Abstract
Faithful renewal of aging and damaged tissues is central to organismal lifespan. Stem cells (SCs) generate the cellular progeny that replenish adult tissues across the body but this task becomes increasingly compromised over time. The age related decline in SC-mediated tissue maintenance is a multifactorial event that commonly affects genome integrity. The presence of DNA damage in SCs that are under continuous demand to divide poses a great risk for age-related disorders such as cancer. However, performing analysis of SCs with genomic instability and the DNA damage response during tissue renewal present significant challenges. Here we introduce an alternative experimental system based on the planaria flatworm Schmidtea mediterranea to address at the organismal level studies intersecting SC-mediated tissue renewal in the presence of genomic instability. Planaria have abundant SCs (neoblasts) that maintain high rates of cellular turnover and a variety of molecular tools have been developed to induce DNA damage and dissect how neoblasts respond to this stressor. S. mediterranea displays high evolutionary conservation of DNA repair mechanisms and signaling pathways regulating adult SCs. We describe genetically induced-DNA damage models and highlight body-wide signals affecting cellular decisions such as survival, proliferation, and death in the presence of genomic instability. We also discuss transcriptomic changes in the DNA damage response during injury repair and propose DNA repair as key component of tissue regeneration. Additional studies using planaria will provide insights about mechanisms regulating survival and growth of cells with DNA damage during tissue renewal and regeneration.
Keywords: DNA damage, DNA repair, DNA double strand breaks, planaria, neoblasts, regeneration, invertebrates, stem cells
1. Introduction
Preserving genomic integrity is essential to life. However, DNA is under constant threat from multiple sources, which include errors during DNA replication, products of intrinsic cellular reactions (e.g. reactive oxygen species) and environmental factors such as UV radiation, chemical exposure, etc (Figure 1A) [1–5]. In humans, for example, these persistent insults generate about 105 DNA lesions per cell every 24 hours [6–8]. If left unchecked, DNA damage can be transmitted to cellular progeny and potentially compromise tissue integrity and function [3, 6, 9–14]. Indeed, about 90% of cancer-related deaths worldwide originate from abnormalities in tissues that are constantly renewed by stem cells (SCs) [5, 15–17]. Genomic instability (i.e. higher rate of genomic changes per cell division) is a major trait in almost all cancers, but the basic mechanisms regulating survival and growth of cells with DNA damage during tissue renewal remain a puzzling biomedical problem.
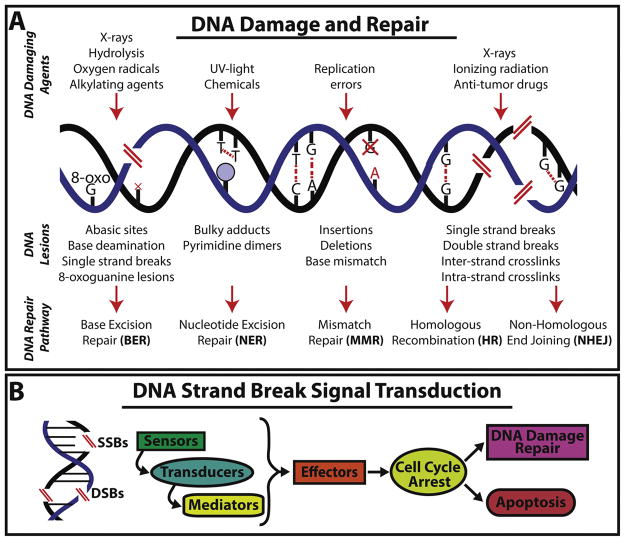
Figure 1. DNA damage responses and stranded break signal transduction.
(A) The illustration summarizes different types of DNA damaging agents, the genomic lession they produce and the specialized DNA repair pathway deployed. For example, expossure to ionizing radiation may lead to DNA single and/or double-stranded breaks and crosslinks that can be repaired through HR or NHEJ (B) Different molecules mediate the repair of DNA strand breaks. Commonly, a signal transduction cascade involving sensors, transducers, mediators, and effector molecules that together influence cellular fate decisions to repair the damage or undergo programmed cell death.
Exposure to DNA damaging agents generally lead to lesions that are common to all living organisms (Figure 1A) [6, 18, 19]. Thus, highly conserved mechanisms of DNA repair have evolved to preserve genetic information and proper cellular function [19–23]. DNA damage response (DDR) sensors and effectors are continuously deployed and are mediated by a specific set of proteins with the goal of re-establishing genomic integrity (Figure 1B) [19]. Importantly, the timely deployment of DDR is synchronized with cellular responses leading to critical decisions that may involve cell cycle arrest, apoptosis, senescence and DNA repair (Figure 1B) [18, 24]. Altogether, the varying responses to DNA damage aim at preventing exhaustion and abnormal transformation of SC pools while maintaining their ability to mediate tissue homeostasis.
The process of DNA damage and its cellular response have been widely documented by in vitro studies and organ specific experimental models. However, the field has benefited less by simultaneous analysis of DNA damage and SC-mediated tissue renewal at the organismal level. We believe that studies merging the cellular response to DNA damage, while attending body demands of cellular turnover may bring important insights about intercellular crosstalk that affects cellular fate decisions in the adult body. For example, there are patterns of regional differences of cell proliferation along the anteroposterior (AP) body axis that affect the fate of SCs and their progeny during tissue renewal, regeneration and carcinogenesis [25–31]. These regional differences are evolutionarily conserved across different species and recent studies have shown that the fate of cells with DNA damage are susceptible to regional signals [22, 23]. In an attempt to complement studies of DDR/DNA repair during tissue renewal and cancer formation, we introduce a simplified model system represented by the planaria flatworm. Planaria possess high rates of cellular turnover and tissue regeneration is driven by adult SCs called neoblasts, which facilitate studies about SC-mediated tissue renewal and DNA damage [32–37]. We present a brief description of the DNA lesions and the molecules involved in repair with special emphasis on double-strand breaks (DSBs), the most dangerous form of DNA damage [8, 20, 38, 39]. We highlight the role of DNA damage during tissue renewal and its possible impact in aging and discuss how recent experimental planaria models associated with DNA damage may provide insights about the SC response during adult tissue maintenance and regeneration. In addition, we identify through data mining of various transcriptomic datasets that the DDR is a critical component of the large-scale tissue homeostasis and regeneration in planaria. Ultimately, we propose the use of planaria as a convenient model to address evolutionarily conserved mechanisms of DDR and DNA repair during tissue repair and regeneration in the adult body.
2. Tissue renewal and stem cell response to DNA damage
Organismal lifespan relies on faithful renewal of aging and damaged tissues [40–46]. SCs generate cellular progeny to maintain adult tissues and in humans, this is a daunting process that requires daily demand of billions of cells that could span over a century [47–50]. Tissue renewal is extremely complex, fulfilling different dynamics of cellular turnover that appear unsynchronized among tissues. For example, the small intestinal epithelium is renewed in about 5 days, while epidermal cells in the skin are replaced every 10–30 days, let alone cells within blood tissue have different renewal rates from 1 day to several months [51–54]. Despite its relevance to physiology and disease, it remains poorly understood how this large scale renewal process is coordinated and how it becomes liable as organisms grow older.
Both tissue maintenance and the capacity to preserve genomic integrity decline with age. Intriguingly, there is positive correlation between DNA repair and lifespan in a variety of organisms [40, 41, 55, 56]. For example, longer-lived species such as humans and naked mole rats consistently display higher expression of genes associated with DNA repair, which supports the idea that preservation of genomic integrity is paramount to the longevity of an organism [41]. The mechanistic process by which genome integrity declines with age is not well understood. Nonetheless, several lines of evidence imply continued exposure of SCs to DNA damage plays a major role in age-related dysfunctions such as cancer and degenerative diseases [1, 3, 9, 16, 57–62]. Eventually, unrepaired genetic lesions may result in SC attrition, cellular transformation and aberrant differentiation that could lead to defective tissue renewal [1, 3, 9, 10, 61–63]. It is unclear whether the increased genome alterations are due to individual or combined effects of: (i) impaired DDR, (ii) increased levels of DNA insults, (iii) epigenetic modifications and telomere shortening with age, (iv) higher susceptibility to damaging agents in SC and progenitor populations (e.g. quiesent, cycling), and/or (v) defects in cell fate decision mechanisms upon demands of cellular turnover. In reality, this list could be more extensive when cellular turnover is considered in the complexity of the whole organism. Systemic factors associated with inflammation, oxidative stress, metabolism, etc, which also have inputs on decisions of cellular proliferation and apoptosis during tissue turnover [1, 3, 10, 61, 63–67]. Collectively, DNA damage greatly influences the ultimate fate of the cell. Nevertheless, it is less clear how cellular decisions are prioritized when physiological demands of tissue turnover are in play and how cancer and degenerative diseases evolve from defects in these cellular decisions.
Increasing evidence demonstrates that preservation of genomic integrity and systemic reduction in DNA damage could be enhanced by physiological or pharmacological manipulations. Specifically, treatments aimed at replenishing the coenzyme NAD+ (nicotinamide adenine dinucleotide) appear to reduce the decline in adult tissue maintenance with age by enhancing DNA repair in animal models, which altogether lead to improvement in lifespan and healthspan [68–70]. These results strongly suggest that it is possible to alter the fate of both DNA repair and cellular turnover with therapeutic interventions. However, many questions still remain and additional model organisms are needed to simultaneously analyze and integrate process of tissue renewal and DNA repair in the complexity of the whole organism. We believe the planaria model system could provide important insights in this regard.
3. Planaria as a model to study DNA damage and tissue renewal
Planaria are members of the phylum Platyhelminthes (flatworms) and are classically known for their robust regenerative capabilities [71]. Planaria display constant cell renewal and undergo repair upon injury to their tissues and organs (e.g. digestive, nervous, muscle, etc.) [34]. The planaria Schmidtea mediterranea, which is the most common species used worldwide to study aspects of tissue homeostasis, contains a large pool of SCs called neoblasts. Neoblasts are recognized as the only cells with capacity to proliferate in S. mediterranea and therefore, serve as the sole source of new cells that support the dozens of different tissues types [32, 34, 71–73]. The neoblast diversity is only beginning to be elucidated and so far, four subpopulations have been described (e.g. Sigma, Gamma, Zeta and Nu) that display restricted potential to generate and maintain tissues [72, 74, 75]. This diversity within planaria neoblasts allows for the integration of local and environmental stimuli throughout its lifespan to maintain tissue homeostasis.
Similar to SCs in other organisms, neoblasts are in constant crosstalk with their surroundings and are influenced by local and systemic signals involving metabolic status, neural inputs, tissue integrity, etc. In the presence of nutrients, planaria increase the body size by incorporation of new cells. Conversely, starvation conditions lead to reduction in animal size by elimination of cells that maintain body proportion [76–78]. Neoblasts also sense and respond to tissue injury by mounting a multi-step proliferative response that mediates the regrowth of missing and damaged parts [79–81].
The capacity to regulate SC division in response to physiological demands and injury has been attributed to the conserved tumor suppressors and oncogenes within the planaria (e.g. PTEN, AKT, p53, Rb) [82–86]. Planaria rarely develop cancer but can be forced to undergo cellular transformation after treatment with carcinogenic compounds or manipulation of tumor suppressor genes [83, 87–89]. In line with previously described organisms, preservation of DNA integrity is paramount for tissue homeostasis and extended lifespan in planaria. Recent interest in dissecting mechanisms of DNA repair in planaria revealed the evolutionary conservation of key regulators such as Rad51, p53, Rb, Ubc9, Brca2, and Rad54B that are activated in response to endogenous and exogenous environmental insults [22, 23, 84, 90–92]. Functional studies of DNA repair pathways identified patterns of SC exhaustion and tissue renewal defects similar to those observed in mammals [10]. These planaria features confer unique advantages to analyze critical parameters in response to DNA damage in the context of the whole body. Thus, we propose the use of S. mediterranea as a simplified platform to address cell fate decisions in the presence of genomic instability during large-scale SC-mediated cellular turnover and tissue repair.
4. Evolutionarily conserved DNA damage repair mechanisms exist in planaria
Older specimens of S. mediterranea are phenotypically indistinguishable from younger ones, which highlight the efficient mechanisms of unlimited cellular renewal in planaria [34]. This also implies that planaria contain efficient DNA repair mechanisms to combat endogenous and exogenous insults that normally deplete SCs in adult tissues, thus preventing aging and cancer-like phenotypes [87]. Indeed, high-throughput query on genomic resources [93, 94] have allowed us to identify a wide range of DSB recognition and repair homologs in planaria (Figure 2A). Specifically, we uncovered components of signaling pathways involved in DNA damage recognition, signaling transduction and effector outcome (e.g. cell cycle arrest, cell death and DNA repair).
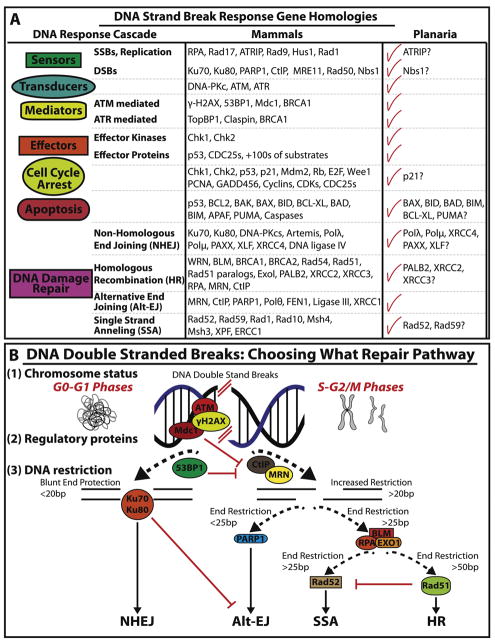
Figure 2. Planaria display evolutionary conservation of the mechanisms involved in DNA double strand break (DSB).
(A) Gene homologies for sequences regulating DDR and signal transduction mechanisms are found in planaria. For simplicity the attention is mostly focused on mediators of DSB. Checkmarks indicate conservation between the planarian species S. mediterranea and humans. However, some genes remain unfound in the planarian genome indicated by a question mark. (B) DSB repair is mediated by four different pathways (e.g. NHEJ, Alt-EJ, SSA and HR) however the choice of these pathways are determined by three mechanisms. (1) The status of chromosome homology which is dependent on what phases of the cell cycle DSBs occur (e.g. homology during S-G2-M phases which favors HR). (2) The abundance of regulatory proteins in each cell cycle phase. For example, increased expression of ATM, Mdc1 and 53BP1 have been shown to favor NHEJ in the G0/G1 phase of the cell cycle by suppressing MRN and CtIP expression needed for HR-mediated repair. (3) Lastly, the restriction of DNA blunt ends. The length of DSB nucleotide base pairs give rise to blunt end protection or a series of end restrictions. (e.g. <20bp for NHEJ and >50bp for HR mediated repair).
Though DNA damage can affect cells in numerous ways, DSBs represent the most severe form of DNA damage as they occur when both strands of the DNA double helix are broken in close proximity. Two DSBs within a cell are capable of forming chromosomal translocations and some estimates establish that 10–50 DSBs occur per cell per cycle [8, 39, 95, 96]. Generally, DSBs can be repaired through: non-homologous end joining (NHEJ), homologous recombination (HR) and their alternative pathways: alternative end joining (alt-EJ) and single strand annealing (SSA) (Figure 2B). The selection of DSB repair pathways is determined by three independent variables: cell cycle phase (e.g. chromosome status), the abundance of regulatory proteins in each cell cycle phase and the resection of DNA blunt ends (Figure 2B). These mechanisms of DSBs repair have been extensively reviewed elsewhere [3, 5, 20, 96–99].
Detailed evaluation of the planaria genome revealed an important molecular conservation of the DDR mediator γ-H2AX and DSB repair protein RAD51 (e.g. ~65% and ~81%, respectively) (Figure 3A, B). Further analysis also showed the presence of key signatures of their phosphorylation sites (e.g. S-Q motif the signature for γ-H2AX) or binding domain activity (e.g. Rad51’s Walker A/B, L1/2 and BRC domains) (Figure 3A, B). The molecular conservation of these molecules in planaria also facilitates the possibility of using commercial antibodies to evaluate the spatial distribution of DDR proteins at cellular and organismal levels (Figure 3C) [22, 23, 100]. DSBs can be induced in planaria through ionizing radiation (IR), RNA interference (RNAi) and through drug exposure such as methyl methanesulfonate (MMS) [22, 23, 89]. In order to gauge the effects of these DSB inducing strategies, three key strategies have been optimized (e.g. TUNEL, COMET assay and karyotyping) [22, 23]. The COMET assay, under alkaline conditions (pH > 13), is commonly used to monitor DNA integrity with focus on DSBs. In addition, karyotyping protocols have been optimized to assay abnormalities found in neoblast-specific chromosomes [22, 101]. FACS protocols have been implemented to monitor neoblast populations, cell cycle progression and cellular apoptosis [102–105] (Figure 3D).
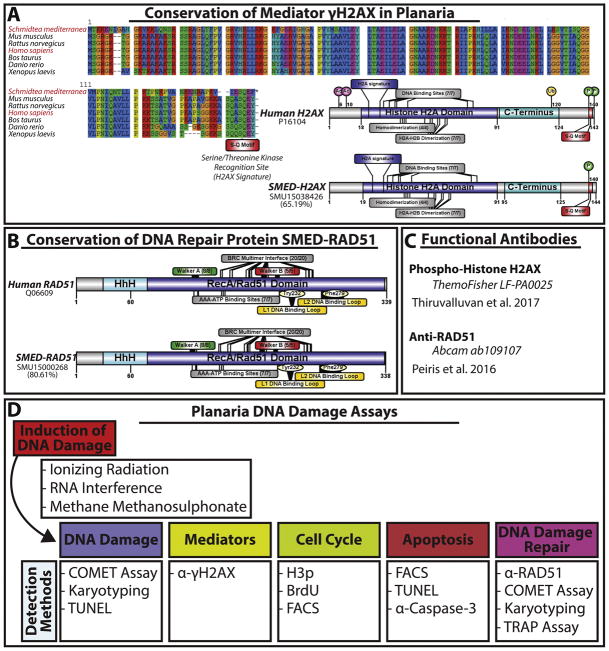
Figure 3. Molecular conservation of key proteins regulating DNA double strand break repair and DNA damage assays in planaria.
(A) Protein alignment of planaria γH2AX and homologs indicate conservation of γH2AX ‘s S-Q motif among species The S. mediterranea mediator γH2AX scores 65.19% similar to the human homolog. Sequence alignment was obtained by CLUSTALW. Using Illustrator for biological sequences (IBS version 1.0) we were able to create diagrams for both human and SMED-γH2AX protein sequences. Human protein length is 143 amino acids (aa) versus 144aa in SMED-γH2AX. Within this protein both the planaria and human sequences contain Histone H2A domain (dark blue, 73aa long) and a C-terminus (light blue, 30aa long). Within the Histone H2A domain both contain the H2A signature (6aa long), DNA binding sites (7/7 sites) homodimerization sites (4/4 sites) and H2A-H2B dimerization sites (7/7 sites). Importantly, both contain the S-Q motif (red domain) phosphorylation on Ser-139 site in the S-Q motif. However, Human γH2AX contains acetylation sites (6aa and 10aa), ubiquination site at 120aa and another phosphorylation site at 143aa. (B) The S. mediterranea RAD51 sequence shares 80.61% similarity with the human homolog and contains all the documented characteristic binding domains. Both contain a helix-hairpin-helix (HhH) motif (light blue, 48aa long) with a large RecA/Rad51 domain (dark blue, 277aa long). Within the RecA/Rad51 domain there two key features Walker A (green and 8/8 sites) and Walker B (red, 5/5 sites) binding regions. Other features of the protein are the BRC multimer interface (20/20 sites), (AAA-ATP bindng sites (7/7 sites), L1/L2 DNA binding loops (yellow) which contain a Tyr 232 and Phe 279, respectively. (C) Details of commercial antibodies that recognize the S. mediterranea RAD51 and γH2AX homologs. SMED-RAD51 anitbody generated by the Sánchez-Alvarado/Hawley labs (Xiang et al., 2014) recognizes similar peptide size as the commercial human RAD51 (Thiruvalluvan et al 2017). (D) General assays and detection methods for inducing and quantifying the response and outcome of DNA damage in native stem cells in the planarian model.
Planaria tolerate relatively high doses of IR, far surpassing the thresholds of exposure that are known to be lethal in mammals [106, 107]. Thus, the DDR in planaria can be analyzed by exposing animals to IR. For example, exposing planaria to IR above 3,000 rad irreversibly eliminates neoblasts, abolishes regeneration, and leads to animal death in about three weeks [37, 71, 90, 108–110]. Lethal doses of IR has been traditionally used as a tool to identify neoblast-associated markers and mechanisms of neoblast repopulation during tissue transplantation and irradiation [37, 71, 90, 108–110]. However, data mining of recent transcriptional data involving samples from a seven day time course upon lethal IR [111] revealed a persistent upregulation of genes involved in DNA damage sensing and signal transduction of DSBs throughout the time course. Interestingly, there was high expression of ATM but reduced gene expression of DNA-PKcs, required for NHEJ, throughout the time course (Figure 4A). Similar effects are observed in primary human fibroblasts cell lines during chronic exposure to IR [112]. Because lethal IR exposure irreversibly eliminates neoblasts, the results confirmed that key regulators of cell cycle progression and markers of proliferating neoblasts were nearly abolished upon IR (e.g. PCNA and cyclinB). Furthermore, the irreversible elimination of neoblasts may also result from a gradual increase in the expression of the inhibitor of growth protein (ING) (Figure 4A). Members of the ING family have been found to negatively regulate EGFR/PI3K/Akt signaling pathway, which is central to planaria neoblast repopulation post IR [90, 113]. Thus, the increased amount of IR-induced DNA damage together with upregulation of ING expression may act together to eliminate neoblast and prevent residual cell proliferation post-IR. These findings also indicate that most components of the DDR are associated with neoblasts.
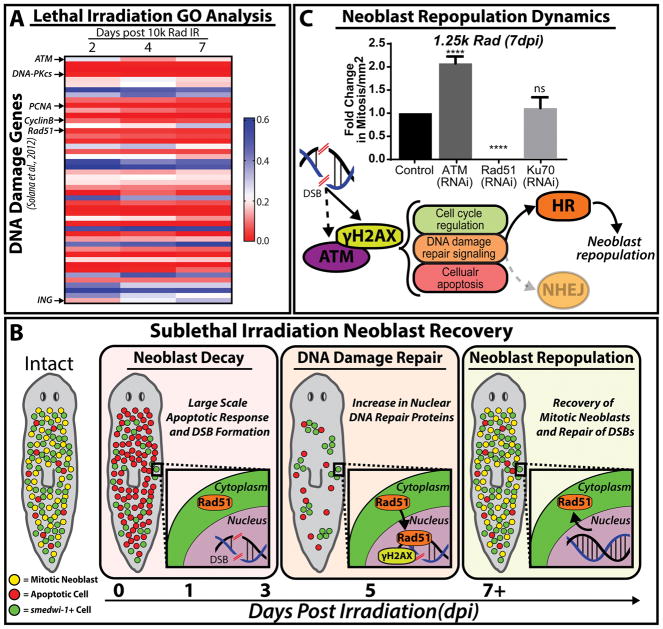
Figure 4. Neoblast response and repopulation post irradiation is attributed to functional DNA damage detection and repair.
(A) Lethal irradiation of planaria (10k rad) irreversibly depletes neoblast populations. Upon analysis of GO term annotations published by Solana et al., 2012, genes involved in DDR and repair along a 7 day time course were selected. The heat map shows changes in gene expression at different times post-irradiation with color red indicating reduction and blue increase in gene expression (scale bar to the right). DNA damage response triggers upregulation of ATM gene expression accompanied by a decrease in cell cycle and DNA repair gene expression (e.g. PCNA, CyclinB and Rad51, respectively). Note that the inhibitor of growth (ING) gene expression is upregulated after 2 hpi. (B) Cartoon depiction of neoblast response to sublethal irradiation. Upon sublethal irradiation, 1-3dpi, neoblast decay arises from a large scale apoptotic response and a lack of RAD51 nuclear translocation accompanies increases of DSBs. Secondly, the remainder of smedwi-1+ neoblast clusters begin to slowly expand and cells exhibit a peak of DDR and DNA repair proteins at 5dpi (e.g. γH2AX and RAD51, respectively); marking the DNA damage repair response. Lastly, neoblast repopulation occurs 7+dpi with increases of smedwi-1+ cells, recovery of mitotic neoblasts and a decrease in DSBs. Cellular events are depicted by the following: mitotic neoblasts (yellow), apoptotic cells (red) and smedwi-1+ cells (green) (C) Fold change in mitosis in RNAi and mock control animals is quantified 7dpi after 1.25k rad (sub-lethal dose). Notice the increase in mitotic events upon ATM(RNAi) and the inability for neoblasts to recover post Rad51(RNAi). Underneath, proposed mechanism of neoblast repopulation post sublethal IR show a possible role of Smed-ATM functioning as a transducer of DSB signal in tandem with γH2AX phosphorylation. Further, this model implies that HR signaling is a key player in neoblast repopulation as RNAi of NHEJ did not affect neoblast repopulation. Dotted arrows and shaded colors imply uncertainties within the model and further experiments are required to validate these assumptions and interactions. ****p < 0.0001; one-way-ANOVA
Exposure to sub-lethal doses of IR (i.e. 1000–1750 rad) leads to a partial elimination of neoblasts, which allow for studies of DDR, SC repopulation and recovery during adult tissue renewal [90, 114] (Figure 4B). The re-establishment of mitotic activity post sub-lethal IR has been attributed to EGF signaling and active DNA repair mechanisms (e.g Rad51 and Rad54B) [90, 115]. Sub-lethal IR depletes neoblast mitotic activity within 24hrs, accompanied by a significant spike in apoptosis and DSBs. There is a gradual increase in DSB repair that peaks at five days post IR as determined by RAD51 gene/protein expression and RAD51 nuclear translocation (Figure 4B). Neoblasts uniquely express the gene smedwi-1 (piwi-1, henceforth) [37, 75, 116]. piwi-1 expression is currently used as the gold standard to recognize the presence of neoblasts and their distribution. piwi-1+ cells clusters are severely reduced during the first 7 days post-irradiation (dpi, 1250 rad) and begin to expand after 9 dpi [90, 114]. However, mitotic activity is detectable after 7 dpi (Figure 4B). Neoblast repopulation depends on EGF signaling that requires active DNA repair mediated by ATM, Rad51, Ku70 and Rad54B [90, 115]. Interestingly, functional disruption of ATM with RNA-interference (RNAi) leads to an accelerated re-establishment of mitotic activity 7dpi [115] (Figure 4C). ATM is an important player of the DDR that influences cellular decisions upon IR through regulation of p53/p21 axis to facilitate cell cycle checkpoint arrest [112, 117]. Thus, we can postulate that Smed-ATM may be key to facilitating an appropriate cellular response to DSBs (e.g. detection and cell cycle arrest). Further experiments will be required to identify the role of Smed-ATM upon IR in cell cycle regulation and determine if Smed-ATM(RNAi) hyper-proliferative neoblasts are gnomically stable; altogether validating the conservation of this protein in planaria. The interplay of DNA repair and neoblast repopulation is only beginning to be understood but the recent evidence suggest that mechanisms of HR are the predominant repair pathway in planaria [23, 90, 115]. This is further supported by results demonstrating that RNAi of Rad51 and Rad54B in sub-lethally irradiated animals fail to repopulate piwi-1+ cells and mitotic activity, resulting in lethality. Conversely, dynamics of mitotic repopulation in Ku70(RNAi) sub-lethally irradiated animals are indistinguishable from untreated control group [22] (Figure 4C). All together, these results imply that Smed-ATM is a key upstream regulator of cell fate in response to IR-induced DSBs and HR is the dominant pathway used in repairing damaged DNA in planaria.
5. Genetic Models of DNA Damage in S. mediterranea
Two independent DNA damage models have been developed in planaria by disrupting gene function via RNAi of Rad51 and Ubc9 [22, 23]. Planaria homologs of Rad51 and Ubc9 show high evolutionary conservation with higher organisms and their disturbance revealed important patterns of regional defects along the AP axis (Figure 5A–D). Furthermore, RNAi of Rad51 and Ubc9 display loss of genomic integrity, specifically by the accumulation of DSB throughout the planaria body [22, 23].
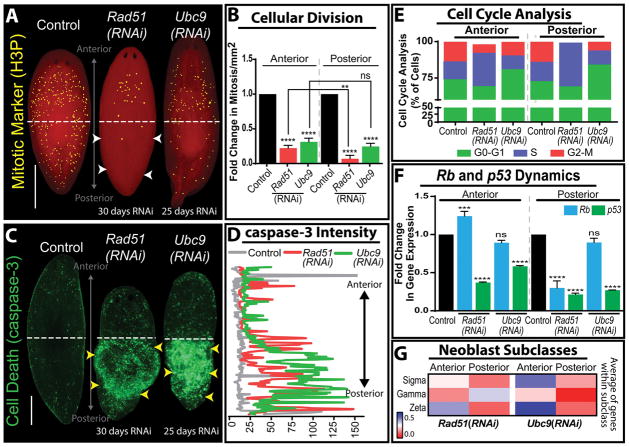
Figure 5. Rad51 and Ubc9 inhibition in planaria yields high levels of DSB and region-specific neoblast responses.
(A) Whole mount immunostaining of mitotic neoblasts marked by Histone3 phosphorylated (H3P) in mock control, Rad51(RNAi) and Ubc9(RNAi) models. Note the difference in mitotic cells along the anteroposterior axis in the Rad51(RNAi) group; white arrow heads indicate a severe decrease in mitosis in the posterior versus the anterior. (B) Levels of mitotic cells across the anteroposterior axis, confirms the asymmetric distribution of mitoses is specific to Rad51(RNAi) but not to animals subjected to Ubc9(RNAi). (C) Whole mount immunostaining of cell death marked by the caspase-3 antibody. Note the increase of apoptotic events (green signal) is concentrated in the posterior region of both Rad51(RNAi) and Ubc9(RNAi) animals (yellow arroheads). (D) Quantification of the fluorescent intensity signal from caspase-3 stained Rad51(RNAi) and Ubc9(RNAi) animals confirming the increased cell death in the posterior regions compared to the anterior and control (yellow arrowheads coincide with the increase in cell death). Intensity readings were obtained by tracing a line in the middle of the animal from the anterior to the posterior region using ImageJ software. (E) Cell cycle analysis using flow cytometry in cells dissociated from the anterior and posterior regions in animals subjected to Rad51(RNAi) and Ubc9(RNAi). Different phases of the cell cycle are color coded and show that Rad51(RNAi) and Ubc9(RNAi) animals display different effects on cell cycle repsented by important reduction in M-phase in the posterior region of Rad51(RNAi), while Ubc9(RNAi) animals tend to halt cell cycle in G0/G1 phases but some cells still continue to divide. (F) Fold change in gene expression of Rb and p53 relative to the control. Rb expression is asymmetrically upregulated in the anterior of Rad51(RNAi) but it does not appear affected in the same manner after Ubc9(RNAi). (G) Heat map, representing fold change in the average of gene expression of neoblast subclass (sigma, gamma, zeta) markers in the anterior and posterior regions. Red indicates diminished gene expression and blue increased gene expression. **p < 0.01; ***p < 0.001; ****p < 0.0001; two-way-ANOVA. Scale bars: 200μm.
Rad51 is required for nucleofilament formation and without functional RAD51 protein the HR repair complex cannot form [20, 118, 119]. Full knockout of Rad51 results in embryonic lethality in mammals [120, 121]. Nonetheless, it is possible to knockdown the HR pathway but it is challenging to evaluate organismal SC response and track their progeny in an environment of genomic instability. Thus, the planaria model system offers unique opportunities to overcome these limitations by enabling the possibility of disrupting HR while SC attend systemic demands of cellular turnover and repair. On the other hand, SUMOylation is a dynamic and reversible post-translational modification that requires the cooperation of a host of proteins [122]. Critical to this pathway is UBC9, which determines protein SUMOylation [122]. SUMO attachment regulates protein function as it can affect protein localization, stability, protein–protein interaction, cause conformational changes or act as a hub to form multi-protein complexes [123]. Mounting evidence suggests SUMOylation plays a critical role in the regulation of DSB repair [124–128]. It does so in three ways: (1) regulates protein stability, DNA binding ability and localization of sensors and effectors of DSB repair, including both NHEJ and HR; (2) leads to creation of an open chromatin state more amenable to repair by controlling epigenetic modification through modulation of various methylases and acetylases; (3) orchestrates successful DDR response by coordinating multiple types of post-translational modifications, most notably stubl-mediated ubiquitination [129–138].
5.1 Functional Disruption of Rad51 and Ubc9 Affects Tissue Homeostasis in Planaria
The unique feature of the Rad51 and Ubc9 knockdown models is the prevalence of DNA damage, especially DSBs that is present throughout the planaria body [22, 23]. Molecular analysis in both models revealed a transient presence of DNA DSBs and chromosomal abnormalities that progressively increased over time. This is consistent with the role Rad51 plays in DSBs repair through HR. However, the mechanism driving this phenomenon in the Ubc9 phenotype was less evident. Two observations were critical to relate SUMOylation to DNA damage: (i) the regional defects in animals subjected to Ubc9(RNAi) were similar to those observed in Rad51 phenotype and (ii) late stages of the Ubc9 phenotype display increase in RAD51 and γ-H2AX protein expression, which appeared in clusters along the AP axis. Additional analysis revealed that DNA damage in Ubc9(RNAi) is due to the inability of RAD51 to translocate from the cytoplasm to the nucleus to repair DSBs [23]. This finding links the two models together and additionally explains phenotypic similarities, which altogether supports the idea that HR is the prominent pathway for repair of DSBs in planaria.
The induction of DNA damage after Ubc9 and Rad51(RNAi) results in a cascade of cell fate decisions led by cell cycle arrest [22, 23]. Cell cycle analysis revealed that while most cells in the Rad51(RNAi) were arrested in S phase, cells in Ubc9(RNAi) animals were primarily arrested in the G1 phase (Figure 5E). p53 and Rb commonly regulate neoblast fate decisions (i.e. apoptosis, proliferation andcell cycle arrest) during tissue renewal and regeneration. This is also the case in the presence of DNA damage in planaria but intriguingly; we found that p53 gene expression is downregulated across the AP axis in both RNAi groups (Figure 5F). However, there were stark differences in Rb expression. Specifically, there is an increase in Rb expression in the anterior region of Rad51(RNAi) animals whereas there is no significant change in Ubc9(RNAi) group (Figure 5F). Although our knowledge of Rb dynamics relies on gene expression data, it is tempting to link increased Rb expression with cell cycle arrest. Canonically, Rb is thought to be an important regulator of the G1/S checkpoint and studies suggest that overexpression of Rb can increase rates of cellular survival and predispose cells to become more cancerous [139, 140]. Furthermore, the genomic instability driven cell cycle arrest in both lead to interesting changes in tissue homeostasis and cellular turnover, specifically in terms of cell survival and death [22, 23]. While both models coincide in a significant decrease in the cycling neoblasts, the Rad51 model reveals a remarkably difference across the AP axis, specifically loss of survival in the posterior region. This is likely due to the differential expression of cell fate regulators p53 and Rb. On the other hand, both models show a massive increase in cell death in the tail region with significantly less cells dying in the anterior. It is possible that Rb is acting as switch for allowing cell survival in the anterior but not the posterior. Another explanation as derived from experimentation in the Ubc9(RNAi) model, where the cell death is partially attributed to attenuation of Hedgehog signaling, which is known to be an important regulator of posterior polarity in planaria. Whether the same mechanism is driving cellular decisions in the Rad51 phenotype requires further experimentation.
A remarkable finding from these studies is that some SCs in the anterior region are able to overcome surveillance mechanisms and continue proliferating with genomic instability [22, 23]. In the Rad51(RNAi), this is in part due to increased expression of Rb and neural inputs in the anterior region. Ectopic introduction of brain tissue in the posterior region, induce neoblast proliferation with DSB. These findings highlight the possibility of intercellular effects, whereby neural signals alter fate decisions of neoblasts with DSBs. Likewise, these results also prompt future studies about possible neural regulation of Rb signaling that facilitate proliferation of neoblasts with DSBs. Alternatively, it is possible that a subset of neoblasts is endowed with proliferative capacity to give rise to cancer-like cells in the anterior. Multiple neoblast subtypes have been characterized [72, 74, 75]. We found that gene expression of markers associated with zeta neoblasts are increased in the anterior for both Ubc9 and Rad51(RNAi) animals (Figure 5G). Recent research demonstrates the intriguing possibility that inhibition of Hippo signaling trigger dedifferentiation of postmitotic progenitors in planarians [141]. It is tantalizing to speculate that the increasing load of genomic instability may act as a switch for zeta nebolasts to leave their lineage-restricted state and try to fill the niche left behind by sigma cells that cannot survive increasing DNA damage. It is also possible that persistent demands of cellular turnover override fate decisions to promote exit of cell cycle arrest has been noted in hematopoietic SCs [63]. Some of these cells may have weaker sensors and effectors of the DDR, allowing them to more easily circumvent these checks and balances and ultimately evolve into cancer-like cells. Additional experiments are required to dissect the actual mechanisms driving cells to withstand excessive DNA damage and continue to proliferate. Nonetheless, the results obtained with Rad51 and Ubc9 downregulation supports the notion that cellular decisions in the presence of DNA damage are also influenced by regional signals that may involve crosstalk among tissues and organs. This is an important finding as a more comprehensive focus on the regional signals driving proliferation of cells with genomic instability may help in understand the mechanisms facilitating cancer formation and progression.
5.2 DNA Damage and Repair are Essential Components of the Regenerative Response
Cell death and proliferation are not only instrumental during tissue renewal but also in the process of regenerating missing or injured body parts. Upon amputation, planaria undergo an orchestrated series of localized and systemic cascades of cellular proliferation and programmed cell death (e.g. ~4–6hrs and ~48hrs). Recent research have have greatly furthered our understanding of the genetic and molecular cascades required for tissue repair and regeneration [79, 80, 142]. The initial peaks of systemic cell division and localized cell death events were found to be accompanied by a genetic response called the generic wound response that happen during the first 24 hours post-injury/amputation (hpa). This is followed by a wave of specific gene expression representing the regeneration response (24–70 hpa) that includes the second molecular peak of mitosis and apoptosis. Finally, the differentiation phase is attributed to neoblast progeny mediated differentiation and specialization of the blastema at +70hpa [79, 80] (Figure 6A).
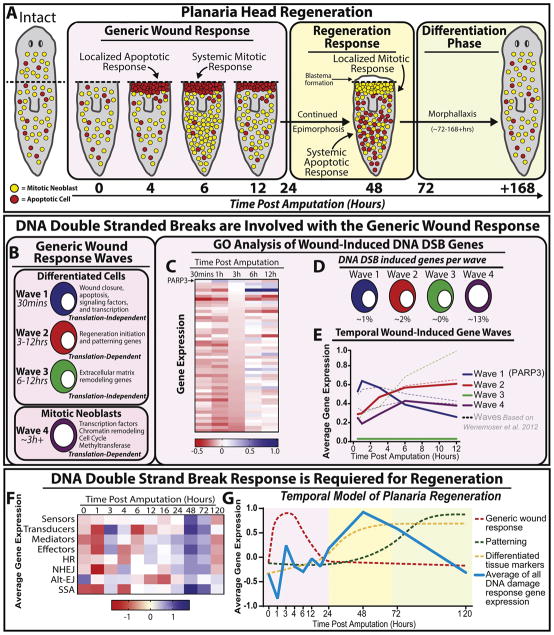
Figure 6. DNA damage repair (DDR) is activated during the initiation of tissue regeneration.
(A) Schematic representation of the general cellular events taking place during the first week post-amputation (hpa) of the anterior region in planaria (color coded boxes). During the generic wound response, a localized apoptotic response occurs ~4hpa, accompanied by a systemic burst of mitotic neoblasts at 6 hpa that gradually reduces ~12hpa. Next, the regeneration response is followed by a localized increase in cell proliferation and a systemic wave of death. After 72hpa the tissue will continue restructuring newly formed tissue through differentiation of stem cells and progenitors. (B) Waves of gene expression found in the generic wound response within the first 12hpa. (C) Heat map representing GO term analysis of wound induced DDR gene expression derived from Wenemoser et al. 2012. (D) Percent of DNA damage genes associated within each wave. (E) Temporal wound-induced gene expression waves. Solid lines indicate averages of DDR specific wave response and dotted lines indicate waves according to Wenemoser et al. 2012. (F) Heat map of average DDR gene expression post amputation. At 48hpa the average expression of all DDR genes is upregulated following wounding. (G) Graph depicts the temporal model of planaria regeneration including the average expression for genes involved in the DDR. The average of all DNA damage gene expression was obtained from the heat map in (F), (blue line) is plotted against the established temporal model of regeneration (dotted lines) according to Wenemoser et al., 2012 and is color coded as shown in (A).
The ability to adjust cell proliferation during simultaneous demands of tissue renewal and injury highlight the faithful mechanisms used by planaria to regulate cell number. Since injury repair relies on cell proliferation and consequently DNA replication, we argue that the DDR is an active player that preserves genome integrity during regeneration. In other words, an increase in cell division is accompanied by DNA replication that is carefully monitored by DNA repair mechanisms. Indeed, recent studies have demonstrated that key components of DNA replication and repair (e.g. p53, Rb, Rad51 and Ubc9) are critical for the regenerative process and without them; planaria fail to regenerate [23, 84, 86, 115]. In addition, cell death is necessary for proper regeneration. The TUNEL assay, which detects cellular apoptosis induced by DSB-nicked ends, is commonly used to evaluate cell death in planaria [104]. After amputation, two peaks of cell death are known to happen at ~4hrs and ~48hrs. However, it remains unclear whether these stereotypical patterns of TUNEL+ cells are derived from the stressful environment of regeneration or actual DNA damage, specifically DSBs.
We were prompted to reanalyze the possible role DNA damage response play during the early and late phases of tissue regeneration based on published transcriptomic data [79, 80]. Transcriptomic changes during the first half of the generic response in planaria offer an interesting resource to discern the role of DDR [79]. This process involves four waves of gene expression found within differentiated tissues (waves 1–3) and neoblasts (wave 4) (Figure 6B). Using published RNAseq data from Wenemoser et al. 2012, we were able to identify genes involved in DNA damage response by GO term analysis. We found that all waves except wave 3 contained genes involved in DDR (Figure 6C–E). Interestingly, we found PARP-3 as the only DDR gene involved in wave 1 and the only DDR-specific gene with a peak in gene expression within the first hour post amputation. At the decline of PARP-3 expression at 3hrs, both wave 2 and wave 4 DDR specific gene expression increased and peaked at 6hrs, which coincide with the system wide mitotic response (Figure 6C). These results suggest that expression of genes in the DDR follow similar transcriptomic changes of the generic wound response except in wave 3. It also implies that PARP family genes may prime the DDR during early regenerative events (Figure 6C–E). However, the question still remains whether the first peak of TUNEL+ cells starting ~4hrs is linked to DNA damage.
PARP-3 catalyzes post-translational modifications of proteins involved in transcription silencing, cell death and interacts with PARP-1 during DDR to accelerate NHEJ [143–146]. In addition, PARP-3 has been shown to act independently of DNA damage and mediate centrosome stability, G1/S cell cycle progression and telomerase activity [144, 147, 148]. For instance, PARP-3 expression has an inverse relationship with telomerase activity. Lung cancer cells depleted of PARP-3 displayed an increase in telomerase activity [148]. During planaria regeneration, telomerase activity is upregulated and accompanies neoblast proliferation [148]. Thus, the possible role of PARP-3 expression during the first 3hrs of regeneration may be to restrict both neoblast cell cycle progression and telomerase activity. The decline in PARP-3 after 3hrs may allow the priming of transcription of both cell cycle and DNA repair proteins that are involved during initial mitotic burst at 6hrs. The data analysis shows that the initial localized peak of TUNEL+ cells may not be in response to DNA damage but to the harsh environment at the amputation site to facilitate wound closure and there is a possibility that PARP-3 may be mediating the post-translational modifications associated with cell death.
Regenerating planaria exhibit a localized mitotic peak and a systemic increase in TUNEL+ cells resulting in the formation of a regenerative blastema starting at 48hrs post amputation [79, 81, 104]. Gene expression levels associated with DNA damage response and repair are relatively low within the first 24hrs of regeneration (Figure 6F). Strikingly, at 48hrs, there was a dramatic increase in the expression of genes involved in DNA damage signal transduction and DDR (Figure 6F). Thus, by plotting the average of DDR gene expression values onto the established temporal model of planaria regeneration [149], we observed that the second wave of DDR is much more significant than that of the generic phase and remains a key feature in the regeneration and early differentiation phases (Figure 6G). Consistently, our recent work strongly supports the idea that DDR is required for large-scale tissue regeneration. Without key molecules such as Rad51, BRCA2, Ubc9 animals, fail to regenerate and replace lost tissues [23, 115]. Furthermore, according to Wurtzel et al., 2015 [80]; expression levels of genes involved in HR (e.g. Rad51-A,-B,-C and BRCA2) were highly active compared to the decrease in NHEJ (e.g. Ku70, Ku80, and Ligase IV) related genes. However, gene expression analysis revealed that HR and NHEJ were at their peaks at 48hrs post amputation. Delving into the molecular dynamics of mitotic events during regeneration in Rad51 and Ubc9(RNAi) animals, we observe that at 6hrs post-amputation both groups respond with a slight increase in mitoses but fail to elicit a second mitotic peak at 48hrs [23, 115]. These results together provide further evidence that DDR is a crucial component of the overall regenerative response in planaria.
6. Final Remarks
Preserving genomic integrity over recurrent tissue renewal is an important challenge that wanes with age. DNA damage and the cellular responses associated with it are at the center of efficient cellular turnover. Thus, we advocate for studies of SCs in their natural environment as they attend demands of tissue renewal and repair. The analyses integrating SCs, DNA damage and tissue renewal in the complexity of the whole body present fresh opportunities to the field and has the potential to inform about cellular crosstalk and regulation of signaling pathways (e.g. tumor suppressors, oncogenes) that control cellular decisions in the face of DNA damage (Figure 7A). The evolutionary and functional conservation of the DDR and mechanisms of SC function in planaria offers a simplified paradigm, in which pharmacological or genetic screens can be performed rapidly and cost-effectively.
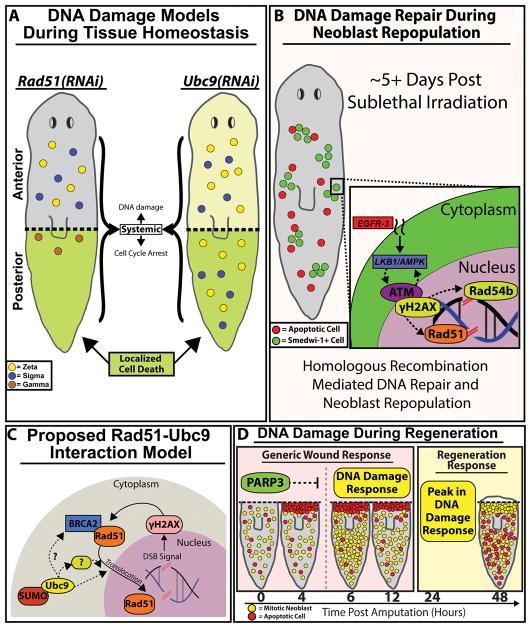
Figure 7. DNA damage repair (DDR) pathways are critical components of the cell proliferation required for tissue regeneration.
(A) Rad51(RNAi)(gray) and Ubc9(RNAi) (light-yellow) planaria share similar increases in DNA damage along the AP axis accompanied by varying levels of cell cycle arrest. Localized cell death (green) is observed in the posterior region of both RNAi conditions. Survival of neoblasts/sub-types were determined from gene expression data from figure 5G; neoblast subtypes: zeta (yellow), sigma (blue) and gamma (brown). (B) 5 days post sublethal irradiation, remaining neoblasts will repair DSBs by homologous recombination and neoblasts will repopulate the entire animal. We propose a hypothetical model whereby EGFR signaling integrates with DDR to facilitate neoblast repopulation post-irradiation (indicated with dotted lines). (C) Proposed RAD51-UBC9 interaction model. Presence of DNA DSB breaks triggers upregulation of RAD51 and γ-H2AX that together participate in DNA repair. However, after Ubc9(RNAi) the RAD51 protein accumulates in the cytoplasm as SUMOylation is required for RAD51 translocation to the nucleus. We propose that Ubc9 may interact with intermediate regulators of Rad51 for DDR. (D) Model of DNA damage during large-scale tissue regeneration. PARP3 expression peaks during the first 4hpa and may be suppressing DDR gene expression within that timeframe. Upon the decline of PARP3 expression, DDR expression at +6hpa is observed. A substantial peak in DDR observed in parallel with the regeneration responses that begins +24hpa. The robust responses of DDR pathways during wound repair could be necessary to drive DNA repair during neoblast proliferation that is required in tissue regeneration.
The capacity to induce different levels/types of DNA damage with pharmacological compounds, IR and genetic manipulations paves the way for additional studies aimed at understanding the biology of the DDR and possible alternatives for therapeutic applications (Figure 7B). The genetic models using loss of function of Rad51 and Ubc9 enable in situ analysis of tissue renewal in response to DNA damage that uniquely allow us to address the molecular basis controlling regional differences in the adult body (Figure 7A). Furthermore, the activation of regional cell survival and death in response to the systemic presence of DSBs is a great resource to address mechanisms of SC survival with defective DNA. This example prompts the possibility of studying intrinsic and extrinsic cues that may favor or restrict the growth of cancer initiating cells (Figure 7C, D). What are the signals facilitating survival of SCs with DSB in the anterior, whereas in the posterior surveillance mechanisms remain active and effectively eliminate damaged cells? Our results suggest that neural inputs may influence dynamics of cell cycle in neoblasts carrying DSBs. Thus, future experiments will address the source and the molecules mediating nervous signals that facilitate cell survival in an environment of genomic instability. Since neoblasts in the anterior region tend to survive with DSBs, it brings interesting opportunities to dissect potential differences among SCs and their susceptibility and advantages to DNA damage. Likewise, future studies will also integrate positional cues along the AP axis [150, 151] with DNA repair to inform about potential interactions that can enhance or reduce the integrity of the genome. Answering these fundamental questions will enable the field to identify early markers of cellular transformation, mechanisms regulating tumor suppressors, and to define means by which transformed cells survive and form tumors. Finally, we believe that planaria traditionally recognized as model for tissue regeneration, may also represent a fresh alternative to understand and manipulate DNA damage and its effects in the adult body.
Highlights.
Planarian model as an alternative to dissect DNA damage repair during tissue renewal
DNA repair mechanisms are evolutionarily conserved in planarians
Genetic models of DNA damage have been developed in planarians
Regional signals affect cellular decision in the presence of DNA Damage
DNA damage repair plays a central role during regeneration of tissues
Acknowledgments
Funding: We acknowledge support from the University of California Cancer Research Coordinating Committee (award CRR-18-525108), the National Cancer Institute and National Institute of General Medical Sciences of the National Institute of Health, awards CA176114 and GM109372 to NJO.
We thank members of the Oviedo Lab for discussion and comments on the manuscript.
Footnotes
Competing interest: The authors declare no competing or financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267) doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual review of biochemistry. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Vitale I, Manic G, Maria R, Kroemer G, Galluzzi L. DNA Damage in Stem Cells. Mol Cell. 2017;66(3) doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Haber JE. DNA recombination: the replication connection. Trends Biochem Sci. 1999;24(7):271–5. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 5.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer research. 1998;58(2):222–5. [PubMed] [Google Scholar]
- 8.Saul RL, Ames BN. Background levels of DNA damage in the population. Basic life sciences. 1986;38:529–35. doi: 10.1007/978-1-4615-9462-8_55. [DOI] [PubMed] [Google Scholar]
- 9.Adams PD, Jasper H, Rudolph LK. Aging-Induced Stem Cell Mutations as Drivers for Disease and Cancer. Cell stem cell. 2015;16(6):601–612. doi: 10.1016/j.stem.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nature Reviews Molecular Cell Biology. 2007;8(9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 11.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132(4):681–96. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Fox EJ, Prindle MJ, Loeb LA. Do mutator mutations fuel tumorigenesis? Cancer metastasis reviews. 2013;32(3–4):353–61. doi: 10.1007/s10555-013-9426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakem R. DNA-damage repair; the good, the bad, and the ugly. The EMBO journal. 2008;27(4):589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeggo PA, Lobrich M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. The Biochemical journal. 2015;471(1):1–11. doi: 10.1042/BJ20150582. [DOI] [PubMed] [Google Scholar]
- 15.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer. Journal international du cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 16.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci U S A. 2013;110(6):1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A.C. Society. Cancer Facts & Figures 2013. 2013 http://www.cancer.org/research/cancerfactsfigures/cancerfactsfigures/cancer-facts-figures-2013.
- 18.Iyama T, Wilson DM., 3rd DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 2013;12(8):620–36. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual review of biochemistry. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 20.Haber JE. A Life Investigating Pathways That Repair Broken Chromosomes. Annu Rev Genet. 2016;50:1–28. doi: 10.1146/annurev-genet-120215-035043. [DOI] [PubMed] [Google Scholar]
- 21.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120(4):497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 22.Peiris TH, Ramirez D, Barghouth PG, Ofoha U, Davidian D, Weckerle F, Oviedo NJ. Regional signals in the planarian body guide stem cell fate in the presence of genomic instability. Development. 2016;143(10):1697–709. doi: 10.1242/dev.131318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiruvalluvan M, Barghouth PG, Tsur A, Broday L, Oviedo NJ. SUMOylation controls stem cell proliferation and regional cell death through Hedgehog signaling in planarians. Cell Mol Life Sci. 2017 doi: 10.1007/s00018-017-2697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann WK. Cell cycle checkpoints and DNA repair preserve the stability of the human genome. Cancer metastasis reviews. 1995;14(1):31–41. doi: 10.1007/BF00690209. [DOI] [PubMed] [Google Scholar]
- 25.Auerbach R, Auerbach W. Regional differences in the growth of normal and neoplastic cells. Science. 1982;215(4529):127–34. doi: 10.1126/science.7053564. [DOI] [PubMed] [Google Scholar]
- 26.Auerbach R, Morrissey LW, Kubai L, Sidky YA. Regional differences in tumor growth: studies of the vascular system. International journal of cancer. Journal international du cancer. 1978;22(1):40–6. doi: 10.1002/ijc.2910220110. [DOI] [PubMed] [Google Scholar]
- 27.Auerbach R, Morrissey LW, Sidky YA. Regional differences in the incidence and growth of mouse tumors following intradermal or subcutaneous inoculation. Cancer research. 1978;38(6):1739–44. [PubMed] [Google Scholar]
- 28.Auerbach R, Morrissey LW, Sidky YA. Gradients in tumour growth. Nature. 1978;274(5672):697–9. doi: 10.1038/274697a0. [DOI] [PubMed] [Google Scholar]
- 29.Dispersio L. Regional growth differences of human tumor xenografts in nude mice. Laboratory Animals. 1981;15:179–180. doi: 10.1258/002367781780959044. [DOI] [PubMed] [Google Scholar]
- 30.Kubai L, Auerbach R. Regional differences in the growth of skin transplants. Transplantation. 1980;30(2):128–31. doi: 10.1097/00007890-198008000-00010. [DOI] [PubMed] [Google Scholar]
- 31.Prehn RT, Karnik V. Differential susceptibility of the axilla and groin of the mouse to chemical oncogenesis. Nature. 1979;279(5712):431–3. doi: 10.1038/279431a0. [DOI] [PubMed] [Google Scholar]
- 32.Aboobaker AA. Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 2011;21(5):304–11. doi: 10.1016/j.tcb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 33.King RS, Newmark PA. The cell biology of regeneration. The Journal of cell biology. 2012;196(5):553–62. doi: 10.1083/jcb.201105099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellettieri J, Sanchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- 35.Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–57. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 36.Sánchez Alvarado A. Planarian regeneration: its end is its beginning. Cell. 2006;124(2):241–5. doi: 10.1016/j.cell.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 38.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405(6787):697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 40.Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proceedings of the National Academy of Sciences of the United States of America. 1974;71(6):2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacRae SL, Croken M, Calder RB, Aliper A, Milholland B, White RR, Zhavoronkov A, Gladyshev VN, Seluanov A, Gorbunova V, Zhang ZD, Vijg J. DNA repair in species with extreme lifespan differences. Aging. 2015;7(12):1171–1182. doi: 10.18632/aging.100866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464(7288):529–35. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapetanaki MG, Mora AL, Rojas M. Influence of age on wound healing and fibrosis. The Journal of pathology. 2013;229(2):310–22. doi: 10.1002/path.4122. [DOI] [PubMed] [Google Scholar]
- 44.Lang T, Streeper T, Cawthon P, Baldwin K, Taaffe DR, Harris TB. Sarcopenia: etiology, clinical consequences, intervention, and assessment. Osteoporosis international: a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA. 2010;21(4):543–59. doi: 10.1007/s00198-009-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307(5710):720–4. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- 47.Reed JC. Dysregulation of apoptosis in cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1999;17(9):2941–53. doi: 10.1200/JCO.1999.17.9.2941. [DOI] [PubMed] [Google Scholar]
- 48.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145(6):851–62. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 49.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nature reviews. Genetics. 2008;9(2):115–28. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 50.Hsu CH, Stedeford T, Okochi-Takada E, Ushijima T, Noguchi H, Muro-Cacho C, Holder JW, Banasik M. Framework analysis for the carcinogenic mode of action of nitrobenzene. Journal of environmental science and health. Part C, Environmental carcinogenesis & ecotoxicology reviews. 2007;25(2):155–84. doi: 10.1080/10590500701399234. [DOI] [PubMed] [Google Scholar]
- 51.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 52.Gonzales KAU, Fuchs E. Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev Cell. 2017;43(4):387–401. doi: 10.1016/j.devcel.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuchs E. Epithelial Skin Biology: Three Decades of Developmental Biology, a Hundred Questions Answered and a Thousand New Ones to Address. Curr Top Dev Biol. 2016;116:357–74. doi: 10.1016/bs.ctdb.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pang WW, Schrier SL, Weissman IL. Age-associated changes in human hematopoietic stem cells. Semin Hematol. 2017;54(1):39–42. doi: 10.1053/j.seminhematol.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Hyun M, Lee J, Lee K, May A, Bohr VA, Ahn B. Longevity and resistance to stress correlate with DNA repair capacity in Caenorhabditis elegans. Nucleic Acids Res. 2008;36(4):1380–1389. doi: 10.1093/nar/gkm1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lans H, Lindvall JM, Thijssen K, Karambelas AE, Cupac D, Fensgård Ø, Jansen G, Hoeijmakers JHJ, Nilsen H, Vermeulen W. DNA damage leads to progressive replicative decline but extends the life span of long-lived mutant animals. Cell Death Differ. 2013;20(12) doi: 10.1038/cdd.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Signer R, Morrison SJ. Mechanisms that Regulate Stem Cell Aging and Life Span. Cell stem cell. 2013;12(2):152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355(6331):1330–1334. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landais S, D’Alterio C, Jones DL. Persistent replicative stress alters polycomb phenotypes and tissue homeostasis in Drosophila melanogaster. Cell Rep. 2014;7(3):859–70. doi: 10.1016/j.celrep.2014.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nat Cell Biol. 2011;13(5):506–12. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brosh RM, Jr, Bellani M, Liu Y, Seidman MM. Fanconi Anemia: A DNA repair disorder characterized by accelerated decline of the hematopoietic stem cell compartment and other features of aging. Ageing research reviews. 2017;33:67–75. doi: 10.1016/j.arr.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 63.Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC, Moehrle B, Brocks D, Bayindir I, Kaschutnig P, Muedder K, Klein C, Jauch A, Schroeder T, Geiger H, Dick TP, Holland-Letz T, Schmezer P, Lane SW, Rieger MA, Essers MAG, Williams DA, Trumpp A, Milsom MD. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 64.Zlatanova I, Pinto C, Silvestre JS. Immune Modulation of Cardiac Repair and Regeneration: The Art of Mending Broken Hearts. Frontiers in cardiovascular medicine. 2016;3:40. doi: 10.3389/fcvm.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamada M, Fujino N, Ichinose M. Inflammatory responses in the initiation of lung repair and regeneration: their role in stimulating lung resident stem cells. Inflammation and regeneration. 2016;36:15. doi: 10.1186/s41232-016-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jasper H, Jones DL. Metabolic regulation of stem cell behavior and implications for aging. Cell metabolism. 2010;12(6):561–5. doi: 10.1016/j.cmet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Houston BJ, Nixon B, Martin JH, Iuliis G, Bromfield EG, McEwen KE, Aitkena RJ. Heat exposure induces oxidative stress and DNA damage in the male germ line. Biology of reproduction. 2018 doi: 10.1093/biolre/ioy009. [DOI] [PubMed] [Google Scholar]
- 68.Fang EF, Lautrup S, Hou Y, Demarest TG, Croteau DL, Mattson MP, Bohr VA. NAD + in Aging: Molecular Mechanisms and Translational Implications. Trends in Molecular Medicine. 2017;23(10):899–916. doi: 10.1016/j.molmed.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Bonkowski MS, Moniot S, Zhang D, Hubbard BP, Ling AJY, Rajman LA, Qin B, Lou Z, Gorbunova V, Aravind L, Steegborn C, Sinclair DA. A conserved NAD + binding pocket that regulates protein-protein interactions during aging. Science. 2017;355(6331):1312–1317. doi: 10.1126/science.aad8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshino J, Baur JA, Imai SI. NAD(+) Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell metabolism. 2017 doi: 10.1016/j.cmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annual Review of Cell and Developmental Biology. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 72.Zhu SJ, Pearson BJ. (Neo)blast from the past: new insights into planarian stem cell lineages. Current opinion in genetics & development. 2016;40:74–80. doi: 10.1016/j.gde.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 73.Rink JC. Stem cell systems and regeneration in planaria. Dev Genes Evol. 2013;223(1–2):67–84. doi: 10.1007/s00427-012-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molinaro AM, Pearson BJ. In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol. 2016;17:87. doi: 10.1186/s13059-016-0937-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15(3):326–39. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.González-Estévez C, Felix DA, Rodríguez-Esteban G, Aboobaker AA. Decreased neoblast progeny and increased cell death during starvation-induced planarian degrowth. The International Journal of Developmental Biology. 2012;56(1–3):83–91. doi: 10.1387/ijdb.113452cg. [DOI] [PubMed] [Google Scholar]
- 77.Oviedo NJ, Newmark PA, Sanchez Alvarado A. Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn. 2003;226(2):326–33. doi: 10.1002/dvdy.10228. [DOI] [PubMed] [Google Scholar]
- 78.Newmark PA, Alvarado AS. NOT YOUR FATHER’S PLANARIAN: A CLASSIC MODEL ENTERS THE ERA OF FUNCTIONAL GENOMICS. Nature Reviews Genetics. 2002;3(3):210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- 79.Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Genes & Development. 2012;26(9):988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW. A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. Dev Cell. 2015;35(5):632–45. doi: 10.1016/j.devcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344(2):979–91. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oviedo NJ, Pearson BJ, Levin M, Sánchez Alvarado A. Planarian PTEN homologs regulate stem cells and regeneration through TOR signaling. Dis Model Mech. 2008;1(2–3):131–43. doi: 10.1242/dmm.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pearson BJ, Sanchez Alvarado A. Regeneration, stem cells, and the evolution of tumor suppression. Cold Spring Harbor symposia on quantitative biology. 2008;73:565–72. doi: 10.1101/sqb.2008.73.045. [DOI] [PubMed] [Google Scholar]
- 84.Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development (Cambridge, England) 2010;137(2):213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peiris TH, Ramirez D, Barghouth PG, Oviedo NJ. The Akt signaling pathway is required for tissue maintenance and regeneration in planarians. BMC developmental biology. 2016;16:7. doi: 10.1186/s12861-016-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu SJ, Pearson BJ. The Retinoblastoma pathway regulates stem cell proliferation in freshwater planarians. Developmental Biology. 2013;373(2):442–452. doi: 10.1016/j.ydbio.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 87.Oviedo NJ, Beane WS. Regeneration: The origin of cancer or a possible cure? Semin Cell Dev Biol. 2009;20(5):557–64. doi: 10.1016/j.semcdb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oviedo NJ, Pearson BJ, Levin M, Sánchez Alvarado A. Planarian PTEN homologs regulate stem cells and regeneration through TOR signaling. Disease Models & Mechanisms. 2008;1(2–3):131–143. doi: 10.1242/dmm.000117. discussion 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stevens AS, Wouters A, Ploem JP, Pirotte N, Van Roten A, Willems M, Hellings N, Franken C, Koppen G, Artois T, Plusquin M, Smeets K. Planarians customize their stem cell responses following genotoxic stress as a function of exposure time and regenerative state. Toxicol Sci. 2017 doi: 10.1093/toxsci/kfx247. [DOI] [PubMed] [Google Scholar]
- 90.Lei K, Thi-Kim Vu H, Mohan RD, McKinney SA, Seidel CW, Alexander R, Gotting K, Workman JL, Sánchez Alvarado A. Egf Signaling Directs Neoblast Repopulation by Regulating Asymmetric Cell Division in Planarians. Developmental Cell. 2016;38(4):413–429. doi: 10.1016/j.devcel.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chinone A, Matsumoto M. DrRad51 is required for chiasmata formation in meiosis in planarian Dugesia ryukyuensis. Mol Reprod Dev. 2014;81(5):409–21. doi: 10.1002/mrd.22308. [DOI] [PubMed] [Google Scholar]
- 92.Xiang Y, Miller DE, Ross EJ, Sanchez Alvarado A, Hawley RS. Synaptonemal complex extension from clustered telomeres mediates full-length chromosome pairing in Schmidtea mediterranea. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(48):E5159–68. doi: 10.1073/pnas.1420287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brandl H, Moon H, Vila-Farre M, Liu SY, Henry I, Rink JC. PlanMine--a mineable resource of planarian biology and biodiversity. Nucleic acids research. 2016;44(D1):D764–73. doi: 10.1093/nar/gkv1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robb SMC, Gotting K, Ross E, Sánchez Alvarado A. SmedGD 2.0: The Schmidtea mediterranea genome database. Genesis (New York, NY: 2000) 2015;53(8):535–546. doi: 10.1002/dvg.22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(22):12871–6. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends in Cell Biology. 2016;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends in biochemical sciences. 1998;23(7):247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 99.Shibata A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat Res. 2017;803–805:51–55. doi: 10.1016/j.mrfmmm.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 100.Yin S, Huang Y, Zhangfang Y, Zhong X, Li P, Huang J, Liu D, Songyang Z. SmedOB1 is Required for Planarian Homeostasis and Regeneration. Sci Rep. 2016;6:34013. doi: 10.1038/srep34013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo L, Accorsi A, He S, Guerrero-Hernandez C, Sivagnanam S, McKinney S, Gibson M, Sanchez Alvarado A. An adaptable chromosome preparation methodology for use in invertebrate research organisms. BMC biology. 2018;16(1):25. doi: 10.1186/s12915-018-0497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peiris TH, Garcia-Ojeda ME, Oviedo NJ. Alternative flow cytometry strategies to analyze stem cells and cell death in planarians. Regeneration. 2016;3(2):123–35. doi: 10.1002/reg2.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48(6):371–80. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- 104.Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Developmental Biology. 2010;338(1):76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bender CE, Fitzgerald P, Tait SW, Llambi F, McStay GP, Tupper DO, Pellettieri J, Sanchez Alvarado A, Salvesen GS, Green DR. Mitochondrial pathway of apoptosis is ancestral in metazoans. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(13):4904–9. doi: 10.1073/pnas.1120680109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seung E, Iwakoshi N, Woda BA, Markees TG, Mordes JP, Rossini AA, Greiner DL. Allogeneic hematopoietic chimerism in mice treated with sublethal myeloablation and anti-CD154 antibody: absence of graft-versus-host disease, induction of skin allograft tolerance, and prevention of recurrent autoimmunity in islet-allografted NOD/Lt mice. Blood. 2000;95(6):2175–82. [PubMed] [Google Scholar]
- 107.Mole RH. The LD50 for uniform low LET irradiation of man. Br J Radiol. 1984;57(677):355–69. doi: 10.1259/0007-1285-57-677-355. [DOI] [PubMed] [Google Scholar]
- 108.Bardeen CR, Baetjer FH. The inhibitive action of the Roentgen rays on regeneration in planarians. J Exp Zool. 1904;1:191–195. [Google Scholar]
- 109.Guedelhoefer OC, Sánchez Alvarado A. Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development (Cambridge, England) 2012;139(19):3510–3520. doi: 10.1242/dev.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abnave P, Aboukhatwa E, Kosaka N, Thompson J, Hill MA, Aboobaker AA. Epithelial-mesenchymal transition transcription factors control pluripotent adult stem cell migration in vivo in planarians. Development. 2017;144(19):3440–3453. doi: 10.1242/dev.154971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Solana J, Kao D, Mihaylova Y, Jaber-Hijazi F, Malla S, Wilson R, Aboobaker A. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biology. 2012;13(3):R19. doi: 10.1186/gb-2012-13-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cao L, Kawai H, Sasatani M, Iizuka D, Masuda Y, Inaba T, Suzuki K, Ootsuyama A, Umata T, Kamiya K, Suzuki F. A novel ATM/TP53/p21-mediated checkpoint only activated by chronic γ-irradiation. PloS One. 2014;9(8):e104279. doi: 10.1371/journal.pone.0104279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu X-L, Zhang X-T, Meng J, Zhang H-F, Zhao Y, Li C, Sun Y, Mei Q-B, Zhang F, Zhang T. ING5 knockdown enhances migration and invasion of lung cancer cells by inducing EMT via EGFR/PI3K/Akt and IL-6/STAT3 signaling pathways. Oncotarget. 2017;8(33):54265–54276. doi: 10.18632/oncotarget.17346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science (New York, NY) 2011;332(6031):811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peiris TH, Ramirez D, Barghouth PG, Ofoha U, Davidian D, Weckerle F, Oviedo NJ. Regional signals in the planarian body guide stem cell fate in the presence of genomic instability. Development (Cambridge, England) 2016;143(10):1697–1709. doi: 10.1242/dev.131318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332(6031):811–6. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Herzog KH, Chong MJ, Kapsetaki M, Morgan JI, McKinnon PJ. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science (New York, NY) 1998;280(5366):1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- 118.Taylor MRG, Spirek M, Chaurasiya KR, Ward JD, Carzaniga R, Yu X, Egelman EH, Collinson LM, Rueda D, Krejci L, Boulton SJ. Rad51 Paralogs Remodel Pre-synaptic Rad51 Filaments to Stimulate Homologous Recombination. Cell. 2015;162(2):271–286. doi: 10.1016/j.cell.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nature structural & molecular biology. 2010;17(10):1260–2. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(13):6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Molecular and Cellular Biology. 1996;16(12):7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annual review of biochemistry. 2013;82:357–85. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 123.Pichler A, Fatouros C, Lee H, Eisenhardt N. SUMO conjugation - a mechanistic view. Biomol Concepts. 2017;8(1):13–36. doi: 10.1515/bmc-2016-0030. [DOI] [PubMed] [Google Scholar]
- 124.Garvin AJ, Morris JR. SUMO, a small, but powerful, regulator of double-strand break repair. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2017;372(1731) doi: 10.1098/rstb.2016.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morris JR, Garvin AJ. SUMO in the DNA Double-Stranded Break Response: Similarities, Differences, and Cooperation with Ubiquitin. Journal of Molecular Biology. 2017;429(22):3376–3387. doi: 10.1016/j.jmb.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 126.Nie M, Boddy MN. Cooperativity of the SUMO and Ubiquitin Pathways in Genome Stability. Biomolecules. 2016;6(1) doi: 10.3390/biom6010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sarangi P, Zhao X. SUMO-mediated regulation of DNA damage repair and responses. Trends in biochemical sciences. 2015;40(4):233–242. doi: 10.1016/j.tibs.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang Z, Zhu W-G, Xu X. Ubiquitin-like modifications in the DNA damage response. Mutation Research. 2017;803–805:56–75. doi: 10.1016/j.mrfmmm.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 129.Bologna S, Altmannova V, Valtorta E, Koenig C, Liberali P, Gentili C, Anrather D, Ammerer G, Pelkmans L, Krejci L, Ferrari S. Sumoylation regulates EXO1 stability and processing of DNA damage. Cell Cycle (Georgetown, Tex) 2015;14(15):2439–2450. doi: 10.1080/15384101.2015.1060381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hendriks IA, Treffers LW, Verlaan-de Vries M, Olsen JV, Vertegaal ACO. SUMO-2 Orchestrates Chromatin Modifiers in Response to DNA Damage. Cell Reports. 2015 doi: 10.1016/j.celrep.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hendriks IA, Vertegaal ACO. SUMO in the DNA damage response. Oncotarget. 2015;6(18):15734–15735. doi: 10.18632/oncotarget.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang C, Cheng J, Bawa-Khalfe T, Yao X, Chin YE, Yeh ET. SUMOylated ORC2 Recruits a Histone Demethylase to Regulate Centromeric Histone Modification and Genomic Stability. Cell Rep. 2016;15(1):147–57. doi: 10.1016/j.celrep.2016.02.091. [DOI] [PubMed] [Google Scholar]
- 133.Wu C-S, Ouyang J, Mori E, Nguyen HD, Maréchal A, Hallet A, Chen DJ, Zou L. SUMOylation of ATRIP potentiates DNA damage signaling by boosting multiple protein interactions in the ATR pathway. Genes & Development. 2014;28(13):1472–1484. doi: 10.1101/gad.238535.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Molecular Cell. 2013;49(5):795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 135.Lee B, Muller MT. SUMOylation enhances DNA methyltransferase 1 activity. The Biochemical journal. 2009;421(3):449–461. doi: 10.1042/BJ20090142. [DOI] [PubMed] [Google Scholar]
- 136.Qin Y, Xu J, Aysola K, Begum N, Reddy V, Chai Y, Grizzle WE, Partridge EE, Reddy ESP, Rao VN. Ubc9 mediates nuclear localization and growth suppression of BRCA1 and BRCA1a proteins. Journal of Cellular Physiology. 2011;226(12):3355–3367. doi: 10.1002/jcp.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shima H, Suzuki H, Sun J, Kono K, Shi L, Kinomura A, Horikoshi Y, Ikura T, Ikura M, Kanaar R, Igarashi K, Saitoh H, Kurumizaka H, Tashiro S. Activation of the SUMO modification system is required for the accumulation of RAD51 at sites of DNA damage. Journal of Cell Science. 2013;126(Pt 22):5284–5292. doi: 10.1242/jcs.133744. [DOI] [PubMed] [Google Scholar]
- 138.Vyas R, Kumar R, Clermont F, Helfricht A, Kalev P, Sotiropoulou P, Hendriks IA, Radaelli E, Hochepied T, Blanpain C, Sablina A, van Attikum H, Olsen JV, Jochemsen AG, Vertegaal ACO, Marine JC. RNF4 is required for DNA double-strand break repair in vivo. Cell Death and Differentiation. 2013;20(3):490–502. doi: 10.1038/cdd.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Davis JN, McCabe MT, Hayward SW, Park JM, Day ML. Disruption of Rb/E2F pathway results in increased cyclooxygenase-2 expression and activity in prostate epithelial cells. Cancer research. 2005;65(9):3633–42. doi: 10.1158/0008-5472.CAN-04-3129. [DOI] [PubMed] [Google Scholar]
- 140.Thomas S, Balan A, Balaram P. The expression of retinoblastoma tumor suppressor protein in oral cancers and precancers: A clinicopathological study. Dental research journal. 2015;12(4):307–14. doi: 10.4103/1735-3327.161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Sousa N, Rodriguez-Esteban G, Rojo-Laguna JI, Salo E, Adell T. Hippo signaling controls cell cycle and restricts cell plasticity in planarians. PLoS Biol. 2018;16(1):e2002399. doi: 10.1371/journal.pbio.2002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Owlarn S, Klenner F, Schmidt D, Rabert F, Tomasso A, Reuter H, Mulaw MA, Moritz S, Gentile L, Weidinger G, Bartscherer K. Generic wound signals initiate regeneration in missing-tissue contexts. Nature communications. 2017;8(1):2282. doi: 10.1038/s41467-017-02338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Research. 2009;37(11):3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou J-M, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rouleau M, McDonald D, Gagné P, Ouellet ME, Droit A, Hunter JM, Dutertre S, Prigent C, Hendzel MJ, Poirier GG. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. Journal of Cellular Biochemistry. 2007;100(2):385–401. doi: 10.1002/jcb.21051. [DOI] [PubMed] [Google Scholar]
- 146.Rulten SL, Fisher AEO, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Molecular Cell. 2011;41(1):33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 147.Augustin A, Spenlehauer C, Dumond H, Ménissier-De Murcia J, Piel M, Schmit A-C, Apiou F, Vonesch J-L, Kock M, Bornens M, De Murcia G. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. Journal of Cell Science. 2003;116(Pt 8):1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 148.Fernández-Marcelo T, Frías C, Pascua I, de Juan C, Head J, Gómez A, Hernando F, Jarabo J-R, Díaz-Rubio E, Torres A-J, Rouleau M, Benito M, Iniesta P. Poly (ADP-ribose) polymerase 3 (PARP3), a potential repressor of telomerase activity. Journal of experimental & clinical cancer research: CR. 2014;33:19. doi: 10.1186/1756-9966-33-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW. A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. Developmental Cell. 2015;35(5):632–645. doi: 10.1016/j.devcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stuckemann T, Cleland JP, Werner S, Thi-Kim Vu H, Bayersdorf R, Liu SY, Friedrich B, Julicher F, Rink JC. Antagonistic Self-Organizing Patterning Systems Control Maintenance and Regeneration of the Anteroposterior Axis in Planarians. Developmental cell. 2017;40(3):248–263 e4. doi: 10.1016/j.devcel.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 151.Wurtzel O, Oderberg IM, Reddien PW. Planarian Epidermal Stem Cells Respond to Positional Cues to Promote Cell-Type Diversity. Developmental cell. 2017;40(5):491–504. e5. doi: 10.1016/j.devcel.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]