Abstract
L1s are transposable elements that move by a copy-and-paste mechanism that continuously increases their copy number in the genome, such that each genome has a record of the L1 history in that host lineage. They make up about 20% of the genomes of eutherian mammals and have played a major role in shaping genome evolution. Chiroptera has the lowest average genome size among mammalian orders and the only documented case of L1 extinction affecting an entire mammalian family. Herein, L1 activity and extinction are characterized in all families of the order Chiroptera using a method that enriches for the youngest lineages of L1s in the genome. In addition to the previously reported L1 extinction in Pteropodidae, L1 extinction was documented to occur in Mormoops blainvilli, but this event did not affect all species of Mormoopidae. Further, there was no evidence of concordance between the evolution of L1s and their chiropteran host. There were two L1 lineages present before the divergence of all extant bats. Both lineages are extinct in the Pteropodidae. One or the other L1 lineage is extinct in almost all bat families, but Taphozous melanopogon maintains active members of both. Most intriguingly, some families within the Rhinolophoidea retain one active L1 lineage whereas other families retain the other, creating a deep discontinuity between L1 phylogeny and chiropteran phylogeny. These results indicate that there have been numerous losses of active L1 lineages over the history of chiropteran evolution, but that all chiropteran families except Pteropodidae have retained L1 activity.
Keywords: bat, Chiroptera, evolution, L1, LINE-1, phylogeny, retrotransposons, transposable elements
Introduction
L1 retrotransposons (LINE-1; Long INterspersed Element-1) have played a major role in shaping mammalian genomes (de Koning et al. 2011; Platt et al. 2018). In addition to retrotransposing their own sequence to new sites in the genome, L1s can provide the molecular machinery to move SINEs (Short INterspersed Elements) and processed pseudogenes (Dewannieux et al. 2003; Dewannieux and Heidmann 2005). Any of these sequences can cause mutations by inserting into genes, and retrotransposition can also move flanking sequences (Kazazian et al. 1988; Goodier et al. 2000; Ostertag and Kazazian 2001).
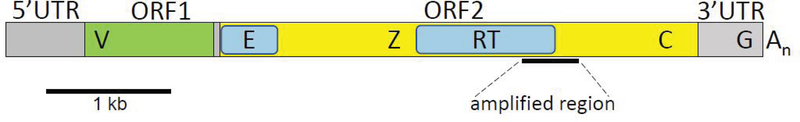
In mammals, full-length L1 elements are 6.5 to 7 kb and are made up of four major segments (Fig. 1): 5′ UTR, ORF1, ORF2, and 3′ UTR (Furano 2000). The 5′ UTR (untranslated region) includes the promoter; this region has been swapped out by recombination many times during mammalian evolution, so it is often non-orthologous between species and even for different subfamilies within a species (Boissinot and Sookdeo 2016). The ORF1 (open reading frame 1) segment encodes a nucleic acid binding protein that is associated with the L1 transcript as part of the retrotransposition complex. It has a hypervariable region (V) near the 5′ end that is either very rapidly evolving or also has been swapped out over the evolutionary history of the element. The ORF2 segment has four conserved domains: endonuclease (E), an octapeptide-containing sequence (Z), reverse transcriptase (RT), and a RNase-H-like zinc finger (C). The 3’ UTR segment contains a G-rich polypurine tract and terminates with a poly-A tail. The proteins encoded by ORF1 and ORF2, along with host proteins, are responsible for retrotransposition. Sequences generally are inserted into the genome starting at the 3′ end and most insertions are truncated, so there are relatively few full length L1s in the genome (Furano 2000).
Figure 1.
Structure of a typical mammalian L1. Full-length elements are ~7 kb in length and have four major segments: 5′ and 3′ untranslated regions (UTRs) and two open reading frames (ORFs). ORF1 has a 5′ hypervariable region (V) and ORF2 contains four conserved domains: endonuclease (E), an octapeptide-containing sequence (Z), reverse transcriptase (RT) and a RNase-H-like zinc finger (C). A G-rich polypurine tract (G) resides in the 3′ UTR and elements terminate with a poly-A tail. The region cloned for this study straddles the RT domain in ORF2 and was isolated by PCR with degenerate primers.
Whole-genome sequencing has greatly expanded what is known about the evolution of mammalian L1s. These studies provide a broad overview of L1 evolution. L1s have persisted in the mammalian genome since before the divergence of placental mammals from marsupials, but are not found in monotremes (Ivancevic et al. 2016). Given the presence of multiple active elements retrotransposing in the genome at any given time, one would expect that over the course of evolutionary history the active elements would have diverged such that they form a bush-like phylogeny within each host species (Clough et al. 1996). Although this is true of other vertebrates that have retrotransposons related to LINE-1—fish, reptiles, and amphibians (Platt et al. 2018)—mammalian L1s from a given species generally form a pectinate tree with a single trunk, indicating that the active elements found in the genome (at any point in their history) within the host lineage are very closely related. The mechanism behind this unique mode of evolution within a genome is not well understood, but it is thought to indicate an ongoing arms race where the genome evolves to suppress retrotransposition and the L1 elements evolve to escape this control (Platt et al. 2018). Occasionally, multiple well-diverged L1 lineages persist over evolutionary time. For example, the deer mouse Peromyscus has two active lineages (Casavant et al. 1996), but these lineages arose subsequent to the origin of Peromyscus (Casavant et al. 1998) and are not found in all species of the genus.
Previously, aPCR-based approach was developed to enrich for relatively young L1 pseudogenes if they are present in the genome (Cantrell et al. 2000). If young elements are not present, older L1 pseudogenes are amplified. Using this technique, a comprehensive screen for L1 activity across all families of Chiroptera was conducted. In all species examined with active L1s, they evolve as one or two persistent lineages. In addition to the extinction event previously documented for the family Pteropodidae (Cantrell et al. 2008), an L1 extinction event was identified in Mormoops blainvilli, however, in this case it did not affect the entire family Mormoopidae.
Methods
Specimens examined
Genomic DNA from a total of 57 species of bats was examined by a PCR-based method that enriches for a conserved region of recently active L1s (Cantrell et al. 2000). Specimens examined and sources of material are provided in Table 1.
Table 1.
Specimens examined in this study. Tissues with TK numbers were acquired from the Museum of Texas Tech University, gE numbers from the New Zealand Department of Conservation, and CT18 from University College Dublin, Belfield. L1 Activity indicates which ancestral L1 lineage is active within each species examined. An asterisk (*) indicates species that are included in the trees in Figure 3.
| Family | Genus, Species | Tissue ID | L1 Activity |
|---|---|---|---|
| Pteropodidae | *Cynopterus sphinx | TK21250 | none |
| Rhinolophidae | *Rhinolophus eloquens | TK33101 | Lineage 2 |
| Hipposideridae | *Hipposideros armiger | TK21147 | Lineage 2 |
| Megadermatidae | *Megaderma lyra | TK21292 | Lineage 1 |
| Craseonycteridae | *Craseonycteris thonglongyai | CT18 | Lineage 1 |
| Rhinopomatidae | *Rhinopoma hardwickei | TK40884 | Lineage 1 |
| Nycteridae | *Nycteris thebaica | TK33153 | Lineage 2 |
| Emballonuridae | *Rhynchonycteris naso | TK15108 | Lineage 2 |
| Emballonuridae | *Taphozous melanopogon | TK21446 | Lineages 1, 2 |
| Phyllostomidae | *Artibeus jamaicensis | TK27682 | Lineage 2 |
| Phyllostomidae | *Tonatia saurophila bakeri | TK104519 | Lineage 2 |
| Mormoopidae | *Mormoops blainvilli | TK32173 | none |
| Mormoopidae | *Pteronotus quadridens | TK9497 | Lineage 2 |
| Noctilionidae | *Noctilio albiventris | TK17633 | Lineage 2 |
| Furipteridae | *Furipterus horrens | TK17149 | Lineage 2 |
| Thyropteridae | *Thyroptera discifera | TK104577 | Lineage 2 |
| Mystacinidae | *Mystacina tuberculata | gE266 | Lineage 2 |
| Myzopodidae | *Myzopoda aurita | gE172 | Lineage 2 |
| Vespertilionidae | *Antrozous pallidus | TK44027 | Lineage 2 |
| Vespertilionidae | *Myotis velifer | TK44032 | Lineage 2 |
| Molossidae | *Tadarida brasiliensis | TK44001 | Lineage 2 |
| Natalidae | *Natalus stramineus | TK15661 | Lineage 2 |
| Pteropodidae | Dobsonia moluccensis | TK20261 | none |
| Pteropodidae | Hypsignathus monstrosus | TK21542 | none |
| Pteropodidae | Macroglossus sp. | TK 20305 | none |
| Pteropodidae | Megaerops niphanae | TK21085 | none |
| Pteropodidae | Megaloglossus woermanni | TK21565 | none |
| Pteropodidae | Melonycteris melanops | TK20071 | none |
| Pteropodidae | Nyctimene albiventer | TK20056 | none |
| Pteropodidae | Pteropus hypomelanus | TK20059 | none |
| Pteropodidae | Pteropus macrotis | TK20310 | none |
| Pteropodidae | Rousettus amplexicaudatus | TK20031 | none |
| Phyllostomidae | Ametrida centurio | TK17743 | Lineage 2 |
| Phyllostomidae | Anoura geoffroyi | TK19385 | Lineage 2 |
| Phyllostomidae | Ardops nichollsi | TK15576 | Lineage 2 |
| Phyllostomidae | Artibeus cinereus | TK19226 | Lineage 2 |
| Phyllostomidae | Artibeus lituratus | TK104427 | Lineage 2 |
| Phyllostomidae | Artibeus planirostris | TK15011 | Lineage 2 |
| Phyllostomidae | Artibeus schwartzi | TK82838 | Lineage 2 |
| Phyllostomidae | Carollia perspicillata | TK104347 | Lineage 2 |
| Phyllostomidae | Choeroniscus godmani | TK40021 | Lineage 2 |
| Phyllostomidae | Choeronycteris mexicana | TK27013 | Lineage 2 |
| Phyllostomidae | Desmodus rotundus | TK40368 | Lineage 2 |
| Phyllostomidae | Diphylla ecaudata | TK13508 | Lineage 2 |
| Phyllostomidae | Glossophaga soricina | TK9251 | Lineage 2 |
| Phyllostomidae | Glyphonycteris sylvestris | TK10454 | Lineage 2 |
| Phyllostomidae | Hylonicteris underwoodi | TK20540 | Lineage 2 |
| Phyllostomidae | Lionycteris spurrelli | TK22524 | Lineage 2 |
| Phyllostomidae | Lonchophylla thomasi | TK17580 | Lineage 2 |
| Phyllostomidae | Lonchorhina aurita | TK20560 | Lineage 2 |
| Phyllostomidae | Macrotus waterhousii | TK27889 | Lineage 2 |
| Phyllostomidae | Micronycteris minuta | TK15174 | Lineage 2 |
| Phyllostomidae | Micronycteris nicefori | TK25119 | Lineage 2 |
| Phyllostomidae | Plathyrrhinus helleri | TK14577 | Lineage 2 |
| Phyllostomidae | Rhinophylla pumilio | TK10130 | Lineage 2 |
| Phyllostomidae | Sturnira lucovici | TK34856 | Lineage 2 |
| Phyllostomidae | Trachops cirrhosus | TK19132 | Lineage 2 |
Degenerate PCR, L1 cloning, and colony screening
A 575 bp region of L1 (Fig. 1) ORF2 homologous to bases 4989–5563 of a full-length Mus L1 (GenBank accession number M13002) was amplified and cloned from each species as described previously (Cantrell et al. 2000). This technique uses degenerate primers to regions that are highly conserved based on a previous alignment of reverse transcriptases from viruses and transposable elements plus alignments of L1s from a broad range of mammalian species. The primers also contain 5′ clamps to increase specificity and introduce two restriction sites at each end of the amplified elements. Restriction digestion after amplification is followed by ligation into a modified lacZ reporter vector, pKSW, that was engineered such that the PCR product is cloned in-frame and in the sense orientation. Insertion of an L1 fragment from an element that has transposed so recently that it still contains an ORF results in production of an Ll/β-galactosidase fusion protein. Insertion of an L1 region that has suffered stop mutations in the normal reading frame blocks production of the fusion protein. Thus, blue colonies are enriched for recently inserted L1 sequences that maintain ORFs, whereas white colonies generally have indels and stop codons.
For initial characterization of each species, clones were sequenced from both blue and white colonies. If identical clones were found, only one was included in the final dataset. Potential recombinants were detected as described previously (Cantrell et al. 2008) and were removed from the dataset. If primarily truncated ORFs were found due to internal restriction sites, PCR products were cloned with alternate enzymes. For each species, a minimum of 20 sequences was included in the final data set, generally from the first 10 blue and first 10 white colonies isolated except where unavailable. All L1 sequences isolated from species analyzed for Figures 2 and 3 of this study were deposited in GenBank (accession numbers EF437602–EF437898 and MK991326–MK991766).
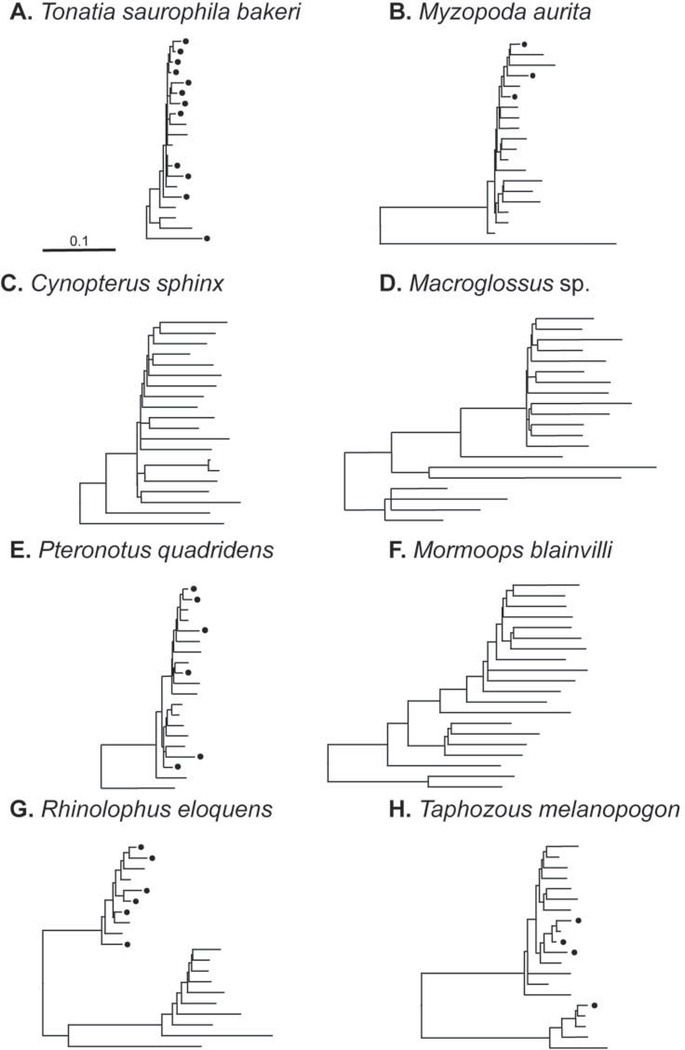
Figure 2.
Example L1 phylogenies of 20 elements from eight bat species. Taxa names have been removed; a black dot represents an L1 with an open reading frame across the region of analysis, indicating recent L1 activity. Terminal branch lengths reflect relative time since insertion. The trees demonstrate the variation in bat L1 evolutionary dynamics: single and multiple lineages as well as cessation of activity.
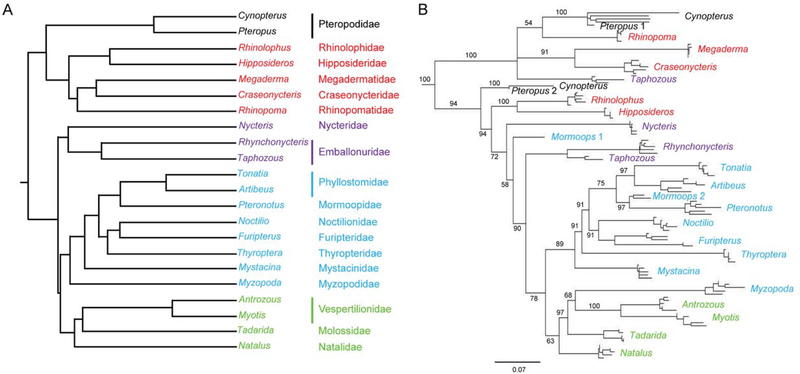
Figure 3.
Comparison of phylogénies of bat families and bat L1 lineages. Taxa from all 18 bat families are included. Colors indicate families and genera within superfamilial groups: Rhinolophoidea, red; Emballonuroidea, purple; Noctillonoidea, blue; and, Verpertillonoidea, green. A. Family tree of Chiroptera derived from Teeling et al. Figure 2 (2005), from a maximum-likelihood analysis of a 13.7 kb concatenated data set. B. Maximum likelihood free of L1s from the same genera as in tree A. Five L1s with open reading frames from each species plus reconstructed ancestral L1s from extinct lineages in Pteropus (Pteropus 1, Pteropus 2) and Mormoops (Mormoops 1 and Mormoops 2) are included in the analysis. Numbers at the nodes indicate majority rule bootstrap support values.
Species were designated as having recently active L1s if at least two sequences were found with intact reading frames and in the correct reading frame across the entire length of the amplified region. In cases where this criterion was not met, additional clones were sequenced in an attempt to detect elements containing ORFs.
Phylogenetic analysis
For each species, 20 L1 sequences (usually 10 from blue colonies and 10 from white colonies) were aligned by the ClustalW algorithm (Thompson et al. 1994). Two young L1s from the most closely related sister taxon were included as outgroup. Alignments were adjusted manually. Phylogenetic analysis was carried out under maximum-likelihood criteria in PAUP* version 4.0bl0 (Swofford 2003). To select the most appropriate model of evolution, the alignments were subjected to an iterative search strategy that estimated the parameters of16 alternative maximum-likelihood models from an initial neighbor joining tree. The relative fit of the models was assessed using the χ2-approximation to the null distribution as a likelihood-ratio test (Yang 1994). Heuristic searches with 100 replicate random addition sequences and tree bisection-reconnection branch swapping were then conducted under likelihood criteria with the fully defined, best-fit model, which was either HKY+G or GTR+G for all species. The trees were subsequently rooted with the outgroup and the taxa names and outgroup branches were removed for ease of viewing. Examples of species-specific L1 trees are shown in Figure 2 (see Results). Tree size was adjusted so that the height and scale bars were uniform. Black dots were added to indicate L1s with ORFs. To be considered an element with an ORF, the sequence was required to be full length, with intact reading frames maintaining the correct reading frame across the entire length of the amplified region. The same methods were used to build an L1 phylogeny representing all families of Chiroptera except that fewer sequences were used for each species, as described under Results.
Results
A 575 bp region of L1 ORF2 (Fig. 1) was amplified, cloned, sequenced, and analyzed from 57 species of Chiroptera (Table 1). All families of bats were sampled and, when possible, the same genera used by Teeling (Teeling et al. 2005) to construct a phylogeny of all chiropteran families were included. Phylogenetic analysis was carried out on elements from each species separately and as well as collectively on species representing all families of Chiroptera. L1s for each of the 57 individual species were analyzed to determine if there was evidence of recent L1 activity and to assess the number of active L1 lineages. For the combined analysis of L1 from the order Chiroptera, one or two species were included for each family. Pteropodidae and Phyllostomidae were sampled more extensively (Table 1).
The targeted region was cloned in frame with lacZ such that a fusion protein was produced in clones where the reading frame of the 575 bp region was maintained, giving rise to blue colonies when clones were plated on β-galactosidase. This technique is extremely effective at enriching for young elements even in the presence of a vast excess of old L1 pseudogenes in the genome. To assess the sensitivity of the technique, DNA from Rousettus amplexicaudatus, a species of Pteropodidae with long extinct L1s, was seeded with quantities of a cloned mouse L1 element equivalent to 1, 3, 10, 100, or 1,000 young L1 copies per haploid genome. Using this PCR-based enrichment technique, no mouse L1 clones were found among 16 sequenced from the sample spiked with mouse L1 equivalent to 1 copy per haploid genome, but samples spiked with 3, 10, 100, or 1,000 copies per haploid genome yielded 25, 38, 94, and 100% mouse L1 clones, respectively (Cantrell et al. 2008). This reconstruction experiment suggested two points of interest: 1) young L1 copies were enriched even at far lower numbers than would be expected in a typical genome; and 2) the resulting phylogenies ofLl elements identified by this technique were more reflective of recent retrotransposition than of the complete history of L1 in that host species. The PCR relies on primers to conserved regions of L1 ORF2 and, thus, PCR amplified relatively young elements more readily than old degenerate elements. The colorimetric assay provides further enrichment for young elements by identifying elements with intact reading frames in the amplified region. The recent activity of L1s can be deduced from the structure of their phylogenetic trees. For example, if L1s have had recent bursts of retrotransposition in a species, this is reflected by the short terminal branch lengths and abundance of open reading frames (ORFs) on the tree. Alternatively, if L1 activity is scant or absent, the past activity is revealed, and branch lengths tend to be longer and ORFs few or absent.
L1 activity within species
As expected, species L1 trees tended to have a pectinate appearance with one or sometimes two lineages evident. Alternative L1 topologies in bats are shown in Figure 2. Single lineages are evident (Fig. 2A, B, and E), but a range of L1 activity can be implied in these species, from very active in Tonatia saurophila bakeri to low levels of recent activity in Myzopoda aurita. Extinction of L1 in megabats was reported previously (Cantrell et al. 2008) and is evident in these L1 phylogenies by the long terminal branch lengths and lack of ORFs in the two Pteropodidae (Fig. 2C and D). An independent L1 extinction event was evident in Mormoops blainvilli (Fig. 2F). Multiple lineages are evident in both L1 extinction events. Multiple lineages also are evident in species with active L1s. For example, Rhinolophus eloquens (Fig. 2G) had one active lineage and one extinct lineage, while T. melanopogan (Fig. 2H) had two very divergent active lineages. No L1 extinction events were found among the 27 species of Phyllostomidae examined, although some families possessed low levels of activity. As previously shown, L1 is extinct in all species of Pteropodidae (Cantrell et al. 2008).
L1 activity in Chiroptera
To compare the evolution of L1s in Chiroptera to the phylogeny of their hosts, young L1s from genera examined by Teeling (Teeling et al. 2005) were analyzed. Five L1s with intact open reading frames from each species were included in the analysis; where multiple lineages were present, representatives from each L1 lineage were included. Five elements that lack intact reading frames from Cynopterus sphinx were included to represent the Pteropodidae. The reconstructed ancestors from both extinct Pteropodidae lineages (Pteropus 1, Pteropus 2) and from both extinct Mormoops lineages (Mormoops 1 and Mormoops 2) also were included.
Although there was an overall similarity between the L1 phylogeny and the bat phylogeny proposed by Teeling at al. (2005), there were many differences (Fig. 3). None of the superfamilies were conserved on the L1 phylogeny. Rhinolophidae and Hipposideridae clustered with the Yangochiroptera rather than the Yinpterochiroptera. Among the Yangochiroptera, L1s from Myzopodidae were sister to those from Vespertilionidae. The relationships among the Noctilionidae, Furipteridae, and Thyropteridae differed, and Nycteridae was not sister to Emballonuridae. Taphozous also was exceptional because of its two extremely divergent L1 active lineages (see below for further discussion of these lineages). One lineage clustered where expected with L1s from the other emballonurid, Rhynchonycteris. The other active L1 lineage in Taphozous clustered with L1s from the Yinpterochiroptera, and that lineage was the more active one in Taphozous. Although there were no active lineages in M. blainvilli, one of the two extinct lineages clustered with L1s from Pteronotus quadridens, consistent with its expected placement among the Mormoopidae.
There were two active L1 lineages present before the divergence of the families of bats. However, there must have been multiple extinctions within both ancestral L1 lineages over the course of chiropteran evolution, irrespective of which recently proposed chiropteran phylogeny is used for comparison. For example, one proposed phylogeny that supports the Yinptero- and Yangochiroptera groupings (Teeling et al. 2005) would require seven independent extinctions of L1 lineage 1 or lineage 2 to account for the active lineages observed in this study, whereas an alternative phylogeny (Van den Bussche and Hoofer 2004) would require eight L1 independent extinction events. The evolution of L1 in Chiroptera also was compared to phylogenies that support the monophyly of all microbats; this relationship required either seven (Jones et al. 2002) or nine (Agnarsson et al. 2011) independent extinction events. An example of mapping extinctions of L1 lineages onto the Teeling bat phylogeny is shown in Figure 4. Minimizing the number of lineage extinction events would require splitting the superfamily Rhinolophoidea so that 1) Megadermatidae, Craseonycteridae, and Rhinopomatidae were members of a clade with Pteropidae, and 2) Rhinolophidae and Hipposideridae were members of a clade with the Emballonuroidea, Noctillonoidea, and Verpertillonoidea (see Fig. 3B). This arrangement does not appear to be consistent with any proposed chiropteran phylogeny.
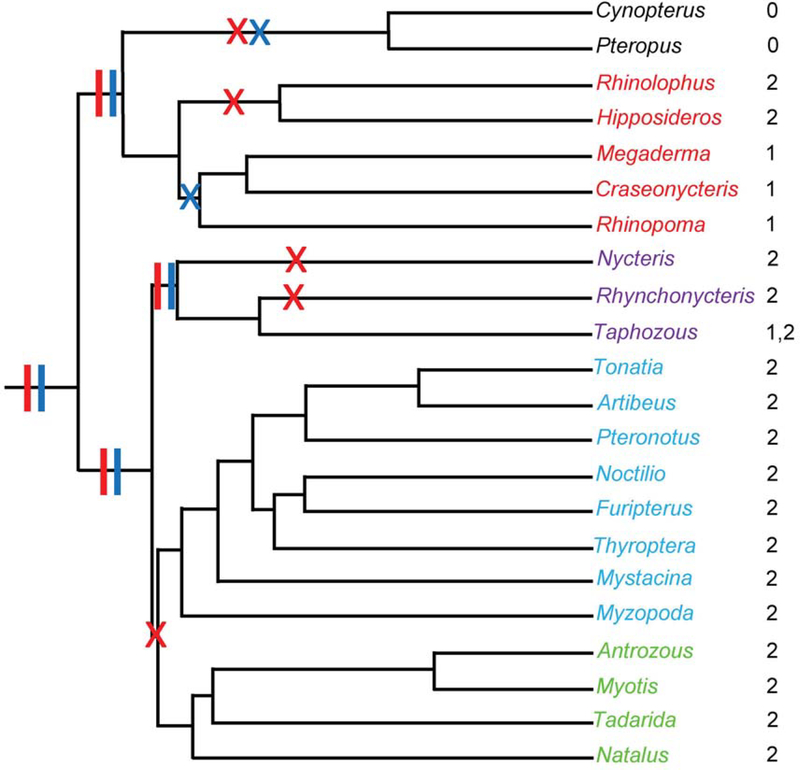
Figure 4.
An example of activity and extinction of two ancient L1 lineages mapped onto the phylogeny of Chiroptera. Colors of taxa names indicate families and genera within superfamilial groups: Rhinolophoidea, red; Emballonuroidea, purple; Noctillonoidea, blue; and, Verpertillonoidea, green. Vertical bars represent activity of L1 lineages in common ancestors and Xs indicate extinction events: lineage 1, red; lineage 2, blue. Numbers to the right of the taxa are the lineage(s) active in the corresponding genus.
Discussion
Persistence and extinction of L1s
Persistence of L1 requires ongoing retrotransposition so that new active copies are inserted before debilitating mutations inactivate the minute fraction of L1s capable of replication; L1 lineages that do not replicate eventually will become extinct. Finding evidence of recent activity has not always been straightforward. Ancient L1s persist in the genome as molecular fossils that obscure the small subset of elements that are products of recent retrotransposition (Deininger et al. 1992; Deininger and Batzer 1993; Furano 2000). The method employed for this study is very sensitive for finding recently transposed L1s (Cantrell et al. 2000; Cantrell et al. 2008), but it does not uncover the complete history of L1s within a species because old elements generally are amplified only in the absence of younger elements. Although this can be partially mitigated using the blue-white screening technique to enrich for clones both with and without intact reading frames over the region of interest, the phylogenies produced by this method should be considered a history of the most recent L1 activity rather than a complete history.
Occasionally, the active L1 lineages go extinct within a mammalian clade so that all subsequently derived species lack active L1s (Casavant et al. 2000; Cantrell et al. 2008; Sookdeo et al. 2018). Such extinctions may be underestimated because recognizing them requires that L1 copies remaining in the genome have acquired enough mutation to be clearly identifiable as inactive. Deeper extinctions are readily identifiable both because the fossil copies have accumulated more mutations and because cladogenesis after an L1 extinction event gives rise to more taxa that also lack active L1s. Why, then, have so few mammalian clades been discovered that lack active L1s? Certainly, sufficient mammalian clades to identify all L1 extinctions have not yet been examined, but among those mammals examined in this study, most were found to have active L1s. It is possible that this is just a historical accident—that L1 extinctions have occurred throughout mammalian evolution, but by chance few of those lineages gave rise to major mammalian radiations. This would make those extinction events harder to find because it would be necessary to locate one of a few species instead of one of many. For example, one could find the L1 extinction in Pteropodidae by looking at any one of the ~65 species in the family, but Mormoopidae contains only eight species and it is known that some of those still have active L1s. This study was very “lucky” to find the L1 extinction event in M. blainvilli.
Although only two complete extinctions of L1 activity were detected in Chiroptera, one in all Pteropodidae and one in M. blainvilli, a surprising number of L1 lineage extinctions in the group were identified. Additional sampling will be required to completely document the number of L1 lineage extinctions, but it seems likely that there have been at least seven independent deep extinctions (Fig. 4), as well as a number of more recent L1 lineage extinctions. For example, two lineage extinctions occurred in M. blainvilli to give rise to complete loss of L1 activity. Lineage extinction without loss of L1 activity likely occurred in several species where there was evidence of one active linage and one inactive one, such as Hipposideros armiger and R. eloquens. For reasons mentioned above, the methods used in this study likely underestimate the number of these extinctions. However, these lineage extinctions highlight what could be a major problem with using L1 phylogeny to reconstruct host phylogeny.
L1 activity and genome size in bats
Among mammals, the genomes of Chiroptera are particularly interesting because average genome size is the lowest among mammalian orders—2.35 picograms in Chiroptera versus 3.5 picograms among all mammals (Smith et al. 2013). Although their small genome size seems exceptional, this has not hindered their evolutionary diversification. The order Chiroptera includes 20% of all extant species of placental mammals, second only to rodents (Wilson and Reeder 2005). Small genome size in both bats and birds has been proposed to be adaptive for flight (Hughes and Hughes 1995). Previous work has concluded that the reduced size of the chiropteran genome is due to extensive DNA loss due to deletions, rather than reduced gains due to retrotransposition (Kapusta et al. 2017). However, Pteropodidae have even smaller genomes than other bats—2.2 picograms—so lack of retrotransposition in these bats likely plays some role in restraining genome size (Smith et al. 2013).
Do L1s provide a function for the host?
Transposable elements are viewed widely as selfish parasites, but the long-term and widespread persistence of L1s has fueled speculation that they may provide a function for their mammalian hosts. Specific proposed functions include a role in chromosomal repair (Hutchison III et al. 1989; Morrish et al. 2002), X chromosome inactivation (Lyon 1998), modulating gene expression (Han et al. 2004; Elbarbary et al. 2016), and neuronal differentiation (Singer et al. 2010). However, if L1 elements play an essential function in their mammalian host, one must account for how that function would be maintained after the extinction of L1s, and that has not yet been documented for any of these proposed functions.
Whether L1s provide an essential function for the host is not known, but it may be that losing L1s could be deleterious in the long run. L1s account not only for their own retrotransposition but also for the movement of SINEs and processed pseudogenes, so losing the major source of retrotransposition in the genome may be akin to drastically lowering the point mutation rate. In the short run, there may be no deleterious effect of losing L1 activity, and, in fact, the loss could be beneficial. But in the long run, the ability of species to evolve could be constrained by the reduction in the amount and type of genetic variation available. The central role of L1 in generating specific types of variation could be replaced by another retrotransposon. For example, sigmodontine rodents that lack active L1s have mysTR, a very active family of endogenous retroviruses (Erickson et al. 2011), but no such driver of retrotransposition has been found in the megabats.
L1s and their parasitic SINEs as phylogenetic markers
“The only homoplasy-free phylogenetic marker is the new one” (Robert J. Baker)—meaning that each newly discovered phylogenetic marker is assumed to be homoplasy free, until sufficient data are generated that show otherwise. Given their vast representation in the genome, L1s and SINEs would seem to be ideal markers for reconstructing the history of their hosts. There are at least two ways by which retrotransposons might be used as phylogenetic markers for their mammalian hosts. First, the history of the L1s or SINEs can be reconstructed. At speciation events the active lineage will diverge and accumulate changes independently in the derived species (Sookdeo et al. 2018). Changes that accumulate in the active L1s can be used as markers to reconstruct the history of their hosts (Verneau et al. 1997; Casavant et al. 1998; Verneau et al. 1998). Second, individual insertions of L1s, SINEs, or other retrotransposons can be used as presence-or-absence characters that can be detected by PCR with flanking single copy primers (Shedlock and Okada 2000). Because there are so many L1 and SINE inserts in the genome, there is an almost unlimited supply of potential markers across a wide range of ages.
Neither of these approaches is completely homoplasy free. First, both may be subject to lineage sorting, as are all phylogenetic markers. As seen here, this may be more serious when reconstructing L1 (or SINE) history because multiple active lineages can coexist, and active lineages can go extinct in patterns that do not recapitulate species histories. It might be assumed that this would not be a problem when using individual insertions as presence-or-absence characters, but same-site insertions do occur. For example, a study of insertions sites of mys retrotransposons in the Peromyscus genome revealed both lineage sorting (Lee et al. 1996) and same-site insertions (Cantrell et al. 2001). One ancient mys insertion had accumulated 12 independent insertions of other retroelements among 13 alleles examined. At two sites, the insertions used identical initial nick sites to insert, but were clearly different events; in one case, two SINEs from different families inserted into the same site, and in another case, the insertions were resolved differently at the 5′ insertion site (Cantrell et al. 2001). Although allele size differences would have been detectable between some alleles in a presence-or-absence PCR assay, some alleles containing different insertions would have appeared to be the same size. It is unknown how common such insertional ‘hot spots’ are in mammalian genomes, but these findings caution against using a small number of insertion sites for phylogenetic reconstruction of the host. However, studies of millions of Alu SINE insertions in primates found that 0.01% or less exhibited homoplasy (Doronina et al. 2018). Phylogenies based on a large number of retrotransposon insertions sites distributed across the genome should be more phylogenetically robust than either studies based on single nucleotide polymorphisms (SNPs) or comparison of retrotransposons phylogenies to host phylogenies.
It was not the intent of this study to use L1s to reconstruct chiropteran phylogeny. Instead, the chiropteran phylogeny was used to better understand the biology of L1 elements. The findings of the study suggest that there may have been extensive lineage sorting of L1 elements in bats, along with a number of cases of multiple, highly diverged active lineages. It appears that the order began its history with two active lineages that were already ~27% divergent at the time of their extinction in the Pteropodidae. These two lineages gave rise to the active lineages in all Chiroptera, but through a lineage sorting process that did not result in L1 phylogeny recapitulating chiropteran phylogeny. Both lineages survived in at least one species, Taphozous melanopogon, where the two clades now differ by ~33%. The two complete extinctions of L1 activity in the order, along with the numerous extinctions of L1 lineages over time, may reflect the intensity of the ongoing arms race between L1 for its survival and strong selection on genome size in Chiroptera.
Acknowledgments
We thank the following sources for the bat tissues acquired for this study: Field Museum of Natural History, New Zealand Department of Conservation, University College Dublin, Belfield, and especially the Museum of Texas Tech University and H. J. Garner, Curator of Collections at the Natural Science Research Laboratory. We appreciate the Wichman Lab researchers who contributed to this study by acquiring bat L1 sequence data: M. A. Cantrell, K. Bush, L. Guerra, C. Simpson Beery, L. Bronson, R. Hailey Emerson, I. K. Erickson, B. Lund, J. Millstein and P. D. Vise. Robert J. Baker was involved in the planning of this study, providing research material and insights about chiropteran phylogeny, and in discussing results. Research reported in this publication was supported by grants from the National Institutes of Health: R01GM38727, P20RR016448, and P20GM104420. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Holly A. Wichman, Center for Modeling Complex Interactions & Department of Biological Sciences, University of Idaho, MS 1122, Moscow, ID 83844 -3051 USA
LuAnn Scott, Department of Biological Sciences, University of Idaho, Moscow, ID 83844-3051 USA.
Eric K. Howell, ÅF AB, Frösundaleden 2A, 169 99, Solna, Stockholm, Sweden
Armando R. Martinez, Environmental Compliance Division, City of Nampa, 340 W. Railroad St. Nampa, ID 83687 USA
Lei Yang, Pacific Northwest Research Institute, 720 Broadway, Seattle, WA 98122 USA.
Robert J. Baker, Department of Biological Sciences and the Museum, Texas Tech University, Lubbock, TX 79409-3131 USA
Literature Cited
- Agnarsson I, Zambrana-Torrelio CM, Flores-Saldana NP, and May-Collado LJ 2011. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLoS Currents 3:RRN1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, and Sookdeo A 2016. The evolution of LINE-1 in vertebrates. Genome Biology and Evolution 8:3485–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell MA, et al. 2001. An ancient retrovirus-like element contains hot spots for SINE insertion. Genetics 158:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell MA, Grahn RA, Scott L, and Wichman HA 2000. Isolation of markers from recently transposed LINE-1 retrotransposons. Biotechniques 29:1310–1316. [DOI] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, and Wichman HA 2008. Loss of LINE-1 activity in the megabats. Genetics 178:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant NC, Lee RN, Sherman AN, and Wichman HA 1998. Molecular evolution of two lineages of L1 (LINE-1) retrotransposons in the california mouse, Peromyscus californicus. Genetics 150:345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant NC, Scott L, Cantrell MA, Wiggins LE, Baker RJ, and Wichman HA 2000. The end of the LINE?: Lack of recent L1 activity in a group of South American rodents. Genetics 154:1809–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant NC, Sherman AN, and Wichman HA 1996. Two persistent LINE-1 lineages in Peromyscus have unequal rates of evolution. Genetics 142:1289–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough JE, Foster JA, Barnett M, and Wichman HA 1996. Computer simulation of transposable element evolution: Random template and strict master models. Journal of Molecular Evolution 42:52–58. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, and Pollock DD 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genetics 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, and Batzer MA 1993. Evolution of retroposons. Evolutionary Biology 27:157–196. [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA 3rd, and Edgell MH 1992. Master genes in mammalian repetitive DNA amplification. Trends in Genetics 8:307–311. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, and Heidmann T 2003. LINE-mediated retrotransposition of marked Alu sequences. Nature Genetics 35:41–48. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, and Heidmann T 2005. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. Journal of Molecular Biology 349:241–247. [DOI] [PubMed] [Google Scholar]
- Doronina L, Reising O, Clawson H, Ray DA, and Schmitz J 2018. True homoplasy of retrotransposon insertions in primates. Systematic Biology 68:482–493. [DOI] [PubMed] [Google Scholar]
- Elbarbary RA, Lucas BA, and Maquat LE 2016. Retrotransposons as regulators of gene expression. Science 351:aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson IK, Cantrell MA, Scott L, and Wichman HA 2011. Retrofitting the genome: L1 extinction follows endogenous retroviral expansion in a group of muroid rodents. Journal of Virology 85:12315–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano AV 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Progress in Nucleic Acid Research and Molecular Biology 64:255–294. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, and Kazazian HH Jr. 2000. Transduction of 3’-flanking sequences is common in L1 retrotransposition. Human Molecular Genetics 9:653–657. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, and Boeke JD 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429:268–274. [DOI] [PubMed] [Google Scholar]
- Hughes AL, and Hughes MK 1995. Small genomes for better flyers. Nature 377:391. [DOI] [PubMed] [Google Scholar]
- Hutchison CA III, Hardies SC, Loeb DD, Shehee WR, and Edgell MH 1989. LINEs and related retroposons: long interspersed repeated sequences in the eucaryotic genome. Pp. 593–617 in Mobile DNA (Berg DE and Howe MM, eds.). American Society for Microbiology, Washington, DC. [Google Scholar]
- Ivancevic AM, Kortschak RD, Bertozzi T, and Adelson DL 2016. LINEs between species: Evolutionary dynamics of LINE-1 retrotransposons across the eukaryotic tree of life. Genome Biology and Evolution 8:3301–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Purvis A, MacLarnon A, Bininda-Edmonds ORP, and Simmons NB 2002. A phylogenetic supertree of bats (Mammalia: Chiroptera). Biological Reviews 77:223–259. [DOI] [PubMed] [Google Scholar]
- Kapusta A, Suh A, and Feschotte C 2017. Dynamics of genome size evolution in birds and mammals. Proceedings of the National Academy of Science USA 114:E1460–E1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr., Wong C, Youssoufian H, Scott AF, Phillips DG, and Antonarakis SE 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332:164–166. [DOI] [PubMed] [Google Scholar]
- Lee RN, Jaskula JC, van den Bussche RA, Baker RJ, and Wichman HA 1996. Retrotransposon Mys was active during evolution ofthe Peromyscus leucopus-maniculatus complex. Journal of Molecular Evolution 42:44–51. [DOI] [PubMed] [Google Scholar]
- Lyon MF 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenetics and Cell Genetics 80:133–137. [DOI] [PubMed] [Google Scholar]
- Morrish TA, et al. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genetics 31:159–165. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, and Kazazian HH Jr. 2001. Biology of mammalian L1 retrotransposons. Annual Review of Genetics 35:501–538. [DOI] [PubMed] [Google Scholar]
- Platt RN II, Vandewege MW, and Ray DA 2018. Mammalian transposable elements and their impacts on genome evolution. Chromosome Research 26:25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock AM, and Okada N 2000. SINE insertions: powerful tools for molecular systematics. Bioessays 22:148–160. [DOI] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, and Gage FH 2010. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends in Neuroscience 33:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Bickham JW, and Gregory TR 2013. Patterns of genome size diversity in bats (order Chiroptera). Genome 56:457–472. [DOI] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, and Boissinot S 2018. Contrasted patterns of evolution of the LINE-1 retrotransposon in perissodactyls: the history of a LINE-1 extinction. Mobile DNA 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O’Brien SJ, and Murphy WJ 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, and Gibson TJ 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bussche RA, and Hoofer SR 2004. Phylogenetic relationships among recent Chiropteran families and the importance of choosing appropriate out-group taxa. Jounal of Mammalogy 85:321–330. [Google Scholar]
- Verneau O, Catzeflis F, and Furano AV 1997. Determination of the evolutionary relationships in (Rodentia : Muridae) using L1 (LINE-1) amplification events. Journal of Molecular Evolution 45:424–436. [DOI] [PubMed] [Google Scholar]
- Verneau O, Catzeflis F, andFurano AV 1998. Determining and dating recent rodent speciation events by using L1 (LINE-1) retrotransposons. Proceedings ofthe National Academy of Science USA 95:11284–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DE, and Reeder DM 2005. Mammal Species of the World. Johns Hopkins University Press, Baltimore. [Google Scholar]
- Yang Z 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. Journal of Molecular Evolution 39:306–314. [DOI] [PubMed] [Google Scholar]