Abstract
Cartilaginous fish (e.g., sharks) are derived from the oldest vertebrate ancestor having an adaptive immune system, and thus are key models for examining MHC evolution. Previously, family studies in two shark species showed that classical class I (UAA) and class II genes are genetically linked. In this study, we show that proteasome genes LMP2 and LMP7, shark-specific LMP7-like, and the TAP1/2 genes are linked to class I/II. Functional LMP7 and LMP7-like genes, as well as multiple LMP2 genes or gene fragments, are found only in some sharks, suggesting that different sets of peptides might be generated depending upon inherited MHC haplotypes. Cosmid clones bearing the MHC-linked classical class I genes were isolated and shown to contain proteasome gene fragments. A non-MHC-linked LMP7 gene also was identified on another cosmid, but only two exons of this gene were detected, closely linked to a class I pseudogene (UAA-NC2); this region probably resulted from a recent duplication and translocation from the functional MHC. Tight linkage of proteasome and class I genes, in comparison with gene organizations of other vertebrates, suggests a primordial MHC organization. Another nonclassical class I gene (UAA-NC1) was detected that is linked neither to MHC nor to UAA-NC2; its high level of sequence similarity to UAA suggests that UAA-NC1 also was recently derived from UAA and translocated from MHC. These data further support the principle of a primordial class I region with few class I genes. Finally, multiple paternities in one family were demonstrated, with potential segregation distortions.
Major histocompatibility complex class I and class II molecules are fundamental components of the adaptive immune system. Antigenic peptides, bound and presented to T cells by class I molecules, are produced by the cytosolic proteasome. In mammalian cells stimulated by IFN-γ, three components of the constitutive catalytic proteasome core, X/PSMB5/β5, Y/δ/PSMB6/β1, and Z/PSMB7/β2, are replaced by LMP7/PSMB8/β5i, LMP2/PSMB9/β1i, and MECL-1/PSMB10/β2i, respectively, to produce immunoproteasomes, which are believed to generate peptides suitable in length and sequence for binding to class I molecules (2–4). Peptides transported into the endoplasmic reticulum via a heterodimer of TAP proteins are loaded onto class I molecules, which then are competent to be expressed at the cell surface (5).
In eukaryotes, one of the first examples of genetic linkage between functionally related, yet structurally unrelated loci was shown for the class I Ag pathway genes, LMP2, LMP7, TAP1, and TAP2, which are closely linked and encoded in the MHC of all vertebrates studied to date (6–17). In mammals, these genes are located in the class II region, while both classical and nonclassical class I genes are encoded in another region of the MHC. The MHC organization of chickens (11) and all teleost fish (9, 13–16) examined is quite different from mouse and human: classical class I genes are closely linked to proteasome and transporter genes (except in chicken, the inducible proteasome elements have been deleted from the MHC and perhaps from the genome entirely). Such close linkage among class I and the functionally related proteasome and transporter genes has been proposed to be a true class I region, which may allow coevolution of these components (9–11). The class I region in teleosts is further highlighted because class II genes are found in another region of the genome entirely, so that there are actually (at least) two MHCs in the bony fish (14, 18). Because class I is closely linked to proteasome/transporter genes in all nonmammalian species studied to date, it is likely that this MHC organization is primitive.
In addition to differences in MHC genomic organization in birds and teleosts, suggesting coevolution of class I pathway genes, there are lineages of TAP alleles that have coevolved to function with sets of class I alleles in the rat (19), hamster (20), and probably the chicken (11, 21). In the frog, Xenopus allelic lineages of TAP and LMP7 are linked to particular class I lineages (22–24). Coevolution of the proteases, transporters, and presenting molecules in the class I Ag presentation pathway may be the selecting force to preserve linkage of the genes in many vertebrate species. Consistent with this idea is the relatively low number of class I genes that are linked to the MHC proper in most nonmammalian vertebrates (excluding the cichlid (25, 26), the axolotl (27, 28), and perhaps the cod (29)), i.e., a stable class I region.
Cartilaginous fish (sharks, skates, rays, and chimeras) are derived from the oldest vertebrate ancestor with an adaptive immune system, and as such are important models for study of the MHC (7). Previously, we have shown that, in contrast to teleost fish, class I and class II genes are closely linked in sharks (1). In addition, previous work demonstrated the existence of TAP1, TAP2 (30), LMP7 homologues (31), and a proteasome gene unique to the cartilaginous fish, called LMP7-like (31). Based on differences in their catalytic sites, it was suggested that proteasomes containing LMP7 or LMP7-like might provide a diverse repertoire of peptides for class I molecules, consistent with the idea that certain class I alleles could bind peptides preferentially produced by one or the other enzyme. In this study, we begin to focus on whether the class I region found in all teleosts and chickens (and probably amphibians) exists in cartilaginous fish by investigating the linkage between class I and class I-related genes (LMP2, LMP7, LMP7-like, TAP1, and TAP2). We also analyze the number of class I genes and their linkage to each other and to other MHC-encoded genes.
Materials and Methods
Animals
A pregnant nurse shark (Ginglymostoma cirratum, order Orectolobiformes) was captured off Little Torch Key, FL, in October 1998, and 39 pups were delivered by Caesarean section (1). Nurse sharks are ovoviviparous, i.e., maintain their young within the uterus, where they are nourished by egg yolk; the size of the yolk sac was an indication of the pups’ ages (32). Shark Yellow used for preparation of the cosmid library is maintained in an outdoor facility in Miami, Florida, with running seawater.
LMP2 isolation
PCR via 5′ RACE with a degenerate primer was performed to isolate the LMP2 gene. The primer 5′-GCRTCNACRTWNCCRTADAT-3′, in which R is A or G, N is A or G or C or T, W is A or T, D is G or A or T (aa position 135–141 (33), Fig. 1A), and the T3 primer on the Uni-ZAP phage vector (Stratagene, La Jolla, CA) were used with a horn shark (Heterodontus francisci) spleen cDNA library as template (34). PCR was performed for 4 min at 94°C, followed by 35 cycles of 94°C for 30 s, 45°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 15 min. A single 600-bp fragment was amplified, cloned into the pCR2.1 TA cloning vector (Invitrogen, San Diego, CA), and sequenced.
FIGURE 1.
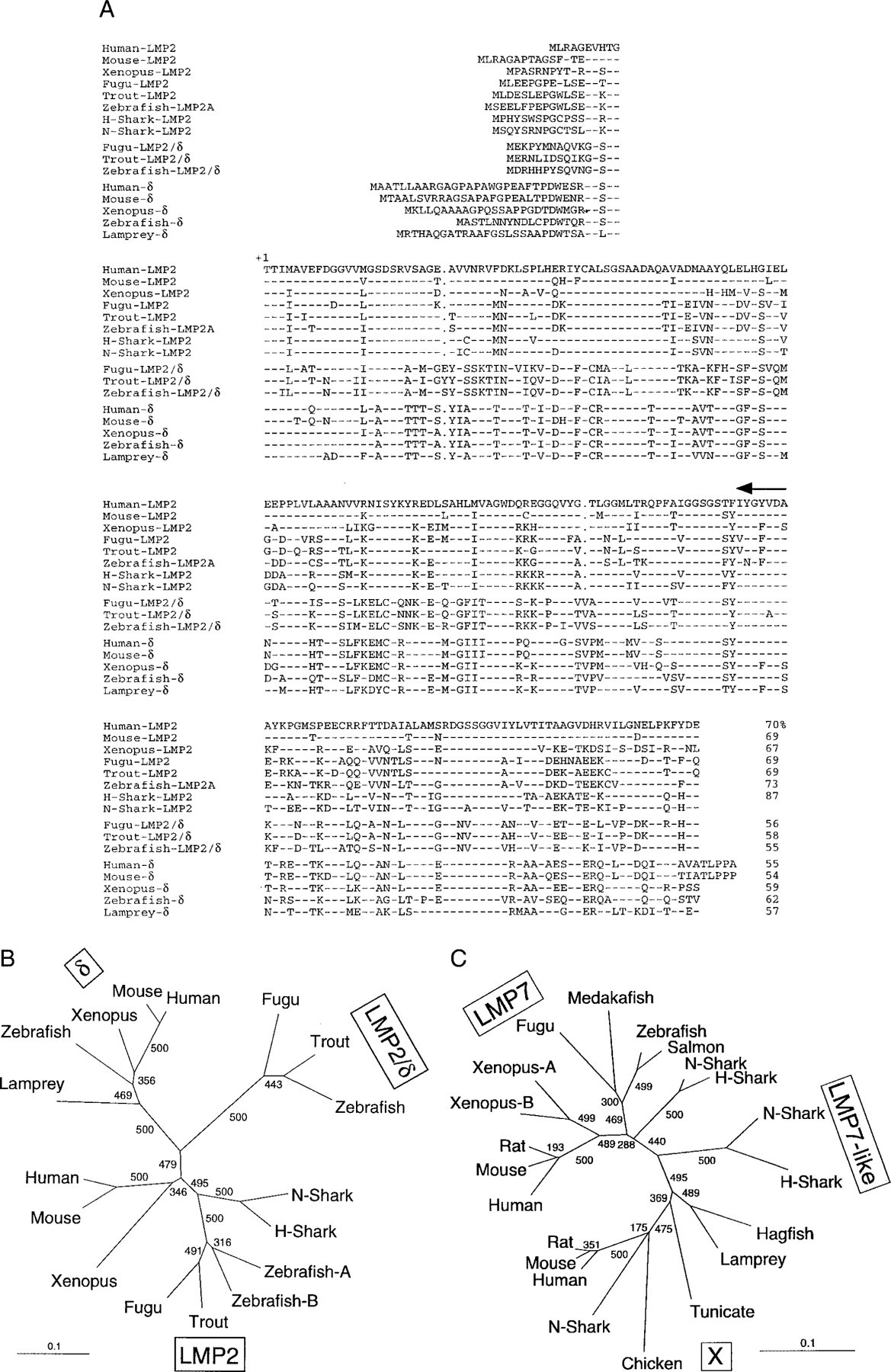
A, Comparison of the amino acid sequence of nurse shark (N-shark) (AF357920) and horn shark (H-shark) (AF357921) LMP2 with LMP2 from human (U01025), mouse (D43620), Xenopus (D87687), trout (AF115541), Fugu (AJ271723), and zebrafish (LMP2A: AF155579, LMP2B: AF155580); LMP2/δ from Fugu (AJ271723), trout (AF115540), and zebrafish (AF155576); and with δ from human (D29012), mouse (U13394), Xenopus (D87689), zebrafish (AF032392), and lamprey (D87690). Dashes and dots show sequences identical to human LMP2 and gaps, respectively. The mature protein begins at +1, and the primer position used for 5′ RACE is indicated with an arrow above the sequence. The percentage of similarity to nurse shark LMP2 is shown at the end of each sequence. B, LMP2 phylogenetic tree using the mature protein. The scale shows the genetic distance. Bootstrap values were obtained with a total of 500 runs and are shown at the branches. C, LMP7/X/LMP7-like phylogenetic tree using the mature protein. GenBank accession numbers are human (Z14982), mouse (U22033), rat (P28064), Fugu (AJ271723), zebrafish (AF032390), N-shark (D64057), salmon (AF184938), Xenopus7B (I51537), Xenopus7A (I51536), H-shark (AF363583) LMP7, N-shark (D64056), H-shark (AF363582) LMP7-like, hagfish (D64054), lamprey (D64055), medakafish (D89725), tunicate (X97729), nurse shark (D64058), human (P28074), mouse (O55234), rat (P28075), and chicken (X57210) X. Scale as in B.
cDNA library screen
The 600-bp LMP2 PCR fragment was used to screen the horn shark spleen cDNA library. This horn shark library was also screened with the full-length nurse shark LMP7 probe (accession D64057). A full-length LMP2 insert was used to probe a nurse shark spleen cDNA library prepared from siblings 34 and 37 (35). This nurse shark spleen cDNA library was also screened with horn shark TAP2 (AF108387, AF108386) and TAP1 (AF108385) (30) to obtain the orthologous nurse shark cDNAs, and with nurse shark nonclassical class I (UAA-NC1, 1) using a probe encoding the α1 and α2 domains (AF220360) to obtain full-length clones. All library screenings were done under high stringency conditions (34).
Sequence alignment and phylogenetic tree
The mature LMP2 and LMP7 protein sequences were aligned using Clustal W with minor adjustments, and an unrooted tree was made using the Tree-View 1.5 program (36).
Southern blotting
A total of 1–3 ml blood was collected from each pup from the caudal vein, and DNA was prepared from nucleated erythrocytes, as described previously (37). Genomic DNA (5 μg) was digested with various restriction enzymes (RE)3 to obtain useful restriction fragment-length polymorphism (RFLP). Full-length nurse shark cDNA inserts were used as probes, except for TAP2 (it lacked the region encoding the N-terminal 160 aa (30)) and UAA-NC1 (the α1 domain exon was used), and all hybridizations were performed under high stringency conditions.
Cosmid library screening
The nurse shark erythrocyte genomic DNA cosmid library prepared from shark Yellow was custom-made (SuperCos1 cosmid library; Stratagene) and has average insert sizes of ~40 kb. Membranes from library lifts were prepared per the manufacturer’s recommendation with minor modifications (38), and ~800,000 clones were screened with the full-length nurse shark LMP7 (D64057 (31)) or class Ia probes (UAA01: AF220063 (1)) under high stringency conditions.
Restriction mapping and subcloning
The cosmid clone 32 was digested with BamHI, SacI, ScaI, XbaI, and all possible combinations of two enzymes, and ClaI, SmaI, BamHI, and ScaI were used for digestion of cosmids 19 and 65. Gels were stained with ethidium bromide to visualize DNA fragments and then blotted onto nitrocellulose membranes. Membranes were sequentially hybridized with either 32P-labeled double-strand probes (labeled with the random-primed labeling method; Boehringer Mannheim, Indianapolis, IN) or end-labeled exon-specific oligonucleotides to identify the digested fragments. Cosmid DNAs were also partially digested with RE and hybridized with end-labeled T3 or T7 oligomers to construct restriction maps. The LMP7-positive BamHI-XbaI 4.5-kb fragment (cosmid 32) and the ClaI-SmaI 1.9-kb fragment (cosmid 65) were subcloned into the pBluescript cloning vector and sequenced to identify the exons. For comparison with hybridizing genomic DNA fragments, 10 μg of cosmid DNA was digested with 15 U of RE for 3 h, and digests were checked on a 1% agarose gel and diluted to convenient concentrations; ~50 ng of cosmid DNA was loaded onto the gel compared with 5 μg of genomic DNA. A class I α1 domain-specific primer (5′-TCTCACAGTCTCCGGTATTT-3′) and a highly conserved α3 domain primer (5′-CCATCGTGATCCACCACACA-3′) were used to amplify the class I sequence from the cosmid 32 DNA; PCR was performed for 4 min at 94°C, followed by 35 cycles of 94°C for 1 min, 56°C for 1 min, 72°C for 3 min, and a final extension at 72°C for 15 min. A 1.5-kb fragment was amplified, cloned into pCR2.1 TA cloning vector, and sequenced.
PCR of genomic DNA
To obtain the transmembrane/cytoplasmic region genomic sequence for the UAA-NC1 gene, a UAA/UAA-NC1 exon 5 primer, 5′-TGAACCAAAGACCAGCTC-3′, and a UAA-NC1-specific exon 6 primer, 5′-TGGATTGTAGCCAGTCTC-3′, were used. PCR was performed at 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 3 min, with final extension of 72°C for 15 min using 2–500 ng “mother” genomic DNA as template. A 1-kb product was cloned into pCR2.1 TA cloning vector (Invitrogen) and sequenced. To obtain the UAA transmembrane/cytoplasmic genomic sequence, PCR was performed using cosmid 65 DNA as template. The same exon 5 primer was used as the forward primer with a UAA-specific reverse primer: 5′-AAGCTTAAATCAGCAGGT-3′. PCR was performed as above. The 2.5-kb product was cloned into the pCR2.1 TA cloning vector and sequenced. The PCR-amplified fragment for UAA exon 5 and intron 5 of cosmid 19 was also sequenced.
Northern blotting
RNA was isolated from various tissues using the TRIzol reagent (Life Technologies, Rockville, MD). Total RNA (20 μg) was electrophoresed in 1% agarose gels using MOPS as running buffer (Quality Biologicals, Gaithersburg, MD), and then transferred to an Optitran nitrocellulose membrane (Schleicher & Schuell, Keene, NH). The membranes were hybridized with UAA 3′ untranslated region (UTR), UAA-NC1 (α1 domain and intron 2), and nucleoside diphosphate kinase probes (39) on the tissue distribution blot. The 3′ UTR of LMP7 and LMP7-like were used as probes with RNA made from PBL of 8 sibling sharks, shark Yellow, and 11 wild animals. All blotting was done under high stringency conditions (34).
RT-PCR
cDNA was prepared from 5 μg of total RNA from shark Yellow PBL with the Superscript preamplification system for first-strand cDNA synthesis kit (Life Technologies). PCR was then performed by 3′ RACE using a UAA α1 domain forward primer, 5′-TCTCACAGTCTCCGGTATTT-3′, and the RACE reverse primer, 5′-CTCGAGAAGCTTGAATTCGGATCC-3′ at 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 4 min, and 72°C for 15-min final extension. A 1.7-kb single band was cloned into pCR2.1 TA cloning vector (Invitrogen) and sequenced.
Results
Isolation of shark LMP2 and horn shark LMP7-like
A 600-bp PCR product was amplified by 5′ RACE from a horn shark spleen cDNA library with a primer specific for LMP2 genes (33). The sequence of this product showed high similarity to all known LMP2 genes, and it was used to screen the same library to isolate LMP2 cDNA clones. The longest horn shark clone (1044 bp, with a 654-bp open reading frame, 304-bp 5′ UTR, and 86-bp 3′ UTR, Fig. 1A) was then used to screen a nurse shark spleen cDNA library. One full-length clone was sequenced and consisted of a 119-bp 5′ UTR, 61-bp 3′ UTR, and 654-bp open reading frame encoding 217 aa (Fig. 1A). The horn and nurse shark clones are 81% identical at the nucleotide level, and the deduced mature nurse shark protein sequence (start at +1 in Fig. 1A) is 87% identical to horn shark LMP2, 67–73% to LMP2 of other species, and 54–62% to proteasome δ (δ throughout the text). Nurse and horn shark LMP2 group with other species’ LMP2, whereas LMP2/δ (an intermediate found only in teleosts (14, 15, 40, 41)) and δ form separate clusters (Fig. 1B).
The nurse shark LMP7-like gene was described previously (31). To determine whether this gene is expressed by other elasmobranchs, we screened a horn shark spleen cDNA library and isolated the orthologous LMP7-like. Rather like the teleost LMP2/δ, the sequence of the resulting horn shark clone forms a cluster with nurse shark LMP7-like in phylogenetic trees, which is equidistant from the LMP7 and X clusters (Fig. 1C). This position of the LMP7-like cluster in the tree suggests that the gene arose not long after the divergence of LMP7 and X, and indeed it may be present in all extant cartilaginous fish.
Linkage studies of TAP, LMP, and class I
In the family of 39 nurse sharks, 13 segregating groups (a–m) were identified previously from classical class I (UAA) RFLP typing (1). Now we have extended the linkage analysis for LMP2, LMP7 (31), LMP7-like (31), TAP1, and TAP2 (30). Representative Southern blots of 20 of these siblings are shown in Fig. 2, and summaries of all experiments are shown in Tables I and II. TAP1 provided limited RFLP and formed only three groups (denoted a–c under the TAP1 blot in Fig. 2, a = U/L, b = L, c = U; U and L designate the upper and lower bands, respectively, in Tables I and II), but each group matched perfectly to the class I groups. RFLP for LMP2, LMP7 (EcoRI), and TAP2 were found only for paternal MHC haplotypes within groups d, e, g, and h (marked with dashes at the side of each blot, and see Table II). One weakly hybridizing LMP7 band was shown not to be MHC linked (BamHI digest in Fig. 2).
FIGURE 2.
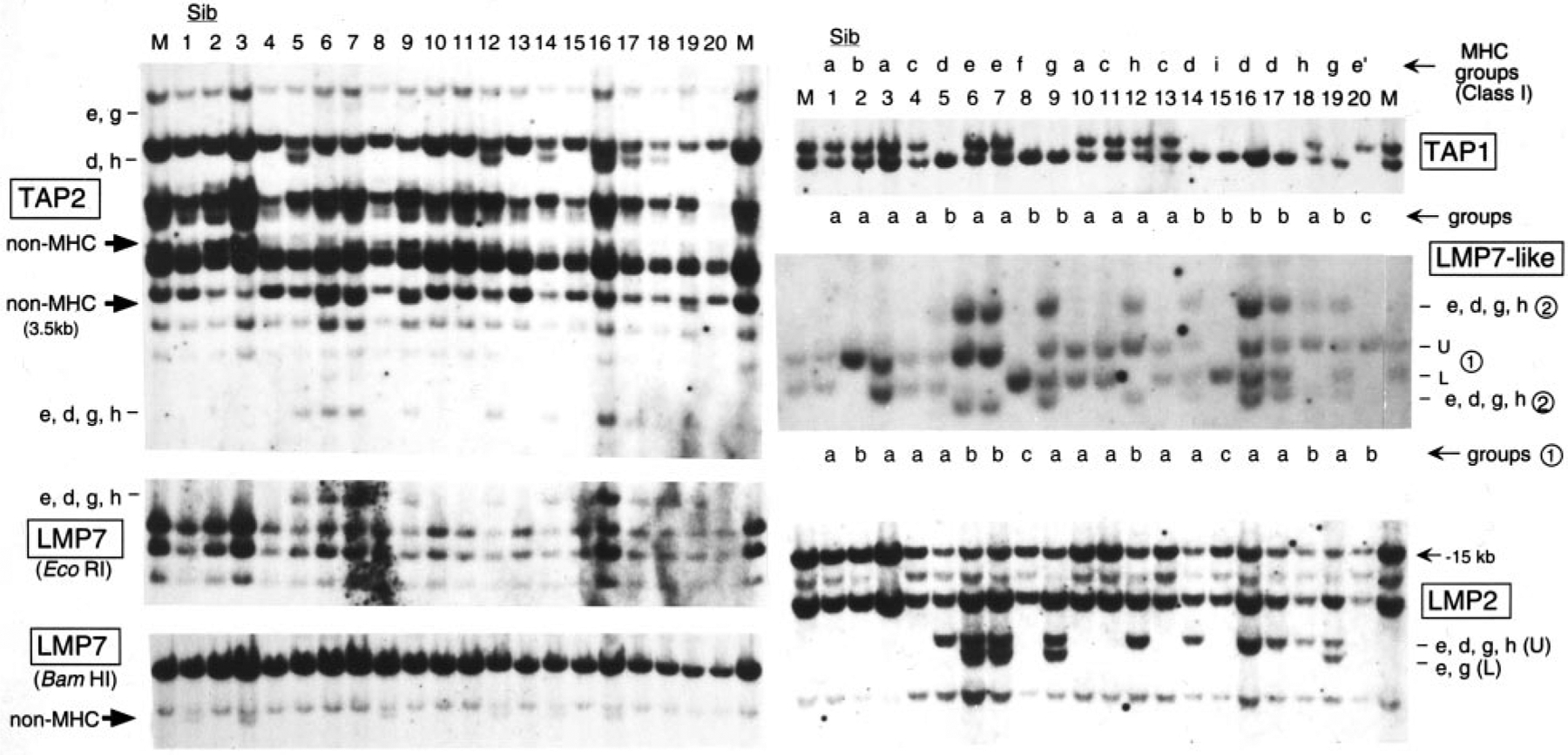
Linkage of LMP7, LMP2, LMP7-like, TAP1, and TAP2 to the nurse shark MHC. Southern blot analyses with 20 siblings from the family with 39 offspring (1). RE were: BamHI for TAP1; BamHI/EcoRI for LMP7-like; PvuII for TAP2; and LMP2, EcoRI, and BamHI for LMP7. Lower case letters at the side of each blot indicate MHC groups, and large arrows indicate the non-MHC-linked bands. Two loci revealed on the LMP7-like blot are indicated as ➀ and ➁, and groups are shown under the blot. Segregants sorted with previous UAA typing (1) are shown above the TAP1 blot as MHC groups. TAP1 groups are shown under TAP1 blot. M, mother; Sib, siblings (numbers indicate individuals from Ref. 1). Band sizes are as follows: TAP2: e, g, 9 kb; d, h, 6.5 kb. Non-MHC: 5, 3.5 kb; e, d, g, h, 2.5 kb. LMP7: EcoRI, 3 kb; BamHI, 9 kb. TAP1: 10, 9 kb. LMP2: e, d, g, h, 6.6 kb; e, g, 6 kb. LMP7-like: ➀, 5.3 and 4.8 kb; ➁, 6.3 and 4 kb.
Table I.
Summary of maternal RFLP in mother and individual siblings
| Siblings | Mother | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sexa | F | F | F | M | F | M | M | M | F | F | F | F | F | M | F | M | M | F | M | F |
| Yolk sac diameter (cm) | 2.5 | – | 2.5 | – | 2.5 | 1.2 | 0.6 | 2.5 | – | 1.8 | – | 2.5 | 0.6 | 2.5 | 0.6 | 1.8 | 1.2 | 1.2 | – | |
| Body length (cm) | 23 | 27 | 23 | 28 | 23 | 25.5 | 28 | 23 | 31 | 24 | 29 | 27 | 25.5 | 27 | 27 | 25.5 | 25.5 | 25.5 | 28 | |
| MHC groupb | a | b | a | c | d | e | e | f | g | a | c | h | c | d | i | d | d | h | g | |
| Maternal haplotype | m1/2 | m1 | m1 | m1 | m2 | m2 | m1 | m1 | m2 | m2 | m1 | m2 | m1 | m2 | m2 | m2 | m2 | m2 | m1 | m2 |
| UAA α1b | L/U | L | L | L | U/L | U | L | L | U | U/L | L | U/L | L | U/L | U | U | U | U | L | U/L |
| TAP1 | U/L | U/L | U/L | U/L | U/L | L | U/L | U/L | L | L | U/L | U/L | U/L | U/L | L | L | L | L | U/L | L |
| LMP7-like ➀ | U/L | U/L | U | U/L | U/L | U/L | U | U | U/L | U/L | U/L | U/L | U | U/L | U/L | U/L | U/L | U/L | U | U/L |
| TAP2 non-MHC (3.5 kb) | − | − | − | − | − | − | + | + | − | + | − | − | − | − | − | − | − | − | − | + |
| LMP7non-MHC (BamHI) | − | + | − | + | − | − | − | − | + | − | − | − | + | − | + | − | + | − | − | − |
| UAA-NC1 | U/L | L | U/L | U | U/L | U | L | U/L | L | U/L | U | L | U | U/L | U | L | L | U/L | U | L |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | |
| Sexa | F | F | M | M | M | M | F | M | M | F | F | M | F | F | F | F | F | M | F | F |
| Yolk sac diameter (cm) | 3.1 | 0.6 | 0.6 | - | 0.8 | 0.6 | 1.8 | 1.8 | 0.6 | 1.2 | - | 0.3 | 0.6 | 0.6 | 2.5 | 2.5 | 0.6 | 0.3 | 0.3 | 0.3 |
| Body length (cm) | 24 | 27 | 27 | 28 | 25.5 | 28 | 24 | 24 | 25.5 | 25.5 | 28 | 28 | 25.5 | 25.5 | 23.5 | 25.5 | 25.5 | 27 | 25.5 | 28 |
| MHC groupb | e′ | j | h | k | d | j | d | d | g′ | d | i | j | e′ | j | h | l | e′ | m | h | j |
| Maternal haplotype | m1 | m1 | m1 | m2 | m2 | m1 | m2 | m2 | m2 | m2 | m2 | m1 | m1 | m1 | m1 | m1 | m1 | m1 | m1 | m1 |
| UAA α1b | L | L | L | U | U | L | U | U | U/L | U | U | L | L | L | L | L | L | L | L | L |
| TAP1 | U | U/L | U/L | L | L | U/L | L | L | U/L | L | L | U/L | U | U/L | U/L | U | U | U | U/L | U/L |
| LMP7-like ➀ | U | U/L | U | U/L | U/L | U/L | U/L | U/L | U/L | U/L | U/L | U/L | U | U/L | U | U/L | U | U | U | U/L |
| TAP2 non-MHC (3.5 kb) | − | − | + | + | − | − | + | − | − | − | − | − | − | − | + | + | − | − | + | − |
| LMP7non-MHC (BamHI) | − | − | + | − | − | − | + | + | − | + | − | − | − | − | − | − | − | − | + | − |
| UAA-NC1 | U/L | L | U | U/L | U/L | L | U | U | U/L | L | U/L | U/L | U/L | U/L | U/L | L | U/L | U | U | L |
F, female; M, male.
Data taken from Ref. 1.
Table II.
Summary of paternal RFLP and haplotypes
| No. of Siblings | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MHC Groupa | UAA α1/Leadera | LMP2 | LMP7 (EcoRI) | LMP7-like ➁ | TAP1 | TAP2 (9, 6.5, 2.5 kb) | Paternal-Maternal Haplotype | LMP7 non-MHC | Total | Yolk Sac Average Diameter (cm) | Body Length Range (cm) | Predicted Father |
| a | UU/U | − | − | − | L | −−− | p1-m1 | 2 | 3 | 2.3 | 23–24 | 1 |
| f | UU/U | − | − | − | L | −−− | p1-m2 | 1 | 1 | 2.5 | 23 | 1 |
| d | LL/L | U | + | + | L | −++ | p2-m2 | 5 | 8 | 1.7 | 23–27 | 1 |
| h | LL/L | U | + | + | L | −++ | p2-m1 | 3 | 5 | 1.4 | 23.5–27 | 1 |
| i | L/L | − | − | − | L | −−− | p3-m2 | 0 | 2 | 0.3 | 27–28 | 2 |
| j | L/L | − | − | − | L | −−− | p3-m1 | 0 | 5 | 0.5 | 25.5–28 | 2 |
| e | L/U | U, L | + | + | L | +−+ | p4-m1 | 0 | 2 | 0.9 | 25.5–28 | 3 |
| g | L/U | U, L | + | + | L | +−+ | p5-m2 | 0 | 2 | 0 | 28–31 | |
| e′ | L/U | − | − | − | U | −−− | p6-m1 | 0 | 3 | 1.4 | 24–25.5 | |
| b | L/Lb | − | − | − | L | −−− | p7-m1 | 0 | 1 | 0 | 27 | |
| k | L/L | U, L | + | + | L | −−+ | p8-m2 | 0 | 1 | 0 | 28 | |
| l | ML/U | L | + | + | U | −−+ | p9-m1 | 0 | 1 | 2.5 | 25.5 | |
| m | L/U | L | − | − | U | −−? | p10-m1 | 0 | 1 | 0.3 | 27 | |
| c | L/U | − | − | − | U | −−− | p11-m2 | 0 | 3 | 0.2 | 25.5–29 | |
| g′ | L/U | − | − | − | U | −−− | p12-m2 | 0 | 1 | 0.6 | 25.5 | |
Data taken from Ref. 1.
The group b α1/leader band migrates slightly faster than groups i and j.
On the LMP7-like blot, there are two sets of segregating loci (indicated as ➀ and ➁). The three groups of ➀ bands (a = U/L, b = U, c = L, shown on the bottom of the blot) matched the class I MHC groups perfectly, and the presence vs absence of the ➁ set of bands precisely matched paternal haplotypes in groups d, e, g, and h; thus, both ➀ and ➁ LMP7-like genes are MHC linked, but only one set of bands is found in all haplotypes (and see later).
RFLP of all 39 siblings are summarized in Table I, in which results with the maternal UAA class I α1 domain are shown to illustrate the inheritance of maternal haplotypes identified definitively from siblings having distinctive paternal bands (1). The predicted maternal haplotype m1 was in 18 offspring and is L (UAA), U (TAP1), and U (LMP7-like ➀); m2 was in 21 offspring and is U (UAA), L (TAP1), and L (LMP7-like ➀); neither maternal haplotype contains the LMP7-like ➁ set of bands. Unfortunately, no LMP2, LMP7, or TAP2 RFLP were detected for the maternal haplotypes; nevertheless, as mentioned above, several paternal haplotypes had distinctive RFLP mapping to MHC for all of these genes.
Sorting out paternal haplotypes
Twelve paternal haplotypes were detected (p1 through p12), demonstrating that there were at least six fathers of this family (Table II). Besides the MHC-linked genes, we used the non-MHC-linked LMP7 RFLP (Fig. 2) and other nonlinked genes (data not shown), as well as body and yolk sac size, to determine haplotype pairs for each father. The p1 and p2 haplotypes had RFLP for the nonlinked LMP7, strongly suggesting that they were derived from one father (designated father 1, responsible for 17 of 39 offspring). In addition, siblings belonging to the four groups, a, f, d, and h, were younger than the others, having larger yolk sacs and smaller body sizes. Unfortunately, no other haplotypes produced RFLP, or yolk sac and body size were not informative enough to discriminate the pairs (the e and g groups had the same paternal RFLP for all genes tested; however, yolk sac and body length in these two groups are distinct, suggesting different fathers, or the same father after a long refractory period). Interestingly, there is an allelic preference for father 1 since 76% (13 of 17) of the offspring have the p2 haplotype. We observed the same phenomenon in another family of 18 nurse shark pups: one haplotype was found in six siblings, and the other haplotype in a maximum of three pups (1).
Cosmid clones bearing class I and proteasome genes
A genomic cosmid library was prepared from shark Yellow to study physical linkage of the shark MHC genes. Three class I-positive clones (cosmids 19, 32, 65) were isolated that hybridized weakly to the full-length LMP7 probe, with cosmid 32 displaying the highest intensity signal. Comparison of BamHI digests of cosmid 32 and shark Yellow genomic DNA resulted in comigration of the strongly hybridizing band (the weakly hybridizing band is due to cross-hybridization to the vector) in the cosmid clone and the weakly hybridizing, non-MHC-linked band in genomic DNA (Figs. 3A and 2, LMP7 BamHI blot). Two LMP7-positive fragments were identified in cosmid 32, and sequence of the XbaI-BamHI fragment revealed LMP7 exon 4 (Fig. 3C, accession AF357928). Exon 5 was identified by Southern blotting and PCR with exon-specific primers and sequencing of the product (data not shown). An exon 6 (3′ UTR)-specific probe did not hybridize to cosmid 32; furthermore, none of the fragments upstream from exon 4 hybridized with the full-length LMP7 probe, and since no other non-MHC-linked bands were detected on the genomic Southern blot with the probe, this gene must be truncated. PCR was then performed with cosmid 32 as template and primers designed to amplify the UAA α1 to α3 domain coding regions. The sequence of this class I gene in cosmid 32 (UAA-NC2) has two stop codons in the exon encoding the α2 domain and one in the α3 domain-encoding exon (Fig. 4), and the intron length and sequence between the α1 and α2 domain-encoding exons are also not conserved compared with UAA, UAA-NC2 having 326 bp rather than 108 bp in UAA. In summary, this cosmid clone contains a pseudogene related to class Ia and an incomplete LMP7 gene, and this syntenic cluster is not linked to the functional MHC.
FIGURE 3.
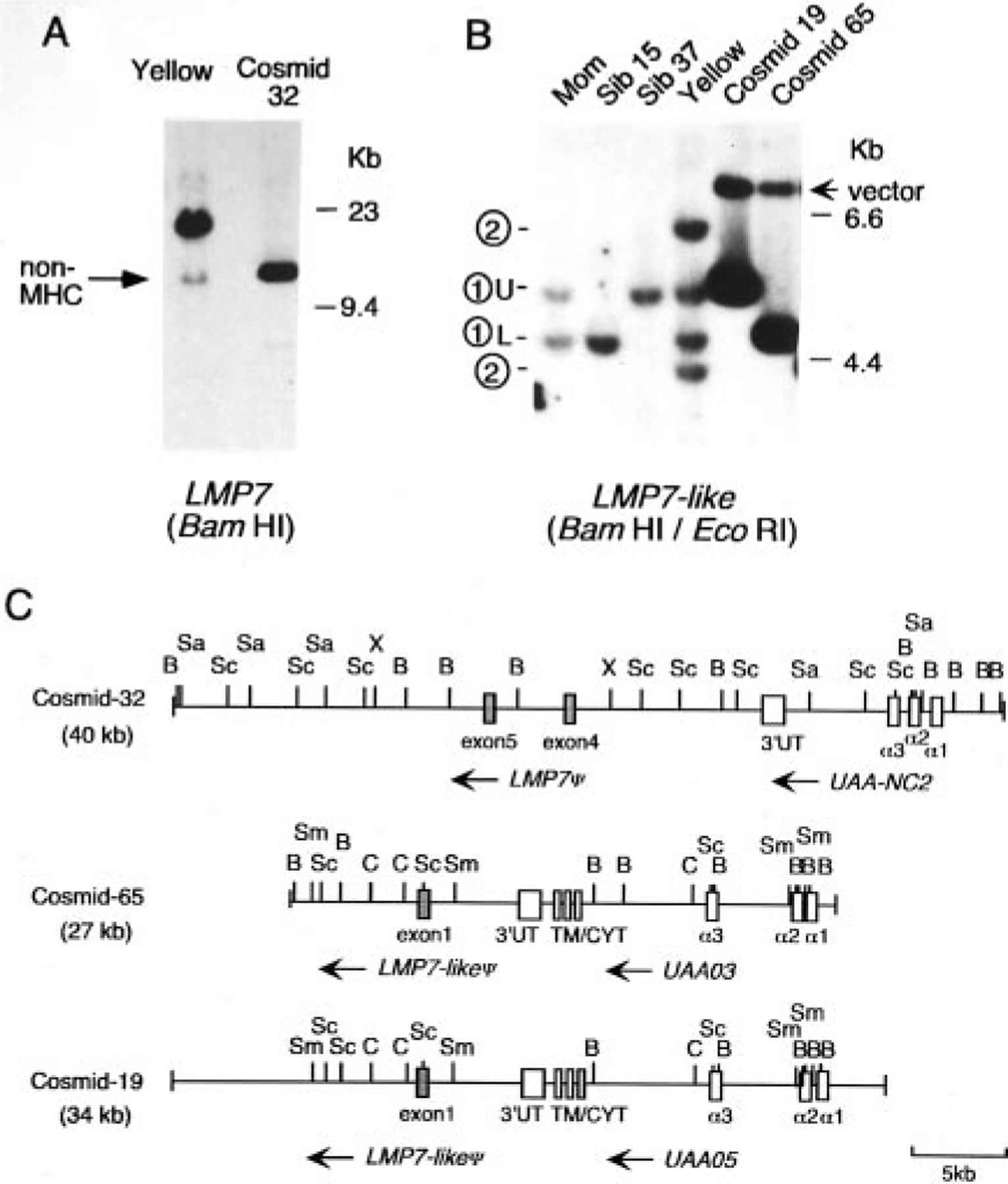
Analysis of class I+ nurse shark cosmid clones reveals closely linked inducible proteasome genes. A, Southern blot comparing shark genomic DNA and cosmid 32. A total of 5 μg of BamHI-digested DNA from shark Yellow and 50 ng of cosmid DNA were loaded side by side and hybridized with the full-length LMP7 probe. Non-MHC-linked bands are indicated by arrows on the left. HindIII-digested λ DNA used as size standard is noted on the right. B, Southern blot comparing genomic DNA and cosmids 19 and 65. A total of 5 μg of mother, siblings 15 and 37, shark Yellow genomic DNA, and 50 ng of cosmid DNA was loaded and hybridized with the full-length LMP7-like probe. BamHI/EcoRI double digests were used to identify band ➀. The 7-kb band in the cosmid DNA lanes was due to cross-hybridization to the SuperCos 1 vector (arrow). C, Restriction mapping of the class I+ cosmid clones. Insert sizes are shown in parentheses. Exons are shown as open boxes and identified with the exon number or domain names; mammalian numbering for the LMP7 exons was used (58). The transcriptional orientation is shown with arrows, and the 5-kb scale is shown on the bottom right. The following RE were used for mapping. B, BamHI; Sa, SacI; Sc, ScaI; X, XbaI; C, ClaI; Sm, SmaI.
FIGURE 4.

Amino acid comparison of all nurse shark class I and human HLA-Cw3. GenBank accession numbers are: UAA01 (AF220063), UAA02 (AF220358), UAA09 (AF220359), UAA05 (AF357925), UAA03 (AF357924), UAA-NC1 (AF220360), UAA-NC2 from cosmid 32 (AF357926), UCA01 (AF028557), and HLA-cw3 (M84172). Dashes and dots show sequences identical to UAA01 and gaps, respectively. Predicted peptide-binding residues and NK cell receptor interaction sites (48) are indicated as p and k above and under the sequences, respectively. 79R is in boldface, and the regions that differ dramatically between the two class Ia alleles are bracketed. Three stop codons in the cosmid 32 class I clone are shown as *.
As mentioned above, we previously showed that there is only one class Ia gene per MHC haplotype in the nurse shark (1). Two cosmid clones (clones 19 and 65) were mapped (Fig. 3C) and shown to bear class Ia genes with sequences precisely matching the two expressed UAA cDNAs expressed by shark Yellow, the presumed alleles (UAA03 and UAA05, Fig. 4). To identify the weakly hybridizing LMP7 band in each cosmid, the 1.9-kb ClaI/SmaI DNA fragment was subcloned, and the sequence revealed an LMP7-like exon 1 (accession AF357927). Double digestions of the cosmid clones with BamHI and EcoRI demonstrated that the LMP7-like genes on cosmid clones 19 and 65 are the ➀ U and ➀ L bands, respectively, detected on the genomic blot in Fig. 2 (Fig. 3B). As expected from mapping of the cosmids (the polymorphic BamHI site downstream of LMP7-like exon 1), shark Yellow, like the mother of the family shown in Fig. 2, has both bands (➀ U/L, Fig. 3B). The ➁ bands present in shark Yellow and in some of the fathers of the family encode at least one complete LMP7-like gene (there are additional ➁ bands not shown in Figs. 2 and 3B). As with the LMP7 gene in cosmid 32, LMP7-like ➀ is a pseudogene, as only exon 1 was identified from sequencing analysis. In summary, the cosmid clones bearing the nurse shark MHC-linked classical class I genes were isolated, and such genes are closely linked to proteasome gene fragments.
To prove conclusively that some animals lacked a complete LMP7-like gene, we performed Northern blots with PBL RNA from eight siblings, three positive (groups d and h) and five negative for the ➁ set of bands (Fig. 2 and Table II), and with shark Yellow, which is also positive (Fig. 3B). Only those animals that were band ➁ positive on the Southern blot expressed LMP7-like transcripts, while all nine animals expressed LMP7 (Fig. 5A).
FIGURE 5.
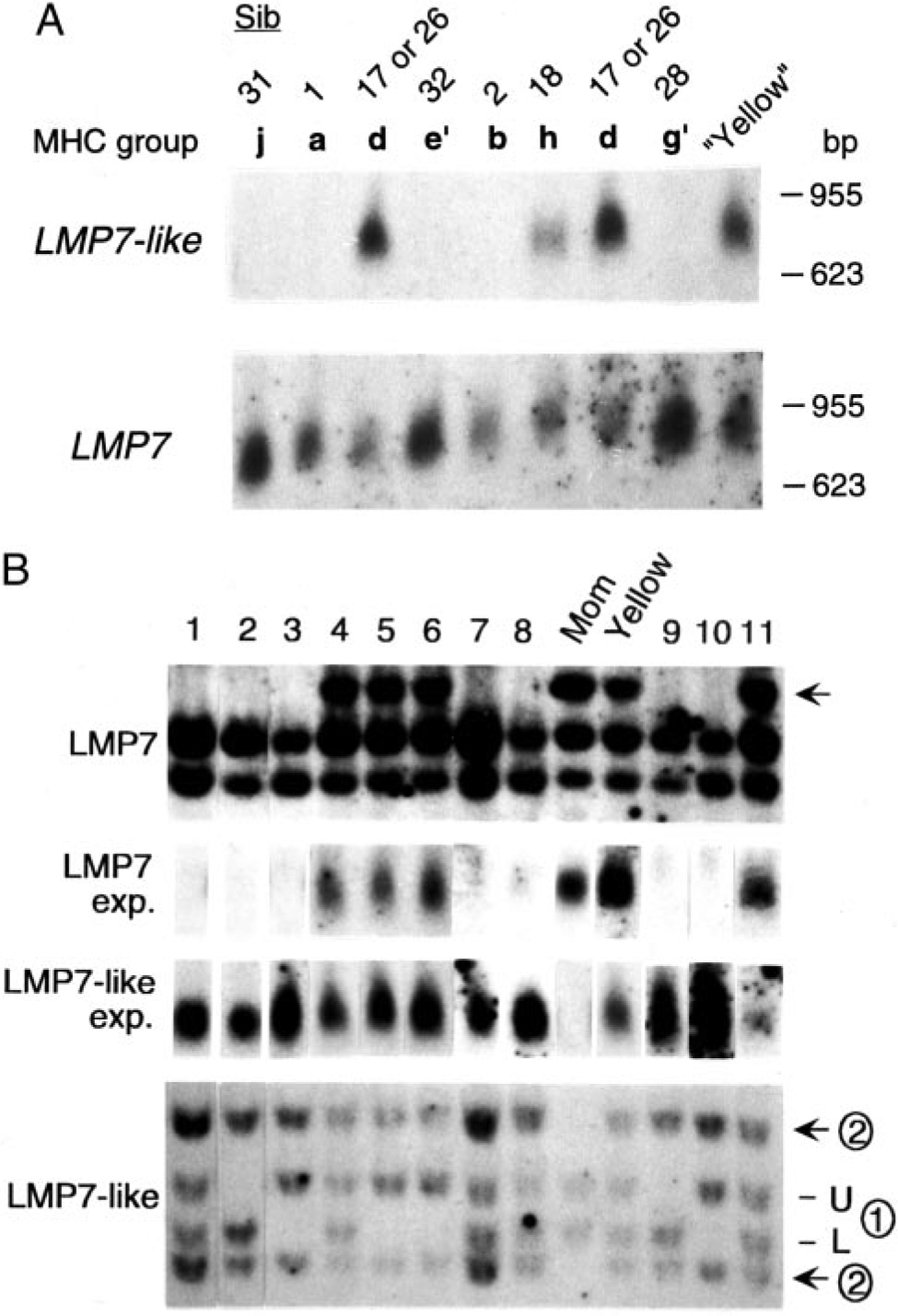
Northern blot analysis shows that some animals lack expression of either LMP7 or LMP7-like. A, Total PBL RNA from eight siblings and shark Yellow were compared. The number of siblings and their MHC groups were shown above blots. RNA size marker (Promega, Madison, WI) is shown on the right. B, Southern and Northern blots from 11 wild nurse sharks (noted as 1–11 above the blots) as well as the mother of the family and shark Yellow with LMP7 and LMP7-like probes. The LMP7 Southern band correlating positively with LMP7 expression is noted with an arrow, and the two LMP7-like loci are designated as ➀ and ➁ (see Fig. 2). BamHI and EcoRI were used for the Southern blots, and band sizes were as follows: LMP7, 2.8, 2.5, and 2.3 kb; LMP7-like ➀, 5.3 and 4.8 kb; ➁, 6.3 and 4 kb.
To further investigate expression of the LMP7 and LMP7-like genes, we performed Northern blots of 11 unrelated (to our knowledge) nurse sharks. As expected, the mother of the family lacking the ➁ set of LMP7-like bands detected by Southern blotting (Figs. 2 and 5B) did not express this gene (Fig. 5B). However, we were surprised to find that only four of the wild animals expressed LMP7, correlating with the presence of a 2.8-kb RFLP detected on the LMP7 Southern blot (arrow in Fig. 5B). Thus, from the family study and examination of the wild animals, functional LMP7 and LMP7-like behave like alleles (pseudoalleles), but LMP7 and LMP7-like gene fragments are nevertheless present in all haplotypes (Figs. 2, 5B, 9, and 10). The presence of two LMP7 alleles with quite different sequences is reminiscent of the situation in Xenopus, in which ancient LMP7 lineages have been detected (23) (see below).
FIGURE 9.
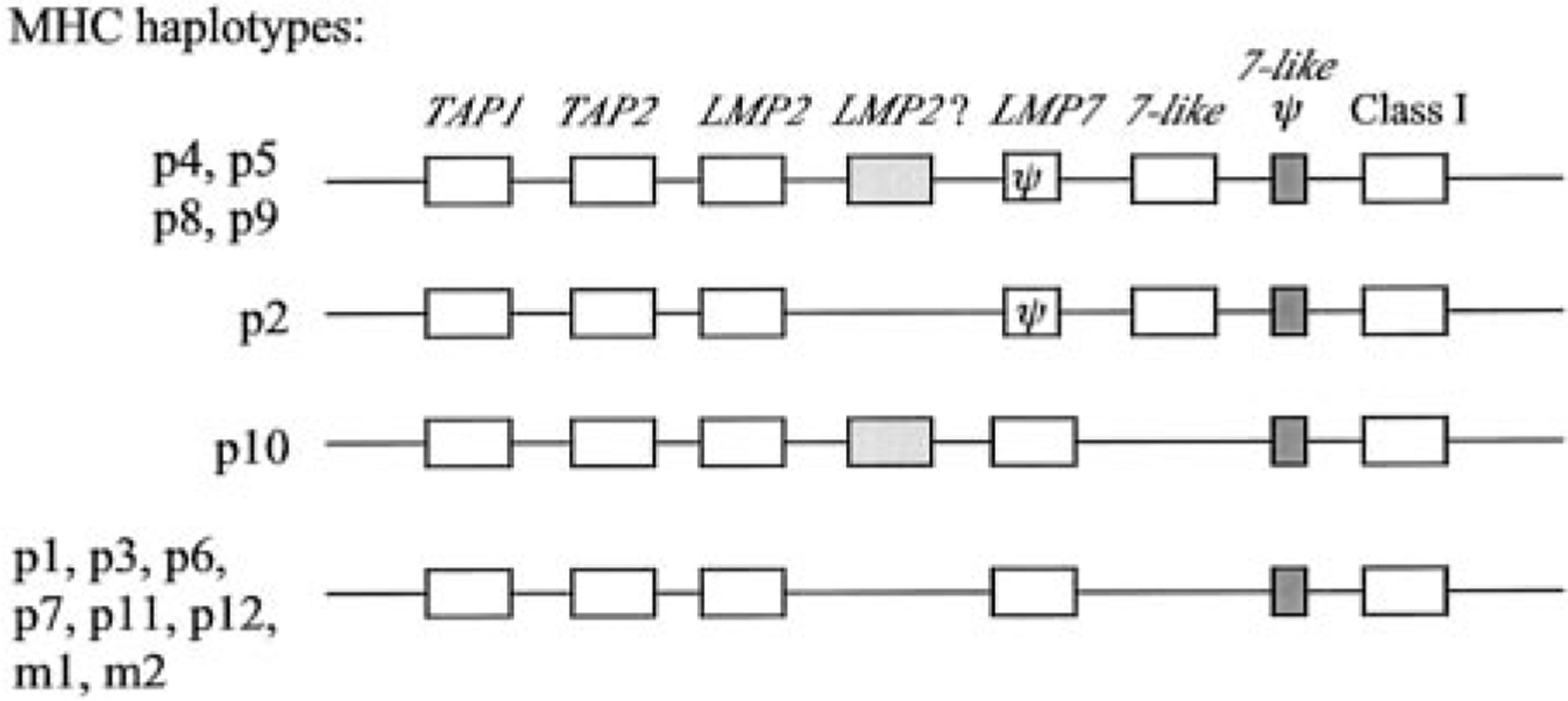
Hypothetical organization of nurse shark MHC haplotypes. All genes identified in this report and previous reports are shown in boxes, and LMP7-like and LMP7 pseudogenes are shown as dark-shaded boxes and ψ, respectively. The second LMP2 gene (fragment) is shown as a lightly shaded box. The order of genes and intergenic distances are not to scale (and are unknown). TAP2, LMP7, and LMP2 were definitively shown to map to MHC only in haplotypes from the P2, P4, and P5 groups.
FIGURE 10.
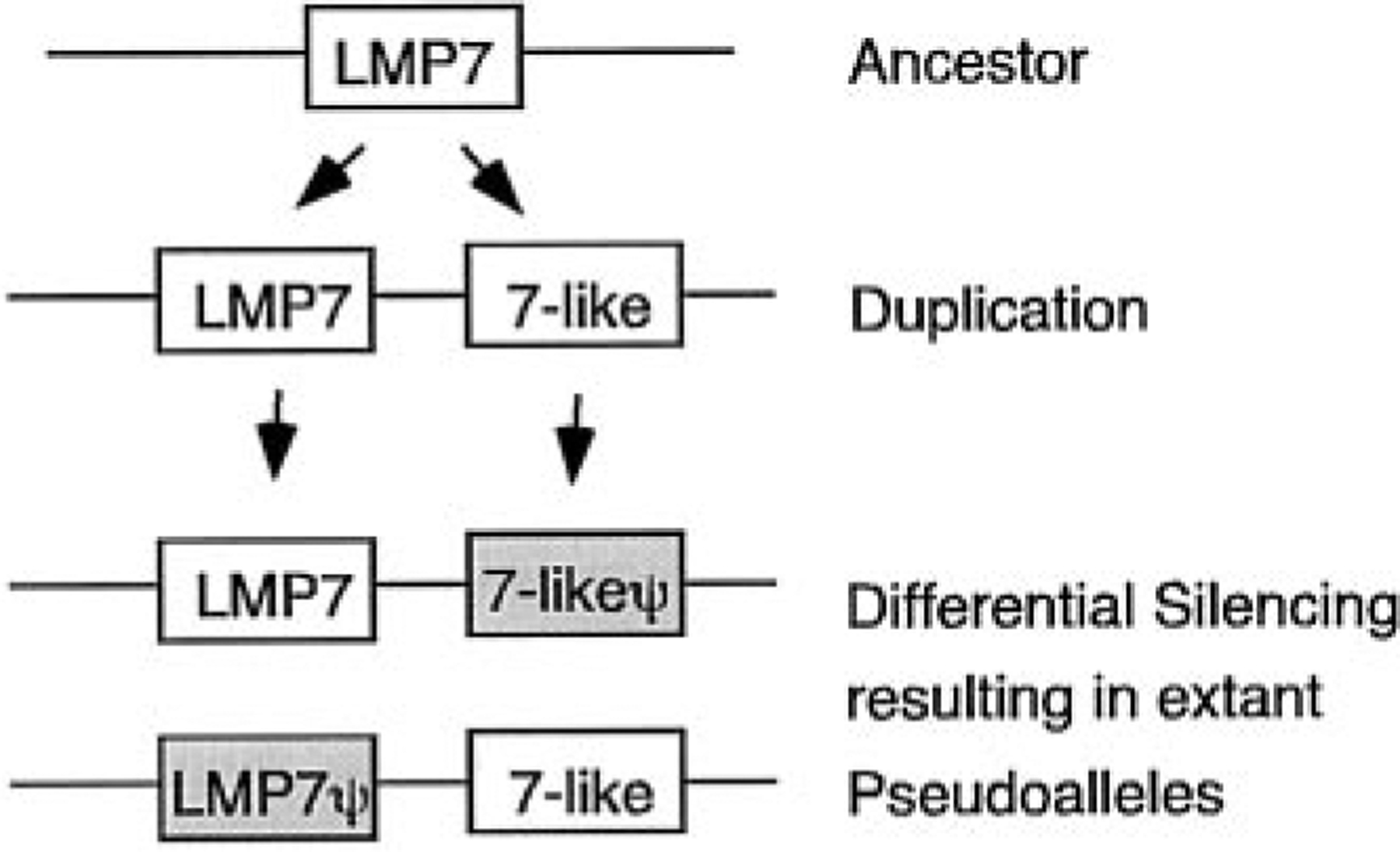
Postulated evolutionary progression of the pseudoallelic nature of LMP7 and LMP7-like. This model is based upon the expression analyses (Fig. 5) and the fact that LMP7 and LMP7-like gene fragments (i.e., incomplete genes) are detected in MHC haplotypes from which the functional, homologous gene is not present.
Other nonclassical class I
In our previous study, we isolated a nonclassical class I gene (UAA-NC1, accession AF220360) by PCR with the primer set used to amplify UAA from genomic DNA (1). UAA-NC1-specific bands segregating in the family were detected (Fig. 6), but the UAA-NC1 groups matched neither the MHC groups nor the non-MHC-linked TAP2 and LMP7 RFLP (Fig. 2 and Table I). To further understand the UAA-NC1 protein structure, the nurse shark spleen cDNA library was screened, resulting in isolation of a 2072-bp clone. Amino acid identity to Gici-UAA is 81% in leader, 61% in α1, 75% in α2, and 89% in α3 domains (Fig. 4). There is a 186-bp short interspersed elements retrotransposon element in the 3′ UTR not detected in UAA, and this element may have been involved in movement of UAA-NC1 out of the MHC (42). The beginning of the transmembrane domain is highly conserved, but there is a 49-nt deletion compared with UAA, resulting in a downstream frameshift (UAA-NC1a, Fig. 7). In addition, a clone that was 3 nt shorter at the start of exon 6 was isolated from the same cDNA library (UAA-NC1b). Since the deletion occurred near the exon-intron boundary, PCR was performed to amplify the region encompassing exons 5 through 6 from genomic DNA. The 1-kb fragment so amplified revealed that all the missing nucleotides were indeed present in UAA-NC1 genomic DNA, and the intron sequence was highly conserved between UAA and UAA-NC1. However, 3 nt were deleted from the middle of exon 5 (shown as a bracket above the sequence), and there are some nucleotide replacements (asterisks above the sequence) that might promote alternative splicing of UAA-NC1 transcripts (Fig. 7).
FIGURE 6.
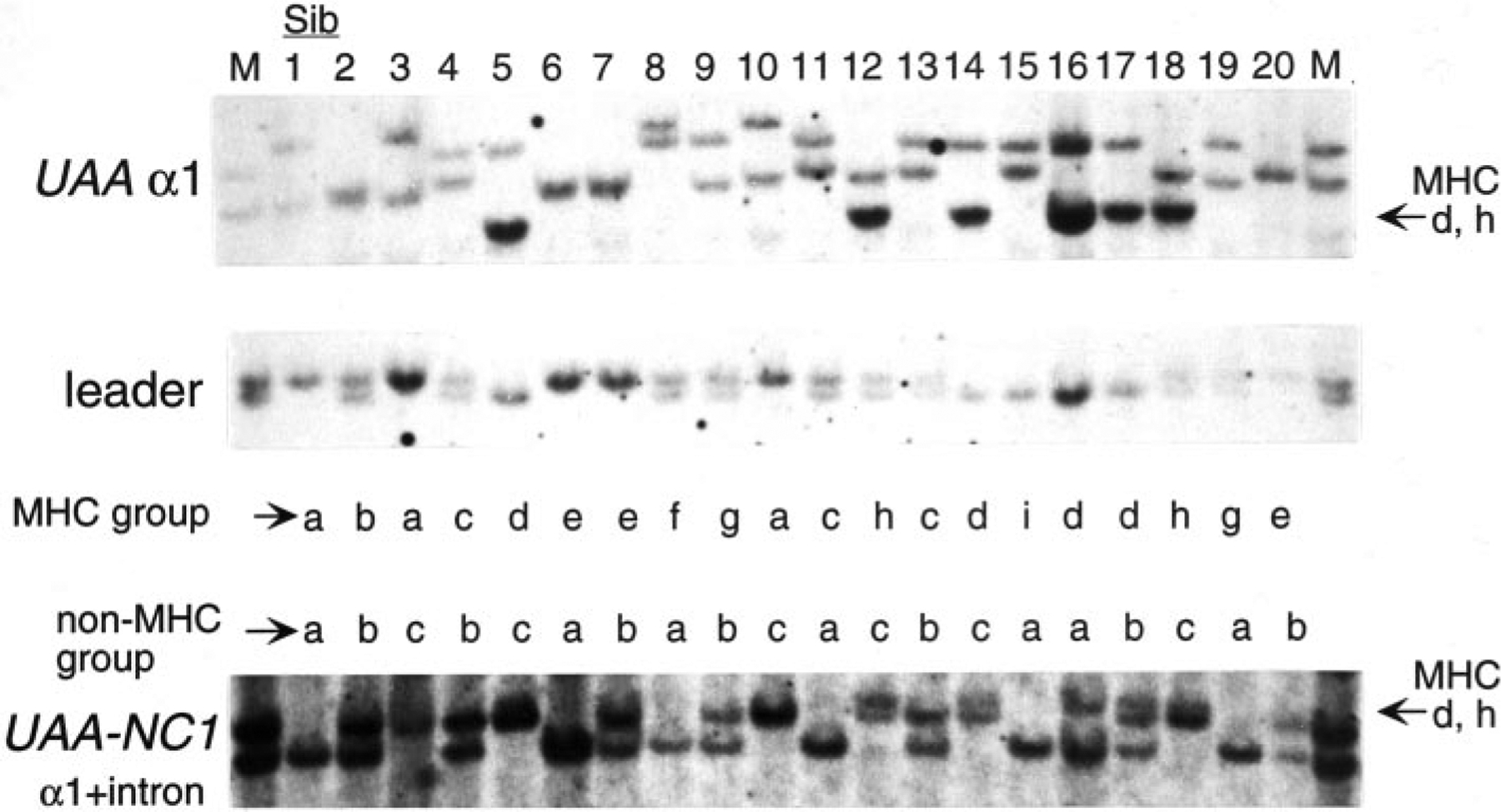
UAA-NC1 is not linked to the MHC. Southern blot analysis of 20 siblings in the family (same individuals in Fig. 2). RE used were: HindIII for UAA α1 and leader and BamHI for UAA-NC1. The probes and sibling numbers are indicated on the left side and above the blots, respectively. MHC and UAA-NC1 groupings are indicated below and above the blots, respectively (and in Table I). The cross-hybridization of UAA-NC1 to UAA α1 of the d and h groups is indicated on the right side of the UAA-NC1 blot. Band sizes are: UAA α1, d, h, ~14 kb; leader, ~4.5, 4 kb; UAA-NC1, ~13, 9.4 kb.
FIGURE 7.

Alternative splice sites in the nonclassical class I gene UAA-NC1. The UAA-NC1 genomic sequence was compared with two alternatively spliced cDNA clones (UAA-NC1a (AF357922) and b (AF357923)). Splice signal sites (GT… AG) are noted in boldface. Possible alternative splice sites are also marked in bold and underlined. Nucleotide differences between UAA and UAA-NC1 are marked as * above the sequence. Polymorphic nucleotides are marked as # underneath the sequence. The classical class I allele UAA03 was aligned above to highlight the 3-nt deletion (bracketed). The end of exon 5 and the beginning of exon 6 are indicated with arrows.
UAA-NC1 is expressed at much lower levels than UAA: only 4 UAA-NC1 clones (two of each splice variant) were isolated from library screenings compared with 42 UAA clones. The tissue distribution, however, is similar for both genes (Fig. 8). The 49-bp deletion resulting in premature termination in UAA-NC1a provides a transcript encoding the first 10 (of 24) hydrophobic aa in the putative transmembrane region. Thus, perhaps like proteins translated from differentially spliced class I transcripts in mammals (43, 44), different forms of the shark protein might be found.
FIGURE 8.
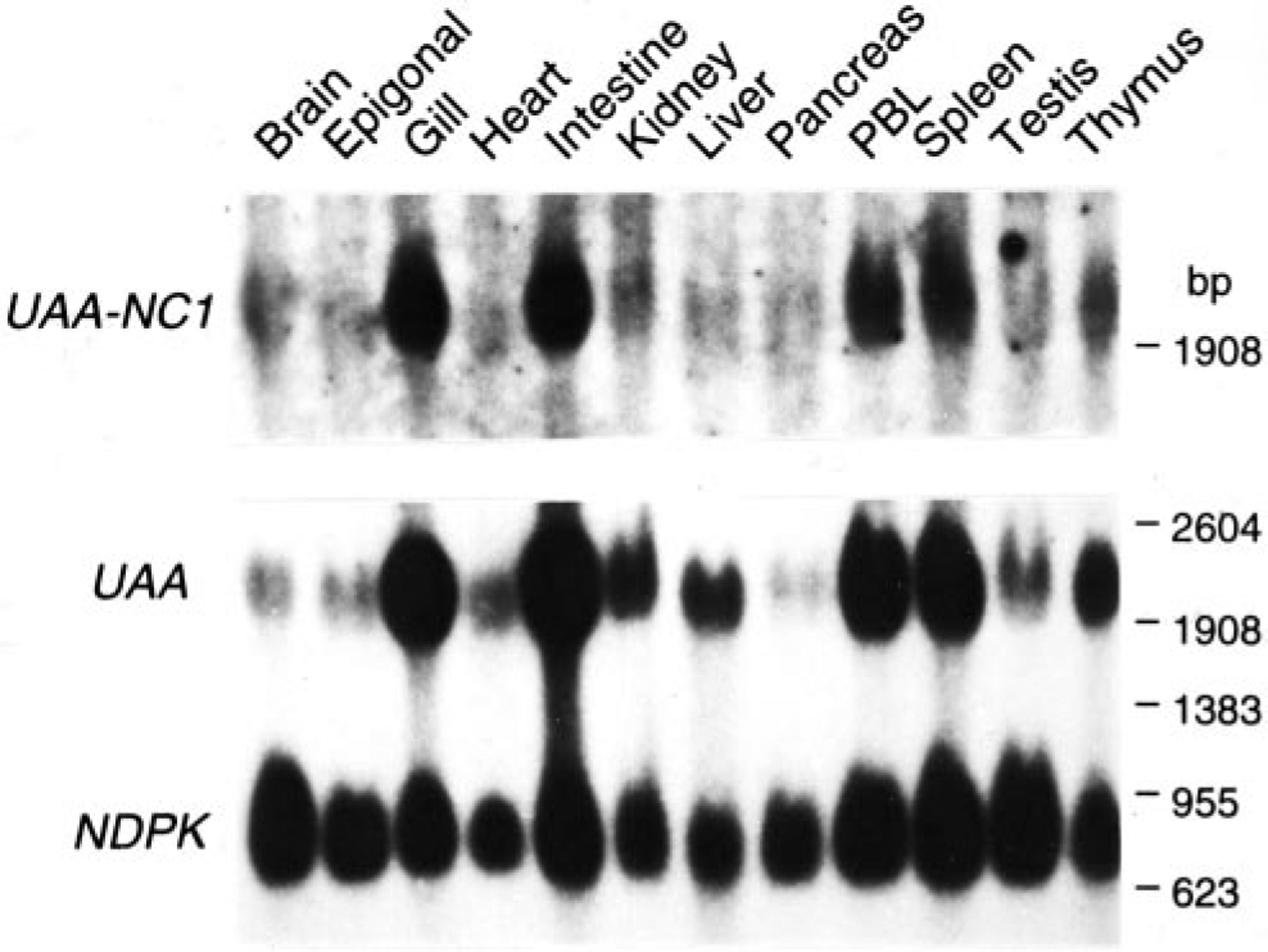
UAA and UAA-NC1 expression in various nurse shark tissues. The ubiquitously expressed nucleoside diphosphate kinase was used as the positive control (39). Size markers shown on the right.
Discussion
In this study, we show that TAP1, TAP2, LMP7, LMP7-like, and LMP2 are linked to classical class I and class II genes in the nurse shark MHC. One set of haplotypes in 19 of 39 siblings has a functional, expressed LMP7-like gene and apparently more than one LMP2 gene or gene fragment (Fig. 9). In groups d, e, g, h, k, l, and m, the LMP2 U band seems to segregate with the very top band (~15 kb in Fig. 2, e.g., compare siblings 4 and 5 in which this uppermost band is approximately half of the intensity in sib 5), but the LMP2 L band is found only in groups e, g, k, l, and m, suggesting the presence of additional genes or gene fragments. Whether there is a second expressed LMP2 gene in elasmobranchs, and whether this gene is related to the novel teleost LMP2 (PMSB11) gene (14, 15, 40, 41) requires further study.
The LMP7-like gene, previously identified in nurse sharks (31), now has been found in horn sharks as well, and, based on the phylogenetic tree analysis (Fig. 1C), this gene arose early in evolution and may be found in all elasmobranchs. Despite its apparent early emergence, a complete LMP7-like gene is not found in all haplotypes, and the functional gene is likely to be allelic with LMP7 (Figs. 5, 9, and 10). It was already recognized that shark LMP7 and LMP7-like proteins differ in residues near their respective catalytic sites (31), implying that the spectrum of peptides might be altered in animals that express both LMP7 genes compared with those with only one. Fig. 11 displays stereoimages of LMP7, LMP7-like, and the constitutive enzyme LMPX, modeled onto the crystal structure of yeast LMPX (45). Residues in red include the active site Thr-1, the obligatory (proton acceptor?) Lys33, and Met45, which is believed to coordinate chymotryptic-like cleavages (after hydrophobic residues) in this active site (46). Five residues in positions 28 (green), 29 (yellow), 30 (blue), 31 (black), and 32 (purple) (Ser-Gln-Thr-Val-Lys, Ser-Val-Asp-Ala-Asn, and Thr-Gln-Met-Phe-Lys in LMPX, LMP7, and LMP7-like, respectively) are diverse among the three proteins (31), and their positions relative to the active site are displayed. Note especially the proximity of the bulky Phe31 (black) to Met45 in LMP7-like as compared with Ala31 in LMP7 and Val31 in LMPX. We think that it would be surprising if these differences in the homologous catalytic subunits did not alter the spectrum of peptides produced by proteasomes containing them. Amazingly, for species separated from each other by ~500 million years, the two old allelic lineages of Xenopus LMP7 genes also differ from each other primarily in the vicinity of the catalytic site, one lineage having Phe31 and the other Ala31 (Ref. 23 and Y. Ohta, S. J. Powis, and M. F. Flajnik, manuscript in preparation). Although we are just beginning our analyses of the inducible proteasome genes in nonmammalian vertebrates, based upon 1) the very old gene/allelic lineages for several MHC genes (22–24, 47); 2) different proteasome genes, in particular MHC haplotypes (23; this study); and 3) the close linkage of proteasome genes to classical class I (6, 11, 13, 15, 17), one can propose that the generation of diverse sets of peptides by immunoproteasomes may be more crucial for adaptive responses in these nonmammalian species than in mice or humans.
FIGURE 11.
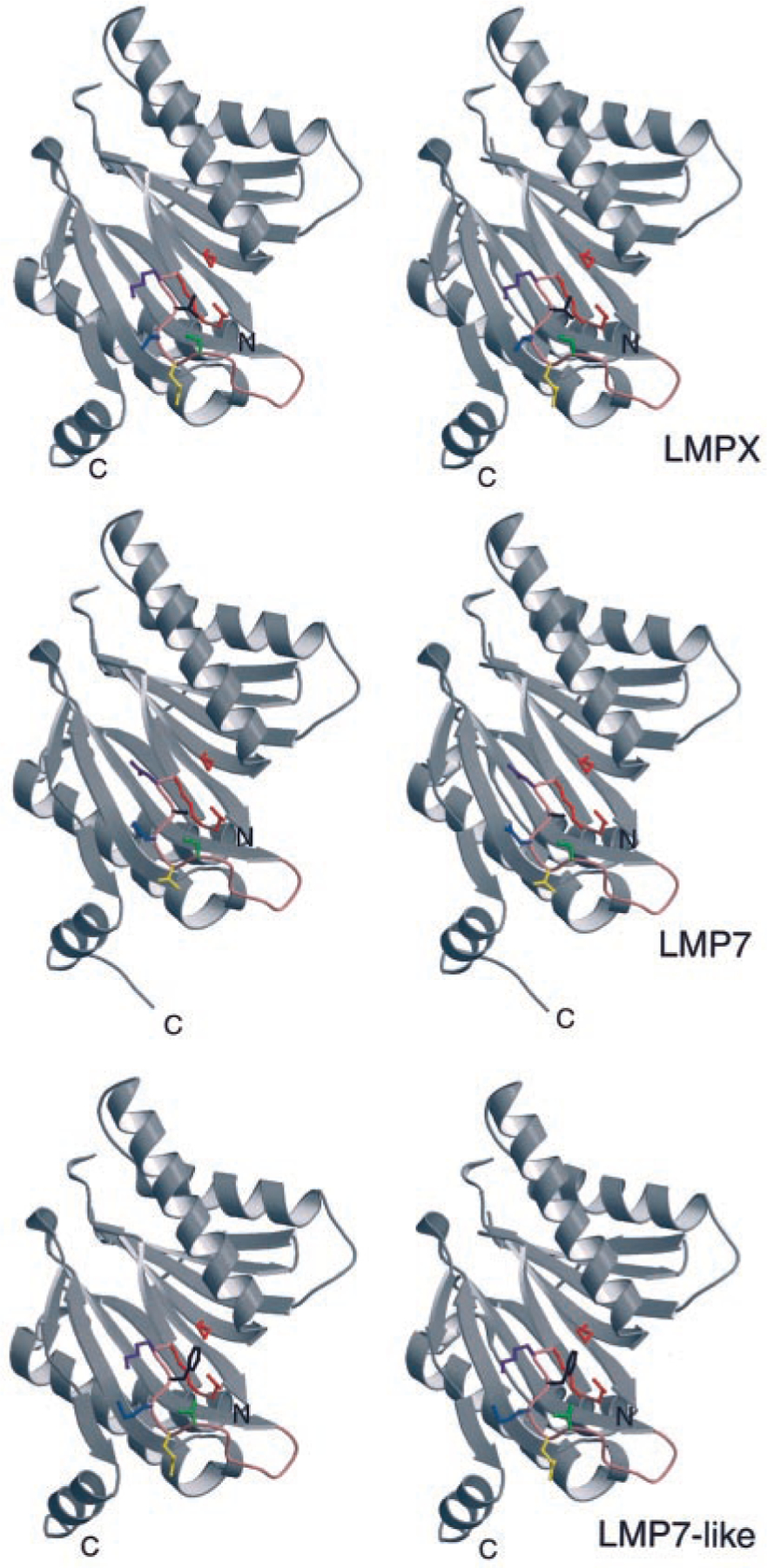
Stereoimage models of shark LMPX, LMP7, and LMP7-like proteins, based upon the crystal structure of the yeast orthologue of LMPX (45). PDB files were created in the program Homology Modeling (www.embi.kun.nl:1100/WIWWWI/). The models were generated in the three-dimensional imaging program MolScript (http://www.avatar.se/molscript/). See text for details of designations for the particular amino acid residues.
In addition to having more genes or gene fragments, RFLP for TAP2, LMP2, LMP7, and LMP7-like were more apparent in the group d, e, g, h, k, and l paternal haplotypes. One possible explanation for these findings is that there are only two major nurse shark MHC allelic lineages and only one lineage generates RFLP with the restriction enzymes that we used. To date, our analysis has shown two distinct class Ia molecules in all nurse sharks tested that are very different at the C terminus of the α1 helix, one having 77DILSK and the other 77KTALE (bracketed in Fig. 4). This polymorphic region itself could be recognized by different sets of NK cell receptors (e.g., see Ref. 48 for NK receptor interactions with a human killer inhibitory receptor), or perhaps different arrays of peptides generated, depending upon the constellation of inherited proteasome genes, may alter the orientation of this class I helical region recognized by TCR and/or NK cell receptors. It appears that the p1 and p2 haplotypes were inherited from one father, but the p2 haplotype was more successful than p1 in being perpetuated; the p2 haplotype in the d and h groups had RFLP for LMP2, LMP7, and TAP2, and contains a functional LMP7-like gene. Perhaps such a haplotype provided an advantage to offspring through expression of the LMP7-like gene, the particular class I allele, or other genes in the haplotype, so that survival was enhanced, but there are many other explanations for this finding that must be explored.
To date, four class I genes have been isolated from the nurse shark, one classical gene, and three nonclassical class I genes. The nonclassical UCA (formerly Gici-11 (34)) was the first gene to be cloned, and it is expressed in a similar fashion to UAA; unfortunately, we have not been able to map this class I gene because of its low level of polymorphism. UAA-NC1, isolated by a PCR “accident” with primers designed for UAA, was shown in this study not to map to MHC, despite its high sequence similarity to UAA. Some UAA alleles are more similar to UAA-NC1 than others (seen as cross-hybridization in groups d and h in Fig. 6), suggesting that UAA-NC1 arose from one particular UAA lineage. The third nonclassical class I UAA-NC2 was found in a cosmid clone bearing a truncated LMP7 gene. This pseudogene complex was probably the result of recent duplication event, which indicates that MHC class Ia was closely linked to LMP7 in the MHC. In fact, all of the class I genes found on the cosmid clones isolated to date are linked to an LMP7 (or 7-like) gene fragment. In the teleosts Fugu (13, 40) and zebrafish (15, 41), class Ia genes are also physically linked to LMP7 genes (or gene fragments), suggesting that this close physical linkage is ancient.
Based upon Southern blotting analyses with a variety of probes under low stringency conditions, UAA may be the only class I gene linked to MHC. The same feature is found for the Xenopus MHC, in which only one class I gene of the classical type is MHC linked (49), while a large set of linked nonclassical class I genes is found in another gene complex half a chromosome away (50, 51). In chickens, two class I genes (one highly expressed) sandwich the TAP1 and TAP2 genes, and Kaufman and colleagues (11, 21, 52) have proposed that the tight linkage has allowed the particular class I and TAP alleles to coevolve; furthermore, like in frogs, several nonclassical chicken class I genes are found a great distance away from the true MHC on the same chromosome (53). In the banded houndshark Triakis scyllium, there are one or two classical class I genes depending on the haplotype examined (54), and the majority of teleost species examined rigorously (except cod (29) and cichlid (25, 26)) have relatively few class I genes in the class I region. Previously, others and we have suggested that close linkage of class I/proteasome/TAP genes constitutes a class I region, in which the genes involved in class I processing and presentation coevolve (10, 21). A large number of class I genes are not optimal in this situation, as this would probably result in rapid evolution of class I by gene conversion or other recombinations known to occur in mammals (55). The nurse shark seems to have carried this feature to the extreme, with even recently duplicated class I genes being exiled from the MHC proper. Thus, we predict that the shark class I genes will also be closely linked to and coevolving with TAP and proteasome genes in a true class I region, consistent with our preliminary analysis of the cosmid clones.
Other cosmid clones containing TAP and proteasome genes have been isolated, and we are currently sequencing and analyzing such clones (in collaboration with T. Shiina and H. Inoko, Tokai University, Isehara, Japan). Examination of these clones will be helpful in comparing with other species to infer the primordial MHC organization. For example, recent work has shown that the third inducible proteasome gene MECL-1, while not linked to the MHC in mammals, is found in the teleost MHC class I region (13, 40, 56), consistent with predictions of large scale duplications being involved in the genesis of MHC (57); linkage of this gene, or other proteasome genes, to shark MHC will further test the duplication model. Since shark classical class II genes are linked to class I, it will be interesting as well to contrast the organization of all shark MHC regions with those of teleosts, in which the classical class II genes have been expelled to another region of the genome (14, 18). Finally, we may uncover genes in the shark MHC involved in Ag presentation that are no longer found in MHCs of more recently derived vertebrates.
Acknowledgments
We thank Robyn Stanfield for generating the proteasome models, Chris Rackley for helping us capture the nurse sharks, and Rebecca Lohr and Lori Clow for discussions and review of this work. We also are grateful to our colleagues Masaru Nonaka and Masanori Kasahara for being the first to uncover the interesting differences between the different proteasome subunits.
This work was supported by National Institutes of Health Grant AI27877.
Footnotes
Abbreviations used in this paper: RE, restriction enzyme; RFLP, restriction fragment-length polymorphism; UTR, untranslated region.
References
- 1.Ohta Y, Okamura K, McKinney EC, Bartl S, Hashimoto K, and Flajnik MF. 2000. Primitive synteny of vertebrate major histocompatibility complex class I and class II genes. Proc. Natl. Acad. Sci. USA 97:4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driscoll J, Brown MG, Finley D, and Monaco JJ. 1993. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature 365:262. [DOI] [PubMed] [Google Scholar]
- 3.Rammensee HG, Friede T, and Stevanoviic S. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178. [DOI] [PubMed] [Google Scholar]
- 4.Kloetzel P-M 2001. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol 2:179. [DOI] [PubMed] [Google Scholar]
- 5.Margulies DH 1999. Fundamental Immunology. Paul WE, ed. Lippincott, Philadelphia, p. 263. [Google Scholar]
- 6.Flajnik MF, Ohta Y, Namikawa-Yamada C, and Nonaka M. 1999. Insight into the primordial MHC from studies in ectothermic vertebrates. Immunol. Rev 167:59. [DOI] [PubMed] [Google Scholar]
- 7.Du Pasquier L, and Flajnik MF. 1999. Fundamental Immunology. Paul WE, ed. Lippincott, Philadelphia, p. 605. [Google Scholar]
- 8.The MHC Sequencing Consortium. 1999. Complete sequence and gene map of the human major histocompatibility complex. Nature 410:921. [DOI] [PubMed] [Google Scholar]
- 9.Takami K, Zaleska-Rutcynska Z, Figueroa F, and Klein J. 1997. Linkage of LMP, TAP, and RING3 with MHC class I rather than class II genes in the zebrafish. J. Immunol 159:6052. [PubMed] [Google Scholar]
- 10.Nonaka M, Namikawa C, Kato Y, Sasaki M, Salter-Cid L, and Flajnik MF. 1997. Major histocompatibility complex gene mapping in the amphibian Xenopus implies a primordial organization. Proc. Natl. Acad. Sci. USA 94:5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufman J, Milne S, Göbel WF, Walker BA, Jacob JP, Auffray C, Zoorob R, and Beck S. 1999. The chicken B locus is a minimal essential major histocompatibility complex. Nature 401:923. [DOI] [PubMed] [Google Scholar]
- 12.Shiina T, Shimizu C, Oka A, Teraoka Y, Imanishi T, Gojobori T,Hanzawa K, Watanabe S, and Inoko H. 1999. Gene organization of the quail major histocompatibility complex (MhcCoja) class I gene region. Immunogenetics 49:384. [DOI] [PubMed] [Google Scholar]
- 13.Clark MS, Shaw L, Kelly A, Snell P, and Elgar G. 2001. Characterization of the MHC class I region of the Japanese pufferfish (Fugu rubripes). Immunogenetics 52:174. [DOI] [PubMed] [Google Scholar]
- 14.Hansen JD, Strassburger P, Thorgaard GH, Young WP, and Du Pasquier L. 1999. Expression, linkage, and polymorphism of MHC-related genes in rainbow trout, Oncorhynchus mykiss. J. Immunol 163:774. [PubMed] [Google Scholar]
- 15.Michalova V, Murray BW, Sultmann H, and Klein J. 2000. A contig map of the MHC class I genomic region in the zebrafish reveals ancient synteny. J. Immunol 164:5296. [DOI] [PubMed] [Google Scholar]
- 16.Graser R, Vincek V, Takami K, and Klein J. 1998. Analysis of zebrafish MHC using BAC clones. Immunogenetics 47:318. [DOI] [PubMed] [Google Scholar]
- 17.Flajnik MF, and Kasahara M. 2001. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15:351. [DOI] [PubMed] [Google Scholar]
- 18.Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M, Postlethwait JH, and Klein J. 1997. Mapping of MHC class I and class II regions to different linkage groups in the zebrafish Danio rerio. Immunogenetics 46:129. [DOI] [PubMed] [Google Scholar]
- 19.Powis SJ, Young LL, Joly E, Barker PJ, Richardson L, Brandt RP, Melief CJ, Howard JC, and Butcher GW. 1996. The rat cim effect: TAP allele-dependent changes in a class I MHC anchor motif and evidence against C-terminal trimming of peptides in the ER. Immunity 4:159. [DOI] [PubMed] [Google Scholar]
- 20.Lobigs M, Mullbacher A, Blanden RV, Hämmerling GJ, and Momburg F. 1999. Antigen presentation in Syrian hamster cells: substrate selectivity of TAP controlled by polymorphic residues in TAP1 and differential requirements for loading of H2 class I molecules. Immunogenetics 49:931. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman J 1999. Co-evolving genes in MHC haplotypes: the “rule” for nonmammalian vertebrates? Immunogenetics 50:228. [DOI] [PubMed] [Google Scholar]
- 22.Flajnik MF, Ohta Y, Greenberg AS, Salter-Cid L, Carrizosa A,Du Pasquier L, and Kasahara M. 1999. Two ancient allelic lineages at the single classical class I locus in the Xenopus MHC. J. Immunol 163:3826. [PubMed] [Google Scholar]
- 23.Nonaka M, Yamada-Namikawa C, Flajnik MF, and Du Pasquier L. 2000. Trans-species polymorphism of the major histocompatibility complex-encoded proteasome subunit LMP7 in an amphibian genus, Xenopus. Immunogenetics 51:186. [DOI] [PubMed] [Google Scholar]
- 24.Ohta Y, Powis SJ, Coadwell WJ, Haliniewski DE, Liu Y, Li H, and Flajnik MF. 1999. Identification and genetic mapping of Xenopus TAP2 genes. Immunogenetics 49:171. [DOI] [PubMed] [Google Scholar]
- 25.Sato A, Klein D, Sultmann H, Figueroa F, O’hUigin C, and Klein J. 1996. Class I MHC genes of cichlid fishes: identification, expression, and polymorphism. Immunogenetics 46:63. [DOI] [PubMed] [Google Scholar]
- 26.Murray BW, Nilsson P, Zaleska-Rutczynska Z, Sultmann H, and Klein J. 2000. Linkage relationships and haplotype variation of the major histocompatibility complex class I A genes in the cichlid fish. Oreochromis niloticus. Mar. Biotechnol 2:437. [DOI] [PubMed] [Google Scholar]
- 27.Sammut B, Laurens V, and Tournefier A. 1997. Isolation of MHC class I cDNAs from the axolotl Ambystoma mexicanum. Immunogenetics 45:285. [DOI] [PubMed] [Google Scholar]
- 28.Sammut B, Du Pasquier L, Ducoroy P, Laurens V, Marcuz A, and Tournefier A. 1999. Axolotl MHC architecture and polymorphism. Eur. J. Immunol 29:2897. [DOI] [PubMed] [Google Scholar]
- 29.Persson AC, Stet RJ, and Pilström L. 1999. Characterization of MHC class I and β(2)-microglobulin sequences in Atlantic cod reveals an unusually high number of expressed class I genes. Immunogenetics 50:49. [DOI] [PubMed] [Google Scholar]
- 30.Ohta Y, Haliniewski DE, Hansen JD, and Flajnik MF. 1999. Isolation of transporter associated with antigen processing genes, TAP1 and TAP2, from the horned shark Heterodontus francisci. Immunogenetics 49:981. [DOI] [PubMed] [Google Scholar]
- 31.Kandil E, Namikawa C, Nonaka M, Greenberg A, Flajnik MF, Ishibashi T, and Kasahara M. 1996. Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates: implications for the origin of MHC class I-restricted antigen presentation. J. Immunol 156:4245. [PubMed] [Google Scholar]
- 32.Castro JI 2000. The biology of the nurse shark, Ginglymostoma cirratum, off the Florida east coast and the Bahama Islands. Environ. Biol. Fishes 58:1. [Google Scholar]
- 33.Nonaka M, Namikawa-Yamada C, Sasaki M, Salter-Cid L, and Flajnik MF. 1997. Evolution of proteasome subunits δ and LMP2: complementary DNA cloning and linkage analysis with MHC in lower vertebrates. J. Immunol 159:734. [PubMed] [Google Scholar]
- 34.Bartl S, Baish MA, Flajnik MF, and Ohta Y. 1997. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J. Immunol 159:6096. [PubMed] [Google Scholar]
- 35.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, and Flajnik MF. 2001. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc. Natl. Acad. Sci. USA 98:1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page RD 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci 12:357. [DOI] [PubMed] [Google Scholar]
- 37.Wong WM, Au DMY, Lam VHS, Tam JWO, and Cheng LYL. 1990. A simplified and improved method for the efficient double-stranded sequencing of mini-prep plasmid DNA. Nucleic Acids Res. 18:5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nizetic D, Drmanac R, and Lehrach H. 1991. An improved bacterial colony lysis procedure enables direct DNA hybridization using short (10, 11 bases) oligonucleotides to cosmids. Nucleic Acids Res. 9:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasahara M, Canel C, McKinney EC, and Flajnik MF. 1991. Molecular cloning of nurse shark cDNAs with high sequence similarity to nucleotide diphosphate kinase genes In NATO ASI Series: Molecular Evolution of the Major Histocompatibility Complex, Vol. H59 Klein D and Klein J, eds. Springer-Verlag, Berlin, p. 491. [Google Scholar]
- 40.Clark MS, Pontarotti P, Gilles A, Kelly A, and Elgar G. 2000. Identification and characterization of a β proteasome subunit cluster in the Japanese pufferfish (Fugu rubripes). J. Immunol 165:4446. [DOI] [PubMed] [Google Scholar]
- 41.Murray BW, Sultmann H, and Klein J. 1999. Analysis of a 26-kb region linked to the MHC in zebrafish: genomic organization of the proteasome component β/transporter associated with antigen processing-2 gene cluster and identification of five new proteasome β subunit genes. J. Immunol 163:2657. [PubMed] [Google Scholar]
- 42.Ogiwara I, Miya M, Ohshima K, and Okada N. 1999. Retropositional parasitism of SINEs on LINEs: identification of SINEs and LINEs in elasmobranchs. Mol. Biol. Evol 16:1238. [DOI] [PubMed] [Google Scholar]
- 43.Kress M, Cosman D, Khoury G, and Jay G. 1983. Secretion of a transplantation-related antigen. Cell 34:189. [DOI] [PubMed] [Google Scholar]
- 44.Carosella ED, Paul P, Moreau P, and Rouas-Freiss N. 2000. HLA-G and HLA-E: fundamental and pathophysiological aspects. Immunol. Today 21:532. [PubMed] [Google Scholar]
- 45.Groll M, Ditzel L, Stock D, Bochtler M, Bartunik HD, and Huber R. 1997. Structure of 20S proteasome from yeast at 2.4 angstrom resolution. Nature 386: 463. [DOI] [PubMed] [Google Scholar]
- 46.Bochtler M, Ditzel L, Groll M, Hartmann C, and Huber R. 1999. The proteasome. Annu. Rev. Biophys. Biomol. Struct 28:295. [DOI] [PubMed] [Google Scholar]
- 47.Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring RB, Stet RJ, Secombes C, and Parham P. 2001. Modes of salmonid MHC class I and II evolution differ from the primate paradigm. J. Immunol 166:3297. [DOI] [PubMed] [Google Scholar]
- 48.Boyington JC, Motyka SA, Schuck P, Brooks AG, and Sun PD. 2000. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 405:537. [DOI] [PubMed] [Google Scholar]
- 49.Shum BP, Avila D, Du Pasquier L, Kasahara M, and Flajnik MF. 1993. Isolation of a classical MHC class I cDNA from an amphibian: evidence for only one class I locus in the Xenopus MHC. J. Immunol 151:5376. [PubMed] [Google Scholar]
- 50.Flajnik MF, Kasahara M, Shum BP, Salter-Cid L, Taylor E, and Du Pasquier L. 1993. A novel type of class I gene organization in vertebrates: a large family of non-MHC-linked class I genes is expressed at the RNA level in the amphibian Xenopus. EMBO J. 12:4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Courtet M, Flajnik MF, and Du Pasquier L. 2001. Major histocompatibility complex and immunoglobulin loci visualized by in situ hybridization on Xenopus chromosomes. Dev. Comp. Immunol 25:149. [DOI] [PubMed] [Google Scholar]
- 52.Kaufman J, Jacob J, Shaw I, Walker B, Milne S, Beck S, and Salomonsen J. 1999. Gene organization determines evolution of function in the chicken MHC. Immunol. Rev 167:101. [DOI] [PubMed] [Google Scholar]
- 53.Miller MM, Goto RM, Taylor RL, Zoorob R, Auffray C, Briles RW, Briles WE, and Bloom SE. 1996. Assignment of Rfp-Y to the chicken MHC/NOR microchromosome and evidence for high-frequency recombination associated with the nucleolar organizer region. Proc. Natl Acad. Sci. USA 93:3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamura K, Ototake M, Nakanishi T, Kurosawa Y, and Hashimoto K. 1997. The most primitive vertebrate with jaws possess highly polymorphic MHC class I genes comparable to those of humans. Immunity 7:777. [DOI] [PubMed] [Google Scholar]
- 55.Martinsohn JT, Sousa AB, Guethlein LA, and Howard JC. 1999. The gene conversion hypothesis of MHC evolution: a review. Immunogenetics 50: 168. [DOI] [PubMed] [Google Scholar]
- 56.Sultmann H, Murray BW, and Klein J. 2000. Identification of seven genes in the major histocompatibility complex class I region of the zebrafish. Scand. J. Immunol 51:577. [DOI] [PubMed] [Google Scholar]
- 57.Kasahara M 1999. The chromosomal duplication model of the major histocompatibility complex. Immunol. Rev 167:17. [DOI] [PubMed] [Google Scholar]
- 58.Meinhardt T, Graf U, and Hämmerling GJ. 1993. Different genomic structure of mouse and human Lmp7 genes: characterization of MHC-encoded proteasome genes. Immunogenetics 38:373. [DOI] [PubMed] [Google Scholar]


