Abstract
Proteinase-activated receptor (PAR)-4 is a member of the proteolytically-activated PAR family of G-protein–coupled receptors (GPCR) that represents an important target in the development of anti-platelet therapeutics. PARs are activated by proteolytic cleavage of their receptor N terminus by enzymes such as thrombin, trypsin, and cathepsin-G. This reveals the receptor-activating motif, termed the tethered ligand that binds intramolecularly to the receptor and triggers signaling. However, PARs are also activated by exogenous application of synthetic peptides derived from the tethered-ligand sequence. To better understand the molecular basis for PAR4-dependent signaling, we examined PAR4-signaling responses to a peptide library derived from the canonical PAR4-agonist peptide, AYPGKF-NH2, and we monitored activation of the Gαq/11-coupled calcium-signaling pathway, β-arrestin recruitment, and mitogen-activated protein kinase (MAPK) pathway activation. We identified peptides that are poor activators of PAR4-dependent calcium signaling but were fully competent in recruiting β-arrestin-1 and -2. Peptides that were unable to stimulate PAR4-dependent calcium signaling could not trigger MAPK activation. Using in silico docking and site-directed mutagenesis, we identified Asp230 in the extracellular loop-2 as being critical for PAR4 activation by both agonist peptide and the tethered ligand. Probing the consequence of biased signaling on platelet activation, we found that a peptide that cannot activate calcium signaling fails to cause platelet aggregation, whereas a peptide that is able to stimulate calcium signaling and is more potent for β-arrestin recruitment triggered greater levels of platelet aggregation compared with the canonical PAR4 agonist peptide. These findings uncover molecular determinants critical for agonist binding and biased signaling through PAR4.
Keywords: G-protein–coupled receptor (GPCR), bioluminescence, bioluminescence resonance energy transfer (BRET), mitogen-activated protein kinase (MAPK), arrestin, docking, platelet, biased signaling, G-protein, peptide, protease-activated receptor-4 (PAR4)
Introduction
G-protein–coupled receptors (GPCRs)4 are the largest family of cell-surface receptors and regulate a host of important physiological responses (1, 2). GPCRs respond to a variety of extracellular signals and regulate cellular behavior through engaging intracellular effector molecules to activate various cell-signaling pathways (3, 4). In drug discovery, GPCRs are a valuable and highly-tractable class of drug targets with over 30% of all currently-approved drugs acting on GPCRs (5). GPCR activation occurs typically through binding of a soluble ligand and conformational changes that enable engagement of G-protein–dependent or β-arrestin–mediated signal transduction. The proteinase-activated receptors (PARs) are a four-member family of GPCRs with a distinct mechanism of activation that involves limited proteolysis and unmasking of a receptor-activating motif called the tethered ligand. PARs were first discovered in an effort to identify the cellular receptors responsible for actions of thrombin that were independent from its role in the coagulation cascade (6). Various proteinases have now been described as activators of the different members of the PAR family, but canonically PAR1 and PAR4 are described as thrombin-activated receptors, whereas PAR2 is activated by trypsin and other trypsin-like serine proteinases. PAR4 can also be activated by trypsin. Whether PAR3 signals independently is unclear, although thrombin can cleave the PAR3 N terminus to reveal a tethered ligand (7, 8). Following the discovery of PARs, it was quickly realized that two receptors, PAR1 and PAR4, serve as the thrombin receptors on human platelets (9, 10). PAR1 and PAR4 both trigger platelet activation and aggregation. Given that anti-platelet agents are an important class of drugs for the treatment of various cardiovascular diseases, there has been a concerted effort aimed at understanding the role of these receptors in platelet activation and in identifying molecules that can act as PAR1 and PAR4 antagonists. A PAR1 antagonist was recently identified and approved for clinical use; however, significant bleeding side effects were noted in patients administered this drug (11–13). Although bleeding is a frequently-encountered side effect of therapeutically-targeting pathways involved in blood coagulation, it was hoped that targeting PAR1 would not present such liability. Much attention has now shifted to understanding and targeting the other thrombin receptor PAR4. Although both PAR1 and PAR4 are activated by thrombin, there are some key differences reported in the activation of these two receptors on platelets that argue for PAR4 being a better anti-platelet drug target than PAR1. PAR1 possesses a hirudin-like binding site that enables it to bind thrombin with relatively high affinity (14, 15). In lacking this thrombin-binding site, PAR4 can only be activated at much higher concentrations of thrombin, such as may be encountered on a growing platelet thrombus. There are also reported differences in the kinetics of PAR1- and PAR4-dependent calcium signaling, with PAR4 causing a delayed but more sustained signal compared with PAR1 (16). This is thought to be due to dual proline residues in the PAR4 exodomain that provide low-affinity interactions with the thrombin-active site and an anionic cluster that slows dissociation from thrombin (17). Overall, PAR1 is thought to initiate platelet activation in response to low concentrations of thrombin, whereas PAR4 is engaged at higher concentrations to further consolidate and stabilize the clot.
Given the liabilities seen with targeting PAR1 as a platelet thrombin receptor, there has been renewed interest in developing small molecule PAR4 antagonists (18–20). Recently, small molecule antagonists of PAR4 have been described with efficacy in inhibiting injury-induced thrombosis in both nonhuman primates and humans (21–23). Despite these promising advances, there remains the question of whether a complete inhibition of PAR4 might also lead to bleeding liability. Our efforts have therefore focused on examining biased signaling through PAR4 in order to identify pathways that are responsible for platelet aggregation and that could be selectively manipulated for therapeutic efficacy. PAR4 can activate multiple signaling pathways through coupling to Gαq/11, Gα12/13, and β-arrestin (24–26). In previous work, we and others (26, 27) showed that pepducins targeting different intracellular motifs in PAR4 could be employed to differentially inhibit platelet activation by blocking G-protein–coupled or β-arrestin-dependent signaling. The ability to selectively activate or inhibit GPCR signaling in a biased manner is of great interest and has been proposed as a strategy for obtaining therapeutically superior drugs for a number of conditions (28–30). In the case of the finely-balanced signaling systems involved in coagulation and platelet activation, such subtle perturbations may be key to obtaining drugs that do not result in bleeding liability.
Here, we seek to gain an understanding of the molecular interactions responsible for agonist docking to PAR4 and to identify interactions that can be manipulated to obtain biased ligands for PAR4. By screening a peptide library derived from the most widely used PAR4 agonist peptide, AYPGKF-NH2, we have identified key residues that confer selective activation of Gαq/11-coupled calcium signaling and/or β-arrestin recruitment. Through in silico docking and confirmatory mutagenesis experiments, we further identify an extracellular loop 2 residue (Asp230) that is critical for receptor activation and signaling. We also examine platelet activation by peptides that show differential activation of PAR4 signaling. Overall, these studies advance our understanding of PAR4 activation and signaling and will help guide future drug discovery efforts targeting this receptor.
Results
To understand the rules governing agonist peptide activation of PAR4, we synthesized 37 hexapeptides through modification to the synthetic PAR4 agonist peptide AYPGKF-NH2. A number of different modifications were examined at each of the six positions, including alanine, d-isomer, and N-methyl substitutions, as well as other modifications aimed at further probing each of the six agonist peptide positions. Henceforth, the parental PAR4 agonist peptide for this study, AYPGKF-NH2 (Ala1–Tyr2–Pro3–Gly4–Lys5–Phe6–NH2; superscript denotes position from the N terminus), will be referred to as PAR4-AP. Gαq/11-coupled calcium signaling, β-arrestin-1/2 recruitment (β-arr-1–rluc and β-arr-2–rluc, respectively), and p44/42 mitogen-activated protein kinase (MAPK) signaling by each of these peptides was monitored and compared with responses triggered by PAR4-AP (Table 1). Whereas PAR4-AP-triggered Gαq/11 signaling reached a clear plateau over the range of concentrations tested (0.3–300 μm), β-arrestin recruitment did not, and therefore, the effective concentration required to achieve half-maximal response (EC50) values could not be accurately assessed. In contrast, with the substituted peptides, Gαq/11 signaling concentration–effect curves in some cases did not clearly plateau, whereas β-arrestin recruitment did. Where possible, EC50 values are compared between PAR4-AP and substituted peptides. However, because clear maxima were not always obtained, comparisons of signal magnitude throughout this study are presented as the response elicited by a given compound at 100 μm, shown as a percentage of responses elicited by PAR4-AP at the same concentration. Because the concentration–effect curve for PAR4-AP exhibits a saturating calcium signal, peptides that failed to saturate calcium signaling responses are depicted as having EC50 values that were “not determined” (ND). Maximal calcium signal responses are shown as a percentage of the response obtained with the calcium ionophore A23187 (Table 1, Maximum % of A23187 calcium ionophore) and are referred to throughout the “Results” as a change in maximal calcium signaling as a measure of the efficacy of the peptides. Throughout the “Results,” β-arrestin recruitment values are presented as a percentage (%) of the PAR4-AP–induced recruitment at 100 μm concentration. For ease of comparison, concentration–effect curves for PAR4-AP–mediated Gαq/11 calcium signaling and β-arrestin recruitment are displayed as a repeated reference value in Figs. 1–7.
Table 1.
Summary table of all calcium, β-arrestin, and p44/42 data
Calcium data are displayed as EC50 or not determined (ND) if EC50 values could not be calculated. Maximum calcium response achieved with each peptide is shown as a percentage of the maximum calcium response achievable in a given experiment revealed by calcium ionophore (% of A23187 calcium ionophore). Statistically significant shifts in EC50 values compared with PAR4-AP, AYPGKF-NH2, were found using the F-statistic (*, p < 0.05). Given that saturation did not always occur, all peptides are compared with the parental PAR4-AP, AYPGKF-NH2, and shown as a percent of the response obtained with PAR4-AP at a given concentration (100 μm). Finally, fold increase over unstimulated baseline is shown for phosphorylation of p44/42 MAPK activation. Statistical significance for comparisons at 100 μm and p44/42 phosphorylation are calculated using one-way ANOVA of response compared to PAR4-AP (*, p < 0.05).
Figure 1.
Calcium and β-arrestin recruitment in response to alanine (A–C); d-isomer (D–F), or N-methylation (G–I) substitutions in the PAR4-AP. A, D, and G, agonist-stimulated calcium signaling. Calcium signaling is shown as a percentage of the maximum calcium signaling obtained by application of calcium ionophore (A23187, 3 μm). (n = 3–7). Agonist-stimulated recruitment of β-arrestin-1 (B, E, and H) and β-arrestin-2 (C, F, and I) to PAR4 is shown. Concentration effect curves of BRET values from cells treated by increasing concentrations of peptide are presented as normalized net BRET ratio (n = 3).
Figure 2.
Calcium and β-arrestin recruitment in response to additional substitutions to the PAR4-AP by position (A, D, G, and J) for agonist-stimulated calcium signaling is shown. Calcium signaling is shown as a percentage of the maximum calcium signaling obtained by application of calcium ionophore (A23187, 3 μm) (n = 3–7). Agonist-stimulated recruitment of β-arrestin-1 (B, E, H, and K) and β-arrestin-2 (C, F, I, and L) to PAR4. Concentration effect curves of BRET values from cells treated increasing concentrations of peptide are presented as normalized net BRET ratio (n = 3).
Figure 3.
PAR4-mediated phosphorylation of p44/42(ERK) is attenuated upon stimulation of PAR4 with peptides unable to stimulate PAR4-dependent calcium signaling. HEK-293 cells stably expressing PAR4-YFP were stimulated with AYPGKF-NH2 (30 μm) or peptide (30 μm) from a library of compounds and Western blotting for p44/42 phosphorylation (p-p44/42). Data are shown as normalized fold increase over unstimulated control cells. Dashed baseline shows mean of unstimulated baseline for comparison. Dotted baseline shows mean response achieved following AYPGKF-NH2 stimulation. (#, p > 0.05 AYPGKF-NH2 compared with untreated control; *, p > 0.05 peptide agonists compared with AYPGKF-NH2; one-way ANOVA). Peptides that were unable to stimulate calcium signaling were also generally unable to stimulate phosphorylation of p44/42. Interestingly, several peptides were able to significantly increase phosphorylation of p44/42 compared with AYPGKF-NH2. (Representative blots are shown in Fig. S4; n = 4–5.)
Figure 4.
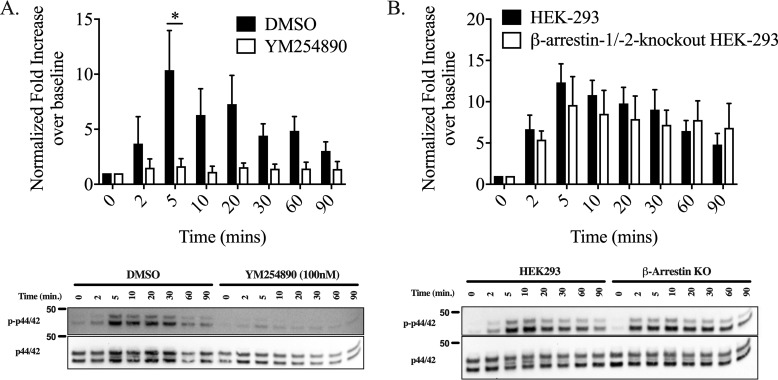
PAR4-mediated phosphorylation of p44/42 (ERK) is Gαq/11-dependent and β-arrestin–independent. To evaluate the contribution of Gαq/11 and β-arrestin to p44/42 phosphorylation, p-p44/42 was monitored by Western blotting following stimulation with AYPGKF-NH2 (30 μm) for various time points. A, to evaluate the contribution of Gαq/11-signaling, cells were incubated with vehicle control (0.001% DMSO) or the Gαq/11-inhibitor, YM254890 (100 nm), prior to agonist stimulation. B, to determine the contribution of β-arrestin–signaling to p44/42 phosphorylation, MAPK activation in HEK-293 and β-arrestin-1/2 knockout cells was analyzed following agonist stimulation. Values are expressed as fold increase over unstimulated 0-min time ((p-p44/22/total p44/42)/(baseline p-p44/42/total p44/42)). We find that Gαq/11 inhibition with YM254890 results in a significant reduction of p44/42 phosphorylation. We observed no statistically significant difference in ERK activation between HEK-293 and β-arrestin-1/2 knockouts HEK-293 cells. Data were analyzed by two-way ANOVA (* indicates p < 0.05). p44/42 MAPK activation did remain sustained up to 90 min in the β-arrestin-1/2 knockout HEK-293 cells whereasp44/42 MAPK phosphorylation in HEK-293 cells decrease toward the baseline by 90 min (5 min compared with the 90-min time point is significantly different in HEK-293 cells, p < 0.05, but not in β-arrestin-1/2 knockout HEK-293, p = not significant, two-tailed t test). (Representative blots are shown; n = 4.)
Figure 5.
Asp230 is essential for PAR4 activation with both PAR4-AP and tethered ligand (revealed by thrombin cleavage) activation. A, frequency table of PAR4/AYPGKF-NH2 interactions as predicted by in silico peptide docking (GalaxyPepDock). Numbers represent the frequency with which an atom in a PAR4-AP residue was predicted to interact with an atom of a PAR4 residue. B, representation of the most highly-predicted AYPGKF-NH2–binding site (Asp230, shown as sticks) interacting (yellow dashed lines) with position 5 (Lys5; shown as ball and sticks) of AYPGKF-NH2. Interaction of Lys5 and Asp230 was highly predicted in 7/10 models (B). Alignment of the receptor backbone (shown as a transparent cartoon) reveals similar binding poses adopted by the side-chain position of Asp230 and the position of Lys5 of AYPGKF-NH2. AYPGKF-NH2- and tethered ligand-stimulated (thrombin revealed) calcium signaling and β-arrestin-1/2 recruitment in PAR4 extracellular loop 2 mutants. Agonist-stimulated calcium signaling: calcium signaling in response to AYPGKF-NH2 (C) or thrombin (F) was recorded in HEK-293 cells transiently expressing PAR4-YFP, single site-directed mutant PAR4-YFP (PAR4H229A-YFP and PAR4D230A-YFP, PAR4Q242A-YFP), or double site-directed mutant PAR4-YFP (PAR4H229A, D230A-YFP and PAR4H229D, A231D-YFP). Data are shown as a percentage of maximum response obtained in cells treated with the calcium ionophore A23187 (3 μm) for each transient cell line, per experimental day. AYPGKF-NH2 concentration is in mass, and thrombin concentration is shown as units (n = 3–5). Agonist-stimulated β-arrestin-1/2 recruitment: HEK-293 cells were transiently transfected with BRET pair of either β-arrestin-1-rluc or -2-rluc and PAR4-YFP or site-directed mutant PAR4-YFP (described above), and agonist-stimulated arrestin recruitment to PAR4 was monitored. Cells were incubated with increasing concentration of either AYPGKF-NH2 (D and E) or thrombin (G and H) for 20 min, and BRET values were recorded. BRET values from cells treated with HBSS were also collected to normalize the data. Renilla luciferase substrate coelenterazine-h (5 μm) was added 10 min prior to data collection. Data are presented as the normalized net BRET ratio (eYFP/rluc − baseline (HBSS control)). AYPGKF-NH2 concentration is in mass, and thrombin concentration is shown as units (n = 4).
Figure 6.

PAR4 agonist-mediated platelet aggregation. Both thrombin (1 unit/ml; shown in black) and AYPGKF-NH2 (100 μm; shown in gray) initiated PAR4-dependent rat platelet aggregation. We observed that stimulation of washed platelet samples with a β-arrestin biased peptide that is unable to stimulate calcium signaling, AyPGKF-NH2 (100 μm; shown in yellow), did not stimulate platelet aggregation. Furthermore, stimulation of washed platelets with the peptide agonist AYPGRF-NH2 (100 μm; shown in purple) caused a robust platelet aggregation comparable with the response with 1unit/ml thrombin response (n = 2–4; representative trace shown).
Effect of alanine substitution on PAR4-AP–mediated signaling responses
Our first set of changes involved alanine substitutions at positions 2–6 of the PAR4-AP (calcium EC50 = 12.1 ± 3.1 μm; β-arrestin-1 recruitment at 100 μm for comparison is considered 100 ± 3.9 and 100 ± 3.5% for β-arrestin-2) (Fig. 1, A–C, and Table 1). Potency was decreased with respect to calcium signaling when Tyr2 was substituted with alanine and failed to reach a plateau over the concentration range tested (AAPGKF-NH2;5 EC50 = ND). Correspondingly, β-arrestin recruitment was reduced compared with PAR4-AP at the same concentration (β-arr-1 47.3 ± 3.6%; β-arr-2 46.4 ± 3.2%). Substitution of Pro3 with alanine resulted in a significant decrease in potency for calcium signaling (AYAGKF-NH2 EC50 = 63.7 ± 20.8 μm) and yielded reduced β-arrestin recruitment (β-arr-1, 48.5 ± 7.9%; β-arr-2, 43.4 ± 7.1%). Either Gly4 or Phe6 substitution to alanine resulted in modest decreases in maximal calcium signaling compared with PAR4-AP at the same concentration, with no significant effect on potency (EC50 = 21.2 ± 6.6 μm; EC50 = 17.3 ± 5.0 μm, respectively). Additionally, alanine substitutions in these positions resulted in significantly-reduced β-arrestin recruitment to levels approximately half of those observed with PAR4-AP (AYPAKF-NH2: β-arr-1, 56.4 ± 5.6%; β-arr-2, 54.6 ± 5.1%; AYPGKA-NH2: β-arr-1, 47.9 ± 3.8%; β-arr-2, 52.6 ± 3.5%). Interestingly, we observed that Lys5 to alanine substitution had no appreciable impact on potency with respect to calcium signaling (AYPGAF-NH2 EC50 = 9.8 ± 2.9 μm) and modestly-reduced recruitment of β-arrestin-1 (59.3 ± 7.2%) and -2 (70.8 ± 6.5%). Collectively, alanine substitutions of positions 2–6 resulted in decreased β-arrestin recruitment compared with the parental peptide (Fig. 1, B and C, and Table 1). Interestingly, we observe differential effects on calcium signaling with significant detriment to potency attributed to tyrosine substitution (Fig. 1A). Additionally, substitution of glycine and phenylalanine moderately decreased maximal calcium-signaling responses, perhaps indicating that these residues are important for full agonism of this pathway compared with that achieved with the parental peptide. Notably, all single alanine substitutions of positions 2–6 of the PAR4-AP retained some agonist activity indicating that signaling is not wholly reliant on any single residue alone.
Effect of d-isomer amino acid substitutions on PAR4-AP–mediated signaling responses
To assess the impact of stereochemical inversion at each residue position, we generated five peptides with single amino acid substitutions from l- to their respective d-isomers (Fig. 1, D–F, and Table 1). Glycine possesses only one isomeric form and thus is not substituted. Both l-alanine to d-alanine(aYPGKF-NH2)6 and l-lysine to d-lysine (AYPGkF-NH2) substitutions resulted in significant losses of potency in calcium-signaling assays compared with PAR4-AP. Consistent with calcium data, we observed a significant decrease in β-arrestin recruitment compared with the parental PAR4-AP (aYPGKF-NH2: β-arr-1, 48.7 ± 2.6%; β-arr-2, 47.5 ± 2.4%; AYPGkF-NH2: β-arr-1, 47.4 ± 1.0%; β-arr-2, 36.5 ± 0.9%). Interestingly, both d-tyrosine (AyPGKF-NH2) and d-proline (AYpGKF-NH2) substitutions completely abolished calcium signaling. Comparably, β-arrestin recruitment was significantly reduced with stimulation by either peptide (AyPGKF-NH2: β-arr-1, 50.7 ± 4.6%; β-arr-2, 46.4 ± 4.2%; AYpGKF-NH2: β-arr-1, 45.2 ± 10.5%; β-arr-2, 38.1 ± 9.5%). In keeping with what we observed with phenylalanine to alanine substitution (Fig. 1, A–C), d-phenylalanine substitution in position 6 (AYPGKf-NH2) resulted in less efficacious agonism of both calcium (EC50 = 22.2 ± 4.7 μm) and β-arrestin recruitment (β-arr-1, 80.3 ± 6.6%; β-arr-2, 74.1 ± 6.0%). Overall, d-isomers substitutions of amino acids Ala1, Tyr2, Pro3, and Lys5 resulted in significant reduction of (d-alanine and d-lysine) or abolition of (d-tyrosine and d-proline) calcium signaling while causing significant reductions in β-arrestin recruitment. Substitution of l-Phe6 for d-Phe was the only d-isomer substitution that is somewhat tolerated, resulting in modest reductions in potency with respect to both calcium signaling and β-arrestin recruitment. These data reveal that the side-chain stereochemistry of residues within the AYPGKF-NH2 peptide performs a role in agonism.
Effect of N-methylation on PAR4-AP–mediated signaling responses
We generated a series of peptides to investigate the pharmacological impact of N-methylation along the peptide backbone (Fig. 1, G–I, and Table 1). In contrast to the parental PAR4-AP, all but one of the N-methylated peptides evaluated yielded clear plateaus and discernible EC50 values in β-arrestin recruitment assays, and where applicable, these are indicated below along with their relative responses compared with PAR4-AP at 100 μm. Substitution of Ala1 with N-methylalanine ((N-Me-A-YPGKF-NH2) almost completely abolished calcium signaling (EC50 = ND). Interestingly, this peptide was able to initiate β-arrestin-1/2 recruitment comparable with levels obtained with PAR4-AP (β-arr-1, 119.2 ± 11.9%; EC50 = 6.8 ± 2.6 μm; β-arr-2, 80.6 ± 10.8%; EC50 = 2.0 ± 0.7 μm). These results suggest that increasing steric bulk and hydrophobicity of the backbone at amide positions through the addition of an N-terminal methyl group drives PAR4 signaling toward β-arrestin pathways and causes only modest activation of calcium signaling. Substitution of Tyr2 for N-methyltyrosine (A-(N-Me-Y)-PGKF-NH2) abolished calcium signaling up to 300 μm (EC50 = ND). Consistent with calcium signaling, we observed decreased signal magnitude in β-arrestin recruitment assays compared with PAR4-AP (β-arr-1, 71.4 ± 10.8%; EC50 = 14.2 ± 8.6 μm; β-arr-2, 61.5 ± 9.8%; EC50 = 11.8 ± 5.7 μm). Substitution of the PAR4-AP Gly4 with sarcosine (Sar, also known as N-methylglycine; AYP-Sar-KF-NH2) resulted in significantly reduced calcium potency (EC50 = ND) and significantly reduced β-arrestin-1/2 recruitment (35.9 ± 4.3 and 36.1 ± 3.9%, respectively). N-Methylation of the α-amine of Lys5 (AYPG-(Nα-Me-K)-F-NH2) resulted in significantly reduced calcium signaling (EC50 = ND) with no significant effect on maximal β-arrestin recruitment (β-arr-1, 126.7 ± 18.6%; EC50 = 12.2 ± 4.3 μm; β-arr-2, 77.1 ± 16.9%; EC50 = 2.5 ± 1.0 μm). Substitution of Phe6 with N-methylphenylalanine (AYPGK-(N-Me-F)-NH2) was equipotent for calcium signaling compared with PAR4-AP (EC50 = 7.6 ± 1.9 μm). Interestingly, this peptide showed increased recruitment of both β-arrestins (β-arr-1, 136.9 ± 18.0%; EC50 = 10.4 ± 3.3 μm; β-arr-2, 96.6 ± 16.4%; EC50 = 15.3 ± 4.6 μm). Together, these data reveal that N-methylation of positions 1, 2, 4, and 5 are deleterious to calcium signaling.
We were excited to observe evidence of β-arrestin–biased peptides within this group, all of which showed decreased calcium signaling and/or increased β-arrestin recruitment. Moreover, the leftward shifts in their associated β-arrestin concentration–effect profiles imply that substitution of N-methylated Ala1, Tyr2, Lys5, or Phe6 residues increases peptide potency with respect to G-protein–independent signaling. Only N-methylation of Gly4 (Sar) resulted in a reduced overall ability to activate PAR4, with relative calcium and β-arrestin signals being reduced by ∼90 and 60%, respectively.
Effect of N-terminal and position 1 modifications on PAR4-AP–mediated signaling responses
To further investigate the impact of N-terminal alterations to the PAR4-AP, we examined the impact of N-terminal acetylation and substitutions of position 1 (Fig. 2, A–C, and Table 1). Previously, it was demonstrated that protonation of the N terminus of the PAR1-tethered ligand (SFLLRN) is critical for agonist function (31). Additionally, substitution of glycine for mercaptopropionic acid (Mpr), which lacks an N-terminal protonated amine found in the native tethered ligand-mimicking peptide GYPGQV, resulted in complete loss of agonist activity (32). Given the evidence for N-terminal protonation in PAR agonism, we sought to determine the impact of N-terminal acetylation on PAR4-AP activity. N-terminal acetylation of the PAR4-AP (Ac-AYPGKF-NH2) negatively impacted both calcium signaling (EC50 = ND) as well as the magnitude of β-arrestin recruitment to PAR4 as expected (β-arr-1, 47.3 ± 3.1%; EC50 = 15.0 ± 4.3 μm; β-arr-2, 45.5 ± 2.8; EC50 = 26.7 ± 5.6 μm). In contrast to the previously reported complete loss of activity with Mpr-YPGQV peptide, we observed partial agonism with the loss of protonation through acetylation of PAR4-AP. This could also be influenced by the additional methyl group of the Ala in Ac-AYPGKF-NH2, which is not present in the Mpr-YPGQV peptide.
In both the native human PAR4-tethered-ligand sequence and the PAR4-AP, position 1 is occupied by small (tethered ligand − glycine, PAR4-AP − alanine) and nonpolar (PAR4-AP − alanine) residues. To investigate the role of position 1 in the PAR4-AP, we generated seven peptides with position 1 substitutions.
Introduction of steric bulk and increased hydrophobicity through substitution of Ala1 for valine resulted in partial agonism of both calcium signaling and β-arrestin recruitment (VYPGKF-NH2 EC50 = 22.9 ± 8.3 μm; β-arr-1, 73.6 ± 13.0%; EC50 = 33.8 ± 13.7 μm; β-arr-2, 55.2 ± 11.8%; EC50 = 13.7 ± 8.2 μm). Additionally, increasing hydrophobic bulk at position 1, through substitution with norleucine, completely abolished calcium signaling (Nle-YPGKF-NH2, EC50 = ND) and resulted in partial agonism of β-arrestin recruitment (β-arr-1, 40.2 ± 9.7%; β-arr-2, 36.5 ± 8.8%). Together, these data indicate that the precise steric bulk of the side chain in position 1 is important for PAR4 agonism.
Next, we evaluated the impact of steric bulk relocation in position 1. Relocation of the methyl group in alanine to the N terminus and loss of side chain through substitution with sarcosine (Sar-YPGKF-NH2) resulted in poor agonism at both calcium signaling (EC50 = ND) and β-arrestin recruitment (β-arr-1, 35.6 ± 2.0%; β-arr-2, 39.8 ± 1.8%). Furthermore, the addition of an α-methyl group through substitution of Ala1 with 2-aminoisobutyric acid (Aib-YPGKF-NH2) resulted in a significantly less potent and partial agonist for calcium signaling (EC50 = 50.2 ± 15.3 μm) and also resulting in a significant reduction in β-arrestin recruitment (β-arr-1, 38.6 ± 3.9%; β-arr-2, 37.3 ± 3.6%). Similarly, replacement with N-methylserine ((N-Me-S)-YPGKF-NH2) also caused a significant attenuation of calcium signaling (EC50 = ND), while also revealing a decrease in β-arrestin recruitment compared with PAR4-AP (β-arr-1, 78.1 ± 6.9%; β-arr-2, 51.7 ± 6.3%). Thus, it is apparent that the precise position of the side chain in position 1 is important for agonism.
To further probe the importance of charge localization at the N-terminal region of the peptide, we made several additional substitutions to position 1. Substitution of alanine with either β-alanine or isonipecotic acid (Inp) significantly reduced the potency and activity of the PAR4-AP, similar to the results with PAR4-AP N-terminal acetylation (βAla-YPGKF-NH2 calcium EC50 = ND, β-arr-1, 25.7 ± 4.8%; β-arr-2, 31.8 ± 4.4%; Inp-YPGKF-NH2 calcium EC50 = ND, β-arr-1, 32.0 ± 3.3%; β-arr-2, 26.9 ± 3.0%). Together, these data suggest a role for N-terminal charge localization in agonist activity.
Effect of additional substitutions in positions 2–4 on PAR4-AP–mediated signaling
Previously, structure–activity studies with PAR4-activating peptides revealed an apparent requirement for an aromatic residue in position 2 (tyrosine) and found that alterations to position 3 (proline) and position 4 (glycine) were not well-tolerated (32). In this study, we observed that substitution of either tyrosine, proline, or glycine with alanine reduced calcium-signaling potency and β-arrestin recruitment; however, none of these substitutions completely abolished the activity of the peptide (Fig. 1, A–C). Interestingly, substitution of either l-tyrosine or l-proline with the respective d-isomer significantly reduced the activity of the PAR4-AP (Fig. 1, D–F, and Table 1). Together, these data suggested that although no individual residue is wholly required for agonist activity, the side chains of these residues and their positioning are important for agonist function. To further probe the contribution of these residues to PAR4-AP activity at PAR4, we generated peptides with alterations in the aromatic side chain of position 2, the backbone conformation of position 3, or position 4 (Fig. 2, D–F, and Table 1).
Peptides with substitution of Tyr2 with either 4-fluoro-l-phenylalanine (A-F(4-fluoro)-PGKF-NH2) or O-methyltyrosine (A-Tyr(Me)-PGKF-NH2) retained calcium signaling and β-arrestin recruitment with no significant changes in potency compared with the PAR4-AP. These data may suggest that the side chain of tyrosine is a site in which further modifications can be made to the peptide without affecting agonism; however, by comparison with AAPGKF-NH2, the presence of an aromatic ring in position 2 is of importance. Given the results obtained with alanine or d-proline substitution of Pro3 (Fig. 1, A–C and D–F, respectively), we furthered probed conformational contributions of this residue to the parental PAR4-AP. Proline was substituted with pipecolic acid (2-piperidinecarboxylic acid; Pip), which is structurally similar to proline but has an increased steric bulk and backbone elongation provided by a six-membered ring compared with the five-membered ring of proline. We observed partial agonism of calcium signaling in response to AY-Pip-GKF-NH2 stimulation with no significant difference in agonist potency (EC50 = 30.7 ± 4.9 μm) and with a correlating partial agonism of β-arrestin recruitment (β-arr-1, 38.8 ± 3.0%; β-arr-2, 44.0 ± 2.7%). Thus, it appears that increasing steric bulk/backbone length may affect efficacy but still results in agonism. To further investigate the role of backbone conformation provided at position 3, proline was substituted with nipecotic acid (3-piperidinecarboxylic acid; Nip) which is similar to pipecolic acid; however, it results in change of the peptide backbone direction (AY-Nip-GKF-NH2). With this modification, both calcium signaling (EC50 = ND) and β-arrestin recruitment (β-arr-1, 32.9 ± 4.9%; β-arr-2, 30.9 ± 4.5%) were significantly decreased in response to PAR4 stimulation. Elongation of the backbone in position 4 (glycine) by substitution with β-alanine (AYP-βAla-KF-NH2) was also significantly detrimental to both calcium signaling (EC50 = ND) and β-arrestin recruitment (β-arr-1,42.5 ± 1.8%; β-arr-2, 24.9 ± 1.6%). Together, these data suggest an important role for backbone conformation contributed by Pro3 and Gly4 compactness in the PAR4-AP. As well, these data suggest peptide backbone length at Pro3 and Gly4 has an important role, consistent with what we have observed with backbone elongation modifications at position 1 (Inp-YPGKF-NH2 and βAla-YPGKF-NH2; Fig. 2, A–C, and Table 1).
Effect of additional substitutions in position 5 on PAR4-AP–mediated signaling responses
Previously, investigations into glutamine substitutions in the native tethered ligand-mimicking peptide (GYPGQV) revealed that agonism is maintained when this position is substituted with a charged residue such as arginine or ornithine (32). In this study, we have identified that substitution of position 5 (lysine) of the PAR4-AP (AYPGKF-NH2) with alanine does not significantly affect the agonist capacity of the peptide (Fig. 1, A–C). Furthermore, we identified that substitution of l-lysine with d-lysine significantly decreases agonist stimulation of calcium signaling and β-arrestin recruitment (Fig. 1, D–F). To further probe the effect of positively-charged residues in position 5, we generated several PAR4-AP analogues with Lys5 substitutions by either arginine or ornithine (Fig. 2, G–I, and Table 1). Substitution of Lys5 with arginine resulted in a partial agonist with respect to calcium signaling but increased β-arrestin recruitment compared with the parental peptide (AYPGRF-NH2 calcium EC50 = 4.0 ± 1.1 μm, β-arr-1, 154.2 ± 10.3%; β-arr-2, 140.1 ± 9.4%). This substitution delocalizes the positive charge of the side chain compared with lysine and increases steric bulk. Interestingly, substitution with ornithine, which is similarly charged but has a shortened distance between the charge and the peptide backbone compared with lysine, also resulted in partial agonism of PAR4-mediated calcium signaling accompanied by a modest increase in the recruitment of β-arrestins (AYPGOF-NH2 calcium EC50 = 4.2 ± 1.1 μm, β-arr-1, 121.7 ± 8.5%; β-arr-2, 112.1 ± 7.7%). To further probe whether steric bulk in the side chain of position 5 affects PAR4-AP potency, we increased steric bulk through methylation of the ϵ-amine of lysine (AYPG-(Nϵ-Me-K)-F-NH2). This substitution yielded a full agonist for calcium signaling (EC50 = 6.7 ± 1.9 μm) and a modestly more efficacious agonist for the recruitment of β-arrestins (β-arr-1, 122.5 ± 12.4%; β-arr-2 116.7 ± 11.2%). All of these substitutions maintain a positive charge and are able to act as agonists for PAR4. To determine whether charge in position 5 is required, we substituted lysine with citrulline, which maintains a similar structure to arginine and lysine while removing a positive charge (AYPG-Cit-F-NH2). Interestingly, we observed partial agonism with respect to calcium signaling (EC50 = 5.4 ± 0.8 μm) with equipotent β-arrestin recruitment compared with the PAR4-AP, consistent with the alanine scan data for this position. Thus, positively charged residues in position 5 seem to contribute to agonism. Interestingly, we observed similar increases in β-arrestin recruitment and equipotent calcium signaling when stericity was either increased (AYPGRF-NH2) or decreased (AYPGOF-NH2), suggesting that the positive charge in this position is more important than the size of the side chain. Finally, although both the charge and size of the size chain have an effect, all modifications to position 5, with the exception of d-lysine (Fig. 1, D–F, and Table 1) and Nα-methyl-lysine (Fig. 1, G–I, Table 1) modifications, were generally well-tolerated, and there appears to be modest freedom in the side chain of this position. This makes the side chain a good candidate for further investigations of structure–activity relationships as well as a good region to label with an imagine modality.
Effect of additional substitutions in position 6 on PAR4-AP–mediated signaling responses
Through our initial investigations into position 6 substitutions, we observed that replacement of Phe6 with either alanine, d-phenylalanine, or N-methyl-phenylalanine had varied effects on PAR4-AP potency and efficacy. Previous investigations into position 6 substitutions in the native tethered ligand (GYPGQV) revealed the requirement for an aromatic residue, as substitution with nonaromatic residues such as lysine or ornithine significantly decreased or abolished the activity of the peptide (32). In addition to being an aromatic residue, phenylalanine is a hydrophobic residue, so we investigated the impact of altering the hydrophobicity/hydrophilicity and stericity while maintaining an aromatic residue (Fig. 2, J–L, and Table 1).
We investigated the effect of polar changes that decrease hydrophobicity. Substitution of Phe6 with tyrosine results in the addition of a hydroxyl group to position 6 (AYPGKY-NH2). We observed partial agonism of calcium signaling with no significant change in potency compared with the PAR4-AP (AYPGKY-NH2, EC50 = 5.2 ± 1.0 μm). Interestingly, the peptide was able to recruit β-arrestins at least as well as the PAR4-AP (β-arr-1, 120.5 ± 12.5%; β-arr-2, 109.9 ± 11.4%). Furthermore, replacing the tyrosine hydroxyl group with an electron withdrawing fluorine, by means of substitution with 4-fluoro-l-phenylalaine, caused significant impairment of agonist activity, both decreasing calcium signaling and significantly decreasing β-arrestin recruitment (AYPGK-F(4-fluoro)-NH2, calcium EC50 = ND; β-arr-1, 25.2 ± 6.5%; β-arr-2, 41.3 ± 5.9%). The results of these two substitutions suggest that altering the polarity, while maintaining aromaticity, in position 6 reduces efficacy (33). Because decreasing hydrophobicity did not increase PAR4-AP activity, we next investigated alterations that increase stericity and hydrophobicity. Substitution with 4-methyl-l-phenylalanine reduced the efficacy of the PAR4-AP to a partial agonist for both calcium signaling and β-arrestin recruitment (AYPGK-F(4-Me)-NH2 calcium EC50 = 9.4 ± 3.0 μm, β-arr-1, 56.4 ± 1.2%; β-arr-2, 63.2 ± 1.1%). Given these results, we wanted to assess the pharmacological impact of substituting phenylalanine with (1-naphthyl)-l-alanine (1Nal) and (2-naphthyl)-l-alanine (2Nal), which not only increases hydrophobicity but also has a modest increase in steric bulk and π-stacking potential. Substitution of Phe6 with (1-naphthyl)-l-alanine (1Nal; AYPGK-1Nal-NH2) generated a partial agonist for both calcium signaling (EC50 = 9.6 ± 1.9 μm) and β-arrestin recruitment (β-arr-1, 77.5 ± 2.3%; β-arr-2, 64.1 ± 2.0%) compared with PAR4-AP. Notably, substitution to (2-naphthyl)-l-alanine (2Nal; AYPGK-2Nal-NH2), which differs in orientation compared with (1-naphthyl)-l-alanine such that the alkyl chain is connected to position 2 of the naphthyl group (instead of position 1 as in 1Nal), restored potency of the peptide to the levels observed with PAR4-AP. AYPGK-2Nal-NH2 was an equipotent agonist for calcium signaling (EC50 = 11.1 ± 2.3 μm) and β-arrestin recruitment (β-arr-1, 77.0 ± 7.0%; β-arr-2, 91.6 ± 6.4%). Together, these results indicate that hydrophobicity, stericity, and side chain positioning of the C-terminal residue all contributes to agonist peptide activity at PAR4.
Probing antagonism of PAR4-AP–stimulated calcium signaling by peptides unable to stimulate calcium signaling
The failure of some substituted peptides to stimulate calcium signaling could reflect either a failure to bind or a loss of efficacy. To determine whether such peptides can act as antagonists of PAR4-AP–stimulated calcium signaling, we preincubated HEK-293 cells stably expressing PAR4-YFP prior to stimulation with PAR4-AP. Pretreatment and competition with 100 μm of peptides unable to stimulate PAR4-dependent calcium signaling were largely unable to affect the calcium response elicited by 30 μm PAR4-AP (AYPGKF-NH2) compared with vehicle-treated controls. Interestingly, two peptides with substitution in position 3 or 4, which were unable to stimulate calcium signaling, were able to decrease calcium signals elicited by PAR4-AP. Calcium signaling in response to AYPGKF-NH2 (30 μm) was significantly reduced following pretreatment with AY-Nip-GKF-NH2 (10.6 ± 0.9% of A23187; p < 0.05) or AYP-Sar-KF-NH2 (13.0 ± 1.5% of A23187; p < 0.05) compared with nonpretreated control (20.8 ± 1.3% of A23187; p < 0.05) (Fig. S2).
PAR4-dependent calcium signaling is Gαq/11-dependent
To verify that PAR4-dependent calcium signaling was Gαq/11-dependent, HEK-293 cells stably expressing PAR4-YFP were stimulated with 100 μm AYPGKF-NH2 following preincubation with selective Gαq/11-inhibitor YM254890 (34). YM254890 inhibited AYPGKF-NH2–stimulated calcium signaling in a concentration-dependent manner, with an IC50 of 16.5 ± 6.1 nm and complete inhibition observed by 100 nm YM254890. The control vehicle (0.001% DMSO) had no effect on calcium signaling (Fig. S3).
Peptides unable to stimulate PAR4-dependent calcium signaling do not activate MAPK signaling
Activation of G-protein and β-arrestin pathways downstream of GPCRs often converge on the activation of the p44/42 MAPK–signaling pathway (35). We therefore monitored phosphorylation of p44/42 MAPK in response to the 37 AYPGKF-NH2 derivative peptides. HEK-293 cells, stably expressing PAR4-YFP, were incubated with control vehicle (0.001% DMSO), 100 μm AYPGKF-NH2, or derivative peptides (100 μm) for 10 min. Data were normalized as a ratio of phosphorylated p44/42 (p-p44/42) to total p44/42 (p44/42). Interestingly, we find that most peptides that were unable to stimulate calcium signaling were also unable to activate p44/42 MAPK signaling (i.e. AyPGKF-NH2, AYpGKF-NH2, (N-Me-S)-YPGKF-NH2, Inp-YPGKF-NH2, Sar-YPGKF-NH2, AY-Nip-GKF-NH2, and AYP-Sar-KF-NH2). Similarly, peptides that perform as partial agonists of Gαq/11-mediated calcium signaling were also partial agonists for p44/42 phosphorylation. Several peptides were able to cause a significantly greater increase in phosphorylation compared with the parental peptide (i.e. Nle-YPGKF-NH2, AYPGOF-NH2, AYPG-(Nϵ-Me-K)-F-NH2, AYPG-Cit-F-NH2, AYPGKY-NH2, AYPGK-1Nal-NH2,) (Fig. 3 and Table 1; representative blots in Fig. S4).
PAR4-mediated MAPK signaling is Gαq/11-dependent and β-arrestin–independent
To further probe the contribution of different pathways downstream of PAR4 to p44/42 phosphorylation, we examined the effect of blocking Gαq/11 signaling with the specific antagonist YM254890 and examined the role of β-arrestins using a β-arrestin-1/2 double knockout HEK-293 cell derived by CRISPR/Cas9-mediated targeting (Fig. S5). Pretreatment of PAR4-YFP HEK-293 cells with 100 nm YM254890 for 20 min abolished PAR4-AP (30 μm)-stimulated activation of MAPK signaling and subsequent phosphorylation of p-p44/42 at 5 min (Fig. 4A). In contrast, there was no significant difference in the phosphorylation of p44/42 MAPK between WT HEK-293 and β-arrestin-1/2 knockout HEK-293 cells stimulated with 30 μm PAR4-AP for 0–90 min (two-way ANOVA). p44/42 MAPK did remain sustained up to 90 min in the β-arrestin-1/2 knockout HEK-293 cells, whereas p44/42 MAPK phosphorylation in HEK-293 cells began to decrease toward baseline by 90 min (5 min compared with a 90-min stimulation within cell line HEK-293 cells, p < 0.05, β-arrestin-1/2 knockout HEK-293, p = n.s., two-tailed t test; Fig. 4B).
Homology modeling of PAR4 and in silico docking of AYPGKF-NH2
To gain a better understanding of the ligand-binding pocket in PAR4, we turned to homology modeling and in silico docking experiments. A homology model of PAR4 was generated using known coordinates of the thermostabilized, antagonist-bound (AZ8838) human PAR2 crystal structure (2.8 Å resolution, PDB code 5NDD (36)). Sequences of human PAR4 were aligned with the structure-resolved residues of PAR2 using Clustal Omega (37, 38). We generated a homology model of WT PAR2 on the PAR2 structure, which contained modifications necessary for crystallization. Twenty human PAR4 homology models, using the WT human PAR2 model as a template, were generated in MODELLER (Webb and Sali (69)). The PAR4 model with the lowest discrete optimized protein energy (DOPE) score was utilized for in silico analyses.
Docking AYPGKF-NH2 to the PAR4 homology model (GalaxyPepDock; Lee et al. (39)) revealed a number of highly-predicted peptide–receptor interaction sites (Fig. 5A; Ser67, Tyr157, His229, Asp230, Leu232, Leu234, Asp235, Ala238, Gln242, His306, Tyr307, Pro310, Ser311, Ala314, Gly316, Tyr319, and Tyr322). Of these, the most frequently predicted interactions were receptor residues His229, Asp230, and Gln242. In peptide-docked models, Gln242 made contact with alanine, tyrosine, and lysine, with the most frequent prediction being with phenylalanine. His229 was predicted to interact with lysine and phenylalanine. Asp230 was frequently predicted to interact with lysine and phenylalanine, and contacts with lysine appeared in 60% of predictions (Fig. 5B). Interestingly, when visualizing receptor residues predicted to interact with the peptide, we found that a large number of these residues were localized on extracellular loop 2 (ECL2).
Calcium signaling and β-arrestin recruitment to predicted peptide-binding site mutant PAR4
We experimentally examined in silico binding predictions through site-directed mutagenesis introducing alanine substitutions at the three most highly-predicted interacting residues, His229, Asp230, and Gln242. Additionally, we generated a double mutation, PAR4H229A,D230A, to determine whether combined loss of His229 and Asp230 would result in additive detriment to receptor activation. We evaluated the functional consequences of these mutations by recording calcium signaling in response to AYPGKF-NH2 with all mutants. We observed no calcium response in cells expressing PAR4D230A-YFP or PAR4H229A,D230A-YFP mutant receptors up to 300 μm AYPGKF-NH2 (Fig. 5C). Similarly, peptide-induced β-arrestin-1/2 recruitment to PAR4D230A-YFP and PAR4H229A,D230A-YFP was significantly reduced compared with the WT PAR4. Both PAR4H229A and PAR4Q242A mutations were fully able to recruit β-arrestin-1/2 to the same levels observed with WT PAR4-YFP (Fig. 5, D and E).
To investigate whether these sites are important for tethered ligand activation of the receptor, we stimulated cells expressing WT and mutant constructs with thrombin. Thrombin cleavage of the receptor reveals the native tethered ligand, GYPGQV (40). Comparable with activation with AYPGKF-NH2, we observed significantly diminished calcium signaling in HEK-293 cells expressing PAR4D230A-YFP and PAR4H229A, 230A-YFP mutants, even up to 10 units/ml thrombin (Fig. 5F). Additionally, β-arrestin-1/2 recruitment was also impaired by PAR4D230A and PAR4H229A, D230A mutants (Fig. 5, G and H). We observed no appreciable difference in calcium signaling or β-arrestin recruitment when PAR4H229A-YFP and PAR4Q242A-YFP mutants were stimulated with thrombin, with the exception of a modest decrease in β-arrestin-1 recruitment to the PAR4Q242A-YFP mutant receptor. Thus, Asp230 interactions with either PAR4-AP or the tethered ligand are critical for receptor activation.
To examine whether differences in cell-surface expression of the mutant receptors could underlie the defect in signaling, we employed confocal microscopy to examine receptor expression and localization. We did not find any differences in expression levels and found appropriate cell-surface localization of the mutant receptors (Fig. S6).
Platelet aggregation response to differential activation of PAR4 signaling
To determine the effects of targeted signaling observed with several peptides, we investigated PAR4-mediated platelet aggregation in response to the β-arrestin–biased, calcium-signaling null peptide AyPGKF-NH2, as well as a modestly more potent calcium agonist AYPGRF-NH2 (compared with AYPGKF-NH2). Thrombin (1 unit/ml) and 100 μm AYPGKF-NH2 were used as controls to ensure successfully-washed platelet preparations. In response to thrombin stimulation, we observed a characteristic aggregation trace resulting in 100% aggregation at 1 unit/ml. In agreement with previously published data, AYPGKF-NH2 (100 μm) stimulation resulted in ∼50% platelet aggregation (26). We observed that platelet aggregation was not stimulated by 100 μm AyPGKF-NH2. Interestingly, when platelet preparations were stimulated with AYPGRF-NH2, in which we observed a modest increase in potency in calcium-signaling assays, enhanced β-arrestin recruitment, and an increased phosphorylation of p44/42, we observed platelet aggregation response comparable with levels observed with thrombin (1 unit/ml) (Fig. 6).
Discussion
PAR4 is emerging as a novel anti-platelet drug target. Here, we examine structure–activity requirements for PAR4 activation of calcium signaling, β-arrestin recruitment, and MAPK activation. PAR cleavage by a number of different enzymes has been reported (41). Canonically, thrombin has been described as a PAR1 and PAR4 activator, and trypsin is described as a PAR2 and PAR4 activator. Although PAR3 can be cleaved by thrombin, its ability to act as a signaling receptor remains ambiguous. In each case, cleavage by serine proteases occurs at an arginine–serine site, which reveals a unique tethered ligand specific to each PAR (42)–human PAR1-SFLLRNP… (6, 15), human PAR2-SLIGKV… (43, 44), and human PAR4-GYPGQV… (40, 45). Although these tethered-ligand sequences are diverse, they retain some highly-conserved properties, including a highly-conserved charged residue in position 5 of the revealed tethered ligands (arginine in PAR1; lysine in PAR2). Additionally, PAR2 and PAR4 have a conserved glycine in position 3 and valine in position 6 of their tethered ligands. The similarities of the proteolytically-revealed tethered ligands are also observed in the optimized tethered ligand-mimicking synthetic agonist peptides. Hydrophobic residues leucine, isoleucine, and proline are preferred in position 3 of PAR1, PAR2, and PAR4 agonist peptides, respectively. Additionally, there is a basic residue in position 5 of the tethered ligand peptide in all three receptors (arginine in PAR1 and PAR2 and lysine in PAR4 activating peptides). These conserved residue properties highlight some of the key requirements that a PAR4 agonist peptide must meet. Previous structure–activity work starting with the proteolytically-revealed sequences of human (GYPGQV) and murine (GYPGKF) PAR4 identified the hexapeptide AYPGKF-NH2 or SYPGKF-NH2 to be potent and selective agonists for PAR4 (32). In this study, we started with the hexapeptide AYPGKF-NH2 as our parental peptide agonist and incorporated various modifications at each of the six positions in this peptide to investigate determinants of agonist-induced PAR4-coupling to Gαq/11, MAPK, and β-arrestin-1/2 recruitment to guide development of biased agonists of PAR4.
Previously, it was demonstrated that substitution of alanine or serine in position 1 of the GYPGKF-NH2–activating peptide retained the ability to trigger tritiated inositol 1,4,5-triphosphate release to a level comparable with that observed with thrombin activation and much higher than the release caused by the GYPGKF-NH2 (32). Interestingly, substitution of position 1 with threonine was reported to significantly reduce the potency of the activating peptide (32). In keeping with results from previous studies, we find a requirement for small aliphatic side chains in position 1 of PAR4-activating peptides, with substitutions that increased bulk in this position consistently resulting in poor agonists for both calcium signaling and β-arrestin recruitment (N-Me-A Fig. 1, G–I; N-Me-S, Aib, Inp, V, Nle in Fig. 2, A–C; and Table 1). Additionally, we observed that both stereochemical inversion of l-alanine to d-alanine (d-alanine, Fig. 1, D–F) and backbone elongation (βAla, Fig. 2, A–C) were detrimental to both calcium signaling and β-arrestin recruitment. Additionally, we observed that capping of the N-terminal charge via acetylation significantly decreases agonist potency. Together, these results indicate that a positive charge and the localization of that charge in addition to precise side-chain positioning at position 1 are critical for effective PAR4-agonism.
The aromatic side chain, and to a lesser extent the hydroxyl group, of the Tyr2 residue of the parental peptide also proved to be important for peptide activity with alanine or d-isomer substitutions resulting in decreased activity of all pathways tested. These results are consistent with previous reports that replacement of tyrosine with other aromatic residues, such as phenylalanine, significantly reduced both the selectivity and the potency of the activating peptide (32). Interestingly, substituting l-Tyr2 for either d-tyrosine (Figs. 1D and 3) or N-methyltyrosine (Figs. 1G and 3) resulted in significant decreases in calcium and p44/42 signaling, while having relatively modest effects on β-arrestin recruitment (Fig. 1, E–F, H, and I, respectively, and Table 1). Also, substitution of tyrosine with either 4-fluorophenylalanine or O-methyltyrosine did not significantly affect calcium transients, p44/42 phosphorylation, or β-arrestin recruitment compared with AYPGKF-NH2 (Figs. 2, D–F and 3, and Table 1). Our findings, together with previously reported data (32), point to the necessity of an aromatic residue in position 2 to maintain agonist potency. Furthermore, our data reveal that backbone orientation in position 2 is a key determinant of agonist potency. Future experiments fine-tuning the aromatic substituents may be beneficial for peptide analogue development.
Our data reveal that alterations affecting proline backbone conformation or glycine residue compactness are not well-tolerated, as these decreased calcium signaling, p44/42 MAPK signaling, and β-arrestin recruitment to PAR4. Calcium signaling has previously been reported to be negatively impacted by alterations to either of these two residues proline and glycine (32). Both proline and glycine residues are frequently found in β-turns, in which proline provides a characteristic kink, and the available data support the notion that the conformation(s) (Pro3 and Gly4) and rigidity (Pro3) of PAR4 agonists permitted by these residues is/are important for receptor activation. To test this hypothesis further, we made several substitutions in position 3 (proline) aimed at evaluating favored backbone conformations. We observed that a return to a predominantly trans-amide conformation by substitution of proline with alanine (Fig. 1, A–C), results in decreased potency in all pathways studied. Interestingly, this substitution did not abolish signaling but did significantly decrease agonist potency, which suggests that the backbone conformation of proline may be necessary for receptor binding but not receptor activation. Substitution of proline to pipecolic acid (Pip., Fig. 2, D–F), which has a similar peptide backbone orientation to proline with a six-membered aromatic ring, decreased β-arrestin recruitment with relatively modest effects for calcium signaling activation. Interestingly, substitution of proline with nipecotic acid (Nip., Fig. 2, D–F) abolished both calcium signaling and p44/42 phosphorylation while also significantly reducing β-arrestin recruitment. Nipecotic acid is similar to pipecolic acid with the exception that nipecotic acid changes the direction of the peptide backbone compared with that adopted by proline, whereas pipecolic acid favors a similar backbone directionality and conformation similar to proline. Interestingly, we observed a similar pharmacological profile observed with AY-Nip-GKF-NH2 when we substituted d-proline in position 3, which also changes the backbone orientation from that of the parent peptide. These findings demonstrate that the backbone conformation provided by proline in position 3 of the parental peptide is necessary for agonism of PAR4-signaling pathways. Furthermore, substitution of a Pro3 with alanine (AYAGKF-NH2, Fig. 1, A–C) significantly and detrimentally altered the pharmacological profile compared with the parental peptide PAR4-AP, suggesting the possibility that agonist competency contributed by position 3 is largely conformational and not chemical. Substitution of sarcosine or β-alanine at position 4 also negatively affected all signaling pathways, whereas substitution of Gly4 with alanine resulted in a decrease in potency with calcium signaling as well as a reduction of β-arrestin recruitment. Together, these substitutions reveal the requirement for a small flexible residue in position 4, consistent with previous reports (32).
It has been previously demonstrated that substitution of position 5 of glutamine in the native tethered ligand (GYPGQV) with arginine or ornithine results in decreased potency in the PAR4 agonist peptide for calcium-signaling pathways (32). However, there is a basic residue found in position 5 of PAR1- (arginine, TFLLR), PAR2- (arginine, SLIGRL), and PAR4 (lysine, AYPGKF)-tethered ligand-mimicking peptides. Given the conservation of charged residues in position 5 of PAR-tethered ligand-mimicking peptides, we generated PAR4-AP–derivative peptides with Lys5 substitutions that maintained or altered charge in this position to determine whether this is a requirement for activity at PAR4. Substitution of Lys5 with alanine had no appreciable effect on calcium or MAPK signaling, while resulting in decreased β-arrestin recruitment (Fig. 1, A–C). Substitution of l-Lys5 with d-lysine, however, significantly reduced potency and efficacy in calcium signaling and β-arrestin recruitment assays and furthermore resulted in a loss of stimulation of MAPK signaling (Figs. 1, D–F, and 3). We continued our exploration of this position with an Nα-methyl-lysine substitution, which increases steric bulk through the addition of a methyl group to the backbone while also removing a hydrogen bond donor and increasing the amount of peptide in the cis-transformation of the amide bond between positions 4 and 5. We found this substitution detrimental to calcium signaling but advantageous for β-arrestin recruitment, resulting in comparable maximal levels of recruitment to those observed with PAR4-AP but at lower concentrations (Fig. 1, G–I). Thus, backbone conformation and bulk at position 5 are important for agonism of calcium signaling and MAPK activation, while possibly being an important site for modification in the search for β-arrestin–biased ligands.
We next turned our attention to analogues that resemble the side chain of Lys5, with a specific interest on structure–activity in addition to ascertaining the importance of charge in this residue. In contrast to the observation with similar substitutions to the tethered ligand-mimicking peptide (GYPGQV) (32) wherein signaling was decreased compared with PAR4-AP, we found that both calcium signaling and β-arrestin recruitment are only modestly impacted by position 5 substitution to ornithine in the PAR4-AP, which decreases steric bulk and shortens the distance between the backbone and the positively charged amine compared with lysine (AYPGOF-NH2; Fig. 2, G–I). Interestingly, however, we observed significantly increased p44/42 phosphorylation in response to AYPGOF-NH2 stimulation (Fig. 3). This is somewhat puzzling because we found p44/42 phosphorylation to be Gαq/11-dependent (Fig. 4A); however, it is important to note that MAPK and other cellular signaling pathways downstream of receptor activation are subject to amplification/deamplification processes within the cell (46), and these observations may point to additional differences in signaling stimulated by this peptide that needs to be examined. We further probed stericity in this position through increasing bulk with substitution of Lys5 with arginine (AYPGRF-NH2). Interestingly, this substitution also results in a delocalization of charge compared with the charge on the Lys5 side chain. This modification provided a similar calcium-signaling profile compared with that observed with ornithine substitution; however, it also resulted in a significant increase of β-arrestin recruitment compared with the parental PAR4-AP. Interestingly, there was no significant change in p44/42 phosphorylation unlike what was observed with ornithine substitution. Another peptide with increased steric bulk, Nϵ-methyl-lysine substitution in position 5, improved calcium-signaling potency, modestly improved β-arrestin recruitment, and significantly increased p44/42 phosphorylation (AYPG-(Nϵ-Me-K)-F-NH2, Fig. 2, G–I). We further investigated whether charge was a significant determinant of agonism by substituting Lys5 with citrulline (AYPG-Cit-F-NH2), which lacks charge but retains a similar structure compared with arginine and, to a lesser extent, lysine. We observed decreased calcium, equivalent β-arrestin recruitment, and significantly increased p44/42 phosphorylation compared with the parental peptide (Figs. 2, G–I, and 3). Together, these data reveal that changes to side-chain charge, bond donors/acceptors, and steric bulk result in varied pharmacological responses that may be exploited to bias signaling responses in favor of β-arrestin recruitment (AYPGRF-NH2) or p44/42 phosphorylation (AYPGOF-NH2; AYPG-(Nϵ-Me-K)-F-NH2) with modest improvements to potency in calcium signaling. Interestingly, we observed that charge is not wholly required for receptor activation as both alanine and citrulline substitutions resulted in agonism of PAR4.
Finally, our structure–activity investigation turned to position 6, phenylalanine, of the parental peptide. Given the significant impact that alterations to position 2 (tyrosine) had on agonist potency, we predicted that there may be a π–π stacking interaction between tyrosine and phenylalanine, which is supported by the backbone kink provided by proline in position 3. Substitution of position 6 with alanine or d-phenylalanine reduced calcium-signaling potency while reducing β-arrestin recruitment and p44/42 phosphorylation. These alterations suggest that phenylalanine in position 6 is not essential for PAR4 activation by agonist peptide but is important for agonist potency and full activation of all signaling pathways. Methylation of the backbone nitrogen in phenylalanine (AYPGK-(N-Me-F)-NH2) resulted in improved activation of calcium signaling, β-arrestin recruitment, and p44/42 phosphorylation compared with AYPGKF-NH2 (Fig. 3). Addition of either methyl (AYPGK-F(4-Me)-NH2), fluorine (AYPGK-F(4-fluoro)-NH2), or hydroxyl (AYPGKY-NH2) (Fig. 2, J–L) off of the para-position of the aromatic side chain of phenylalanine was deleterious to all signaling pathways studied. Substitution of unnatural residues that increase the size of steric bulk and π-stacking potential yielded mixed results based on the orientation of the naphthyl. Substitution of phenylalanine with (1-naphthyl)-l-alanine (1Nal) increased p44/42 phosphorylation but decreased both calcium signaling and β-arrestin recruitment (Fig. 2, J–L). Changing the alkyl chain connection from position 1 to position 2 of the naphthyl (AYPGK-2Nal-NH2; Fig. 2, J–L) resulted in activity similar to AYPGKF-NH2. With (2-naphthyl)-l-alanine showing some flexibility in the steric bulk of position 6, it would be interesting for future work to investigate increasing the π-stacking potential at position 2 while simultaneously increasing the π-stacking potential at position 6, because we hypothesize that these two residues' aromatic regions of the PAR4-AP may interact.
To determine whether any of the peptides that failed to stimulate PAR4-dependent calcium signaling were able to act as antagonists, cells were pretreated with these peptides and subsequently stimulated with AYPGKF-NH2 to see whether the parental peptide was able to still elicit a signal. We observed that both AY-Nip-GKF-NH2 and AYP-Sar-KF-NH2 were both able to significantly decrease the calcium signal elicited by PAR4-AP (Fig. S2). Interestingly, each of these substitutions resulted in ablation of both calcium and p44/42 signaling as well as significantly decreasing β-arrestin recruitment. These results may indicate that targeting PAR4 with small molecules mimicking positions 3 and 4 dipeptides may provide a starting place for the design of novel PAR4 antagonists. Additionally, peptides that further explore modifications to positions 3 and 4 could be investigated for potent PAR4 antagonists.
In platelets, it has previously been demonstrated that PAR4-mediated calcium mobilization induces activation and aggregation (24, 47). Both thrombin and AYPGKF-NH2 initiated PAR4-dependent platelet aggregation to levels comparable with those previously reported in the literature in the same rat platelet system (26). As was hypothesized, we observed that stimulation of washed platelet samples with a β-arrestin–biased peptide (AyPGKF-NH2; 100 μm) that is unable to stimulate PAR4-dependent calcium signaling did not stimulate platelet aggregation (Fig. 6). Furthermore, stimulation of washed platelets with the agonist peptide, AYPGRF-NH2 (Fig. 2, G–I), enhanced platelet aggregation to levels comparable with those observed with thrombin activation of PAR4 (Fig. 6). AYPGRF-NH2 stimulates calcium signaling to levels comparable with those seen with PAR4-AP but triggers more β-arrestin-1 recruitment than the parental peptide. These data suggest that calcium signaling is essential for triggering platelet activation, but β-arrestin recruitment may also play a role.
Having ascertained some of the governing agonist peptide residue characteristics enabling PAR4 activation and validated their signaling consequences in an ex vivo system, we turned our attention to investigating the ligand-binding site of the receptor. The extracellular loops of many GPCRs have been demonstrated to be important facilitators of ligand entry into the orthosteric site, with ECL2 possessing the most diversity in sequence and structure (48, 49). Analysis of common mechanisms of GPCR-ligand interaction derived from crystal structure and biochemical experiments reveals a possible role of ECL2 secondary loop in binding and facilitating entry, whereas the diversity of residue sequences across class A GPCRs is thought to confer their ligand specificity (48, 49). In many class A GPCRs, a disulfide bond between ECL2 and a transmembrane 3 cysteine (Cys3.25, Ballesteros-Weinstein numbering) is thought to stabilize the extracellular topology such that ligand recognition at ECL2 and subsequent orthosteric binding can occur (50). All PAR receptors possess this canonical TM3 Cys3.25 (PAR1 Cys-1753.25, PAR2 Cys-1483.25, PAR3 Cys-1663.25, and PAR4 Cys-1493.25), and residues adjacent to this conserved cysteine are important for ligand binding and receptor activation. Investigations into the role of PAR ECL2 domains have previously demonstrated that all PAR receptors possess a homologous CHDXL motif (…CHDVL…-PAR1C254-L258 and PAR2C226-L230; …CHDVH… PAR3C245-L249…CHDAL; … PAR4C226-L230) in their ECL2 (40, 51). Given that the activating peptides of PAR1 and PAR2 (51, 52) are similar (SFLLRN, PAR1; SLIGRL, PAR2), additional studies were conducted that revealed that ligand specificity is conferred by acidic residues distal to the CHDXL motif (PAR1, 259NETL262; PAR2 231PEQL234 (52–54). Interestingly, the equivalent human PAR4 ECL2 residues distal to the CHDAL domain, 233PLDA236, share sequence homology with those distal to the human PAR2 CHDVL domain (231PEQL234), both of which contain an acidic residue. Al-Ani et al. (51) demonstrated that glutamic acid residues in the rat PAR2 ECL2 (PEEVL) conferred specificity to PAR2 ligands. In previous studies, a series of aspartic acids in the PAR4 extracellular loop 2 (ECL2) and N-terminal (N)-tail were proposed to be key for thrombin binding to PAR4 (55–57). Mutations of ECL2 aspartic acid residues D224A, D230A/D235A (double mutation), and D22A/D230A/D235A mutants were all found to decrease thrombin cleavage of PAR4 and activation in yeast models (57). Furthermore, the activity of the PAR4 agonist peptide AYPGKF-NH2 was also abolished in PAR4 mutants containing the ECL2 single mutant D224A, double mutant D230A/D235A, or triple mutant D224A/D230A/D235A aspartic acid mutations to alanine which indicate that these aspartic acid residues may be important for both agonist peptide and tethered ligand bindings and activation of PAR4. The D230A/D235A mutation to alanine removed two acidic residues in the highly-conserved …CHD230AL domain (PAR1 Asp256; PAR2 Asp228) as well as the homologous PLD235A… specificity-conferring domain observed in PAR1 (Glu260) and PAR2 (Glu232) thus making it difficult to assess whether the loss of cleavage and activation is due to the CHD230AL or PLD235A aspartic acid.
In this study, in silico docking to a homology model of PAR4 revealed 17 predicted sites of interaction with AYPGKF-NH2 (Ser67, Tyr157, His229, Asp230, Leu232, Leu234, Asp235, Ala238, Gln242, His306, Tyr307, Pro310, Ser311, Ala314, Gly316, Tyr319, Tyr322; Fig. 5 A). Interestingly, these predicted sites reveal residues that have been shown or predicted to have a role in PAR4 activation by thrombin or agonist peptide. These sites include the aspartic acid residue Asp230 found in the highly-conserved CHDXL domain in PARs as well as Asp235 in the downstream region that is homologous to the specificity-conferring motif found in PAR1 and PAR2 (57). Additionally, previous in silico docking revealed a role for aspartic acid and histidine in this motif in binding of AYPGKF-NH2 to PAR4 (58). We generated mutations of the most frequently-predicted interaction sites (His229, Asp230, Gln242, and His229/Asp230) to alanine, which revealed that Asp230 is essential for productive interaction of the peptide with the receptor, as mutation to alanine abolished calcium signal potentiation as well as β-arrestin recruitment. Interestingly, this loss of activity was observed with both peptide agonist AYPGKF-NH2 as well as with thrombin-stimulated activation of PAR4, which agrees with previous reports of the importance of these sites while also elucidating the importance of the conserved CHD230AL motif in PAR4 activation, consistent with the role of this domain in PAR1 and PAR2 (51–54, 57, 59). Another frequently predicted residue belonging to the CHDAL domain, His229, when mutated to alanine did not alter the ability of either AYPGKF-NH2 or the tethered ligand as revealed by thrombin to activate calcium signaling or β-arrestin recruitment pathways. Furthermore, we observed that a double mutation of both of these sites to alanine (H229A/D230A) did not result in any additive detriment to PAR4 activation; therefore, we conclude that Asp230 interaction with PAR4 peptide and the tethered ligand agonist performs an essential role in activation of PAR4.
Finally, in the context of platelet activation, we find that our modified peptides that are able to cause a greater level of calcium signaling and β-arrestin recruitment than PAR4-AP could also trigger greater platelet aggregation. In our hands, PAR4-AP was unable to elicit greater than 50% of the aggregation observed in response to 1 unit/ml thrombin, whereas the peptide AYPGRF-NH2 was able to trigger aggregation comparable with that seen with thrombin. A role for PAR4-dependent calcium signaling is well-established in platelet aggregation (47, 60, 61), and consistent with this, AyPGKF-NH2, which is unable to stimulate calcium signaling, failed to induce aggregation. A role for β-arrestin–mediated PAR4 signaling in platelets has also been demonstrated (26, 62). Together, these data suggest that calcium signaling is critical for initiating PAR4-mediated platelet aggregation, but the precise role and hierarchy of the different PAR4-signaling pathways in platelet responses remains to be fully established.
In summary, this study identifies key mechanisms involved in PAR4 activation and signaling. We also present and characterize a novel toolkit of PAR4 agonist peptides that could be used to study biased signaling through PAR4 in platelets as well as other physiological systems. We further show that platelet activation by PAR4 critically depends on the Gαq/11 pathway, and selective targeting of this pathway might yield a useful anti-platelet agent. Overall, this study advances our understanding of agonist binding to PAR4 and will support future efforts aimed at defining signaling contribution in homeostatic signaling and the discovery of novel therapeutics targeting PAR.
Experimental procedures
Chemicals and reagents
All chemicals were purchased from Millipore-Sigma, Thermo Fisher Scientific (Hampton, NH), or BioShop Canada, Inc. (Burlington, Ontario, Canada), unless otherwise stated. AYPGKF-NH2 was purchased from and synthesized by Genscript (Piscataway). Thrombin from human plasma was obtained from Millipore-Sigma (catalogue no. 605195, 5000 units).
Molecular cloning and constructs
The plasmid encoding PAR4-YFP was cloned and validated as described previously (26). The use of an in-frame, C-terminal eYFP fusion with PAR4 has been utilized in several published studies, and such modification does not significantly alter signaling downstream of PAR4 (26, 62–64). Plasmid DNA mutations in the predicted orthosteric binding site of PAR4 were generated using the QuikChange XL multisite-directed mutagenesis kit (Agilent Technologies, Mississauga, Ontario, Canada) to generate all mutants described in this study. All constructs were verified by Sanger sequencing (Robarts DNA Sequencing Facility, University of Western Ontario). Confocal microscopy of HEK-293 cells transfected with PAR4-YFP and PAR4-YFP mutants generated by site-directed mutagenesis was employed to ensure proper localization of receptors to the plasma membrane (Fig. S6). Confocal microscopy was conducted on an Olympus FV1000 (×60 magnification) following fixation of samples with 4% methanol-free formaldehyde and nuclear staining with 4′,6-diamidino-2-phenylindole dihydrochloride (1:1000). Renilla luciferase-tagged β-arrestin-1/2 constructs were a kind gift from Michel Bouvier (Université de Montréal). PX458 vector for CRISPR/Cas9 targeting was a kind gift from Feng Zhang (Massachusetts Institute of Technology; Addgene catalogue no. 48138).
Site-directed mutagenesis of predicted PAR4 receptor–peptide interaction sites
Site-directed mutagenesis using Agilent QuikChange-XL was undertaken to generate mutations in highly-predicted receptor residues to validate the GalaxyPepDock model and to determine key binding-site residues for the native PAR4-tethered ligand and PAR4 agonist peptide AYPGKF-NH2. Residues were mutated by single-nucleotide substitutions replacing native residues with alanine. Successful mutations were confirmed by DNA sequencing (London Regional Genomics Centre, London, Ontario, Canada).
Cell lines and culture conditions
All media and cell culture reagents were purchased from Thermo Fisher Scientific. HEK-293 cells (ATCC; Manassas, VA) and CRISPR/Cas9 HEK-293 β-arrestin double knockout (β-arr. knockout) cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% sodium pyruvate, and 1% streptomycin/penicillin. Cells stably transfected with PAR4-YFP vector were routinely cultured in the above media supplemented with 600 μg/ml G418 sulfate (Geneticin; Thermo Fisher Scientific, Waltham, MA). Because trypsin activates PAR4, cells were routinely subcultured using enzyme-free isotonic phosphate-buffered saline (PBS), pH 7.4, containing 1 mm EDTA followed by centrifugation to remove PBS/EDTA, and plated in appropriate culture plates for further experimentation. Cells were transiently transfected with appropriate vectors using a modified calcium phosphate transfection (nuclease-free water, 2.5 m CaCl2, and 2× HEPES-buffered saline). Transient transfections were rinsed with PBS 24 h post-transfection and replaced with growth media. Transiently transfected cells were assayed or imaged at 48 h post-transfection.
Generation of β-arrestin-1/2 double knockout HEK-293 cells using CRISPR/Cas9
A β-arrestin-1/2 double knockout HEK-293 cell line was generated using CRISPR/Cas9-mediated gene targeting. Guide sequences targeted to β-arrestin-1 or β-arrestin-2 were designed using a CRISPR guide design tool (72, 73). Gene-targeting guides (β-arrestin 1, TGTGGACCACATCGACCTCG, and β-arrestin 2, GCGGGACTTCGTAGATCACC) were ligated into the PX458 vector (kind gift from Dr. Feng Zhang, MIT, Addgene plasmid no. 48138), verified by direct sequencing, and transfected in HEK-293 cells via calcium phosphate transfection (65, 66). GFP-expressing cells were single-cell–selected by flow cytometry (FACSAria III; London Regional Flow Cytometry Facility) into wells of a 96-well plate. Clonal cell populations were expanded and screened by Western blotting to identify successful knockout (anti-β-arrestin-1/2 antibody, rabbit mAb D24H9, Cell Signaling Technologies, Danvers, MA; anti-β-actin antibody, mouse mAb 8H10D10, Cell Signaling Technologies; Fig. S5).
General methods for peptide synthesis
All reagents were purchased from Sigma, ChemImpex, or Thermo Fisher Scientific and used without further purification. Peptides were synthesized using standard Fmoc–solid-phase peptide synthesis (SPPS) on Rink-amide 4-Methylbenzhydrylamine (MBHA) resin (256 mg, 0.1 mmol, and 0.39 mmol/g) with a Biotage® SyrowaveTM automated peptide synthesizer (0.4 mmol of O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HCTU), 0.4 mmol of Fmoc-amino acids, and 0.6 mmol of DIPEA, 1 h coupling). Some peptides also involved additional manual synthesis (see procedures below). Following automated and manual synthesis, peptides were cleaved off the resin by being shaken in a cleavage mixture solution of 95% TFA, 2.5% TIPS, and 2.5% H2O (4 ml, 5 h), precipitated in ice-cold tert-butyl methyl ether, lyophilized, purified by preparative RP-HPLC, and further lyophilized to obtain a dry powder. Purity was assessed by analytical RP-HPLC and characterized by mass spectrometry (MS). The analytical RP-HPLC was performed on a system consisting of an analytical Agilent Zorbax SB-C8 column (4.6 × 150 mm, 5 μm), Waters 600 controller, Waters in-line degasser, and Waters Masslynx software (version 4.1). Two mobile phases were used: eluent A (0.1% TFA in acetonitrile) and eluent B (0.1% TFA in MilliQ water). The flow rate was set at 1.5 ml min−1 over 10 min with an additional 5-min wash (95% solvent A in solvent B). A Waters 2998 Photodiode array detector (200–800 nm) and an ESI-MS (Waters Quattro Micro API mass spectrometer) were used to monitor the column eluate. The preparative RP-HPLC used the same system, eluents, and detection method as mentioned above for the analytical RP-HPLC, except that a preparative Agilent Zorbax SB-C8 column (21.2 × 150 mm, 5 μm) at a flow rate of 20 ml min−1 was used. The mass spectra for all peptides were determined in positive mode using an ESI ion source on either a Bruker micrOToF II mass spectrometer or a Xevo QToF mass spectrometer (Table S1). All peptides had purity >95% as determined by analytical HPLC (see Fig. S1).
Synthesis of Ac-AYPGKF-NH2
The hexapeptide (AYPGKF) was synthesized using the standard automated Fmoc-SPPS discussed under “General methods for peptide synthesis.” While still on resin, with all of the side chains protected and with the N-terminal Fmoc-deprotection completed, the N terminus of this peptide was acetylated by being shaken in a solution of acetic anhydride and DMF (v/v 1:4, 5 ml, 30 min). Following this, the peptide was cleaved, purified, and analyzed as mentioned under “General methods for peptide synthesis.”
Synthesis of peptides involving N-methylation of the peptide backbone
Peptides ((N-Me-A)-YPGKF-NH2, (N-Me-S)-YPGKF-NH2, A-(N-Me-Y)-PGKF-NH2, AYPG-(Nα-Me-K)-F-NH2, and AYPGK-(N-Me-F)-NH2) were synthesized using the above-mentioned automated Fmoc-SPPS up to the desired amino acid that was N-methylated (e.g. KF-resin for AYPG-(Nα-Me-K)-F-NH2). Following Fmoc deprotection of the resin-bound N-terminal amino acid (e.g. lysine for AYPG-(Nα-Me-K)-F-NH2), site-selective N-methylation of the N-terminal amino acid was completed as follows. First, protection of the primary amine was completed to favor monomethylation by adding 2-nitrobenzenesulfonyl chloride (110.8 mg, 0.5 mmol) and 2,4,6-trimethylpyridine (132 μl, 1.0 mmol) in NMP (4 ml) to the on-resin peptide (0.1 mmol) and shaken (15 min). Following completion of the protection reaction, the resin was washed three times with NMP (three times with 5 ml) and dry THF (three times with 5 ml). Second, N-methylation was completed through Mitsunobu conditions by adding triphenylphosphine (131.2 mg, 0.5 mmol), diisopropyl azodicarboxylate (98 μl, 0.5 mmol), and methanol (41 μl, 1.0 mmol) in dry THF (4 ml) to the on-resin peptide and shaken under N2 (10 min). Following completion of the N-methylation reaction, the resin was washed three times with dry THF (three times with 5 ml) and NMP (three times with 5 ml). Third, the methylated N-terminal amine was then deprotected by adding 2-mercaptoethanol (70 μl, 1.0 mmol) and 1,8-diazabicyclo(5.4.0)undec-7-ene (75 μl, 0.5 mmol) in NMP (4 ml) to the on-resin peptide and shaken (5 min). The solution turned bright yellow as expected. The solution was removed, and then this third step was repeated, followed by being washed five times with NMP (five times with 5 ml) and three times with DMF (three times with 5 ml). Addition of the next sequential amino acid (except for inapplicable peptides, i.e. (N-Me-A)-YPGKF-NH2 and (N-Me-S)-YPGKF-NH2) was added manually (0.3 mmol of O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU), 0.3 mmol of Fmoc-amino acid (e.g. Fmoc-Gly-OH for AYPG-(Nα-Me-K)-F-NH2), 0.6 mmol of DIPEA, 4 ml of DMF, and 3 h coupling). Following completion of the sequential amino acid reaction, the resin was washed five times with DMF (five times with 5 ml) and then the next sequential amino acid was added (on applicable peptides i.e. AYPG-(Nα-Me-K)-F-NH2 and AYPGK-(N-Me-F)-NH2) using the automated procedure mentioned under “General methods for peptide synthesis.” Following completion of the synthesis, the peptide was cleaved, purified, and analyzed as mentioned under “General methods for peptide synthesis.” The amino acid N-methylglycine, commonly known as sarcosine, was obtained from a commercial source, and therefore, N-methylglycine–containing peptides (i.e. Sar-YPGKF-NH2 and AYP-Sar-KF-NH2) was synthesized solely by the standard automated Fmoc-SPPS discussed under “General methods for peptide synthesis.”
Synthesis of AYPG-(Nϵ-Me-K)-F-NH2
The hexapeptide (AYPGKF) was synthesized using the standard automated Fmoc-SPPS discussed under “General methods for peptide synthesis,” except that Fmoc-Lys(Alloc)-OH was used in place of standard Fmoc-Lys(Boc)-OH. While still on resin, with all of the side chains protected, and with the N-terminal Fmoc-deprotection completed, the N terminus of this peptide was Boc-protected by adding Boc-ON (98.5 mg, 0.4 mmol) and DIPEA (70 μl, 0.4 mmol) in DMF (4 ml) and shaken (4 h). The solution was removed, and this Boc protection was repeated overnight (20 h) to ensure reaction completion. The resin was washed three times with DMF (three times with 5 ml) and dry DCM (three times for 5 ml) and was placed under N2 for site-selective Alloc deprotection of the on-resin lysine side chain. Phenylsilane (296 μl, 2.4 mmol) in dry DCM (2 ml) was added to the resin. Tetrakis(triphenylphosphine) palladium(0) (23.1 mg, 20 μmol) was dissolved in dry DCM (1 ml) and added to the resin. The peptide column was flushed with N2 (2 min) followed by shaking (5 min). The resin was washed three times with dry DCM (three times with 5 ml). The procedure was repeated and shaken again (30 min). The resin was washed four times with DCM (four times with 5 ml) and DMF (four times with 5 ml). Following this N-terminal Boc protection and subsequent side chain Alloc deprotection, N-methylation of the on-resin lysine side chain was completed through the site-selective N-methylation procedure mentioned above. Following completion of the synthesis, the peptide was cleaved, purified, and analyzed as discussed under “General methods for peptide synthesis.”
SPPS reaction monitoring
Three methods were used to monitor SPPS reactions. Two methods (the Kaiser test and the small-scale resin cleavage) were used to monitor general peptide synthesis. Three methods (the Kaiser test, the small-scale resin cleavage, and the Chloranil test) were used to monitor synthesis of peptides involving N-methylation. For the Kaiser test, several resin beads were placed in a test tube followed by the addition of 42.5 mm phenol in ethanol (50 μl), 20 μm potassium cyanide in pyridine (50 μl), and 280.7 mm ninhydrin in ethanol (50 μl). The mixture was then heated (100 °C, 5 min), and a positive test indicates the presence of a free primary amine. For the small-scale resin cleavage method, several beads and cleavage mixture (500 μl) were shaken and worked up as per the full cleavage of the resin procedure described above, and the desired peptide was confirmed through HPLC-MS. For the Chloranil test, several resin beads were placed in a test tube followed by the addition of 357.8 mm acetaldehyde in DMF (50 μl) and 81.3 mm p-chloranil in DMF (50 μl) and allowed to stand at room temperature (5 min), where a positive test indicates the presence of a free secondary amine.
Calcium-signaling assay
HEK-293 cells or HEK-293 cells stably expressing PAR4-YFP were loaded with the calcium-sensitive dye (Fluo-4 NW, F36206, Life Technologies, Inc., and Thermo Fisher Scientific) and aliquoted into cuvettes with Hanks' buffered salt solution (HBSS containing Ca2+ and Mg2+). Samples were excited at 480 nm, and emission was recorded at 530 nm. Baseline fluorescence from PAR4-YFP excitation remains constant throughout the experiment. Following baseline recording, agonist-induced changes in fluorescence were recorded. Change in fluorescence from baseline to maximum response at a given agonist concentration (Δ) were expressed as a percentage of maximum cellular response elicited by the calcium ionophore (A23187, Sigma).
Calcium-signaling assay with PAR4 orthosteric site-directed mutants
HEK-293 cells were transiently transfected with 2 μg of WT YFP tagged PAR4 or PAR4 mutant receptors (described above) using the calcium phosphate transfection protocol as described above. Calcium signaling was investigated as described above for the WT receptor.
β-Arrestin recruitment–bioluminescence resonance energy transfer (BRET)
HEK-293 cells were co-transfected with either WT or mutant PAR4-YFP and β-arrestin-1- or -2–rluc (0.2 μg) using modified calcium phosphate transfection. Cells were re-plated (24 h post-transfection) into white 96-well plates (Corning). 48 h post-transfection, agonist-stimulated β-arrestin recruitment to PAR4 was detected by measuring BRET following 20 min of agonist stimulation. Coelenterazine (5 μm; Nanolight Technology, Pinetop, AZ) was added 10 min prior to BRET collection using a Berthold Mithras LB 940 plate reader.
Testing peptide library specificity for PAR4
Previous studies have pointed to the possibility of PAR agonist peptides cross-activating other members of this family. For example, the PAR1-tethered ligand peptide SFLLRN is able to activate PAR2, and substitution of a phenylalanine residue in certain PAR4 agonist peptides confers PAR1 activity (32, 52, 67). Activating peptides typically requires an aromatic side chain in position 2 (Phe in PAR1), and we wondered whether substitution of tyrosine for phenylalanine may decrease selectivity at PAR4 and increase cross-talk with PAR1. We examined whether substitutions made to AYPGKF-NH2 affected their selectivity using a calcium-signaling assay in HEK-293 cells that endogenously express PAR1 and PAR2. None of the peptides tested elicited a signaling greater than 5% of the calcium ionophore A23187 showing that they retain selectivity toward PAR4 (Fig. S7). Additionally, we investigated specificity using BRET for β-arrestin-2 recruitment to cells expressing PAR4-YFP or PAR2-YFP. None of the peptides were able to induce β-arrestin recruitment in PAR2-YFP–expressing HEK-293 cells (net BRET of 0.05 and over) with the exception of AYPGK-1Nal-NH2 (Fig. S8).
PAR4-stimulated MAPK signaling
HEK-293 and CRISPR/Cas9 β-arrestin-1/2 double knockout HEK-293
Agonist-stimulated MAPK signaling in CRISPR/Cas9 HEK-293 β-arr. knockout and WT HEK-293 cells transfected with PAR4-YFP were analyzed by Western blotting. Cells were placed in serum-free media (serum-free DMEM, Gibco) for 3 h before beginning the experiment. Cells were then stimulated with 30 μm AYPGKF-NH2 for varying time periods (0, 2, 5, 10, 20, 30, 60, and 90 min) at 37 °C. Treated cells were placed on ice following the stimulation period. Total protein was extracted by adding kinase lysis buffer with phosphatase and protease inhibitors (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 0.5% Nonidet P-40, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 25 mm NaF, 1 mg/ml leupeptin, 1 mg/m aprotinin, 1 mm phenylmethylsulfonyl fluoride, and 1 mm DTT) for 20 min, and cell membranes were cleared by centrifugation (13,300 × g for 3 min). Protein concentrations were measured using the Pierce BCA protein assay (Thermo Fisher Scientific). Protein samples were heat-denatured at 98 °C for 8 min in denaturing Laemmli buffer (containing 2-mercaptoethanol) and resolved on 4–12% gradient Invitrogen Bolt BisTris Plus gels (Thermo Fisher Scientific). The resolved proteins were transferred via wet transfer in buffer (1× Tris-glycine, 20% methanol) to a polyvinylidene difluoride membrane and blocked in TBST buffer (TBS with 0.1% (v/v) Tween 20) supplemented with 1% ECL Advance Blocking Agent (GE Healthcare) for 40 min at room temperature. p44/42 (Thr-202/Tyr-204) phosphorylation was detected with specific antibodies (Cell Signaling Technology, Danvers, MA; diluted 1:1000 in TBST) overnight at 4 °C. Phospho-p44/42 immunoreactivity was detected using the horseradish peroxidase-conjugated anti-rabbit secondary antibody (Cell Signaling Technology; 1:10,000 in TBST for 1 h). After washing the membrane with TBST (three times with 10-min washes), the peroxidase activity was detected with the chemiluminescence reagent ECL Advance (GE Healthcare) on iBright image Station (Invitrogen). Membranes were then stripped with blot-stripping buffer (Thermo Fisher Scientific) at room temperature and blocked in TBST with 1% ECL Advance Blocking Agent before incubation with the p44/42 (1:1000 in TBST with 1% ECL Advance Blocking Agent) overnight at 4 °C. Following incubation with primary antibody, blots were rinsed in TBST and incubated with appropriate horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody (1:10,000 in TBST for 1 h). Membranes were washed and imaged as described above. Band intensities representing activated MAPK were quantified using FIJI (68). Phospho-kinase levels were normalized for differences in protein loading by expressing the data as a ratio of the corresponding total p44/42 signal. Data are expressed as a fold increase over unstimulated baseline control.
PAR4-stimulated MAPK signaling in HEK-293 +/− YM254890
Agonist-stimulated MAPK signaling in WT HEK-293 cells transfected with PAR4-YFP was analyzed by Western blotting. Cells were placed in serum-free media (serum-free DMEM, Gibco) for 3 h before beginning the experiment. Prior to agonist stimulation, the cells were treated with either control (0.001% DMSO) or YM254890 (100 nm) for 20 min. Cells were then stimulated with 30 μm AYPGKF-NH2 for varying time periods (0, 2, 5, 10, 20, 30, 60, and 90 min) at 37 °C. Cells were lysed and protein-quantified, and samples were run and imaged as described previously.
MAPK signaling in PAR4-YFP stable cells with peptide library
Agonist-stimulated MAPK signaling in HEK-293 cells stably expressing PAR4-YFP were analyzed by Western blotting following stimulation with PAR4 peptide agonists. Cells were placed in serum-free media (serum-free DMEM, Gibco) for 3 h before beginning the experiment. Cells were stimulated with either control (no agonist), AYPGKF-NH2, or agonist peptides described at 100 μm concentration. Cells were stimulated for 10 min with agonist and lysed, quantified, and blotted as described above.
Preparation of washed platelets and light-transmission aggregometry
Animal care and experimental procedures were carried out under a protocol (AUP no. 2018-047) that was reviewed and approved by the University of Western Ontario Animal Care Committee in accordance with the Canadian Council on Animal Care (CCAC) standards and guidelines. Blood was drawn from the abdominal aorta or heart of Sprague-Dawley rats into trisodium citrate (3.4% w/v). The blood was centrifuged at 200 × g for 15 min at 22 °C to isolate platelet-rich plasma (PRP). PRP was transferred to a 14-ml polypropylene round-bottom tube (Falcon) and centrifuged at 1800 rotations per min (594 × g) for 10 min at 22 °C to pellet platelets and obtain platelet-poor plasma (PPP). PPP was decanted and used for 100% light transmission control. To obtain the washed platelet sample, platelets were washed twice in Tyrode's buffer (12 mm NaHCO3, 127 mm NaCl, 5 mm KCl, 0.5 mm NaH2PO4, 1 mm MgCl2, 5 mm glucose, and 10 mm HEPES; Ramachandran et al. (26)) and resuspended to a volume equivalent to that of the initial blood draw. Washed platelet sample was equilibrated for 1 h at ambient temperature following the addition of 1 m CaCl2 (1 μl/ml). 400 μl of this washed platelet suspension was used for light transmission aggregometry on a platelet aggregometer (Model 700; Chrono-Log Corp, Havertown, PA).
Structural homology modeling of PAR4
Residues resolved in the PAR2 crystal (Protein Data Bank (PDB) code 5NDD) were used to generate a homology model of WT human PAR2. Homology model of WT human PAR2 was then aligned with the WT human PAR4 amino acid sequence (National Center for Biotechnology Information (NCBI) accession number NM_003950) using Clustal Omega (37, 38). Only residues resolved in the crystal structure of PAR2 were used as template coordinates for PAR4 homology modeling. Twenty human PAR4 homology models were generated using the human PAR2 crystal structure coordinates (PDB code 5NDD) (36) as a template in MODELLER (version 9.16) (69). The model with the lowest DOPE score was taken as the best structure and visualized using PyMOL (version 1.7.4, Schrödinger, LLC). Electrostatic surface potential was calculated after assignment of partial charges by using PDB2PQR and by using the Adaptive Poisson-Boltzmann Solver plugin in PyMOL (70).
In silico docking of AYPGKF-NH2 to the PAR4 homology model
In silico docking of the PAR4 agonist peptide, AYPGKF-NH2, to the PAR4 homology model was undertaken using GalaxyPepDock (39). The lowest energy PAR4 homology model PDB file was loaded into GalaxyPepDock along with a text file of the PAR4 agonist peptide sequence, which GalaxyPepDock uses to predict a peptide structure for docking analyses (Ala-Tyr-Pro-Gly-Lys-Phe; amidation is not included in the structure prediction). Ten models of PAR4-AP docking were generated. Frequency of receptor–peptide residue interactions, taken as atom–atom distances of less than 3 Å, was quantified manually to determine highly recurrent interactions.
Data analysis and statistical tests
Data are shown as mean ± S.E. Calcium signaling and β-arrestin-1/2 recruitment concentration–effect curves were calculated using nonlinear regression curve fitting (three parameters; Prism7). EC50 values are reported in Table 1 where the signal has saturated. Where signaling is not saturated, the EC50 is reported for calcium as not determined (ND). The F-statistic calculated to compare EC50 values was calculated using the extra sum-of-squares analysis (71). For Table 1, values comparing percentage of signaling or recruitment at 100 μm to values obtained from the parental PAR4-AP, AYPGKF-NH2, are shown. Significance was determined for comparisons with 100 μm AYPGKF-NH2 and p44/42 phosphorylation by one-way ANOVA. Western blotting protein densitometry was quantified using FIJI. Transformation and analysis were conducted on raw data, and significance is indicated by * (p < 0.05).
Author contributions
P. E. T., J. C. L., L. G. L., and R. R. conceptualization; P. E. T., J. C. L., and R. R. data curation; P. E. T., P. B. S., and R. R. formal analysis; P. E. T. validation; P. E. T., J. C. L., D. A., M. F., P. B. S., and R. R. investigation; P. E. T. and R. R. visualization; P. E. T., J. C. L., and R. R. methodology; P. E. T., J. C. L., L. G. L., and R. R. writing-original draft; P. E. T. and R. R. project administration; P. E. T., J. C. L., D. A., M. F., P. C., P. B. S., L. G. L., and R. R. writing-review and editing; P. B. S., L. G. L., and R. R. supervision; P. B. S., L. G. L., and R. R. funding acquisition; L. G. L. and R. R. resources.
Supplementary Material
Acknowledgments
We thank Dr. Michel Bouvier for providing the Renilla luciferase-tagged β-arrestin-1/2 constructs and Dr. Feng Zhang for providing the PX458 vector for CRISPR/Cas9 targeting.
This work was supported by a Canadian Institutes of Health Research (CIHR) grant 376560 (to R. R.) and by grants from the Natural Sciences and Engineering Research Council of Canada (to L. G. L. and P. B. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Table S1.
Throughout the text boldface indicates amino acid differing from parental PAR4 agonist peptide.
Lowercase letter indicates d-amino acid throughout.
- GPCR
- G-protein–coupled receptor
- PAR4
- proteinase activated receptor-4
- PAR4-AP
- parental PAR4 agonist peptide
- PAR4-AP
- parental PAR4 agonist peptide; AYPGKF-NH2
- ANOVA
- analysis of variance
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- BRET
- bioluminescence resonance energy transfer
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- DCM
- dichloromethane
- DIPEA
- N,N-diisopropylethylamine
- DMF
- N,N-dimethylformamide
- HBSS
- Hanks' buffered salt solution
- Sar
- sarcosine
- β-arr
- β-arrestin
- Mpr
- mercaptopropionic acid
- eYFP
- enhanced yellow fluorescent protein
- SPSS
- solid-phase peptide synthesis
- PRP
- platelet-rich plasma
- PPP
- platelet-poor plasma
- Pip
- 2-piperidinecarboxylic acid
- Nip
- 3-piperidinecarboxylic acid
- PDB
- Protein Data Bank
- rluc
- Renilla luciferase
- ESI
- electrospray ionization
- Boc
- t-butoxycarbonyl
- Cit
- citrulline
- N-Me-S
- N-methylserine
- Aib
- 2-aminoisobutyric acid
- Nal
- naphthyl
- Nle
- norleucine
- Alloc
- allyloxycarbonyl
- TIPS
- triisopropylsilane
- Inp
- isonipecotic acid.
References
- 1. Fredriksson R., and Schiöth H. B. (2005) The repertoire of G-protein–coupled receptors in fully sequenced genomes. Mol. Pharmacol. 67, 1414–1425 10.1124/mol.104.009001 [DOI] [PubMed] [Google Scholar]
- 2. Pierce K. L., Premont R. T., and Lefkowitz R. J. (2002) Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 10.1038/nrm908 [DOI] [PubMed] [Google Scholar]
- 3. Erlandson S. C., McMahon C., and Kruse A. C. (2018) Structural basis for G protein–coupled receptor signaling. Annu. Rev. Biophys. 47 10.1146/annurev-biophys-070317-032931 [DOI] [PubMed] [Google Scholar]
- 4. Hilger D., Masureel M., and Kobilka B. K. (2018) Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12 10.1038/s41594-017-0011-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sriram K., and Insel P. A. (2018) G-protein–coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol. Pharmacol. 93, 251–258 10.1124/mol.117.111062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vu T. K., Hung D. T., Wheaton V. I., and Coughlin S. R. (1991) Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell 64, 1057–1068 10.1016/0092-8674(91)90261-V [DOI] [PubMed] [Google Scholar]
- 7. Adams M. N., Ramachandran R., Yau M.-K., Suen J. Y., Fairlie D. P., Hollenberg M. D., and Hooper J. D. (2011) Structure, function and pathophysiology of protease activated receptors. Pharmacol. Ther. 130, 248–282 10.1016/j.pharmthera.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 8. Ishihara H., Connolly A. J., Zeng D., Kahn M. L., Zheng Y. W., Timmons C., Tram T., and Coughlin S. R. (1997) Protease-activated receptor 3 is a second thrombin receptor in humans. Nature 386, 502–506 10.1038/386502a0 [DOI] [PubMed] [Google Scholar]
- 9. Coughlin S. R. (2005) Protease-activated receptors in hemostasis, thrombosis and vascular biology. J. Thromb. Haemost. 3, 1800–1814 10.1111/j.1538-7836.2005.01377.x [DOI] [PubMed] [Google Scholar]
- 10. Coughlin S. R. (1999) How the protease thrombin talks to cells. Proc. Natl. Acad. Sci. U.S.A. 96, 11023–11027 10.1073/pnas.96.20.11023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morrow D. A., Braunwald E., Bonaca M. P., Ameriso S. F., Dalby A. J., Fish M. P., Fox K. A., Lipka L. J., Liu X., Nicolau J. C., Ophuis A. J., Paolasso E., Scirica B. M., Spinar J., Theroux P., et al. (2012) Vorapaxar in the secondary prevention of atherothrombotic events. N. Engl. J. Med. 366, 1404–1413 10.1056/NEJMoa1200933 [DOI] [PubMed] [Google Scholar]
- 12. Morrow D. A., Scirica B. M., Fox K. A., Berman G., Strony J., Veltri E., Bonaca M. P., Fish P., McCabe C. H., Braunwald E., and TRA 2°P-TIMI 50 Investigators. (2009) Evaluation of a novel antiplatelet agent for secondary prevention in patients with a history of atherosclerotic disease: design and rationale for the thrombin-receptor antagonist in secondary prevention of atherothrombotic ischemic events (TRA 2°P)-TIMI 50 trial. Am. Heart J. 158, 335–341.e3 10.1016/j.ahj.2009.06.027 [DOI] [PubMed] [Google Scholar]
- 13. Scirica B. M., Bonaca M. P., Braunwald E., De Ferrari G. M., Isaza D., Lewis B. S., Mehrhof F., Merlini P. A., Murphy S. A., Sabatine M. S., Tendera M., Van de Werf F., Wilcox R., Morrow D. A., and TRA 2°P-TIMI 50 Steering Committee Investigators. (2012) Vorapaxar for secondary prevention of thrombotic events for patients with previous myocardial infarction: a prespecified subgroup analysis of the TRA 2°P-TIMI 50 trial. Lancet 380, 1317–1324 10.1016/S0140-6736(12)61269-0 [DOI] [PubMed] [Google Scholar]
- 14. Jacques S. L., LeMasurier M., Sheridan P. J., Seeley S. K., and Kuliopulos A. (2000) Substrate-assisted catalysis of the PAR1 thrombin receptor. Enhancement of macromolecular association and cleavage. J. Biol. Chem. 275, 40671–40678 10.1074/jbc.M004544200 [DOI] [PubMed] [Google Scholar]
- 15. Liu L. W., Vu T. K., Esmon C. T., and Coughlin S. R. (1991) The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J. Biol. Chem. 266, 16977–16980 [PubMed] [Google Scholar]
- 16. Shapiro M. J., Weiss E. J., Faruqi T. R., and Coughlin S. R. (2000) Protease-activated receptors 1 and 4 are shut off with distinct kinetics after activation by thrombin. J. Biol. Chem. 275, 25216–25221 10.1074/jbc.M004589200 [DOI] [PubMed] [Google Scholar]
- 17. Jacques S. L., and Kuliopulos A. (2003) Protease-activated receptor-4 uses dual prolines and an anionic retention motif for thrombin recognition and cleavage. Biochem. J. 376, 733–740 10.1042/bj20030954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rwibasira Rudinga G., Khan G. J., and Kong Y. (2018) Protease-activated receptor 4 (PAR4): a promising target for antiplatelet therapy. Int. J. Mol. Sci. 19, E573 10.3390/ijms19020573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu C. C., Huang S. W., Hwang T. L., Kuo S. C., Lee F. Y., and Teng C. M. (2000) YD-3, a novel inhibitor of protease-induced platelet activation. Br. J. Pharmacol. 130, 1289–1296 10.1038/sj.bjp.0703437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Young S. E., Duvernay M. T., Schulte M. L., Nance K. D., Melancon B. J., Engers J., Wood M. R., Hamm H. E., and Lindsley C. W. (2010) A novel and selective PAR4 antagonist: ML354. in Probe Reports from the NIH Molecular Libraries Program, National Center for Biotechnology Information, Bethesda, MD: [PubMed] [Google Scholar]
- 21. Miller M. M., Banville J., Friends T. J., Gagnon M., Hangeland J. J., Lavalée J.-F., Martel A., O'Grady H., Rémillard R., Ruediger E., Tremblay F., Posy S. L., Allegretto N. J., Guarino V. R., Harden D. G., et al. (2019) Discovery of potent protease-activated receptor 4 antagonists with in vivo antithrombotic efficacy. J. Med. Chem. 62, 7400–7416 10.1021/acs.jmedchem.9b00186 [DOI] [PubMed] [Google Scholar]
- 22. Wilson S. J., Ismat F. A., Wang Z., Cerra M., Narayan H., Raftis J., Gray T. J., Connell S., Garonzik S., Ma X., Yang J., and Newby D. E. (2018) PAR4 (protease-activated receptor 4) antagonism with BMS-986120 inhibits human ex vivo thrombus formation. Arterioscler. Thromb. Vasc. Biol. 38, 448–456 10.1161/ATVBAHA.117.310104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong P. C., Seiffert D., Bird J. E., Watson C. A., Bostwick J. S., Giancarli M., Allegretto N., Hua J., Harden D., Guay J., Callejo M., Miller M. M., Lawrence R. M., Banville J., Guy J., et al. (2017) Blockade of protease-activated receptor-4 (PAR4) provides robust antithrombotic activity with low bleeding. Sci. Transl. Med. 9, eaaf5294 10.1126/scitranslmed.aaf5294 [DOI] [PubMed] [Google Scholar]
- 24. Holinstat M., Voss B., Bilodeau M. L., McLaughlin J. N., Cleator J., and Hamm H. E. (2006) PAR4, but not PAR1, signals human platelet aggregation via Ca2+ mobilization and synergistic P2Y12 receptor activation. J. Biol. Chem. 281, 26665–26674 10.1074/jbc.M602174200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li D., D'Angelo L., Chavez M., and Woulfe D. S. (2011) Arrestin-2 differentially regulates PAR4 and ADP receptor signaling in platelets. J. Biol. Chem. 286, 3805–3814 10.1074/jbc.M110.118018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramachandran R., Mihara K., Thibeault P., Vanderboor C. M., Petri B., Saifeddine M., Bouvier M., and Hollenberg M. D. (2017) Targeting a proteinase-activated receptor 4 (PAR4) carboxyl-terminal motif to regulate platelet function. Mol. Pharmacol. 91, 287–295 10.1124/mol.116.106526 [DOI] [PubMed] [Google Scholar]
- 27. Covic L., Misra M., Badar J., Singh C., and Kuliopulos A. (2002) Pepducin-based intervention of thrombin-receptor signaling and systemic platelet activation. Nat. Med. 8, 1161–1165 10.1038/nm760 [DOI] [PubMed] [Google Scholar]
- 28. Bar-Shavit R., Maoz M., Kancharla A., Jaber M., Agranovich D., Grisaru-Granovsky S., and Uziely B. (2016) Protease-activated receptors (PARs) in cancer: novel biased signaling and targets for therapy. Methods Cell biol. 132, 341–358 10.1016/bs.mcb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 29. Hollenberg M. D., Mihara K., Polley D., Suen J. Y., Han A., Fairlie D. P., and Ramachandran R. (2014) Biased signaling and proteinase-activated receptors (PARs): targeting inflammatory disease. Br. J. Pharmacol. 171, 1180–1194 10.1111/bph.12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seyedabadi M., Ghahremani M. H., and Albert P. R. (2019) Biased signaling of G protein coupled receptors (GPCRs): molecular determinants of GPCR/transducer selectivity and therapeutic potential. Pharmacol. Ther. 200, 148–178 10.1016/j.pharmthera.2019.05.006 [DOI] [PubMed] [Google Scholar]
- 31. Scarborough R. M., Naughton M. A., Teng W., Hung D. T., Rose J., Vu T. K., Wheaton V. I., Turck C. W., and Coughlin S. R. (1992) Tethered ligand agonist peptides. Structural requirements for thrombin receptor activation reveal mechanism of proteolytic unmasking of agonist function. J. Biol. Chem. 267, 13146–13149 [PubMed] [Google Scholar]
- 32. Faruqi T. R., Weiss E. J., Shapiro M. J., Huang W., and Coughlin S. R. (2000) Structure–function analysis of protease-activated receptor 4 tethered ligand peptides. Determinants of specificity and utility in assays of receptor function. J. Biol. Chem. 275, 19728–19734 10.1074/jbc.M909960199 [DOI] [PubMed] [Google Scholar]
- 33. Hoffmann W., Langenhan J., Huhmann S., Moschner J., Chang R., Accorsi M., Seo J., Rademann J., Meijer G., Koksch B., Bowers M. T., von Helden G., and Pagel K. (2019) An intrinsic hydrophobicity scale for amino acids and its application to fluorinated compounds. Angew. Chem. Int. Ed. Engl. 58, 8216–8220 10.1002/anie.201813954 [DOI] [PubMed] [Google Scholar]
- 34. Takasaki J., Saito T., Taniguchi M., Kawasaki T., Moritani Y., Hayashi K., and Kobori M. (2004) A novel Gαq/11-selective inhibitor. J. Biol. Chem. 279, 47438–47445 10.1074/jbc.M408846200 [DOI] [PubMed] [Google Scholar]
- 35. Jean-Charles P.-Y., Kaur S., and Shenoy S. K. (2017) GPCR signaling via β-arrestin-dependent mechanisms. J. Cardiovasc. Pharmacol. 70, 142–158 10.1097/FJC.0000000000000482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng R. K. Y., Fiez-Vandal C., Schlenker O., Edman K., Aggeler B., Brown D. G., Brown G. A., Cooke R. M., Dumelin C. E., Doré A. S., Geschwindner S., Grebner C., Hermansson N.-O., Jazayeri A., Johansson P., et al. (2017) Structural insight into allosteric modulation of protease-activated receptor 2. Nature 545, 112–115 10.1038/nature22309 [DOI] [PubMed] [Google Scholar]
- 37. Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A. R. N., Potter S. C., Finn R. D., and Lopez R. (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee H., Heo L., Lee M. S., and Seok C. (2015) GalaxyPepDock: a protein–peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 43, W431–W435 10.1093/nar/gkv495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu W. F., Andersen H., Whitmore T. E., Presnell S. R., Yee D. P., Ching A., Gilbert T., Davie E. W., and Foster D. C. (1998) Cloning and characterization of human protease-activated receptor 4. Proc. Natl. Acad. Sci. U.S.A. 95, 6642–6646 10.1073/pnas.95.12.6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soh U. J., Dores M. R., Chen B., and Trejo J. (2010) Signal transduction by protease-activated receptors. Br. J. Pharmacol. 160, 191–203 10.1111/j.1476-5381.2010.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hollenberg M. D. (2003) Proteinase-activated receptors: tethered ligands and receptor-activating peptides. Drug Dev. Res. 59, 336–343 10.1002/ddr.10301 [DOI] [Google Scholar]
- 43. Al-Ani B., Wijesuriya S. J., and Hollenberg M. D. (2002) Proteinase-activated receptor 2: differential activation of the receptor by tethered ligand and soluble peptide analogs. J. Pharmacol. Exp. Ther. 302, 1046–1054 10.1124/jpet.302.3.1046 [DOI] [PubMed] [Google Scholar]
- 44. Nystedt S., Emilsson K., Wahlestedt C., and Sundelin J. (1994) Molecular cloning of a potential proteinase activated receptor. Proc. Natl. Acad. Sci. U.S.A. 91, 9208–9212 10.1073/pnas.91.20.9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kahn M. L., Zheng Y. W., Huang W., Bigornia V., Zeng D., Moff S., Farese R. V. Jr., Tam C., and Coughlin S. R. (1998) A dual thrombin receptor system for platelet activation. Nature 394, 690–694 10.1038/29325 [DOI] [PubMed] [Google Scholar]
- 46. Chidiac P. (2016) RGS proteins destroy spare receptors: effects of GPCR-interacting proteins and signal deamplification on measurements of GPCR agonist potency. Methods 92, 87–93 10.1016/j.ymeth.2015.08.011 [DOI] [PubMed] [Google Scholar]
- 47. Edelstein L. C., Simon L. M., Lindsay C. R., Kong X., Teruel-Montoya R., Tourdot B. E., Chen E. S., Ma L., Coughlin S., Nieman M., Holinstat M., Shaw C. A., and Bray P. F. (2014) Common variants in the human platelet PAR4 thrombin receptor alter platelet function and differ by race. Blood 124, 3450–3458 10.1182/blood-2014-04-572479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Venkatakrishnan A. J., Deupi X., Lebon G., Tate C. G., Schertler G. F., and Babu M. M. (2013) Molecular signatures of G-protein–coupled receptors. Nature 494, 185–194 10.1038/nature11896 [DOI] [PubMed] [Google Scholar]
- 49. Woolley M. J., and Conner A. C. (2017) Understanding the common themes and diverse roles of the second extracellular loop (ECL2) of the GPCR super-family. Mol. Cell. Endocrinol. 449, 3–11 10.1016/j.mce.2016.11.023 [DOI] [PubMed] [Google Scholar]
- 50. Wheatley M., Wootten D., Conner M. T., Simms J., Kendrick R., Logan R. T., Poyner D. R., and Barwell J. (2012) Lifting the lid on GPCRs: the role of extracellular loops. Br. J. Pharmacol. 165, 1688–1703 10.1111/j.1476-5381.2011.01629.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Al-Ani B., Saifeddine M., Kawabata A., and Hollenberg M. D. (1999) Proteinase activated receptor 2: role of extracellular loop 2 for ligand-mediated activation. Br. J. Pharmacol. 128, 1105–1113 10.1038/sj.bjp.0702834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lerner D. J., Chen M., Tram T., and Coughlin S. R. (1996) Agonist recognition by proteinase-activated receptor 2 and thrombin receptor. Importance of extracellular loop interactions for receptor function. J. Biol. Chem. 271, 13943–13947 10.1074/jbc.271.24.13943 [DOI] [PubMed] [Google Scholar]
- 53. Gerszten R. E., Chen J., Ishii M., Ishii K., Wang L., Nanevicz T., Turck C. W., Vu T. K., and Coughlin S. R. (1994) Specificity of the thrombin receptor for agonist peptide is defined by its extracellular surface. Nature 368, 648–651 10.1038/368648a0 [DOI] [PubMed] [Google Scholar]
- 54. Nanevicz T., Ishii M., Wang L., Chen M., Chen J., Turck C. W., Cohen F. E., and Coughlin S. R. (1995) Mechanisms of thrombin receptor agonist specificity. Chimeric receptors and complementary mutations identify an agonist recognition site. J. Biol. Chem. 270, 21619–21625 10.1074/jbc.270.37.21619 [DOI] [PubMed] [Google Scholar]
- 55. Nieman M. T. (2008) Protease-activated receptor 4 uses anionic residues to interact with α-thrombin in the absence or presence of protease-activated receptor 1. Biochemistry 47, 13279–13286 10.1021/bi801334s [DOI] [PubMed] [Google Scholar]
- 56. Nieman M. T., and Schmaier A. H. (2007) Interaction of thrombin with PAR1 and PAR4 at the thrombin cleavage site. Biochemistry 46, 8603–8610 10.1021/bi700597p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sánchez Centellas D., Gudlur S., Vicente-Carrillo A., Ramström S., and Lindahl T. L. (2017) A cluster of aspartic residues in the extracellular loop II of PAR 4 is important for thrombin interaction and activation of platelets. Thromb. Res. 154, 84–92 10.1016/j.thromres.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 58. Moschonas I. C., Kellici T. F., Mavromoustakos T., Stathopoulos P., Tsikaris V., Magafa V., Tzakos A. G., and Tselepis A. D. (2017) Molecular requirements involving the human platelet protease-activated receptor-4 mechanism of activation by peptide analogues of its tethered ligand. Platelets 28, 812–821 10.1080/09537104.2017.1282607 [DOI] [PubMed] [Google Scholar]
- 59. Blackhart B. D., Emilsson K., Nguyen D., Teng W., Martelli A. J., Nystedt S., Sundelin J., and Scarborough R. M. (1996) Ligand cross-reactivity within the protease-activated receptor family. J. Biol. Chem. 271, 16466–16471 10.1074/jbc.271.28.16466 [DOI] [PubMed] [Google Scholar]
- 60. French S. L., and Hamilton J. R. (2016) Protease-activated receptor 4: from structure to function and back again. Br. J. Pharmacol. 173, 2952–2965 10.1111/bph.13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tourdot B. E., Stoveken H., Trumbo D., Yeung J., Kanthi Y., Edelstein L. C., Bray P. F., Tall G. G., and Holinstat M. (2018) Genetic variant in human PAR (protease-activated receptor) 4 enhances thrombus formation resulting in resistance to antiplatelet therapeutics. Arterioscler. Thromb. Vasc. Biol. 38, 1632–1643 10.1161/ATVBAHA.118.311112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khan A., Li D., Ibrahim S., Smyth E., and Woulfe D. S. (2014) The physical association of the P2Y12 receptor with PAR4 regulates arrestin-mediated Akt activation. Mol. Pharmacol. 86, 1–11 10.1124/mol.114.091595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Leger A. J., Jacques S. L., Badar J., Kaneider N. C., Derian C. K., Andrade-Gordon P., Covic L., and Kuliopulos A. (2006) Blocking the protease-activated receptor 1–4 heterodimer in platelet-mediated thrombosis. Circulation 113, 1244–1254 10.1161/CIRCULATIONAHA.105.587758 [DOI] [PubMed] [Google Scholar]
- 64. Vanderboor C. M., Thibeault P. E., Nixon K. C., Gros R., Kramer J., and Ramachandran R. (2019) Proteinase-activated receptor 4 (PAR4) activation triggers cell membrane blebbing through RhoA and β-arrestin. bioRxiv 10.1101/746677 [DOI] [PubMed]
- 65. Ferguson S. S., and Caron M. G. (2004) Green fluorescent protein-tagged β-arrestin translocation as a measure of G-protein–coupled receptor activation. Methods Mol. Biol. 237, 121–126 10.1385/1-59259-430-1:121 [DOI] [PubMed] [Google Scholar]
- 66. Graham F. L., and van der Eb A. J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52, 456–467 10.1016/0042-6822(73)90341-3 [DOI] [PubMed] [Google Scholar]
- 67. Boitano S., Flynn A. N., Schulz S. M., Hoffman J., Price T. J., and Vagner J. (2011) Potent agonists of the protease activated receptor 2 (PAR2). J. Med. Chem. 54, 1308–1313 10.1021/jm1013049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Webb B., and Sali A. (2017) Protein Structure Modeling with MODELLER. Functional Genomics, pp. 39–54, Humana Press, New York, NY, New York, NY [Google Scholar]
- 70. Dolinsky T. J., Nielsen J. E., McCammon J. A., and Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim B. H., Wang F. I., Pereverzev A., Chidiac P., and Dixon S. J. (2019) Toward defining the pharmacophore for positive allosteric modulation of PTH1 receptor signaling by extracellular nucleotides. ACS Pharmacol. Transl. Sci. 2, 155–167 10.1021/acsptsci.8b00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., Li Y., Fine E. J., Wu X., Shalem O., Cradick T. J., Marraffini L. A., Bao G., and Zhang F. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.