Abstract
Many genetic markers have been associated with variations in treatment response to analgesics, but none have been assessed in the context of combination therapies. In this study, the treatment effects of nortriptyline and morphine were tested for association with genetic markers relevant to pain pathways. Treatment effects were determined for single and combination therapies. A total of 24 functional Single Nucleotide Polymorphisms (SNPs) were tested within the gene loci of OPRM1, ABCB1, CYP2C19 and CYP2D6, COMT and HTR2A. Genotyping was performed in a population of neuropathic pain patients that previously participated in a clinical trial. For monotherapy, neither nortriptyline nor morphine responses were associated with SNPs. However, for nortriptyline + morphine combination therapy, the SNP rs1045642, within the drug efflux pump ABCB1 transporter significantly predicted analgesic response. The presence of the C allele accounted for 51% of pain variance in this subgroup in response to combination treatment. The T-allele homozygotes demonstrated only 20% improvement in pain scores, while the C-allele homozygotes 88%. There was no significant contribution of rs1045642 to the medication side-effects under all treatment conditions. The UK Biobank dataset was then used to validate this genetic association. Here, patients receiving similar combination therapy (opioid + tricyclic antidepressant) carrying the C allele of rs1045642 displayed 33% fewer body pain sites than patients without that allele, suggesting better pain control. In all, our results show a robust effect of the rs1045642 polymorphism on response to chronic pain treatment with a nortriptyline + morphine combination.
Introduction
Chronic neuropathic pain is a debilitating condition that affects 7 to 10% of the general population and requires challenging multidisciplinary treatment strategies.[11,22,87] Because first line medications usually provide only partial relief, an effort has been made to determine if combination therapies deliver more effective symptom control.[8,22] However, the great variability amongst individual patient response to analgesics has been a limiting factor in predicting treatment response and individualizing therapy. One important cause of this variability is related to the genetic variants associated with pain pathways and the metabolism of analgesic drugs.[1,63,84,85,94]
A recent crossover clinical trial involving patients being treated for neuropathic pain compared a morphine + nortriptyline combination to either drug alone, and found improved outcomes with combination therapy versus monotherapy.[24] Even though patients receiving combination therapy had significant improvement in pain scores overall, there were patients who responded very well and those who demonstrated little to no response. A question remains whether it is possible to distinguish which patients could potentially respond better to combination drug therapy. The current study is a secondary analysis of the results from this clinical trial, in which the treatment effects are examined in the context of different genetic variants.
Many genetic markers have been associated with variation in treatment response to opioids or tricyclic antidepressants. These mainly consist of single nucleotide polymorphisms (SNPs) within the mu opioid receptor (OPRM1) gene locus, Cytochrome P450 gene family (CYP2C19 and CYP2D6), catecholamine inactivator Catechol–O-Methyl Transferase (COMT), ATP-Binding Cassette B1 Transporter (ABCB1), and serotonin receptor 2A (HTR2A).[28,39,85,94] From this group of candidate markers, the OPRM1 SNP rs1799971 has demonstrated strong evidence linking it to opioid dose requirements, risk of dependence and pain intensity in conditions such as diabetic neuropathy and low back pain.[9,10,27,35,44,49] Genetic markers in drug metabolizing enzymes such as CYP2C19 and CYP2D6 have also been associated with altered drug plasma levels, treatment effect and incidence of side effects with medications including amitriptyline, nortriptyline and duloxetine. However, this evidence was obtained from studies focusing on psychiatric treatment response rather than pain.[30,31,80] Genetic variations in the COMT gene have been implicated in the risk of developing pain conditions, and in treatment response to analgesics, including opioids and antidepressants.[14,43,59,82] SNPs within the COMT and HTR2A genes have also been implicated in the incidence of chronic pain conditions and the risk of depression associated with pain.[13,28,45,77,93] The ABCB1 gene codes for a transporter (ABC) of a wide variety of drugs, and polymorphisms within its gene have been shown to have an effect on morphine concentrations in the central nervous system, and incidence of side effects and treatment effects with amitriptyline and nortriptyline.[33,36,65,68] Despite this growing awareness of the clinical implications of genetic variability, most genetic markers have not been tested in the setting of combination therapies or in relation to treatments in chronic pain. In this study, we describe a novel effect of the ABCB1 gene SNP rs1045642 in the treatment response to nortriptyline and morphine combination therapy for patients with neuropathic pain.
Materials and Methods
The neuropathic pain discovery cohort
The secondary genetic analysis was performed in a cohort of neuropathic pain patients from a double-blind crossover clinical trial by Gilron et al.[24] The study was conducted with the approval of the Queen’s University Health Sciences & Affiliated Teaching Hospitals Research Ethics Committee and all study subjects consented to participate. In brief, participants were recruited from January 2010 to May 2014, and randomized, using a double-dummy design, to receive a sequence of oral study medications: nortriptyline, morphine, and combination nortriptyline/morphine. The trial started with a 7-day baseline period with no antidepressant or opioid treatment, and then each of the three 6 week treatment periods consisted of a 3.5 week titration phase in which the study medication was titrated to reach the maximum tolerated dose (MTD), then a maintenance treatment phase where participants were treated with the MTD of the medication for 7 days, and finally a 7-day dose taper followed by a 4-day washout period prior to starting the next treatment period. Outcomes documented in this trial included, amongst others, the average daily pain intensity at MTD (0-10 numerical rating scale), percentage change in average daily pain from baseline to MTD, the Stanford Sleepiness Scale (SSS), the Patient Assessment of Constipation Symptoms (PAC-SYM), and postural changes in heart rate and blood pressure.[19,32,38]
The 36 participants who completed all three treatment periods from the original clinical trial were contacted and all consented to this secondary genetic analysis study. Thirty-two participants returned the saliva sample kit (Oragene DNA OG-500, DNA Genotek, Ontario, Canada) as requested. Four participants did not return the saliva sample kit by mail and were lost to follow up. These four participants were eliminated from the study. A total of 27 DNA samples were successfully obtained for genetic analysis, with five deemed not adequate for DNA extraction (inadequate volume, damaged container). DNA purification was performed in accordance with the manufacturer recommendation (DNA Genotek, Ontario, Canada) and genotyping for a total of 34 SNPs was performed by the Genome Quebec Innovation Center with Sequenom IPLEX Gold technology and using pre-designed TaqMan® assays (Applied Biosystems; Foster City, CA). Allele-specific fluorescence signals were distinguished by measuring endpoint 6-FAM or VIC fluorescence intensities at 508 nm and 560 nm, respectively, and genotypes were generated using Genotyper® Software V 1.3 (Applied Biosystems; Foster City, CA). The DNA Elution Buffer was used as a negative control, and K562 Cell Line DNA (Promega Corporation; Madison, WI) was included in each batch of samples tested as positive control. Results of two participants were further removed from the analysis because their analgesic responses were missing for morphine and combination therapy medications, resulting in a final sample of 25 participants.
Genetic Variant Selection
The following genetic variants were selected for our analysis based on previous literature linking genetic polymorphisms with analgesia/pain and drug metabolism (Table 1). This method has been chosen over the potential functionality of SNPs (or gene sequencing approach) because of the small sample size of the discovery cohort. We decided to apply the smallest list of candidate SNPs that might show association with the highest probability to reduce the burden of multiple testing correction. At the OPRM1 locus, we tested the single functional non-synonymous C>T variation at rs1799971. For the COMT locus, a set of four COMT SNPs (rs6269, rs4633, rs4818 and rs4680) were grouped to form three pain haplotypes: low pain sensitive (LPS), average pain sensitive (APS) and high pain sensitive (HPS) as previously described.[14,56,88] Then, the number of LPS alleles expressed by each subject was counted, creating three different risk groups: highly pain responsive group (0 LPS alleles), medium pain responsive group (1 LPS allele) and low pain responsive group (2 LPS alleles) following the method previously described by Tchivileva et al.[82] For the serotonin receptor pathway, rs6313 and rs7997012 were selected because they were shown to be associated with pain and with antidepressant response.[3,77] Two SNPs, rs1045642 and rs2032582 were selected from the ABCB1 gene. Finally, a functional grouping was used to assess variants within CYP2D6 and CYP2C19. Nortriptyline is metabolized in the liver by CYP2D6. Genetic variations of the gene coding for this enzyme have been shown to alter the drugś bioavailability depending on the level of activity of the enzyme [29]. CYP2C19 metabolism is a secondary path for nortriptyline, and thus it was included as a gene variant of interest as it is commonly associated with therapeutic and side effect profile of tricyclic antidepressants in general [66]. SNPs within the CYP2C19 and CYP2D6 gene locus were tested as diplotypes denoting enzymatic metabolic rate activity (PharmGKB), based on which patients can be categorized as poor (PM), intermediate (IM), extensive (EM) and ultra-rapid (UM) metabolizers. Among 25 patients, with respect to CYP2C19 genotypes, 3 were identified as IM, 11 as EM and 11 as UM. With respect to CYP2D6 genotypes, one patient was identified as PM, one patient was identified as IM, and the remaining 23 as EM. Thus, CYP2D6 genotypes have been removed from subsequent analyses due to low number of genetic variants carriers.
Table 1.
SNPs selected for the genetic association analysis.
| Gene or metabolic pathway | rs number | Location | Functional change | Gene symbol | Gene name | Reference |
|---|---|---|---|---|---|---|
| OPRM1 gene | rs1799971 | Exon 1 | Asn40Asp | OPRM1 | Opioid Receptor Mu 1 | [4,8–10,24,27,35,44,49] |
| COMT gene | rs6269 | Intron 2 | COMT | Catechol-O-Methyl Transferase | [14,43,59,82] | |
| rs4633 | Codon 62 | |||||
| rs4818 | Exon 4 | |||||
| rs4680 | Exon 4 | Val158Met | ||||
| Serotonin receptor | rs6313 | Exon 2 | T102C | HTR2A | 5-Hydroxytryptamine Receptor 2A | [28,45,77,94] |
| rs7997012 | 13:46837850 | |||||
| ABCB1 gene | rs1045642 | Exon 26 | C3435T | ABCB1 | ATP Binding Cassette Subfamily B | [33,36,65] |
| rs2032582 | Exon 21 | Ala893Thr | Member 1 | |||
| CYP2C19 metabolism | rs12248560 | Promoter | CYP2C19 | Cytochrome P450 Family 2 | [25,30,31,71,80] | |
| rs4244285 | Exon 5 | Subfamily C Member 19 | ||||
| rs4986893 | Exon 4 | |||||
| rs28399504 | Exon 1 | |||||
| rs56337013 | Exon 9 | |||||
| rs72552267 | Exon 3 | |||||
| rs72558186 | Intron 5 | |||||
| rs41291556 | Exon 3 | |||||
| CYP2D6 metabolism | rs1065852 | 22:42130692 | P34S | CYP2D6 | Cytochrome P450 Family 2 | [25,30,31,80] |
| rs201377835 | 22:42129910 | Subfamily D Member 6 | ||||
| rs5030862 | 22:42130668 | G42R | ||||
| rs72549357 | 22:42526657 | |||||
| rs28371706 | 22:42129770 | T107I | ||||
| rs1080985 | 22:42132375 | |||||
| rs72549354 | 22:42128815 | |||||
| rs59421388 | 22:42127608 | V338M | ||||
| rs35742686 | 22:42128242 | 259Frameshift | ||||
| rs769258 | 22:42130761 | |||||
| rs3892097 | 22:42128945 | Splicing defect | ||||
| rs28371725 | Intron 6 | Splicing defect | ||||
| rs1135840 | 22:42126611 | S486T | ||||
| rs5030655 | 22:42129084 | 118Frameshift | ||||
| rs5030867 | 22:42127856 | H324P | ||||
| rs5030865 | 22:42129033 | G169R | ||||
| rs5030656 | 22:42128174 | K281del | ||||
Validation analysis using the UK Biobank
The UK Biobank (UKBB) data (UKBB application number 20802) was used to validate our results from the neuropathic pain cohort. This validation step was considered for significant associations found in the discovery cohort. Thus, only the rs1045642 effect on combination treatment response was tested. The side effect profile or associations with other genotypes such as metabolic enzyme activity were not tested in the UK Biobank cohort. The UKBB is a database with genetic and phenotypic information of 40 to 69 year old people living in the United Kingdom [81]. A total of 502,000 participants were recruited between 2006 and 2010 and informed consent was obtained from all.
For the UKBB validation analysis, data were extracted from the data-field 20003, which captured the patient’s answers provided at an assessment center through a verbal interview with a trained nurse, regarding which prescription medications they were taking on a regular basis (not including short term medication taken for less than 1 week). Participants were divided into the following treatment groups: Treatment Group 1, consisting of patients taking amitriptyline or nortriptyline monotherapy; Treatment Group 2, consisting of participants taking morphine or fentanyl monotherapy; or Treatment Group 3, consisting of participants taking a combination of one of the tricyclic antidepressants (amitriptyline or nortriptyline) along with one of the opioids (morphine or fentanyl). The second set of data was obtained using a touchscreen question delivered during the assessment center visit: “In the last month have you experienced any of the following that interfered with your usual activities?” (UKBB data-field 6159). Participants could choose all that applied from the following options: headache, facial pain, neck or shoulder pain, back pain, stomach or abdominal pain, hip pain, knee pain, pain all over the body, none of the above, or prefer not to answer. From these options, a score between 0 and 8 was calculated with one point allocated to each painful site. Participants that answered “pain all over the body” were given the maximum score of 8, and the ones that marked “none of the above” or “prefer not to answer” were given a score of 0. Lastly, genotyping data that passed quality control was available for 487,409 participants.
Data analysis
The discovery analyses focused on genetic factors associated with better response to combination pain therapy versus mono- pain therapy. We selected the primary outcome, for the purposes of this secondary genetic analysis, as the “% reduction in pain”, defined as “change in pain/treatment period baseline” ×100% by Gilron et al.[24] The rationale for using % change from baseline rather than using actual pain ratings is that this controls for the wide variation of start-of-treatment and end-of-treatment pain scores across all trial patients. The “% reduction in pain” during each study period was calculated as the difference between treatment period baseline and average pain during MTD. We fitted a linear mixed-effects model to the “% reduction in pain” with age, sex, treatment type, genetic factor and an interaction between drug and genetic factor as the fixed effects and patient ID as the random effect. Under this model, we evaluated an association between the genetic factor and “% reduction in pain” under specific therapy type by assessing statistical significance of a contrast, H0 : βgf + βgf:tr = 0, where βgf represents the baseline effect of the genetic factor and βgf:tr represents the interaction term between genetic factor and treatment type. To determine if a genetic factor was associated with an increased risk of side effects, we fit a linear mixed effects model with the same set of predictors but with the self-reported side effect as an outcome. To determine if a genetic factor was associated with an increased risk of side effects, we fit a linear mixed effects model with the same set of predictors but with the self-reported side effect as an outcome. All analyses were performed using R statistical software (version 3.5.1). Linear mixed-effects models were fit using the “lme4” package [3] and linear contrasts were evaluated using the “multcomp” package. [34]
For the UKBB replication analyses of genetic factors associated with benefits of either mono- or combination therapy, an overdispersed Poisson model was fit to the total number of painful body sites from UKBB data as the outcome with age, sex, genetic factor, treatment group, an interaction between treatment group and genetic factor, and 40 first principal components as linear predictors. The main predictors were chosen to be the same as in the discovery analysis. We included all 40 principal components provided by Bycroft et al. [6] as co-variates to correct for UKBB population structure, self-reported sex, and genotyping array. However, since a majority (88.26%) of participants in the UKBB cohort report their ethnic background as “British,” within the broader-level group “white,” one can assume that this cohort has relatively homogeneous ancestry. Therefore, we repeated our analysis with adjustment for only the first 3 principal components instead of 40 (results are not presented here). The estimated coefficients for the key variables (i.e., genetic factor and treatment groups) and their corresponding P-values did not differ appreciably between the two models.
Results
Association analysis in the discovery cohort
A set of 34 preselected SNPs were used for the discovery association analysis. This selection was based on previous reports demonstrating an association with morphine or nortriptyline response or those that have been shown to modify levels of morphine, serotonin, or norepinephrine (Table 1). As the test cohort was small (n=25), the number of genotyped SNPs was conservative (n=34). Furthermore, when possible, SNPs were grouped based on their known functionality (see Materials and Methods). With this grouping, we now performed eight independent tests, bringing our correction for multiple testing threshold to 6.25x10−3.
All twenty-five genotyped participants (7 females; 18 males) were Caucasian, with average age of 67±9 years old (mean ± standard deviation) and average weight of 90±16 kg. In line with the previous report [28], the effect of combination therapy in this subgroup (n=25) of genotyped participants demonstrated an average % reduction in pain with nortriptyline monotherapy that was 10% lower than with combination therapy (P = 0.085) and 14% lower with morphine monotherapy than with combination therapy (P = 0.015) (Figure 1).
Figure 1. Pain score characteristics in the discovery cohort.
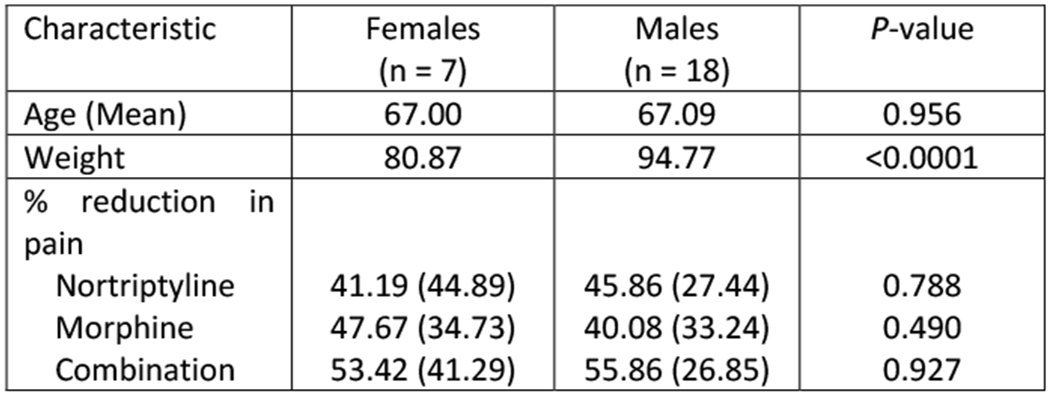
(A) Participant demographics and percent change in pain scores characteristics by sex. None of the observed differences were statistically significant. (B) Percent change in pain score under mono- (i.e., nortriptyline or morphine) or combination therapies. Dots represent individual observations, bars represent means, and error bars represent standard deviations.
We then examined whether SNPs were associated with the % reduction in pain and thus underpin changes in the benefits of mono- or combination therapy. Thirty-four SNPs were combined into eight genetic test groups based on functionality (Table 2). The association between the C allele of ABCB1 rs1045642 (C/T genotype) with the percent change in pain was the only association that reached statistical significance (P = 4.89x10−5) and continued to be significant after Bonferroni correction for multiple testing. In the mixed-effects model, the presence of the C allele accounted for 21% of the variance in pain score, while only 7% of the pain score variance was accounted for by the rest of the predictors. The C allele homozygotes showed an improved response under all pain treatment conditions but this improvement had the greatest magnitude (and was the only one that reached statistical significance) under the combination therapy treatment. Specifically, in the combination treatment group only, the C allele of rs1045642 accounted for 51% of the variance in pain scores. Moreover, in the combination therapy group, the homozygotes for the T allele demonstrated only 20% improvement in pain scores, while the homozygotes for the C allele demonstrated an 88% improvement (Figure 2). Furthermore, the probability of having more than 30% improvement in pain rating given that a subject was taking the combination therapy and had the C allele was 88% (i.e., positive predictive values (PPV)), while that among subjects under combination therapy with the T allele was 54%. As none of the other genotyped SNPs demonstrated an association, our results suggest that the C allele of ABCB1 rs1045642 may significantly improve neuropathic pain treatment management and specifically with morphine/nortriptyline combination therapy.
Table 2.
Association results between the selected SNP/metabolic pathway and % reduction in pain by therapy type in the discovery cohort.
| Gene or metabolic Pathway | SNP rsID | Genotype frequency | Morphine | Nortriptyline | Drug Combination | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| f(AA) | f(Aa) | f(aa) | β (SE) | P-value | β (SE) | P-value | β (SE) | P-value | ||
| OPRM1 gene | rs1799971 | 0.77 | 0.07 | 0.16 | −0.97 (9.18) | 0.999 | 2.15 (9.14) | 0.988 | −5.51 (9.18) | 0.854 |
| COMT metabolism | 0 LPS (n = 5) | −7.13 (42.9) | 0.973 | −0.33 (42.9) | 1.000 | 5.03 (42.9) | 0.989 | |||
| 1 LPS (n = 17) | −32.2 (41.9) | 0.504 | −28.9 (41.8) | 0.555 | −18.3 (41.9) | 0.737 | ||||
| 2 LPS (n = 3) | −35.8 (49.3) | 0.600 | −46.6 (49.3) | 0.507 | −28.4 (49.3) | 0.705 | ||||
| Serotonin receptor | rs6313 | 0.35 | 0.38 | 0.27 | −0.88 (8.14) | 0.999 | 2.12 (7.96) | 0.985 | −2.33 (8.15) | 0.981 |
| rs7997012 | 0.54 | 0.42 | 0.04 | 2.92 (11.5) | 0.986 | −2.38 (11.5) | 0.992 | 8.26 (11.5) | 0.786 | |
| ABCB1 gene | rs1045642 | 0.23 | 0.54 | 0.23 | 14.5 (8.33) | 0.193 | 18.2 (8.13) | 0.065 | 32.5 (8.33) | 4.89e-05* |
| rs2032582 | 0.35 | 0.50 | 0.15 | −13.4 (9.11) | 0.307 | −14.4 (9.10) | 0.253 | −21.2 (9.11) | 0.052 | |
| CYP2C19 metabolism | PM (n = 0) | NA | NA | NA | NA | NA | NA | |||
| IM (n = 3) | −30.4 (46.7) | 0.586 | −14.6 (46.7) | 0.838 | −8.50 (46.7) | 0.939 | ||||
| EM (n = 11) | −0.54 (51.1) | 1.000 | 0.46 (51.1) | 1.000 | 5.74 (51.1) | 0.993 | ||||
| UM (n = 11) | −27.2 (44.8) | 0.624 | −34.5 (44.6) | 0.512 | −20.8 (44.8) | 0.729 | ||||
| CYP2D6 metabolism | PM (n = 1) | 25.8 (46.9) | 0.833 | 19.8 (46.9) | 0.911 | −1.90 (46.9) | 1.000 | |||
| IM (n = 1) | 23.2 (53.2) | 0.886 | −20.1 (53.2) | 0.919 | −3.46 (53.2) | 0.999 | ||||
| EM (n = 23) | −35.7 (44.9) | 0.473 | −45.0 (44.9) | 0.355 | −50.6 (44.9) | 0.295 | ||||
| UM (n = 0) | NA | NA | NA | NA | NA | NA | ||||
The results are based on linear mixed-effect models fitted to % reduction in pain with age, sex, treatment type, genetic factor, and an interaction between treatment type and genetic factor. A: major allele, a: minor allele. LPS: Low Pain Sensitivity; PM= poor metabolizers; IM= intermediate metabolizers; EM= extensive metabolizers and UM= ultra-rapid metabolizers.
Statistical significant set at α=6.25e-03 level.
Figure 2. Percent reduction in pain stratified by C allele of ABCB1 rs1045642 and treatment type in the discovery cohort.
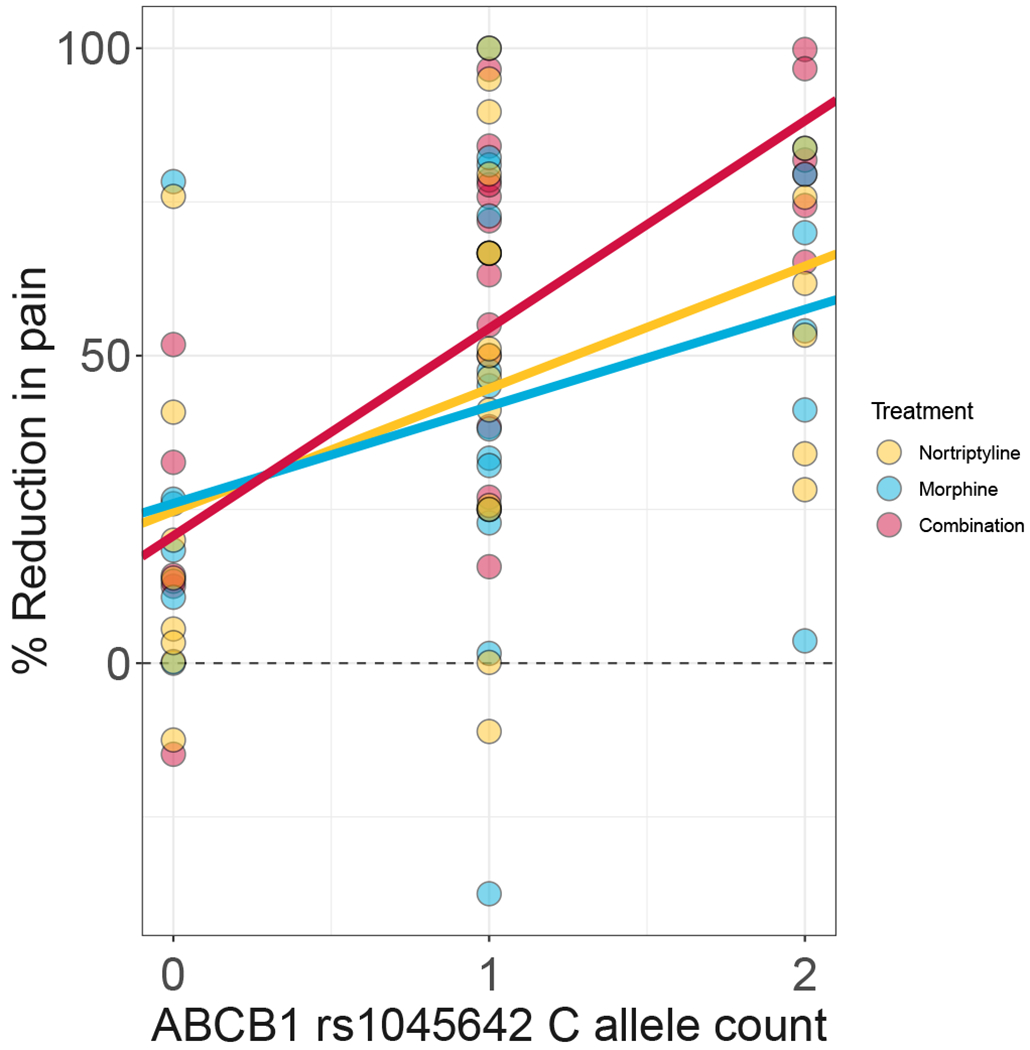
Dots represent individual observations and lines represent fitted values from a linear mixed-effect model. Percent values highlight differences in treatment type efficacy between the homozygotes for the T allele and the homozygotes for the C allele. Under combination therapy, the homozygotes for C allele, on average, reported 88% improvement for pain reduction while the homozygotes for T allele reported 20% improvement for pain reduction. The improvement for pain reduction for C allele homozygotes was 64% in nortriptyline treatment group and 57% morphine treatment group.
Association of ABCB1 rs1045642 with adverse drug events
The original study by Gilron et al. collected self-reported side effects, such as The Stanford Sleepiness Scale, Patient Assessment of Constipation Symptoms (PAC_SYM) and blood pressure changes during combination treatment of neuropathic pain.[19,24,32] Here, we tested the association of ABCB1 rs1045642 with side effects in the three treatment groups described above. We questioned if the increased therapeutic efficacy of C allele carriers of rs1045642 was also associated with stronger side effects. There was no statistically significant contribution of ABCB1 rs1045642 with the observed side-effects in any of the treatment groups (Table 3). These results suggest that only the therapeutic effects described herein but not the observed side effects of morphine-nortriptyline combination therapy are related to ABCB1 genotype.
Table 3.
Association results for SNP rs1045642 and side effects by treatment group in the discovery cohort.
| Side effect | Morphine | Nortriptyline | Drug Combination | |||
|---|---|---|---|---|---|---|
| β (SE) | P-value | β (SE) | P-value | β (SE) | P-value | |
| Epworth Sleepiness Scale (1-7) | 0.07 (0.26) | 0.988 | −0.08 (0.24) | 0.974 | −0.11 (0.26) | 0.956 |
| Patient Assessment of Constipation Symptoms (0-4) | 0.02 (0.19) | 0.999 | −0.02 (0.19) | 0.999 | 0.07 (0.19) | 0.970 |
| Systolic Blood Pressure | 3.54 (3.68) | 0.657 | 0.55 (3.58) | 0.997 | −0.26 (3.67) | 0.999 |
| Diastolic Blood Pressure | −4.17 (3.27) | 0.475 | 2.68 (3.27) | 0.783 | −0.49 (3.38) | 0.998 |
The results are based on linear mixed-effect models fitted to the side effects with age, sex, treatment type, SNP, and an interaction between treatment type and SNP. P-values are for contrast coding that evaluate statistical significance of a genetic factor on a side effect under identified treatment group.
Validation analysis in the UK Biobank
Next, we analyzed the UKBB dataset to validate associations found between ABCB1 rs1045642 and therapeutic effect based in the discovery cohort findings. In the UKBB, we included participants who were also taking amitriptyline and fentanyl in order to increase sample size due to the extensive use of these drugs and their similarities to nortriptyline and morphine respectively, leading to identical interactions with the ABCB1 transporter.
There were 1,624 participants taking amitriptyline or nortriptyline (Treatment Group 1), 689 taking morphine or fentanyl (Treatment Group 2), and 37 taking a combination of amitriptyline or nortriptyline along with morphine or fentanyl (Treatment Group 3). There were no significant sex differences, except there were more females than males taking amitriptyline or nortriptyline (Treatment Group 1), and more males than females taking morphine or fentanyl (Treatment Group 2, Table 4). The ABCB1 rs1045642 genotype was also obtained for each study participant. We used the number of painful body sites as a proxy for clinically relevant chronic pain intensity, as a strong correlation between these two phenotypes has been reported previously. [16,17,67,74,79,89] (Table 4)
Table 4.
Participant demographics in the UK biobank.
| Females (n = 1569) | Males (n = 792) | P-value | |
|---|---|---|---|
| Age in years, mean (S.E.) | 58 (7) | 59 (8) | 0.266 |
| Treatment | <0.0001 | ||
| Group 1* | 1165 | 472 | |
| Group 2* | 378 | 314 | |
| Group 3 | 26 | 12 | |
| Number of pain sites | 0.478 | ||
| 0 | 174 | 74 | |
| 1 | 265 | 143 | |
| 2 | 278 | 129 | |
| 3 | 287 | 151 | |
| 4 | 179 | 99 | |
| 5 | 104 | 51 | |
| 6 | 43 | 13 | |
| 7 | 6 | 3 | |
| 8 | 222 | 129 | |
Treatment Group 1 - amitriptyline or nortriptyline; Treatment Group 2 - morphine or fentanyl; Treatment Group 3 – combination of amitriptyline or nortriptyline along with morphine or fentanyl.
Significant difference between females and males (P < 0.05). A two-sample t-test was used to assess differences between continues variables and a chi-square test was used to compare counts.
Our primary outcome of interest was the relationship between the C allele of rs1045642 and the total number of painful body sites (Figure 3). Subjects in Treatment Group 3 (combination therapy) reported the highest number of painful body sites (4.04 ±1.11). The number of pain sites were 2% lower (P = 0.896) in Treatment Group 2 (a proxy for nortriptyline monotherapy) and 29% lower (P = 0.003) in Treatment Group 1 (a proxy for morphine monotherapy) relative to group 3 (Figure 3A).
Figure 3. The number of pain sites by treatment group in the UK Biobank cohort (UKBB).
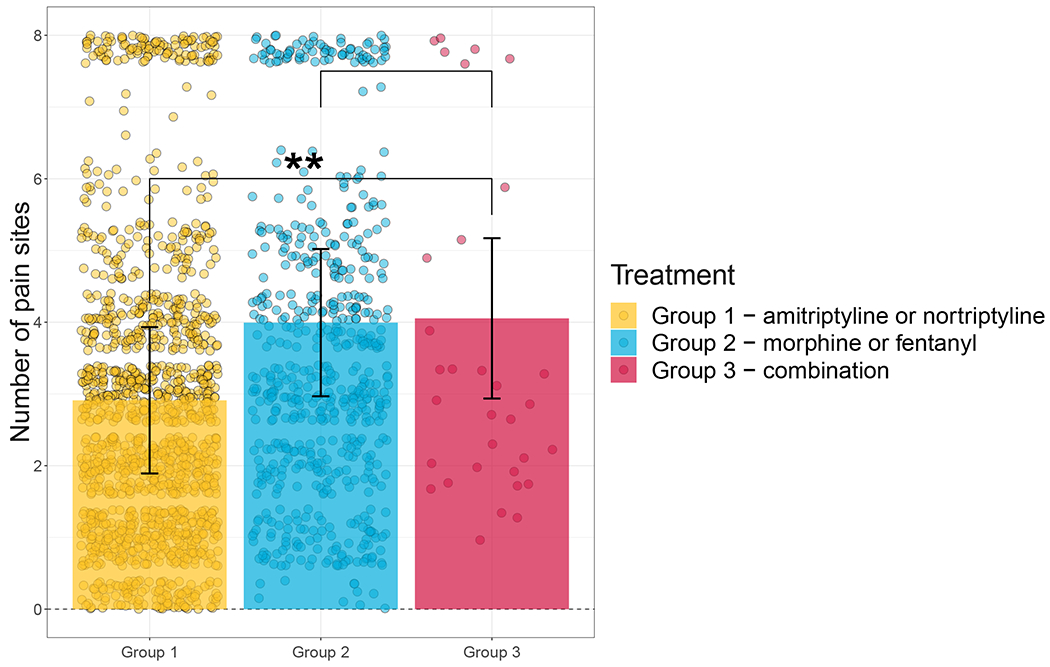
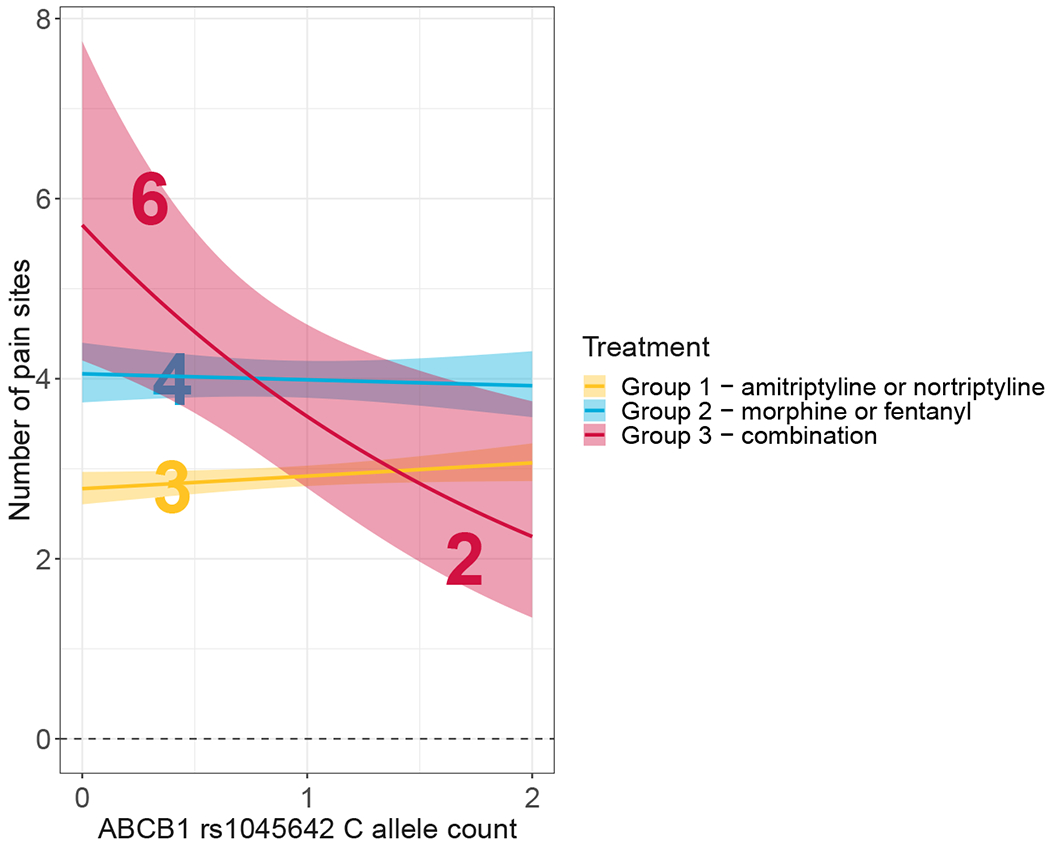
(A) The number of total painful body sites reported by subject in the UKBB cohort stratified by treatment group. Dots represent individual observations, bars represent means, error bars represent standard errors. (B) The reduction in number of reported pain sites stratified by C allele of rs1045642 and treatment group. Under combination therapy, the homozygotes for T allele, on average, reported 6 painful sites while the homozygotes for C allele reported 2 painful sites. The number of painful sites did not vary appreciably between the homozygotes for C or T alleles under either monotherapy treatment.
As with the discovery cohort analysis, the results of the Poisson model, fitted to the number of painful sites, suggest that rs1045642 of ABCB1 is associated with reduced pain in the therapy group (Treatment Group 3, P = 0.022) but not either of the monotherapies (Treatment Group 1, P = 0.138; Treatment Group 2, P = 0.530) (Table 5). We considered one-sided P-values at this replication stage with a prior effect direction based on the discovery cohort (Treatment Group 3, P = 0.011). The target SNP, rs1045642, showed a concordant direction of effect size in the UKBB data. Specifically, in the combination therapy group (Treatment Group 3), the T/T homozygotes demonstrated, on average, 6 painful sites, whereas the C/C homozygotes averaged 2 painful sites (Fig. 3B). Furthermore, the probability of having 8 pain sites given that a subject was taking the combination therapy (Treatment Group 3) and had the C allele was 13.8% (i.e., PPV), while that among subjects under combination therapy with the T allele was 37.2%; the probability of having more than 3 pain sites among people with the C allele and combination therapy was 20.68%, while that among people with the T allele was 51.1%. The presence of the C allele of rs1045642 in the Poisson model explained about 2% of the variance in the number of painful sites, while the rest of the predictors (including 40 first principal components) accounted for an additional 7% of the variance. The weaker effect of rs1045642 genotype on the treatment effect in the replication cohort has several possible explanations. It may be due to an overestimation of the effect in the discovery cohort and/or a difference in the quality of the phenotypic information collected in the discovery cohort through the randomized control clinical trial versus self-report in the UKBB with a proxy measure for pain levels. Another explanation may be the inclusion of fentanyl as a proxy for morphine. While morphine and fentanyl interact with ABCB1 in a similar manner, they are metabolized differently.
Table 5.
Association results for functional SNPs within the ABCB1 gene locus and the number of pain sites by therapy group in the UK Biobank.
| ABCB1 SNP | Genotype frequencies | Morphine | Nortriptyline | Drug Combination | |||||
|---|---|---|---|---|---|---|---|---|---|
| f(AA) | f(Aa) | f(aa) | β (SE) | P-value | β (SE) | P-value | β (SE) | P-value | |
| rs10248420 | 0.71 | 0.26 | 0.03 | −0.02(0.05) | 0.643 | −0.02 (0.04) | 0.511 | −0.52(0.28) | 0.062 |
| rs1045642 | 0.30 | 0.48 | 0.22 | −0.02 (0.04) | 0.530 | 0.04 (0.03) | 0.138 | −0.37 (0.16) | 0.022* |
| rs1128503 | 0.32 | 0.49 | 0.19 | 0.02 (0.04) | 0.595 | −0.05 (0.03) | 0.061 | 0.11 (0.17) | 0.500 |
| rs11983225 | 0.77 | 0.21 | 0.02 | −0.007 (0.06) | 0.891 | −0.04 (0.04) | 0.367 | −0.51 (0.32) | 0.109 |
| rs12720067 | 0.77 | 0.21 | 0.02 | −0.0003 (0.06) | 0.995 | −0.05 (0.04) | 0.258 | −0.51 (0.32) | 0.109 |
| rs2032583 | 0.77 | 0.21 | 0.02 | −0.007 (0.06) | 0.891 | −0.04 (0.04) | 0.367 | −0.51 (0.32) | 0.109 |
| rs2235040 | 0.77 | 0.21 | 0.02 | −0.007 (0.06) | 0.891 | −0.04 (0.04) | 0.406 | −0.51 (0.32) | 0.109 |
| rs2235067 | 0.77 | 0.21 | 0.02 | −0.001 (0.05) | 0.983 | −0.03 (0.04) | 0.431 | −0.52 (0.32) | 0.105 |
| rs4148739 | 0.77 | 0.21 | 0.02 | −0.007 (0.05) | 0.891 | −0.03 (0.04) | 0.367 | −0.51 (0.32) | 0.109 |
| rs4148740 | 0.77 | 0.21 | 0.02 | −0.007 (0.05) | 0.894 | −0.04 (0.04) | 0.320 | −0.51(0.31) | 0.109 |
| rs7787082 | 0.71 | 0.26 | 0.03 | −0.02 (0.05) | 0.633 | −0.03 (0.04) | 0.481 | −0.52 (0.28) | 0.062 |
The results are based on a Poisson model fit to the number of pain sites with age, sex, SNP, treatment group, an interaction between SNP and treatment group, and the first 40 principal components as linear predictors.
Statistically significant result at α = 0.05 level.
Associations between other ABCB1 functional SNPs and the number of pain sites in the UK Biobank
As ABCB1 has a number of functional polymorphisms and several are in linkage disequilibrium (LD) [42], we tested whether the rs1045642 polymorphism is driving the efficacy of combination therapy as described above, or if it is a marker for a different functional allele. We re-fitted Poisson regression models to the number of pain sites with other functional ABCB1 SNPs as genetic predictors. The results of these additional analyses are summarized in Table 5 and unequivocally demonstrate that other ABCB1 polymorphisms do not appreciably alter the efficacy of the three treatments considered.
Discussion
Responses to the evolving opioid crisis in many parts of the world have emphasized tremendous caution around the use and prescribing of opioids for chronic noncancer pain, and this has included recommendations to limit the daily opioid dose prescribed. [5,15] In addition to careful screening, intensive education, and close follow-up of patients receiving opioids for chronic pain, strategies to limit prescribed opioid doses may include combination pharmacotherapy whereby additive or synergistic analgesic interactions may facilitate an opioid sparing effect. [5,21,24,51] Thus, understanding the genetic basis of therapeutic responses to various analgesics may serve to better guide new pain management strategies.
In this manuscript, we selected a group of SNPs to be tested as potential predictors of treatment response in neuropathic pain patients. Despite convincing findings on their role in opioid or tricyclic antidepressant pharmacology [14,29,35,39,85,94], little has been published of their value as markers of treatment response for chronic pain conditions, such as neuropathic pain, and in the setting of mixed medication regimens. From all the genetic variations tested, the ABCB1 SNP rs1045642, but none of the other ABCB1 polymorphisms, demonstrated a replicable association with the efficacy of nortriptyline/morphine combination treatment as compared to that of either nortriptyline or morphine as monotherapy.
The ABCB1 gene, also known as the MDR1 or multidrug resistance 1 gene, encodes for P-glycoprotein, which is a transmembrane ATP dependent efflux pump that is expressed in many tissues, including the renal tubules, small and large intestinal epithelium, hepatocytes, in the endothelial cells of the blood-brain barrier, and spinal cord.[12,70,72,83] P-glycoprotein recognizes and transports a broad variety of structurally different and functionally unrelated drugs, including tricyclic antidepressants amitriptyline and doxepin, selective serotonin reuptake inhibitors (SSRIs) such as citalopram, and opioids such as morphine and fentanyl.[36,50,57] . Relevant to our study, nortriptyline has been classified as a moderate affinity substrate of P-glycoprotein, with a transmembrane transport ratio determined between 0.9 to 1.1[58]. On the other hand morphine is classified as a weak P-glycoprotein substrate with an efflux: influx ratio of 1.2 to 1.5[55]. Pharmacokinetic interactions exist among drugs transported by P-glycoproteins [78,91]. Combination treatments with drugs that either inhibit or induce P-glycoproteins might alter the amount of drug available in the brain.
Polymorphisms in the ABCB1 gene have been associated with altered P-glycoprotein activity and expression levels, which have been demonstrated to alter plasma drug levels, side effect profiles and clinical response.[33,41,76] Although the first report by Hoffmeyer et al. showed that the T allele is associated with reduced mRNA expression of P-glycoprotein, subsequent studies on the role of ABCB1 polymorphisms SNP rs1045642 on P- glycoprotein functionality and drug response are controversial, with evidence on both sides.[33,47] When we extend this to drug concentrations and resulting plasma levels, once again, there is evidence to support higher plasma levels and, therefore, greater efflux for both CC [61] and TT [93] carriers.
There is also equivocal evidence surrounding the downstream effect of the SNP rs1045642 on opioid response.[47] A study by Campa et al. identified an increased drug effect in patients treated with morphine carrying the T allele as opposed to C. [7] However, in some studies, there is no reported effect of the SNP on response to opioids, including fentanyl [69], morphine [46,73], or oxycodone. [95] On the other hand, individuals with TT genotype as part of a haplotype required more methadone. [48] In the same study, the TT genotype individually was associated with the higher methadone dose requirement, although the association did not reach statistical significance.[48] Specifically for nortriptyline, rs1045642 has been associated with changes in the incidence of postural hypotension as a side effect, in which the T homozygote population is more at risk of postural hypotension compared to the C homozygote, but reports have been conflicting and no changes have been found in its therapeutic effect.[37,65]
Although a mechanism to explain the functional effect of this SNP in our study still needs to be demonstrated, several hypotheses can be offered. Importantly, this is the first published evidence to demonstrate the genotypic effect of the ABCB1 transporter variant for combination therapy of two analgesics, but neither morphine nor nortriptyline separately, so this effect can be different from what is expected based on the single drug response versus combination. Furthermore, existing controversy on the functional effect of rs1045642 at molecular genetic level may result from a complex epistatic interaction of different allelic variants of ABCB1. It has recently been reported that the rs1045642 polymorphism affects mRNA stability as part of haplotype, but this may not necessarily translate directly to reduced protein levels.[90] There is often an inverse correlation between secondary-structure dependent RNA stability and translation efficiency, and this translation efficiency is often tissue-dependent. [56,86] Furthermore, there is evidence to suggest the haplotype of ABCB1 tagged by rs1045642 controls the translation efficiency, and, more importantly, co-translational folding and insertion of P- glycoprotein into the membrane, thereby altering substrate specificity. [36] Thus, first, a better understanding of the impact of rs1045642 polymorphism on P-glycoprotein protein expression and its specificity to nortriptyline and morphine should be established.
Furthermore, it is well documented that a decreased expression of P-glycoprotein increases bioavailability of many drugs in plasma and in the CNS through the blood brain barrier. [33,70,72,83] From this perspective, we should expect that the C allele of rs1045642 should be associated with decreased expression of P-glycoprotein protein. However, there is also evidence of a reverse effect in which P-glycoprotein in the choroid plexus increases the transport of molecules from the circulation into the cerebrospinal fluid [64] and hence could augment the influx of drugs from the choroid plexus into the CSF. The degree to which these mechanisms specifically influence the concentration and pharmacologic effect of morphine or nortriptyline in the CNS has yet to be determined.
Finally, transporters are known to be dynamic structures and their substrate specificity is influenced by a series of factors, including the size of the molecules to be transported and their chemical composition.[76] The effect of a single genetic variant in the ABCB1 gene can be tissue specific, and compensatory transport pathways may also be activated. These compensatory transport pathways may have a greater impact on transporter activity when two or more drugs with different mechanisms of action are combined.[33,54,92]
An obvious limitation of this study is the number of subjects that were available for genetic testing. Despite the small sample, the controlled settings of the clinical trial allowed the detection of an effect of ABCB1 rs1045642 in treatment response. The detection was possible in a small sample size due to the strong effect of the C allele. To determine if this association was relevant in more heterogeneous populations, the secondary analysis was pursued using UKBB data. The phenotypic outcome selected for the study was the number of self-reported painful sites within the past month, and even though the scale of characterization in the UKBB is not at the level of a controlled clinical trial for chronic pain, the number of painful sites is well understood to have a direct relationship with perceived intensity of pain, and with other pain-related outcomes, such as risk of pain chronicity, sleep disorders and limited functional capacity.[16,18,26,60,79] The drugs studied in this secondary analysis were expanded to include amitriptyline, which is a tricyclic antidepressant structurally similar to nortriptyline, and fentanyl, an opioid like morphine, both of which are substrates of ABCB1.[40,58,62] The results from our analysis show that rs1045642 is related to a decrease in the number of painful sites in participants that take nortriptyline or amitriptyline in combination with morphine or fentanyl. Thus, although the UK Biobank cohort used for validation phenotypically different than the discovery clinical trial cohort, and the drug combination is extended rather than direct replication, the genetic association results are congruent.
In summary, our results suggest that the rs1045642 polymorphism has a strong clinical effect on combination treatment responses and may represent one example of the utility of genetic testing in the effective treatment of chronic pain. The genetic effect size of the C allele on treatment response is impressive and has little precedence on the pharmacokinetics field. [52] The increase in therapeutic efficacy of C allele carriers of rs1045642 does not correlate with increased adverse drug events, which suggest this combination of morphine and nortriptyline may be of benefit for neuropathic patients with this genotype. The results of this study increase the understanding of molecular genetic pathophysiology of outcomes in combination therapies, and the potential development of genetically targeted treatments for neuropathic pain. The standard implementation of genetic testing to future clinical trials studying treatment effects will speed the translation of knowledge, and fast-track the control of symptoms in chronic pain patients.
Acknowledgments
Financial support:
Canada Excellence Research Chairs Program (Grant # CERC08) to Luda Diatchenko
Alan Edwards Fellowship in clinical pain research to Rodrigo Benavides
Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to Dmitri Zaykin
Clinical trial funding: Canadian Institutes of Health Research (CIHR) Grant #MCT-94187, Ethypharm (morphine study drug) and Apotex (nortriptyline study drug) to Ian Gilron
Gilron research salary support: CIHR-Pfizer Rx&Dc Collaborative Research Investigator Program (CIHR Grant #MSH-55041) and the Queen’s University Department of Anesthesiology & Perioperative Medicine to Ian Gilron. Ian Gilron has received industry support from Adynxx, Biogen, Eupraxia, Novaremed and Teva.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- [1].Ablin JN; Buskila D, Personalized treatment of pain. Curr Rheumatol Rep 2013, 15 (1), 298. [DOI] [PubMed] [Google Scholar]
- [2].Bates D, Machler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software 2015;67(1):1–48 [Google Scholar]
- [3].Benedetti F; Barbini B; Bernasconi A; Fulgosi MC; Colombo C; Dallaspezia S; Gavinelli C; Marino E; Pirovano A; Radaelli D; Smeraldi E, Serotonin 5-HT2A receptor gene variants influence antidepressant response to repeated total sleep deprivation in bipolar depression. Prog Neuropsychopharmacol Biol Psychiatry 2008, 32 (8), 1863–6. [DOI] [PubMed] [Google Scholar]
- [4].Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc. Nat. Acad. Sci 1998; 95: 9608–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Busse JW, Craigie S, Juurlink DN, Buckley DN, Wang L, Couban RJ, Agoritsas T, Akl EA, Carrasco-Labra A, Cooper L, Cull C, da Costa BR, Frank JW, Grant G, Iorio A, Persaud N, Stern S, Tugwell P, Vandvik PO, Guyatt GH. Guideline for opioid therapy and chronic noncancer pain. CMAJ : Canadian Medical Association journal = journal de l’Association medicale canadienne 2017;189(18):E659–e666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562(7726):203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Campa D, Gioia A, Tomei A, Poli P, Barale R. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clinical pharmacology and therapeutics 2008;83(4):559–566 [DOI] [PubMed] [Google Scholar]
- [8].Chaparro LE; Wiffen PJ; Moore RA; Gilron I, Combination pharmacotherapy for the treatment of neuropathic pain in adults. Cochrane Database Syst Rev 2012, (7), Cd008943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cheng KI; Lin SR; Chang LL; Wang JY; Lai CS, Association of the functional A118G polymorphism of OPRM1 in diabetic patients with foot ulcer pain. J Diabetes Complications 2010, 24 (2), 102–8. [DOI] [PubMed] [Google Scholar]
- [10].Chou WY; Yang LC; Lu HF; Ko JY; Wang CH; Lin SH; Lee TH; Concejero A; Hsu CJ, Association of mu-opioid receptor gene polymorphism (A118G) with variations in morphine consumption for analgesia after total knee arthroplasty. Acta Anaesthesiol Scand 2006, 50 (7), 787–92. [DOI] [PubMed] [Google Scholar]
- [11].Colloca L; Ludman T; Bouhassira D; Baron R; Dickenson AH; Yarnitsky D; Freeman R; Truini A; Attal N; Finnerup NB; Eccleston C; Kalso E; Bennett DL; Dworkin RH; Raja SN, Neuropathic pain. Nat Rev Dis Primers 2017, 3, 17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cordon-Cardo C; O’Brien JP; Boccia J; Casals D; Bertino JR; Melamed MR, Expression of the multidrug resistance gene product (P-glycoprotein) in human normal and tumor tissues. J Histochem Cytochem 1990, 38 (9), 1277–87. [DOI] [PubMed] [Google Scholar]
- [13].Diatchenko L; Fillingim RB; Smith SB; Maixner W, The phenotypic and genetic signatures of common musculoskeletal pain conditions. Nat Rev Rheumatol 2013, 9 (6), 340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Diatchenko L; Nackley AG; Slade GD; Bhalang K; Belfer I; Max MB; Goldman D; Maixner W, Catechol-O-methyltransferase gene polymorphisms are associated with multiple pain-evoking stimuli. Pain 2006, 125 (3), 216–24. [DOI] [PubMed] [Google Scholar]
- [15].Dowell D, Haegerich TM, Chou R. CDC Guideline for Prescribing Opioids for Chronic Pain - United States, 2016. MMWR Recommendations and reports : Morbidity and mortality weekly report Recommendations and reports 2016;65(1):1–49 [DOI] [PubMed] [Google Scholar]
- [16].Dragioti E; Larsson B; Bernfort L; Levin LA; Gerdle B, A cross-sectional study of factors associated with the number of anatomical pain sites in an actual elderly general population: results from the PainS65+ cohort. J Pain Res 2017, 10, 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Finney A; Dziedzic KS; Lewis M; Healey E, Multisite peripheral joint pain: a cross-sectional study of prevalence and impact on general health, quality of life, pain intensity and consultation behaviour. BMC Musculoskelet Disord 2017, 18 (1), 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Forssell H; Kauko T; Kotiranta U; Suvinen T, Predictors for future clinically significant pain in patients with temporomandibular disorder: A prospective cohort study. Eur J Pain 2017, 21 (1), 188–197. [DOI] [PubMed] [Google Scholar]
- [19].Frank L; Kleinman L; Farup C; Taylor L; Miner P Jr., Psychometric validation of a constipation symptom assessment questionnaire. Scand J Gastroenterol 1999, 34 (9), 870–7. [DOI] [PubMed] [Google Scholar]
- [20].Frye R; Mathews SE, Effect of digoxin-like immunoreactive factor on the TDx digoxin II assay. Clin Chem 1987, 33 (4), 629–30. [PubMed] [Google Scholar]
- [21].Gilron I, Bailey JM, Tu D, Holden RR, Weaver DF, Houlden RL. Morphine, gabapentin, or their combination for neuropathic pain. The New England journal of medicine 2005;352(13):1324–1334 [DOI] [PubMed] [Google Scholar]
- [22].Gilron I; Baron R; Jensen T, Neuropathic pain: principles of diagnosis and treatment. Mayo Clin Proc 2015, 90 (4), 532–45.2 [DOI] [PubMed] [Google Scholar]
- [23].Gilron I, Jensen TS, Dickenson AH. Combination pharmacotherapy for management of chronic pain: from bench to bedside. The Lancet Neurology 2013;12(11):1084–1095 [DOI] [PubMed] [Google Scholar]
- [24].Gilron I; Tu D; Holden RR; Jackson AC; DuMerton-Shore D, Combination of morphine with nortriptyline for neuropathic pain. Pain 2015, 156 (8), 1440–8. [DOI] [PubMed] [Google Scholar]
- [25].Goh LL, Lim CW, Sim WC, Toh LX, Leong KP. Analysis of Genetic Variation in CYP450 Genes for Clinical Implementation. PLoS One. 2017;12(1):e0169233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Grimby-Ekman A; Gerdle B; Bjork J; Larsson B, Comorbidities, intensity, frequency and duration of pain, daily functioning and health care seeking in local, regional, and widespread pain - a descriptive population-based survey (SwePain). BMC Musculoskelet Disord 2015, 16, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hasvik E; Iordanova Schistad E; Grovle L; Julsrud Haugen A; Roe C; Gjerstad J, Subjective health complaints in patients with lumbar radicular pain and disc herniation are associated with a sex - OPRM1 A118G polymorphism interaction: a prospective 1-year observational study. BMC Musculoskelet Disord 2014, 15, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Helton SG; Lohoff FW, Serotonin pathway polymorphisms and the treatment of major depressive disorder and anxiety disorders. Pharmacogenomics 2015, 16 (5), 541–53. [DOI] [PubMed] [Google Scholar]
- [29].Hicks JK; Sangkuhl K; Swen JJ; Ellingrod VL; Muller DJ; Shimoda K; Bishop JR; Kharasch ED; Skaar TC; Gaedigk A; Dunnenberger HM; Klein TE; Caudle KE; Stingl JC, Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hicks JK; Swen JJ; Thorn CF; Sangkuhl K; Kharasch ED; Ellingrod VL; Skaar TC; Muller DJ; Gaedigk A; Stingl JC, Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther 2013, 93 (5), 402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hiemke C; Baumann P; Bergemann N; Conca A; Dietmaier O; Egberts K; Fric M; Gerlach M; Greiner C; Grunder G; Haen E; Havemann-Reinecke U; Jaquenoud Sirot E; Kirchherr H; Laux G; Lutz UC; Messer T; Muller MJ; Pfuhlmann B; Rambeck B; Riederer P; Schoppek B; Stingl J; Uhr M; Ulrich S; Waschgler R; Zernig G, AGNP Consensus Guidelines for Therapeutic Drug Monitoring in Psychiatry: Update 2011. Pharmacopsychiatry 2011, 44 (6), 195–235. [DOI] [PubMed] [Google Scholar]
- [32].Hoddes E; Zarcone V; Smythe H; Phillips R; Dement WC, Quantification of sleepiness: a new approach. Psychophysiology 1973, 10 (4), 431–6. [DOI] [PubMed] [Google Scholar]
- [33].Hoffmeyer S; Burk O; von Richter O; Arnold HP; Brockmoller J; Johne A; Cascorbi I; Gerloff T; Roots I; Eichelbaum M; Brinkmann U, Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A 2000, 97 (7), 3473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hothorn T, Bretz F, Westfall P. Simultaneous Inference in General Parametric Models. Biometrical Journal 2008;50(3):346–363 [DOI] [PubMed] [Google Scholar]
- [35].Hwang IC; Park JY; Myung SK; Ahn HY; Fukuda K; Liao Q, OPRM1 A118G gene variant and postoperative opioid requirement: a systematic review and meta-analysis. Anesthesiology 2014, 121 (4), 825–34. [DOI] [PubMed] [Google Scholar]
- [36].Ieiri I; Takane H; Otsubo K, The MDR1 (ABCB1) gene polymorphism and its clinical implications. Clin Pharmacokinet 2004, 43 (9), 553–76. [DOI] [PubMed] [Google Scholar]
- [37].Jensen BP; Roberts RL; Vyas R; Bonke G; Jardine DL; Begg EJ, Influence of ABCB1 (P-glycoprotein) haplotypes on nortriptyline pharmacokinetics and nortriptyline-induced postural hypotension in healthy volunteers. Br J Clin Pharmacol 2012, 73 (4), 619–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jensen MP; Karoly P; Braver S, The measurement of clinical pain intensity: a comparison of six methods. Pain 1986, 27 (1), 117–26. [DOI] [PubMed] [Google Scholar]
- [39].Kapur BM; Lala PK; Shaw JL, Pharmacogenetics of chronic pain management. Clin Biochem 2014, 47 (13–14), 1169–87. [DOI] [PubMed] [Google Scholar]
- [40].Kharasch ED; Hoffer C; Altuntas TG; Whittington D, Quinidine as a probe for the role of p-glycoprotein in the intestinal absorption and clinical effects of fentanyl. J Clin Pharmacol 2004, 44 (3), 224–33. [DOI] [PubMed] [Google Scholar]
- [41].Kimchi-Sarfaty C; Marple AH; Shinar S; Kimchi AM; Scavo D; Roma MI; Kim IW; Jones A; Arora M; Gribar J; Gurwitz D; Gottesman MM, Ethnicity-related polymorphisms and haplotypes in the human ABCB1 gene. Pharmacogenomics 2007, 8 (1), 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kimchi-Sarfaty C; Oh JM; Kim IW; Sauna ZE; Calcagno AM; Ambudkar SV; Gottesman MM, A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007, 315 (5811), 525–8. [DOI] [PubMed] [Google Scholar]
- [43].Kocabas NA, Catechol-O-methyltransferase (COMT) pharmacogenetics in the treatment response phenotypes of major depressive disorder (MDD). CNS Neurol Disord Drug Targets 2012, 11 (3), 264–72. [DOI] [PubMed] [Google Scholar]
- [44].Kreek MJ; Levran O; Reed B; Schlussman SD; Zhou Y; Butelman ER, Opiate addiction and cocaine addiction: underlying molecular neurobiology and genetics. J Clin Invest 2012, 122 (10), 3387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lebe M; Hasenbring MI; Schmieder K; Jetschke K; Harders A; Epplen JT; Hoffjan S; Kotting J, Association of serotonin-1A and -2A receptor promoter polymorphisms with depressive symptoms, functional recovery, and pain in patients 6 months after lumbar disc surgery. Pain 2013, 154 (3), 377–84. [DOI] [PubMed] [Google Scholar]
- [46].Lee MG, Kim HJ, Lee KH, Choi YS. The Influence of Genotype Polymorphism on Morphine Analgesic Effect for Postoperative Pain in Children. The Korean journal of pain 2016;29(1):34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Leschziner GD, Andrew T, Pirmohamed M, Johnson MR. ABCB1 genotype and PGP expression, function and therapeutic drug response: a critical review and recommendations for future research. The pharmacogenomics journal 2007;7(3):154–179 [DOI] [PubMed] [Google Scholar]
- [48].Levran O, O’Hara K, Peles E, Li D, Barral S, Ray B, Borg L, Ott J, Adelson M, Kreek MJ. ABCB1 (MDR1) genetic variants are associated with methadone doses required for effective treatment of heroin dependence. Human molecular genetics 2008;17(14):2219–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liu YC; Wang WS, Human mu-opioid receptor gene A118G polymorphism predicts the efficacy of tramadol/acetaminophen combination tablets (ultracet) in oxaliplatin-induced painful neuropathy. Cancer 2012, 118 (6), 1718–25. [DOI] [PubMed] [Google Scholar]
- [50].Loscher W; Potschka H, Drug resistance in brain diseases and the role of drug efflux transporters. Nat Rev Neurosci 2005, 6 (8), 591–602. [DOI] [PubMed] [Google Scholar]
- [51].Mao J, Gold MS, Backonja MM. Combination drug therapy for chronic pain: a call for more clinical studies. J Pain 2011;12(2):157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Maranville JC; Cox NJ, Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenomics J 2016, 16 (4), 388–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Meineke I; Freudenthaler S; Hofmann U; Schaeffeler E; Mikus G; Schwab M; Prange HW; Gleiter CH; Brockmoller J, Pharmacokinetic modelling of morphine, morphine-3-glucuronide and morphine-6-glucuronide in plasma and cerebrospinal fluid of neurosurgical patients after short-term infusion of morphine. Br J Clin Pharmacol 2002, 54 (6), 592–603. [DOI] [PubMed] [Google Scholar]
- [54].Meissner K; Jedlitschky G; Meyer zu Schwabedissen H; Dazert P; Eckel L; Vogelgesang S; Warzok RW; Bohm M; Lehmann C; Wendt M; Cascorbi I; Kroemer HK, Modulation of multidrug resistance P-glycoprotein 1 (ABCB1) expression in human heart by hereditary polymorphisms. Pharmacogenetics 2004, 14 (6), 381–5. [DOI] [PubMed] [Google Scholar]
- [55].Mercer SL, Coop A. Opioid analgesics and P-glycoprotein efflux transporters: a potential systems-level contribution to analgesic tolerance. Current topics in medicinal chemistry 2011;11(9):1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nackley AG; Shabalina SA; Tchivileva IE; Satterfield K; Korchynskyi O; Makarov SS; Maixner W; Diatchenko L, Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 2006, 314 (5807), 1930–3. [DOI] [PubMed] [Google Scholar]
- [57].Nielsen LM; Olesen AE; Branford R; Christrup LL; Sato H; Drewes AM, Association Between Human Pain-Related Genotypes and Variability in Opioid Analgesia: An Updated Review. Pain Pract 2015, 15 (6), 580–94. [DOI] [PubMed] [Google Scholar]
- [58].O’Brien FE; Dinan TG; Griffin BT; Cryan JF, Interactions between antidepressants and P-glycoprotein at the blood-brain barrier: clinical significance of in vitro and in vivo findings. Br J Pharmacol 2012, 165 (2), 289–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Oertel B; Lotsch J, Genetic mutations that prevent pain: implications for future pain medication. Pharmacogenomics 2008, 9 (2), 179–94. [DOI] [PubMed] [Google Scholar]
- [60].Ohrbach R; Fillingim RB; Mulkey F; Gonzalez Y; Gordon S; Gremillion H; Lim PF; Ribeiro-Dasilva M; Greenspan JD; Knott C; Maixner W; Slade G, Clinical findings and pain symptoms as potential risk factors for chronic TMD: descriptive data and empirically identified domains from the OPPERA case-control study. J Pain 2011, 12 (11 Suppl), T27–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Owen A, Goldring C, Morgan P, Chadwick D, Park BK, Pirmohamed M. Relationship between the C3435T and G2677T(A) polymorphisms in the ABCB1 gene and P-glycoprotein expression in human liver. British journal of clinical pharmacology 2005;59(3):365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Park HJ; Shinn HK; Ryu SH; Lee HS; Park CS; Kang JH, Genetic polymorphisms in the ABCB1 gene and the effects of fentanyl in Koreans. Clin Pharmacol Ther 2007, 81 (4), 539–46. [DOI] [PubMed] [Google Scholar]
- [63].Peiro AM; Planelles B; Juhasz G; Bagdy G; Libert F; Eschalier A; Busserolles J; Sperlagh B; Llerena A, Pharmacogenomics in pain treatment. Drug Metab Pers Ther 2016, 31 (3), 131–42. [DOI] [PubMed] [Google Scholar]
- [64].Rao VV, Dahlheimer JL, Bardgett ME, Snyder AZ, Finch RA, Sartorelli AC, Piwnica-Worms D. Choroid plexus epithelial expression of MDR1 P glycoprotein and multidrug resistance-associated protein contribute to the blood-cerebrospinal-fluid drug-permeability barrier. Proc Natl Acad Sci. 1999; 96: 3900–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Roberts RL; Joyce PR; Mulder RT; Begg EJ; Kennedy MA, A common P-glycoprotein polymorphism is associated with nortriptyline-induced postural hypotension in patients treated for major depression. Pharmacogenomics J 2002, 2 (3), 191–6. [DOI] [PubMed] [Google Scholar]
- [66].Rudorfer MV, Potter WZ. Metabolism of tricyclic antidepressants. Cellular and molecular neurobiology 1999;19(3):373–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Rundell SD; Patel KV; Krook MA; Heagerty PJ; Suri P; Friedly JL; Turner JA; Deyo RA; Bauer Z; Nerenz DR; Avins AL; Nedeljkovic SS; Jarvik JG, Multisite Pain Is Associated with Long-term Patient-Reported Outcomes in Older Adults with Persistent Back Pain. Pain Med 2019. [DOI] [PubMed] [Google Scholar]
- [68].Sadhasivam S; Chidambaran V; Zhang X; Meller J; Esslinger H; Zhang K; Martin LJ; McAuliffe J, Opioid-induced respiratory depression: ABCB1 transporter pharmacogenetics. Pharmacogenomics J 2015, 15 (2), 119–26. [DOI] [PubMed] [Google Scholar]
- [69].Saiz-Rodriguez M, Ochoa D, Herrador C, Belmonte C, Roman M, Alday E, Koller D, Zubiaur P, Mejia G, Hernandez-Martinez M, Abad-Santos F. Polymorphisms associated with fentanyl pharmacokinetics, pharmacodynamics and adverse effects. Basic & clinical pharmacology & toxicology 2019;124(3):321–329 [DOI] [PubMed] [Google Scholar]
- [70].Schinkel AH; Wagenaar E; Mol CA; van Deemter L, P-glycoprotein in the blood-brain barrier of mice influences the brain penetration and pharmacological activity of many drugs. J Clin Invest 1996, 97 (11), 2517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Scott SA, Sangkuhl K, Shuldiner AR, Hulot JS, Thorn CF, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 19. Pharmacogenet Genomics. 2012;22(2):159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sharom FJ, The P-glycoprotein multidrug transporter. Essays Biochem 2011, 50 (1), 161–78. [DOI] [PubMed] [Google Scholar]
- [73].Sia AT, Sng BL, Lim EC, Law H, Tan EC. The influence of ATP-binding cassette sub-family B member -1 (ABCB1) genetic polymorphisms on acute and chronic pain after intrathecal morphine for caesarean section: a prospective cohort study. International journal of obstetric anesthesia 2010;19(3):254–260 [DOI] [PubMed] [Google Scholar]
- [74].Sibille KT; Chen H; Bartley EJ; Riley J 3rd; Glover TL; King CD; Zhang H; Cruz-Almeida Y; Goodin BR; Sotolongo A; Petrov ME; Herbert M; Bulls HW; Edberg JC; Staud R; Redden D; Bradley LA; Fillingim RB, Accelerated aging in adults with knee osteoarthritis pain: consideration for frequency, intensity, time, and total pain sites. Pain Rep 2017, 2 (3), e591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Sissung TM, Baum CE, Deeken J, Price DK, Aragon-Ching J, Steinberg SM, Dahut W, Sparreboom A, Figg WD. ABCB1 genetic variation influences the toxicity and clinical outcome of patients with androgen-independent prostate cancer treated with docetaxel. Clinical cancer research : an official journal of the American Association for Cancer Research 2008;14(14):4543–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sissung TM; Troutman SM; Campbell TJ; Pressler HM; Sung H; Bates SE; Figg WD, Transporter pharmacogenetics: transporter polymorphisms affect normal physiology, diseases, and pharmacotherapy. Discov Med 2012, 13 (68), 19–34. [PMC free article] [PubMed] [Google Scholar]
- [77].Slade GD; Smith SB; Zaykin DV; Tchivileva IE; Gibson DG; Yuryev A; Mazo I; Bair E; Fillingim R; Ohrbach R; Greenspan J; Maixner W; Diatchenko L, Facial pain with localized and widespread manifestations: separate pathways of vulnerability. Pain 2013, 154 (11), 2335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Solhaug V, Molden E. Individual variability in clinical effect and tolerability of opioid analgesics - Importance of drug interactions and pharmacogenetics. Scandinavian journal of pain 2017;17:193–200 [DOI] [PubMed] [Google Scholar]
- [79].Staud R; Price DD; Robinson ME; Vierck CJ Jr., Body pain area and pain-related negative affect predict clinical pain intensity in patients with fibromyalgia. J Pain 2004, 5 (6), 338–43. [DOI] [PubMed] [Google Scholar]
- [80].Stingl JC; Brockmoller J; Viviani R, Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol Psychiatry 2013, 18 (3), 273–87. [DOI] [PubMed] [Google Scholar]
- [81].Sudlow C; Gallacher J; Allen N; Beral V; Burton P; Danesh J; Downey P; Elliott P; Green J; Landray M; Liu B; Matthews P; Ong G; Pell J; Silman A; Young A; Sprosen T; Peakman T; Collins R, UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015, 12 (3), e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Tchivileva IE; Lim PF; Smith SB; Slade GD; Diatchenko L; McLean SA; Maixner W, Effect of catechol-O-methyltransferase polymorphism on response to propranolol therapy in chronic musculoskeletal pain: a randomized, double-blind, placebo-controlled, crossover pilot study. Pharmacogenet Genomics 2010, 20 (4), 239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Thiebaut F; Tsuruo T; Hamada H; Gottesman MM; Pastan I; Willingham MC, Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci U S A 1987, 84 (21), 7735–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ting S; Schug S, The pharmacogenomics of pain management: prospects for personalized medicine. J Pain Res 2016, 9, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Trescot AM; Faynboym S, A review of the role of genetic testing in pain medicine. Pain Physician 2014, 17 (5), 425–45. [PubMed] [Google Scholar]
- [86].Tsao D; Shabalina SA; Gauthier J; Dokholyan NV; Diatchenko L. Disruptive mRNA folding increases translational efficiency of catechol-O-methyltransferase variant. Nucleic Acids Res. 2011; 39(14), 6201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].van Hecke O; Austin SK; Khan RA; Smith BH; Torrance N, Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain 2014, 155 (4), 654–62. [DOI] [PubMed] [Google Scholar]
- [88].Vargas-Alarcon G; Fragoso JM; Cruz-Robles D; Vargas A; Lao-Villadoniga JI; Garcia-Fructuoso F; Ramos-Kuri M; Hernandez F; Springall R; Bojalil R; Vallejo M; Martinez-Lavin M, Catechol-O-methyltransferase gene haplotypes in Mexican and Spanish patients with fibromyalgia. Arthritis Res Ther 2007, 9 (5), R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Vivaldi D; Di Giosia M; Tchivileva IE; Jay GW; Slade GD; Lim PF, Headache attributed to TMD Is Associated With the Presence of Comorbid Bodily Pain: A Case-Control Study. Headache 2018, 58 (10), 1593–1600. [DOI] [PubMed] [Google Scholar]
- [90].Wang D, Johnson AD, Papp AC, Kroetz DL, Sadee W . Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genom 2005; 15: 693–704 [PubMed] [Google Scholar]
- [91].Wang JS, DeVane CL, Gibson BB, Donovan JL, Markowitz JS, Zhu HJ. Population pharmacokinetic analysis of drug-drug interactions among risperidone, bupropion, and sertraline in CF1 mice. Psychopharmacology 2006;183(4):490–499 [DOI] [PubMed] [Google Scholar]
- [92].Zhou L; Schmidt K; Nelson FR; Zelesky V; Troutman MD; Feng B, The effect of breast cancer resistance protein and P-glycoprotein on the brain penetration of flavopiridol, imatinib mesylate (Gleevec), prazosin, and 2-methoxy-3-(4-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)phenyl)propanoic acid (PF-407288) in mice. Drug Metab Dispos 2009, 37 (5), 946–55. [DOI] [PubMed] [Google Scholar]
- [93].Zhu D, Taguchi-Nakamura H, Goto M, Odawara T, Nakamura T, Yamada H, Kotaki H, Sugiura W, Iwamoto A, Kitamura Y. Influence of single-nucleotide polymorphisms in the multidrug resistance-1 gene on the cellular export of nelfinavir and its clinical implication for highly active antiretroviral therapy. Antiviral therapy 2004;9(6):929–935 [PubMed] [Google Scholar]
- [94].Zorina-Lichtenwalter K; Parisien M; Diatchenko L, Genetic studies of human neuropathic pain conditions: a review. Pain 2018, 159 (3), 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zwisler ST, Enggaard TP, Mikkelsen S, Verstuyft C, Becquemont L, Sindrup SH, Brosen K. Lack of association of OPRM1 and ABCB1 single-nucleotide polymorphisms to oxycodone response in postoperative pain. Journal of clinical pharmacology 2012;52(2):234–242 [DOI] [PubMed] [Google Scholar]


