To prevent chromosome missegregation, a metaphase checkpoint is activated when topoisomerase II is catalytically inhibited and DNA catenations persist. Pandey et al. dissect the key molecular events triggering this regulatory system.
Abstract
Topoisomerase II (Topo II) is essential for mitosis since it resolves sister chromatid catenations. Topo II dysfunction promotes aneuploidy and drives cancer. To protect from aneuploidy, cells possess mechanisms to delay anaphase onset when Topo II is perturbed, providing additional time for decatenation. Molecular insight into this checkpoint is lacking. Here we present evidence that catalytic inhibition of Topo II, which activates the checkpoint, leads to SUMOylation of the Topo II C-terminal domain (CTD). This modification triggers mobilization of Aurora B kinase from inner centromeres to kinetochore proximal centromeres and the core of chromosome arms. Aurora B recruitment accompanies histone H3 threonine-3 phosphorylation and requires Haspin kinase. Strikingly, activation of the checkpoint depends both on Haspin and Aurora B. Moreover, mutation of the conserved CTD SUMOylation sites perturbs Aurora B recruitment and checkpoint activation. The data indicate that SUMOylated Topo II recruits Aurora B to ectopic sites, constituting the molecular trigger of the metaphase checkpoint when Topo II is catalytically inhibited.
Introduction
Type II DNA topoisomerases are universal enzymes that play crucial roles in mitosis due to their unique strand passage reaction (SPR). The SPR is a multistep action involving large conformational changes and using ATP hydrolysis (Dong and Berger, 2007; Wang, 2007). A dimeric Topoisomerase II (Topo II) holoenzyme introduces a double-strand break into a bound DNA helix. A second, intact DNA helix is passed through the break, which is then religated. This catalytic cycle has been well studied, because widely used anticancer drugs target the SPR (Nitiss, 2009b).
Previous studies showed that yeast Topo II mutants with a low rate of ATP hydrolysis activate the metaphase checkpoint (Andrews et al., 2006; Furniss et al., 2013). However, yeast Topo II mutants defective at the initiation step of the SPR do not. This suggests that the checkpoint is activated only when the SPR is impaired at specific stages, requiring ATP hydrolysis, and not due to a defect in SPR initiation.
The catalytic Topo II inhibitor ICRF-193 acts at the step of ATP hydrolysis and thus chemically mimics the genetic effects of the yeast mutants with a slow rate of ATP hydrolysis (Nitiss, 2009b). Human cells treated with ICRF-193 also activate a metaphase checkpoint (Clarke et al., 2006; Skoufias et al., 2004; Toyoda and Yanagida, 2006). However, it remains unclear how disruption of the Topo II SPR, particularly as late as the ATP hydrolysis stage, can induce a metaphase checkpoint. Recent studies provided a hint toward the molecular mechanism. HeLa cells treated with ICRF-187 (which inhibits Topo II using the same mechanism as ICRF-193) up-regulate small ubiquitin-like modifier 2/3 (SUMO2/3) modification of Topo IIα on mitotic chromosomes (Agostinho et al., 2008). Another Topo II inhibitor, merbarone, that blocks an early step of the SPR, did not up-regulate SUMO2/3 modification. SUMOylation is important for error-free chromosome segregation in many eukaryotes (Biggins et al., 2001; Hari et al., 2001; Mukhopadhyay and Dasso, 2017; Takahashi et al., 2006; Zhang et al., 2008). These observations indicate that catalytic inhibition of Topo IIα at the ATP hydrolysis step leads to SUMO2/3-modified Topo IIα and that this biochemical event may play a role in metaphase checkpoint activation.
Supporting this notion, we reported that Topo IIα C-terminal domain (CTD) SUMOylation regulates Aurora B at mitotic centromeres (Edgerton et al., 2016; Yoshida et al., 2016). Aurora B is the kinase component of the chromosome passenger complex (CPC) that controls the metaphase-to-anaphase transition. In Xenopus laevis egg extracts (XEEs), SUMOylated Topo IIα CTD interacts with Claspin (Ryu et al., 2015), which binds to Chk1 kinase; Chk1 can activate Aurora B via phosphorylation of S331 in human cells (Petsalaki et al., 2011). Further, SUMOylated Topo IIα CTD binds to Haspin kinase and promotes Aurora B recruitment to inner centromeres via phosphorylation of histone H3 threonine 3 (H3T3p; Dai and Higgins, 2005; Dai et al., 2005; Kelly et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). This Topo II SUMOylation-dependent mechanism of Aurora B recruitment to mitotic centromeres is conserved in yeast and XEEs (Edgerton et al., 2016; Yoshida et al., 2016).
Here, we provide evidence that the metaphase checkpoint accompanies SUMOylation-dependent activation of Aurora B kinase in XEE and cultured cells. Checkpoint activation requires Aurora B and Haspin, both of which are recruited to novel chromosomal positions upon Topo II catalytic inhibition. Aurora B and H3T3p are depleted from their normal residence at inner centromeres: ectopic phosphorylation of H3T3 is induced at kinetochore proximal centromeres (KPCs) and chromosome arms; Aurora B is recruited to those same locales. We propose that upon detection of a stalled SPR, SUMOylation of the Topo II CTD triggers Aurora B activation to induce a metaphase delay. The data have implications for cancer therapies that may use Aurora B and Topo II inhibitors.
Results
Topo II catalytic inhibition increases Topo IIα SUMOylation on mitotic chromosomes in XEE
SPR defects at the step of ATP hydrolysis activate a metaphase checkpoint in yeast and human cells (Clarke et al., 2006; Furniss et al., 2009). We found that Topo IIα SUMOylation stimulates Aurora B recruitment to centromeres in yeast and XEE (Edgerton et al., 2016; Yoshida et al., 2016), and Aurora B is known to regulate anaphase onset. Thus, we postulated that SPR stalling at the ATP hydrolysis step leads to SUMOylation of Topo II that recruits Aurora B to mitotic centromeres.
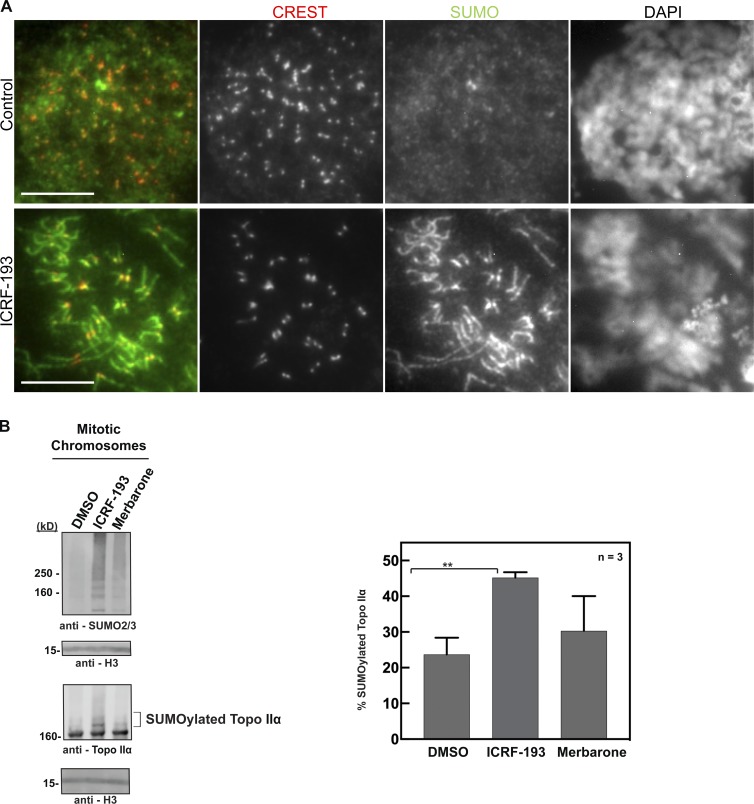
We first asked if ICRF-193, which inhibits ATP hydrolysis by Topo II, induces Topo II SUMOylation. In the XEE cell-free system, we observed chromosome condensation under the microscope, then immediately added ICRF-193 or merbarone (a Topo II inhibitor that does not act at the step of ATP hydrolysis). Adding the inhibitors after condensation eliminates indirect effects due to disruption of Topo IIα activity required for chromosome condensation (Fig. 1 A). After 10-min incubation with the inhibitors, chromosomes were isolated and subjected to Western blotting. Both inhibitors increased chromosomal mitotic SUMOylation, but ICRF-193 had a much greater effect (Fig. 1, B and C). Increased mitotic SUMOylation was observed prominently for large molecular weight proteins, consistent with the molecular weight of SUMOylated Topo IIα. Indeed, anti-Topo IIα antibodies revealed that ICRF-193–treated chromosomes had more SUMOylated Topo IIα than control and merbarone-treated samples (Fig. 1, B and D). Quantification revealed a significant increase in SUMOylated Topo IIα in ICRF-193–treated extracts, ∼1.2 times more than the DMSO control, whereas merbarone-treated extracts showed only a slight increase. Increased SUMOylation of PARP1, another major SUMOylated protein on mitotic chromosomes in XEE (Ryu et al., 2010a), was not observed (Fig. 1, B and E). Notably, increased chromosomal SUMOylation with ICRF-193 was significantly reduced in the presence of a dominant-negative E2 SUMO-conjugating enzyme, UbC9 (dnUbC9), indicating that the normal SUMOylation machinery was used for ICRF-193–mediated SUMOylation of Topo IIα (Fig. 1 B). Because we observed a large increase in overall chromosomal SUMO2/3 modification, we examined if the SUMOylation machinery itself is affected by ICRF-193. Addition of ICRF-193 to in vitro SUMOylation assays using recombinant Topo IIα-CTD as a substrate did not have a measurable effect on SUMOylation efficiency (Fig. S1). Thus, the SUMOylation machinery was not targeted directly by ICRF-193. This is consistent with a specific effect of ICRF-193 on Topo IIα SUMOylation, not on PARP1 SUMOylation, on chromosomes in XEE. The results demonstrate that the Topo II inhibitor ICRF-193, which inhibits ATP hydrolysis by Topo II, specifically increases Topo IIα SUMOylation, and that merbarone, which blocks initiation of the SPR, has a much weaker effect. Thus, Topo IIα at a specific stage in the SPR cycle could be especially susceptible to SUMOylation.
Figure 1.
Topo II inhibitor addition increases mitotic SUMOylation on mitotic chromosomes in XEEs. (A) Schematic representation for preparation of mitotic replicated chromosomes treated with inhibitors from XEEs (see Materials and methods). (B) Inhibitor-treated mitotic chromosomes were isolated from XEEs as shown in A with (+dnUbc9) and without (control) dnUbc9 and subjected to Western blotting. Histone H4 was probed as a loading control for the mitotic chromosomes. (C–E) Quantification of mitotic SUMOylation in the inhibitor-treated mitotic chromosomes relative to DMSO-treated chromosomes, percentage SUMOylation of Topo IIα and PARP1 as seen in B, from four independent experiments (n = 4). Error bars, standard deviation. *, P value from Student’s t test. **, Statistically significant difference, P ≤ 0.01.
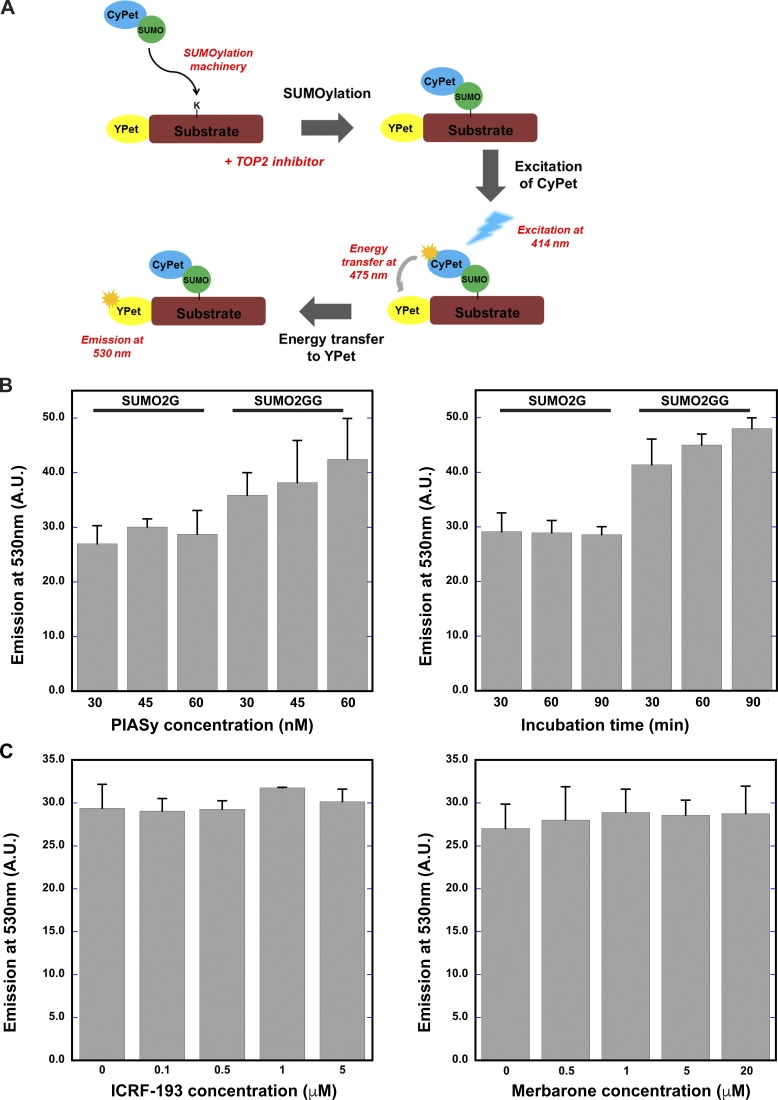
Figure S1.
Topo II inhibitor treatment does not affect SUMOylation machinery. (A) A schematic representation of FRET-based in vitro SUMOylation assay. YPet-tagged Topo IIα CTD was used as a substrate, along with CyPet-tagged SUMO2G and SUMO2 GG isoforms in the reaction. (B) In vitro SUMOylation was performed with different PIASy concentrations and at different time points. a.u., arbitrary unit. (C) In vitro SUMOylation was performed for 30 min with different ICRF-193 and merbarone concentrations. The emission spectrum was recorded at 530 nm from three different experiments (n = 3), and error bars represent standard deviation. Paired t test was performed comparing the reaction with inhibitors and without inhibitors in C. None of the inhibitor-containing conditions showed statistically significant differences from the conditions without inhibitors.
Topo II catalytic inhibition induces SUMOylation of the physiologically relevant CTD residues
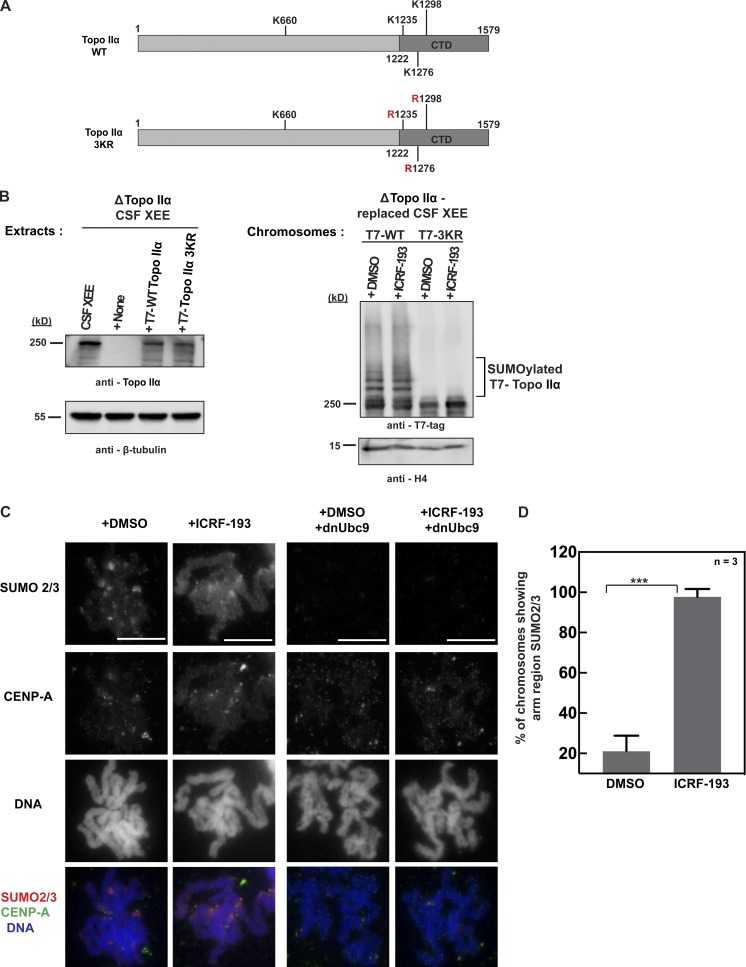
Next, we asked if ICRF-193 induces Topo IIα SUMOylation at the physiologically relevant residues. We had identified all SUMO acceptor sites in Xenopus Topo IIα, which is exclusively modified with SUMO2/3 during mitosis (Ryu et al., 2015). Three sites are located in the CTD, and one is in the DNA gate domain. To ask if ICRF-193–mediated Topo IIα SUMOylation occurs at the native lysines in the CTD, we prepared mitotic chromosomes after immunodepletion of endogenous Topo IIα from XEEs and addition of recombinant T7-tagged WT Topo IIα or the 3KR mutant where all three CTD lysines are mutated to arginine (Fig. 2, A and B). Since Topo IIα depletion prevents proper chromosome formation in replicated chromatin, we used unreplicated chromosomes for this analysis, which have less SUMOylated Topo IIα than replicated mitotic chromosomes (Azuma et al., 2003). Still, the mitotic chromosomes with recombinant WT Topo IIα exhibited increased SUMOylation with ICRF-193, although the SUMOylation increase was not as clear as in replicated chromosomes (see Fig. 1). Importantly, the mitotic chromosomes with recombinant Topo IIα 3KR did not show Topo IIα SUMOylation even with ICRF-193 (Fig. 2 B). Thus, ICRF-193 increased Topo IIα SUMOylation on the native SUMO acceptor residues in the CTD.
Figure 2.
Topo II inhibitor ICRF-193 increases Topo IIα SUMOylation at CTD and up-regulates SUMOylation on mitotic centromeres and chromosome arms in XEEs. (A) Schematic representation for the primary structure of WT X. laevis Topo IIα and Topo IIα 3KR mutant. The three lysine residues indicated in the CTD were mutated to arginine, which inhibits SUMO2/3 conjugation of the CTD. (B) Endogenous Topo IIα in CSF XEEs was depleted using affinity-purified anti-Topo IIα antibody and replaced with recombinant full-length WT T7-tagged Topo IIα or Topo IIα 3KR (left). β-Tubulin, loading control for Topo IIα levels in CSF XEEs. The inhibitor-treated mitotic chromosomes were isolated from Topo IIα–replaced CSF XEEs and probed for Topo IIα SUMOylation using T7 antibody by Western blotting (right). Histone H4 is the loading control for the mitotic chromosomes. (C) DMSO- and ICRF-193–treated mitotic replicated chromosomes were isolated from XEEs as shown in Fig. 1 A with or without dnUbc9 (control). The mitotic chromosomes were subjected to immunofluorescence staining using the indicated antibodies, and DNA was stained with Hoechst 33342. Bars, 10 µm. (D) Quantification of mitotic chromosomes showing arm region SUMO2/3 signals. The mitotic chromosomes with arm region SUMO2/3 signal were counted for 30 chromosomes from three independent experiments (n = 3). Error bars, standard deviation. *, P value from Student’s t test. ***, Statistically significant difference, P ≤ 0.001.
Previous work revealed that SUMOylated proteins are mainly confined to mitotic centromeres during mitosis in XEE. Thus, we asked if increased SUMOylation with ICRF-193 occurs at centromeres. Immunostaining of mitotic chromosomes from ICRF-193–treated XEEs showed increased SUMO2/3 at centromeres as well as chromosome arms (Fig. 2, C and D). Consistent with the Western blot analysis, SUMOylation on chromosome arms with ICRF-193 was diminished by addition of dnUbC9 (Fig. 2 C). The ICRF-193–mediated increase in SUMO2/3 on chromosome arms was observed in 97% of treated chromosomes (Fig. 2 D). Interestingly, we also observed spreading of the inner-kinetochore protein CENP-A after ICRF-193 treatment, which did not alter the overall chromosome morphology but could affect recruitment of centromeric components (Fig. 2 C). In contrast, the localization of Topo IIα throughout the chromosome axis and with enrichment at the mitotic centromere was not affected by ICRF-193 or merbarone (Fig. 3, A and B). The data show that ICRF-193 induces increased mitotic SUMOylation (primarily at the CTD lysine residues) of Topo IIα located on chromosome arms and at centromeres.
Figure 3.
Aurora B is recruited to chromosome arms in response to ICRF-193 addition in XEEs. (A and B) Mitotic replicated chromosomes were prepared from XEEs with the inhibitor treatment as in Fig. 1 A. Isolation of fixed chromosomes was performed as described in Materials and methods for immunostaining using the indicated antibodies. DNA was stained with Hoechst 33342. (C) Immunostaining of a single pair of sister chromatids showed Aurora B recruitment to chromosome arms with ICRF-193 addition. Inhibition of SUMOylation by dnUbc9 addition reduced Aurora B signals at chromosome arms. Bars, 10 µm. (D) Quantification of the mitotic chromosomes showing arm region Aurora B signals. The mitotic chromosomes showing arm region Aurora B foci were counted for >25 chromosomes from four independent experiments (n = 4). Error bars, standard deviation. *, P value from Student’s t test. *, P ≤ 0.05.
ICRF-193–induced Topo IIα SUMOylation recruits Aurora B to mitotic chromosomes
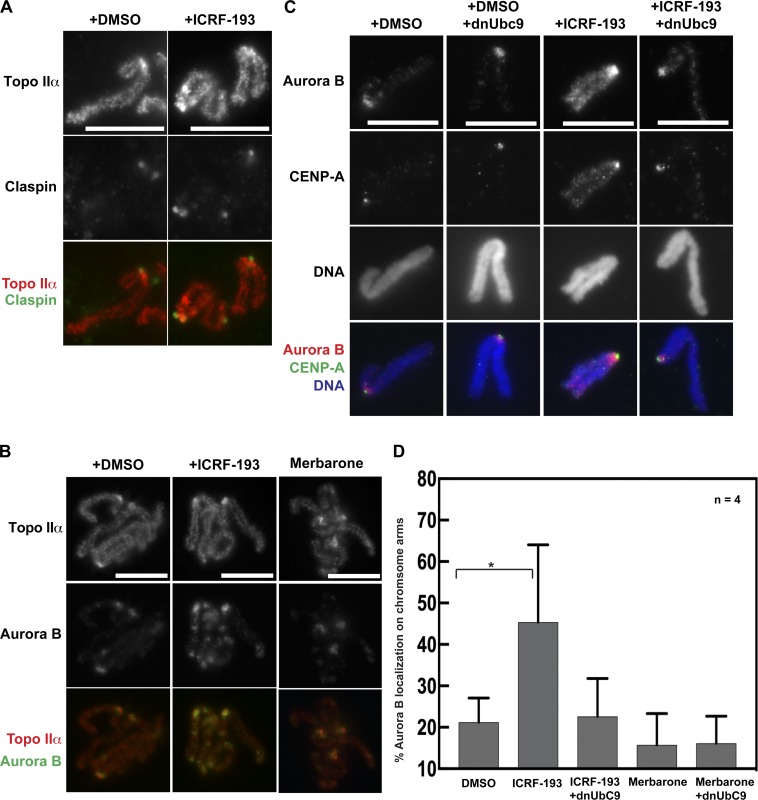
Previously, we demonstrated a role for Topo IIα CTD SUMOylation in mediating protein interactions in XEEs (Ryu et al., 2015; Yoshida et al., 2016). Mass spectrometry identified Claspin and Haspin as SUMOylated CTD binding proteins, and both were recruited to mitotic centromeres dependent on CTD SUMOylation. Since we observed ICRF-193–mediated up-regulation of Topo IIα SUMOylation, we asked if Topo IIα SUMOylation-dependent binding proteins are recruited to chromosomes in the context of Topo II inhibition. First, we examined Claspin localization on mitotic chromosomes isolated from ICRF-193–treated XEEs. Claspin was enriched at mitotic centromeres and colocalized with Topo IIα foci (Fig. 3 A), but this pattern did not change after ICRF-193 treatment. Next, we examined Aurora B, which is recruited to centromeres in part by SUMOylated Topo IIα via Haspin-mediated H3T3p (Edgerton et al., 2016; Yoshida et al., 2016). In contrast to Claspin, Aurora B became more abundant at centromeres after ICRF-193 treatment and was also recruited to chromosome arms (Fig. 3 B). The chromosome arm foci were striking, because Aurora B normally remains restricted to inner centromeres in metaphase. In contrast, Aurora B remained localized at centromeres and did not move to chromosome arms after merbarone treatment (Fig. 3 B). Also consistent with Topo II SUMOylation inducing the recruitment of Aurora B to these novel chromosome sites, addition of dnUbc9 eliminated the Aurora B signals on chromosome arms with ICRF-193 treatment (Fig. 3 C). The ICRF-193–induced and SUMOylation-dependent chromosome arm localization of Aurora B was observed in 45% of mitotic chromosomes, and addition of dnUbc9 reduced (by about half, ∼22.6%) the number of chromosomes showing arm-region Aurora B signals. However, after merbarone treatment, the number of mitotic chromosomes with arm-region Aurora B signals was less than the DMSO control and was not affected by dnUbc9 (Fig. 3 D). Together, the results suggest that Topo IIα SUMOylation on chromosomes arms induced by ICRF-193 triggers recruitment of Aurora B. On the other hand, Claspin remained restricted to the mitotic centromeres, suggesting that the chromatin association of Claspin is not solely dependent on the SUMO/SUMO-interacting motif interaction between Claspin and the SUMOylated Topo II CTD.
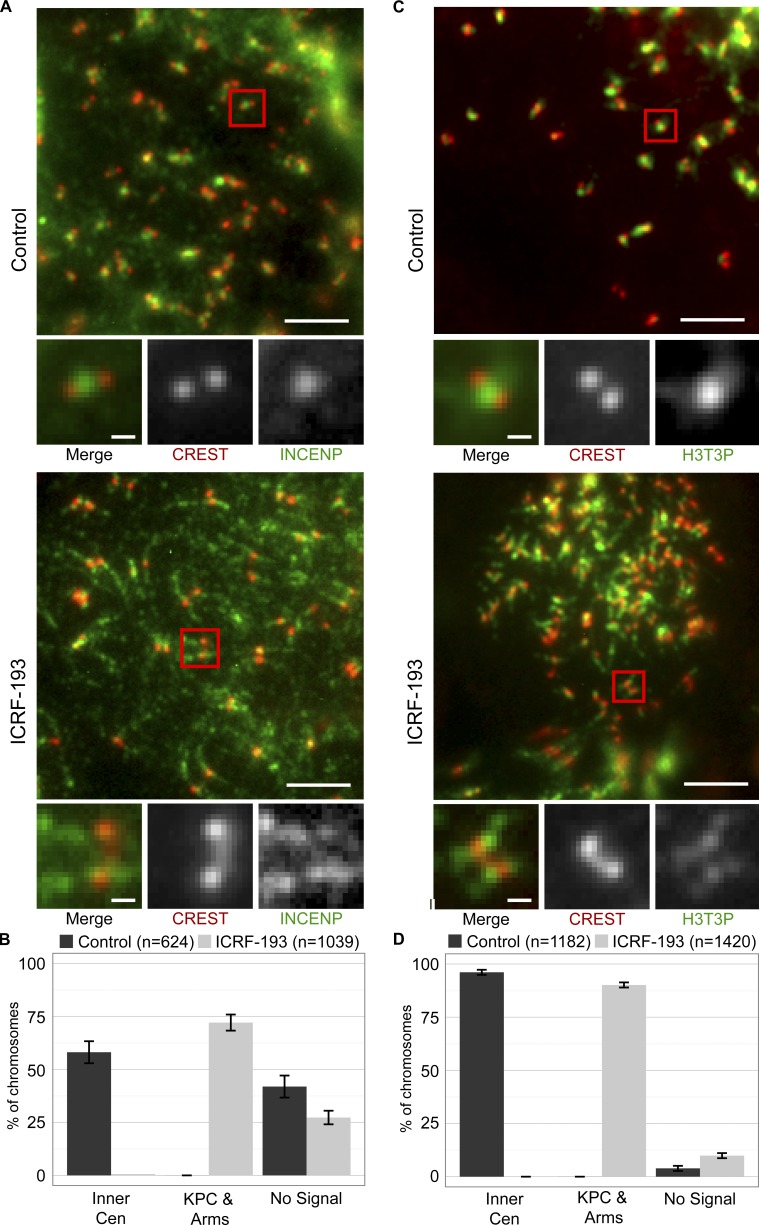
Aurora B recruitment to chromosome arms is conserved in human cells
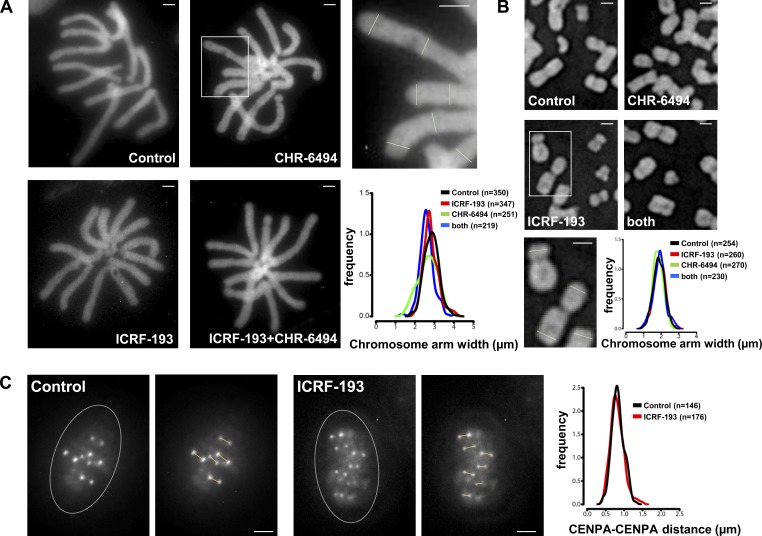
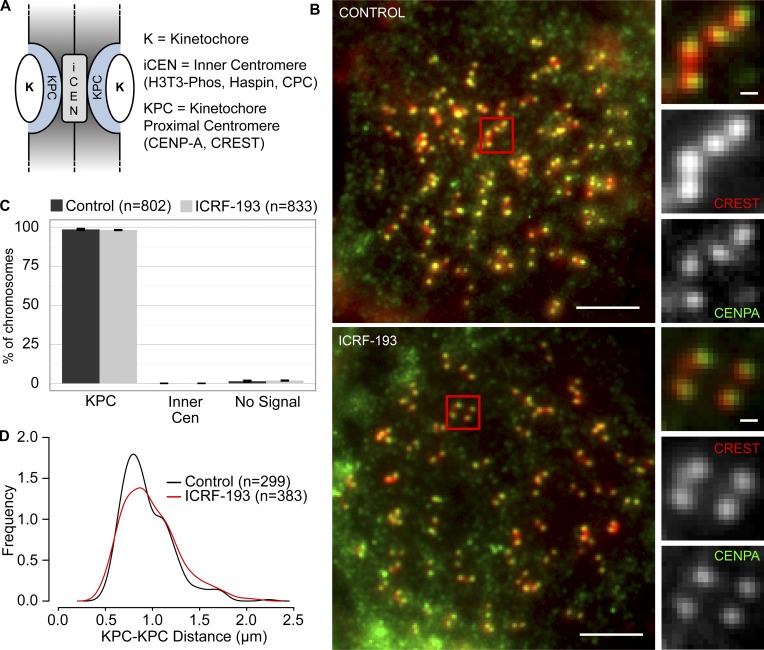
To find out if Aurora B recruitment is conserved, we treated mammalian cells with nocodazole to give a pure population of pseudometaphases with condensed chromosomes, and then incubated a further 45 min with or without ICRF-193. As in XEE, we avoided effects that may arise due to Topo II inhibition during chromosome condensation. To validate that this approach and the ICRF-193 treatment did not grossly affect chromosome structure, we measured widths of chromosome arms in live HeLa and Muntiacus muntjak cells expressing H2B-GFP. This revealed a similar distribution of arms widths with or without ICRF-193 (Fig. S2, A and B). Then, we defined the positions of the KPCs by immunostaining with anti–CENP-A antibodies and CS1058 CREST serum (Fig. 4 A). The KPCs are distinct from the inner centromere of a chromatid pair, lying more peripherally at the base of the kinetochores, (Cheeseman et al., 2002, Hindriksen et al., 2017). In control and ICRF-193–treated HeLa cells, chromosomes were indistinguishable in terms of these colocalizing KPC epitopes (Fig. 4, B and C). The distribution of KPC-to-KPC distances was similar (Fig. 4 D). Thus, ICRF-193 treatment did not measurably alter overall centromere architecture. Finally, we examined KPC–KPC distances without nocodazole, in live cells (Fig. S2 C). The KPC–KPC distances observed in controls were consistent with previous reports (Smith et al., 2016) and were not altered by ICRF-193 treatment.
Figure S2.
Chromosome morphologies in unfixed cells after Topo II and Haspin inhibitor treatments in mitosis. (A) Muntjac chromosomes in live cells expressing H2B-GFP, arrested with nocodazole, treated for 45 min as indicated, and then gently spun onto glass slides and imaged immediately. Bars, 4 µm. Histogram plot shows the distribution of chromosome arm widths measured as indicated by lines in the magnified image above (corresponding to the boxed region). (B) HeLa chromosomes prepared and analyzed as in A. Bars, 2 µm. (C) CENP-A-GFP in live metaphase HT-1080 (human) cells, treated for 45 min as indicated, then gently spun onto glass slides and imaged immediately (without nocodazole). Bars, 2 µm. Histogram plot shows the distribution of KPC-to-KPC distances measured as indicated by lines in the images on the right of each example (corresponding to the boxed region). Circled region indicates the position of the metaphase plate. Data were collected from three independent experiments.
Figure 4.
CENP-A and CREST colocalize regardless of ICRF-193 treatment in HeLa cells. (A) Cartoon depicts centromere/kinetochore regions. (B) Representative immunofluorescent-stained images of pseudometaphase HeLa cells (nocodazole arrested) ± ICRF-193 treatment for 45 min. CREST, red; CENP-A, green. Bars, 10 µm (insets, 1 µm). (C) Quantification of immunofluorescent staining at centromeres (Cen). Error bars, standard deviation. (D) Distribution plot of KPC-to-KPC distances. Error bars, standard deviation. Data were collected from three independent experiments.
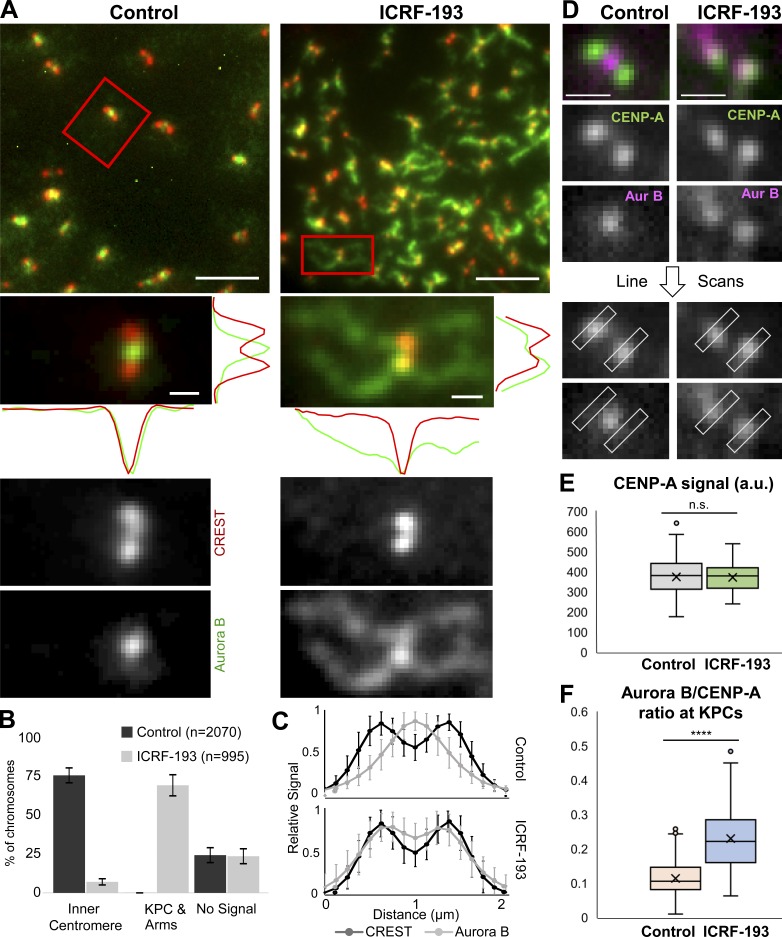
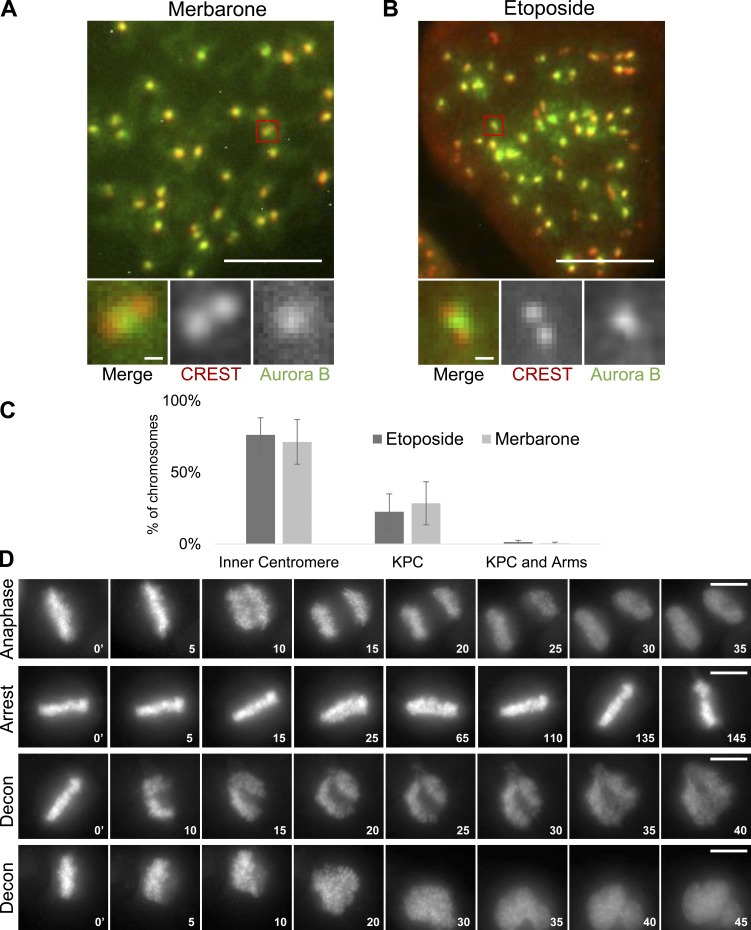
Having established that ICRF-193 treatment did not grossly affect condensed mammalian chromosome morphology, we immunostained with anti-Aurora B antibodies. As observed previously in control cells, Aurora B localized to a discrete focus at the inner centromere of each chromatid pair (Adams et al., 2000; Bischoff et al., 1998; Gassmann et al., 2004; Hindriksen et al., 2017; Nozawa et al., 2010; Terada et al., 1998; Vagnarelli and Earnshaw, 2004; Fig. 5 A). In contrast, ICRF-193 treatment induced recruitment of Aurora B to chromosome arms, similar to the observations in XEE. Aurora B was diminished at inner centromeres and instead was enriched at KPCs. To quantify this, we first categorized chromosomes based on localization of Aurora B to inner centromeres versus chromosome arms and KPCs (Fig. 5 B). Second, we performed line scans across pairs of KPCs and averaged the positions of the KPCs relative to the Aurora B signal (Fig. 5 C). Both analyses revealed a pronounced recruitment of Aurora B to chromosome arms and a redistribution from inner centromere to the KPCs induced by ICRF-193. Finally, we analyzed metaphase cells (not synchronized with nocodazole). Here, we made line scans across KPCs perpendicular to the kinetochore–kinetochore axis (Fig. 5 D). Quantification of CENP-A and Aurora B signals revealed that ICRF-193 treatment did not affect the CENP-A signal (Fig. 5 E), but significantly increased the Aurora B:CENP-A ratio at KPCs (Fig. 5 F). Altogether, the data reveal that catalytic inhibition of Topo II induced recruitment of Aurora B to KPCs and chromosome arms. Consistent with the lack of Topo II SUMOylation induced by merbarone, merbarone did not induce recruitment of Aurora B to chromosome arms and did so only weakly to KPCs (Fig. S3, A and C). Similar effects were observed upon treatment with etoposide, a Topo II poison that traps Topo II in a covalent complex with broken DNA (Fig. S3, B and C).
Figure 5.
Aurora B recruitment to KPCs and chromosome arms during ICRF-193 treatment in HeLa cells. (A) Representative immunofluorescent-stained images of pseudometaphase cells (nocodazole arrested) ± ICRF-193 treatment for 45 min. CREST, red; Aurora B, green. Line scans were done in the x and y axis. Bars, 10 µm (insets, 1 µm). (B) Quantification of immunofluorescent staining at centromeres/chromosome arms. Error bars, standard deviation. (C) Averaged plots of Aurora B and CREST signal intensities in line scans across centromeres in cells treated as in A. Each scan was normalized to the highest value. Control, n = 52 chromosomes from 25 cells; ICRF, n = 50 chromosomes from 31 cells. (D) Images showing line scans of Aurora B and CENP-A perpendicular to the kinetochore–kinetochore axis in metaphase cells (no nocodazole). Bars, 1 µm. (E) Quantification of peak CENP-A signals from D. Control, n = 15 cells and n = 124 kinetochores; ICRF-193, n = 12 cells and n = 44 kinetochores; P = 0.85. Data collected from three independent experiments. P values from Student’s t test. n.s., not significant. (F) Quantification of Aurora B/CENP-A ratios from D. Control, n = 40 cells and n = 204 kinetochores; ICRF-193, n = 42 cells and n = 172 kinetochores; ****, P = 2 × 10−39. Data collected from at least three independent experiments. P values from Student’s t test. All error bars, standard deviation.
Figure S3.
Topo II inhibitors etoposide and merbarone only weakly affect Aurora B localization in mitotic chromosomes. (A and B) Representative immunofluorescent-stained images of pseudometaphase HeLa cells (nocodazole arrested) after merbarone (200 µM) or etoposide (10 µM) treatment for 45 min. Bars, 10 µm (insets, 1 µm). CREST, red; Aurora B, green. (C) Quantification of immunofluorescent staining at centromeres/chromosome arms. Error bars, standard deviation. Etoposide, n = 873 chromosomes; merbarone, n = 683 chromosomes. Data were collected from at least three independent experiments. (D) Representative examples of live-cell imaging showing normal anaphase onset, metaphase arrest, and decondensation (Decon) of chromatin following attempted anaphase when Topo II is inhibited and chromosomes cannot segregate. Bars, 10 µm.
Aurora B is required for metaphase arrest upon Topo II catalytic inhibition
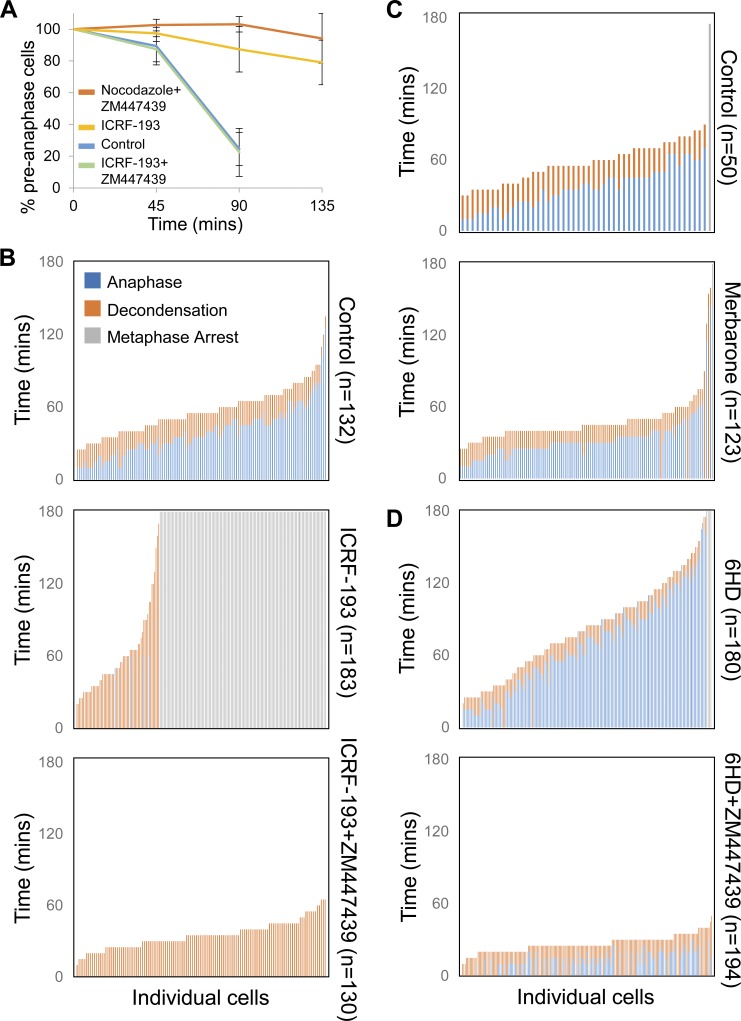
In metaphase, Aurora B is largely restricted to inner centromeres. Metaphase arrest can be induced by experimentally targeting Aurora B to KPCs (Liu et al., 2009). Since ICRF-193 recruited Aurora B to KPCs, we asked if Aurora B is required for metaphase arrest induced by ICRF-193. First, we collected pseudometaphases via nocodazole synchrony. After washing, we seeded them into medium with or without ICRF-193 and the Aurora B inhibitor ZM447439 and collected cells at intervals to make chromosome spreads. We used a modified spreading method that retains chromosome positions on the mitotic spindle, allowing accurate assessment of anaphase onset (Giménez-Abián et al., 2005). Control cells initiated anaphase after 45–90 min (Fig. 6 A). ICRF-193–treated cells delayed in metaphase as expected, but inhibition of Aurora B completely abolished this response. With ZM447439 and nocodazole, Aurora B inhibition was not able to immediately bypass the spindle assembly checkpoint, as reported previously (Ditchfield et al., 2003).
Figure 6.
Aurora B inhibition bypasses the metaphase checkpoint induced by Topo II catalytic inhibitors. (A) Quantification of percentage preanaphase cells following nocodazole arrest and release. Cells were treated with various combinations of drugs after release from nocodazole arrest, and preanaphase cells were counted at each time point to assess mitotic progression. For each treatment and time point, n = 400 cells. Data collected from three independent experiments. Error bars, standard deviation. (B–D) Live single-cell analysis of mitotic progression. Quantitation of time to anaphase and decondensation during drug treatments. Each vertical bar represents one cell, with each cell being scored for time to anaphase and decondensation from the start of the time course. Error bars, standard deviation. Data collected from three independent experiments.
In a second approach, we used live-cell analysis. HeLa cells were grown and imaged in normal culture conditions in an environmental chamber housed around an inverted microscope (example images are presented in Fig. S3 D). ICRF-193 was added and images were recorded at 5-min intervals to provide enough temporal resolution for accurate estimates of metaphase duration. We analyzed only cells that were in metaphase at the time of ICRF-193 and/or ZM447439 addition, to exclude effects of Topo II and Aurora B inhibition before metaphase. Finally, we included two other inhibitors of Topo II: merbarone, which did not induce Topo II SUMOylation, and 6-hydroxydaidzein (6HD), a naturally occurring catalytic inhibitor abundant in plants (Baechler et al., 2014). In controls, the interval from metaphase to anaphase was on average 39.2 min (Fig. 6 B). ICRF-193 addition in metaphase substantially delayed the onset of anaphase, and most cells remained arrested for the duration of the experiments. Consistent with the lack of Topo II SUMOylation after merbarone treatment, this inhibitor did not arrest cells in metaphase (Fig. 6 C). 6HD did induce metaphase arrest, although more weakly than ICRF-193 (Fig. 6 D). With both ICRF-193 and 6HD treatment, inhibition of Aurora B abolished the metaphase arrest (Fig. 6, B and D). Similarly, an alternative Aurora B inhibitor had the same effect (Fig. S4 A). Thus, the data are consistent with recruitment of Aurora B upon Topo II catalytic inhibition promoting metaphase arrest.
Figure S4.
Aurora B or Haspin inhibition bypasses the metaphase checkpoint induced by Topo II catalytic inhibitors. (A) Live single-cell analysis of mitotic progression. Quantitation of time to anaphase and decondensation during drug treatments. Hesperadin (1 µM), an Aurora B inhibitor (Hauf et al., 2003), and 5-ITu (10 µM), a Haspin inhibitor (De Antoni et al., 2012), were added to metaphase cells 10 min after ICRF-193 was added. Each vertical bar represents one cell, with each cell being scored for time to anaphase and decondensation from the start of the time course. Cells that remained arrested in metaphase for the duration of the experiment are indicated by a red bar. (B) Representative immunofluorescent-stained images of pseudometaphase HeLa cells (nocodazole arrested) treated with ICRF-193 ± 5-ITu for 45 min. CREST, red; Aurora B, green. After ICRF-193 treatment, Aurora B localizes to the KPCs and chromosome core (right). After ICRF-193 + 5-ITu treatment (left), Aurora B typically localizes diffusely on chromatin and is present on some KPCs. The histogram plot shows classification of Aurora B staining pattern. Error bars, standard deviation. Bars, 10 µm. Data were collected from at least three independent experiments.
H3T3p recruits Aurora B to KPCs and chromosome arms
Aurora B was recruited to KPCs and chromosome arms upon Topo II catalytic inhibition and is required for the coincident metaphase arrest. To understand the mechanism of Aurora B recruitment, we asked if it is recruited with the inner centromere protein (INCENP) subunit of the CPC to which Aurora B binds directly. In control pseudometaphase cells, INCENP localized to inner centromeres, as expected (Fig. 7, A and B). However, similar to Aurora B, INCENP was recruited to KPCs and chromosome arms after ICRF-193 treatment. This indicates that Aurora B is mobilized as a component of the CPC, where Aurora B binds to INCENP and bridging molecules tether INCENP to chromatin. One such bridging molecule is Survivin, which binds to INCENP and interacts with H3T3p via its BIR domain (Kelly et al., 2010; Wang et al., 2010; Jeyaprakash et al., 2011). In metaphase, H3T3p is enriched at inner centromeres and, in part, accounts for the specificity of CPC localization to inner centromeres (Hindriksen et al., 2017). Thus, we asked if de novo phosphorylation of H3T3 might account for Aurora B recruitment with ICRF-193. As expected, we detected H3T3p in control pseudometaphase cells between the KPCs of sister chromatids (Fig. 7, C and D). With ICRF-193, H3T3p localized to ectopic sites corresponding to KPCs and chromosome arms, similar to Aurora B and INCENP.
Figure 7.
INCENP and H3T3p at KPCs and chromosome arms after ICRF-193 treatment in mitosis. (A and C) Representative immunofluorescent-stained images of pseudometaphase HeLa cells (nocodazole arrested) ± ICRF-193 treatment for 45 min. Bars, 10 µm (insets, 1 µm). (A) CREST, red; INCENP, green. (C) CREST, red. H3T3p, green. (B and D) Quantification of immunofluorescent staining at centromeres/chromosome arms. Error bars, standard deviation. Data collected from three independent experiments.
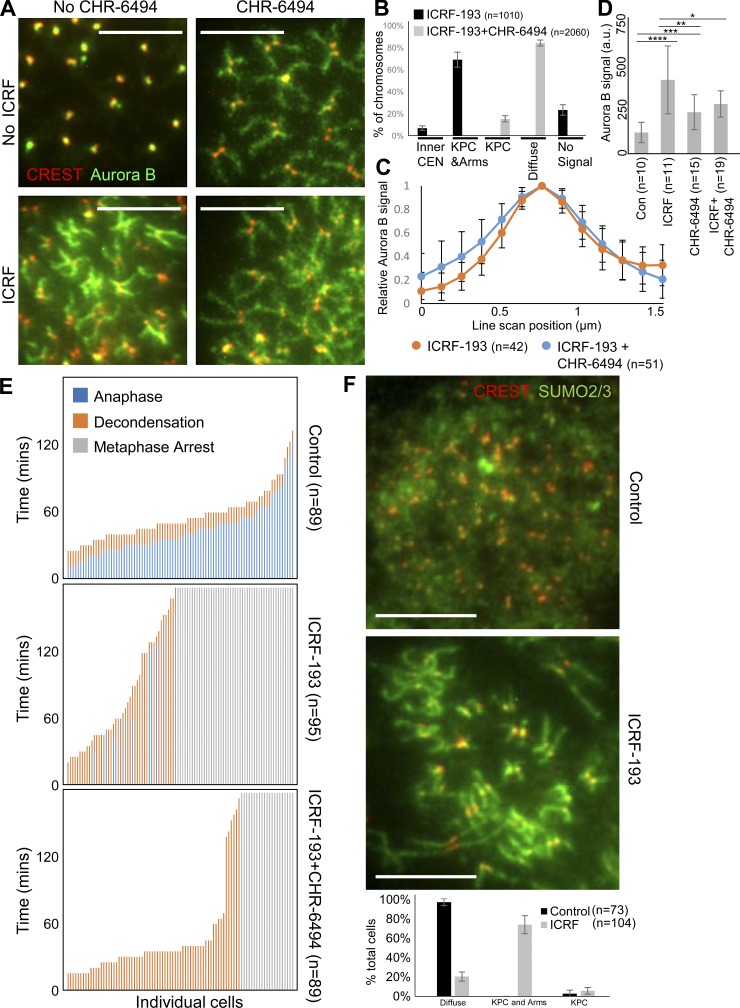
An explanation for these localization patterns is that Topo II catalytic inhibition induced H3T3p within nucleosomes at KPCs and chromosome arms, leading to CPC recruitment. To test this, we asked if Haspin, which phosphorylates H3T3 (Dai and Higgins, 2005; Wang et al., 2010), is required for recruitment of Aurora B to KPCs and chromosome arms. We combined ICRF-193 treatment with Haspin inhibitors, CHR-6494 (Fig. 8) or 5-ITu (Fig. S4 B). In pseudometaphases treated with ICRF-193 and the Haspin inhibitors, there was a qualitative difference in Aurora B localization. Compared with ICRF-193 treatment alone, Aurora B appeared more diffusely dispersed on chromatin, as observed previously after Haspin inhibition (Bekier et al., 2015; De Antoni et al., 2012; Kelly et al., 2010; Wang et al., 2010, 2012; Yamagishi et al., 2010). Categorization of cells based on this phenotype revealed a clear difference between cells treated with ICRF-193 alone and with ICRF-193 plus Haspin inhibitor (Figs. 8 B and S4 B). To gain quantitative information, we analyzed chromosomes based on two features observed to distinguish the samples. First, we measured the distribution of Aurora B across the width of chromosome arms, because inhibition of Haspin appeared to spread the Aurora B laterally. An averaged plot of Aurora B intensity indeed revealed a small difference in distribution (Fig. 8 C). Second, we measured the abundance of Aurora B on the chromosome arms. This revealed a significant reduction in Aurora B recruitment in the presence of CHR-6494 (Fig. 8 D). Together, the data are consistent with Haspin inhibition restricting the ICRF-193–induced recruitment of Aurora B to chromosomes. Thus, H3T3p likely contributes to Aurora B recruitment upon Topo II catalytic inhibition.
Figure 8.
Haspin promotes Aurora B localization to KPCs and chromosome arms during ICRF-193 treatment in HeLa cells. (A) Representative immunofluorescent-stained images of pseudometaphase HeLa cells (nocodazole arrested) ± ICRF-193 ± CHR-6494 treatment for 45 min. CREST, red; Aurora B, green. (B) Classification of Aurora B staining pattern. (C) Quantification of average Aurora B signal intensity spanning chromosome arms. Each scan was normalized to the highest value. ICRF, n = 42 chromosomes from 35 cells; Haspin, n = 51 chromosomes from 27 cells. (D) Quantification of Aurora B signal intensity on chromatin. P values from Student’s t test. *, P = 0.04; **, P = 0.01; ***, P = 0.001; ****, P = 0.0004. a.u., arbitrary unit. (E) Live single-cell analysis of mitotic progression. Quantitation of time to anaphase and decondensation during drug treatments. Each vertical bar represents one cell, with each cell being scored for time to anaphase and decondensation from the start of the time course. (F) Representative immunofluorescent-stained images of pseudometaphase HeLa cells (nocodazole arrested) ± ICRF-193 for 45 min. CREST, red; SUMO2/3, green. The individual channels are provided for reference in Fig. S5 A. The plot shows classification of SUMO2/3 staining pattern. Bars, 10 µm. Error bars, standard deviation. The images are representative examples of those collected from three independent experiments.
Haspin activity is required for ICRF-193–induced metaphase arrest
If the mechanism of metaphase arrest upon Topo II catalytic inhibition involves activation of Haspin at KPCs and chromosome arms, which mobilizes Aurora B to these sites, then Haspin ought to be required for the metaphase arrest. To test this, we used live-cell analysis, adding CHR-6494 at the same time as ICRF-193. Metaphase cells treated with CHR-6494 alone behaved similar to controls, spending on average 42.5 min in metaphase (not depicted). However, in the presence of ICRF-193, CHR-6494 bypassed the metaphase arrest in most cells (Fig. 8 E). The Haspin inhibitor 5-ITu more completely abolished the checkpoint response (Fig. S4 A). Together, the data indicate that Haspin is required for the metaphase arrest, presumably due to its ability to generate H3T3p at KPCs and chromosome arms for CPC binding.
Aurora B and SUMO2/3 are recruited to KPCs and the chromosome core upon Topo II catalytic inhibition
In XEE, Haspin binds specifically to SUMOylated Topo II (Yoshida et al., 2016). To find out if Topo II SUMOylation in human cells might induce Aurora B recruitment to KPCs and chromosome arms, we characterized SUMO2/3 localization in mitotic chromosomes after ICRF-193 treatment. In control pseudometaphases, weak diffuse signals were observed in the nucleoplasm after staining with anti-SUMO2/3 antibodies (Fig. 8 F). After ICRF-193 treatment, SUMO2/3 became prominently localized to chromosome arms and KPCs in most cells, similar to Aurora B localization (Fig. 8 F). This provides circumstantial evidence for a link between Topo IIα SUMOylation and Aurora B recruitment upon Topo II catalytic inhibition.
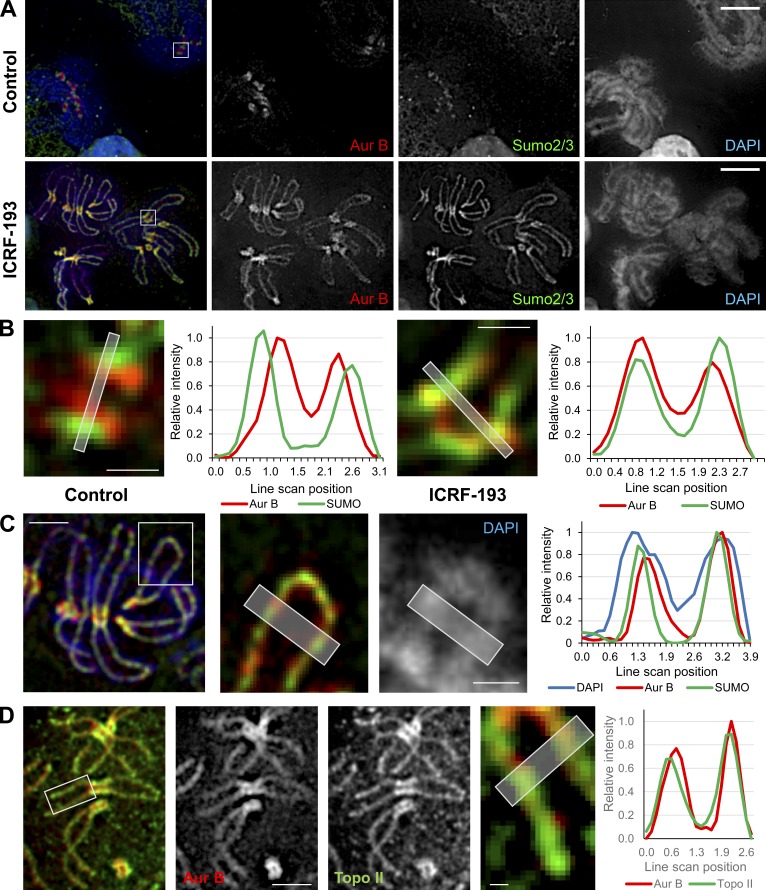
Although the patterns of Aurora B, INCENP, and SUMO2/3 staining after ICRF-193 treatment were similar, the small size of human chromosomes limited the accuracy with which this could be evaluated. To circumvent this, we examined mitotic chromosomes in M. muntjak cells, which have the largest chromosomes found in mammals. This allows higher-resolution observations of chromosome arms and centromeres. Pseudometaphase M. muntjak cells were prepared in the same manner as the HeLa cells, by synchrony with nocodazole, mitotic shake-off, and then a 45-min incubation in nocodazole alone or with the addition of ICRF-193. Similar results to HeLa mitotic chromosomes were observed in controls, where Aurora B and SUMO2/3 were enriched at centromeres, with Aurora B typically adopting a bilobed position internal to the SUMO2/3 (Fig. 9, A and B). After ICRF-193, the Aurora B and SUMO2/3 at centromeres were positioned more similarly (Fig. 9, A and B), suggesting relocalization of the Aurora B to a more lateral position. Strikingly, after ICRF-193 treatment, SUMO2/3 and Aurora B accumulated on chromosome arms (Fig. 9, A and C). The dimensions of M. muntjak chromosomes permitted analysis of sub–chromosome arm distribution. Line scans across the width of the arms revealed that, compared with the entire chromosome width, Aurora B and SUMO2/3 were restricted, occupying the central axial core regions of chromosome arms (Fig. 9 C). Costaining of Aurora B and SUMO2/3 confirmed that they adopted similar distributions along the chromosome arms and enriched at the axial core (Fig. 9, A–C). This was intriguing, because Topo II is known to isolate biochemically within the chromosome axial core (Earnshaw et al., 1985, Adachi et al., 1989). We therefore costained SUMO2/3 and Topo II, which revealed that they indeed have a similar restricted pattern consistent with localization to the axial core (Fig. 9 D). Thus, mobilization of Aurora B observed in XEE and human cells upon Topo II catalytic inhibition is conserved in M. muntjak cells, and Aurora B and SUMO2/3 are recruited to the central core regions of chromosome arms, where Topo II is abundant in mitosis.
Figure 9.
Aurora B and SUMO2/3 are recruited to the chromosome core upon Topo II catalytic inhibition. Representative immunofluorescent-stained images of M. muntjak cells arrested in mitosis with nocodazole then treated ± ICRF-193 for 45 min. The experiment was performed at least three times, and the images shown are a true representation of the staining patterns that were consistently observed. (A) Aurora B, red; SUMO2/3, green; DAPI (DNA), blue. Bars, 10 µm. (B) Magnification of kinetochores from boxed regions in A. Bars, 1 µm. (C) Right: Magnified chromosome arms from the boxed region in the image on the left. Bar, 4 µm (inset, 2 µm). (D) Aurora B, red; Topo IIα, green. Bars, 5 µm (inset, 0.5 µm). In B–D, the histogram plots show average signal intensities of a 5-pixel-wide line scan across the planes indicated in the magnified images. The line scans are representative examples from three independent experiments.
Evidence that Topo II CTD SUMOylation induces recruitment of Aurora B and promotes metaphase arrest
The immunostaining results could not provide a definitive link between Topo IIα CTD SUMOylation, Aurora B recruitment to chromosomes, and metaphase checkpoint activation. To test these relationships directly, we exchanged endogenous human Topo IIα with a mutant lacking the three conserved SUMOylation sites in the CTD. We used a system previously characterized to insert a single copy of a doxycycline (Dox)-inducible mCherry-Topo IIα or a mCherry-3KR-Topo IIα mutant allele into the 5q31.3 locus using Flp recombinase (Lane et al., 2013). We achieved efficient depletion of endogenous Topo IIα using siRNAs targeting the 3′ UTR, and we simultaneously induced the mCherry-Topo IIα proteins with Dox (Fig. 10 A). We then synchronized the cells with nocodazole and, after mitotic shake-off, incubated them for a further 45 min with or without ICRF-193. Without ICRF-193 treatment, immunostaining revealed that mCherry-Topo IIα or mCherry-3KR Topo IIα cells had similar patterns of Aurora B localization, largely restricted to inner centromeres (Fig. 10, B and C). After ICRF-193 treatment, the mCherry-Topo IIα cells efficiently recruited Aurora B to chromosome arms. However, there was a statistically significant defect in the ability of the mCherry-3KR Topo IIα cells to recruit Aurora B (an approximate sevenfold increase in the percentage of cells that were unable to recruit Aurora B to the chromosome arms and KPC; P ≤ 0.03). This suggests that Topo IIα CTD SUMOylation at these conserved residues promotes Aurora B recruitment.
Figure 10.
Topo II CTD SUMOylation promotes Aurora B recruitment and metaphase checkpoint activation. (A) Western blot showing endogenous Topo II depletion achieved after RNAi in HeLa EM2-11ht cell lines and induction (Dox) of exogenous mCherry-Topo II and mCherry-TopoII-3KR. (B) Classification of Aurora B localization in cells treated as in A, with endogenous Topo II depleted and exogenous Topo II expressed. (C) Representative examples of Aurora B localization categories as in B. (D and E) Analysis of metaphase duration in live HeLa cell lines in A, after depletion of endogenous Topo II and expression of exogenous alleles. (D) Quantification of the mCherry-Topo II signal at the first time point captured for each of the cells analyzed in E. a.u., arbitrary unit. Bars, 10 µm. Error bars, standard deviation. Data collected from three independent experiments.
Next, we used the same strategy to test if the 3KR mutant had a functional checkpoint. After depletion of endogenous Topo IIα and induction of the exogenous Topo IIα alleles, cells in metaphase were analyzed immediately following addition of ICRF-193. Mitotic progression in these single cells using time-lapse microscopy was followed by directly observing the exogenous mCherry-Topo IIα or mCherry-3KR Topo IIα proteins. Thus, we could quantify the level of the exogenous Topo IIα proteins in the same cohort of cells analyzed to determine metaphase duration. Most single cells did not vary more than twofold in their levels of exogenous Topo IIα expression, and the distributions were similar (Fig. 10 D). However, exogenous mCherry-Topo IIα–expressing cells had a substantially more proficient checkpoint response in the presence of ICRF-193 than did the mCherry-3KR-Topo IIα–expressing cells (Fig. 10 E). These experiments therefore provide direct support to the hypothesis that SUMOylation of the conserved Topo IIα CTD acceptor lysine residues constitutes a molecular signal that recruits Aurora B to KPCs and the chromosome axial core and triggers activation of the metaphase checkpoint.
Discussion
The SPR has been studied for decades because it has extensive roles in cell division and is the target of major classes of anticancer drugs (Nitiss, 2009a,b; Wang, 2002). A yeast top2 mutant, top2-B44, which hydrolyzes ATP slowly, triggers activation of the metaphase checkpoint (Andrews et al., 2006; Furniss et al., 2013). Thus, progression through mitosis and Topo II activity are coupled. However, molecular insight is lacking into the mechanism of activation of the metaphase checkpoint when Topo II is perturbed.
Studies in mammalian cells revealed that this checkpoint may be conserved (Clarke et al., 2006; Skoufias et al., 2004). Analogous to the top2-B44 mutant, catalytic Topo II inhibitors (e.g., ICRF-193) inhibit ATP hydrolysis and activate a metaphase checkpoint in human cells (Clarke et al., 2006; Skoufias et al., 2004; Toyoda and Yanagida, 2006). In both cases, impaired ATP hydrolysis delays the enzyme in a particular structural conformation, within the SPR. This suggests that cells might detect the persistence of that Topo II conformer. Here we have provided evidence that SUMOylation of the Topo II CTD triggers the checkpoint, perhaps because the conformation of the enzyme is specifically recognized by SUMO ligases when ATP hydrolysis is slow.
In yeast, the checkpoint is transduced by the CTD of the enzyme, which possesses conserved SUMOylation sites (Bachant et al., 2002). Inhibition of Topo II with ICRF-193 in mitotic XEE induced Topo II CTD SUMOylation on these physiologically relevant sites (Figs. 1 and 2). This SUMOylation was conserved in mitotic HeLa cells treated with ICRF-193, which activated the metaphase checkpoint, but was not observed after treatment with merbarone, which did not activate the checkpoint (Figs. 6 and S5 B). How might Topo II CTD SUMOylation activate the downstream checkpoint factors? In both budding yeast and XEE, Topo II CTD SUMOylation can regulate Haspin kinase activity to promote H3T3 phosphorylation that recruits the CPC to mitotic centromeres (Edgerton et al., 2016; Yoshida et al., 2016). This mechanism relies on binding of Haspin to the SUMO2/3 moieties on the Topo II CTD. Thus, it is predicted that direct binding of Haspin to SUMOylated Topo II occurs upon ICRF-193 treatment in human cells.
Figure S5.
Topo II inhibitor ICRF-193 up-regulates Topo IIα SUMOylation and induces recruitment of SUMO2/3 to KPCs and chromosome arms in HeLa cells. (A) Representative images of pseudometaphase HeLa cells (nocodazole arrested) ± ICRF-193 for 45 min and then immunostained with CREST serum and anti-SUMO2/3 antibody. DAPI stain for DNA. These are individual channels from Fig. 8 F. Bars, 10 µm. The images are representative examples of those collected from at least three independent experiments. (B) DMSO-, ICRF-193–, and merbarone-treated mitotic chromosomes were isolated from HeLa cells and subjected to Western blotting. The mitotic SUMOylation and Topo SUMOylation were probed using the indicated antibodies. Histone H3 was probed as a loading control for the mitotic chromosomes. Percentage SUMOylation of Topo IIα was calculated from three independent experiments (n = 3). Error bars, standard deviation. *, P value from Student’s t test. **, Statistically significant difference, P ≤ 0.01.
When we mutated the three conserved SUMOylated sites in the Topo II CTD, activation of the checkpoint and recruitment of Aurora B were abrogated but not abolished (Fig. 10). This may indicate that additional CPC recruitment pathways are activated by ICRF-193 (Zhou et al., 2017; Goto et al., 2017). Another possibility is that additional SUMOylated CTD lysine residues contribute to Aurora B recruitment and activation. A recent study provided important insight into as many as 11 lysine residues that can be SUMOylated, as well as their functions in Topo II localization in mitosis (Antoniou-Kourounioti et al., 2019). Together with data presented here, the study made it clear that CTD SUMOylation serves at least two mitotic functions: Topo II localization and recruitment of Aurora B. These functions could be mediated by different SUMOylation sites, or the same SUMOylated residues may be capable of providing the protein–protein interactions needed for both functions.
In XEE, ICRF-193 simultaneously induced CTD SUMOylation and mobilization of Aurora B. This was conserved in human cells, where Aurora B was recruited away from inner centromeres to the KPC and chromosome arms and the metaphase checkpoint was activated. Aurora B has been experimentally targeted to the KPC using several approaches, including expression of a fusion between INCENP and the DNA binding domain of CENP-B (Hengeveld et al., 2017). This leads to forced activation of the metaphase checkpoint. Moreover, a pool of Aurora B is revealed near kinetochores after inhibition of Haspin, which causes Aurora B to be dispersed from chromatin (Bekier et al., 2015). Thus, when Topo II is catalytically inhibited, it is possible that cells similarly induce recruitment of Aurora B to the KPC to activate the checkpoint. However, another study reported that Aurora B kinase activity at the KPC is diminished upon Topo II inhibition, based on reduced phosphorylation of CENP-A (Coelho et al., 2008). This may indicate that Aurora B recruitment to the chromosome arms activates the checkpoint. It will be interesting to test if Aurora B recruitment to either arms or KPCs is sufficient for checkpoint activation.
A body of work has revealed that restriction of the CPC to inner centromeres in mitosis is based on feedback cycles (Hindriksen et al., 2017). Factors that recruit Aurora B are themselves regulated by Aurora B, e.g., phosphorylation by Aurora B enhances Haspin kinase activity (Ghenoiu et al., 2013; Wang et al., 2011; Zhou et al., 2014). Conversely, phosphorylation of Repo-Man by Aurora B protects the centromeric H3T3p from PP1γ-Repo-Man (Qian et al., 2011, 2013). Clustering of Aurora B promotes its maximum activity by inducing autophosphorylation (Kelly et al., 2007, Bishop and Schumacher, 2002). It is perhaps striking, then, that catalytic Topo II inhibition is able to break this cycle, release Aurora B from inner centromeres, and allow recruitment to KPCs and chromosome arms. It will be interesting to establish if H3T3p is sufficient for Aurora B recruitment to KPCs and chromosome arms. Previous work has established that Pds5 functions to recruit Aurora B by facilitating localization of Haspin to inner centromeres (Goto et al., 2017; Zhou et al., 2017). It will be important to test if Pds5 is recruited to KPCs and chromosome arms upon Topo II catalytic inhibition.
The ICRF-193–mediated metaphase checkpoint requires Aurora B activity, suggesting that up-regulation of Aurora B activity at the KPC and/or chromosome arms induces mitotic arrest. What molecules are phosphorylated by Aurora B to activate this checkpoint? If Aurora B at the KPC is responsible for checkpoint activation, we anticipate the downstream targets are Ndc80/Hec1, Dsn1, and CENP-E (Cheeseman et al., 2006; DeLuca et al., 2006; Welburn et al., 2010), and in that case, checkpoint activation likely occurs via microtubule detachment from kinetochores. If recruitment to the chromosome arms is required for checkpoint activation, Aurora B may have novel substrates.
In summary, we propose a model in which Topo II catalytic inhibition activates a metaphase checkpoint via Aurora B. Checkpoint activation involves Topo II CTD SUMOylation as an initial signaling step, which recruits Haspin kinase as a direct downstream mediator of Aurora B recruitment via H3T3p. We anticipate that this mechanism contributes to the timing of mitosis so that cells can monitor catenations and stalled SPR cycles. Studies revealed that Topo II catalytic inhibition in G2 cells delays entry into mitosis (Damelin and Bestor, 2007; Deming et al., 2001; Downes et al., 1994), but that this cell cycle response is frequently diminished in cancer cells (Brooks et al., 2014; Deiss et al., 2019; Franchitto et al., 2003; Jain et al., 2015). Intriguingly, a deficiency in the G2 checkpoint may make the metaphase checkpoint critical for survival of cancer cells (Brownlow et al., 2014; Deiss et al., 2019). Targeting the mechanism of metaphase arrest upon Topo II catalytic inhibition may differentially kill cancer cells.
Materials and methods
Plasmid construction, site-directed mutagenesis, recombinant protein expression and purification, and XEE antibodies and inhibitors
For full-length recombinant WT Topo IIα and 3KR Topo IIα, the coding sequence was subcloned into a modified pPIC3.5Kb vector, which had a calmodulin-binding protein–T7 tag sequence for N-terminal fusion (Ryu et al., 2010b; Yoshida et al., 2016). The recombinant proteins were expressed in GS 115 strain of Pichia pastoris yeast and purified. Frozen yeast cells were ground with dry ice in a coffee grinder followed by lysis using lysis buffer (150 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 30 mM Hepes, pH 7.8, 1 mM DTT, 0.1% Triton X-100, 5% glycerol, and 10 mM PMSF). The lysed samples were centrifuged at 25,000 g for 40 min. To capture the calmodulin-binding protein–tagged protein, supernatant was mixed with calmodulin-sepharose resin and rotated for 90 min at 4°C. The resins were washed with lysis buffer and eluted with 10 mM EGTA (GE Healthcare). The eluted fractions were checked by Coomassie staining and further purified by Q-sepharose anion-exchange chromatography (GE Healthcare). For 3KR Topo IIα, lysine-to-arginine substitution was done by site-directed mutagenesis using QuikChange II kit (Agilent) according to the manufacturer’s protocol.
Both guinea pig and rabbit polyclonal anti-SUMO2/3 antibodies were generated using full-length human SUMO2 as an antigen. Human SUMO2 cDNA was subcloned into pGEX4T-1 or pET28a using BamHI/XhoI sites. Expressed proteins were purified using either glutathione-sepharose (GST) affinity resin (GE Healthcare) or hexa histidine affinity resin (Talone Metal; Clontech/Takara). GST moiety on GST-SUMO2 was cleaved by thrombin (T7513; Sigma-Aldrich) to obtain full-length untagged SUMO2 for antigen. Hexa histidine–tagged SUMO2 was conjugated to N-hydroxysuccinimidyl (NHS)–sepharose (GE Healthcare) for antigen affinity column. cDNA fragments encoding Xenopus Topo IIα (C-terminus region, aa 1,358–1,579) and Xenopus PARP1 (N-terminus region, aa 1–150) were amplified and subcloned in pGEX4T-1 (GE Healthcare) and pET28a (Novagen). The recombinant proteins were expressed in Escherichia coli and purified by hexa histidine tag–based purification or glutathione affinity tag–based purification followed by ion-exchange chromatography. Polyclonal antibodies for Topo IIα and PARP1 were generated in rabbit by injecting hexa histidine–tagged recombinant proteins. The rabbit antisera were collected, and antibodies were affinity purified using NHS-Sepharose columns with their covalently bound GST-tagged antigens. In-house-made antibodies were used for Western blotting at 1:1,000 dilution. Commercial primary antibodies used in this study and their dilution for Western blotting were mouse monoclonal anti-histone H4-HRP (197517, 1:1,000; Abcam), mouse monoclonal anti-β-tubulin (T4026-2ML, 1:1,000; Sigma-Aldrich), mouse monoclonal anti-histone H3 (IB1B2, 1:2,000; Cell Signaling), and rabbit polyclonal anti-T7 tag (ab19291, 1:1,000; Abcam). Secondary antibodies included Licor-specific goat anti-rabbit IRDye 680RD (925-68071, 1:20,000), goat anti-rabbit IRDye 800CW (925-32211, 1:20,000), goat anti-mouse IRDye 800CW (925-32210, 1:20,000), and goat anti-rat IRDye 680 RD (925-68076, 1:20,000).
For immunofluorescence staining, anti-SUMO2/3 guinea pig polyclonal antibody was used at a 1:500 dilution. For CENP-A antibody, a synthesized peptide of the N-terminus of Xenopus CENP-A was used as an antigen, and then antibody was purified with an antigen affinity column with the antigen peptide conjugated to NHS-sepharose. Anti-Aurora B rabbit polyclonal antibody (1:500) was prepared using full-length X. laevis Aurora B kinase as an antigen, and anti-Claspin rabbit polyclonal antibody was prepared using a His6-T7–tagged N-terminus region of Claspin (aa 1–271). For Topo IIα staining, a commercial anti-Topo IIα/β mouse monoclonal antibody (M052-3, 1:500, MBL International) was used. The fluorescence labeled secondary antibodies goat anti-mouse IgG Alexa Fluor 568 (A11031, 1:500; Invitrogen), goat anti-rabbit IgG Alexa Fluor 568 (A11036, 1:500; Invitrogen), goat anti-rabbit IgG Alexa Fluor 488 (A11034, 1:500; Invitrogen), goat anti-guinea pig IgG Alexa Fluor 647 (A21450, 1:500; Invitrogen), and goat anti-chicken IgG Alexa Fluor 488 (A11039, 1:750; Invitrogen) were used for visualization in experiments using XEE.
The Topo II inhibitors used in this study, ICRF-193 (BML-GR332; Enzo Life Sciences), merbarone (445800; Calbiochem), and 6-hydroxydaidzein(4′,6,7-trihydroxyisoflavone; CAS#17817-31-1; Carbosynth), were dissolved in DMSO and stored at −20°C.
XEE assays
Xenopus demembraned sperm nuclei and low-speed cytostatic factor (CSF) arrested XEEs were prepared according to standard protocols (Kornbluth and Evans, 2001; Murray, 1991). In detail, sperm was isolated from dissected testis then treated with detergent (lysophosphatidylcholine, L4129; Sigma-Aldrich) containing buffer to obtain demembraned sperm nuclei. Corrected frog eggs were dejellied with cysteine-containing buffer, and then crushed by centrifugation in buffer containing EGTA to preserve CSF activity. For mitotic replicated chromosome isolation, CSF extracts were driven into interphase with the addition of 0.6 mM CaCl2. Demembraned sperm nuclei were added to interphase extract at either 4,000 sperm nuclei/µl (for Western blotting analysis) or 800 sperm nuclei/µl (for immunofluorescence analysis), then incubated for ∼45 min. Equal amounts of CSF XEEs were used to induce mitosis. To inhibit the mitotic SUMOylation, dnUbC9 was added to XEEs at 150 ng/µl at the onset of mitosis induction by addition of CFS XEEs. After incubating for 60∼90 min, followed by microscopic analysis of condensed mitotic chromosomes, Topo II inhibitors (ICRF-193 1 µM and merbarone 10 µM) were added into XEEs for 10 min.
For isolation of inhibitor-treated mitotic chromosomes, CSF XEEs were diluted with three times their volume of 0.5× CSF-XB containing 18 mM β-glycerophosphate, 0.25% Triton X-100, 10 mg/ml protease inhibitors (leupeptin, pepstatin, and chymostatin; EMD Millipore), and 0.2 µM okadaic acid (EMD Millipore). Diluted XEEs were layered on top of a 40% glycerol cushion and centrifuged at 10,000 g for 5 min at 4°C. Isolated mitotic chromosomes were boiled in SDS-PAGE sample buffer. Samples were resolved on 8–16% gradient gels and subjected to immunoblotting with the indicated antibodies. Signals were acquired using a LI-COR OdysseyFc digital imager, and quantification was performed using Image Studio Lite software.
Relative SUMOylation levels were calculated for each sample and normalized to the loading control histone H4. The mean and standard deviation of four independent experiments were calculated. Statistical analysis for each sample was calculated by Student’s t test of the mean. Percentage Topo IIα and PARP1 SUMOylation were calculated for each sample with respect to total protein levels. For the immunofluorescence staining, XEEs were diluted with three times their volume in immunofluorescence dilution buffer (0.5× CSF-XB containing 18 mM β-glycerophosphate and 250 mM sucrose) and fixed with an equal volume of fixation buffer (immunofluorescence dilution buffer containing 4% p-formaldehyde) for 6-min incubation at RT. Fixed chromosome samples were loaded onto 8 ml of a 40% glycerol cushion with coverslips at the bottom and spun at 6,000 g for 20 min at RT. Precipitated chromosomes on coverslips were postfixed with 1.6% p-formaldehyde in PBS, blocked with PBS containing 5% BSA, and processed for immunostaining with the indicated antibodies. For DNA staining, Hoechst 33342 dye (EMD Millipore) was used, and for mounting the specimen, Vectashield H-1000 medium (Vector Laboratories) was used. Images were acquired at 20°C using the Plan Apo 100×/1.4 objective lens on a Nikon TE2000-U microscope with a Retiga SRV charge-coupled device camera (QImaging) operated by Volocity imaging software (PerkinElmer). Adobe Photoshop (CS6) software was used to process the images for signal intensities and size according to journal policy. For immunofluorescence quantification, ImageJ software (National Institutes of Health) was used. 30 mitotic chromosomes for arm region SUMO2/3 signal were counted from three independent experiments. The statistical analysis for each sample was calculated by Student’s t test of the mean. For Aurora B mislocalization, distinct Aurora B foci at chromosome arms were counted for >25 chromosomes for four independent experiments. Statistical analysis for each sample was calculated by Student’s t test of the mean.
The immunodepletion/add-back experiment and chromosome isolation were performed exactly as previously described (Yoshida et al., 2016). Specifically, Topo IIα antibodies (1 mg/ml) were captured on protein A–conjugated magnetic beads (Thermo Fisher Scientific) at a 1:1 ratio. Anti-Topo IIα-Protein A beads were blocked with 5% BSA containing CSF-XB (100 mM KCl, 0.1 mM CaCl2, 2 mM MgCl2, 5 mM EGTA, 50 mM sucrose, and 10 mM Hepes, pH 7.8). To obtain >99% Topo IIα depletion, we used anti-Topo IIα-Protein A beads at a ratio of 1.1 µl to 1 µl CSF XEEs. Protein A Dynabeads and XEEs were incubated at RT for 15 min followed by 15-min incubation on ice, repeated twice. For add-back, purified recombinant proteins were added to immunodepleted extracts similar to endogenous Topo IIα, which was confirmed by immunoblotting. For mitotic chromosome isolation, demembraned sperm nuclei were added to CSF XEEs and incubated for 60 to ∼90 min. Chromosomes were isolated as described above.
HeLa cell Topo IIα SUMOylation assay
For the analysis of mitotic chromosomes, HeLa cells were synchronized with 2 mM thymidine for 18 h. Cells were released from thymidine into fresh medium, and 6 h later, 0.1 µg/ml nocodazole was added to cells. 4 h after nocodazole addition, mitotic cells were collected by mitotic shake-off and released with 7 µM ICRF-193–containing fresh medium for 20 min. For mitotic chromosome isolation, cells were lysed with lysis buffer (250 mM sucrose, 20 mm Hepes, 100 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 0.2% Triton X-100, 1:2,000 lysophosphatidylcholine, and 15 mM iodoacetic acid), and lysed cells were layered on a 40% glycerol cushion as for chromosome isolation from XEEs. Later, isolated mitotic chromosomes were boiled in SDS-PAGE sample buffer, resolved on 8–16% gradient gels, and subjected to immunoblotting with the indicated antibodies. Signals were acquired using a LI-COR Odyssey Fc machine.
In vitro SUMOylation Förster resonance energy transfer (FRET) assay
The CyPet-tagged SUMO2G, CyPet-SUMO2GG, and YPet-tagged Topo IIα CTDs were subcloned in pET28a with His6-tag. CyPet and YPet coding sequences were amplified by PCR from pH2B-CyPet-FHA2-AuroraB Substrate-YPet-IRES-puro2b obtained through Addgene (Wurzenberger et al., 2012). Recombinant proteins were expressed in Rosseta2(DE3) strain, and then purified using Cobalt affinity beads (Talon Beads; Takara Bio). The imidazole-eluted His6-tag proteins were further purified using ion-exchange columns. The enzymes used for in vitro SUMOylation FRET assays, E1 complex (Aos1/Uba2 heterodimer), PIASy, Ubc9, and the dominant-negative form of Ubc9 (dnUbc9-C93S/L97S), were expressed in BL21 (DE3) or Rosetta2 (DE3) bacteria (Azuma et al., 2003, 2005; Yoshida et al., 2016). E1 complex and heterodimer of Aos1 and Uba2 were expressed in E. coli using pRSF plasmid. Hexa histidine tag was fused to the N-terminus of Uba2. The E1 complex was purified by hexa histidine affinity resin followed by SUMO affinity column and Q-sepharose anion-exchange column. Ubc9-containing bacterial lysate was initially fractionated by DEAD-sepharose equilibrated with buffer containing 100 mM NaCl. The unbound fraction containing Ubc9 was further purified by SP-sepharose cation-exchange column followed by S-100 gel filtration column. Hexa histidine–tagged PIASy proteins were purified by hexa histidine affinity resin, and then further cleaned by Q-sepharose and SP-sepharose column chromatography.
In vitro SUMOylation FRET reactions were performed at 25°C for 30 min using 30 nM E1 Uba2/Aos1, 60 nM E2 Ubc9, 60 nM PIASy, 2.5 µM Cypet-tagged SUMO2G and SUMO2GG, 0.5 µM Ypet-tagged Topo IIα, 2.5 mM ATP, 1 µM ICRF-193, and 10 µM merbarone. The reactions were stopped by addition of 10 mM EDTA. There were different concentrations for PIASy and inhibitors and different time points used for time course experiments, as indicated in Fig. S2. Reaction buffer consisted of 20 mM Hepes, pH 7.8, 100 mM NaCl, 5 mM MgCl2, 5% glycerol, 0.05% Tween 20, and 1 mM DTT. The emission spectrum at 530 nM was measured using a fluorescence spectrophotometer (Varian Cary Eclipse) with excitation at 414 nM. The emission spectrum was recorded at 530 nM for three independent experiments. Standard deviation was calculated for statistical analysis, and error bars represent standard deviation.
Cell fixation and staining
Mammalian cells were grown to ∼70–80% confluence on a 10-cm dish and arrested for 3 h in 100 ng/ml nocodazole. Cells were then treated with 14 µM ICRF-193 and/or 2 µM CHR-6494 for 45 min before isolating mitotic cells by mitotic shake-off into 500 µl of PBS. Cells were treated for 5 min with 1 ml of water to hypotonic shock the cells for better chromosome spreading. 500 µl of the hypotonic mixture was added to a cytology funnel attached to a glass slide with a Cytospin (Thermo Fisher Scientific) clip. The assembly was spun for 5 min at 2,000 rpm with maximum acceleration. Working quickly so that the cells did not fully dry out, glass slides were removed from the clip, and a ring was drawn around the deposited cells with a Super-PAP pen to form a hydrophobic barrier to help keep the fixing and staining reagents on the cells. There is a fine balance between letting the Super-Pap solution dry (∼30–60 s) and not letting the cells dry out. When the Super-PAP dried sufficiently, the cells were fixed with 3.8% PFA for 5 min. Cells were permeated with 0.05% Triton X-100 in PBS for 5 min. Cells were then treated with 50 mM ammonium chloride in PBS for 2 min to quench PFA. Cells were washed with PBS-0.01% Triton X-100 and then blocked with 0.1% casein in PBS-0.01% Triton X-100. Cells were stained overnight at 4°C with primary antibodies. Following primary antibody staining, cells were washed three times with PBS-0.01% Triton X-100 and stained with secondary antibody for 1 h. Following secondary antibody staining, cells were washed twice with PBS-0.01% Triton X-100 and once with PBS-DAPI and mounted with ProLong Gold (Invitrogen). The following primary antibodies were used for staining: CREST antibody (human, CS1058, 1:200; Cortex Biochem), anti-Aurora B antibody (mouse, 611083, 1:200; BD Transduction), anti–CENP-A antibody (rabbit, 2186S, 1:200; Cell Signaling Technology), anti-INCENP antibody (rabbit, I5283, 1:2,000; Sigma-Aldrich), and anti-H3T3-Phos antibody (rabbit, ab78351, 1:1,000; Abcam). Secondary antibodies used were goat anti-human (A11013, Alexa Fluor 488 [green], 1:200; Thermo Fisher Scientific), goat anti-rabbit (A11011, Alexa Fluor 568, 1:200; Thermo Fisher Scientific), goat anti-mouse (A11004, Alexa Fluor 568, 1:200; Thermo Fisher Scientific), goat anti-guinea pig (A21450, Alexa Fluor 647, 1:200; Thermo Fisher Scientific), and goat anti-mouse (405322, Alexa Fluor 647, 1:200; BioLegend).
Image analysis of HeLa cells
To assess the localization patterns of Aurora B (Figs. 5, 8, 10, and S3), H3T3p (Fig. 7), INCENP (Fig. 7), and SUMO2/3 (Fig. 8) at centromeres and chromosome arms, individual chromosomes were assigned to categories based on the following criteria: (a) inner centromere, the signal was restricted to between the two CENP-A/CREST foci; (b) KPC, the signal overlapped with the two CENP-A/CREST foci; (c) chromosome arms, the signal was enriched at the core of each chromatid rather than across the entire chromatid width; and (d) diffuse, the signal was not localized as described in categories a–c, but was present homogeneously on the chromatin.
To quantify the amount of Aurora B at KPCs (Fig. 5, D–F), metaphase HeLa cells (no nocodazole) were treated with ICRF-193 or DMSO for 45 min, fixed, and stained immediately using antibodies against CENP-A and Aurora B. Images were analyzed by quantifying the signal in line scans across the KPCs perpendicular to the kinetochore–kinetochore axis. The minimum and maximum values were extracted from the resulting histogram plots, and background-subtracted maximum signal intensities were calculated as maximum minus minimum. The Aurora B signal was then expressed relative to the maximum minus minimum CENP-A signal as a ratio.
Somatic cell microscopy
Cells were imaged using a DeltaVision microscope system (Applied Precision) based on an Olympus IX-71 inverted microscope and either an Olympus UPLSAPO 100×, 1.40-NA, oil objective for stained images (mounted with ProLong Gold, Invitrogen) or an Olympus LUCPLFLN 20×, 0.45-NA, air objective (with CO2-independent culture medium; Thermo Fisher Scientific) for live-cell imaging. A CoolSNAP HQ2 camera (Photometrics) was used to capture images. Soft Worx (version 6.1.3; Applied Precision) software was used to acquire images. Images were cropped and contrast enhanced using ImageJ (Fiji) and Photoshop (Adobe). All imaging was done at 37°C. Fluorochromes used are listed for each secondary antibody in the Cell fixation and staining section.
Quantification of metaphase checkpoint duration
For nocodazole arrest and release time courses, Hela cells were grown to ∼80% confluence in a 15-cm dish. Cells were then arrested in 100 ng/ml nocodazole for 3 h. After arrest, cells were washed once with warm medium, and then mitotic cells were shaken off and aliquoted into 14 3-cm dishes and treated with various drugs. At each 45-min time point, cells were trypsinized and then fixed for preparation of chromosome spreads (Giménez-Abián and Clarke, 2009; Giménez-Abián et al., 2005). Briefly, cells were washed with PBS and then resuspended in 2 ml of PBS in a 15-ml conical tube. To hypotonic shock the cells, 3 ml of water was added for 5 min. To the hypotonic mixture, 10 ml of freshly made Carnoy’s fixation solution (25% glacial acetic acid and 75% methanol) was added to the cells, and the mixture was spun at 1,000 g for 5 min. After the spin, the supernatant was carefully aspirated off the pellet, taking care not to disturb the delicate pellet. Cells were then washed three times with 15 ml Carnoy’s solution, taking care to gently but thoroughly resuspend cells during each wash. After the three washes, the cells were incubated overnight at RT in 15 ml Carnoy’s solution. The next day, the cells were spun down and resuspended in 0.2 ml Carnoy’s solution and dropped onto a glass microscope slide. After the slides dried, they were stained with 5% Giemsa solution for 7 min and then mounted with Entellan (Merck).
For the live-cell imaging time courses, HeLa cells with H2B-GFP were plated on a 4-chamber, 3-cm glass-bottom plate and grown overnight to a confluence of ∼80%. Medium was replaced with CO2-independent medium. Cells were then treated with various drugs and quickly transferred to the heated Delta Vision microscope chamber. Cells were imaged for 3 h, and cells that were in metaphase at the beginning of the time course were scored for time to anaphase and chromosome decondensation. In the presence of Topo II inhibitors, chromatids are unable to separate. In this case, metaphase checkpoint bypass is observed as a failed attempt at chromosome segregation in anaphase, followed by decondensation of the chromosomes in telophase. Additional examples of these phenotypes have been provided previously (Giménez-Abián and Clarke, 2009). Inhibitors were used at the following concentrations: ZM447439 (8 µM), ICRF-193 (14 µM), merbarone (200 µM), and 6HD (120 µg/ml).
Endogenous Topo IIα knockdown and mutant expression
The S2F-IMCg-F-mCherry-Topo IIα plasmid (Lane et al., 2013) was mutagenized using QuikChange II XL (200521; Agilent) to generate S2F-IMCg-F-mCherry-3KR-Topo IIα (K1240R, K1267R, K1287R). The latter was then inserted into the Flp site at the 5q31.3 locus of the HeLa EM2-11ht cell line, as follows. Cells were transfected with S2F vector, FLP recombinase vector, and puromycin vector in a 1:2:1 ratio by weight using Lipofectamine 2000. After 24 h, transfected cells were selected by treating the cells with 3.3 µg/ml of puromycin overnight. Cells that had properly inserted the S2F vector were selected by treating the cells with 100 µM ganciclovir, with medium changes every 2–3 d until colonies developed. The inserted Topo IIα was induced with 250 ng/ml Dox for 24 h before experiments. Endogenous Topo IIα was knocked down using the hs.Ri.TOP2A.13.1 (Integrated DNA Technologies) oligonucleotide (5′-GGUGUUUUUAGUACAAGAUUUGAUGUCUUGUACUAA-3′) targeting the 3′ UTR. Cells were grown to ∼40% confluence in a 3-cm dish to account for cells doubling during the RNAi treatment. The oligonucleotide was prepared for transfection with 50 pmol of oligonucleotide, 2 µl Lipofectamine RNAiMAX Reagent (Life Technologies), and 150 µl OptiMEM as carrier for the oligonucleotide and RNAiMAX per manufacturer’s protocol. Cells were treated for 24 h before experiments.
Online supplemental material
Fig. S1 presents data from a FRET-based in vitro SUMOylation assay using recombinant Topo II CTD as substrate. Fig. S2 shows chromosome morphologies after inhibition of Topo II or Haspin in mitosis. Fig. S3 shows effects of merbarone and etoposide on Aurora B localization and examples of images from live-cell microscopy. Fig. S4 shows metaphase checkpoint bypass with Aurora B and Haspin inhibitors. Fig. S5 shows individual channels from the image in Fig. 8 F and presents Western blotting results that show increased SUMOylation of Topo II after treating HeLa cells with ICRF-193 but not merbarone.
Acknowledgments
We thank the Clarke, Gardner, Titus, and Courtemanche laboratories at the University of Minnesota and the Azuma laboratory at the University of Kansas for discussions. We thank the University of Minnesota Imaging Center, Guillermo Marquez, and Mark Sanders for help with microscopy.
This study was funded by National Institutes of Health grants R01GM112793 and R01GM130858; the University of Minnesota Foundation; the University of Minnesota Office of the Vice President for Research; and University of Kansas Strategic Initiative Grant (INS0073115).
The authors declare no competing financial interests.
Author contributions: N. Pandey and D. Keifenheim performed the majority of the experiments and contributed to experimental design and manuscript preparation. M.M. Yoshida initiated the study and performed preliminary experiments. V. Hassebroek performed the experiments in Fig. S5 B. C. Soroka performed the experiments in Fig. S1. D.J. Clarke performed the experiments in Figs. 6 A, 9 F, and S2. Y. Azuma and D.J. Clarke directed the research in each laboratory and cowrote the manuscript.
References
- Adachi Y., Käs E., and Laemmli U.K.. 1989. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 8:3997–4006. 10.1002/j.1460-2075.1989.tb08582.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.R., Wheatley S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., and Earnshaw W.C.. 2000. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10:1075–1078. 10.1016/S0960-9822(00)00673-4 [DOI] [PubMed] [Google Scholar]
- Agostinho M., Santos V., Ferreira F., Costa R., Cardoso J., Pinheiro I., Rino J., Jaffray E., Hay R.T., and Ferreira J.. 2008. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 68:2409–2418. 10.1158/0008-5472.CAN-07-2092 [DOI] [PubMed] [Google Scholar]
- Andrews C.A., Vas A.C., Meier B., Giménez-Abián J.F., Díaz-Martínez L.A., Green J., Erickson S.L., Vanderwaal K.E., Hsu W.S., and Clarke D.J.. 2006. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 20:1162–1174. 10.1101/gad.1367206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou-Kourounioti M., Mimmack M.L., Porter A.C.G., and Farr C.J.. 2019. The Impact of the C-Terminal Region on the Interaction of Topoisomerase II Alpha with Mitotic Chromatin. Int. J. Mol. Sci. 20:1238 10.3390/ijms20051238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Arnaoutov A., and Dasso M.. 2003. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 163:477–487. 10.1083/jcb.200304088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Arnaoutov A., Anan T., and Dasso M.. 2005. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24:2172–2182. 10.1038/sj.emboj.7600700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J., Alcasabas A., Blat Y., Kleckner N., and Elledge S.J.. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 9:1169–1182. 10.1016/S1097-2765(02)00543-9 [DOI] [PubMed] [Google Scholar]
- Baechler S.A., Schroeter A., Walker J., Aichinger G., and Marko D.. 2014. Oxidative metabolism enhances the cytotoxic and genotoxic properties of the soy isoflavone daidzein. Mol. Nutr. Food Res. 58:1269–1281. 10.1002/mnfr.201300531 [DOI] [PubMed] [Google Scholar]
- Bekier M.E., Mazur T., Rashid M.S., and Taylor W.R.. 2015. Borealin dimerization mediates optimal CPC checkpoint function by enhancing localization to centromeres and kinetochores. Nat. Commun. 6:6775 10.1038/ncomms7775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D.L., and Murray A.W.. 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 159:453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff J.R., Anderson L., Zhu Y., Mossie K., Ng L., Souza B., Schryver B., Flanagan P., Clairvoyant F., Ginther C., et al. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052–3065. 10.1093/emboj/17.11.3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J.D., and Schumacher J.M.. 2002. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277:27577–27580. 10.1074/jbc.C200307200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks K., Chia K.M., Spoerri L., Mukhopadhyay P., Wigan M., Stark M., Pavey S., and Gabrielli B.. 2014. Defective decatenation checkpoint function is a common feature of melanoma. J. Invest. Dermatol. 134:150–158. 10.1038/jid.2013.264 [DOI] [PubMed] [Google Scholar]
- Brownlow N., Pike T., Zicha D., Collinson L., and Parker P.J.. 2014. Mitotic catenation is monitored and resolved by a PKCε-regulated pathway. Nat. Commun. 5:5685 10.1038/ncomms6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Drubin D.G., and Barnes G.. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199–203. 10.1083/jcb.200201052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Clarke D.J., Vas A.C., Andrews C.A., Díaz-Martínez L.A., and Giménez-Abián J.F.. 2006. Topoisomerase II checkpoints: universal mechanisms that regulate mitosis. Cell Cycle. 5:1925–1928. 10.4161/cc.5.17.3200 [DOI] [PubMed] [Google Scholar]
- Coelho P.A., Queiroz-Machado J., Carmo A.M., Moutinho-Pereira S., Maiato H., and Sunkel C.E.. 2008. Dual role of topoisomerase II in centromere resolution and aurora B activity. PLoS Biol. 6:e207 10.1371/journal.pbio.0060207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., and Higgins J.M.. 2005. Haspin: a mitotic histone kinase required for metaphase chromosome alignment. Cell Cycle. 4:665–668. 10.4161/cc.4.5.1683 [DOI] [PubMed] [Google Scholar]
- Dai J., Sultan S., Taylor S.S., and Higgins J.M.. 2005. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19:472–488. 10.1101/gad.1267105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M., and Bestor T.H.. 2007. The decatenation checkpoint. Br. J. Cancer. 96:201–205. 10.1038/sj.bjc.6603537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Maffini S., Knapp S., Musacchio A., and Santaguida S.. 2012. A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. J. Cell Biol. 199:269–284. 10.1083/jcb.201205119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss K., Lockwood N., Howell M., Segeren H.A., Saunders R.E., Chakravarty P., Soliman T.N., Martini S., Rocha N., Semple R., et al. 2019. A genome-wide RNAi screen identifies the SMC5/6 complex as a non-redundant regulator of a Topo2a-dependent G2 arrest. Nucleic Acids Res. 47:2906–2921. 10.1093/nar/gky1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Deming P.B., Cistulli C.A., Zhao H., Graves P.R., Piwnica-Worms H., Paules R.S., Downes C.S., and Kaufmann W.K.. 2001. The human decatenation checkpoint. Proc. Natl. Acad. Sci. USA. 98:12044–12049. 10.1073/pnas.221430898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., and Taylor S.S.. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280. 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K.C., and Berger J.M.. 2007. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 450:1201–1205. 10.1038/nature06396 [DOI] [PubMed] [Google Scholar]
- Downes C.S., Clarke D.J., Mullinger A.M., Giménez-Abián J.F., Creighton A.M., and Johnson R.T.. 1994. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells/. Nature. 372:467–470. 10.1038/372467a0 [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Halligan B., Cooke C.A., Heck M.M., and Liu L.F.. 1985. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100:1706–1715. 10.1083/jcb.100.5.1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton H., Johansson M., Keifenheim D., Mukherjee S., Chacón J.M., Bachant J., Gardner M.K., and Clarke D.J.. 2016. A noncatalytic function of the topoisomerase II CTD in Aurora B recruitment to inner centromeres during mitosis. J. Cell Biol. 213:651–664. 10.1083/jcb.201511080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A., Oshima J., and Pichierri P.. 2003. The G2-phase decatenation checkpoint is defective in Werner syndrome cells. Cancer Res. 63:3289–3295. [PubMed] [Google Scholar]
- Furniss K., Vas A.C., Lane A., and Clarke D.J.. 2009. Assaying topoisomerase II checkpoints in yeast. Methods Mol. Biol. 582:167–187. 10.1007/978-1-60761-340-4_14 [DOI] [PubMed] [Google Scholar]
- Furniss K.L., Tsai H.J., Byl J.A., Lane A.B., Vas A.C., Hsu W.S., Osheroff N., and Clarke D.J.. 2013. Direct monitoring of the strand passage reaction of DNA topoisomerase II triggers checkpoint activation. PLoS Genet. 9:e1003832 10.1371/journal.pgen.1003832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Carvalho A., Henzing A.J., Ruchaud S., Hudson D.F., Honda R., Nigg E.A., Gerloff D.L., and Earnshaw W.C.. 2004. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166:179–191. 10.1083/jcb.200404001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghenoiu C., Wheelock M.S., and Funabiki H.. 2013. Autoinhibition and Polo-dependent multisite phosphorylation restrict activity of the histone H3 kinase Haspin to mitosis. Mol. Cell. 52:734–745. 10.1016/j.molcel.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Abián J.F., and Clarke D.J.. 2009. Cytological analysis of chromosome structural defects that result from topoisomerase II dysfunction. Methods Mol. Biol. 582:189–207. 10.1007/978-1-60761-340-4_15 [DOI] [PubMed] [Google Scholar]
- Giménez-Abián J.F., Díaz-Martínez L.A., Wirth K.G., Andrews C.A., Giménez-Martín G., and Clarke D.J.. 2005. Regulated separation of sister centromeres depends on the spindle assembly checkpoint but not on the anaphase promoting complex/cyclosome. Cell Cycle. 4:1561–1575. 10.4161/cc.4.11.2146 [DOI] [PubMed] [Google Scholar]
- Goto Y., Yamagishi Y., Shintomi-Kawamura M., Abe M., Tanno Y., and Watanabe Y.. 2017. Pds5 Regulates Sister-Chromatid Cohesion and Chromosome Bi-orientation through a Conserved Protein Interaction Module. Curr. Biol. 27:1005–1012. 10.1016/j.cub.2017.02.066 [DOI] [PubMed] [Google Scholar]
- Hari K.L., Cook K.R., and Karpen G.H.. 2001. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15:1334–1348. 10.1101/gad.877901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengeveld R.C.C., Vromans M.J.M., Vleugel M., Hadders M.A., and Lens S.M.A.. 2017. Inner centromere localization of the CPC maintains centromere cohesion and allows mitotic checkpoint silencing. Nat. Commun. 8:15542 10.1038/ncomms15542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindriksen S., Lens S.M.A., and Hadders M.A.. 2017. The Ins and Outs of Aurora B Inner Centromere Localization. Front. Cell Dev. Biol. 5:112 10.3389/fcell.2017.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C.K., Roychoudhury S., and Majumder H.K.. 2015. Selective killing of G2 decatenation checkpoint defective colon cancer cells by catalytic topoisomerase II inhibitor. Biochim. Biophys. Acta. 1853:1195–1204. 10.1016/j.bbamcr.2015.02.021 [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A.A., Basquin C., Jayachandran U., and Conti E.. 2011. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure. 19:1625–1634. 10.1016/j.str.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Sampath S.C., Maniar T.A., Woo E.M., Chait B.T., and Funabiki H.. 2007. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 12:31–43. 10.1016/j.devcel.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., and Funabiki H.. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S., and Evans E.K.. 2001. Analysis of apoptosis using Xenopus egg extracts. Curr. Protoc. Cell Biol. Chapter 11:Unit 11.12. [DOI] [PubMed] [Google Scholar]
- Lane A.B., Giménez-Abián J.F., and Clarke D.J.. 2013. A novel chromatin tether domain controls topoisomerase IIα dynamics and mitotic chromosome formation. J. Cell Biol. 203:471–486. 10.1083/jcb.201303045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., and Lens S.M.. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353. 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., and Dasso M.. 2017. The SUMO Pathway in Mitosis. Adv. Exp. Med. Biol. 963:171–184. 10.1007/978-3-319-50044-7_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605. 10.1016/S0091-679X(08)60298-8 [DOI] [PubMed] [Google Scholar]
- Nitiss J.L. 2009a DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 9:327–337. 10.1038/nrc2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J.L. 2009b Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 9:338–350. 10.1038/nrc2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R.S., Nagao K., Masuda H.T., Iwasaki O., Hirota T., Nozaki N., Kimura H., and Obuse C.. 2010. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12:719–727. 10.1038/ncb2075 [DOI] [PubMed] [Google Scholar]
- Petsalaki E., Akoumianaki T., Black E.J., Gillespie D.A., and Zachos G.. 2011. Phosphorylation at serine 331 is required for Aurora B activation. J. Cell Biol. 195:449–466. 10.1083/jcb.201104023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Lesage B., Beullens M., Van Eynde A., and Bollen M.. 2011. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr. Biol. 21:766–773. 10.1016/j.cub.2011.03.047 [DOI] [PubMed] [Google Scholar]
- Qian J., Beullens M., Lesage B., and Bollen M.. 2013. Aurora B defines its own chromosomal targeting by opposing the recruitment of the phosphatase scaffold Repo-Man. Curr. Biol. 23:1136–1143. 10.1016/j.cub.2013.05.017 [DOI] [PubMed] [Google Scholar]
- Ryu H., Al-Ani G., Deckert K., Kirkpatrick D., Gygi S.P., Dasso M., and Azuma Y.. 2010a PIASy mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J. Biol. Chem. 285:14415–14423. 10.1074/jbc.M109.074583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Furuta M., Kirkpatrick D., Gygi S.P., and Azuma Y.. 2010b PIASy-dependent SUMOylation regulates DNA topoisomerase IIalpha activity. J. Cell Biol. 191:783–794. 10.1083/jcb.201004033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Yoshida M.M., Sridharan V., Kumagai A., Dunphy W.G., Dasso M., and Azuma Y.. 2015. SUMOylation of the C-terminal domain of DNA topoisomerase IIα regulates the centromeric localization of Claspin. Cell Cycle. 14:2777–2784. 10.1080/15384101.2015.1066537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., Lacroix F.B., Andreassen P.R., Wilson L., and Margolis R.L.. 2004. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell. 15:977–990. 10.1016/j.molcel.2004.08.018 [DOI] [PubMed] [Google Scholar]
- Smith C.A., McAinsh A.D., and Burroughs N.J.. 2016. Human kinetochores are swivel joints that mediate microtubule attachments. eLife. 5:e16159 10.7554/eLife.16159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Yong-Gonzalez V., Kikuchi Y., and Strunnikov A.. 2006. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 172:783–794. 10.1534/genetics.105.047167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Tatsuka M., Suzuki F., Yasuda Y., Fujita S., and Otsu M.. 1998. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17:667–676. 10.1093/emboj/17.3.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda Y., and Yanagida M.. 2006. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell. 17:2287–2302. 10.1091/mbc.e05-11-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., and Earnshaw W.C.. 2004. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 113:211–222. 10.1007/s00412-004-0307-3 [DOI] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., and Higgins J.M.. 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 330:231–235. 10.1126/science.1189435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., Daum J.R., Patnaik D., Kateneva A.V., Gorbsky G.J., and Higgins J.M.. 2012. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J. Cell Biol. 199:251–268. 10.1083/jcb.201205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., van der Waal M.S., Patnaik D., Lens S.M., and Higgins J.M.. 2011. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr. Biol. 21:1061–1069. 10.1016/j.cub.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430–440. 10.1038/nrm831 [DOI] [PubMed] [Google Scholar]
- Wang J.C. 2007. Unlocking and opening a DNA gate. Proc. Natl. Acad. Sci. USA. 104:4773–4774. 10.1073/pnas.0701070104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R. III, Lampson M.A., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392. 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C., Held M., Lampson M.A., Poser I., Hyman A.A., and Gerlich D.W.. 2012. Sds22 and Repo-Man stabilize chromosome segregation by counteracting Aurora B on anaphase kinetochores. J. Cell Biol. 198:173–183. 10.1083/jcb.201112112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., and Watanabe Y.. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243. 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yoshida M.M., Ting L., Gygi S.P., and Azuma Y.. 2016. SUMOylation of DNA topoisomerase IIα regulates histone H3 kinase Haspin and H3 phosphorylation in mitosis. J. Cell Biol. 213:665–678. 10.1083/jcb.201511079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.D., Goeres J., Zhang H., Yen T.J., Porter A.C., and Matunis M.J.. 2008. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell. 29:729–741. 10.1016/j.molcel.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Liang C., Chen Q., Zhang Z., Zhang B., Yan H., Qi F., Zhang M., Yi Q., Guan Y., et al. 2017. The N-Terminal Non-Kinase-Domain-Mediated Binding of Haspin to Pds5B Protects Centromeric Cohesion in Mitosis. Curr. Biol. 27:992–1004. 10.1016/j.cub.2017.02.019 [DOI] [PubMed] [Google Scholar]
- Zhou L., Tian X., Zhu C., Wang F., and Higgins J.M.. 2014. Polo-like kinase-1 triggers histone phosphorylation by Haspin in mitosis. EMBO Rep. 15:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]