Abstract
The hippocampus is well established as an essential brain center for learning and memory. Within the hippocampus, recent studies show that area CA2 is important for social memory, and is an anomaly compared to its better understood neighboring region, CA1. Unlike CA1, CA2 displays a lack of typical synaptic plasticity, enhanced calcium buffering and extrusion, and resilience to cell death following injury. Although recent studies have identified multiple molecular markers of area CA2, the proteins that mediate the unique physiology, signaling, and resilience of this region are unknown. Using a transgenic GFP-reporter mouse line that expresses eGFP in CA2, we were able to perform targeted dissections of area CA2 and CA1 for proteomic analysis. We identified over 100 proteins with robustly enriched expression in area CA2 compared to CA1. Many of these proteins, including RGS14 and NECAB2 have already been shown to be enriched in CA2 and important for its function, while many more merit further study in the context of enhanced expression in this enigmatic brain region. Furthermore, we performed a comprehensive analysis of the entire data set (>2300 proteins) using a weighted protein co-expression network analysis (WPCNA). This identified eight distinct co-expressed patterns of protein co-enrichment associated with increased expression in area CA2 tissue (compared to CA1). The novel data set we present here reveals a specific CA2 hippocampal proteome, laying the groundwork for future studies and a deeper understanding of area CA2 and the proteins mediating its unique physiology and signaling.
Keywords: CA2, CA1, hippocampus, RGS14, proteome, calcium, synaptic plasticity, systems biology, protein-protein interaction network, actin
Graphical Abstract
INTRODUCTION
The hippocampus is well established as serving essential roles in the processing of learning and memory. Anatomically, this brain region is composed of the Cornu Ammonis (CA), a strip of pyramidal neurons, and the dentate gyrus (DG), which is composed of granule cells. The pyramidal cell-containing CA regions can be further divided into distinct areas CA1, CA2, and CA3, based on anatomical differences. The original, anatomical definition of CA2 indicated a portion of the CA between CA1 and CA3 contained large pyramidal neurons like those in CA3, but that lacked thorny excrescences indicative of mossy fiber synapses on CA3 neurons from the DG1. However, the extent of DG axon projection into the CA regions appears to be unreliable for distinguishing CA3 from CA2 as it differs among species2–4. Indeed, in rodents, DG neurons have been shown to project not only to CA3, but also to CA2 neurons2. Thus a more reliable marker of the distinction between CA3 and CA2 has been the expression of proteins specific to CA25. Many of these mRNAs and/or proteins have been identified including regulator of G protein signaling 14 (RGS14)6–8, amigo-29, Purkinje cell protein 4 (PCP4)3, 10, alpha-actinin 2 (ACTN2)11, 12, and striatal-enriched protein-tyrosine phosphatase (STEP)13. These proteins have overlapping expression and many have been used to reliably identify a large proportion of CA2 neurons in functional studies examining physiology14 and behavior15 associated with CA2.
Far and away, the largest number of studies focusing on long-term potentiation (LTP) and synaptic plasticity in the hippocampus have done so looking at CA3>CA1 synapses16. Notably, until recently, CA2 was largely ignored in many functional studies of the hippocampus. Like neurons in CA1, neurons in CA2 also receive synaptic input from CA3, but oddly, LTP could not be induced at these synapses under the same conditions that elicit LTP in CA16, 14 This lack of LTP in CA2 has been shown to be due to the selective expression of RGS146, the unusually robust calcium buffering and extrusion properties of CA2 pyramidal cells17, and the presence of perineuronal nets containing aggrecan18, though how these mechanisms may interact are unclear. Interestingly, LTP could be induced in the distal dendrites of CA2 neurons, which receive input from the entorhinal cortex19. One of the most notable differences between CA2 and surrounding regions is the resistance of CA2 neurons to cell death following a variety of insults. Area CA2 is resistant to cell loss in temporal lobe epilepsy as well as status epilepticus20, 21. CA2 neurons are resistant to cell death following ischemia22–24 and traumatic brain injury25, 26. The mechanism(s) by which area CA2 resists injury following such insults is unknown, but likely some of the same properties that make CA2 synapses resistant to LTP may also contribute to CA2 neurons being resistant to cell death.
In order to better understand the differences between CA2 and its neighboring hippocampal regions, we initiated studies to define differences in protein expression between CA2 and CA1. Multiple proteins identified to be enriched in CA2 have already been shown to mediate unique aspects of CA2 signaling and physiology including aggrecan18 and RGS146 which each serve roles in the suppression of LTP in CA2. Additionally, both agonists of the vasopressin 1b receptor27 and antagonists of the adenosine A1 receptor28, 29 have been shown to allow or induce potentiation of postsynaptic responses in CA2, respectively, albeit through different mechanisms. To further our understanding of area CA2, it is vital to know what proteins are preferentially expressed in this region. Although multiple studies have focused on differences between the CA3 and CA1 regions of the hippocampus30, 31, to date, there has been no published study focused on either the transcriptome or proteome of the CA2 region.
Here we provide a differential proteomic analysis focused on proteins enriched in the CA2 region over CA1. Using mice expressing enhanced green fluorescent protein (eGFP) under control of the amigo-2 promoter as a guide, we dissected out dorsal CA2 and CA1 and performed mass spectrometry (MS)-based proteomics using both label-free and isobaric tandem mass tag approaches. Of the 7000+ proteins surveyed, we identified over 100 proteins with consistently and robustly higher expression in CA2 (vs. CA1), including established markers of area CA2. Many of these differentially expressed proteins are associated with calcium ion binding and synaptic plasticity, providing possible targets for study in respect to the resistance of CA2 neurons to cell death following injury as well. Furthermore, we identify eight separate co-expressed modules enriched in CA2 (vs. CA1). These modules contain proteins already known to be expressed highly in CA2 as well as new targets that merit further study. By combining the differential expression data with co-enrichment (co-expression) network analysis, we reveal a specific CA2 (vs. CA1) proteome that can be used by us and other interested researchers to identify functional protein networks that contribute to the unique physiology and signaling of CA2.
METHODS
Mouse model for hippocampus dissections
C57B6 mice heterozygous for GFP expression under the CA2-enriched amigo-2 promoter were used for the experiment. Mice were deeply anesthetized with Fatal-Plus before brains were removed and dissected on dry ice. Hippocampi were isolated and the dorsal portion of the hippocampus was used to collect tissue from areas CA1 and CA2, with CA2 being identified through GFP fluorescence. The dissection method for experiments 1 and 3 (n=8 each) was pooling of CA2 or CA1-enriched coronal slabs, while dissection for experiment 2 (n=6) relied upon dissection along the whole length of the hippocampus. Experiment 2 mice were 4-month old females whereas the mice used for experiments 1 and 3 were 2-month old males.
Tissue homogenization and digestion
Procedures for tissue homogenization were performed essentially as described32. In total, 22 samples for either of the two regions of hippocampus were analyzed; all 11 CA1 samples were matched to the corresponding 11 CA2-derived tissue samples. Approximately 100 mg (wet tissue weight) of brain tissue was homogenized in 8 M urea lysis buffer (8 M urea, 100 mM NaHPO4, pH 8.5) with HALT protease and phosphatase inhibitor cocktail (ThermoFisher) using a Bullet Blender (NextAdvance). Each Rino sample tube (NextAdvance) was supplemented with ~100 μL of stainless steel beads (0.9 to 2.0 mm blend, NextAdvance) and 500 μL of lysis buffer. Frozen samples were then placed into the bullet blender (in 4 °C cold room). The samples were homogenized for 2 full 5 min cycles and the lysates were transferred to new Eppendorf Lobind tubes. Each sample was then sonicated for 3 cycles consisting of 5 s of active sonication at 30% amplitude followed by 15 s on ice. Samples were then centrifuged for 5 min at 15 000 g and the supernatant was transferred to a new tube. Protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce). For protein digestion, 100 μg of each sample was aliquoted and volumes normalized with additional lysis buffer. Samples were reduced with 1 mM dithiothreitol (DTT) at room temperature for 30 min, followed by 5 mM iodoacetamide (IAA) alkylation in the dark for another 30 min. Samples were then diluted 8-fold with 50 mM triethylammonium bicarbonate (TEAB). Lysyl endopeptidase (Wako) at 1:100 (w/w) was added and digestion continued overnight. Trypsin (Promega) was then added at 1:50 (w/w) and digestion was carried out for another 12 h. The peptide solutions were acidified to a final concentration of 1% formic acid (FA) and 0.1% triflouroacetic acid (TFA) and desalted with a C18 Sep-Pak column (Waters). Each Sep-Pak column was activated with 1 mL of methanol, washed with 1 mL of 80% acetonitrile, and equilibrated with 2×1 mL 0.1% TFA. The samples were then loaded and each column was washed with 2×1 mL 0.1% TFA. Elution was performed with 2 rounds of 400 μL of 50% acetonitrile. Peptides were recovered by drying eluates under vacuum with a chilled vapor trap (Labconco).
Liquid-chromatography coupled to mass spectrometry (LC-MS/MS) for label free quantitation (LFQ) experiments
For experiments 1 and 2, tryptic peptides (2 μg) from CA1 and CA2-specific samples were resuspended in peptide loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile) containing 0.2 pmol of isotopically labeled peptide calibrants (ThermoFisher, #88321). Peptide mixtures were separated on a self-packed C18 (1.9 um Dr. Maisch, Germany) fused silica column (25 cm x 75 μM internal diameter; New Objective, Woburn, MA) by a NanoAcquity UHPLC (Waters, Milford, FA) and monitored on a Q-Exactive Plus mass spectrometer (ThermoFisher Scientific, San Jose, CA). Elution was performed over a 140-minute gradient at a rate of 300 nL/min with buffer B ranging from 1% to 65% in experiment 1 and from 3% to 60% in experiment 2 (buffer A: 0.1% formic acid and 5% DMSO in water, buffer B: 0.1 % formic and 5% DMSO in acetonitrile). The mass spectrometer cycle was programmed to collect one full MS scan followed by 10 data dependent MS/MS scans. The MS scans [m/z ranges 400–1600 (experiment 1) and 400–1500 (experiment 2), 400,000 AGC, 50 ms maximum ion collection time] were collected at a resolution of 120,000 at m/z 200 in profile mode and the MS/MS spectra [0.7 m/z isolation width, 30% HCD collision energy, 10,000 AGC target, with maximum ion time of 35 ms (experiment 1) and 60 ms (experiment 2)] were acquired at a resolution of 15,000 (experiment 1) or 30,000 (experiment 2) at m/z 200. Dynamic exclusion was set to exclude previous sequenced precursor ions and their isotopes for 30 seconds within a 10 ppm window. Precursor ions with +1, and +8 or higher charge states were excluded from sequencing. Experiment 1 RAW files are provided [Data Link 1: SAGE Bionetworks https://dx.doi.org/10.7303/syn16805615 ]; as are experiment 2 RAW files [Data Link 2: SAGE Bionetworks https://dx.doi.org/10.7303/syn16805616 ]. One technically replicated RAW file derived from peptide re-injection in experiment 2 (ca1_31_B.RAW) was generated and analyzed in downstream MaxQuant analysis, but not considered further.
MaxQuant for label-free proteome quantification
Experiment 1 and 2 RAW data files for the 14 samples plus one technical replicate were analyzed using MaxQuant v1.5.2.8 with Thermo Foundation 2.0 for RAW file reading capability. Raw files for each experiment were analyzed and summarized in separate runs of MaxQuant, but with the same database and search parameters as follows. The search engine Andromeda was used to build and search a concatenated target-decoy UniProt Knowledgebase (UniProtKB) peptide database containing both Swiss-Prot and TrEMBL mouse reference protein sequences (54,490 target sequences), plus 245 contaminant proteins included as a parameter for Andromeda search within MaxQuant33. Methionine oxidation (+15.9949 Da) (up to 5 allowed each per peptide), and protein N-terminal acetylation (+42.0106 Da) were variable modifications; cysteine was assigned a fixed carbamidomethyl modification (+57.0215 Da). Only fully tryptic peptides were considered with up to 2 miscleavages in the database search. A precursor mass tolerance of ±10 ppm was applied prior to mass accuracy calibration and ±4.5 ppm after internal MaxQuant calibration. Other search settings included a maximum peptide mass of 6,000 Da, a minimum peptide length of 6 residues, 0.05 Da tolerance for high resolution MS/MS scans. The false discovery rate (FDR) for peptide spectral matches, proteins, and site decoy fraction were all set to 1 percent. Quantification settings were as follows: re-quantify with a second peak finding attempt after protein identification has completed; match full MS1 peaks between runs; a 0.7 min retention time match window was used after an alignment function was found with a 20 min retention time search space. The label free quantitation (LFQ) normalization algorithm in MaxQuant34, 35 was used for protein quantitation. The quantitation method only considered razor and unique peptides for protein level quantitation. All tab-separated text MaxQuant outputs, parameters, and summary files are available for experiment 1 [Data Link 3: SAGE Bionetworks https://dx.doi.org/10.7303/syn16805840 ], and experiment 2 [Data Link 4: SAGE Bionetworks https://dx.doi.org/10.7303/syn16805857 ]. The FASTA UniprotKB database used for all 3 experiments is provided [Data Link 5: SAGE Bionetworks https://dx.doi.org/10.7303/syn17056501.1 ].
Tandem mass tag (TMT) peptide labeling
For experiment 3, complete digestion of all samples was assumed and an aliquot equivalent to 20 μg was taken from each of the coronal slab-dissected, pooled samples and combined to make a global internal standard (GIS) for mixed CA1 and CA2 regions of mouse hippocampus. All peptide mixtures were dried down under vacuum. One batch using the 10-plex TMT kit (ThermoFisher) was used to label the 8 samples and 2 GIS mixture replicates. Sample arrangement by reporter channel is as follows: 127N: CA1_WT; 127C: CA1_540; 128N: CA1_542; 128C: CA1_545; 129N: CA2_WT; 129C: CA2_540; 130N: CA2_542; 130C: CA2_545. TMT channels 126 and 131 were used to label global internal standard (GIS) replicates. Information about age and sex of mice used is provided in Supplemental Table 3. Labeling was performed according to manufacturer’s protocol. Briefly, each sample (80 μg of peptides per sample/channel) was resuspended in 100 μL of 100 mM TEAB buffer. The TMT labeling reagents were equilibrated to room temperature and 41 μL anhydrous acetonitrile was added to each reagent channel and softly vortexed for 5 min. Peptide suspensions were transferred to the corresponding TMT channels and incubated for 1 h at room temperature. The reaction was quenched with 8 μl of 5% hydroxylamine. To ensure complete labeling, select channels from each batch were analyzed by LC-MS/MS according to previously published methods36. All 10 channels were then combined and dried by vacuum to~500 μL. Sep-Pak desalting was performed and the elution was dried to completeness.
Electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) fractionation for 10-plex TMT
Dried samples were resuspended in 100 μL of ERLIC buffer A (90% acetonitrile with 0.1% acetic acid) and loaded onto a PolyWAX LP column (20 cm by 3.2 mm packed with 300 Å 5 μm beads from PolyLC Inc) as reported previously37. An Agilent 1100 HPLC system consisting of a degasser, a binary pump, an autosampler, a microflow UV detector, and a fraction collector was used to carry out the fractionation. The gradient was from 0 to 50% ERLIC buffer B (30% ACN with 0.1% FA) over 45 min. A total of 44 fractions were collected and then combined to 22 fractions. Final fractions 1 to 21 consisted of alternating combinations (1 and 21, 2 and 22, etc.) and fraction 22 consisted of the last fractions (43 to 44) similar to the method previously described37.
LC-MS/MS and TMT data acquisition on an Orbitrap Fusion mass spectrometer
Assuming equal distribution of peptide concentration across all 22 ERLIC fractions, 40 μL of loading buffer (0.1% TFA) was added to each of the fractions and 2 μL (2 μg equivalent) was separated on 25 cm long 75 μm internal diameter fused silica columns (New Objective, Woburn, MA) packed in-house with 1.9 μm Reprosil-Pur C18-AQ resin (Dr. Maisch). The LC-MS platforms consisted of a Dionex RSLCnano UPLC coupled to an Orbitrap Fusion mass spectrometer with a Flex nano-electrospray ion source (ThermoFisher). Sample elution was performed over a gradient of 3 to 30% Buffer B (0.1% formic acid in ACN) over 105 min, from 30 to 60% B over 20 min, and from 60 to 99% B over 5 min at 300 nL/min. The column was reconditioned with 99% B for 10 min at a flow rate of 500 nl/min and equilibrated with 1% B for 15 min at a flow rate of 350 nl/min. The Orbitrap Fusion (Thermo Scientific) was operated in positive ion data-dependent mode with synchronous precursor selection (SPS)-MS3 analysis for reporter ion quantitation. The full scan was performed in the range of 380–1500 m/z at nominal resolution of 120 000 at 200 m/z and AGC set to 2×105, followed by selection of the most intense ions above an intensity threshold of 5,000 for collision-induced dissociation (CID)-MS2 fragmentation in the linear ion trap with 35% normalized collision energy and 30 000 resolution. The isolation width was set to 1.5 m/z with a 0.5 m/z offset. Each of the top 10 fragment ions for each peptide MS2 was notched out with an isolation width of 2 m/z and co-fragmented to produce MS3 scans analyzed in the Orbitrap at a nominal resolution of 60,000 after higher-energy collision dissociation (HCD) fragmentation at a normalized collision energy of 65%. All resulting raw files (n=22) are provided [Data Link 6: SAGE Bionetworks https://dx.doi.org/10.7303/syn16805617 ].
TMT Protein identification and quantification
Raw data files from the Orbitrap Fusion were processed using Proteome Discover (version 2.1). MS/MS spectra were searched against the same UniProtKB mouse proteome database as used for LFQ (54,489 total sequences plus EGFP). We chose to include both Swiss-Prot and TrEMBL sequences in the database as the additional depth provided by ERLIC fractionation enables the sequencing of rare protein isoforms (i.e., proteoforms) that may not be appreciated in mouse brain. The respective FASTA database used in this study is deposited on Synapse (https://www.synapse.org). SEQUEST parameters were specified as: trypsin enzyme, two missed cleavages allowed, minimum peptide length of 6, TMT tags on lysine residues and peptide N-termini (+ 229.162932 Da) and carbamidomethylation of cysteine residues (+ 57.02146 Da) as fixed modifications, oxidation of methionine residues (+ 15.99492 Da) and deamidation of asparagine and glutamine (+0.984 Da) as a variable modification, precursor mass tolerance of 20 ppm, and a fragment mass tolerance of 0.6 Daltons. Peptide spectral match (PSM) error rates were determined using the target-decoy strategy coupled to Percolator38 modeling of true and false matches. Reporter ions were quantified from MS3 scans using an integration tolerance of 20 ppm with the most confident centroid setting. An MS2 spectral assignment false discovery rate (FDR) of less than 1% was achieved by applying the target-decoy strategy. Following spectral assignment, peptides were assembled into proteins and were further filtered based on the combined probabilities of their constituent peptides to a final FDR of 1%. In cases of redundancy, shared peptides were assigned to the protein sequence with the most matching peptides, thus adhering to principles of parsimony. The search results and TMT quantification are provided [Data Link 7: SAGE Bionetworks https://dx.doi.org/10.7303/syn16805770 ].
Statistics for comparison of CA2 to CA1 quantification across three experiments
LFQ or TMT normalized abundances summarized for protein-level quantification based on parsimoniously assembled razor plus unique peptides in MaxQuant or Proteome Discoverer, respectively, were considered for statistical comparisons of CA1 to CA2 sample abundances within experiment. For the TMT normalized abundances, the ratio of the sample channel normalized reporter abundance to that of the average for the two GIS channels was calculated prior to log2-transformation. Likewise, LFQ abundances were log2-transformed. Then Student’s T-test was performed for each of the three within-experiment comparisons using two tails and assuming equal variance for the two sample groups. Log2(CA1 mean / CA2 mean) was also calculated for each experiment. To get a meta-analysis statistic across all three experiments, Fisher’s negative log sum/Chi2 was calculated in R using the survcomp::combine.test function with method=”fisher” parameter. The alternate method for calculating meta-p, “z.transform”, was also used separately (Supplemental Table 1, master summary of 3-experiment meta-analysis). Meta-p was only calculated if all available experiments’ measurements of log2(CA2)-log2(CA1) mean log2 abundance ratio were equivalent in sign, or direction of change, and at least two of the three experiments measured the protein. Fisher’s meta-p was minus-log10-transformed and used for volcano calculations. Of 2,947 proteins measured in at least 2 experiments, 1,625 (55%) were consistent in directionality; and more importantly, the standard deviation from mean of 0.000 for inconsistent (sign of change) proteins was only 0.107 log2 units, whereas for consistently measured proteins, the SD was 0.306; SD increased further to 0.384 for the 740 Fisher meta-p significant (meta-p < 0.05) proteins measured consistently in at least 2 experiments. Thus, the volcano fold-change cutoff for proteins with a meta-p calculation was set to <−0.3219 or >0.3219, which corresponds to a 25% fold enrichment minimum in CA1 or CA2, and which is more than 1.96xSD for inconsistently measured proteins, and more than 1.96xSD of all null TMT measurements, SD[log2(GIS2/GIS1)]=0.137. Greater than 1.96-fold of these SD represent a probability of <0.05 that Gaussian population tails overlap with markers deemed reliable, defining an upper bound on the false positive rate for the CA2-enriched markers determined by Fisher’s meta-p. Finally, 134 additional proteins only measured in the deeper TMT experiment were found significantly CA2-enriched by Student’s T-test and are summarized in Supplemental Table 2.
Chord plots for selected brain-relevant gene ontologies (e.g. not cardiac) were selected from GO Elite 1.2.5 output for CA2 and CA1-enriched proteins (min enrichment 1.25-fold and min 2-experiment metaP < 0.01, enriched versus the background of all 2+ experiment identified gene product proteins. GOplot R package was used to visualize the results.
Data Normalization and Batch Correction for Weighted protein Co-expression Network Analysis (WPCNA)
A 3,758-protein by 22-sample matrix was assembled for all common Uniprot identifiers across the data sets representing proteins with 10 or fewer (<50%) missing measurements. LFQ abundance data was downsampled, dividing by the lowest non-missing LFQ abundance in both experiment 1 and experiment 2. After, LFQ intensities were on average 5-fold greater than TMT reporter intensities in our 3 experiments (2 LFQ, and 1 TMT); thus, prior to robust regression of batch effects, we set the means of all abundances to be roughly equivalent, further dividing all LFQ abundances by 5. Then all normalized abundances including TMT normalized abundances were log2-transformed, prior to R impute::impute.knn imputation, and sva::ComBat Bayesian batch effect removal, modelling hippocampus region as a common, protected variable, and removing inter-experiment systematic variance. Then originally missing values were returned to the matrix following ComBat.
WPCNA Networks and Analyses of Batch-Normalized Abundance
A protein network was built using the above corrected log2-abundance matrix after finding scale free toplogy at power Beta=6.6 using WPCNA::pickSoftThreshold function. WPCNA::blockwiseModules was used to automate network building with the following parameters, mergeCutHeight=0.07, replaceMissing=TRUE, minimum module size 17, deepSplit=4, corType=”bicor”, networkType=” signed”, PAMstage=TRUE, pamRespectsDendro=TRUE, reassignThreshold=0.05, and maxBlockSize greater than the number of proteins in the network. Resulting module eigenproteins were then correlated to hippocampus region as a binary trait, and selected module structures were drawn as iGraphs as previously published32, and mapping edges which had been curated in the BioGRID interactome database previously, much as previously published39.
The R boxplot() function was used to generate boxplots of CA1 and CA2 marker genes determined by meta-significance analysis using the same normalized abundance data generated from LFQ and TMT abundances for WGCNA. Boxes extend from the twenty-fifth to 75th percentiles of intra-group variance, with whiskers at 5th and 95th percentiles, excluding outliers.
RESULTS
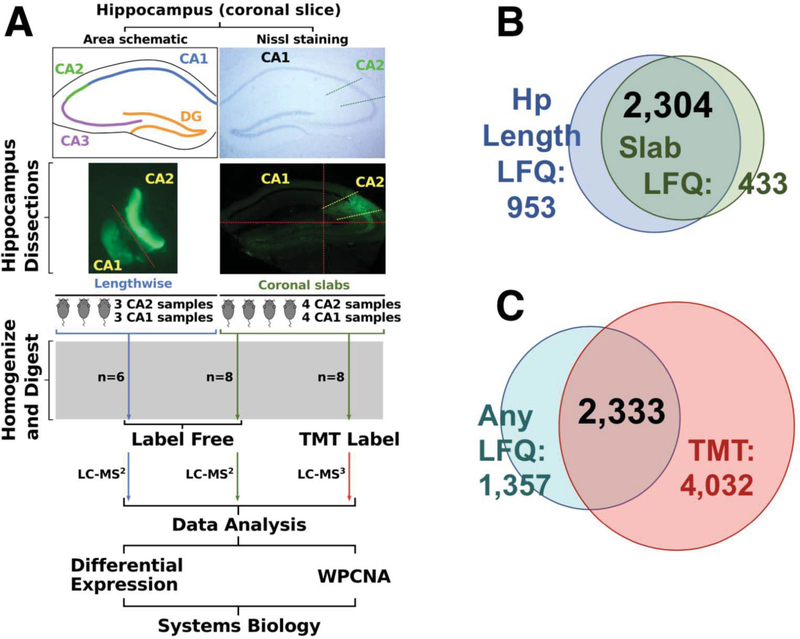
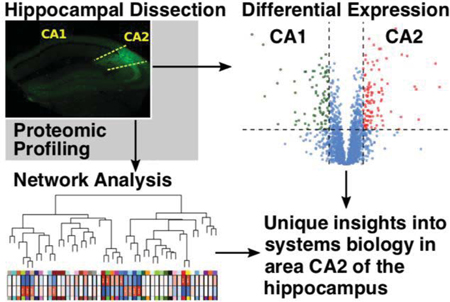
Previous studies have identified amigo-2 as a marker for the CA2 region of the hippocampus5, 15, 40, 41. Using mice expressing eGFP under the amigo-2 promoter, we isolated dorsal CA2 and CA1 from mouse hippocampus using two different approaches. One approach isolated the hippocampal regions by cutting along the length of the hippocampus while the other approach cut the hippocampus into slabs resembling coronal slices, pooling the CA2 enriched and CA1 enriched tissue fragments from the slabs for each sample (Fig. 1A). CA2 and paired CA1 tissue samples were homogenized, digested, and analyzed by label free mass spectrometry. Following LFQ, 3,257 unique proteins were well-quantified across samples obtained by dissection method 1 and 2,737 proteins were quantified in the samples obtained by dissection method 2, with 2,304 proteins overlapping between the two dissection methods (Fig. 1B). To dive deeper into these isolated tissue preparations, the samples collected via the hippocampal slab method (dissection method 2) were subjected to tandem mass tag (TMT) labeling, followed by extensive orthogonal prefractionation, and a batch of LC-MS3 runs. This allowed us to identify more peptides in the samples and therefore, a more extensive proteome. In the TMT-labeled samples we identified 6,365 proteins compared to 3,690 total proteins identified in any of the LFQ samples with 2,333 proteins overlapping (Fig. 1C).
Figure 1. Dissection and analysis workflow.
(A) A schematic and Nissl stained section of the hippocampus highlight the location of area CA2. Areas CA2 and CA1 of dorsal sections of excised mouse hippocampi were isolated with the aid of eGFP expression in area CA2 under the amigo-2 promoter. Cuts made for the dissections are approximated by red dashed lines. Tissue dissected out along the length of the hippocampus was collected from 3 mice (3 CA2 samples, 3 CA1 samples, n=6) was processed and CA1 and CA2 proteomes were identified via LC-MS/MS. Coronal slab samples (4 CA2 samples, 4 CA1 samples, n=4) were not only analyzed by LC-MS/MS but also LC-MS3 following tandem mass tag (TMT) labeling. Following quantification samples were analyzed through differential expression as well as weighted protein-protein correlation analysis (WPCNA), a systems biology approach examining networks of co-expressed proteins. (B) A comparison of dissection methods of areas CA1 and CA2 show that dissection via coronal slabs led to quantification of 2737 proteins with dissection along the length of the hippocampus yielding 3257 with 2304 overlapping proteins. (C) TMT labeling and orthogonal prefractionation of coronal slab samples followed by LC-MS3 quantified 6365 proteins compared to 3690 in any of the label free quantification (LFQ) samples with 2333 proteins overlapping.
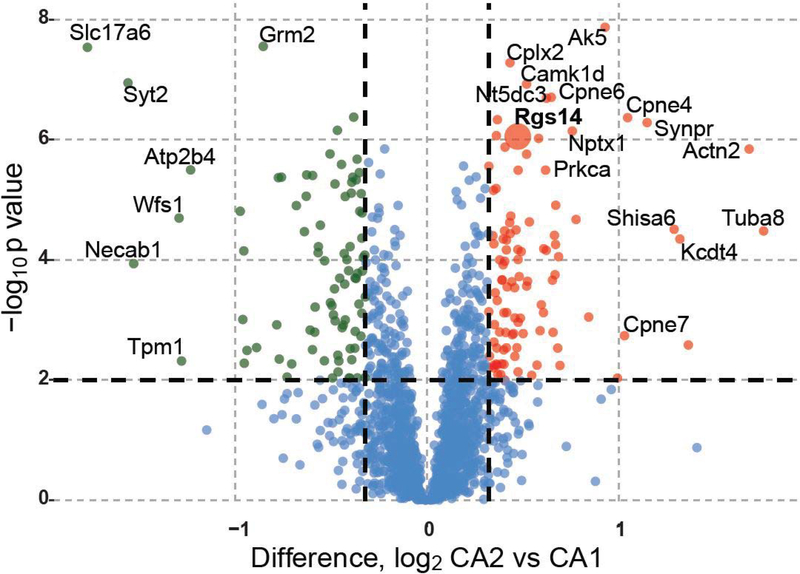
Comparing the proteins found in areas CA2 and CA1 from both dissection methods and both the LFQ and TMT quantification strategies, we identified 101 proteins enriched in CA2 and 85 proteins in CA1 that met the criteria of having a meta p-value less than 0.01 and a greater than 1.25-fold change in expression. These proteins are shown in a volcano plot (Fig. 2) with CA2 enriched proteins on the right and CA1 enriched proteins on the left. Alternatively, focusing on “consensus” changed proteins in Supplemental Table 1 (master proteomics quantification and statistics), upon filtering for a minimum of 3 experiments all with the same change in direction and a Fisher’s meta-p combined_p_value of <0.05, and minimum absolute 3-experiment mean fold change of at least 25% (i.e., a log2(CA1/CA2) fold change of log2(1.25)=0.3219), 128 targets are identifiable as CA2 enriched or depleted (68/128 are enriched at least 25%, while 60 are depleted at least 25%).
Figure 2. Three-experiment meta-analyis of differential expression reveals distinct CA1 and CA2 proteome.
Volcano plot comparing differential expression of proteins in areas CA2 and CA1 across all dissection methods and mass spectrometry modalities. 101 proteins are enriched in CA2 (red) and 85 in CA1 (green) based on the criteria of a Fisher’s meta p-value across all experiments < 0.01 and greater than 1.25-fold change in expression for each protein.
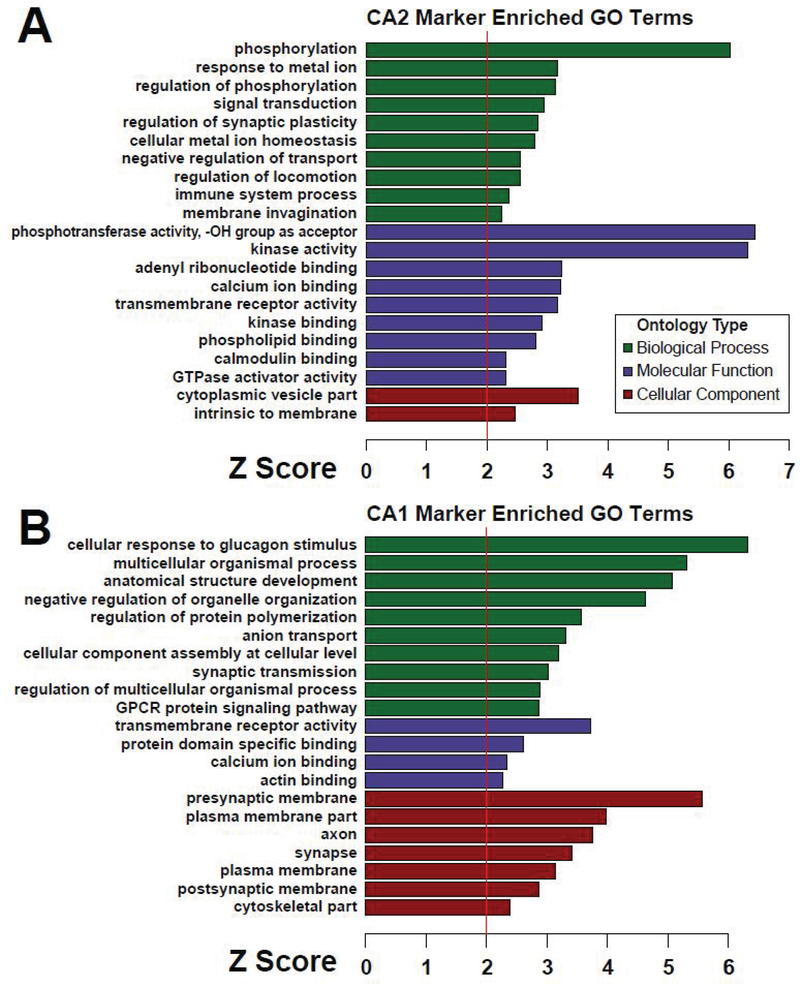
The CA2-enriched proteins in the meta-analysis volcano include well-known markers for area CA2, including RGS146–8 and α-actinin 211, 42. A gene ontology (GO) analysis of the CA2 enriched proteins is shown as a bar graph with terms grouped by biological process, molecular function, and cellular component (Fig. 3A). Notably, the gene ontology terms “regulation of synaptic plasticity” and “calcium ion binding” are associated with proteins enriched in CA2, which is known to be distinctly different from CA1 in these functions6, 14, 17, 43, 44 Based on these known differences and to gain insight into the specific proteins contributing to the ontologies that set area CA2 apart from CA1, we created chord plots from selected ontologies in Figure 3 (Fig. 4A).
Figure 3. Gene ontology (GO) analysis of CA2- and CA1-enriched proteins.
(A) Gene ontologies enriched in the 100 CA2-enriched gene products relative to 2,947 gene products identified an d quantified in a minimum of two out of three experiments are shown grouped by biological process, molecular function, and cellular component. (B) Gene ontologies likewise enriched in the 85 CA1-enriched gene product proteins are shown grouped by biological process, molecular function, and cellular component. A Z-score of 1.96 represents significant overrepresentation (p < 0.05) relative to the background list of 2,947 gene products.
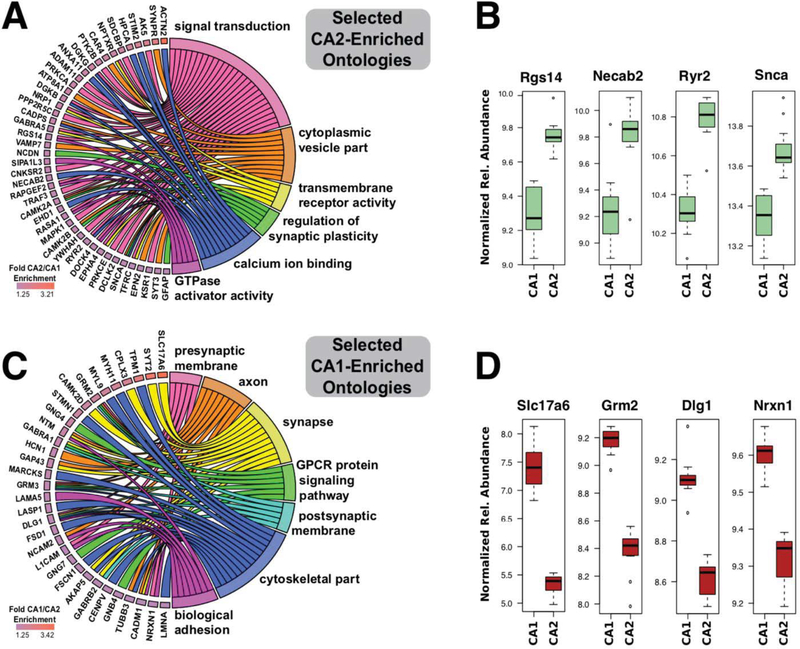
Figure 4. A circular representation of specific proteins contributing to selected ontologies linked to unique physiology in areas CA2 and CA1.
(A) Chord plot linking specific gene product proteins to associated gene ontology terms derived from Figure 3 for proteins enriched in area CA2. (B) Box plots for individual proteins selected from CA2-enriched ontologies comparing differential expression across samples between areas CA1 and CA2. (C) Chord plot linking specific gene product proteins to associated gene ontology terms for proteins enriched in area CA1. (D) Box plots for individual proteins selected from CA1-enriched ontologies comparing differential expression across samples between areas CA1 and CA2. For (B) and (D) relative abundance units (following normalization across the 3 experiments) are all log2 transformed.
We also created a bar graph and chord plot displaying CA1 enriched ontologies (Fig. 3B) as well as selected CA1 enriched proteins linked to their respective gene ontology terms (Fig. 4C). To provide some context on variability of expression between CA2 and CA1 for individual proteins, we created box plots showing relative expression of representative proteins from areas CA2 (Fig. 4B) and CA1 (Fig. 4D) that also contribute to the ontologies in the chord plots. Box plots comparing differential protein expression between CA1 and CA2 samples as well as all three mass spectrometry experiments comparing CA1 and CA2 for each individual protein in the chord plots (Fig. 4 A and C) can be found in Supplemental Figure 1 and 2 respectively.
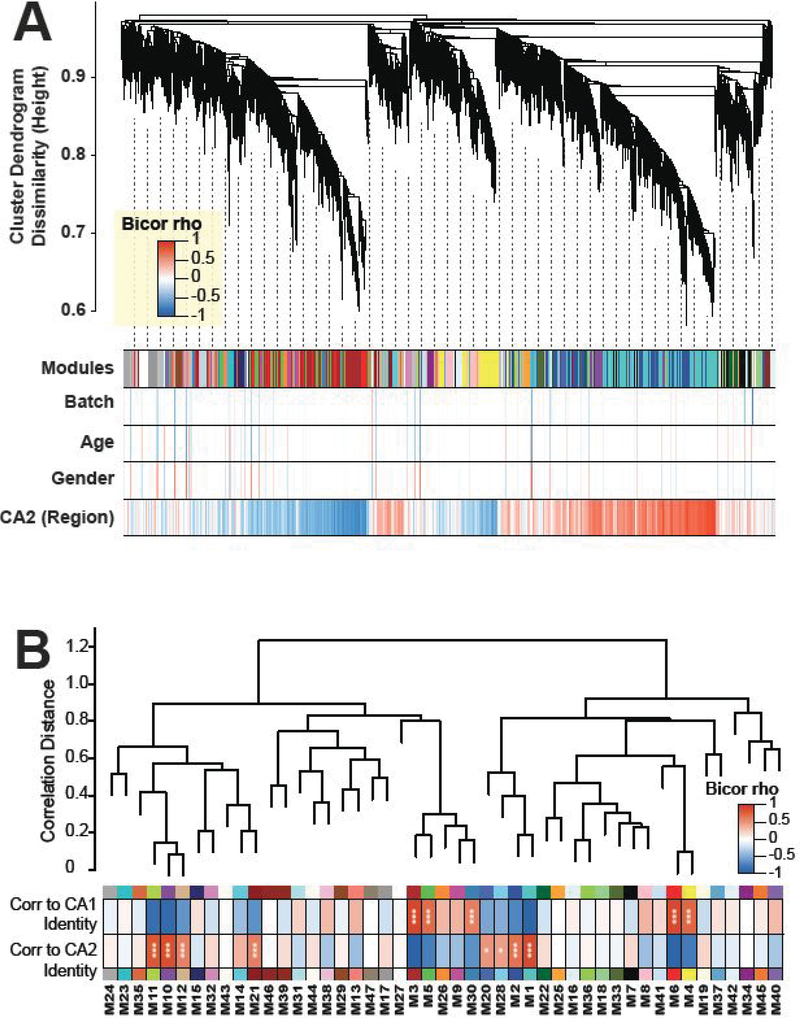
While examining differential expression is common and useful when comparing two proteomes, many proteins of interest are excluded due to variance in expression across samples or small fold-differences between area CA1 and CA2. To fully take advantage of our data, we performed a meta-analysis taking into account the expression of all proteins quantified across all of our samples. In order to identify proteins that have coherent patterns of abundance across the 22 CA1- and CA2- specific dissection samples, we performed weighted protein co-expression network analysis (WPCNA) following batch correction of the abundances from the 3 replicate experiments (Fig. 5A). WPCNA groups proteins based on correlated expression and can be used to identify communities of proteins with shared biological function. Out of 47 distinct modules of co-expressed proteins, we found eight distinct co-expression patterns (module eigenproteins) that were significantly enriched in samples from area CA2 (vs. CA1) (M11, M10, M12, M21, M20, M28, M2, and M1) (Fig. 5B). Of these modules, Fisher exact one-tailed test for overlap of module members with differentially enriched proteins in CA2 identified M1 (BH-corrected FDR 1.32×10−8), M11 (BH-corrected FDR 3.49×10−7), and M10 (BH-corrected FDR 5.76×10−5) as modules significantly overrepresenting differentially expressed CA2 markers we identified by meta-p and log2(fold change) analysis (Supplemental Table 4). The reason that only three of the eight CA2-enriched modules (as indicated by significance of a Student’s correlation p value) show significance by overlap with differentially expressed CA2 markers is that thresholding for differential expression may be less sensitive to change across regions compared to co-enrichment (co-expression) analysis, thus providing justification for implementation of the more sensitive co-enrichment approach.
Figure 5. WPCNA reveals protein modules enriched in CA2.
(A). Weighted protein correlation network module detection by dissimilarity measure (1 minus topology overlap matrix) grouping proteins into dendrogram branches of correlated abundance patterns across the 22 CA2 and CA1 samples across 3 experiments. Heatmap of bicor correlation (rho) for individual proteins in the network is given to traits including batch, age, gender, and region. (B) A dendrogram displays relationships between protein co-expression network modules. Out of 47 distinct modules of co-expressed proteins, eight co-expression patterns (module eigenproteins) that were significantly enriched in CA2 (vs. CA1) (M11, M10, M12, M21, M20, M28, M2, and M1), via two-sided Student bicor correlation p values (*, p<0.05; ***, p<0.002).
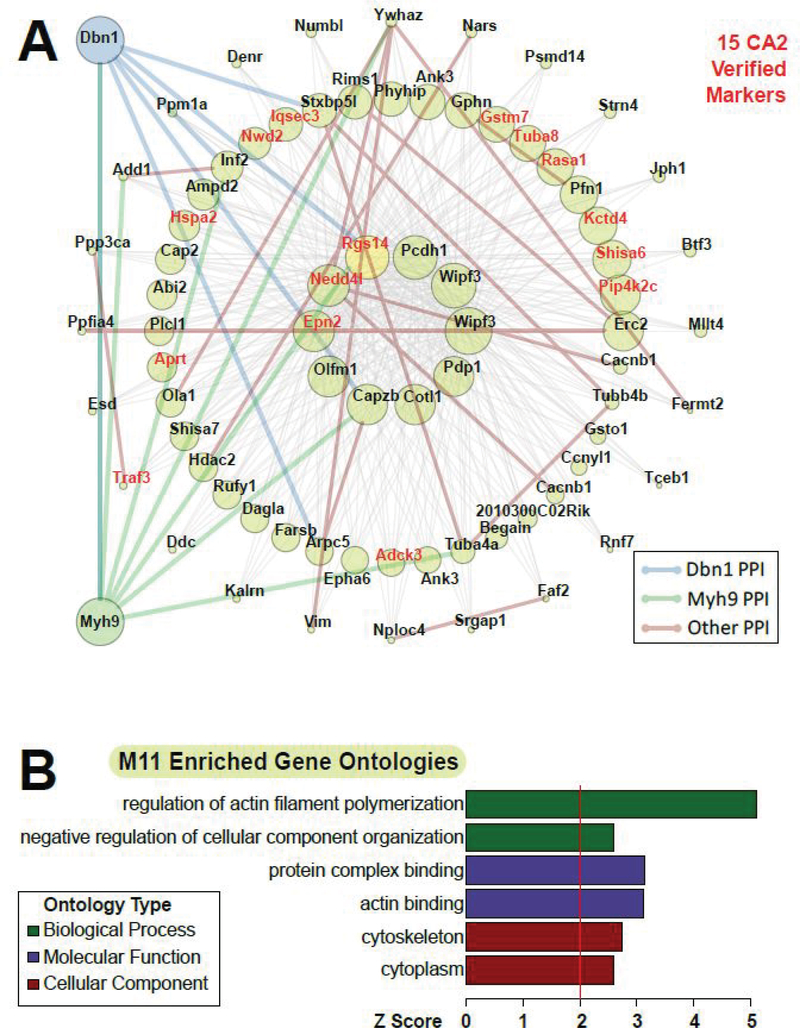
One network of particular interest is M11 (Fig. 6A). This module contains 15 out of the 101 differentially expressed CA2 (vs. CA1) markers; these markers verified across 3 experiments, and distinctly co-express across the 22 samples with strong enrichment of the M11 module eigenprotein in CA2 samples (differentially expressed gene symbols highlighted in red). Proteins closest to the center of the module are the hub proteins that are most correlated and representative of the expression pattern of all of the proteins in the module as a whole. Among these hub proteins is RGS14 (highlighted yellow node), a known marker of CA2 that regulates synaptic plasticity in CA2 pyramidal cells6, 43. Examining GO terms associated with proteins in this module, we find that this module is associated with regulation of actin filament polymerization and actin binding (Fig. 6B). Relevant to this finding, RGS14 was recently shown to exist in a complex with multiple actin binding proteins and myosin motor proteins in mouse brain44.
Figure 6. WPCNA module implicating key CA2 biology and PPIs, with gene ontology analysis.
(A) Gene symbols for protein isoforms we found to be CA2 markers by meta-p<0.01 are highlighted in red in network M11. One of these proteins, RGS14, has been shown to interact with Dbn1 and Myh9, which were added along with bold, colored edges for all known PPIs among visualized nodes of the network (Dbn1 interactions: blue; Myh9 interactions: green; all other interactions: brown). (B) M11-enriched gene ontologies are shown grouped by biological process, molecular function, and cellular component.
To gain insight into functional protein-protein interactions (PPIs) between the RGS14-driven subnetwork of CA2-enriched markers and the multiple actin binding proteins in this module, known PPIs from the BioGRID database and from our own RGS14 interactome44 were added as edges in the subnetwork graph. We also checked for PPIs among M11 members and actin binding proteins we had previously shown to interact with RGS14 and found that drebrin 1 (Dbn1), and myosin IIA (Myh9) PPIs were present among M11 members (Fig. 6A, edges connecting to added blue and green nodes), but not among CA1-specific modules, including M4, M8, and M9, which had other myosins as high correlation-ranked members (M4: Myo5a, Myh10; M8: Myo6; M9: Myl6). These data point to drebrin 1 and myosin IIA as potential functional interactors with RGS14, expediting targeted efforts to examine the cellular signaling and physiology mediated by these interactions in particular. Our network/systems approach has thus identified more (and less) CA2-relevant actin-binding protein PPIs that connect the M11 hub, RGS14, and other co-expressed proteins’ functions and that merit further study.
To further validate our findings, we compiled a list of proteins we observed here in our analysis that also have been previously observed to be preferentially expressed in area CA2 over CA1 (Table 1). A function for each protein in brain is also listed to provide context. Out of 101 proteins shown to be enriched in CA2, 18 already have been shown to be enriched in CA2 compared to CA1 either by mRNA and/or protein levels. These 18 proteins include RGS14 and alpha-actinin 2, known markers for the CA2 region6–8, 11, 42 that are not highly enriched in any other CA region of the hippocampus.
Table 1. Externally validated CA2-enriched proteins.
Here, we list expression (mRNA and protein) and known function of proteins that we found to have robustly enriched expression in CA2 (vs. CA1). These 18 proteins have been previously shown in the literature to be enriched in CA2 through examination of mRNA and/or protein expression and provide external validation of the methods used to isolate and analyze areas CA1 and CA2 for this study.
| Gene Symbol | Protein name | Validation by mRNA | Validation by Protein | Known Function in neurons |
|---|---|---|---|---|
| DGKG | Diacylglycerol kinase gamma | Yes71 | N/A | N/A |
| MCU | Mitochondrial Calcium Uniporter | Yes71 | N/A | Main component of the protein complex that mediates mitochondrial calcium uptake56 |
| NECAB2 | N-terminal EF-hand calcium-binding protein 2 | Yes45 | Yes45 | Interacts with and modulates the activity of the adenosine 2A receptor72 and metabotropic glutamate receptor 573 |
| ACTN2 | Alpha-actinin 2 | N/A | Yes11, 42 | Mediates spine morphology and assembly of the post-synaptic density in hippocampal neurons,52 anchoring PSD-95 at post-synaptic sites53 |
| SMS | Spermine synthase | Yes71 | N/A | Converts spermidine to spermine, a polyamine which has multiple effects54 including modulation of NMDA receptor function55 |
| PRKCE | Protein kinase C ε | N/A | Yes74 | Activation and increased expression of this kinase mediates neuroprotection following oxygen and glucose deprivation62 |
| SLC25A22 | Mitochondrial glutamate carrier 1 | Yes71 | N/A | Transports glutamate into the inner mitochondrial membrane75 |
| RGS14 | Regulator of G protein signaling 14 | Yes71 | Yes6–8 | Natural suppressor of LTP in the CA2 region of the hippocampus6 through modulation of calcium signaling43· 44 |
| OSTF1 | Osteoclast-stimulating factor 1 | Yes71 | Yes76 | N/A |
| CADPS | Calcium-dependent secretion activator 1 | Yes71 | N/A | Essential factor for synaptic vesicle priming in hippocampal neurons60 |
| GPC1 | Glypican-1 | Yes71 | N/A | N/A |
| PPFIA2 | Liprin-alpha 2 | No77 | Yes78 | Controls synaptic output by regulating synaptic vesicle pool size59 |
| KCTD4 | Potassium channel tetramerization domain containing 4 | Yes71 | N/A | N/A |
| RYR2 | Ryanodine receptor 2 | No71 | Yes79· 80 | Mediates calcium-induced calcium release from intracellular calcium stores57, affecting cognitive function and hippocampal synaptic plasticity in chronically stressed mice81 |
| ATP8A1 | ATPase phospholipid transporting 8A1 | Yes71 | N/A | Deficiency in this flippase is associated with externalization of phosphatidylserine in hippocampal neurons, delaying hippocampal-dependent learning63 |
| NRP1 | Neuropilin 1 | Yes71 | N/A | Interacts with semaphoring 3 and VEGF to mediate axon guidance and angiogenesis respectively58 |
| ANXA11 | Annexin A11 | Yes71 | N/A | Participates in ER-to-Golgi trafficking, modulating the early secretory pathway82 |
| CLVS2 | Clavesin 2 | Yes71 | N/A | Regulates late endosome/lysosome morphology83 |
DISCUSSION
Here, we provide evidence of a CA2-enriched proteome distinct from that of CA1 mouse hippocampus. Of the more than 7000 proteins surveyed, we identified over 100 proteins with consistently and robustly higher expression in CA2 (vs. CA1) including established markers of area CA2, such as RGS146–8 and NECAB245. Gene ontology (GO) terms associated with these CA2-enriched proteins include functions for “regulation of neuronal synaptic plasticity” and “calcium ion binding,” both of which have been shown to be different between CA2 from CA1 in the current literature6, 14, 17, 41, 46. Our findings identify eight distinct co-expression networks enriched in CA2 (vs. CA1). These networks contain proteins already known to be expressed highly in CA2, as well as newly validated proteins that can highlight potentially novel functions which contribute to area CA2’s unique physiology and signaling roles.
Although area CA1 of the hippocampus has been extensively studied, much less effort has been directed toward understanding area CA25. This is because CA2 had long been thought to be more of a transition zone between CA1 and CA3, and perhaps because CA2 is small compared with the surrounding CA regions with undefined boundaries, making it more difficult to study and dissect by traditional means. Recently identified CA2-specific marker proteins5 have helped to make this hippocampal region more accessible to investigators, including our use of amigo2-GFP here combined with manual dissection. Fluorescence-guided laser capture microdissection is an alternate approach that allows for more discrete dissection, but yields much less tissue and protein for LC-MS/MS analysis, prompting us to utilize a manual dissection method (Fig. 1A). Physiologically, the CA1 region has been of interest to researchers in part because it displays LTP at synapses projecting from CA3, which is thought to underlie the basis of some forms of hippocampal-dependent learning and memory47. Notably, LTP is absent in CA2 pyramidal neurons under the same conditions in which it is elicited in CA114, 41. This lack of LTP can be overcome through select interventions including knockout of RGS146, 41 and increasing extracellular calcium to overcome the robust calcium buffering and extrusion properties in CA2 pyramidal cells17, 41.
In relation to disease, the hippocampus has been of interest due to its susceptibility to damage from a variety of insults including temporal lobe epilepsy21, 48, traumatic brain injury25, 26, and ischemia22–24. However, area CA2 is uniquely resistant to cell death following each of these forms of damage compared to area CA121–26, 48. Although the mechanism of this protection of the CA2 region is unknown, the process may be related to the resistance to LTP, as both are known to require calcium. Our data showing the enrichment of calcium-regulating proteins in CA2 support this idea.
We identified over 100 proteins that are robustly and reliably enriched in CA2 (vs. CA1). Although many of these proteins have not been previously reported to be enriched in CA2, we identified 18 proteins that had already been shown to be preferentially expressed in CA2 (vs.CA1) via either mRNA and/or protein expression levels (Table 1). Considering these proteins are validated not only by our results here, but also by outside findings, many of these targets merit further research in the context of CA2 function.
Two of these proteins, MCU and RGS14, are of particular interest due to their ties to multiple aspects of physiology that are unique to area CA2 within the hippocampus. RGS14 is a known suppressor of LTP in the CA2 region of the hippocampus6. Furthermore, this activity has been attributed, at least in part, to RGS14’s capacity to bind calcium regulated proteins calmodulin (CaM) and CaMKII44 and its suppression of postsynaptic calcium transients during structural plasticity induction41. It is quite possible that RGS14’s ties to enhanced calcium buffering and suppression of LTP in area CA2 may be tied to CA2’s resistance to neuronal cell death, and future studies should further investigate this connection. Additionally, we find that MCU, the mitochondrial calcium uniporter responsible for mitochondrial calcium import, is highly expressed in area CA2 (vs. CA1). MCU has already been shown to mediate cytoplasmic calcium sequestration in other cell types by importing calcium into the mitochondria49. MCU has also been shown to be involved not only in NMDA receptor-mediated excitotoxicity50 but also LTP outside of the hippocampus51. The role of MCU in the CA2 region of the hippocampus merits further study and could potentially reveal a link between enhanced calcium buffering and decreased cell death and plasticity in this enigmatic brain reigon.
Many other proteins that have been previously identified to be enriched in area CA2 (vs. CA1) likely play functional roles in this region of the hippocampus as well. Alpha-actinin 2, for example, has been used as a marker for area CA2, and has been shown to mediate spine morphology and assembly of the post-synaptic density in hippocampal neurons52, 53, which are known to affect post-synaptic plasticity. Notably, alpha-actinin 2 is calcium insensitive, providing a mechanism by which CA2 neurons may be able to control spine morphology under conditions in which calcium levels are limited. Spermine synthase may also be involved in post-synaptic plasticity in CA2. This enzyme converts spermidine to spermine, which can modulate the activity of multiple ion channels including inwardly rectifying potassium channels, NMDA receptors, AMPA receptors, and kainate receptors54, 55. All of these ion channels have been shown to play a role in synaptic plasticity, and higher levels of spermine in CA2 due to spermine synthase may play a role in CA2’s relative lack of plasticity compared to area CA1. MCU, a mitochondrial calcium ion uniporter, and ryanodine receptor 2 each control intracellular calcium levels56, 57 Both may participate in post-synaptic calcium handling properties, with MCU already having been shown to mediate cytoplasmic calcium sequestration in other cell types by importing calcium into mitochondria49.
Many of the proteins we found to be enriched in CA2 (vs. CA1) are reported to control presynaptic signaling or function, and include neuropilin 1 (Nrp1)58, liprin-alpha 2 (Ppfia2)59, and calcium-dependent secretion activator 1 (Cadps) or CAPS-160. Although these proteins have been shown to be enriched in CA2 by other methods, it is worth noting that these and other presynaptic proteins may be enriched in area CA2 due to their expression in other areas that project to the CA2 region including CA3 6,14, the DG 2, and the entorhinal cortex (EC)19, 40. Also, CA2 neurons project into CA1 potentially confusing identification of additional presynaptic proteins preferentially expressed in CA2. These limitations potentially could be addressed by future studies using RNA sequencing to examine differential expression in CA2 and CA1. However mRNA also is known to be transported throughout the axon as well61.
Area CA2 is also noteworthy for its remarkable resistance to cell loss following injury. Neurons in CA2 are resistant to damage from temporal lobe epilepsy, ischemia, and traumatic brain injury12, 21–26, 48. Among the proteins we identified as enriched in CA2 (vs. CA1) and that also have been previously shown to be high in CA2 (Table 1), are protein kinase C-ε (PRKCE) and ATPase phospholipid transporter 8A1 (ATP8A1), both of which are known to have neuroprotective effects. Protein kinase C-ε activation is reported to mediate neuroprotection following oxygen and glucose deprivation as is the case during ischemia62. ATP8A1 deficiency in hippocampal neurons is associated with externalization of phosphatidylserine in hippocampal neurons63, which is associated with apoptosis64. Enhanced ATP8A1 expression in CA2 pyramidal cells may decrease phosphatidylserine externalization and subsequent programmed cell death following injury, providing a mechanism of neuroprotection. These proteins with known neuroprotective effects merit further study in the context of area CA2 along with proteins such as RGS14 and MCU that appear to play a role in suppression of high intracellular calcium levels linked to excitotoxicity caused by excessive glutamate stimulation.
Beyond these individual proteins that show preferential enrichment in CA2 (vs. CA1), we have identified entire networks of proteins with correlated expression, and eight of these networks’ principle component patterns of expression are enriched in samples derived from CA2 versus samples from CA1, i.e., these network eigenproteins significantly correlate with CA2 sample identity (Fig. 5B). Furthermore, one of these networks (M11) contains 15 proteins we find to be enriched in CA2 (vs. CA1) via differential expression and includes RGS14 as a hub protein of the module. By incorporating previously reported brain co-immunoprecipitation data for the RGS14 interactome44 into our network analysis, we identify drebrin 1 and myosin IIA as likely functional interactors. Related to RGS14’s known role as a suppressor of LTP in area CA26, 41, drebrin 1 has been shown in numerous studies to be important for spine formation and plasticity65–68. Drebrin forms a stable structure with actin in dendritic spines, but following NMDA receptor activation, drebrin exits spines, presumably allowing F-actin polymerization associated with plasticity68. Furthermore, this removal of drebrin from spines is mediated by myosin II68, which has been shown to be necessary for spine maturation69 and maintenance of LTP70. Based on the known functions of drebrin and myosin II, as well as the network enrichments for PPIs that implicate these proteins as relevant RGS14 interactions in CA2, RGS14 appears likely to interact functionally with these new targets. RGS14 may act on either drebrin 1 or myosin II, or both, to prevent their interaction to disrupt actin dynamics and suppress LTP in CA2 pyramidal neurons, providing an example of how this network analysis can be used to narrow focus for CA2-relevant PPIs and subnetworks in future studies.
Our findings here identify over 100 proteins enriched in CA2 that are involved in synaptic signaling and plasticity. Most of these proteins have never been reported to be highly expressed in CA2 (vs. CA1) and merit further research to determine how their selective expression may affect the unique physiology and signaling observed in this hippocampal subregion. In addition to a wealth of newly identified proteins of interest, we have uncovered networks of proteins with correlated expression, wherein eight of these networks are significantly enriched in CA2 samples. Here we performed a comprehensive “deep dive” into one WPCNA module (M11) to illustrate the power of this data set. While practical contraints prevent us from doing a comprehensive dive here into all 47 networks, this comprehensive network data set is now available for study by all interested researchers, and can be used to provide context for expression of proteins within the same module, and help to identify and sharpen focus on potential functional interactions unique to CA2, as we have shown with RGS14. Future studies will focus on newly identified CA2-enriched (vs. CA1) proteins with known functions that appear to explain aspects of area CA2’s unique physiology and signaling.
Supplementary Material
Supplemental Figure 1 – Box plots showing differential expression between CA1 and CA2 for proteins included in chord plots
Supplemental Figure 2 – Box plots showing differential expression between CA1 and CA2 across all mass spectrometry experiments for proteins included in chord plots
Supplemental Table 1 – Meta-analysis of quantitative proteomics
Supplemental Table 2 – Additional CA2 markers in TMT proteomics
Supplemental Table 3 – Information on dissected mouse samples
Supplemental Table 4 – Significance of differentially expressed proteins in modules
Acknowledgements
Grant funding was as follows: NINDS 1R21 NS087488 (for JRH, NTS, and EBD), and NINDS R01 NS037112 (for JRH and NTS), NINDS F31NS098648 and T32 GM008602 for KJG. Proteomics analysis and LC-MS/MS were subsidized through an additional grant for the NINDS Emory Neuroscience Core (P30NS055077). This research was funded in part by the Intramural Research Program of the NIEHS (ES100221 for SMD). We thank Daniel Lustberg for assistance with the mice and Georgia Alexander for image collection.
Footnotes
SUPPORTING INFORMATION
The following supporting information is available free of charge at ACS website http://pubs.acs.org.
References
- 1.Lorente de No R, Studies on the structure of the cerebral cortex. II. Continuation of the study of the ammonic system. J Psychol Neurol 1934, (46), 113–177. [Google Scholar]
- 2.Kohara K; Pignatelli M; Rivest AJ; Jung HY; Kitamura T; Suh J; Frank D; Kajikawa K; Mise N; Obata Y; Wickersham IR; Tonegawa S, Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat Neurosci 2014, 17 (2), 269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San Antonio A; Liban K; Ikrar T; Tsyganovskiy E; Xu X, Distinct physiological and developmental properties of hippocampal CA2 subfield revealed by using anti-Purkinje cell protein 4 (PCP4) immunostaining. J Comp Neurol 2014, 522 (6), 1333–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurberg S; Zimmer J, Aberrant hippocampal mossy fibers in cats. Brain Res 1980, 188 (2), 555–9. [DOI] [PubMed] [Google Scholar]
- 5.Dudek SM; Alexander GM; Farris S, Rediscovering area CA2: unique properties and functions. Nat Rev Neurosci 2016, 17 (2), 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SE; Simons SB; Heldt SA; Zhao M; Schroeder JP; Vellano CP; Cowan DP; Ramineni S; Yates CK; Feng Y; Smith Y; Sweatt JD; Weinshenker D; Ressler KJ; Dudek SM; Hepler JR, RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc Natl Acad Sci U S A 2010, 107 (39), 16994–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans PR; Lee SE; Smith Y; Hepler JR, Postnatal developmental expression of regulator of G protein signaling 14 (RGS14) in the mouse brain. J Comp Neurol 2014, 522 (1), 186–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squires KE; Gerber KJ; Pare JF; Branch MR; Smith Y; Hepler JR, Regulator of G protein signaling 14 (RGS14) is expressed pre- and postsynaptically in neurons of hippocampus, basal ganglia, and amygdala of monkey and human brain. Brain Struct Funct 2018, 223 (1), 233–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laeremans A; Nys J; Luyten W; D’Hooge R; Paulussen M; Arckens L, AMIGO2 mRNA expression in hippocampal CA2 and CA3a. Brain Struct Funct 2013, 218 (1), 123–30. [DOI] [PubMed] [Google Scholar]
- 10.Lein ES; Callaway EM; Albright TD; Gage FH, Redefining the boundaries of the hippocampal CA2 subfield in the mouse using gene expression and 3-dimensional reconstruction. J Comp Neurol 2005, 485 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- 11.Wyszynski M; Kharazia V; Shanghvi R; Rao A; Beggs AH; Craig AM; Weinberg R; Sheng M, Differential regional expression and ultrastructural localization of alpha-actinin-2, a putative NMDA receptor-anchoring protein, in rat brain. J Neurosci 1998, 18 (4), 1383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer A; Trigg HL; Thomson AM, Characterization of neurons in the CA2 subfield of the adult rat hippocampus. J Neurosci 2007, 27 (27), 7329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulanger LM; Lombroso PJ; Raghunathan A; During MJ; Wahle P; Naegele JR, Cellular and molecular characterization of a brain-enriched protein tyrosine phosphatase. J Neurosci 1995, 15 (2), 1532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao M; Choi YS; Obrietan K; Dudek SM, Synaptic plasticity (and the lack thereof) in hippocampal CA2 neurons. J Neurosci 2007, 27 (44), 12025–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hitti FL; Siegelbaum SA, The hippocampal CA2 region is essential for social memory. Nature 2014, 508 (7494), 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malenka RC; Bear MF, LTP and LTD: an embarrassment of riches. Neuron 2004, 44 (1), 5–21. [DOI] [PubMed] [Google Scholar]
- 17.Simons SB; Escobedo Y; Yasuda R; Dudek SM, Regional differences in hippocampal calcium handling provide a cellular mechanism for limiting plasticity. Proc Natl Acad Sci U S A 2009, 106 (33), 14080–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carstens KE; Phillips ML; Pozzo-Miller L; Weinberg RJ; Dudek SM, Perineuronal Nets Suppress Plasticity of Excitatory Synapses on CA2 Pyramidal Neurons. J Neurosci 2016, 36 (23), 6312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevaleyre V; Siegelbaum SA, Strong CA2 pyramidal neuron synapses define a powerful disynaptic cortico-hippocampal loop. Neuron 2010, 66 (4), 560–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corsellis JA; Bruton CJ, Neuropathology of status epilepticus in humans. Adv Neurol 1983, 34, 129–39. [PubMed] [Google Scholar]
- 21.Sloviter RS, Permanently altered hippocampal structure, excitability, and inhibition after experimental status epilepticus in the rat: the “dormant basket cell” hypothesis and its possible relevance to temporal lobe epilepsy. Hippocampus 1991, 1 (1), 41–66. [DOI] [PubMed] [Google Scholar]
- 22.Kirino T, Delayed neuronal death in the gerbil hippocampus following ischemia. Brain Res 1982, 239 (1), 57–69. [DOI] [PubMed] [Google Scholar]
- 23.Sadowski M; Wisniewski HM; Jakubowska-Sadowska K; Tarnawski M; Lazarewicz JW; Mossakowski MJ, Pattern of neuronal loss in the rat hippocampus following experimental cardiac arrest-induced ischemia. J Neurol Sci 1999, 168 (1), 13–20. [DOI] [PubMed] [Google Scholar]
- 24.Yang G; Kitagawa K; Ohtsuki T; Kuwabara K; Mabuchi T; Yagita Y; Takazawa K; Tanaka S; Yanagihara T; Hori M; Matsumoto M, Regional difference of neuronal vulnerability in the murine hippocampus after transient forebrain ischemia. Brain Res 2000, 870 (1–2), 195–8. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell WL; Dhillon K; Harper L; Espin J; MacIntosh TK; Smith DH; Graham DI, There is differential loss of pyramidal cells from the human hippocampus with survival after blunt head injury. J Neuropathol Exp Neurol 2003, 62 (3), 272–9. [DOI] [PubMed] [Google Scholar]
- 26.Kotapka MJ; Graham DI; Adams JH; Gennarelli TA, Hippocampal pathology in fatal non-missile human head injury. Acta Neuropathol 1992, 83 (5), 530–4. [DOI] [PubMed] [Google Scholar]
- 27.Pagani JH; Zhao M; Cui Z; Avram SK; Caruana DA; Dudek SM; Young WS, Role of the vasopressin 1b receptor in rodent aggressive behavior and synaptic plasticity in hippocampal area CA2. Mol Psychiatry 2015, 20 (4), 490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons SB; Caruana DA; Zhao M; Dudek SM, Caffeine-induced synaptic potentiation in hippocampal CA2 neurons. Nat Neurosci 2012, 15 (1), 23–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz MD; Solis JM, Characterisation of the mechanisms underlying the special sensitivity of the CA2 hippocampal area to adenosine receptor antagonists. Neuropharmacology 2019, 144, 9–18. [DOI] [PubMed] [Google Scholar]
- 30.Gozal E; Gozal D; Pierce WM; Thongboonkerd V; Scherzer JA; Sachleben LR Jr.; Brittian KR; Guo SZ; Cai J; Klein JB, Proteomic analysis of CA1 and CA3 regions of rat hippocampus and differential susceptibility to intermittent hypoxia. J Neurochem 2002, 83 (2), 331–45. [DOI] [PubMed] [Google Scholar]
- 31.von Ziegler LM; Selevsek N; Tweedie-Cullen RY; Kremer E; Mansuy IM, Subregion-Specific Proteomic Signature in the Hippocampus for Recognition Processes in Adult Mice. Cell Rep 2018, 22 (12), 3362–3374. [DOI] [PubMed] [Google Scholar]
- 32.Seyfried NT; Dammer EB; Swarup V; Nandakumar D; Duong DM; Yin L; Deng Q; Nguyen T; Hales CM; Wingo T, A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s disease. Cell systems 2017, 4 (1), 60–72. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J; Neuhauser N; Michalski A; Scheltema RA; Olsen JV; Mann M, Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. Journal of proteome research 2011, 10 (4), 1794–1805. [DOI] [PubMed] [Google Scholar]
- 34.Cox J; Hein MY; Luber CA; Paron I; Nagaraj N; Mann M, Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Molecular & Cellular Proteomics 2014, 13 (9), 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luber CA; Cox J; Lauterbach H; Fancke B; Selbach M; Tschopp J; Akira S; Wiegand M; Hochrein H; O’Keeffe M; Mann M, Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 2010, 32 (2), 279–89. [DOI] [PubMed] [Google Scholar]
- 36.Pichler P; Köcher T; Holzmann J; Mazanek M; Taus T; Ammerer G; Mechtler K, Peptide Labeling with Isobaric Tags Yields Higher Identification Rates Using iTRAQ 4-Plex Compared to TMT 6-Plex and iTRAQ 8-Plex on LTQ Orbitrap. Analytical Chemistry 2010, 82 (15), 6549–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wingo TS; Duong DM; Zhou M; Dammer EB; Wu H; Cutler DJ; Lah JJ; Levey AI; Seyfried NT, Integrating next-generation genomic sequencing and mass spectrometry to estimate allele-specific protein abundance in human brain. Journal of proteome research 2017, 16 (9), 3336–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Käll L; Canterbury JD; Weston J; Noble WS; MacCoss MJ, Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature methods 2007, 4 (11), 923. [DOI] [PubMed] [Google Scholar]
- 39.Umoh ME; Dammer EB; Dai J; Duong DM; Lah JJ; Levey AI; Gearing M; Glass JD; Seyfried NT, A proteomic network approach across the ALS-FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Molecular Medicine 2018, 10 (1), 48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivas KV; Buss EW; Sun Q; Santoro B; Takahashi H; Nicholson DA; Siegelbaum SA, The Dendrites of CA2 and CA1 Pyramidal Neurons Differentially Regulate Information Flow in the Cortico-Hippocampal Circuit. J Neurosci 2017, 37 (12), 3276–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans PR; Parra-Bueno P; Smirnov MS; Lustberg DJ; Dudek SM; Hepler JR; Yasuda R, RGS14 Restricts Plasticity in Hippocampal CA2 by Limiting Postsynaptic Calcium Signaling. eNeuro 2018, 5 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratzliff AD; Soltesz I, Differential immunoreactivity for alpha-actinin-2, an N-methyl-D-aspartate-receptor/actin binding protein, in hippocampal interneurons. Neuroscience 2001, 103 (2), 337–49. [DOI] [PubMed] [Google Scholar]
- 43.Evans P; Hepler J, Regulator of G protein signaling 14 (RGS14) interacts with calmodulin (CaM) in a calcium-dependent manner. Society for Neuroscience 2012. [Google Scholar]
- 44.Evans PR; Gerber KJ; Dammer EB; Duong DM; Goswami D; Lustberg DJ; Zou J; Yang JJ; Dudek SM; Griffin PR; Seyfried NT; Hepler JR, Interactome Analysis Reveals Regulator of G Protein Signaling 14 (RGS14) is a Novel Calcium/Calmodulin (Ca(2+)/CaM) and CaM Kinase II (CaMKII) Binding Partner. J Proteome Res 2018, 17 (4), 1700–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmermann B; Girard F; Meszar Z; Celio MR, Expression of the calcium binding proteins Necab-1,−2 and −3 in the adult mouse hippocampus and dentate gyrus. Brain Res 2013, 1528, 1–7. [DOI] [PubMed] [Google Scholar]
- 46.Matsuzaki M; Honkura N; Ellis-Davies GC; Kasai H, Structural basis of long-term potentiation in single dendritic spines. Nature 2004, 429 (6993), 761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicoll RA, A Brief History of Long-Term Potentiation. Neuron 2017, 93 (2), 281–290. [DOI] [PubMed] [Google Scholar]
- 48.Haussler U; Rinas K; Kilias A; Egert U; Haas CA, Mossy fiber sprouting and pyramidal cell dispersion in the hippocampal CA2 region in a mouse model of temporal lobe epilepsy. Hippocampus 2016, 26 (5), 577–88. [DOI] [PubMed] [Google Scholar]
- 49.Drago I; De Stefani D; Rizzuto R; Pozzan T, Mitochondrial Ca2+ uptake contributes to buffering cytoplasmic Ca2+ peaks in cardiomyocytes. Proc Natl Acad Sci U S A 2012, 109 (32), 12986–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu J; Tan YW; Hagenston AM; Martel MA; Kneisel N; Skehel PA; Wyllie DJ; Bading H; Hardingham GE, Mitochondrial calcium uniporter Mcu controls excitotoxicity and is transcriptionally repressed by neuroprotective nuclear calcium signals. Nat Commun 2013, 4, 2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HY; Lee KY; Lu Y; Wang J; Cui L; Kim SJ; Chung JM; Chung K, Mitochondrial Ca(2+) uptake is essential for synaptic plasticity in pain. J Neurosci 2011, 31 (36), 12982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hodges JL; Vilchez SM; Asmussen H; Whitmore LA; Horwitz AR, alpha-Actinin-2 mediates spine morphology and assembly of the post-synaptic density in hippocampal neurons. PLoS One 2014, 9 (7), e101770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matt L; Kim K; Hergarden AC; Patriarchi T; Malik ZA; Park DK; Chowdhury D; Buonarati OR; Henderson PB; Gokcek Sarac C; Zhang Y; Mohapatra D; Horne MC; Ames JB; Hell JW, alpha-Actinin Anchors PSD-95 at Postsynaptic Sites. Neuron 2018, 97 (5), 1094–1109 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pegg AE, The function of spermine. IUBMB Life 2014, 66 (1), 8–18. [DOI] [PubMed] [Google Scholar]
- 55.Hackos DH; Hanson JE, Diverse modes of NMDA receptor positive allosteric modulation: Mechanisms and consequences. Neuropharmacology 2017, 112 (Pt A), 34–45. [DOI] [PubMed] [Google Scholar]
- 56.Baughman JM; Perocchi F; Girgis HS; Plovanich M; Belcher-Timme CA; Sancak Y; Bao XR; Strittmatter L; Goldberger O; Bogorad RL; Koteliansky V; Mootha VK, Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 2011, 476 (7360), 341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tully K; Treistman SN, Distinct intracellular calcium profiles following influx through N- versus L-type calcium channels: role of Ca2+-induced Ca2+ release. J Neurophysiol 2004, 92 (1), 135–43. [DOI] [PubMed] [Google Scholar]
- 58.Parker MW; Guo HF; Li X; Linkugel AD; Vander Kooi CW, Function of members of the neuropilin family as essential pleiotropic cell surface receptors. Biochemistry 2012, 51 (47), 9437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spangler SA; Schmitz SK; Kevenaar JT; de Graaff E; de Wit H; Demmers J; Toonen RF; Hoogenraad CC, Liprin-alpha2 promotes the presynaptic recruitment and turnover of RIM1/CASK to facilitate synaptic transmission. J Cell Biol 2013, 201 (6), 915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jockusch WJ; Speidel D; Sigler A; Sorensen JB; Varoqueaux F; Rhee JS; Brose N, CAPS-1 and CAPS-2 are essential synaptic vesicle priming proteins. Cell 2007, 131 (4), 796–808. [DOI] [PubMed] [Google Scholar]
- 61.Sahoo PK; Smith DS; Perrone-Bizzozero N; Twiss JL, Axonal mRNA transport and translation at a glance. J Cell Sci 2018, 131 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohan CH; Stradecki-Cohan HM; Morris-Blanco KC; Khoury N; Koronowski KB; Youbi M; Wright CB; Perez-Pinzon MA, Protein kinase C epsilon delays latency until anoxic depolarization through arc expression and GluR2 internalization. J Cereb Blood Flow Metab 2017, 37 (12), 3774–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levano K; Punia V; Raghunath M; Debata PR; Curcio GM; Mogha A; Purkayastha S; McCloskey D; Fata J; Banerjee P, Atp8a1 deficiency is associated with phosphatidylserine externalization in hippocampus and delayed hippocampus-dependent learning. J Neurochem 2012, 120 (2), 302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fadok VA; Bratton DL; Rose DM; Pearson A; Ezekewitz RA; Henson PM, A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 2000, 405 (6782), 85–90. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi H; Sekino Y; Tanaka S; Mizui T; Kishi S; Shirao T, Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J Neurosci 2003, 23 (16), 6586–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi H; Mizui T; Shirao T, Down-regulation of drebrin A expression suppresses synaptic targeting of NMDA receptors in developing hippocampal neurones. J Neurochem 2006, 97Suppl 1, 1105. [DOI] [PubMed] [Google Scholar]
- 67.Koganezawa N; Hanamura K; Sekino Y; Shirao T, The role of drebrin in dendritic spines. Mol Cell Neurosci 2017, 84, 85–92. [DOI] [PubMed] [Google Scholar]
- 68.Mizui T; Sekino Y; Yamazaki H; Ishizuka Y; Takahashi H; Kojima N; Kojima M; Shirao T, Myosin II ATPase activity mediates the long-term potentiation-induced exodus of stable F-actin bound by drebrin A from dendritic spines. PLoS One 2014, 9 (1), e85367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koskinen M; Bertling E; Hotulainen R; Tanhuanpaa K; Hotulainen P, Myosin IIb controls actin dynamics underlying the dendritic spine maturation. Mol Cell Neurosci 2014, 61, 56–64. [DOI] [PubMed] [Google Scholar]
- 70.Ryu J; Liu L; Wong TP; Wu DC; Burette A; Weinberg R; Wang YT; Sheng M, A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron 2006, 49 (2), 175–82. [DOI] [PubMed] [Google Scholar]
- 71.Lein ES; Hawrylycz MJ; Ao N; Ayres M; Bensinger A; Bernard A; Boe AF; Boguski MS; Brockway KS; Byrnes EJ; Chen L; Chen L; Chen TM; Chin MC; Chong J; Crook BE; Czaplinska A; Dang CN; Datta S; Dee NR; Desaki AL; Desta T; Diep E; Dolbeare TA; Donelan MJ; Dong HW; Dougherty JG; Duncan BJ; Ebbert AJ; Eichele G; Estin LK; Faber C; Facer BA; Fields R; Fischer SR; Fliss TP; Frensley C; Gates SN; Glattfelder KJ; Halverson KR; Hart MR; Hohmann JG; Howell MP; Jeung DP; Johnson RA; Karr PT; Kawal R; Kidney JM; Knapik RH; Kuan CL; Lake JH; Laramee AR; Larsen KD; Lau C; Lemon TA; Liang AJ; Liu Y; Luong LT; Michaels J; Morgan JJ; Morgan RJ; Mortrud MT; Mosqueda NF; Ng LL; Ng R; Orta GJ; Overly CC; Pak TH; Parry SE; Pathak SD; Pearson OC; Puchalski RB; Riley ZL; Rockett HR; Rowland SA; Royall JJ; Ruiz MJ; Sarno NR; Schaffnit K; Shapovalova NV; Sivisay T; Slaughterbeck CR; Smith SC; Smith KA; Smith BI; Sodt AJ; Stewart NN; Stumpf KR; Sunkin SM; Sutram M; Tam A; Teemer CD; Thaller C; Thompson CL; Varnam LR; Visel A; Whitlock RM; Wohnoutka PE; Wolkey CK; Wong VY; Wood M; Yaylaoglu MB; Young RC; Youngstrom BL; Yuan XF; Zhang B; Zwingman TA; Jones AR, Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445 (7124), 168–76. [DOI] [PubMed] [Google Scholar]
- 72.Canela L; Lujan R; Lluis C; Burgueno J; Mallol J; Canela EI; Franco R; Ciruela F, The neuronal Ca(2+) -binding protein 2 (NECAB2) interacts with the adenosine A(2A) receptor and modulates the cell surface expression and function of the receptor. Mol Cell Neurosci 2007, 36 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- 73.Canela L; Fernandez-Duenas V; Albergaria C; Watanabe M; Lluis C; Mallol J; Canela EI; Franco R; Lujan R; Ciruela F, The association of metabotropic glutamate receptor type 5 with the neuronal Ca2+-binding protein 2 modulates receptor function. J Neurochem 2009, 111 (2), 555–67. [DOI] [PubMed] [Google Scholar]
- 74.Kaasinen SK; Goldsteins G; Alhonen L; Janne J; Koistinaho J, Induction and activation of protein kinase C delta in hippocampus and cortex after kainic acid treatment. Exp Neurol 2002, 176 (1), 203–12. [DOI] [PubMed] [Google Scholar]
- 75.Fiermonte G; Palmieri L; Todisco S; Agrimi G; Palmieri F; Walker JE, Identification of the mitochondrial glutamate transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem 2002, 277 (22), 19289–94. [DOI] [PubMed] [Google Scholar]
- 76.Vermeren M; Lyraki R; Wani S; Airik R; Albagha O; Mort R; Hildebrandt F; Hurd T, Osteoclast stimulation factor 1 (Ostf1) KNOCKOUT increases trabecular bone mass in mice. Mamm Genome 2017, 28 (11–12), 498–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zurner M; Mittelstaedt T; tom Dieck S; Becker A; Schoch S, Analyses of the spatiotemporal expression and subcellular localization of liprin-alpha proteins. J Comp Neurol 2011, 519 (15), 3019–39. [DOI] [PubMed] [Google Scholar]
- 78.Spangler SA; Jaarsma D; De Graaff E; Wulf PS; Akhmanova A; Hoogenraad CC, Differential expression of liprin-alpha family proteins in the brain suggests functional diversification. J Comp Neurol 2011, 519 (15), 3040–60. [DOI] [PubMed] [Google Scholar]
- 79.Shimizu H; Fukaya M; Yamasaki M; Watanabe M; Manabe T; Kamiya H, Use-dependent amplification of presynaptic Ca2+ signaling by axonal ryanodine receptors at the hippocampal mossy fiber synapse. Proc Natl Acad Sci U S A 2008, 105 (33), 11998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaya L; Meissner B; Riedl MC; Muik M; Schwarzer C; Ferraguti F; Sarg B; Lindner H; Schweigreiter R; Knaus HG; Romanin C; Bandtlow CE, Direct association of the reticulon protein RTN1A with the ryanodine receptor 2 in neurons. Biochim Biophys Acta 2013, 1833 (6), 1421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu X; Betzenhauser MJ; Reiken S; Meli AC; Xie W; Chen BX; Arancio O; Marks AR, Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell 2012, 150 (5), 1055–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shibata H; Kanadome T; Sugiura H; Yokoyama T; Yamamuro M; Moss SE; Maki M, A new role for annexin A11 in the early secretory pathway via stabilizing Sec31A protein at the endoplasmic reticulum exit sites (ERES). J Biol Chem 2015, 290 (8), 4981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katoh Y; Ritter B; Gaffry T; Blondeau F; Honing S; McPherson PS, The clavesin family, neuron-specific lipid- and clathrin-binding Sec14 proteins regulating lysosomal morphology. J Biol Chem 2009, 284 (40), 27646–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 – Box plots showing differential expression between CA1 and CA2 for proteins included in chord plots
Supplemental Figure 2 – Box plots showing differential expression between CA1 and CA2 across all mass spectrometry experiments for proteins included in chord plots
Supplemental Table 1 – Meta-analysis of quantitative proteomics
Supplemental Table 2 – Additional CA2 markers in TMT proteomics
Supplemental Table 3 – Information on dissected mouse samples
Supplemental Table 4 – Significance of differentially expressed proteins in modules