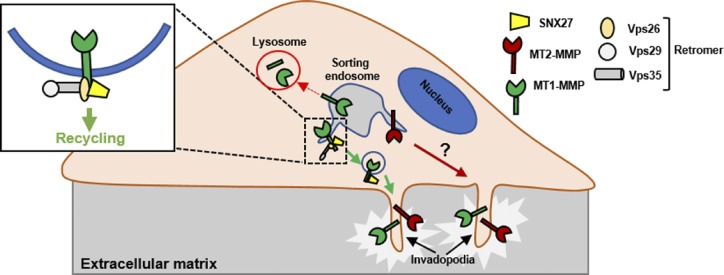
Recycling of MT-MMPs to actin-rich membrane-protrusive structures promotes breast cancer invasion. This study shows that SNX27–retromer, an endosomal sorting and recycling machinery, interacts with MT1-MMP and regulates its transport to the cell surface, thus promoting matrix invasive activity of the breast cancer cells.
Abstract
A variety of metastatic cancer cells use actin-rich membrane protrusions, known as invadopodia, for efficient ECM degradation, which involves trafficking of proteases from intracellular compartments to these structures. Here, we demonstrate that in the metastatic breast cancer cell line MDA-MB-231, retromer regulates the matrix invasion activity by recycling matrix metalloprotease, MT1-MMP. We further found that MT2-MMP, another abundantly expressed metalloprotease, is also invadopodia associated. MT1- and MT2-MMP showed a high degree of colocalization but were located on the distinct endosomal domains. Retromer and its associated sorting nexin, SNX27, phenocopied each other in matrix degradation via selectively recycling MT1-MMP but not MT2-MMP. ITC-based studies revealed that both SNX27 and retromer could directly interact with MT1-MMP. Analysis from a publicly available database showed SNX27 to be overexpressed or frequently altered in the patients having invasive breast cancer. In xenograft-based studies, SNX27-depleted cell lines showed prolonged survival of SCID mice, suggesting a possible implication for overexpression of the sorting nexin in tumor samples.
Graphical Abstract
Introduction
MT1-MMP–mediated ECM degradation
Metastasis to a secondary site is among the major causes of tumor relapse and cancer-associated fatalities around the world. Invasive cancer cells form actin-rich plasma membrane protrusions called invadopodia that facilitate breaching of the underlying basement membrane. Invadopodia act like molecular scissors, where various proteases are continuously delivered to degrade ECM (Linder et al., 2011). Membrane type 1 matrix metalloproteinase (MT1-MMP), a member of the MMP family, is a well-studied invadosome-associated protease (Jabłońska-Trypuć et al., 2016; Itoh, 2015; Holmbeck et al., 2003; Jiang et al., 2006; Sodek et al., 2007; Qiang et al., 2019; Artym et al., 2006; Poincloux et al., 2009). It was initially characterized as an interstitial collagenase which degraded ECM by directly cleaving its substrate and activating a secretory matrix metalloprotease, MMP-2 (Ohuchi et al., 1997; Sato et al., 1994; Al-Raawi et al., 2011; Clark et al., 2007). Although other membrane-type metalloproteases are also involved in cancer metastasis, their molecular role in ECM remodeling is less explored (Wells et al., 2015; Tatti et al., 2011, 2015; Shen et al., 2017; Yip et al., 2017; Huang et al., 2009; Wu et al., 2017; Wang et al., 2015; Jiang et al., 2017). MT2-MMP is overexpressed in breast cancer and is involved in basement membrane transmigration in breast cancer (Kousidou et al., 2004; Benson et al., 2013; Hotary et al., 2006; Ota et al., 2009), but the molecular mechanism governing its role in cancer metastasis is unexplored.
A rapid and tightly regulated recycling of MT1-MMP to invadopodia conforms with the dynamicity of these ECM remodeling structures, which is exclusively governed by intracellular trafficking (Jacob and Prekeris, 2015; Poincloux et al., 2009; Linder, 2015; Frittoli et al., 2011; Castro-Castro et al., 2016). Some of the components of recycling circuitries, including Exocyst complex, SNAREs, and Rabs, have been identified to play a crucial role in transport of MT1-MMP to invadopodia (Monteiro et al., 2013; Steffen et al., 2008; Williams and Coppolino, 2011; Williams et al., 2014; Macpherson et al., 2014; Wiesner et al., 2013; Frittoli et al., 2014; Kajiho et al., 2016). All these studies collectively imply that disruption of the protease-recycling axis has a pronounced impact on the invasive properties of the cancer cell.
Retromer complex in endosomal sorting and recycling
Retromer, an evolutionarily well-conserved complex, plays a vital role in the sorting and recycling of various transmembrane cargoes (Seaman et al., 1997; Burd and Cullen, 2014; Cullen and Steinberg, 2018). It was discovered in yeast to recycle Vps10, a sorting receptor for vacuolar carboxypeptidase Y, from endosomes to the TGN (Seaman et al., 1998; Arighi et al., 2004; Seaman, 2004; Johannes and Popoff, 2008). Retromer is a heteromeric protein complex comprising vacuolar protein sorting (Vps) subunits, i.e., Vps35, Vps26, and Vps29, that form the core. The trimer is unable to get recruited on the endosomal membrane on its own, which is prerequisite for its cargo-retrieval activity. To accomplish this, core retromer components are shown to be associated with small GTPase Rab7A and some of the members of the sorting nexin (SNX) family (Gallon and Cullen, 2015; Rojas et al., 2007; Cullen and Korswagen, 2011; Seaman et al., 2009, 1998; Rojas et al., 2008; Harrison et al., 2014; Wassmer et al., 2009). The retromer-mediated cargo recycling is crucial for lysosomal functioning, nutrient uptake, and Drosophila melanogaster wing development, maintaining apical polarity and neuronal functions (Burd and Cullen, 2014; Seaman et al., 1997; Cui et al., 2019; Vardarajan et al., 2012; Harterink et al., 2011). Thus, perturbation of retromer is linked to pathologies like metabolic myopathy, neurodegenerative disorders, etc. (Wang and Bellen, 2015; Small and Petsko, 2015). Retromer also interacts with WASH (Wiskott-Aldrich syndrome protein and Scar homologue), an endosomal actin nucleator, to facilitate cargo sorting (Harbour et al., 2010; Derivery et al., 2009; McGough et al., 2014). It is now well established that, in metazoans, retromer plays a central role in plasma membrane recycling of various cargoes (Seaman et al., 2013; Chamberland and Ritter, 2017; Burd and Cullen, 2014). This expanded function is dependent on its association with WASH and SNX27, a PDZ (PSD95, Dlg1, and zo-1) domain-containing member of the SNX family. Glucose transporter (GLUT1), β-adrenergic receptor (β-AR), and parathyroid-receptor hormone are among the well-studied cargoes that are directed to the plasma membrane by retromer and its associated SNX27 (Steinberg et al., 2013; Temkin et al., 2011; Lauffer et al., 2010; McGarvey et al., 2016). The loss of SNX27 or retromer trimer resulted in missorting and lysosomal degradation of these cargoes. Temkin et al. (2011) have shown that SNX27 associates with the cargo and mediates its entry to retromer-decorated tubules, thus giving new insight into endosomal sorting by SNX27 and retromer. A plethora of cargoes has been found comprising of a PDZ-binding motif in their cytoplasmic tail. This motif is required for the recognition of these cargoes by SNX27 (Steinberg et al., 2013; Lunn et al., 2007; Gallon et al., 2014). Lauffer et al. (2010) have shown that mutation of a single conserved His residue in the PDZ domain of SNX27 significantly reduced the recycling of β2-AR, which has PDZ-ligand in its cytoplasmic tail. Thus, these studies have established the role of SNX27 protein in cargo selection and acting as a cargo adaptor.
In the present study, we investigate the sorting and recycling mechanism of MT1-MMP and MT2-MMP, abundant membrane-type proteases in the metastatic breast cancer cell line. We hypothesized that retromer might be facilitating the recycling of these MT-MMPs conferring ECM-degrading abilities to the breast tumor cells. Retromer preferentially recycled MT1-MMP to the cell surface. Further, among retromer-associated SNXs, SNX27 was found to assist retromer for recycling of the proteases. Perturbation of either retromer or SNX27 abrogated the ECM degradation activity of the cancer cells. The mechanism governing sorting of the proteases by the retromer–SNX27 assembly is studied using microscopy and biochemical assays. Finally, the in vivo implication of our observations was tested in a metastatic breast cancer xenograft model.
Results
Retromer contributes to matrix degradation and invasion in breast cancer cells
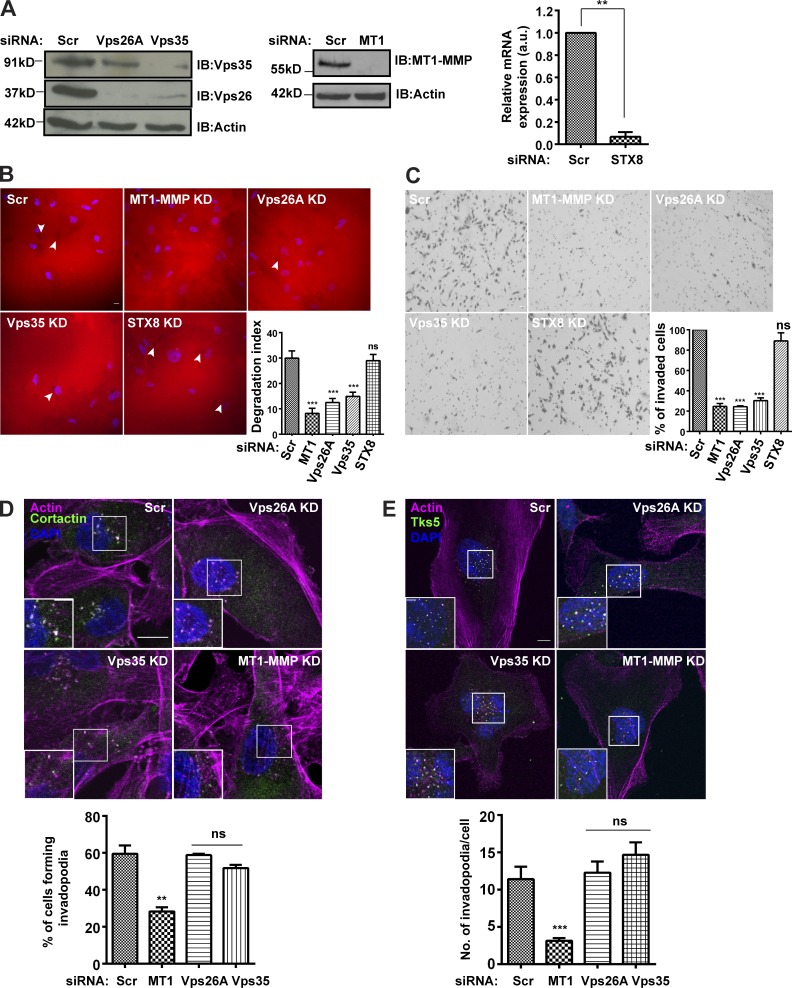
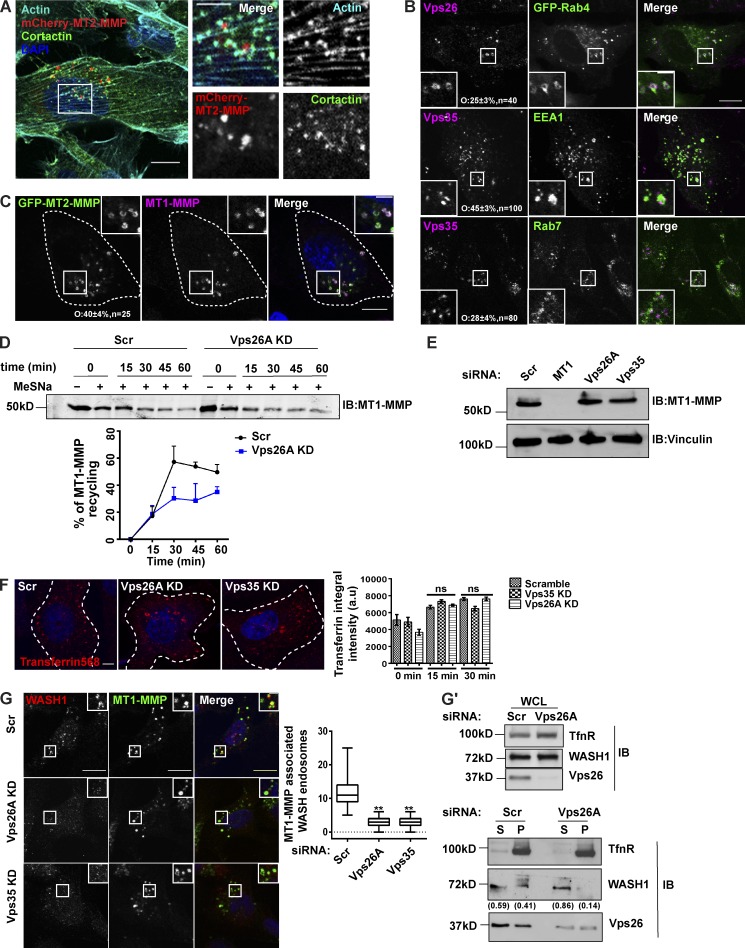
Targeted trafficking and recycling of the cargoes is critical for the life cycle/turnover of invadopodia. Thus, we sought to investigate the consequences of disrupting retromer, a key player of endosomal protein sorting and recycling (Klinger et al., 2015; Burd and Cullen, 2014), on the invasive properties of MDA-MB-231. We used ON-TARGETplus SMARTpools for targeted siRNA-mediated downregulation of Vps26A, Vps35, MT1-MMP, and syntaxin 8 (STX8). The knockdown (KD) efficiency was measured at the protein or mRNA level (Fig. 1 A). Subsequently, the cells were seeded on fluorescently labeled gelatin or Matrigel-coated cell inserts and allowed to degrade or invade, after which they were fixed and processed. The percentage of invaded cells or degradation index were quantified for control and siRNA-treated cells. Control cells showed matrix degradation and Matrigel invasion, thus confirming the formation of functional invadopodia, whereas depletion of Vps subunits (retromer) significantly abrogated matrix degradation and cellular ability to invade Matrigel-coated inserts (Fig. 1, B and C). A similar phenotype was observed for the cells with reduced expression of MT1-MMP. However, cells depleted of STX8, a member of the SNARE family, which are crucial machinery for the endosomal trafficking route (Chen and Scheller, 2001; Jahn and Scheller, 2006; Risselada and Grubmüller, 2012), could still degrade the matrix similar to that of control cells (Fig. 1, B and C). This suggests that STX8 might not be involved in intracellular trafficking of the cargoes required for the degradative activity of invadopodia.
Figure 1.
Retromer contributes to the invasive properties of breast cancer cells. (A) The KD efficiency of the respective genes was confirmed by Western blot or quantitative PCR (qPCR; n = 3). Values in the graph represent means ± SEM. Two-tailed Student’s t test, ** P < 0.01. (B) Retromer, MT1-MMP, or STX8 were depleted via siRNA and cells were seeded on Alexa Fluor 568–labeled gelatin-coated coverslips for 12 h. Degradation activity was quantified as degradation index, described in the Materials and methods (N = 4, n = 300; scale bar = 10 µm). Arrowheads represent degradation spots. One-way ANOVA, *** P < 0.001. The graph represents means ± SEM. (C) Cells depleted for indicated molecules were seeded on Matrigel-coated cell inserts and allowed to invade for 20 h. Invasive cells were counted and the percentage of invaded cells were plotted (N = 3, scale bar = 30 µm). One-way ANOVA, *** P < 0.001. The graph represents means ± SEM. (D and E) MDA-MB-231 cells treated with indicated siRNA were immunostained for invadopodia markers cortactin (D) or Tks5 (E) and stained with phalloidin to label actin. Images were analyzed on the confocal microscope and the percentage of cells forming invadopodia was quantified and plotted (N = 3, n = 400; scale bar = 10 µm, inset = 4 µm). The inset represents the actin-cortactin– or Tks5-actin–rich invadopodia. Invadopodia (Tks5/actin dots) per cell were counted (N = 3, n = 50). The graph represents means ± SEM. One-way ANOVA, ** P < 0.01, *** P < 0.001. N, number of experimental repeats.
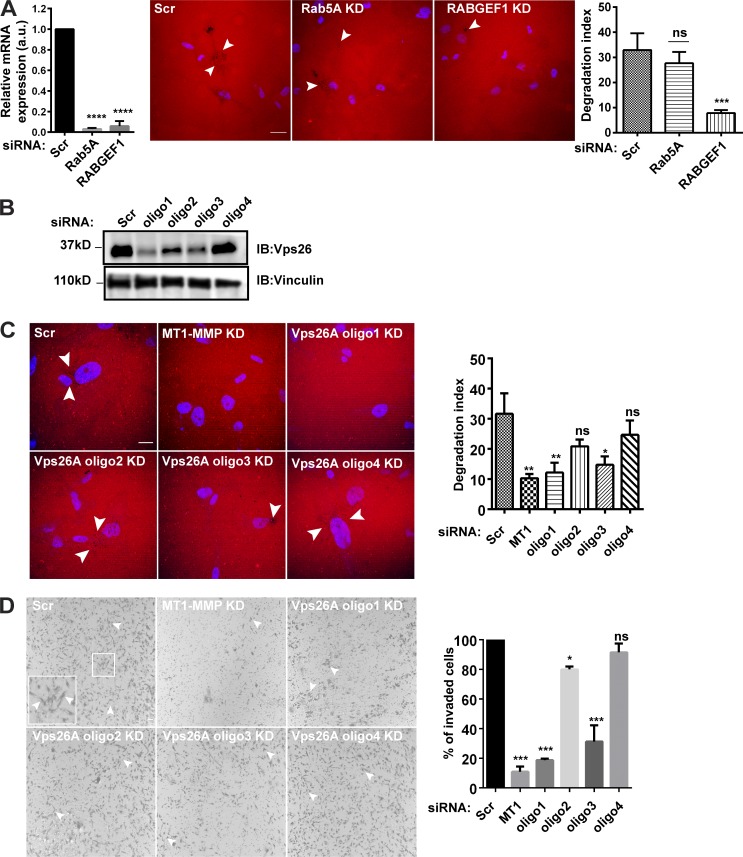
Additionally, the depletion of Rab5A isoforms did not affect the degradation index. Previously, it has been shown that Rab5 isoforms show redundancy in their function, and cells depleted of a single isoform could degrade the matrix (Frittoli et al., 2014). However, the downregulation of its activator (guanine nucleotide exchange factor [GEF] for Rab5A/B/C), RABGEF1, reduced the matrix degradation (Fig. S1 A). These observations demonstrated the sensitivity of this assay.
Figure S1.
Retromer-mediated abrogation of the invasive potential of MDA-MB-231 is a gene-specific effect. Gelatin degradation assay is a sensitive assay to measure matrix degradation activity of the cancer cells. (A) Cells were transfected with siRNA against Rab5A and RABGEF1. After 72 h, mRNA was isolated and qPCR was performed to measure the efficacy of the KD. Subsequently, similarly treated cells were seeded on fluorescent gelatin for 12 h, followed by fixation and DAPI staining to label nuclei. Cells were imaged with a confocal microscope and the degradation index was analyzed. Arrowheads represent the degradation dots. (N = 3, n = 100; scale bar = 10 µm). The graphs represent means ± SEM. One-way ANOVA, *** P < 0.001, **** P < 0.0001. (B) To ensure the target specificity by siRNA, individual oligos were used to deplete Vps26A. Cells were transfected with the four oligos targeting different sites on Vps26A and subjected to immunoblotting. Of the four, oligo1 and oligo3 could efficiently deplete the Vps26A. (C and D) Similarly treated cells were subjected to gelatin degradation (C) or Matrigel invasion (D), where cells were seeded on fluorescent gelatin or Matrigel-coated cell inserts, respectively. The degradation index and percentage of invaded cells were analyzed and plotted. (N = 3, scale bars = 30 pixels). Values in the graphs represent means ± SEM. One-way ANOVA, * P < 0.05, ** P < 0.001, *** P < 0.001. N, number of experimental repeats.
Further, the observed phenotype for retromer was cross-validated using four individual oligos. Among these, oligo1 and oligo3 showed maximum KD of Vps26A (Fig. S1 B) and thus impaired the gelatin degradation and Matrigel invasion activity (Fig. S1, C and D). Validation using individual oligos also helped us in ruling out the off-target effect, if any occurred, due to the use of SMARTpool siRNA.
Next, we asked whether the downregulation of retromer hampered invadopodia formation. Retromer-depleted cells were immunostained for well-known invadopodia markers Tks5 or cortactin. The cells could form invadopodia, as marked by actin-cortactin– or actin-Tks5–positive dots, comparable to that of control (Fig. 1, D and E). On the contrary, cells suppressed for MT1-MMP showed a reduced ability to form invadopodia, which is in line with the previous reports (Clark et al., 2007; Steffen et al., 2008). Altogether, these results suggested that the endosomal sorting and recycling pathways are crucial for the invadopodia maturation, perhaps via their role in the trafficking of some of the maturation factors like proteases.
Retromer preferentially recycles MT1-MMP but not MT2-MMP from endosomes to the cell surface
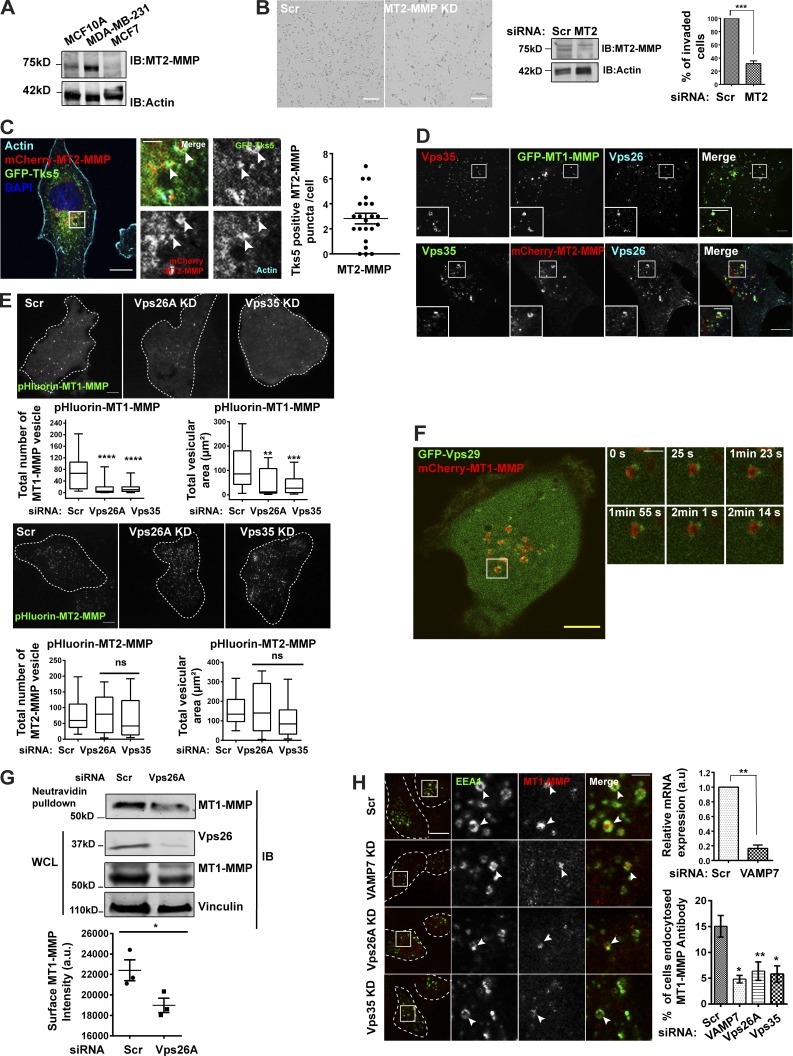
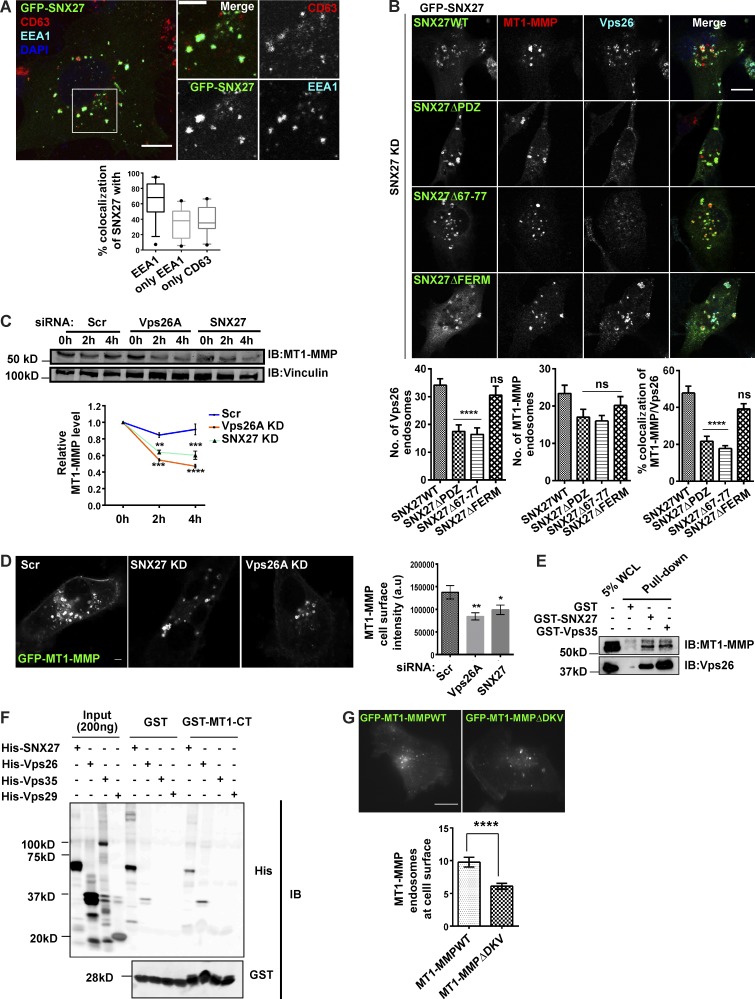
Next, we asked which cargo might be transported via retromer to invadopodia. Since retromer-depleted cells formed nonfunctional invadopodia, we focused on investigating the recruitment of the proteases on these structures. MT1-MMP is known to be overexpressed in various cancers, including breast cancer, and promotes tumor invasion (Sato et al., 1994; Okada et al., 1995; Jiang et al., 2006; Perentes et al., 2011; Al-Raawi et al., 2011; Rossé et al., 2014; Li et al., 2015). Also, elevated transcript levels of MT2-MMP, a member of the MT-MMP family that shares 73.9% similarity with MT1-MMP, were reported in the MDA-MB-231 cell line (Hotary et al., 2006; Ota et al., 2009). We performed a comparative expression analysis using an antibody against MT2-MMP and found its overexpression in the invasive MDA-MB-231 cell line, compared with nontransformed MCF10A or poorly invasive MCF7 cell lines (Fig. 2 A). To understand the functional relevance of the overexpression of the protease in MDA-MB-231, we performed a Matrigel invasion assay in the cells depleted of MT2-MMP and observed a significant reduction in the percent of invaded cells (Fig. 2 B). This was consistent with the previous reports, where this protease is shown to facilitate basement membrane degradation (Hotary et al., 2006; Ota et al., 2009). Interestingly, MT2-MMP was also found to be invadopodia associated since it showed colocalization with GFP-Tks5 and cortactin, as revealed by fixed-cell (Fig. 2 C and Fig. S2 A) and live-cell imaging (Video 1). We decided to focus on investigating the role of retromer in the recycling of MT1-MMP and MT2-MMP. Consistent with the previous reports, retromer was found to be distributed on endosomes positive for EEA1 (45 ± 3%, n = 100), Rab7 (28 ± 4%, n = 80), and GFP-Rab4 (25 ± 3%, n = 40) in PFA-fixed MDA-MB-231 cells (Fig. S2 B; Seaman, 2004; Rojas et al., 2008; Temkin et al., 2011).
Figure 2.
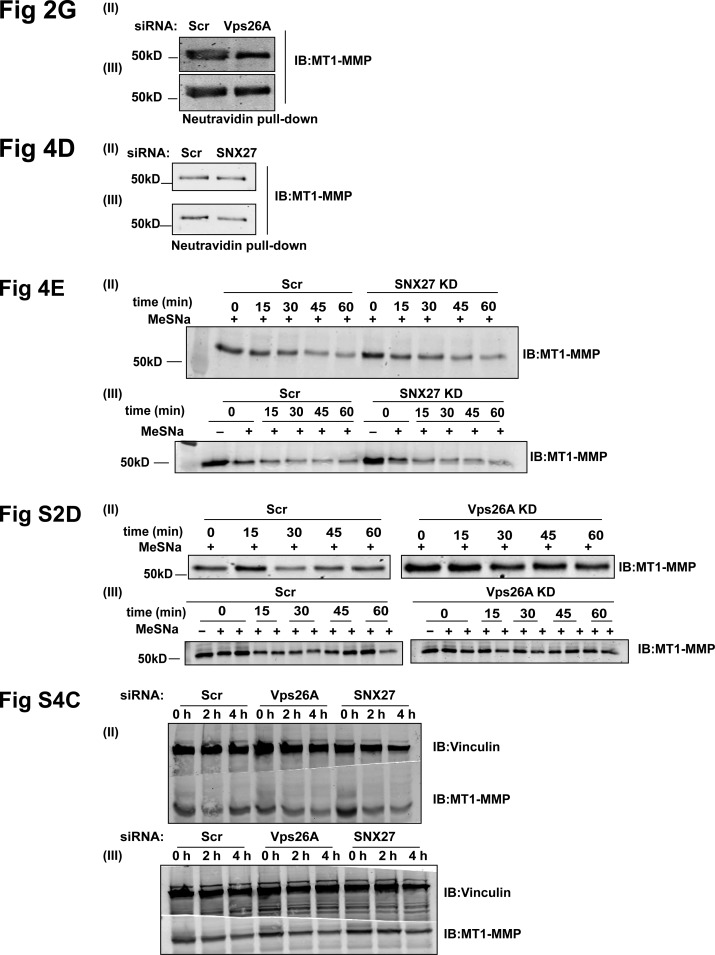
Retromer mediates cell surface recycling of MT1-MMP but not MT2-MMP. (A–C) MT2-MMP is overexpressed and invadopodia associated in MDA-MB-231 cells. (A) Indicated cells were lysed and immunoblotted with anti–MT2-MMP and anti-actin antibody. (B) Control and MT2-MMP–depleted cells were lysed and immunoblotted to determine KD levels. Subsequently, cells were seeded on Matrigel-coated cell inserts and allowed to invade for 20 h. The number of invasive cells was quantified and plotted (N = 3, scale bars = 50 µm). Values in the graph represent means ± SEM. Two-tailed Student’s t test, *** P < 0.001. (C) MDA-MB-231 cells transiently expressing mCherry–MT2-MMP and GFP-Tks5 were stained with phalloidin and DAPI and analyzed with a confocal microscope. The number of MT2-MMP endosomes positive for Tks5 was counted and plotted (N = 3, n = 45; scale bar = 10 µm, inset = 4 µm). The insets show the MT2-MMP vesicles positive for actin and Tks5. (D) Confocal images of MDA-MB-231 cells that were stably expressing GFP–MT1-MMP or transfected with mCherry–MT2-MMP and immunostained to label Vps26 and Vps35 (N = 3, n = 40; scale bars = 10 µm, inset = 4 µm). The insets show Vps35 and Vps26 localizing on the MT1-MMP or MT2-MMP endosomes. (E) MDA-MB-231 cells depleted of Vps26A and Vps35 were transfected with pHluorin–MT1-MMP or pHluorin–MT2-MMP and analyzed by TIRF microscopy. The number of vesicles and their total area was quantified and plotted (N = 3, n = 40; scale bars = 10 µm). Values are means ± SEM. One-way ANOVA, ** P < 0.01, *** P < 0.001, **** P < 0.0001. (F) mCherry–MT1-MMP and GFP-Vps29 were cotransfected in MDA-MB-231 cells and subjected to live-cell confocal imaging (Scale bars = 5 µm, inset = 4 µm). The insets show the multiple events over time where retromer is associated with the MT1-MMP endosome. (G) Surface levels of MT1-MMP were measured in control and Vps26A-depleted cells. Biotin labeling was done at 4°C for 45 min, followed by quenching and lysing the cells (refer to Materials and methods). The lysate was allowed to bind with Neutravidin beads followed by immunoblotting. Quantification of immunoblots was done to measure intensity of MT1-MMP (refer to Materials and methods; N = 3). Additional blots in Fig. S5. Values in the graph are means ± SEM. Two-tailed Student’s t test, * P < 0.05. WCL, whole cell lysate. (H) The KD efficiency of VAMP7 was measured by qPCR (N = 3). Two-tailed Student’s t test. MDA-MB-231 cells treated with indicated siRNA were surface labeled with MT1-MMP antibody at 4°C for 1 h, shifted to 37°C, fixed at 15 min, and immunostained to label with EEA1. Cells positive for MT1-MMP and EEA1 were counted and the percentage of cells that endocytosed MT1-MMP antibody was calculated. The insets represent the MT1-MMP–positive EEA1 endosome. The arrowheads in the enlarged insets show the internalized MT1-MMP antibody inside the vesicles positive for EEA1. (N = 4, n = 300; scale bars = 10 µm, inset = 4 µm). The graphs represent three independent experiments; values are means ± SEM. One-way ANOVA, * P < 0.05, ** P < 0.01. N, number of experimental repeats.
Figure S2.
Retromer is distributed on early and late endosomes carrying MT1-MMP and recruits WASH on these endosomes in MDA-MB-231 cells. MT2-MMP is invadopodia associated. (A) mCherry–MT2-MMP–expressing cells were fixed and labeled for endogenous cortactin and phalloidin. Images were acquired using a confocal microscope. The insets show a magnification of the boxed region where MT2-MMP endosomes are localizing on actin-cortactin–rich invadopodia. (N = 3, n = 40; scale bars = 10 µm, inset = 4 µm). Retromer localization on Rab4-, Rab7-, and EEA1-positive endosomes. (B) GFP-RAB4–expressing cells were fixed and labeled with endogenous Vps26. Also, untransfected MDA-MB-231 cells were fixed and immunolabeled using antibodies against Vps35 and EEA1 or Vps35 and Rab7. Images captured by confocal microscope were analyzed and percentage of colocalization was calculated. (N = 3, n = 50; scale bars = 10 µm, inset = 4 µm). (C) MDA-MB-231 cells stably expressing GFP–MT2-MMP were fixed and labeled with endogenous MT1-MMP. Cells were imaged with a confocal microscope and analyzed for percentage of colocalization. The insets show the colocalization of MT1-MMP and the MT2-MMP endosomes. (N = 3, n = 40; scale bars = 10 µm, inset = 4 µm). (D) Cells treated with Scr siRNA (control) and siRNA targeting Vps26A were labeled with biotin at 4°C after 72 h of treatment. Cells were then shifted to 37°C for 30 min to allow endocytosis and then reshifted to 4°C and treated with or without MeSNa (0 min + or −, respectively) to remove noninternalized biotin. They were again shifted to 37°C, followed by MeSNa treatment to measure the recycling of biotin-labeled surface protein, and were harvested at indicated recycling time points (15, 30, 45, and 60 min). Lysates were incubated with Neutravidin beads; bound protein was eluted and separated on SDS gel. Immunoblotting was performed and the level of MT1-MMP intensity was quantified by densitometric analysis of Western blots. For each time point, percentage of recycling was analyzed by calculating the relative loss of MT1-MMP intensity measured at 0 min, followed by MeSNa-treated recycling time points. See Materials and methods for details. Three independent experiments were performed (N = 3; additional blots in Fig. S5). (E) MDA-MB-231 cells were transfected with the siRNA against MT1-MMP, Vps26A, and Vps35. After 72 h, cells were lysed and subjected to immunoblotting to measure levels of MT1-MMP. Vinculin was used as a loading control. (F) Vps26A- or Vps35-depleted cells were serum-starved for 2 h and labeled with transferrin at 4°C for 30 min. Unbound transferrin was removed by PBS washing and cells were shifted to 37°C and fixed at indicated time points. Transferrin uptake was calculated by measuring the integral intensity of the transferrin (N = 3, n = 300; scale bar = 10 µm). The graph represents means ± SEM. One-way ANOVA. (G) Cells treated with indicated siRNA were fixed 72 h after transfection and immunostained with antibodies against MT1-MMP and WASH1, followed by imaging and quantification of the number of WASH-associated MT1-MMP endosomes. The insets show the WASH1 puncta on the MT1-MMP endosomes. (N = 4, n = 400; scale bar = 10 µm, inset = 4 µm). The graph represents means ± SEM. One-way ANOVA, ** P < 0.01. (G′) After 72-h treatment with indicated siRNA, cells were lysed and proceeded for immunoblotting to detect WASH1 expression. Transferrin receptor was used as a control. Similarly, treated cells were snap-frozen to isolate the membrane and soluble fraction. Protein fractions were separated on SDS gel, followed by immunoblotting against the transferrin receptor and WASH antibody. Transferrin receptor was used as a control to ensure the quality of the membrane fractionation. The distribution of WASH1 in membrane and supernatant was quantified by comparing the ratio of supernatant to total protein, i.e., Sup+pellet, or to that of pellet to total protein for each condition. The numbers between the blots represent the quantified values. N, number of experimental repeats.
Video 1.
MDA-MB-231 cells cotransfected with mCherry–MT2-MMP and GFP-Tks5. Cells were analyzed on a TIRF microscope (Fig. 2 C). Scale bars = 10 µm. 1,000 ms/frame.
Additionally, retromer showed colocalization with both MT1-MMP (40 ± 6%, n = 80) and MT2-MMP (22 ± 4%, n = 52; Fig. 2 D). Notably, the two proteases showed a high percentage of colocalization (40 ± 4%, n = 60), suggesting that they share a common endosomal pool (Fig. S2 C). Next, we investigated if the perturbation of retromer affects the cell surface population of the proteases. Cells with reduced expression of retromer were allowed to express pHluorin-tagged MT1-MMP or MT2-MMP. pHluorin is a pH-sensitive GFP mutant that is often used to study exocytic events at the cell surface (Monteiro et al., 2013; Kajiho et al., 2016). The total internal reflection fluorescence (TIRF) microscopy–based analysis revealed that KD of either Vps26A or Vps35 subunits of retromer significantly reduced the cell surface population of MT1-MMP, whereas MT2-MMP remained unaffected (Fig. 2 E and Videos 2 and 3). This result indicated that retromer might be specifically involved in the sorting and recycling of MT1-MMP. Therefore, from here onwards, we focused on understanding the retromer-mediated cell surface recycling of MT1-MMP.
Video 2.
MDA-MB-231 cells transfected with indicated siRNA (Scr or Vps26A KD) for 60 h were transfected with pHluorin–MT1-MMP. Cells were plated on a gelatin-coated imaging dish and videos were captured using a TIRF microscope (Fig. 2 E). Scale bar = 10 µm. 500 ms/frame.
Video 3.
siRNA transfected cells (Scr or Vps26A) for 60 h, were transfected with pHluorin–MT2-MMP, and 12 h after transfection imaging was performed using a TIRF microscope. Scale bar = 10 µm (Fig. 2 E). 500 ms/frame.
In the live-cell confocal microscopy, dynamic MT1-MMP endosomes were found to be cotransported with Vps29, which was associated as a distinct puncta on these endosomes (Fig. 2 F and Video 4). This cotrafficking indicated that retromer carries MT1-MMP from the sorting endosomes and ensures its recycling. Next, we measured the surface levels of MT1-MMP by performing biotinylation assay, which has been used extensively to measure the surface proteome over a large population (Steinberg et al., 2013; Qiang et al., 2019). Controls and the cells with reduced expression of Vps26A of retromer were incubated with noncleavable biotin, EZ-link Sulfo-NHS-Biotin, and lysed. The labeled proteins were captured using Neutravidin beads and analyzed by immunoblotting. Compared with controls, these cells showed a significant reduction in the levels of MT1-MMP at the plasma membrane (Fig. 2 G). This observation prompted us to measure the recycling kinetics of MT1-MMP in these cells. For this, proteins at the cell surface were labeled with the cleavable biotin, i.e., EZ-link Sulfo-NHS-SS-Biotin, followed by treatment with 2-mercaptoethanesulfonic acid (MeSNa) to remove biotin from the recycled population of protein, as described previously (Remacle et al., 2003). The depletion of retromer slowed down the recycling of MT1-MMP without perturbing its rate of endocytosis (Fig. S2 D). On the contrary, total levels of the protease were unaffected in cells suppressed for retromer (Fig. S2 E), suggesting that retromer is vital for replenishing the surface population of MT1-MMP.
Video 4.
MDA-MB-231 cells expressing GFP-Vps29 and mCherry–MT1-MMP. After 12 h of transfection, live-cell confocal microscopy was performed (Fig. 2 F). Scale bar = 5 µm. 800 ms/frame.
Further, these observations were supported by MT1-MMP antibody uptake assay (Remacle et al., 2003) in cells depleted of retromer or VAMP7. VAMP7 was included as a control since it is an established regulator of MT1-MMP recycling (Steffen et al., 2008; Williams et al., 2014; Castro-Castro et al., 2016). Image analysis revealed a substantial reduction in the uptake of antibodies, thus suggesting that the cells had a reduced surface pool of the protease to begin with (Fig. 2 H). Further, to ensure that the reduced uptake is not because of a defect in endocytosis, we performed the transferrin uptake assay in retromer-depleted cells. The quantification of the intensity of the internalized transferrin revealed that the cells could endocytose transferrin similar to that of control (Fig. S2 F), thus suggesting that reduced MT1-MMP uptake was because of the low abundance of protease on the surface and not due to its reduced rate of endocytosis in retromer-depleted cells.
WASH, a heteropentameric complex, generates endosomal actin patches and facilitates protein sorting (Derivery et al., 2009; Gomez and Billadeau, 2009). It is also known that retromer subunit Vps35 interacts with the FAM21 subunit of the WASH complex, thus facilitating its recruitment on the endosomal membrane (Harbour et al., 2010, 2012; McGough et al., 2014). In agreement with these studies, we found that retromer depletion led to a reduction in WASH1 punctae on the MT1-MMP endosomes in MDA-MB-231 cells (Fig. S2 G). Further, the biochemical assay also revealed lower levels of WASH1 in the membrane fractions of Vps26A-depleted cells (Fig. S2 G′).
SNX27 contributes to matrix degradation and is overexpressed in patients having invasive breast cancer
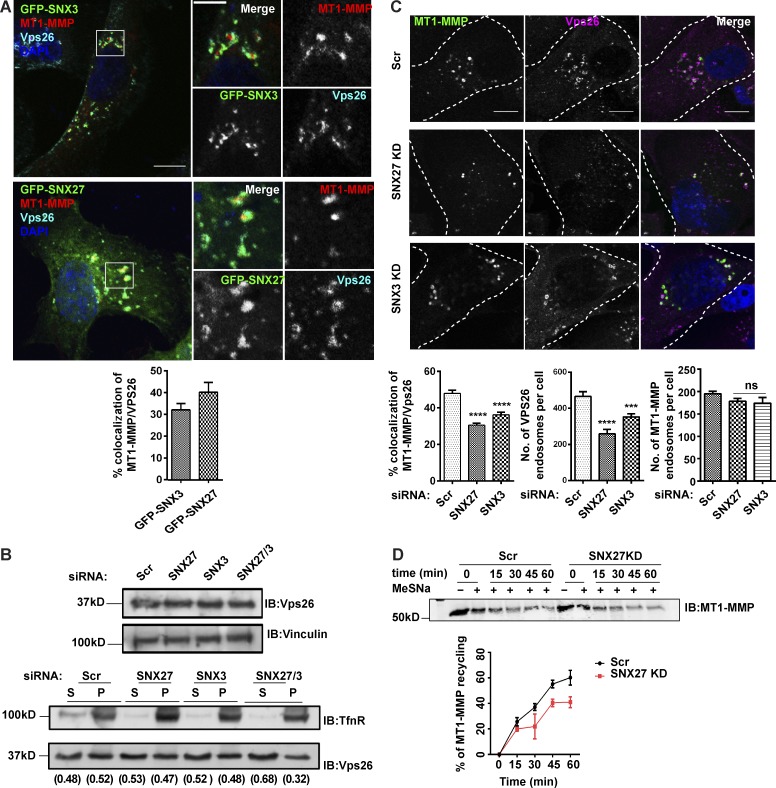
Association of retromer with some of the SNXs members, which include SNX3, SNX27, SNX1/SNX2, and SNX5/SNX6, is crucial for the efficient retrieval of cargoes (Harterink et al., 2011; Steinberg et al., 2013; Wassmer et al., 2007; Rojas et al., 2007). Next, we sought to identify the retromer-associated SNX that would facilitate endosomal sorting and recycling of MT1-MMP. Using siRNA-mediated KD, we screened a few of the retromer-associated SNX proteins (SNX1, SNX2, SNX5, SNX6, SNX3, and SNX27) for their effect on gelatin degradation activity in MDA-MB-231 cells. SNX1 and SNX2 were codepleted since they showed functional redundancy among themselves (Schwarz et al., 2002; Rojas et al., 2007). Likewise, SNX5 and SNX6 are also functionally similar and thus were cosuppressed (Wassmer et al., 2007). Analysis of degradation indices revealed that the silencing of SNX3 and SNX27 remarkably reduced matrix degradation activity (Fig. 3 A). Also, we observed colocalization of these two SNXs with MT1-MMP and retromer-positive endosomes (Fig. S3 A). Further, in line with the previous reports, KD of SNX3 affected the membrane association of retromer as revealed by the biochemical membrane fractionation assay (Fig. S3 B; Harterink et al., 2011; Vardarajan et al., 2012). A similar effect was also observed for SNX27 depletion. However, this was pronounced in SNX3/SNX27 codepletion. In support of the biochemical observation, immunofluorescence (IF) microscopy analysis showed a significant reduction in the number of retromer-positive MT1-MMP endosomes upon SNX3 or SNX27 depletion (Fig. S3 C).
Figure 3.
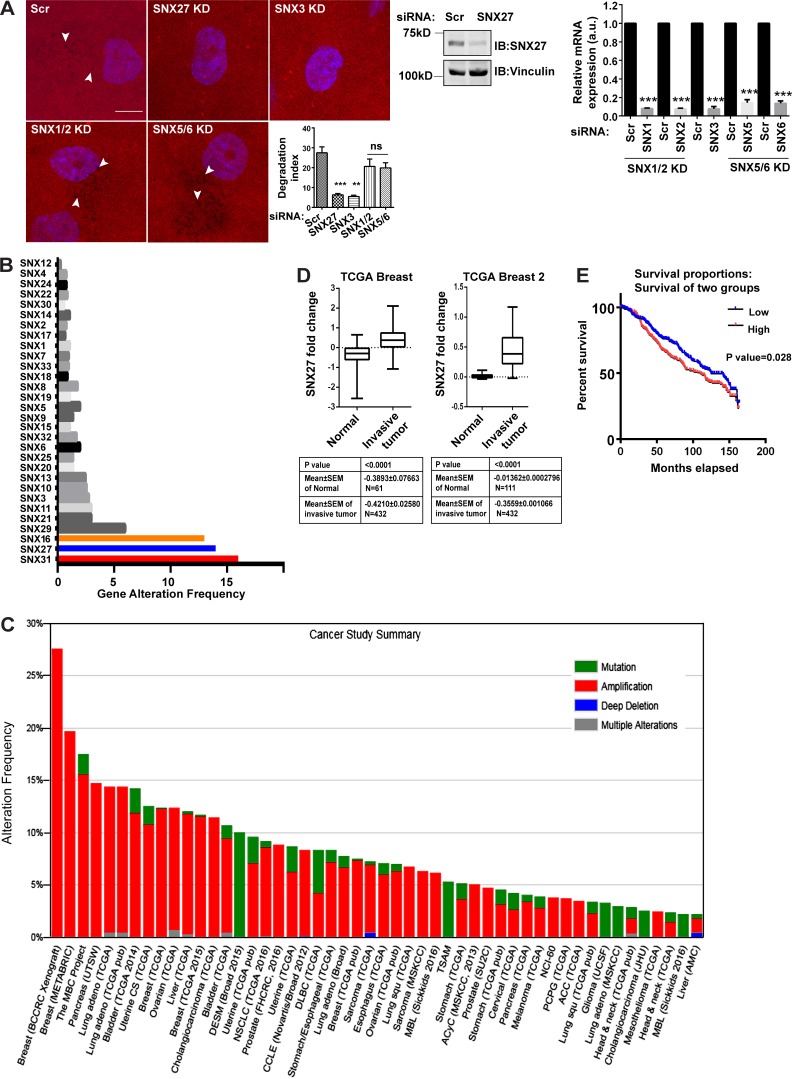
SNX27 contributes to MDA-MB-231 matrix degradation and is overexpressed in invasive breast tumor patients. (A) MDA-MB-231 cells were treated with indicated siRNA for SNX27 and SNX3 and codepleted of SNX1/2 or SNX5/6 and efficacy of gene silencing was detected by immunoblotting, where vinculin was used as a loading control, or qPCR (N = 3). Two-tailed Student’s t test, *** P < 0.001. Subsequently, gelatin degradation assay was performed as mentioned above and the degradation index was analyzed (N = 3, n = 250; scale bar = 10 µm). The graph represents three independent experiments; values are means ± SEM. One-way ANOVA, ** P < 0.01, *** P < 0.001. Arrowheads represent degradation dots. (B) From the publicly available database cBio Cancer Genomics Portal, various breast tumor datasets were analyzed for alteration frequency among SNX family members. (C) The SNX27 alteration frequency, types of alteration, availability of mutation analysis, and copy number alteration in the various tumor cohorts are shown. (D) SNX27 expression in normal versus invasive breast carcinoma reported in two different TCGA datasets was analyzed for fold change. (E) In the Curtis dataset, samples were separated for high and low SNX27 expression populations (n = 534). The Kaplan-Meier survival curve was plotted among these separated populations. N, number of experimental repeats.
Figure S3.
SNX3 and SNX27, retromer-associated SNXs, contribute to matrix degradation in MDA-MB-231 and endosomal recruitment of retromer. (A) GFP-SNX3– or GFP-SNX27–expressing MDA-MB-231 cells were fixed and immunolabeled with an antibody against Vps26 and MT1-MMP. Images were acquired on the confocal microscope and analyzed for percentage of colocalization of Vps26 and MT1-MMP with SNX3 or SNX27. Magnified boxed regions show SNX3 or SNX27 localizing on MT1-MMP and Vps26 endosomes (N = 3, n = 46; scale bar = 10 µm, inset = 4 µm). (B) Cells depleted of SNX3 or SNX27 or codepleted of SNX3 and SNX27 were lysed at 72 h after transfection and immunoblotted for Vps26. Similarly, siRNA-treated cells were lysed to separate membrane and soluble fraction and immunoblotted for Vps26 and transferrin receptor (TfnR), which is used as a control. The membrane distribution of Vps26 was analyzed by calculating the ratio of supernatant:pellet fraction for each condition independently. The numbers on the blot signify the ratio of supernatant to total protein, i.e., Sup+pellet, or to that of pellet to total protein for each condition. (C) MDA-MB-231 cells were transfected with control and indicated siRNAs. IF was proceeded 72 h after transfection using antibodies against Vps26 and MT1-MMP, imaged on a confocal microscope, and percentage of colocalization of MT1-MMP with Vps26 was calculated. Also, the number of Vps26 or MT1-MMP endosomes were counted and plotted. (N = 4, n = 300; scale bars = 10 µm). Values in the graphs represent means ± SEM. One-way ANOVA, *** P < 0.001, **** P < 0.0001. (D) Cells transfected with siRNA targeting SNX27 were labeled with cleavable biotin and biotinylation was performed (as described earlier) to measure MT1-MMP recycling. MT1-MMP recycling percentage was analyzed by quantifying the relative loss of MT1-MMP at each time point with respect to the total endocytosed population captured at 0 min. Three independent experiments were performed (N = 3; additional blots in Fig. S5). N, number of experimental repeats.
Next, we set out to study the clinical relevance for SNX in breast cancer invasion. Information was mined from the publicly available database cBio Cancer Genomics Portal (http://www.cbioportal.org/public-portal/). Eight published datasets were selected that had gene expression information from patients diagnosed with invasive breast carcinoma (sample size = 4,431). Studies with overlapping data to that of the selected datasets were discarded. We analyzed the alterations in the expression of all the members of the human SNX family, as well as the components of the Vps subcomplex. While SNX27 was found to be the second-most frequently altered (14%) among all the SNX family members, preceded by SNX31 (16%; Fig. 3 B), expression of Vps subunits was minimally altered (Vps35, 2.3%; Vps26A, 1%; Vps29, 0.2%). We also checked the genetic alteration frequency of SNX27 across tumors associated with different organs. It showed >25% alterations in breast cancers, with amplification being the major representative of alterations (Fig. 3 C).
Further, we investigated the expression pattern of SNX27 in two of The Cancer Genome Atlas datasets: TCGA Breast and TCGA Breast 2, available on Oncomine, a cancer microarray database (https://www.oncomine.org). Both the datasets revealed significantly higher SNX27 expression in the invasive breast tumor compared with the normal breast tissue (Fig. 3 D). Interestingly, using an independent breast dataset (Curtis breast cancer dataset, 534 samples), we also observed that the higher expression of SNX27 is anticorrelated to patient survival (P = 0.028; Fig. 3 E).
SNX27 plays a crucial role in cell surface transport of MT1-MMP in MDA-MB-231 cells
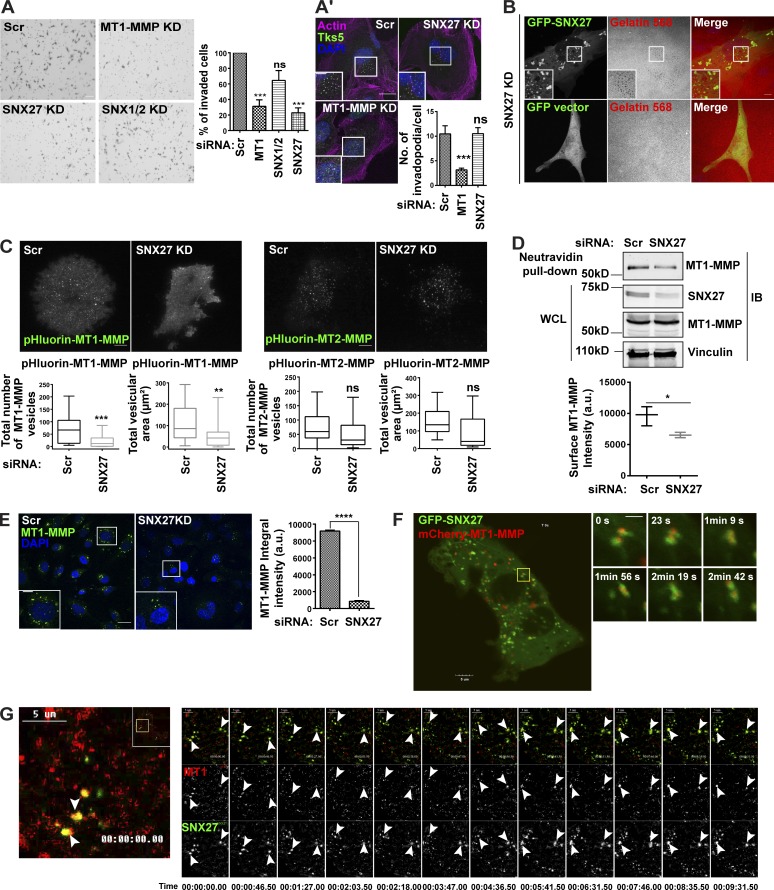
To understand the cellular implication of SNX27 overexpression in the invasive breast tumor samples, we performed Matrigel invasion assay in SNX27-depleted MDA-MB-231 cells. Downregulation of SNX27 significantly reduced 3D Matrigel invasion, whereas double depletion of SNX1/2 showed no effect (Fig. 4 A). This supported the previous observation, where SNX27-depleted cells exhibited reduced gelatin degradation (Fig. 3 A). Also, cells with reduced expression of SNX27 could form Tks5-enriched actin dots, suggesting their retained ability to form invadopodia (Fig. 4 A′). To further validate these findings and rule out the off-target effect, we conducted a rescue experiment where siRNA-resistant GFP-SNX27 was overexpressed in the SNX27 KD background. The cells overexpressing GFP-SNX27 could recover gelatin degradation activity, ensuring the gene-specific effect (Fig. 4 B) of KD.
Figure 4.
SNX27 carries MT1-MMP to the cell surface and contributes to its recycling. (A) Matrigel invasion assay was performed (as described earlier) in MDA-MB-231 cells transfected with control siRNA and siRNA targeting indicated molecules. The number of invasive cells was counted. One-way ANOVA, *** P < 0.001. The graph represents means ± SEM. (A′) Cells depleted for SNX27 were fixed after 72 h and immunostained with Tks5. Phalloidin was used to label actin. Images were acquired with a confocal microscope and the number of Tks5/actin (invadopodia) per cell were counted (N = 3, n = 30; scale bars = 10 µm, inset = 4 µm). The insets display the Tks5/actin-rich structures. One-way ANOVA, *** P < 0.001. The graph represents means ± SEM. (B) Cells were treated with siRNA against SNX27 and 60 h after treatment were transfected with GFP vector or GFP-SNX27WT. After 12 h, cells were fixed and imaged using a confocal microscope (N = 3, n = 60; scale bars = 10 µm, inset = 4 µm). The insets show the degradation spots. (C) MDA-MB-231 cells were transfected with control siRNA and siRNA targeting SNX27. 60 h after transfection, pHluorin–MT1-MMP and pHluorin–MT2-MMP were transfected. Cells were allowed to adhere to the gelatin-coated imaging dishes for 6 h and then subjected to TIRF microscopy. The number of vesicles and their total area was quantified and plotted. (N = 3, n = 30; scale bars = 10 µm). Two-tailed Student’s t test, **P < 0.01, *** P < 0.001. The graphs represent means ± SEM. (D) Cells were treated with control and SNX27 siRNA and subjected to biotinylation to measure only the surface MT1-MMP levels. Cells were labeled with noncleavable biotin and lysed, followed by binding with Neutravidin beads (refer to Materials and methods). The graph represents the quantification from the three individual blots for surface MT1-MMP levels (N = 3; additional blots in Fig. S5). Two-tailed Student’s t test, * P < 0.05. (E) Cells were treated with control and SNX27 siRNA. After 72 h, cells were incubated with antibodies against MT1-MMP at 4°C for 1 h, then shifted to 37°C for 15 min. Cells were fixed, stained with DAPI, and imaged with a confocal microscope. The total intensity of the MT1-MMP vesicles per frame was calculated and plotted (N = 3, n = 3,000; scale bars = 10 µm, inset = 4 µm). The insets show the MT1-MMP antibody uptake by the cells. Two-tailed Student’s t test, **** P < 0.0001. The graph represents means ± SEM. (F) MDA-MB-231 cells were cotransfected with GFP-SNX27 and mCherry–MT1-MMP and 12 h after transfection were imaged by a live confocal microscope (N = 3, n = 25; scale bars = 5 µm, inset = 4 µm). The insets represent various events from different time points of the live-cell imaging, where SNX27 was associated with MT1-MMP vesicles. (G) MDA-MB-231 cells transfected with mCherry–MT1-MMP and GFP-SNX27 were subjected to TIRF microscopy, and endosome dynamics near the cell surface were analyzed using a TIRF microscope. The inset panel represents a series of frames (along with the time point) from a captured video (inset scale bars = 5 µm). Arrowheads show vesicles positive for SNX27 and MT1-MMP near the cell surface (N = 3, n = 12). N, number of experimental repeats; WCL, whole cell lysate.
Previously, a quantitative surface proteomics study combined with interactome analysis has demonstrated that depletion of either SNX27 or retromer in HeLa cells led to a significant reduction in various cell surface cargoes, including nutrient transporters, suggesting their role in endosomal recycling itineraries (Steinberg et al., 2013). The authors also found MT1-MMP as one of the cargoes enriched in the SNX27 interactome. In the present study, SNX27 phenocopied retromer (Figs. 3 A and 4 A) in matrix degradation and invasion. Also, we demonstrated that retromer recruits WASH on MT1-MMP endosomes and facilitates recycling of the protease (Fig. S2 G). Interestingly, SNX27 is also known to interact with the WASH complex (Lee et al., 2016). Based on these observations, we then asked whether SNX27 takes part in retromer-mediated recycling of MT1-MMP. We performed TIRF microscopy and found a reduced cell surface population of MT1-MMP in the SNX27 depleted cells, whereas MT2-MMP remained unaffected (Fig. 4 C and Videos 5 and 6).
Video 5.
MDA-MB-231 cells transfected with control and siRNA targeting SNX27 for 60 h and transfected again with pHluorin–MT1-MMP. Cells were plated on a gelatin-coated imaging dish and videos were captured on the TIRF microscope (Fig. 4 C). Scale bar = 10 µm. 500 ms/frame.
Video 6.
Cells treated, as described in Video 5, for Scr and SNX27 KD and transfected with pHluorin–MT2-MMP (Fig. 4 C). Scale bar = 10 µm. 500 ms/frame.
Further, to measure the protein levels of MT1-MMP at the cell surface, we performed surface biotinylation by labeling cells with the noncleavable biotin as described in the previous section. Immunoblotting analysis showed that, compared to control, there was a marginal but significant reduction in the MT1-MMP surface population in SNX27-depleted cells. However, SNX27 depletion did not affect the total levels of MT1-MMP (Fig. 4 D).
Next, we determined the effect of reduced SNX27 expression on the recycling kinetics of MT1-MMP. Surface proteins were labeled with cleavable biotin and lysates were collected at different time points (as described earlier) for immunoblot analysis. SNX27-depleted cells showed a reduction in MT1-MMP recycling compared with the control cells, although the effect was subtle (Fig. S3 D). To further confirm this observation and ensure the effect over a large population, we performed an MT1-MMP uptake assay. An extensive analysis was done over a large set of images collected from control (∼3,000 cells) and SNX27-depleted cells (∼4,400 cells). An automated image analysis program was used to measure the integral intensity of the MT1-MMP puncta. Indeed, SNX27-depleted cells showed a significant reduction in the MT1-MMP uptake, thus supporting our previous observations (Fig. 4 E). Since SNX27 colocalized with endosomes positive for endogenous MT1-MMP and Vps26 (Fig. S3 A), we asked whether SNX27 traffics MT1-MMP from the endosomal population. Using live-cell confocal microscopy, we visualized numerous mCherry–MT1-MMP and GFP-SNX27 endosomes, which were cotransported. Also, as observed for retromer, GFP-SNX27 was associated as a puncta on the MT1-MMP endosomes (Fig. 4 F and Video 7). Further, we found them associated near the cell surface when imaged using TIRF microscopy (Fig. 4 G and Video 8). Collectively, these data demonstrated the contribution of SNX27 to the cancer cell invasion by facilitating the trafficking of MT1-MMP from endosomes to the plasma membrane.
Video 7.
MDA-MB-231 cells transfected with mCherry–MT1-MMP and GFP-SNX27. Live-cell imaging was performed using a confocal microscope 12 h after transfection (Fig. 4 F). Scale bar = 5 µm. 800 ms/frame.
Video 8.
Cells were cotransfected as described in Video 7, and live-cell imaging was performed using a TIRF microscope. The video represent the inset from an observed cell. Inset scale bar = 3 µm (Fig. 4 G). 500 ms/frame.
SNX27–retromer assembly localizes on endosomal domains and facilitates matrix degradation by recycling MT1-MMP
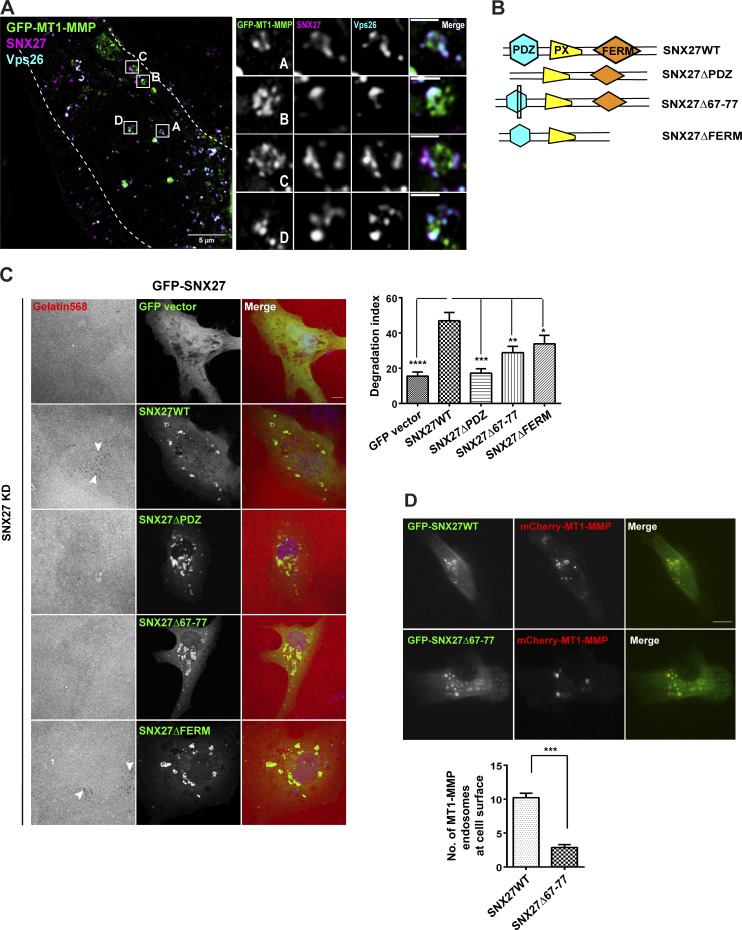
SNX27 is shown to be localized on early/sorting endosomes that are enriched in phosphatidylinositol 3-phosphate (Lunn et al., 2007; Rincón et al., 2007). Consistent with these observations, we also found that SNX27 primarily localized on EEA1- and CD63-positive endosomes (Fig. S4 A). In the present study, the depletion of SNX27 reduced the recycling of MT1-MMP to the cell surface and thus phenocopied retromer (Fig. 4, C–F). Therefore, we investigated their proximity to the MT1-MMP endosomes in MDA-MB-231 cells. GFP–MT1-MMP–expressing cells were fixed and labeled for endogenous Vps26 and SNX27. Analysis by super-resolution 3D-SIM (structured illumination microscopy) showed retromer and SNX27 punctae on the subdomains of MT1-MMP endosomes (Fig. 5 A), perhaps indicating their physical association. These observations prompted us to hypothesize that the SNX27–retromer complex recycles MT1-MMP from discrete subdomains on the endosomes to the cell surface.
Figure S4.
Retromer associates with SNX27 on MT1-MMP endosomes and prevents its lysosomal degradation. SNX27 largely localizes on EEA1- and CD63-marked endosomes in MDA-MB-231. (A) GFP-SNX27 overexpressing cells were fixed and IF was performed where antibodies against EEA1 and CD63 were used. Cells were imaged with a confocal microscope and the percentage of colocalization was calculated. Insets show the SNX27 puncta associated with CD63 and/or EEA1 endosomes. (N = 3, n = 60; scale bar = 10 µm, inset = 4 µm). (B) Cells were treated with siRNA against SNX27 and indicated siRNA-resistant, GFP-tagged SNX27 constructs were expressed. Cells were fixed and labeled with antibodies against Vps26 and MT1-MMP, imaged with a confocal microscope, and the number of MT1-MMP or Vps26 endosomes, as well as the percentage of colocalization of MT1-MMP with Vps26, were analyzed. (N = 4, n = 200; scale bar = 10 µm). Values in the graphs represent means ± SEM. One-way ANOVA, **** P < 0.0001. (C) Vps26A and SNX27 expression were downregulated by transfecting MDA-MB-231 cells with the respective siRNA. 68 h after transfection, media with the cycloheximide was added to cells at a working concentration of 10 μg μl−1 and the lysate was collected at indicated time points. MT1-MMP protein levels were detected by immunoblotting with antibodies against MT1-MMP, and vinculin was used as a loading control (N = 3). For additional blots, see Fig. S5. Values in the graph represent means ± SEM. One-way ANOVA, ** P < 0.01, *** P < 0.001, **** P < 0.0001. (D) GFP–MT1-MMP stably expressing MDA-MB-231 cells were treated with siRNA against Vps26A and SNX27. Live-cell imaging was performed and MT1-MMP surface intensity was calculated (N = 3, n = 35; scale bar = 10 µm). Values in the graph represent means ± SEM. One-way ANOVA, * P < 0.05, ** P < 0.01. (E) Lysates from MDA-MB-231 cells allowed to bind recombinantly expressed and purified GST-fused SNX27 and Vps35 bound to sepharose beads. The protein complex was eluted and run on denatured SDS gel. Immunoblotting was performed against antibodies for MT1-MMP and Vps26. (F) Purified GST and the cytoplasmic tail of MT1-MMP (GST-MT1-CT) was incubated with indicated His-tagged proteins and pull-down was performed using glutathione sepharose beads. Precipitates were immunoblotted with an anti-His antibody. (G) GFP–MT1-MMPWT and ΔDKV mutants were expressed in MDA-MB-231 cells, and 8 h after transfection were monitored by TIRF microscope (N = 2, n = 35; scale bar = 10 µm). Values in the graph represent means ± SEM, **** P < 0.0001. N, number of experimental repeats; WCL, whole cell lysate.
Figure 5.
SNX27 and retromer reside on the endosomal subdomains of MT1-MMP to facilitate its recycling and matrix degradation activity. (A) MDA-MB-231 cells expressing GFP–MT1-MMP were fixed, immunostained with an antibody against SNX27 and Vps26, and analyzed by super-resolution microscopy 3D-SIM. Boxed regions are labeled with the corresponding enlarged view of the inset on the right. These insets represent the multiple events of SNX27–retromer on MT1-MMP endosomal subdomains (N = 2, n = 6; scale bars = 5 µm, inset = 2 µm). (B) Diagrammatic representation of SNX27WT and its different deletion mutants used in the study. (C) MDA-MB-231 cells transfected with control siRNA or siRNA against SNX27 for 48 h and transfected again with siRNA-resistant GFP-fused SNX27WT and mutants. Cells were seeded on labeled gelatin and were fixed and stained with DAPI before the effect of gene suppression diminished. The degradation index was calculated as described earlier. Arrows represent the degradation dots. (N = 4, n = 200; scale bar = 10 µm). One-way ANOVA, * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. The graph represents means ± SEM. (D) GFP-SNX27WT and GFP-SNX27Δ67–77 mutants were cotransfected along with mCherry–MT1-MMP. 12 h after transfection, cells were analyzed with a TIRF microscope. The number of MT1-MMP endosomes was counted (N = 3, n = 45; scale bar = 10 µm). Values in the graph represent mean ± SEM. Two-tailed Student’s t test, *** P < 0.001. N, number of experimental repeats.
To understand the mechanistic details of SNX27-mediated MT1-MMP recycling, we asked two crucial questions. First, which domain of SNX27 is involved in the MT1-MMP recycling and second, how retromer–SNX27 assembly is contributing to this recycling axis. To address these, we used different deletion mutants of SNX27, i.e., SNX27 lacking PDZ (SNX27ΔPDZ) or FERM domains (SNX27ΔFERM). We also used a retromer binding-deficient mutant of SNX27, lacking residues in the PDZ domain crucial for its interaction with Vps26 (SNX27Δ67–77; Fig. 5 B). These mutants were employed in earlier studies where the role of SNX27 and retromer was found to be crucial in the transport of the associated cargoes, like β-AR (Lauffer et al., 2010; Temkin et al., 2011) and GLUT1 (Steinberg et al., 2013; McGough et al., 2014). We investigated whether these mutants could rescue the gelatin degradation activity of MDA-MB-231 cells lacking SNX27. siRNA-resistant SNX27WT or its mutants were overexpressed in the SNX27-depleted cells. As shown in Fig. 5 C, while SNX27WT could rescue the degradative activity, the SNX27ΔPDZ or SNX27Δ67–77 mutant failed to do so. On the contrary, cells expressing SNX27ΔFERM could degrade fluorescent gelatin, almost comparable with the WT.
Furthermore, TIRF analysis showed a significantly reduced cell surface population of MT1-MMP in the SNX27Δ67–77 mutant compared with SNX27WT-expressing cells (Fig. 5 D). These observations possibly suggest that the association of SNX27 and retromer is required for recycling of the protease.
Next, we examined whether these mutants affect the endosomal population of retromer or proteases and thus perturb the recycling axis. We attempted to overexpress the siRNA-resistant WT and mutant constructs in SNX27-depleted cells and immunostained for endogenous Vps26 and MT1-MMP. Cells expressing SNX27WT or SNX27ΔFERM showed a punctate localization of the retromer, and it could colocalize with the MT1-MMP endosomes (Fig. S4 B). The SNX27ΔPDZ or SNX27Δ66–77 mutant–expressing cells showed a substantial reduction in retromer punctae. Contrarily, the MT1-MMP endosomes were unaffected in these cells, suggesting that SNX27 does not have an impact on the intracellular pool of the protease. Collectively, these results supported our hypothesis that retromer association with the PDZ domain of SNX27 is indispensable for MT1-MMP recycling and thus its matrix degradation activity at the cell surface.
Direct interaction of SNX27 with Vps26 is reported to be crucial to divert cargoes from lysosomal degradation by directing them to the cell surface (Steinberg et al., 2013). Defects in cargo recycling should ideally enhance its lysosomal degradation. To confirm whether it is valid for MT1-MMP, we determined the turnover kinetics of endogenous MT1-MMP upon depletion of retromer or SNX27. We blocked protein translation using cycloheximide and depleted either Vps26A or SNX27 (Fig. S4 C). Indeed, compared with the control cells, retromer- or SNX27-depleted cells showed significantly faster lysosomal degradation of endogenous MT1-MMP, suggesting a possible role of the retromer–SNX27 in MT1-MMP recycling. In MDA-MB-231, overexpressed MT1-MMP also showed membrane localization on the entire cell surface as revealed by live-cell imaging (Fig. S4 D). We studied whether depletion of retromer and SNX27 also affects peripheral cell surface MT1-MMP. We measured the cell surface intensity of GFP–MT1-MMP at the entire periphery of the cell and could find a significant reduction in the protease at the membrane upon perturbation of retromer or SNX27 (Fig. S4 D).
MT1-MMP interacts with SNX27 and the Vps26 subunit of the retromer
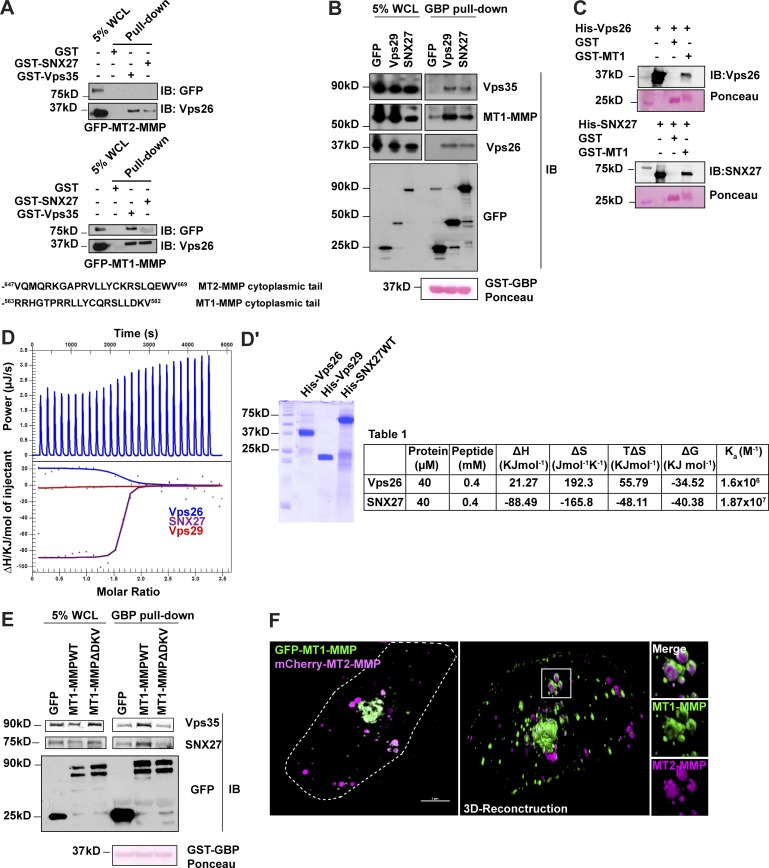
Sorting of MT1-MMP from a pool of various endosomal cargoes requires the selective recognition of the protease by the retromer–SNX complex. This recognition could either be direct or via an adaptor molecule. We used biochemical approaches to investigate if the retromer–SNX27 complex is associated with the protease. GST pull-down assay was performed using lysate from the MDA-MB-231 cells stably expressing GFP–MT1-MMP or GFP–MT2-MMP. While the recombinantly purified GST-Vps35 or GST-SNX27 could pull down endogenous Vps26 and GFP–MT1-MMP, they failed to associate with GFP–MT2-MMP (Fig. 6 A). A similar observation was also made for endogenous MT1-MMP in MDA-MB-231 cells (Fig. S4 E). The lack of MT2-MMP association might be because of the distinct amino acid residues in its cytoplasmic tail.
Figure 6.
Interaction analysis of SNX27 and retromer with MT1-MMP. (A) Lysates were prepared from MDA-MB-231 cells stably expressing GFP–MT1-MMP or GFP–MT2-MMP and allowed to bind recombinant GST-fused SNX27 and Vps35 immobilized on the glutathione sepharose beads. The protein complex was eluted and run on denatured SDS gel. Immunoblotting was performed using anti-GFP and anti-Vps26 antibodies. Amino acid sequences represent the cytoplasmic tail sequences of MT2-MMP and MT1-MMP. (B) GBP pull-down was performed with lysates from MDA-MB-231 cells expressing GFP vector, GFP-Vps29, and GFP-SNX27. Purified GST–GBP was incubated with glutathione sepharose beads and allowed to bind with the respective lysates for 1 h. Beads were washed and boiled, samples were run on SDS gel, and analysis was done by Western blotting. (C) Purified GST or GST-MT1 tails bound to glutathione sepharose beads were incubated with His-Vps26 or SNX27 in the binding buffer (see Materials and methods). Immunoblotting was done using an anti-Vps26 or anti-SNX27 antibody. (D) Calorimetric titrations of the MT1-MMP–CT peptide (400 µM) with His-tagged SNX27, Vps26, and Vps29 were performed. The upper panel shows raw ITC data for the Vps26 (40 µM) obtained from the 25 automated injections (2 µl in each injection). The lower panel shows integrated peak areas fitted using a 1:1 independent model of binding. (D′) The quality of proteins used in ITC was determined by running on SDS gel. Table 1 represents the analyzed thermodynamic parameters for the measured protein–peptide interaction. In all the performed experiments, the heat of ligand dilution was subtracted from the raw data to obtain normalized integrated heats. All ITC titrations were performed at 17°C. (E) MDA-MB-231 cells expressing the GFP–MT1-MMP or MT1-MMPΔDKV mutant were lysed and subjected to GBP pull-down as described above. Immunoblotting was performed using an antibody against Vps35, SNX27, and GFP. (F) Cells cotransfected with GFP–MT1-MMP and mCherry–MT2-MMP were fixed and analyzed by super-resolution microscopy, 3D-SIM. Images were obtained after 3D reconstruction (scale bar = 5 µm). The insets show the localization of MT1-MMP and MT2-MMP on discrete endosomal domains. WCL, whole cell lysate.
To validate the above observation, we performed a GFP-trap pull-down assay, also called the GBP pull-down. This assay is a modified immunoprecipitation where the GFP antibody is replaced by GFP-binding protein (GBP). GBP is used to efficiently isolate GFP-tagged proteins from the cell lysate, similar to anti-GFP antibodies (Rothbauer et al., 2006, 2008). GST–GBP–bound glutathione sepharose beads were incubated with the lysates prepared from the cells overexpressing GFP-tagged proteins, i.e., GFP-Vps29 or GFP-SNX27, or GFP vector. The complex formed between GST–GBP and GFP-tagged protein was eluted, separated on SDS-PAGE gel, and analyzed by Western blotting (Gujrati et al., 2019). We detected endogenous MT1-MMP, Vps26, and Vps35 from the above lysates (Fig. 6 B) being pulled down together with GFP-Vps29 and GFP-SNX27. Thus, this confirmed our observations from the GST pull-down and showed that the SNX27–retromer assembly interacts with MT1-MMP in vivo.
Often, the sequence motifs recognized by the sorting machinery are present in the cytoplasmic tail of transmembrane cargo molecules. SNX27 and retromer are known to directly interact with their associated cargoes via specific motifs in their cytoplasmic tails. MT1-MMP’s cytoplasmic tail is shown to be crucial for its internalization and localization on invadopodia (Lehti et al., 2000; Uekita et al., 2004; Yudowski et al., 2009; Qiang et al., 2019). We next probed whether MT1-MMP directly interacts with the individual retromer components or SNX27. We employed a GST pull-down assay where the GST tag fused a 20-amino acid–long cytoplasmic tail to the MT1-MMP (GST-MT1-CT) was used as bait. His-tagged SNX27, as well as retromer components Vps26, Vps35, and Vps29, were expressed and purified from Escherichia coli and probed as potential binding partners for MT1-MMP. Immunoblotting using an anti-His antibody showed that both SNX27 and the Vps26 subunit of retromer could interact with the MT1-MMP tail (Fig. S4 F). However, we could also detect a measurable amount of signal for SNX27 where only GST protein was used as a bait. To ensure if there is genuine interaction between GST-MT1 tail and Vps26 or SNX27, the pull-down assay was reperformed with a modified protocol. In the binding buffer, 0.5% BSA was added to block nonspecific binding and protein-specific antibodies were used for immunoblotting. A clear binding was observed on the MT1 tail with SNX27 and Vps26 (Fig. 6 C).
To further validate these results and determine the affinity of the interactions, we set out to perform isothermal titration calorimetry (ITC). We used a 20-mer peptide derived from MT1-MMP cytosolic tail (MT1-MMP-CT) and measured its interaction with SNX27 and Vps26. While SNX27 and Vps26 interacted with the peptide with a Ka of 1.87 × 107 and 1.6 × 106, respectively, Vps29 did not show any interaction (Fig. 6, D and D′). Importantly, the analysis of the obtained thermodynamic parameters revealed that different guiding factors are contributing to these interactions. While Vps26 binding is entropically favored, the interaction to SNX27 is driven by enthalpic factors.
These results prompted us to identify the sequence motif in the MT1-MMP cytoplasmic tail that might be governing retromer–SNX27 interaction. A previous study performed in MDCK cells suggested a role for the last three amino acids of the MT1-MMP cytoplasmic tail, DKV, in the recycling of the protease (Wang et al., 2004). Interestingly, a study has shown that the DKV motif is required for the association of MT1-MMP with the LIM kinase, which possesses a PDZ domain (Lagoutte et al., 2016). We generated a MT1-MMPΔDKV mutant and proceeded with the GBP pull-down. Compared with the full-length MT1-MMP, the ΔDKV mutant showed weak interaction with retromer and SNX27 (Fig. 6 E). Significantly, this recycling-deficient MT1-MMP mutant also showed less MT1-MMP population near the cell surface as measured by TIRF microscopy (Fig. S4 G). These observations suggest the role of retromer–SNX27, possibly by associating to DKV motif, which is unique to MT1-MMP, and thus mediates the proteases’ cell surface recycling. The absence of interaction of MT2-MMP with the assembly perhaps also explains why the cell surface population of MT2-MMP is unaffected by depletion of retromer or SNX27 as shown in (Fig. 2 E and 4 C). Corroborating with this observation, 3D-SIM revealed that MT1-MMP and MT2-MMP are located on spatially distinct domains on the same endosomes (Fig. 6 F and Video 9). TIRF analysis revealed that the motile MT1-MMP vesicles were not colocalizing to MT2-MMP (Video 10). Altogether, these results suggest that retromer–SNX27 specifically interacts with endosomal MT1-MMP and helps in its sorting from the endosomal pool for cell surface recycling. Additionally, MT1-MMP and MT2-MMP possibly follow distinct trafficking routes to be delivered to invadopodia.
Video 9.
MDA-MB-231 cells cotransfected with GFP–MT1-MMP and mCherry–MT2-MMP were fixed with PFA 12 h after transfection. Images were acquired by SIM. Z slices were set for 0.2 µm. The video represents 3D reconstruction along the x-y and x-z plane (Fig. 6 F).
Video 10.
MDA-MB-231 cells were plated on unlabeled gelatin and cotransfected with pHluorin–MT1-MMP and mCherry–MT2-MMP. 12 h after transfection, live-cell video was captured with a TIRF microscope. Scale bar = 3 µm. 500 ms/frame.
SNX27 depletion leads to delayed metastasis and prolonged survival in vivo
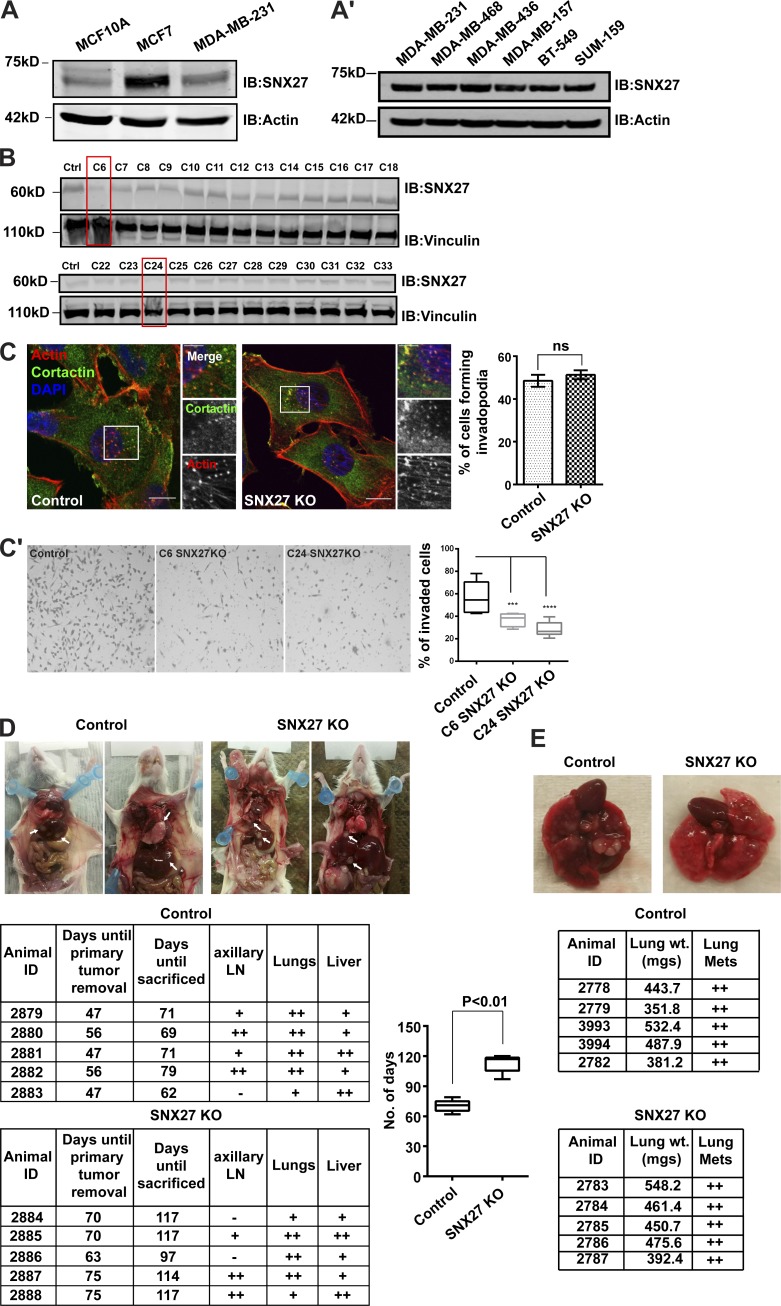
So far, our study focused on MDA-MB-231, as it is a well-studied cell line model, to understand the metastatic property of breast cancer. Therefore, we set out to perform immunoblot analysis to determine the SNX27 expression in MDA-MB-231 and compared it with that in MCF7 (poorly invasive cell line) and nontransformed MCF10A cell lines (Fig. 7 A). While the expression was high in MDA-MB-231 compared with MCF10A, it was even higher in MCF7. Further, we checked whether this expression level is similar in other breast cancer cell lines, i.e., metastatic BT-549 and SUM-159, as well as MDA-MB–derived cell lines. Indeed, we found that SNX27 protein levels were comparable across these cell lines, indicating it as a breast tumor-associated protein (Fig. 7 A′). This was in line with our tumor sample-based analysis (Fig. 3 D).
Figure 7.
SNX27 depletion prolonged animal survival in a xenograft model. (A and A′) For comparative analysis of SNX27 expression, indicated cell lines were lysed and subjected to immunoblotting using anti-SNX27 and anti-actin antibodies (loading control). (B) Different clones of MDA-MB-231 SNX27KO were obtained after cells were transfected with Cas9 protein complexed with either control sgRNA or sgRNA-targeted SNX27. The knockout efficiency was determined by immunoblotting for SNX27 and vinculin (loading control). (C) Control sgRNA transfected MDA-MB-231 cells and SNX27KO clones were seeded on gelatin-coated coverslips, and after fixation immunostained with the anti-cortactin antibody. DAPI and phalloidin were used to label the nucleus and actin, respectively. Images were acquired with a confocal microscope, and the number of cells forming invadopodia were plotted. The inset is representing the actin-cortactin–rich invadopodia. (N = 3, n = 80; scale bars = 10 µm, inset = 4 µm). The graph represents means ± SEM. Two-tailed Student’s t test. (C′) Control MDA-MB-231 cells and two clones of SNX27KO MDA-MB-231 were plated on Matrigel-coated inserts. The cellular invasion was measured and plotted as described earlier (N = 3). The graph represents means ± SEM. One-way ANOVA, *** P < 0.001, **** P < 0.0001. (D and E) An equal number of control MDA-MB-231 cells and SNX27KO cells were injected in the mammary fat pad (D) or intravenous (E) of the mice. (D) 7 wk after primary tumor removal, animals were sacrificed upon developing secondary tumors, as determined by the general health condition of the animals. The number of days for which animals survived was plotted (n = 5 mice/group). Arrows indicate the presence of secondary metastatic tumors in the lungs and liver. (E) Intravenous injection of control MDA-MB-231 cells and SNX27KO cells was evaluated in terms of the appearance of lung metastasis and survival. Tables in D and E use a scoring system based on the level of tumor infiltration: +, at least one tumor nodule present; ++, multiple (three or more) nodules present. Two-tailed Student’s t test was used to determine significance. LN, lymph node; Mets, metastasis; N, number of experimental repeats.
Using the CRISPR-Cas9–mediated gene depletion strategy, we generated an SNX27 knockout (SNX27KO) to study its effect on tumor metastasis in vivo. Various clones were screened and downregulation of gene expression was confirmed at the protein level (Fig. 7 B). Although the SNX27KO clones C6 and C24 could form invadopodia (Fig. 7 C), their invasive potential was markedly impaired as measured by Matrigel invasion assay (Fig. 7 C′).
The established SNX27KO cell lines were used in vivo where an equal number of the control (as mentioned earlier) and SNX27KO cells (clone C24) were xenografted into the mammary fat pad of immunocompromised SCID mice. Animal survival was accounted for as a major readout for tumor metastasis since once the cancer cells spread out of the primary site of implantation, animals suffer from multiple organ failure and hence show severe symptoms of sickness. After 7 wk, the primary tumor was removed and animals were kept alive until they showed signs of any illness as we have previously shown (Tanei et al., 2016; Liang et al., 2016). While control animals displayed significantly earlier signs of sickness and reduced survival, most likely as the result of metastasis to local and distal organs, those engrafted with the SNX27KO cells displayed prolonged survival (i.e., control: 70.4 ± 5.4 d versus SNX27KO: 112.4 ± 7.8 d; P < 0.01; Fig. 7 D). This further correlates with our analysis of the breast cancer patients’ survival index, where patients with low SNX27 expression showed a marginally longer life span (Fig. 3 E). Thus, the extended survival in SNX27KO-injected mice may suggest a correlation between survival and delayed onset of metastasis. However, to conclude the role of SNX27 in the progression of tumor metastasis, one needs to track the growth and development of secondary tumors in real-time using bioluminescence or fluorescence-based imaging assays in vivo.
Next, we sought to also test this hypothesis and to establish the effect of SNX27 in tumor colonization. An equal number of either control or SNX27KO cells were injected intravenously via the tail vein. After 70–80 d, both groups of animals were euthanized and investigated for the appearance of tumor formation. Animals in both groups showed the presence of tumors, although estimation of the percentage of tumor infiltration in lungs suggested no differential effects on lung colonization (measurement indicated as the percentage of total organ involvement; control: 83 ± 9.7% versus SNX27KO: 84 ± 6.5%; Fig. 7 E). However, there was a marginal delay in the growth of the primary tumor in mice injected with the SNX27KO cell line. This might be due to reduced proliferation and hence suggests the possible role of SNX27 in cellular proliferation as reported in recent studies (Yang et al., 2018; Zhang et al., 2019).
Together, these results suggest the involvement of SNX27 in facilitating metastasis, although it appears not to be required for tumor colonization while establishing at a new site. Also, these findings provide evidence supporting the notion that SNX27 deletion may improve survival rate in breast cancer xenograft models by delaying the onset of metastasis.
Discussion
Invadopodia are actin-rich membrane protrusions involved in ECM remodeling, which contributes to cancer cell invasion and tumor dissemination (Paterson and Courtneidge, 2018; Linder et al., 2011). Along with a host of proteins, proteases are recruited on invadopodia where they act as molecular scissors for efficient matrix degradation (Weaver, 2006; Watanabe et al., 2013; Beaty and Condeelis, 2014; Poincloux et al., 2009; Murphy and Courtneidge, 2011). MT1-MMP is a key enzyme associated with invadopodia and is required for degrading the matrix (Jacob and Prekeris, 2015; Castro-Castro et al., 2016). MT2-MMP, the other member of the MT-MMP family, is also overexpressed in various cancers. It is involved in cancer cell transmigration via ECM, thus promoting tumor invasion (Chen et al., 2014; Nakada et al., 1999; Gilles et al., 1996; Jiang et al., 2006; Asano et al., 2008; Ito et al., 2010; Hotary et al., 2006). In the current study, we are able to demonstrate a high level of MT2-MMP expression in the MDA-MB-231 cell line at the protein level (Fig. 2 A). Previously, an in vitro assay-based study has reported the broad spectrum of substrate activity for MT1- and MT2-MMP (d’Ortho et al., 1997). The authors demonstrated that both of the proteases cleaved fibronectin and tenascin, whereas only MT2-MMP could degrade laminin. Also, MT1-MMP is well-characterized as an interstitial collagenase that could digest type I, II, and III collagens (Ohuchi et al., 1997; Holmbeck et al., 2003; Itoh et al., 2006). In cell line–based assays, MT1-MMP and MT2-MMP were shown to activate proMMP2, thereby contributing to the remodeling of the basement membrane (Morrison et al., 2001; Nishida et al., 2008). Also, two independent studies showed that depletion of endogenous MT1-MMP or MT2-MMP reduced the basement membrane invasive activity in the MDA-MB-231 cell line (Hotary et al., 2006; Ota et al., 2009). Further, MT2-MMP is shown to degrade E-cadherin by its proteolytic activity, leading to epithelial–mesenchymal transition, and thereby facilitates cell invasion (Liu et al., 2016). Consistent with these reports, we observed that depletion of MT2-MMP led to abrogation of Matrigel invasion activity (Fig. 2 B).
The present study reveals that MT2-MMP mediates focalized matrix degradation by associating with invadopodia (Fig. 2 C and Fig. S2 A). Since invadopodia are the site of action for these proteases, we hypothesized that MT2-MMP facilitates cancer invasion along with MT1-MMP by getting recruited on these membrane protrusions. These proteases colocalized in the endosomal storage pools, where they get accumulated and are available for recycling. However, they were localized on distinct endosomal subdomains. This corroborated with our observations where retromer or SNX27 depletion perturbed the population of MT1-MMP at the cell surface, but MT2-MMP remained unaffected (Figs. 2 E and 4 C). Subsequently, MT1-MMP but not MT2-MMP was found to be associated with retromer and SNX27 (Fig. 6 A). The unique sorting signals in the cytoplasmic tail of the transmembrane proteins are recognized by sorting molecules to mediate cargo trafficking (Gallon and Cullen, 2015). MT1-MMP has a DKV motif, distinct from MT2-MMP, at its C-terminal tail (Fig. 6 A). Our interaction studies from the cytosolic extracts revealed that MT1-MMP lacking the DKV motif failed to associate with the retromer–SNX27 assembly compared with the WT protease (Fig. 6 E), thus explaining the reduced cell surface transport of this mutant protease (Fig. S4 G). The dissimilarity in cytosolic tail sequences of these two proteases unveils the possibility of MT2-MMP sorting from the endosomal subdomains by associating with distinct trafficking machinery.
WASH, an endosomal Arp2/3 activator, was shown to interact with the exocyst complex to facilitate MT1-MMP exocytosis from the late endosomes to invadopodia (Monteiro et al., 2013). It is also required for the cell surface recycling of transferrin receptor, α5β1–integrin complex, β2AR, and GLUT1 (Derivery et al., 2009; Zech et al., 2011; Puthenveedu et al., 2010; Piotrowski et al., 2013). Retromer is known to regulate membrane recruitment of the WASH1 complex. We also found reduced membrane recruitment of WASH upon retromer depletion (Fig. S2, G and G′); however, it was not completely abolished, suggesting a possible role of additional players. Recently, the ESCRT-0 component of HRS has been shown to recruit WASH in a retromer-independent pathway and regulate cargo recycling (MacDonald et al., 2018). Additionally, retromer directly interacts with VARP (Vps9-ankyrin repeat protein), a Rab32 effector, and recruits it on the endosomal membranes (Hesketh et al., 2014). This interaction governs the retromer, VARP, and VAMP7-mediated cell surface transport (Hesketh et al., 2014). VARP is also required for the directed movement of VAMP7 vesicles (Burgo et al., 2012). Moreover, the recycling of MT1-MMP from the late endosomes is known to be mediated by VAMP7 (Steffen et al., 2008; Williams and Coppolino, 2011; Williams et al., 2014). In light of these facts, there is a possibility that retromer-mediated exocytosis of MT1-MMP involves WASH, VARP, and VAMP7.
For the recycling of transmembrane cargoes, one of the crucial steps is their association with sorting machinery. Our in vitro interaction studies have demonstrated that both Vps26 or SNX27 could directly interact with the cytoplasmic tail of MT1-MMP (Fig. 6, C and D). However, the results from the rescue-based functional assays and TIRF microscopy revealed that association of Vps26 with SNX27 is crucial for the recycling of the protease (Fig. 5, C and D). Thus, the exact mode of binding of the protease with retromer–SNX27 in the cellular environment remains elusive. Previously, Gallon et al. (2014) demonstrated that the binding affinity of SNX27 for its cognate cargoes was allosterically increased when it was bound to Vps26. They also found that the SNX27 mutant, deficient in binding to retromer, showed abrogation of this observed cooperative effect. It is possible that the association of Vps26 with SNX27 enhances the latter’s affinity for the protease.
Alternatively, the protease might bind at the interface between Vps26 and SNX27. This mode of binding of retromer, SNX proteins, to the cargo is already known in the literature (Lucas et al., 2016). In a structure-based study, the authors have shown that the binding of the recycling motif of divalent metal transporter 1-isoform II (DMT1-II), a retrograde cargo, required its coincident interaction with retromer and SNX3 (Lucas et al., 2016).
It is already established that retromer recognizes cargoes, including cation independent mannose-6-phosphate receptor (CIMPR) and sortilin-related receptor (SorLA) via F/W-L-M/V and FANSHY motifs, respectively (Seaman, 2007; Fjorback et al., 2012). However, it has also been shown recently that SNX5 and SNX6 could recognize CIMPR via WLM motif in the tail, independent of retromer (Simonetti et al., 2017; Kvainickas et al., 2017). SNX27, the only member of the SNX family with a PDZ domain, binds to PDZ binding motif (PDZbm) present in the C-terminal region of the PDZ ligands such as β2AR, Kir3 (potassium channel inwardly rectifying), and SDC2 (syndecan2; Cao et al., 1999; Lauffer et al., 2010; Lunn et al., 2007; Steinberg et al., 2013). Nonetheless, the cytoplasmic tail of MT1-MMP does not harbor any of these PDZbm’s. However, it is interesting to note that DKV motif in its tail possesses the features of ClassIII PDZbm, i.e., X[DE]Xϕ (Nourry et al., 2003). Our results suggest that the SNX27–retromer assembly binds to MT1-MMP via its DKV motif (Fig. 6 E).
Further delineation of the molecular mode of binding of MT1-MMP with retromer–SNX27 assembly would require extensive biophysical and structural studies.
Taken altogether, our study establishes that the two major abundant membrane-type metalloproteases localize on the distinct endosomal domains in the metastatic cancer cell line. Retromer and SNX27 selectively recognize MT1-MMP by direct interaction with its tail and promote cell surface recycling of the protease, thereby mediating ECM degradation (Fig. 8). Our xenograft model-based studies, as well as expression analysis from the invasive breast tumor samples, suggest that abundance of SNX27 might play an important role in tumor metastasis. Accordingly, the elevated expression of SNX27 could be related to the marginally shorter survival of the patients.
Figure 8.
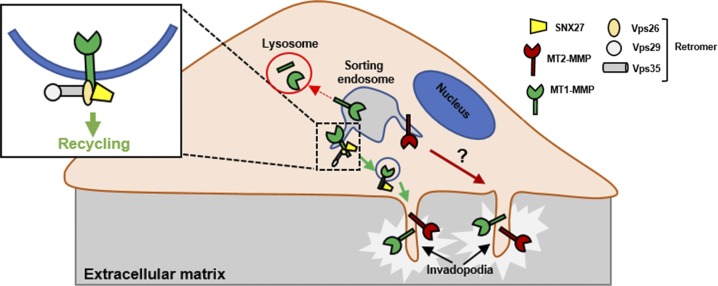
The proposed model showing SNX27–retromer assembly directly interacts with MT1-MMP and mediates its recycling to invadopodia. MT1-MMP and MT2-MMP reside on distinct endosomal domains. Retromer and SNX27 localize on the subdomains of the sorting endosomes carrying MT1-MMP and MT2-MMP. This sorting machinery selectively recycles MT1-MMP and recognizes the cargo by directly interacting with its cytoplasmic tail.
Materials and methods
Plasmids, antibodies, and reagents
The following antibodies were kind gifts and used at the mentioned dilutions: α-rabbit EEA1 antibody from Professor M. Zerial used at 1:1,000 (IF); α-rabbit WASH antibody from Professor Alexis Gautreau, used at 1:1,000 (immunoblotting [IB]) and 1:300 (IF); α-rabbit GST antibody from Dr. Ram Kumar Mishra (Indian Institute of Science Education and Research, Bhopal, India) used at 1:500 (IB). The following antibodies were purchased commercially: α-mouse MT1-MMP (Millipore, MAB3328), 1:1,000 (IB) and 1:400 (IF); α-rabbit MT2-MMP (Abcam, ab135562); α-mouse SNX27 (Abcam, ab77799), 1:1,000 (IB) and 1:400 (IF); α-mouse Vps35 (Santa Cruz, sc-374372), 1:500 (IB) and 1:300 (IF); α-rabbit Vps26 (Abcam, ab23892), 1:1,000 (IB and IF); α-mouse vinculin (Sigma, V9131), 1:1,000 (IB); α-mouse cortactin (Millipore, 05–180), 1:300 (IF); α-mouse transferrin receptor (Invitrogen), 1:1,000 (IB); α-rabbit actin (Sigma, A2066), 1:1,000 (IB); α-mouse GFP (Roche, 11814460001), 1:2,000 (IB); α-rabbit Rab7 (CST, D95F2), 1:200 (IF); α-mouse Rab5 (BD Biosciences, 610724), 1:300 (IF); α-mouse CD107a (LAMP1; BD Biosciences, 555798), 1:300 (IF); α-mouse His (ThermoFischer Scientific), 1:1,000 (IB).
MT2-MMP gene was amplified from MT2-MMP pCR3.1 (Professor Stephen Weiss’s laboratory) and cloned into pEGFP-N1 and pmCherry-N1 mammalian expression vectors with enzyme sites XhoI/EcoRI using the following primers: Fwd, 5′-AAACTCGAGATGGGCAGCGAC-3′ and Rev, 5′-CCCGAATTCGACCCACTCCTG-3′. For the generation of MT2-MMP pHluorin construct, MT2-MMP was first cloned into pcDNA 3.1 plasmid with enzyme sites NheI/EcoRI using the following primers: Fwd, 5′-TTTAAGCTTATGGGCAGCGACCCGAGC-3′ and Rev, 5′-GAATTCTCAAGCGTAATCTGGAACATCGTATGGGTAGACCCACTCCTGCAG-3′. Chimeric pHluorin–MT2-MMP was then generated in this construct by Mutagenex, Inc., wherein the tag was inserted between the hemopexin domain and the transmembrane domain of the protein after residue number 1863 in the gene sequence. The mCherry–C1-SNX27 construct was generated by amplifying SNX27 from pEGFP–C1-SNX27 (Professor Peter Cullen) with enzyme sites HindIII/BamHI using the following primers: Fwd, 5′-GAAAGCTTATGGCGGACGAGGACG-3′ and Rev, 5′-CTGGATCCCTAGTATTCCTCTTTTCTCC-3′. A pEGFP–N1–MT1-MMP mutant lacking DKV motif is generated by PCR amplifying MT1-MMP lacking DKV encoding nucleotides using the following primers: Fwd, 5′-TCGAATTCGGTGGTCTCGGACCATGTCT-3′ and Rev, 5′-CCGCGGCAGCAGGGAACGCTGGCAGTA-3′. Amplified product was digested with enzymes EcoRI/SacII and ligated into the pEGFP-N1 vector.
Cell culture and transient transfection
MDA-MB-231 (HTB-26), MCF10A (CRL-10317), and MCF7 (HTB-22) cell lines were purchased from ATCC. MDA-MB-231 cells were maintained in L-15 (Invitrogen) media with 10% FBS, 100 µg ml−1 penicillin, and 100 µg ml−1 streptomycin at 37°C in CO2-free conditions. MCF10A cells were cultured in DMEM/F12 Ham’s Mixture (Invitrogen) supplemented with 5% horse serum, 20 ng ml−1 EGF (Sigma), 10 µg ml−1 insulin (Sigma), 0.5 mg ml−1 hydrocortisone (Sigma), and 100 ng ml−1 cholera toxin (Sigma), and cells were maintained at 37°C in 5% CO2. MCF7 cells were grown in DMEM high glucose (Invitrogen) supplemented with 10% FBS, 100 μg ml−1 penicillin, and streptomycin. Cells were maintained in an incubator at 37°C and 5% CO2.
The MDA-MB-231 stable cell line was grown in L-15, supplemented with 10% FBS, 100 µg ml−1 penicillin, 100 µg ml−1 streptomycin, and 500 µg ml−1 G418. For transient transfection, cells were grown on coverslips in 24-well plates 1 d before the experiment. Cells were transfected at 50–60% confluency with 0.5 µg of plasmid DNA using LTX/Plus transfection reagent (Life Technologies).
Generation of stable cell lines
To generate a stable cell line, 2 × 106 cells per ml were seeded in a 30-mm dish and after 24 h were transfected with GFP–MT1-MMP or GFP–MT2-MMP using Lipofectamine LTX. 48 h after transfection, the antibiotic selection was proceeded by adding G418 to the transfected cells at a concentration of 800 µg ml−1 in L-15 complete media supplemented with 10% FBS. The media was changed every fourth day since the antibiotic degrades at 37°C. Cells that survived antibiotic stress proliferated and formed colonies. Several such clones were collected and grown. Each clone was detected for GFP expression by performing immunoblotting. Clones with high GFP expression were further grown in 150-mm dishes, followed by passing through a cell sorter (FACS). Different cell populations with high, medium, and low GFP expressions were collected and grown separately. After sorting, cells were grown in L-15 media with G418 concentration at 500 µg ml−1. In all the experiments, cells with medium GFP expression were used.
siRNA transfection
Cells were seeded 24 h before performing siRNA transfection. Standard protocol, available at Dharmacon (https://dharmacon.horizondiscovery.com/uploadedFiles/Resources/basic-dharmafect-protocol.pdf), was used for transfection using Dharmafect as the transfection reagent. SMARTpool siRNAs targeting the gene of interest were purchased from Dharmacon. All siRNA were used at the working concentration of 10–20 nM. MDA-MB-231 cells were transfected with negative control, Scr siRNA, and SMARTpool siRNA against MT1-MMP, MT2-MMP, VAMP7, Vps35, Vps26A, SNX27, SNX1, SNX2, SNX3, SNX5, and SNX6. A set of four individual oligos was used at a working concentration of 30 nM against Vps26A. The sequences of oligonucleotides are oligo1, 5′-GCUAGAACACCAAGGAAUU-3′; oligo2, 5′-UAAAGUGACAAUAGUGAGA-3′; oligo3, 5′-UGAGAUCGAUAUUGUUCUU-3′; and oligo4, 5′-CCACCUAUCCUGAUGUUAA-3′. The sequences of SMARTpool siRNAs are provided in Table S1.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from the cells using RNA easy kit (Qiagen, Cat. 74104), and cDNA was prepared using the High Capacity RNA-to-cDNA kit (Life Technologies, Cat. 4387406). Real-time qPCR reactions were performed using the SYBR Green Kit and corresponding primers on Applied Biosystems 7300 Real-Time PCR System.
IF microscopy and colocalization analysis
Cells were fixed in 4% PFA for 18 min at room temperature. Indirect IF was performed on fixed cells permeabilized with 0.1% Triton X-100 for 15 min at room temperature, followed by blocking with 5% FBS in PBS before staining with respective antibodies. The coverslips were mounted using Mowiol (Calbiochem, Cat. 475904) on glass slides and imaged using Zeiss LSM 780 laser-scanning confocal microscope with the Zen 2010 software. Data from three independent experiments were subjected to analysis by the automated image analysis program, Motion Tracking (http://motiontracking.mpi-cbg.de; Rink et al., 2005; Collinet et al., 2010). The cells were randomly selected for imaging. At least 15 images were acquired for each condition in a given experimental setup, and all the images have been processed together for quantification. The objects were identified as vesicles in each channel based on their size, fluorescence intensity, and other parameters by Motion Tracking software (Rink et al., 2005; Collinet et al., 2010). Objects detected in two different channels were considered to be colocalized if the relative overlap of respective areas was >35%. The apparent colocalization value was calculated as the ratio of integral intensities of colocalized objects to the integral intensities of all objects carrying a given marker and varied from 0.0 to 1.0. The colocalization-by-chance (random colocalization) was estimated by random permutation of objects localization in different channels. The apparent colocalization was corrected for random colocalization.
MT1-MMP antibody uptake assay
MDA-MB-231 cells were trypsinized, counted, and 30,000 cells per well were seeded on the gelatin-coated coverslips for 24 h and transfected with siRNA. After 72 h of transfection, antibody uptake was performed. Cells were incubated at 4°C with 10 µg ml−1 MT1-MMP antibody diluted in L-15 complete media for 30 min. The unbound antibody was removed after washing with 1× PBS. L-15 complete media was added, and cells were shifted to 37°C to allow internalization of the surface-bound antibody. At the 15-min time point, cells were fixed by adding 4% PFA, followed by permeabilization, and were immunolabeled with anti-EEA1 antibody.
Gelatin degradation assay
MDA-MB-231 cells after 60 h of transfection were trypsinized and 50,000 cells were plated on Alexa Fluor 568–labeled fluorescent gelatin for 12 h. Cells were then fixed and stained with DAPI. Images were captured using Zeiss LSM 780 confocal laser-scanning microscope with 40× oil immersion objective, 1.4 NA. Images were processed in Motion Tracking software, where degradation area was identified as an object and degradation index was calculated using the following formula:
The mean background intensity is the intensity of the gelatin normalized to the frame size of the image (512 × 512 pixels).
Matrigel invasion assay
Matrigel invasion assay was performed by transfecting MDA-MB-231 cells with 20 nM siRNA targeting genes of interest. After 60 h of transfection, cells were trypsinized and counted, and 50,000 cells per well were resuspended in serum-free media and seeded on BD Transwell cell inserts (8-µm pore size) coated with 0.5 mg ml−1 of Matrigel (BD Biosciences, Cat. 354277). Complete media with 10% FBS was added to the lower chamber. Transwell plates were kept at 37°C for 16–18 h. Cells on the inserts were fixed with 4% PFA for 20 min, washed three times with 1× PBS, and permeabilized by adding 100% methanol for 15 min. Staining was done by adding 1× Giemsa dye (from 20× stock) and noninvaded cells were removed using a cotton swab. Inserts were dried for 6 h at room temperature and imaged using an inverted epifluorescence microscope using 10× objective at bright field mode. Cells were counted using ImageJ software.
Live-cell imaging
Cells were cotransfected with GFP and mCherry-fused proteins for 12 h, followed by trypsinization, and were seeded on glass-bottom dishes coated with gelatin. Cells in L-15 complete media were incubated for 4 h at 37°C in an incubator and further imaged with an Olympus FV3000 confocal laser-scanning microscope with a 60× Plan Apo N objective (oil, 1.42 NA) on an inverted stage. Dual-color sequential imaging was performed using 488-nm and 561-nm lasers to excite GFP and mCherry-fusion proteins, respectively. Images were acquired and processed using FV31S-SW software.
SIM
Sample preparation was similar to that for confocal imaging. High-resolution imaging was performed using a Nikon N-SIM with a 100× Apochromat objective (oil, 1.49 NA) using the 561-nm and 633-nm laser lines and an Andor iXon 897 electron multiplying charge-coupled device camera. To obtain super-resolution images, optimal intensity settings were maintained. Images acquired were processed for reconstruction using Nikon NIS-Elements software version 4.30.
TIRF microscopy
To capture events specifically near the cell surface, images were captured with a Nikon Eclipse Ti inverted microscope equipped with TIRF setup and a temperature controller. MDA-MB-231 cells cotransfected with GFP- and mCherry-fusion proteins were plated on gelatin-coated, glass-bottom dishes filled with L-15 complete media. Sequential dual-color imaging was done using a 100× Apochromat objective (oil, 1.49 NA), and an Agilent laser combiner MCL400 (Agilent Technologies) with an electron multiplying charge-coupled device camera (Photometrics Inc.). Image acquisition was done with Micro-Manager software version 1.4.23. To excite GFP and mCherry, 488-nm and 562-nm lasers were used, respectively. Bandpass filters were selected to control fluorescent emission. Cells were transfected with siRNA followed by pHluorin-fused MT1-MMP or MT2-MMP and images were acquired at an exposure time of 500 ms. Objects were identified using Motion Tracking software, and the density of the proteases was calculated.
Western blotting
Cells were harvested and lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM DTT, 1% Triton X-100, and 1 mM EDTA) supplemented with 10 µg ml−1 protease inhibitor cocktail (PIC). Cell debris was removed by centrifuging at 16,000 g for 15 min at 4°C. Protein concentration was estimated using the Bradford assay. Samples were prepared by adding 1× SDS loading dye and separated by running on SDS-PAGE, followed by transfer to a nitrocellulose membrane (0.45 µm; GE Healthcare, Cat. 10600002). The membrane was blocked with 5% BSA then incubated for 1 h at room temperature with the respective primary antibody. To detect the bound antibody signal, the membrane was incubated with HRP-conjugated or fluorescently labeled secondary antibody, and protein bands were detected using chemiluminescence-based ECL Western blot detection reagent (Bio-Rad, Cat. 1705060) or on a Li-Cor Odyssey Infrared Scanning System, respectively.
MT1-MMP surface biotinylation
To measure surface population, MDA-MB-231 cells (9 × 104 cells per ml) were grown in a 35-mm plate for 24 h. Cells were then transfected with siRNA. At the 72-h posttransfection time point, biotinylation was performed. The culture medium was removed and the cells were washed carefully with 2 ml PBS (6.7 mM NaHPO4, 3.3 mM NaH2PO4, and 140 mM NaCl, pH 7.2) and incubated with noncleavable biotin and EZ-link Sulfo-NHS-Biotin (Invitrogen, Cat. 21217) at 4°C for 45 min. Further, to quench unreacted biotin, cells were treated with a quenching solution (50 mM Tris-HCl, pH 8.0 ). After quenching, cells were washed with 1× PBS and solubilized in lysis buffer. The lysate was incubated with Neutravidin beads for 1 h at room temperature. Biotin-labeled proteins were eluted from the beads and subjected to immunoblotting.
The intensity of the biotinylated MT1-MMP was quantified for Scrambled (Scr) and siRNA-treated samples for Vps26A and SNX27 using ImageJ software. The scanned Western blot image was converted to grayscale for its densiometric analysis. Using the square selection tool, a square box was plotted on the lanes that represented MT1-MMP bands and another box of the exact same dimension was plotted to measure the background for the corresponding lanes (normalized intensity of MT1-MMP in Scr = ScrMT1-MMP intensity − Scr background; normalized intensity of MT1-MMP in Vps26A KD = Vps26A KDMT1-MMP intensity − Vps26A KD background; normalized intensity of MT1-MMP in SNX27 KD = SNX27 KDMT1-MMP intensity − SNX27 KD background).
The normalized intensities of MT1-MMP in Scr were compared and plotted to those of Vps26A KD or SNX27 KD.
To measure recycling kinetics, cells were transfected with siRNA as mentioned above, and surface proteins were labeled with cleavable biotin and EZ-Link Sulfo-NHS-SS-Biotin (0.2 mg ml−1; Invitrogen, 21331) for 45 min at 4°C. Unreacted biotin was quenched with the quenching solution. To measure internalization, cells were allowed to endocytose biotinylated proteins at 37°C for 30 min and then shifted to 4°C. After 30 min of endocytosis (0 min, MeSNa−), cells were immediately lysed to measure the total biotinylated population. However, other cells were washed and given MeSNa treatment, i.e., 50 mM MeSNa in MeSNa buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.2 wt/vol BSA, pH 8.6) to remove noninternalized biotin-labeled proteins, followed by quenching. After 30 min of endocytosis (0 min, MeSNa+), cells were solubilized in the lysis buffer. Recycling was measured by reshifting the remaining cells at 37°C and incubating for different time points (15, 30, 45, and 60 min). At each recycling time point, cells were shifted to 4°C, washed, MeSNa treated, quenched, and solubilized. The biotin-labeled proteins (lysate) were incubated with Neutravidin beads (Pierce, Cat. 29200) for 1 h and eluted by boiling the beads in 1× SDS loading dye. Immunoblotting was performed to detect protein levels.
The percentage of recycling was calculated as
where time t = 0, intensity = A, t = t1, and intensity = B.
Assay to measure membrane-associated proteins
This was performed as demonstrated earlier (Seaman et al., 2009; Breusegem and Seaman, 2014). An equal number of cells for control and KD were grown in 6-well plate tissue culture dishes 1 d before the experiment. siRNA transfection for SNX27 and Vps26A was performed. After 72 h of transfection, media was removed and cells were washed with ice-cold PBS, snap-frozen using liquid nitrogen, and then thawed at room temperature. The cells were scraped off the dish using lysis buffer containing 0.1 M MES-NaOH, pH 6.5, 1 mM magnesium acetate, 0.5 mM EGTA, 200 µM sodium orthovanadate, 0.2 M sucrose, and PIC. These were centrifuged at 10,000 g for 10 min and the supernatant was collected in a fresh tube. The pellet was then solubilized in lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% SDS, followed by centrifugation at 10,000 g for 10 min. This supernatant comprises of all the membrane-associated proteins. Collected samples were run on SDS-PAGE and proteins detected by immunoblotting.
Recombinant protein purification
To express and purify the recombinant proteins, both GST (pGEX-6P-1) and His (pET28a) gene constructs were transformed into E. coli BL21 (DE3) cells. The bacterial culture was grown at 37°C and induced at an OD600 of 0.6 by adding 200 µM IPTG. The induced bacterial culture was grown at 16°C (for Vps26, 500 µM IPTG at 18°C) overnight. The freshly grown culture was harvested and cells were resuspended in lysis buffer, 1× PBS containing 1.25 mM NaCl, 5 mM DTT, 1 mM EDTA, 0.5% Tween 20, 10 µg ml−1 PIC, and 200 mM PMSF for GST-tagged proteins, and 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 10 mM β-mercaptoethanol, 10 mM imidazole, 0.5% Triton X-100, 10 µg ml−1 PIC, and 200 mM PMSF for His-tagged protein, respectively. The lysed bacterial culture was centrifuged at a speed of 15,000 rpm for 45 min at 4°C and separated into pellet and supernatant. The supernatant was incubated for 1 h with Glutathione Sepharose 4B (GE Healthcare), and for 15 min with Ni-NTA beads (Qiagen) for GST- and His-tagged proteins, respectively. Both glutathione sepharose and Ni-NTA agarose beads were washed with the wash buffers containing 20 mM Hepes and 1 M NaCl, and 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 10 mM imidazole, respectively. Finally, GST- and His-tagged proteins were eluted with 10 mM reduced glutathione dissolved in 50 mM Tris-HCl, pH 8.0, and 200 mM imidazole dissolved in 20 mM Tris-HCl, pH 8.0, and 200 mM NaCl, respectively.
GST pull-down assay
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM DTT, 1% Triton X-100, 1 mM EDTA, and 10 µg ml−1 PIC) and centrifuged at 19,000 g for 15 min at 4°C to remove cell debris. The recombinant proteins were quantified. For each reaction, 0.5 µM of GST-tagged bait was added to the glutathione sepharose beads and incubated for 1 h at 4°C on the rotamer. Unbound bait was removed by washing beads three times with 1× PBS while spinning at 4°C and 600 g for 3 min. Bait bound with beads were incubated with the cell lysate followed by washes with high salt (0.5 M NaCl, 1% Triton X-100) and low salt (0.1 M NaCl, 0.5% Triton X-100) buffers. To elute protein complexes, beads were boiled in 1× SDS loading dye for 5 min at 95°C. The samples were run on 4–12% SDS gradient gel and proceeded for immunoblotting.
In vitro pull-down assay
In the pull-down assay, 0.5 µM GST-tagged MT1-MMP cytoplasmic tail was immobilized on glutathione sepharose beads and incubated with an equimolar concentration of His-tagged SNX27, Vps26, Vps35, or Vps29 in the binding buffer composed of 50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1% Triton X-100, and 0.5% BSA. Subsequently, the beads were washed twice with both high salt (0.5 M NaCl, 1% Triton X-100) and low salt (0.1 M NaCl, 0.5% Triton X-100) buffers. Bound proteins were eluted by boiling in 1× SDS loading dye, separated on 4–12% SDS gradient gel and detected by immunoblotting.
ITC
To understand the thermodynamic basis of protein–peptide interactions, ITC experiments were performed on a Nano ITC instrument (TA Instruments). His-tagged SNX27, Vps26, and Vps29 proteins were purified using two-step purification, i.e., affinity chromatography followed by size-exclusion chromatography. In this study, 40 µM His-tagged proteins were titrated against the synthetic peptide for the MT1-MMP cytoplasmic tail region (GenScript). All the solutions were filtered and degassed before the ITC run. 400 µM peptide was injected into the ITC cell containing 40 µM His-tagged proteins in 25 automated injections (2 µl in each injection at 200 rpm) at an interval of 180 s and mixed homogeneously. The heat of interaction between both of the components was measured after subtracting the heat of dilution. The data were processed to obtain thermodynamic parameters using the NanoAnalyze software.
GFP-trap pull down
GBP clone was purchased from Addgene and subcloned into the pGEX-6P1 vector to express GST-tagged GBP. GFP vector or GFP fusion proteins were overexpressed in 6-well format in the MDA-MB-231 cell line. 16 h after transfection, cells were lysed in 20 mM Tris-HCl, pH 8.0, 137 mM NaCl, 10% glycerol, 2 mM EDTA, and 1% NP-40, and cleared by centrifuging at 16,000 g at 4°C for 20 min. Cleared lysates were mixed with 15 µg of GST–GBP and bound to the glutathione sepharose beads. The beads bound with GBP–GFP protein complexes were then washed three times with PBS, followed by elution using 1× SDS sample loading buffer. Samples were then separated on 4–12% SDS gradient gel followed by Western blotting.
Degradation experiment
Cells were plated for 24 h before experimenting. 30,000 cells per well were seeded in 24-well plates and transfected with control and siRNA targeting molecule of interest. After 68 h of transfection, the media was removed and cells were washed with sterile 1× PBS. L-15 complete media with cycloheximide at a working concentration of 10 µg ml−1 were added in cells. At different time points, cells were lysed by adding lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM DTT, 1% Triton X-100, and 1 mM EDTA) supplemented with PIC. Cell debris was removed by spinning lysate at 16,000 g for 15 min at 4°C. Protein estimation was done by Bradford assay, and samples were prepared by adding 1× SDS sample loading buffer and boiling at 95°C. Proteins were separated on SDS gel and proceeded for Western blotting.
Generation of SNX27KO cell line using CRISPR/Cas9
Using an online CRISPR design tool (https://www.thermofisher.com/in/en/home/life-science/genome-editing/geneart-crispr/crispr-libraries/trueguide-grnas.html) from ThermoFisher Scientific, a 20-nucleotide guide sequence was designed against human SNX27 isoform 2. The targeted DNA sequence was 5′-GTGGTGGACCTGATTCGAGC-3′ in exon2 with protospacer-adjacent motif sequence AGG. A negative control, nontargeting single gRNA (sgRNA) 5′-AAAUGUGAGAUCAGAGUAAU-3′, that does not recognize any specific sequence was also used. 1,000 cells per well were seeded for 24 h before transfection in a 96-well plate. Cells were transfected with Cas9–sgRNA complex at a ratio of 250 ng/1.5 pmol per protein using lipofectamine Cas9 plus (Invitrogen, Cat. A36496) and lipofectamine CRISPRMAX (Invitrogen, Cat. CMAX00003) reagents. 48 h after transfection, cells were serially diluted and grown until they reached 80% confluency. CRISPR knockout was confirmed by Western blotting.
Xenograft studies
All the present study protocols involving mice followed the standard regulations and were approved by the Houston Methodist Research Institute Animal Care and Use Committee.
Mammary fat pad tumor metastasis development
Low-passage control MDA-MB-231 and SNX27KO–MDA-MB-231 cells were cultured under standard conditions (37°C) in CO2-independent L-15 media (as recommended by ATCC) until they were 80–90% confluent, after which they were trypsinized and the number of viable cells was determined by counting after Trypan blue staining. A cell suspension was prepared by mixing with Matrigel at a 1:1 ratio. Under aseptic conditions, the cell suspension containing 5 × 106 cells 0.1 ml−1 was injected into the mammary fat pad (near the ninth nipple) of 8–10-wk-old female NSG mice anesthetized with isoflurane. Tumor growth was monitored weekly using calipers and tumor volume was calculated according to the formula L × W2 / 2 = mm3. Once primary tumors approached ∼1,000 mm3 vol, they were partially removed in order to provide a source for developing a secondary tumor and the incision was closed. Animals were maintained until they showed signs of sickness, indicative of potential metastasis.
Lung colonization assay
Intravenous injection of experimental cell lines (5 × 105 cells 0.2 ml−1 per mouse) was performed through the mouse tail vein. Briefly, conscious animals were placed in an appropriate restrainer after which tails were warmed in a water bath to dilate the vein, disinfected with an alcohol pad, and injected with the corresponding cell line. Animals were then transferred to a clean cage.
Statistical analysis
For the statistical evaluation of the datasets from quantitative Western blotting and quantitative image analysis, unpaired two-tailed Student’s t test or one-way ANOVA was performed using GraphPad Prism6 software version 6.01. A P value <0.05 was considered as statistically significant. Datasets are expressed as means ± SEM. Data distribution was assumed to be normal but this was not formally tested. The number of samples, images, and experiments are mentioned in the respective figure legends.
Online supplemental material
Fig. S1 shows that the depletion of retromer via individual oligos abrogates gelatin degradation and Matrigel invasion in breast cancer cells (related to Fig. 1). Fig. S2 shows that MT2-MMP is associated with invadopodia marker cortactin, localization of retromer on various endosomal compartments in MDA-MB-231 cells, decreased recycling kinetics of MT1-MMP upon depletion of retromer, and that retromer recruits WASH on the endosomes in breast cancer cells (related to Fig. 2). Fig. S3 shows that SNX3 or SNX27 localizes to retromer and MT1-MMP, that these SNXs recruit retromer on the endosomes, and SNX27 is required for recycling of MT1-MMP on the cell surface, measured by biotinylation (related to Figs. 3 and 4). Fig. S4 shows endosomal localization of retromer in cells expressing various SNX27 mutants; retromer and SNX27 rescues MT1-MMP from the lysosomal degradation; MT1-MMP is associated with retromer or SNX27 in breast cancer cells; MT1-MMP cytoplasmic tails do not interact with Vps29 or Vps35; and DKV motif in the cytoplasmic tail of MT1-MMP is required for its recycling at the cell surface (related to Figs. 5 and 6). Fig. S5 shows all the additional immunoblots used for the quantification (Figs. 2 G and 4, D and E; and Figs. S2 D and S4 C). Table S1 lists oligonucleotides sequences of SMARTpool siRNA used in this study. Video 1 shows the dynamics of MT2-MMP on invadopodia marked by Tks5. Videos 2 and 3 show MT1-MMP or MT2-MMP endosomes at the cell surface in control or retromer-depleted cells, respectively. Video 4 shows the cotransport of retromer with MT1-MMP. Videos 5 and 6 show MT1-MMP or MT2-MMP endosomes at the cell surface in control or SNX27-depleted cells, respectively. Video 7 and 8 show cotransport of SNX27 with MT1-MMP endosomes in the endosomal compartments or at the cell surface, respectively. Video 9 shows the spatial distribution of MT1-MMP and MT2-MMP on the endosomal subdomains. Video 10 shows that both the proteases are carried to the cell surface via distinct endosomes.
Figure S5.
Additional Western blots that were used for quantification are provided, along with their relevant figure numbers.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Varadharajan Sundaramurthy (National Center for Biological Sciences [NCBS], Bangalore, India) for providing us the SIM and TIRF imaging facility at NCBS. We thank the Nikon imaging facility at NCBS and Mr. Suparno Gupta for helping with the SIM imaging. We thank Dr. Thomas van Zanten (NCBS) for helping with the TIRF imaging. We thank Professor Stephen Weiss (University of Michigan, Ann Arbor, MI) for sharing MT2-MMP pCR3.1 construct; Professor Philippe Chavrier (Centre National de la Recherche Scientifique [CNRS], Paris, France) for sharing pCDNA3.1 MT1-MMP pHluorin construct; Professor Alexis Gautreau (CNRS) for providing WASH1 antibody; Professor Marino Zerial (Max Planck Institute, Dresden, Germany) for sharing EEA1 antibody and pEGFPC1 Rab4 construct; Professor Marc Coppolino (University of Guelph, Guelph, Canada) for sharing pEGFP–MT1-MMP and mCherry–MT1-MMP constructs; Professor Peter Cullen (University of Bristol, Bristol, UK) for providing all SNX27WT and mutant constructs; Professor Craig Blackstone (National Institutes of Health, Bethesda, MD) for sharing GST-SNX27 construct; and Professor Mark A. McNiven (Mayo Clinic, Rochester, MN) for sharing GST-MT1-MMP-CT construct. We thank Cassy Anselme (Houston Methodist Research Institute, Houston, TX) for helping with the survival plot analysis and Jui Anil Shinde (Indian Institutes of Science, Education, and Research [IISER], Bhopal, India) for initiating biochemical experiments. We are thankful to Professor Mark von Zastrow (University of California, San Francisco, San Francisco, CA), Dr. Boris Simonetti (University of Bristol), Dr. Subba Rao Gangi Setty (Indian Institute of Science, Bangalore, India) and Dr. Amulya Priya (Institute Curie, Paris, France) for their helpful suggestions and discussions. We deeply appreciate Dr. Merlyn Emmanuel (Vanderbilt University, Nashville, TN) for her critical comments and insightful suggestions that improved the manuscript. We are grateful to JCS for providing the travel grant to visit Dr. Jenny Chang’s laboratory for performing xenograft-based studies. We are thankful to the Central Instrumentation Facility at IISER Bhopal that was used to perform ITC and confocal microscope-based experiments. We also acknowledge the FIST facility at IISER Bhopal by DST for providing facility to conduct live-cell imaging based experiments.
This work is part of the doctoral thesis of P. Sharma and was supported by the following projects: Department of Science and Technology (EMR/2016/00340 and SR/FST/LSI-643/2015 (C)), the Council of Scientific and Industrial Research 27(0326/17/EMR-II7), and Department of Biotechnology (102/IFD/SAN/1941/2018-2019). P. Sharma is funded by the Ministry of Human Resources Development, India, and was awarded JCS International travel grant number 170809.
The authors declare no competing financial interests.
Author contribution: P. Sharma performed all the experiments. P. Sharma and S. Datta conceptualized the study, designed the experiments performed analyses, and wrote the manuscript. S. Parveen performed biotinylation experiments. L.V. Shah generated MT2-MMP constructs and performed MT2-MMP functional studies. M. Mukherjee helped in performing ITC experiments and data analysis. Y. Kalaidzidis helped with the TIRF image analysis. A.J. Kozielski, R. Rosato, and J.C. Chang designed, performed, and analyzed xenograft-based experiments.
References
- Al-Raawi D., Abu-El-Zahab H., El-Shinawi M., and Mohamed M.M.. 2011. Membrane type-1 matrix metalloproteinase (MT1-MMP) correlates with the expression and activation of matrix metalloproteinase-2 (MMP-2) in inflammatory breast cancer. Int. J. Clin. Exp. Med. 4:265–275. [PMC free article] [PubMed] [Google Scholar]
- Arighi C.N., Hartnell L.M., Aguilar R.C., Haft C.R., and Bonifacino J.S.. 2004. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165:123–133. 10.1083/jcb.200312055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., and Mueller S.C.. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043. 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Asano T., Tada M., Cheng S., Takemoto N., Kuramae T., Abe M., Takahashi O., Miyamoto M., Hamada J., Moriuchi T., and Kondo S.. 2008. Prognostic values of matrix metalloproteinase family expression in human colorectal carcinoma. J. Surg. Res. 146:32–42. 10.1016/j.jss.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Beaty B.T., and Condeelis J.. 2014. Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 93:438–444. 10.1016/j.ejcb.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson C.S., Babu S.D., Radhakrishna S., Selvamurugan N., and Ravi Sankar B.. 2013. Expression of matrix metalloproteinases in human breast cancer tissues. Dis. Markers. 34:395–405. 10.3233/DMA-130986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breusegem S.Y., and Seaman M.N.J. Conn, P.M., editor. 2014. Image-based and biochemical assays to investigate endosomal protein sorting. Methods in Enzymology. Vol. 534. Elsevier, New York. [DOI] [PubMed] [Google Scholar]
- Burd C., and Cullen P.J.. 2014. Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6:a016774 10.1101/cshperspect.a016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgo A., Proux-Gillardeaux V., Sotirakis E., Bun P., Casano A., Verraes A., Liem R.K.H., Formstecher E., Coppey-Moisan M., and Galli T.. 2012. A molecular network for the transport of the TI-VAMP/VAMP7 vesicles from cell center to periphery. Dev. Cell. 23:166–180. 10.1016/j.devcel.2012.04.019 [DOI] [PubMed] [Google Scholar]
- Cao T.T., Deacon H.W., Reczek D., Bretscher A., and von Zastrow M.. 1999. A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature. 401:286–290. 10.1038/45816 [DOI] [PubMed] [Google Scholar]
- Castro-Castro A., Marchesin V., Monteiro P., Lodillinsky C., Rossé C., and Chavrier P.. 2016. Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu. Rev. Cell Dev. Biol. 32:555–576. 10.1146/annurev-cellbio-111315-125227 [DOI] [PubMed] [Google Scholar]
- Chamberland J.P., and Ritter B.. 2017. Retromer revisited: Evolving roles for retromer in endosomal sorting. J. Cell Biol. 216:3433–3436. 10.1083/jcb.201708111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.A., and Scheller R.H.. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98–106. 10.1038/35052017 [DOI] [PubMed] [Google Scholar]
- Chen L., Zhou Q., Xu B., Liu J., Shi L., Zhu D., Wu C., and Jiang J.. 2014. MT2-MMP expression associates with tumor progression and angiogenesis in human lung cancer. Int. J. Clin. Exp. Pathol. 7:3469–3477. [PMC free article] [PubMed] [Google Scholar]
- Clark E.S., Whigham A.S., Yarbrough W.G., and Weaver A.M.. 2007. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67:4227–4235. 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Collinet C., Stöter M., Bradshaw C.R., Samusik N., Rink J.C., Kenski D., Habermann B., Buchholz F., Henschel R., Mueller M.S., et al. 2010. Systems survey of endocytosis by multiparametric image analysis. Nature. 464:243–249. 10.1038/nature08779 [DOI] [PubMed] [Google Scholar]
- Cui Y., Carosi J.M., Yang Z., Ariotti N., Kerr M.C., Parton R.G., Sargeant T.J., and Teasdale R.D.. 2019. Retromer has a selective function in cargo sorting via endosome transport carriers. J. Cell Biol. 218:615–631. 10.1083/jcb.201806153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., and Korswagen H.C.. 2011. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat. Cell Biol. 14:29–37. 10.1038/ncb2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., and Steinberg F.. 2018. To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19:679–696. 10.1038/s41580-018-0053-7 [DOI] [PubMed] [Google Scholar]
- d’Ortho M.P., Will H., Atkinson S., Butler G., Messent A., Gavrilovic J., Smith B., Timpl R., Zardi L., and Murphy G.. 1997. Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur. J. Biochem. 250:751–757. 10.1111/j.1432-1033.1997.00751.x [DOI] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J.J., Lombard B., Loew D., and Gautreau A.. 2009. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell. 17:712–723. 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Fjorback A.W., Seaman M., Gustafsen C., Mehmedbasic A., Gokool S., Wu C., Militz D., Schmidt V., Madsen P., Nyengaard J.R., et al. 2012. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J. Neurosci. 32:1467–1480. 10.1523/JNEUROSCI.2272-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frittoli E., Palamidessi A., Disanza A., and Scita G.. 2011. Secretory and endo/exocytic trafficking in invadopodia formation: the MT1-MMP paradigm. Eur. J. Cell Biol. 90:108–114. 10.1016/j.ejcb.2010.04.007 [DOI] [PubMed] [Google Scholar]
- Frittoli E., Palamidessi A., Marighetti P., Confalonieri S., Bianchi F., Malinverno C., Mazzarol G., Viale G., Martin-Padura I., Garré M., et al. 2014. A RAB5/RAB4 recycling circuitry induces a proteolytic invasive program and promotes tumor dissemination. J. Cell Biol. 206:307–328. 10.1083/jcb.201403127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallon M., and Cullen P.J.. 2015. Retromer and sorting nexins in endosomal sorting. Biochem. Soc. Trans. 43:33–47. 10.1042/BST20140290 [DOI] [PubMed] [Google Scholar]
- Gallon M., Clairfeuille T., Steinberg F., Mas C., Ghai R., Sessions R.B., Teasdale R.D., Collins B.M., and Cullen P.J.. 2014. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. USA. 111:E3604–E3613. 10.1073/pnas.1410552111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles C., Polette M., Piette J., Munaut C., Thompson E.W., Birembaut P., and Foidart J.M.. 1996. High level of MT-MMP expression is associated with invasiveness of cervical cancer cells. Int. J. Cancer. 65:209–213. [DOI] [PubMed] [Google Scholar]
- Gomez T.S., and Billadeau D.D.. 2009. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 17:699–711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujrati M., Mittal R., Ekal L., and Mishra R.K.. 2019. SUMOylation of periplakin is critical for efficient reorganization of keratin filament network. Mol. Biol. Cell. 30:357–369. 10.1091/mbc.E18-04-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M.E., Breusegem S.Y.A., Antrobus R., Freeman C., Reid E., and Seaman M.N.J.. 2010. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J. Cell Sci. 123:3703–3717. 10.1242/jcs.071472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M.E., Breusegem S.Y., and Seaman M.N.J.. 2012. Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem. J. 442:209–220. 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- Harrison M.S., Hung C.-S., Liu T.T., Christiano R., Walther T.C., and Burd C.G.. 2014. A mechanism for retromer endosomal coat complex assembly with cargo. Proc. Natl. Acad. Sci. USA. 111:267–272. 10.1073/pnas.1316482111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M., Port F., Lorenowicz M.J., McGough I.J., Silhankova M., Betist M.C., van Weering J.R.T., van Heesbeen R.G.H.P., Middelkoop T.C., Basler K., et al. 2011. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat. Cell Biol. 13:914–923. 10.1038/ncb2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh G.G., Pérez-Dorado I., Jackson L.P., Wartosch L., Schäfer I.B., Gray S.R., McCoy A.J., Zeldin O.B., Garman E.F., Harbour M.E., et al. 2014. VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev. Cell. 29:591–606. 10.1016/j.devcel.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbeck K., Bianco P., and Birkedal-Hansen H.. 2003. MT1-mmp: a collagenase essential for tumor cell invasive growth. Cancer Cell. 4:83–84. 10.1016/S1535-6108(03)00196-X [DOI] [PubMed] [Google Scholar]
- Hotary K., Li X.Y., Allen E., Stevens S.L., and Weiss S.J.. 2006. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 20:2673–2686. 10.1101/gad.1451806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-H., Yang W.-H., Chang S.-Y., Tai S.-K., Tzeng C.-H., Kao J.-Y., Wu K.-J., and Yang M.-H.. 2009. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia. 11:1371–1382. 10.1593/neo.91326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E., Yana I., Fujita C., Irifune A., Takeda M., Madachi A., Mori S., Hamada Y., Kawaguchi N., and Matsuura N.. 2010. The role of MT2-MMP in cancer progression. Biochem. Biophys. Res. Commun. 393:222–227. 10.1016/j.bbrc.2010.01.105 [DOI] [PubMed] [Google Scholar]
- Itoh Y. 2015. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 44-46:207–223. 10.1016/j.matbio.2015.03.004 [DOI] [PubMed] [Google Scholar]
- Itoh Y., Ito N., Nagase H., Evans R.D., Bird S.A., and Seiki M.. 2006. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol. Biol. Cell. 17:5390–5399. 10.1091/mbc.e06-08-0740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabłońska-Trypuć A., Matejczyk M., and Rosochacki S.. 2016. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J. Enzyme Inhib. Med. Chem. 31(sup1):177–183. 10.3109/14756366.2016.1161620 [DOI] [PubMed]
- Jacob A., and Prekeris R.. 2015. The regulation of MMP targeting to invadopodia during cancer metastasis. Front. Cell Dev. Biol. 3:4 10.3389/fcell.2015.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., and Scheller R.H.. 2006. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Jiang W.G., Davies G., Martin T.A., Parr C., Watkins G., Mason M.D., and Mansel R.E.. 2006. Expression of membrane type-1 matrix metalloproteinase, MT1-MMP in human breast cancer and its impact on invasiveness of breast cancer cells. Int. J. Mol. Med. 17:583–590. [PubMed] [Google Scholar]
- Jiang C., Wang J., Dong C., Wei W., Li J., and Li X.. 2017. Membranous type matrix metalloproteinase 16 induces human prostate cancer metastasis. Oncol. Lett. 14:3096–3102. 10.3892/ol.2017.6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L., and Popoff V.. 2008. Tracing the retrograde route in protein trafficking. Cell. 135:1175–1187. 10.1016/j.cell.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Kajiho H., Kajiho Y., Frittoli E., Confalonieri S., Bertalot G., Viale G., Di Fiore P.P., Oldani A., Garre M., Beznoussenko G.V., et al. 2016. RAB2A controls MT1-MMP endocytic and E-cadherin polarized Golgi trafficking to promote invasive breast cancer programs. EMBO Rep. 17:1061–1080. 10.15252/embr.201642032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger S.C., Siupka P., and Nielsen M.S.. 2015. Retromer-mediated trafficking of transmembrane receptors and transporters. Membranes (Basel). 5:288–306. 10.3390/membranes5030288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousidou O.C., Roussidis A.E., Theocharis A.D., and Karamanos N.K.. 2004. Expression of MMPs and TIMPs genes in human breast cancer epithelial cells depends on cell culture conditions and is associated with their invasive potential. Anticancer Res. 24:4025–4030. [PubMed] [Google Scholar]
- Kvainickas A., Jimenez-Orgaz A., Nägele H., Hu Z., Dengjel J., and Steinberg F.. 2017. Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J. Cell Biol. 216:3677–3693. 10.1083/jcb.201702137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagoutte E., Villeneuve C., Lafanechère L., Wells C.M., Jones G.E., Chavrier P., and Rossé C.. 2016. LIMK Regulates Tumor-Cell Invasion and Matrix Degradation Through Tyrosine Phosphorylation of MT1-MMP. Sci. Rep. 6:24925 10.1038/srep24925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffer B.E.L., Melero C., Temkin P., Lei C., Hong W., Kortemme T., and von Zastrow M.. 2010. SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J. Cell Biol. 190:565–574. 10.1083/jcb.201004060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Chang J., and Blackstone C.. 2016. FAM21 directs SNX27-retromer cargoes to the plasma membrane by preventing transport to the Golgi apparatus. Nat. Commun. 7:10939 10.1038/ncomms10939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti K., Valtanen H., Wickström S.A., Lohi J., and Keski-Oja J.. 2000. Regulation of membrane-type-1 matrix metalloproteinase activity by its cytoplasmic domain. J. Biol. Chem. 275:15006–15013. 10.1074/jbc.M910220199 [DOI] [PubMed] [Google Scholar]
- Li Y., Cai G., Yuan S., Jun Y., Li N., Wang L., Chen F., Ling R., and Yun J.. 2015. The overexpression membrane type 1 matrix metalloproteinase is associated with the progression and prognosis in breast cancer. Am. J. Transl. Res. 7:120–127. [PMC free article] [PubMed] [Google Scholar]
- Liang D.H., Choi D.S., Ensor J.E., Kaipparettu B.A., Bass B.L., and Chang J.C.. 2016. The autophagy inhibitor chloroquine targets cancer stem cells in triple negative breast cancer by inducing mitochondrial damage and impairing DNA break repair. Cancer Lett. 376:249–258. 10.1016/j.canlet.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S. 2015. MT1-MMP: Endosomal delivery drives breast cancer metastasis. J. Cell Biol. 211:215–217. 10.1083/jcb.201510009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., Wiesner C., and Himmel M.. 2011. Degrading devices: invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 27:185–211. 10.1146/annurev-cellbio-092910-154216 [DOI] [PubMed] [Google Scholar]
- Liu Y., Sun X., Feng J., Deng L.-L., Liu Y., Li B., Zhu M., Lu C., and Zhou L.. 2016. MT2-MMP induces proteolysis and leads to EMT in carcinomas. Oncotarget. 7:48193–48205. 10.18632/oncotarget.10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M., Gershlick D.C., Vidaurrazaga A., Rojas A.L., Bonifacino J.S., and Hierro A.. 2016. Structural Mechanism for Cargo Recognition by the Retromer Complex. Cell. 167:1623–1635.e14. 10.1016/j.cell.2016.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn M.L., Nassirpour R., Arrabit C., Tan J., McLeod I., Arias C.M., Sawchenko P.E., Yates J.R. III, and Slesinger P.A.. 2007. A unique sorting nexin regulates trafficking of potassium channels via a PDZ domain interaction. Nat. Neurosci. 10:1249–1259. 10.1038/nn1953 [DOI] [PubMed] [Google Scholar]
- MacDonald E., Brown L., Selvais A., Liu H., Waring T., Newman D., Bithell J., Grimes D., Urbé S., Clague M.J., and Zech T.. 2018. HRS-WASH axis governs actin-mediated endosomal recycling and cell invasion. J. Cell Biol. 217:2549–2564. 10.1083/jcb.201710051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson I.R., Rainero E., Mitchell L.E., van den Berghe P.V.E., Speirs C., Dozynkiewicz M.A., Chaudhary S., Kalna G., Edwards J., Timpson P., and Norman J.C.. 2014. CLIC3 controls recycling of late endosomal MT1-MMP and dictates invasion and metastasis in breast cancer. J. Cell Sci. 127:3893–3901. 10.1242/jcs.135947 [DOI] [PubMed] [Google Scholar]
- McGarvey J.C., Xiao K., Bowman S.L., Mamonova T., Zhang Q., Bisello A., Sneddon W.B., Ardura J.A., Jean-Alphonse F., Vilardaga J.-P., et al. 2016. Actin-Sorting Nexin 27 (SNX27)-Retromer Complex Mediates Rapid Parathyroid Hormone Receptor Recycling. J. Biol. Chem. 291:10986–11002. 10.1074/jbc.M115.697045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I.J., Steinberg F., Jia D., Barbuti P.A., McMillan K.J., Heesom K.J., Whone A.L., Caldwell M.A., Billadeau D.D., Rosen M.K., and Cullen P.J.. 2014. Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol. 24:1670–1676. 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D., et al. 2013. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203:1063–1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C.J., Butler G.S., Bigg H.F., Roberts C.R., Soloway P.D., and Overall C.M.. 2001. Cellular activation of MMP-2 (gelatinase A) by MT2-MMP occurs via a TIMP-2-independent pathway. J. Biol. Chem. 276:47402–47410. 10.1074/jbc.M108643200 [DOI] [PubMed] [Google Scholar]
- Murphy D.A., and Courtneidge S.A.. 2011. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada M., Nakamura H., Ikeda E., Fujimoto N., Yamashita J., Sato H., Seiki M., and Okada Y.. 1999. Expression and tissue localization of membrane-type 1, 2, and 3 matrix metalloproteinases in human astrocytic tumors. Am. J. Pathol. 154:417–428. 10.1016/S0002-9440(10)65288-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Miyamori H., Thompson E.W., Takino T., Endo Y., and Sato H.. 2008. Activation of matrix metalloproteinase-2 (MMP-2) by membrane type 1 matrix metalloproteinase through an artificial receptor for proMMP-2 generates active MMP-2. Cancer Res. 68:9096–9104. 10.1158/0008-5472.CAN-08-2522 [DOI] [PubMed] [Google Scholar]
- Nourry C., Grant S.G.N., and Borg J.-P.. 2003. PDZ domain proteins: plug and play! Sci. STKE 2003:RE7. 10.1126/stke.2003.179.re7 [DOI] [PubMed]
- Ohuchi E., Imai K., Fujii Y., Sato H., Seiki M., and Okada Y.. 1997. Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 272:2446–2451. 10.1074/jbc.272.4.2446 [DOI] [PubMed] [Google Scholar]
- Okada A., Bellocq J.P., Rouyer N., Chenard M.P., Rio M.C., Chambon P., and Basset P.. 1995. Membrane-type matrix metalloproteinase (MT-MMP) gene is expressed in stromal cells of human colon, breast, and head and neck carcinomas. Proc. Natl. Acad. Sci. USA. 92:2730–2734. 10.1073/pnas.92.7.2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota I., Li X.-Y., Hu Y., and Weiss S.J.. 2009. Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc. Natl. Acad. Sci. USA. 106:20318–20323. 10.1073/pnas.0910962106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson E.K., and Courtneidge S.A.. 2018. Invadosomes are coming: new insights into function and disease relevance. FEBS J. 285:8–27. 10.1111/febs.14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perentes J.Y., Kirkpatrick N.D., Nagano S., Smith E.Y., Shaver C.M., Sgroi D., Garkavtsev I., Munn L.L., Jain R.K., and Boucher Y.. 2011. Cancer cell-associated MT1-MMP promotes blood vessel invasion and distant metastasis in triple-negative mammary tumors. Cancer Res. 71:4527–4538. 10.1158/0008-5472.CAN-10-4376 [DOI] [PubMed] [Google Scholar]
- Piotrowski J.T., Gomez T.S., Schoon R.A., Mangalam A.K., and Billadeau D.D.. 2013. WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. Mol. Cell. Biol. 33:958–973. 10.1128/MCB.01288-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R., Lizárraga F., and Chavrier P.. 2009. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 122:3015–3024. 10.1242/jcs.034561 [DOI] [PubMed] [Google Scholar]
- Puthenveedu M.A., Lauffer B., Temkin P., Vistein R., Carlton P., Thorn K., Taunton J., Weiner O.D., Parton R.G., and von Zastrow M.. 2010. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 143:761–773. 10.1016/j.cell.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L., Cao H., Chen J., Weller S.G., Krueger E.W., Zhang L., Razidlo G.L., and McNiven M.A.. 2019. Pancreatic tumor cell metastasis is restricted by MT1-MMP binding protein MTCBP-1. J. Cell Biol. 218:317–332. 10.1083/jcb.201802032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle A., Murphy G., and Roghi C.. 2003. Membrane type I-matrix metalloproteinase (MT1-MMP) is internalised by two different pathways and is recycled to the cell surface. J. Cell Sci. 116:3905–3916. 10.1242/jcs.00710 [DOI] [PubMed] [Google Scholar]
- Rincón E., Santos T., Ávila-Flores A., Albar J.P., Lalioti V., Lei C., Hong W., and Mérida I.. 2007. Proteomics identification of sorting nexin 27 as a diacylglycerol kinase ζ-associated protein: new diacylglycerol kinase roles in endocytic recycling. Mol. Cell. Proteomics. 6:1073–1087. 10.1074/mcp.M700047-MCP200 [DOI] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Risselada H.J., and Grubmüller H.. 2012. How SNARE molecules mediate membrane fusion: recent insights from molecular simulations. Curr. Opin. Struct. Biol. 22:187–196. 10.1016/j.sbi.2012.01.007 [DOI] [PubMed] [Google Scholar]
- Rojas R., Kametaka S., Haft C.R., and Bonifacino J.S.. 2007. Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol. Cell. Biol. 27:1112–1124. 10.1128/MCB.00156-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R., van Vlijmen T., Mardones G.A., Prabhu Y., Rojas A.L., Mohammed S., Heck A.J.R., Raposo G., van der Sluijs P., and Bonifacino J.S.. 2008. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183:513–526. 10.1083/jcb.200804048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossé C., Lodillinsky C., Fuhrmann L., Nourieh M., Monteiro P., Irondelle M., Lagoutte E., Vacher S., Waharte F., Paul-Gilloteaux P., et al. 2014. Control of MT1-MMP transport by atypical PKC during breast-cancer progression. Proc. Natl. Acad. Sci. USA. 111:E1872–E1879. 10.1073/pnas.1400749111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M.C., and Leonhardt H.. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 3:887–889. 10.1038/nmeth953 [DOI] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., and Leonhardt H.. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289. 10.1074/mcp.M700342-MCP200 [DOI] [PubMed] [Google Scholar]
- Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., and Seiki M.. 1994. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 370:61–65. 10.1038/370061a0 [DOI] [PubMed] [Google Scholar]
- Schwarz D.G., Griffin C.T., Schneider E.A., Yee D., and Magnuson T.. 2002. Genetic analysis of sorting nexins 1 and 2 reveals a redundant and essential function in mice. Mol. Biol. Cell. 13:3588–3600. 10.1091/mbc.e02-03-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J. 2004. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165:111–122. 10.1083/jcb.200312034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J. 2007. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J. Cell Sci. 120:2378–2389. 10.1242/jcs.009654 [DOI] [PubMed] [Google Scholar]
- Seaman M.N.J., Marcusson E.G., Cereghino J.L., and Emr S.D.. 1997. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 137:79–92. 10.1083/jcb.137.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J., McCaffery J.M., and Emr S.D.. 1998. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142:665–681. 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J., Harbour M.E., Tattersall D., Read E., and Bright N.. 2009. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122:2371–2382. 10.1242/jcs.048686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N.J., Gautreau A., and Billadeau D.D.. 2013. Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 23:522–528. 10.1016/j.tcb.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Wang X., Yu X., Zhang Y., and Qin L.. 2017. MMP16 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Oncotarget. 8:72197–72204. 10.18632/oncotarget.20060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti B., Danson C.M., Heesom K.J., and Cullen P.J.. 2017. Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J. Cell Biol. 216:3695–3712. 10.1083/jcb.201703015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S.A., and Petsko G.A.. 2015. Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat. Rev. Neurosci. 16:126–132. 10.1038/nrn3896 [DOI] [PubMed] [Google Scholar]
- Sodek K.L., Ringuette M.J., and Brown T.J.. 2007. MT1-MMP is the critical determinant of matrix degradation and invasion by ovarian cancer cells. Br. J. Cancer. 97:358–367. 10.1038/sj.bjc.6603863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A., Le Dez G., Poincloux R., Recchi C., Nassoy P., Rottner K., Galli T., and Chavrier P.. 2008. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol. 18:926–931. 10.1016/j.cub.2008.05.044 [DOI] [PubMed] [Google Scholar]
- Steinberg F., Gallon M., Winfield M., Thomas E.C., Bell A.J., Heesom K.J., Tavaré J.M., and Cullen P.J.. 2013. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15:461–471. 10.1038/ncb2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanei T., Choi D.S., Rodriguez A.A., Liang D.H., Dobrolecki L., Ghosh M., Landis M.D., and Chang J.C.. 2016. Antitumor activity of Cetuximab in combination with Ixabepilone on triple negative breast cancer stem cells. Breast Cancer Res. 18:6 10.1186/s13058-015-0662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatti O., Arjama M., Ranki A., Weiss S.J., Keski-Oja J., and Lehti K.. 2011. Membrane-type-3 matrix metalloproteinase (MT3-MMP) functions as a matrix composition-dependent effector of melanoma cell invasion. PLoS One. 6:e28325 10.1371/journal.pone.0028325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatti O., Gucciardo E., Pekkonen P., Holopainen T., Louhimo R., Repo P., Maliniemi P., Lohi J., Rantanen V., Hautaniemi S., et al. 2015. MMP16 Mediates a Proteolytic Switch to Promote Cell-Cell Adhesion, Collagen Alignment, and Lymphatic Invasion in Melanoma. Cancer Res. 75:2083–2094. 10.1158/0008-5472.CAN-14-1923 [DOI] [PubMed] [Google Scholar]
- Temkin P., Lauffer B., Jäger S., Cimermancic P., Krogan N.J., and von Zastrow M.. 2011. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13:715–721. 10.1038/ncb2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uekita T., Gotoh I., Kinoshita T., Itoh Y., Sato H., Shiomi T., Okada Y., and Seiki M.. 2004. Membrane-type 1 matrix metalloproteinase cytoplasmic tail-binding protein-1 is a new member of the Cupin superfamily. A possible multifunctional protein acting as an invasion suppressor down-regulated in tumors. J. Biol. Chem. 279:12734–12743. 10.1074/jbc.M309957200 [DOI] [PubMed] [Google Scholar]
- Vardarajan B.N., Bruesegem S.Y., Harbour M.E., Inzelberg R., Friedland R., St George-Hyslop P., Seaman M.N.J., and Farrer L.A.. 2012. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol. Aging. 33:2231.e15–2231.e30. 10.1016/j.neurobiolaging.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., and Bellen H.J.. 2015. The retromer complex in development and disease. Development. 142:2392–2396. 10.1242/dev.123737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ma D., Keski-Oja J., and Pei D.. 2004. Co-recycling of MT1-MMP and MT3-MMP through the trans-Golgi network. Identification of DKV582 as a recycling signal. J. Biol. Chem. 279:9331–9336. 10.1074/jbc.M312369200 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yu S.-J., Li Y.-X., and Luo H.-S.. 2015. Expression and clinical significance of matrix metalloproteinase-17 and -25 in gastric cancer. Oncol. Lett. 9:671–676. 10.3892/ol.2014.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer T., Attar N., Bujny M.V., Oakley J., Traer C.J., and Cullen P.J.. 2007. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J. Cell Sci. 120:45–54. 10.1242/jcs.03302 [DOI] [PubMed] [Google Scholar]
- Wassmer T., Attar N., Harterink M., van Weering J.R.T., Traer C.J., Oakley J., Goud B., Stephens D.J., Verkade P., Korswagen H.C., and Cullen P.J.. 2009. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell. 17:110–122. 10.1016/j.devcel.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Hoshino D., Koshikawa N., Seiki M., Suzuki T., and Ichikawa K.. 2013. Critical role of transient activity of MT1-MMP for ECM degradation in invadopodia. PLOS Comput. Biol. 9:e1003086 10.1371/journal.pcbi.1003086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A.M. 2006. Invadopodia: specialized cell structures for cancer invasion. Clin. Exp. Metastasis. 23:97–105. 10.1007/s10585-006-9014-1 [DOI] [PubMed] [Google Scholar]
- Wells J.M., Gaggar A., and Blalock J.E.. 2015. MMP generated matrikines. Matrix Biol. 44-46:122–129. 10.1016/j.matbio.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner C., El Azzouzi K., and Linder S.. 2013. A specific subset of RabGTPases controls cell surface exposure of MT1-MMP, extracellular matrix degradation and three-dimensional invasion of macrophages. J. Cell Sci. 126:2820–2833. 10.1242/jcs.122358 [DOI] [PubMed] [Google Scholar]
- Williams K.C., and Coppolino M.G.. 2011. Phosphorylation of membrane type 1-matrix metalloproteinase (MT1-MMP) and its vesicle-associated membrane protein 7 (VAMP7)-dependent trafficking facilitate cell invasion and migration. J. Biol. Chem. 286:43405–43416. 10.1074/jbc.M111.297069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.C., McNeilly R.E., and Coppolino M.G.. 2014. SNAP23, Syntaxin4, and vesicle-associated membrane protein 7 (VAMP7) mediate trafficking of membrane type 1-matrix metalloproteinase (MT1-MMP) during invadopodium formation and tumor cell invasion. Mol. Biol. Cell. 25:2061–2070. 10.1091/mbc.e13-10-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Ma C., Shan S., Zhou L., and Li W.. 2017. High expression of matrix metalloproteinases 16 is associated with the aggressive malignant behavior and poor survival outcome in colorectal carcinoma. Sci. Rep. 7:46531 10.1038/srep46531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Follett J., Kerr M.C., Clairfeuille T., Chandra M., Collins B.M., and Teasdale R.D.. 2018. Sorting nexin 27 (SNX27) regulates the trafficking and activity of the glutamine transporter ASCT2. J. Biol. Chem. 293:6802–6811. 10.1074/jbc.RA117.000735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C., Foidart P., Somja J., Truong A., Lienard M., Feyereisen E., Schroeder H., Gofflot S., Donneau A.-F.F., Collignon J., et al. 2017. MT4-MMP and EGFR expression levels are key biomarkers for breast cancer patient response to chemotherapy and erlotinib. Br. J. Cancer. 116:742–751. 10.1038/bjc.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudowski G.A., Puthenveedu M.A., Henry A.G., and von Zastrow M.. 2009. Cargo-mediated regulation of a rapid Rab4-dependent recycling pathway. Mol. Biol. Cell. 20:2774–2784. 10.1091/mbc.e08-08-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Calaminus S.D.J., Caswell P., Spence H.J., Carnell M., Insall R.H., Norman J., and Machesky L.M.. 2011. The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J. Cell Sci. 124:3753–3759. 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li K., Zhang Y., Lu R., Wu S., Tang J., Xia Y., and Sun J.. 2019. Deletion of sorting nexin 27 suppresses proliferation in highly aggressive breast cancer MDA-MB-231 cells in vitro and in vivo. BMC Cancer. 19:555 10.1186/s12885-019-5769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.