Abstract
The immune-regulatory compound histamine is involved in the metabolism of the essential skin component hyaluronan (HA). We previously reported that histamine up-regulates the expression of HYBID (hyaluronan-binding protein involved in hyaluronan depolymerization, also called CEMIP or KIAA1199), which plays a key role in HA degradation. However, no information is available about histamine's effects on HA synthase (HAS) expression, the molecular sizes of HA species produced, and histamine receptors and their signaling pathways in skin fibroblasts. Moreover, histamine's effects on photoaged skin remain elusive. Here, we show that histamine increases HA degradation by up-regulating HYBID and down-regulating HAS2 in human skin fibroblasts in a dose- and time-dependent manner and thereby decreases the total amounts and sizes of newly produced HA. Histamine H1 blocker abrogated the histamine effects on HYBID up-regulation, HAS2 suppression, and HA degradation. Histamine H1 agonist exhibited effects on HA levels, composition, and breakdown similar to those of histamine. Of note, blockade of protein kinase Cδ or PI3K–Akt signaling abolished histamine-mediated HYBID stimulation and HAS2 suppression, respectively. Immunohistochemical experiments revealed a significant ∼2-fold increase in tryptase-positive mast cells in photoaged skin, where HYBID and HAS2 expression levels were increased and decreased, respectively, compared with photoprotected skin. These results indicate that histamine controls HA metabolism by up-regulating HYBID and down-regulating HAS2 via distinct signaling pathways downstream of histamine receptor H1. They further suggest that histamine may contribute to photoaged skin damage by skewing HA metabolism toward degradation.
Keywords: fibroblast, skin, hyaluronan, mast cell, histamine, cell signaling, catabolism, hyaluronan synthase, KIAA1199/HYBID, photoaging
Introduction
Histamine is an important mediator of many physiological and pathological processes such as gastric acid secretion, neuromodulation, cell proliferation and differentiation, inflammation, and regulation of immune function (1). Mast cells store histamine in the granules and release it upon their activation, i.e. degranulation of mast cells (1). Increased numbers of mast cells are commonly observed in various tissues under pathological conditions such as tumors and inflammation (2). One example of these conditions is photoaged skin, which is induced by chronic exposure to sun light. This is characterized clinically by wrinkle formation of the skin and histologically by mild infiltrations of inflammatory cells composed of lymphocytes, macrophages and mast cells and degenerated extracellular matrix (ECM)3 molecules such as hyaluronan (HA), collagen, and elastin (3). In photoaged skin, mast cells are often located close to fibroblasts, suggesting that fibroblasts stimulated by mast cell-derived mediators may contribute to dermal changes of the photoaged skin (4).
HA, a nonsulfated linear glycosaminoglycan composed of repeating disaccharide units of β-(1,3)-linked d-glucuronic acid and β-(1,4)-linked N-acetyl-d-glucosamine, is distributed in the dermis as an essential component of the skin and contributes to hydration of the skin and regulation of cell proliferation and migration in the dermis (5). Although HA is synthesized by three different HA synthases, i.e. HAS1, HAS2, and HAS3 (6), our previous study demonstrated that among them, HAS1 and HAS2 play a key role in HA production in normal human skin fibroblasts (7). In addition, we have disclosed that HYBID (hyaluronan-binding protein involved in hyaluronan depolymerization), alias CEMIP and KIAA1199, plays an essential role in HA degradation in skin fibroblasts independently of HYAL2 (hyaluronidase 2) and a cell-surface HA receptor CD44 (8, 9). Because HYBID was identified by microarray analysis among the genes that were up-regulated by histamine and down-regulated by transforming growth factor β1 (TGF-β1), histamine is known to enhance HA degradation by up-regulating the HYBID expression in skin fibroblasts (8, 9). However, there have been no studies about the effect of histamine on the HA synthesis by HAS1, HAS2, and/or HAS3. Histamine effects are mediated by four types of histamine receptors, i.e. HRH1, HRH2, HRH3, and HRH4, all of which belong to the G protein–coupled receptors (1). Little information is available for the histamine receptors and their downstream signaling pathways involved in the expression of HYBID and HAS genes in skin fibroblasts. We recently reported that in the photoaged skin of Japanese and Caucasian women, reduction in content and molecular size of HA in the dermis caused by HYBID overexpression and decreased expression of HAS1/2 is related to facial skin wrinkling (10, 11). However, involvement of mast cells and/or histamine in the expression of HYBID and HAS genes in photoaged skin remains elusive.
In the present study, we tested dose- and time-dependent effects of histamine on the expression of the HYBID and HAS genes and studied the newly synthesized HA species in human skin fibroblasts under stimulation with different concentrations of histamine. We also investigated the histamine receptors and their downstream signaling pathways responsible for the histamine-induced HYBID and HAS expression. In addition, we examined distribution of tryptase-positive mast cells, a major source of histamine, and HYBID-positive cells in the photoaged skin samples, which showed HYBID overexpression and HAS2 down-regulation as compared with the photoprotected skin. Our data in the present study provide, to the best of our knowledge, the first evidence that histamine contributes to the production of lower-molecular-weight HA through up-regulation of HYBID and down-regulation of HAS2 in skin fibroblasts and suggest that mast cell–derived histamine may play a role in aberrant HA metabolism in photoaged skin.
Results
Dose- and time-dependent expression of HYBID and HAS in human skin fibroblasts under stimulation with histamine
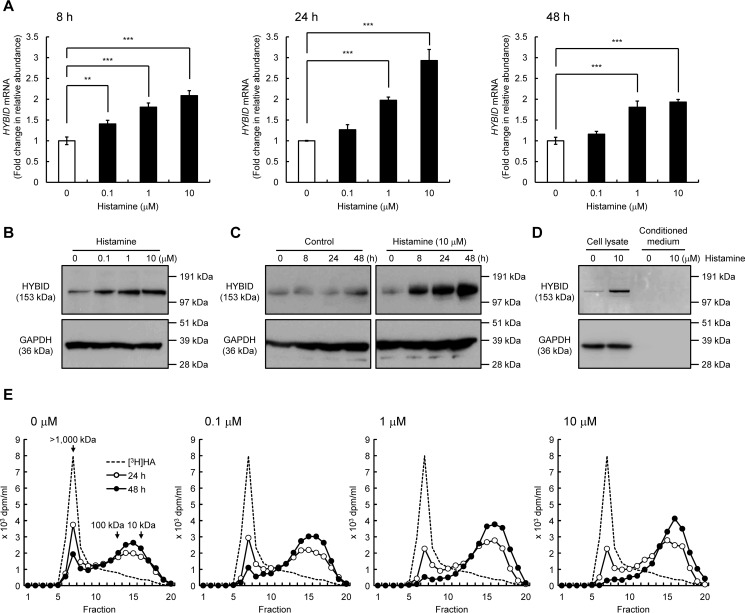
We first examined the dose- and time-dependent effects of histamine on the HYBID expression in normal human embryonic skin fibroblasts (Detroit 551 cells). As shown in Fig. 1 (A and B), histamine dose-dependently enhanced the HYBID expression, showing the highest up-regulation at a concentration of 10 μm, i.e. 2.9- and 3.0-fold increases in the mRNA and protein expression, respectively. Time-course experiments showed that histamine-induced up-regulation of the HYBID mRNA and protein expression begins at 8 h, reaches maximum levels at 24 h, and remains increased at 48 h after the stimulation (Fig. 1, A and C). HYBID protein was detected only in the cell lysates, but a negligible amount of HYBID was present in conditioned medium from control or histamine-treated Detroit 551 cells (Fig. 1D). In accordance with the HYBID up-regulation by histamine, degradation activity of high-molecular-weight [3H]HA was dose-dependently increased (Fig. 1E). When fluoresceinamine-labeled HA (FA-HA) was added to the culture media of control or histamine-treated Detroit 551 cells, FA-HA was almost completely recovered in the media without intracellular accumulation (Table 1). These results suggest that histamine-stimulated HA depolymerization occurs in a cell-associated manner, and HA fragments are released from the cells to extracellular milieu.
Figure 1.
Effects of histamine on the expression of HYBID in human normal skin fibroblasts (Detroit 551 cells). A, the dose-dependent and time-course expression of the HYBID mRNA. Detroit 551 cells were treated with histamine (0, 0.1, 1, or 10 μm), and the mRNA levels were measured by quantitative real-time PCR at 8, 24, or 48 h after the histamine treatment. The values are expressed as means ± S.D. (n = 3) and shown as fold increases in mRNA expression relative to control cells. Control, cells treated with vehicle alone. The Williams' test was used for statistical analysis. ***, p < 0.001; **, p < 0.01. B, the dose-dependent expression of HYBID protein in Detroit 551 cells. They were treated with 0, 0.1, 1, or 10 μm of histamine for 24 h, and the extracted proteins were subjected to immunoblotting with anti-HYBID antibody. GAPDH, loading control. C, the time-course expression of the HYBID protein. Detroit 551 cells were treated with histamine (0 or 10 μm) for 0, 8, 24, or 48 h, and the proteins extracted were analyzed by immunoblotting using anti-HYBID antibody. D, HYBID protein in the cell lysates and conditioned media from Detroit 551 cells treated with histamine (0 or 10 μm) for 24 h. They were subjected to immunoblotting with antibodies against HYBID or GAPDH. GAPDH, loading control for the cell lysates. E, HA depolymerization in Detroit 551 cells treated with histamine. The cells in culture were treated with 0, 0.1, 1, or 10 μm of histamine in the presence of [3H]HA for 24 h (open circle) or 48 h (closed circle), and HA fragments in the media were examined by size-exclusion chromatography.
Table 1.
Distribution of FA-HA in extracellular and cell-associated compartments after addition to histamine-stimulated Detroit 551 cells
FA-HA was incubated with Detroit 551 cells for 24 h, and fluorescence activity in the extracellular and cell-associated fractions was measured as described under “Experimental procedures.” Recovery is expressed as percentages of the total fluorescence activity. The values represent means ± S.D. (n = 3).
| 24 h | ||
|---|---|---|
| Extracellular | Cell-associated | |
| Control | 99.52 ±2.95 | 0.48 ±0.42 |
| Histamine | 99.47 ±4.30 | 0.53 ±0.10 |
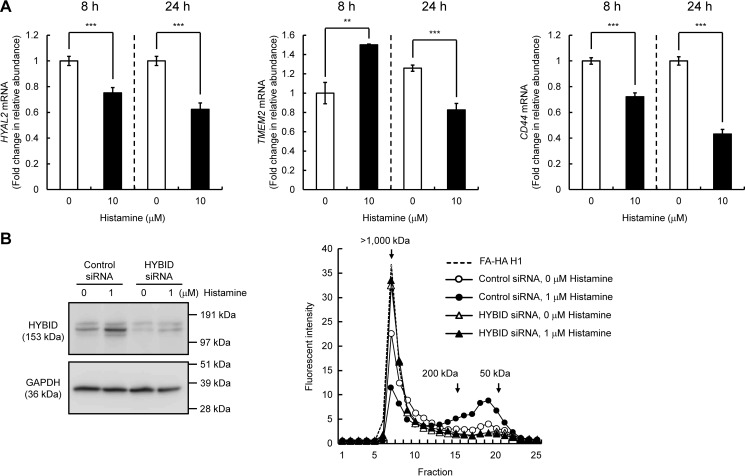
Previous studies reported that two hyaluronidases HYAL2 and transmembrane protein 2 (TMEM2) and a cell-surface HA receptor CD44 play a role in HA degradation (12, 13). Thus, we examined their mRNA expression in histamine-treated Detroit 551 cells and found that they are significantly down-regulated by histamine at 24 h after stimulation (Fig. 2A). To further study whether HYBID is directly involved in the histamine-mediated production of low-molecular-weight HA, we assessed HA degrading activity in siRNA-mediated HYBID knocked-down cells. As shown in Fig. 2B, knockdown of HYBID expression abrogated production of low-molecular-weight HA in Detroit 551 cells under either stimulated or unstimulated conditions. The data suggest that the histamine-mediated production of low-molecular-weight HA is derived from the action of up-regulated HYBID by histamine.
Figure 2.
Effects of histamine on the expression of HYAL2, TMEM2, and CD44 in human normal skin fibroblasts (Detroit 551 cells) and HA degradation by HYBID knocked-down Detroit 551 cells stimulated with histamine. A, expression levels of HYAL2, TMEM2, and CD44 mRNA. Detroit 551 cells were treated with histamine (0 or 10 μm), and the mRNA levels were measured by quantitative real-time PCR at 8 or 24 h after the histamine treatment. The values are expressed as means ± S.D. (n = 3) and shown as fold increases in mRNA expression relative to control cells. Control, cells treated with vehicle alone. The Student's t test was used for statistical analysis. ***, p < 0.001; **, p < 0.01. B, HA depolymerization in HYBID knocked-down Detroit 551 cells treated with histamine. HYBID was knocked down by treating Detroit 551 cells with siRNAs to HYBID. As for controls, the cells were transfected with control nonsilencing siRNA (Control siRNA). The expression levels of HYBID protein in control or HYBID knocked-down cells stimulated with 0 or 1 μm of histamine at 24 h after the stimulation were evaluated by immunoblotting. GAPDH was used as a loading control. Representative data for two siRNAs are shown. HA-degrading activity in control (circles) or HYBID (triangles) siRNA-treated Detroit 551 cells was detected by size-exclusion chromatography. siRNA-treated Detroit 551 cells were stimulated with 0 (open) or 1 μm (closed) of histamine in the presence of fluoresceinamine-labeled HA H1 for 48 h, and HA fragments in the media were subjected to chromatography.
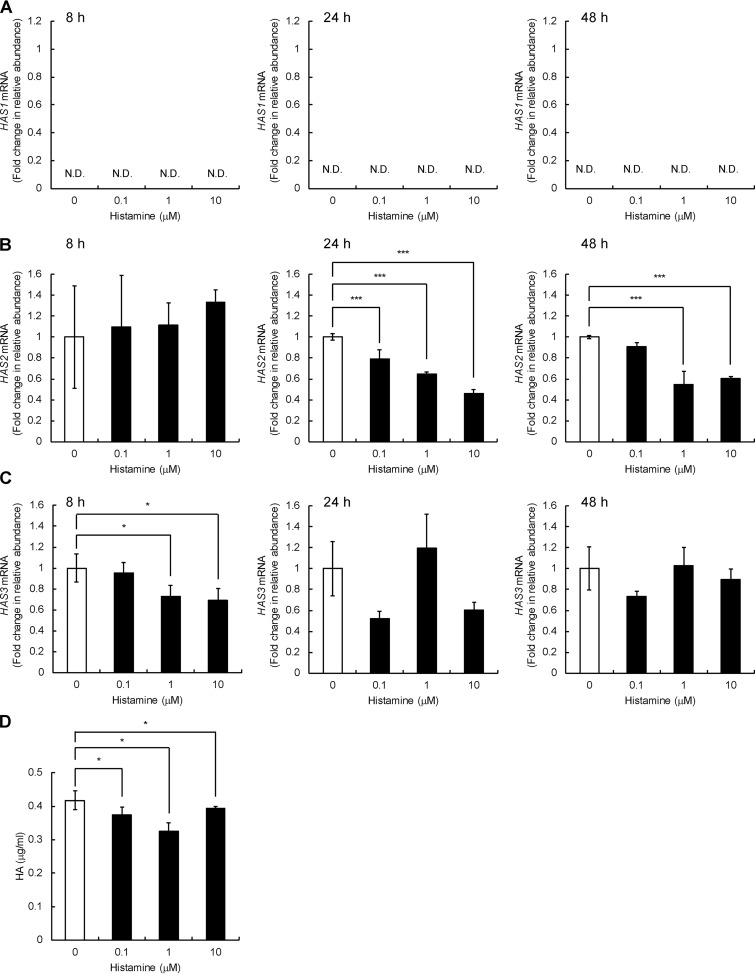
We next examined the expression of HAS1, HAS2, and HAS3 in Detroit 551 cells and found that HAS2 and HAS3 are expressed, whereas HAS1 mRNA expression is negligible under both basal and histamine-stimulated conditions (Fig. 3, A–C). As shown in Fig. 3B, histamine dose-dependently down-regulated the HAS2 mRNA expression, and its maximal effect was exerted at a dose of 10 μm, showing down-regulation up to 46% of the original level at 24 h after the treatment. Differently from the HYBID mRNA expression, a significant decrease in the HAS2 mRNA expression was detected at 24 h, but not at 8 h, after histamine treatment (Fig. 3B). HAS3 mRNA expression was dose-dependently down-regulated by histamine at 8 h after the treatment, but the down-regulation was not maintained at 24 and 48 h (Fig. 3C). HA production levels were slightly but significantly reduced in the cells treated with 0.1, 1.0, or 10 μm histamine compared with the control cells without histamine treatment (Fig. 3D).
Figure 3.
Effects of histamine on the expression of the HAS1–3 mRNA and HA production in human normal skin fibroblast Detroit 551 cells. A–C, the dose-dependent and time-course expression of the HAS1 (A), HAS2 (B), and HAS3 (C) mRNA. Detroit 551 cells were treated with histamine (0, 0.1, 1, or 10 μm), and the mRNA levels were measured by quantitative real-time PCR at 8, 24, or 48 h after the histamine treatment. The values are expressed as means ± S.D. (n = 3) and shown as fold increases in mRNA expression relative to control cells. Control, cells treated with vehicle alone. The Williams' test was used for statistical analysis. ***, p < 0.001; *, p < 0.05. D, HA content in the culture media of Detroit 551 cells stimulated with histamine. The cells were cultured in the presence of 0, 0.1, 1, or 10 μm of histamine for 24 h. The HA concentrations in the media were quantified by ELISA. The values represent means ± S.D. (n = 3). The Williams' test was used for statistical analysis. *, p < 0.05. N.D., not detected.
Similar expression profiles of HYBID and HAS1–3 were observed with normal human neonatal skin fibroblasts (HS27 cells) (Fig. S1). Histamine significantly ∼2-fold up-regulated the mRNA and protein expression of HYBID (Fig. S1, A and B) and down-regulated the HAS2 mRNA expression to ∼65% of the original level in HS27 cells (Fig. S1D). However, no changes were seen in the HAS3 mRNA expression (Fig. S1E). HA production was significantly reduced by the histamine treatment (Fig. S1F). Normal human adult skin fibroblasts (NHDF-Ad cells) also showed the up-regulation of mRNA and protein expression of HYBID, up to an ∼2-fold increase compared with the original level, after treatment with histamine (Fig. S2, A and B), whereas neither down-regulation of the HAS2 and HAS3 expression nor HA production was seen by histamine treatment (Fig. S2, D–F). These data suggest that histamine commonly up-regulates HYBID expression, but histamine-mediated HAS down-regulation may be cell type–specific.
Production of lower-molecular-weight HA by skin fibroblasts under stimulation with histamine
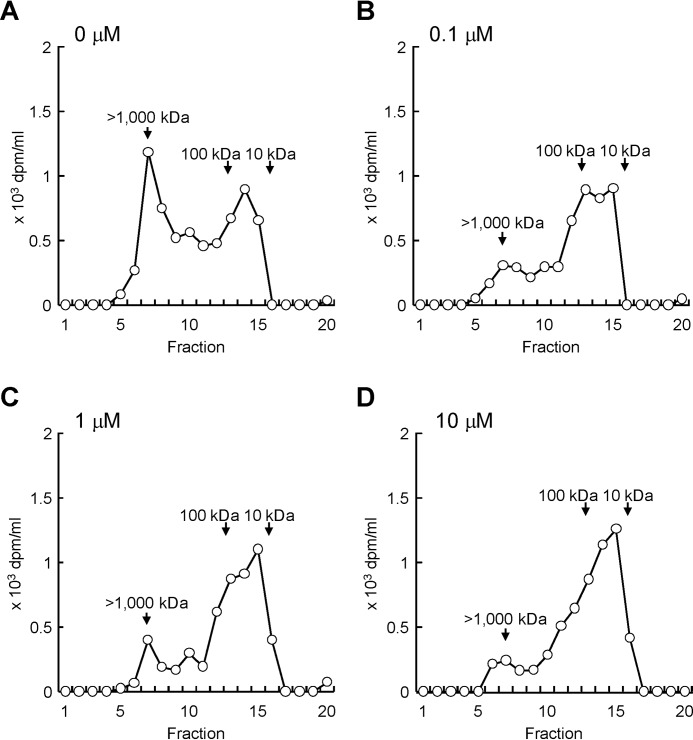
We then analyzed molecular weight distribution of newly produced HA in Detroit 551 cells under stimulation with different concentrations of histamine. The cells were cultured with [3H]glucosamine in the presence of histamine (0, 0.1, 1.0, or 10 μm) for 48 h, and metabolically labeled HA in the media was subjected to the gel filtration chromatography. As shown in Fig. 4A, nonstimulated cells produced HA species with two main fractions: i.e. high-molecular-weight [3H]HA of >1000 kDa and lower-molecular-weight [3H]HA of 10–100 kDa. Because HAS2 protein synthesizes only high-molecular-weight HA (6), lower-molecular-weight [3H]HA was considered to be a degradation product from high-molecular-weight [3H]HA. By contrast, treatment with histamine dose-dependently decreased high-molecular-weight [3H]HA and increased lower-molecular-weight [3H]HA (Fig. 4, B–D). These data suggest that histamine stimulation leads to the accumulation of lower-molecular-weight HA via increase in HYBID-mediated HA degradation and decrease in HAS2-mediated HA production.
Figure 4.
Effect of histamine on the size distribution of newly produced HA. A–D, detroit 551 cells were metabolically labeled with 10 μCi/ml of [3H]glucosamine in the presence of 0 (A), 0.1 (B), 1 (C), or 10 (D) μm of histamine for 48 h. The radiolabeled HA was isolated from the conditioned media and subjected to size-exclusion chromatography.
Identification of histamine receptor(s) for the regulation of HYBID and HAS2 expression in skin fibroblasts under stimulation with histamine
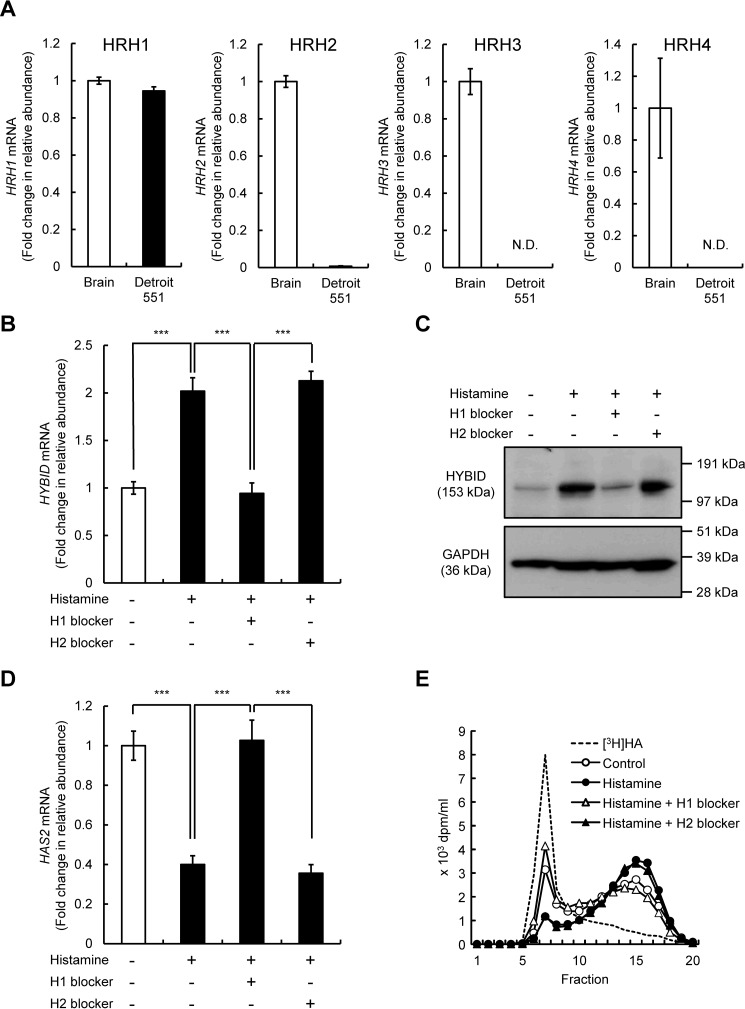
All four histamine receptors, i.e. HRH1, HRH2, HRH3, and HRH4, are expressed in normal human brain (1, 14). Therefore, using the brain tissue as a positive control, we examined mRNA expression of the receptors in Detroit 551 cells. As shown in Fig. 5A, HRH1 mRNA was abundantly expressed in Detroit 551 cells, whereas only low-level expression of HRH2 mRNA and negligible or no expression of HRH3 and HRH4 mRNA were seen. Therefore, we then examined the involvement of HRH1 and HRH2 in the HYBID and HAS2 expression by using pharmacological histamine receptor antagonists and agonists. As shown in Fig. 5 (B–D), HRH1 antagonist (H1 blocker; chlorpheniramine maleate) completely abrogated the histamine-mediated up-regulation of HYBID mRNA and protein expression and the down-regulation of HAS2 mRNA expression. However, HRH2 antagonist (H2 blocker; cimetidine) exhibited no effects on the histamine-mediated HYBID or HAS2 expression (Fig. 5, B–D). Consistent with the results of HYBID expression, H1 blocker attenuated the histamine-mediated HA degradation, whereas H2 blocker showed no similar effect (Fig. 5E). We also studied the effects of HRH1 and HRH2 agonists on the expression of HYBID and HAS2. As shown in Fig. 6 (A–C), HRH1 agonist (2-pyridylethylamine dihydrochloride) up-regulated mRNA and protein expression of HYBID and down-regulated the HAS2 expression, whereas HRH2 agonist (amthamine dihydrobromide) showed no changes in the expression of HYBID or HAS2. In agreement with these results, HRH1 agonist up-regulated HA degradation activity at a similar level to that of histamine, but HA degradation activity was not altered by HRH2 agonist (Fig. 6D). All these data strongly suggest that among the four histamine receptors, HRH1 is responsible for the action of histamine on the regulation of HYBID and HAS2 expression in skin fibroblasts.
Figure 5.
Expression of HRH1–4 mRNAs in normal human skin fibroblast Detroit 551 cells and effects of histamine H1 and H2 blockers on the expression of HYBID and HAS2 mRNA and HA degradation. A, expression levels of HRH1–4 mRNAs in the brain tissue (positive control) and Detroit 551 cells. The expression levels of HRH1–4 mRNAs in the brain tissue and Detroit 551 cells were measured by quantitative real-time PCR. The values are expressed as means ± S.D. (n = 3). N.D., not detected. B and C, effects of histamine H1 and H2 blockers on the HYBID mRNA and protein expression. Detroit 551 cells were cultured for 1 h in the absence or presence of 10 μm of chlorpheniramine maleate (histamine H1 blocker) or 10 μm of cimetidine (histamine H2 blocker) prior to stimulation with 10 μm of histamine for 24 h. The mRNA and protein expression of HYBID was measured by quantitative real-time PCR and immunoblotting, respectively. The values are expressed as means ± S.D. (n = 3) and shown as fold increases in mRNA expression relative to control cells in the absence of histamine. The Tukey's test was used for statistical analysis. ***, p < 0.001. D, effects of histamine H1 and H2 blockers on the HAS2 mRNA expression. Detroit 551 cells were treated with 10 μm of chlorpheniramine maleate (histamine H1 blocker) or 10 μm of cimetidine (histamine H2 blocker) prior to stimulation with 10 μm of histamine for 24 h, followed by analysis by quantitative real-time PCR for HAS2 as shown in B. E, HA depolymerization by Detroit 551 cells treated with histamine blockers. Detroit 551 cells were incubated for 1 h in the absence or presence of 10 μm of chlorpheniramine maleate (histamine H1 blocker) (open triangle) or 10 μm of cimetidine (histamine H2 blocker) (closed triangle), followed by additional incubation for 24 h with [3H]HA without (control; open circle) or with (closed circle) 10 μm of histamine, and HA fragments in the culture media were examined by size-exclusion chromatography.
Figure 6.
Effects of histamine H1 and H2 agonists on the expression of HYBID and HAS2 mRNA and HA degradation. A and B, effects of histamine H1 and H2 agonists on the HYBID mRNA and protein expression. Detroit 551 cells were cultured for 24 h in the absence or presence of 10 μm of 2-pyridylethylamine dihydrochloride (histamine H1 agonist) or 10 μm of amthamine dihydrobromide (histamine H2 agonist). The mRNA and protein expression of HYBID was measured by quantitative real-time PCR and immunoblotting, respectively. The values are expressed as means ± S.D. (n = 3) and shown as fold increases in mRNA expression relative to control cells. The Tukey's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01. C, effects of histamine H1 and H2 agonists on the HAS2 mRNA expression. Detroit 551 cells were treated cultured for 24 h in the absence or presence of 10 μm of 2-pyridylethylamine dihydrochloride (histamine H1 agonist) or 10 μm of amthamine dihydrobromide (histamine H2 agonist), followed by quantitative real-time PCR for HAS2 as shown in B. D, HA depolymerization by Detroit 551 cells treated with histamine agonists. Detroit 551 cells were incubated for 24 h in the absence (control; open circle) or presence of 10 μm of histamine (closed circle), 10 μm of 2-pyridylethylamine dihydrochloride (histamine H1 agonist) (open triangle), or 10 μm of amthamine dihydrobromide (histamine H2 agonist) (closed triangle) with [3H]HA, and HA fragments in the culture media were examined by size-exclusion chromatography.
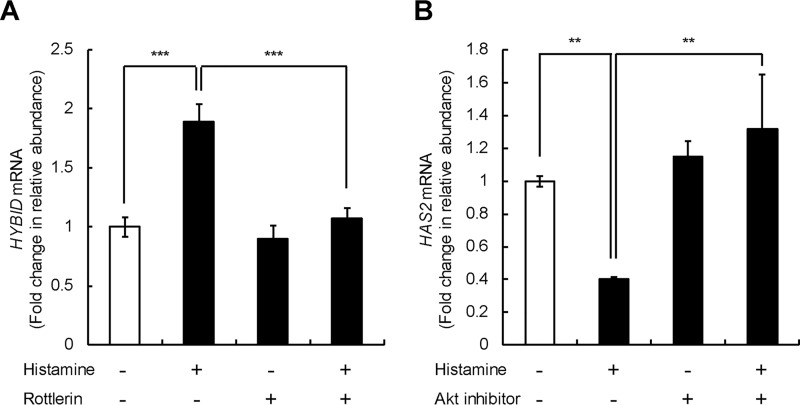
Differential regulation of HYBID and HAS2 gene expression by histamine through the protein kinase Cδ (PKCδ) or PI3K–Akt pathway in skin fibroblasts
We further investigated the signaling pathways downstream of HRH1 for histamine-mediated HYBID and HAS2 expression in skin fibroblasts (Detroit 551 cells) and found that rottlerin, a protein kinase Cδ inhibitor, completely suppresses the histamine-mediated up-regulation of HYBID but not the basal HYBID expression (Fig. 7A). Akt inhibitor V significantly attenuated the histamine-mediated down-regulation of HAS2 but not the basal HAS2 expression (Fig. 7B). These results suggest that the histamine-mediated gene expression of HYBID and HAS2 is regulated by distinct signaling pathways via the PKCδ pathway for HYBID and the PI3K–Akt signaling pathway for HAS2.
Figure 7.
Effect of PKCδ and PI3K–Akt inhibitors on the expression of HYBID and HAS2 mRNA. A and B, effects of rottlerin on HYBID expression (A) and Akt inhibitor V on HAS2 (B) mRNA expression. Detroit 551 cells were incubated for 1 h in the absence (control) or presence of 3 μm of rottlerin (PKCδ inhibitor) or 20 μm of Akt inhibitor V (Akt inhibitor) prior to stimulation with 10 μm of histamine for 24 h. The expression levels of HYBID and HAS2 mRNAs were measured by quantitative real-time PCR. The values are expressed as means ± S.D. (n = 3) and shown as fold increases in mRNA expression relative to control cells in the absence of histamine. The Tukey's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01.
Increased number of mast cells in the dermis of photoaged skin that is characterized by enhanced HYBID and decreased HAS2 expression
Real-time PCR analysis of the photoaged and photoprotected skin samples indicated that the expression of HYBID and HAS2 is significantly enhanced and reduced, respectively, in the photoaged skin compared with the photoprotected skin (Fig. 8A), confirming our previous data (11). Because mast cells are a major source of histamine, we performed immunohistochemistry using an antibody specific to tryptase, a selective marker of mast cells, in the skin specimens. As shown in Fig. 8B, many tryptase-positive mast cells were observed in the dermis of the photoaged skin, whereas they were sparsely present in the photoprotected skin. Negligible staining was obtained with control nonimmune IgG (Fig. 8B). By counting numbers of tryptase-positive mast cells, we demonstrated that the mean number of mast cells is significantly ∼2-fold higher in the photoaged skin than in the photoprotected skin (Fig. 8C). Interestingly, mast cells in the photoaged skin occasionally showed weak diffuse staining around mast cells, which suggests degranulation (15) (Fig. 8B, insets). Immunofluorescence staining of HYBID or tryptase in serial sections of the photoaged skin tissue demonstrated that HYBID is positively immunostained in the area where many tryptase-positive mast cells are present (Fig. 8D). Negligible staining was obtained with control nonimmune IgG (Fig. 8D). These data suggest that histamine produced by the increased mast cells may be involved in increased HYBID expression and decreased HAS2 expression in photoaged skin.
Figure 8.
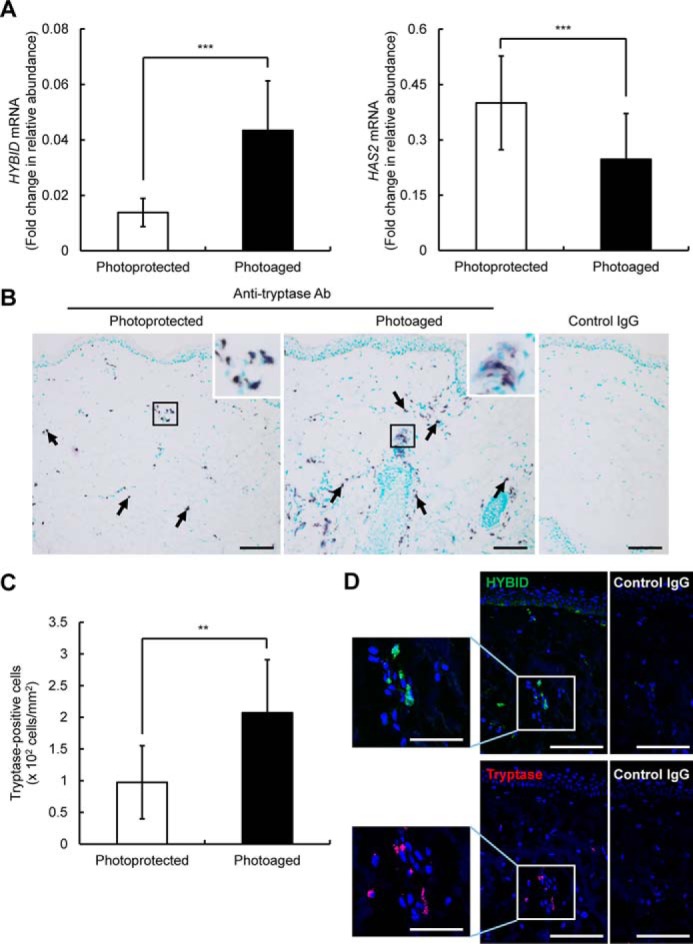
Expression levels of HYBID and HAS2 mRNA and immunohistochemistry of HYBID and tryptase and quantification of tryptase-immunoreactive cells in the dermis of photoprotected and photoaged skin. A and B, expression levels of HYBID and HAS2 mRNA in the photoprotected and photoaged skin specimens. The values are presented as the means ± S.D. (n = 10). The paired Student's t test was used for statistical analysis. ***, p < 0.001. B, immunohistochemistry of tryptase in the skin biopsy specimens. Representative images from the photoprotected and photoaged skin specimens are shown. Frozen skin sections were immunostained with anti-tryptase antibody (Ab) or control mouse IgG and counterstained with methylgreen. Arrows, purple-stained tryptase-positive cells. Scale bars, 100 μm. Insets, high-power view of boxed areas. C, the numbers of tryptase-positive cells in the photoprotected and photoaged skin specimens. The values represent means ± S.D. The paired Student's t test was used for statistical analysis. **, p < 0.01. D, immunofluorescence staining of HYBID and tryptase. Representative images from the photoaged skin specimens are shown. Sequential frozen skin sections were immunofluorescence stained with anti-HYBID antibody (green) or control rat IgG, and anti-tryptase antibody (red) or control mouse IgG, followed by counterstaining with DAPI (blue). Boxed areas are shown at higher magnification to the left. Scale bars, 100 μm. Insets, 50 μm.
Discussion
In the present study, to the best of our knowledge, we have demonstrated for the first time that histamine decreases total amount and molecular size of newly produced HA by the increased HYBID-mediated HA degradation and the suppressed HAS2-mediated HA synthesis in human skin fibroblasts. Although we previously reported that histamine up-regulates the HYBID expression and enhances HA degradation in human skin fibroblasts (8), the present study has further demonstrated the histamine-induced down-regulation of the HAS2 expression together with HYBID up-regulation in two different types of fibroblasts. We have also shown that unlike HYBID, the expression of HYAL2, TMEM2, and CD44, all of which may be involved in HA degradation (12, 13), is down-regulated by histamine treatment in Detroit 551 cells and that knockdown of the HYBID expression abrogates the production of low-molecular-weight HA under stimulation with histamine. Therefore, it seems likely that HYBID is responsible for histamine-stimulated HA depolymerization in skin fibroblasts.
Several growth factors including TGF-β1, basic fibroblast growth factor, epidermal growth factor, and platelet-derived growth factor BB are known to act as up-regulators for HAS2 expression (9). However, limited information is available for suppressors of the HAS2 expression, and to the best of our knowledge, only glucocorticoids are reported to suppress the HAS2 expression in skin fibroblasts and keratinocytes (16). Thus, our data in the present study indicate that histamine should be added to a list of down-regulators of the HAS2 gene and suggest that newly produced lower-molecular-weight HA under stimulation with histamine is ascribed to degradation of high-molecular-weight HA through the action of overexpressed HYBID, as we observed with the above-mentioned growth factors (9).
The diverse biological effects of histamine are mediated through the interaction with the histamine receptors, i.e. HRH1, HRH2, HRH3, and HRH4, expressed on the cells within the tissues (17). Therefore, expression profile of histamine receptors is considered to be an important determinant for histamine effects. Accumulating lines of evidence have demonstrated that HRH1 is expressed by a broad range of cell types such as neurons, smooth muscle cells, endothelial cells, and inflammatory and immune cells, whereas HRH2 is commonly co-expressed with HRH1 in most cell types (18). Conversely, the expression of HRH3 is confined to neural cells in the central and peripheral nervous system, and HRH4 is expressed mainly in cells of hematopoietic origin particularly dendritic cells, mast cells, eosinophils, monocytes, basophils, and T cells (17). Although information was limited about histamine receptors in skin fibroblasts (17, 19), our data in the present study have demonstrated that the effects of histamine on the HYBID and HAS2 expression are mediated mainly through HRH1 in skin fibroblasts.
In various pathophysiological conditions, metabolism of ECM macromolecules such as collagens and HA is controlled by the action of factors, e.g. grown factors and cytokines. TGF-β1 is one of the representative anabolic factors that are implicated in the overproduction of ECM molecules (20). Concerning the TGF-β1–mediated metabolism of collagens in skin fibroblasts, TGF-β1 appears to act through the same PI3K–Akt signaling pathway to down-regulate the MMP-1 (collagenase-1) gene and up-regulate the COL1A1/2 (collagen type I, α1 and α2 chains) gene (20). However, our previous study indicated that the different signaling pathways are used for TGF-β1–induced HA metabolism: the HYBID down-regulation via the PI3K–Akt signaling pathway and the HAS1/2 up-regulation through Smad3, ERK1/2, and/or p38-MAPK pathway (9). In the present study, we have provided data of the different signaling pathways for histamine-induced HYBID and HAS2 expression, showing that the histamine/HRH1 signal is transduced through the PKCδ pathway for the HYBID up-regulation and the PI3K–Akt pathway for the HAS2 down-regulation. Ligand binding to G protein–coupled receptors promotes association of the receptors with G proteins, which consist of α, β, and γ subunits and causes activation of the classical and nonclassical intracellular signaling pathways (18). Histamine binding to HRH1 activates the Gαq subunit and leads to increased cytosolic calcium levels and generation of diacyl glycerol, both of which activate PKC in the classical signaling pathway (18). Conversely, the nonclassical pathway by the Gβγ subunits of histamine-stimulated HRH1 is known to activate PI3K, leading to activation of Akt (18, 21). Therefore, both the classical and nonclassical routes through the distinct intracellular signaling pathways are involved in histamine-induced HA metabolism. Taken together, our data on HYBID and HAS expression suggest the possibility that the HA metabolism by catabolic and anabolic enzymes may be regulated by the divergent signaling pathways of the stimulatory factors, although further detailed studies are needed to clarify the hypothesis.
The alteration in number of mast cells during photoaging is controversial, although previous studies suggested that mast cells are tended to be increased in photoaged skin (4, 22–26). It seems likely that numbers of mast cells in the dermis are dependent on anatomical locations and severity of photoaging of the skin examined. In addition, gender, age, and ethnicity of the subjects may affect numbers of mast cells in photoaged skin. In the present study, we have shown that number of mast cells in the dermis is significantly ∼2-fold higher in the photoaged skin (crow's-foot areas) compared with the photoprotected skin (inner forearms), both of which were obtained by punch biopsy from the photoaged and photoprotected areas of the same Japanese female individuals. As shown in the previous (11) and present studies, the photoaged skin was characterized by increased HYBID expression and decreased HAS2 expression compared with the photoprotective skin. Mast cells in the photoaged skin are known to commonly localize near fibroblasts, showing their frequent contact each other (4). UV irradiation of human skin is reported to induce degranulation of mast cells in the dermis (15), and actually our immunohistochemical study of the photoaged skin specimens showed the finding suggestive of degranulation. In addition, HYBID-positive cells and tryptase-positive mast cells were closely located in the dermis of the photoaged skin. Taken together, it is plausible to speculate that in the photoaged skin, histamine released from mast cells stimulates fibroblasts to overexpress HYBID and down-regulate HAS2, leading to the reduction in amount and molecular size of HA in the photoaged skin (11). However, future experimental work is definitely necessary to demonstrate these processes.
In summary, we have demonstrated that in human skin fibroblasts, histamine contributes to the reduction in molecular sizes and amounts of newly produced HA by increasing the HYBID expression and decreasing the HAS2 expression using different histamine–HRH1 signaling pathways. We have also provided data that mast cell–derived histamine may be implicated in enhanced HYBID expression and decreased HAS2 expression in photoaged skin. Although high-molecular-weight HA is anti-inflammatory, and anti-angiogenic, low-molecular-weight HA fragments are potently pro-inflammatory and pro-angiogenic and promote cell migration (27–29). Therefore, in the photoaged skin, the HA fragments generated by the action of histamine-induced HYBID may be involved in angiogenesis and chronic inflammatory cell infiltration, which are commonly observed in photoaged skin (30). To demonstrate our hypothesis, however, detailed analyses of the histamine–HYBID axis in the photoaged skin are definitely necessary.
Experimental procedures
Cell cultures
Normal human skin fibroblasts including Detroit 551 (American Type Culture Collection), HS27 (American Type Culture Collection), and NHDF-Ad (Takara Bio) cells were cultured in Eagle's minimum essential medium (MP Biomedicals) supplemented with nonessential amino acids, 1 mm sodium pyruvate, and 10% (v/v) fetal bovine serum (JRH Biosciences) at 37 °C in a humidified atmosphere containing 5% CO2.
Quantitative real-time PCR
Total RNA was isolated from skin fibroblasts by the RNeasy mini kit (Qiagen), and that from normal human brain was purchased from Biochain. They were reverse-transcribed to cDNA using a high-capacity cDNA archive kit (Applied Biosystems). Expression of the target mRNAs was quantitatively analyzed using the cDNA templates in a TaqMan real-time PCR assay (Applied Biosystems; StepOnePlusTM real-time PCR system) according to the manufacturer's protocol. The relative quantification values of HYBID, HAS1, HAS2, HAS3, HYAL2, TMEM2, CD44, HRH1, HRH2, HRH3, and HRH4 were normalized to endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for in vitro study or β-actin for in vivo study.
Antibodies
A rat mAb against HYBID was developed and characterized previously (8). Antibodies against GAPDH and tryptase were purchased from Santa Cruz Biotechnology and DAKO, respectively. Anti-rat IgG–Alexa Fluor® 594 and anti-mouse IgG–Alexa Fluor® 647 were purchased from Invitrogen. Control rat IgG and control mouse IgG were purchased from Thermo Scientific and R&D Systems, respectively.
Immunoblotting
Cell homogenate supernatants and conditioned media were subjected to SDS–polyacrylamide gel by electrophoresis on NuPAGE 4–12% Bis-Tris gels (Invitrogen), and the proteins resolved were transferred onto polyvinylidene difluoride membranes. The membranes were reacted with the antibodies and then incubated with the horseradish peroxidase–conjugated secondary antibodies: donkey anti-rat IgG antibody for HYBID (Jackson ImmunoResearch) and goat anti-rabbit IgG antibody for GAPDH (DAKO). Immunoreactive bands were detected using the SuperSignal West Pico chemiluminescent substrate system (Thermo Scientific).
Assay for cellular depolymerization
High-molecular-weight 3H-labeled HA ([3H]HA) of >1000 kDa was prepared as described previously (8). Cellular HA depolymerization was assayed by culturing confluent cells in medium containing [3H]HA (40,000 dpm/ml) and applying the media to a Sepharose CL-2B (GE Healthcare) column (1 × 60 cm) equilibrated with 0.5 m NaCl. The flow rate was 0.65 ml/min, and fractions of 2.5 ml were collected. The radioactivity of each fraction was measured by a scintillation counter (Aloka; LSC-6100).
Cellular FA-HA H1 (1,760 kDa; peak top kDa) depolymerization was assayed by culturing confluent cells in medium containing FA-HA H1 (10 μg/ml) and applying the harvested media to a Sepharose CL-2B (GE Healthcare) column (1 × 60 cm) equilibrated with 0.5 m NaCl. The flow rate was 0.8 ml/min, and fractions of 1.6 ml were collected. Depolymerized FA-HA H1 was detected at an excitation wavelength of 490 nm using an emission wavelength of 525 nm.
The column was calibrated with the FA-HA species: H1 (1,760 kDa; peak top kDa), M1 (907 kDa), L1 (197 kDa), S1 (56 kDa), T1 (28 kDa), and U1 (9.8 kDa) (PG Research). An excitation wavelength of 490 nm and an emission wavelength of 525 nm were used for the detection of fluoresceinamine.
Assay for HA production
Culture media were harvested at 24 h after culturing skin fibroblasts in the presence of histamine (0, 0.1, 1, or 10 μm) (WAKO). The HA content in the media was determined using a QnE HA ELISA (Biotech Trading Partners, Inc.) according to the manufacturer's protocol. The assay detected HA molecular mass forms as small as 20–30 kDa.
RNA interference
Knockdown experiments were performed using 25-nucleotide siRNA duplexes chemically synthesized for HYBID and control nonsilencing siRNA by Invitrogen. The siRNA oligonucleotide sequences were as follows: HYBID-1, 5′-AAACAUUGAAAUAUUCGCCAUGCUC-3′; and HYBID-2, 5′-UUGACAAGGAGGCCAAGACAGUGGU-3′. These siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen), and the knocked-down cells were used for the experiments at 24 h after transfection.
Molecular size distribution of synthesized HA
Detroit 551 cells were cultured for 48 h in culture media containing 10 μCi/ml of d-[1,6-3H(N)]glucosamine hydrochloride (American Radiolabeled Chemicals) in the presence of histamine (0, 0.1, 1, or 10 μm). The harvested culture media were treated with 30 μg/ml Pronase (Merck) and then divided into two parts: one was digested with 0.1 unit/ml Streptomyces hyaluronidase (WAKO) in 0.5 m MES buffer, pH 6.0, containing 0.15 m sodium acetate at 37 °C overnight, whereas the other was left undigested by the enzyme. They were applied to a PD-10 desalting column equilibrated with 0.5 m NaCl to remove unincorporated radiolabel, and each sample was fractionated in a Sepharose CL-2B (GE Healthcare) column. The chromatography profiles depicted only the hyaluronidase-sensitive activity in each fraction plotted against the fraction number by subtracting the hyaluronidase-resistant counts from the radioactivity of the hyaluronidase-undigested samples.
Determination of extracellular and cell surface-associated HA
Detroit 551 cells were cultured in 6-well plates in the medium containing FA-HA (10 μg/ml) (PG research) for 24 h and washed with ice-cold PBS. The medium and wash were combined and designated as “extracellular” fraction. The cells were removed from the well by incubation with AccumaxTM (Innovative Cell Technologies). The cell suspension was centrifuged at 4 °C, and the resulting supernatant was designated as “cell surface-associated” fraction. An excitation wavelength of 490 nm and an emission wavelength of 525 nm were used for the detection of FA.
Chemicals
Histamine H1 receptor antagonist chlorpheniramine maleate and H2 receptor antagonist cimetidine were purchased from WAKO. Histamine H1-receptor agonist 2-pyridylethylamine dihydrochloride and H2-receptor agonist amthamine dihydrobromide were obtained from R&D Systems and Sigma–Aldrich, respectively. Rottlerin, a PKCδ inhibitor, and Akt inhibitor V were from Millipore and Merck, respectively.
Human tissue samples
Human skin tissues were taken by punch biopsy (3 mm in diameter) from the inner arm (photoprotected area) and the corner of the eye (photoaged area) of 10 Japanese female volunteers (age range, 65–72 years; mean, 69.1 years) at Yaesu Nihonbashi skin Clinic (Tokyo, Japan). The skin tissue collection was conducted according to the principles expressed in the Declaration of Helsinki and approved by the Institutional Review Board of Tokyo-Eki Center-building Clinic (Tokyo, Japan) and Kao Corporation (Tokyo, Japan), and informed consent was obtained from all of the volunteers before surgery.
Immunohistochemistry
Tryptase staining was performed on frozen skin sections. Endogenous peroxidase activity and nonspecific binding were blocked by incubating them with 0.3% (v/v) H2O2 in methanol for 15 min and 1% (v/v) BSA in PBS for 30 min, respectively. They were incubated with the mouse mAb against human mast cell tryptase (DAKO) or control mouse IgG (R&D Systems) for 18 h at 4 °C. After washing in PBS, they were incubated with EnVision+ Dual Link (DAKO) for 30 min at room temperature. Color was developed by reacting with the Vector VIP kit (Vector Laboratories). Tryptase-positive cells were counted in the dermal areas 500 μm below the epidermal–dermal junction (counting field area, 0.05 mm2) by observing at ×200 magnification. The quantification of 16 randomly selected areas per each skin sample was performed.
Immunofluorescence study
Serial frozen sections of the photoaged skin tissue were subjected to immunostained with rat anti-HYBID mAb or control rat IgG (Thermo Scientific), or mouse anti-tryptase mAb (DAKO) or control mouse IgG (R&D Systems), followed by incubation with goat anti-rat IgG–Alexa Fluor® 594 (Invitrogen) or goat anti-mouse IgG-Alexa Fluor® 647 (Invitrogen). The nuclei were stained with DAPI (Vector Laboratories), and autofluorescence was blocked with Vector® TrueVIEWTM autofluorescence quenching kit (Vector Laboratories). The images were obtained with a confocal microscope LSM710 (Zeiss).
Statistics
Statistical significance was assessed by Student's t test, paired Student's t test, analysis of variance, Williams' test, and Tukey's test using EXSUS version 8.0.0 (CAC EXICARE Corporation). A probability of p < 0.05 was considered significant.
Author contributions
H. Y. and S. S. conceptualization; H. Y. resources; H. Y., M. A., A. K., Y. E., K. K., T. N., and S. S. data curation; H. Y. and S. S. formal analysis; H. Y., T. S., and Y. T. supervision; H. Y. and S. S. investigation; H. Y. methodology; H. Y. and Y.O. writing-original draft; H. Y. project administration; S. S., T. S., Y. O., and Y. T. writing-review and editing.
Supplementary Material
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- ECM
- extracellular matrix
- FA
- fluoresceinamine-labeled
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HA
- hyaluronan
- HAS
- hyaluronan synthase
- HR
- histamine receptor
- HYAL2
- hyaluronidase 2
- PKC
- protein kinase C
- TGF
- transforming growth factor
- TMEM2
- transmembrane protein 2.
References
- 1. Dong H., Zhang W., Zeng X., Hu G., Zhang H., He S., and Zhang S. (2014) Histamine induces upregulated expression of histamine receptors and increases release of inflammatory mediators from microglia. Mol. Neurobiol. 49, 1487–1500 10.1007/s12035-014-8697-6 [DOI] [PubMed] [Google Scholar]
- 2. Mantovani A., Allavena P., Sica A., and Balkwill F. (2008) Cancer-related inflammation. Nature 454, 436–444 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- 3. Iddamalgoda A., Le Q. T., Ito K., Tanaka K., Kojima H., and Kido H. (2008) Mast cell tryptase and photoaging: possible involvement in the degradation of extra cellular matrix and basement membrane proteins. Arch. Dermatol. Res. 300, S69–S76 [DOI] [PubMed] [Google Scholar]
- 4. Toyoda M., Nakamura M., Nakada K., Nakagawa H., and Morohashi M. (2005) Characteristic alterations of cutaneous neurogenic factors in photoaged skin. Br. J. Dermatol. 153, 13–22 10.1111/j.1365-2133.2005.06965.x [DOI] [PubMed] [Google Scholar]
- 5. Laurent T. C., and Fraser J. R. (1992) Hyaluronan. FASEB J. 6, 2397–2404 10.1096/fasebj.6.7.1563592 [DOI] [PubMed] [Google Scholar]
- 6. Itano N., and Kimata K. (2002) Mammalian hyaluronan synthases. IUBMB Life 54, 195–199 10.1080/15216540214929 [DOI] [PubMed] [Google Scholar]
- 7. Sugiyama Y., Shimada A., Sayo T., Sakai S., and Inoue S. (1998) Putative hyaluronan synthase mRNA are expressed in mouse skin and TGF-β upregulates their expression in cultured human skin cells. J. Invest. Dermatol. 110, 116–121 10.1046/j.1523-1747.1998.00093.x [DOI] [PubMed] [Google Scholar]
- 8. Yoshida H., Nagaoka A., Kusaka-Kikushima A., Tobiishi M., Kawabata K., Sayo T., Sakai S., Sugiyama Y., Enomoto H., Okada Y., and Inoue S. (2013) KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc. Natl. Acad. Sci. U.S.A. 110, 5612–5617 10.1073/pnas.1215432110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nagaoka A., Yoshida H., Nakamura S., Morikawa T., Kawabata K., Kobayashi M., Sakai S., Takahashi Y., Okada Y., and Inoue S. (2015) Regulation of hyaluronan (HA) metabolism mediated by HYBID (hyaluronan-binding protein involved in HA depolymerization, KIAA1199) and HA synthases in growth factor-stimulated fibroblasts. J. Biol. Chem. 290, 30910–30923 10.1074/jbc.M115.673566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshida H., Komiya A., Ohtsuki R., Kusaka-Kikushima A., Sakai S., Kawabata K., Kobayashi M., Nakamura S., Nagaoka A., Sayo T., Okada Y., and Takahashi Y. (2018) Relationship of hyaluronan and HYBID (KIAA1199) expression with roughness parameters of photoaged skin in Caucasian women. Skin Res. Technol. 24, 562–569 10.1111/srt.12467 [DOI] [PubMed] [Google Scholar]
- 11. Yoshida H., Nagaoka A., Komiya A., Aoki M., Nakamura S., Morikawa T., Ohtsuki R., Sayo T., Okada Y., and Takahashi Y. (2018) Reduction of hyaluronan and increased expression of HYBID (KIAA1199) correlate with clinical symptoms in photoaged skin. Br. J. Dermatol. 179, 136–144 10.1111/bjd.16335 [DOI] [PubMed] [Google Scholar]
- 12. Bourguignon L. Y., Singleton P. A., Diedrich F., Stern R., and Gilad E. (2004) CD44 interaction with Na+–H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J. Biol. Chem. 279, 26991–27007 10.1074/jbc.M311838200 [DOI] [PubMed] [Google Scholar]
- 13. Yamamoto H., Tobisawa Y., Inubushi T., Irie F., Ohyama C., and Yamaguchi Y. (2017) A mammalian homolog of the zebrafish transmembrane protein 2 (TMEM2) is the long-sought-after cell-surface hyaluronidase. J. Biol. Chem. 292, 7304–7313 10.1074/jbc.M116.770149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H., Burkhardt C., Heinrich U. R., Brausch I., Xia N., and Förstermann U. (2003) Histamine upregulates gene expression of endothelial nitric oxide synthase in human vascular endothelial cells. Circulation 107, 2348–2354 10.1161/01.CIR.0000066697.19571.AF [DOI] [PubMed] [Google Scholar]
- 15. Kim M. S., Kim Y. K., Lee D. H., Seo J. E., Cho K. H., Eun H. C., and Chung J. H. (2009) Acute exposure of human skin to ultraviolet or infrared radiation or heat stimuli increases mast cell numbers and tryptase expression in human skin in vivo. Br. J. Dermatol. 160, 393–402 10.1111/j.1365-2133.2008.08838.x [DOI] [PubMed] [Google Scholar]
- 16. Gebhardt C., Averbeck M., Diedenhofen N., Willenberg A., Anderegg U., Sleeman J. P., and Simon J. C. (2010) Dermal hyaluronan is rapidly reduced by topical treatment with glucocorticoids. J. Invest. Dermatol. 130, 141–149 10.1038/jid.2009.210 [DOI] [PubMed] [Google Scholar]
- 17. Thurmond R. L., Gelfand E. W., and Dunford P. J. (2008) The role of histamine H1 and H4 receptors in allergic inflammation: the search for new antihistamines. Nat. Rev. Drug Discov. 7, 41–53 10.1038/nrd2465 [DOI] [PubMed] [Google Scholar]
- 18. O'Mahony L., Akdis M., and Akdis C. A. (2011) Regulation of the immune response and inflammation by histamine and histamine receptors. J. Allergy Clin. Immunol. 128, 1153–1162 10.1016/j.jaci.2011.06.051 [DOI] [PubMed] [Google Scholar]
- 19. Johnson C. L., Johnson C. G., Bazan E., Garver D., Gruenstein E., and Ahluwalia M. (1990) Histamine receptors in human fibroblasts: inositol phosphates, Ca2+, and cell growth. Am. J. Physiol. 258, C533–C543 10.1152/ajpcell.1990.258.3.C533 [DOI] [PubMed] [Google Scholar]
- 20. Bujor A. M., Pannu J., Bu S., Smith E. A., Muise-Helmericks R. C., and Trojanowska M. (2008) Akt blockade downregulates collagen and upregulates MMP1 in human dermal fibroblasts. J. Invest. Dermatol. 128, 1906–1914 10.1038/jid.2008.39 [DOI] [PubMed] [Google Scholar]
- 21. Mettler S. E., Ghayouri S., Christensen G. P., and Forte J. G. (2007) Modulatory role of phosphoinositide 3-kinase in gastric acid secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G532–G543 10.1152/ajpgi.00138.2007 [DOI] [PubMed] [Google Scholar]
- 22. Bosset S., Bonnet-Duquennoy M., Barré P., Chalon A., Kurfurst R., Bonté F., Schnébert S., Le Varlet B., and Nicolas J. F. (2003) Photoageing shows histological features of chronic skin inflammation without clinical and molecular abnormalities. Br. J. Dermatol. 149, 826–835 10.1046/j.1365-2133.2003.05456.x [DOI] [PubMed] [Google Scholar]
- 23. Bhawan J., Oh C. H., Lew R., Nehal K. S., Labadie R. R., Tsay A., and Gilchrest B. A. (1992) Histopathologic differences in the photoaging process in facial versus arm skin. Am. J. Dermatopathol. 14, 224–230 10.1097/00000372-199206000-00008 [DOI] [PubMed] [Google Scholar]
- 24. Lavker R. M., and Kligman A. M. (1988) Chronic heliodermatitis: a morphologic evaluation of chronic actinic dermal damage with emphasis on the role of mast cells. J. Invest. Dermatol. 90, 325–330 10.1111/1523-1747.ep12456193 [DOI] [PubMed] [Google Scholar]
- 25. Bhawan J., Andersen W., Lee J., Labadie R., and Solares G. (1995) Photoaging versus intrinsic aging: a morphologic assessment of facial skin. J. Cutan. Pathol. 22, 154–159 10.1111/j.1600-0560.1995.tb01399.x [DOI] [PubMed] [Google Scholar]
- 26. Rojas I. G., Martínez A., Pineda A., Spencer M. L., Jiménez M., and Rudolph M. I. (2004) Increased mast cell density and protease content in actinic cheilitis. J. Oral. Pathol. Med. 33, 567–573 10.1111/j.1600-0714.2004.00242.x [DOI] [PubMed] [Google Scholar]
- 27. West D. C., Hampson I. N., Arnold F., and Kumar S. (1985) Angiogenesis induced by degradation products of hyaluronic acid. Science 228, 1324–1326 10.1126/science.2408340 [DOI] [PubMed] [Google Scholar]
- 28. Rooney P., Kumar S., Ponting J., and Wang M. (1995) The role of hyaluronan in tumour neovascularization (review). Int. J. Cancer 60, 632–636 10.1002/ijc.2910600511 [DOI] [PubMed] [Google Scholar]
- 29. Sugahara K. N., Murai T., Nishinakamura H., Kawashima H., Saya H., and Miyasaka M. (2003) Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J. Biol. Chem. 278, 32259–32265 10.1074/jbc.M300347200 [DOI] [PubMed] [Google Scholar]
- 30. Toyoda M., Nakamura M., Luo Y., and Morohashi M. (2001) Ultrastructural characterization of microvasculature in photoaging. J. Dermatol. Sci. 27, S32–S41 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.