Abstract
Background
Circulating tumour DNA from colorectal cancer (CRC) is a biomarker for early detection of the disease and therefore potentially useful for screening. One such biomarker is the methylated SEPT9 (mSEPT9) gene, which occurs during CRC tumourigenesis. This systematic review and meta-analysis aims to establish the sensitivity, specificity and accuracy of mSEPT9 tests for the early diagnosis of CRC.
Methods
A systematic search of the relevant literature was conducted using Medline and Embase databases. Data were extracted from the eligible studies and analysed to estimate pooled sensitivity, specificity and diagnostic test accuracy.
Results
Based on 19 studies, the pooled estimates (and 95% CIs) for mSEPT9 to detect CRC were: sensitivity 69% (62–75); specificity 92% (89–95); positive likelihood ratio 9.1 (6.1–13.8); negative likelihood ratio 0.34 (0.27–0.42); diagnostic OR 27 (15–48) and area under the curve 0.89 (0.86–0.91). The test has a positive predictive value of 2.6% and negative predictive value of 99.9% in an average risk population (0.3% CRC prevalence), and 9.5% (positive predictive value) and 99.6% (negative predictive value) in a high-risk population (1.2% CRC prevalence).
Conclusion
The mSEPT9 test has high specificity and moderate sensitivity for CRC and is therefore a potential alternative screening method for those declining faecal immunochemical test for occult blood (FIT) or other screening modalities. However, it is limited by its poor diagnostic performance for precancerous lesions (advanced adenomas and polyps) and its relatively high costs, and little is known about its acceptability to those declining to use the FIT.
Keywords: colorectal cancer, screening, cancer epidemiology
Summary box.
-
What is already known about this subject?
The methylation of the SEPT9 (mSEPT9) test has shown promise as a screening test for early detection of colorectal cancers.
-
What are the new findings?
This updated meta-analysis estimates the sensitivity to be 69% and specificity of 92% and adds to current evidence by including new studies, evaluating the covariate effects and how these interact with pooled estimates of diagnostic performance.
While diagnostic performance seems promising for colorectal cancers, the mSEPT9 test has poorer performance for detecting precancerous lesions (advanced adenomas and polyps) and is more expensive.
-
How might it impact on clinical practice in the forseeable future?
The clinically relevant findings of this study posits the potential use of an alternative screening method for those that decline the faecal immunochemical test (FIT) to provide cover to individuals who would otherwise miss out on the benefits offered by colorectal cancer screening.
Further studies are needed to test whether mSEPT9 is acceptable for those that decline the FIT and whether this strategy would be cost-effective.
Introduction
Colorectal cancer (CRC) survival is highly dependent on stage and therefore methods for early detection are critical to reduce mortality. In the USA, only 40% of CRC cases are diagnosed at an early stage; the 5-year relative survival for stage 1 is 89.9%, stage 2 is 71.1%, stage 3–4 is 14.2%.1
Liquid biopsy screening tests are blood-based tests for early detection of cancer. They examine peripheral blood for circulating tumour DNA (ctDNA), which can be present in the earliest stages of tumourigenesis.2 ctDNA is released into the bloodstream from malignant cells during apoptosis. These fragments of DNA are representative of cancer cells of the tumour. Commonly, the DNA of tumour cells and therefore the ctDNA is methylated at the 5’-Cytosine-phosphate-Guanine-3’ sites.2 Therefore, detection of methylated ctDNA is used as a biomarker for the detection of asymptomatic cancer including CRC.
For the early detection of CRC, methylation of the SEPT9 (mSEPT9) gene located on chromosome 17 at q25.33 has shown promise as a ctDNA biomarker as CRC tumours have an increased likelihood of exhibiting methylation of the promoter region of the SEPT9 gene.3 For this test, a purified DNA sample obtained from peripheral blood is evaluated using duplex real-time PCR to identify methylation in the v2 region of the SEPT9 gene,3 which is suggestive of CRC.
A previous meta-analysis of diagnostic performance of mSEPT94 reported estimates of the sensitivity of mSEPT9 for CRC ranging from 37% up to 88%. However, most of the studies within this meta-analysis have not assessed the effect of covariates on sensitivity and specificity (such as the study sample size, region of recruitment of participants, total number of CRC cases and the distribution of stage). Additionally, four studies included in this meta-analysis were assessed as having a high probability of patient selection bias and could not include research reported since 2017. Two other meta-analyses5 6 did not conduct a comprehensive analysis of diagnostic accuracy of mSEPT9 tests: the first study5 considered covariates to assess sources of heterogeneity but provided limited evidence of the effect of those covariates on the diagnostic efficacy of mSEPT9. The second study6 only provided estimates on sensitivity and specificity. Finally, few studies have estimated the accuracy of the newer and commonly used mSEPT9 test, Epi-ProColon V.2.0 to the commonly used screening method for CRC, the faecal immunochemical tests (FIT), thereby limiting understanding of its clinical applicability. Only one previous meta-analysis4 assessed the impact of the threshold level for the number of PCR results that constitute a positive test, which is a major issue affecting the accuracy of mSEPT9 test for CRC.7
The aim of this systematic review was to conduct an updated meta-analysis stratified by the region of recruitment, sample size and stage, and to evaluate the accuracy of a range of positivity thresholds for the mSEPT9 test.
Methods
This systematic review and meta-analysis was conducted as per Guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis.8
Search strategy
Medline and Embase databases were searched for relevant literature. The search was divided into three phases. All articles included in these databases from January 1946 to May 2018 were considered. The search terms for the first phase were: (‘colon’ or ‘colorectal’) and (‘cancer’ or ‘carcinoma’) and (‘serum’ or ‘plasma’) and ‘sensitivity’ and ‘specificity’. The search terms for the second phase were: (‘SEPT9’ or ‘SEPT9’ or ‘Septin-9’ or ‘mSEPT9’) and (‘colon’ or ‘colorectal’) and (‘methylated’ or ‘methylation’). The search terms for the third phase were: ‘Epi procolon’ and ‘SEPT9’ and ‘diagnosis’. The articles from all three phases were combined and duplicate articles removed. A detailed search strategy is provided in online supplementary file 1.
bmjgast-2019-000355supp001.pdf (179.1KB, pdf)
Eligibility criteria
The inclusion criteria for the papers were: full-text available, English language, human studies, case-control study design, CRC diagnosis either prior or following mSEPT9 test, report of sufficient data to enable construction of 2×2 tables of mSEPT9 result and CRC status, and the use of a mSEPT9 single gene assay for both cases and controls. Exclusion criteria were: articles that were meta-analysis, conference and meeting abstracts, studies that used a multigene ctDNA panel; and studies with <20 CRC case participants.
Study selection
Endnote X9 was used to manage citations. Each article was assigned to one or more of the following reference categories: ‘articles with SEPT9 information’, ‘potential candidates for meta-analysis’, ‘irrelevant to this study’ and ‘selected studies’ by the following method. Articles that matched the inclusion criteria, identified by the three search phases were assigned to ‘article with SEPT9 information’. The abstracts of these papers were scanned for validity. Those meeting all inclusion criteria were assigned to ‘potential candidates for meta-analysis’. The full-text of these articles were evaluated, and the applicable studies for the meta-analysis were assigned to ‘selected studies’. Articles such as meeting abstracts, conference notes, meta-analysis and those studies which matched one exclusion criteria were assigned to ‘irrelevant for this study’.
Quality assessment of individual studies
The two authors (RH and MJ) independently carried out quality assessment of each study to assess risk of bias and applicability concerns using the Quality Assessment of Diagnostic Accuracy Studies V.2.0 tool.9 Risk of bias and applicability concerns of the study methodology was assessed under four domains: ‘patient selection’, ‘index test (mSEPT9)’, ‘reference standard (histopathology or colonoscopy to define CRC)’. The ‘flow and timing’ domain was omitted when assessing ‘applicability concerns’ as instructed by the tool. Each domain comprised two or three signalling questions, for which each reviewer answered ‘yes’, ‘no’ or ‘unclear’. Based on this, each study was graded ‘high’, ‘low’ or ‘unclear’ in each of the four domains, and the assessment results were then graphed (see online supplementary table 1, online supplementary figure 1). Inter-rater reliability/agreeability was measured using Cohen’s kappa statistic.
bmjgast-2019-000355supp002.pdf (678.2KB, pdf)
Data extraction
From each of the articles, the following results were abstracted: the number of true positives (tp)=participants who had CRC at the time of testing and were positive for the mSEPT9 test; true negatives (tn)=participants who did not have CRC at the time of testing and were negative for the mSEPT9 test; false positives (fp)=participants who did not have CRC and were positive for the mSEPT9 test and false negatives (fn)=participants who had CRC and were negative for the mSEPT9 test. All data were extracted by the author of this systematic review and was doubled checked to ensure accuracy.
Statistical analysis
Calculations of the following measures of accuracy of mSEPT9 were conducted: sensitivity [tp/(tp+fn)], specificity [tn/(tn+fp)], positive likelihood ratio [sensitivity/(1−specificity)], negative likelihood ratio [(1−sensitivity)/specificity] and diagnostic ORs [(tp/fn)/(fp/tn)] as well as their respective 95% CIs. As a secondary measure, positive predictive value (PPV) (tp/tp+fp) and negative predictive value (NPV) (tn/tn+fn) was also calculated assuming a CRC prevalence of 0.3% for an average risk population (consistent with having no first-degree relatives with CRC) and a CRC prevalence of 1.2% for a high-risk population (consistent with having two first-degree relatives with CRC). The range of threshold levels signifying a positive test were considered using either “one positive count out of three PCR results (1/3 algorithm)’ or ‘two positive counts out of three PCRs (2/3 algorithm)’,7 for the stratified subgroup analysis and meta-regression.
To investigate the relationship between performance indicators and the covariates that affect them, we used mixed effect bivariate and hierarchical statistical models.10 We estimated accuracy of diagnostic tests, how this accuracy varies with alterations in clinical and methodological practices and the comparison of accuracy of two or more tests.10
Forest plots were synthesised for the pooled sensitivity and specificity estimates. A stratified analysis (and subgroup meta-regression) was conducted based on country of recruitment, sample size, recruiting more stage 1 over stage 4 CRC participants and threshold algorithm. Heterogeneity from the pooled estimates of sensitivity and specificity was evaluated via the I2 statistic. To determine whether varying threshold levels to qualify as a positive test was a potential source for heterogeneity, an overall Spearman’s coefficient was estimated between sensitivity and specificity and respectively for studies sorted by region of recruitment and CRC case sample sizes (online supplementary table 2). A positive correlation between sensitivity and specificity is indicative of heterogeneity due to varying thresholds for positivity chosen for each study.
To assess the possibility of publication bias, Deek’s asymmetry test was conducted,11 by plotting the log of diagnostic ORs against 1/effective sample size1/2.11
A hierarchical summary operator receiver curve (HSROC) model was used to evaluate the effects of covariates simultaneously on the pooled estimates of mSEPT9.10 The HSROC plot provides estimates of diagnostic performance around a summary point and is suited to evaluate the effects on accuracy and positive test cut-off thresholds. The area under the curve (AUC) of the HSROC was calculated to provide evidence for diagnostic validity of the mSEPT9 tests. Pooled positive likelihood ratio and pooled negative likelihood ratio was used to assess diagnostic test accuracy. A high pooled positive likelihood ratio combined with a low pooled negative likelihood ratio provides evidence of validation of mSEPT9 tests as an early detection test for CRC. To illustrate the performance of the test in terms of predicting presence of CRC, a Fagan Nomogram was generated to convert a pretest probability of CRC to post-test probability of CRC depending on whether the mSEPT9 test was positive or negative. All statistical analysis were computed with STATA V.13.1.
Results
Literature search
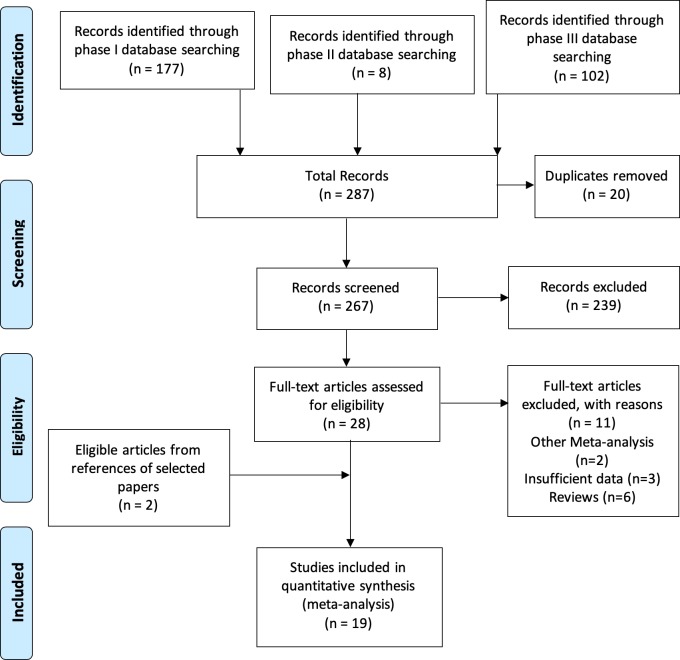
Based on the selected search terms 287 articles were identified, of which 20 were duplicates, leaving a total of 267 (figure 1). Of these, 239 studies were excluded after screening the abstracts. The full text of the 28 papers selected were read and the references from each article screened for other eligible studies. From the pool of 28 eligible studies, 11 studies were excluded (2 meta-analyses, 3 insufficient information to produce and 6 reviews). In the references of the eligible studies, 2 more eligible studies were identified, leaving a final total of 19 eligible studies (table 1).
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analysis flow chart of the selection of eligible literature.8
Table 1.
Eligible studies and the characteristics of participants included in the study
| Author | Year | Country | Participants | Stage | Reference | |||||||
| Total | Cases (CRC):controls (no CRC) | 0 | 1 | 2 | 3 | 4 | Unknown | |||||
| 1 | Grützmann et al | 2008 | USA | 309 | 126:183 | 0 | 22 | 36 | 54 | 11 | 3 | 12 |
| 2 | Lofton-Day et al | 2008 | USA | 312 | 133:179 | 0 | 20 | 32 | 47 | 31 | 3 | 23 |
| 3 | deVos et al | 2009 | Germany | 245 | 90:155 | 0 | 19 | 40 | 27 | 4 | 0 | 24 |
| 4 | Tänzer et al | 2010 | Germany | 67 | 33:34 | 5 | 6 | 0 | 1 | 14 | 7 | 13 |
| 5 | Ahlquist et al | 2012 | USA | 79 | 30:49 | 0 | 7 | 7 | 8 | 8 | 0 | 25 |
| 6 | Tóth et al | 2012 | Hungary | 184 | 92:92 | 0 | 25 | 14 | 36 | 18 | 0 | 14 |
| 7 | Lee et al | 2013 | South Korea | 197 | 101:96 | 0 | 26 | 30 | 28 | 17 | 0 | 26 |
| 8 | Liu et al | 2013 | Singapore | 52 | 26:26 | 0 | 26 | 0 | 0 | 0 | 0 | 27 |
| 9 | Church et al | 2014 | USA | 1510 | 53:1457 | 0 | 22 | 14 | 12 | 5 | 0 | 15 |
| 10 | Johnson et al | 2014 | USA | 301 | 101:200 | 2 | 26 | 20 | 23 | 13 | 17 | 16 |
| 11 | Potter et al | 2014 | USA | 1544 | 44:1500 | 0 | 39 | 5 | 0 | 17 | ||
| 12 | Su et al | 2014 | China | 234 | 172:62 | 0 | Unclear | 0 | 28 | |||
| 13 | Tóth et al | 2014 | Hungary | 58 | 34:24 | 0 | 6 | 11 | 11 | 5 | 1 | 18 |
| 14 | Jin et al | 2014 | China | 226 | 135:91 | 0 | Unclear | 0 | 19 | |||
| 15 | Ørntoft et al | 2015 | Denmark | 278 | 128:150 | 0 | 35 | 35 | 30 | 28 | 0 | 20 |
| 16 | Wu et al | 2016 | China | 586 | 291:295 | 4 | 37 | 66 | 82 | 33 | 62 | 21 |
| 17 | Song et al | 2017 | China | 978 | 388:590 | 0 | 52 | 129 | 162 | 19 | 0 | 7 |
| 18 | Song et al | 2017 | China | 409 | 85:324 | 0 | 9 | 38 | 31 | 7 | 0 | 22 |
| 19 | Chen et al | 2017 | Taiwan | 60 | 51: 9 | 0 | 16 | 27 | 0 | 29 | ||
CRC, colorectal cancer.
Quality assessment of individual studies
Of the 19 studies,127 12–22 were screening studies (blood sample for mSEPT9 test taken prior to CRC diagnosis) and 723–29 were clinical studies (blood sample for mSEPT9 test after CRC diagnosis). As the mSEPT9 test is being evaluated for its screening ability, the majority being screening studies is a strength of this meta-analysis. Fifteen studies7 12–19 21–24 26 27 collected plasma samples from participants for the mSEPT9 test prior to starting neoadjuvant therapy, adjuvant chemotherapy, radiotherapy or surgical resection and 17 studies,7 12–24 26 27 29 confirmed CRC status by gold standard histopathology.
Three studies were assessed as having a high risk of bias due to patient selection; six studies were assessed as having ‘unclear’ risk of bias due to the conduct of the index test and seven studies were assessed as ‘unclear’ risk of bias due to flow and timing. Five studies were assessed as having ‘high’ applicability concerns due to patient selection, and three studies were assessed as having ‘high’ applicability concerns due to the index test. One study was assessed as having high applicability concerns and two ‘unclear’ for the reference standard domain. There was good inter-rater agreeability for risk of bias (k=0.75) and excellent agreeability (k=1) for applicability concerns between the quality assessors.
Participant characteristics
The 19 selected studies had a total of 7629 cases and controls from 7 countries, in North America, Europe and East/South-East Asia. All CRC cases were diagnosed and staged before treatment and controls were individuals with no evidence of CRC, most (94.3%) confirmed by prior colonoscopy.
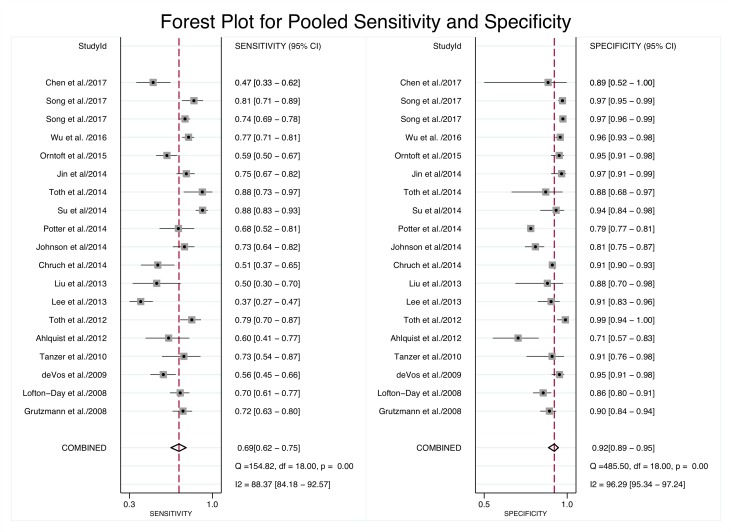
The pooled sensitivity of mSEPT9 in detecting CRC was 0.69 (95% CI 0.62 to 0.75) and the pooled specificity was 0.92 (95% CI 0.89 to 0.95) (table 2). There was considerable heterogeneity for both sensitivity (I2=88.37%) and specificity (I2=96.29%) (figure 2). The pooled positive likelihood ratio of 9.1 (95% CI 6.1 to 13.8) indicates that CRC cases were ninefold more likely to have a positive mSEPT9 test compared with controls (table 2). The negative likelihood ratio of 0.33 (95% CI 0.22 to 0.50) indicates that CRC cases were one-third less likely to have a negative mSEPT9 test compared with controls (table 2). The overall pooled diagnostic OR for mSEPT9 was 27 (95% CI 15 to 48)(table 2), which indicates that a case had 27-fold greater odds of being positive than a control, or to put it another way, a positive mSEPT9 result was associated with a 27-fold increase in odds of the person having CRC. Pooled specificity and positive likelihood ratio of North American studies was lower compared with Asian and European studies (p<0.001).
Table 2.
Pooled performance indicators for all eligible studies and subgroup analysis based on country of recruitment, mSEPT9 test cut-off level (threshold) and study sample size
| Pooled sensitivity (95% CI) |
Pooled specificity (95% CI) |
Pooled positive likelihood ratio (95% CI) |
Pooled negative likelihood ratio (95% CI) |
Pooled diagnostic OR (95% CI) |
|
| Overall | 0.69 (0.62 to 0.75) | 0.92 (0.89 to 0.95) | 9.1 (6.1 to 13.8) | 0.34 (0.27 to 0.42) | 27 (15 to 48) |
| Country of recruitment | |||||
| Asian | 0.68 (0.55 to 0.79) | 0.96 (0.93 to 0.97) | 16 (8.9 to 28.6) | 0.33 (0.22 to 0.50) | 48 (19 to 126) |
| European | 0.71 (0.58 to 0.81) | 0.95 (0.92 to 0.97) | 15.3 (9.3 to 25.1) | 0.30 (0.20 to 0.45) | 51 (25 to 104) |
| North American | 0.68 (0.62 to 0.74) | 0.85 (0.79 to 0.89) | 4.4 (3.3 to 5.9) | 0.38 (0.31 to 0.45) | 12 (8 to 17) |
| Threshold level | |||||
| Algorithm | |||||
| 1/3 | 0.68 (0.57 to 0.78) | 0.88 (0.83 to 0.92) | 5.7 (3.7 to 8.8) | 0.36 (0.25 to 0.51) | 16 (8 to 32) |
| 2/3 | 0.69 (0.60 to 0.76) | 0.95 (0.92 to 0.97) | 14.6 (8.5 to 24.8) | 0.33 (0.25 to 0.43) | 44 (21 to 91) |
| Study sample size | |||||
| ≥100 | 0.71 (0.61 to 0.79) | 0.93 (0.89 to 0.96) | 10.4 (6.3 to 17) | 0.31 (0.23 to 0.61) | 33 (16 to 66) |
| <100 | 0.66 (0.57 to 0.75) | 0.92 (0.85 to 0.95) | 8.0 (4.2 to 15.3) | 0.37 (0.27 to 0.49) | 22 (9 to 52) |
Figure 2.
Forest plot of pooled sensitivity and specificity from meta-analysis and heterogeneity values based on the I2 statistic.
The 2/3 algorithm for positivity threshold appeared to have the same sensitivity as the 1/3 threshold (approximately 0.69), but had a higher specificity and thereby a higher positive likelihood ratio and diagnostic OR, with a lower negative likelihood ratio. Sensitivity and specificity appeared similar for large (>100 participants) and small (<100 participants) studies.
The Spearman’s correlation coefficient for all studies was 0.19 (p=0.4) indicating no evidence that the heterogeneity was due to differences in the definition of threshold used by each study (online supplementary table 2) or of a monotonic relationship between sensitivity and specificity. The effect investigated here was the algorithm chosen by the study as 2/3 PCR results. While some studies clearly stated the use of the 1/3 algorithm, other studies which did not explicitly state this were considered within the 1/3 PCR result subgroup, as this is the minimum threshold to establish a positive result (online supplementary table 2). There was some evidence that the threshold used contributed to the heterogeneity of Asian studies (Spearman’s coefficient 0.793, p=0.02), but no such evidence was observed for European or North American studies or large or small studies (online supplementary table 2).
There was marginal evidence (p=0.06) for the heterogeneity in sensitivity being attributable to differences in sensitivity in North American studies and studies that recruited more cases with stage 1 CRC than cases with stage 4 CRC (p=0.06) (see online supplementary table 3, online supplementary figure 2). There was strong evidence (p<0.01) that the heterogeneity in specificity was largely affected by size of study, and whether it was conducted in Asian (p<0.01) or North American (p<0.001) regions. Studies with a high proportion of stage 1 CRCs had a lower sensitivity (p=0.06) than those with a high proportion of stage 4 CRCs (see online supplementary table 3, online supplementary figure 2).
There was marginal evidence of publication bias (p=0.05) (see online supplementary figure 3).
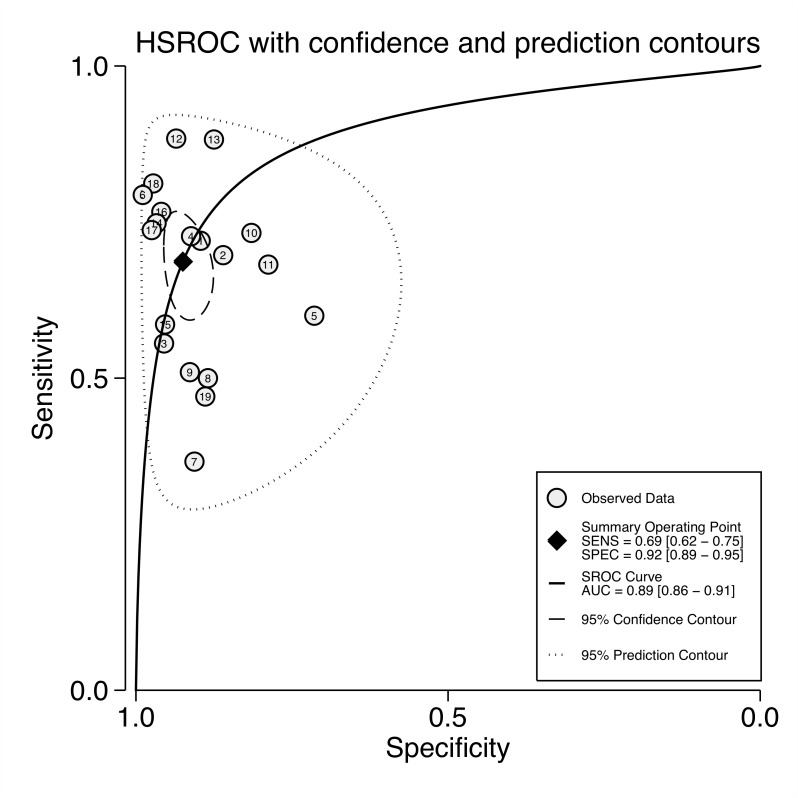
Hierarchical summary operator receiver curve
The 95% prediction contour of the HSROC encompassed all eligible studies indicating the likely region of sensitivity and specificity of mSEPT9 (figure 3). The AUC of 0.89 (95% CI 0.86 to 0.91) confirms the mSEPT9 test as being diagnostically valid with moderate-to-high accuracy. The β-coefficient of the HSROC, which describes a symmetrical distribution of data points (studies), is not significantly different from zero (0.37; 95% CI −0.25 to 0.79) and therefore the lambda coefficient is a reasonable measure of the test’s accuracy; 3.01 (95% CI 2.45 to 3.73). The theta coefficient of −0.65 (95% CI –1.09 to –0.202) represents the positivity threshold cut-off on a log scale (0.522 on the linear scale).
Figure 3.
Hierarchical summary operator receiver curve (HSROC) plot for all studies, the summary operating point with confidence contours and prediction contours for future tests. AUC, area under the curve; SENS, sensitivity; SPEC, specificity.
The sensitivity and specificity was more efficient for the positivity threshold chosen in the Asian and European studies compared with the North American studies (see online supplementary figure 4a–c). The diagnostic performance for European studies was similar to Asian studies, both with AUC 0.96 (95% CI 0.92 to 0.97), which were higher compared with North American studies 0.80 (95% CI 0.76 to 0.83).
Visual assessment of pretest and post-test probability and likelihood ratios
The summary points from this meta-analysis (pooled positive likelihood ratio=9.4 and cut-off exclusion level or negative likelihood ratio=0.3) falls just below the cut-off confirmation of diagnostic validity, defined as a test with a positive likelihood ratio >10 (confirmatory role) and negative likelihood ratio of <0.1 (exclusory role) (see online supplementary figure 5a–c).
Positive and negative predictive values
A mSEPT9 test with sensitivity of 69% and specificity of 92% would accurately diagnose 21 of 30 CRC cases present in a sample of 10 000 people at average risk. For this sample, the PPV is 2.6% and NPV is 99.9%.
For the same sensitivity and specificity, a mSEPT9 test would accurately diagnose 83 of 120 CRC cases in a sample of 10 000 people at high risk. For this sample, the PPV is 9.5% and NPV is 99.6%.
Discussion
In this meta-analysis, mSEPT9 has an estimated sensitivity of 0.69 (95% CI 0.62 to 0.75) and specificity of 0.92 (95% CI 0.89 to 0.95), AUC of 0.89 (95% CI 0.86 to 0.91) and positive and negative likelihood ratios of 9.1 and 0.33, respectively. For a screened population of 10 000 people, of which 0.3% (30 cases) have CRC, that is, typical of an average risk population aged 50–74 years, 21 of the 30 CRC cases will be detected by the mSEPT9 test and 9172 of the 9970 healthy individuals would be correctly classified by the test as not having CRC. A person who tests positive for the mSEPT9 test has a 2.6% chance of having CRC (PPV). A negative mSEPT9 test virtually rules out CRC with an NPV of 99.9%. For a population at high risk, for example, with a strong family history putting them at four-fold increased risk, the corresponding PPV and NPV would be 9.5% and 99.6%.
A previous meta-analysis of mSEPT9 in 2017 (which included 14 of the 19 studies we included in this meta-analysis) reported a pooled sensitivity of 0.71, specificity of 0.92 and AUC 0.88, which are in line with previous diagnostic test performance indicators.4 Previous meta-analyses reported that estimates of sensitivity and specificity were highly heterogeneous, probably due to differences in test performance by geographic region of recruitment of participants. We also observed marginal evidence that some of heterogeneity could be due to varying ctDNA positivity thresholds inferring that test accuracy depended on the positivity algorithm, with the optimum threshold level being the 2/3 algorithm. The North American studies had overall lower pooled diagnostic accuracy indicators potentially due to different positivity threshold used in that region. As reported by Nian et al,4 we found marginal evidence for the sensitivity being lower for studies with a higher proportion of early stage disease is expected given earlier stage CRC would be expected to produce lower amounts of circulating mSEPT9 given fewer cancer cells at the site of the original tumour and no metastases.
Limitations of this meta-analysis included inability to investigate other non-threshold effects on heterogeneity or variations in PCR methodology and technology.
Many CRC screening programmes use the FIT to detect occult blood in faeces. The FIT has a sensitivity of 79% (95% CI 69 to 86) and specificity of 94% (95% CI 92 to 95),30 that is, is more sensitive and specific than mSEPT9. For a screened population of 10 000 people, at average CRC risk, 24 of the 30 CRC cases will be detected by FIT and 9372 of the 9970 healthy individuals would be correctly classified by FIT as not having CRC. The PPV for this population would be 3.9% and NPV would be 99.9%. In the same screened population, for individuals at high risk of CRC, the corresponding PPV and NPV would be 13.8% and 99.7%. Therefore, on a purely performance-based comparison, mSEPT9 is not a replacement test for FIT. However, as with all cancer screening programmes, there is incomplete uptake of screening by FIT, particularly so for some countries where less than half of the eligible population are participating in population-based organised programmes, for example, Australia, France, Ireland, Italy, Republic of Korea and Singapore.31 One reason (of many) for this non-participation being some people find talking about and collecting/storing faecal samples unpleasant and embarrassing.32 For these people, a blood test may be more appealing and result in higher participation. To support this strategy, studies need to be done on decliners of FIT screening to determine their uptake in an alternative screen via the mSEPT9 test. Such studies would indicate whether decliners of the FIT might be amenable to an alternative test based on a blood sampled test such as mSEPT9, and therefore mSEPT9 might be a viable alternative screening test for those who would otherwise miss out on the protection that CRC screening affords. A recent study reported that 94% of average risk patients who declined a FIT test, opted for mSEPT9 testing; however, the study sample only included people who attended a screening clinic and therefore does not represent all decliners of FIT screening.33 One argument against the strategy of offering mSEPT9 as an alternative to FIT screening is the high cost of the mSEPT9 test (€15015), which means that screening an average risk population via mSEPT9 is twice as expensive as screening via FIT, even under the assumption of equivalent sensitivity and specificity of the tests.34 Another argument against this strategy is that mSEPT9 might not be sensitive for precancerous lesions: advanced adenomas (15%) and polyps (5%).4 Because of these limitations of mSEPT9, investment in strategies to facilitate participation in FIT remains important.
Conclusion
This analysis of data from 19 studies finds that the mSEPT9 test has potential for a population screening of CRC with the ability to detect approximately two-thirds of CRC cases and ruling out 92% of non-cases. While mSEPT9 has diagnostic performance similar (although lower) to the FIT, it is currently unsuitable as its replacement in screening programmes. However, it may be a valid alternative to the FIT, when considering those unwilling to participate in the home faecal test. Further screening studies are required to assess the uptake of mSEPT9 tests when offered to FIT decliners in lieu of FIT as well as an economic evaluation that includes the costs of the test.
Footnotes
Twitter: @markaj64
Contributors: RH conceptualised the study, carried out literature search and selected the studies, provided quality assessment, extraction of data, conducted data analysis, conceptualised the design and draft of the manuscript and approved the final manuscript. MJ contributed to the conception of the paper, supported in selection of papers and provided quality assessment, supported in data analysis and design and draft of the manuscript, proofread and approved the final manuscript.
Funding: This study was funded by National Health and Medical Research Council (Grant number: APP1117611).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information. All data for this analysis are included in the article or uploaded supplementary material.
References
- 1.Howlader N, Noone A, Krapcho M, et al. . Seer cancer statistics review, 1975–2016. Bethesda, MD: National Cancer Institute, 2016. https://seer.cancer.gov/csr/1975_2016/ [Google Scholar]
- 2.Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics 2017;15:59–72. 10.1016/j.gpb.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molnár B, Tóth K, Barták BK, et al. . Plasma methylated septin 9: a colorectal cancer screening marker. Expert Rev Mol Diagn 2015;15:171–84. 10.1586/14737159.2015.975212 [DOI] [PubMed] [Google Scholar]
- 4.Nian J, Sun X, Ming S, et al. . Diagnostic accuracy of methylated SEPT9 for blood-based colorectal cancer detection: a systematic review and meta-analysis. Clin Transl Gastroenterol 2017;8:e216–e16. 10.1038/ctg.2016.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, He Y, Zhang X, et al. . A pooled analysis of the diagnostic efficacy of plasmic methylated septin-9 as a novel biomarker for colorectal cancer. Biomed Rep 2017;7:353–60. 10.3892/br.2017.970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song L, Yu H, Jia J, et al. . A systematic review of the performance of the SEPT9 gene methylation assay in colorectal cancer screening, monitoring, diagnosis and prognosis. CBM 2017;18:425–32. 10.3233/CBM-160321 [DOI] [PubMed] [Google Scholar]
- 7.Song L, Jia J, Yu H, et al. . The performance of the mSEPT9 assay is influenced by algorithm, cancer stage and age, but not sex and cancer location. J Cancer Res Clin Oncol 2017;143:1093–101. 10.1007/s00432-017-2363-0 [DOI] [PubMed] [Google Scholar]
- 8.Moher D, Liberati A, Tetzlaff J, et al. . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiting PF, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529–36. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 10.Macaskill P, Gastonis C, Deeks J. Chapter 10: Analysing and Presenting Results : Deeks JJ BP, Gatsonis C, Cochrane Handbook for systematic reviews of diagnostic test accuracy version 1. The Cochran Collaboration, 2010. [Google Scholar]
- 11.van Enst WA, Ochodo E, Scholten RJPM, et al. . Investigation of publication bias in meta-analyses of diagnostic test accuracy: a meta-epidemiological study. BMC Med Res Methodol 2014;14:70 10.1186/1471-2288-14-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grützmann R, Molnar B, Pilarsky C, et al. . Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One 2008;3:e3759–8. 10.1371/journal.pone.0003759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tänzer M, Balluff B, Distler J, et al. . Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One 2010;5:e9061 10.1371/journal.pone.0009061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tóth K, Sipos F, Kalmár A, et al. . Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One 2012;7:e46000 10.1371/journal.pone.0046000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Church TR, Wandell M, Lofton-Day C, et al. . Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317–25. 10.1136/gutjnl-2012-304149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson DA, Barclay RL, Mergener K, et al. . Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS One 2014;9:e98238–8. 10.1371/journal.pone.0098238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter NT, Hurban P, White MN, et al. . Validation of a real-time PCR–Based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem 2014;60:1183–91. 10.1373/clinchem.2013.221044 [DOI] [PubMed] [Google Scholar]
- 18.Tóth K, Wasserkort R, Sipos F, et al. . Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One 2014;9:e115415–9. 10.1371/journal.pone.0115415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin P, Kang Q, Wang X, et al. . Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015;30:830–3. 10.1111/jgh.12855 [DOI] [PubMed] [Google Scholar]
- 20.Ørntoft M-BW, Nielsen HJ, Ørntoft TF, et al. . Performance of the colorectal cancer screening marker SEPT9 is influenced by age, diabetes and arthritis: a nested case-control study. BMC Cancer 2015;15:819 10.1186/s12885-015-1832-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu D, Zhou G, Jin P, et al. . Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn 2016;18:535–45. 10.1016/j.jmoldx.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 22.Song L, Peng X, Li Y, et al. . The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics 2017;9:599–610. 10.2217/epi-2016-0146 [DOI] [PubMed] [Google Scholar]
- 23.Lofton-Day C, Model F, DeVos T, et al. . Dna methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414–23. 10.1373/clinchem.2007.095992 [DOI] [PubMed] [Google Scholar]
- 24.deVos T, Tetzner R, Model F, et al. . Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 2009;55:1337–46. 10.1373/clinchem.2008.115808 [DOI] [PubMed] [Google Scholar]
- 25.Ahlquist DA, Taylor WR, Mahoney DW, et al. . The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol 2012;10:272–7. 10.1016/j.cgh.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HS, Hwang SM, Kim TS, et al. . Circulating methylated septin 9 nucleic acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol 2013;6:290–4. 10.1593/tlo.13118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Tham CK, Ong SYK, et al. . Serum methylation levels of TAC1. SEPT9 and EYA4 as diagnostic markers for early colorectal cancers: a pilot study. Biomarkers 2013;18:399–405. 10.3109/1354750X.2013.798745 [DOI] [PubMed] [Google Scholar]
- 28.Su XL, Wang YF, Li SJ, et al. . High methylation of the SEPT9 gene in Chinese colorectal cancer patients. Genet Mol Res 2014;13:2513–20. 10.4238/2014.January.17.5 [DOI] [PubMed] [Google Scholar]
- 29.Chen C-H, Yan S-L, Yang T-H, et al. . The relationship between the methylated Septin-9 DNA blood test and stool occult blood test for diagnosing colorectal cancer in Taiwanese people. J Clin Lab Anal 2017. [Epub ahead of print: 08 Jul 2016]. 10.1002/jcla.22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JK, Liles EG, Bent S, et al. . Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med 2014;160:171–71. 10.7326/M13-1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.IARC Colorectal cancer screening. IARC Handb Cancer Prev 2019;17:1–300http://publications.iarc.fr/573 [Google Scholar]
- 32.Honein-AbouHaidar GN, Kastner M, Vuong V, et al. . Systematic review and Meta-study synthesis of qualitative studies evaluating facilitators and barriers to participation in colorectal cancer screening. Cancer Epidemiol Biomarkers Prev 2016;25:907–17. 10.1158/1055-9965.EPI-15-0990 [DOI] [PubMed] [Google Scholar]
- 33.Ioannou S, Sutherland K, Iyengar R, et al. . Su1667 – increasing uptake of colon cancer screening in a medically underserved population with the addition of blood-based testing. Gastroenterology 2019;156:S-604–4. 10.1016/S0016-5085(19)38405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth JA, deVos T, Ramsey SD. Clinical and budget impact of increasing colorectal cancer screening by blood-and stool-based testing. American Health & Drug Benefits 2019;12. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjgast-2019-000355supp001.pdf (179.1KB, pdf)
bmjgast-2019-000355supp002.pdf (678.2KB, pdf)