Summary
A central aspect of aging research concerns the question as to when individuality in lifespan arises1. We have now discovered that a transient increase in reactive oxygen species (ROS), which occurs naturally during early development in a subpopulation of synchronized Caenorhabditis elegans, sets processes into motion that increase stress resistance, improve redox homeostasis and ultimately prolong lifespan in those animals. We find that these effects are linked to the global ROS-mediated decrease in developmental histone H3K4me3 levels. Studies in HeLa cells confirmed that global H3K4me3 levels are ROS-sensitive, and that depletion of H3K4me3 levels increases stress resistance in mammalian cell cultures. In vitro studies identified the Set1/MLL histone methyltransferase as the redox sensitive unit of the H3K4-trimethylating COMPASS complex. Our findings imply a novel link between early-life events, ROS-sensitive epigenetic marks, stress resistance and lifespan.
Genetic effects are estimated to account for only 10 – 25% of the observed differences in human lifespan1. However, the remaining differences are not entirely attributable to environmental factors. Even when isogenic animals, such as Caenorhabditis elegans, are cultivated under identical environmental conditions, individual lifespans can vary by over 50-fold. These results suggest that other, more stochastic factors account for lifespan variations. Previous studies in C. elegans revealed that as early as day 1 of adulthood, subpopulations of longer-lived animals emerge2. We thus focused on the concept that specific fluctuating signals during development might differentially affect processes that determine lifespan. We explored the idea that reactive oxygen species (ROS)3, might serve as early lifespan determining modulators in C. elegans. This hypothesis was premised by C. elegans studies, which showed i) that significant lifespan extension occurs upon pharmacologically generated ROS in young adults4 or during development5, ii) that exposure of nematodes to non-lethal concentrations of ROS leads to increased stress resistance and longevity, a phenomenon termed mitohormesis4,6 and iii) that larvae of a synchronized wild-type population exhibit large inter-individual variations in endogenous ROS levels7.
Early-Life ROS affect adult redox states
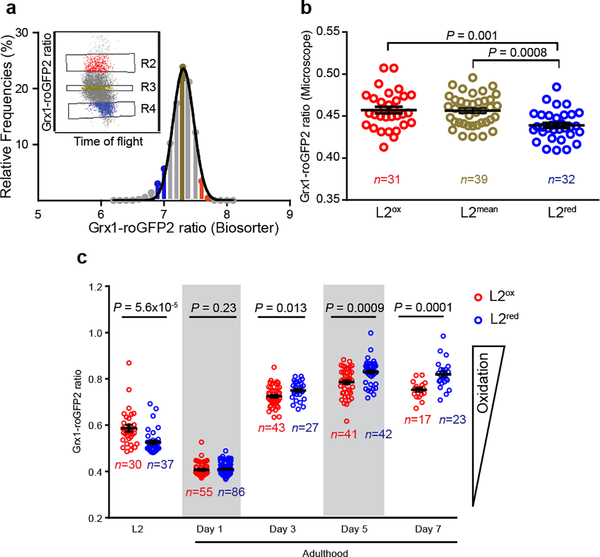
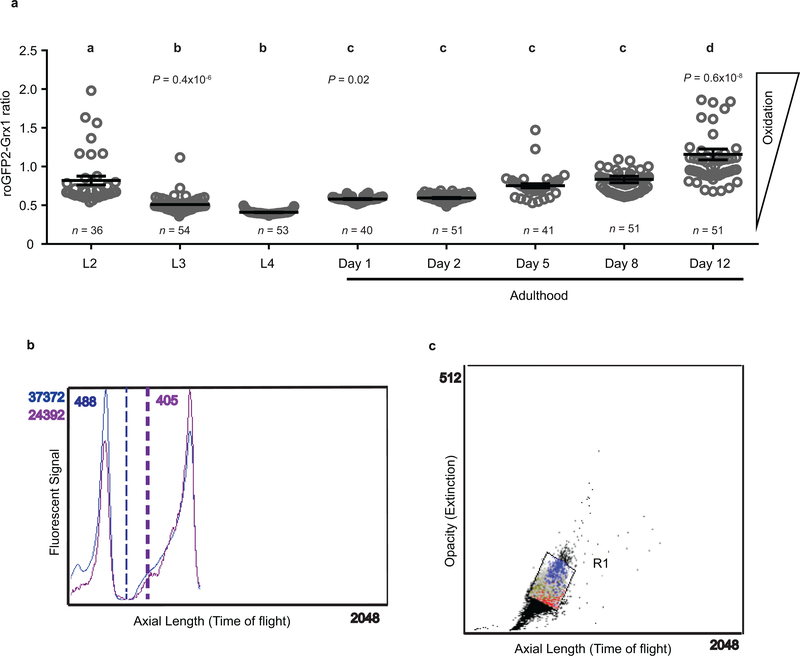
To investigate if and how developmental ROS levels affect C. elegans later in life, we used wild-type N2 worms that ubiquitously express the integrated redox sensing protein Grx1-roGFP2, which faithfully responds to the cellular ratio of oxidized and reduced glutathione (GSSG:GSH)8. Consistent with previous peroxide measurements7, L2 larvae revealed a significantly more oxidizing redox environment and substantially higher inter-individual differences than young adults, which appeared maximally reduced with little inter-individual differences (Extended Data Fig. 1a). With age, the average redox state became more oxidizing and inter-individual redox differences re-emerged. Subsequent analysis of about 16,000 age-synchronized L2 larvae using a reconfigured large particle BioSorter (Extended Data Fig. 1b) confirmed our microscopic studies and showed that the GSSG/GSH ratio varies widely among individuals (Fig. 1a). We sorted and binned L2 worms with redox states 2~3 standard deviations above (L2ox) or below (L2red) the mean population (L2mean) (Fig. 1a, Extended Data Fig. 1c), and confirmed their different redox states by fluorescence microscopy (Fig. 1b, Extended Data Fig. 2a–d). Importantly, L2ox and L2red worms did not differ significantly in size, reproductive activity, mitochondrial respiratory chain function or glycolytic flux (Extended Data Fig. 3a–f), excluding that more extreme early-life redox states affect development or other relevant physiological parameters. Subsequent redox analysis of sorted L2ox and L2red-worms showed that all animals become similarly reduced in young adulthood and become more oxidized as they age (Fig. 1c). By day 7 of adulthood, however, the L2ox-worms were significantly more reduced than the L2red worms. At this point, we know neither what event(s) trigger the transient increase in GSSG:GSH ratios during early development, nor what mechanisms cause the observed switch in endogenous redox states during adulthood. However, our results demonstrate that a synchronized population of C. elegans larvae contains subpopulations with redox environments that imprint information relevant later in life.
Figure 1. Endogenous redox state in an age-synchronized population of C. elegans larvae.
(a) Distribution of Grx1-roGFP2 ratios of a L2-staged N2jrIs2[Prpl-17::Grx1-roGFP2] population. L2 worms with Grx1-roGFP2 ratios between 2 and 3 standard deviations above (red line, insert: R2, red, L2ox) or below (blue line, insert: R4, blue, L2red) the mean were sorted and compared to animals with mean Grx1-roGFP2 ratios (green line, insert: R3, green, L2mean). Insert: axes are redrawn to scale. n = 15,599 animals. (b) Representative microscopic analysis of the Grx1-roGFP2 ratio of individual worms (symbols) of the L2ox, L2mean and L2red subpopulations. n = animals; One-way ANOVA with Tukey correction. The experiment was repeated 4 more times with similar results (see Extended Data Figure 2a–d). (c) Longitudinal analysis of the redox state. L2ox and L2 red-sorted were cultivated at 20°C and the Grx1-roGFP2 ratio of animals (symbol) in each subpopulation was determined microscopically at the indicated time points. n = animals; Mann-Whitney U test, two - sided. Data in (b) and (c) represent mean ± SEM.
Early-life ROS extend lifespan
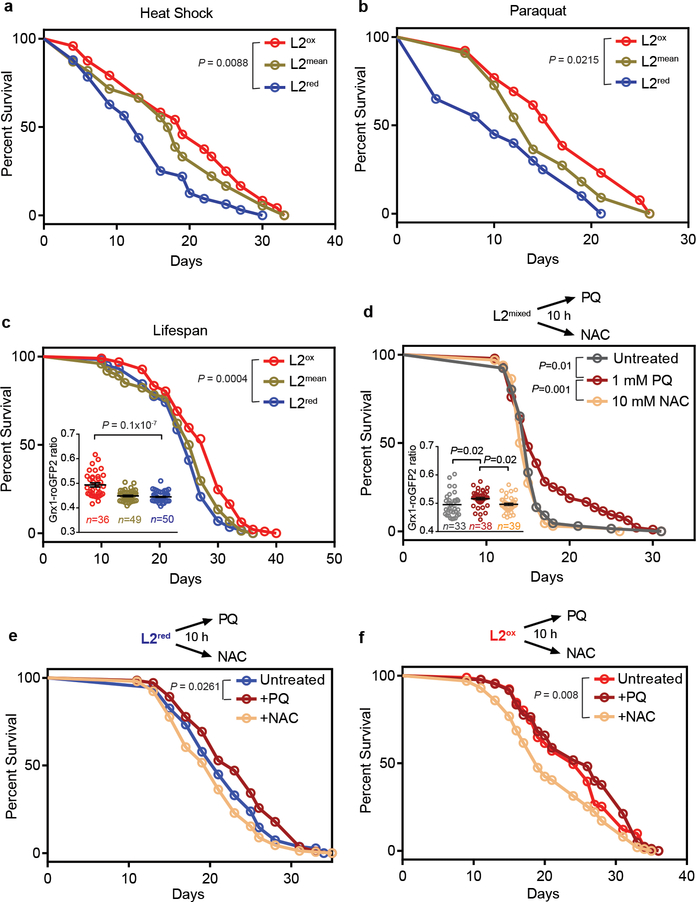
To investigate potential downstream effects of the observed variations in developmental redox levels, we compared stress resistance and lifespan of the sorted worms. We found that compared to L2red-worms, L2ox-worms were significantly more heat shock resistant (Extended Data Fig. 3g), about 30% longer-lived after the initial heat shock treatment (Fig. 2a), and substantially longer-lived when grown in the presence of oxidants, such as paraquat (PQ) (Fig. 2b) or juglone (Extended Data Table 1). Moreover, L2ox-worms displayed an up to 18% increase in median lifespan and a 1 – 4 day increase in maximal lifespan (Fig. 2c; Extended Data Figure 2e–i, Extended Data Table 2). This clear correlation between increased GSSG:GSH ratios, stress resistance, and lifespan suggested that a subpopulation of synchronized worms undergo a naturally occurring hormesis event during early development. Indeed, when we generated a more oxidizing environment in an entire population of worms by exposing them to 1 mM PQ for 10-h during their L2-stage, the entire population became longer-lived (Fig. 2d with insert). Treatment of L2 worms with 10 mM NAC for 10 h did not substantially alter the redox state of the population and had no significant lifespan effect. By conducting the same experiments with previously sorted subpopulations, however, we found that a 10h-NAC treatment decreased the lifespan of L2ox-worms but not of the already reduced L2red-worms, whereas a 10h-PQ exposure increased the lifespan of L2red-worms but not of the already oxidized L2ox-worms (Fig. 2e, f and Extended Data Table 3). These results provide evidence that transient changes in the redox-environment during early development are sufficient to positively affect the lifespan of C. elegans.
Figure 2. Oxidized L2 subpopulations show increased stress resistance and longer lifespan.
Experiments were performed with N2jrIs2[Prpl-17::Grx1-roGFP2] animals sorted into L2ox, L2mean and L2red subpopulations. (a) Representative survival curves of sorted worms that survived heat shock treatment. (b) Representative survival curves of sorted worms cultivated on NGM plates supplemented with 2 mM PQ. (c) Representative survival curves of sorted worms. See Extended Data Fig. 2e–i for repetitions. (Insert) The Grx1-roGFP2 ratio of individual worms (symbol) after sorting is shown. n = animals; unpaired t-test, two sided. (d) Representative survival curves of a non-sorted (mixed) worm population treated at the L2 stage with either nothing, 1 mM PQ or 10 mM NAC for 10 hours. (Insert) The Grx1-roGFP2 ratio of individual worms (symbol) after treatment is shown. n animals; one-way ANOVA with Tukey correction. Data in inserts represent mean ± SEM. (e-f) Representative survival curves of a L2red (e) or L2ox subpopulation (f) after a 10 h-treatment with either nothing, 1 mM PQ or 10 mM NAC. The specific sorting events, number of individuals, repetitions and statistical analysis (log-rank) for each of the data sets shown in this figure can be found in Ext. Data Tables 1–3.
Early-life ROS reduce H3K4me3 levels
To gain insights into the mechanism(s) by which a transient increase in the cellular redox state during development causes an increase in stress resistance and lifespan, we first conducted quantitative RT-PCR, testing for mRNA levels of commonly assessed heat shock genes (Fig. 3a) and oxidative stress-related genes (Extended Data Fig. 4a). Unexpectedly, L2ox and L2red worms did not significantly differ in the steady-state expression levels of any of these genes. However, upon exposure to heat shock conditions, L2ox-worms showed a significantly increased capacity to upregulate heat shock gene expression compared to L2red-worms (Fig. 3a). Changes in transcript levels of heat shock factor HSF-1 were not significantly different, suggesting that the transcriptional stimulation in L2ox-worms is either due to specific changes in HSF-1 activity, its subcellular localization, or heat shock promoter accessibility9.
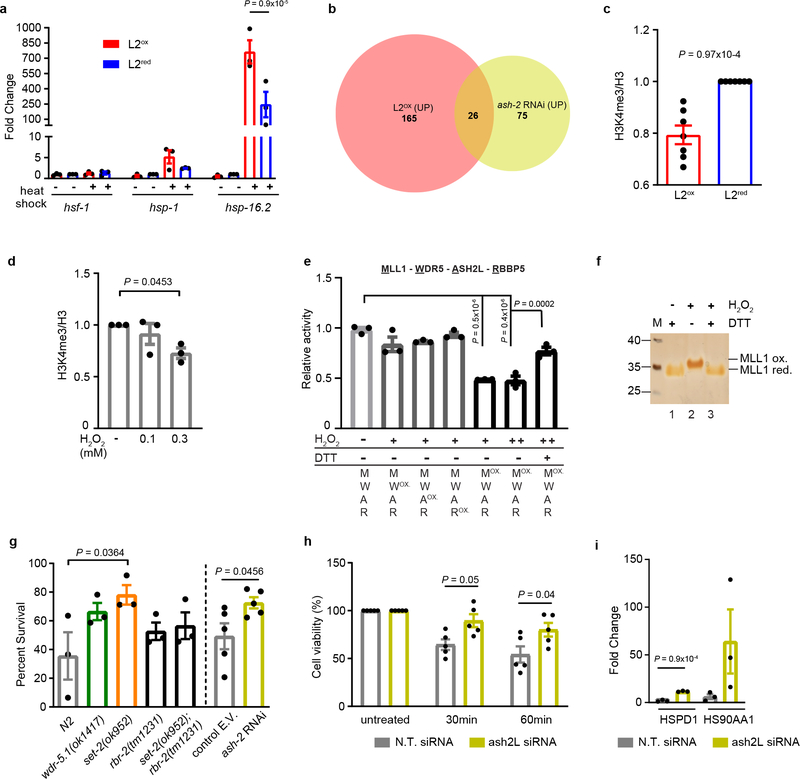
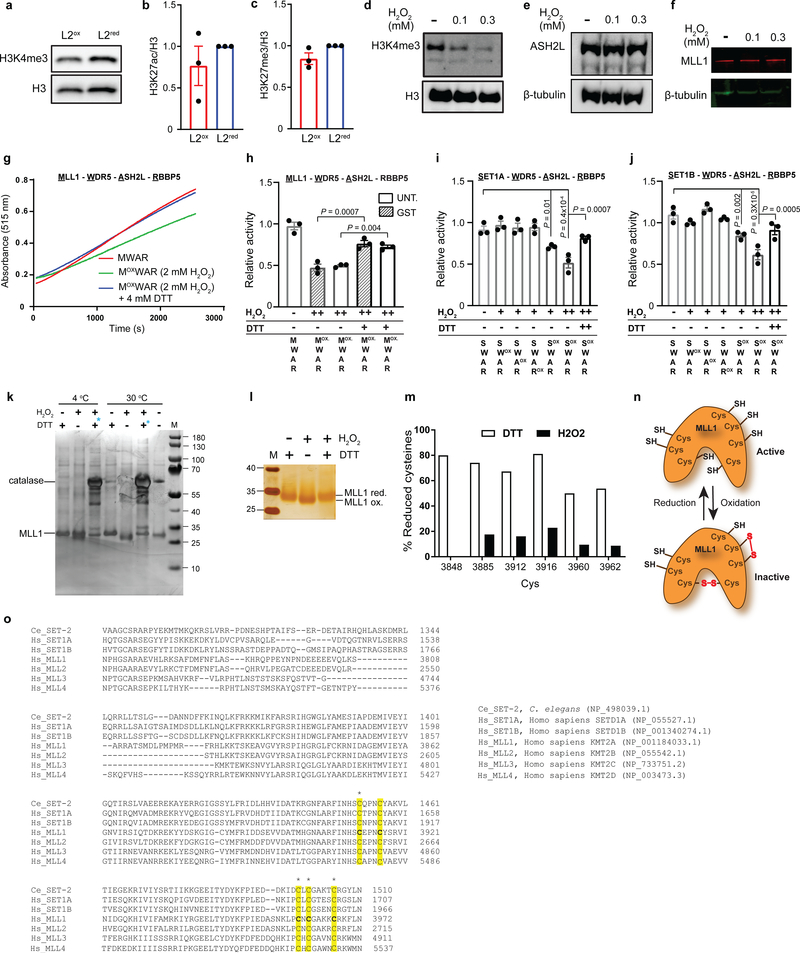
Figure 3. H3K4me3 - A redox sensitive histone modification involved in stress gene expression and resistance.
(a) Heat shock gene transcripts in sorted subpopulations before and after heat shock. n = 3 independent sorting experiments; two-way ANOVA with Tukey correction. (b) Venn diagram of upregulated genes in L2ox and ash-2 RNAi worms 10 (complete list in Supplementary Table 1). (c) Quantification of global H3K4me3 levels in L2ox and L2red worms. n = 7 independent sorting experiments; unpaired t-test, two-sided. (d) Quantification of H3K4me3 levels in HeLa cells before and after H2O2 treatment. n = 3 independent experiments; one-way ANOVA with Dunnett correction. (e) In vitro histone methyltransferase assays of core HMC members; ox: pre-treated with 1 mM (+) or 2 mM (++) H2O2 for 30 min prior to activity assay. Thiol-reducing agent dithiothreitol (DTT) was added after the H2O2 treatment. n = 3 independent experiments; one-way ANOVA with Sidak correction. (f) Reverse thiol trapping of oxidized and reduced MLL1SET. A 500-Da mass increase per oxidized thiol can be detected on non-reducing SDS-PAGE. (g) Heat shock survival of C. elegans N2 wild-type, wdr-5, set-2, rbr-2 and set2/rbr-2 mutants after 48 hours (n = 3 independent experiments; one-way ANOVA with Dunnett correction) or N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with ash-2 or control RNAi after 24 hours (n = 5 independent experiments; unpaired t-test, two-sided. E.V. empty vector). (h) Heat shock survival of ASH2L-siRNA treated HeLa cells. n = 5 independent experiments; two-way ANOVA with Tukey correction. (i) Transcript levels of heat shock genes after 30 min heat stress treatment of ASH2L siRNA treated HeLa cells. N.T., non-targeting. n = 3 independent experiments; unpaired t-test, two-sided. Data in (a), (c), (d), (e) and (g-i) represent mean ± SEM. For blot and gel source images, see Supplementary Fig. 1–3.
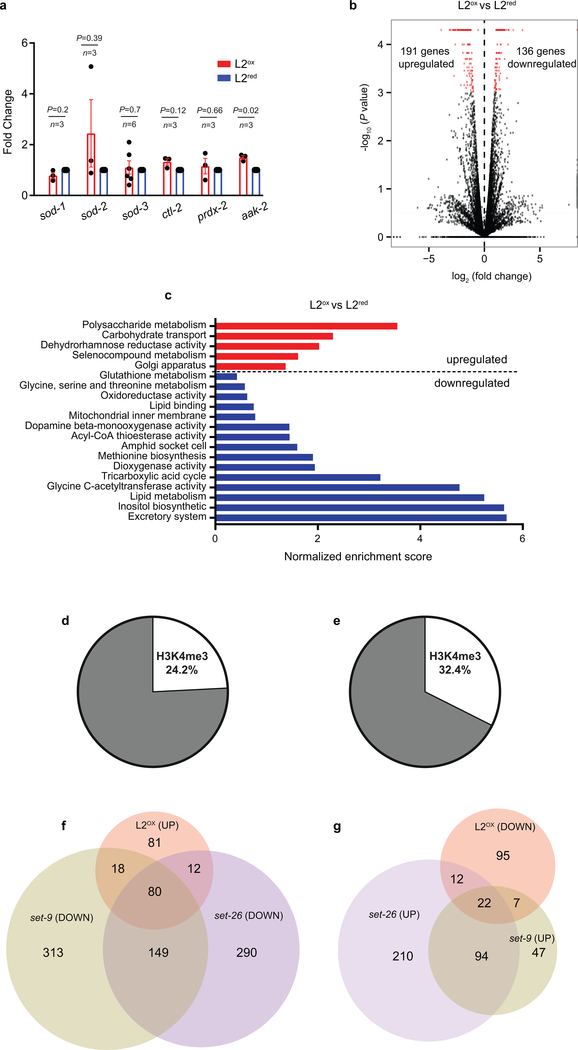
Subsequent RNA-seq analysis from four independent large-scale sorting experiments identified 191 up-regulated and 136 downregulated genes in L2ox-worms compared to L2red-worms (Extended Data Fig. 4b). We were unable to draw any clear connection between the differentially expressed genes (DEGs) and previously identified sets of stress or longevity-related genes (Extended Data Fig. 4c and Supplementary Table 1). To our surprise, however, 26 of the 191 up-regulated genes in L2ox-worms overlapped with a set of 101 genes, previously shown to be up-regulated in worms lacking the Absent Small Homeodisc protein ASH-210 (expected overlap if no correlation <1; P = 1.7×10−22, hypergeometric probability) (Fig. 3b and Supplementary Table 1). ASH-2 is a component of the highly conserved histone methylation complex COMPASS, which, together with a member of the SET-1/MLL (mixed lineage leukemia) histone methyltransferase family and WDR-5 causes trimethylation of lysine 4 in histone H3 (H3K4me3)11. H3K4me3 is primarily found at transcriptional start sites (TSS), where the modification is thought to mark and maintain transcriptionally active genes12. Intriguingly, recent studies in C. elegans revealed that TSS-associated H3K4me3 marks are set during early development, and remain stable throughout life13. Indeed, analysis of published ChIP-data revealed that about 25% of DEGs that we identified in L2ox-worms associate with H3K4me3 marks that appear to be set during development (Extended Data Fig. 4d, e). Furthermore, we found a highly significant overlap between DEGs in L2ox-worms and DEGs in strains lacking the H3K4me3-readers SET-9 or SET-2614, similar to the overlap between ash-2 knockdown and set-9 or set-26 deletion strains (Extended Data Fig. 4f, 3g). Based on these results, we decided to analyze the global H3K4me3 abundancy in L2ox and L2red-worms by western blot using antibodies against H3K4me3. We tested the sorted subpopulations of seven independent sorting experiments and found a clear and highly reproducible >25% reduction in global H3K4me3 levels in L2ox-worms (Fig. 3c, Extended Data Fig. 5a). In contrast, other marks, such as H3K27ac or H3K27me3, were not significantly different between the two subpopulations (Extended Data Fig. 5b, c). These results strongly suggest that we discovered a connection between endogenous ROS levels, H3K4 trimethylation levels, and gene regulation.
H3K4me3 - A redox-sensitive histone mark
RNA-seq analysis of L2ox and L2red-worms did not reveal any transcriptional changes in components of the COMPASS complex. This result suggested that the activity rather than the level of the H3K4me3 complex is affected by the redox environment. To directly test whether the H3K4me3 machinery is ROS sensitive, we attempted to purify the C. elegans proteins for in vitro methylation assays. Yet, this turned out to be impossible due to their instability. In contrast, the mammalian members of the COMPASS have been previously used in H3K4 methylation assays in vitro15. To first ascertain that H3K4 trimethylation is a redox sensitive process in mammalian cells as well, we subjected HeLa cells to non-lethal H2O2 treatment, and investigated global H3K4me3 levels. Consistent with our findings in C. elegans, we observed a >30% global decrease in H3K4me3 levels within <30 min of peroxide treatment (Fig. 3d and Extended Data Fig. 5d), without detectable changes in steady-state levels of two of the main COMPASS components (Extended Data Fig. 5e, f). We subsequently purified the SET domain of MLL1 (C.e. SET-2), which catalyzes the lysine-directed histone methylation16, ASH2L, WDR5, and RBBP5 (ASH-2, WDR-5.1, and RBBP-5 in C.e.). We individually treated the four proteins with peroxide for 30 min, removed the oxidant, combined the proteins and tested for in vitro histone methylation activity. Notably, only one protein appeared to be reproducibly peroxide-sensitive; the SET-domain of MLL1 (Fig. 3e and Extended Data Fig. 5g, h). Importantly, incubation of the peroxide-inactivated SET-domain with thiol-reducing dithiothreitol restored the original activity, strongly suggesting that thiol oxidation is responsible for the reversible inactivation (Fig. 3e and Extended Data Fig. 5g, h). Analysis of the SET-domain of other SET1/MLL-family members17, including SET1A, the most closely related human homologue of C. elegans SET-218 (Extended Data Fig. 5i), SET1B (Extended Data Fig. 5j) and a version of MLL1 lacking the cysteine-containing GST-tag used for purification (Extended Data Fig. 5h) demonstrated that sensitivity towards peroxide is a universal feature for SET1/MLL family members. To investigate which of the seven cysteines in the MLL1-SET domain might be sensitive to reversible thiol oxidation, we conducted direct (Extended Data Fig. 5k, l) and reverse thiol trapping (Fig. 3f) on oxidized and reduced MLL1SET using the 500-Da thiol-reactive compound 4-acetamido-4’maleimidylstilbene-2,2’-disulfonic acid (AMS), followed by SDS PAGE19. Analysis of the migration behavior of thiol-trapped oxidized versus reduced MLL1SET suggested that peroxide-treatment leads to the formation of two intramolecular disulfide bonds. Subsequent mass spectrometric analysis of in vitro modified cysteines confirmed these results and revealed that at least four of the five absolutely conserved cysteines in the SET domain are highly oxidation-sensitive (Extended Data Fig. 5m–o). To our knowledge, this makes H3K4me3 the first histone methylation mark and MLL1 the first histone methyltransferase known to be redox-regulated.
H3K4me3 modulates stress resistance
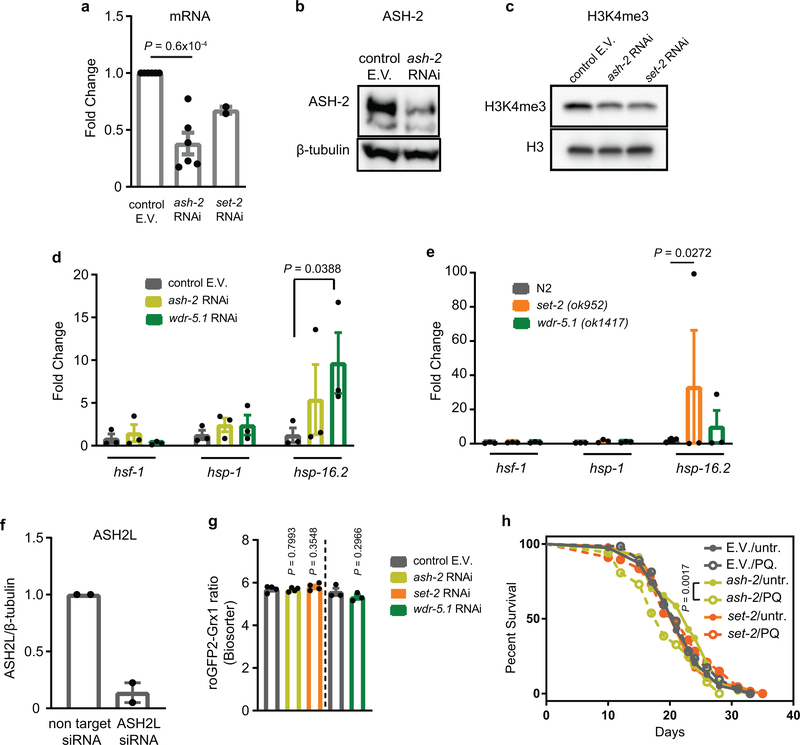
Our studies raised the intriguing possibility that redox-mediated inactivation of the lone SET-1 homologue in the oxidized subpopulation of C. elegans (i.e., SET-2) leads to a reduction in global H3K4me3 levels, which causes increased stress resistance and longevity. This theory was supported by recent studies, which showed that deleting or knocking down components of the COMPASS complex in C. elegans increase lifespan10. Indeed, strains either deleted (i.e., set-2, wdr-5) or depleted (i.e., ash-2) for members of the COMPASS complex were significantly more heat stress resistant than respective control strains or a strain deficient in H3K4me3-demethylase RBR-2 (Fig. 3g). In addition, and comparable to results obtained with L2ox-worms, we found that H3K4me3-deficient worms (Extended Data Fig. 6a–c) showed a substantially increased transcriptional response upon heat shock (Extended Data Fig. 6d, e). We obtained very similar results with ASH2L-depleted HeLa cells (Extended Data Fig. 6f), which were more heat stress resistant (Fig. 3h) and demonstrated an augmented transcriptional response to heat stress compared to control RNAi cells (Fig. 3i). These results strongly suggested that the downstream effects of H3Kme3 depletion on stress resistance and longevity are conserved.
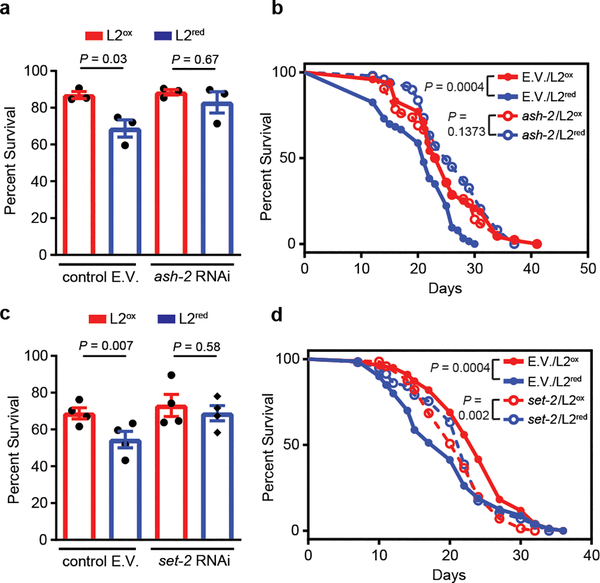
To finally test whether down-regulation of H3K4me3 levels is sufficient to increase heat shock resistance and lifespan in L2ox-worms, we generated ash-2 or set-2 RNAi knockdown worms expressing the Grx1-roGFP2 sensor protein and sorted the synchronized L2 population as before. We detected no significant difference in the relative distribution or range of GSSG:GSH ratios between mutant and control RNAi worms (Extended Data Fig. 6g). Yet, the sorted L2ox and L2red subpopulations of ash-2 and set-2 RNAi worms no longer exhibited any difference in heat shock sensitivity (Fig. 4a, b) or lifespan (Fig. 4c, d; Extended Data Tables 2 and 4). Similarly, no life-prolonging effect was observed when we treated ash-2 or set-2-RNAi-worms with PQ for 10h-treatment at the L2 larval state (Extended Data Fig. 6h and Extended Data Table 4). These results imply that down-regulation of H3K4me3 levels is both necessary and sufficient to increase heat shock resistance and lifespan in the oxidized subpopulation of worms.
Figure 4. An intrinsically oxidizing environment confers increased stress resistance via down-regulation of global H3K4me3 levels.
Survival of N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with ash-2 (a) or set-2 (b) RNAi, sorted into L2ox and L2red and measured 24 hours after heat shock. n = 3 (a) and n = 4 (b) independent experiments; two-way ANOVA with Tukey correction. Data represent mean ± SEM. Representative survival curves of N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with ash-2 (c) or set-2 (d) RNAi and sorted into L2ox and L2red. For n numbers, repetitions and statistics (log-rank) in (c) and (d), see Extended Data Tables 2 and 4.
Conclusions
In 2010, Cynthia Kenyon wrote in the journal Nature: “It is possible that a stochastic event — a metabolic insult or noise in the expression of a regulatory gene — flips an epigenetic switch or sets in motion a chain of events that promotes ageing”20. Our studies have identified that variations in endogenous ROS during development, potentially caused by locally different growth conditions, contribute to the lifespan variation observed in synchronized populations of C. elegans. Animals that accumulate high levels of ROS during development apparently undergo an endogenous hormesis event, which, as previously observed upon exogenous ROS treatment21, increases stress resistance and lifespan. This might serve as a bet-hedging strategy to provide subpopulations of worms with improved survival during stress. Our studies uncovered the underlying mechanism of ROS-mediated hormesis by demonstrating that global H3K4me3 levels are redox-sensitive, and decrease in response to oxidative stress. Based on the findings that reduction of global H3K4me3 levels increase stress resistance and C. elegans lifespan10, we now postulate that we have indeed identified one stochastic event, the respective epigenetic switch and the chain of events that are set into motion during early development to increase lifespan.
Recent studies in C. elegans demonstrated that H3K4me3 marks within gene bodies are set during adulthood and change with age13. In contrast, H3K4me3 that are enriched at transcriptional start sites and thought to provide the memory of actively transcribed genes, are set during development and remain stable throughout the lifespan13. This result likely explains how transient redox-mediated changes in H3K4me3 levels during development are sufficient to exert long-lasting effects despite the dramatic changes in the redox environment during adulthood. Our finding that organisms with reduced levels of H3K4me3 show an increased transcriptional response to stress conditions is initially counterintuitive, given that H3K4me3 is widely considered an activating mark11. However, our results are fully consistent with yeast studies, which showed that reduction of H3K4me3 levels causes substantially more robust gene expression changes upon stress treatment22. The extent to which this increase in transcriptional capacity of stress-related genes is linked to the observed lifespan extension, however, remains to be determined. A significant number of genes involved in lipid metabolism were found to be downregulated in L2ox-worms (Extended Data Fig. 4c, Supplementary Table 1). This is of note since increased lipid storage and altered lipid signaling have been previously linked to increased lifespans of a variety of different organisms23, and found to play a role in the lifespan extension of worms globally deficient in H3K4me3 levels24. Future work is needed to reveal how the transient downregulation of H3K4me3 levels selectively during development can elicit similarly profound life-altering effects. Our ability to change the lifespan of an entire population by a simple 10-hour exposure to ROS during development suggests that we have identified a window in time and a mechanism that helps to individualize lifespan in animals. This study will be forming the groundwork for future work in mammals where very early and transient metabolic events in life seem to have equally profound impacts on lifespan25.
ONLINE METHODS
C. elegans strains, maintenance and lifespan assays
The following C. elegans strains were used in this study: PB020: N2jrIs2[Prpl-17::Grx-1-roGFP2], N2: Wild-type Bristol isolate, RB1304: wdr-5.1(ok1417), RB1025: set-2(ok952), ZR1: rbr-2(tm1231) and ABR9: set-2(ok952);rbr-2(tm1231). If not stated otherwise, worms were cultured at 20°C. Standard procedures were followed for C. elegans strain maintenance 26. Synchronization was performed using alkaline hypochlorite solution; eggs were allowed to hatch by overnight incubation in M9 medium during gentle shaking. Newly hatched, arrested L1 larvae were transferred onto standard nematode growth medium (NGM) plates seeded with live E. coli OP50. Lifespan studies were performed at 20°C in the presence of FuDR. Survival was scored every 2 days, and worms were censored if they crawled off the plate, hatched inside, or lost vulva integrity during reproduction. The first day of adulthood was set as t = 0. Lifespan of unsorted worm populations were performed in a lifespan machine according to 27. Survival plots were generated using GraphPad Prism. Lifespan data were analyzed for statistical significance with log-rank (Mantel-Cox) or Gehan-Breslow-Wilcoxon test.
Reconfiguration of BioSorter for ratiometric sorting
405 and 488 nm lasers were used to excite the Grx1-roGFP2 sensor protein. Since the protein possesses a single emission maximum (~520 nm), the two lasers in the BioSorter (Union Biometrica) were realigned to sequentially illuminate single L2-staged worms as they pass through the flow cell, without emitting overlapping signals. This enabled collection of signals from 405 and 488nm lasers separately, from two photon multipliers tubes (PMTs). As result, data were displayed as two groups of peaks (Extended Data Fig. 1b). Using the partial profiling feature (pp) of the FlowPilot-Pro™ software, we mapped the peaks corresponding to each laser that trace the fluorescent intensity and extinction signals. The extinction signal from the 488 nm laser was used to initially gate worms at the L2 stage larva (R1 gate, see Extended Data Fig.1c). Oxidized, mean and reduced L2 worms were sorted from R2, R3 and R4 gates respectively, based on the peak 405 and 488 fluorescent intensities (insert in Fig. 1a).
Microscopy
Worms were mounted on objective slides using 4 μl thermoreversible CyGEL (BioStatus; Fisher Scientific) and 2 μl of 50 mM levamisole for immobilization. Fluorescence and DIC images were acquired with an upright microscope equipped with a Photometrics Coolsnap HQ2 cooled CCD camera, a UPlan S-Apo 20x objective (NA 0.75) and a X-CITE® exacte light source equipped with a closed feedback-loop. For Grx1-roGFP2 fluorescence, an external filter wheel was used with excitation filters 420/40x, 500/20x, dual bandpass dichroic T515LPXR and a single emission filter 535/30x. Image analysis was performed in Metamorph (Molecular Devices, Inc) using a custom script. Briefly, an intensity threshold was chosen by the user. Pixels above this threshold constitute regions of interest. Regions with very high signal in any channel (e.g., fluorescent particles) were identified by applying an over-saturation threshold and excluded from regions of interest. Mean ratiometric values (Ex420Em535/Ex500Em535) of regions of interest are calculated after subtraction of background. Acquisition parameters were kept identical across all samples. For body length measurements, worms were measured from the nose to the tail tip and analysis was performed with ImageJ.
Brood size
L4-staged worms were transferred onto NGM plates and incubated at 15°C. The parental animals were transferred daily to individual NGM plates until the end of the reproductive period. The progeny of each animal was counted at the L2 or L3 stage.
Cellular respiration
Real-time oxygen consumption rates (OCR) and extra-cellular acidification rate (ECAR) were measured with a Seahorse XFe96 Analyzer (Seahorse bioscience Inc., North Billerica, MA, USA) as described28. Briefly 100 L2–staged worms were sorted directly into individual wells of 96-well Seahorse utility plates at a final volume of 200 ul of 10% M9. Acute effects of pharmacological inhibitors FCCP (ETC accelerator) and sodium azide (NaN3, Complex IV and V inhibitor) were evaluated by injecting them during the run at final concentrations of 20 uM and 40 mM respectively.
Heat shock treatment
Heat shock was performed on solid OP50-seeded NGM plates wrapped in parafilm and submerged in a pre-heated water bath. For thermotolerance assays, worms were heat-shocked for 45 min at 38°C. Survival was scored after 24 hours and then until the death of the last worm by absence of touch response or pharyngeal pumping. For transcriptional response assays, worms were heat-shocked for 30 min at 35°C. After 1 hour recovery at 20°C, worms were harvested and snap-frozen in liquid nitrogen. All heat shock treatments were applied to worms at the L2 stage.
Treatments with N-acetyl cysteine (NAC) and paraquat (PQ)
For survival assays, worms were cultivated on solid OP50-seeded NGM plates, supplemented with the indicated concentrations of NAC or PQ. Survival was determined by absence of touch response or pharyngeal pumping. For transient exposure to compounds, worms were transferred into M9-media supplemented with OP50 and the indicated concentrations of NAC or PQ for 10 hours. Worms were harvested, washed three times with M9 and transferred onto regular OP50-seeded NGM plates.
RNA extraction and real-time qPCR
3,000–5,000 worms at the L2 stage (whole population or after sorting) were grounded in Trizol reagent (Life Technologies) with sea sand and pestle. After filtering of the sand, samples were vigorously shaken with chloroform, allowed to stand for 3 minutes at room temperature, and then centrifuged at 16,000 g at 4°C. The aqueous phase was then collected and RNA was purified using QIAGEN RNeasy RNA extraction columns as per manufacturers’ recommendations. HeLa cells were directly lysed in the culture dish by adding Trizol, as per manufacturers’ recommendations. For RNA isolation, after addition of ethanol, the lysate was loaded onto QIAGEN RNeasy RNA extraction columns. cDNA synthesis was performed using PrimeScript™ 1st strand cDNA Synthesis Kit (Takara) and real-time quantitative PCR was performed using Radiant™ Green Lo-ROX qPCR Kit (Alkali Scientific), as per manufacturers’ recommendations, in an Eppendorf Mastercycler epgradient S realplex2 detection system. Relative expression was calculated from Cycle threshold values using the 2−ΔΔCt method and the expression of genes of interest were normalized to housekeeping genes cdc-42, pmp-3, panactin) and/or spiked-in luciferase (10 pg/ml Trizol).
Primers used were cdc-2: 5’-AGCCATTCTGGCCGCTCTCG-3’ and 5’-GCAACCGCCTTCTCGTTTGGC-3’; pmp-3: 5’-TTTGTGTCAATTGGTCATCG-3’ and 5’-CTGTGTCAATGTCGTGAAGG-3’; panactin: 5’-TCGGTATGGGACAGAAGGAC-3’ and 5’-CATCCCAGTTGGTGACGATA-3’; sod-1: 5’-AAAATGTGGAACCGTGCTG-3’ and 5’-TGAACGTGGAATCCATGAA-3’; sod-2: 5’-GATTTGGAGCCTGTAATCAGTC-3’ and 5’-GAAGAGCGATAGCTTCTTTGAC-3’; sod-3: 5’-CACTATTAAGCGCGACTTCGG-3’ and 5’-CAATATCCCAACCATCCCCAG-3’; ctl-2: 5’-ATCCCAACATGATCTTTGA-3’ and 5’-TGAGATTCTTCACTGGTTG-3’; prdx-2: 5’-CGACTCTGTCTTCTCTCAC-3’ and 5’-GAAGATCATTGATGGTGAT-3’; aak-2: 5’-AAGTCTGGAGTTGGGAATACG-3’ and 5’-GTATGCACTTCTTTGTGGAACC-3’; hsf-1: 5’-TCCGTATAAGAATGCGACTAGG-3’ and 5’-TAGCTTCTGATGTGGTTGAAGG-3’; hsp-1: 5’-GGACGTCTTTCCAAGGATGA-3’ and 5’-TCAAGATCTCGTCGACTTG-3’; hsp-16.2: 5’-CTGTGAGACGTTGAGATTGATG-3’ and 5’-CTTTACCACTATTTCCGTCCAG-3’; ash-2: 5’-CGATCGAAACACGGAACGA-3’ and 5’-TGCCGGAATCTGCAGTTTTT-3’, set-2: 5’-TCGAAGATTGAAGGTGAAGAGAG-3’ and 5’-ATCATCTTTTTGCGGAACTGTAA-3’; HSPD1: 5’-TGCTGAGTTTTGAATGAGCAA-3’ and 5’-CAATCTGCTCTCAAATGGACA-3’; Hsp90AA1: 5’-GAAATCTGTAGAACCCAAATTTCAA-3’ and 5’-TCTTTGGATACCTAATGCGACA-3’; Luciferase: 5’- ACGTCTTCCCGACGATGA-3’ and 5’-GTCTTTCCGTGCTCCAAAAC-3’.
RNA-seq analysis
Total RNA from 4 biological replicates of worms sorted at the L2 stage (extracted as described above) was assessed for quality using the TapeStation (Agilent). Samples were prepared using the Illumina TruSeq Stranded Total RNA Library Prep kit (Illumina). 100 ng of total RNA was rRNA-depleted using Ribo-Gone (Takara Bio USA). The rRNA-depleted RNA was then fragmented and copied into first strand cDNA using reverse transcriptase and random primers. The products were purified and enriched by PCR (15 cycles) to create the final cDNA library. The 3’ prime ends of the cDNA were adenylated and ligated to adapters, including a 6-nt barcode unique for each sample. Final libraries were checked for quality and quantity by TapeStation and qPCR using Kapa’s library quantification kit for Illumina Sequencing platforms (Kapa Biosystems,Wilmington MA). The samples were pooled, clustered on an Illumina cBot and sequenced on one lane of an Illumina HiSeq4000 flow cell, as paired-end 50 nt reads. The quality of the raw reads data for each sample (e.g. low-quality scores, over-represented sequences, inappropriate GC content) was checked using FastQC (version v0.11.3). The Tuxedo Suite software package was used for the computational analysis of the RNA sequencing 29,30. Briefly, reads were aligned to the reference genome WS220 using TopHat (version 2.0.13) and Bowtie2 (version 2.2.1.). Cufflinks/CuffDiff (version 2.1.1) was used for expression quantitation, normalization, and differential expression analysis, using reference genome WS220. For this analysis, we used parameter settings: “--multi-read-correct” to adjust expression calculations for reads that map in more than one locus, as well as “--compatible-hits-norm” and “--upper-quartile–norm” for normalization of expression values. Diagnostic plots were generated using the CummeRbund R package. Genes and transcripts were identified as being differentially expressed based on three criteria: test status = “OK”, FDR ≤0.05, and fold change ≥±1.5. The Bioconductor Package GSA was used to perform enrichment test analysis. The algorithm was modified from the original “Gene Set Enrichment Analysis” (GSEA) 31, for better power. Genesets were downloaded from sources indicated in Supplementary Table 1. All FDR corrected P-values in this result are extremely significant (FDR~0).
Western blot
Standard methods for western blotting were used for the detection of proteins from worm lysates. Briefly, 3,000–5,000 L2-staged worms were collected in 20 μl of M9 buffer and snap frozen in liquid nitrogen. Laemmli loading buffer was added to the worm pellet (1:1 volume) and the samples were boiled for 5 min, separated by SDS-PAGE and transferred to PVDF membranes. Blots were blocked for 1 hour with 5% milk in PBS and probed with anti-H3 (Abcam, ab1791; 1:2,000), anti-H3K4me3 (Abcam, ab8580; 1:1,000), anti-H3K27ac (Abcam, ab4729; 1:1000), anti-H3K27me3 (Millipore, #07–449; 1:1000), anti-ASH-2 (Abmart, X3-G5EFZ3, 1:1,000;) or anti-β-tubulin (Santa Cruz, sc-5274; 1:2,000) primary antibodies overnight at 4 °C. For the extraction of mammalian proteins, HeLa cells were treated with trypsin (Invitrogen, 25200056) washed twice with PBS and collected in lysis buffer (RIPA + 1 mM PMSF + protease inhibitor cocktail + 1 mM EDTA/0.5 ml lysis buffer per 5 × 106 cells). Samples were incubated for 45 min at 4 °C with constant agitation. Lysates were spun down (4 °C, 20 min, 12,000 rpm) and snap frozen in liquid nitrogen. Laemmli loading buffer was added to the lysates (1:1 volume) and the samples were boiled for 5 min, separated by SDS-PAGE and transferred to PVDF membranes. Blots were blocked for 1 hour with 5% milk in PBS and probed with anti-H3 (Abcam, ab1791; 1:2000), anti-H3K4me3 (Abcam, ab8580; 1:1,000), anti-ASH2L (Bethyl laboratories, polyclonal, A300–489A; 1:1,000), anti-MLL1 (Bethyl laboratories, polyclonal, A300–374A; 1:500) or anti-β-tubulin (Santa Cruz, sc-5274; 1:2,500) primary antibodies overnight at 4°C. HRP conjugated anti-rabbit (ThermoScientific, 31460) and anti-mouse (ThermoScientific, 31430) secondary antibodies were used at 1:5,000 dilution for 1 h at room temperature. Proteins were detected using Clarity ECL Western blotting substrate (Biorad) and signal was captured using a BioRad ChemiDoc Touch imaging system.
C. elegans RNAi
Escherichia coli HT115 (DE3) strains transformed with vectors expressing dsRNA of the genes of interest (ash-2, set-2, wdr-5.1, empty pL4440) were obtained from the Ahringer library (a gift from G. Csankovszki), sequence-verified and grown at 37 °C as per manufacturer’s recommendations. L1 worms obtained from synchronized populations were placed onto NGM plates containing ampicillin (100 mg/ml−1) and IPTG (0.4 mM) seeded with the respective bacteria. Worms were cultivated on either RNAi or the empty vector control bacteria for two generations.
Mammalian cell culture and H2O2 treatment
HeLa (EM-2–11ht) cells (a gift from J. Nandakumar and authenticated by STR) were cultured in DMEM (Life Technologies, 11995–065), supplemented with 10% Fetal Bovine Serum (Sigma-Aldrich, F4135) and 1% Penicillin-Streptomycin (Gibco, 15140–122) at 5% CO2. At 80% confluency, cells were washed with PBS (Life Technologies, 10010023) and treated with HBSS (Life Technologies, 14025–092) supplemented with 0.1 mM or 0.3 mM H2O2 and incubated at 37°C for 30 min.
Mammalian siRNA and heat shock treatment
8,000 cells were transfected with 4.8 pmoles of ASH2L siRNA (Dharmacon, M-019831–01-0005) or non-targeting siRNA (Dharmacon, D-001210–02-05) using Lipofectamine RNAiMax (Invitrogen, 13778–150) in OPTI-MEM I Reduced serum medium (Gibco, 31985–062). For heat shock treatment, cells were washed with PBS 72 hours after siRNA transfection and placed in HBSS. Plates were wrapped with parafilm and submerged in a pre-heated water bath at 43°C for the indicated time points. Viability was determined using the CellTiter-Glo Kit (Promega) as per manufacturer’s recommendations. Luminescence was monitored on a FLUOstar Omega microplate reader (BMG Labtech).
Histone methyltransferase activity assays
SET domains of MLL/SET family proteins (MLL1, SET1A, SET1B), RBBP5 (full length), ASH2L (full length), and WDR5 (full length) were purified as previously described 32. The purified proteins were diluted to 10 μM and incubated with 1 mM or 2 mM H2O2 at 4°C for one hour in the buffer 25 mM Tris-HCl, pH 8.0. For DTT recovery, 4 mM DTT was added after H2O2 treatment and was incubated at 4°C for 60 min. After oxidation, excess H2O2 was removed by ultrafiltration. Methyltransferase assays were performed using H3 peptides (residues 1–20) with one additional Tyr-residue at C-terminus for accurate quantification of peptides. An enzyme-coupled continuous spectrophotometric assay system was employed to monitor the time course of the reaction33,34. This assay system, which monitors the appearance of the cofactor product (SAH) at an absorbance of 515 nm (i.e., OD515) contained the following components: 25 mM Tris (pH 8.0), 320 nM AdoHcy nucleosidase, 480 nM adenine deaminase, 40 U/L xanthine oxidase, 20,000 U/L horseradish peroxidase, 4.5 mM 3,5-dicholoro-2-hydroxybenzenesulfonic acid, 0.894 mM 4-aminophena-zone, 40 μM MnCl2, 2.25 μM K4Fe(CN)6·3H2O, 200 μM S-Adenosyl-Methionine and 1 μM of the four mammalian proteins that constitute the minimal H3K4-methylating complex. All components were mixed in 30 μl volume in 384-well plate at RT, and the reaction was initiated by adding 400 μM H3 peptide substrate. The OD515 was monitored using a Synergy Neo Multi-Mode Reader (Bio-Tek) for 1 hour at 28°C. The slope of OD515 vs. time from the first 20 minutes linear range was converted into reaction rates. The relative activity for each complex without any H2O2 pretreatment was set to 1. A buffer control was used to determine the baseline.
In vitro protein oxidation and thiol trapping
Purified MLL1SET (15 μM) was treated with either 2 mM DTT or 2 mM H2O2 for 30 min at 4°C or 30°C. To stop the reaction, the H2O2-treated samples were mixed with catalase (0.5 mg/ml). To reduce reversible thiol modifications the oxidized sample were treated with 4 mM DTT for 30 min at 30°C. The reduced cysteines were blocked with 20 mM NEM prior to SDS-PAGE analysis. For reverse thiol trapping experiments, the samples were resuspended in a denaturing thiol-trapping buffer (2.3 M urea, 0.2% SDS, 10 mM EDTA, 200 mM Tris-HCl, pH 8.5) supplemented with 20 mM NEM for 30 min at 25°C. Proteins were precipitated with 10% trichloracetic acid (TCA). After centrifugation, the pellets were washed with 10% TCA and 5% TCA and re-dissolved in the denaturing thiol-trapping buffer supplemented with 4 mM DTT to reduce reversible thiol modifications. After 45 min of incubation at 30°C, all new cysteine thiols were labeled with 25 mM AMS for 5 min at 25°C. Proteins were analyzed on SDS-PAGE under non-reducing conditions and visualized using silver staining. For the mass spectrometric analysis of the cysteine-containing peptides, iodoacetamide (IAM) was used instead of AMS to label reversibly oxidized cysteines. After SDS-PAGE under non-reducing conditions and Coomassie staining, protein bands were cut out, trypsin-digested, and analyzed by nano LC-MS/MS (MS Bioworks).
Statistical analysis
The Prism software package (GraphPad Software 7) and the Microsoft Office 2010 Excel software package (Microsoft Corporation) were used to carry out statistical analyses. Information about statistical tests, P values and n numbers are provided in the respective figures and figure legends.
DATA AVAILABILITY STATEMENT
All relevant data are available and/or included with the manuscript as source data or Supplementary Information. RNA-sequencing data have been uploaded to the Gene Expression Omnibus (GEO) database with accession number GSE138502.
Extended Data
Extended Data Figure 1. In vivo read-out of endogenous redox states at different stages during C. elegans lifespan and sorting parameters of oxidized and reduced subpopulations.
(a) Microscopic analysis of the Grx1-roGFP2 ratio of individual N2jrIs2[Prpl-17::Grx1-roGFP2] worms (symbol) cultivated at 15 °C and imaged at the indicated time points. Means (bars) of every two sequential time points that are not significantly different from each other (P > 0.05) share the same letter. Data represent mean ± SEM; n, number of animals; one-way ANOVA with Tukey correction. (b) The roGFP2 ratio (405/488) was calculated using the partial profiling feature (pp) configured to analyze extinction and emission data from each 488 nm and 405 nm lasers that sequentially excited each worm. (c) A population of N2jrIs2[Prpl-17::Grx1-roGFP2] at the L2 stage separated based on their opacity (extinction) and length (time of flight) was gated as R1.
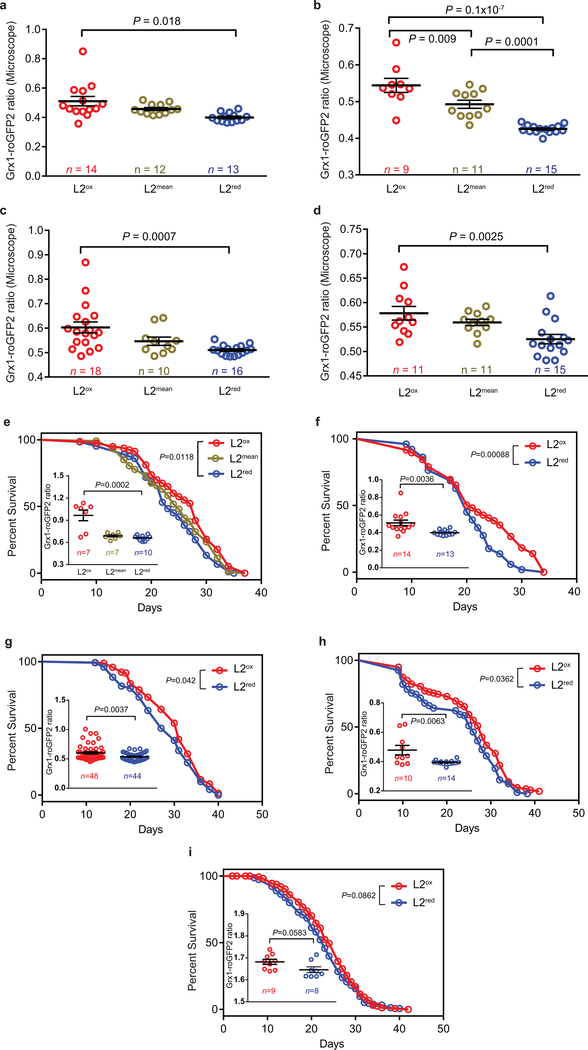
Extended Data Figure 2. Sorting efficiency and lifespan of L2ox and L2red subpopulations.
(a-d) Microscopic analysis of the Grx1-roGFP2 ratio of individual worms (symbol) previously sorted into L2ox, L2mean and L2red subpopulations. n = animals; one-way ANOVA with Tukey correction. (e-i) Survival curves of sorted L2ox, L2mean and L2red worms. For n numbers, P values (log rank test) see Extended Table 2. (Inserts) The Grx1-roGFP2 ratio of individual worms (symbol), as assessed by fluorescence microscopy after sorting is shown. n = animals. For P values (unpaired t-test, two sided) see Extended Table 2.
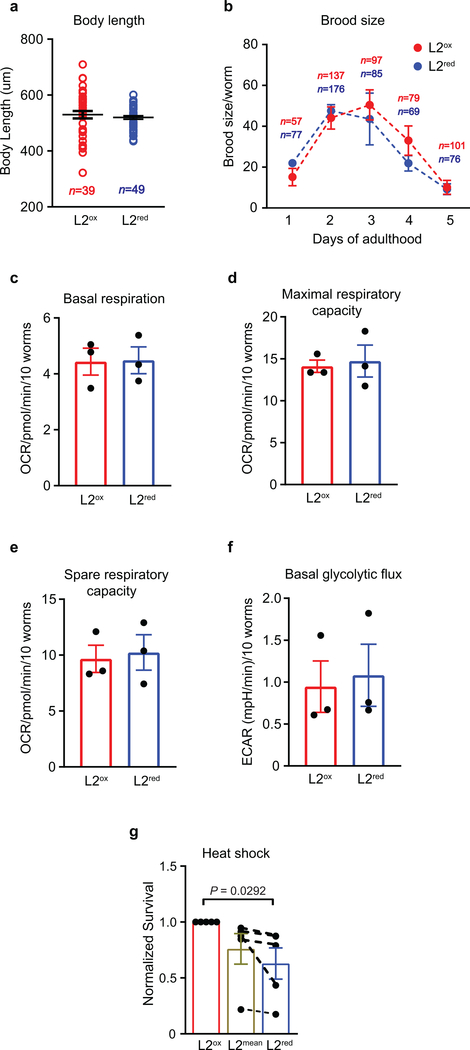
Extended Data Figure 3. Physiological properties of L2ox and L2red sorted worms.
(a) Length measurements of L2ox and L2red worms (symbol) from nose to tail tip immediately after sorting. No significant difference; P = 0.4735 (unpaired t-test, two-sided). (b) Brood size of L2ox and L2red worms, measured at the indicated time points. n = animals. No significant difference within a single age; P = 0.6532 (two – way ANOVA). (c) Basal respiration, (d) maximal and (e) spare respiratory capacity and (f) basal rates of flux through glycolysis (ECAR) of L2ox and L2red worms. n = 3 independent sorting experiments. P = 0.9469 (c), P = 0.7784 (d), P = 0.7904 (e), P = 0.7925 (f); unpaired t-test, two-sided. (g) Survival of L2ox and L2red worms 20 hours after heat shock. n = 5 independent sorting experiments; unpaired t-test, two-sided. The data points connected represent data from the same sorting experiment. The survival of L2ox is set to 1. All data represent mean ± SEM.
Extended Data Figure 4. Gene expression profiles of L2ox and L2red.
(a) Steady-state transcript levels of selected oxidative stress-related genes in L2ox and L2red worms. n, independent sorting experiments; unpaired t-test, two sided. Data represent mean ± SEM. (b) Volcano plot showing fold changes versus P values for the transcriptomes of L2ox and L2red subpopulations. Differentially expressed genes (DEGs, P ≤ 0.05, changes ≥ log2 ± 0.6) are represented by red dots (see methods for statistical definition of DEGs). Data were collected from 4 independent sorting experiments. (c) Gene Set Enrichment Analysis of the 327 DEGs. Normalized enrichment scores (see methods for calculation) are represented by the bar graph. Terms (for summary, see Supplementary Table 1) indicating origin/process/phenotype associated with genes known to play a role in the process are shown on the left. Some terms (*) have been merged and are represented as a single category bar for simplicity (for detailed values, see Supplementary Table 1). (d, e) Percentage of DEGs identified in L2ox that intersect with H3K4me3 peak signals within their 5’ region (500 bp upstream and downstream from the transcription start site). The H3K4me3 ChIP data sets were generated from L3-staged N2 worms (ChIP–chip, GEO entry: GSE30789) in (a) and ChIP-seq, GEO entry: GSE28770) in (b), indicating that these marks are set during larval development. Hypergeometric probability for (a): P = 0.064 and (b): P = 2.786 × 10-6. (c) and (d) Venn diagrams show the overlap among up-regulated (c) or down-regulated (d) gene sets in L2ox ]and down – or up – regulated set-9(rw5) and set-26(tm2467) gene sets (GEO entry: GSE100623). See Supplementary Table 1 for data sets in (a) – (d).
Extended Data Figure 5. Redox sensitivity of in vivo H3K4m3e3 levels and in vitro histone methyltransferase complex activity.
(a) Global H3K4me3 levels in the L2ox and L2red sorted worms. A representative westernblot using antibodies against H3K4me3 is shown. Quantification of global H3K27ac (b) and H3K27me3 (c) levels by westernblot. n = 3 independent sorting experiments. P = 0.3793 (b) and P = 0.0905 (c); unpaired t-test, two-sided. Data represent mean ± SEM. Global H3K4me3 (d), ASH2L (e) and MLL1 (f) levels in HeLa cells before and after H2O2 treatment, as assessed by western blot. (g) Time course of the in vitro methyltransferase reaction for core HMC (MLL1-WDR5-ASH2L-RBBP5). Reaction rates were derived from the first 20 min of the linear range. In vitro histone methyltransferase assays of core HMC members, consisting of purified GST-WDR5 (WDR5), GST-ASH2L (ASH2L), GST-RBBP5 (RBBP5) and either GST-MLL1SET or untagged MLL1SET (h), GST-SET1ASET (i) or GST-SET1BSET (j). Superscript OX indicates that the protein was pre-treated with either 1 mM (+) or 2 mM (++) H2O2 for 30 min prior to the activity assay. DTT was added after the H2O2 treatment. n = 3 independent experiments; one - way ANOVA with Sidak correction. Data represent mean ± SEM. (k) MLL1SET was treated with either 2 mM DTT, 2 mM H2O2 or 2 mM H2O2 followed by 4 mM DTT-treatment. Catalase was used to quench the H2O2. The proteins were denatured and thiols were modified with NEM prior to loading onto non-reducing SDS-PAGE to prevent non-specific thiol oxidation. The proteins were visualized by silver staining. M, marker. (l) MLL1SET treated with either 2 mM DTT, 2 mM H2O2 or 2 mM H2O2 followed by 4 mM DTT-treatment was analyzed. All reduced protein thiols were then labeled with the 500 Da thiol-reactive AMS, causing a 500-Da mass decrease per oxidized thiol detectable on reducing SDS-PAGE. (m) Cysteine oxidation state in MLL1SET after treatment with either 2 mM DTT or 2 mM H2O2 followed by NEM labeling as assessed by LC-MS/MS. The peptide containing Cys3967 could not be detected. (n) Schematic representation of the redox sensitivity of the MLL1SET. For blot and gel source images, see Supplementary Fig. 1 and 3. o) Sequence alignment of the SET-domain. All cysteines present in MLL1 are shown in bold, and the five absolutely conserved cysteines are highlighted in yellow. Cysteines shown to be involved in zinc coordination are marked with an asterisk. NCBI protein blast and Clustal Omega Multiple Sequence Alignment, Clustal O (1.2.4) were used.
Extended Data Figure 6. Effects of H3K4me3 down-regulation on heat shock response and endogenous redox state.
(a) ASH-2 and SET-2 transcript levels of N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with ash-2 or set-2 RNAi for 2 generations. n = 6 (ash-2) and n = 2 (set-2) independent experiments; unpaired t-test, two-sided. E.V., empty vector. (b) ASH-2 protein levels in N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with control RNAi or ash-2 RNAi for 2 generations using westernblot analysis. (c) H3K4me3 levels in N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with control RNAi, ash-2 RNAi or set-2 RNAi for 2 generations. (d) Transcript levels of selected heat shock genes after heat shock treatment of N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with the indicated RNAi. n = 3 independent experiments; one-way ANOVA with Bonferroni correction. (e) Transcript levels of selected heat shock genes in set-2 or wdr-5.1 mutants before and after heat shock treatment. n = 3 independent experiments; one-way ANOVA with Bonferroni correction. (f) ASH2L levels following ash2L siRNA treatment of HeLa cells. n = 2 independent experiments. (g) Grx1-roGFP2 ratios of L2 larval worms treated with ash-2 RNAi, set-2 RNAi, wdr-5.1 RNAi or the empty vector were measured using the BioSorter. n = 4 (ash-2, set-2) and n = 3 (wdr-5.1) independent sorting experiments; unpaired t-test, two-sided. (h) Representative survival curves of N2jrIs2[Prpl-17::Grx1-roGFP2] worms treated with ash-2 or set-2 RNAi for 2 generations, and treated with 1 mM PQ for 10 hours at the L2 larval stage. For n numbers, repetitions and statistics (log-rank), see Extended Data Table 4. Data in (a), (d), (e-g) represent mean ± SEM. For blot source images, see Supplementary Fig. 1 and 3.
Extended Data Table 1. Lifespan assays of L2ox, L2mean and L2red subpopulations following heat shock treatment or in the continuous presence of paraquat or juglone.
Worms were sorted onto NGM plates and exposed to heat shock treatment or the indicated compounds. P values for Kaplan Meier survival analysis were calculated based on the log-rank (Mantel-Cox) method. Samples were compared to L2ox subpopulations. Maximum lifespan was defined as the 10% of last survival population.
| Condition | Experiment | Sorted population | Mean | Max | Dead/censored worms | P value | % change | Figure on text | Ratio BioSorter‡ | P value (BioS.) |
|---|---|---|---|---|---|---|---|---|---|---|
| Heat shock (long term) | 1 | L2ox | 19 | 32.5 | 24 | Figure 2a | 7.44±0.01(n=238) | |||
| L2mean | 17 | 31.5 | 19/4 | 0.639 | 10.5%* | 7.37±0.02(n=135) | 0.002 | |||
| L2red | 13 | 27.3 | 32/1 | 0.0088 | 31.6%* | 7.29±0.02(n=132) | 0.2×10−8 | |||
| 2 | L2ox | 17.5 | 25 | 29/13 | 7.06±0.01(n=698) | |||||
| L2red | 12 | 21 | 20/3 | 0.0077 | 31.4%* | 7.01±0.012(n=319) | 0.002 | |||
| 3 | L2ox | 18 | 25.9 | 96/2 | 7.15±0.007(n=893) | |||||
| L2red | 15 | 24.2 | 77/1 | 0.0153 | 16.7%* | 7.12±0.012(n=914) | 0.009 | |||
| Paraquat (2 mM) | 1 | L2ox | 17 | 26 | 13 | Figure 2b | 7.09±0.02(n=116) | |||
| L2mean | 14 | 26 | 11 | 0.4456 | 17.6%* | 6.99±0.033(n=113) | 0.006 | |||
| L2red | 10 | 21 | 20 | 0.0215 | 41.2%* | 6.67±0.045(n=108) | 0.2×10−13 | |||
| 2 | L2ox | 13 | 18.5 | 38 | 7.35±0.017(n=158) | |||||
| L2red | 11 | 18.5 | 40 | 0.0456 | 15.4%* | 7.14±0.011(n=277) | 0.1×10−19 | |||
| 3 | L2ox | 13 | 18.6 | 47 | 6.71±0.02(n=139) | |||||
| L2red | 12 | 15.5 | 38 | 0.0037 | 16.7%† | 6.33±0.06(n=100) | 0.8×10−7 | |||
| Juglone (250 uM) | 1 | L2ox | 23 | 32.4 | 72 | 6.59±0.012(n=428) | ||||
| L2red | 19 | 32.4 | 73 | 0.0033 | 17.4%* | 6.55±0.012(n=513) | 0.007 | |||
| 2 | L2ox | 18 | 33.3 | 86 | 7.43±0.01(n=186) | |||||
| L2red | 18 | 29.5 | 59 | 0.0202 | 11,4%† | 7.27±0.015(n=161) | 0.1×10−16 | |||
percentage change based on mean lifespan.
percentage change based on max lifespan.
Grx1-roGFP2 ratio of L2ox and L2red subpopulations, based on BioSorter analysis during sorting. Data represent mean ± SEM. n, number of worms. Unpaired t-test, two sided.
Extended Data Table 2. Lifespan assays of L2ox, L2mean and L2red subpopulations.
Worms were sorted onto NGM plates and assessed for lifespan. P values for Kaplan Meier survival analysis were calculated based on the log-rank (Mantel-Cox) method unless noted otherwise. Samples were compared to L2ox. Maximum lifespan was defined as the 10% of last survival population. E.V., empty vector (control RNAi conditions for experiments in Ext. Data Table 4).
| Exp. | Sorted population | Mean | Max | Dead/censored worms | P value (survival) | % change | Figure on text | Ratio BioSorter§ | P value (BioS.) | Ratio Microscope∥ | P value (Micr.) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L2ox | 30 | 35.8 | 97 | Fig. 2c | 6.88±0.02(n=134) | 0.49±0.008(n=36) | ||||
| L2mean | 26 | 33.5 | 74 | 0.007 | 13.3%* | 6.69±0.02(n=143) | 0.1×10−8 | 0.45±0.003(n=49) | 0.1×10−6 | ||
| L2red | 25 | 32.3 | 58 | 0.0004 | 17%* | 6.62±0.04(n=131) | 0.4×10−6 | 0.44±0.003(n=50) | 0.1×10−7 | ||
| 2 | L2ox | 28 | 35.1 | 78/2 | 7.14±0.02(n=127) | 0.96±0.07(n=7) | |||||
| L2mean | 25 | 34.3 | 105/3 | 0.1018 | 10.7%* | 7.11±0.03(n=133) | 0.532 | 0.69±0.01(n=7) | 0.0027 | ||
| L2red | 23 | 33.5 | 63/1 | 0.0118 | 17.8%* | 6.94±0.07(n=136) | 0.0101 | 0.66±0.01(n=10) | 0.0002 | ||
| 3 | L2ox | 21 | 34 | 94/2 | 7.45±0.02(n=139) | 0.51±0.03(n=14) | |||||
| L2red | 20 | 30 | 51 | 0.0088 | 12%† | 7.35±0.01(n=716) | 0.0003 | 0.45±0.01(n=13) | 0.0036 | ||
| 4 | L2ox | 31 | 39.4 | 162/3 | 6.61±0.02(n=170) | 0.6±0.02(n=48) | |||||
| L2red | 28.5 | 38.8 | 120 | 0.042 | 8%* | 6.55±0.02(n=161) | 0.0456 | 0.54±0.01 (n=44) | 0.0037 | ||
| 5 | L2ox | 28 | 36 | 114/4 | 7.29±0.01(n=191) | 0.48±0.03(n=10) | |||||
| L2red | 26 | 35.4 | 106 | 0.0362 | 7%* | 7.12±0.02(n=88) | 0.3×10−9 | 0.39±0.004(n=14) | 0.0063 | ||
| 6¶ | L2ox | 24 | 34 | 164/7 | 6.19±0.02(n=275) | 1.68±0.01 (n=9) | |||||
| L2red | 23 | 34 | 155/9 | 0.0862‡ | 4%* | 6.03±0.02(n=273) | 0.5×10−7 | 1.65±0.01 (n=8) | 0.0583 | ||
| 7 | E.V.ox | 24 | 34 | 46 | Fig. 4d | 5.55±0.04(n=114) | |||||
| E.V.red | 21 | 28.3 | 63 | 0.0004 | 12.5%* | 4.49±0.067(n=114) | 0.5×10−28 | ||||
| 8 | E.V.ox | 24 | 33 | 77/3 | Fig. 4e | 6.37±0.035(n=60) | |||||
| E.V.red | 20 | 31 | 80 | 0.002 | 17%* | 5.85±0.076(n=63) | 0.1×10−7 | ||||
| 9 | E.V.ox | 20 | 33 | 85 | 6.08±0.046(n=133) | ||||||
| E.V.red | 17 | 30.7 | 77 | 0.0264 | 15%* | 5.87±0.052(n=107) | 0.0032 | ||||
| 10 | E.V.ox | 18 | 28.7 | 89/1 | 6.03±0.022(n=225) | ||||||
| E.V.red | 18 | 25.4 | 85 | 0.0077 | 11.5%† | 5.10±0.038(n=226) | 0.3×10−62 | ||||
percentage change based on mean lifespan.
percentage change based on max lifespan.
p value for survival analysis calculated based on the Gehan-Breslow-Wilcoxon method.
Grx1-roGFP2 ratio of L2ox and L2red subpopulations, based on BioSorter analysis during sorting. Data represent mean ± SEM. n, number of worms. Unpaired t-test, two sided.
Grx1-roGFP2 ratio of L2ox and L2red subpopulations based on microscopic, following sorting. Data represent mean ± SEM. n, number of worms. Unpaired t-test, two sided.
Lifespan conducted with experimenter blind to the type of worms assayed.
Extended Data Table 3. Lifespan assays of L2ox and L2red subpopulations following transient exposure to oxidizing or reducing conditions.
Worms were sorted into liquid medium, exposed to paraquat (PQ) or N-acetylcysteine (NAC) for 10 hours and then returned to NGM plates for lifespan assessment. P values for Kaplan Meier survival analysis were calculated based on the log-rank (Mantel-Cox) method unless noted otherwise. Samples were compared to L2ox subpopulations or L2mixed. Maximum lifespan was defined as the 10% of last survival population.
| Experiment | Sorted population | Mean | Max | Dead/censored worms | P value | % change | Figure on text | Ratio BioSorter§ | P value (BioS.) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | L2ox | 24 | 34.4 | 91 | Figure 2f | 7.07±0.018(n=317) | |||
| 1 mM PQ | 26 | 33.7 | 85 | 0.4364 | 7.7%* | ||||
| 10 mM NAC | 19 | 32.9 | 99 | 0.008 | 20.8%* | ||||
| L2red | 21 | 31.2 | 109 | Figure 2e | 7.±0.011(n=402) | 0.00074 | |||
| 1 mM PQ | 23 | 31.7 | 139/1 | 0.0261 | 8.7%* | ||||
| 10 mM NAC | 21 | 29.4 | 157 | 0.0546 | - | ||||
| 2 | L2ox | 16 | 23.3 | 86 | 7.09±0.007(n=800) | ||||
| L2red | 15 | 19 | 54 | 0.0002 | 18.4%† | 7.05±0.009(n=1218) | 0.00034 | ||
| L2red + 1 mM PQ | 15.5 | 25.6 | 64 | 0.0002∥ | 9%† | ||||
| 3 | L2ox | 23 | 29.8 | 89 | |||||
| 10 mM NAC | 21 | 30.4 | 50 | 0.05‡ | 8.7%* | ||||
| 4¶ | L2Mixed | 15 | 21.1 | 66 | Figure 2d | ||||
| 0.1 mM PQ | 15 | 20.6 | 107 | 0.5137 | 2.4%† | ||||
| 1 mM PQ | 15 | 27.8 | 96 | 0.0175 | 24.1%† | ||||
| 10 mM NAC | 14.5 | 19.5 | 66 | 0.3667 | 7.6%† | ||||
| 5¶ | L2Mixed | 12 | 16 | 84 | |||||
| 0.1 mM PQ | 12 | 20.7 | 101 | 0.003 | 22.7%† | ||||
| 1 mM PQ | 12 | 29.7 | 88 | 0.0023 | 46.1%† | ||||
| 10 mM NAC | 11 | 18.7 | 101 | 0.1809 | 14.4%† | ||||
percentage change based on mean lifespan.
percentage change based on max lifespan.
p value for survival analysis calculated based on the Gehan-Breslow-Wilcoxon method.
Grx1-roGFP2 ratio of L2ox and L2red subpopulations at the L2 stage, based on BioSorter analysis during sorting. Data represent mean ± SEM. n, number of worms. Unpaired t-test, two sided.
Sample compared to the reduced subpopulation.
lifespan assays performed in the lifespan machine (see methods).
Extended Data Table 4. Lifespan assays upon H3K4me3-targeting RNAi treatment.
Worms were treated with the indicated RNAi and sorted into L2ox and L2red at the F2 generation (Exp. 1–4).
| Exp. | Sorted population | Mean | Max | Dead/censored worms | P value | % change | Figure on text | Ratio BioSorter‡ | P value (BioS.) |
|---|---|---|---|---|---|---|---|---|---|
| 1& | ash-2 RNAi ox | 24 | 33.3 | 40/2 | Figure 4d | 5.00±0.043(n=95) | |||
| ash-2 RNAi red | 25 | 34 | 45/4 | 0.1373 | 4.2%* | 4.66±0.046(n=85) | 0.1×10−6 | ||
| 2& | set-2 RNAi ox | 22 | 32 | 57/1 | Figure 4e | 6.17±0.047(n=52) | |||
| set-2 RNAi red | 22 | 30 | 72/1 | 0.7739 | - | 5.64±0.03(n=224) | 0.2×10−14 | ||
| 3& | ash-2 RNAi ox | 30 | 39.1 | 70 | 6.12±0.03(n=186) | ||||
| ash-2 RNAi red | 30 | 37.1 | 79 | 0.5721 | - | 5.8±0.039(n=216) | 0.9×10−9 | ||
| set-2 RNAi ox | 20 | 34 | 76 | 6.13±0.019(n=259) | |||||
| set-2 RNAi red | 20 | 33 | 55 | 0.7339 | - | 5.67±0.026(n=295) | 0.1×10−38 | ||
| 4& | ash-2 RNAi ox | 25 | 30.2 | 87 | 6.17±0.021(n=223) | ||||
| ash-2 RNAi red | 25 | 30.4 | 79 | 0.9133 | - | 5.56±0.046(n=223) | 0.5×10−26 | ||
| set-2 RNAi ox | 18 | 25.7 | 76 | 6.19±0.025(n=214) | |||||
| set-2 RNAi red | 18 | 27 | 85 | 0.8589 | 4.8%† | 5.29±0.041(n=217) | 0.1×10−52 | ||
| 5 | control E.V. untr. | 21 | 30 | 78 | Ext. Data Fig. 6h | ||||
| control E.V. PQ | 21 | 31.7 | 64 | 0.5073 | - | ||||
| ash-2 RNAi untr. | 23 | 30.4 | 70 | ||||||
| ash-2 RNAi PQ | 19 | 27.6 | 52 | 0.0017 | 17.4%* | ||||
| set-2 RNAi untr. | 21 | 31.3 | 59 | ||||||
| set-2 RNAi PQ | 21 | 32.1 | 67 | 0.4515 | - | ||||
| 6 | control E.V. untr. | 20 | 29.5 | 58 | |||||
| control E.V. PQ | 18 | 32.3 | 43 | 0.7952 | 10%* | ||||
| ash-2 RNAi untr. | 23 | 31.4 | 46 | ||||||
| ash-2 RNAi PQ | 17 | 32.3 | 30 | 0.3873 | 26%* | ||||
| set-2 RNAi untr. | 20 | 28.8 | 47 | ||||||
| set-2 RNAi PQ | 23 | 28.7 | 39 | 0.3225 | 13%* | ||||
The empty-vector controls for experiments 1–4 correspond are found in Extended Data Table 1, experiments 7–10. Worms were treated with 1 mM PQ for 10 h at the L2 stage following RNAi treatment with ash-2 or set-2 (Exp. 5 and 6). P values for Kaplan Meier survival analysis were calculated based on the log-rank (Mantel-Cox) method. Samples were compared to the respective L2ox or untreated control. Maximum lifespan was defined as the 10% of last survival population.
percentage change based on mean lifespan.
percentage change based on max lifespan.
Grx1-roGFP2 ratio of L2ox and L2red subpopulations, based on BioSorter analysis during sorting. Data represent mean ± SEM. n, number of worms. Unpaired t-test, two sided.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to M. Malinouski and T. Mullins from Union Biometrica for their invaluable help and assistance with the reconfiguration of the Biosorter. We thank G. Csankovszki for antibodies, C. elegans RNAi feeding clones and comments, B. Braeckman for the N2jrIs2[Prpl-17::Grx-1-roGFP2] strain, J. Nandakumar for HeLa (EM-2–11ht) cells, the Caenorhabditis Genetics Center (funded by National Institutes of Health Infrastructure Program P40 OD010440) for C. elegans strains, the DNA Sequencing Core (BRCF), R. Tagett, W. Wu and the Bioinformatics Core of University of Michigan for RNA sequencing, K. Wan for protein purification and Jakob laboratory members for comments on the manuscript. We also thank R. Sawarkar and J. Labbadia for important suggestions, and J. Bardwell for critically reading the manuscript. Mass spectrometry was performed by MS Bioworks. This work was supported by NIH grants GM122506 and AG046799 as well as the Priority Program SPP 1710 of the Deutsche Forschungsgemeinschaft (Schw823/3–2) to U.J., a NIH T32 Career Training in the Biology of Aging grant to D.B., a NIH T32 Career Training in the Biology of Aging grant and a Bright Focus ADR Fellowship (A2019250F) to B.J.O., and the National Natural Science Foundation of China (31470737) to Y.C.
Footnotes
The authors declare no competing financial interests.
SUPPLEMENTARY INFORMATION
See SI guide
REFERENCES
- 1.Finch CE & Tanzi RE Genetics of aging. Science (New York, N.Y 278, 407–411 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Rea SL, Wu D, Cypser JR, Vaupel JW & Johnson TE A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nature genetics 37, 894–898 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmstrom KM & Finkel T Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature reviews 15, 411–421 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Schulz TJ et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell metabolism 6, 280–293 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Dillin A et al. Rates of behavior and aging specified by mitochondrial function during development. Science (New York, N.Y 298, 2398–2401 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Ristow M & Schmeisser S Extending life span by increasing oxidative stress. Free radical biology & medicine 51, 327–336 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Knoefler D et al. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Molecular cell 47, 767–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutscher M et al. Real-time imaging of the intracellular glutathione redox potential. Nature methods 5, 553–559 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Labbadia J & Morimoto RI Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Molecular cell 59, 639–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greer EL et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466, 383–387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shilatifard A The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annual review of biochemistry 81, 65–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guenther MG, Levine SS, Boyer LA, Jaenisch R & Young RA A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Back P et al. Exploring real-time in vivo redox biology of developing and aging Caenorhabditis elegans. Free radical biology & medicine 52, 850–859 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Wang W et al. SET-9 and SET-26 are H3K4me3 readers and play critical roles in germline development and longevity. eLife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou Y et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature structural & molecular biology 13, 713–719 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Southall SM, Wong PS, Odho Z, Roe SM & Wilson JR Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Molecular cell 33, 181–191 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Patel A, Dharmarajan V, Vought VE & Cosgrove MS On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. The Journal of biological chemistry 284, 24242–24256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T & Kelly WG A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet 7, e1001349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leichert LI et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proceedings of the National Academy of Sciences of the United States of America 105, 8197–8202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenyon CJ The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Yang W & Hekimi S A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS biology 8, e1000556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiner A et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS biology 10, e1001369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen M, Flatt T & Aguilaniu H Reproduction, fat metabolism, and life span: what is the connection? Cell metabolism 17, 10–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han S et al. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544, 185–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Sadighi Akha AA, Miller RA & Harper JM Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. The journals of gerontology. Series A, Biological sciences and medical sciences 64, 711–722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 26.Brenner S The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroustrup N et al. The Caenorhabditis elegans Lifespan Machine. Nature methods 10, 665–670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koopman M et al. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nature protocols 11, 1798–1816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Pachter L & Salzberg SL TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng SB et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature genetics 42, 790–793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorgan KM et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Analytical biochemistry 350, 249–255 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Southall SM, Cronin NB & Wilson JR A novel route to product specificity in the Suv4–20 family of histone H4K20 methyltransferases. Nucleic acids research 42, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available and/or included with the manuscript as source data or Supplementary Information. RNA-sequencing data have been uploaded to the Gene Expression Omnibus (GEO) database with accession number GSE138502.