SUMMARY
While interferon (IFN) responses are critical for mammalian antiviral defense, induction of antiviral RNA interference (RNAi) is evident. To date, individual functions of the mammalian RNAi and micro RNA (miRNA) effector proteins Argonautes 1–4 (AGO1–AGO4) during virus infection remain undetermined. AGO2 was recently implicated in mammalian antiviral defense, so we examined antiviral activity of AGO1, AGO3, or AGO4 in IFN-competent immune cells. Only AGO4-deficient cells are hyper-susceptible to virus infection. AGO4 antiviral function is both IFN dependent and IFN independent, since AGO4 promotes IFN but also maintains antiviral capacity following prevention of IFN signaling or production. We identified AGO-loaded virus-derived short interfering RNAs (vsiRNAs), a molecular marker of antiviral RNAi, in macrophages infected with influenza or influenza lacking the IFN and RNAi suppressor NS1, which are uniquely diminished without AGO4. Importantly, AGO4-deficient influenza-infected mice have significantly higher burden and viral titers in vivo. Together, our data assign an essential role for AGO4 in mammalian antiviral defense.
In Brief
Functional specificities for the four mammalian RNAi/miRNA effector proteins AGO1–AGO4 remain enigmatic. Adiliaghdam et al. demonstrate a surprising and essential role for AGO4, but not AGO1 or AGO3, in the defense against multiple viral types in immune cells and in vivo.
Graphical Abstract

INTRODUCTION
Almost 20 years after the discovery of RNA interference (RNAi) (Fire et al., 1998) and following the recent approval of an RNAi based therapy by the Food and Drug Administration, decrypting the roles of individual effector RNAi proteins remains enigmatic. This is especially true for the Argonaute (AGO) proteins that are responsible for execution of small-RNA-mediated silencing and are consequently highly conserved throughout the three domains of life (Czech and Hannon, 2011; Meister, 2013). The number of encoded AGOs varies among species, but there is always preservation of at least one catalytically active (slicing-competent) AGO (Czech and Hannon, 2011; Meister, 2013). Mammals possess four AGO proteins (AGO1–AGO4, also known as Eif2c1–Eif2c4) in somatic cells that bind all known short interfering RNAs (siRNAs) or microRNAs (miRNAs) and form the core of the RNA-induced silencing complex (RISC). AGO2 is the sole catalytically active AGO in mice and humans and is essential for small RNA processing (Cheloufi et al., 2010; Yang et al., 2010; O’Carroll et al., 2007) or siRNA-mediated RNAi (Liu et al., 2004; Meister et al., 2004). These activities of AGO2 are fundamental to mammalian development, survival (O’Carroll et al., 2007; Cheloufi et al., 2010), and protection from virus infection (Li et al., 2016; Qiu et al., 2017). The other three AGOs (AGO1, AGO3, and AGO4) are linked within a single ~190-kb locus and despite possessing a catalytic PIWI domain with >90% homology to AGO2 (Meister, 2013), and can load siRNAs (Dueck et al., 2012; Ruda et al., 2014), they are thought to have lost their intrinsic endonuclease activity (Liu et al., 2004; Meister et al., 2004). However, AGO1, 3 and 4 can acquire catalytic activity under certain conditions (Hauptmann et al., 2013, 2014; Schürmann et al., 2013; Bridge et al., 2017; Ruda et al., 2014). Nonetheless, this activity is superfluous for AGO-miRNA-mediated silencing that occurs through mRNA destabilization and inhibition of translation (Eichhorn et al., 2014; Pillai et al., 2005). It is presumed that AGO1, AGO3, and AGO4 have redundant functions by binding identical RNAs (Liu et al., 2004; Wang et al., 2012; Meister et al., 2004; Landthaler et al., 2008), although individual RNA preferences have been demonstrated (Azuma-Mukai et al., 2008; Dueck et al., 2012). Moreover, distinct expression patterns, protein binding partners, and post-translational modifications of mammalian AGO1–AGO4 (Bridge et al., 2017; Li et al., 2014) may dictate different quantitative or qualitative functions. Finally, a unique physiological role for AGO4 has been reported in the context of meiotic sex chromosome inactivation (Modzelewski et al., 2012). Therefore, functional specificities of mammalian AGO1–AGO4 can occur but remain elusive in most biological contexts. Whether AGO1–AGO4 have distinct roles in mammalian host-virus interactions, the ancestral function of RNAi, is not established.
RNAi is the major antiviral defense strategy in plants and invertebrates (Guo et al., 2019). In these organisms, viral RNA is sensed by the RNase III endonuclease Dicer that “dices” viral RNAs into ~22-nt siRNAs that bind AGO proteins for complementary virus silencing. In mammals, however, long double-stranded or single-stranded hairpin viral RNA primarily induces type I interferon (IFN) production via a variety of pattern recognition receptors (PRRs) (Iwasaki and Pillai, 2014) such as retinoic-acid-inducible gene I (RIG-I)-like receptors (RLRs), RIG-I, and melanoma differentiation-associated protein 5 (MDA-5), both of which are helicases closely related to Dicer (Zou et al., 2009). The key RNAi effector proteins Dicer and AGOs are retained in mammals and functional, acting to process host miRNA or RNAi responses driven by exogenously introduced siRNAs or short hairpin RNAs (shRNAs) (Liu et al., 2004; Bernstein et al., 2001). However, the ubiquity and effectiveness of the PRR/RLR-mediated antiviral response have questioned the utility of RNAi for antiviral defense in mammals (Pare and Sullivan, 2014; Cullen et al., 2013). Indeed, Dicer processing of long double-stranded RNAs (dsRNAs) is suboptimal in mammalian somatic cells (Flemr et al., 2013), and mammals lack the RNA-dependent RNA polymerase that amplifies viral-derived siRNAs in some, but not all, organisms that utilize antiviral RNAi (Guo et al., 2019). Furthermore, since viruses are obligate intracellular parasites that rely on the host for replication, many viruses that infect mammalian cells have employed a myriad of counter-defense strategies that suppress RNAi (Fabozzi et al., 2011; Kakumani et al., 2013; Li et al., 2002, 2004, 2013, 2016; Qiu et al., 2017; Wang et al., 2006). Nevertheless, RNAs of viral origin with canonical features of siRNAs have been detected in virus-infected mammalian cells (Qiu et al., 2017; Kennedy et al., 2015; Parameswaran et al., 2010; Schopman et al., 2012; Li et al., 2013, 2016), but their function in host antiviral defense, if any (Backes et al., 2014; Tsai et al., 2018), or their dependence on the single mammalian Dicer for their production remained unclear (Otsuka et al., 2007; Matskevich and Moelling, 2007; Shapiro et al., 2014; Bogerd et al., 2014). Dicer-dependent virus-derived small RNAs were detected in embryonic stem cells (ESCs) but reduced upon cell differentiation (Maillard et al., 2013), and masking of antiviral RNAi by IFN is evident (Maillard et al., 2016; van der Veen et al., 2018), leading to the hypothesis that antiviral RNAi may be confined to undifferentiated cells with attenuated IFN (Pare and Sullivan, 2014). However, stem cells, including mouse ESCs, robustly express intrinsic IFN-stimulated genes (ISGs) that autonomously protect from virus infection (Wu et al., 2018). Moreover, exogenously introduced siRNAs or viruses engineered to self-target by RNAi can suppress virus in IFN-sufficient cells (Benitez et al., 2015); AGO-loaded, Dicer-dependent virus-derived siRNAs (vsiRNAs) are detected in human IFN-sufficient somatic cells (Kennedy et al., 2015; Li et al., 2016; Qiu et al., 2017); and AGO2 catalytic activity is antiviral in the presence of IFN (Li et al., 2016; Qiu et al., 2017). Thus, while IFN restrains antiviral RNAi, and conversely, RNAi components regulate IFN (Kok et al., 2011; Seo et al., 2013), how IFN and RNAi (and other antiviral systems) cooperate, complement, or compensate for each other (Nish and Medzhitov, 2011) in various cell types infected by constantly evolving viruses is unclear. Pertinent to this is that few studies have examined RNAi components in vivo or in physiologically relevant immune cells that respond to virus infection. This is first and foremost because depletion of most RNAi components, including Dicer, Drosha, or AGO2, results in embryonic lethality (Bernstein et al., 2003; Cheloufi et al., 2010; Liu et al., 2004; Chong et al., 2010), leading most studies to examine antiviral RNAi in immortalized cell lines with minimal relevance to virus infection. Second, depletion of RNAi components also disrupts miRNA populations, which can repress host antiviral pathways or promote or limit virus directly (Luna et al., 2015; Scheel et al., 2016), making interpretation of antiviral phenotypes difficult. However, single AGO1-, AGO3-, and AGO4-deficient mice are viable (O’Carroll et al., 2007; Van Stry et al., 2012), and their loss does not disrupt miRNA populations (O’Carroll et al., 2007). Given this, we sought to examine if AGO1, AGO3, or AGO4 performs an antiviral task in mammalian immune cells. Herein, we report that the RNAi/miRNA machinery component AGO4 has a unique antiviral role over AGO1 or AGO3 in mammalian innate immune cells and in vivo. This fundamental requirement of AGO4 for the optimal resistance from virus infection is not only IFN and RLR signaling independent but also ostensibly IFN promoting. Consistently, these functions rendered AGO4 ineffective at controlling influenza virus lacking its key virulence factor, nonstructural protein 1 (NS1), that acts to repress both cognate siRNAs for antiviral RNAi (Li et al., 2016; Qiu et al., 2017) and the IFN response (Ayllon and García-Sastre, 2015).
RESULTS
Unique Antiviral Role for AGO4 in Mammalian Cells
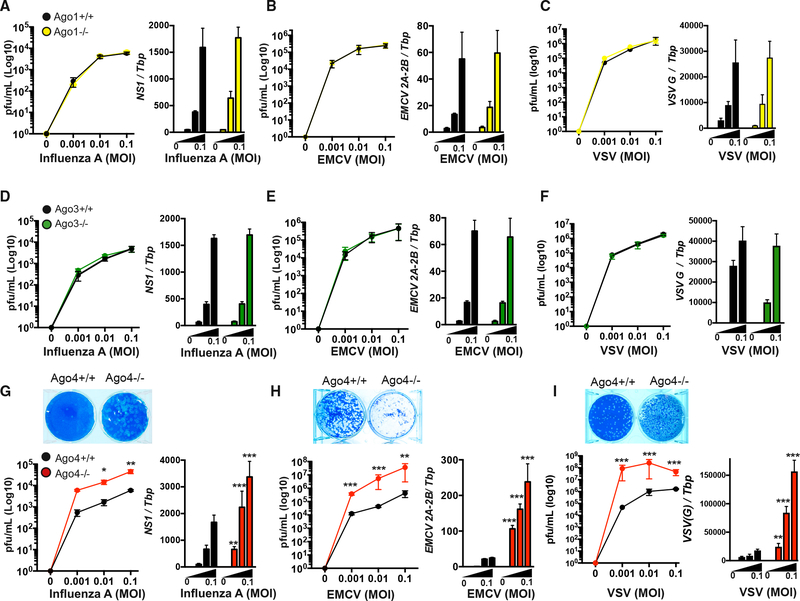
Quantitative differences in AGO function have been attributed to differential expression (Wang et al., 2012). However, there is a gap in knowledge regarding AGO expression, particularly in immune cells that respond to virus infection. We examined Ago1–Ago4 transcript expression and found Ago4, reported to be absent in many somatic cell lines and high in the male germline (Petri et al., 2011; Modzelewski et al., 2012), was the most abundant AGO in IFN-producing innate immune cells such as macrophages, monocytes, dendritic cells, and granulocytes (Figure S1). Conversely, Ago1 and Ago2 were low in these cells. Interestingly, Ago1, Ago2, and Ago3 were highest in adaptive immune cells (Figure S1). We bred AGO1, AGO3, or AGO4 heterozygote parents and isolated bone marrow from AGO1, AGO3, or AGO4 homozygous null progeny or their littermate wild-type (WT) controls. Single AGO deficiency and a lack of compensatory expression of other AGOs were confirmed (Figure S1). Furthermore, AGO1-, AGO3-, or AGO4-deficient bone marrow was matured to macrophages and no evidence of compromised maturation was observed (Figure S1). To determine whether macrophages deficient in AGO1, AGO3, or AGO4 could mount a normal antiviral response to influenza, we infected homozygous null (Ago1−/−, Ago3−/−, or Ago4−/−) macrophages with influenza (H1N1, A/Puerto Rico/8/34) and quantified virus levels. Single AGO1 deficiency yielded no differences in viral levels following influenza infection (Figure 1A). Furthermore, AGO1-deficient macrophages yielded no difference virus levels when infected with the positive-strand RNA virus encephalomyocarditis (EMCV; Figure 1B) or another negative-strand RNA virus, vesicular stomatitis virus (VSV; Figure 1C). Similarly, single AGO3-deficient macrophages yielded no differences in viral levels following infection with influenza, EMCV, or VSV (Figures 1D–1F). Hence, individually, AGO1 and AGO3 do not contribute to the containment of virus levels. Our data are in agreement with previous studies examining Influenza-infected AGO1/AGO3 double-knockout mice that displayed similar viral titers to wild-type counterparts, despite increased mortality following infection (Van Stry et al., 2012). In contrast, however, AGO4-deficient macrophages displayed marked hyper-susceptibility to influenza infection with significantly elevated viral titers and viral RNA levels following infection (Figure 1G). Furthermore, there was a significant increase in EMCV and VSV levels in AGO4-deficient macrophages (Figures 1H and 1I). We also confirmed an antiviral role for AGO4 in bone-marrow-derived dendritic cells, as well as mouse embryonic fibroblasts (Figure S2). Hence, AGO4 has a unique antiviral role against a range of RNA viruses
Figure 1. Argonaute 4 (AGO4) Is a Unique and Essential Antiviral Mediator.
Viral titers and viral RNA of (A) influenza-A-infected (PR/8/1934), (B) encephalomyocarditis (EMCV)-infected, or (C) vesicular stomatitis virus (VSV)-infected (indicated multiplicities of infection [MOIs] for 16 h) bone-marrow-derived macrophages (BMDMs) from Ago1+/+ (black) and Ago1−/− (yellow), (D–F) Ago3+/+ (black) and Ago3−/− (green), or (G–I) Ago4+/+ (black) and Ago4−/− (red) littermate mice. Representative plaque assay images are shown. Influenza Ns1, EMCV 2A-2B, or VSV G RNA levels were quantified by qPCR relative to TATA box protein (Tbp). All data are from two or three biological replicates performed in triplicate and combined. Errors bars represent SEM. *p < 0.05, **p < 0.01, and ***p < 0.001, as measured by two-way ANOVA with Bonferroni multiple comparison test.
IFN-Dependent and IFN-Independent Antiviral Role for AGO4
The characteristic antiviral response in mammals is type I IFN mediated, and macrophages are the major source of type I IFN following RNA virus infection in vivo (Kumagai et al., 2007). Furthermore, there is much evidence for IFN and RNAi bi-directional crosstalk (Maillard et al., 2016; van der Veen et al., 2018; Kok et al., 2011; Seo et al., 2013). We thus determined if AGO1, AGO3, or AGO4 deficiency altered IFN induction in macrophages. AGO1- or AGO3-deficient macrophages had IFN induction equivalent to their WT counterparts. Conversely, AGO4 knockout cells displayed a significant reduction in IFN-β following infection with a range of RNA viruses or stimulation with viral ligands (Figures 2A and 2B). Thus, AGO4 promotes IFN following activation of antiviral pathways. To determine if AGO4 knockout macrophages were hyper-susceptible to virus infection because of compromised IFN, we infected AGO4 knockout macrophages in the presence of an IFN receptor blocking antibody (αIFNAR) that significantly decreased the ISG Isg15 (Figure 2C). As expected, αIFNAR increased viral titers, but AGO4 deficiency further increased viral titers in the absence of IFN signaling (Figures 2D and 2E), demonstrating that AGO4 can elicit antiviral defense independently of, and in addition to, IFN. Additionally, we created AGO4/mitochondrial antiviral-signaling protein (MAVS) double-knockout mice to assess AGO4 with impaired RLR signaling and IFN production (Figure 2F). AGO4 maintained antiviral activity in the absence of MAVS, as significantly elevated virus levels and virus-induced cytopathy were observed in AGO4/MAVS double-deficient macrophages compared to WT, single AGO4-deficient, or single MAVS-deficient macrophages (Figures 2G–2J). Interestingly, AGO4 and MAVS displayed a similar individual contribution to antiviral defense in macrophages (Figures 2G–2J). However, AGO4/MAVS double-knockout macrophages displayed significantly more virus levels compared to either single MAVS-deficient or single AGO4-deficient cells (Figures 2G–2J), demonstrating an independent and additive contribution of AGO4 and MAVS to mammalian antiviral defense.
Figure 2. Antiviral Activity of AGO4 Is Interferon (IFN) Dependent and Independent.
(A) Ifnb1 in VSV-infected BMDMs as measured by qPCR.
(B) IFNβ ELISA of poly(I:C)-treated or virus-infected BMDMs.
(C) VSV-induced Isg15 levels in the presence or absence of 10 μg/mL anti-IFNAR (MAR-5A3), as measured by qPCR.
(D and E) VSV levels at indicated MOIs for 16 h in the presence or absence of 10 μg/mL anti-IFNAR (MAR-5A3) antibody as quantified by plaque assays (D) or qPCR (E) of VSV-G RNA.
(F) Ifnb1 induction in influenza-infected BMDMs as measured by qPCR.
(G) Representative flow cytometry plots of influenza-GFP-infected BMDMs (MOI 1, 24 h post-infection [p.i.]). GFP percentages of Annexin V− (non-apoptotic) or Annexin V+ (apoptotic) cells are indicated.
(H) Percentage of influenza-GFP+ BMDMs at indicated MOIs for 24 h.
(I and J) Influenza Ns1 RNA levels as determined by qPCR (I) or influenza titers (J) as determined by plaque assay.
Data are from three combined biological replicates performed in triplicate. Errors bars represent SEM. *p < 0.05, **p < 0.01, and ***p < 0.001, as measured by an unpaired t test or one-way or two-way ANOVA with Bonferroni multiple comparison test. N.S., not significant.
Overexpression of Ago4 Suppresses Virus Levels
To determine if increasing AGO4 abundance could enhance the antiviral capacity of mammalian cells, we ectopically expressed increasing doses of AGO4 in HEK293T cells and infected with influenza. Indeed, overexpression of AGO4, but not AGO1 or AGO3, suppressed influenza in a dose-dependent manner (Figures 3A–3E). Moreover, given that overexpression of AGO1 or AGO3 could not enhance virus suppression despite expression levels equal to AGO4 (Figure 3D), it is unlikely that quantitative differences alone dictate the unique antiviral function of AGO4. In vitro analyses have not identified slicing capacity of AGO4 toward miRNA or siRNA substrates (Meister et al., 2004; Schürmann et al., 2013). However, AGO4 can be converted to a catalytically activate AGO (Hauptmann et al., 2014). We wondered if AGO4 could become catalytically active in the presence of virus and if AGO4 antiviral capacity was due to this activity, so we mutated each predicted AGO4 amino acid that comprises the catalytic triad (D587, G671, and R809) to alanine. Expression of AGO4D587A, AGO4G671A, or AGO4R809A repressed influenza to the same degree as AGO4, suggesting that these individual sites do not contribute to the antiviral function of AGO4 (Figure 3E). Furthermore, we transfected siRNAs complementary to the influenza genome into cells deficient in each AGO and found that siRNAs only failed to suppress influenza in the absence of the known slicing AGO2,or in AGO2 catalytic-dead (Ago2D597A) cells but could successfully suppress virus titers in the absence of AGO1, AGO3, or AGO4 (Figure 3F). Thus, AGO4 is unlikely antiviral through siRNA loading to RISC or siRNA-mediated slicing mechanisms.
Figure 3. Ago4 Overexpression Reduces Viral Load.
(A) Flow cytometry of HEK293T transfected with empty vector (EV) or indicated concentrations of Ago4-Myc for 24 h prior to infection with influenza-GFP at MOI 1 for 24 h. GFP percentages of 7-AAD− (non-apoptotic) cells are indicated. Right: mean and SEM of percentage of Flu-GFP relative to EV control.
(B and C) Ago4 and influenza Ns1 RNA expression in HEK293T transfected with EV or 10 μg of Ago4-Myc and infected with Influenza A (MOI 1, 24 h p.i.).
(D) Myc immunoblot of Ago1, Ago3, or Ago4 transfected HEK293T cells.
(E) Percentage of Flu-GFP+ cells in HEK293T cells transfected with 10 μg of the indicated plasmid and infected at MOI 1 for 24 h. 7-AAD−, relative to EV.
(F) Influenza viral titers of mouse embryonic fibroblasts (MEFs) transfected with a negative control (NC) siRNA or vsiRNA against NS1.
Data are from three biological replicates performed in triplicate and combined. Errors bars represent SEM. *p < 0.05, **p < 0.01, and ***p < 0.001, as measured by an unpaired t test or one-way ANOVA with Bonferroni multiple comparison test.
Identification of AGO-Loaded Influenza-Derived RNAs in Mature Macrophages
To understand if the IFN-independent antiviral role of AGO4 is related to its function in executing RNAi, we first determined if canonical viral siRNAs are present in virus-infected macrophages. The RNAi process is efficient in that a few RNA molecules can trigger inactivation of a continuously transcribed mRNA target for prolonged periods of time (Nishikura, 2001). Thus, vsiRNAs are optimally identified by AGO immunoprecipitation (IP) (Li et al., 2016; Qiu et al., 2017; Tsai et al., 2018). To date, AGO-loaded vsiRNAs have not been detected in primary immune cells, only mouse or human cell lines or suckling mice. To this end, we infected bone-marrow-derived macrophages with influenza A or with influenza A lacking the IFN and RNAi suppressor protein NS1 (influenza delNS1) and used a pan-AGO antibody (recognizing all four AGOs) to immunoprecipitate AGO-bound RNAs after UV crosslinking. UV crosslinking of protein-RNA allows stringent washing and minimizes the recovery of non-specific RNA through reassociation of RNA-binding proteins to RNA after cell lysis (Mili and Steitz, 2004). Sequencing of RNA revealed influenza-derived RNAs stringently bound to AGO in influenza-infected macrophages. AGO-loaded influenza derived RNAs sized 18–40 nucleotides (Figure 4A) were observed which are likely a combination of siRNAs, siRNA precursors and/or complementary target influenza RNAs. However, the canonical 21–23 nucleotide length vsiRNAs were a more prominent proportion of the AGO-loaded RNAs following infection with influenza lacking NS1 (Figure 4A; Tables S1 and S2).
Figure 4. Reduction in AGO-Loaded Influenza-Derived RNAs in AGO4-Deficient Macrophages.
(A) Size distribution and percentage of AGO-bound reads mapped to influenza following infection of BMDMs with influenza or influenza del NS1 (MOI 1, 24h.p.i).
(B) Immunoblot of pan-AGO or AGO2 of cell lysates or (C) pan-AGO of immunoprecipitates.
(D) Amount of AGO-loaded influenza RNA in influenza-infected BMDMs, relative to WT controls.
(E) Quantity of AGO-loaded influenza-derived RNA at indicated nucleotide lengths.
(F) Origin of AGO-bound RNA that mapped to the influenza A genome. Genomic segments presented from the 3′ end (left) to the 5′ end (right), and negative (−) or positive (+) strand and scale are indicated. Boxes highlight bona fide vsiRNAs (Li et al., 2016; Qiu et al., 2017).
(G) Percentage and size distribution of AGO co-immunoprecipitated influenza RNA in BMDMs infected with influenza delNS1 (MOI 1, 24 h p.i.).
(H) Relative influenza HA RNA as quantified by qPCR in MEFs infected with influenza or influenza delNS1 in the presence or absence of 10 μg/mL anti-IFNAR.
Data are the mean of two or three biological replicates. Error bars represent SEM. *p < 0.05, **p < 0.01, and ***p < 0.001, as determined by an unpaired t test or one-way ANOVA with a Bonferroni multiple comparisons test.
Reduced AGO-Loaded Influenza RNAs in AGO4-Deficient Macrophages
To determine the contribution of AGO4 to the abundance of AGO-loaded virus-derived RNAs, we used the same pan-AGO antibody to standardize the efficiency of IP and performed IP in AGO1-, AGO3-, or AGO4-deficient macrophages (Figures 4B and 4C). Despite increased virus titers in AGO4-deficient macrophages, we found a significant reduction in AGO-loaded influenza-derived RNA in the absence of AGO4, but not AGO1 or AGO3, (Figure 4D; Tables S1 and S2). Interestingly, the loss of AGO-bound influenza RNAs with AGO4 deficiency was of all lengths (Figure 4E), suggesting no preferential loss of canonical siRNAs over precursor or complementary target RNAs. We aligned the AGO-loaded influenza RNA to the eight segments of the influenza genome and found that AGO-loaded RNA was derived from both positive and negative strands and prominently from the neuraminidase (NA) genomic strand (Figure 4F). In AGO4-deficient macrophages, we found that the majority of AGO-loaded influenza-derived RNAs were lost at the terminal 5′ or 3′ ends if the influenza genome (Figure 4F), previously described as hotspots of vsiRNAs (Li et al., 2016; Maillard et al., 2013). Moreover, the proportion of AGO-loaded influenza-derived canonical siRNAs size (21–23 nt), seen predominantly after infection with influenza delNS1, was markedly reduced in AGO4-deficient macrophages (Figure 4G; Table S2). Importantly, the amount of pan AGO, as well as slicing-capable AGO2, was equal in WT and AGO4 knockout macrophages (Figure 4B), as was the amount of pan-AGO protein immunoprecipitated (Figure 4C). Thus, AGO4 appears to be essential for the generation of vsiRNAs or stability of the AGO-influenza RNA complex.
Genetic Rescue of Influenza delNS1 in AGO4-Deficient Cells
Demonstration of antiviral RNAi entails genetic rescue of viral suppressor of RNAi (VSR)-deficient viruses in RNAi-compromised host cells, so we next examined virus levels in AGO4-deficient cells infected with influenza lacking NS1. Indeed, infection of AGO4-deficient cells with influenza delNS1 resulted in significantly enhanced virus levels (Figure 4H). This enhancement was significantly more than observed for influenza with intact NS1 (Figure 4H), demonstrating genetic rescue of influenza delNS1 in AGO4-deficient cells. Importantly, NS1 suppresses both IFN and RNAi (Ayllon and García-Sastre, 2015; Li et al., 2016; Qiu et al., 2017), yet AGO4 deficiency was capable of enhancing influenza delNS1 even when IFNAR was blocked (Figure 4H), attributing the genetic rescue to IFN-independent mechanisms.
AGO4 Is an Antiviral Factor In Vivo
We next wanted to establish if AGO4 is capable of autonomously defending against virus infection in vivo. We infected AGO4 knockout mice or littermate controls intranasally with influenza A virus and found that AGO4 knockout mice exhibited significantly more burden of infection. AGO4 knockout mice lost significantly more weight than WT littermate controls (Figure 5A). In addition, increased leukocyte infiltration and lung damage in AGO4 knockout mice was apparent (Figure 5B), and flow cytometry revealed a significant elevation in inflammatory CD11bhi monocytes infiltrating the lung of AGO4 knockout mice (Figure 5C). To determine if increased burden of infection was due to elevated virus titers, we infected AGO4 knockout mice with influenza in which NS1 is tagged with green fluorescent protein (influenza-GFP) (Manicassamy et al., 2010) and found increased GFP in the lungs of AGO4-deficient mice compared to littermate controls (Figure 5D). We also isolated bronchoalveolar lavage fluid (BALF) and found significantly elevated influenza titers in AGO4 knockout mice (Figure 5E). Moreover, we detected elevated influenza NP protein by immunoblot (Figure 5F) and influenza RNA in the lung (Figures 5G and 5H). Thus, AGO4 is an essential mammalian antiviral factor in vivo, controlling both viral titers and virus-induced pathology.
Figure 5. AGO4 Is an Essential Antiviral Mediator In Vivo.
(A) Weight loss over time following intranasal infection with influenza A (PR/8/1934, 105 plaque-forming units [pfu]/mouse, n = 10 per group).
(B) Hematoxylin and eosin staining of lungs 4 days p.i. Scale bar represents 0.2 μm.
(C) Proportion of monocyte and dendritic cell populations infiltrating lungs 4 days p.i., as assessed by flow cytometry.
(D) Confocal microscopy of influenza-GFP (green) or wheat germ agglutinin (blue) of lungs 72 h p.i. (105 pfu/mouse). Scale bar represents 350 μm.
(E–H) Viral titers of bronchoalveolar lavage fluid (BALF) measured by plaque assay (E), immunoblot of influenza NP protein (F), or influenza PA (G) or NP (H) RNA expression in lungs measured by qPCR 4 days p.i.
*p < 0.05, **p < 0.01, and ***p < 0.001, as determined by an unpaired t test.
DISCUSSION
All small RNA types, regardless of their origin, mediate their regulatory functions through RISC-containing AGO proteins, which are remarkably conserved proteins in all domains of life (Czech and Hannon, 2011). Individual functions of the four mammalian AGOs in somatic cells, particularly AGO1, AGO3, and AGO4, in any biological context, is largely unresolved. Here, we have identified that the small RNA machinery protein AGO4 has a unique function in the context of virus infection of mammalian immune cells and in vivo. This function was divergent from other AGOs (AGO1 and AGO3) that are considered to be non-catalytic and was independent of, and complementary to, IFN- or RLR-dependent pathways.
Several observations support our conclusion. First, AGO4 uniquely restricted influenza, VSV, and EMCV in macrophages, dendritic cells, and mouse embryonic fibroblasts. Second, we found that AGO4 deficiency disrupted IFN production, but antiviral activity was maintained following blockade of IFN signaling or in the absence of the RLR signaling adaptor protein MAVS, demonstrating this role is independent and additive to the RLR/IFN-based antiviral arsenal. Future experiments will understand how AGO4 may promote the IFN response. Third, overexpression of AGO4, but not AGO1 or AGO3, suppressed virus levels. Fourth, we revealed the presence of AGO-loaded influenza-derived RNAs, the hallmark of antiviral RNAi, in primary macrophages, which were reduced in the absence of AGO4, but not AGO1 or AGO3. We also demonstrated a genetic rescue of influenza lacking the RNAi and IFN suppressor protein NS1 in AGO4-deficient cells when IFNAR was blocked, suggesting that the role of AGO4 in antiviral defense is RNAi dependent. There is active siRNA-mediated silencing in the absence of AGO4, indicating the function of AGO4 is not at the level of siRNA loading or target slicing, at least in the context of experimental RNAi. Moreover, these results established that AGO2 is functional in the absence of AGO4. However, to prove beyond doubt that the IFN-independent antiviral function of AGO4 we have identified is RNAi mediated, future experiments should include an analysis of AGO4 function in Dicer-deficient cells. Furthermore, a demonstration that the AGO-bound influenza-derived RNAs reduced in AGO4 knockouts are functionally capable of silencing influenza is needed. Fifth, and most importantly, we demonstrated an indispensable antiviral role for AGO4 against influenza in vivo. There was a greater burden of infection, and AGO4 deficiency resulted in elevated virus titers, which is in contrast to AGO1/AGO3-deficient mice (Van Stry et al., 2012).
It is reasonable to assume that the same RNAi pathway is activated in response to exogenous dsRNA and viruses. However, it is appealing to speculate that the unique function of AGO4 we have identified is exclusive to virus-infected cells. In C. elegans, the Dicer-related helicase DRH-1 is essential for silencing a replicating viral replicon, but not a homologous cellular mRNA targeted by the same pool of siRNAs (Guo et al., 2013). Similarly, in D. melanogaster, a loquacious-independent siRNA pathway is dedicated to the antiviral response, while Dcr-2, AGO2, and r2d2 are required for silencing triggered by both exogenous dsRNA and viruses (Marques et al., 2013). Furthermore, since there is a precedence for the RNAi machinery to silence virus via translational repression, AGO4 may also act in this manner. Ago1–Ago27 mutant A. thaliana have defective translational repression and normal slicing yet elevated virus levels (Guo et al., 2019). Likewise, prevention of the VSR protein during flock house virus (FHV) replication in mammalian cells induced translational inhibition of viral RNAs independent of protein kinase R or other translational repression mechanisms (Petrillo et al., 2013). Moreover, the unique IFN-independent antiviral role of AGO4 could involve miRNAs. AGO4-deficient cells displayed a small loss of AGO-loaded miRNAs, and certain RNA viruses use cellular miRNAs as a resource to promote their replication (Scheel et al., 2016) that AGO4 may uniquely control.
Indisputably, the primary mode of mammalian antiviral defense culminates in IFN production and many aspects of the IFN antiviral system act to suppress RNAi mechanisms (Maillard et al., 2016; van der Veen et al., 2018). This inhibition has been thought to primarily preserve or promote the IFN antiviral response. However, while there is no obvious evolutionary reason for mammals to eliminate a functional host defense system in the host-pathogen arms race, it is appealing to speculate that since RNAi machinery is fundamental to maintenance of host mRNA transcripts, including induced antiviral transcripts, using RNAi components primarily for antiviral defense may pose a greater fitness cost to the host than employing IFN-based strategies. Critical to our understanding of the intersection of these antiviral pathways for optimal defense, future studies that functionally characterize vsiRNAs and RNAi pathway members must be examined under conditions that do not compromise the essential functions of cellular miRNAs and in physiologically relevant immune cells or epithelial cells with intact IFN responses.
In summary, we ascribe a previously unidentified and unique IFN-promoting and IFN-independent function for AGO4 in the restriction of influenza and other pathogenic RNA viruses in mature immune cells and in vivo. Taken together, our data add to the emerging evidence that RNAi-related mechanisms are a functional mode of antiviral defense in mammals and assign a specific and unique role for AGO4 in that context. Our work also reveals that individual AGOs can have unique functions in mammalian cells, at least during host-virus interactions. Specific promotion of AGO4 function in mammalian cells may be an effective antiviral strategy.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kate L. Jeffrey (KJeffrey@mgh.harvard.edu).
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
AGO1, AGO3, and AGO4 conventionally deleted mice have been previously described (O’Carroll et al., 2007) and were obtained from A. Tarakhovsky (The Rockefeller University). AGO mice have been backcrossed to C57BL/6 at least 10 times and all experiments were performed with littermate controls from heterozygote breedings. MAVS knockout mice were previously described (Sun et al., 2006) and obtained from The Jackson Laboratory. Mice were housed under specific pathogen-free conditions and experimental protocols were performed according to Massachusetts General Hospital and Harvard Medical School Animal Care and Use (IACUC).
Cells and reagents
Bone marrow-derived macrophages (BMDMs) were produced from AGO deficient mice as described (Mehta et al., 2017) Briefly, bone marrow was flushed from tibia and fibia and allowed to adhere to a non-treated tissue culture plate for 1 day. Non-adherent cells were then differentiated to macrophages in DMEM containing 10% FBS, 1% L-Glutamine, 1% penicillin-streptomycin, 0.1% β-mercaptoethanol, 5 ng/mL of interleukin (IL)-3 (Peprotech) and Macrophage Colony Stimulating Factor M-CSF (Peprotech) for 7 days. Macrophage maturity was assessed by surface expression of CD11b and F4/80 with Flow Cytometry. Deletion for AGO1,3,4 and lack of transcriptional upregulation of other Agos was confirmed with qPCR. AGO1 protein deletion was confirmed with available antibodies (anti-AGO1 from Abcam). For the generation of dendritic cells, bone marrow was seeded in non-treated 10-cm dishes, cultured in RPMI plus 10% fetal bovine serum, 2% penicillin/streptomycin, and 1% of 200 mM glutamine containing 20 ng/ml GM-CSF and 20 ng/ml IL-4, and incubated at 37°C/5% CO2. Medium was replenished on days 3, 5, and 7. At day 8, non-adherent cells were collected and adherent cells were discarded. These cells were immature DCs, and purity was confirmed by FACS analysis using anti-CD11c mAb (BD PharMingen). AGO1, 3 and 4 deficient mouse embryonic fibroblasts (MEFs) were kindly provided by A. Tarakhovsky and cultured in DMEM with 15% FBS, L-Glutamine, and penicillin-streptomycin, β-mercaptoethanol. Polyinosinic:polycytidylic acid (PolyI:C, used at 10 μg/mL) was purchased from Sigma. Monoclonal antibody to Interferon A Receptor (anti-IFNAR, MAR103, used at 10 μg/mL) was purchased from eBiosciences.
Viruses
Influenza A virus (A/Puerto Rico/8/34), A/PR/8/1934 delNS1, A/PR/8/1934 NS1-GFP, and anti-NP antibody were kindly provided by A. García-Sastre (Mount Sinai School of Medicine). Encephalomyocarditis (EMCV) was purchased from ATCC. Vesicular Stomatitis Virus (VSV, Indiana strain) and VSV-GFP were kindly provided by C. Rice and M. MacDonald (The Rockefeller University).
METHOD DETAILS
In vitro virus infection
Bone marrow-derived macrophages were infected in 12 well plates with 100 μL of virus for the indicated multiplicity of infection (MOI) diluted in phosphate buffered saline (PBS) supplemented with 1% FBS, calcium and magnesium for 1 h at 37°C, rocking every 15 minutes. Virus was removed, cells were washed once with PBS and replaced with macrophage media This was considered time 0. To determine influenza titers in macrophage supernatants, plaque assays were performed on canine kidney (MDCK) cells using 0.6% BSA, 2% Oxoid Agarose, 1 mg/mL Trypsin-TPCK, 0.6% NaHCO3, 2% sodium pyruvate, 1% penicillin//streptomycin, 50 mM HEPES, 4 mM L-glutamine in DMEM for plaque formation and plaques counted 2–3 days post infection (dpi). For quantification of EMCV titers, plaque assays were performed on Vero cells with 1.5% LE agarose and plaques counted at 24 hpi. To determine VSV titers, plaque assays were performed on Baby Hamster Kidney (BHK-21) cells using 1.5% LE agarose with DMEM for 16 hours post infection (hpi). All cells were fixed with 7% formaldehyde followed by crystal violet staining.
In vivo influenza infection
Age and sex-matched (8–12 weeks, female) littermate controls were anesthetized by i.p. injection of Tribromoethanol (Avertin) and challenged with 30 μL (15 μL per nostril) of influenza A/PR/8/1934 diluted in PBS (105 pfu) intranasally. Lungs were removed and snap frozen in liquid N2 and RNA extracted (QIAGEN) 4 days post-infection. Bronchoalveolar lavage fluid (BALF) was recovered by cannulation with 1 mL of PBS after terminal exsanguination. Body weight was measured twice daily. Mice showing more than 25% of body weight loss were considered to have reached the experimental end point and were humanely killed.
Flow cytometry
Single-cell suspensions of mouse lung were prepared using collagenase/DNase treatment. Cells were incubated in Fc block for 20 minutes at 4°C and then stained with the following fluorescent-conjugated antibodies: APC-conjugated anti-CD11c (HL3; 550261), FITC-conjugated anti-F4/80 (6F12; 552958), PE-conjugated anti-CD11b (M1/70; 553311). All antibodies were obtained from BD PharMingen. Cells were analyzed on an LSRII flow cytometer (BD Bioscience) and analyzed by FlowJo (Tree Star).
Identification virus-derived RNA bound to AGO in Influenza A-infected macrophages
AGO IP was carried out as previously described using a pan AGO antibody (2A8, Millipore) that recognizes all four endogenous AGOs (Li et al., 2016; Qiu et al., 2017). In summary, 2–3 biological replicates each of Influenza A or Influenza delNS1-infected (MOI 1, 24 hours) bone marrow-derived macrophages from WT, Ago1, Ago3 or Ago4 knockout mice were UVB-irradiated three times with 200 mJ/cm2 in UV Stratalinker 2400 from Stratagene on ice to form covalent bonds between protein and RNA. 10 million cells were lysed in 1 mL of PXL buffer (PBS, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40) with protease inhibitors (Roche) and DNA digested with DNase. Protein A Dynabeads (Invitrogen) were washed with PBS/0.02%Tween and incubated with a bridging antibody (Rabbit anti-mouse IgG, Jackson Immunoresearch) to increase avidity for 45 minutes at room temperature, washed and then incubated with AGO antibody for 1.5 hours at room temperature. AGO bound RNA was immunoprecipitated overnight at 4 degrees and washed with high stringency buffers and RNA extracted from beads with Trizol. Libraries of small RNAs were constructed by a method depending on the 5′monophosphate of small RNAs, with a NEBNext Multiplex Small RNA Library Prep Set (Illumina) and sequenced on an Illumina HiSeq System.
Processing of AGO IP RNA sequencing data
TrimGalore (v0.4.2) was used to remove Illumina sequencing adaptors, using a quality score cutoff of 20, stringency set to 3 and minimum read length of 18 (https://github.com/FelixKrueger/TrimGalore). Reads were first aligned to mouse genome build UCSC mm10 using Tophat2 (v2.1.1 with Bowtie2 v2.2.9) (Kim et al., 2013) with a maximum of two mismatches. Reads that were unmapped to mm10 were then aligned to the H1N1 influenza (PR/8/1934) genome using Tophat2, with maximum multihit of one. Read length distributions were obtained using custom python scripts that extracted read length counts from the aligned reads to mouse or influenza genomes, these scripts are provided as Figure S3.
Viral siRNA transfections
Mouse embryonic fibroblasts were plated 12–16h prior to transfection to achieve 70%–80% confluency. vsiRNAs (Integrated DNA Technologies) were transfected to a final concentration of 100 nM in a total volume of 500 μL antibiotic-free media using Lipofectamine 2000 (Life Technologies) as per the manufacturer’s protocol. Cells were then incubated at 37°C for 6h followed by a 1h adsorption with Influenza A-GFP, MOI 1 (virus was adsorbed in a manner identical to that described previously for macrophages). Following infection, cells were once again incubated at 37°C for 6h and then transfected again. Viral supernatants were collected at 24h and 48h and cells were lysed for RNA extraction using RLT buffer (QIAGEN) at 48h from the point of infection.
RNA extraction and quatitative reverse transcription-PCR (RT-PCR)
RNA was isolated with the RNeasy mini kit (QIAGEN). RNA was reverse transcribed using Reverse Transcription Supermix (Biorad). qPCR was performed using SYBR green (BioRad) and PCR products were quantified with a standard curve. Gene expression was displayed relative to the housekeeping gene, TATA box protein, Tbp.
Western Blotting
2 million macrophages were lysed in 200 μL of RIPA buffer (Tris-HCl, pH 7.4, 50 mM, NaCl 150mM, NP-40 1%, sodium deoxycholate 0.5%, SDS 0.1%) with a cocktail of protease and phosphatase inhibitors (Roche). 20 μg of protein was loaded per lane and separated on a 4%–12% SDS/PAGE gel and transferred to PVDF. Ponceau stain (Sigma) determined protein transfer and membrane was blocked for 1 h with 5% skim milk in TBS/0.1% Tween at 25°C, and then incubated with the indicated primary antibody in 3% BSA TBS/0.1% Tween overnight at 4°C. Specific antibodies Influenza NP protein (A. Sastre-Garcia) were used to quantify virus protein in the lung tissue. β-actin (Sigma) was used as a loading control. Anti-pan-AGO for immunoblotting (7G1.1) was from the Darnell laboratory (Rockefeller University), anti-AGO1 antibody was from Abcam.
Enzyme-linked immunosorbent assay
IFNβ levels in cell supernatants was determined with a mouse IFNβ ELISA (PBL Interferon Source).
Plasmids
FLAG/HA-tagged Ago1–4 were obtained from Addgene (#10820, 10822, 10823, 10824) and the the FLAG/HA was replaced with a Myc tag. Mutations were introduced into Ago4 by site-directed mutagenesis using the QuickChange Kit (Stratagene).
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism was used for all statistical analysis. Total sample size was determined based on the previous studies with similar genetically modified mice with comparable functional defects. All experiments were repeated at least three times. Statistical analysis was carried out by two-way or one-way analysis of variance (ANOVA), or by an unpaired t test of biological replicates. P value < 0.05 was considered statistically significant.
DATA AND CODE AVAILABILITY
Flow Cytometry data were analyzed using FlowJo (Treestar, USA) software. Graphics and statistics were generated with GraphPad PRISM software 7.0 (GraphPad Software, USA).
The RNA sequencing data reported in this paper are archived at the Gene Expression Omnibus (GEO): GSE128228 (https://www.ncbi.nlm.nih.gov/geo).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-conjugated anti-CD11c | BD PharMingen | Cat# HL3;550261; RRID:AB_398460 |
| FITC-conjugated anti-F4/80 | BD PharMingen | Cat# 6F12;552958; RRID:AB_394533 |
| PE-conjugated anti-CD11b | BD PharMingen | Cat# M1/70;553311; RRID:AB_394775 |
| Rabbit anti-mouse IgG antibody | Abcam | Ab190475; RRID:AB_2827162 |
| Anti-pan-AGO (7G1.1) used for immunoblotting | R. Darnell (Rockefeller University) (Chi et al., 2009) |
DSHB 7G1.1; RRID:AB_2827163 |
| Anti-pan Ago Antibody, clone 2A8 | Millipore | Cat# MABE56; RRID:AB_11214388 |
| Bridging antibody (Rabbit anti-mouse IgG) | Jackson Immunoresearch | Cat# 315001008; RRID:AB_2340032 |
| Anti-Ago1 antibody | Abcam | Ab5070; RRID:AB_2277644 |
| Anti-Ago2 antibody | A. Tarakhovsky | N/A |
| Influenza anti-NP antibody | A. Sastre-Garcia | N/A |
| Anti β-actin | Sigma | Cat# NB600-501; RRID:AB_10077656 |
| Anti Myc | Abcam | Ab9106; RRID:AB_307014 |
| Bacterial and Virus Strains | ||
| Influenza A virus (A/Puerto Rico/8/34) MacDonald (The Rockefeller University). | A. García-Sastre (Mount Sinai School of Medicine). | N/A |
| A/PR/8/1934 ΔNS1 | A. García-Sastre (Mount Sinai School of Medicine). | N/A |
| A/PR/8/1934 NS1-GFP | A. García-Sastre (Mount Sinai School of Medicine). | N/A |
| Encephalomyocarditis (EMCV) | ATCC | Cat# VR-1479 |
| Vesicular Stomatitis Virus (VSV, Indiana strain) | C. Rice (Rockefeller University) | N/A |
| VSV-GFP | C. Rice (Rockefeller University) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Polyinosinic:polycytidylic acid (PolyI:C) | Sigma | Cat# P1530 |
| IFNAR1 monoclonal Antibody | eBiosciences | Cat# MAR103 |
| Halt Protease and phosphatase inhibitors cocktail | Roche | Cat# 78440 |
| Protein A Dynabeads | Invitrogen | Cat# 10001D |
| Lipofectamine 2000 | Life Technologies | Cat# 11668027 |
| Recombinant murine Interleukin (IL)-3 | Peprotech | Cat# 213-13 |
| Recombinant murine Macrophage Colony Stimulating Factor M-CSF |
Peprotech | Cat# 315-02 |
| Critical Commercial Assays | ||
| RNeasy mini kit | QIAGEN | Cat# 74104 |
| Reverse Transcription Supermix | Biorad | Cat# 1708840 |
| SYBR green | Biorad | Cat# 1725275 |
| mouse IFNβ ELISA kit | PBL Interferon Source | Cat# 42400 |
| NEBNext Multiplex Small RNA Library Prep Set | Illumina | Cat# E7300L |
| Deposited Data | ||
| https://www.ncbi.nlm.nih.gov/geo/ | GEO: GSE128228 | |
| Experimental Models: Cell Lines | ||
| AGO4 +/+ Mouse embryonic fibroblasts (MEFs) | A. Tarakhovsky | N/A |
| AGO4 −/− Mouse embryonic fibroblasts (MEFs) | A. Tarakhovsky | N/A |
| Baby Hamster Kidney (BHK-21) cell line | ATCC | Cat# CCL-10; RRID:CVCL_1915 |
| Vero cell line | ATCC | Cat# CCL-81; RRID:CVCL_0059 |
| MDCK cell line | ATCC | Cat# CCL-34; RRID:CVCL_0422 |
| HEK293T cell line | ATCC | Cat# CRL-3216; RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse: AGO4 −/− | A. Tarakhovsky | O’Carroll et al., 2007 |
| Mouse: MAVS −/− | The Jackson Laboratory | 008634; RRID:IMSR_JAX:008634 |
| Oligonucleotides | ||
| Table S3 | ||
| Recombinant DNA | ||
| Plasmid FLAG/HA-tagged Ago1 | Thomas Tuschl | Addgene #10820; RRID:Addgene_10820 |
| Plasmid FLAG/HA-tagged Ago2 | Thomas Tuschl | Addgene #10822; RRID:Addgene_10822 |
| Plasmid FLAG/HA-tagged Ago3 | Thomas Tuschl | Addgene #10823; RRID:Addgene_10823 |
| Plasmid FLAG/HA-tagged Ago4 | Thomas Tuschl | Addgene #10824; RRID:Addgene_10824 |
| Plasmid Myc-tagged Ago1 | This paper | N/A |
| Plasmid Myc-tagged Ago3 | This paper | N/A |
| Plasmid Myc-tagged Ago4 | This paper | N/A |
| Plasmid Myc-tagged Ago4D587A | This paper | N/A |
| Plasmid Myc-tagged Ago4G671A | This paper | N/A |
| Plasmid Myc-tagged Ago4R809A | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism v5.02 | GraphPad Software | https://www.graphpad.com/; RRID:SCR_002798 |
| FlowJo (Tree Star) | FlowJo |
https://www.flowjo.com/solutions/flowjo/; RRID:SCR_008520 |
Highlights.
Individual roles for the RNAi/miRNA effector proteins AGO1, AGO3, and AGO4 are elusive
AGO4 is uniquely antiviral in mammalian immune cells
Mammalian AGO4 has evolved to both elicit antiviral RNAi and boost IFN
AGO4 is indispensable for control of influenza in vivo
ACKNOWLEDGMENTS
The authors wish to thank Massachusetts General Hospital (MGH) next-generation sequencing, Center for Comparative Medicine, and Flow Cytometry Cores. We would like to sincerely thank A. Tarakhovsky for AGO knockout mice and mouse embryonic fibroblast (MEF) lines and invaluable scientific discussions; A. Garcia-Sastre, C. Rice, and M. McDonald for virus reagents; and H.-C. Reinecker for confocal microscopy assistance. We thank R. Xavier and R.M. Anthony for critical reading of the manuscript and all members of the Jeffrey lab for valuable input. This work was supported by MGH Executive Committee on Research (ECOR) funds, NIH grant U19 AI082630, NIH grant R01 AI107087 (K.L.J.), a Helmsley Postdoctoral Fellowship (D.H.), and Starr Cancer Consortium grant I4-A447 (N.P). The RNA sequencing data reported in this paper are archived at the Gene Expression Omnibus (GEO): GSE128228 (https://www.ncbi.nlm.nih.gov/geo).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.01.021.
REFERENCES
- Ayllon J, and García-Sastre A (2015). The NS1 protein: a multitasking virulence factor. Curr. Top. Microbiol. Immunol 386, 73–107. [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, and Siomi MC (2008). Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. USA 105, 7964–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S, Langlois RA, Schmid S, Varble A, Shim JV, Sachs D, and tenOever BR (2014). The Mammalian response to virus infection is independent of small RNA silencing. Cell Rep 8, 114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez AA, Spanko LA, Bouhaddou M, Sachs D, and tenOever BR (2015). Engineered mammalian RNAi can elicit antiviral protection that negates the requirement for the interferon response. Cell Rep. 13, 1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, and Hannon GJ (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, and Hannon GJ (2003). Dicer is essential for mouse development. Nat. Genet 35, 215–217. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KL, Putnam N, Barrows NJ, Sherry B, et al. (2014). Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol 88, 8065–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge KS, Shah KM, Li Y, Foxler DE, Wong SCK, Miller DC, Davidson KM, Foster JG, Rose R, Hodgkinson MR, et al. (2017). Argonaute utilization for miRNA silencing is determined by phosphorylation-dependent recruitment of LIM-domain-containing proteins. Cell Rep. 20, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, and Hannon GJ (2010). A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, and Darnell RB (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, and Littman DR (2010). Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 24, 1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR, Cherry S, and tenOever BR (2013). Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell Host Microbe 14, 374–378. [DOI] [PubMed] [Google Scholar]
- Czech B, and Hannon GJ (2011). Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet 12, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck A, Ziegler C, Eichner A, Berezikov E, and Meister G (2012). Micro-RNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 40, 9850–9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, and Bartel DP (2014). mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabozzi G, Nabel CS, Dolan MA, and Sullivan NJ (2011). Ebolavirus proteins suppress siRNA effects by direct interaction with the mammalian RNAi pathway. J. Virol 85, 2512–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, and Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, and Svoboda P (2013). A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155, 807–816. [DOI] [PubMed] [Google Scholar]
- Guo X, Zhang R, Wang J, Ding SW, and Lu R (2013). Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc. Natl. Acad. Sci. USA 110, 16085–16090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Li Y, and Ding SW (2019). Small RNA-based antimicrobial immunity. Nat. Rev. Immunol 19, 31–44. [DOI] [PubMed] [Google Scholar]
- Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, and Meister G (2013). Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol 20, 814–817. [DOI] [PubMed] [Google Scholar]
- Hauptmann J, Kater L, Löffler P, Merkl R, and Meister G (2014). Generation of catalytic human Ago4 identifies structural elements important for RNA cleavage. RNA 20, 1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, and Pillai PS (2014). Innate immunity to influenza virus infection. Nat. Rev. Immunol 14, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumani PK, Ponia SS, S RK, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, and Bhatnagar RK (2013). Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J. Virol. 87, 8870–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Whisnant AW, Kornepati AV, Marshall JB, Bogerd HP, and Cullen BR (2015). Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc. Natl. Acad. Sci. USA 112, E6945–E6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok KH, Lui PY, Ng MH, Siu KL, Au SW, and Jin DY (2011). The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9, 299–309. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, Aozasa K, Kawai T, and Akira S (2007). Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27, 240–252. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, and Tuschl T (2008). Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14, 2580–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Basavappa M, Jinfeng L, Dong S, Cronkite DA, Prior JT, Reinecker H-C, Hertzog P, Han Y, Li W-X, Cheloufi S, Karginov F, Ding S-W, and Jeffrey KL (2016). Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nature Microbiology 2, 16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, and Ding SW (2002). Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321. [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, García-Sastre A, Ball LA, et al. (2004). Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101, 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu J, Han Y, Fan X, and Ding SW (2013). RNA interference functions as an antiviral immunity mechanism in mammals. Science 342, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Fu B, Berman MA, Diallo A, and Dorf ME (2014). TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc. Natl. Acad. Sci. USA 111, 6970–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, and Hannon GJ (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. [DOI] [PubMed] [Google Scholar]
- Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP, et al. (2015). Hepatitis C virus RNA functionally sequesters miR-122. Cell 160, 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, and Voinnet O (2013). Antiviral RNA interference in mammalian cells. Science 342, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Van der Veen AG, Deddouche-Grass S, Rogers NC, Merits A, and Reis e Sousa C (2016). Inactivation of the type I interferon pathway reveals long double-stranded RNA-mediated RNA interference in mammalian cells. EMBO J. 35, 2505–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, and García-Sastre A (2010). Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. USA 107, 11531–11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Wang JP, Wang X, de Oliveira KP, Gao C, Aguiar ER, Jafari N, and Carthew RW (2013). Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog. 9, e1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matskevich AA, and Moelling K (2007). Dicer is involved in protection against influenza A virus infection. J. Gen. Virol 88, 2627–2635. [DOI] [PubMed] [Google Scholar]
- Mehta S, Cronkite DA, Basavappa M, Saunders T, Adiliaghdam F, Amatullah H, Morrison S, Pagan J, Anthony R, Tonerre P, et al. (2017). Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. Science Immunology 2, eaag3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G (2013). Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet 14, 447–459. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, and Tuschl T (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197. [DOI] [PubMed] [Google Scholar]
- Mili S, and Steitz JA (2004). Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, and Cohen PE (2012). AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev. Cell 23, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nish S, and Medzhitov R (2011). Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity 34, 629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K (2001). A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107, 415–418. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, and Tarakhovsky A (2007). A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 21, 1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. (2007). Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27, 123–134. [DOI] [PubMed] [Google Scholar]
- Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. (2010). Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6, e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, and Sullivan CS (2014). Distinct antiviral responses in pluripotent versus differentiated cells. PLoS Pathog. 10, e1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri S, Dueck A, Lehmann G, Putz N, Rüdel S, Kremmer E, and Meister G (2011). Increased siRNA duplex stability correlates with reduced offtarget and elevated on-target effects. RNA 17, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo JE, Venter PA, Short JR, Gopal R, Deddouche S, Lamiable O, Imler JL, and Schneemann A (2013). Cytoplasmic granule formation and translational inhibition of nodaviral RNAs in the absence of the double-stranded RNA binding protein B2. J. Virol 87, 13409–13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, and Filipowicz W (2005). Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309, 1573–1576. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Xu Y, Zhang Y, Zhou H, Deng YQ, Li XF, Miao M, Zhang Q, Zhong B, Hu Y, et al. (2017). Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity 46, 992–1004.e5. [DOI] [PubMed] [Google Scholar]
- Ruda VM,Chandwani R,Sehgal A,Bogorad RL,Akinc A,Charisse K,Tarakhovsky A, Novobrantseva TI, and Koteliansky V (2014). The roles of individual mammalian argonautes in RNA interference in vivo. PLoS ONE 9, e101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel TK, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, Auray G, Gerber M, Fak J, Keller I, et al. (2016). A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe 19, 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, Berkhout B, and Haasnoot J (2012). Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 40, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann N, Trabuco LG, Bender C, Russell RB, and Grimm D (2013). Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol 20, 818–826. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, and Sullivan CS (2013). Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JS, Schmid S, Aguado LC, Sabin LR, Yasunaga A, Shim JV, Sachs D, Cherry S, and tenOever BR (2014). Drosha as an interferon-independent antiviral factor. Proc. Natl. Acad. Sci. USA 111, 7108–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, and Chen ZJ (2006). The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24, 633–642. [DOI] [PubMed] [Google Scholar]
- Tsai K, Courtney DG, Kennedy EM, and Cullen BR (2018). Influenza A virus-derived siRNAs increase in the absence of NS1 yet fail to inhibit virus replication. RNA 24, 1172–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche-Grass S, Borg A, Kjær S, Snijders AP, and Reis e Sousa C (2018). The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 37, e97479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Stry M, Oguin TH 3rd, Cheloufi S, Vogel P, Watanabe M, Pillai MR, Dash P, Thomas PG, Hannon GJ, and Bix M (2012). Enhanced susceptibility of Ago1/3 double-null mice to influenza A virus infection. J. Virol 86, 4151–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, and Omata M (2006). Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology 130, 883–892. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O’Loughlin E, Lee T, Houel S, O’Carroll D, Tarakhovsky A, Ahn NG, and Yi R (2012). Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 26, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, Wang Y, Silva LAV, Sarbanes S, Sun T, et al. (2018). Intrinsic immunity shapes viral resistance of stem cells. Cell 172, 423–438.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS,Maurin T,Robine N,Rasmussen KD,Jeffrey KL,Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, and Lai EC (2010). Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 107, 15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Chang M, Nie P, and Secombes CJ (2009). Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Flow Cytometry data were analyzed using FlowJo (Treestar, USA) software. Graphics and statistics were generated with GraphPad PRISM software 7.0 (GraphPad Software, USA).
The RNA sequencing data reported in this paper are archived at the Gene Expression Omnibus (GEO): GSE128228 (https://www.ncbi.nlm.nih.gov/geo).