Abstract
Design parameters for microphysiological systems (MPS) are driven by the need for new tools to answer questions focusing on human physiology in a robust and reliable manner. Within this perspective, engineering benchmarks and principles are identified to guide the construction of new devices in the MPS field, with emphasis placed on the design principles common to all tissues, as well as those unique to a subset of tissues. Leading organ replica technologies that recapitulate various functions of the brain, heart, intestine, and lung are highlighted as examples that meet the identified benchmarks and standards, with current barriers for large scale production and commercialization discussed. To reach their full potential and achieve widespread use, MPS will have to be recognized officially by government agencies, and toward this end, considerations of MPS as a potential regulatory tool are presented.
Keywords: microphysiological system, organ-on-a-chip, high-throughput, microfabrication, primary cell culture
Introduction
Microphysiological systems (MPS), which include organs-on-chips and organ replicas, have undergone rapid development since their inception [1], driven by the need to replace animal models and recapitulate human physiology, thus paving the way for rapid discoveries in basic and pharmaceutical sciences, as well as personalized medicine [2]. The development of a single, modular and universal microphysiological platform for all organs or tissues represents a tantalizing goal; however, each carries a distinct set of requirements that are dependent on the tissue under study. Beyond ensuring that engineered platforms are capable of hosting healthy populations of cultured cells, engineers must also consider whether devices are scalable, manufacturable, reproducible, and predictive. Within this perspective, engineering benchmarks and design principles relevant to MPS are presented, and technologies that push current limitations of scalability and throughput are highlighted. Ultimately, new systems should be widely accessible and enable acquisition of high-content, reproducible information in a reliable format [3,4]. Moreover, medical and regulatory decisions are expected to benefit from this suite of technologies, in particular where endpoints of significance to human health can be obtained and validated across multiple laboratories.
Engineering benchmarks and design principles
In designing platforms that are not only physiologically relevant, but also scalable in manufacture and usage, it is helpful to distill each organ down into representative tissue subunits, from which dominant features can be identified and engineered into the organ-specific tissue replica. For example, the crypt or crypt/villus is the primary tissue subunit of the intestinal epithelium with its polarized architecture supported by in vivo biochemical gradients that regulate epithelial cell self-renewal and differentiation. The alveoli of the lung, in which an air-liquid interface is present, is responsible for gas exchange and is continuously subjected to rhythmic stretching that modulates cellular physiology. Likewise, the nephron of the kidney, sinusoids and lobules of the liver, and electrically connected, anisotropic tissue layers of the heart could be considered as the smallest derivative subunits of their respective organ that support its major function. Identifying these major subunits and their critical microenvironment features will enable design, construction, and operation of these tissue replicas to yield complex organ-level behavior and enable predictive experiments and assays. Additionally, these tissue subunits while recapitulating organ-level physiology do so with out the high construction complexity required to build a full organ.
Cultured cells are a core component of microphysiological systems, and thus, the choice of appropriate cell source should be a primary consideration during design (Fig. 1). The cells must be competent to interface with neighboring cells and/or tissue subunit components, in order to develop complex behaviors and display interactions that are not present in simpler culture systems. Human primary cells can retain a normal genome and physiology that closely mimic the in vivo phenotypes and functions of the original tissue, and thus are the preferred tissue source [5–8]. For many organs (e.g., blood, skin, intestine), primary cells are readily available from biopsies of healthy and disease state donors. However, in the case of organs in which biopsies are more difficult to obtain (e.g., brain, kidney, liver), primary cells can be isolated from surgically resected tissue or cadaveric donor organs. To address the finite lifespan of end-differentiated primary cells, tissue-specific, adult stem/progenitor cells are often used as the they have ability to renew themselves indefinitely under ideal growth conditions, and can be differentiated to yield some or all of the major specialized cell types of the in vivo tissue. Alternatively, primary somatic cells can be used to generate induced pluripotent stem cells (iPSCs), which thereafter may be differentiated into tissue-specific cell types. All primary cells require highly specialized growth conditions, which may include specific cytokines, metabolites, and growth factors that must be incorporated into the microdevice. Given that many stem cells can be long lived in culture, with some capable of being banked and frozen, genetically diverse biobanks can be established for population screens [9]. These attributes make stem cells a very attractive source to build microphysiological systems; however, robust protocols for stem cell maintenance and differentiation need to be established [10,11]. Despite the many advantages of using primary cells, it is acknowledged that immortalized cell lines can be used to build microphysiological systems, benefitting from their predictability, ease of culture, and low handling costs, despite having lost the complexities of the original tissue from which they were isolated.
Figure 1.
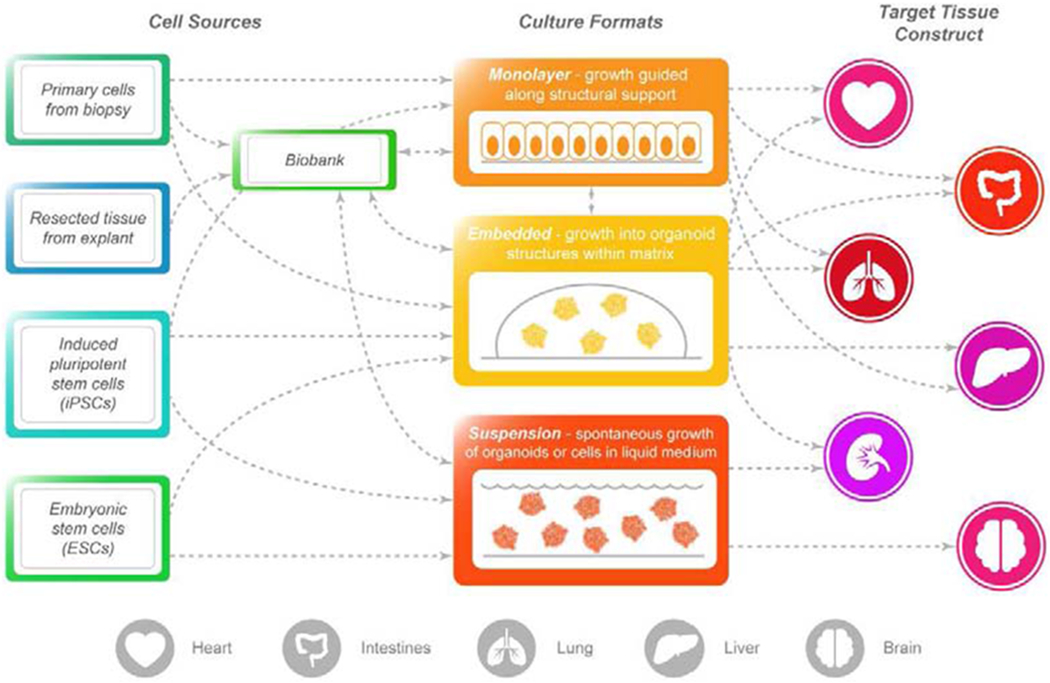
Sources of tissue and example culture schemes to expand the primary cells. Diagrams indicate potential routes that can be taken for expansion of primary cells for culture within microphysiological systems that recapitulate functions of specific organs.
A second consideration for the development of in vitro tissue replicas is the organization of cell types into distinct regions that are reminiscent of in vivo tissue structure. Porous membranes are often used to place different cell types at opposing boundaries, thus generating an artificial tissue boundary analogous to that found in vivo. Microfabrication technologies such as 3D printing have the potential to build a variety of cell types with precise 3D architecture. To maintain cell viability, nutrient/oxygen supply and waste removal should be specifically considered for cell culture in microphysiological systems as they are usually in miniaturized formats with reduced media volume and non-convective geometries. Microfluidic perfusion [12], frequent medium replenishes [13,14], or integrations of mechanical mixers and gas reservoirs [15] have been used to facilitate the rates of nutrient/oxygen delivery and waste removal.
For adherent cells, extracellular matrix (ECM) is an important component to consider. Beyond providing binding sites for cell adhesion, material properties such as stiffness, viscoelasticity, and porosity should be considered to correctly recapitulate the cell phenotypes, gene expression and key functions of the target tissue. Tumor-cell-derived ECMs (e.g., Matrigel) are frequently used for culture of 3D organoids; however, the composition of cell-derived ECMs are not well defined and vary batch to batch [16,17]. Purified protein or polysaccharide ECMs (e.g., collagens, hyaluronic acids, alginate, and chitosan) are also used as matrices for 2D and 3D culture and are less costly compared to Matrigel. Moreover, as the biochemical properties and molecular structures of these purified, naturally-derived ECMs are well defined, the material properties are easier to tune than cell-derived ECMs, with preservation of cell compatibility. Synthetic polymer ECMs are also used in 2D and 3D culture when there is a necessity to tailor the material and structural properties of the matrix with the highest precision [18]. Dynamic modification such as photoreactive crosslinking [19,20], physical and chemical crosslinking [21], degradation [22], or enzyme driven modification [23,24] can be exploited to alter the mechanical properties [19,20] or create 3D microstructures [14,25]. However, protein coatings or chemical conjugation of cell binding moieties may be necessary to promote cell adhesion. Synthetic polymers in general are reproducibly manufactured, economical, and free of endotoxin or nucleases, but the cell behaviors including proliferation and cell phenotypes cannot always be fully replicated.
Implementing physiological features of the tissue (e.g., shear forces, 3D microstructures, dynamic strains) or in situ sensing units (e.g., electrical, mechanical, biochemical) for large scale use require materials and methods that are compatible with manufacturing methods (Fig. 2). Since the key features for successful tissue subunit recapitulation depend on the organ, material selection ideally should be target-tissue specific. However, there are manufacturing considerations that are universal across all in vitro tissue systems. All materials used that contact cells should be biocompatible, reproducibly manufacturable, and exhibit minimal variation within a batch or between batches. Absorption of small molecules should be mitigated for drug screening applications by choice of material or surface treatments and coatings. Compatibility with the standardized multiwell plates is advantageous for automation, and to fit into existing workflows and assay platforms. Optical transparency and low fluorescence background are required if the tissue properties will be evaluated by microscopic imaging. Examples of common chemical and physical attributes for tissue subunits to guide microphysiological device design are highlighted in Fig. 3.
Figure 2.
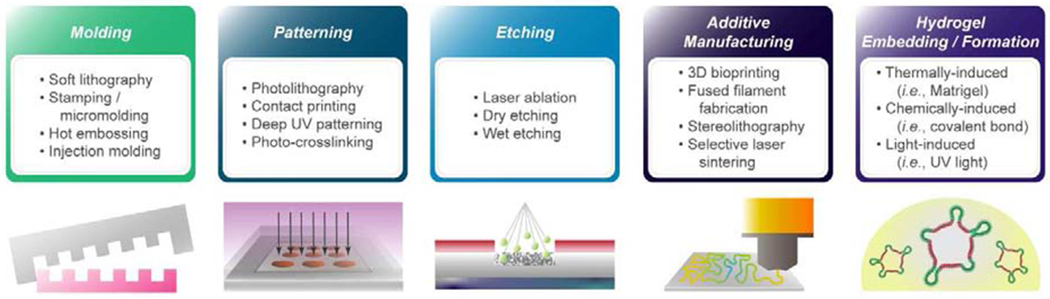
Example manufacturing categories and techniques for microphysiological systems.
Figure 3.
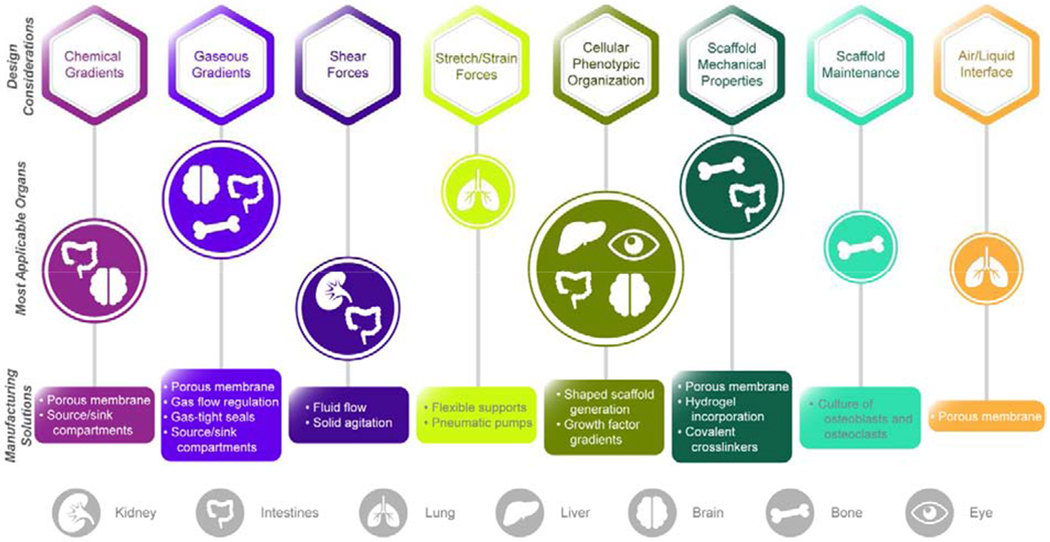
Design considerations and commonly utilized solutions for the manufacturing of microphysiological systems, with representative organs pertinent to each category highlighted in the central row.
Highlights of leading organ replica technologies
Hundreds of MPS devices exist throughout published literature, with a handful of commercial products under development. Within this section, we highlight several technologies developed in academic laboratories that push the limits of manufacturing and analytical scalability. Specifically, these technologies exhibit exceptional promise for automation of fabrication, tissue culture, and/or analytical assays, ultimately rendering the collection of high content information accessible to a greater number of end-users.
Miniaturized Bioreactors for Brain Organoids.
Brain organoids have shown potential for high throughput compatibility with in vivo-like tissue complexity. Typically, generation of brain organoids begins by forming an embryonic body from iPSC or ESC, followed by neuroectoderm induction and expansion, and finally differentiation leading to mature brain organoids. The initial embryonic body can be formed in a high throughput manner (i.e., multiwell plates). However, as the organoids expand in size and cell number over time, static culture in multiwell plates often fails to provide efficient transport of oxygen and nutrients, leading to cell death in the long-term [26]. Qian and colleagues developed miniaturized spinning reactors, SpinΩ, to enable long term brain organoid culture (Fig. 4A). Representative regions include forebrain organoids containing all 6 cortical layers of the human fetal brain, as well as midbrain and hypothalamus organoids [27]. This system offers a cost-efficient and high-throughput compatible platform for delivering nutrients and oxygen, which may have applications for other cell and tissue types.
Figure 4.
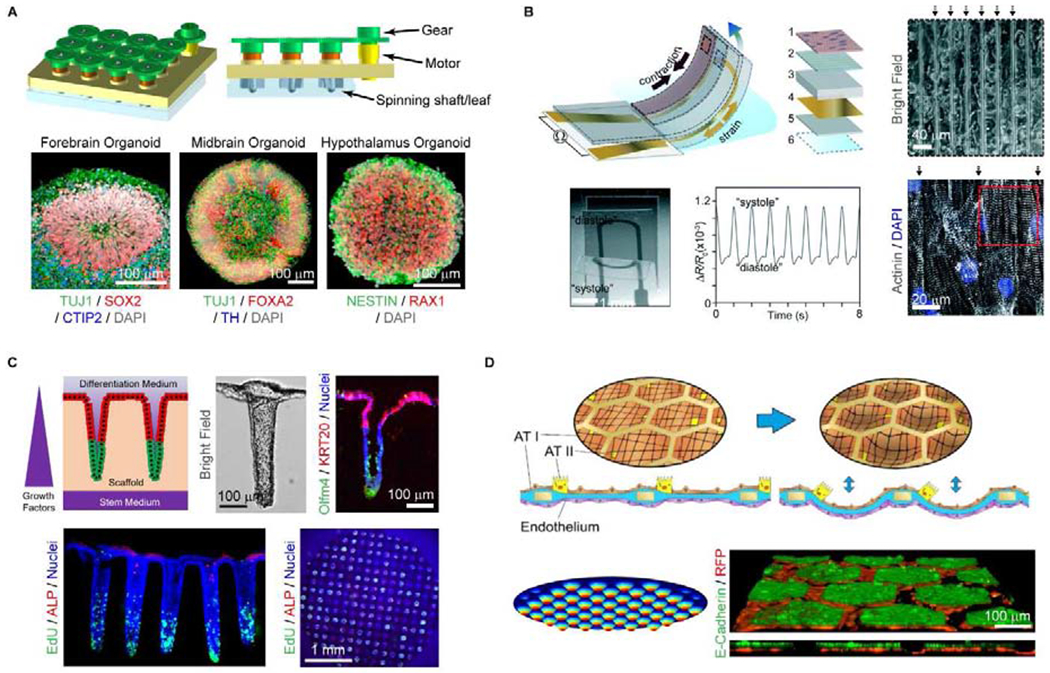
Recent technologies pushing current limitations of scalability and throughput. A) Diagram of SpinΩ bioreactor used for the formation of brain organoids. Region-specific organoids grown within the bioreactor are displayed via confocal microscopy in lower panels. Reproduced from Ref. [27] with permission from Elsevier. B) Schematic of instrumented cardiac microphysiological device, capable of measuring cardiac stresses within cell incubator environments in real-time. Reproduced from Ref. [29] with permission from The Royal Society of Chemistry. C) Generation of in vitro human colonic crypt arrays by application of chemical gradients across shaped collagen hydrogels. Reproduced from Ref. [11] with permission from Elsevier. D) Formation of lung alveoli replicas over a collagen/elastin membrane suspended within a gold mesh. The segregated membrane areas, with primary co-cultured lung epithelial (E-cadherin+) and endothelial (RFP+) cells, can be stretched and analyzed in parallel. Reproduced from P Zamprogno, et al., bioRxiv doi: 10.1011/608919, with permission from the corresponding author.
3D Printing of Cardiac Muscle Subunits.
Lind et. al. developed a cardiac MPS using a novel multimaterial three-dimensional (3D) printing method [28,29]. The group integrated multiple functional inks with different properties to grow multiple tissue constructs on a multiwell device with embedded sensors (Fig. 4B). The ability to print grooved micro-architectures using soft polydimethylsiloxane (PDMS) within each culture site facilitated the self-guided assembly of progenitor cells into electrically anisotropic layers of cardiac tissue. Multimaterial 3D printing incorporated electrically conductive, soft strain gauge sensors for a continuous, instantaneous, non-invasive readout. The cardiac tissue layers displayed physiologic functions, including contraction, in response to appropriate stimuli and increased or decreased contractility upon application of cardiac-active drugs. An advantage of this platform was the wide variety of printable polymer-based materials employed on a single, programmable fabrication platform and in particular the highly parallelizable embedded electric sensors to provide a continuous, near-instantaneous readout.
Hydrogel Molding for In Vitro Intestinal Crypts.
The development of organoid and monolayer culture methods for intestinal epithelial stem cells (IESCs) has enabled primary cell-derived, physiologically relevant, in vitro models of the intestines [13,14,30–37]. To create high-fidelity replicas of the microarchitecture and asymmetry of human colonic epithelium, arrays of crypt-like invaginations shaped from crosslinked collagen hydrogels mimicked the shape, size and density of human colon crypts [14] (Fig. 4C). Application of a growth factor gradient was sufficient to polarize the crypts so that the stem/proliferative cells were restricted at the base of the crypts (forming a stem cell niche) while various differentiated cell types were located at the luminal surface. The crypts responded appropriately to the luminal application of short-chain fatty acids or basal application of inflammatory cytokines. Platforms such as these enable replication of the intestinal microarchitecture, epithelial cell compartmentalization and polarity as well as key intestinal features such as chemical and gas gradients. Each cassette, containing <120 patterned crypts, exhibits great potential as a high-content and physiologically relevant cellular assay tool. While currently in 12-well format, innovations in automated scaffold fabrication strategies and 3D microscopy methods will maximize throughput.
Stretchable Thin Films to Mimic Lung Alveoli.
Generation of scalable models of the lung epithelium has presented a challenge in that appropriate mechanical forces must be simultaneously applied across all arrayed elements to simulate the rhythmic stretching and relaxation that occurs in vivo [38,39]. A lung-on-a-chip platform in which primary lung epithelial and endothelial cells were supported over a biomimetic collagen/elastin membrane covering the arrayed hexagonal holes of an electron microscopy grid, replicated lung alveoli both in the architecture and physical forces during breathing (P Zamprogno et al., bioRxiv doi: 10.1011/608919, Fig. 4D). Application of a cyclic negative pressure below the membrane enabled the in vitro alveoli to deflect in concert, experiencing radial strains similar to those found in vivo. Given that the dimensions of each hexagonal area (i.e., 225 μm dia.) were nearly identical to those of in vivo lung alveoli (160 – 200 μm), the platform represented an impressive scalable mimic of its in vivo counterpart.
Considerations for commercialization
Though MPS carries tremendous potential to advance the areas of personalized medicine and drug development, there are significant hurdles that the field must overcome in order for MPS to be commercially viable and executable across laboratories [40]. Certain device features could make a technically capable method great for academic research and prototyping, yet hinder scale-up and widespread use. Concepts of scale often vastly differ between academic and industrial settings, with fabrication of 1 – 100 units considered sufficient for feasibility studies and/or peer-reviewed publication, though 100,000+ units potentially required for launch of a commercial product [41]. While the considerations below are not intended to be exhaustive, it is important that developers of MPS technology begin studying whether these issues are present and how to overcome them.
Sourcing of raw materials. Raw materials that are to be included in the physical device or cassette, which may include plastics, polymers, and hydrogels, must not only be sourced for analytical assay compatibility, yet must also be done so at a large volume that meets good manufacturing practice (GMP) constraints, such as batch-to-batch reproducibility and the ability to apply robust standard operating procedures (SOP) during fabrication and assembly [42,43]. A commonly used material for organ-on-chip microfluidic devices is poly(dimethylsiloxane) (PDMS), which is biocompatible, widely accessible, and easy-to-use. However, attempted scaling of PDMS based devices reveals hidden issues and costs, including the need for large-volume fabrication of master replicas, and variabilities in obtaining high-resolution features during polymer molding [3,44]. Moreover, due to the high propensity of PDMS to absorb hydrophobic small molecules and proteins [45], antifouling solutions must be optimized and validated for each application and device.
Cell selection and validation. Cells cultured within MPS are also subject to GMP compliance to provide assurance that any conclusions drawn are reliable and predictive, and thus must be selected, cultured, and validated accordingly. Geraghty et al. have provided a general protocol for acquisition, naming, and usage of new cell lines, with recommendations for avoiding pitfalls that could result in contamination or unreliable results [46]. Methods for scaled-up primary stem cell manufacturing are under active investigation [47], and it is generally accepted that all culture conditions should be fully defined and free of animal-derived substances [48–50]. With the above in mind, studies must be performed to determine the biological impact of donor-to-donor and passage-to-passage differences, which may inform decisions on whether multiple donors must be included for every assay and suitable “passage windows” in which these cells can be reliably utilized. While karyotyping can be a robust and informative method for quality control (QC), it is becoming clear that more in-depth characterizations are required for assessment of genetic drift of stem cells throughout culture, which may include short tandem repeat profiling, copy number variation mapping, or whole-genome sequencing [46,51].
Assembly and quality control. Given the complexity of most MPS, which integrate both living and non-biological components/mechanics, it is expected that some assembly will be required by the end-user (e.g., securing of all device components and seeding of cells), preventing a complete QC assessment of each assembled device by the manufacturer. Instead, QC will likely have to be based on thorough benchmarking of the separate components (e.g., genetic drift assessment of cells, size tolerance of machined components, coefficients of variation for embedded sensor responses), in accordance with the considerations for raw materials and cells outlined above. Further, this will necessitate providing detailed and unambiguous protocols for final MPS assembly and usage to the end-user, which will require continuous optimization and validation by the manufacturer.
Journey to the end-user. Shipping of cell-based platforms over large distances and varying environmental conditions may present several concerns. What is the shelf life of each constituent? Must the devices and cells be shipped in an expedited fashion, under separate conditions, and are there federal transport agreements that must be agreed upon ahead of time? Once the device reaches the end-user, storage of each component much be clearly delineated, and/or device packaging must be designed in such a way that it is easy to use and store appropriately.
As suggested by these points, to reach their full potential these devices will ultimately have to meet regulatory guidelines where ruggedness and inter-laboratory repeatability are critical. The extensive testing that will be required may potentially need to be facilitated through public-private partnerships [52].
Outlook: Regulatory approval and broad adoption
In the United States, regulatory science is “the science of developing new tools, standards, and approaches to assess the safety, efficacy, quality, and performance of all FDA-regulated products” [53]. Since regulatory science is used to make decisions that can have life-and-death consequences for large numbers of people, it employs consensus and time-proven methods. Within this context, the FDA has developed innovative approaches to advance regulatory science, such as its Advancing Regulatory Science Initiative and Critical Path Initiative. Section #1 of the FDA’s “Strategic Plan for Regulatory Science” addresses the need to modernize toxicology to enhance product safety [53]. One of the aims of MPS to-date has been to supplement or even replace in vivo toxicology testing. FDA’s “Predictive Toxicology Roadmap” also discusses MPS as a promising new technology [54]. FDA, DARPA and NIH-NCATS have made significant contributions to these efforts through a collaborative “Tissue Chip” program with the ambitious goal of developing a “human-body-on-a-chip” for drug screening [55]. Outside of the United States, high enthusiasm exists for advancing MPS technology, with the European Union (EU) funded Project ORCHID (Organ-on-Chip In Development) advocating for increased awareness of organ-on-chip technology and providing a roadmap for organ-on-chip adoption throughout the EU [56,57].
MPS have potential applications in regulatory science that go far beyond toxicology testing. In particular, the ability of MPS to recapitulate a significant functional aspect of an organ or system can benefit preclinical performance and safety testing of medical devices, reducing or replacing the burden of animal testing [58]. With the potential for application to so many different medical products across multiple government agencies, a useful thought experiment is to ask what qualities MPS should embody to be highly successful tools for regulatory science. All regulatory tools have basic requirements such as precision, reproducibility, and robustness [59,60], but what are some particular areas that could be challenging or problematic for MPS that deserve further research and attention early in development? A few to consider are:
Determining which biological signals are clinically important for human health. There are many possible endpoints that can be measured in an MPS, and it will be important to conclude which ones can consistently relate to effects in humans.
Challenges of representing the whole with a small sample size. Micro-sized samples often do not contain a large enough population to be representative of the distribution of all cell behaviors, etc. in an organ or system. This would likely be caught in reproducibility testing, but may only show up as a problem when certain test variables are employed which select for sub-populations.
Challenges in comparing data with large numbers of variables that impact the outcome. We have found that the more input variables are involved in determining the outcome of an in vitro test, the less likely that it is useful across multiple companies or products because of the inability to reproduce every one of those inputs in testing. With a small number of variables some assumptions and corrections can be made, but with MPS, which are highly complex systems, it will be essential to have very fine control over many parameters. This stresses the urgent nature of having consensus guidelines for competing technologies with similar measurement goals.
It will be important for MPS to make the transition from answering research driven questions to satisfying requirements pertaining to regulatory decision making. Mature, robust MPS platforms with a significant public health benefit will catalyze continuing public-private collaborations and facilitate future regulatory application.
Acknowledgements
We gratefully acknowledge financial support from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health (R01 DK109559). Furthermore, the authors thank Prof. Christopher E. Sims and Dr. Kristy S. McKeating for their insights on manufacturing and commercialization.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: Disclaimer
The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services. The findings and conclusions in this Perspective have not been formally disseminated by the US Food and Drug Administration and should not be construed to represent any agency determination or policy.
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Nancy Allbritton and Yuli Wang have a financial interest in Altis Biosystems, Inc. The remaining authors declare no competing financial interests.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Bhatia SN, Chen CS: Tissue engineering at the micro-scale. Biomed Microdevices 1999, 2:131–144. [Google Scholar]
- 2.Reardon S: ‘Organs-on-chips’ go mainstream. Nature 2015, 523:266. [DOI] [PubMed] [Google Scholar]
- 3.Probst C, Schneider S, Loskill P: High-throughput organ-on-a-chip systems: Current status and remaining challenges. Curr Opin Biomed Eng 2018, 6:33–41. [Google Scholar]
- 4.Wikswo JP, Block FE, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, et al. : Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng 2013, 60:682–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon K, Clouaire T, Bao XX, Kemp SE, Xenophontos M, de Las Heras JI, Stancheva I: Immortality, but not oncogenic transformation, of primary human cells leads to epigenetic reprogramming of DNA methylation and gene expression. Nucleic Acids Res. 2014, 42:3529–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M: Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics 2009, 8:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou AH, Voorhoeve PM, Lan WW, Tin MQ, Tong L: Comparison of gene expression profiles in primary and immortalized human pterygium fibroblast cells. Exp. Cell Res 2013, 319:2781–2789. [DOI] [PubMed] [Google Scholar]
- 8.Herbst-Kralovetz MM, Quayle AJ, Ficarra M, Greene S, Rose WA, Chesson R, Spagnuolo RA, Pyles RB: Quantification and comparison of toll-like receptor expression and responsiveness in primary and immortalized human female lower genital tract epithelia. Am. J. Reprod. Immunol 2008, 59:212–224. [DOI] [PubMed] [Google Scholar]
- 9.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J,Taylor-Weiner A, Kester L, et al. : Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161:933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews PW, Benvenisty N, McKay R, Pera MF, Rossant J, Semb H, Stacey GN, Initia SCISC: The international stem cell initiative: Toward benchmarks for human embryonic stem cell research. Nat. Biotechnol 2005, 23:795–797. [DOI] [PubMed] [Google Scholar]
- 11.Di Nardo P, Parker GC: Stem cell standardization. Stem Cells Dev 2011, 20:375–377. [DOI] [PubMed] [Google Scholar]
- *12.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE: Reconstituting organ-level lung functions on a chip. Science 2010, 328:1662–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lung epithelial and endothelial cells were cultured on opposite sides of a porous membrane within a poly(dimethylsiloxane) microfluidic device, to which stretch-strain forces could be applied. This is one of the first organ-on-a-chip systems replicating organ-level physiology and has since advanced to market.
- 13.Wang Y, Gunasekara DB, Reed MI, DiSalvo M, Bultman SJ, Sims CE, Magness ST, Allbritton NL: A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 2017, 128:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Wang Y, Kim R, Gunasekara DB, Reed MI, DiSalvo M, Nguyen DL, Bultman SJ, Sims CE, Magness ST, Allbritton NL: Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell. Mol. Gastroenterol. Hepatol. 2018, 5:113–130. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scaffolds of covalently crosslinked collagen were shaped into the microscale architecture of the large intestinal epithelium, over which human patient-derived intestinal epithelial cells were grown and subjected to various biochemical gradients known to exist in vivo. This platform benefits from high sampling efficiency, analytical assay compatibility, and the ability to co-culture the different cell types of the intestine in an organized and physiologically accurate manner.
- 15.Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming G-l: Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc 2018, 13:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serban MA, Prestwich GD: Modular extracellular matrices: Solutions for the puzzle. Methods 2008, 45:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen EH, Daly WT, Le NNT, Farnoodian M, Belair DG, Schwartz MP, Lebakken CS, Ananiev GE, Saghiri MA, Knudsen TB, et al. : Versatile synthetic alternatives to matrigel for vascular toxicity screening and stem cell expansion. Nat Biomed Eng 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordonez-Moran P, Clevers H, Lutolf MP: Designer matrices for intestinal stem cell and organoid culture. Nature 2016, 539:560–564. [DOI] [PubMed] [Google Scholar]
- 19.Jeon O, Bouhadir KH, Mansour JM, Alsberg E: Photocrosslinked alginate hydrogels with tunable biodegradation rates and mechanical properties. Biomaterials 2009, 30:2724–2734. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell CD, Zhang B, Onofrillo C, Duchi S, Blanchard R, Quigley A, Bourke J, Gambhir S, Kapsa R, Di Bella C, et al. : Tailoring the mechanical properties of gelatin methacryloyl hydrogels through manipulation of the photocrosslinking conditions. Soft Matter 2018, 14:2142–2151. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Sun Q, Li Q, Kawazoe N, Chen G: Functional hydrogels with tunable structures and properties for tissue engineering applications. Front Chem 2018, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarem M, Arya N, Heizmann M, Neffe AT, Barbero A, Gebauer TP, Martin I, Lendlein A, Shastri VP: Interplay between stiffness and degradation of architectured gelatin hydrogels leads to differential modulation of chondrogenesis in vitro and in vivo. Acta Biomater. 2018, 69:83–94. [DOI] [PubMed] [Google Scholar]
- 23.Schweller RM, Wu ZJ, Klitzman B, West JL: Stiffness of protease sensitive and cell adhesive PEG hydrogels promotes neovascularization in vivo. Ann. Biomed. Eng 2017, 45:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruz-Acuña R, Quirós M, Farkas AE, Dedhia PH, Huang S, Siuda D, Garcia-Hernández V, Miller AJ, Spence JR, Nusrat A, et al. : Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol 2017, 19:1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira RF, Bártolo PJ: 3D bioprinting of photocrosslinkable hydrogel constructs. J. Appl. Polym. Sci 2015, 132. [Google Scholar]
- 26.Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA: Cerebral organoids model human brain development and microcephaly. Nature 2013, 501:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Qian X, Nguyen Ha N, Song Mingxi M, Hadiono C, Ogden Sarah C, Hammack C, Yao B, Hamersky Gregory R, Jacob F, Zhong C, et al. : Brain-region-specific organoids using minibioreactors for modeling ZIKV exposure. Cell 2016, 165:1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miniaturized spinning bioreactor inserts were designed and 3D printed to enable scalable and reproducible culture of brain-region-specific organoids from human induced pluripotent stem cells within standardized 12-well plates. Forebrain, midbrain, and hypothalamus organoids were successfully generated within this system, and the efficacies of selected Zika antiviral drugs were tested.
- **28.Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan HY, Yadid M, Park SJ, Kotikian A, Nesmith AP, Campbell PH, et al. : Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater 2017, 16:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]; Multimaterial three-dimensional printing was applied to generate cardiac microphysiological devices with integrated sensors that are capable of monitoring tissue contractile stresses within incubator environments in real-time. Beyond confirmation of systole and diastole measurements within the device, applications for drug monitoring and contractile development characterization were demonstrated.
- 29.Lind JU, Yadid M, Perkins I, O’Connor BB, Eweje F, Chantre CO, Hemphill MA, Yuan HY, Campbell PH, Vlassak JJ, et al. : Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab Chip 2017, 17:3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. : Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459:262–265. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, DiSalvo M, Gunasekara DB, Dutton J, Proctor A, Lebhar MS, Williamson IA, Speer J, Howard RL, Smiddy NM, et al. : Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell. Mol. Gastroenterol. Hepatol 2017, 4: 165–182.e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutton JS, Hinman SS, Kim R, Wang Y, Allbritton NL: Primary cell-derived intestinal models: Recapitulating physiology. Trends Biotechnol. 2019, 37:744–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, et al. : Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci Rep 2018, 8:2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, Hamilton GA, Targan SR, Svendsen CN, Barrett RJ: Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterol. Hepatol 2017, 5:669–677.e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim R, Wang Y, Hwang S-HJ, Attayek PJ, Smiddy NM, Reed MI, Sims CE, Allbritton NL: Formation of arrays of planar, murine, intestinal crypts possessing a stem/proliferative cell compartment and differentiated cell zone. Lab Chip 2018, 18:2202–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim R, Attayek PJ, Wang Y, Furtado KL, Tamayo R, Sims CE, Allbritton NL: An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 2019, DOI: 10.1088/1758-5090/ab446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinman SS, Wang Y, Allbritton NL: Photopatterned membranes and chemical gradients enable scalable phenotypic organization of primary human colon epithelial models. Anal Chem 2019, 91:15240–15247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stucki AO, Stucki JD, Hall SR, Felder M, Mermoud Y, Schmid RA, Geiser T, Guenat OT: A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip 2015, 15:1302–1310. [DOI] [PubMed] [Google Scholar]
- 39.Stucki JD, Hobi N, Galimov A, Stucki AO, Schneider-Daum N, Lehr CM, Huwer H, Frick M, Funke-Chambour M, Geiser T, et al. : Medium throughput breathing human primary cell alveolus-on-chip model. Sci. Rep 2018, 8:14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang BY, Radisic M: Organ-on-a-chip devices advance to market. Lab Chip 2017, 17:2395–2420. [DOI] [PubMed] [Google Scholar]
- 41.Kumar AA: Creating novel medical diagnostics with a design for manufacturing. Transl Mater Res 2018, 5:024001. [Google Scholar]
- 42.International Organization for Standardization: ISO 13485:2016: Medical devices – Quality management systems -- Requirements for regulatory purposes. https://www.iso.org/standard/59752.html, 2016. (accessed December 10, 2019).
- 43.Electronic Code of Federal Regulations: Title 21, Chapter I, Subchapter H, Part 820. https://www.ecfr.gov/cgi-bin/text-idx?SID=cbf078d98db332db6af97029d2030bbe&mc=true&tpl=/ecfrbrowse/Title21/21cfr820_main_02.tpl, 2019. (accessed December 10, 2019).
- 44.Novak R, Didier M, Calamari E, Ng CF, Choe Y, Clauson SL, Nestor BA, Puerta J, Fleming R, Firoozinezhad SJ, et al. : Scalable fabrication of stretchable, dual channel, microfluidic organ chips. J Vis Exp 2018, e58151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toepke MW, Beebe DJ: PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6:1484–1486. [DOI] [PubMed] [Google Scholar]
- 46.Geraghty RJ, Capes-Davis A, Davis JM, Downward J, Freshney RI, Knezevic I, Lovell-Badge R, Masters JRW, Meredith J, Stacey GN, et al. : Guidelines for the use of cell lines in biomedical research. Brit J Cancer 2014, 111:1021–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VC, Couture LA, Gold J: Application of the suspension culture system for scale-up manufacture of hPSCs and hPSC-derived cardiomyocytes. Card Vasc Biol 2017, doi: 10.1007/978-3-319-56106-6_7:145–161. [DOI] [Google Scholar]
- 48.Serra M, Brito C, Correia C, Alves PM: Process engineering of human pluripotent stem cells for clinical application. Trends Biotechnol 2012, 30:350–359. [DOI] [PubMed] [Google Scholar]
- 49.Unger C, Skottman H, Blomberg P, Dilber MS, Hovatta O: Good manufacturing practice and clinical-grade human embryonic stem cell lines. Hum Mol Genet 2008, 17:R48–R53. [DOI] [PubMed] [Google Scholar]
- 50.Lei YG, Schaffer DV: A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc Natl Acad Sci USA 2013, 110:E5039–E5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes P, Marshall D, Reid Y, Parkes H, Gelber C: The costs of using unauthenticated, overpassaged cell lines: How much more data do we need? Biotechniques 2007, 43:575–584. [DOI] [PubMed] [Google Scholar]
- *52.Livingston CA, Fabre KM, Tagle DA: Facilitating the commercialization and use of organ platforms generated by the microphysiological systems (tissue chip) program through public-private partnerships. Comput Struct Biotec 2016, 14:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review provides details of the Tissue Chip Program, organized by the National Center for Advancing Translation Science (NCATS), that are relevant to academic labs seeking to make their technology widely available. Among other topics, the role of public-private partnerships, regulatory considerations, and routes toward commercialization are thoroughly detailed.
- 53.U.S. Food and Drug Administration, Advancing regulatory science at FDA: A strategic plan. https://www.fda.gov/science-research/advancing-regulatory-science/strategic-plan-regulatory-science, 2011. (accessed September 21, 2019).
- 54.U.S. Food and Drug Administration, FDA’s predictive toxicology roadmap. https://www.fda.gov/media/109634/download, 2019. (accessed September 21, 2019).
- 55.National Center for Advancing Translational Sciences, How the tissue chip program works. https://ncats.nih.gov/tissuechip/about/operations, 2018. (accessed September 21, 2019).
- 56.Mastrangeli M, Millet S, Orchid Partners T, Van den Eijnden-van Raaij J: Organ-on-chip in development: Towards a roadmap for organs-on-chip. ALTEX 2019, 36:650–668. [DOI] [PubMed] [Google Scholar]
- 57.Mastrangeli M, Millet S, Mummery C, Loskill P, Braeken D, Eberle W, Cipriano M, Fernandez L, Graef M, Gidrol X, et al. : Building blocks for a european organ-on-chip roadmap. ALTEX 2019, 36:481–492. [DOI] [PubMed] [Google Scholar]
- 58.Guan A, Hamilton P, Wang Y, Gorbet M, Li ZY, Phillips KS: Medical devices on chips. Nat. Biomed. Eng 2017, 1:0045. [Google Scholar]
- 59.Van Arnum P, Robustness in analytical methods outlined. http://www.pharmtech.com/robustness-analytical-methods-outlined, 2007. (accessed September 21, 2019).
- 60.Hendricks MMWB, Boer JHd, Smilde AK: Robustness of analytical chemical methods and pharmaceutical technological products. Amsterdam ; New York: Elsevier; 1996. [Google Scholar]


