Abstract
Background:
Alzheimer’s disease is the most common form of brain dementia characterized by gradual loss of memory followed by further deterioration of other cognitive function. Large-scale genome-wide association studies have identified and validated more than 20 AD risk genes. However, how these genes are related to the brain-wide breakdown of structural connectivity in AD patients remains unknown.
Methods:
We used the genotype and DTI data in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. After constructing the brain network for each subject, we extracted three types of link measures, including fiber anisotropy, fiber length and density. We then performed a targeted genetic association analysis of brain-wide connectivity measures using general linear regression models. Age at scan and gender were included in the regression model as covariates. For fair comparison of the genetic effect on different measures, fiber anisotropy, fiber length and density were all normalized with mean as 0 and standard deviation as one.We aim to discover the abnormal brain-wide network alterations under the control of 34 AD risk SNPs identified in previous large-scale genome-wide association studies.
Results:
After enforcing the stringent Bonferroni correction, rs10498633 in SLC24A4 were found to significantly associated with anisotropy, total number and length of fibers, including some connecting brain hemispheres. With a lower level of significance at 5e-6, we observed significant genetic effect of SNPs in APOE, ABCA7, EPHA1 and CASS4 on various brain connectivity measures.
Keywords: brain connectivity, imaging genetics association, Alzheimer’s disease
Background
Alzheimer’s disease is the most common form of brain dementia characterized by gradual loss of memory followed by further deterioration of other cognitive function. It has become one of the leading cause of death and is still increasingly affecting the aging population nationwide[1]. Due to the lack of effective treatment, number of deaths due to Alzheimer’s between 2000 and 2014 has significantly increased [1].
Neuroimaging has been a major approach to study AD which allows mapping of structural, functional and molecular AD pathology inside brain [2]. Accumulating evidence from previous studies suggests abnormal imaging patterns in AD patients, such as cortical and subcortical atrophy [3], cortical amyloid deposition[4], grey matter atrophy[5], and functional cortical disconnection [6]. Recently, there is a growing interest in structural brain connectivity captured through diffusion tensor imaging (DTI)in AD. AD patients and those in a mild stage were found to have loss of inter-hemisphere connectivities [7] and increased diffusion anisotropy [8].
On the other hand, genetic factors play an essential role in AD. Results from large-scale twin studies suggests the heritability of AD to reach 70%-80% [9, 10]. APOE is the most well-known gene associated with increased AD risk and remains as the risk gene with the greatest known impact. Later, large-scale genome-wide association studies (GWASs) identified and validated twenty novel risk genetic loci [11, 12, 13, 14, 15, 16]. These risk variants have been recently found to differentially regulate brain amyloidosis across different disease stages [17]. However, how they exert effect on the brain-wide breakdown of structural brain connectivity has not been studied yet.
Leveraging the genotype and DTI data in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) [18, 19], in this paper, we perform a targeted genetic association analysis of brain-wide connectivity measures to discover the brain network alterations under the control of AD risk SNPs. We focus our analysis on link level measures, including fiber anisotropy, fiber length and density. In addition, to avoid potential bias introduced in imaging processing pipeline, we repeat our imaging processing pipeline and evaluate the reliability of all connectivity measures. Only high quality measures will be considered for the further imaging genetics association analysis. Using age at scan and gender as covariates, we employ general linear regression models to investigate the association between each pair of candidate SNP and connectivity measure. After enforcing the stringent Bonferroni correction, SNPs rs10498633 in SLC24A4 were found to be significantly associated with anisotropy, total number and length of fibers including some connecting hemispheres, which is consistent with existing findings. With a lower level of significance at 5e-6, we observed significant genetic effect of SNPs in APOE, ABCA7, EPHA1 and CASS4 on various brain connectivity measures.
Methods
ADNI Cohort
Data used in this study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database [19, 18]. Involving researchers from more than 50 sites in the USA and Canada, the ADNI is a multi-site study to track the progression of AD in the human brain by collecting longitudinal neuroimaging, biochemical, and genetic biological data. A key aim of the ADNI is to provide the opportunity to combine genetics with multiple types of imaging (e.g. magnetic resonance imaging (MRI) and positron emission tomography (PET)) and clinical data to help investigate mechanisms of the disease. For up-to-date information, see www.adni-info.org. For the proposed brain-wide connectomics genetics association analysis, we downloaded the structural MRI (sMRI) scans, DTI scans, GWAS genotype and demographic data from the ADNI website. Written informed consent was obtained at the time of enrollment and/or genetic sample collection and protocols were approved by each participating study and sites’ Institutional Review Board.
Brain Connectivity Measures
The DTI data was first denoised and corrected for motion and distortion using the approach described in a previous study [20]. Tractography was performed in Camino [21] based on white matter fiber orientation distribution function (ODF). Streamlines were modeled with a multi-tensor modeling approach, where voxels will fit up to two fiber orientations. Second, the sMRI images were registered to the b0 volume of DTI data using the FNIRT toolbox [22] and 278 brain regions of interest (ROIs) were extracted following [23]. The final networks between 278 brain ROIs were constructed using fibers going through white matter and connecting ROIs. In this project, we focus on the link level measures and calculated the fiber anisotropy (FA), length of fibers (LOF) and number of the fibers (NOF) connecting each pair of ROIs. Considering that the number of fibers are partially dependent on the surface area of their connected ROIs, we derived a new measure, the fiber density (FD), for the following association analysis, which is the fraction between number of fibers and the average surface of grey-matter regions i and j.
Reliability Test
To avoid potential bias introduced in imaging processing pipeline, We further evaluated the reliability of all brain connectivity measures. Since each subject only have one DTI scan in ADNI, we repeated our imaging processing pipeline three times and quantified the reliability of FA, FD and LOF measures by calculating their intraclass correlation coefficients (ICC) across three runs.
Genotype Data
Genotyping was performed using the Illumina HumanOmni Express BeadChip for all participants included. We first performed standard sample and SNP quality control procedures as described previously [24]. The un-genotyped SNPs were imputed using MACH and minimac in a two-stage procedure following a previous study [25]. The pilot 1 data of the 1000 Genomes Project were used as a reference panels for inferring missing genotypes. Minimac produced the posterior probabilities of the imputed genotypes at un-genotyped marker loci for each individual. To assure the quality of imputation, an r2 value equal to 0.30 was imposed as the threshold to filter the imputed genotypes. In this project, since we are particularly interested in the brain network alterations under the control of AD, rs429358 in APOE and 33 AD risk SNPs used in [17] were included for the association analysis (Table.1).
Table 1.
34 AD risk loci included in the brain-wide connectomics genetics association.
| SNP | Gene | MAF | SNP | Gene | MAF |
|---|---|---|---|---|---|
| rs3752246 | ABCA7 | 0.17 | rs6701713 | CR1 | 0.25 |
| rs3764650 | ABCA7 | 0.2 | rs11767557 | EPHA1 | 0.2 |
| rs4147929 | ABCA7 | 0.18 | rs11771145 | EPHA1 | 0.43 |
| rs6733839 | BIN1 | 0.39 | rs17125944 | FERMT2 | 0.11 |
| rs744373 | BIN1 | 0.36 | rs35349669 | INPP5D | 0.21 |
| rs7561528 | BIN1 | 0.2 | rs190982 | MEF2C | 0.22 |
| rs7274581 | CASS4 | 0.09 | rs610932 | MS4A6A | 0.45 |
| rs9349407 | CD2AP | 0.19 | rs983392 | MS4A6A | 0.23 |
| rs10948363 | CD2AP | 0.19 | rs2718058 | NME8 | 0.34 |
| rs3865444 | CD33 | 0.21 | rs3851179 | PICALM | 0.31 |
| rs10838725 | CELF1 | 0.26 | rs10792832 | PICALM | 0.31 |
| rs11136000 | CLU | 0.38 | rs561655 | PICALM | 0.34 |
| rs1532278 | CLU | 0.26 | rs28834970 | PTK2B | 0.32 |
| rs9331896 | CLU | 0.38 | rs10498633 | SLC24A4/RIN3 | 0.15 |
| rs12034383 | CR1 | 0.41 | rs1131497 | SORL1 | 0.37 |
| rs3818361 | CR1 | 0.25 | rs1476679 | ZCWPW1 | 0.21 |
| rs6656401 | CR1 | 0.07 | rs429358 | APOE | 0.15 |
Brain Connectomics Genetics Association
We performed a targeted genetic association analysis of brain-wide connectivity measures by employing a general linear model (GLM) approach in R. GLMs were developed using age at scan and gender as independent variables. For fair comparison of genetic effect on different measures, FA, LOF and FD measures were normalized to have zero mean and standard deviation as one. Following analysis of brain connectivity to examine the association between candidate SNPs and brainwide connectivity measures, correction for multiple comparisons was enforced using the Bonferroni correction method at a 0.05 level of significance. We also examined the associations showing trend-level significance (uncorrected p ≤ 5e-6).
Results
Subject
All the subjects included in this study are participants from the ADNI-2 and ADNI-GO stages. Among all 273 subjects with DTI scans, 214 of them without missing values in sMRI scans, genotype of 34 risk SNPs and demographic information were kept for the association analysis. In total, the study population is consisted of 34 healthy controls (HC), 26 individuals with subjective memory complain (SMC), 59 individuals with early mild cognitive impairment (EMCI), 23 individuals with late MCI (LMCI) and 36 individuals with AD. Shown in Table. 2 is the detailed demographic information for all 214 subjects.
Table 2.
Demographic information of all participants.
| HC | SMC | EMCI | LMCI | AD | |
|---|---|---|---|---|---|
| Number | 34 | 26 | 59 | 23 | 36 |
| Gender(M/F) | 19/15 | 18/8 | 34/25 | 16/7 | 22/14 |
| Age(Mean±std) | 72.97±5.94 | 73.5±5.22 | 72.9±7.59 | 71.39±8.1 | 75.06±8.94 |
Reliability Test
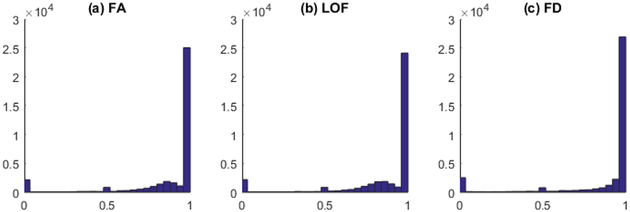
Among all brain connectivity measures, including FA, LOF and FD, about one third of the measures show inconsistency across three runs with ICC smaller than 0.9. More specifically, 26886 out of 38503 FA measures (69.83%) passed the reliability test with ICC greater than 0.9 [26]. For LOF and FD measures, there are 25644 (66.6%) and 29791 (77.37%) passing the same threshold. Shown in Figure. 1 is the ICC distribution for FA, LOF and FD respectively. In this paper, we only included those measures with excellent reliability (ICC ≥ 0.9) for further analysis.
Figure 1. ICC distribution for all measures.
(a) Fiber anisotropy, (b) Length of fibers, (c) Fiber density.
Genetic effect of AD risk genes on connectivity measures
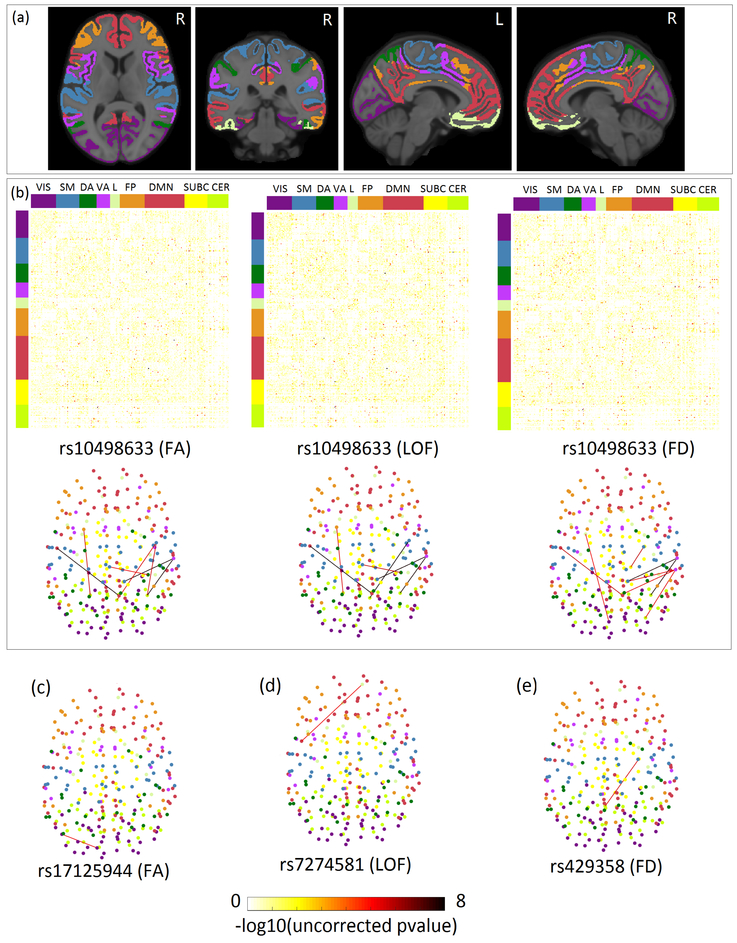
We identified several significant association between 34 AD risk loci and three types of connectivity measures with excellent reliability (Table. 3). For better interpretation of results, we mapped 278 brain ROIs to Yeo parcellation with seven groups (3rd and 4th columns in Table. 3), including Visual (VIS), Somato-Motor (SM), Dorsal Attention (DA), Ventral Attention (VA), Limbic system (L), Fronto-Parietal (FP), and Default Mode Network (DMN). Shown in Figure. 2(a) is the brain map of Yeo atlas we used in MNI 152 space. We also added subcortical regions (SUBC) and cerebellum (CER) to complement Yeo atlas using the strategy previously described in [27].
Table 3.
Significant associations between AD risk SNPs and brain connectivity measures.
| Data Type | SNP | ROI1 | ROI2 | Beta | p(uncorrected) | ICC |
|---|---|---|---|---|---|---|
| Fiber Anisotropy | rs10498633 | DMN | DMN | 0.89 | 1.44E-08 | 1.00 |
| rs10498633 | VA | CER | 0.88 | 1.68E-08 | 1.00 | |
| rs10498633 | VA | SUBC | 0.88 | 1.73E-08 | 1.00 | |
| rs10498633 | CER | FP | 0.76 | 1.52E-06 | 1.00 | |
| rs10498633 | CER | SM | 0.75 | 2.08E-06 | 1.00 | |
| rs10498633 | CER | SM | 0.75 | 2.72E-06 | 1.00 | |
| rs11771145 | CER | SM | 0.58 | 4.08E-06 | 1.00 | |
| rs17125944 | VS | VS | 0.48 | 4.35E-06 | 0.92 | |
| rs10498633 | DA | SM | 0.72 | 4.89E-06 | 1.00 | |
| Length of Fibers | rs10498633 | CER | SM | 0.90 | 8.73E-09 | 1.00 |
| rs10498633 | DMN | DMN | 0.90 | 9.06E-09 | 1.00 | |
| rs10498633 | VA | CER | 0.88 | 1.68E-08 | 1.00 | |
| rs10498633 | VA | SUBC | 0.88 | 1.68E-08 | 1.00 | |
| rs10498633 | DA | SM | 0.76 | 1.54E-06 | 1.00 | |
| rs10498633 | CER | FP | 0.75 | 2.56E-06 | 1.00 | |
| rs7274581 | L | DMN | 0.53 | 4.49E-06 | 1.00 | |
| Fiber Density | rs10498633 | VA | CER | 0.88 | 1.76E-08 | 1.00 |
| rs10498633 | VA | SUBC | 0.88 | 2.20E-08 | 1.00 | |
| rs4147929 | SUBC | CER | 0.68 | 1.04E-06 | 1.00 | |
| rs10498633 | SM | SUBC | 0.77 | 1.40E-06 | 0.95 | |
| rs10498633 | DMN | DMN | 0.76 | 1.48E-06 | 1.00 | |
| rs429358 | SUBC | CER | 0.64 | 2.71E-06 | 1.00 | |
| rs10498633 | VA | CER | 0.74 | 3.37E-06 | 1.00 | |
| rs10498633 | VA | CER | 0.74 | 3.62E-06 | 1.00 | |
| rs10498633 | VA | SUBC | 0.73 | 4.15E-06 | 1.00 | |
| rs10498633 | VS | L | 0.73 | 4.56E-06 | 1.00 |
Figure 2. Heatmap of all SNP-connectivity associations and brain map of selected connectivities with uncorrected p ≤ 5e-6.
(a) Brain map of Yeo parcellation with 7 groups in MNI 152 space. (b) Top panel: Heatmap showing the association of rs10498633 in SLC24A4 with three different types of connectivity measures. Rows and columns are reordered to form 7 groups corresponding to Yeo parcellation. Top and side colorbasr indicate the corresponding Yeo parcellation of each ROI. The last two groups, subcortical (SUBC) and Cerebellum (CER), are added to complement the Yeo atlas. (c) Brain map of the association between rs17125944 and FA measures. (d) Brain map of the association between rs7274581 and LOF measures. (e) Brain map of the association between rs429358 and FD measures. All the links in the brain connectivity map share the same colormap with the dots in the heatmap. Yeo parcellation in (a), top and side colorbars in (b), and all the nodes in the brain connectivity map share the same color scheme.
For FA measure, after enforcing the stringent Bonferroni correction based on the total number of FA measures and risk SNPs, the genetic effect of rs10498633 in SLC24A4 achieved brain-wide significance and showed association with the anisotropy of fibers connecting Ventral Attention and Subcortical, Ventral Attention and Cerebellum, and within Default Mode Network respectively. rs10498633 in SLC24A4 is also found to be significantly associated with the length and density of fibers connecting Cerebellum and Somato-Motor, Ventral Attention and Cerebellum, Ventral Attention and Subcortical, and within Default Mode Network etc. After lowering the level of significance to 5e-6 (uncorrected), other SNPs such as rs11771145 and rs10498633 are found to be nominally associated with the fibers connecting Cerebellum and Somato-Motor, Dorsal Attention and Somato-Motor respectively. In addition, rs17125944 in FERMT2 is associated with the internal connectivity anisotropy inside Default Mode Network with a nominal significance. rs429358 in top AD risk gene APOE only shows nominal significance of association with the density of fibers between Subcortical and Cerebellum.
Shown in the top panel of Figure. 2(b) is the heatmap of association results between rs10498633 and three types of connectivity measures. Each row and column were reordered based on Yeo atlas. The bottom panel is the corresponding brain connectivity map showing only links with ICC ≥ 0.9 and uncorrected p leq 5e-6. Links in the bottom panel are corresponding to the dots in the top panel. Different node colors in the brain connectivity map indicate the Yeo group information of each ROI. Among all the fibers affected by those risk genes, we observed that some of them associated with rs10498633 connect two hemispheres. This is consistent with previous findings that AD patients show loss of inter-hemispheric connectivity. Figure. 2(c)-(e) are example brain maps of three associations with nominal significance.
Discussion
To the best of our knowledge, this is the first comprehensive analysis to test the association of the top AD risk variants with brain connectivity measures. By performing a targeted genetic association of brain-wide connectivity, we were able to replicate the previous findings such as abnormal inter-hemispheric connectivity patterns and fiber anisotropy in AD. rs10498633 in SLC24A4 consistently shows significant genetic effect on all three type of brain connectivity measures, including some inter-hemispheric connectivity which is a known abnormal pattern in AD patients.
SLC24A4 encodes a member of the potassium-dependent sodium/calcium exchanger protein family. It has been previously reported to have significant association with gray matter density and brain metabolism [28]. According to the Allen Human Brain Atlas (AHBA) (www.brain-map.org), SLC24A4 is highly expressed in part of the cerebellum and some subcortical regions, including Amygdala, Puta-men, etc. Results of a large-scale eQTL analysis (www.braineac.org) show that rs10498633 is an eQTL in Thalamas, Putamen and white matter. Taken together, these evidence gives strong support to our connectomics-genetics findings. It also suggests potential role of rs10498633 in regulating the brain connectivity by mediating the expression of certain genes including SLC24A4.
Conclusion
We performed a targeted genetic association analysis of brain-wide connectivity measures to investigate the effect of AD risk genes on brain networks. We identified several significant genetics-connectomics associations. Particularly, rs10498633 in SLC24A4 shows significant genetic effect on the anisotropy, length and density of fibers, some of which connect two hemispheres. Since rs10498633 is an eQTL locus in multiple brain regions, the regulation role of rs10498633 in brain network may be achieved by mediating the expression of certain genes including SLC24A4.
Acknowledgements
This research was supported by NIH grants R01 AG053993, R01 EB022574, R01 LM011360, R01MH108467, R01 AG019771, P30 AG010133 and K01 AG049050. This project was also funded, in part, with support from the Indiana Clinical and Translational Sciences Institute funded, in part by Grant Number UL1TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- 1.Association A, et al. : 2017 alzheimer’s disease facts and figures. Alzheimer’s & Dementia 13(4), 325–373 (2017) [Google Scholar]
- 2.Teipel S, Drzezga A, Grothe MJ, Barthel H, Chételat G, Schuff N, Skudlarski P, Cavedo E, Frisoni GB, Hoffmann W, et al. : Multimodal imaging in alzheimer’s disease: validity and usefulness for early detection. The Lancet Neurology 14(10), 1037–1053 (2015) [DOI] [PubMed] [Google Scholar]
- 3.Fox NC, Schott JM: Imaging cerebral atrophy: normal ageing to alzheimer’s disease. The Lancet 363(9406), 392–394 (2004) [DOI] [PubMed] [Google Scholar]
- 4.Villemagne VL, Ong K, Mulligan RS, Holl G, Pejoska S, Jones G, O’Keefe G, Ackerman U, Tochon-Danguy H, Chan JG, et al. : Amyloid imaging with 18f-florbetaben in alzheimer disease and other dementias. Journal of Nuclear Medicine 52(8), 1210–1217 (2011) [DOI] [PubMed] [Google Scholar]
- 5.Thompson PM, Hayashi KM, De Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, et al. : Dynamics of gray matter loss in alzheimer’s disease. Journal of neuroscience 23(3), 994–1005 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pievani M, de Haan W, Wu T, Seeley WW, Frisoni GB: Functional network disruption in the degenerative dementias. The Lancet Neurology 10(9), 829–843 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Wang J, Zhang H, Mchugh R, Sun X, Li K, Yang QX: Interhemispheric functional and structural disconnection in alzheimer’s disease: a combined resting-state fmri and dti study. PLoS One 10(5), 0126310 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douaud G, Jbabdi S, Behrens TE, Menke RA, Gass A, Monsch AU, Rao A, Whitcher B, Kindlmann G, Matthews PM, et al. : Dti measures in crossing-fibre areas: increased diffusion anisotropy reveals early white matter alteration in mci and mild alzheimer’s disease. Neuroimage 55(3), 880–890 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingo TS, Lah JJ, Levey AI, Cutler DJ: Autosomal recessive causes likely in early-onset alzheimer disease. Archives of neurology 69(1), 59–64 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sleegers K, Lambert J-C, Bertram L, Cruts M, Amouyel P, Van Broeckhoven C: The pursuit of susceptibility genes for alzheimer’s disease: progress and prospects. Trends in Genetics 26(2), 84–93 (2010) [DOI] [PubMed] [Google Scholar]
- 11.Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, et al. : Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for alzheimer’s disease. Nature genetics 45(12), 1452 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. : Common variants at abca7, ms4a6a/ms4a4e, epha1, cd33 and cd2ap are associated with alzheimer’s disease. Nature genetics 43(5), 429 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P: Genome-wide association study identifies variants at clu and cr1 associated with alzheimer’s disease. Nat Genet (2009) [DOI] [PubMed] [Google Scholar]
- 14.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J: Genome-wide association study identifies variants at clu and picalm associated with alzheimer’s disease. Nat Genet (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. : Common variants at ms4a4/ms4a6e, cd2ap, cd33 and epha1 are associated with late-onset alzheimer’s disease. Nature genetics 43(5), 436 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carrasquillo MM, Lambert JC, et al. : Genome-wide analysis of genetic loci associated with alzheimer disease. Jama 303(18), 1832–1840 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apostolova LG, Risacher SL, Duran T, Stage EC, Goukasian N, West JD, Do TM, Grotts J, Wilhalme H, Nho K, et al. : Associations of the top 20 alzheimer disease risk variants with brain amyloidosis. JAMA neurology (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiner MW, Aisen PS, et al. : The alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimers Dement 6(3), 202–117 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, Ramanan VK, Foroud TM, Faber KM, Sarwar N, et al. : Genetic studies of quantitative mci and ad phenotypes in adni: Progress, opportunities, and plans. Alzheimer’s & dementia: the journal of the Alzheimer’s Association 11(7), 792–814 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manjón JV, Coupé P, Concha L, Buades A, Collins DL, Robles M: Diffusion weighted image denoising using overcomplete local pca. PloS one 8(9), 73021 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook P, Bai Y, Nedjati-Gilani S, Seunarine K, Hall M, Parker G, Alexander D: Camino: open-source diffusion-mri reconstruction and processing In: 14th Scientific Meeting of the International Society for Magnetic Resonance in Medicine, vol. 2759 (2006). Seattle WA, USA [Google Scholar]
- 22.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM: Fsl. Neuroimage 62(2), 782–790 (2012) [DOI] [PubMed] [Google Scholar]
- 23.Shen X, Tokoglu F, Papademetris X, Constable RT: Groupwise whole-brain parcellation from resting-state fmri data for network node identification. Neuroimage 82, 403–415 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nho K, Corneveaux J, Kim S, Lin H, Risacher S, Shen L, Swaminathan S, Ramanan V, Liu Y, Foroud T, et al. : Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Molecular psychiatry 18(7), 781 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nho K, Kim S, Risacher SL, Shen L, Corneveaux JJ, Swaminathan S, Lin H, Ramanan VK, Liu Y, Foroud TM, et al. : Protective variant for hippocampal atrophy identified by whole exome sequencing. Annals of neurology 77(3), 547–552 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koo TK, Li MY: A guideline of selecting and reporting intraclass correlation coefficients for reliability research. Journal of chiropractic medicine 15(2), 155–163 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amico E, Marinazzo D, Di Perri C, Heine L, Annen J, Martial C, Dzemidzic M, Kirsch M, Bonhomme V, Laureys S, et al. : Mapping the functional connectome traits of levels of consciousness. NeuroImage 148, 201–211 (2017) [DOI] [PubMed] [Google Scholar]
- 28.Stage E, Duran T, Risacher SL, Goukasian N, Do TM, West JD, Wilhalme H, Nho K, Phillips M, Elashoff D, et al. : The effect of the top 20 alzheimer disease risk genes on gray-matter density and fdg pet brain metabolism. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring 5, 53–66 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]