decay of the MS2 signal. When nascent transcript levels were low, we typically observed loss of the transcript signal right after transgene–speckle separation, while when nascent transcript levels were high, a complete loss of transcript signal appeared to require a larger separation. Indeed, we could observe creation of a connecting “bridge” of MS2-tagged transcripts lying between the transgene and speckle periphery. We also could observe deformation of the speckle shape toward the transcripts (Fig. 4 B and Video 4). These transcript bridges sometimes elongated as the transgene moved away from the speckle until the transgene had moved far enough away to break contact. Once the transgene dissociated with the speckle, the nascent transcript signal did not increase without new speckle association (Fig. 4 C). Plotting speckle–transgene distances and nascent transcript levels, averaged over tens of movies, as a function of time after HS (Fig. 5 A, left), after speckle association (Fig. 5 A, middle), or after speckle disassociation (Fig. 5 A, right) reveals that these typical examples, discussed above, represent predominant patterns for these three categories of speckle-transgene dynamics.
Video 3.
Maximum-intensity 2D projections of 3D image stacks calculated for each 1-min time point. Time given in minutes. Scale bar = 1 µm; 12 fps. Temperature increase begins at 1 min, reaching 42°C at ∼5 min. Decrease and disappearance of MS2-tagged nascent transcripts (red) after disassociation of transgene (green) from nuclear speckle (lighter green; see also Fig. 4 A).
Video 4.
Maximum-intensity 2D projections of 3D image stacks calculated for each 1-min time point. Time given in minutes. Scale bar = 1 µm; 12 fps. Temperature increase begins at 1 min, reaching 42°C at ∼5 min. Delayed decay of MS2-tagged nascent transcripts (red) after transgene (green) disassociation from nuclear speckle (lighter green). This delay is associated with a nascent transcript accumulation between transgene and speckle that elongates and appears to physically connect the nuclear speckle with the transgene even after the transgene moves away from the speckle (see also Fig. 4 B).
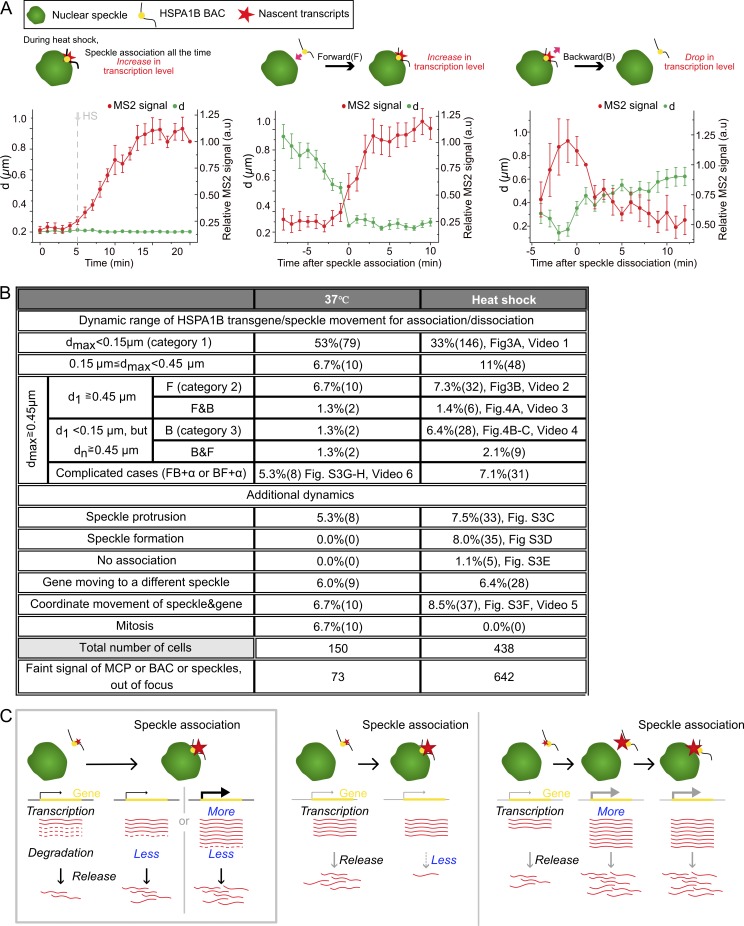
Figure 5.
Statistics for classifications of relative movements between BAC transgene and nuclear speckle. (A) Cartoons of representative dynamics (categories 1–3, top) during HS and corresponding averaged plots of live-cell data (bottom, mean ± SEM, n = 21–47 movies). Distance (d, green) of HSPA1B transgene from closest speckle and nascent RNA levels (red) in arbitrary units. Category 1 (left): stable speckle association of HSPA1B transgene and transcription increase after temperature reaches 42°C, 47 movies; category 2: transcription increase as function of time after transgene association with speckle (t = 0), 24 movies; category 3: transcription decrease as function of time after transgene dissociation (t = 0) from speckle, 21 movies. (B) Numbers for categories of dynamics observed for 20 to ∼25 min with or without HS. dmax, maximum distance between transgene locus and speckle during imaging; d1, distance between transgene locus and speckle at first time point; dn, distance between transgene locus and speckle at any time point during imaging; F, forward relative motion bringing transgene and speckle closer; B, backward relative motion moving transgene and speckle apart; +α, additional movements. (C) Three models for correlation of nascent transcript increase with speckle association: nuclear speckle (green), gene (yellow), and nascent transcripts (red; solid lines, intact; dashed lines, degraded). Left: Current working model. Nascent transcripts increase after speckle association through a decreased, exosome-mediated nascent transcript degradation rate, and possibly an increased transcription rate. Middle: Alternative model 1. Nascent transcripts adjacent to transcription site increase due to a reduced transcript release rate; at steady state, this would not change the rate of mRNA production and is inconsistent with the increased mRNA count 15 min after HS. Right: Alternative model 2. Nascent transcript levels increase before speckle association, causing subsequent speckle association; this is inconsistent with live-cell imaging temporal ordering of events.
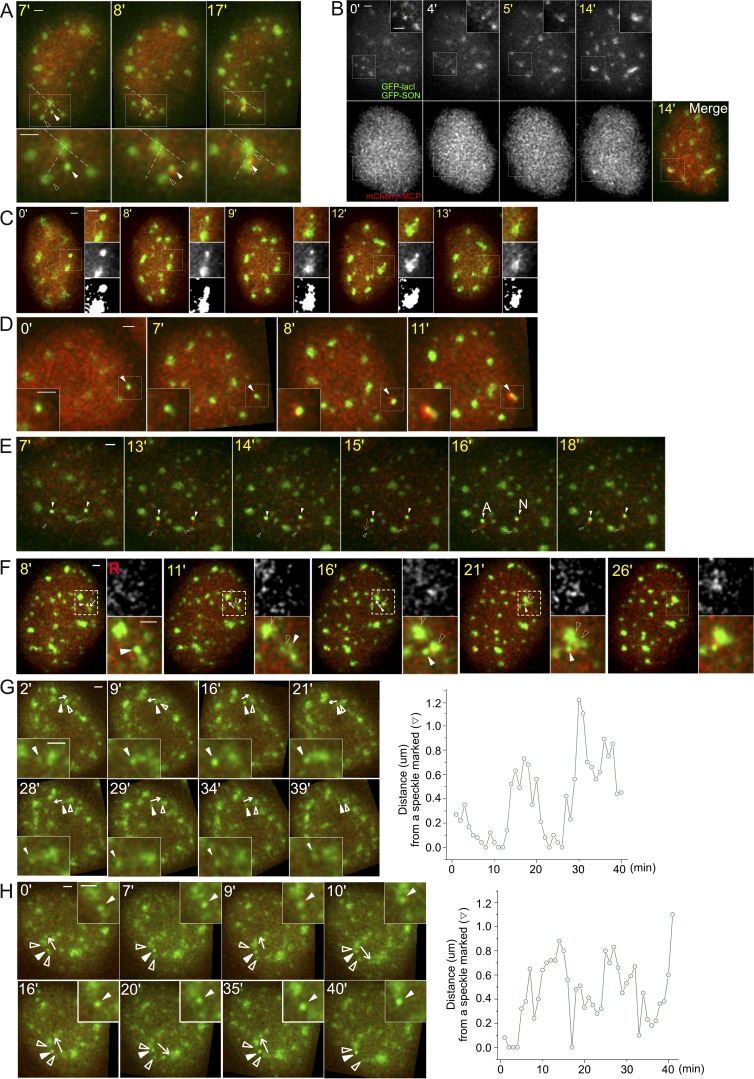
More complicated dynamics classifications were also seen (Figs. 5 B and S3). Although transgenes and speckles usually associated by transgenes moving to the speckle, occasionally speckles moved to the transgenes (Fig. S3, A–D). Additional dynamics include speckle protrusion to a transgene (7.5%, Fig. S3 C), nucleation of a new speckle (8%, Fig. S3 D), and movement of the transgene from one speckle to another (6.4%). In 5 of 438 cells (1.1%), although the transgene did not associate with any speckles, we observed small transient increases for no longer than one time point of the MS2-tagged nascent transcript signal above the background (Fig. S3 E). These transient increases above background are consistent with fluctuations in the low-level transcript levels seen by smRNA FISH for nonassociated HSPA1B transgenes (Fig. 1) but mostly missed in our live-cell imaging due to the mCherry-MCP background. In all other cases, appearance of a MS2 signal above background was coupled to nuclear speckle association, which in contrast yielded substantially higher frequency and longer duration of increased transcript signals. A physical connection between the BAC transgene, its nascent transcripts, and the associated nuclear speckle is demonstrated by their coordinated movements (8.5%; Fig. S3 F and Video 5). These rarer classifications shed additional light on the functional significance of nuclear speckle association to HSPA1B transcription.
Figure S3.
Additional examples of speckle and transgene dynamics. Time stamp during HS (yellow), BAC transgene (white arrowheads, bright green), nuclear speckle (empty arrowheads, lighter green), and RNA MS2-tagged transcripts (red, mCherry-MCP). All scale bars = 1 µm. (A) Combined speckle and transgene movements after speckle movement toward and association with the transgene (category: F, Fig. 5 B). Dotted lines connect to relatively stationary speckles. (B) Movement of two speckles toward the transgene locus (category: F, Fig. 5 B). (C) Speckle protrusion toward the active transgene locus (category: speckle protrusion, Fig. 5 B), with the transgene then moving closer to speckle. (D) Speckle formation at the active transgene (white arrowhead) after HS at 7 min, with nascent transcripts appearing above background at 8 min (category: speckle formation, Fig. 5 B). (E) No persisting transcription for nonassociating locus (category: no association, Fig. 5 B). Arrow (15 min) shows direction of transgene movement. Transcriptional bursting is observed at both nonassociating transgene loci at 13 min. These bursting signals are observed again at 15 min at both loci. At 16 min, one transgene (left) associates (A) with a speckle and now maintains elevated transcript signal during the rest of the observation time, whereas the other transgene that is not associated (N) with speckle does not maintain an elevated transcript signal. (F) Coordinated movement of speckle and gene (category: coordinate movement of speckle and gene, Fig. 5 B). White arrows mark the direction of gene movements. Nuclear speckle and associated BAC transgene (white arrowhead) move together as a single unit before merging with a different speckle, suggesting a stable attachment of transgene and speckle (Video 5). (G and H) Movements of Hsp70 BAC transgene away from and back to nuclear speckles at 37°C. White arrows mark the direction of gene movements. BAC transgene shows two (G) or four (H) long-range movements away from and then back toward nuclear speckle. Category in G: FB + α, Fig. 5 B and Video 6; category in H: gene moving to a different speckle, Fig. 5 B.
Video 5.
Maximum-intensity 2D projections of 3D image stacks calculated for each 1-min time point. Time given in minutes. Scale bar = 1 µm; 12 fps. Temperature increase begins at 1 min, reaching 42°C at ∼5 min. Coordinated movement of transgene (green) and nuclear speckle (light green) suggesting stable physical attachment of transgene with speckle during HS (see also Fig. S3 F). Red, MS2-tagged nascent transcripts.
Interestingly, at both HS and normal temperatures, we observed long-range transgene movements relative to nuclear speckles (Fig. 5 B). These movements included repetitive oscillations in which transgenes moved large distances away from speckles and then back to the same speckle (Fig. S3 G and Video 6) or sometimes to a different speckle (Fig. S3 H). Based on our results, we anticipate that such oscillations should produce significant variations in the transcriptional levels of the four genes flanking the Hsp70 locus and, during HS, of the Hsp70 genes.
Video 6.
Maximum-intensity 2D projections of 3D image stacks calculated for each 1-min time point. Time given in minutes. Scale bar = 1 µm; 12 fps. Oscillating movement of transgene (green) relative to nuclear speckle (light green) at 37°C (see also Fig. S3 G). Red, MS2-tagged nascent transcripts.
In summary, our results suggest a new phenomenon of “gene expression amplification” for Hsp70 genes and four genes flanking the Hsp70 locus, whereby association with nuclear speckles is associated with a severalfold boost in gene expression. This gene expression amplification phenomenon appears to be distinct from the now well-described phenomenon of transcriptional bursting, in which genes pulse on and off for extended time periods.
More specifically, we showed that whereas all Hsp70 alleles turn on within several minutes after HS, the amount of nascent transcripts measured adjacent to Hsp70 transgene and endogenous genes is several-fold higher for alleles that are associated with nuclear speckles. At 15 min after HS, at least, this integrated nascent transcript signal correlates with the actual levels of gene expression measured by counting total numbers of Hsp70 mRNAs per cell. At later times after HS, we assume that this nascent transcript signal remains proportional to the net rate of transcript production and nuclear export. Moreover, by live-cell imaging, we showed a tight temporal order of events whereby increased/decreased nascent transcripts, labeled by the MS2 repeats, strictly follow speckle association/disassociation. This temporal ordering of events is consistent with a causal relationship between speckle association and increased net production of MS2-tagged transcripts.
Our results currently support a working model (Fig. 5 C, left) in which speckle association leads to decreased nascent transcript degradation via exosomes, possibly combined with an increased rate of transcription, leading to a net increased rate of transcript production and gene expression. Alternative model 1 (Fig. 5 C, middle), in which speckle association results in a reduced release rate of nascent transcripts from the transcription site, would also lead to an observed increased nascent transcript accumulation with speckle association. However, this would not cause an increased net production of transcripts and is therefore inconsistent with our observation of increased gene expression measured by total cellular mRNA count 15 min after HS. Alternative model 2 (Fig. 5 C, right) would be that increased transcription and nascent transcript accumulation occur first, causing increased speckle association shortly afterward. While this second model would explain the correlation between speckle association and increased nascent transcript levels, it is inconsistent with our observed tight temporal ordering of events in which increased (decreased) nascent transcript levels always follows rather than precedes speckle association (disassociation). Further mechanistic studies will be needed to dissect the exact contributions of changes in transcriptional initiation, pause-release, elongation, and/or RNA degradation rates to the increased nascent Hsp70 transcript accumulation and net increase in mRNA production after 15-min HS that we observe with speckle association.
Our results parallel our previous findings of the appearance of Hsp70 nascent transcripts after speckle association of a very large plasmid transgene array. However, this plasmid array was unusually heterochromatic, and its activation dynamics after HS were greatly delayed and abnormal compared with the endogenous heat-shock locus. Our new results now extend this earlier work by placing these observations in a more physiological context relevant to gene regulation of endogenous gene loci. Moreover, our results now strongly suggest that increased Hsp70 gene expression results from increased expression per allele and not to changes in the percentage of expressing alleles. The tight temporal correlation between speckle association/disassociation and increased/decreased nascent transcripts suggests that expression amplification follows contact with the nuclear speckle periphery, through direct contact between transgene and speckle and/or through the bridging of nascent transcripts. An actual physical linkage between transgene and speckle through the bridging nascent transcripts is further suggested by live-cell videos showing elongation of the MS2 signal during transgene movement away from speckles (Fig. 4 B and Video 4) and coordinated movements of speckle, transcript, and transgene (Fig. S3 F and Video 5).
Overall, our results support the concept of nuclear speckles as a gene expression hub (Hall et al., 2006), capable of increasing the net production of transcripts of associated genes. This expression amplification function of nuclear speckles can add to previously suggested nuclear speckle functions, including modulation of posttranscriptional processing activities such as splicing and nuclear export (Galganski et al., 2017; Spector and Lamond, 2011). We propose speckle association as another possible mechanism of stochastic gene expression for a potentially large subset of active genes. Future work will be aimed at determining the prevalence of expression amplification mediated by nuclear speckle contact and its underlying molecular mechanism.
Materials and methods
Cell culture and establishment of cell lines
CHOK1 cells were grown in Ham’s F12 medium (Cell Media Facility, University of Illinois at Urbana-Champaign) with 10% FBS (Sigma-Aldrich, F2442) at 37°C in a 5% CO2 incubator. To generate stable CHOK1 cell lines, we performed a series of DNA transfections followed by selection of stable colonies with four different DNA constructs in the following order: 92G8+3′MS2+GKREP_C26 Hsp70 BAC (G418, 400 µg/ml), p3′SS-EGFP-dlacI (hygromycin, 200 µg/ml; Robinett et al., 1996), EGFP-SON-Zeo BAC (zeocin, 200 µg/ml), and pUb-MS2bp-mCherry (puromycin, 400 µg/ml; Khanna et al., 2014). Modifications of the original 92G8 Hsp70 BAC (Invitrogen) to 92G8+3′MS2+GKREP_C26 included insertion of a lacO repeat and Kan/Neo selectable marker cassette (Hu et al., 2009), deletion of 8 kb containing the HSPA1A and HSPA1L genes (Hu et al., 2010), and insertion of MS2 repeats into the 3′ UTR of HSPA1B (Khanna et al., 2014). The original SON BAC (Invitrogen, 165J2) was modified by adding GFP to the NH2 terminus of the SON coding region to generate EGFP-SON-Zeo BAC (Khanna et al., 2014). The final CHOK1 cell clone we used for live-cell imaging in these studies was C7MCP. C7MCP cells were maintained in complete medium with 200 µg/ml G418, 100 µg/ml hygromycin, 100 µg/ml zeocin, and 200 µg/ml puromycin. When cells were seeded on coverslips or glass-bottom dishes 2 d before cell imaging, medium was changed to complete medium without any pH indicator and with no G418, hygromycin, zeocin, or puromycin added. The Hsp70_38 CHO DG44 stable cell clone (Hu et al., 2009) contains the HSPA1B BAC, modified with the lacO repeat and Kan/Neo selectable marker, integrated at low copy number at a single chromosomal site. The parental CHO DG44 cell line used for the generation of this clone stably expressed EGFP-Lac repressor. Hsp70_38 cells were grown in Ham’s F12 medium with 10% FBS, 100 µg/ml hygromycin, and 200 µg/ml G418.
smRNA FISH probe design
smRNA FISH was performed using Stellaris probes and the Stellaris protocol (Biosearch Technologies). Probes were designed with the Stellaris Probe Designer using 1.5 to ∼2.5 kb of the 5′ end of the coding sequence. FISH probe sets for each gene consisted of ∼33 20mer DNA oligonucleotides, complementary to the target RNA. A probe set for MS2 repeats consisted of eight 20mer oligonucleotides. Probe sets are listed in Table 1.
Table 1. Probe sets for smRNA FISH.
| Sequence (5′–3′) | ||||||
|---|---|---|---|---|---|---|
| Hsp70 for CHO | Hsp70 | MSH5 | VARS | LSM | C6orf48 | MS2 repeats |
| CACGCACGAGTAGGTGGTG | GGGAGTCACTCTCGAAAGAC | CCACGAGCCTGCAAAAGGA | ACACCCCTGAGCACGACG | GAGCGCAAGCTGGGTAGAG | GAGCCCACTTCGCAAAAAG | CATGTTTTCTAGAGTCGACC |
| CTTCCGTGCTGGAACACG | CACAGGTTCGCTCTGGAAAG | CGCTACAGGTGGGAGAACG | CGCGGACGCAGGACGAGA | CAAGCGCTGACGGGCAAAG | AACTCTCATACTGCCAACC | ATACTGCAGACATGGGTGAT |
| TGGTCGTTGGCGATGATCT | AACGCCGGAAACTCAACACG | CGCCTTTTCAGTAACCTGA | CCGGTCTCACGAGGAACA | GCGGGAAGCGACGCAGAAA | CACTCAACAGTCGGGCCAT | CCTTAGGATCTAATGAACCC |
| GTCGGTGAAGGCCACGTAG | CGACAAGAGCTCAGTCCTT | TCACGCGCTTATCTTCCTC | CTTTGTGACAGGGAGCGT | CAGGTCTGGGGAAACCGAA | TGCGCAAAGGCAGCGCAAG | TGATCCTCATGTTTTCTAGG |
| CGTCGAAACACGGTGTTCTG | TGAGACTGGGGGCTGGAAAC | GAGTCGTGCACGTCTTATG | TAGTTCCCTAAGATCGCC | GGAAGACAGCAGGGTGCTG | CAACGGTAGTTCACCCAAC | AGAGTCGACCTGCAGACATG |
| CGAACTTGCGGCCGATCAG | TGGTCGTTGGCGATGATCTC | GAAGGAAGGGGTCTGAGGG | AGTCGAGCGGGCAGAGAC | CTTTTGACGTCACGGTACC | AGACACCAGAAACTCCAGT | ATGGGTGATCCTCATGTTTT |
| GATCTCCTCGGGGTAGAAG | TTCGCGTCAAACACGGTGTT | TCATTCCTGTCACGCGGAG | AATCCACCTCACAGCCAG | GGTACAAAGGCCAGATCCC | GAGCTAGGTCAGTTCCAAG | AATGAACCCGGGAATACTGC |
| CATCTTCGTCAGCACCATG | TTGTCTCCGTCGTTGATCAC | ATTGTGGGAAACTCCACGC | AGATGGTCAGACTGGGCC | CTCCTCAATGAACCTGAGA | TTGTTTTGAGTCAGCAGGG | AGGCAATTAGGTACCTTAGG |
| TGATCACCGCGTTGGTCAC | AGATCTCCTCGGGGTAGAAT | GGTGAATTCTCGGGTATTT | CGCGTGTACGTACTGGAG | ATATCCCATTTGTTCTCAG | CCATTTTGCAATCACTCGC | |
| GAGTCGTTGAAGTAGGCGG | ATCTCCTTCATCTTGGTCAG | TGGAGAGCTGTGGACACAG | TCGGCGTCTGGTTGGATG | TTTTTTCCCTCATCATGGA | ATCATCACTCCCTTCCTAT | |
| CGTGGGCTCGTTGATGATC | CTGCGAGTCGTTGAAGTAGG | TACTTCTGCTACAGGGCTG | GGAGTGTGGAAGGCTCTC | TCCTCACTGAATCTCTCTC | CACTCAAGAGTTACCTGGG | |
| CCAGGTCGAAGATGAGCAC | ATGATCCGCAGCACGTTGAG | TGGCGCGCAGAATGCAAAG | AACTATCCACCATCGCGG | AGGTCTAACTTTTCCGTCT | TCAGAGCGCTGCGGTGATG | |
| ATGGACACGTCGAACGTGC | TCAGGATGGACACGTCGAAG | ACGATTCACAGAGGAGGCC | GGGGAGTTCCTGGGGAAG | AAGCTTGCCTGGCAGAGAA | AAATGCTGGACCGAGGGGG | |
| GAAGATGCCGTCGTCGATC | TCGAAGATGCCGTCGTCGAT | AAAAGTGAGGGCGGTTCGG | TCTCAGAGGGGCAGTGTC | AGAAAATATCCCACCCGCA | CCATGAACTCGTTGAGCCT | |
| CACGAAGTGGCTCACCAGC | CCCTCAAACAGGGAGTCGAT | CATGAGCTTGGAGGCTCTG | CTGTCAGGAGCCGAGGAC | CCCACTCCTTTCAATGAAT | TTACAACTCCTAACGGGGA | |
| TGGACGACAGGGTCCTCTT | GGTGATGGACGTGTAGAAGT | CTTGGGTTCGCTCCTAAG | CTGAGGGTCTGACCAGGC | CTGTGCTCTCTCAGTCGAC | GGATAACACGGCGATGAAC | |
| CCTCGAACAGGGAGTCGAT | TTCGGAACAGGTCGGAGCAC | CTGGGGAAGCCGGAGGAG | AGACACCCGAGTCCCATA | AAGTCCCAGAGAGACTCTG | CCTAATACTCACCTTTACT | |
| CGCCGTGATGGACGTGTAGA | AGGACCAGGTCGTGAATCTG | TTCTACTCCCCTCAGAGAC | CTCACGGGAGCTCCTTCG | AAGGCTGGAGCCCAAATTA | AGGCATTCAAAAGGCTCTC | |
| GGAACAGGTCCGAGCACAG | TTGAAGAAGTCCTGCAGCAG | CCATCAACTCTCCATTCAA | GGTCCTATGTTTGAGTAG | CATCCCTGACAGTTCTCAA | TTGGGAACAAAGCTTTCCG | |
| CTTCTGCACCTTGGGGATG | TTGATGCTCTTGTTCAGGTC | CCAACCCTCTTTTATTCTA | GAAGACTGCGGGATCGAG | TACTGGTATTGTGAACCCC | ATGCCCATGAACGAAAGCT | |
| GTTGAAGAAGTCCTGCAGC | TGCACGTTCTCGGACTTGTC | GATCTGTCCAGCAAGGAAG | AGGTCCGAACGAAGTGGA | GGAGTGTCCTTTGACAGTA | ACTCCTATTTTGCAGTAGA | |
| GATGCTCTTGTTGAGGTCG | TTGATCAGGGCAGTCATCAC | ATTCCACAGCACACACAGA | GGGAGACGTAGAGGGTGG | TGTGGGAAGGAGCATGGTA | GGACGTTAGAAAGGGAGGA | |
| CTGCACGTTCTCAGACTTG | TCGTACACCTGGATCAGCAC | TAGGCAATGCCCAAGTATC | CTGGGGAAGGCATCTGGG | GGGAGAGAGGGGGAAAACC | AGGGAAGCTCTTCTGGAAA | |
| TTGATGAGCGCCGTCATCA | CAGATTGTTGTCTTTCGTCA | GTGGAGTCACTAGTATCAT | TAGCGAGCGGCTATGAGG | TAGTTCCACGACCACATCC | CCAATTAGGGAGATCTGGA | |
| GAGTAGGTGGTGAAGGTCT | ATGTCGAAGGTCACCTCGAT | GCATCTGGCATGAAGTGGA | GTGGCTGGAGACAGATGC | GTACCTACCTCAGGTCATT | AGCATCGGAGACTCTAGTC | |
| CGTACACCTGGATCAGCAC | TGACGTTCAGGATGCCGTTG | GAGAAGCTTGAGGCTCTCG | GTTGGACAAGGACAGCCG | CCCAACAGACTTGTTGGAA | GTTCCCTAAATGAGTCAGA | |
| GATGCCGCTGAGCTCGAAG | TCATGTTGAAGGCGTAGGAC | GGAATTCATGGTTCCATCC | CGTGTCGGCGTAACTGAC | TGCTTTGTATTGTTTTCCA | CTTCTGGAGACCCAAGTAT | |
| ATCGATGTCGAAGGTCACC | TGTCCAGAACCTTCTTCTTG | CATCTGCAATCCCAGAGAG | CACAGGCAGCTGGTATTA | AAAGAGGTTCCAGGGCCC | TGGGCTTCCAGAGTTCATT | |
| TGACGTTCAGGATGCCGTT | CACGAGATGACCTCTTGACA | CGAGCTTCGGAGTCCCAG | GTACATCTTCCACCTCGC | CTCTGTGAAGGTGCATTGT | ||
| TAGGACTCGAGCGCGTTCT | TTGTGCTCAAACTCGTCCTT | AGGCACAAGGCATTTATTT | CCTTCACTCAGATTAGTGC | |||
| GCGCTCTTCATGTTGAAGG | CTGATGATGGGGTTACACAC | ACTGCGCCTGACCTGTATT | GAGGGGGAGATTCCAAACC | |||
| AGGAGATGACCTCCTGACA | ACTAAAGAACAAAGGCCCCT | GCCATACAAAGCTTCTCTC | ||||
| CACGAACTCCTCCTTGTCG | AAGTCCTTGAGTCCCAACAG | |||||
| CTAATCCACCTCCTCGATG | CCATCAGGTTACAACTTAAC | |||||
| ACACCGGGAGAGCAAGCAG | ||||||
| AGGGCTAACTAACCCTGAC | ||||||
smRNA FISH procedure
Each oligonucleotide contained an amino group at the 3′ end for fluorophore coupling using either Cy5 N-hydroxysuccinimide (NHS) ester (GE Healthcare, PA15102) or rhodamine NHS ester (Thermo Fisher Scientific, 46406). NHS esters and probes were incubated overnight in 0.1 M sodium bicarbonate solution (pH 8.0) at RT, and purified using Bio-Spin P6 columns (Bio-Rad, 7326221) according to the manufacturer’s protocol. Purified, pooled probe concentrations were ∼50–100 µM in Tris buffer, pH 7.4.
Cells were seeded on coverslips (Thermo Fisher Scientific) 2 d before experiments. For HS, a well-plate or dish containing the coverslip and medium was sealed using parafilm and incubated in a 42°C water bath. Cells were fixed using freshly prepared 3.6% PFA (Sigma-Alrich, P6148-500G) in PBS for 15 min at RT. After washing for 5 min three times in PBS, cells were permeabilized using 0.5% Triton X-100 (Thermo Fisher Scientific, 28314) for 10 min in diethyl pyrocarbonate (DEPC)-treated PBS. For DEPC treatment, 1 ml fresh DEPC was added to 1 liter PBS or water, incubated >15 h at RT, and autoclaved for 25 min to inactivate the remaining DEPC. After rinsing three times in DEPC-treated PBS, the permeabilized cells were equilibrated in wash buffer (10% formamide [Sigma-Aldrich, F9037] and 2× SSC) for 30 min. The cells were incubated in hybridization buffer with smRNA FISH probes (final concentration, ∼300–500 nM) for 15 h at 37°C. The hybridization buffer contained 2× SSC, 10% formamide, 10% wt/vol dextran sulfate (Sigma-Aldrich, D8906), 1 mg/ml Escherichia coli tRNA (Sigma-Aldrich, R8759), 2 mM ribonucleoside vanadyl complex (NEB, S1402), and 0.02% RNase-free BSA (Ambion, AM2618) in DEPC-treated water. The cells were then washed twice for 30 min at 37°C in wash buffer and mounted in a Mowiol-DABCO antifade medium (Harlow and Lane, 1988).
smRNA FISH in HAP1 cells was followed by immunostaining against SON. After smRNA FISH, cells were fixed again with 3.6% PFA in DEPC-treated PBS for 15 min and washed three times for 5 min. Cells were incubated in blocking buffer (0.5% Triton X-100, 1% wt/vol RNase-free BSA, and 20 µM ribonucleoside vanadyl complex). Cells were incubated with primary antibody against SON (Sigma-Aldrich, HPA 023535) at a 1:300 dilution in DEPC-treated PBS for 1 h in a humid chamber at RT, washed three times for 5 min in DEPC-treated PBS, and incubated with Alexa Fluor 488–labeled, goat anti-rabbit IgG secondary antibody (Invitrogen, A11008) at a 1:300 dilution in DEPC-treated PBS for 1 h in a humid chamber at RT. All antibody solutions were supplemented with 0.4 U/µl RNase inhibitors (Lucigen, E0126). For smRNA FISH against endogenous transcripts in WT CHOK1 cells, immunostaining against SC-35 was done before smRNA FISH. We followed a similar immunostaining procedure as described above, but with primary antibody against SC35 (1:300 in PBS; Abcam, ab11826) for 12 h at 4°C and with Alexa Fluor 488–labeled, secondary goat anti-mouse IgG antibody (Invitrogen, A11029) at a 1:300 dilution in DEPC-treated PBS. After washing three times for 5 min in DEPC-treated PBS, slides were mounted in a Mowiol-DABCO antifade medium.
Exosome knockdown
C7MCP cells were seeded on coverslips (Thermo Fisher Scientific) and transfected the next day with 20 nM of scrambled or EXOSC3 (Dharmacon) siRNA using RNAiMax (Life Technologies) as per the manufacturer’s instructions. The sequence of siRNA for RRP40 knockdown was 5′-GGCAUAGUGAUUGCUAAAUUU-3′. Scrambled siRNA were commercially available, ON-TARGETplus non-targeting pool (Dharmacon, D-001810-01-05). Cells were used for either immunoblotting or RNA-immunoFISH after 96 h (two rounds of transfection). For HS, dishes containing the coverslips were sealed with parafilm and placed in a water bath at 42°C for 1 h and immediately fixed using freshly prepared 4% PFA. smRNA FISH for Hsp70 and immunostaining was performed as described previously using a 1:250 dilution of rabbit monoclonal EXOSC3 antibody (Abcam, ab190689) and 1:100 dilution of secondary anti-rabbit Alexa Fluor 405 antibody (Jackson Immunoresearch Laboratories, 711-475-152). Cell lysates were prepared using Laemmli buffer and resolved on a 10% polyacrylamide gel. Proteins were transferred onto a Immobilon polyvinylidene fluoride membrane (Millipore). The blot, after blocking in 5% skim milk in 0.1% Triton X-100 in PBS, was incubated overnight with either anti-EXOSC3 (1:10,000 dilution) or anti-Lamin B1 (Abcam, ab16048) antibody in 5% skim milk in 0.1% Triton X-100 in PBS at 4°C overnight. The blot was washed three times with 0.1% Triton X-100 in PBS and incubated with HRP-conjugated goat anti-rabbit antibody (1:10,000 dilution in 5% skim milk in 0.1% Triton X-100 in PBS) at RT for 1 h, and then washed three times with 0.1% Triton X-100 in PBS. Using luminol as substrate, the blot was visualized by chemiluminescence using the Immobilon Western Chemiluminescent HRP substrate (Millipore, WBKLS0100) and iBright CL1000 Imaging system (Invitrogen).
RNA polymerase staining
Immunostaining used a 1:1,000 dilution of rabbit anti-RNA polII COOH-terminal domain repeat YSPTSPS (phospho S2) antibody (Abcam, ab193468) followed by a 1:500 dilution of secondary anti-rabbit Alexa Fluor 647 (Invitrogen, A-21244). To measure Ser2P accumulation adjacent and over the transgene, projected images of the Z-stacks showing the transgene were generated from conservative, iterative deconvolved (Softworx, GE Healthcare) images. Using ImageJ (National Institutes of Health), integrated intensity measurements were made within a manually defined region of interest surrounding the Ser2P foci near the transgene. Background integrated intensity from a nearby region of the same area was subtracted from these measurements.
Microscopy and data analysis of fixed samples
For fixed cells, we used a Personal Delta Vision microscope (GE Healthcare) equipped with a Coolsnap HQ camera and Plan Apo N 60×/1.42-NA oil-immersion objective (Olympus). Sections were spaced every 200 nm in z. Pixel size was 67 nm. 3D z-stacks were processed using the “enhanced” version of the iterative, nonlinear deconvolution algorithm provided by the Softworx software (GE Healthcare; Agard et al., 1989) and projected in the x-y plane using a maximum-intensity algorithm. The distance between a gene and a nuclear speckle was measured from the maximum-intensity projection of the 3D dataset (the edge of the speckle was defined as where the nuclear speckle intensity fell to 40% of its intensity maximum), and the distance was measured to the center of the BAC transgene or the edge of a FISH signal. Measurements were made manually using the line profile function in ImageJ. smRNA FISH signal intensities over nascent transcripts were measured by manually selecting the nascent RNA FISH area in 2D summed projections of the 3D raw dataset, summing the pixel intensities, and then subtracting the background intensity estimated from a same-size area adjacent to the nascent transcript signal. Measurements were made using ImageJ. For counting the number of mature mRNA spots, we used the StarSearch software program, as described elsewhere (Levesque et al., 2013; Shaffer et al., 2013). Figs. 1 E and S2 D (but not line scans) were scaled using a gamma value <1, applied uniformly across all images. This reduced the intensity dynamic range, allowing simultaneous visualization of both intense nascent transcript signals and dimmer, less intense nascent transcript signals and cytoplasmic mRNAs with the same scaling.
Live-cell imaging and analysis
Cells were plated on 35-mm dishes with a #1.5-thickness glass coverslip bottom (MatTek, P35G-1.5-14-C) 48 h before imaging. For rapid wide-field live-cell imaging, we used an OMX V4 microscope (GE Healthcare) equipped with a U Plan S-Apo 100×/1.40-NA oil-immersion objective (Olympus), two Evolve electron-multiplying charge-coupled device cameras (Photometrics), a live cell incubator chamber (GE Healthcare) with separate temperature controllers for the objective lens and the incubator heater, and a humidified CO2 supply. MatTek dishes were placed on the microscope, and temperatures for both the objective lens and incubator chamber were maintained at 37°C for ∼1–3 h before data acquisition. Temperatures were set to 44°C on both temperature controllers immediately after the first time frame was taken, with ramping from 37°C to 44°C requiring ∼5 min. Empirically, this temperature setting of 44°C for the controllers produced a temperature of 42°C in the medium inside the Matek dish, and a similar transcriptional induction of the Hsp70 BAC transgene MS2 signal on the microscope as seen off the microscope with a 42°C HS.
3D images (z-spacing = 200 nm) were acquired once every minute, typically using the solid-state illumination with 1% transmittance, 10–15-ms exposure for each z-slice for 477 ± 16-nm excitation (GFP) and 2 to ∼5% transmittance and 10–20-ms exposure for each z-slice for 572 ± 10-nm excitation (mCherry). Typically, 3D stacks from each of 20–25 fields of view were taken during each 1-min interval using the point-visiting function of the Softworx image acquisition software, with each 3D data stack acquired over ∼1 s or less. 3D z-stacks were processed using the enhanced version of the iterative, nonlinear deconvolution algorithm provided by Softworx (Agard et al., 1989; GE Healthcare), and projected into the x-y plane using a maximum-intensity algorithm. Using a custom Matlab program, each cell in the projected live-cell videos (512 × 512-pixel area per frame) was tracked and cropped into a 256 × 256-pixel area per frame with the cell translated to the center of the area and saved as a single file. Custom Matlab scripts written for data analysis described here are publicly available on Github: https://doi.org/10.5281/zenodo.2559675. If necessary, a rigid body registration (ImageJ plugin “StackReg”) was applied to correct for any x-y nuclear rotation and/or translational displacement between sequential time points. Each of these individual cell video files was then visually inspected and sorted into different categories of dynamics for further analysis.
Online supplemental material
Fig. S1 shows positioning of HSPA1B BAC transgene or endogenous Hsp70 genes relative to nuclear speckles before or after HS. Fig. S2 contains various supporting data: transcription induction kinetics in different clones; cell images corresponding to Fig. 1 D; exosome knockdown and HSPA1B transcription levels; intensity of Ser2P foci as function of speckle association; and speckle association of Hsp70 flanking genes. Fig. S3 shows additional examples of speckle and transgene dynamics. Video 1 shows transcription increase after HS for stably associated transgene (Fig. 3 A). Video 2 shows transcription increases after speckle association (Fig. 3 B). Video 3 shows transcription decreases after disassociation from speckle (Fig. 4 A). Video 4 shows dissociation of transgene from speckle with transcripts bridging transgene and speckle (Fig. 4 B). Video 5 shows coordinated movement of attached transgene and speckle (Fig. S3 F). Video 6 shows transgene oscillations away from and back toward nuclear speckle (Fig. S3 G).
Acknowledgments
A.S. Belmont acknowledges support from National Institutes of Health grant R01 GM58460 and from its Common Fund 4D Nucleome Program (U54 DK107965).
The authors declare no competing financial interests.
Author contributions: J. Kim and A.S. Belmont conceived of and designed the study; J. Kim performed most experiments and analyzed data; N. Chivukula Venkata performed exosome and RNA polII Ser2p experiments; N. Chivukula Venkata and G.A. Hernandez Gonzalez helped with RNA FISH experimental replicates and controls; J. Kim and A.S. Belmont wrote the manuscript; N. Khanna provided a stable cell line.
References
- Agard D.A., Hiraoka Y., Shaw P., and Sedat J.W.. 1989. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30:353–377. 10.1016/S0091-679X(08)60986-3 [DOI] [PubMed] [Google Scholar]
- Brown J.M., Green J., das Neves R.P., Wallace H.A., Smith A.J., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R., et al. 2008. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 182:1083–1097. 10.1083/jcb.200803174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang Y., Wang Y., Zhang L., Brinkman E.K., Adam S.A., Goldman R., van Steensel B., Ma J., and Belmont A.S.. 2018. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 217:4025–4048. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Kuai B., Wang K., Wang L., Wang Y., Wu X., Chi B., Li G., and Cheng H.. 2018. mRNAs are sorted for export or degradation before passing through nuclear speckles. Nucleic Acids Res. 46:8404–8416. 10.1093/nar/gky650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galganski L., Urbanek M.O., and Krzyzosiak W.J.. 2017. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 45:10350–10368. 10.1093/nar/gkx759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.L., Smith K.P., Byron M., and Lawrence J.B.. 2006. Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288:664–675. 10.1002/ar.a.20336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., and Lane D.. 1988. Antibodies. A laboratory manual. Cold Spring Harbor Laboratory, New York. [Google Scholar]
- Hu Y. 2010. Changes in large-scale chromatin structure and dynamics of BAC transgenes during transcriptional activation. University of Illinois, Urbana, IL. [Google Scholar]
- Hu Y., Kireev I., Plutz M., Ashourian N., and Belmont A.S.. 2009. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J. Cell Biol. 185:87–100. 10.1083/jcb.200809196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Plutz M., and Belmont A.S.. 2010. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J. Cell Biol. 191:711–719. 10.1083/jcb.201004041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N., Hu Y., and Belmont A.S.. 2014. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. 24:1138–1144. 10.1016/j.cub.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Han K.Y., Khanna N., Ha T., and Belmont A.S.. 2019. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 132:jcs226563 10.1242/jcs.226563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J., Pagie L., Ortabozkoyun H., Boyle S., de Vries S.S., Janssen H., Amendola M., Nolen L.D., Bickmore W.A., and van Steensel B.. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 153:178–192. 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Kölbl A.C., Weigl D., Mulaw M., Thormeyer T., Bohlander S.K., Cremer T., and Dietzel S.. 2012. The radial nuclear positioning of genes correlates with features of megabase-sized chromatin domains. Chromosome Res. 20:735–752. 10.1007/s10577-012-9309-9 [DOI] [PubMed] [Google Scholar]
- Lamond A.I., and Spector D.L.. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605–612. 10.1038/nrm1172 [DOI] [PubMed] [Google Scholar]
- Levesque M.J., Ginart P., Wei Y., and Raj A.. 2013. Visualizing SNVs to quantify allele-specific expression in single cells. Nat. Methods. 10:865–867. 10.1038/nmeth.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz S.A., Ollikainen N., Tabak B., Palla A., Schmidt J.M., Detmar E., Lai M.M., Shishkin A.A., Bhat P., Takei Y., et al. 2018. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 174:744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., and Aiden E.L.. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159:1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., and Belmont A.S.. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700. 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer S.M., Wu M.T., Levesque M.J., and Raj A.. 2013. Turbo FISH: a method for rapid single molecule RNA FISH. PLoS One. 8:e75120 10.1371/journal.pone.0075120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland L.S., Johnson C.V., Byron M., McNeil J., and Lawrence J.B.. 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J. Cell Biol. 162:981–990. 10.1083/jcb.200303131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silla T., Karadoulama E., Mąkosa D., Lubas M., and Jensen T.H.. 2018. The RNA Exosome Adaptor ZFC3H1 Functionally Competes with Nuclear Export Activity to Retain Target Transcripts. Cell Reports. 23:2199–2210. 10.1016/j.celrep.2018.04.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., and Lamond A.I.. 2011. Nuclear speckles. Cold Spring Harb. Perpect. Biol. 3:a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T., Meaburn K.J., and Misteli T.. 2008. The meaning of gene positioning. Cell. 135:9–13. 10.1016/j.cell.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasan I., Sustackova G., Zhang L., Kim J., Sivaguru M., HamediRad M., Wang Y., Genova J., Ma J., Belmont A.S., and Zhao H.. 2018. CRISPR/Cas9-mediated knock-in of an optimized TetO repeat for live cell imaging of endogenous loci. Nucleic Acids Res. 46:e100 10.1093/nar/gky501 [DOI] [PMC free article] [PubMed] [Google Scholar]