Abstract
Purpose:
In this cross-sectional pilot study we set out to discover a non-invasive biomarker that could distinguish steroid-resistant nephrotic syndrome (SRNS) from steroid-sensitive nephrotic syndrome (SSNS).
Experimental design:
Urine and clinical data were collected from patients with idiopathic nephrotic syndrome and healthy controls. Using SELDI-TOF-MS, we identified an 11-fold upregulated 13.8 kDa fragment of α 1-B glycoprotein (A1BG) in urine in SRNS. To validate our findings, A1BG was detected by Western blot. Creatinine was measured and transformed to glomerular filtration rate (GFR) by the new Schwartz formula and classified to chronic kidney disease (CKD) stage. p-Values were determined by unpaired t-test and Mann–Whitney rank sum test. Microalbumin was also measured to determine albumin/creatinine ratios.
Results:
The 13.8 kDa A1BG was present in 7 of 19 patients with SRNS; but absent in all SSNS (n = 15) and controls (n = 10). The A1BG+ patients had lower GFR than A1BG− patients (p<0.009) and tended to have higher CKD stage.
Conclusion and clinical relevance:
The 13.8 kDa A1BG fragment had a high discriminatory power for steroid resistance in pediatric nephrotic syndrome, but is only present in a subset of patients. Additional longitudinal studies are required to determine the usefulness of this biomarker as a non-invasive predictive marker of therapeutic response.
Keywords: Biomarker, Focal segmental glomerulosclerosis, Minimal change disease, Nephrotic syndrome, Steroid resistance
1. Introduction
Idiopathic nephrotic syndrome (INS) is the most common glomerular disease of childhood, characterized by the presence of proteinuria, edema, hypoalbuminemia and hypercholesterolemia. Glucocorticoids are the first-line treatment to achieve remission targeting a reduction in urine protein levels. Long-term prognosis is directly related to steroid responsiveness. Steroid-resistant nephrotic syndrome (SRNS) and biopsy-proven focal segmental glomerulosclerosis (FSGS) are significantly associated with poor outcome [1-4]. FSGS is a pathologic diagnosis that is steroid resistant (SRNS) in approximately 70% of cases, [5] and is substantially increasing in incidence [6-8]. The 30% of FSGS patients who respond to steroids generally have better prognosis. FSGS is the most common primary glomerular disease leading to end-stage renal disease (ESRD) in children [9]. An additional complication in patients with FSGS leading to ESRD is the high rate of recurrence (30–40%) following transplant [10].
There are currently no diagnostic tests that accurately predict steroid responsiveness in pediatric nephrotic syndrome or distinguish SRNS from steroid-sensitive nephrotic syndrome (SSNS). The initial prolonged course of daily, high-dose corticosteroid therapy thus serves both as a diagnostic and therapeutic maneuver. Renal biopsy and histological evaluation remain as the standard for providing the pathologic diagnosis in adults. Histological features, such as degree of interstitial fibrosis and variants of FSGS, help predict clinical courses and renal outcomes [11-14]. Pediatric patients are typically not biopsied at initial presentation, but are assumed to have minimal change disease (MCD) and are treated as such, based on earlier studies demonstrating that up to 78% of new onset nephrotic syndrome in children is MCD [15]. It should also be noted that single biopsies are thought to under diagnose FSGS in children because of the focal nature of the lesion, and the fact that there are generally few glomeruli present in pediatric needle biopsies. Therefore, identification of urinary biomarkers that predict steroid responsiveness or differentiate SR/FSGS from SS/MCD would benefit patients with SRNS by avoiding their exposure to high-dose corticosteroids.
A few studies have investigated differential biomarkers for FSGS and MCD in urine. For instance, the viable podocytes were detected only in the urine of FSGS but not in MCD and healthy controls [16], and the urinary cytokine TGF-β(1) was identified as a marker associated with FSGS, but failed to predict steroid responsiveness [17].
Advances in proteomics have hastened the discovery of urinary biomarkers, which may lead to early diagnosis, identify risk factors, and predict course and outcome in kidney diseases [18-22]. Urine proteomics is emerging as a potential rich source of non-invasive biomarkers of drug responsiveness in NS. Urine proteomic profiling studies of pediatric nephrotic syndrome using gel-based analysis or performing SELDI-TOF-MS has been published [23-26]. Some potential biomarkers have been discovered but thorough validation has not been conducted.
In this pilot study we identified a candidate biomarker for pediatric SRNS using SELDI, coupled with MALDI-TOF-MS, and conducted initial validation studies using a qualitative Western blot analysis in an expanded patient population to determine the value of this candidate biomarker in differentiating steroid sensitivity from resistance in idiopathic pediatric nephrotic syndrome.
2. Materials and methods
2.1. Study design, patient groups and sample collection
This is a cross-sectional study approved by the Institutional Review Board of the Cincinnati Children’s Hospital Medical Center, OH, USA. Written informed consent was obtained from each patient or legal guardian before enrollment. Thirty-four patients between the ages of 2–19-y old diagnosed with nephrotic syndrome were enrolled in this pilot study. Normal age-matched healthy controls were recruited from family members of colleagues. Urine was collected as part of a routine clinic visit, spun for 5 min at 2000 × g, and stored at −80°C. Demographic data, diagnosis regarding steroid responsiveness, renal biopsy results, immunosuppression and serum creatinine at the time of sampling were obtained (Table 1). Creatinine was transformed to glomerular filtration rate (GFR) by the new Schwartz formula [27] and classified to chronic kidney disease (CKD) stage [28].
Table 1.
Patient demographics
| Variable | SRNS (n = 19) | SSNS (n = 15) | Control (n = 10) | p-Value |
|---|---|---|---|---|
| Age (y; mean±SD) | 12.8±5.1 | 6.5±3.7 | 10.5±4.5 | 0.001a) |
| Sex: male:female | 11:8 | 10:5 | 8:2 | 0.49 |
| Race (%) | ||||
| African American | 47.4 (9) | 46.7 (7) | 50 (5) | 0.76 |
| Caucasian | 47.4 (9) | 40 (6) | 50 (5) | |
| Other | 5.2 (1) | 13.3 (2) | 0 (0) | |
| Hypertension (%) | 8 (42.1) | 2 (13.3) | 0.12 | |
| Immunosuppressant | ||||
| Steroid | 26.3 (5) | 73.3 (11) | NA | 0.003a) |
| CNI | 42.1 (8) | 13.3 (2) | NA | |
| MMF | 36.8 (7) | 0 (0) | NA | |
| ACEI/ARB | 8 (42.1) | 0 (0) | NA |
Denotes significant difference.
SRNS was defined when a patient failed to respond to a standard steroid therapy (2 mg/kg/day) for at least 8 wk. SSNS was defined as the ability to achieve remission in response to the steroid treatment within 8 wk after initial diagnosis as evidenced by normalization of urine protein to a negative reading on urine dipstick. Normal controls were children who did not have any history or evidence of renal disease. Nineteen patients with SRNS, 15 patients with SSNS and 10 healthy controls were included.
Fresh mid-stream urine samples were collected (approximately 5–10 mL) in a standard urine container without preservatives and protease inhibitors. The samples were initially placed at 4°C (for not more than 24 h), then centrifuged at 2000 × g for 5 min at room temperature (RT), and aliquoted supernatants were stored at −80°C. Not more than two freeze–thaw cycles were permissible for each sample.
2.2. Urine proteomics with SELDI-TOF-MS
Initial samples (nine SRNS and seven SSNS samples) were subjected to SELDI-TOF-MS and analyzed using the ProteinChip SELDI System Enterprise Edition (Bio-Rad Laboratories, Hercules, CA, USA). Four types of ProteinChips with different chromatographic surfaces were pre-equilibrated in the following buffers: weak cation exchange (CM10), 0.1 M sodium acetate, pH 4.0; hydrophobic/reverse phase (H50), 10% ACN, 0.1% TFA; metal-affinity (IMAC30) arrays charged with copper sulfate, 0.1 M sodium phosphate, 0.5 M sodium chloride, pH 7.0; Normal phase (NP20), 0.1% sodium phosphate, pH 6.0. Equal volumes (20 μL) of urine were diluted 1:5 in a chip-specific buffer and incubated on spot for 60 min. Spots were washed with the chip-specific buffer and distilled water as per the manufacturer’s instructions. Bound proteins were co-crystalized on spot with 50% saturated sinapinic acid (SPA) (2 × 1 μL) prepared in 50% ACN, 0.5% TFA. Low mass spectra (0–20 000 Da) were obtained with a laser intensity of 2500 nJ for CM10, IMAC30, H50 and NP20 chips. High mass spectra (20 000–200 000 Da) were obtained with laser intensities of 4000 nJ for all Proteinchips. The resulting spectra were calibrated using All-In-One Peptide/Protein Standards (Bio-Rad Laboratories).
2.3. Isolation of candidate biomarker
The biomarker that displayed the largest difference between SRNS and SSNS groups was selected for further investigation. A pooled urine sample from two SRNS patients with a high intensity value for the candidate peak was used for subsequent identification steps. Briefly, 400 μL of urine was centrifuged at 4000 rpm through a Microcon Ultracel YM-50 50kDa molecular weight filter (Millipore, Billerica, MA, USA) to remove albumin and the flow through was collected. The filter was then washed with 400 μL of 10, 15, 20 and 25% ACN to elute any small proteins that may be bound to albumin and the flow through was collected. The fractions were lyophilized in a vacuum centrifuge, for 2.5 h at RT and reconstituted with a Laemmli sample buffer (Bio-Rad Laboratories) for SDS-PAGE and boiled for 10 min. Gel electrophoresis was performed by using 12% Tris-glycine gels with molecular weight standard markers (Invitrogen, Carlsbad, CA, USA). Proteins were visualized with MS compatible silver stain (Pierce Silver Stain for MS, Thermo Fisher Scientific, Rockford, IL, USA). The 20% ACN eluate lane had a very distinct band in the appropriate mass range. This band was selected for identification by MS/MS (Supporting Information).
2.4. Protein identification by tandem mass spectrometry
Proteins were identified in the University of Cincinnati Proteomics Laboratory (UC-PL) using an Applied Biosystems 4800 MALDI-TOF/TOF instrument (running the Explorer 4000 Series operation software, ver 3.5.28) from peptides obtained by in-gel trypsin digestion of silver-stained protein bands as described in detail elsewhere [29]. MALDI-TOF spectra were collected from peptides spotted in 2.5 mg/mL CHCA matrix in positive-ion reflector mode across a range of 875–4000 m/z. The top 15 peptide signals (excluding known auto-digestion peptides of trypsin) were further fragmented in positive-ion TOF/TOF mode for protein identification. The fragmentation spectra were searched against 232 509 protein entries annotated as “human” on an in-house 4 processor MASCOT (ver. 2.2.07 from Matrix Science) cluster. The annotated “human” sequences were parsed from the entire NCBInr database dated July 8, 2010. Specific search criteria were limited to trypsin digestion with up to two missed cleavages, peptide mass tolerance of 25 ppm, MS/MS fragmentation tolerance of 0.25 Da, variable modifications including oxidized methionine and deamidation of glutamine and asparagine, and a fixed modification of carbamidomethyl-cysteine due to the reduction and alkylation of cysteine with iodoacetamide prior to trypsin digestion [29]. Protein identification criteria included a minimum of two fragmentation spectra and a protein significance threshold of p<0.005. The results are presented as Supporting Information with the total MASCOT score, the protein sequence coverage, the individual peptide sequences (with ion scores) and one fragmentation spectrum for each protein.
2.5. Initial validation
Qualitative study by Western blot analysis was performed in an expanded patient set with 19 samples of patients with SRNS, 15 with SSNS and 10 healthy controls. We used the full-length recombinant α 1-B glycoprotein (A1BG) protein (67.14 kDa) (P01, Abnova, Walnut, CA, USA), as the positive control. MagicMarlc™ XP and SeeBlue® Prestained Standard (Invitrogen) were used as the molecular weight markers. Thirty microliters of urine sample from each patient and control were subjected to electrophoresis on 12% SDS-polyacrylamide gels and transferred to PVDF membranes (Millipore, Bedford, MA, USA). The membranes were incubated with monoclonal mouse anti-human A1BG (A1BG antibody [51A6], Abcam, Cambridge, MA, USA) at 4°C for overnight, and followed by the secondary antibody, peroxidase-conjugated goat anti-mouse IgG (Millipore), at RT for 1 h. The protein and fragmentations were detected by ECL (GE Healthcare, Chalfont, UK).
2.6. Urine α 1-B glycoprotein
We developed an ELISA for A1BG to determine total urine A1BG levels. Urine creatinine was measured and A1BG concentrations expressed as ng/mg creatinine. We applied a direct ELISA protocol by using the same standard protein and primary antibody as the Western blot. Briefly, 100 μL full-length A1BG recombinant protein in various concentrations using twofold serial dilutions (1000, 500, 250, 125, 62.5, 31.25, 15.6, 7.8, 3.9, 1.95 ng/mL and blank) and 100 μL urine samples were added to a 96-well, Immulon 4HBX ELISA plate (Thermo Fisher Scientific, Waltham, MA, USA) and coated overnight at 4°C. The plate was washed three times with 1 × PBS and blocked with 1% BSA in buffer “A” (0.5 M NaCl, 3 mM KCl, 1 × Na2HPO4/KH2PO4 pH 7.2 and 1% Triton X-100) for 2-h incubation. After washing three times with buffer A, the plate was incubated with monoclonal mouse anti-human A1BG (1 μg/mL in blocking buffer) at RT for 2h, then washed three times and incubated with 2 μg/mL biotinylated goat anti-mouse IgG (ab6788, Abcam) in blocking buffer at RT for 1 h. The plate was washed three times and 50 μL avidin–HRP (1:2000; Dako, Glostrup, Denmark, UK) was added at RT for 1 h. Plates were washed three times and 50 μL TMB substrate reagent (BD Biosciences, Franklin Lakes, NJ, USA) was added and the plates were protected from light. The optical density at 450 nm was measured by microtiter plate spectrophotometer (Benchmark Plus, Bio-Rad) at RT for 20, 25 and 30 min. A standard curve was created using a cubic regression and a log (concentration)-linear (absorbance) transformation. The urine A1BG level was averaged from the 3-time readings. The precision values of the intra-assay and inter-assay were also determined as the coefficient of variance (CV). The mean intra-CV values were 5.92% and the inter-assay CV was 11%.
2.7. Urine microalbumin and creatinine
Urine microalbumin was measured by immunoturbidimetry using MALB flex reagent on a Siemens Dimension Xpand Plus with HM clinical analyzer (Siemens Healthcare, New York, NY, USA). Intra- and inter-assay CV values for the assay are 2.3 and 5.9%, respectively. Creatinine values were obtained using a colorimetric creatinine kit based on the Jaffe reaction (Enzo Life Sciences, Plymouth Meeting, PA, USA). Intra- and inter-assay CVs for the creatinine assay are 2.4 and 3.15%, respectively.
2.8. Statistics
For SELDI analysis, spectra were analyzed with ProteinChip Data Manager Software 3.0.7 (Bio-Rad Laboratories). Peak intensity was normalized to total ion current. Spectra were baseline subtracted and clustered using default settings. Peaks with a signal-to-noise ratio of > 5 in a mass window of 0.3% found in at least 20% of spectra were identified as clusters. To measure the sensitivity and specificity for a protein peak in distinguishing steroid responsiveness, receiver-operating characteristic (ROC) curves were generated and the area under the curve (AUC) calculated. Peaks demonstrating a 10-fold difference in intensity between groups, with a p<0.05 using a Mann–Whitney rank sum analysis and an AUC greater than 0.7 were considered statistically significant and were subjected to further analysis.
SigmaStat 3.5 was used for the analysis of clinical variables and demographics. p-Values were determined by Chi-square, unpaired t-test or Mann–Whitney rank sum analysis.
3. Results
3.1. Discovery and Identification of a 13.8 kDa fragment of α 1-B glycoprotein
A comparison of SELDI spectra from SSNS and SRNS patient urines revealed ten significantly different (p<0.05) peaks with a minimum threefold change in intensity that could distinguish the two groups, with an ROC of at least 0.70 (Table 2). The most significantly different peak was at 13.8 kDa on the H50 surface (p = 0.039, AUC 0.73), which was upregulated 11-fold in SRNS compared with SSNS (Fig. 1). When the data for this marker were normalized to urine creatinine, the differences were even greater (41-fold increase in SRNS, p = 0.01 by Mann–Whitney rank sum analysis). The wide margin of difference between the groups led us to believe that this could be a potential biomarker for distinguishing steroid responsiveness; therefore, we decided to further analyze the protein to determine its identity. It should be noted that a peak of approximately the same size was also found on the IMAC30 surface and was upregulated 3.6-fold (p = 0.003; AUC 0.76). This peak exhibited the same differential behavior between groups and had an average mass of 13841 Da, compared with 13827 Da on the H50 surface. However, the mass resolution of the SELDI instrument is only 0.1% and peak clustering occurs in a 0.3% mass window, thus these could not be confidently identified as separate putative biomarkers. Two patient samples with high intensity values for the 13.8 kDa peak were pooled, lyophilized, processed to remove albumin and the 13.8 kDa peak was isolated by PAGE. Proteins were visualized by MS compatible silver stain. The peak was subjected to in-gel trypsin digestion, c18ZipTip extraction and identified by MALDI-TOF/TOF as A1BG, a member of the immunoglobulin superfamily.
Table 2.
SELDI peaks with >threefold change in expression
| Surface | p-Value | AUC | m/z | Fold change |
|---|---|---|---|---|
| IMAC | 0.002 | 0.88 | 4358 | 8 |
| 0.003 | 0.76 | 13841 | 3.6 | |
| H50 | 0.013 | 0.84 | 6984 | 3 |
| 0.016 | 0.84 | 6442 | 4 | |
| 0.028 | 0.22 | 4740 | −3.8 | |
| 0.028 | 0.76 | 15810 | 3.3 | |
| 0.039 | 0.73 | 13827 | 11 | |
| CM10 | 0.007 | 0.84 | 4030 | 3.3 |
| 0.013 | 0.84 | 7048 | 6.3 | |
| 0.047 | 0.76 | 7505 | 4.3 |
Figure 1.
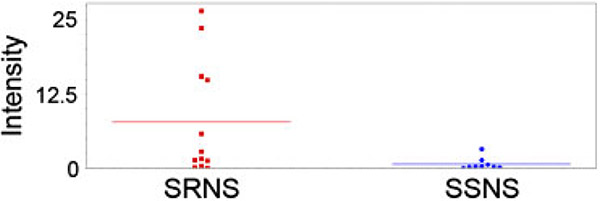
Scatterplot of peak intensity of the 13.8 kDa peak in SRNS vs. SSNS. The horizontal lines represent the mean value and each dot represents the intensity of the peak in one patient sample. The mean intensity is 11-fold higher (p<0.039) in SRNS patients than in SSNS patients with an AUC of 0.73.
3.2. Initial validation
The expanded validation set included 19 patients with SRNS, 15 patients with SSNS and 10 healthy controls. In conjunction with renal pathology, there were 14 SRNS with biopsy-proven FSGS (SR/FSGS), 1 SRNS with MCD, 2 SSNS with FSGS and the remainder had no biopsy. On Western blot, there were multiple fragments of A1BG protein in several urine samples. The representative appearance of A1BG bands on Western blot among controls, SSNS and SRNS patients can be seen in Fig. 2. The 13.8 kDa band was the fragment of interest. It was present in seven patients with SRNS, but never appeared in patients with SSNS or in healthy controls. The sensitivity of 13.8 kDa A1BG fragment to differentiate SRNS from SSNS was 37%, specificity was 100% and positive discriminatory value was 100%. Six of the seven SRNS presenting the fragment were SR/FSGS. The remaining A1BG+ patient was steroid resistant and presented with clinical symptoms in line with FSGS pathology, but has not yet been biopsied for definitive diagnosis.
Figure 2.
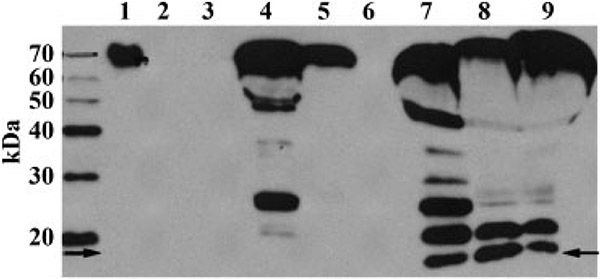
Western blot of A1BG. Lane 1 is 50 ng recombinant full-length human A1BG. This lane was moved in the image from the left side of the MW markers to its current location, but was otherwise not manipulated. Lanes 2 and 3 are healthy controls that demonstrate a lack of protein relative to the disease samples. Lanes 4–6 are from SSNS patients and lanes 7–9 are from SRNS patients. Note the appearance of the lower band (arrows) at approximately 14 kDa. This band is present in the SRNS samples, which display a greater degree of fragmentation, but is absent in the SSNS samples.
Demographic data, clinical and laboratory parameters of patients with nephrotic syndrome who had the 13.8 kDa fragment (A1BG+) and patients who did not have the fragment (A1BG−) are listed in Table 3.
Table 3.
Demographic and clinical variables in patients with and without the 13.8 kDa fragment of A1BG
| Variable | A1BG+ (n = 7) | A1BG− (n = 27) | p-Value |
|---|---|---|---|
| Age (y; mean±SD) | 12.7±2.8 | 9.3±5.8 | 0.149 |
| Sex: male:female | 5:2 | 16:11 | 0.68 |
| Race (%) | 0.56 | ||
| African American | 42.9 (3) | 48.1 (13) | |
| Caucasian | 57.1 (4) | 40.7 (11) | |
| Other | 0 (0) | 11.1 (3) | |
| Pathology (%) | |||
| FSGS | 85.7 (6) | 37 (10) | |
| MCD | 0 (0) | 3.7 (1) | |
| No biopsy | 14.3 (1) | 59.3 (16) | |
| Hypertension (%) | 57.1 (4) | 22.2 (6) | 0.157 |
| Immunosuppressant (%) | 0.005a) | ||
| Steroid | 14.3 (1) | 56 (15) | |
| CNI | 71.4 (5) | 22.2 (6) | |
| MMF | 71.4 (5) | 7.4 (2) | |
| ACEI/ARB (%) | 57.1 (4) | 14.8 (4) | 0.037 |
| Cr (mg/dL) | 2.1±1.9 | 0.8±1.1 | 0.01a) |
| GFR (mL/min/1.73m2) | 54.7±36.3 | 97.7±36.4 | 0.009a) |
| CKD stage | 2.7±1.50 | 1.67±0.92 | 0.06 |
| A1BG level (ng/mL) | 1831.95±1593.68 | 1814.58±1748.17 | 0.96 |
| A1BG/Cr (ng/mg) | 1333.66±622.39 | 3350.53±3515.39 | 0.33 |
| ACR (mg/g) | 17 451.07±6284.66 | 18 433.78±33 381.01 | 0.125 |
Denotes significant difference.
Between A1BG+ and A1BG− groups, there were no significant differences in age, gender, race and the prevalence of hypertension, although the patients in A1BG+ were on average older and the proportion of patients who developed hypertension was higher, but failed to reach significance. The pattern of immunosuppressive administration was considerably different (p = 0.005). The patients in the A1BG+ group were prescribed corticosteroids in the lower proportion than in A1BG− patients, while calcineurin inhibitors (CNI) and mycophenolate mofetil (MMF) were prescribed substantially more to A1BG+ patients. Angiotensin-converting enzyme inhibitors (ACEIs) and angiotensin receptor blockers (ARBs) were also more likely to be used in A1BG+ patients (p = 0.037).
In terms of renal function, the patients with the 13.8 kDa A1BG fragment had significant lower renal function than the patients without the fragment (p<0.05). Serum creatinine levels were 2.1±1.9 and 0.8±1.1 mg/dL in A1BG+ and A1BG− groups, respectively. The eGFR of A1BG+ patients (54.7±36.3mL/min/1.73m2) was lower than in A1BG− patients (97.7±36.4mL/min/1.73m2; p<0.009). A1BG+ was more likely to have a higher stage of CKD, but this did not reach statistical significance (p = 0.06).
To quantitate urine A1BG, we developed a direct ELISA using the same primary antibody we used for Western blot. Furthermore, Spearman correlation for urine A1BG, A1BG per creatinine ratio (A1BG/Cr) (ng/mg) and U.ACR (mg/g) were determined. There was a good correlation between urine A1BG/Cr (ng/mg) and U.ACR (mg/g) (r = 0.42, p = 0.01), but not for U.A1BG (ng/mL) vs U.ACR (mg/g) (r = 0.19, p = 0.27). In spite of the difference in the fragmentation of A1BG, there were not significant differences in the level of urine A1BG and A1BG/Cr between A1BG+ and A1BG− groups (p = 0.96 and 0.33, respectively). Urine A1BG and A1BG/Cr between SRNS and SSNS were also not significantly different (p = 0.7 and 0.256, respectively). For urine albumin/creatinine ratios (ACR), there were no significant differences between A1BG+ and A1BG− groups (p = 0.125), or between SRNS and SSNS patients (p = 0.266, data not shown).
3.3. Fragmentation pattern of α 1-B glycoprotein
The Western blot was performed for each sample of 19 patients with SRNS, 15 patients with SSNS (9 relapsed and 6 remission) and 10 healthy controls. We used the full length of A1BG recombinant protein as the protein of reference. The Western blot revealed A1BG bands for each urine sample at the same molecular weight of the protein standard. In addition, there were multiple fragments of A1BG protein present in several urine samples. The degradation was shown in 17 SRNS (89.5%), including all SRNS with 13.8 kDa fragment and 7 SSNS (46.7%). No SSNS patients with remission or healthy controls presented this fragmentation pattern.
4. Discussion
SRNS and FSGS are significantly associated with poor outcome. Of the various methods of urine proteomic technology, SELDI-TOF-MS provides the advantage of rapid high-throughput urinary protein profiling. Recent studies have reported potential biomarkers of steroid resistance by using SELDI-TOF-MS. One study performed SELDI-TOF-MS analysis using Biomarker Pattern Software™ and the boosting algorithm to determine a protein of mass 4.144 kDa as a candidate biomarker [25]. Another study revealed a group of five peaks distinguishing SRNS. They identified one peak as B2-microglobulin (B2M). However, there has been no follow up of clinical validation study in these groups of patients.
This study was intended to apply SELDI-TOF-MS to discover urinary biomarkers that would be able to differentiate steroid resistance in pediatric INS and then to validate any candidate biomarker for its discriminatory ability in an extended number of patients. We identified a protein peak associated with steroid resistance and identified it as a 13.8 kDa fragment of A1BG. A1BG is an approximately 63 kDa plasma glycoprotein, containing a single peptide of 474 amino acid residues with four glucosamine oligosaccharides [30]. It is a member of the immunoglobulin superfamily, but the function is not known. The 13.8 kDa fragment of A1BG was present in 7/19 SRNS patients, 6 of whom were SR/FSGS. The 7th patient presented with clinical symptoms of FSGS and steroid resistance, but did not have a biopsy and therefore cannot be labeled as FSGS. Although the fragment appeared only in a subset ofpatients with SRNS, the presence of the fragment in these samples was indicative of steroid resistance 100% of the time and was associated with significantly reduced renal function. The reduction of renal function represents greater severity, or chronicity, of the disease in these patients. FSGS is a diagnosis that can represent differing pathologies that are often classified as variants. A limitation of this study is that our pathology reports did not specify FSGS variants. It is possible that this subset of fragment positive patients represents a specific variant of FSGS, but we cannot make that pathologic determination in this cohort. A potential confounding variable that needs to be explored in greater depth in future studies is the correlation of this fragmentation pattern with the prescription of potential nephrotoxic drugs to control the disease in these patients, especially CNI, ACEI and ARB.
Despite the difference in the presence of the 13.8 kDa fragment, the levels of total urine A1BG and A1BG/Cr were not significantly different between A1BG+ and A1BG− groups. Therefore, the total levels of A1BG and the A1BG/Cr ratio were not related with the appearance of the fragment and do not differentiate steroid resistance and/or SR/FSGS. Nevertheless, there was a good correlation between total urine A1BG/Cr and ACR. This is probably the result of increased levels of urine protein, and not specific to A1BG.
In nephrotic syndrome the glomerular permeability barrier is altered resulting in proteinuria. The high incidence of recurrence of FSGS after renal transplantation has lead to the idea of circulating factor(s) contributing to pathology, but this is not clearly defined. Recently, a urine proteomic study in a different INS has demonstrated that not only intact albumin, but also numerous albumin fragments were detected in nephrotic urine [26]. The albumin fragmentation was also found in plasma. Another recent study found that patients with active FSGS or with post-transplantation recurrence had oxidized plasma albumin, suggesting that massive oxidation was playing a role in the pathogenesis of the proteinuria associated with active FSGS [31]. In our study, multiple fragments of A1BG protein were detected in urine as well. These were displayed in 90% of patients with SRNS and less than half in patients with SSNS. The degradation was not seen in patients with SSNS who were in remission or in healthy controls. It is probably that the degradation of urine A1BG is the result of active nephrotic syndrome, and might be explained by a similar pathogenesis proposed to albumin fragmentation. It might be argued that the use of protease inhibitors post-collection could assist in determining if the degradation was specific to the disease state, but several recent reports have generally found that the absence of protease inhibitors in urine proteomics do not alter their findings [32, 33]. Additionally, it can be detrimental to use protease inhibitors during the discovery phase of urine biomarker research because they can interfere with results obtained by SELDI analysis and mask potentially important differences. While it may be true that A1BG fragmentation occurs post-collection, it appears very specific nonetheless, and could reflect a pathological increase in the protease activity.
This study is not without limitations. This was a single-center cross-sectional study with a limited number of patients. It would be helpful to design a large prospective multi-center study to further investigate what makes the subset of patients that present the 13.8 kDa fragment of A1BG different than those that do not. Also, it will be important to design a longitudinal study in which patient samples are collected prior to steroid treatment to determine if this fragment can predict therapeutic response. In this pilot study the results suggest that the appearance of the 13.8 kDa A1BG fragment in nephrotic syndrome differentiates steroid resistance associated with unfavorable renal function, but since FSGS is a diagnostic pathology, as opposed to a unique disease entity, there could be underlying mechanistic differences resulting in differential A1BG fragmentation that will require a larger patient population and more detailed mechanistic studies to decipher.
Supplementary Material
Clinical Relevance.
INS is the most common glomerular disorder of childhood. Pathology on invasive biopsy remains the diagnostic method of choice for nephrotic syndrome in adults. Biopsies are used less often in children and are seen as less reliable for predicting steroid responsiveness. Prognosis correlates with steroid responsiveness, from sensitive (SSNS) to resistant (SRNS). SRNS is the most common acquired cause of ESRD in children. Non-invasive biomarkers that could predict steroid responsiveness would help patients to avoid unnecessary exposure to high-dose corticosteroids and help to tailor treatments with alternative drugs that are more likely to be beneficial for steroid-resistant patients. In this pilot study we have discovered and performed an initial validation on a candidate biomarker that differentiates steroid resistance in a subset of patients with nephrotic syndrome and correlates with poor renal function. Further analysis of this biomarker in future longitudinal studies may yield valuable insight into steroid-resistant pathological mechanisms of nephrotic syndrome and could potentially be useful as a predictive marker of steroid responsiveness.
Acknowledgments
Dr. Devarajan is funded in part by NIDDK R01 DK069749. Dr. Czech is supported by an NRSA fellowship grant F32 DK079545-01.
Abbreviations:
- A1BG
α 1-B glycoprotein
- ACR
albumin/creatinine ratio
- CKD
chronic kidney disease
- ESRD
end-stage renal disease
- FSGS
focal segmental glomerulosclerosis
- GFR
glomerular filtration rate
- INS
idiopathic nephrotic syndrome
- MCD
minimal change disease
- NS
nephrotic syndrome
- RT
room temperature
- SRNS
steroid-resistant nephrotic syndrome
- SSNS
steroid-sensitive nephrotic syndrome
5 References
- [1].Cattran DC, Rao P, Long-term outcome in children and adults with classic focal segmental glomerulosclerosis. Am. J. Kidney Dis 1998. 32, 72–79. [DOI] [PubMed] [Google Scholar]
- [2].Hari P, Bagga A, Jindal N, Srivastava RN, Treatment of focal glomerulosclerosis with pulse steroids and oral cyclophosphamide. Pediatr. Nephrol 2001, 16, 901–905. [DOI] [PubMed] [Google Scholar]
- [3].Gipson DS, Chin H, Presler TP, Jennette C et al. , Differential risk of remission and ESRD in childhood FSGS. Pediatr. Nephrol 2006, 21, 344–349. [DOI] [PubMed] [Google Scholar]
- [4].Roberti I, Vyas S, Long-term outcome of children with steroid-resistant nephrotic syndrome treated with tacrolimus. Pediatr. Nephrol 2010, 25, 1117–1124. [DOI] [PubMed] [Google Scholar]
- [5].Gipson DS, Gibson K, Gipson PE, Watkins S et al. , Therapeutic approach to FSGS in children. Pediatr. Nephrol 2007, 22, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bonilla-Felix M, Parra C, Dajani T, Ferris M et al. , Changing patterns in the histopathology of idiopathic nephrotic syndrome in children. Kidney Int. 1999, 55, 1885–1890. [DOI] [PubMed] [Google Scholar]
- [7].Dragovic D, Rosenstock JL, Wahl SJ, Panagopoulos G et al. , Increasing incidence of focal segmental glomerulosclerosis and an examination of demographic patterns. Clin. Nephrol 2005, 6, 1–7. [DOI] [PubMed] [Google Scholar]
- [8].Boyer O, Moulder JK, Somers MJ, Focal and segmental glomerulosclerosis in children: a longitudinal assessment. Pediatr. Nephrol 2007, 22, 1159–1166. [DOI] [PubMed] [Google Scholar]
- [9].Smith JM, Stablein DM, Munoz R, Hebert D et al. , Contributions of the transplant registry: the 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr. Transplant 2007, 11, 366–373. [DOI] [PubMed] [Google Scholar]
- [10].Korbet SM, Clinical picture and outcome of primary focal segmental glomerulosclerosis. Nephrol. Dial. Transplant 1999, 14, 68–73. [DOI] [PubMed] [Google Scholar]
- [11].Schwartz MM, Korbet SM, Rydell J, Borok R et al. , Primary focal segmental glomerular sclerosis in adults: prognostic value of histologic variants. Am. J. Kidney Dis 1995, 25, 845–852. [DOI] [PubMed] [Google Scholar]
- [12].Alexopoulos E, Stangou M, Papagianni A, Pantzaki A et al. , Factors influencing the course and the response to treatment in primary focal segmental glomerulosclerosis. Nephrol. Dial. Transplant 2000, 15, 1348–1356. [DOI] [PubMed] [Google Scholar]
- [13].Thomas DB, Franceschini N, Hogan SL, Ten Holder S et al. , Clinical and pathologic characteristics of focal segmental glomerulosclerosis pathologic variants. Kidney Int. 2006, 69, 920–926. [DOI] [PubMed] [Google Scholar]
- [14].Silverstein DM, Craver R, Presenting features and short-term outcome according to pathologic variant in childhood primary focal segmental glomerulosclerosis. Clin. J. Am. Soc. Nephrol 2007, 2, 700–707. [DOI] [PubMed] [Google Scholar]
- [15].A Report of the International Study of Kidney Disease in Children, Primary nephrotic syndrome in children: clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. Kidney Int. 1981, 20, 765–771. [DOI] [PubMed] [Google Scholar]
- [16].Nakamura T, Ushiyama C, Suzuki S, Hara M et al. , The urinary podocyte as a marker for the differential diagnosis of idiopathic focal glomerulosclerosis and minimal-change nephrotic syndrome. Am. J. Nephrol 2000, 20, 175–179. [DOI] [PubMed] [Google Scholar]
- [17].Woroniecki RP, Shatat IF, Supe K, Du Z et al. , Urinary cytokines and steroid responsiveness in idiopathic nephrotic syndrome of childhood. Am. J. Nephrol 2008, 28, 83–90. [DOI] [PubMed] [Google Scholar]
- [18].Bennett MR, Ravipati N, Ross G, Nguyen MT et al. , Using proteomics to identify preprocedural risk factors for contrast induced nephropathy. Proteomics Clin. Appl 2008, 2, 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nguyen MT, Ross GF, Dent CL, Devarajan P, Early prediction of acute renal injury using urinary proteomics. Am. J. Nephrol 2005, 25, 318–326. [DOI] [PubMed] [Google Scholar]
- [20].Parikh CR, Jani A, Mishra J, Ma Q et al. , Urine NGAL and IL-18 are predictive biomarkers for delayed graft function following kidney transplantation. Am. J. Transplant 2006, 6, 1639–1645. [DOI] [PubMed] [Google Scholar]
- [21].Hinze CH, Suzuki M, Klein-Gitelman M, Passo MH et al. , Neutrophil gelatinase-associated lipocalin is a predictor of the course of global and renal childhood-onset systemic lupus erythematosus disease activity. Arthritis Rheum. 2009, 60, 2772–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stangou M, Alexopoulos E, Papagianni A, Pantzaki A et al. , Urinary levels of epidermal growth factor, interleukin-6 and monocyte chemoattractant protein-1 may act as predictor markers of renal function outcome in immunoglobulin A nephropathy. Nephrology (Carlton) 2009, 14, 613–620. [DOI] [PubMed] [Google Scholar]
- [23].Magistroni R, Ligabue G, Lupo V, Furci L et al. , Proteomic analysis of urine from proteinuric patients shows a proteolitic activity directed against albumin. Nephrol. Dial. Transplant 2009, 24, 1672–1681. [DOI] [PubMed] [Google Scholar]
- [24].Khurana M, Traum AZ, Aivado M, Wells MP et al. , Urine proteomic profiling of pediatric nephrotic syndrome. Pediatr. Nephrol 2006, 21, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Woroniecki RP, Orlova TN, Mendelev N, Shatat IF et al. , Urinary proteome of steroid-sensitive and steroid-resistant idiopathic nephrotic syndrome of childhood. Am. J. Nephrol 2006, 26, 258–267. [DOI] [PubMed] [Google Scholar]
- [26].Candiano G, Musante L, Bruschi M, Petretto A et al. , Repetitive fragmentation products of albumin and alpha1-antitrypsin in glomerular diseases associated with nephrotic syndrome. J. Am. Soc. Nephrol 2006, 17, 3139–3148. [DOI] [PubMed] [Google Scholar]
- [27].Schwartz GJ, Munoz A, Schneider MF, Mak RH et al. , New equations to estimate GFR in children with CKD. J. Am. Soc. Nephrol 2009, 20, 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Eknoyan G, Levin NW, K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am. J. Kidney Dis 2002, 39, S1–S266. [PubMed] [Google Scholar]
- [29].Eismann T, Huber N, Shin T, Kuboki S et al. , Peroxir-edoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am. J. Physiol. Gastrointest. Liver Physiol 2009, 296, G266–G274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ishioka N, Takahashi N, Putnam FW, Amino acid sequence of human plasma alpha 1B-glycoprotein: homology to the immunoglobulin supergene family. Proc. Natl. Acad. Sci. USA 1986, 83, 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Musante L, Candiano G, Petretto A, Bruschi M et al. , Active focal segmental glomerulosclerosis is associated with massive oxidation of plasma albumin. J. Am. Soc. Nephrol 2007, 18, 799–810. [DOI] [PubMed] [Google Scholar]
- [32].Afkarian M, Bhasin M, Dillon ST, Guerrero MC et al. , Optimizing a proteomics platform for urine biomarker discovery. Mol. Cell. Proteomics 2010, 9, 2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Havanapan PO, Thongboonkerd V, Are protease inhibitors required for gel-based proteomics of kidney and urine? J. Proteome Res 2009, 8, 3109–3117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


