ABSTRACT
Antibodies from B-cell clonal lineages share sequence and structural properties as well as epitope specificity. Clonally unrelated antibodies can similarly share sequence and specificity properties and are said to be convergent. Convergent antibody responses against several antigens have been described in humans and mice and include different classes of shared sequence features. In particular, some antigens and epitopes can induce convergent responses of clonally unrelated antibodies with restricted heavy (VH) and light (VL) chain variable region germline segment usage without similarity in the heavy chain third complementarity-determining region (CDR H3), a critical specificity determinant. Whether these V germline segment-restricted responses reflect a general epitope specificity restriction of antibodies with shared VH/VL pairing is not known. Here, we investigated this question by determining patterns of antigen binding competition between clonally unrelated antigen-specific rat antibodies from paired-chain deep sequencing datasets selected based solely on VH/VL pairing. We found that antibodies with shared VH/VL germline segment pairings but divergent CDR H3 sequences almost invariably have restricted epitope specificity indicated by shared binding competition patterns. This epitope restriction included 82 of 85 clonally unrelated antibodies with 13 different VH/VL pairings binding in 8 epitope groups in 2 antigens. The corollary that antibodies with shared VH/VL pairing and epitope-restricted binding can accommodate widely divergent CDR H3 sequences was confirmed by in vitro selection of variants of anti-human epidermal growth factor receptor 2 antibodies known to mediate critical antigen interactions through CDR H3. Our results show that restricted epitope specificity determined by VH/VL germline segment pairing is a general property of rodent antigen-specific antibodies.
KEYWORDS: B cell lineages, binning, convergence, redundancy, next-Gen Sequencing, NGS
Introduction
Sequence and structural diversity in immunoglobulin variable regions enable antibodies to bind a virtually unlimited number of antigenic structures. Sequence diversity is generated somatically during the process of B cell maturation by recombination of germline-encoded heavy and light chain germline gene segments into full-length functional variable region exons.1 Light chain variable region sequence diversity is generated by recombination of VL and JL gene germline segments along with relatively limited junctional diversity generated by nucleotide nibbling and incorporation between these segments. By contrast, heavy chain variable region sequence diversity is generated by recombination of three germline segments, VH, DH and JH, which, along with significant nucleotide nibbling and incorporation, generates high sequence and length diversity in the third complementarity-determining region of the heavy chain (CDR H3).2,3 The CDR H3 region is located centrally in the interface between antibody and antigen and usually provides critical contacts with the antigen. The CDR H3 region is considered to be a major determinant of antibody specificity due to its high sequence diversity and central role in antigen binding.1,4
Immune repertoires, while highly diverse, have a significant degree of redundancy in specificities. This redundancy is mostly due to expansion of B cell clones harboring antigen-specific B cell receptors.5 Somatic mutation and clonal selection results in antibody clones with differing affinities for antigen and sometimes differing fine specificity for homologous antigens. However, these clonally related antibodies have essentially the same binding mode, defined as overall occluded antigen surface and binding geometry. Another level of redundancy is generated by what has been termed clonal convergence, in which antibodies from different B cell lineages share sequence elements that allow, or are presumed to allow, binding to the same antigen and epitope with similar binding modes.6-9 Sequence features that are conserved in convergent antibodies vary and include shared VH and/or VL chain germline segment use, CDR H3 sequences or a combination of these. Convergent antibody responses have been observed in human and rodent immune responses to specific antigens and epitopes, including proteins, bacterial polysaccharides and viral antigens, allergens, blood group antigens, and haptens.6,8,10-24 Antibody convergence is now a major principle behind rational human immunodeficiency virus type 1 vaccine design.25
A relatively common convergence class includes groups of clonally unrelated antibodies with recurrent VH and VL germline segment pairing without CDR H3 sequence similarity or length conservation that bind certain antigens or epitopes.8,10-18 These convergences have been described as VH/VL germline segment usage restrictions as a function of antigen or epitope specificity.12-18 Whether these convergent responses are special cases in which certain V region germline segments are particularly suited for binding to certain epitopes independently of CDR H3 diversity or are examples of more widespread VH/VL germline segment convergences within antigen-specific repertoires is not clear. Here, we show that multiple epitopes in an antigen can induce antibodies with convergent VH/VL germline segment pairing in immunized rats. In addition, we show that shared VH/VL germline segment use in clonally unrelated antigen-specific antibodies is sufficient to predict shared epitope specificity with high confidence regardless of CDR H3 sequence and length, indicating that VH/VL germline segment pairing strongly restricts epitope specificity. Finally, we show by means of in vitro selection that the CDR H3 sequence and functional redundancy implied by widespread VH/VL germline segment pairing convergences can include clones with critical antigen contacts mediated by CDR H3. Our results extend our understanding of the role of V germline segment pairing and CDR H3 sequence diversity in determining epitope specificity in natural immune responses and provide insights about the organization of natural immune repertoires beyond clonal expansion.
Results
Germline segment pairing convergences are readily observed in a large anti-chicken ovalbumin antibody panel
A previously described panel of rat antibodies specific for chicken ovalbumin (OVA) generated by single-cell B-cell receptor sequencing (scBCR-seq) of antigen-sorted IgMneg B cells pooled from 3 immunized rats was used in this study.26 This panel includes 93 OVA-specific clones from different clonal lineages selected without regard for germline segment use, CDR sequence or length. As previously reported, the monovalent equilibrium dissociation constant (KD) of the clones by surface plasmon resonance (SPR) ranged from less than 10 pM to 30 nM.26 The scBCR-seq dataset includes 670 additional uncharacterized lineages, each lineage defined here as clones with the same heavy and light chain V and J germline segments, the same CDR H3 length, and at least 80% nucleotide identity within CDR H3 (IMGT® CDR H3 definition27) between clones. We initially searched for correlations between sequence features and epitope specificity assessed by cross-competition binding experiments in an SPR format. In this assay, binding competition between two antibodies is determined by testing the binding of a “probe” antibody to immobilized antigen bound to a “capture” antibody (Suppl. Fig. 1). A subset of 18 clones showed cross-competition with several antibodies in different epitope groups (Suppl. Fig. 2), probably caused by recognition of alternative OVA conformational states.28 These antibodies did not share VH/VL pairing with any other clones or amongst themselves and were excluded from further analysis. Six antibodies did not show detectable binding in this assay and were also excluded (data not shown).
Competition data was obtained for 71 of the clones, with at least 28 epitope groups identified for clones in competition assays (Suppl. Fig. 2). Eighteen of these clones could be classified in 7 groups of antibodies with recurring VH/VL germline segment pair use, ranging from 2 to 4 clones per group (Figure 1 and Suppl. Fig. 2, color-coded OVA clones). Within 6 of the 7 VH/VL pair groups, all clones cross-competed with each other in 4 epitope groups and, with some exceptions, behaved similarly in cross-competitions with other clones in the panel (Figure 1, Suppl. Fig. 2), indicating similar epitope specificity within each group. The exceptions were within the range of uncertainty for this technique, which may occasionally disagree for nearly identical clonally related antibodies, usually observed as patterns discordant between reciprocal competitions (e.g., clones OVA-80 and OVA-96, OVA-70 and OVA-13). Only one pair of clones with shared VH/VL germline segment pairing, clones OVA-36 and OVA-46, had cross-competition patterns with other antibodies clearly different from each other (Figure 1 and Suppl. Fig. 2, black background). We refer to clones with shared VH/VL pairing and epitope specificity as “parallel lineages” for simplicity.
Figure 1.
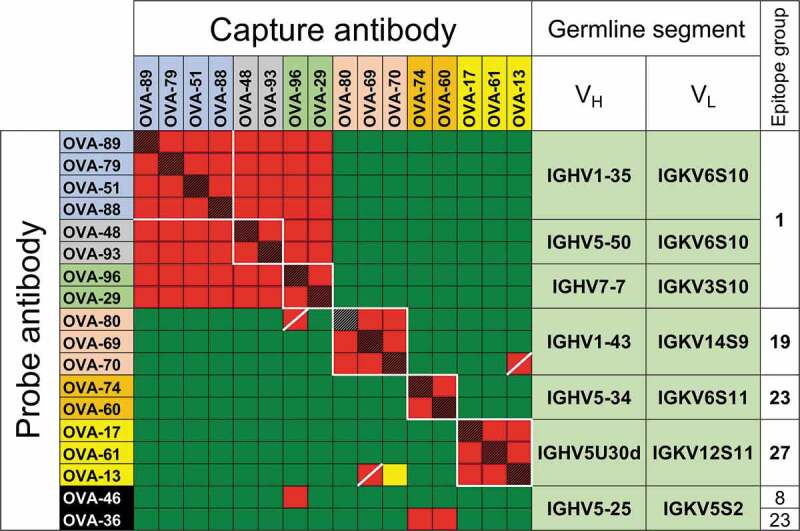
Competitive binding between anti-OVA antibodies. Capture and probe antibodies refer to antibodies in solid phase used to capture OVA and antibodies in solution phase in the assay used to determine cross-competition, respectively. Red and red/black cross-hatched squares indicate no binding of the probe antibody. Green squares indicate binding of the probe antibody. Yellow squares indicate weak binding of probe antibody. OVA-36 and 46, highlighted in black, share IGHV5-25 and IGKV5S2 germline segment use but have distinct binding competition patterns. Groups of squares highlighted with white lines indicate competition of clones with the same VH/VL germline segment pairing. Names of antibodies within each VH/VL germline segment group are color-coded as in Suppl. Figure 2. Only antibodies with competition data as both capture and probe reagents as well as OVA-36 and 46 are shown, with the full dataset shown in Suppl. Figure 2.
Sequences of antibodies within the parallel lineages were analyzed in detail to confirm different clonal origins. As expected, since these clones were originally selected to represent unique B-cell lineages, no significant CDR H3 sequence similarity was observed between clones within groups, with pairwise amino acid identities between clones with the same CDR H3 length below 67% (Figure 2). The independent clonal origin of the antibodies with the same CDR H3 length within groups were also confirmed by junctional DNA analysis (Suppl. Fig. 3). Our results show that convergences in the form of parallel lineages are readily apparent in a sufficiently large antibody panel.
Figure 2.
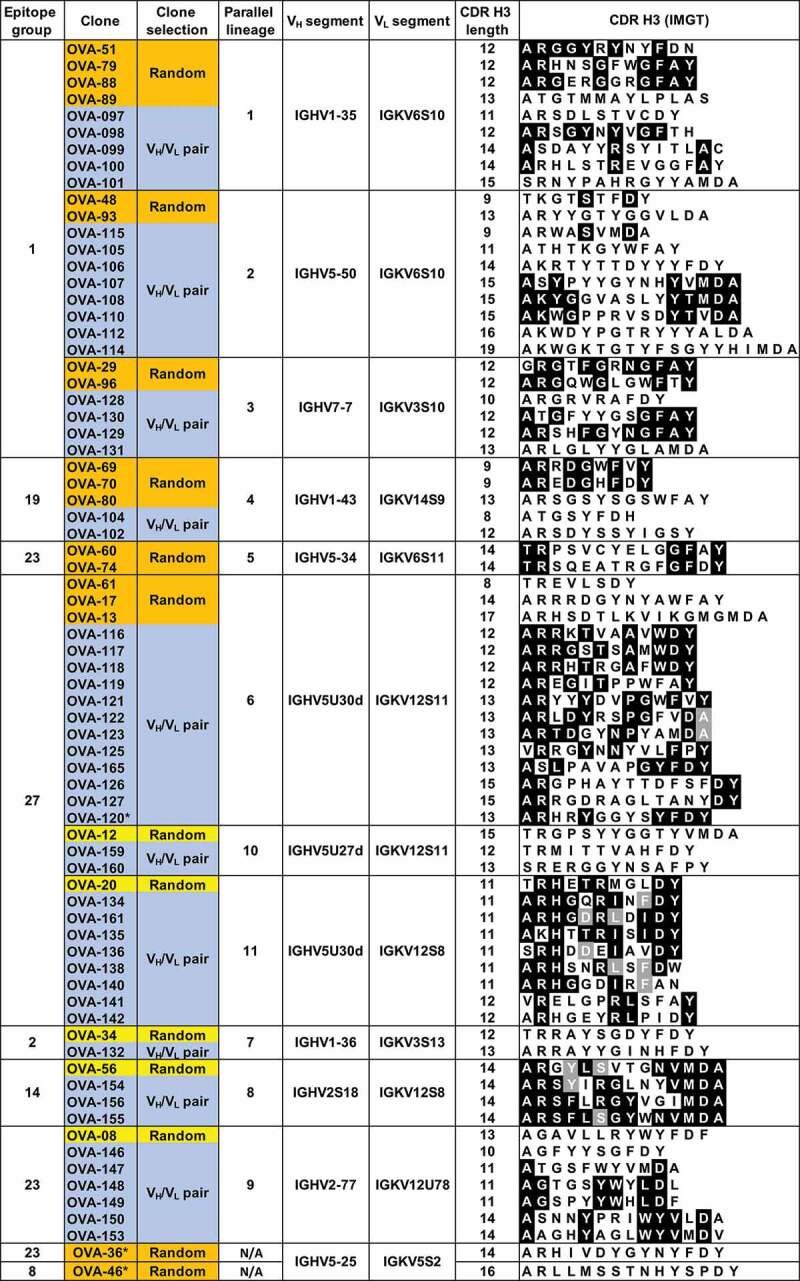
VH and VL germline segments and CDR H3 sequences of anti-OVA clones in parallel lineages. Amino acid identities between 2 or more clones within the same parallel lineage with the same CDR H3 length are shown in black and gray backgrounds. Randomly selected clones that were and were not part of parallel lineages in the initial antibody panel are highlighted in orange and yellow, respectively. Clones selected based on VH/VL germline segment pair use are highlighted in blue.
VH/VL germline segment pairing restricts epitope specificity of anti-OVA antibodies
The observation that most clones with shared VH/VL germline segment pairing recognize the same epitope suggests epitope specificity is restricted by VH/VL germline segment use. However, although unlikely, it is possible that the epitope restriction observed in the analyzed panel of 93 clones is due to chance. To test whether epitope restriction is determined by VH/VL germline segment pairing, additional clonally unrelated antibodies were sampled from the scBCR-seq dataset based solely on VH and VL germline segment use. Two classes of clones were selected. The first class includes clones with the same VH/VL germline segment pairings as clones in 5 of the 6 parallel lineages identified above. A second class of clones had the same VH/VL pairings as clones that were not part of parallel lineages in the initial panel (Figure 2, orange and yellow backgrounds, respectively, and Suppl. Fig. 2). These included clones in 2 epitope groups (2 and 14) not recognized by antibodies from parallel lineages. The only information used for clone selection from the scBCR-seq dataset was VH and VL germline segment use and independent clonal lineage. A total of 51 clonally unrelated OVA-specific antibodies covering 6 epitope groups and 10 VH/VL germline segment combinations were selected (Figure 2, blue background). Clones within the same parallel lineage with identical CDR H3 length had pairwise CDR H3 amino acid sequence identity of less than 72% (Suppl. Fig. 4). Clones with the same VH/VL germline segment pairing, CDR H3 length and JH and JL germline segments were confirmed to belong to different lineages by CDR H3 sequence identity and junctional analysis (Figure 2 and Suppl. Fig. 3). Affinities of selected clones ranged from 30 pM to 78 nM, within the same range as the clones picked randomly (Suppl. Fig. 5).
Forty-one of the clones were tested in two-way competitions with clones in other parallel lineages, as well as with clones selected to differentiate between similar epitope groups (Figure 3). The other 10 clones were tested in one- or two-way competitions with a subset of the clones in the same or other parallel lineages (Suppl. Fig. 6A and B). All clones within each of the putative parallel lineages showed restricted epitope specificity indicated by identical or nearly identical cross-competition patterns with antibodies in different epitope groups. As above, the observed discrepancies were limited to one-way patterns, typical of noise in panels of this size. Of the 56 antibodies within putative parallel lineages (51 selected plus 5 clones from the original panel not initially known to be in parallel lineages), only 5 clones showed patterns in two-way competitions that differed from other clones within the same parallel lineages. Four of these clones, OVA-100, OVA-105, OVA-104 and OVA-117, showed differences limited to 4 cross-competition data points relative to other members of their parallel lineages in two-way competitions with antibodies in other parallel lineages (Suppl. Fig. 7). However, clone OVA-120 showed patterns of cross-competitions different from other clones in its putative parallel lineage, although many of the discrepancies were only observed in one-way competition patterns (Figure 3). The atypical cross-competition pattern of clone OVA-120 was not related to polyspecificity; score of OVA-120 in a baculovirus virus particle ELISA (BV ELISA) to detect polyspecificity was 1.82, lower than the score of 5 for polyspecific antibodies.29 Combined with the reference antibodies in the initial panel, 67 of the 74 (91%) anti-OVA antibodies with repeated VH/VL germline segment pairs showed virtually identical cross-competition patterns within each parallel lineage, 4 clones (5%) showed minor intra-group differences and only 3 clones (4%) showed competition patterns incongruent with VH/VL pairing.
Figure 3.

Binding competitions between anti-OVA antibodies in parallel lineages. Red and red/black cross-hatched squares indicate no binding of the probe antibody. Green squares indicate binding of the probe antibody. Yellow and gray squares indicate weak and indeterminate binding of probe antibody, respectively. Squares with diagonal bars indicate discordant one-way competition results within parallel lineages. Clones are highlighted in orange, yellow and blue as in Figure 2. Clones with white background are not part of parallel lineages and were used to discriminate between epitope groups. Clone OVA-164 is a clonal variant of OVA-64. Competition patterns for reference antibodies of parallel lineages 8, 9 and 10 are shown in Suppl. Fig. 6B.
VH/VL germline segment pairing restricts epitope specificity of antibodies against hen egg-white lysozyme
We extended our findings to a second antigen, hen egg-white lysozyme (HEL). An anti-HEL scBCR-seq dataset was obtained by similar methods used to derive the anti-OVA dataset. Significant nonspecific binding of labeled HEL was observed in sorting of B cells prior to sequencing (data not shown), resulting in only 20 of 96 selected clones showing binding to HEL by ELISA (Suppl. Fig. 8 and Suppl. File 1). Fourteen of these antibodies, along with 6 previously described anti-HEL mouse clones with known epitopes,8,30-33 were tested in SPR-based competition assays. Reference mouse clones competed with each other as expected (Figure 4(a,b)). The HEL-specific rat clones derived from the scBCR-seq dataset bound in 11 overlapping epitope groups (Figure 4(a)). No recurring VH/VL germline segment pairs were observed in this relatively small panel of antibodies (Suppl. Table 1).
Figure 4.
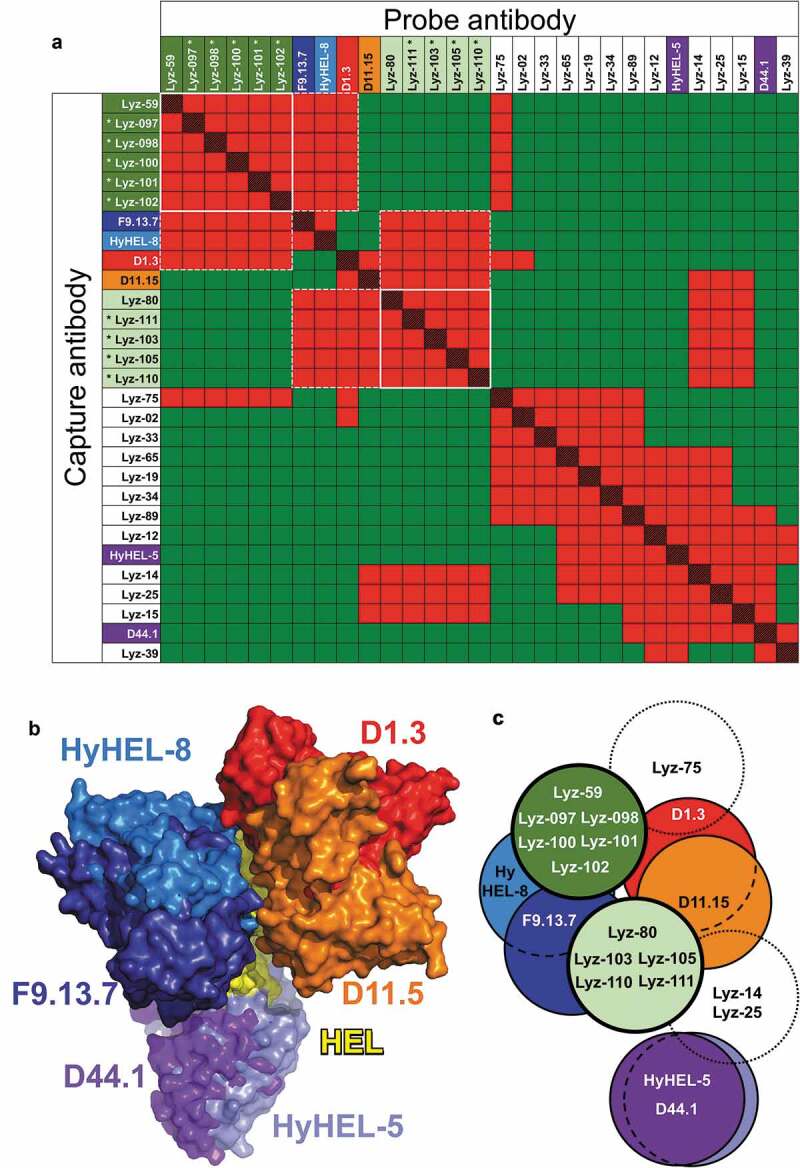
Epitope mapping of anti-HEL antibodies. (a) Competition patterns of anti-HEL clones tested. Red and red/black cross-hatched squares indicate no binding of the probe antibody. Green squares indicate binding of the probe antibody. Mouse antibodies of known epitope specificity are highlighted in shades of blue, orange, red and purple. Anti-HEL clones selected based on VH/VL pairing are shown with asterisks. White lines and dotted lines indicate cross-competition with other clones in parallel lineages and mouse anti-HEL antibodies. (b) Composite structure of HEL bound to reference mouse antibodies (PDB 1YQV, 1FDL, 1FBI, 1MLC, 1NDG and 1JHL). Variable regions of antibodies are color-coded as in panel a. (c) Graphical representation of epitope groups of antibodies in parallel lineages, mouse anti-HEL antibodies and additional selected rat anti-HEL antibodies. Antibodies and parallel lineages colored as in panels a and b.
Two antibodies with different VH/VL germline segment pairs, Lyz-59 and Lyz-80, were selected to predict additional candidates binding the same epitopes based solely on VH/VL pairing data. Both antibodies compete with F9.13.7 and Hy-HEL-8. While Lyz-80 competes with both D11.15 and D1.3, which have closely overlapping epitopes, Lyz-59 does not compete with D11.15 (Figure 4(a–c)). Thus, subtle differences in competition between these two antibodies provide a more precise prediction of their cross-competition properties. A total of 5 and 4 lineages with HEL-specific antibodies with the same VH/VL germline segment pairing as clones Lyz-59 and Lyz-80, respectively, were identified, and one clone was selected from each lineage (Suppl. Table 1, Suppl. File 1). CDR H3 lengths ranged from 14 to 20 and 10 to 15 residues for clones in the Lyz-59 and Lyz-80 groups, respectively (Suppl. Table 1). Amino acid identity between 3 pairs of clones with the same CDR H3 length (Lyz-097/098, Lyz-100/101 and Lyz-80/111) ranged from 33% to 53%. These were also confirmed to belong to unique B cell lineages by junctional analysis (data not shown). Clones Lyz-80 and Lyz-111 shared an amino acid motif (TMGI) encoded by a rat DH segment but shifted by one position, and some clones in the Lyz-59 group shared some similarities created by junctional variations of IGHD1-12 and IGHJ4, with IGHD1-12 in different registers (Suppl. Fig. 3 and Suppl. Table 1). All 9 clones within each parallel lineage showed identical patterns of cross-competition as the Lyz-59 and Lyz-80 clones (Figure 4(a,c)). This included identical competition patterns of the Lyz-59 group of clones with the reference D1.3 and D11.15 mouse antibodies with closely overlapping epitopes (Figure 4(b,c)). Our results indicate that restricted epitope specificity of antibodies sharing VH/VL germline segment use occurs for different antigens.
Synthetic parallel lineages of clones mediating critical interactions with antigen through CDR H3
The fact that convergences are readily detectable in large antibody panels in the form of parallel lineages, and the apparent epitope restriction of antibodies with shared VH/VL use, suggest that CDR H3 in parallel lineage clones may not play a critical role in antigen binding, allowing a wide range of possible CDR H3 sequences compatible with epitope binding. Alternatively, the diverse CDR H3 sequences may represent structurally distinct solutions for critical antigen interactions with a given epitope and binding mode. The latter possibility would imply that antibodies known to mediate critical antigen interactions through CDR H3 may accept distinct CDR H3 sequences for epitope binding while retaining epitope specificity.
We tested this with two humanized anti-human epidermal growth factor receptor 2 (Her2) antibodies binding different epitopes, hu4D5 and hu2C4, that are known to mediate critical antigen interactions through CDR H3.34-37 The CDR H3 surface area buried in complex structures is 187 Å2 for hu4D5 and 201 Å2 for hu2C4. In hu2C4, mutation of residues Asn-108, Leu-109, Pro-111 or Tyr-114 (numbering according to the International ImMunoGeneTics information system®, IMGT®27) to Ala individually results in reductions of about 100-fold in binding.35 In hu4D5, the Trp-107 and Tyr-113 residue side-chains contact Her2 through hydrophobic interactions.36 Both residues make large contributions to the free energy of binding, with ΔΔG values of 5.9 and 5.6 kcal/mol when individually mutated to Ala.34
Variants of hu4D5 and hu2C4 were obtained from phage display libraries with randomized CDR H3 sequences, followed by testing of epitope specificity and affinity of variants with diverse CDR H3 sequences. Two sets of libraries were made for each clone. One design randomized CDR H3 sequences with NNK codons whereas the other used a mixture of trinucleotide codons (Suppl. Fig. 6). Both libraries were designed to retain the CDR H3 length of each parental clone. Libraries were panned on immobilized Her2 under low-stringency conditions to maximize sequence diversity in the output clones. Clones retaining antigen binding in ELISA and SPR after reformatting to IgG were selected for sequence analysis and functional characterization.
Despite the differences between randomization schemes and the relatively limited sequence space coverage in most libraries (Suppl. Fig. 9), results were comparable between the two libraries for each antibody and are thus presented in combination. Clones were grouped by sequence types, arbitrarily prioritizing patterns similar to the parental CDR H3 sequence that may reflect similar antigen interactions (Figure 5, Suppl. Fig. 10, Suppl. File 1). Two major consensus sequence groups were identified for hu4D5-derived clones. One group of 301 unique clones (hu4D5-g1) retained at least some of the critical contact residues of the parental antibody, substituting His or Tyr for the critical Trp in position 107 in some NNK-derived clones and all clones from the trinucleotide-based library (Figure 5(a)). Group hu4D5-g2 included 62 unique clones with strong consensus features within the group that did not retain recognizable similarity with the parental sequence (Figure 5(a)). An additional 98 unique clones in group hu4D5-g3 had limited consensus patterns (Figure 5(a)). A group of 13 clones in group hu4D5-g4 with CDR H3 sequences were one residue shorter relative to the parental clone (Figure 5(a)). Three major groups of consensus sequences were obtained for randomized hu2C4 clones. A major group of 79 unique clones (group hu2C4-g1) retained Ile/Leu in position 108, perhaps analogous to the Leu-108 residue making direct antigen contact in hu2C4,37 but otherwise without recognizable similarity with the parental clone except perhaps Gly in position 109 (Figure 5(b)). Group hu2C4-g2 with 7 unique sequences had no apparent sequence similarity with the parental hu2C4 clone (Figure 5(b)). In addition, group hu2C4-g3 included 72 unique clones resembling the group hu2C4-g1 consensus, but with a CDR H3 length of 11 rather than 12 amino acid residues as in wild-type hu2C4 and group hu2C4-g1 clones (Figure 5(b)).
Figure 5.
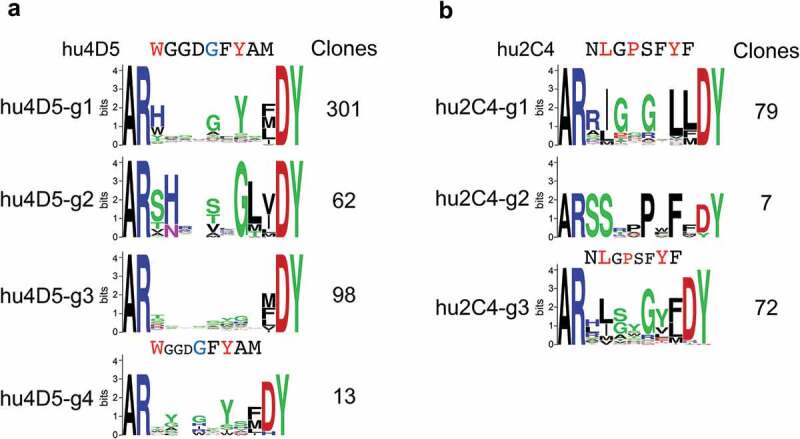
CDR H3 consensus sequences of unique anti-Her2 hu4D5 (a) and hu2C4 (b) variant clones. Parental CDR H3 sequence regions randomized in libraries are shown above the sequence logos, highlighting residues contacting Her2 through side-chains in red and through main-chain in blue. The number of unique clones for each group are shown.
A total of 63 variants with higher affinity in SPR assays representing different consensus groups and unique, non-consensus patterns derived from hu4D5 and hu2C4 were expressed as human IgG1 and tested in competition assays with each of the parental antibodies. Not unexpectedly, as all clones retain non-CDR H3 residues that were optimized for binding in the parental antibodies, all variants competed only with the cognate parental antibody in SPR-based competition assays (data not shown), confirmed by competition assays with a subset of higher affinity clones from different sequence groups (Suppl. Fig. 11), indicating that epitope specificity remained unchanged in the variants relative to parental antibodies. The 63 variants were screened for affinity to Her2 and the best variants of each major consensus or unique group except the wild type-like hu4D5-g1 group were selected for detailed affinity analysis. Despite the almost complete absence of CDR H3 sequence similarity with parental antibodies, the best variants of many groups had affinities that were comparable to the parental antibodies (Table 1). Remarkably, these included hu4D5 variants that have small residues instead of Trp in position 107 and mostly non-aromatic residues in place of Tyr-113, the two CDR H3 residues that make critical direct hydrophobic contacts with Her2 through side-chains and that are critical for binding. In addition, the strongly selected hu2C4 variants with shorter CDR H3, which are a minor fraction of the library caused by inefficiencies in trinucleotide incorporation, were among the best hu2C4 variants, with affinities similar to the parental clone. The results indicate that significant CDR H3 sequence diversity is allowed in antibodies known to mediate critical interactions with antigen through this region while retaining epitope specificity and robust antigen binding.
Table 1.
CDR H3 sequence and binding kinetics of best anti-Her2 variants in different consensus groups.
| Clonea | CDR H3b | ka (M−1s−1) | kd (s−1) | KD (nM) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hu4D5 | A | R | W | G | G | D | G | F | Y | A | M | D | Y | 7.2 × 105 [1.06]c | 1.1 × 10−4 [1.04] | 0.13 [1.10] |
| hu4D5-g2.1 | A | R | S | H | K | N | V | M | G | L | V | D | Y | 1.6 × 106 [1.12] | 3.5 × 10−4 [1.09] | 0.27 [1.13] |
| hu4D5-g2.2 | A | R | S | H | A | L | V | Y | G | L | I | D | Y | 1.8 × 106 [1.21] | 4.3 × 10−4 [1.08] | 0.33 [1.27] |
| hu4D5-g3.1 | A | R | G | H | Y | S | S | G | H | P | F | D | Y | 9.6 × 105 [1.09] | 3.2 × 10−4 [1.03] | 0.39 [1.10] |
| hu4D5-g3.2 | A | R | G | I | G | N | G | A | E | G | M | D | Y | 6.6 × 105 [1.05] | 1.1 × 10−3 [1.11] | 1.8 [1.17] |
| hu2C4 | A | R | N | L | G | P | – | S | F | Y | F | D | Y | 5.5 × 104 [1.04] | 2.2 × 10−4 [1.04] | 4.6 [1.14] |
| hu2C4-g3.1 | A | R | L | M | S | Y | – | – | G | Y | F | D | Y | 6.3 × 104 [1.17] | 4.0 × 10−4 [1.01] | 6.4 [1.16] |
| hu2C4-g3.2 | A | R | L | L | A | Y | – | – | G | Y | L | D | Y | 5.8 × 104 [1.15] | 5.7 × 10−4 [1.09] | 9.9 [1.07] |
| hu2C4-g1.1 | A | R | A | I | A | G | – | G | V | L | L | D | Y | 4.8 × 104 [1.16] | 1.2 × 10−3 [1.03] | 25 [1.13] |
| hu2C4-g1.2 | A | R | A | I | T | N | – | E | V | L | L | D | Y | 4.9 × 104 [1.10] | 1.4 × 10−3 [1.01] | 28 [1.07] |
| hu2C4-g2.1 | A | R | S | S | T | P | – | P | F | F | M | D | Y | 3.9 × 104 [1.15] | 9.6 × 10−4 [1.03] | 25 [1.11] |
| hu2C4-g2.2 | A | R | S | S | R | P | – | P | W | F | F | D | Y | 6.5 × 104 [1.23] | 1.5 × 10−3 [1.01] | 23 [1.25] |
a Clone name suffixes indicate groups according to Figure 5.
b IMGT® positions 105 to 117 shown. Underlined boldface residues contact antigen through side-chains in parental antibodies. Highlighted hu4D5 residues are IMGT® W107 and Y113 (Kabat W95 and Y100a). Highlighted hu2C4 residues are IMGT® L108, P110 and Y114 (Kabat L96, P98 and Y100a). Residues shown in italics were not randomized. Dashes introduced to conform all clones to the IMGT® numbering system.
c Values shown indicate geometric means and geometric standard deviation factors (in brackets) of experiments performed four times (one n = 1 and one n = 3 experiment).
Antibodies with shared VH/VL germline segment use and different antigen specificities have different binding modes
A possible basis for the restricted epitope specificity of antibodies with shared VH/VL germline segment pairs is that these antibodies bind antigen through stereotyped interactions, using a defined subset of residues in the binding interface. We addressed this question by analyzing antigen-binding interfaces in complex structures of antibodies with shared VH/VL germline segment use but different antigen or epitope specificities, including two previously described convergent human antibodies specific for the respiratory syncytial virus F (RSV F) glycoprotein and a human antibody with the same IGHV3-21/IGLV1-40 germline segment pairing that binds the unrelated proprotein convertase subtilisin-kexin type 9 (PCSK9).11,38-41 All three antibodies have similar CDR H3 lengths and occur in a germline or near-germline configuration (Suppl. Fig. 12), allowing direct comparison of contacts by germline residues in the binding interface. Given the role of CDRs in antigen binding, several non-CDR H3 interface residues are shared between the anti-RSV F and anti-PCSK9 antibodies as determined by buried surface area per position (Suppl. Fig. 13). However, while the interface profiles of both anti-RSV F antibodies are nearly identical outside CDR H3 and the junctional region of light chain CDR3, significant differences are observed between the anti-RSV F and anti-PCSK9 buried surface area profiles outside CDR H3, especially in the light chain and CDR H2. Thus, the overall binding mode is not necessarily conserved between antibodies with shared VH/VL germline segment pairs binding different epitopes.
Discussion
Numerous reports have described convergent responses in human and mouse antibody repertoires.6,8,10-24 One particular convergence class has been described as a restriction in VH and VL germline segment usage determined by antigen or epitope specificity.12-18 Collectively, these reports show that a number of epitopes or antigens can induce convergent immune responses with recurring germline segments. Repertoire deep sequencing studies suggest that convergences in CDR H3 sequences with or without VH germline segment recurrence, which are different from parallel lineages, may be prevalent.21,42-46 However, with two exceptions,21,42 there is no data to confirm the functional significance of these convergences. For VH/VL convergences, descriptions have been based on incidental findings of antibody characterization rather than systematic searches for these convergences.10-23 Thus, while convergences have been described with many antigens, no report has described the extent by which different parallel lineage convergences can occur for a single antigen or, to our knowledge, predicted this convergence class to be pervasive in immune repertoires. In addition, whether VH/VL germline segment pairing also has a restrictive role on epitope specificity has not been previously addressed.
Here, we show, based on systematic analysis of convergences in antibody repertoires derived by high-throughput paired-chain sequencing, that VH/VL pairing parallel lineage convergences are common within a single antigen and that VH/VL pairing restricts epitope specificity. Of note, epitope restriction by germline segment use is not a logical consequence of convergent responses, structural principles or biological processes. Epitope restriction of antibodies with recurrent VH/VL pairing is evident because VH/VL convergences are prevalent while, at the same time, antibodies with the same VH/VL germline segment binding different epitopes in the same antigen are rare. The epitope restrictions would be unremarkable in the absence of pervasive parallel lineage responses. The widespread occurrence of parallel lineage convergences in natural repertoires led us to investigate whether highly redundant CDR H3 sequences are compatible with epitope binding in the context of a VH/VL germline segment pair. We show here by means of phage display that functional CDR H3 redundancy includes clones known to mediate critical interactions with antigen through CDR H3, consistent with the observed widespread occurrence of parallel lineages and possibly other types of convergences.
Shared VH/VL germline segment pairing, which may not seem very informative due to the relatively limited number of different possible pairings, is highly predictive of functional redundancy within antigen-specific repertoires. In total, 78 (92%) of 85 clonally unrelated antigen-specific antibodies with 13 different VH/VL germline segment pairings binding in 8 epitope groups in 2 antigens had restricted epitope specificity as determined by competition assays. Four additional antibodies had only minor differences in cross-competition patterns relative to other antibodies in the same parallel lineages, bringing the agreement between VH/VL pairing and epitope specificity to 96%. Importantly, for 64 of these antibodies the epitope restriction was identified prospectively, with clones selected based only on VH/VL germline segment pairing. Of the 64 antibodies, 34 had 7 different VH/VL pairs not known a priori to belong to epitope-restricted groups. In addition, for both antigens, epitope restriction as a function of VH/VL use was observed in the context of a relatively large diversity of epitope specificities in the repertoires. Thus, our dataset represents clones with shared VH/VL germline segment pairing that were selected randomly, except for the availability of repeated VH/VL pairs in the sequence sets. Based on this, we expect our results showing restricted epitope specificity in clonally unrelated antibodies sharing VH/VL germline segment pairing to be largely representative of unsampled antigen-specific clones in the datasets described here and in antigen-specific repertoires for other antigens.
Epitope specificity determination by cross-competition is a relatively low resolution epitope mapping technique. Antibodies that cross-compete can have significantly different overlapping epitopes or cross-compete through constant region bulk. However, the virtually indistinguishable cross-competition patterns of a relatively large number of antibodies with recurring VH/VL germline segment pairs to several epitopes indicates a close epitope similarity among antibodies in parallel lineages. A virtually identical binding geometry for clones within parallel lineages is the most parsimonious interpretation for the generally restricted epitope specificity of antibodies with the same VH/VL germline segment pair. Details in the interactions may differ among clones, especially in CDR H3, which can vary both in sequence and length between antibodies in parallel lineages and may cover slightly different epitope areas. However, overall binding geometry and many of the interactions would be expected to be similar or identical in most or all cases.
The generalized epitope restriction of antibodies with the same VH/VL pair that we observed would not be congruent with binding of these antibodies to partially overlapping epitopes or with significantly different rotational angles relative to the antigen, as these would be effectively as different structurally as non-overlapping epitopes. If that were the case, a high prevalence of non-overlapping epitopes would be expected within sets of antibodies with the same VH/VL pairing, unlike what is observed here. Consistent with our interpretation of conserved binding geometries in parallel lineages, virtually identical binding geometries have been observed in structures of antibodies with the same VH/VL germline segment pairs in complex with protein or peptide antigens.8,11,47,48 It should be pointed out that conserved binding geometry may not necessarily extend to constant regions. This is because angles between variable and constant regions may differ with framework sequences encoded by JH germline segments,49,50 which do not necessarily recur within parallel lineages.
An unexpected implication of our results showing restricted epitope specificity determined by VH/VL germline segment pairing is that the number of distinct epitopes recognized by antibodies from immune repertoires generated in vivo with a particular VH/VL pair is apparently low, probably less than one epitope on average per small- to medium-size antigen with multiple epitope groups (HEL, 14 kDa; OVA, 43 kDa). This is unexpected, as no structural principle requires VH/VL pairs to have significantly restricted epitope specificities in the context of the highly variable CDR H3 region. A more general specificity preference based on light chain CDR 1 length, which correlates with binding of antibodies to different antigen classes due to different overall shape complementarity requirements, was previously described.51 However, the restricted epitope specificity based on VH/VL germline segment use shown here includes antibodies binding the same antigen class, globular proteins, with similar overall epitope shapes. Thus, epitope restriction based on VH/VL germline segment use is not expected based on that precedent.
As currently understood, VH/VL germline segment pairs recur in antibodies specific for different antigens, and thus have a broad range of potential specificities when combined with different CDR H3 sequences. In fact, it has been shown that CDR H3 diversity in the context of limited VH/VL germline segment diversity is sufficient for a relatively broad range of antigen specificities,4 although the number of unique antibodies reported for each antigen is low and epitope coverage in these cases is not known. It is possible that epitope restriction may be only partially valid with larger or more complex antigens such as whole viruses and cells. That is, clones with recurrent VH/VL germline segment pairs may have more than one distinct epitope specificity in more complex antigens. However, our data suggest that these would be restricted to a relatively limited set of epitopes. One point to note is that the epitope restriction by VH/VL germline segment pairing probably happens only in the context of naïve repertoires without somatic diversification prior to antigen stimulation, as it occurs in rodents and humans. Somatic mutation after antigen stimulation is constrained to retain epitope specificity except in cases of secondary responses to cross-reactive epitopes in different antigens, which is unlikely to occur within an antigen. However, in species in which the naïve repertoire undergoes somatic diversification of V germline segment regions prior to antigen stimulation, such as rabbits, the spectrum of potential epitope specificities of VH/VL germline segment pairs in a single antigen may be expanded. This is in fact observed with antibodies obtained from libraries with V regions randomized, or “somatically mutated”, in vitro.52
A second unexpected implication of the apparent widespread occurrence of parallel lineages in repertoires is the functional redundancy of CDR H3. A certain level of sequence similarity is observed between the CDR H3 of clones in some parallel lineages, as might be expected from their shared epitope specificity. Many of these similarities involve lower diversity sequences contributed by JH germline segments as well as the region contributed by VH (IMGT® positions 105 and 106), which are invariant in the absence of nucleotide nibbling or somatic mutation and could be expected with relatively high frequency by chance. In addition, these similarities usually involve only subsets of clones within each parallel lineage, and the positions of amino acid identities in CDR H3 vary in different pairwise comparisons. Thus, the significance of many of these sequence similarities is evident only with the benefit of data indicating functional similarities of a group of clones. Clones in this category include anti-OVA parallel lineages 7 and 10 and subsets of other parallel lineages, as well as some clones in both anti-HEL parallel lineages (Figure 2 and Suppl. Table 1). However, the level of pairwise amino acid identity between clones with the same CDR H3 length is generally low (Suppl. Fig. 4).
The widespread occurrence of restricted epitope specificity in parallel lineages in the context of CDR H3 regions of different lengths and sequences implies that CDR H3 in parallel lineages may not be as important energetically for binding or, alternatively, may provide the necessary free energy of binding and epitope specificity with a range of distinct sequences and lengths. Rather than surveying the binding contribution of CDR H3 in parallel lineage clones, we tested whether the well-characterized hu4D5 and hu2C4 anti-Her2 antibodies known to mediate energetically important interactions with antigen through CDR H3 would accept widely distinct CDR H3 sequences while retaining epitope specificity and robust binding. We found that, contrary to a recently stated expectation that complete randomization of hu4D5 CDR H3 would be unlikely to yield a significant number of Her2 binders,53 CDR H3 sequences that differ significantly from the parental antibodies can accommodate strong binding to the same epitope as determined by binding competition assays. As CDR H3 in these synthetic parallel lineage clones must provide much of the free energy of binding that is contributed by the corresponding parental CDR H3 regions, our results indicate a significant degree of sequence and functional redundancy in CDR H3. By extension, our results indicate that natural parallel lineages may include clones with CDR H3 regions with major energetic contributions to binding, explaining the relatively high frequency of these convergences in repertoires.
Parallel lineages imply a major role of V-germline segment residues in the energetics of binding. In fact, non-CDR H3 regions of natural antibodies have been shown to contribute a large fraction of binding residues and free energy of binding, although the relative contribution of germline segment and somatically mutated residues has not been determined.54,55 Previous descriptions of structures of convergent antibodies in complex with antigen have uncovered some shared mechanisms of binding based on specific germline segment features. This is exemplified by the binding of convergent antibodies against influenza A hemagglutinin and the Staphylococcus aureus virulence factor IsdB with IGHV1-69 or IGHV4-39 germline segments, which make use of hydrophobic germline residues in CDR H2 for antigen binding.22,56 However, other convergent clones against similar influenza A hemagglutinin epitopes using other germline segments lack that signature or any other sequence and structural signatures in common between different convergent groups,23 suggesting different structural mechanisms for different convergent groups. Given the variety of germline segments and heavy and light chain subgroups involved in the convergences shown here, it is unlikely that these share a common structural basis, except for CDR H3 sequence and functional diversity. CDR H3 redundancy was previously examined in specific cases of convergences. In some cases, conservation of specific or chemically similar residues in certain CDR H3 positions was observed.11,23,56 However, in other cases no conserved CDR H3 signatures were observed,11,22 similar to our results with the hu4D5-g2 and hu2C4-g2 group clones and many of the parallel lineages described here.
One difficulty of determining the structural basis for restricted epitope specificity is that the absence of epitope specificities in the same antigen cannot be directly addressed by structural studies. Thus, the structural basis for this restriction may only be inferred. One possibility is that a given VH/VL pair can only interact with different epitopes through specific stereotyped interactions. Our analysis of the binding modes of different antibodies with shared VH/VL germline segment use and different antigen specificities indicates that this is not necessarily the case (Suppl. Fig. 13), which is not unexpected. Determining whether subtle antigen interactions are recurrently used by antibodies with the same VH/VL pairing with different epitopes will require systematic large-scale analyses of complex structures of analogous sets of antibodies with shared VH/VL pairing binding different antigens. However, it is possible that the generally restricted epitope specificity of VH/VL pairs within antigens may not be based on stereotypic antigen binding interactions and that this restriction may be best described statistically. Thus, epitope restriction by VH/VL germline segment pairing within antigens is a repertoire feature that emerges from large-scale functional analyses that, while structurally determined, may not be readily apparent from a strictly structural perspective.
Whether any VH/VL pairs in a set of antigen-specific clones can be potentially part of parallel lineages is not clear. About half of the anti-OVA scBCR-seq dataset, an estimated 96% of which bind OVA,26 is composed of lineages with unique VH/VL germline segment pairs (Suppl. Fig. 14). Whether these are unique due to undersampling, relatively low frequency of compatible CDR H3 sequences, immune regulation or stochastic factors cannot be easily determined. The chemistry of the antigen interactions mediated by CDR H3 may affect the prevalence of CDR H3 sequences meeting the structural requirements for binding and hence parallel lineage formation. However, CDR H3 length does not seem to be a major factor in parallel lineage convergences within species. The CDR H3 lengths of the parallel lineage clones we characterized largely reflect the CDR H3 lengths of clones in the anti-OVA repertoire, which are slightly longer on average than the CDR H3 length of naïve rat IgG repertoires (Suppl. Fig. 15). Similarly, CDR H3 lengths of previously described human and mouse antibodies with convergent parallel lineages range from 15 to 18 and 9 to 17 residues, respectively,10-12,15,17,41 slightly longer than the average CDR H3 length of each species.57 Thus, parallel lineage convergences do not seem to be generally associated with readily apparent CDR H3 length biases within species. Whether different species have different propensities for parallel lineage convergences and germline segment-restricted epitope specificity due to differences in average CDR H3 length remains to be determined.
Our results extend our understanding of the role of VH/VL pairing and CDR H3 diversity in determining epitope specificity. The CDR H3 functional redundancy that results in widespread parallel lineage convergences, combined with the highly predictive epitope restriction of VH/VL germline segment pairings, reveal a logical organization of immune repertoires beyond classical B cell lineages that is determined by structural rather than biological factors, with implications for vaccine development and antibody discovery and engineering.
Materials and methods
Selection of anti-OVA and anti-HEL clones from scBCR-seq datasets
A dataset of anti-chicken ovalbumin clones derived by scBCR-seq with paired heavy and light chain variable regions was previously described.26 A subset of 93 clones selected from the scBCR-seq dataset were shown to be specific for OVA, numbered here as “OVA” followed by a two-digit number. Additional clones, indicated by “OVA” followed by a three-digit number, were selected from different B-cell lineages in scBCR-seq datasets using VH and VL germline segment use data and prioritizing clones within lineages with full-length JH sequence information. Lineage definition for the anti-OVA panel was the same as previously described, using same VH and VL germline segments and at least 80% nucleotide identity within CDR H3, ignoring J germline segment identity, to include clones within the same lineage.26 A similar definition was used for the anti-HEL panel, using 80% amino acid identity within CDR H3 instead as the threshold cutoff. The junctional nucleotide sequences of selected clones within the same lineage and 50% or greater amino acid sequence identity were further analyzed for VH, DH, and JH use and boundaries using an in-house tool and a composite database of IMGT®27 and in-house determined reference germline segments sequences as previously described.26 Clones with the same D and J segments in the same location and similar boundaries were excluded from the dataset as likely clonal variants modified by somatic mutation. All clones were expressed as rat/human chimeric immunoglobulin G after DNA synthesis as previously described.26
Sequencing of rat anti-HEL repertoires
A dataset of B-cell receptor sequences enriched for HEL-specific clones with paired heavy and light chain information was obtained as previously described.26 Briefly, three Sprague Dawley (SD) rats (Charles River, Hollister, CA) were immunized with 100 µg HEL (Sigma-Aldrich, St. Louis, MO) in Complete Freund’s adjuvant (BD, Franklin Lakes, NJ) followed by bi-weekly boosts of 50 µg OVA in incomplete Freund’s adjuvant divided in three sites (intraperitoneally, subcutaneously at base of tail, at nape of neck and in both hocks). Cells from multiple lymph nodes from the 3 rats were pooled, depleted of IgMpos B cells, stained with anti-rat IgM (Clone G53-238, BD Biosciences, San Jose, CA) conjugated to PE Cy7 and HEL conjugated to Alexa Fluor 647 (Thermo Fisher Scientific, Waltham, MA) and sorted for IgMneg/HELpos cells in a FACSAriaIII sorter (BD, Franklin Lakes, NJ). Due to visible nonspecific binding of labeled antigen in control T cells, gating for cell selection was set at a high threshold to enrich for antigen-specific B cells. A total of 49,000 IgMneg/HELpos events were detected from the three rats and processed for scBCR-seq. Sequences were parsed for germline segments and processed for paired VH/VL sequences and clones grouped by putative lineages as described below.
Selection of anti-Her2 CDR H3 variants by phage display
The variable regions of anti-Her2 hu4D5 and hu2C4 were cloned in a monovalent antigen-binding fragment (Fab) phage display vector.58 The CDR H3 region of both clones was replaced by site-directed mutagenesis with stop codons and two sets of libraries with randomized CDR H3 sequences were produced for each clone by oligonucleotide-directed mutagenesis as previously described.52 CDR H3 randomization was achieved by use of NNK (IUPAC code) or trinucleotide codon mixes. The trinucleotide-based randomization was designed to partly mimic human CDR H3 diversity with position and length-specific codon mixtures (Suppl. Fig. 9). Clones were selected on Her2 directly immobilized on ELISA plates (hu4D5) or on biotinylated Her2 immobilized on streptavidin-coated ELISA plates (hu2C4). Libraries were panned for up to 5 rounds as previously described, selecting clones from rounds 3 and 5 that were confirmed to bind Her2 in phage-ELISA for Sanger sequencing.35 Sequence logos were generated using the WebLogo server.59
Sequence coverage of the anti-Her2 variant libraries was calculated based on library randomization scheme, library colony-forming units (cfu) achieved and fraction of successfully mutated clones in the library determined by Sanger sequencing of a library clone sample. Briefly, because the expected frequencies of different sequences in the library are unequal, the sequence coverage was calculated by generating j (104 in this case) sequences in a Monte Carlo simulation using the probabilities of each amino acid being yielded by a given codon and calculating the expected frequency P of each sequence i. This generates a set in which each simulated sequence i represents L/j sequences of identical or very similar expected frequencies, where L is the number of successfully mutated (non-template) clones in the library. The average number of unique sequences C in a library size L is approximated as:
The contribution to C by clones with stop or Cys codons, which are rarely selected by phage display and effectively defective, is defined as zero here. The estimated percentage F of the total theoretical diversity V of clones without stop or Cys codons achieved in a library (functional sequence coverage) is therefore approximately
| (2) |
Note that if Pi is constant, as in an idealized library in which each sequence variant has the same expected frequency Pi = 1/V, equation (1) simplifies to the previously described equation60
| (3) |
Expression and purification of antibodies and Fabs
DNA fragments encoding variable regions of anti-OVA and anti-HEL scBCR-seq sequences were obtained by DNA synthesis and cloned into human immunoglobulin class 1 (IgG1)/kappa mammalian expression vectors as previously described.26 The heavy chain variable region of selected anti-Her2 clones were subcloned into human IgG1 and flag-tagged Fab mammalian expression vectors. Heavy and light chain expression plasmids were co-transfected into Expi293 cells at 1 ml scale and purified in batch mode by protein A chromatography as previously described.61,62 Fabs of hu4D5 variants were transiently expressed in 30 ml Expi293 cells for 7 days and purified by anti-flag-tag and size-exclusion chromatography. Fabs were confirmed to be monomeric by analytical size-exclusion chromatography.
Affinity and binding competition assays
Binding of clones to OVA or HEL was confirmed by ELISA and affinities determined by SPR as previously described.26 Purified hu2C4 variant IgG1 was tested for binding to Her2 by SPR at 25°C using a BIAcore T200 apparatus (GE Life Sciences, Piscataway, NJ), immobilizing IgG in protein A chips and using monomeric Her2 as analyte. The hu4D5 variants were tested by SPR in BIAcore T200 apparatus with Her2 immobilized on CM5 chips, using Fabs as analytes at 25°C. Binding competition assays were performed in Carterra IBIS and LSA instruments as previously described.63 Binding competition results were analyzed in a blinded manner without reference to antibody sequence groups using the Carterra Epitope Binning software following standard vendor guidelines, previously described in detail.64,65
Polyspecificity assay
Polyspecificity of antibody OVA-120 was performed in a BV ELISA assay as previously reported,29 using IgG purified by protein A and size-exclusion chromatography in the assay.
Determination of antigen binding interfaces
Buried surface area of antibody regions and residues in the interface with antigen (excluding crystal contacts) were determined using the PDBePISA server at http://www.ebi.ac.uk/pdbe/pisa.66 PDB files with accession numbers 1N8Z (hu4D5), 1S78 (hu2C4), 6APD (anti-RSV F ADI-19425), 5U68 (anti-RSV F MPE8) and 3H42, 4K8R and 5OCA (anti-PCSK9) were used in the analyses.
Abbreviations
- CDR H3
Heavy chain third complementarity determining region
- cfu
Colony-forming units
- HEL
Hen egg-white lysozyme
- IgG1
Human immunoglobulin class 1
- IMGT®
The International ImMunoGeneTics information system®
- OVA
Chicken ovalbumin
- PCSK9
Proprotein convertase subtilisin-kexin type 9
- RSV F
Respiratory syncytial virus F glycoprotein
- scBCR-seq
Single-cell B-cell receptor sequencing
- SD
Sprague Dawley
- SPR
Surface plasmon resonance
- VH
Heavy chain variable region germline segment
- VL
Light chain variable region germline segment
Acknowledgments
We thank Bijay S. Jaiswal and Jeremy Stinson for project support and Jack Bevers III for assistance with Carterra-based epitope competition assays.
Disclosure of potential conflicts of interest
All authors except A.A. are current or former employees of Genentech and may be Roche shareholders. A.A. is an employee of SciGenom Labs. S.S. is affiliated with SciGenom Research Foundation.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Schroeder HW Jr., Cavacini L.. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125:S41–14. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM.. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. [DOI] [PubMed] [Google Scholar]
- 3.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/S0092-8674(02)00675-X. [DOI] [PubMed] [Google Scholar]
- 4.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/S1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 5.Finney J, Yeh CH, Kelsoe G, Kuraoka M. Germinal center responses to complex antigens. Immunol Rev. 2018;284:42–50. doi: 10.1111/imr.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imkeller K, Wardemann H. Assessing human B cell repertoire diversity and convergence. Immunol Rev. 2018;284:51–66. doi: 10.1111/imr.2018.284.issue-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev I, Altae-Tran H, Chuang G-Y, Joyce M, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–58. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Li H, Yang F, Smith-Gill SJ, Mariuzza RA. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nat Struct Biol. 2003;10:482–88. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- 9.Dunand CJH, Wilson PC. Restricted, canonical, stereotyped and convergent immunoglobulin responses. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140238. doi: 10.1098/rstb.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croote D, Darmanis S, Nadeau KC, Quake SR. High-affinity allergen-specific human antibodies cloned from single IgE B cell transcriptomes. Science. 2018;362:1306–09. doi: 10.1126/science.aau2599. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin E, Gilman MSA, Wrapp D, Chen M, Ngwuta JO, Moin SM, Bai P, Sivasubramanian A, Connor RI, Wright PF, et al. Infants infected with respiratory syncytial virus generate potent neutralizing antibodies that lack somatic hypermutation. Immunity. 2018;48:339–49 e5. doi: 10.1016/j.immuni.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert R, Lefranc MP, Ghochikyan A, Agadjanyan MG, Cribbs DH, Van Nostrand WE, Wark KL, Dolezal O. Restricted V gene usage and VH/VL pairing of mouse humoral response against the N-terminal immunodominant epitope of the amyloid beta peptide. Mol Immunol. 2010;48:59–72. doi: 10.1016/j.molimm.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Sanchez A, Garcia-Ocana M, de los Toyos JR. Mouse monoclonal antibodies to pneumococcal C-polysaccharide backbone show restricted usage of VH-DH-JH gene segments and share the same kappa chain. Immunol Lett. 2009;123:125–31. [DOI] [PubMed] [Google Scholar]
- 14.Persson H, Flicker S, Sadegh MK, Greiff L, Valenta R, Ohlin M. A common idiotype in IgE and its relation to recognition of the grass pollen allergen Phl p 2. Mol Immunol. 2008;45:2715–20. doi: 10.1016/j.molimm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 15.McLean GR, Olsen OA, Watt IN, Rathanaswami P, Leslie KB, Babcook JS, Schrader JW. Recognition of human cytomegalovirus by human primary immunoglobulins identifies an innate foundation to an adaptive immune response. J Immunol. 2005;174:4768–78. doi: 10.4049/jimmunol.174.8.4768. [DOI] [PubMed] [Google Scholar]
- 16.Smith-Gill SJ, Mainhart CR, Lavoie TB, Rudikoff S, Potter M. VL-VH expression by monoclonal antibodies recognizing avian lysozyme. J Immunol. 1984;132:963–67. [PubMed] [Google Scholar]
- 17.Zhang M, Zharikova D, Mozdzanowska K, Otvos L, Gerhard W. Fine specificity and sequence of antibodies directed against the ectodomain of matrix protein 2 of influenza A virus. Mol Immunol. 2006;43:2195–206. doi: 10.1016/j.molimm.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 18.Thomson CA, Bryson S, McLean GR, Creagh AL, Pai EF, Schrader JW. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. Embo J. 2008;27:2592–602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen PS, Haahr-Hansen M, Coljee VW, Hinnerfeldt FR, Varming K, Bregenholt S, Haurum JS. Extensive restrictions in the VH sequence usage of the human antibody response against the Rhesus D antigen. Mol Immunol. 2007;44:412–22. doi: 10.1016/j.molimm.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Setliff I, McDonnell WJ, Raju N, Bombardi RG, Murji AA, Scheepers C, Ziki R, Mynhardt C, Shepherd BE, Mamchak AA, et al. Multi-donor longitudinal antibody repertoire sequencing reveals the existence of public antibody clonotypes in HIV-1 infection. Cell Host Microbe. 2018;23:845–54 e6. doi: 10.1016/j.chom.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Wu X, Zhang B, McKee K, O’Dell S, Soto C, Zhou T, Casazza JP, Mullikin JC, Kwong PD, et al. De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc Natl Acad Sci U S A. 2013;110:E4088–97. doi: 10.1073/pnas.1306262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung YA, Foletti D, Deng X, Abdiche Y, Strop P, Glanville J, Pitts S, Lindquist K, Sundar PD, Sirota M, et al. Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire. Nat Commun. 2016;7:13376. doi: 10.1038/ncomms13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza a viruses. Cell. 2016;166:609–23. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mou Y, Zhou XX, Leung K, Martinko AJ, Yu JY, Chen W, Wells JA. Engineering improved antiphosphotyrosine antibodies based on an immunoconvergent binding motif. J Am Chem Soc. 2018;140:16615–24. doi: 10.1021/jacs.8b08402. [DOI] [PubMed] [Google Scholar]
- 25.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–25. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein LD, Chen YJ, Wu J, Chaudhuri S, Hsiao Y, Schneider K, Hoi KH, Lin Z, Guerrero S, Jaiswal BS, et al. Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun Biol. 2019;2:304. doi: 10.1038/s42003-019-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefranc MP, Ehrenmann F, Ginestoux C, Giudicelli V, Duroux P. Use of IMGT((R)) databases and tools for antibody engineering and humanization. Methods Mol Biol. 2012;907:3–37. [DOI] [PubMed] [Google Scholar]
- 28.Hu HY, Du HN. Alpha-to-beta structural transformation of ovalbumin: heat and pH effects. J Protein Chem. 2000;19:177–83. doi: 10.1023/A:1007099502179. [DOI] [PubMed] [Google Scholar]
- 29.Hotzel I, Theil F-P, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al. A strategy for risk mitigation of antibodies with fast clearance. MAbs. 2012;4:753–60. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chitarra V, Alzari PM, Bentley GA, Bhat TN, Eisele JL, Houdusse A, Lescar J, Souchon H, Poljak RJ. Three-dimensional structure of a heteroclitic antigen-antibody cross-reaction complex. Proc Natl Acad Sci U S A. 1993;90:7711–15. doi: 10.1073/pnas.90.16.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith-Gill SJ, Wilson AC, Potter M, Prager EM, Feldmann RJ, Mainhart CR. Mapping the antigenic epitope for a monoclonal antibody against lysozyme. J Immunol. 1982;128:314–22. [PubMed] [Google Scholar]
- 32.Braden BC, Souchon H, Eisele JL, Bentley GA, Bhat TN, Navaza J, Poljak RJ. Three-dimensional structures of the free and the antigen-complexed Fab from monoclonal anti-lysozyme antibody D44.1. J Mol Biol. 1994;243:767–81. doi: 10.1016/0022-2836(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 33.Boulot G, Eisele JL, Bentley GA, Bhat TN, Ward ES, Winter G, Poljak RJ. Crystallization and preliminary X-ray diffraction study of the bacterially expressed Fv from the monoclonal anti-lysozyme antibody D1.3 and of its complex with the antigen, lysozyme. J Mol Biol. 1990;213:617–19. doi: 10.1016/S0022-2836(05)80248-7. [DOI] [PubMed] [Google Scholar]
- 34.Kelley RF, O’Connell MP. Thermodynamic analysis of an antibody functional epitope. Biochemistry. 1993;32:6828–35. doi: 10.1021/bi00078a005. [DOI] [PubMed] [Google Scholar]
- 35.Vajdos FF, Adams CW, Breece TN, Presta LG, de Vos AM, Sidhu SS. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J Mol Biol. 2002;320:415–28. doi: 10.1016/S0022-2836(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 36.Cho H-S, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW Jr., Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–60. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 37.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–28. doi: 10.1016/S1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 38.Chan JC, Piper DE, Cao Q, Liu D, King C, Wang W, Tang J, Liu Q, Higbee J, Xia Z, et al. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc Natl Acad Sci U S A. 2009;106:9820–25. doi: 10.1073/pnas.0903849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gustafsen C, Olsen D, Vilstrup J, Lund S, Reinhardt A, Wellner N, Larsen T, Andersen CBF, Weyer K, Li J-P, et al. Heparan sulfate proteoglycans present PCSK9 to the LDL receptor. Nat Commun. 2017;8:503. doi: 10.1038/s41467-017-00568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiele F, Park J, Redemann N, Luippold G, Nar H. An antibody against the C-terminal domain of PCSK9 lowers LDL cholesterol levels in vivo. J Mol Biol. 2014;426:843–52. doi: 10.1016/j.jmb.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE Jr., Jardetzky TS. Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol. 2017;2:16272. doi: 10.1038/nmicrobiol.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson KJL, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall E, Gurley T, Moody M, Haynes B, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16:105–14. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parameswaran P, Liu Y, Roskin KM, Jackson KL, Dixit VP, Lee J-Y, Artiles KL, Zompi S, Vargas M, Simen B, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinstein JA, Jiang N, White RA 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–10. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burckert J-P, Dubois A, Faison WJ, Farinelle S, Charpentier E, Sinner R, Wienecke-Baldacchino A, Muller CP. Functionally convergent b cell receptor sequences in transgenic rats expressing a human B cell repertoire in response to tetanus toxoid and measles antigens. Front Immunol. 2017;8:1834. doi: 10.3389/fimmu.2017.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briney B, Inderbitzin A, Joyce C, Burton DR. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. 2019;566:393–97. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho KJ, Hong KW, Kim S-H, Seok JH, Kim S, Lee J-H, Saelens X, Kim KH. Insight into highly conserved H1 subtype-specific epitopes in influenza virus hemagglutinin. PLoS One. 2014;9:e89803. doi: 10.1371/journal.pone.0089803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du J, Wang H, Zhong C, Peng B, Zhang M, Li B, Hou S, Guo Y, Ding J. Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide. Mol Immunol. 2008;45:2861–68. doi: 10.1016/j.molimm.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 49.Landolfi NF, Thakur AB, Fu H, Vasquez M, Queen C, Tsurushita N. The integrity of the ball-and-socket joint between V and C domains is essential for complete activity of a humanized antibody. J Immunol. 2001;166:1748–54. doi: 10.4049/jimmunol.166.3.1748. [DOI] [PubMed] [Google Scholar]
- 50.Koenig P, Lee CV, Walters BT, Janakiraman V, Stinson J, Patapoff TW, Fuh G. Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc Natl Acad Sci U S A. 2017;114:E486–E95. doi: 10.1073/pnas.1613231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raghunathan G, Smart J, Williams J, Almagro JC. Antigen-binding site anatomy and somatic mutations in antibodies that recognize different types of antigens. J Mol Recognit. 2012;25:103–13. doi: 10.1002/jmr.v25.3. [DOI] [PubMed] [Google Scholar]
- 52.Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340:1073–93. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 53.Mason DM, Friedensohn S, Weber CR, Jordi C, Wagner B, Meng S, Gainza P, Correia BE, Reddy ST. Deep learning enables therapeutic antibody optimization in mammalian cells by deciphering high-dimensional protein sequence space. BioRxiv. 2019. doi: 10.1101/617860. [DOI] [Google Scholar]
- 54.Robin G, Sato Y, Desplancq D, Rochel N, Weiss E, Martineau P. Restricted diversity of antigen binding residues of antibodies revealed by computational alanine scanning of 227 antibody-antigen complexes. J Mol Biol. 2014;426:3729–43. doi: 10.1016/j.jmb.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 55.Burkovitz A, Ofran Y. Understanding differences between synthetic and natural antibodies can help improve antibody engineering. Mabs. 2016;8:278–87. doi: 10.1080/19420862.2015.1123365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang C-Y, Cadwell G, et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog. 2014;10:e1004103. doi: 10.1371/journal.ppat.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–49. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 58.Lee CV, Sidhu SS, Fuh G. Bivalent antibody phage display mimics natural immunoglobulin. J Immunol Methods. 2004;284:119–32. doi: 10.1016/j.jim.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patrick WM, Firth AE, Blackburn JM. User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. 2003;16:451–57. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- 61.Bos AB, Luan P, Duque JN, Reilly D, Harms PD, Wong AW. Optimization and automation of an end-to-end high throughput microscale transient protein production process. Biotechnol Bioeng. 2015;112:1832–42. doi: 10.1002/bit.25601. [DOI] [PubMed] [Google Scholar]
- 62.Luan P, Lee S, Arena TA, Paluch M, Kansopon J, Viajar S, Begum Z, Chiang N, Nakamura G, Hass PE, et al. Automated high throughput microscale antibody purification workflows for accelerating antibody discovery. MAbs. 2018;10:624–35. doi: 10.1080/19420862.2018.1445450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vij R, Lin Z, Chiang N, Vernes JM, Storek KM, Park S, Chan J, Meng YG, Comps-Agrar L, Luan P, et al. A targeted boost-and-sort immunization strategy using Escherichia coli BamA identifies rare growth inhibitory antibodies. Sci Rep. 2018;8:7136. doi: 10.1038/s41598-018-25609-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abdiche YN, Yeung AY, Ni I, Stone D, Miles A, Morishige W, Rossi A, Strop P. Antibodies targeting closely adjacent or minimally overlapping epitopes can displace one another. PLoS One. 2017;12:e0169535. doi: 10.1371/journal.pone.0169535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sivasubramanian A, Estep P, Lynaugh H, Yu Y, Miles A, Eckman J, Schutz K, Piffath C, Boland N, Niles RH, et al. Broad epitope coverage of a human in vitro antibody library. MAbs. 2017;9:29–42. doi: 10.1080/19420862.2016.1246096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


