Abstract
The genus Flavivirus in the family Flaviviridae comprises many medically important viruses, such as dengue virus (DENV), Zika virus (ZIKV), and yellow fever virus. The quest for therapeutic targets to combat flavivirus infections requires a better understanding of the kinetics of virus–host interactions during infections with native viral strains. However, this is precluded by limitations of current cell-based systems for monitoring flavivirus infection in living cells. In the present study, we report the construction of fluorescence-activatable sensors to detect the activities of flavivirus NS2B–NS3 serine proteases in living cells. The system consists of GFP-based reporters that become fluorescent upon cleavage by recombinant DENV-2/ZIKV proteases in vitro. A version of this sensor containing the flavivirus internal NS3 cleavage site linker reported the highest fluorescence activation in stably transduced mammalian cells upon DENV-2/ZIKV infection. Moreover, the onset of fluorescence correlated with viral protease activity. A far-red version of this flavivirus sensor had the best signal-to-noise ratio in a fluorescent Dulbecco's plaque assay, leading to the construction of a multireporter platform combining the flavivirus sensor with reporter dyes for detection of chromatin condensation and cell death, enabling studies of viral plaque formation with single-cell resolution. Finally, the application of this platform enabled the study of cell-population kinetics of infection and cell death by DENV-2, ZIKV, and yellow fever virus. We anticipate that future studies of viral infection kinetics with this reporter system will enable basic investigations of virus–host interactions and facilitate future applications in antiviral drug research to manage flavivirus infections.
Keywords: cell death, dengue virus (DENV), flavivirus, fluorescence, protease, cell-based reporter, internal cleavage, NS2B–NS3, plaque assay, Zika virus (ZIKV)
Introduction
The genus Flavivirus in the family Flaviviridae comprises more than 70 species of arthropod-borne viruses (arboviruses) that are transmitted to vertebrates by infected mosquitoes or ticks, producing diseases in animals and humans, including many medically important viruses like West Nile virus (WNV),2 yellow fever virus (YFV), St. Louis encephalitis virus, dengue virus (DENV), Japanese encephalitis virus (JEV), Zika virus (ZIKV), and tick-borne encephalitis virus (TBEV) (1).
The genome of flaviviruses is a positive sense RNA of ∼11 kb that encodes three structural proteins, i.e. capsid (C), membrane precursor (prM), and envelope (E), and seven nonstructural proteins, i.e. NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. These proteins initially form a precursor polyprotein (NH2-C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-COOH) that is cleaved by both cellular and viral proteases to release the mature viral proteins (2). The flavivirus serine protease NS2B–NS3 consists of the N-terminal domain of the NS3 protein associated with the membrane-resident NS2B cofactor to form an active complex. This viral protease cleaves the precursor polyprotein at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, and NS4B/NS5 junctions, as well as at internal sites within C, NS2A, NS3, and NS4A (3–5).
Flaviviruses have continued to emerge in recent years, and together represent a global threat responsible for pandemics associated with encephalitis and hemorrhagic fever diseases for which there are no specific treatments available other than supportive care upon hospitalization (2). Moreover, the development of successful human vaccines seems to be challenging for some flaviviruses. Although YFV, JEV, and TBEV vaccines are highly effective, the development of vaccines for other flaviviruses like WNV and DENV have presented some drawbacks and safety concerns (6–8) This situation partially arises from the limitations of clinical studies, and although there are established animal models for flaviviruses, they do not faithfully reproduce all the clinical manifestations observed in the human host (9, 10). Therefore, post-mortem studies and cell culture models are still an important approach to study flavivirus diseases (11–13), especially for the quest of novel therapeutic targets to combat these infections, either on the virus or on the host (14, 15).
Currently, the identification of flavivirus-infected cells relies on either immunostaining of viral proteins (12), the application of recombinant reporter replicons or viral genomes (16–20), or the use of cell-based molecular reporters of the NS2B–NS3 activity (21–23). Antibody staining techniques require both fixation and permeabilization because of the lack of flavivirus expressed proteins directly on the cell surface of infected cells as a part of the viral replication cycle (2, 24, 25), which precludes their application for live-cell imaging. Reporter replicons and viral genomes allow kinetic studies in living cells but are limited to molecular clones and thus not suitable to study clinical isolates or native virus strains. In this respect, genetically encoded molecular reporters monitoring the flavivirus NS2B–NS3 proteolytic activity upon infection are an advantageous approach that is suitable for live-cell imaging studies of native flavivirus strains.
Previously, we developed a series of caspase-activatable reporters by fusing, via a linker containing the caspase-3/7 cleavage site DEVD, a hydrophobic quenching peptide to the C terminus of a fluorescent protein (26–28). This quenching peptide inhibits the maturation of the chromophore in the fluorescent protein until it is proteolytically removed by an active caspase, fully restoring the fluorescence (26, 27). In the present study, we developed genetically encoded flavivirus molecular reporters by inserting a flaviviral NS2B–NS3 cleavage site into our caspase-activatable (CA) GFP (26) or CA-mNeptune (28), giving rise to the flavivirus-activatable (FlaviA) GFP and FlaviA-mNeptune reporters, respectively. To our knowledge, this is the first fluorescence-activatable molecular reporter system for live-cell imaging of the infection by both reference and native strains of flaviviruses like DENV, ZIKV, and YFV.
Results
Fluorescence-activatable GFP-based reporters of flavivirus NS2B–NS3 protease activity become fluorescent upon cleavage by recombinant DENV-2/ZIKV proteases in vitro
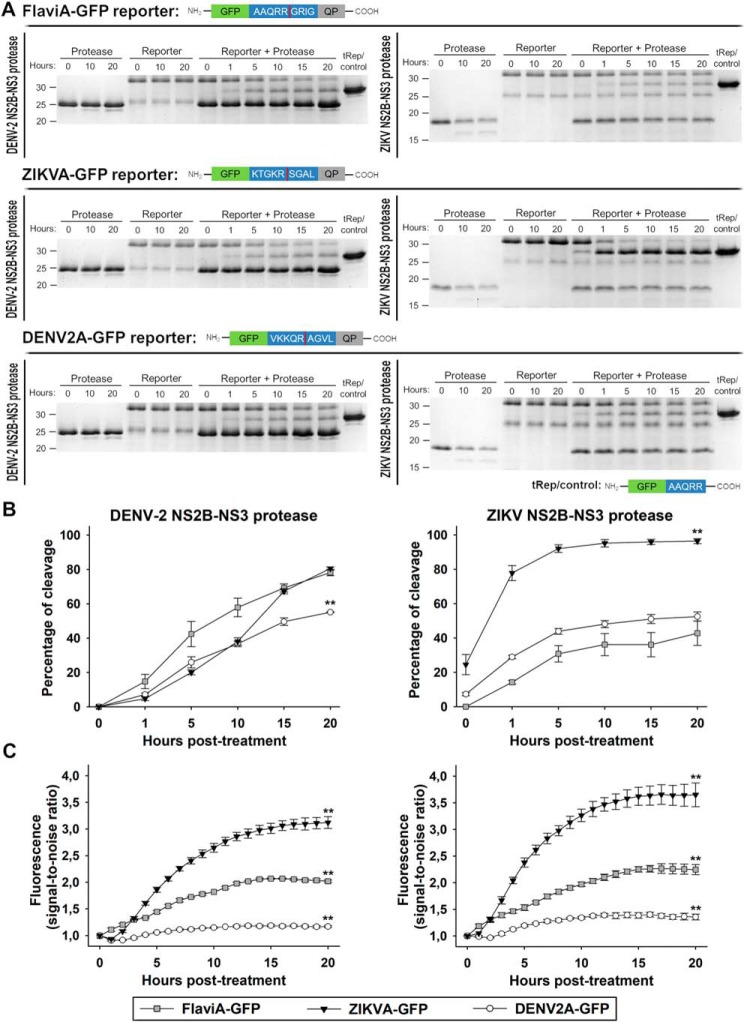
We based the design of a molecular sensor for flavivirus proteases on our previously reported CA-GFP sensor that comprises GFP, a linker for caspase cleavage and a C-terminal quenching peptide (26–28). However, we encountered several limitations for the development of the new sensor, mainly with the linker sequence for the reporter function. This led us to envisage several alternative designs by changing the linker sequence. Indeed, we generated several variants of the reporter that remained uncleaved and/or nonfluorescent upon DENV-2 NS2B–NS3 protease treatment in vitro (Table S1). Therefore, we designed a linker based on previously characterized flavivirus polyprotein cleavage sites (29). After careful analysis and avoiding the formation of cleavage sites for other cellular proteases within the resulting protein sequence of the sensor (http://web.expasy.org/peptide_cutter/),3 we selected the cleavage sequences that define the linker. Three variants of this reporter were constructed by changing the linker sequence: ZIKVA-GFP (ZIKV polyprotein NS2B/NS3 cleavage site linker), DENV2A-GFP (DENV-2 polyprotein NS2B/NS3 cleavage site linker), and FlaviA-GFP with the internal NS3 cleavage site present in many members of the Flavivirus genus (3, 5, 30). Using these variants of the reporter, we verified the cleavage in vitro by Coomassie Blue–stained SDS-PAGE gels (Fig. 1, A and B, and Fig. S1) and the fluorescence activation (Fig. 1C) to evaluate at the protein level the potential of these linkers to be used within reporters of viral protease activity.
Figure 1.
Fluorescence-activatable GFP-based reporters for flavivirus NS2B–NS3 protease activity become fluorescent upon cleavage by DENV-2/ZIKV recombinant proteases in vitro. The flavivirus-activatable GFP reporters contain a quenching peptide (QP) at the C terminus of GFP joined by a linker consisting of a cleavage site for the flavivirus NS2B–NS3 proteases. When the viral proteases cleave the linker, the quenching peptide is removed, and the GFP adopts a conformation promoting chromophore maturation. Three variants of this reporter were developed by changing the linker sequence: ZIKVA-GFP (ZIKV polyprotein NS2B/NS3 cleavage site linker), DENV2A-GFP (DENV-2 polyprotein NS2B/NS3 cleavage site linker), and FlaviA-GFP with the internal NS3 cleavage site linker, which is present in many members of the Flavivirus genus. A, in vitro cleavage kinetics of the flavivirus-activatable GFP reporter. Purified reporter proteins were mixed with purified DENV-2 NS2B–NS3 protease (left panels) or ZIKV NS2B–NS3 protease (right panels) at a molar ratio of 1:1 and incubated for given times. The reactions were quenched by thermal treatment in SDS loading buffer, and samples were analyzed by SDS-PAGE and staining of the gels with Coomassie Blue. tRep/control is an engineered cleaved variant of the FlaviA-GFP protein and was used as size marker of cleaved reporters. Representative cropped images from three independent experiments are shown. B, cleavage efficiency kinetics of the purified flavivirus-activatable GFP reporter proteins treated with purified DENV-2 NS2B–NS3 protease (left panel) and ZIKV NS2B–NS3 protease (right panel). C, time-resolved fluorescence signal-to-noise ratio of the purified flavivirus-activatable GFP reporter proteins treated with purified DENV-2 NS2B–NS3 protease (left panel) and ZIKV NS2B–NS3 protease (right panel). The data are expressed as means ± S.D. of three independent experiments. **, p < 0.001 compared with the other two reporter variants at 20 h post-treatment.
Purified recombinant DENV-2 NS2B–NS3 protease (Fig. 1A, left panels) or ZIKV NS2B–NS3 protease (Fig. 1A, right panels) were added to the three purified FlaviA-GFP reporter proteins. The DENV-2 NS2B–NS3 protease band was observed at 25 kDa, and the ZIKV NS2B–NS3 protease was located below 20 kDa, whereas all three full-length reporter proteins appeared above 30 kDa. To determine the location of the cleaved reporters, we generated a truncated variant of the FlaviA-GFP reporter protein (tRep/control) by inserting a stop codon downstream of the cleavage site in the DNA sequence of the linker. The bands of the cleaved reporters appeared between the 25- and 30-kDa markers. The cleavage kinetics of the reporters can be observed over time for the three variants tested (Fig. 1, A and B).
The intensities of these bands were quantified, and a ratio of the cleaved reporter to the total amount of reporter protein for each time point was calculated. The results are displayed as time-resolved cleavage efficiency (%) to compare among the different variants of the reporter (Fig. 1B). The DENV-2 NS2B–NS3 protease has very similar cleavage kinetics for the three variants with some slight differences. Although the FlaviA-GFP reporter showed an earlier increase, the ZIKVA-GFP reporter also reached ∼80% of cleavage efficiency. The DENV2A-GFP reached only ∼50% efficiency (Fig. 1B, left panel). On the other hand, striking differences are observed for the cleavage efficiency of the three variants of the reporter by the ZIKV NS2B–NS3 protease. The ZIKVA-GFP reporter had a much earlier increase, reaching almost 100% cleavage by 10 h. In contrast, the FlaviA-GFP and the DENV2A-GFP variants reached only 40% of cleavage efficiency after 20 h (Fig. 1B, right panel). These results indicate that the reporters are sensitive to flavivirus protease cleavage as designed, although with different efficiencies and kinetics.
To determine whether these cleavage kinetics correlate with fluorescence activation of the reporters, we monitored the fluorescence signal of each reporter as a function of time and normalized it to the background signal for each construct, obtaining thereby a time-resolved signal-to-noise ratio for the fluorescence of the reporters. All three reporters showed an increased in this signal-to-noise ratio for both protease treatments, indicating that the cleavage of the constructs correlates with the fluorescence increase of the GFP. The ZIKVA-GFP reporter showed the highest increase in fluorescence for both protease treatments, followed by the FlaviA-GFP and the DENV2A-GFP. These results indicate that the increase of the signal-to-noise ratio is a sensitive marker of cleavage, especially for the ZIKVA-GFP reporter (Fig. 1C).
The FlaviA-GFP sensor reports the highest fluorescence increase in stably transduced mammalian cells upon DENV-2/ZIKV infection
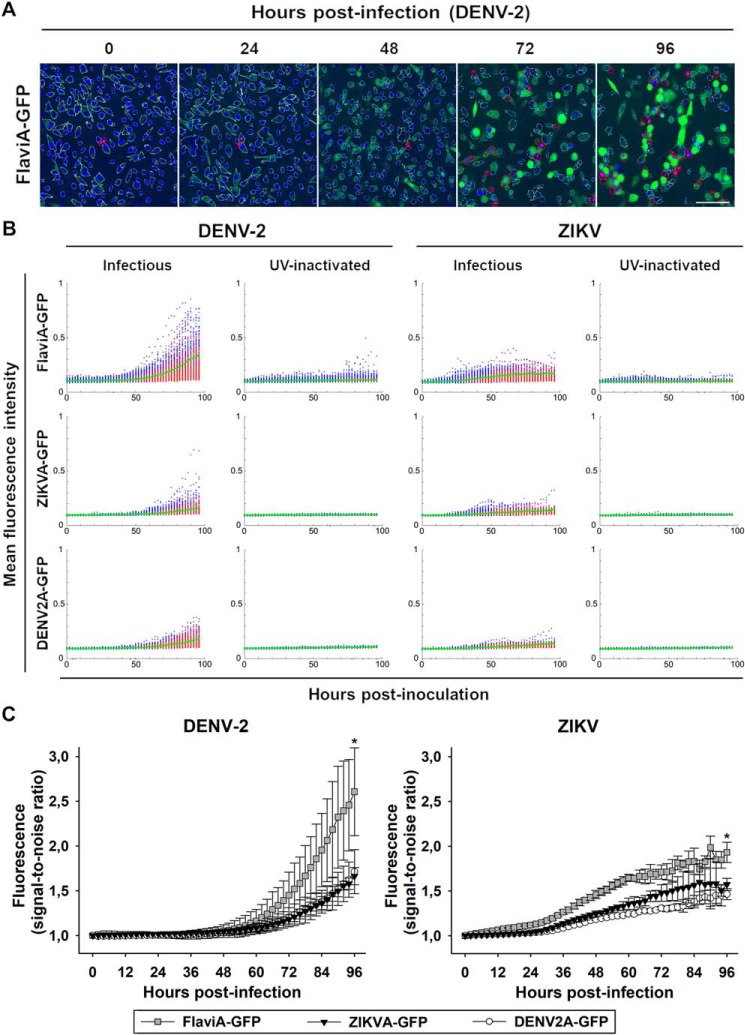
To validate our candidate GFP-based reporters of flavivirus NS2B–NS3 proteases, we generated three BHK-21 stable cell lines expressing each reporter. Upon DENV-2 or ZIKV infection at a low multiplicity of infection (MOI) of 0.25, we monitored the cellular fluorescence as a function of time using live imaging. A qualitative assessment of the images suggested that the cell fluorescence started to increase significantly at 48 h post-infection (Fig. 2A and Fig. S2A). To quantify this increase, we constructed an image analysis pipeline using CellProfiler 2.0 to identify single cells based on their nuclei, recognize their cytoplasms (white outlines), classify them as live (blue dots) or dead cells (red outlines and dots) and quantify the total cell fluorescence (Fig. 2, A and B). Our results showed that the viral-induced cytotoxicity started ∼50–60 h postinfection with DENV-2 and ∼40–50 h post–ZIKV infection (Fig. 2B, red dots). However, the cellular fluorescence started to increase in living cells (Fig. 2B, blue dots) approximately at 48 h postinfection, and we could quantify living cells with increased fluorescence until the end of this time course (96 h). The population mean values for each condition are represented by the green continuous lines. To compare the different variants of the reporter, we monitored the cell population kinetics of the flavivirus-activatable GFP sensor's fluorescence across multiple experiments and confirmed that the flavivirus internal NS3 cleavage site linker (AAQRRGRIG) confers the highest signal-to-noise ratio for reporting the infection of DENV-2 and ZIKV in stably transduced BHK-21 cells (Fig. 2C). These results confirm that our GFP-based fluorescence-activatable reporters of NS2B–NS3 protease activity can be used in living cells and that our variant harboring the flavivirus-conserved linker has the highest sensitivity to report viral infection for both DENV-2 and ZIKV at the single-cell level.
Figure 2.
The FlaviA-GFP sensor reports the highest fluorescence increase in stably transduced mammalian cells upon DENV-2/ZIKV infection. We generated three BHK-21 stable cell lines expressing the flavivirus-activatable GFP reporters, each with one of the previously tested linker sequences. After cell sorting of subpopulations with homogeneous expression of each reporter, the cells were grown and infected with either infectious or UV-inactivated DENV-2 13538/ZIKV CIET-01 at a low MOI of 0.25, for the specified time periods. A, an automated image analysis protocol was constructed in CellProfiler 2.0 for the quantification of live (white outline), dead (red outline), and activated FlaviA-GFP fluorescent cells (green). A representative experiment is shown for the FlaviA-GFP stable cell line infected with DENV-2 (n = three independent experiments, magnification of 200×; scale bar, 100 μm). B, the flavivirus-activatable GFP reporter activation is represented by scatter plots showing the time-resolved fluorescence of the population of single live (blue) and dead (red) reporter cells after the exposure to infectious or UV-inactivated DENV-2 (left panels) or ZIKV (right panels). The population mean values for each condition are represented by the green continuous lines. Representative scatter plots are shown (n = three independent experiments). C, the cell population kinetics of the flavivirus-activatable GFP sensors fluorescence across multiple experiments confirmed that the flavivirus internal NS3 cleavage site linker (AAQRRGRIG) confers the highest signal-to-noise ratio to report the infection with both DENV-2 (left panel) and ZIKV (right panel) in stably transduced BHK-21 cells. The data are expressed as means ± S.D. of three independent experiments. *, p < 0.05 compared with both ZIKVA-GFP and DENV2A-GFP at 96 h postinfection.
The fluorescence activation of the FlaviA-GFP reporter correlates with viral NS3 protease synthesis and activity in the cellular context
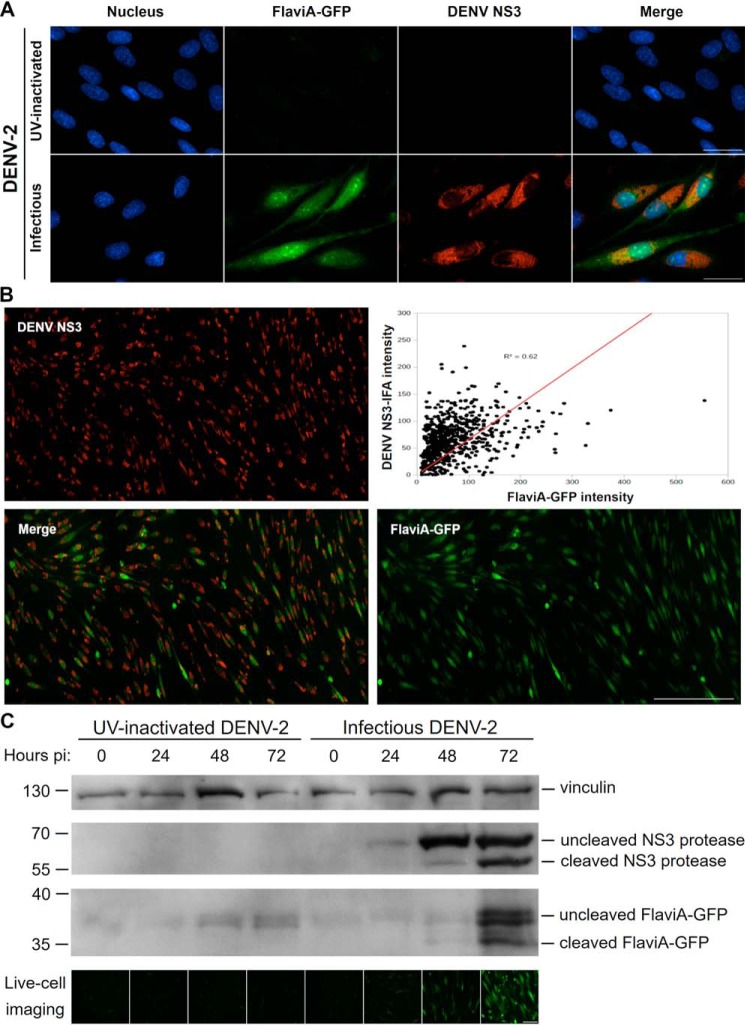
To validate the ability of the FlaviA-GFP construct to report the NS3 protease content in the cellular context, we first monitored the correlation of the reporter fluorescence with an anti-NS3 immunofluorescence staining. BHK-21 stable cells expressing the FlaviA-GFP reporter were infected with DENV-2 13538, and the NS3 protease was revealed by an immunofluorescence assay with an anti-DENV NS3 protein antibody 72 h postinoculation. High magnification images were obtained to study the cellular patterns of both fluorescent signals. The images showed a significant amount of colocalized fluorescence, suggesting a correlation between cellular NS3 amounts and the fluorescent form of the FlaviA-GFP reporter (Fig. 3A). To confirm this correlation, we obtained low magnification images and quantified the total cellular intensity for both fluorescent signals in many single cells using CellProfiler 2.0. This quantification confirmed an important correlation (Fig. 3B), suggesting that the fluorescence of the FlaviA-GFP reporter arises because of the viral-induced synthesis of NS3 protease in the cells. To confirm this hypothesis, we monitored for 72 h the cleavage kinetics of the FlaviA-GFP reporter in BHK-21 stable cells upon DENV-2 infection in parallel to the viral NS3 protease expression by Western blotting. Only the cells incubated with infectious DENV-2 showed a band of uncleaved NS3 protease. This signal was dim at 24 h but became more evident at 48 and 72 h, whereas a band corresponding to the cleaved NS3 protease became visible at 48 h postinfection, because of the autoproteolytic activity of NS3. On the other hand, the band of uncleaved FlaviA-GFP reporter was visible in lysates of cells incubated with UV-inactivated DENV-2, at all the times tested (Fig. 3C). In the presence of the infectious DENV-2 a slight band of cleaved FlaviA-GFP reporter appeared by 48 h, correlating with the onset of autoproteolytic activity of the viral NS3. This band showed a strong increase at 72 h postinfection similar to the increase of cleaved NS3 protease (Fig. 3C). This correlation was expected because both the FlaviA-GFP reporter and the viral NS3 protease contain the same flavivirus internal NS3 cleavage site (AAQRRGRIG). The cleavage of the NS3 protease itself therefore represents an excellent internal control for the NS3 protease activity in this experiment. Together, these results demonstrate that the fluorescence increase of the FlaviA-GFP reporter correlates with its proteolytic cleavage in the cellular context, and this cleavage depends on the presence of active viral NS3 protease.
Figure 3.
The FlaviA-GFP reporter becomes cleaved and fluorescent in stably transduced BHK-21 cells upon DENV-2 infection, which correlates with the viral NS3 protease synthesis and autoproteolysis. Stable BHK-21 cells expressing the FlaviA-GFP reporter were infected with either infectious or UV-inactivated DENV-2 13538 at a low MOI of 0.25, for the specified time periods postinoculation. A, comparison of DENV-2 infection detection by the FlaviA-GFP reporter (green) and immunostaining with an anti-DENV NS3 protease antibody (orange) in stably transduced BHK-21 cells at 72 h postinfection with DENV-2. Images from a representative experiment are shown (n = three independent experiments, magnification of 600×; scale bar, 30 μm). B, correlation of the total cell fluorescence intensity given by the FlaviA-GFP reporter (green) and an immunofluorescence assay (IFA) with an anti-DENV NS3 protease antibody (orange) in stably transduced BHK-21 cells at 72 h postinfection with DENV-2. A representative experiment is shown (n = three independent experiments, magnification of 200×; scale bar, 300 μm). C, cleavage and fluorescence kinetics of the FlaviA-GFP reporter in stable BHK-21 cells upon DENV-2 infection and subsequent viral NS3 protease production and activity. A representative Western blotting kinetics with its corresponding live-cell images set is shown (n = three independent experiments, magnification of 200×; scale bar, 100 μm).
The FlaviA-mNeptune, a far-red version of the flavivirus sensor, reports the best signal-to-noise ratio in a DENV-2/ZIKV fluorescent plaque assay
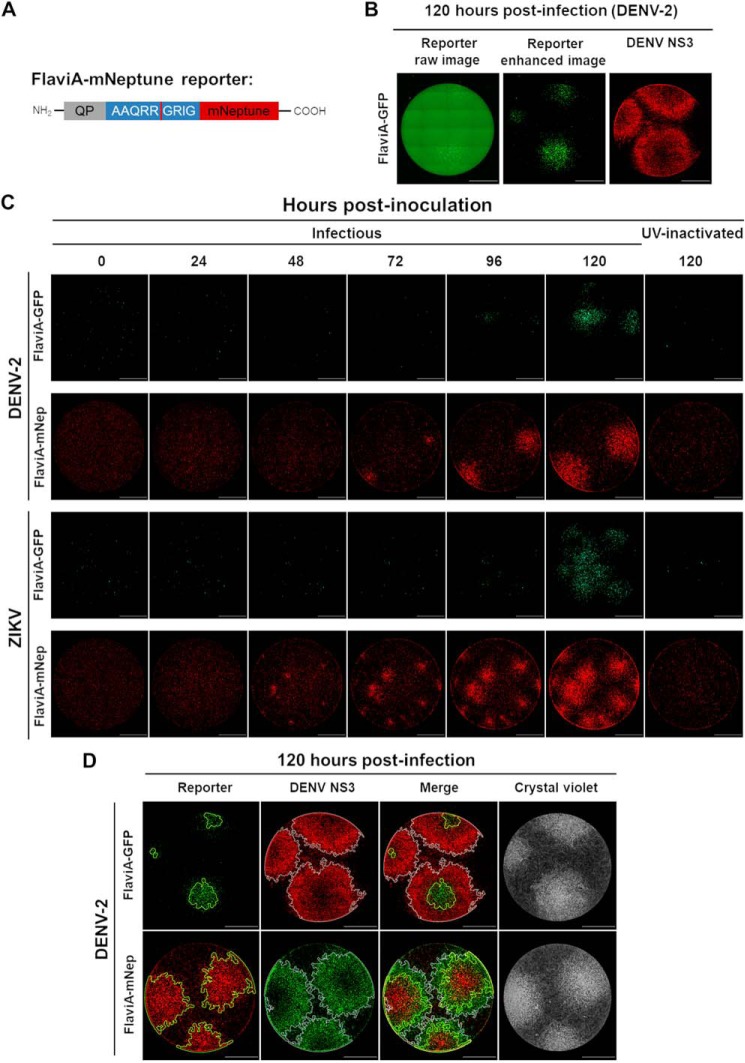
To ascertain the ability of the FlaviA-GFP version of the sensor to report the cell population-based kinetics of infection, we designed an experimental protocol for a Dulbecco's plaque assay in 96-well plates. Briefly, confluent cell monolayers of BHK-21 cells stably expressing the FlaviA-GFP reporter were infected with decimal dilutions of a DENV-2 or ZIKV viral seed and incubated with a medium containing carboxymethylcellulose for 120 h. The plaque-containing wells were completely imaged at 40× magnification, and the whole-well images of cell monolayers were generated with the stitching function of the Gen5 3.0 software (BioTek). However, as shown in Fig. 4B, the raw images of the FlaviA-GFP stable cell monolayers presented high backgrounds at low magnification in the green fluorescence channel, probably because of the autofluorescence of the phenol red and the fetal bovine serum of the culture medium. An alternative medium without phenol red (Gibco, catalog no. 11935-046) was tested but showed an increase in cytotoxicity (data not shown). These images could be enhanced by increasing the contrast but thereby lost a significant fraction of the cellular signal from the fluorescent viral plaques, as shown by the size comparison of the reporter plaques in the enhanced image to the plaques stained with an anti-NS3 antibody 120 h postinfection (Fig. 4B). Therefore, we envisaged a far-red version of our flavivirus reporter (FlaviA-mNeptune, Fig. 4A) based on our previously published CA-mNeptune sensor (28). We generated a new stable cell line with the far-red FlaviA-mNeptune reporter to compare the signal-to-noise ratio between the red fluorescent plaques and the green fluorescent plaques generated in FlaviA-GFP stable cells.
Figure 4.
The FlaviA-mNeptune is a far-red version of the flavivirus reporter that enables earlier detection of the cell infection kinetics in a DENV-2/ZIKV plaque assay. We generated two BHK-21 stable cell lines expressing either a green or a far-red version of the flavivirus reporter. Both versions were compared by plaque assay upon infection with DENV-2 13538 and ZIKV CIET-01 at the specified time periods. A, the FlaviA-mNeptune reporter is a quenched version of the fluorescent protein mNeptune which contains a quenching peptide (QP) at the N terminus joint by a linker consisting of the internal NS3 cleavage site (AAQRRGRIG), which is conserved among many members of the Flavivirus genus. When the flavivirus protease cleaves the linker, the quenching peptide is removed, and the mNeptune adopts its fluorescent structural conformation. B, BHK-21 cells stably transduced with the FlaviA-GFP reporter and infected with DENV-2 required a contrast enhancement procedure to reveal viral plaques at 120 h postinfection but with a poor correlation when compared with the same plaques labeled with an anti-DENV NS3 protease antibody. C, the plaque assay kinetics showed that BHK-21 cells stably transduced with the FlaviA-mNeptune reporter accumulate enough intensity to reveal fluorescent plaques by 48 and 72 h postinfection with ZIKV and DENV-2, respectively, much earlier than their counterparts transduced with the FlaviA-GFP reporter. D, the performance of both GFP and mNeptune reporters was further evaluated by comparing the size of their resulting fluorescent plaques to the signal reported by an anti-DENV NS3 protease antibody and crystal violet staining, confirming that the FlaviA-mNeptune reporter has a better correlation to both the infection front (DENV NS3 immunostaining) and the cytopathic effect (crystal violet staining), compared with the FlaviA-GFP reporter. A representative experiment for each condition is shown (n = three independent experiments, magnification of 40×; scale bar, 1000 μm).
A time-based comparison of the plaque formation kinetics for both DENV-2 and ZIKV indicated that the FlaviA-mNeptune version of the reporter has a higher signal-to-noise ratio, revealing earlier evidence of red fluorescent plaques 48–72 h postinfection compared with the plaques produced by the fluorescence increase of the FlaviA-GFP reporter, which required 96–120 h to become evident (Fig. 4C). To confirm the higher potential of the FlaviA-mNeptune reporter to reveal fluorescent viral plaques, we compared the size of the plaques for both types of reporters with the respective size of the same plaques stained with both an anti-NS3 antibody and crystal violet staining. We also identified the plaque outlines for both reporters using an image analysis protocol constructed in CellProfiler 2.0 based on cell-by-cell neighbor counts and image thresholding (Fig. 5A). The protocol identified much larger plaques for FlaviA-mNeptune–infected cells compared with those of infected FlaviA-GFP cells. In addition, the size of the plaques generated in the FlaviA-mNeptune–infected cells was very similar to the size of the plaques revealed by the NS3 labeling and the crystal violet staining (Fig. 4D). Together, these results demonstrate that the mNeptune version of the flavivirus sensor reports the best signal-to-noise ratio in a Dulbecco's plaque assay, indicating that this reporter is a good marker of viral replication to study cell population-based kinetics of infection by this standard virological technique.
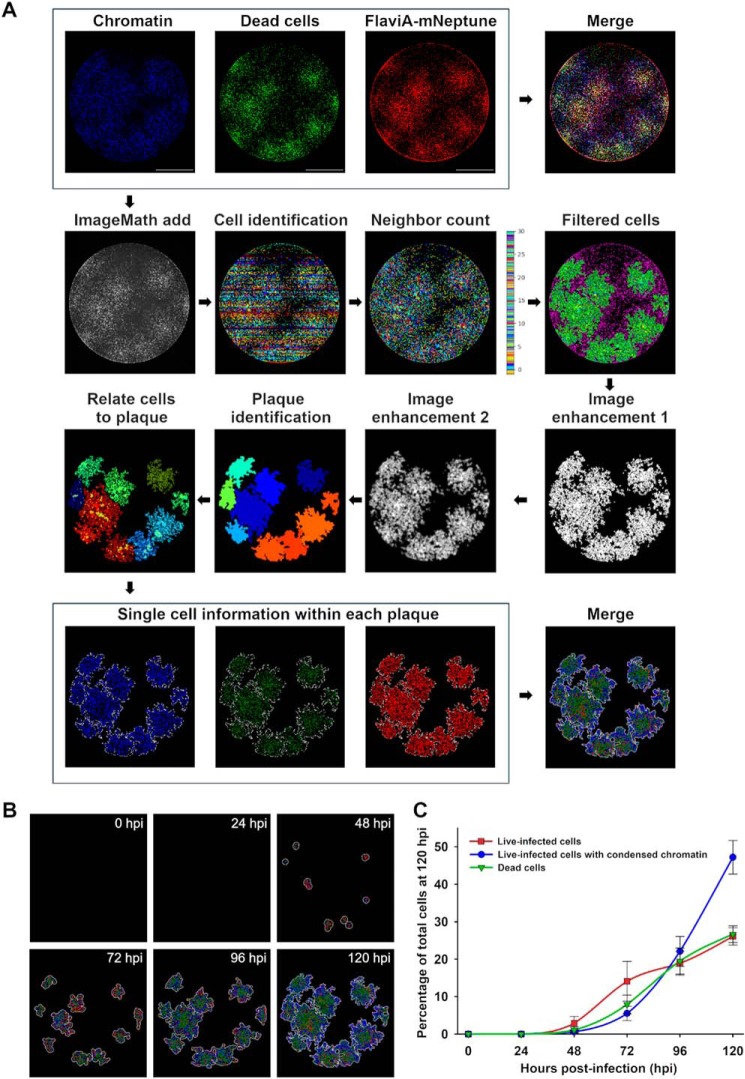
Figure 5.
The FlaviA-mNeptune reporter enables analysis of the kinetics of ZIKV infection and induced cell death in single plaques and single cells. We constructed an image analysis protocol using the software CellProfiler 2.0 to characterize the kinetics of infection and cell death by the quantification of fluorescence features of cells imaged to assess DNA/chromatin condensation (Hoechst 33342), cell death (SYTOX green), and viral infection (FlaviA-mNeptune reporter). A, high resolution images were stitched into a single image of the well for all three fluorescent channels, which intensities were added into a single image to have a redundant single-cell identification. A neighbor count and threshold were performed to filter the cells belonging to plaques and achieve single plaque identification. The single cells previously identified were related to their respective parent plaques to quantify single-cell parameters within each plaque, including chromatin condensation (blue), cell death (green), and the intensity of the FlaviA-mNeptune reporter (red). B, the image analysis protocol was applied to analyze a representative kinetics of ZIKV plaque formation during 120 h postinfection (hpi), achieving the identification of individual plaques and the categorization of single cells within each plaque as live-infected (red), live-infected with chromatin condensation (blue), and dead (green). The 120-h postinfection image was reused from point A, because its generation was used in Fig. 5A to exemplify the image analysis pipeline applied to analyze the viral plaques at every postinfection time depicted in B. C, time-resolved kinetics of ZIKV infection described by parameters of percentage of live-infected cells (red), live-infected cells with chromatin condensation (blue), and dead cells (green) within the viral plaques. Images and data from a representative analyzed experiment are shown (n = three independent experiments, magnification of 40×; scale bar, 1000 μm). The data are expressed as means ± S.D. for the number of plaques identified at each time point depicted.
A multireporter platform to study viral plaques formation at a single-cell resolution reveals differences in cell-population kinetics of infection and cell death induction by several flaviviruses
To further investigate the potential application of our FlaviA-mNeptune reporter system to study the cell population-based kinetics of viral infection and plaque formation, we designed an experimental protocol combining three types of reporters. The FlaviA-mNeptune reporter was used as a surrogate marker of viral replication, Hoechst 33342 was used to stain chromatin condensation as an early marker of an ongoing apoptosis, and SYTOX green was used to label nuclei of cells with permeabilized membranes as a late marker of cell death (Fig. 5A). We constructed an image analysis protocol in CellProfiler 2.0 to identify and characterize each individual plaque at a single-cell level. Briefly, the pipeline adds the images of the three channels into one (ImageMath) to identify all single cells with all possible combinations of the reporters (cell identification). These cells are characterized by the quantification of their neighbors within a specified distance (neighbor count) and filtered based on a minimal number of neighbors per cell (filtered cells) to identify the plaque-forming cells. Then a series of image enhancing steps enable us to perform the identification of plaques as new primary objects (plaque identification). Finally, with the identified plaques we were able to relate the single cells to their corresponding plaques and quantify thereby the number of live-infected cells (red), live-infected cells with chromatin condensation (blue), and dead cells (green) (Fig. 5A). Moreover, the time-based monitoring of these parameters for each fluorescent viral plaque enabled us to study the cell-population kinetics of infection for ZIKV plaque formation until 120 h (Fig. 5B). The quantification of these parameters indicated that the ratios of these three cell subpopulations over time are very similar among different plaques for this specific ZIKV strain, as depicted by the relatively low standard deviations observed (Fig. 5C). These results suggest that with the combination of a FlaviA-mNeptune reporter as a marker of infection with a reporter of chromatin condensation and a reporter for cell death, we can obtain a multireporter platform to characterize the infection kinetics induced by a specific viral strain.
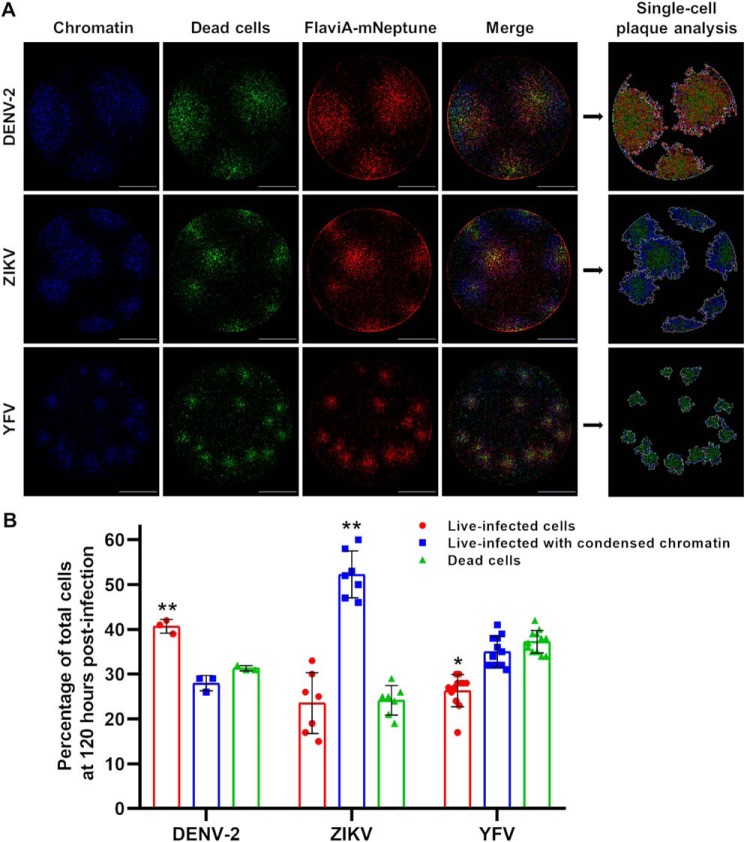
To confirm the potential of this multireporter platform to reveal virus-specific differences in terms of viral replication, chromatin condensation, and cell death, we compared the fluorescent viral plaques generated upon the infection with different flaviviruses including DENV-2, ZIKV, and YFV. YFV represents an additional model to further validate the spectrum of application of our flavivirus reporter (Figs. S2B and S3) and to explore the heterogeneity in replication kinetics and cell death induction across multiple flaviviruses using our multireporter platform. The acquired images at 120 h post-infection with DENV-2, ZIKV, and YFV show qualitative differences (Fig. 6A) that were examined using the image analysis protocol presented above (Fig. 5A). The artificial images generated by the image analysis protocol highlighted those differences (single-cell plaque analysis; Fig. 6A). The identified plaques contained several subpopulations of infected cells: a central core of dead cells (green), surrounded by a ring of cells presenting chromatin condensation (blue), and another ring of live-infected cells (red). Nevertheless, the proportions of these cell subpopulations were very different for DENV-2, ZIKV, and YFV. Finally, the quantification of the percentage of cells corresponding to each subpopulation within each plaque confirmed the qualitative observations, indicating differential proportions of infection and induced cell death among the three viral species tested (Fig. 6B).
Figure 6.
The FlaviA-mNeptune reporter enables the comparative characterization of the infection produced by flaviviruses in terms of viral replication and induced cell death at a single-cell level. We applied our image analysis protocol to characterize the DENV, ZIKV, and YFV infection and induced cell death by the quantification of fluorescence features of cells imaged to assess DNA/chromatin condensation (Hoechst 33342), cell death (SYTOX green), and viral infection (FlaviA-mNeptune reporter). A, the image analysis protocol was applied to achieve the identification of viral plaques produced by DENV, ZIKV, and YFV at 120 h postinfection and the categorization of single cells within each plaque as live-infected (red), live-infected with chromatin condensation (blue), and dead (green). B, DENV, ZIKV, and YFV infection described by parameters of percentage of live-infected cells (red), live-infected cells with chromatin condensation (blue), and dead cells (green) within the viral plaques at 120-h postinfection. The images and data from a representative analyzed experiment are shown (n = three independent experiments, magnification of 40×; scale bar, 1000 μm). The data are expressed as means ± S.D. for the number of plaques generated by each virus. *, p < 0.05; **, p < 0.001 compared with the other two cell populations conforming the plaques of the same viral species.
Next, we asked whether the presence of either type of reporters (FlaviA-GFP and FlaviA-mNeptune) and dyes (Hoechst 33342 and SYTOX green) could affect the viral replication by comparing the pfu determined with the standard crystal violet staining on WT cells with those generated on both reporter cell lines at 96 h postinfection with DENV-2, ZIKV, and YFV. A representative experiment for DENV-2 is shown in Fig. S4A, and the plaque count confirmed that there is no difference in viral replication among all three cell lines infected with all three viral species tested (Fig. S4B). This result validates that the behaviors observed for DENV-2, ZIKV, and YFV arise from intrinsic differences in viral replication kinetics and not from a differential effect of the reporters and dyes in viral replication. Together, these results demonstrate the applicability of our reporter to be used in combination with other molecular sensors to establish a multiparametric characterization of the infection produced by different flaviviruses.
Discussion
The present study reports the construction of fluorescent protein-based sensors of flavivirus NS2B–NS3 serine proteases activity that become fluorescent upon cleavage by recombinant DENV-2/ZIKV proteases in vitro (Fig. 1). Moreover, the variant of this sensor with the internal NS3 cleavage site linker (AAQRRGRIG) reported the highest fluorescence increase in stably transduced mammalian cells upon DENV-2/ZIKV infection, correlating with the viral induced NS3 protease activity in the cellular context (Figs. 2 and 3). A far-red version of this flavivirus sensor reported the best signal-to-noise ratio in a fluorescent Dulbecco's plaque assay, enabling the construction of a multireporter platform to study plaque formation with single-cell resolution (Figs. 4 and 5). Finally, the application of this platform revealed important differences in cell-population kinetics of infection and cell death induced by several flaviviruses (Fig. 6).
As a starting point, we developed this genetically encoded flavivirus reporter system by successfully engineering the linker of our previously published caspase-activatable reporters, CA-GFP and CA-mNeptune (26–28), to be recognized and cleaved by the flavivirus NS2B–NS3 proteases, giving rise to the FlaviA-GFP and FlaviA-mNeptune reporters, respectively. First, we focused on the validation of three proposed linkers in vitro as a proof of concept of the recognition and cleavage of our reporter upon treatment with flavivirus proteases, without the possible effect of other proteases that are present in the cellular context. Our results showed that all three linkers tested were cleaved as soon as 1 h post-treatment with both DENV-2 and ZIKV proteases (Fig. 1, A and B). This fast cleavage validated the amino acid sequences of our linkers as well as the application of “unlinked” versions of both recombinant DENV-2 and ZIKV proteases, in which the NS2B and NS3 regions were produced as independent polypeptides. This choice was based on our own demonstration that the “unlinked” version of DENV-2 protease rests predominantly in a “closed,” active conformation, in contrast with the less active, relaxed conformation, adopted by the frequently used “linked” construct, in which NS2B and NS3 polypeptides are attached by a nine-amino acid linker (31).
Once the biochemical principle behind the preliminary variants of our reporter was confirmed in vitro with recombinant viral proteases, we decided to test its performance upon full flavivirus particle infection in mammalian cells. The highest fluorescence increase to report the infection with DENV-2 and ZIKV in stably transduced BHK-21 cells was obtained with the internal NS3 cleavage site linker (Fig. 2) present in many members of the Flavivirus genus (3, 5, 30). Moreover, a BLASTp search of the NCBI reference proteins database (32) using the query sequence AAQRRGRIG, revealed 100% coverage and 88.9–100% identity to >60 members of the Flavivirus genus. The alignment results included many medically important flaviviruses like WNV, YFV, SEV, DENV, JEV, ZIKV, and TBEV (Fig. S5), among others (1). This finding outlines the potential application of our reporter system to other flaviviruses and supports our hypothesis that the flavivirus internal NS3 cleavage site could represent a conserved viral target for the development of a single antiviral therapy against the members of the Flavivirus genus, as previously suggested for DENV (30). Nevertheless, further work using other flaviviruses is needed to establish the whole spectrum of utility and specificity of this reporter system.
Furthermore, our flavivirus reporter system proved to be a useful method for monitoring the kinetics of DENV-2 and ZIKV infection at a single-cell level by live-cell imaging (Fig. 2) and showed a positive correlation with standard virological approaches like detection of viral NS3 protease synthesis and activity by indirect immunofluorescence and Western blotting (Fig. 3). In this respect, flavivirus reporter replicons have been generated by engineering the viral genomes of reference strains (16–20). However, in recent years special effort has been invested in the development of cell-based molecular reporter systems for live-cell imaging of flavivirus infections (21–23), mainly for their application on the study of native viral strains and clinical isolates. Among the latter, our system shows some advantages in that it is a fluorescence-activatable reporter, which allows an easier interpretation of the results and simplifies the image analysis when compared with those systems based on the relocalization of the fluorescent signal across cellular compartments (21, 23). Moreover, the utility of our fluorescence activation approach arises from its low fluorescent background and the use of a single recombinant construct that codifies for a single fluorescent protein with intramolecular quenching, in contrast with other, difficult to optimize strategies, like the FRET probes (26, 33) and the protease-triggered Cre-mediated reporter system, which rely on the cellular transfection of two different recombinant constructs (22) and could be sensitive to early changes in gene expression. Another great advantage over other systems is that our reporter has been validated with reference strains but also clinical isolates of different flavivirus species like DENV, ZIKV, and YFV, expanding its range of application to more than one flavivirus.
The fluorescence activation principle of our reporter also allowed us to envisage a cell-based fluorescent Dulbecco's plaque assay for flaviviruses (Fig. 4). The plaque assay is one of the “gold standard” virological techniques originally developed by d'Hérelle (34) for titration and isolation of single clones of bacteriophages and later adapted by Dulbecco (35) to animal viruses. Viral plaques arise after genome replication, transcription, translation, virus release from infected cells, and infection of surrounding cells (36). Thus, plaque development constitutes a hallmark of the infection carried out by a specific viral clone. To date, the kinetics of flavivirus plaque formation could only be monitored for transgenic fluorescent viruses made by modifying the genome of reference strains (16–20). However, we applied the far-red version of our flavivirus sensor to establish, for the first time, a mammalian reporter cell line where the plaque assay can be monitored over time with unlabeled native flavivirus strains. This way, we were able to study the kinetics of viral plaque development with the clinical isolate ZIKV CIET-01 (Fig. 4). Considering that viral plaques are clonal lesions of infected cells formed by the cytopathic effect of replicating viruses (36), we decided to incorporate molecular sensors for chromatin condensation (Hoechst 33342) and cell death (SYTOX green) to obtain multiparametric data from single viral plaques. This enabled us to construct a multireporter platform to study flavivirus plaque formation with single-cell resolution (Fig. 5) as a way to obtain a preliminary characterization of the infection produced by a particular flavivirus strain.
Following our approach, the quantification of the percentage of cells within each viral plaque corresponding to live-infected cells, live-infected cells with chromatin condensation, and dead cells indicated differential rates of infection and types of induced cell death for DENV-2, ZIKV, and YFV (Fig. 6). The infection with DENV-2 presents the highest proportion of live-infected cells, indicating that the infected cells take longer time to die upon infection and suggesting that this behavior drives to an increased viral production probably related to a faster infection kinetics leading to the formation of larger plaques (37). The amount of DENV-infected cells with chromatin condensation is very similar to that of dead cells, suggesting a fast type of cell demise once the cell death program is engaged, probably necrosis (38, 39). A similar scenario is shown by YFV that induces a fast type of cell death, probably necrosis, but earlier during the infection, which might limit viral spreading, leading to a lower proportion of live-infected cells and smaller viral plaques (40). On the other hand, ZIKV has a lower proportion of live-infected cells but a very high number of infected cells with chromatin condensation, suggesting that this viral strain induces a delayed type of cell death, probably apoptosis as previously reported (23, 41–43), preserving for a longer time the cell integrity to produce new viral particles and leading to larger plaques compared with YFV infection (44). However, this could be also related to differences in viral permissiveness of the BHK-21 cell line, because it is known that ZIKV virus tends to infect BHK-21 cells poorly compared with YFV, as demonstrated by envelope protein immunostaining (45, 46). Therefore, our multireporter platform serves only as a primary screening of the type of infection exposed by flavivirus strains. A more detailed exploration is needed using virological and cell biology methods to reach strong conclusions about the viral fitness and cell death mechanism induced by a specific viral strain. Nevertheless, our approach would be suitable for the screening of antiviral compounds by a plaque reduction assay, with the advantage of having a readout of the possible cytotoxicity induced by those compounds in a single experiment.
To our knowledge, this is the first fluorescence-activatable cell-based molecular reporter system for live-cell imaging of flavivirus infection suitable to be used in a plaque assay simultaneously with molecular sensors of other cellular or viral-induced processes. Moreover, taking into account the enormous need for preventative and therapeutic treatments for flavivirus infections like dengue and Zika (47), our multireporter platform enables the decoding of viral-specific fingerprints of Dulbecco-plaque formation with a potential future application for antiviral drug research and other studies on viral replication/cell death induction for both native flavivirus strains and clinical isolates.
Experimental procedures
Viruses
Clinical isolate DENV-2 13538 (DENV-2/CR/10066/2007) was provided by Instituto Costarricense de Investigación y Enseñanza en Nutrición y Salud, Cartago, Costa Rica (48). DENV-2 viruses were produced in C6/36 cells from Aedes albopictus (ATCC, Manassas, VA) by inoculating a cellular monolayer at a MOI of 0.01 and incubating for 3 days with RPMI 1640 medium (Gibco) supplemented with 2% fetal bovine serum (FBS; Gibco) at 33 °C in an atmosphere of 5% CO2. Then culture supernatant was collected and centrifuged at 3000 × g for 10 min. Before storage at −80 °C, 23% newborn calf serum (Gibco) was added (49).
Clinical isolate ZIKV CIET-01 (ZIKV/CR/CIET-01/2016) was kindly provided by Claudio Soto-Garita from Universidad de Costa Rica. Vaccine strain YFV 17D (YFV/US/17D/1937) was isolated from the commercial vaccine YF-VAX® (Sanofi Pasteur, Lyon, France). ZIKV and YFV viruses were produced in Vero cells (ATCC) by inoculating cellular monolayers at a MOI of 0.1 and incubating for 5 days with minimum essential medium (MEM, Gibco) supplemented with 2% FBS at 37 °C in an atmosphere of 5% CO2. Culture supernatants were collected, centrifuged at 3000 × g for 10 min, and stored at −80 °C.
Viruses were titrated by plaque assay in BHK-21 cells (ATCC) as previously described (50). Briefly, 10-fold serial dilutions of viruses were added to BHK-21 confluent monolayers. After 2 h of adsorption, the cells were incubated at 37 °C in an atmosphere of 5% CO2 for 5 days with MEM supplemented with 2% FBS and 1% carboxymethylcellulose (Sigma). Plaque numbers were counted after staining with crystal violet. Virus inactivation was carried out by five cycles of UV light (254 nm) exposure at an energy of 400,000 μJ/cm2 in the CL-100 UV Cross-linker (UVP, Upland, CA).
Reporter development and molecular cloning
FlaviA-GFP, ZIKVA-GFP, and DENV2A-GFP reporters were developed by site-directed mutagenesis of the linker in the previously described CA-GFP reporter (26–28) to a sequence coding for the flavivirus NS3 internal cleavage site (AAQRRGRIG), the ZIKV polyprotein NS2B/NS3 cleavage site (KTGKRSGAL), and the DENV-2 polyprotein NS2B/NS3 cleavage site (VKKQRAGVL), respectively, using the QuikChange approach (Agilent, Santa Clara, CA). The tRep/control, a truncated variant of the reporter protein, was generated by inserting a stop codon downstream the cleavage site in the linker of the FlaviA-GFP reporter. Then the genes of the flavivirus-activatable GFP reporters in the plasmid pET21b (Novagen, Madison, WI) were independently amplified by PCR and ligated into the SpeI and XhoI restriction sites of the pLenti-puro vector (a gift from Ie-Ming Shih, Addgene plasmid 39481) (51) to generate the constructs pLenti-ZIKVA-GFP-puro, pLenti-DENV2A-GFP-puro, and pLenti-FlaviA-GFP-puro. Likewise and based on the previously described CA-mNeptune reporter (28), we designed and commercially synthetized (Atum, Newark, CA) the FlaviA-mNeptune reporter gene by changing the linker sequence to codify for AAQRRGRIG. Finally, the FlaviA-mNeptune gene in the plasmid pD2109-CMV (Atum) was PCR-amplified and ligated into the XbaI and SalI restriction sites of the pLenti-CMV-GFP-Puro vector (a gift from Eric Campeau, Addgene plasmid 17448) (52) to produce the plasmid pLenti-CMV-FlaviA-mNeptune-puro. All constructs were confirmed by sequencing (Genewiz, South Plainfield, NJ).
Protein expression and purification
FlaviA-GFP, ZIKVA-GFP, DENV2A-GFP, and tRep/control expression constructs in pET21b vectors were transformed into Escherichia coli strain BL21(DE3). Flasks containing 2 liters of 2X YT media were inoculated with 8 ml of an overnight culture and grown at 37 °C to an A600 of 0.6. The cultures were then induced using 1 mm isopropyl β-d-1-thiogalactopyranoside (Sigma) and incubated at 20 °C for 5 h to allow protein expression. The cells were harvested by centrifugation and disrupted by microfluidization. Clarified lysates were prepared by centrifugation at 15,000 × g for 45 min. The reporter proteins were then purified using Co2+-affinity LC with a 5-ml HiTrap Chelating HP column (GE Healthcare). The column was washed with a buffer composed of 50 mm imidazole, 300 mm NaCl, and 50 mm NaH2PO4, pH 8.0, and the proteins were eluted using a similar buffer with 300 mm imidazole and stored at 4 °C. Proteins purity was assessed in SDS-PAGE gels stained with Coomassie Blue.
DENV-2 protease was expressed as an “unlinked” construct by the cotransformation of BL21(DE3) cells with a pACYDuet plasmid encoding for residues 48–100 of the NS2B cofactor and a pETDuet plasmid comprising amino acids 1–187 of the NS3 protease (a gift from Thomas Keller) (53). ZIKV protease was expressed as an “unlinked” construct by the transformation of BL21(DE3) cells with pET15b vector encoding for amino acids 48–100 of the NS2B cofactor and 1–178 of the NS3 protease (54). The cells harboring either DENV-2 protease or ZIKV protease were grown in 2× YT medium with antibiotics at 37 °C to an A600 of 0.6. Protein expression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside and proceeded for 3 h at 25 °C. The proteases were purified using Ni2+-affinity LC with a 5-ml HiTrap chelating HP column and eluted using a step gradient with 300 mm imidazole. The eluted proteases were further purified using anion exchange with a HiTrap Q HP column (GE Healthcare) with a linear gradient from 10 to 500 mm NaCl and stored at −80 °C. Protein purity was assessed in SDS-PAGE gels stained with Coomassie Blue.
Reporter cleavage and fluorescence assay in vitro
Samples containing 10 μm of either FlaviA-GFP, ZIKVA-GFP, or DENV2A-GFP reporters with and without 10 μm of DENV-2 or ZIKV proteases were prepared with digestion buffer (50 mm Tris buffer, pH 8.5, 0,1% CHAPS, 20% glycerol) in a final volume of 120 μl and added to a costar 96-well black plate. The fluorescence was measured every hour (excitation, 475 nm; emission, 512 nm) for 20 h at 27 °C. Another set of samples was incubated at 27 °C in 30-μl aliquots, to which SDS loading buffer was added at time points of 0, 1, 5, 10, 15, and 20 h. These samples were then run on SDS-PAGE and stained with Coomassie Blue to determine the cleavage kinetics of the flavivirus-activatable GFP reporters by DENV-2 and ZIKV proteases. Gel images were acquired with a ChemiDocTM XRS+ System (Bio-Rad) and analyzed with the ImageJ software (National Institutes of Health, Bethesda, MD) (55) to determine the cleavage efficiency. The fluorescence signal-to-noise ratio was calculated by dividing the signal of the reporter treated with viral protease by the noise gave by the untreated reporter at every time point.
Lentiviral vectors assembly
HEK293T cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% FBS, 1× GlutaMAX (Gibco), 1 mm sodium pyruvate (Gibco), and 1× antibiotic–antimycotic solution (Gibco). Lentiviral particles were generated in 60% confluent HEK293T cell monolayers by triple transfection with polyetilenimide (Polysciences, Warrington, PA) of either pLenti-FlaviA-GFP-puro, pLenti-ZIKVA-GFP-puro, pLenti-DENV2A-GFP-puro, or pLenti-CMV-FlaviA-mNeptune-puro constructs and both packaging plasmids pMD2.G and psPAX2 (a gift from Didier Trono, Addgene plasmids 12259 and 12260). At 72 h post-transfection, virus-containing medium was collected, filtered through a 0.45-μm membrane, supplemented with 5 μg/ml of Polybrene (Sigma), and stored at −80 °C. Finally, 10-fold serial dilutions of each lentiviral seed were added to HEK293T cells, and the biological titers in transducing units/ml were determined by flow cytometry at 48 h post-transduction using a BD AccuriTM C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ), as previously described (56).
Reporter cell lines production and selection
BHK-21 cell monolayers at 80% confluency were stably transduced with lentiviral particles carrying a genetic construct codifying for either FlaviA-GFP, ZIKVA-GFP, DENV2A-GFP, or FlaviA-mNeptune reporters. For cell transduction, lentiviral vector particles were added to the cells at a MOI of 1 and centrifuged for 2 h at 300 × g at 25 °C. At 72 h post-transduction, the cells were selected with 8 μg/ml of puromycin (Sigma) in MEM 10% FBS during 2 days, and then cell populations with homogeneous levels of expression of the constructs were isolated by FACS with a BD FACSJazzTM cell sorter (BD Biosciences) based on the fluorescent basal signal of the reporter proteins. The selected stable cell lines were grown and maintained in MEM supplemented with 10% FBS and 0.5 μg/ml puromycin.
Infection kinetics in reporter cell lines by live-cell imaging
BHK-21 cells stably expressing the FlaviA-GFP, the ZIKVA-GFP or the DENV2A-GFP reporters were seeded on μClear black 96-well plates (Greiner Bio-One, Kremsmünster, Austria) at a density of 15,000 cells/well with MEM supplemented with 2% FBS. After 24 h of incubation at 37 °C with 5% CO2, reporter cells were infected with either infectious or UV-inactivated DENV-2 13538 or ZIKV CIET-01 at a MOI of 0.25 and allowed to adsorb for 2 h at 37 °C. After labeling with 1 μg/ml Hoechst 33342 (Invitrogen) for 10 min, the cells were washed with 1× PBS and incubated for 96 h with FluoroBriteTM DMEM (Gibco) supplemented with 2% FBS and containing 2.5 μg/ml of propidium iodide (Invitrogen) at 37 °C in an atmosphere of 5% CO2. Images were acquired every 2 h with a Cytation 3 cell imaging multimode reader (BioTek, Winooski, VT). The fluorescence signal-to-noise ratio was calculated by dividing the signal of the reporter cells treated with infectious virus by the noise gave by the reporter cells treated with UV-inactivated virus at every time point.
Kinetic plaque assay on reporter cell lines by live-cell imaging
BHK-21 cells stably expressing the FlaviA-mNeptune reporter were seeded on μClear black 96-well plates at a density of 25,000 cells/well with MEM supplemented with 10% FBS. After 24 h of incubation at 37 °C with 5% CO2, reporter cells were infected with 10-fold serial dilutions of either infectious or UV-inactivated DENV-2 13538, ZIKV CIET-01, or YFV 17D. After 2 h of adsorption, the cells were labeled with 1 μg/ml Hoechst 33342 for 10 min, washed once with 1× PBS and incubated for 120 h at 37 °C with 5% CO2 with MEM AutoModTM (Sigma) supplemented with 2% FBS, 1% carboxymethylcellulose, and 500 nm SYTOX green (Invitrogen). Images of the whole well were acquired every 24 h with a Cytation 3 cell imaging multimode reader. Finally, at 120 h postinoculation, viral plaques were confirmed by crystal violet staining.
Indirect immunofluorescence
At 96 and 120 h postinfection with either infectious or UV-inactivated DENV-2 13538, the medium was removed, and cells were fixed for 1 h with 1% paraformaldehyde (Sigma). Later, the cells were permeabilized with 70% ethanol for 15 min and 0,001% Triton X-100 (Sigma) for 10 min at rate temperature. Then cells were incubated with a 1:800 dilution of rabbit polyclonal anti-DENV NS3 antibody (GeneTex, Irvine, CA, GTX124252) for 1 h at 37 °C, washed twice with 1× PBS, and incubated with a 1:400 dilution of goat polyclonal anti-rabbit IgG Alexa Fluor® 594–conjugated antibody (Invitrogen, A11037) for 30 min at 37 °C. After three washes with 1× PBS, FluoroBriteTM DMEM was added, and images of the whole well were acquired with a Cytation 3 cell imaging multimode reader.
Western blotting
BHK-21 cells stably expressing the FlaviA-GFP reporter were seeded on 12-well plates (Greiner Bio-One) at a density of 350,000 cells/well with MEM supplemented with 2% FBS. After 24 h of incubation at 37 °C with 5% CO2, reporter cells were infected with either infectious or UV-inactivated DENV-2 13538 at a MOI of 0.25 and allowed to adsorb for 2 h at 37 °C. Every 24 h during 96 h of incubation, images were acquired with the Cytation 3 cell imaging multimode reader, and the cells were lysed with 2% SDS solution and stored at −20 °C. Later, samples were run on a SDS-PAGE gel, transferred to a PVDF membrane (Millipore), and blotted in a single step with a mixture of 1:100 dilution of rabbit polyclonal anti-GFP antibody (Invitrogen, A-11122), 1:1000 dilution of rabbit polyclonal anti-DENV NS3 antibody, and 1:500 dilution of rabbit monoclonal anti-vinculin antibody (Invitrogen, 700062). Finally, the membrane was treated with a 1:500 dilution of goat polyclonal anti-rabbit IgG HRP-conjugated antibody (Invitrogen, G-21234) and visualized using the Super Signal West Pico Plus chemiluminescent substrate (Thermo Fisher Scientific). The images were acquired with a ChemiDocTM XRS+ system.
Image analysis and statistics
Image analysis was performed with the software CellProfiler 2.0 (http://www.cellprofiler.org; Broad Institute, Cambridge, MA).3 The data are expressed as means ± S.D. of three independent experiments. Statistical significance of the differences between mean values was determined by using one-way analysis of variance followed by a Tukey's post hoc test with the software SigmaPlot 14 (Systat Software Inc., San Jose, CA). The level of significance is denoted in each figure.
Author contributions
J. L. A.-A. and J. A. H. conceptualization; J. L. A.-A. data curation; J. L. A.-A. and R. M.-R. formal analysis; J. L. A.-A., D. J. M., and M. E. H. investigation; J. L. A.-A. and R. M.-R. writing-original draft; J. L. A.-A., J. A. H., and R. M.-R. writing-review and editing; J. L. A.-A., J. A. H. and R. M.-R. resources; J. A. H. and R. M.-R. supervision; J. A. H. and R. M.-R. funding acquisition; J. A. H. and R. M.-R. project administration; J. L. A.-A. and R. M.-R. methodology.
Supplementary Material
Acknowledgments
We thank Francisco Vega-Aguilar for technical assistance with viral strains. We thank Joseph P. Kennedy, Jr., and Derrick P. Feuerstein for support and suggestions during the experimentation. We also thank Ralf Bartenschlager (Heidelberg University) for his scientific advice and critical review of the manuscript. This work is dedicated with regard to Alina I. Arias-Barrantes and Alexander Mora-Solano.
This work was supported by Universidad de Costa Rica Project VI-803-B9–505 (to J. L. A.-A. and R. M.-R.), National Science Foundation Grant NSF CBET1511367 (to J. A. H.), and International Centre for Genetic Engineering and Biotechnology Grant CRP/CRI18-02 (to R. M.-R). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S5.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- WNV
- West Nile virus
- DENV
- dengue virus
- ZIKV
- Zika virus
- YFV
- yellow fever virus
- JEV
- Japanese encephalitis virus
- TBEV
- tick-borne encephalitis virus
- CA
- caspase-activatable
- FlaviA
- flavivirus-activatable
- MOI
- multiplicity of infection
- FBS
- fetal bovine serum
- MEM
- minimum essential medium
- DMEM
- Dulbecco's modified Eagle's medium.
References
- 1. Gould E. A., and Solomon T. (2008) Pathogenic flaviviruses. Lancet 371, 500–509 10.1016/S0140-6736(08)60238-X [DOI] [PubMed] [Google Scholar]
- 2. Lindenbach B., Murray C. L., Thiel H.-J., and Rice C. M. (2013) Flaviviridae. In Fields Virology, pp. 712–746, Lippincott Williams & Wilkins, Philadelphia, Pennsylvania [Google Scholar]
- 3. Teo K. F., and Wright P. J. (1997) Internal proteolysis of the NS3 protein specified by dengue virus 2. J. Gen. Virol. 78, 337–341 10.1099/0022-1317-78-2-337 [DOI] [PubMed] [Google Scholar]
- 4. Lindenbach B. D., and Rice C. M. (2003) Molecular biology of flaviviruses. Adv. Virus Res. 59, 23–61 10.1016/S0065-3527(03)59002-9 [DOI] [PubMed] [Google Scholar]
- 5. Bera A. K., Kuhn R. J., and Smith J. L. (2007) Functional characterization of cis and trans activity of the flavivirus NS2B–NS3 protease. J. Biol. Chem. 282, 12883–12892 10.1074/jbc.M611318200 [DOI] [PubMed] [Google Scholar]
- 6. Ishikawa T., Yamanaka A., and Konishi E. (2014) A review of successful flavivirus vaccines and the problems with those flaviviruses for which vaccines are not yet available. Vaccine 32, 1326–1337 10.1016/j.vaccine.2014.01.040 [DOI] [PubMed] [Google Scholar]
- 7. Scott L. J. (2016) Tetravalent dengue vaccine: a review in the prevention of dengue disease. Drugs 76, 1301–1312 10.1007/s40265-016-0626-8 [DOI] [PubMed] [Google Scholar]
- 8. Normile D. (2017) Safety concerns derail dengue vaccination program. Science 358, 1514–1515 10.1126/science.358.6370.1514 [DOI] [PubMed] [Google Scholar]
- 9. Chan K. W., Watanabe S., Kavishna R., Alonso S., and Vasudevan S. G. (2015) Animal models for studying dengue pathogenesis and therapy. Antiviral Res. 123, 5–14 10.1016/j.antiviral.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 10. Reynolds E. S., Hart C. E., Hermance M. E., Brining D. L., and Thangamani S. (2017) An overview of animal models for arthropod-borne viruses. Comp. Med. 67, 232–241 [PMC free article] [PubMed] [Google Scholar]
- 11. Diamond M. S., Edgil D., Roberts T. G., Lu B., and Harris E. (2000) Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74, 7814–7823 10.1128/JVI.74.17.7814-7823.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balsitis S. J., Coloma J., Castro G., Alava A., Flores D., McKerrow J. H., Beatty P. R., and Harris E. (2009) Tropism of replicating dengue virus in mice and humans defined by viral nonstructural protein 3-specific immunohistochemistry. Am. J. Trop. Med. Hyg. 80, 416–424 10.4269/ajtmh.2009.80.416 [DOI] [PubMed] [Google Scholar]
- 13. Arias-Arias J. L., Vega-Aguilar F., Corrales-Aguilar E., Hun L., Loría G. D., and Mora-Rodríguez R. (2018) Dengue virus infection of primary human smooth muscle cells. Am. J. Trop. Med. Hyg. 99, 1451–1457 10.4269/ajtmh.18-0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Acosta E. G., and Bartenschlager R. (2016) The quest for host targets to combat dengue virus infections. Curr. Opin. Virol. 20, 47–54 10.1016/j.coviro.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 15. de Wispelaere M., Lian W., Potisopon S., Li P. C., Jang J., Ficarro S. B., Clark M. J., Zhu X., Kaplan J. B., Pitts J. D., Wales T. E., Wang J., Engen J. R., Marto J. A., Gray N. S., et al. (2018) Inhibition of flaviviruses by targeting a conserved pocket on the viral envelope protein. Cell Chem. Biol. 25, 1006–1016.e8 10.1016/j.chembiol.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li S. H., Li X. F., Zhao H., Deng Y. Q., Yu X. D., Zhu S. Y., Jiang T., Ye Q., Qin E. D., and Qin C. F. (2013) Development and characterization of the replicon system of Japanese encephalitis live vaccine virus SA14-14-2. Virol. J. 10, 64 10.1186/1743-422X-10-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmid B., Rinas M., Ruggieri A., Acosta E. G., Bartenschlager M., Reuter A., Fischl W., Harder N., Bergeest J. P., Flossdorf M., Rohr K., Höfer T., and Bartenschlager R. (2015) Live cell analysis and mathematical modeling identify determinants of attenuation of dengue virus 2′-O-methylation mutant. PLoS Pathog. 11, e1005345 10.1371/journal.ppat.1005345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie X., Zou J., Shan C., Yang Y., Kum D. B., Dallmeier K., Neyts J., and Shi P. Y. (2016) Zika virus replicons for drug discovery. EBioMedicine 12, 156–160 10.1016/j.ebiom.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tamura T., Fukuhara T., Uchida T., Ono C., Mori H., Sato A., Fauzyah Y., Okamoto T., Kurosu T., Setoh Y. X., Imamura M., Tautz N., Sakoda Y., Khromykh A. A., Chayama K., et al. (2018) Characterization of recombinant Flaviviridae viruses possessing a small reporter tag. J. Virol. 92, e01582–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kümmerer B. M. (2018) Establishment and application of flavivirus replicons. Adv. Exp. Med. Biol. 1062, 165–173 10.1007/978-981-10-8727-1_12 [DOI] [PubMed] [Google Scholar]
- 21. Medin C. L., Valois S., Patkar C. G., and Rothman A. L. (2015) A plasmid-based reporter system for live cell imaging of dengue virus infected cells. J. Virol. Methods 211, 55–62 10.1016/j.jviromet.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsieh M.-S., Chen M.-Y., Hsieh C.-H., Pan C.-H., Yu G.-Y., and Chen H.-W. (2017) Detection and quantification of dengue virus using a novel biosensor system based on dengue NS3 protease activity. PLoS One 12, e0188170 10.1371/journal.pone.0188170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McFadden M. J., Mitchell-Dick A., Vazquez C., Roder A. E., Labagnara K. F., McMahon J. J., Silver D. L., and Horner S. M. (2018) A fluorescent cell-based system for imaging Zika virus infection in real-time. Viruses 10, E95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winkler G., Maxwell S. E., Ruemmler C., and Stollar V. (1989) Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 171, 302–305 10.1016/0042-6822(89)90544-8 [DOI] [PubMed] [Google Scholar]
- 25. Chung K. M., Thompson B. S., Fremont D. H., and Diamond M. S. (2007) Antibody recognition of cell surface-associated NS1 triggers Fc-receptor-mediated phagocytosis and clearance of West Nile virus-infected cells. J. Virol. 81, 9551–9555 10.1128/JVI.00879-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicholls S. B., Chu J., Abbruzzese G., Tremblay K. D., and Hardy J. A. (2011) Mechanism of a genetically encoded dark-to-bright reporter for caspase activity. J. Biol. Chem. 286, 24977–24986 10.1074/jbc.M111.221648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nicholls S. B., and Hardy J. A. (2013) Structural basis of fluorescence quenching in caspase activatable-GFP. Protein Sci. 22, 247–257 10.1002/pro.2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu P., Nicholls S. B., and Hardy J. A. (2013) A tunable, modular approach to fluorescent protease-activated reporters. Biophys. J. 104, 1605–1614 10.1016/j.bpj.2013.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shiryaev S. A., Kozlov I. A., Ratnikov B. I., Smith J. W., Lebl M., and Strongin A. Y. (2007) Cleavage preference distinguishes the two-component NS2B–NS3 serine proteinases of dengue and West Nile viruses. Biochem. J. 401, 743–752 10.1042/BJ20061136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Constant D. A., Mateo R., Nagamine C. M., and Kirkegaard K. (2018) Targeting intramolecular proteinase NS2B/3 cleavages for trans-dominant inhibition of dengue virus. Proc. Natl. Acad. Sci. U.S.A. 115, 10136–10141 10.1073/pnas.1805195115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hill M. E., Yildiz M., and Hardy J. A. (2019) Cysteine disulfide traps reveal distinct conformational ensembles in dengue virus NS2B–NS3 protease. Biochemistry 58, 776–787 10.1021/acs.biochem.8b00978 [DOI] [PubMed] [Google Scholar]
- 32. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 33. Leavesley S. J., and Rich T. C. (2016) Overcoming limitations of FRET measurements. Cytometry A. 89, 325–327 10.1002/cyto.a.22851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. d'Hérelle F. (1926) The bacteriophage and its behavior. Nature 118, 183–185 10.1038/118183a0 [DOI] [Google Scholar]
- 35. Dulbecco R. (1952) Production of plaques in monolayer tissue cultures by single particles of an animal virus. Proc. Natl. Acad. Sci. U.S.A. 38, 747–752 10.1073/pnas.38.8.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yakimovich A., Andriasyan V., Witte R., Wang I. H., Prasad V., Suomalainen M., and Greber U. F. (2015) Plaque2.0: a high-throughput analysis framework to score virus-cell transmission and clonal cell expansion. PLoS One 10, e0138760 10.1371/journal.pone.0138760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goh K. C., Tang C. K., Norton D. C., Gan E. S., Tan H. C., Sun B., Syenina A., Yousuf A., Ong X. M., Kamaraj U. S., Cheung Y. B., Gubler D. J., Davidson A., St. John A. L., Sessions O. M., et al. (2016) Molecular determinants of plaque size as an indicator of dengue virus attenuation. Sci. Rep. 6, 26100 10.1038/srep26100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghosh Roy S., Sadigh B., Datan E., Lockshin R. A., and Zakeri Z. (2014) Regulation of cell survival and death during Flavivirus infections. World J. Biol. Chem. 5, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galluzzi L., Vitale I., Aaronson S. A., Abrams J. M., Adam D., Agostinis P., Alnemri E. S., Altucci L., Amelio I., Andrews D. W., Andrews D. W., Annicchiarico-Petruzzelli M., Antonov A. V., Arama E., Baehrecke E. H., Barlev N. A., et al. (2018) Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liprandi F. (1981) Isolation of plaque variants differing in virulence from the 17D strain of yellow fever virus. J. Gen. Virol. 56, 363–370 10.1099/0022-1317-56-2-363 [DOI] [PubMed] [Google Scholar]
- 41. Liu J., Li Q., Li X., Qiu Z., Li A., Liang W., Chen H., Cai X., Chen X., Duan X., Li J., Wu W., Xu M., Mao Y., Chen H., et al. (2018) Zika virus envelope protein induces G2/M cell cycle arrest and apoptosis via an intrinsic cell death signaling pathway in neuroendocrine PC12 cells. Int. J. Biol. Sci. 14, 1099–1108 10.7150/ijbs.26400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Limonta D., Jovel J., Kumar A., Airo A. M., Hou S., Saito L., Branton W., Ka-Shu Wong G., Mason A., Power C., and Hobman T. C. (2018) Human fetal astrocytes infected with Zika virus exhibit delayed apoptosis and resistance to interferon: Implications for persistence. Viruses 10, E646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Anfasa F., Goeijenbier M., Widagdo W., Siegers J. Y., Mumtaz N., Okba N., van Riel D., Rockx B., Koopmans M. P. G., Meijers J. C. M., and Martina B. E. E. (2019) Zika virus infection induces elevation of tissue factor production and apoptosis on human umbilical vein endothelial cells. Front. Microbiol. 10, 817 10.3389/fmicb.2019.00817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kato F., Tajima S., Nakayama E., Kawai Y., Taniguchi S., Shibasaki K., Taira M., Maeki T., Lim C. K., Takasaki T., and Saijo M. (2017) Characterization of large and small-plaque variants in the Zika virus clinical isolate ZIKV/Hu/S36/Chiba/2016. Sci. Rep. 7, 16160 10.1038/s41598-017-16475-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chan J. F., Yip C. C., Tsang J. O., Tee K. M., Cai J. P., Chik K. K., Zhu Z., Chan C. C., Choi G. K., Sridhar S., Zhang A. J., Lu G., Chiu K., Lo A. C., Tsao S. W., et al. (2016) Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg. Microbes Infect. 5, e93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petrova E., Gracias S., Beauclair G., Tangy F., and Jouvenet N. (2019) Uncovering flavivirus host dependency factors through a genome-wide gain-of-function screen. Viruses 11, E68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zakaria M. K., Carletti T., and Marcello A. (2018) Cellular targets for the treatment of flavivirus infections. Front. Cell Infect. Microbiol. 8, 398 10.3389/fcimb.2018.00398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soto-Garita C., Somogyi T., Vicente-Santos A., and Corrales-Aguilar E. (2016) Molecular characterization of two major dengue outbreaks in Costa Rica. Am. J. Trop. Med. Hyg. 95, 201–205 10.4269/ajtmh.15-0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Medina F., Medina J. F., Colon C., Vergne E., Santiago G. A., and Munoz-Jordan J. L. (2012) Dengue virus: isolation, propagation, quantification, and storage. Curr. Protoc. Microbiol. chapter 15, unit 15D.2 10.1002/9780471729259.mc15d02s27 [DOI] [PubMed] [Google Scholar]
- 50. Morens D. M., Halstead S. B., Repik P. M., Putvatana R., and Raybourne N. (1985) Simplified plaque reduction neutralization assay for dengue viruses by semimicro methods in BHK-21 cells: comparison of the BHK suspension test with standard plaque reduction neutralization. J. Clin. Microbiol. 22, 250–254 10.1128/JCM.22.2.250-254.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guan B., Wang T. L., and Shih I. M. (2011) ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71, 6718–6727 10.1158/0008-5472.CAN-11-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., and Kaufman P. D. (2009) A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529 10.1371/journal.pone.0006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim Y. M., Gayen S., Kang C., Joy J., Huang Q., Chen A. S., Wee J. L., Ang M. J., Lim H. A., Hung A. W., Li R., Noble C. G., Lee L. T., Yip A., Wang Q. Y., et al. (2013) NMR analysis of a novel enzymatically active unlinked dengue NS2B–NS3 protease complex. J. Biol. Chem. 288, 12891–12900 10.1074/jbc.M112.442723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hill M. E., Kumar A., Wells J. A., Hobman T. C., Julien O., and Hardy J. A. (2018) The unique cofactor region of Zika virus NS2B–NS3 protease facilitates cleavage of key host proteins. ACS Chem. Biol. 13, 2398–2405 10.1021/acschembio.8b00508 [DOI] [PubMed] [Google Scholar]
- 55. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tiscornia G., Singer O., and Verma I. M. (2006) Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 10.1038/nprot.2006.37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.