ABSTRACT
Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4, CD152) is a receptor on T cells that inhibits the cell’s functions. Blocking CTLA-4 with an antibody has proven effective for the treatment of cancer patients. Anti-CTLA-4 antibodies currently approved for clinical use can bind to human CTLA-4, but do not cross-react to murine CTLA-4. Here, we report the generation and characterization of a functional humanized antibody, mAb146, against both human and murine CTLA-4. Alanine scanning of CTLA-4 using mammalian cell expression cassette identified the unique epitopes of this novel antibody. In addition to the amino acid residues interacting with ligands CD80 and CD86, an N-glycosylation site on N110, conserved in CTLA-4 of human, monkey, and mouse, was identified as the specific epitope that might contribute to the cross-species binding and function of this antibody. This finding may also contribute to the understanding of the glycosylation of CTLA-4 and its related biologic function. In addition to facilitating preclinical development of anti-CTLA-4 antibodies, mAb146 may be useful as a therapeutic agent.
KEYWORDS: Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), CD152, antibody, cross-species binding, epitope mapping, N-glycosylation
Introduction
Cancer immunotherapy has become an effective approach to treat cancer. Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) is one of the validated targets of immune checkpoints.1 CTLA-4 is a disulfide-linked homodimeric glycoprotein with approximately 75% sequence homology with CD28. Both CTLA-4 and CD28 are members of the Ig superfamily present on T cells. After T-cell activation, CTLA-4 quickly expresses on those T cells, generally within one hour of antigen engagement with T cell receptor. CTLA-4 can inhibit T-cell signaling through competition with CD28. CD28 mediates a well-characterized T-cell co-stimulatory signal by binding to its ligands CD80 (B7-1) and CD86 (B7-2) on antigen-presenting cells, leading to T-cell proliferation by inducing the production of interleukin-2 and anti-apoptotic factors. Due to the much higher affinity binding of CTLA-4 to CD80 and CD86 than that of CD28, CTLA-4 can out-compete with CD28 binding to CD80 and CD86, suppressing T-cell activation. It also is reported that CTLA-4 can capture CD80 and CD86, and subsequently remove these ligands from antigen-presenting cells.2 In addition, CTLA-4 is constitutively expressed on the surface of regulatory T cells (Tregs), suggesting that CTLA-4 may be required for contact-mediated suppression and associated with Tregs-produced immunosuppressive cytokines such as transforming growth factor beta and interleukin-10.3 Recent research indicates that the selective depletion of Tregs in tumor microenvironment is the dominant mechanism of action of anti-CTLA-4-targeted therapies.4 It has been reported that effective anti-CTLA-4-based treatments require Fcγ receptor engagement of anti-CTLA-4 antibody,56 and Fc effector function, such as antibody-dependent cell-mediated cytotoxicity (ADCC) mediated depletion of Tregs, is critical.78
Due to the importance of CTLA-4 function, CTLA-4 blockade has been tested for treatment of cancer in numerous preclinical and clinical studies. A substantial amount of data has been published for two antibodies against CTLA-4, ipilimumab, and tremelimumab. Ipilimumab (MDX-010, BMS-734016) is an immunomodulatory agent that has been approved as monotherapy for treatment of advanced melanoma.9 Combined with an anti-PD-1 antibody, ipilimumab has also been approved for the treatment of advanced melanoma, metastatic colorectal cancer with MMR and MSI-H aberrations and renal cell carcinoma.10 Tremelimumab was evaluated as monotherapy in melanoma and malignant mesothelioma11 and in combination with the anti-PD-L1 antibody durvalumab in multiple cancers.12–15 Since human and mouse CTLA-4 only share approximately 76% amino acid identity, these two anti-CTLA-4 antibodies can only bind to human CTLA-4 (hCTLA-4), but not murine CTLA-4 (mCTLA-4).16 Here, we developed a novel antibody, mAb146, by immunizing rats with both human and mouse CTLA-4 and screening a large number of hybridoma clones. This antibody recognizes not only the MYPPPY motif that interacts with CD80/CD86, but also an N-glycosylated site epitope that is conserved in human, monkey, and murine CTLA-4.
Results
Generation of murine CTLA-4-cross-reactive antibody
Lymphocytes, isolated from spleen and lymph nodes of hCTLA-4 and mCTLA-4 extracellular domain (ECD) alternately immunized Sprague Dawley (SD) rats, were electrically fused with SP2/0 myeloma cells to form hybridoma. Hybridoma clones were screened on binding to human, murine, and monkey CTLA-4 proteins, as well as engineered human CTLA-4-expressing cells. The variable regions of positive clones were isolated, and then humanized using complementary-determining region (CDR)-grafting techniques. After screening and humanization, a monoclonal antibody (mAb) 1.146.19-Z12 (mAb146) with human IgG1 isotype was found that bound to hCTLA-4 with EC50 of 0.03 nM, which is slightly higher than the EC50 of ipilimumab (0.01 nM) (Figure 1a). mAb146 and ipilimumab also bound to monkey CTLA-4 with EC50 of 0.05 nM and 0.03 nM, respectively (Figure 1b). However, only mAb146 bound to mCTLA-4 (EC50 of 0.19 nM) (Figure 1c); ipilimumab did not bind to mCTLA-4.
Figure 1.

Ipilimumab and mAb146 bound to human (a), monkey (b), and murine (c) CTLA-4 measured by ELISA. A 96-well plate was coated with hCTLA-4-6xHis monomer (1.0 μg/mL), cynomolgus monkey CTLA-4-6xHis monomer (0.5 μg/mL) or mouse CTLA-4-6xHis monomer (0.5 μg/mL) at 4◦C. After incubation with the antigens, the binding of Ipilimumab and mAb146 was detected by addition of HRP-conjugated goat anti-human IgG antibody.
The binding kinetics of antibody
The binding kinetics of the antibodies were measured using surface plasmon resonance (SPR). Specifically, we used SPR to measure the on-rate constant (ka) and off-rate constant (kd) of the antibodies to extracellular domain of hCTLA-4, and then determined the affinity constant (KD). mAb146 bound to hCTLA-4 with an affinity (KD=0.477 nM) that is significantly higher than that of ipilimumab (KD = 3.68 nM). The high affinity of mAb146 was mainly due to the slow off rate (kd = 9.82E-05) compared with that of ipilimumab (kd = 3.46E-03). mAb146 could also bind to mCTLA-4 with high affinity (KD = 1.39 nM) (Table 1).
Table 1.
Kinetic of antibody-binding on human/murine CTLA-4 extracellular domain.
| Antigens |
Human CTLA-4 |
Murine CTLA-4 |
|
|---|---|---|---|
| Antibodies | mAb146 | Ipilimumab | mAb146 |
| ka (1/Ms) | 2.06E+05 | 9.42E+05 | 1.72E+05 |
| kd (1/s) | 9.82E-05 | 3.46E-03 | 2.39E-04 |
| KD (M) | 4.77E-10 | 3.68E-09 | 1.39E-09 |
Competition with ligands of antibody
In order to test whether the humanized mAb146 was able to block CTLA-4 binding on CD80 and CD86, we used both enzyme-linked immunosorbent assay (ELISA) and fluorescence-activated cell sorting (FACS). In an ELISA-based competition assay, mAb146 and ipilimumab had similar effects in blocking human CD80 (hCD80) binding to coated hCTLA-4, with IC50 of 0.87 nM and 0.40 nM, respectively (Figure 2a). They also had similar effects in blocking human CD86 (hCD86) binding to hCTLA-4, with IC50 of 0.71 nM and 0.42 nM, respectively (Figure 2b). In a FACS assay, mAb146 could more effectively block CTLA-4/ligand binding than ipilimumab. At the highest concentration used, ipilimumab only blocked 32% of hCTLA-4 binding to hCD80+ cells and 40% of hCTLA-4 binding to hCD86+ cells. In comparison, mAb146 blocked 71% of hCTLA-4 binding on hCD80 and 73% of hCTLA-4 binding on hCD86, significantly higher than that caused by ipilimumab. The IC50 of ipilimumab and mAb146 blocking hCTLA-4 binding to hCD80 were 3.23 nM and 6.60 nM, respectively (Figure 2c), and the blocking of hCTLA-4 binding to hCD86 were 2.52 nM and 5.15 nM, respectively (Figure 2d). The antibodies were also tested for their ability to block mCTLA-4 binding on murine CD80 (mCD80) and murine CD86 (mCD86). As expected, mAb146, but not ipilimumab, could block the binding of mCTLA-4 and mCD80 (Figure 2e) and the binding of mCTLA-4 and mCD86 (Figure 2f), with IC50 of 0.62 nM and 2.32 nM, respectively.
Figure 2.
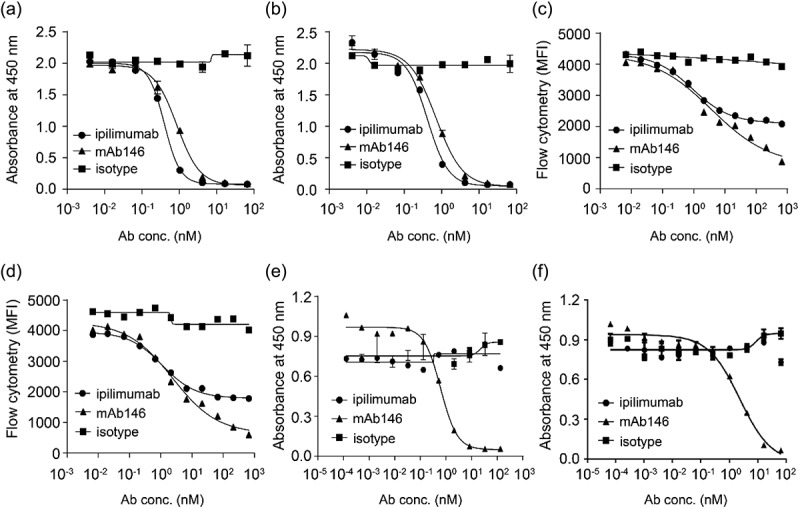
Ipilimumab and mAb146 block ligands human CD80 (a, c) and human CD86 (b, d) binding to hCTLA-4 by ELISA (a–b) and FACS (c–d), and their ability on blocking mouse CD80 (e) and mouse CD86 (f) binding to mCTLA-4 also detected by ELISA (e–f). In ELISA-based competition assays, hCTLA-4-hFc dimer or mCTLA-4-mFc dimer (0.5 μg/mL) were coated on 96-well plates, the antibodies pre-mixed with 0.25 μg/mL of hCD80-6xHis, hCD86-6xHis, or 0.5 μg/ml of mCD80-6xHis, 5 μg/mL of mCD86-6xHis were added. After blocking with 2% BSA, biotinylated anti-His tag antibody was added. The bound ligands were detected by HRP-conjugated streptavidin. In FACS-based competition assay, hCD80- or hCD86-expressing CHO cell lines were used. The details were described in the method section.
ADCC and antibody-dependent cellular phagocytosis effect of the antibody
It has been reported that the Fc of an anti-CTLA-4 antibody is an important factor contributing to anti-tumor efficacy in vivo.5–8 In order to evaluate the possible anti-tumor mechanism of mAb146, ADCC and antibody-dependent cellular phagocytosis (ADCP) assays were performed on engineered human CTLA-4-expressing cells. As shown in Figure 3, mAb146 showed similar ADCC (Figure 3a) and ADCP effects (Figure 3b) as ipilimumab.
Figure 3.
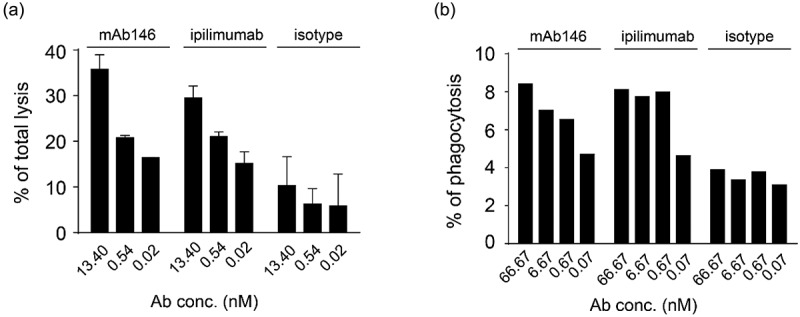
Antibody-dependent cellular cytotoxicity (ADCC) (a) and antibody-dependent cellular phagocytosis (ADCP) (b) of the antibodies against CTLA-4. (a) Human CTLA4-expressing 293F cells were added to 96-well plates at 1 × 104 cells/well, and then the antibodies pre-incubated with 5 × 105 PBMCs were added. The plates were kept at 37°C in a 5% CO2 incubator for 4 h. Lysis of the target cells was determined by the introduction of DELFIA® EuTDA Cytotoxicity Reagents. (b) Human macrophage cells were mixed at 1:1 ratio with CFSE-dyed engineered human CTLA-4 expressing 293F cells in 96-well plates, then antibodies were added and incubated with cells at 37°C in a 5% CO2 incubator for 3 h. After wash, APC-labeled anti-human CD14 antibody was added for flow cytometry detection.
Efficacy study
Due to the cross-reactivity of mAb146 to both human and murine CTLA-4, the anti-tumor efficacy of this antibody could be tested in a syngeneic CT26 mouse model. A functional anti-mCTLA-4 antibody, 9H10, was used as a positive control.1718 As shown in Figure 4, mAb146 significantly inhibited tumor growth in a dose-dependent manner. At 1 mg/kg dose, mAb146 significantly inhibited tumor growth, compared with vehicle control group. At 3 mg/kg and 10 mg/kg dose, mAb146 inhibited tumor growth more effectively than the control anti-mCTLA-4 antibody at 10 mg/kg dose. Moreover, 10 mg/kg mAb146 induced complete tumor regression at the end of the study period.
Figure 4.
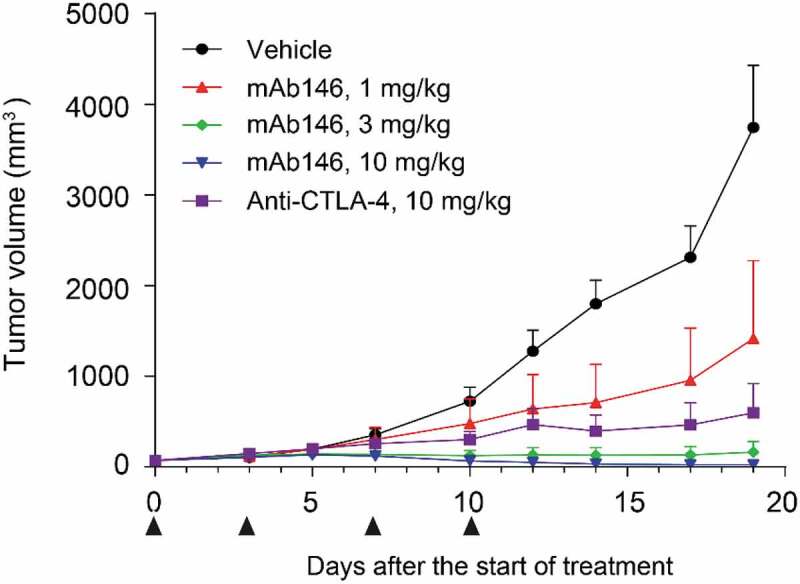
mAb146 significantly inhibited tumor growth in CT-26 syngeneic model. In CT-26 syngeneic model. Female Balb/C mice were inoculated subcutaneously with 1 × 105 tumor cells in 0.1 mL of PBS mixed with 50 μL matrigel. When the average tumor volume reached 60–80 mm3, the animals were randomly grouped (n = 6). The anti-CTLA-4 antibodies and isotype control were used for treatment. The tumor size was measured twice weekly by a vernier caliper, and tumor volume was calculated by the formula a× b2 × π/6 where “a” was length and “b” was width (a > b) of the tumor.
Epitope mapping
To understand mAb146’s cross-reactivity to both human and mouse CTLA-4 and its antagonistic function, we conducted alanine scanning to map their epitopes. In this experiment, hCTLA-4 variants with a single mutation were made by mutating alanine residues on hCTLA-4 to glycine residues, and all other residues to alanine. Three additional mutants were made to test whether the epitope involved an N-glycosylation site: hCTLA-4-N78Q, hCTLA-4-N110Q, and hCTLA-4-N78Q/N110Q. All mutants were transiently expressed in HEK293F/Expi293 cells. A capture ELISA was conducted to test how the mutations affected antibody binding, and binding reduction more than 55% was set as the cutoff. Additionally, a hCTLA-4 crystal structure (PDB code 1AH1) was used to analyze the data of alanine scanning. For example, some amino acid residues (M3, V5, Y25, V36, V38, R40, V49, C50, C94, I114) were identified as buried residues and unlikely to directly contact with the antibodies. The observed binding reductions probably resulted from the instability or conformational change of CTLA-4 structure after alanine substitutions. Details of the final determined epitope are shown in Table 2 and Figure 5. Although some of the contact residues of ipilimumab (Figure 5a) and mAb146 (Figure 5b) overlap, a few residues are unique to mAb146, such as N110 and V96. The overlapped contact residues of the two antibodies mainly involved MYPPPY motif, which has been reported to be the interface on CTLA-4 interacting with the ligands of CTLA-4 (Figure 5c,d, Fig. S1).
Table 2.
List of identified hot spot residues and interface residues from structure complexes on hCTLA-4.
| CTLA4 position | mAb146 epitope* |
Ipilimumab epitope* | mAb146 model interface# |
Ipilimumab interface# [Ref.6] |
Tremelimumab Interface# [Ref.23] |
CD80 Interface# [Ref.13] |
CD86 Interface# [Ref.20] |
Comments |
|---|---|---|---|---|---|---|---|---|
| 3M | x | x | x | x | Buried | |||
| 4H | x | x | x | |||||
| 5V | x | x | Buried | |||||
| 25Y | x | x | Buried | |||||
| 26A | ||||||||
| 27S | ||||||||
| 28P | x | |||||||
| 29G | ||||||||
| 30K | ||||||||
| 31A | x | x | Buried | |||||
| 32T | ||||||||
| 33E | x | x | x | x | x | |||
| 34V | ||||||||
| 35R | x | x | x | x | x | x | x | |
| 36V | x | x | Buried | |||||
| 37T | x | x | x | |||||
| 38V | x | x | Buried | |||||
| 39L | x | x | x | x | x | |||
| 40R | x | x | Buried | |||||
| 41Q | x | x | x | x | Maintain structure | |||
| 42A | ||||||||
| 43D | ||||||||
| 44S | x | x | x | |||||
| 45Q | x | x | x | |||||
| 46V | x | x | x | x | x | |||
| 47T | x | x | x | x | ||||
| 48E | x | x | x | x | ||||
| 49V | x | x | Buried | |||||
| 50C | x | x | Buried | |||||
| 51A | x | x | ||||||
| 52A | ||||||||
| 53T | x | x | x | x | x | |||
| 54Y | x | |||||||
| 55M | x | x | Maintain structure |
|||||
| 56M | ||||||||
| 57G | x | x | Maintain structure | |||||
| 58N | ||||||||
| 59E | ||||||||
| 60L | ||||||||
| 61T | x | |||||||
| 62F | ||||||||
| 63L | x | |||||||
| 64D | ||||||||
| 65D | x | |||||||
| 66S | ||||||||
| 67I | ||||||||
| 68C | x | |||||||
| 90G | x | Maintain structure |
||||||
| 91L | x | x | x | x | Maintain structure |
|||
| 92Y | x | x | Maintain structure |
|||||
| 93I | x | x | x | x | x | |||
| 94C | x | x | Buried | |||||
| 95K | x | x | x | x | x | x | ||
| 96V | x | |||||||
| 97E | x | x | x | x | x | x | x | |
| 98L | x | x | ||||||
| 99M | x | x | x | x | x | x | x | |
| 100Y | x | x | x | x | ||||
| 101P | x | x | x | x | x | |||
| 102P | x | x | x | x | x | x | x | |
| 103P | x | x | x | x | x | x | ||
| 104Y | x | x | x | x | x | x | x | |
| 105Y | x | x | x | x | ||||
| 106L | x | x | x | x | x | x | ||
| 107G | x | x | x | x | ||||
| 108I | x | x | x | x | x | |||
| 109G | x | x | x | x | ||||
| 110N | x | x | x | |||||
| 111G | x | x | x | Buired | ||||
| 112T | x | x | Buried | |||||
| 113Q | x | x | Maintain structure | |||||
| 114I | x | x | Buried |
*: The residues are extracted from ala-scanning experiment data.
#: The residues are extracted from structure complexes using the software COCOMAPS. A maximum distance was set to 6 Å to report atom–atom interactions within this cutoff limit. The mAb146 complex is from homology modeling and docking. The Ipilimumab complex, Tremelimumab complex, CD80 complex, and CD86 complex are from PDB entries 5XJ3, 5GGV, 1I8L, and 1I85, respectively.
Figure 5.
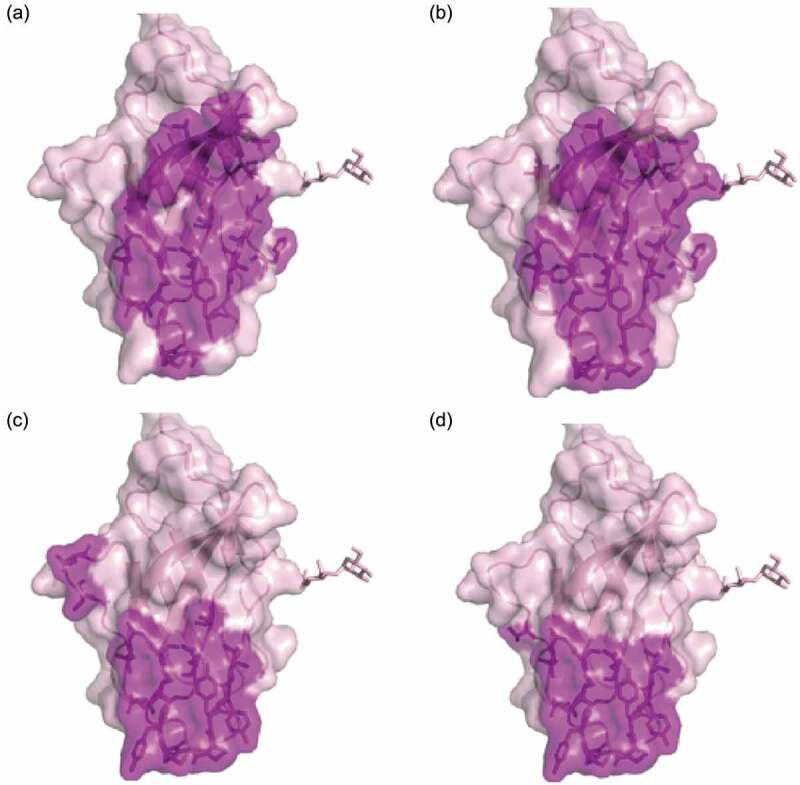
Hot spot residues or ligands binding sites mapped on CTLA-4 structure. Purple-shaded parts were identified as epitopes on human CTLA4. (a) Binding sites of Ipilimumab; (b) binding sites of antibody mAb146, the net indicates the N110 glycosylation site; (c) CD80 binding site (PDB code 1I8L); (d) CD86 binding site (PDB code 1I85).
The impact of N-glycan on antibody cross-species reactivity
The overlapping epitope of ipilimumab and mAb146 did not explain the unique cross-species binding of mAb146. As the N110A mutation on CTLA-4 only affected mAb146 binding to CTLA-4, not affecting ipilimumab, and N110 is predicted as an N-glycosylation site because it is in NGT motif, we further examined the N-glycosylation sites as a potential epitope of mAb146. The two possible N-glycosylation sites N78 and N110 on CTLA-4 were removed by single mutation of N78Q and N110Q and double mutation of N78Q/N110Q. The expression level of the mutants was significantly reduced, especially double mutation of N78Q/N110Q had almost no expression. However, the binding of ipilimumab on mutated CTLA-4 was not significantly changed (Figure 6a), consistent with the data of N110A. In contrast, the binding of mAb146 on mutated CTLA-4 N110Q was significantly reduced, whereas its binding on CTLA-4 N78Q did not change (Figure 6b).
Figure 6.
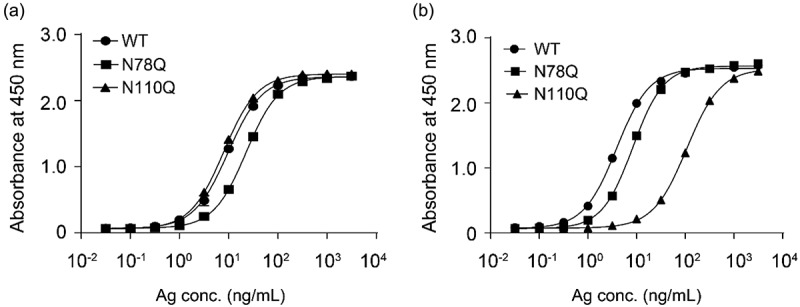
Ipilimumab (a) and mAb146 (b) bound to wide type human CTLA-4 and glycosylation sites mutated (N78Q and N110Q) CTLA-4. The N-glycosylation sites N78 and N110 on CTLA-4 monomer with His tag were removed by single mutation of N78Q, N110Q. These CTLA-4 variants and WT CTLA-4 were incubated with 1 μg/ml of antibodies pre-coated on a 96-well plate. The bound CTLA4 variants were detected by HRP-labeled secondary antibody. The absorbance at 450 nM was measured using a microplate spectrophotometer.
To further investigate whether mAb146 bound to the N110 residue or the glycans on N110, the N-glycan of CTLA-4 was removed using PNGase F, an amidase cleaving between the innermost GlcNAc and asparagine resides of oligosaccharides. Both His-tagged monomeric human CTLA-4 and Fc-tagged dimeric human CTLA-4 were used in this assay. As shown in Figure 7, mAb146 binding to deglycosylated CTLA-4 was significantly reduced, with a 22.3-fold increased EC50 for monomeric CTLA-4 (Figure 7b) and 6.3-fold increased EC50 for dimeric CTLA-4 (Figure 7d). In contrast, the binding of ipilimumab to deglycosylated CTLA-4 did not significantly change (Figure 7a,c).
Figure 7.
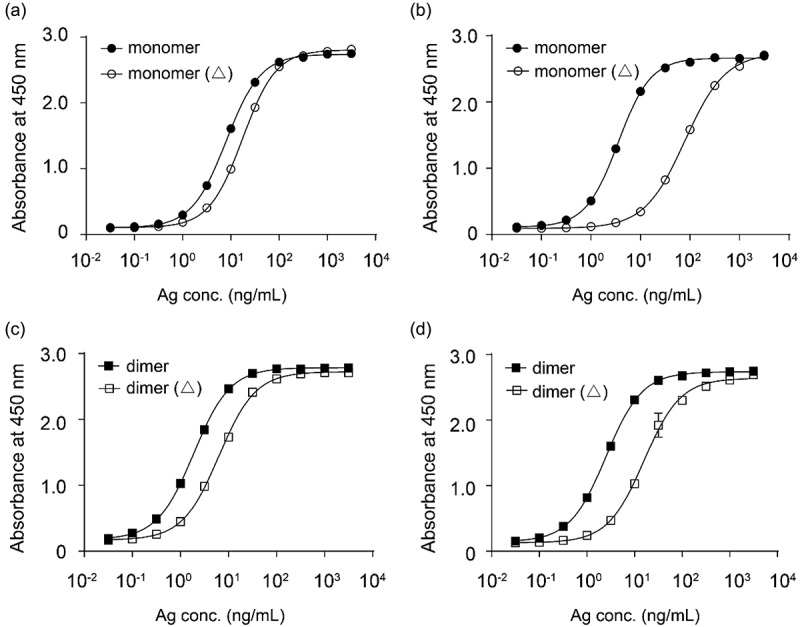
Ipilimumab (a, c) and mAb146 (b, d) bound to monomeric human CTLA-4 with or without deglycosylation (a, b), and dimeric human CTLA-4 with or without deglycosylation (c, d). Each of the antibodies at concentration of 1 μg/ml was coated on 96-well plate overnight for ELISA binding assay. After interacting with untreated or PNase F-treated CTLA-4 protein, HRP-labeled secondary antibody was added as detection antibody. The absorbance at 450 nM was measured using a microplate spectrophotometer.
This set of data confirmed that the N-glycan on N110 of CTLA-4 is part of the epitope of mAb146. Indeed, the N110 residue is conserved in CTLA-4 of cynomolgus monkey and mouse, suggesting the mechanistic cross-species binding of mAb146.
Antibody modeling and antibody-antigen docking
In order to investigate the mAb146-CTLA-4 interaction, modeling of mAb146 was conducted and its docking with hCTLA-4 and mCTLA-4 was examined. The overall structures of the complexes are shown in Figure 8. In this model, the binding interface of hCTLA-4 and mAb146 is formed by residues from the C, C’, F, G strands of the front β-sheet of hCTLA-4 and light chain LCDR 1 and LCDR2 and heavy chain HCDR 1, 2 and 3. One potential interaction involving LCDR3 is the 2.6 Å approach between W101 on LCDR3 and L106 on hCTLA-4. The MYPPPY motif on FG loop of hCTLA-4 interacts with the HCDR2 (Y51, S53 to Y60, L65). The residues (H27, T30 to D34) on HCDR1 form an interface with the C and C’ strands on hCTLA-4. The residues (M99 to Y105) on HCDR3, residues (N31 to N35, Y37) on LCDR1 and residues (V56 to K58) on LCDR2 interact with the G strand of hCTLA-4. The N-glycan on N110 likely inserts into the space between LCDR1 and HCDR3. Based on this complex model, the interactions between mCTLA-4 and mAb146 are very similar to these in the hCTLA-4-mAb146 complex.
Figure 8.
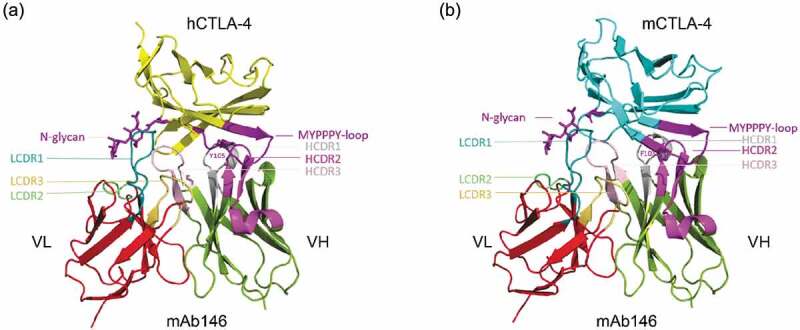
Structure model of the hCTLA-4:mAb146 and mCTLA-4:mAb 146 complexes. (a) The interface of hCTLA-4:mAb 146 complex is mainly composed of hCTHA-4 FG-loop (MYPPPY-loop, dark purple), N-glycan (dark purple) and mAb 146 CDRs. The FG loop mainly interacts with the HCDR2 (light purple). The HCDR1 (gray) mainly interacts with the C and C’ strands on hCTLA-4. The HCDR3 (pink), LCDR1 (cyan) and LCDR2 (light green) interact with the G strand, mainly the N-glycan. (b) The interface of mCTLA-4:mAb 146 is similar to that of hCTLA-4:mAb 146 complex. The color scheme is same as (A).
Overall, about 12 hydrogen bonds and 138 contacts less than 6 Å contribute to the hCTLA-4-mAb146 interface. The total surface area buried at the binding interface is about 1784.0 Å2.
As shown in Figure 8, mAb146 might bind to the MYPPPY motif and the N110 glycan. Details of mAb146’s binding with hCTLA-4 and N110 glycan are shown in Figure 9a,b. The LCDR1, LCDR2, and HCDR3 form a polar pocket and the N110 glycan points into it. P102, H103, Y104 on heavy chain and D33, G34, N35, V56, S57, K58 on light chain may contribute to the interaction.
Figure 9.
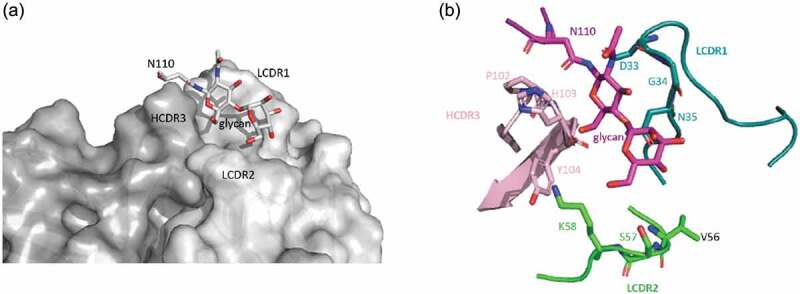
The binding of mAb146 with hCTLA-4 N110 glycan. (a) Surface representation of mAb 146 (heavy chain in dark gray; light chain in light gray) showing the pocket formed by HCDR3, LCDR1, and LCDR2. The N110 glycan inserts into the pocket. (b) Detailed interaction between the N110 glycan (purple) and mAb 146. P102, H103, Y104 on HCDR3 (pink), D33, G34, N35 on LCDR1 (cyan), and V56, S57, K58 on LCDR2 (green).
Comparing the hCTLA-4 and mCTLA-4 in docking models, we found a significant difference: the residue at position 105 is Y on hCTLA-4, but it is F on mCTLA-4. This position has no direct interaction with mAb146, while in the complex of hCTLA-4 and ipilimumab, Y105 directly interacts with LCDR3 (Fig. S2). In mCTLA-4, the residue F with opposite polarity is in this position, which can break its interactions with ipilimumab. This may explain why ipilimumab does not bind to mCTLA-4.
mAb146 specifically binds to CTLA-4, but not its homolog CD28 (data not shown). In order to understand this CTLA-4 specific binding, the structure of hCTLA-4 and human CD28 (hCD28) were superposed (Figure 10). Although the positions corresponding to the hCTLA-4 contact residues on hCD28 are almost the same, the different CC’ loop (HKGLDSAV on hCD28 and LRQADSQVT on hCTLA-4) may have different interactions with mAb146. On hCTLA-4, this loop interacts with mAb146 HCDR1 and HCDR3 as mentioned above. In contrast, a short loop on hCD28 may cause potential clash with mAb146. Another possible reason is that L106 and I108 on hCTLA-4 contribute to hydrophobic interactions with mAb146, while the corresponding residues on hCD28 are D and E, both hydrophilic residues. The physical property of the aforementioned residues may also contribute to the specificity of mAb146.
Figure 10.
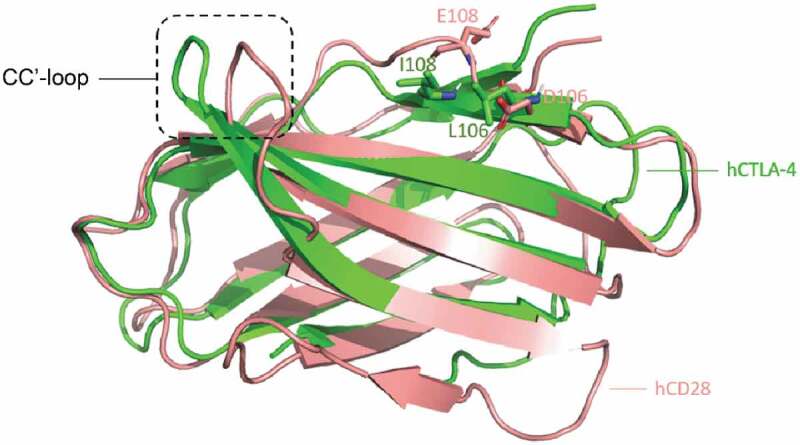
Structure comparison between human CTLA-4 and human CD28. The structures of hCTLA-4 (green) and hCD28 (salmon) are superposed. The main difference between these two is on the CC’-loop. On hCTLA-4, this loop interact with mAb146 HCDR1 and HCDR3, whereas a shorter loop on CD28 may cause potential clash. The L106 and I108 on hCTLA-4 and the corresponding residues D106 and E108 on CD28 are shown in sticks. The physical property of these residues may also contribute to the specificity of mAb146.
Discussion
To facilitate preclinical development of anti-CTLA-4 antibody, including testing different combinations with other immune checkpoint agonists or antagonists, we attempted to generate an antibody that cross-reacts to human, monkey, and murine CTLA-4. Although the extracellular sequence homology between human and murine CTLA-4 is low (67.5%), human CD80/CD86 can bind to both human and murine CTLA-4,19 indicating that hCTLA-4 and mCTLA-4 share structural homology. It has been reported that the proline-rich motif MYPPPY, present in the loops joining the F and G β-strands on both hCTLA-4 and mCTLA-4, is the key interface reacting to CD80 and CD86. After screening a large number of mAbs, we found that only one clone, mAb146, not only cross-reacted to hCTLA-4 and mCTLA-4, but also competed with CD80 and CD86. Furthermore, mAb146 demonstrated anti-tumor efficacy in a mouse tumor model, suggesting it may have potential as a therapeutic agent.
Although cross-reactive to human and murine CTLA-4, mAb146 did not bind to CD28, the homolog of CTLA-4. To understand the specificity and cross-reactivity of mAb146, we did epitope mapping using alanine scanning and protein modeling/docking. In addition to the expected MYPPPY motif, we were surprised to find that mAb146 bound to the N-glycan on N110. Glycosylation of proteins can influence and adjust protein structure and sometimes contribute to ligand recognition and downstream biologic activity. Recent studies have demonstrated that the alteration of antigen conformational equilibrium caused by glycosylation can influence the binding of antibody, and affect the sensitivity of cancer cells to chemotherapeutic agent and growth factors.2021
CTLA-4 is a disulfide-linked homodimeric glycoprotein with two N-linked glycosylation sites on each monomer, i.e., N78 and N110.22 The N78-glycosylation site was located in the E strand of the ABED β-sheet, while the N110 was located in the G strand of the CC’FG β-sheet on the opposite face of the IgV sandwich, and G2FS1 and G2FS2 were the major glycoforms identified by LC-MSE peptide mapping.2324
Different from the MYPPPY loop, which is the key binding interface for the interaction of the CTLA-4 homodimer with the CD80/CD86 ligands, the N-glycosylation sites may play indirect roles in mediating the interaction between CTLA-4 on T cells and CD80/CD86 on antigen-presenting cells. The N-glycosylation may maintain the orientation or spatial organization of CTLA-4.25–27 In a previous report, removal of the N78 glycosylation site in CTLA-4 by N78D mutation led to obvious aggregation and loss of CD80/CD86 ligand binding, while removal of the N110 glycosylation site in CTLA-4 did not significantly affect the CD80 and CD86 ligands binding.27 In our study, mAb146’s binding on N110Q mutant was significantly reduced, whereas its binding on N78Q mutant did not change, indicating that the conserved glycosylation site of N110 is likely part of epitope for mAb146 cross-species binding. When the N-glycan was removed from CTLA-4, we found mAb146 bound to glycan instead of the amino acid residue of N110.
As reported previously, hCTLA-4 can form homodimer in solution or on cell surfaces, which can bridge bivalent CD80 dimer to form a zipper-like oligomerization pattern.25 This could be important for the mechanism of action of CTLA-4. The N110-glycan of CTLA-4 is not on the exact interface between CTLA-4 and CD80/CD86. We suspect this glycan may extend the structure of CTLA-4 homodimer so the two CD80/CD86 binding sites on one CTLA-4 homodimer is long enough to prevent its binding to one CD80/CD86 homodimer. Due to the flexibility of the antibody hinge region, the two binding sites on mAb146 can either bind to one homodimer of CTLA-4 or two units each on two CTLA-4 homodimers. As shown in Figure 2, mAb146 had slightly weaker blocking activity than ipilimumab in ELISA-based competition assays, but mAb146 showed significantly better blocking activity than ipilimumab in FACS-based competition study. A possible reason is that mAb146 could alter the spatial structure of CTLA-4 dimer by binding on the N110 glycosylation, and more effectively break the CTLA-4 directed CD80/CD86 zipper-line-like oligomers. Additionally, mAb146 might inhibit CTLA-4 directed CD80/CD86 trans-endocytosis.
In this study, mAb146 was generated on the backbone of an effector-enabling human IgG1. Therefore, in addition to blockage of the interaction of CD80/CD86 ligands and CTLA-4, ADCC, and ADCP of mAb146 might play important roles in the antibody’s in vivo efficacy. Both human IgG1 and human IgG4 of mAb146 were evaluated in the syngeneic CT26 mouse model. The anti-tumor efficacy of mAb146-human IgG4 was dramatically impaired compared to mAb146-human IgG1 (Fig. S3), indicating that the anti-tumor activity of mAb146 might be mediated by the depletion of Tregs via ADCC and ADCP effects. This observation is consistent with recent studies showing that the anti-tumor efficacy of an anti-mCTLA-4 antibody with hamster IgG2 isotype was also mediated largely by ADCC effects on Tregs.578
The two anti-CTLA-4 antibodies, ipilimumab and tremelimumab, have similar binding properties,16 whereas mAb146 has unique mCTLA-4 cross-reactivity. The epitope comprising glycan on N110 may contribute to this unique binding. MAb146 may be used to facilitate preclinical studies of anti-CTLA-4 using mouse models, but also shed light on the role of N110 glycosylation in CTLA-4 dimerization and biologic function. To our knowledge, mAb146 is the first functional antibody reported to cross-react with human and murine CTLA-4, and, interestingly, target a unique epitope involving N-glycosylation.
Materials and methods
Immunization
The animal handling was conducted under the permission of WuXi Biologics animal care and use committee. hCTLA-4 and mCTLA-4 were used for immunization of SD rats purchased from Beijing Vital River Laboratory Animal Co. Briefly, three SD rats were immunized with 30 μg/animal of human and mouse CTLA-4 ECD protein in adjuvant Titer Max, once a week for 8 weeks. The anti-CTLA-4 titer of immunized serum was measured by ELISA every month. When the antibody titer was sufficiently high, the rat with the highest titer was given a final boost with human and mouse CTLA-4 ECD protein without adjuvant. After 4 days, the spleen and lymph nodes were taken from the rat, and the lymphocytes were separated for hybridoma generation.
Hybridoma generation and screening
The lymphocytes isolated from the lymph node of the immunized rat were mixed with SP2/0 myeloma cells at 1:1 ratio. The cell mixture was then washed and resuspended at 2 × 106 cells/ml in electric fusion solution and the electric cell fusion was conducted using Btx Electro Cell Manipulator (ECM 2001) following the manufacturer’s standard protocol. After fusion, the cell suspension was transferred into 96-well plates at 1 × 104 cells/well for clone formation and the cultural supernatants were collected for screening. Approximately 3,000 hybridoma clones were screened for binding to human, murine, and monkey CTLA-4 proteins, as well as engineered human CTLA-4-expressing cells. The cultural supernatants of selected positive clones were collected for purification and further characterization of the antibodies. The VH and VL genes of the selected hybridoma clones were isolated by RT-PCR or 5ʹ RACE and fused with constant region of human IgG1 for the chimeric antibodies production.
Humanization
CDR-grafting technique was used for humanization. Briefly, the CDRs and framework regions (FRs) of the variable regions of the antibodies were defined using the Kabat system. Based on the sequence homology and structural similarity, the gene of rat region FR1-3 was replaced by humanized region FR1-3, while the rat gene of the FR4 region was replaced by humanized FR4 region derived from JH and JK genes that had the most similar structures. After verifying the template sequence and codon optimization, the heavy chain variable region and light chain variable region were synthesized and cloned into an expression vector, and then used for expression of the humanized antibodies. The humanized antibodies were purified using Protein A chromatography, and were used for further in vitro and in vivo tests.
ELISA assay
A 96-well plate was coated with hCTLA-4-His tag monomer (1.0 μg/mL), cynomolgus monkey CTLA-4-His tag monomer (0.5 μg/mL) or mouse CTLA-4-His tag monomer (0.5 μg/mL) at 4°C for 16–20 h. After 1-h blocking with 2% bovine serum albumin (BSA) in Dulbecco’s phosphate-buffered saline (DPBS), test antibodies, as well as positive and negative control antibodies (prepared in house) were added to the plates and incubated at room temperature for 1 h. The binding of the antibodies to the plates was detected by horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Bethyl Laboratories, A80-119P, 1:5000) with 1-h incubation. The color was developed by dispensing 100 μL of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate, and then stopped by 100 μL of 2 M HCl. The absorbance at 450 nM was measured using a microplate spectrophotometer.
Competition with ligands
In the ELISA-based competition assay, plates were coated with hCTLA-4-hFc tag dimer or mCTLA-4-mFc tag dimer (0.5 μg/mL) at 4°C for 16–20 h. After 1-h blocking with 2% BSA in DPBS, test antibodies, as well as positive and negative control antibodies were pre-mixed with 0.25 μg/mL of hCD80-His tag (Sino Biological, Cat: 10698-H08H) or hCD86-His tag (Sino Biological, Cat: 10699-H08H), or 0.5 μg/ml of mCD80-His tag (Sino Biological, Cat: 50446-M08H) or 5 μg/ml of mCD86-His tag (Sino Biological, Cat: 50068-M08H), and then added to the plates and incubated at room temperature for 1 h. After washing, biotinylated anti-His tag antibody (Genscript, Cat. No. A00613, 1:2000–5000 dilution) was added. The plates were incubated at room temperature for 1 h. The bound ligands were detected by HRP-conjugated streptavidin (Invitrogen, 1:20000). The color was developed by dispensing TMB substrate and then stopped by 2 M HCl. The absorbance at 450 nM was measured using a microplate spectrophotometer.
For FACS-based competition assay, The CD80- and CD86-expressing Chinese hamster ovary cell lines were developed by WuXi Biologics. CD80- or CD86-expressing cells were added to each well of a 96-well plate at 1 × 105 per well. Serial dilutions of test antibodies, and positive and negative controls were mixed with biotinylated human CTLA-4-hFc tag dimer. Then the mixtures of antibody and CTLA-4 were added to the cells and incubated for 1 h at 4°C. The cells were washed two times with 200 μL FACS buffer (DPBS containing 1% BSA). Streptavidin PE (eBioscience) was added to the cells and incubated at 4°C for 1 h. Additional washing steps were performed two times. Finally, the cells were resuspended in 100 μL FACS buffer and fluorescence values were measured by flow cytometry and analyzed by FlowJo.
ADCC and ADCP assays
The ADCC assay was performed based on DELFIA® EuTDA Cytotoxicity Reagents (PerkinElmer-AD0116). Briefly, human CTLA4-expressing 293F cells were added to 96-well plates at 1 × 104 per well, and then various concentrations of antibodies pre-incubated with 5 × 105 peripheral blood mononuclear cells, collected from healthy donors after they provided informed consent, were added to the plates. The plates were kept at 37°C in a 5% CO2 incubator for 4 h. Lysis of the target cells was determined by DELFIA Europium Solution. The europium and the ligand form a highly fluorescent and stable chelate (EuTDA), and then the signal was read using SpectraMax® M5e. In ADCP assay, human monocytes were isolated using Human Monocyte Enrichment Kit (Miltenyi Biotec-130-050-201) and incubated with 100 ng/mL recombinant human M-CSF (R&D-216-MC) for the differentiation of macrophage. Then, macrophage cells were mixed with carboxyfluorescein succinimidyl ester-dyed engineered human CTLA-4 expressing 293F cells at 1:1 ratio; then, various concentrations of antibodies were added and cultured with cells at 37°C in a 5% CO2 incubator for 3 h. After wash, allophycocyanin-labeled anti-human CD14 antibody (1:100, eBioscience-17-0149-42) was added for detection. The phagocytosis rate of antibodies to the cells was tested by flow cytometry.
Animal model
The animal handling was conducted under the permission of WuXi Biologics animal care and use committee, The CT26 tumor cells were maintained in vitro as a monolayer culture in RPMI-1640 medium supplemented with 10% fetal bovine serum at 37°C in an atmosphere of 5% CO2. The tumor cells were routinely subcultured twice weekly after detaching the cells by trypsin-EDTA treatment. The cells growing in an exponential growth phase were harvested and counted for tumor inoculation. Female Balb/C mice were purchased from Beijing Vital River Laboratory Animal Co., Ltd. The mice at age 6–8 weeks with weight approximately 18–22 g were used for the study. Each mouse was inoculated subcutaneously at the right axillaries with 1 × 105 tumor cells in 0.1 mL of PBS mixed with 50 μL matrigel. When the average tumor volume reaches 60–80 mm3, the animals were randomly grouped (n = 6–8). The anti-CTLA-4 antibodies and human IgG1 isotype control were used for treatment. A functional anti-mCTLA-4 antibody, 9H10, purchased from BioXCell (BioXCell-BE0131), was used as a positive control. The tumor size was measured twice weekly by a vernier caliper, and tumor volume was calculated by the formula a× b2 × π/6 where a is length and b is width (a > b).
Epitope mapping
Alanine scanning experiments on hCTLA-4 were conducted and their effects on antibody binding were evaluated. The technical details of epitope mapping have been described previously.28 Briefly, a total of 72 positions on hCTLA-4 were identified. Most of them were located on the side interacting with ligands CD80 or CD86 based on the published structure of CTLA-4, and additional positions that might be buried were selected as control. The 72 mutations on hCTLA-4 monomer with His-tag were made: alanine residues on hCTLA-4 were mutated to glycine residues, and all other residues were mutated to alanine. The binding of these mutants to the antibodies was measured using ELISA.
Study of N-glycosylation as a binding epitope
The N-glycosylation sites N78 and N110 on CTLA-4 monomer with His tag were removed by single mutation of N78Q, N110Q, and double mutation of N78Q/N110Q and expressed in Expi293 cells (Thermo Fisher Scientific). Cells were cultured for 5 days and supernatant was collected for purification using Ni-NTA column (GE Healthcare). PNGase F, an amidase, was used for removing N-linked oligosaccharides from glycosylated CTLA-4 monomer with His tag and also dimer with mFc tag. Briefly, 200–500 μg CTLA-4 protein was incubated with 5 μL PNase F (New England Biolabs) at 37°C for 48 h, and purified using Ni-NTA column and Protein A column (GE Healthcare), respectively.
One μg/ml of each antibody was coated on 96-well plate overnight for ELISA binding assay. After interacting with gradient diluted above mutants or de-glycosylated CTLA-4, HRP-labeled secondary antibody was added as detection antibody. The absorbance at 450 nM was measured using a microplate spectrophotometer.
Antibody modeling and docking
The structure model of mAb146 was generated using the most recently described MODELER29 homology modeling protocol integrated in BIOVIA Discovery Studio. Several templates were chosen from the Protein Databank (PDB). The template for heavy chain was 4LEX, for light chain was 4DTG, and for orientation was 4LEX. Following the MODELER protocol, the loop refinement protocol in Discovery Studio was carried out to improve the modeling of heavy chain CDR3. Default parameters were used in both protocols. We chose the best score model as the mAb146 model.
The hCTLA-4 structure was extracted from a complex structure of ipilimumab-scFV and hCTLA-4 (PDB ID: 5XJ3). The mCTLA-4 structure was selected from PDB entry 5E56. These structures and the generated model of mAb146 were submitted to ZDOCK30 in Discovery Studio to make the molecular complex docking. The alanine scanning experiment data were provided as restraints. The best score docking results were retained and visually checked using Pymol.
A 20ns molecular dynamics (MD) simulation was conducted for further optimizing the mAb146 and hCTLA-4 complex model. The glycosylation parameters for simulation were generated by doGlycans.31 Gromacs32 was used to perform the MD simulation. All input files for MD simulation of the docked conformation were generated by CHARMM-GUI.33 The complex was simulated with CHARMM force field at 300K.
The amino acid residues involved in complex interaction were explored by COCOMAPS.34 A maximum distance was set to 6 Å to report atom–atom interactions within this cutoff limit.
Funding Statement
This work was supported by the Shanghai Rising-Star Program [19QB1406000].
Acknowledgments
We greatly thank Fangfang Luo, Xiaofeng Lu, Yingbo Zhang, Hui Meng, Yuanyuan Ding, Chenghu Wang, Lijun Ge and Jianqing Xu for their technical assistances.
Disclosure of potential conflicts of interest
The authors are the employees of WuXi Biologics. The project was sponsored by Shanghai Rising-Star Program (Grant No. 19QB1406000).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Park J, Kwon M, Shin EC.. Immune checkpoint inhibitors for cancer treatment. Arch Pharm Res. 2016;39:1577–12. doi: 10.1007/s12272-016-0850-5. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi OS, Zheng Y, Nakamura K, Attridge K, Manzotti C, Schmidt EM, Baker J, Jeffery LE, Kaur S, Briggs Z, et al. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science. 2011;332:600–03. doi: 10.1126/science.1202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowshanravan B, Halliday N, Sansom DM. CTLA-4: a moving target in immunotherapy. Blood. 2018;131:58–67. doi: 10.1182/blood-2017-06-741033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang F, Du X, Liu M, Zheng P, Liu Y. Anti-CTLA-4 antibodies in cancer immunotherapy: selective depletion of intratumoral regulatory T cells or checkpoint blockade? Cell Biosci. 2018;8:30. doi: 10.1186/s13578-018-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210:1695–710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingram JR, Blomberg OS, Rashidian M, Ali L, Garforth S, Fedorov E, Fedorov AA, Bonanno JB, Le Gall C, Crowley S, et al. Anti-CTLA-4 therapy requires an Fc domain for efficacy. Proc Natl Acad Sci USA. 2018;115:3912–17. doi: 10.1073/pnas.1801524115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arce Vargas F, Furness AJS, Litchfield K, Joshi K, Rosenthal R, Ghorani E, Solomon I, Lesko MH, Ruef N, Roddie C, et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell. 2018;33:649–63 e4. doi: 10.1016/j.ccell.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romano E, Kusio-Kobialka M, Foukas PG, Baumgaertner P, Meyer C, Ballabeni P, Michielin O, Weide B, Romero P, Speiser DE, et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc Natl Acad Sci USA. 2015;112:6140–45. doi: 10.1073/pnas.1417320112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ipilimumab. Drugs R D. 2010;10:97–110. doi: 10.2165/11584510-000000000-00000. https://www.ncbi.nlm.nih.gov/pubmed/?term=Ipilimumab.+Drugs+R+D.+2010%3B10%3A97%E2%80%93110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J Exp Clin Cancer Res. 2019;38:255. doi: 10.1186/s13046-019-1259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremelimumab. Drugs RD. 2010;10:123–32. doi: 10.2165/11584530-000000000-00000. https://www.ncbi.nlm.nih.gov/pubmed/?term=Tremelimumab.+Drugs+RD.+2010%3B10%3A123%E2%80%9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonia S, Goldberg SB, Balmanoukian A, Chaft JE, Sanborn RE, Gupta A, Narwal R, Steele K, Gu Y, Karakunnel JJ, et al. Safety and antitumour activity of durvalumab plus tremelimumab in non-small cell lung cancer: a multicentre, phase 1b study. Lancet Oncol. 2016;17:299–308. doi: 10.1016/S1470-2045(15)00544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahig H, Aubin F, Stagg J, Gologan O, Ballivy O, Bissada E, Nguyen-Tan F-P, Soulières D, Guertin L, Filion E, et al. Phase I/II trial of durvalumab plus tremelimumab and stereotactic body radiotherapy for metastatic head and neck carcinoma. BMC Cancer. 2019;19:68. doi: 10.1186/s12885-019-5266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fumet JD, Isambert N, Hervieu A, Zanetta S, Guion JF, Hennequin A, Rederstorff E, Bertaut A, Ghiringhelli F. Phase Ib/II trial evaluating the safety, tolerability and immunological activity of durvalumab (MEDI4736) (anti-PD-L1) plus tremelimumab (anti-CTLA-4) combined with FOLFOX in patients with metastatic colorectal cancer. ESMO Open. 2018;3:e000375. doi: 10.1136/esmoopen-2018-000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JY, Kim JW, Lim MC, Kim S, Kim HS, Choi CH, Yi JY, Park S-Y, Kim B-G. A phase II study of neoadjuvant chemotherapy plus durvalumab and tremelimumab in advanced-stage ovarian cancer: a Korean Gynecologic Oncology Group Study (KGOG 3046), TRU-D. J Gynecol Oncol. 2019;30:e112. doi: 10.3802/jgo.2019.30.e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He M, Chai Y, Qi J, Zhang CWH, Tong Z, Shi Y, Yan J, Tan S, Gao GF. Remarkably similar CTLA-4 binding properties of therapeutic ipilimumab and tremelimumab antibodies. Oncotarget. 2017;8:67129–39. doi: 10.18632/oncotarget.18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, Korman AJ. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res. 2013;1:32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- 18.Fiegle E, Doleschel D, Koletnik S, Rix A, Weiskirchen R, Borkham-Kamphorst E, Kiessling F, Lederle W. Dual CTLA-4 and PD-L1 blockade inhibits tumor growth and liver metastasis in a highly aggressive orthotopic mouse model of colon cancer. Neoplasia. 2019;21:932–44. doi: 10.1016/j.neo.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Guo Q, Guo H, Liu T, Zheng Y, Gu P, Chen X, Wang H, Hou S, Guo Y, et al. Versatile characterization of glycosylation modification in CTLA4-Ig fusion proteins by liquid chromatography-mass spectrometry. mAbs. 2014;6:1474–85. doi: 10.4161/mabs.36313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bongers J, Devincentis J, Fu J, Huang P, Kirkley DH, Leister K, Liu P, Ludwig R, Rumney K, Tao L, et al. Characterization of glycosylation sites for a recombinant IgG1 monoclonal antibody and a CTLA4-Ig fusion protein by liquid chromatography-mass spectrometry peptide mapping. J Chromatogr A. 2011;1218:8140–49. doi: 10.1016/j.chroma.2011.08.089. [DOI] [PubMed] [Google Scholar]
- 21.Chikuma S. CTLA-4, an essential immune-checkpoint for T-cell activation. Curr Top Microbiol Immunol. 2017;410:99–126. doi: 10.1007/82_2017_61. [DOI] [PubMed] [Google Scholar]
- 22.Movahedin M, Brooks TM, Supekar NT, Gokanapudi N, Boons GJ, Brooks CL. Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen. Glycobiology. 2017;27:677–87. doi: 10.1093/glycob/cww131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peiris D, Spector AF, Lomax-Browne H, Azimi T, Ramesh B, Loizidou M, Welch H, Dwek MV. Cellular glycosylation affects Herceptin binding and sensitivity of breast cancer cells to doxorubicin and growth factors. Sci Rep. 2017;7:43006. doi: 10.1038/srep43006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C, Sonnen AF, George R, Dessailly BH, Stagg LJ, Evans EJ, Orengo CA, Stuart DI, Ladbury JE, Ikemizu S, et al. Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering. J Biol Chem. 2011;286:6685–96. doi: 10.1074/jbc.M110.182394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamper CC, Zhang Y, Tobin JF, Erbe DV, Ikemizu S, Davis SJ, Stahl ML, Seehra J, Somers WS, Mosyak L, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–11. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 26.Ramagopal UA, Liu W, Garrett-Thomson SC, Bonanno JB, Yan Q, Srinivasan M, Wong SC, Bell A, Mankikar S, Rangan VS, et al. Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc Natl Acad Sci USA. 2017;114:E4223–E32. doi: 10.1073/pnas.1617941114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzler WJ, Bajorath J, Fenderson W, Shaw SY, Constantine KL, Naemura J, Leytze G, Peach RJ, Lavoie TB, Mueller L, Linsey PS. Solution structure of human CTLA-4 and delineation of a CD80/CD86 binding site conserved in CD28. Nat Struct Biol. 1997;4:527–31. doi: 10.1038/nsb0797-527. [DOI] [PubMed] [Google Scholar]
- 28.Li D, Xu J, Wang Z, Gong Z, Liu J, Zheng Y, Li J, Li J. Epitope mapping reveals the binding mechanism of a functional antibody cross-reactive to both human and murine programmed death 1. mAbs. 2017;9:628–37. doi: 10.1080/19420862.2017.1296612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–91. [DOI] [PubMed] [Google Scholar]
- 30.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danne R, Poojari C, Martinez-Seara H, Rissanen S, Lolicato F, Rog T, Vattulainen I. doGlycans-tools for preparing carbohydrate structures for atomistic simulations of glycoproteins, glycolipids, and carbohydrate polymers for GROMACS. J Chem Inf Model. 2017;57:2401–06. doi: 10.1021/acs.jcim.7b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutzner C, Pall S, Fechner M, Esztermann A, de Groot BL, Grubmuller H. More bang for your buck: improved use of GPU nodes for GROMACS 2018. J Comput Chem. 2019. doi: 10.1002/jcc.26011. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12:405–13. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vangone A, Spinelli R, Scarano V, Cavallo L, Oliva R. COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics. 2011;27:2915–16. doi: 10.1093/bioinformatics/btr484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


