Abstract
The yeast vacuolar H+-ATPase (V-ATPase) of budding yeast (Saccharomyces cerevisiae) is regulated by reversible disassembly. Disassembly inhibits V-ATPase activity under low-glucose conditions by releasing peripheral V1 subcomplexes from membrane-bound Vo subcomplexes. V-ATPase reassembly and reactivation requires intervention of the conserved regulator of H+-ATPase of vacuoles and endosomes (RAVE) complex, which binds to cytosolic V1 subcomplexes and assists reassembly with integral membrane Vo complexes. Consistent with its role, the RAVE complex itself is reversibly recruited to the vacuolar membrane by glucose, but the requirements for its recruitment are not understood. We demonstrate here that RAVE recruitment to the membrane does not require an interaction with V1. Glucose-dependent RAVE localization to the vacuolar membrane required only intact Vo complexes containing the Vph1 subunit, suggesting that the RAVE-Vo interaction is glucose-dependent. We identified a short conserved sequence in the center of the RAVE subunit Rav1 that is essential for the interaction with Vph1 in vivo and in vitro. Mutations in this region resulted in the temperature- and pH-dependent growth phenotype characteristic of ravΔ mutants. However, this region did not account for glucose sensitivity of the Rav1-Vph1 interaction. We quantitated glucose-dependent localization of a GFP-tagged RAVE subunit to the vacuolar membrane in several mutants previously implicated in altering V-ATPase assembly state or glucose-induced assembly. RAVE localization did not correlate with V-ATPase assembly levels reported previously in these mutants, highlighting both the catalytic nature of RAVE's role in V-ATPase assembly and the likelihood of glucose signaling to RAVE independently of V1.
Keywords: vacuolar ATPase, vacuole, vacuolar acidification, yeast, organellar pH homeostasis, glucose signaling, protein-protein interaction, proton pump, rabconnectin-3, RAVE complex
Introduction
Tight maintenance of organelle and cytosolic pH is required for cellular homeostasis, and perturbed pH homeostasis is associated with disease states ranging from metabolic acidosis (1) to neurodegeneration (2) and aging (3, 4). V-ATPases2 are highly conserved proton pumps that are responsible for organelle acidification in eukaryotic cells. They are multisubunit complexes composed of two major subcomplexes, a peripheral V1 complex oriented toward the cytosol that contains the sites for ATP hydrolysis and an integral membrane Vo domain that contains the proton pore. Mammalian genomes encode multiple isoforms for several subunits in both V1 and Vo (5). In yeast, however, all subunits except the Vo a-subunit are encoded by single-copy VMA genes. In contrast, the yeast a-subunit consists of Vph1 and Stv1 isoforms that target V-ATPases to the vacuole or Golgi, respectively, and also endow their respective complexes with distinct regulatory properties (6–8).
Reversible disassembly is a conserved, post-translational form of V-ATPase regulation that is able to tune V-ATPase activity to multiple different stimuli (9–12). In yeast, acute glucose deprivation results in separation of the V1 subcomplex and V1 subunit C from the Vo subcomplex, and glucose readdition induces rapid reassembly of both V1 and V1 subunit C with Vo. After disassembly, free V1 subcomplexes lack Mg-ATPase activity and free Vo subdomains are closed to H+ translocation, and activity is restored by reassembly (13–15). The yeast RAVE complex is a V-ATPase–specific chaperone required both for initial biosynthetic assembly of the V-ATPase and for efficient glucose-dependent reassembly of V-ATPases (16, 17). Upon glucose readdition, RAVE catalyzes recruitment of cytosolic V1 and V1 subunit C to the vacuolar membrane and assembly of functional V-ATPases. Yeast RAVE is a heterotrimeric complex comprised of three subunits, Rav1, Rav2, and Skp1 (18). Rav1 is the central component of RAVE and contains N-terminal and C-terminal binding sites for Rav2 and Skp1, respectively (19). Rav1 interacts independently with 1) V1 subcomplexes via one or more of the EG peripheral stalks, 2) V1 subunit C, and 3) the cytosolic N-terminal domain of Vph1 (Vph1NT) of the Vo subcomplex (19). Rav2 interacts with V1 subunit C, making V1 subunit C the only V-ATPase subunit that contains multiple RAVE-binding sites (19). Skp1, a component of many essential cellular complexes, including SCF (Skp1-cullin-F-Box) ubiquitin ligases, does not appear to interact directly with V-ATPase subunits (16).
The rabconnectin-3 (Rbcn3) complex is the structural and functional homologue of RAVE in higher eukaryotes, but it appears to contain only two larger subunits and no Skp1 (20). The Rbcn3 complex was shown to associate with V-ATPase subunits in Drosophila (21), and loss of function compromises organelle acidification in multiple organisms (21–23). The Rbcn3α subunit has regions of homology to the N- and C-terminal domains of Rav1 but is almost twice as large.
It was proposed that RAVE's role in glucose-dependent V-ATPase reassembly is to shuttle cytosolic V1 and V1 subunit C back to Vo at the vacuolar membrane and provide a template for complex assembly, thus improving the efficiency of V-ATPase reassembly (19). However, the cellular levels of Rav1 and Rav2 are only 5–10% of the levels of V1 subunits (24), suggesting that RAVE's role must be catalytic, requiring multiple cycles of binding and release of V1 to achieve V-ATPase reassembly. We recently found that a significant portion of RAVE is present at the vacuolar membrane of glucose-replete cells, released from the membrane by glucose deprivation, and recruited back to the membrane upon glucose readdition (19). This recruitment and release of RAVE is entirely consistent with its role as a glucose-dependent chaperone and assembly factor. However, neither the molecular basis of the glucose-sensitive recruitment of RAVE nor the signaling mechanisms involved are known. The interaction between RAVE and V1 is not intrinsically glucose-sensitive. Rather, RAVE can bind to the V1 subdomain whether it is in the cytosol as a result of glucose deprivation or as a result of Vo mutations that prevent membrane association (17).
In the current study, we find that only Vph1-containing Vo subcomplexes are required for glucose-dependent recruitment of RAVE to the vacuolar membrane. Previous binding data indicated that a fragment consisting of amino acids 679–898 of Rav1 (Rav1 679–898) binds directly to Vph1NT. We identified a six amino acid conserved motif in this region that is required for interaction of Rav1 679–898 with Vph1NT in vitro and for recruitment of RAVE to the vacuolar membrane in vivo. Although this interaction is not intrinsically glucose-sensitive, we tested whether mutations previously associated with altered assembly or reversible disassembly of the V-ATPase would affect steady-state levels of RAVE at the membrane in the presence or absence of glucose. We find that mutations in several glucose-sensing pathways significantly alter RAVE localization. The results indicate that the RAVE complex itself can be recruited to the vacuolar membrane without V1 and that its steady-state localization does not strictly follow that of V1 subcomplexes.
Results
Interactions with the V1 subcomplex are not required for glucose-dependent recruitment of RAVE to the membrane
Levels of RAVE subunits in isolated cytosolic fractions do not change with glucose levels (17), and very little RAVE is isolated with vacuolar membranes containing the V-ATPase. However, using GFP-tagged RAVE subunits, we previously demonstrated that RAVE is reversibly released from the vacuolar membrane upon glucose deprivation and restored to the membrane upon glucose readdition (19). GFP-tagged Rav1 and Rav2 complement the growth phenotypes of the corresponding deletion mutants. We asked whether either subunit could be recruited to membranes in the absence of the other. As shown in Fig. 1, both Rav1-GFP and Rav2-GFP are present at the membrane, as well as the cytosol, in glucose-grown WT cells. In contrast, Rav2-GFP is almost completely cytosolic in rav1Δ cells, and Rav1-GFP is almost completely cytosolic in rav2Δ cells. We conclude that both Rav1 and Rav2 subunits must be present for efficient recruitment of the complex to the vacuolar membrane, and in subsequent experiments, we use Rav2-GFP to track localization of the RAVE complex.
Figure 1.
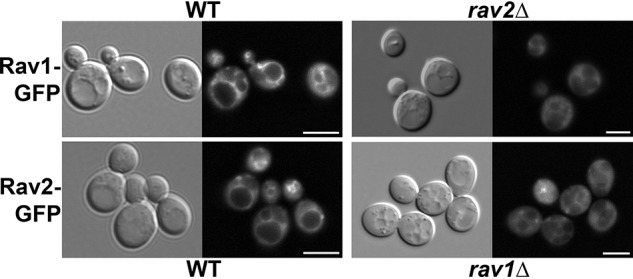
Localization of RAVE subunits Rav1 and Rav2 to the vacuolar membrane requires the presence of both subunits. Top, Rav1 was tagged with GFP and expressed in WT and rav2Δ strain. Bottom, Rav2 was tagged with GFP and expressed in WT and rav1Δ strain. All cells were maintained in glucose. DIC images are shown on the right in each set; the vacuoles are identified as a single or several distinct indentations depending on conditions. Both GFP-tagged subunits were also present in small cytosolic puncta in some cells. The nature of these puncta is not known. Scale bar, 5 μm.
Although V1 subunit C is released from the vacuolar membrane upon glucose deprivation and is proposed to be a key player in reversible disassembly of the yeast V-ATPase, we previously found that the RAVE complex can be recruited to the vacuolar membrane in glucose-grown vma5Δ cells, which lack V1 subunit C. In addition, Rav2-GFP also cycles off and on the membrane with glucose deprivation and readdition in a vma5Δ mutant (19), indicating that V1 subunit C is not required for glucose-sensitive recruitment of RAVE to the vacuolar membrane. V1 assembly is compromised (25), and RAVE-V1 binding is abolished (17), in a vma4Δ mutant, which lacks the V1 subunit E, so we compared Rav2-GFP localization in vma4Δ and WT strains. As shown in Fig. 2A, Rav2-GFP localized to the vacuolar membrane in both the vma4Δ and WT strains in the presence of glucose, suggesting that binding to the V1 subcomplex is not required for vacuolar localization of RAVE. We next tested whether Rav2-GFP was released from the membrane into the cytosol upon glucose deprivation. As shown in both images and line scans in Fig. 2A, vacuolar localization is lost in both vma4Δ and WT cells. However, glucose readdition restores vacuolar localization in both strains. In line scans for high glucose conditions in both strains, there are maxima of fluorescence intensity corresponding to the vacuolar membrane. To compare localization of Rav2-GFP across a larger population of the cells, we performed line scan measurements across 10–11 cells under each condition. The maximum fluorescence intensity for each scan was normalized to the average intensity of the cells maintained in glucose, and the results are plotted in Fig. 2B. For both WT and vma4Δ cells, there is a significant decrease in maximal intensity in the absence of glucose, but no significant difference after glucose readdition. From these data, we conclude that the association of RAVE with V1 is not required for glucose-dependent recruitment of RAVE to the vacuolar membrane.
Figure 2.
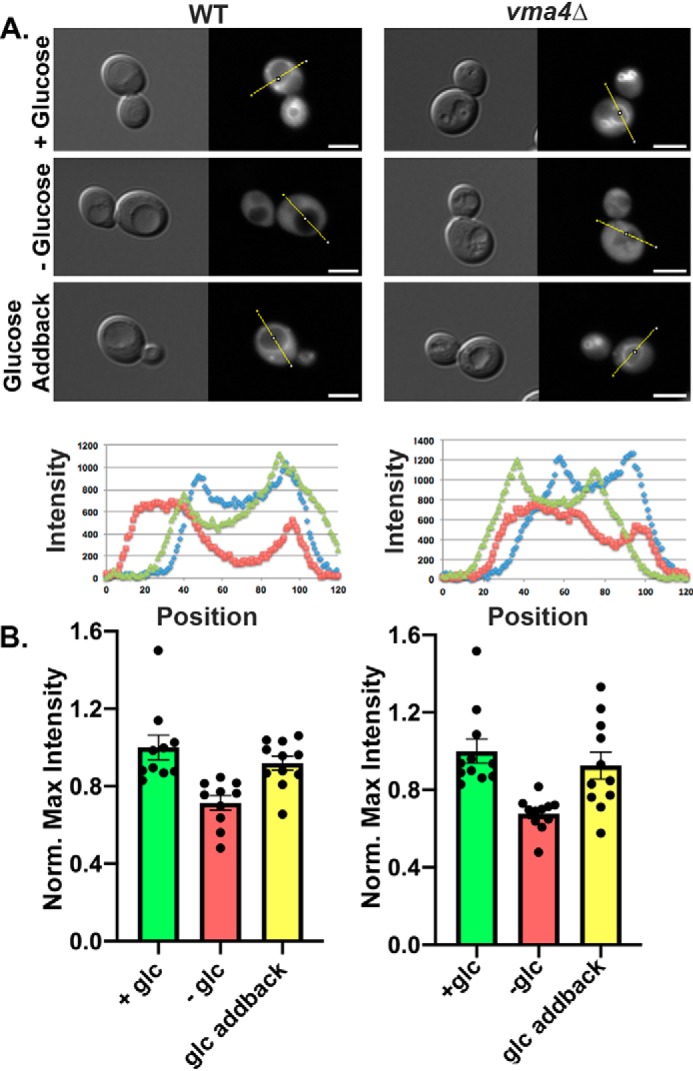
Glucose-dependent recruitment of RAVE to the vacuolar membrane does not require interaction with the V1 subcomplex. A, DIC and Rav2-GFP images were obtained for WT and vma4Δ cells in the presence of glucose (+Glucose), after 15 min of glucose deprivation (−Glucose), and 15 min after the readdition of glucose to glucose-deprived cells (Glucose Addback). The fluorescence intensities on the lines shown in each GFP image are plotted in the graphs below the micrographs: blue lines for +glucose, red lines for −glucose, and green lines for glucose addback. Scale bar, 5 μm. B, line scans were plotted for 10–11 cells from each condition, and maximum intensities along the line scan are shown, after normalization to the average maximum in the +glucose sample for each strain. Bars, mean ± S.E. (error bars); individual data points are also shown.
The results above were somewhat surprising, given that RAVE has been shown to interact with both V1 (17) and V1 subunit C (19, 26) in the cytosol of glucose-deprived cells and to be released from the membrane under conditions of V-ATPase disassembly (19). We hypothesized that recruitment of RAVE to the membrane might solely depend on interaction with Vo subcomplexes and thus would be disrupted in a vma11Δ mutant, which lacks an essential subunit of the proteolipid ring in Vo and fails to assemble or transport Vo subcomplexes to the vacuole (27, 28). Indeed, Rav2-GFP remained cytosolic even in the presence of glucose in a vma11Δ strain (Fig. 3A), indicating that interactions with the Vo subcomplex are required for membrane recruitment of RAVE. We next asked whether Vo complexes needed to be free of V1 for RAVE to bind. A mutation of a critical glutamate (E145L) in VMA11 allows V-ATPase assembly, but abolishes both ATPase activity and proton transport (27). The vma11 E145L mutant was shown to have increased levels of V1 subunits assembled at the vacuole, apparently as a result of assembly of virtually all Vo subcomplexes with V1 (27). We introduced Rav2-GFP into this strain to assess whether RAVE was able to bind to fully assembled, but inactive, V-ATPases. Interestingly, Rav2-GFP was still able to bind to the membrane in presence of glucose and to be reversibly released upon glucose deprivation and readdition (Fig. 3B). These results suggest that the site of RAVE interaction with the Vo subcomplex must be available when the V-ATPase is assembled and suggest that release and recruitment of RAVE do not require catalytic cycling of the V-ATPase.
Figure 3.
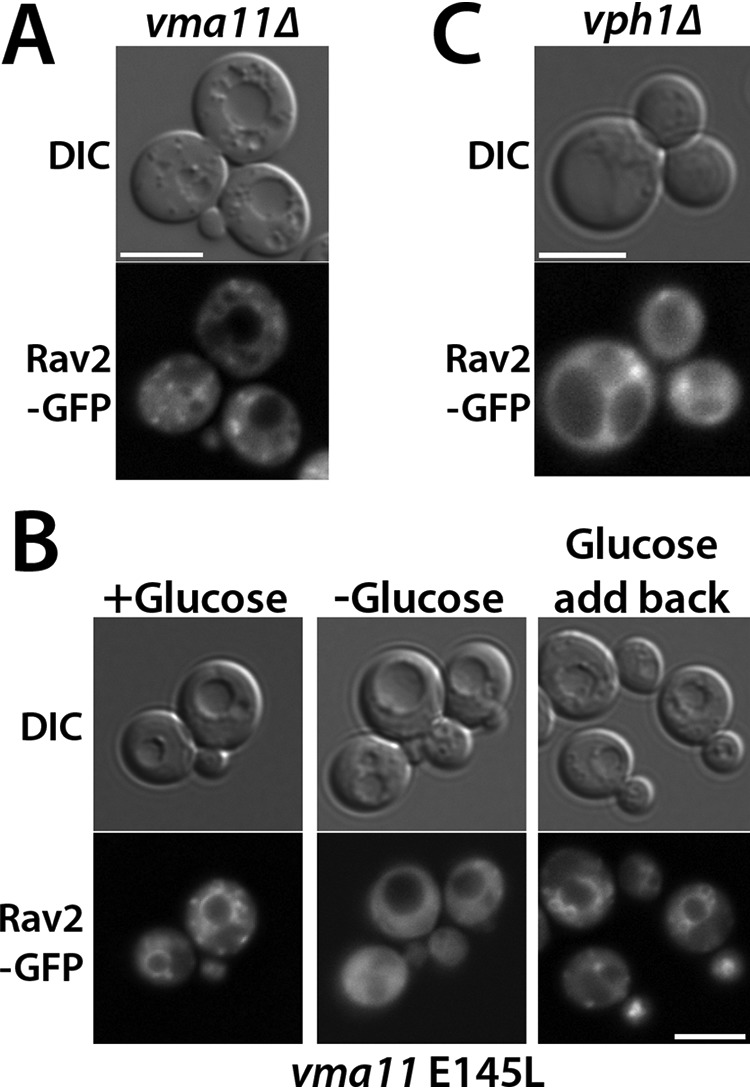
RAVE requires Vph1-containing Vo subcomplexes for association with the vacuolar membrane. A, Rav2-GFP was introduced into a vma11Δ mutant, which abolishes assembly of Vo subcomplexes. Cells that were maintained in glucose were visualized under DIC optics (left) and GFP fluorescence optics (right). B, Rav2-GFP was visualized under varied glucose conditions in a vma11 E145L mutation, reported to contain assembled but inactive V-ATPase complexes at the vacuole that are incapable of disassembly upon glucose deprivation. Glucose treatments were as described in the legend to Fig. 2. C, Rav2-GFP was visualized in a vph1Δ mutant background, which lacks the vacuolar isoform of the Vo a-subunit. The same glucose-grown cells are visualized under DIC (left) and GFP (right) fluorescence optics. Scale bar, 5 μm for all images.
There are two isoforms of the Vo a-subunit, Vph1 and Stv1, but only V-ATPases containing the Vph1 isoform require RAVE for their assembly (29). We also found that the cytosolic domain of Vph1 (Vph1NT) binds directly to Rav1 in vitro (19). Consistent with these observations, we found that Rav2 failed to localize to the vacuolar membrane in a vph1Δ strain, even in the presence of glucose (Fig. 3C). This suggests that Vph1 is likely to be the molecular target for RAVE recruitment to the membrane. In fact, the Rav1-Vph1 interaction appears to be the central glucose-dependent interaction between the RAVE and V-ATPase complexes.
A conserved motif within Rav1 is required for RAVE function, recruitment to vacuolar membranes, interaction with Vph1NT, and V-ATPase assembly
Through a combination of bioinformatics, molecular modeling, and interaction assays (19), we previously predicted that the N-terminal region of yeast Rav1 (Rav1 55–669) is rich in β-sheet structures, likely forming a double β-propeller, and showed that it contains a binding site for Rav2. The C-terminal half of Rav1 (Rav1 840–1357) contains the most highly conserved portion of the protein (Rav1 840–1125), in a region that is predicted to be predominantly α-helical. The C-terminal half of Rav1 contains binding sites for V1, V1 subunit C, and Skp1. We identified a Vph1NT-binding site between the N- and C-terminal regions in a less conserved segment encompassing Rav1 679–898 (19). Sequence analysis of this region revealed the presence of a conserved six amino acid motif (757LPVYHP in yeast; Fig. 4A). To determine whether this region was essential for RAVE function, we generated point mutations targeting conserved residues (rav1 760YH761 mutated to AA (YH-AA) and rav1 K773A). We also deleted the entire six amino acid motif (rav1 6Δ). The mutants were then tested for a Rav1− phenotype by comparing growth on plates buffered to low pH to growth on plates at high pH, with added calcium, at 37 °C (16, 17). rav1Δ and rav2Δ mutants are characterized by a temperature-sensitive Vma− growth phenotype and show WT growth at pH 5 but poor growth on YEPD, pH 7.5, or YEPD, pH 7.5, containing 60 mm CaCl2 at 37 °C (16, 17). Interestingly, the rav1 YH-AA and rav1 6Δ mutations resulted in a Rav− phenotype, whereas the K773A mutation allowed WT growth (Fig. 4B).
Figure 4.
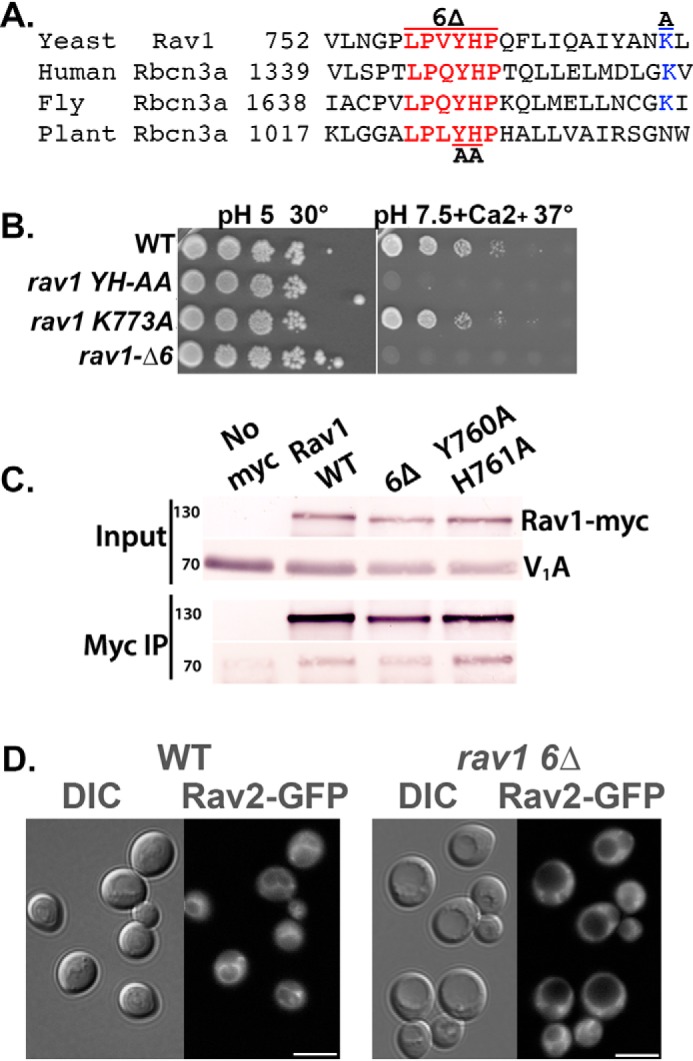
Mutations in a conserved sequence of Rav1 generate a Rav1− growth phenotype and compromise RAVE recruitment to the membrane. A, alignment of the indicated portion of yeast Rav1 with the following rabconnectin-3α homologues in higher eukaryotes: human, Homo sapiens DMXL2 (NCBI locus AAI44540); fly, Drosophila melanogaster rabconnectin-3α (NCBI locus NP_572302); and plant, Arabidopsis thaliana hypothetical protein (NCBI locus AAD20167). Numbers indicate the position of the first amino acid in the protein sequence shown. B, growth phenotype of strains containing WT and mutant RAV1. 10-Fold serial dilutions of log-phase liquid cultures of the indicated strains grown in YEPD, pH 5, were prepared in 96-well plates and then transferred by pinning to YEPD, pH 5, plates or YEPD, pH 7.5, plates containing 60 mm CaCl2. For each plate, the highest concentration of cells is shown on the left, with successive serial dilutions to the right. Growth at 30 °C and 37 °C as indicated. The Rav− mutant phenotype is characterized by sensitivity to high pH and calcium concentrations at 37 °C. C, a C-terminal Myc13 tag was added to Rav1 in all of the strains except the no-Myc control samples (No myc). Cytosolic fractions were prepared from the indicated yeast strains as described under “Experimental procedures,” and a portion was reserved as input. The remainder was combined with mouse anti-Myc mAb followed by Protein A-Sepharose to immunoprecipitate Myc-tagged Rav1 (Myc IP). Both input and immunoprecipitate samples from each strain were separated by SDS-PAGE and transferred to nitrocellulose. The blots were probed was anti-Myc to determine the levels of Rav1-Myc and with mAb 8B1F3 to determine the level of co-precipitated V1A subunit. D, cellular distribution of Rav2-GFP in WT cells and in the rav1 6Δ mutant. DIC images are shown to the left of each set and GFP fluorescence to the right. Scale bar, 5 μm.
The data above suggest that the six amino acid conserved motif is essential for RAVE function, but it is possible that the mutation reduces Rav1p levels or causes overall misfolding of the protein. We tagged both WT Rav1 and the Rav1 6Δ mutant protein with a Myc13 tag and assessed protein stability on immunoblots of whole-cell lysates (Fig. 4C). We observed only a slight decrease in mutant Rav1p levels compared with WT, suggesting that the Rav1− phenotype is not due to loss of Rav1. We next assessed whether the rav1 6Δ mutation altered interaction between Rav1 and V1. Amino acids 840–940 of Rav1 were previously shown to be required for V1 binding (19), so if the rav1 6Δ mutation has a major effect on structure, V1 binding might be lost. We immunoprecipitated the Myc13-tagged WT Rav1 and Rav1 6Δ proteins and assessed co-precipitation of V1 subunits. The rav1 6Δ mutation did not alter the interaction with V1, as indicated by comparable levels of V1 subunit A co-precipitated with Rav1-Myc13 (Fig. 4C). We next hypothesized that the six amino acid conserved motif might be required for RAVE's vacuolar localization. We compared Rav2-GFP localization in the rav1 6Δ mutation to its localization in WT Rav1. As shown in Fig. 4D, Rav2-GFP is largely cytosolic in the rav1 6Δ mutant, even in glucose-replete conditions, indicating that this motif is necessary for membrane recruitment of RAVE.
Because RAVE's vacuolar localization depends on both the presence of glucose and Vph1, we hypothesized that rav1 6Δ mutant might prevent recruitment of RAVE to the vacuolar membrane as the result of a signaling defect, in which RAVE is no longer able to sense glucose, or as the result of a binding defect that prevents RAVE from binding to Vph1. To distinguish between these possibilities, we expressed Vph1NT and Rav1 679–898, with or without the rav1 6Δ mutation, in Escherichia coli and assessed their interaction in vitro. Both WT and mutant Rav1 fragments were tagged with a C-terminal His6 tag. Vph1NT and WT or mutant Rav1 fragments were incubated with TALON resin, which binds to the His6 tag, and co-elution of Vph1NT with the Rav1 fragments at high imidazole concentrations was assessed by immunoblotting. As shown in Fig. 5, whereas Vph1NT eluted with WT Rav1 679–898 (Fig. 5A), Vph1NT showed very little binding to the Rav1 679–898 fragment containing the rav1 6Δ mutation (Fig. 5B) and no binding in the absence of the Rav1 fragment (Fig. 5C). These data indicate that the conserved motif is directly involved in the interaction between Rav1 679–898 and Vph1NT, rather than indirectly affecting the interaction by contributing to glucose sensing.
Figure 5.
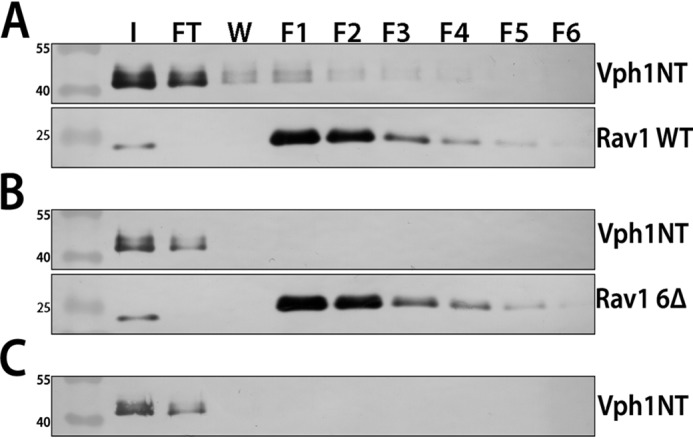
Purified Vph1NT binds to WT Rav1 679–898 in vitro, but not in the presence of the rav1 6Δ mutation. Vph1NT, WT Rav1 679–898-His6, and Rav1 679–898-His6 containing the rav1 6Δ mutation were expressed and purified as described under “Experimental procedures.” Vph1NT was mixed with the WT or mutant Rav1 fragment at a 5:1 molar ratio, and the mixture was added to 300 μl of TALON resin and incubated at 4 °C for 2 h. The resin and protein fragments were transferred to a column and washed with low-imidazole buffer, and bound protein was eluted at high imidazole concentrations. 500-μl fractions (F1–F6) were collected for each sample as described under “Experimental procedures.” Input (I), flow-through (FT), wash (W), and fractions are shown for Vph1NT and WT Rav1 679–898-His6 (Rav1 WT) (A), Vph1NT and Rav1 679–898-His6 containing the 6Δ mutation (Rav1 6Δ) (B), and Vph1NT only (no His-tagged Rav1) (C). For A and B, a single blot that was cut and the upper portion were probed for Vph1NT (10D7 mouse mAb), whereas the lower portion was probed with anti-His6.
We next assessed how the rav1 6Δ mutation affects the assembly of the V-ATPase. We first compared localization of Vma5-GFP (V1 subunit C) to the vacuolar membrane in WT and rav1 6Δ mutant cells. As shown in Fig. 6A, Vma5-GFP is localized to the vacuolar membrane in WT cells but is largely cytosolic in the rav1 6Δ cells, suggesting that V-ATPase assembly is compromised in the mutant. We isolated vacuolar vesicles from WT and rav1 6Δ mutant cells and compared the levels of V-ATPase. As described previously, rav1Δ cells have WT levels of Vo subunit Vph1 in vacuolar vesicles, but reduced levels of V1 subunits at the membrane (26), and the rav1 6Δ exhibits a very similar assembly defect (Fig. 6B). Consistent with their assembly defect, vacuolar vesicles from the rav1 6Δ mutants also have significantly reduced levels of concanamycin-sensitive ATPase activity (Fig. 6C). Taken together, these data indicate that loss of interaction with Vph1 in the rav1 6Δ mutant results in loss of RAVE-dependent V-ATPase assembly and function.
Figure 6.
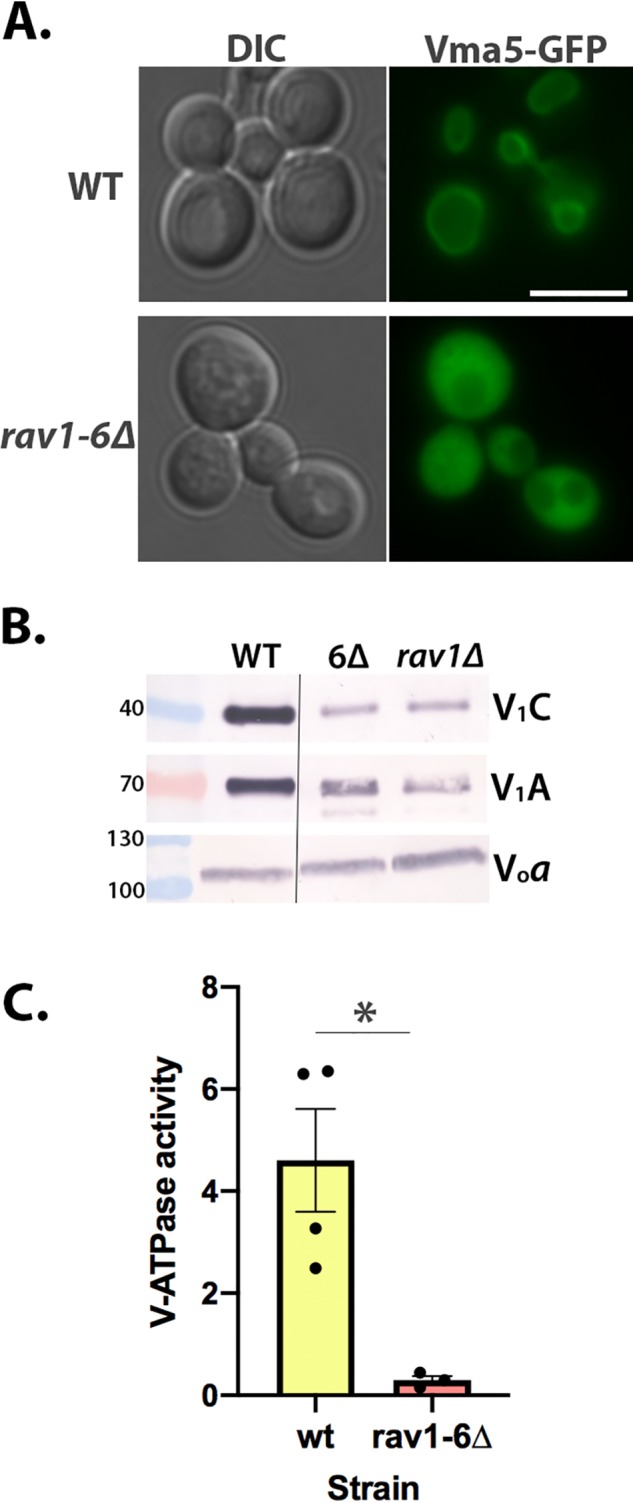
V-ATPase assembly and function are compromised in a rav1 6Δ mutant. A, localization of Vma5-GFP was visualized in WT and rav1 6Δ strain backgrounds (right set of images). DIC images (left set of images) identify the position of vacuoles. Scale bar, 5 μm. B, vacuolar vesicles isolated from WT cells, rav1 6Δ and rav1Δ, were solubilized, and equal amounts of protein from each sample were separated by SDS-PAGE and transferred to nitrocellulose. V1 subunits A and C and Vo subunit a (Vph1) were identified as described under “Experimental procedures.” Molecular masses of markers in kDa are indicated on the left. The line indicates that the samples are from nonadjacent lanes of the same gels. C, ATPase activity sensitive to 200 nm concanamycin A (V-ATPase activity) was measured in 3–4 independent vacuolar vesicles preparations from each strain. Mean activity ± S.E. (error bars) is shown for each strain; *, p < 0.05 by Student's t test.
Cellular factors that contribute to glucose-dependent localization of RAVE
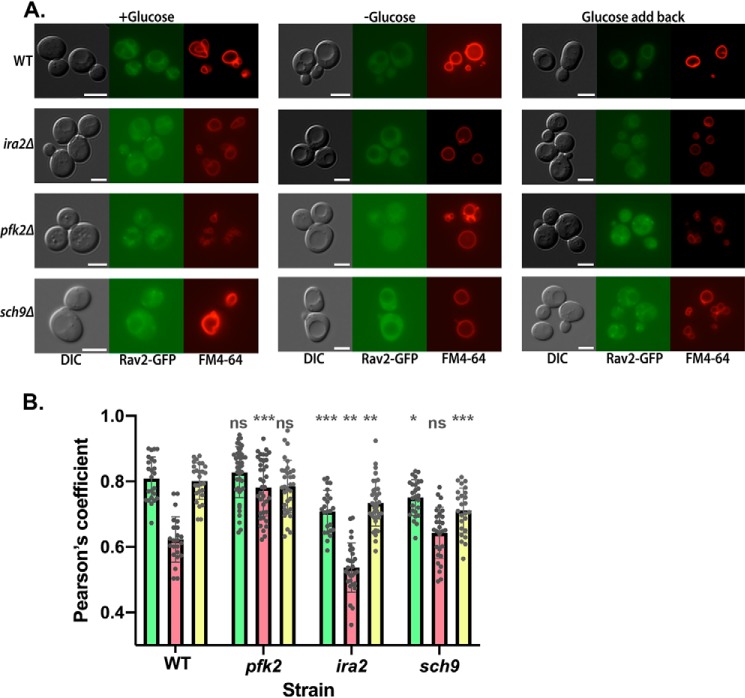
As described above, although RAVE is recruited to the vacuolar membrane by its interaction with Vph1, this interaction is reversed by glucose deprivation. This suggests that either the binding site becomes unavailable, or some other cellular pathway alters the interaction between RAVE and Vph1-containing Vo sectors, directly or indirectly. Several pathways have been associated with signaling the V-ATPase assembly state in yeast, and we hypothesized that one or more of these pathways might affect glucose-dependent association of RAVE with the vacuolar membrane. We tested effects of three mutations previously reported to affect reversible disassembly of the V-ATPase on glucose-dependent RAVE recruitment to the membrane. Deletion of phosphofructokinase subunit Pfk2 (pfk2Δ) was reported to reduce V-ATPase reassembly after glucose readdition and enhance the RAVE-V1 association in cytosolic fractions (30). Deletion of the Ras inhibitor protein Ira2 (ira2Δ) prevented disassembly of the yeast V-ATPase upon glucose deprivation, prompting further investigations that suggest reduced levels of Ras and protein kinase A activity are required to trigger disassembly upon glucose deprivation (31). In contrast, deletion of the master metabolic regulator Sch9 (sch9Δ) increases V-ATPase assembly in both the presence and absence of glucose (32). Rav2 was labeled with GFP in each of the mutant strains, and co-localization of Rav2-GFP with the vacuolar membrane dye FM4-64 was quantitated. This quantitation was necessary because many of the signaling pathways implicated in V-ATPase reversible disassembly give partial effects, so we anticipated that simple visualization of membrane association as described above might be inadequate to show effects.
As shown for the WT cells in Fig. 7, release of Rav2-GFP from the vacuolar membrane upon glucose deprivation results in a corresponding reduction in Pearson coefficient for colocalization with FM4-64, and the Pearson coefficient increases upon glucose readdition. We repeated this analysis for the pfk2Δ, ira2Δ, and sch9Δ mutants, analyzing Rav2-GFP localization in the presence of glucose, 15 min after glucose removal and 15 min after glucose readdition. As shown in Fig. 7, there is significantly less RAVE at the membrane under all conditions in the ira2Δ strain. In contrast, Rav2-GFP localization in the pfk2Δ mutant is significantly different from WT cells only under conditions of glucose deprivation, where pfk2Δ maintained significantly higher levels of Rav2-GFP at the vacuole. The sch9Δ mutant had lower levels of vacuolar Rav2-GFP than WT cells in the presence of glucose (both before and after glucose deprivation) but similar levels of vacuolar Rav2-GFP during glucose deprivation. Interestingly, these results suggest that steady-state localization of RAVE does not always reflect the previously observed localization in V1 in the mutants.
Figure 7.
Glucose-dependent localization of RAVE to the vacuolar membrane in mutants. A, Rav2 was tagged with GFP in the indicated WT and mutant cells. Cells at log phase were labeled with FM4-64 and subjected to a 90-min chase to allow transport of the dye to the vacuole. Each strain was visualized from the glucose growth medium (+Glucose), after a 15-min glucose deprivation (−Glucose) and 15 min after glucose readdition (Glucose add back). For each strain and condition, the same field was visualized under DIC (left), GFP fluorescence (middle), and Texas Red fluorescence (right) optics. Scale bar, 5 μm for all images. Images shown are representative of at least 50 cells imaged from at least two separate experiments. B, Pearson coefficient for colocalization of Rav2-GFP and FM4-64 was calculated for 24–40 cells for each strain and condition, obtained from at least two different experiments. For each strain, individual measurements are superimposed on bars corresponding to the mean Pearson coefficient for glucose-grown cells (green), −glucose cells (red), and glucose addback (yellow). Error bars, S.D. For each condition, the four strains were compared by one-way analysis of variance, and significant differences from the WT strain are indicated: ***, p < 0.0005; **, p < 0.005; *, p < 0.05; ns, p > 0.05.
Discussion
The data above suggest that the information required for glucose-dependent recruitment of RAVE to the vacuolar membrane resides in the interaction of Rav1 with Vph1-containing Vo sectors. This is an important insight, because it suggests that RAVE may play a more active role in V-ATPase reassembly than previously appreciated. Earlier work has established that RAVE can bind to both cytosolic V1 and V1 subunit C and has highlighted RAVE in a passenger role, accompanying V1 subunits to the membrane upon glucose addition and potentially aligning them for reassembly. The results reported here indicate that RAVE can relocalize in response to glucose levels independent of its V1 subunit C and V1 subcomplex partners (Fig. 2) (19). RAVE can even be recruited reversibly to the membrane under conditions where the V-ATPase does not disassemble (the vma11 E145L mutant; Fig. 3). These and previous results support the Rav1-Vph1 interaction as a key, glucose-sensitive interaction and imply that Rav1 can recognize Vph1 even when V1 is bound, consistent with the observed presence of RAVE at the vacuolar membrane in the presence of glucose.
A small, conserved, six amino acid motif located 88 amino acids upstream of the most conserved region of Rav1 is essential for RAVE function, and specifically for interaction of RAVE with Vph1 and recruitment to the vacuolar membrane. Rbcn3α subunits in higher eukaryotes share homology with Rav1 but are much larger. This motif is positioned 130–208 amino acids upstream of the most conserved sequence (corresponding to amino acids 840–1125 of yeast Rav1) in the higher eukaryotic Rbcn3α subunits shown in Fig. 3A, suggesting its position may be relevant for function. It is unlikely, however, that this region is the sole point of contact between Rav1 and Vph1, as we have previously shown that the most conserved fragment of Rav1 (Rav1(840–1125)), which does not contain this motif, also binds to Vph1NT in vitro (29). Nevertheless, it is notable that the rav1 6Δ mutation is sufficient to generate V-ATPase assembly and activity defects comparable with that seen in a complete RAV1 deletion mutant (Fig. 6).
We tested glucose-dependent localization of RAVE in three mutants involved in nutritional signaling, chosen because they have been reported to affect the level of V1 bound to Vo in the presence or absence of glucose (30–32). However, the steady-state localization of GFP-tagged RAVE in these mutants did not parallel the effects of the mutations on V1 localization. Specifically, the two mutants that were reported to induce increased V1-Vo assembly, ira2Δ and sch9Δ, actually had significantly lower levels of RAVE at the vacuolar membrane under most conditions. In contrast, the pfk2Δ mutant, which was reported to slow reassembly of V1 with Vo after glucose deprivation, maintained higher levels of RAVE at the vacuolar membrane during glucose deprivation. Although somewhat unexpected, these results emphasize that RAVE can move independently of V1. This might be expected, because the very different levels of RAVE and V1 subunits in yeast cells suggest that RAVE acts catalytically in V-ATPase assembly, requiring multiple cycles of recruitment from cytosol to vacuole to restore V-ATPase levels after glucose deprivation and readdition.
It is possible that localization of both the RAVE and V1 complexes arises from an equilibrium of release and reassociation with the vacuolar membrane that can be pushed in the disassembly or reassembly direction in the presence or absence of glucose. In WT cells maintained in glucose, the membrane-bound forms of RAVE and V1 predominate, whereas in the absence of glucose, cytosolic levels increase. However, the mutations perturb these equilibria, resulting in different steady-state localizations of RAVE and V1. There may also be states between fully membrane-bound and fully released for both complexes. Such intermediate states could account for the presence of more stably assembled V-ATPase complexes in the presence of high extracellular pH (33) and less stably assembled complexes in the presence of low phosphatidylinositol 3,5-bisphosphate (34). In addition, when viewed by fluorescence microscopy, a higher proportion of GFP-tagged V1 subunits appear to be at or near the vacuolar membrane in the absence of glucose than would be expected from biochemical experiments with lysed cells or isolated vacuoles (35). These V1 complexes may be in an intermediate disassembled state that is easily dissociated during lysis. Very little RAVE complex is present in isolated vacuolar membranes under any condition, also suggesting a fragile attachment. Additional studies of the spatial and temporal behavior of RAVE and V-ATPase subcomplexes, in mutants with varied nutrient signaling capacities, will be needed to address these questions.
Taken together, the data shown here indicate that RAVE can operate much more independently from the V-ATPase and its V1 and Vo domains than previously anticipated. The RAVE complex must also be considered a central player in glucose signaling of V-ATPase assembly state.
Experimental procedures
Materials and growth media
Oligonucleotides were purchased from MWG Operon. Sepharose A beads were purchased from GE Healthcare. Anti-Myc mAb (9E10) was purchased from Santa Cruz Biotechnology, Inc. Anti-FLAG M2 resin, mouse anti-FLAG antibody, and FLAG peptide were purchased from Sigma. Amylose and TALON resin were purchased from New England Biolabs and Clontech, respectively.
Media for yeast and Escherichia coli growth were purchased from Fisher. Yeast were maintained in either yeast extract-peptone-2% dextrose (YEPD) medium, buffered to pH 5.0 with 50 mm potassium phosphate and 50 mm potassium succinate, or in fully supplemented minimal synthetic complete (SC) medium with individual amino acids omitted for selection as indicated. The Rav− phenotype was tested by comparing growth on YEPD plates buffered to pH 5.0 with growth on YEPD plates buffered to pH 7.5 with 50 mm MES and 50 mm MOPS containing 60 mm CaCl2 at 30 °C and 37 °C. E. coli were maintained in either Luria broth (LB) or rich broth (LB supplemented with 2% glucose).
Yeast strain construction
All strains used this in this work were derived from either SF838-5A, BY4741, or BY4742 background. Strain genotypes and their sources are listed in Table 1. Oligonucleotides used to generate strains are listed in Table 2. Strains containing Rav1 or Rav2 C-terminally tagged with GFP or 13 copies of the Myc epitope were generated as described previously (19, 36). Rav1 C-terminally tagged with 13 copies of the Myc epitope was generated as described previously (16, 17).
Table 1.
Genotypes of yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SF838-5A (WT) | MATα ura3-52 leu2-3,112 his4-519 ade6 | Ref. 41 |
| SF838-5A Rav2-GFP | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-kanMX6 | Ref. 19 |
| SF838-5A Rav2-GFP vma4Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-kanMX6 vma4Δ::URA3 | This study |
| SF838-5A Rav2-GFP vma11Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-KanMX6 vma11Δ::LEU2 | This study |
| SF838-5A Rav2-GFP vma11 E145L | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-KanMX6 Nat-vma11 E145L | Ref. 17 |
| SF838-5A Rav2-GFP vph1Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-KanMX6 vph1Δ::LEU2 | This study |
| SF838-5A vma4Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 vma4Δ::URA3 | This study |
| SF838-5A vph1Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 vph1Δ::LEU2 | This study |
| SF838-5A rav1Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 rav1Δ::LEU2 | Ref. 17 |
| SF838-5A Rav1 6Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 | This study |
| SF838-5A Rav1 YH-AA | MATα ura3-52 leu2-3,112 his4-519 ade6 | This study |
| SF838-5A Rav1 K773A | MATα ura-52 leu2-3,112 his4-519 ade6 | This study |
| SF838-5A Rav1 6Δ-Myc | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV1-Myc13-kanMX6 | This study |
| SF838-5A Rav1 YH-AA-Myc | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV1-Myc13-kanMX6 | This study |
| SF838-5A Rav2-GFP rav1Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-KanMX6 rav1Δ::LEU2 | This study |
| SF838-5A Rav2-GFP Rav1 6Δ | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-kanMX6 rav1-6Δ | This study |
| SF838-5A Rav2-GFP Rav1 YH-AA | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-kanMX6 | This study |
| SF838-5A Rav2-GFP Rav1 K773A | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV2-GFP-kanMX6 | This study |
| SF838-5A Rav1-Myc | MATα ura3-52 leu2-3,112 his4-519 ade6 RAV1-Myc13-kanMX6 | Ref. 17 |
| BY4741 Vma5-GFP | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 VMA5-GFP::HIS3 | Ref. 42 |
| BY4741 Vma5-GFP Rav1 6Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 VMA5-GFP::HIS3 rav1-6Δ | This study |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| BY4741 Rav2-GFP | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RAV2-GFP-kanMX6 | This study |
| BY4741 Rav2-GFP pfk2Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RAV2-GFP-kanMX6 pfk2Δ::Nat | This study |
| BY4741 Rav2-GFP ira2Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RAV2-GFP-kanMX6 ira2Δ::Nat | This study |
| BY4742 sch9Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sch9Δ::Nat | Winderickx lab |
| Rav2-GFP sch9Δ | Spore from cross of BY4741 Rav2-GFP::Kan and BY4742 sch9Δ::Nat | This study |
Table 2.
Oligonucleotides used in this study
| Oligonucleotide name | Oligonucleotide sequence (5′ to 3′) |
|---|---|
| Rav1 730–830 Ura For | GCTTCCGGAAATCAATTTTACATCAAGGATAAATCTCTGGATTTGACTGATCCAACCTCTGACACATGCAGC |
| Rav1 730–830 Ura Rev | GTTGTTTTCGTTAGTTGCTCAGTAAGTGCTAGAGACACTGTCTTATTGAAACAGGTATTTCACACCGCAGGGTAA |
| Rav1 YH-AA For | GTGTTTTAAATGGTCCACTTCCAGTTGCCGCTCCACAATTTTTGATTCAAGCCATTTATGCG |
| Rav1 YH-AA Rev | CGCATAAATGGCTTGAATCAAAAATTGTGGAGCGGCAACTGGAAGTGGACCATTTAAAACAC |
| Rav1 6Δ For | CCATTTAAGTAGTGTTTTAAATGGTCCACAATTTTTGATTCAAGCCATTATGCG |
| Rav1 6Δ Rev | CGCATAAATGGCTTGAATCAAAAATTGTGGACCATTTAAAACACTACTTAATGG |
| Rav1 K773A For | GATTCAAGCCATTTATGCGAATGCGTTACAACTTGTCAAGGAGCTG |
| Rav1 K773A Rev | CAGCTCCTTGACAAGTTGTAACGCATTCGCATAAATGGCTTGAATC |
Strains harboring the rav1 6Δ and rav1 YH-AA mutations were constructed by first replacing the sequence for amino acids 730–830 of RAV1 with the URA3 gene and then replacing the URA3 gene with a mutagenized RAV1 fragment. To generate the rav1 730–830Δ::URA3 strain, the URA3 gene was amplified from pRS316 using oligonucleotides Rav1 730–830 Ura For and Rav1 730–830 Ura Rev (Table 2). The RAV1 mutations were introduced by a variation of QuikChange mutagenesis (Agilent), into a fragment encoding Rav1 679–898 cloned into pGEM T-Easy (Promega) (19). PCR amplification with mutagenic primers Rav1 6Δ For and Rav1 6Δ Rev, Rav1 YH-AA For and Rav1 YH-AA Rev, or Rav1 K773A For and Rav1 K773A Rev introduced the rav1 6Δ, rav1 YH-AA, and Rav1 K773A mutations, respectively. The product was treated with DpnI to remove the template plasmid before transformation in E. coli strain DH5α. Plasmids were sequenced to confirm the presence of the desired mutation, digested with EcoRI, and then transformed into the rav1 730–830Δ::URA3 strain. The transformation mixture was plated on YEPD, pH 5, and grown overnight at 30 °C and then replica-plated onto SD−Ura plates containing 5-fluoro-orotic acid (37) to select for replacement of URA3. Integration of the desired mutant allele was verified with PCR and DNA sequencing. Deletion of RAVE or V-ATPase subunits was performed as described (19).
Bacterial expression constructs, protein purification, and protein pulldowns
MBP-Vph1NT and WT MBP-Rav1(679–898)-His6 constructs were generated as described previously (19). Rav1 679–898 was mutagenized and verified as described above. Protein expression of MBP-Vph1NT (amino acids 1–372) and MBP-Rav1(679–898)-His6 was as described (19, 38). Rav1 and Vph1 constructs were inserted into BL-21 or Rosetta cells, respectively, for expression. Protein expression was induced by the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside cells at a density of 0.5–0.6 OD/ml, and growth was continued for 2 h at 37 °C for Rav1 or 16 h at 18 °C for Vph1. MBP-tagged proteins were affinity-purified on amylose columns, as described (19). Peak fractions were pooled and incubated with 10 μg of Prescission protease overnight at 4 °C to cleave off the MBP.
After overnight cleavage, proteins were dialyzed overnight in TALON equilibration buffer (50 mm sodium phosphate, 150 mm NaCl, 5 mm beta-mercaptoethanol, pH 7.4). Rav1 (679–898)-His6 was then mixed with 300 μl of TALON resin in combination with its binding partner, at a ratio of 1:5 Rav1(679–898)-His6 with Vph1-NT, and gently rocked at 4 °C for 2 h. For each sample, 30 μl from a total volume of 1.2 ml was reserved as an input fraction. The reserved input was combined with 30 μl of cracking buffer (50 mm Tris-HCl, pH 6.8, 8 m urea, 5% SDS, 1 mm EDTA), and 2.5 μl of the mixture was loaded for the gels in Fig. 4. The resin-protein mixture was poured into a column and washed with 3 ml of TALON wash buffer (50 mm sodium phosphate, 150 mm NaCl, 5 mm imidazole, pH 7.4), and proteins were eluted with 50 mm sodium phosphate, 150 mm NaCl, 250 mm imidazole, pH 7.4, into 500-μl fractions. Elution fractions were TCA-precipitated and dissolved in 80 μl of cracking buffer; 5 μl from each sample was separated by SDS-PAGE and transferred to nitrocellulose. The resulting blots were cut, and the top was probed with the anti-Vph1 (10D7), and the bottom portion was probed with mouse anti-His6 antibody (GenScript), followed by alkaline phosphatase–conjugated secondary antibody.
Immunoprecipitations and vacuolar purification
Co-immunoprecipitation of RAVE and V1 was based on methods described in Smardon et al. (17). For the experiments described here, cells containing WT or mutant Rav1 tagged with a Myc13 epitope were grown to log phase, and 50 OD600 units were pelleted by centrifugation. The cells were then lysed in cold extraction buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 0.1 mm EDTA) by agitation with glass beads, and cell debris and beads were removed by centrifugation. Sepharose A beads were added to preclear the suspension and then removed by centrifugation. 6 μl of anti-Myc mAb (9E10) was added to the supernatant, and the mixture was rocked at 4 °C for 6 h. Sepharose A beads were then added to the antibody-lysate mixture, which was rocked for another 2 h at 4 °C. After incubation, Sepharose beads were pelleted and washed three times with cold extraction buffer. 80 μl of hot cracking buffer was added directly to the beads, and immunoprecipitated protein was eluted at 95 °C for 10 min. Samples were separated by SDS-polyacrylamide gels, transferred to nitrocellulose, and then immunoblotted with anti-Myc (9E10) or anti-V1A (8B1F3) mAb followed by alkaline phosphatase–conjugated secondary antibody. Vacuolar vesicles were isolated and characterized as described previously (34).
Fluorescence microscopy
Yeast strains expressing Rav1-GFP or Rav2-GFP were grown to log phase in SC medium. Localization of GFP-tagged proteins was visualized by differential interference contrast (DIC) microscopy or fluorescence microscopy using a GFP filter set on a Zeiss Imager.Z1 fluorescence microscope equipped with a Hamamastu CCD camera and AxioVision software. The micrographs were assembled into figures using Adobe Photoshop CC. For all figures, the images shown are representative of at least 50 images obtained in at least two different experiments.
To observe the localization of GFP-tagged proteins under conditions of glucose deprivation and readdition, cells were grown overnight to log phase in SC medium. Cells from the overnight culture were imaged for the “+glucose” condition. For the “−glucose” condition, 1 ml of culture was removed, washed, and resuspended in SC medium lacking glucose and incubated at 30 °C for 15 min before imaging. For the “glucose readdition” condition, glucose was added back to the glucose-deprived cells to a concentration of 2% (w/v), and incubation was continued at 30 °C for an additional 15 min. Cells were then imaged at room temperature for a maximum of 15 min. RAVE subunit fluorescence images in Figs. 1–3 were obtained by importing .zvi files from the microscope into ImageJ, manually setting the brightness and contrast range to 0–3000, and then subtracting background using the “rolling ball radius of 50.0 pixels” setting. For comparison of glucose-dependent localization in WT and vma4Δ cells (Fig. 2), lines were drawn across fluorescence images of cells with well-defined vacuoles, and the fluorescence intensities across the line were determined using the “plot profile” feature. The intensity data were imported into Excel and plotted directly for the single-cell scans shown in Fig. 2A. Maximum values for each line scan were determined and divided by the average value of the +glucose sample to obtain the normalized maximal intensities in Fig. 2B. Data in Fig. 2B were plotted using GraphPad Prism.
For co-localization of Rav2-GFP with the vacuolar membrane, cells were grown overnight to log phase in YEPD. A volume of cells corresponding to 3 OD600 units was pelleted by centrifugation and resuspended in 1 ml of YEPD, and 8 μm FM4-64 was added (39). Tubes were covered in foil and rocked at 30 °C for 30 min. Cells were diluted to 15 ml with YEPD, spun down to remove free FM4-64, and then resuspended in 6 ml of YEPD and rocked at 30 °C for 90 min. Cells were washed twice with 15 ml of SC medium. After washing, cells were resuspended in 1 ml of SC medium, allowed to recover for 15 min, and then imaged. Images were obtained under DIC, GFP, and Texas Red optics in succession, from cells with glucose present, absent, and readded as above. To correct for drift during the relatively long exposure times required for visualizing Rav2-GFP, the resulting micrographs were layered in Photoshop CC, using the vacuole as a guide; the GFP image was placed over the DIC image, where vacuoles are prominent indentations, and then the Texas Red image was placed over GFP.
Co-localization of Rav2-GFP with vacuolar FM4-64 was quantified using Just Another Colocalization Plugin (JACoP) (40) in ImageJ to calculate the Pearson coefficient. Briefly, images were cropped from aligned GFP and FM4-64 micrographs and imported into ImageJ and then JACoP. Pearson coefficients were calculated with the GFP channel serving as Image A and FM4-64 serving as Image B. The depicted Pearson coefficients in Fig. 6 represent an average of at least 24 cells for each strain. Data were plotted using GraphPad Prism, and statistical significance was assessed using a one-way analysis of variance to compare WT and mutant strains under each condition.
Sequence analysis
Sequence analysis and identification of the conserved motif was performed using BLASTP on the NCBI BLAST server.
Author contributions
M. C. J. and P. M. K. conceptualization; M. C. J. and P. M. K. data curation; M. C. J. and P. M. K. formal analysis; P. M. K. supervision; P. M. K. funding acquisition; M. C. J. and P. M. K. validation; M. C. J. and P. M. K. investigation; M. C. J. and P. M. K. visualization; M. C. J. and P. M. K. writing-original draft; P. M. K. project administration; M. C. J. and P. M. K. writing-review and editing.
Acknowledgments
We thank Dr. Joris Winderickx (University of Leuven, Belgium) for the sch9Δ strain, Anne Smardon for construction of Rav2-GFP in vma4Δ, Madeline Clark for construction of the rav1 YH-AA mutant, and Maureen Tarsio for technical support.
This work was supported by National Institutes of Health Grant R01 GM127364 (to P. M. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- V-ATPase
- vacuolar H+-ATPase
- RAVE
- regulator of H+-ATPase of vacuoles and endosomes
- Rbcn3
- rabconnectin-3
- YH-AA
- rav1 760YH761 mutated to AA
- YEPD
- yeast extract-peptone-2% dextrose
- FM4-64
- N-(3-triethylammonium propyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide
- SC
- synthetic complete
- LB
- Luria broth
- MBP
- maltose-binding protein
- OD
- optical density
- DIC
- differential interference contrast.
References
- 1. Breton S., and Brown D. (2013) Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda) 28, 318–329 10.1152/physiol.00007.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Colacurcio D. J., and Nixon R. A. (2016) Disorders of lysosomal acidification: the emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 32, 75–88 10.1016/j.arr.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henderson K. A., Hughes A. L., and Gottschling D. E. (2014) Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife 3, e03504 10.7554/eLife.03504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deprez M. A., Eskes E., Wilms T., Ludovico P., and Winderickx J. (2018) pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 menage-a-trois to stress tolerance and longevity. Microb. Cell 5, 119–136 10.15698/mic2018.03.618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- 6. Manolson M. F., Wu B., Proteau D., Taillon B. E., Roberts B. T., Hoyt M. A., and Jones E. W. (1994) STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H+-ATPase subunit Vph1p. J. Biol. Chem. 269, 14064–14074 [PubMed] [Google Scholar]
- 7. Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., and Stevens T. H. (2001) The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276, 47411–47420 10.1074/jbc.M108310200 [DOI] [PubMed] [Google Scholar]
- 8. Kawasaki-Nishi S., Nishi T., and Forgac M. (2001) Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J. Biol. Chem. 276, 17941–17948 10.1074/jbc.M010790200 [DOI] [PubMed] [Google Scholar]
- 9. Parra K. J., Chan C. Y., and Chen J. (2014) Saccharomyces cerevisiae vacuolar H+-ATPase regulation by disassembly and reassembly: one structure and multiple signals. Eukaryot. Cell 13, 706–714 10.1128/EC.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y., Steinbusch L. K. M., Nabben M., Kapsokalyvas D., van Zandvoort M., Schönleitner P., Antoons G., Simons P. J., Coumans W. A., Geomini A., Chanda D., Glatz J. F. C., Neumann D., and Luiken J. J. F. P. (2017) Palmitate-induced vacuolar-type H+-ATPase inhibition feeds forward into insulin resistance and contractile dysfunction. Diabetes 66, 1521–1534 10.2337/db16-0727 [DOI] [PubMed] [Google Scholar]
- 11. Stransky L. A., and Forgac M. (2015) Amino acid availability modulates vacuolar H+-ATPase assembly. J. Biol. Chem. 290, 27360–27369 10.1074/jbc.M115.659128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGuire C. M., and Forgac M. (2018) Glucose starvation increases V-ATPase assembly and activity in mammalian cells through AMP kinase and phosphatidylinositide 3-kinase/Akt signaling. J. Biol. Chem. 293, 9113–9123 10.1074/jbc.RA117.001327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parra K. J., Keenan K. L., and Kane P. M. (2000) The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J. Biol. Chem. 275, 21761–21767 10.1074/jbc.M002305200 [DOI] [PubMed] [Google Scholar]
- 14. Couoh-Cardel S., Milgrom E., and Wilkens S. (2015) Affinity purification and structural features of the yeast vacuolar ATPase Vo membrane sector. J. Biol. Chem. 290, 27959–27971 10.1074/jbc.M115.662494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma S., Oot R. A., Khan M. M., and Wilkens S. (2019) Functional reconstitution of vacuolar H+-ATPase from Vo proton channel and mutant V1-ATPase provides insight into the mechanism of reversible disassembly. J. Biol. Chem. 294, 6439–6449 10.1074/jbc.RA119.007577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seol J. H., Shevchenko A., Shevchenko A., and Deshaies R. J. (2001) Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3, 384–391 10.1038/35070067 [DOI] [PubMed] [Google Scholar]
- 17. Smardon A. M., Tarsio M., and Kane P. M. (2002) The RAVE complex is essential for stable assembly of the yeast V-ATPase. J. Biol. Chem. 277, 13831–13839 10.1074/jbc.M200682200 [DOI] [PubMed] [Google Scholar]
- 18. Seol J. H., Feldman R. M., Zachariae W., Shevchenko A., Correll C. C., Lyapina S., Chi Y., Galova M., Claypool J., Sandmeyer S., Nasmyth K., Shevchenko A., and Deshaies R. J. (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626 10.1101/gad.13.12.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smardon A. M., Nasab N. D., Tarsio M., Diakov T. T., and Kane P. M. (2015) Molecular interactions and cellular itinerary of the yeast RAVE (regulator of the H+-ATPase of vacuolar and endosomal membranes) complex. J. Biol. Chem. 290, 27511–27523 10.1074/jbc.M115.667634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakisaka T., and Takai Y. (2005) Purification and properties of rabconnectin-3. Methods Enzymol. 403, 401–407 10.1016/S0076-6879(05)03035-1 [DOI] [PubMed] [Google Scholar]
- 21. Yan Y., Denef N., and Schüpbach T. (2009) The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell 17, 387–402 10.1016/j.devcel.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Einhorn Z., Trapani J. G., Liu Q., and Nicolson T. (2012) Rabconnectin3α promotes stable activity of the H+ pump on synaptic vesicles in hair cells. J. Neurosci. 32, 11144–11156 10.1523/JNEUROSCI.1705-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merkulova M., Păunescu T. G., Azroyan A., Marshansky V., Breton S., and Brown D. (2015) Mapping the H+ (V)-ATPase interactome: identification of proteins involved in trafficking, folding, assembly and phosphorylation. Sci. Rep. 5, 14827 10.1038/srep14827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ho B., Baryshnikova A., and Brown G. W. (2018) Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6, 192–205.e3 10.1016/j.cels.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 25. Kane P. M., Tarsio M., and Liu J. (1999) Early steps in assembly of the yeast vacuolar H+-ATPase. J. Biol. Chem. 274, 17275–17283 10.1074/jbc.274.24.17275 [DOI] [PubMed] [Google Scholar]
- 26. Smardon A. M., and Kane P. M. (2007) RAVE is essential for the efficient assembly of the C subunit with the vacuolar H+-ATPase. J. Biol. Chem. 282, 26185–26194 10.1074/jbc.M703627200 [DOI] [PubMed] [Google Scholar]
- 27. Hirata R., Graham L. A., Takatsuki A., Stevens T. H., and Anraku Y. (1997) VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J. Biol. Chem. 272, 4795–4803 10.1074/jbc.272.8.4795 [DOI] [PubMed] [Google Scholar]
- 28. Umemoto N., Ohya Y., and Anraku Y. (1991) VMA11, a novel gene that encodes a putative proteolipid, is indispensable for expression of yeast vacuolar membrane H+-ATPase activity. J. Biol. Chem. 266, 24526–24532 [PubMed] [Google Scholar]
- 29. Smardon A. M., Diab H. I., Tarsio M., Diakov T. T., Nasab N. D., West R. W., and Kane P. M. (2014) The RAVE complex is an isoform-specific V-ATPase assembly factor in yeast. Mol. Biol. Cell 25, 356–367 10.1091/mbc.e13-05-0231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan C. Y., and Parra K. J. (2014) Yeast phosphofructokinase-1 subunit Pfk2p is necessary for pH homeostasis and glucose-dependent vacuolar ATPase reassembly. J. Biol. Chem. 289, 19448–19457 10.1074/jbc.M114.569855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bond S., and Forgac M. (2008) The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast. J. Biol. Chem. 283, 36513–36521 10.1074/jbc.M805232200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilms T., Swinnen E., Eskes E., Dolz-Edo L., Uwineza A., Van Essche R., Rosseels J., Zabrocki P., Cameroni E., Franssens V., De Virgilio C., Smits G. J., and Winderickx J. (2017) The yeast protein kinase Sch9 adjusts V-ATPase assembly/disassembly to control pH homeostasis and longevity in response to glucose availability. PLoS Genet. 13, e1006835 10.1371/journal.pgen.1006835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diakov T. T., and Kane P. M. (2010) Regulation of vacuolar proton-translocating ATPase activity and assembly by extracellular pH. J. Biol. Chem. 285, 23771–23778 10.1074/jbc.M110.110122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li S. C., Diakov T. T., Xu T., Tarsio M., Zhu W., Couoh-Cardel S., Weisman L. S., and Kane P. M. (2014) The signaling lipid PI(3,5)P2 stabilizes V1-Vo sector interactions and activates the V-ATPase. Mol. Biol. Cell 25, 1251–1262 10.1091/mbc.e13-10-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tabke K., Albertmelcher A., Vitavska O., Huss M., Schmitz H. P., and Wieczorek H. (2014) Reversible disassembly of the yeast V-ATPase revisited under in vivo conditions. Biochem. J. 462, 185–197 10.1042/BJ20131293 [DOI] [PubMed] [Google Scholar]
- 36. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 37. Sherman F., Fink G. R., and Hicks J. B. (1982) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 38. Diab H., Ohira M., Liu M., Cobb E., and Kane P. M. (2009) Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J. Biol. Chem. 284, 13316–13325 10.1074/jbc.M900475200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vida T. A., and Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bolte S., and Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 41. Stevens T. H., Rothman J. H., Payne G. S., and Schekman R. (1986) Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J. Cell Biol. 102, 1551–1557 10.1083/jcb.102.5.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]