Abstract
The endoplasmic reticulum (ER) is the site of maturation for roughly one-third of all cellular proteins. ER-resident molecular chaperones and folding catalysts promote folding and assembly of this diverse set of newly synthesized proteins. As these processes are error-prone, all eukaryotic cells have a quality control system in place that constantly monitors the proteins and decides their fate. Proteins with potentially harmful non-native conformations are subjected to assisted-folding or degraded. Persistent folding-defective proteins are distinguished from folding intermediates and targeted for degradation by a specific process involving clearance from the ER. Although the basic principles of these processes appear conserved from yeast to animals and plants, there are distinct differences in the ER-associated degradation of misfolded glycoproteins. The general importance of ER quality control events is underscored by their involvement in biogenesis of diverse cell surface receptors and the crucial function to maintain protein homeostasis under diverse stress conditions.
Keywords: endoplasmic reticulum-associated degradation, ERAD, ERQC, glycoprotein, glycosylation, protein folding
Introduction
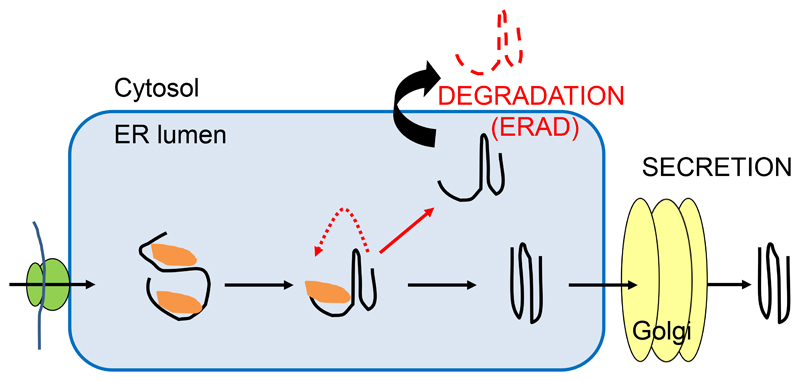
The ER is the site of entry to the secretory pathway and thus serves as major protein folding organelle for a large number of diverse proteins. Protein folding is error-prone and “abnormal” proteins may be the result of different events including genetic mutations, missing post-translational modifications or altered environmental conditions (6; 52). As a consequence, all eukaryotic cells have evolved specific protein quality control processes that recognize non-native protein conformations, promote folding and target terminally misfolded proteins or unassembled proteins for degradation. These mechanisms prevent the accumulation of potentially harmful aberrant proteins and maintain cellular protein homeostasis under different environmental conditions which is especially crucial for sessile organisms. ER-quality control (ERQC) processes including promotion of protein folding and ER-associated degradation (ERAD) as well as ER exit are competing events that must be tightly controlled to avoid an overly stringent quality control that selects functional proteins for degradation as well as an overly lax system that allows targeting of potentially harmful non-functional proteins to other cellular destinations (Figure 1).
Figure 1. Schematic overview of ERQC processes.
As soon as a polypeptide emerges in the lumen of the ER it faces numerous chaperones (shown in orange) and enzymes that promote folding and prevent aggregation. While proper folded proteins with native conformations exit the ER to the Golgi, terminally misfolded proteins are degraded by ER-associated degradation (ERAD). A critical step in ERQC is the distinction between folding intermediates in productive folding cycles and folding-defective polypeptides in futile folding processes.
N-linked glycosylation is the most common co- and posttranslational modification of secretory proteins. N-glycosylation is both directly and indirectly linked with protein folding and plays an important role in quality control and the clearance of misfolded proteins (44). Here, I provide an overview of the current status of the field and discuss the controversial and poorly understood processes with a major focus on glycoproteins and glycan-dependent pathways.
Protein Folding in the ER
Molecular chaperones and factors involved in protein folding in the ER
The majority of cellular proteins that are targeted to the secretory pathway enter the ER by translocation through the Sec61 channel while they are still undergoing synthesis. As soon as the nascent polypeptide chain emerges in the ER lumen, different chaperones, folding factors and enzymes interact with the polypeptide to block aggregation, prevent premature folding and assist with folding towards a native state. Binding protein (BiP), which belongs to the classical heat-shock protein 70 (HSP70) family, is the major molecular chaperone in the ER lumen. BiP binds to newly-synthesized proteins as they enter from the translocation channel and keeps them in a folding-competent state. The molecular chaperone activity of BiP is ATP-dependent and includes cycles of protein binding and ATP hydrolysis. In mammalian cells and in plants, BiP is frequently found in a multiprotein complex (9; 37; 92; 97), which consists of other heat shock proteins, different chaperones and folding enzymes such as protein disulfide isomerase (PDI) and UDP-glucose:glycoprotein glucosyltransferase (UGGT). The presence of UGGT, the folding sensor for misfolded glycoproteins, in this multiprotein complex indicates that glycosylated and non-glycosylated proteins are subjected to folding with the help of a dedicated set of ER chaperones and enzymes that reside next to each other.
In contrast to mammals and yeast, flowering plants contain several BiP proteins (BiP1 to BiP3 in Arabidopsis) (26; 101). BiP1 and BiP2 are almost identical based on their amino acid sequence and are ubiquitously expressed under non-stressed conditions, whereas BiP3, which is more distantly related, is expressed mainly in pollen and highly induced upon ER stress (90; 101). An Arabidopsis bip2 single mutant is more sensitive to tunicamycin treatment and more susceptible to infection with the bacterial pathogen Pseudomonas syringae (140). The bip1 bip2 double mutant is viable, but normal pollen tube growth is decreased and the fusion of polar nuclei in female gametophytes is disrupted in these plants (89; 90). The latter phenotype can be rescued by the expression of BiP3 under the control of the BiP1 promoter, suggesting a comparable chaperone function. The bip1 bip2 bip3 triple mutant is lethal in pollen and causes pollen abortion (90). Not all of the observed phenotypes may be directly related to protein folding processes, as BiPs are involved in other processes in the ER such as protein translocation or activation of the unfolded protein response (UPR) by dissociation from the ER stress sensor bZIP28 (124). BiP3 is implicated in the biogenesis of glycoproteins such as the pattern recognition receptor XA21 (104) and binds to misfolded variants of the growth receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1). Tobacco BiP was also shown to bind to a bean phaseolin domain involved in trimer formation as well as to calreticulin in a larger protein complex (21; 35).
ERdj3, a luminal member of the DNA J protein family, was initially found in mammalian cells in protein binding of the unassembled heavy chain of immunoglobulin G1 (IgG1) (92). Subsequent studies revealed that ERdj3 acts as a co-chaperone of BiP and associates with non-native polypeptides to transfer them to BiP and promote their folding (62). The ER-luminal Arabidopsis proteins ERdj3A, ERdj3B and P58IPK could partially complement the growth phenotype associated with yeast mutants of the corresponding J proteins Scj1p (ERdj3A and ERdj3B homolog) and Jem1p (related to P58IPK) that function in protein folding (145). ERdj3 proteins from Arabidopsis display differences in their expression. ERdj3A is mainly expressed in pollen, whereas ERdj3B is found in many different organs. ERdj3A interacts with BiP1 and BiP3 and stimulates the ATP-hydrolysis that leads to trapping of substrate proteins in the substrate binding domain of BiP (13; 86). In addition, Arabidopsis and rice contain two homologs of ERdj2 (ERdj2A and ERdj2B) and one ERdj5/ERdj7 related protein (102; 145). In mammals, ERdj5 participates in ERAD and promotes misfolded protein degradation by the cleavage of intermolecular disulfide bridges (134). Although this activity is mediated by its CXXC motif, the J domain shows interaction with BiP. ERdj5 also interacts with ER degradation-enhancing α-mannosidase-like (EDEM) proteins, which are key factors for the degradation of misfolded glycoproteins. Arabidopsis ERdj5 is found in the ER but lacks a thioredoxin domain and a CXXC motif, and its involvement in protein folding and the degradation of misfolded glycoproteins remains to be demonstrated (31; 145).
The Arabidopsis HSP90 family consists of seven proteins that are present in the cytosol and different organelles. The single ER-resident HSP90 member in Arabidopsis, called SHEPHERD (SHD; other names include endoplasmin, AtGRP94 or HSP90.7), is required for correct CLAVATA signaling (18; 58; 67). Genetic evidence suggests that SHD is implicated in folding or complex assembly of CLAVATA proteins in the ER, but its mechanism of action is unclear. SHD overexpression in tobacco protoplasts promotes the secretion of α-amylase in a similar albeit less efficient manner as BiP overexpression (67).
In human cells, the homologous ER-resident proteins STROMAL-DERIVED FACTOR-2 (SDF2) and SDF2-LIKE 1 (SDF2L1) associate with ERdj3 and bind to non-native proteins to prevent their aggregation (37). A forward-genetic screen for pathogen-associated molecular pattern (PAMP)-insensitive Arabidopsis mutants identified a T-DNA insertion in the single copy gene coding for the SDF2 homolog from plants (97). Immune complexes from purified SDF2 contained ERdj3B and BiP but not SHD or the lectin chaperone calreticulin (CRT) which is together with its membrane-bound paralogue calnexin (CNX) part of a glycan-dependent folding cycle. In Y2H assays, SDF2 interacted with ERdj3B but not with SHD, BiP1-3, UGGT, CRT2 or CRT3, suggesting that SDF2 may not be a part of the CNX/CRT-UGGT complex. Remarkably, SDF2 deficiency impaired the biogenesis of EFR, the receptor binding to the bacterial PAMP EF-Tu (97). In the sdf2 mutant, EFR carried mainly oligomannosidic N-glycans and its degradation was blocked by the specific ERAD inhibitor kifunensine. More recently, it was shown that SDF2 co-purifies together with other ER-resident proteins such as the rice pattern recognition receptor XA21 (105). XA21 is heavily glycosylated and the co-purified complex contained BiP3, CNX1, CRT3, ERdj3B, PDI and a HSP90 homolog (104; 105). Silencing of SDF2 in rice reduced the XA21 mediated immunity presumably by directly affecting XA21 biogenesis in the ER. Mutations in the genes encoding SDF2, ERdj3b and CRT3 have also been found to suppress the cell death and the constitutive defense response in bir1 plants defective in BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1)-INTERACTING RECEPTOR-LIKE KINASE 1 (BIR1), a negative regulator of plant immunity (129). ERQC involving ERdj3b, CRT3 and presumably SDF2 is required for the efficient biogenesis and maturation of SUPPRESSOR OF BIR1-1 (SOBIR1), a receptor like-kinase with two N-glycosylation sites in its extracellular leucine-rich-repeat domain (38). Another study reported that erdj3b and sdf2 mutants, but not crt3, led to a suppression of BAK1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE 4 (SERK4) silencing-mediated cell death (24). Client proteins of the BAK1/SERK4 silencing-mediated cell death that are subjected to ERQC have not been unequivocally identified. However, candidates such as cysteine-rich receptor-like kinase (CRK4 or CRK5) have several N-glycosylation sites and are glycosylated (24). While mammalian SDF2/SDF2L1 does not distinguish between the glycosylated and non-glycosylated variants of misfolded proteins (37), plant SDF2 may be more specific for folding of glycosylated proteins. Interestingly, determination of the Arabidopsis SDF2 crystal structure revealed the presence of the β-trefoil fold that has is also present in lectins and may act as a carbohydrate binding domain (120).
Co-/posttranslational modifications and protein folding
Protein disulfide isomerases (PDIs) catalyze the formation and rearrangement of disulfide bonds in the ER. The PDI superfamily in Arabidopsis consists of 14 members (147), which can be further divided into 6 distinct subfamilies. They have in common one or more thioredoxin-like domains. Some of the PDIs such as PDI5 or PDI6 – members of subfamily PDI-L, which are categorized by the presence of two thioredoxin domains and two redox-inactive thioredoxin domains - represent classical disulfide oxidoreductases as found in mammals and yeast that have chaperone activity in plants (2). Other PDI proteins display a different domain organization with only one thioredoxin domain (PDI-C subfamily) and an additional domain that is found in proteins implicated in Golgi-to-ER retrograde transport (148). The role of these PDI-C proteins in retrieval of native ER-resident proteins - similar to the well-established KDEL/HDEL receptor - or retrieval of escaped non-native proteins that expose free cysteine residues remains to be demonstrated. In mammalian cells, CNX/CRT are associated with the PDI ERp57 to promote oxidative folding. Yeast PDI forms a complex with the class I α-mannosidase HTM1 for de-mannosylation and initiation of ERAD (39). A misfolded variant of the brassinosteroid receptor BRI1 (BRI1-5) is also retained in the ER by a thiol-based mechanism (47), but the involvement of PDIs is unknown.
Peptidyl-prolyl cis-trans isomerases (PPIases) catalyze the cis-trans isomerization of proline peptide bonds which is a rate-limiting step for folding. PPIase subfamilies include cyclophilins, FK506-binding proteins and parvulins (71). The roles of PPIases in ERQC and functional cyclophilins and parvulins acting in the ER have not been well described in plants (71; 84). The FK506-binding protein TWISTED DWARF1 (FKBP42/TWD1) interacts with BRI1 and has been described as an ER-resident protein (15; 141; 149). However, TWD1 is a type I membrane protein, and the FK506-binding protein domain faces the cytoplasm where it interacts with a cytosolic HSP90 and the cytosolic kinase domain of BRI1. Consistent with a role in BR signaling and not ERQC, the secretion and protein accumulation of BRI1 are unaffected in a tdw1 mutant line (149).
N-glycosylation and carbohydrate-mediated folding
N-glycosylation is a major covalent modification of proteins entering the secretory pathway (3). N-glycosylation is initiated by en bloc transfer of a pre-assembled oligosaccharide (Glc3Man9GlcNAc2) precursor to an asparagine residue within the sequence motif Asn-X-Ser/Thr (X represents any amino acid except proline) on nascent polypeptide chains. For most glycoproteins, the N-glycosylation step is carried out co-translationally and precedes folding (1). N-glycosylation is catalyzed by oligosaccharyltransferase (OST) which is a heteromeric membrane-embedded protein complex consisting of a single catalytic subunit (STT3) and several non-catalytic proteins. In mammalian cells, there is evidence showing that OST, together with Sec61, is an integral component of the native ribosome–translocon complex (25; 70; 108). Although the Sec61 channel and the whole translocon are poorly characterized in plants, it is very likely that the underlying mechanisms of protein translocation into the ER and concomitant glycosylation are conserved. Simultaneous disruption of the two catalytic subunits, STT3A and STT3B, which very likely define two distinct OST complexes, is lethal in Arabidopsis (68). Moreover, a leaky mutant of the non-catalytic OST subunit DGL1 (dgl1-1) displayed a severe underglycosylation defect resulting in a drastic post-embryonic growth defect (73). Together, these data underscore the importance of protein N-glycosylation. In contrast to other eukaryotes, the molecular function of the non-catalytic OST subunits is poorly characterized in plants (126). In yeast and metazoans, they modulate N-glycosylation by regulating the substrate specificity, stability or assembly of the OST complex (64; 93). The yeast OST3 and OST6 subunits, for example, display oxidoreductase activity, whereby a CXXC motif forms mixed disulfides with nascent glycoprotein substrates to prevent their premature folding and keep them in a glycosylation competent state (121). The Arabidopsis OST3/OST6 homolog (known as OST3/6) lacks a CxxC motif, indicating that this OST subunit has evolved a different function or recruited additional thioredoxin-containing proteins, which function as oxidoreductases. Similar to the catalytic OST subunit STT3A, OST3/6 is important for the biogenesis of EFR, and OST3/6-dependent glycosylation affects a yet unknown glycoprotein receptor involved in interspecific pollen tube reception (32; 96). Likewise, underglycosylation, induced by tunicamycin treatment of Arabidopsis seedlings, impaired the accumulation of EFR but had little effect on other heavily glycosylated plant proteins such as the immune receptor FLAGELLIN SENSING 2 (FLS2) and BAK1 (54). Collectively, these studies suggest that protein folding and ERQC of certain types of plant glycoproteins are impaired when N-glycosylation efficiency is reduced.
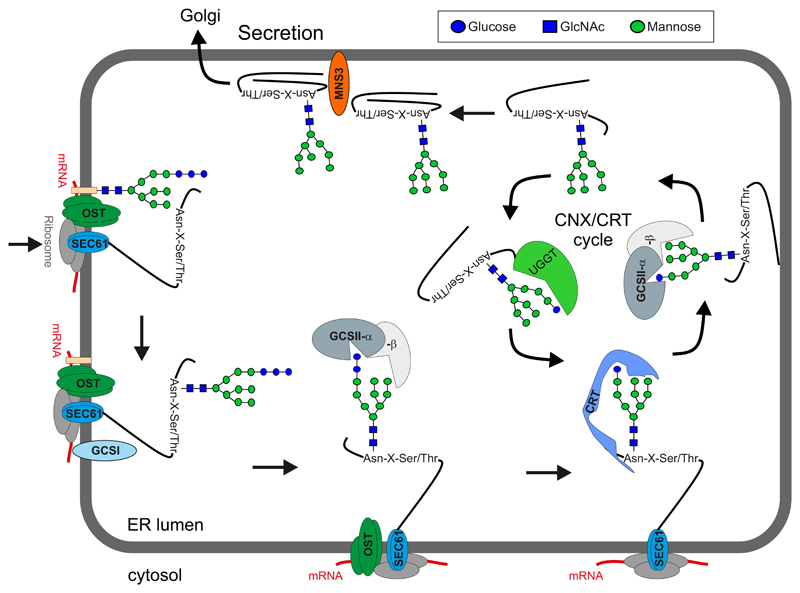
Several mechanisms contribute to the misfolding in the absence of N-glycosylation. Attached N-glycans can directly or indirectly influence the folding of nascent proteins in the lumen of the ER. The direct effect is mainly caused by the hydrophilic nature of the bulky oligosaccharides which stabilize the local protein structures and mask the hydrophobic patches of the polypeptide to prevent aggregation (42). The higher stability of glycosylated compared to non-glycosylated proteins can be the outcome of a less stable unfolded state or a more stable folded state (122). In addition to their direct effects, the covalently attached N-glycans serve as recognition signals for the ER-resident carbohydrate-binding proteins CNX and calreticulin CRT (44). Binding to these specific lectins suppresses aggregation by slowing down folding and prevents trafficking to downstream compartments. These functions are either mediated by intrinsic chaperone functions due to CNX/CRT-polypeptide interactions or by the recruitment of folding factors and enzymes such as the oxidoreductase ERp57 and PPIases. To facilitate the interaction of glycan moieties with CNX and its soluble homolog CRT, the outermost α1,2-linked glucose residue is removed by α-glucosidase I (GCSI) immediately after the transfer of the oligosaccharide (Figure 2). GCSI is a type II transmembrane protein that is associated with the translocon and the OST complex (25). Subsequently, α-glucosidase II (GCSII), a luminal enzyme composed of a catalytic alpha subunit and a beta-subunit which contains a mannose-binding lectin domain with a proposed regulatory function for N-glycan trimming, catalyzes two consecutive cleavage steps. These processing reactions are more regulated than the GCSI-mediated hydrolysis and determine the interaction with CNX/CRT. The first cleavage removes the terminal α1,3-glucose residue, thus generating the mono-glucosylated N-glycan Glc1Man9GlcNAc2 (23), which is optimally suited for binding to the lectin domains from CNX and CRT (69). The second cleavage reaction removes the innermost glucose residue, which liberates the glycoprotein from its association with CNX/CRT. Structural studies of the mammalian enzyme indicate that the glucose-trimming reactions are not consecutive because the substrate must be reoriented and inserted into the active site in the opposite direction (28). This partial separation of GCSII and the substrate may facilitate the CNX/CRT binding.
Figure 2. Glycosylation and productive ERQC steps leading to ER exit and secretion of native glycoproteins.
The nascent polypeptide enters the ER through the SEC61 channel. The oligosaccharyltransferase (OST) complex identifies N-glycosylation sites (Asn-X-Ser/Thr, X can be any amino acid except proline) and transfers the preassembled oligosaccharide to the Asn residue. Glucosidase I (GCSI) removes the ultimate glucose residue and subsequently the heteromeric GCSII cleaves the penultimate glucose. The resulting mono-glucosylated N-glycan is the signal for recognition and binding by the lectins calreticulin (CRT) or the membrane bound calnexin (CNX, not shown here). CNX/CRT promotes folding together with folding catalysts (such as protein disulfide isomerase, not shown). The dynamic interaction between CNX/CRT can be driven towards dissociation by GCSII-mediated de-glucosylation. Incompletely folded glycoproteins are re-glucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT) and subjected to another round of CNX/CRT-mediated folding (CNX/CRT cycle). Properly folded glycoproteins are not re-glucosylated, processed by the plant ER α-mannosidase I (MNS3) and exit the ER to the Golgi.
Upon cleavage of the second glucose, the released glycoprotein is subjected to a quality check by UGGT, which acts post-translationally as the central folding sensor for glycoproteins in the ER and decides their fate. UGGT recognizes minor folding detects as well as permanent misfolding (106; 113; 132) and re-glucosylates N-glycans on maturing proteins. The non-native glycoproteins bearing mono-glucosylated N-glycans can re-enter the CNX/CRT cycle for further folding attempts. Proteins that reach their native conformation are not re-glucosylated by UGGT, do not re-engage with CNX/CRT and are allowed to leave the ER to downstream compartments (Figure 2). By contrast, terminally misfolded glycoproteins or folding intermediates that stay for a prolonged time are retrieved from the CNX/CRT cycle and directed to ERAD.
Complete disruption of GCSI activity is lethal in Arabidopsis (11; 40), but the defect and the resulting phenotype can be rescued by concomitant knockout of ALG10, the glucosyltransferase that adds the outmost glucose residue to the preassembled oligosaccharide precursor (33). Elimination of GCSII activity, by knockout of the alpha subunit has very likely a similar drastic effect leading to lethality in Arabidopsis (123). These findings suggest that early N-glycan trimming steps are essential for ERQC, presumably to facilitate the mono-glucosylated glycan-dependent interaction with CNX/CRT that promotes folding.
A. thaliana contains three CRTs (CRT1, CRT2 and CRT3) and two membrane-bound CNXs (CNX1 and CNX2). Analysis of promoter-GUS fusions showed that CNX1 and CNX2 are differentially expressed with CNX1 displaying higher expression levels in most organs. Promotor swapping and complementation of plant growth defects indicated, however, that both lectins have a redundant molecular function (139). While CRT1/CRT2 display a high sequence identity and are functionally redundant, CRT3 is distinct and has acquired plant-specific functions (107). For example, CRT3 overexpression could not restore the mild growth defect observed for crt1 crt2 seedlings (19). Loss of all CRTs caused increased sensitivity to water stress (65) and additional CNX1 disruption resulted in vegetative growth phenotypes (139). The elimination of all five CNX/CRT encoding genes is not tolerated in Arabidopsis.
Despite the advances in our understanding of the physiologic role of CNX/CRT, we know little about potential client proteins that are subjected to glycan-dependent ERQC. The immune receptor EFR and BRI1-9, another misfolded variant of the brassinosteroid receptor BRI1, displayed a UGGT-dependent interaction with the plant-specific CRT3 (60; 74; 116). Complementation experiments with chimeric fusion proteins and expression of mutated CRT3 variants revealed that the C-terminal subdomain of Arabidopsis CRT3 contains a basic amino acid motif responsible for the ER-retention of BRI1-9 and EFR (48; 80). Quite puzzling, BRI1-9 was also associated with CNX1/CNX2 (61) but only the CRT3 deficiency suppressed the severe growth phenotype of bri1-9 (60). Therefore, CNX1/CNX2 and CRT3 may have indeed different roles with the former involved in protein folding and CRT3 acting as a retention factor to keep the misfolded protein in the ER. Furthermore, misfolded BRI1-5, is cleared by a glycan-dependent ERAD process and found associated with CNX, but in contrast to bri1-9 neither disruption of CRT3 nor of CNX1 and CNX2 suppressed the bri1-5 dwarf phenotype (47) (Table 1).
Table 1. List of A. thaliana ERAD components and their potential client proteins.
| Protein name1 | A. thaliana name | A. thaliana Locus | ERAD substrate | References |
|---|---|---|---|---|
| SEL1L/HRD3 | SEL1L (HRD3A2, EBS5) | At1g18260 | BRI1-5, BRI1-9 MLO-13 SUBEX-C57Y |
(78; 127) (78; 127) (78) (56) |
| HRD1/HRD1 | HRD1A HRD1B4 |
At3g16090 At1g65040 |
BRI1-9 | (127) |
| OS-9, XTP3-B/YOS9 | OS9 (EBS6) | At5g35080 | BRI1-5 BRI1-9 EFR*5 SUBEX-C57Y |
(55; 128) (55; 128) (128) (56) |
| EDEM1-3/HTM1 | MNS4 MNS54 |
At5g43710 At1g27520 |
BRI1-5 BRI1-9 SUBEX-C57Y |
(57) (57) (56) |
| HERP/USA1 | EBS76 | At4G29960 | BRI1-9 BRI1-5 EFR*5 |
(81) (81) (81) |
| UBE2J1/UBC6 | UBC32 | At3g17000 | BRI1-5 BRI1-9 MLO-123 |
(22) (22) (22) |
The names of the mammalian/yeast homologs are given.
There is another SEL1L homologue in the A. thaliana genome (HRD3B), which is a pseudogene and does not play any role in ERAD (78; 127).
MLO-1/MLO-12 are non-glycosylated ERAD-C substrates (95).
HRD1A/HRD1B and MNS4/MNS5 appear functionally redundant.
“*”Highlights that misfolded EFR (e.g. present in uggt or sdf2 mutants) is a glycan-dependent ERAD substrate.
Putative similar function like yeast USA1 and human HERP.
In contrast to mammals where two UGGT isoforms were reported, Arabidopsis harbors a single gene coding for UGGT. Intriguingly, the first reports on deletion of UGGT (alternative names EBS1 or PSL2) did not describe any growth or developmental phenotypes. The first uggt mutants (ebs1) were identified in a genetic screen to identify regulators of the BR signaling pathway. UGGT deficiency suppressed the bri1-9 growth defect, but no alterations of growth or morphology were reported for ebs1 plants when compared to wild-type (61). In additional studies, it was shown that other uggt mutants were impaired in EFR-mediated defense reactions (74; 116). Again no abnormal growth or developmental phenotype was reported for these different uggt alleles suggesting that UGGT function is to a great extent dispensable under non-stressed conditions. UGGT deficiency suppressed the bri1-9 phenotype, but unexpectedly enhanced the bri1-5 phenotype (47; 61). The different nature of the lesion in BRI1-5 and BRI1-9 affects the interaction with ERQC components. BiP binds stronger to BRI1-5, which may compensate for the absence of other chaperones and the BRI1-5 mutation (C69Y) disrupts the formation of a disulfide bond which may contribute to ER-retention of BRI1-5 by mixed disulfide formation with a yet unknown oxidoreductase (47).
Together with the studies for disruption of GCSI, GCSII and CNX/CRT the data from the different uggt alleles imply that a single round of CNX/CRT interaction is essential for the plant development. Re-entrance and association with CNX/CRT that is dependent on UGGT re-glucosylation is required for certain proteins, such as glycoproteins involved in stress response. More recently, a study focused on the physiologic role of Arabidopsis UGGT and investigated the phenotypes of two different uggt alleles (10). The UGGT-deficient plants exhibited a delayed vegetative growth as seen by reduced rosette diameters of 6-week old plants and shorter roots in seedlings. After 60 days the difference between wild-type and mutants were less visible indicating that UGGT activity is important for early growth stages.
Other types of protein glycosylation in the ER
In mammals and other eukaryotes, single sugar residues can be attached to the hydroxyl amino acids serine/threonine (termed O-glycosylation) of nascent polypeptides in the ER. O-mannosylation plays an important role for protein folding and quality control in Saccharomyces cerevisiae (143). The attachment of sugars such as mannose decreases the hydrophobicity and prevents the formation of insoluble aggregates. The involved yeast mannosyltransferases associate with the translocon complex and enable glycosylation prior to folding and competition between N- and O-glycosylation (83; 98). O-mannose or other monosaccharides can also be attached in a later step to terminate the folding of non-native proteins that persist in futile folding cycles for some time and send them for ERAD (142). Some mammalian proteins are also modified with O-mannose in the ER and O-fucosylation, another type of O-glycosylation, has been implicated in protein folding and quality control in mammalian cells and Drosophila. The corresponding O-fucosyltransferase discriminates between folded/unfolded states and contains a chaperon-like functions that is independent of its catalytic activity (137). Similar co- or post-translational modifications that promote folding or prevent aggregation have not been described in plants and they lack clear homologs of yeast O-mannosyltransferases or mammalian O-fucosyltransferases. Recently, Arabidopsis REPRESSOR OF CYTOKININ DEFICIENCY 1 (ROCK1), an ER-resident nucleotide sugar transporter involved in the regulation of the plant response to cytokinin, was identified (99). Transport assays with yeast microsomes expressing ROCK1 showed uptake of the nucleotide sugars UDP-GalNAc and UDP-GlcNAc. Strikingly, ROCK1 deficiency reduced the protein abundance of the cytokinin oxidases/dehydrogenase 1 (CKX1) and suppressed the bri1-9 growth phenotype in a similar way as previously described for uggt mutants and other ERQC components (60; 61; 99). CKX1 and BRI1-9 are both N-glycosylated but GlcNAc-containing N-glycans are unaffected and incorporation of GalNAc to protein linked N-glycans has never been described in plants. Consequently, it is possible that the transported sugars are part of a yet unknown glycosylation reaction that contributes to protein maturation and quality control.
Protein Misfolding and ER-Associated Degradation (ERAD) Pathways
If protein folding attempts fail or the when the native conformation cannot be reached within a certain time, proteins are directed from folding to degradation pathways. Since the ER is not a typical degradation compartment and hosts only a limited set of specific proteases such as the signal peptide peptidase, aberrant proteins, in most cases, are dislocated to the cytosol for proteasomal degradation in a process termed ER-associated degradation (ERAD) (138). Alternatively, misfolded proteins can be eliminated via a proteasome-independent cytosolic pathway or targeted for degradation to the lytic vacuole/lysosome (117). Vacuolar delivery may be required during severe ER stress when proteins aggregate in the ER. Under such conditions, the ERAD capacity is likely insufficient to restore protein homeostasis, leading to the induction of autophagy and delivery of cargo to the vacuole for degradation. In Arabidopsis, it was recently shown that ER stress-inducing agents trigger autophagy by the activation of the UPR in an IRE1b-dependent manner (79). Importantly, the response was dependent on the accumulation of unfolded proteins as chemical chaperones or BiP overexpression reduced the autophagy activation in the presence of ER stress-inducing agents (146). Autophagy was also triggered by the overexpression of constitutively misfolded proteins. ERQC autophagy has also been described in mammals as a mechanism to remove non-aggregated ERAD-resistant misfolded proteins (51). When ERAD was specifically blocked, a normal ERAD substrate was directed towards the ERQC autophagy pathway suggesting a compensation mechanism.
ERAD is a conserved multi-step process involving the recognition of misfolded proteins, transport to the E3 ubiquitin ligase complex, retro-translocation from the ER to the cytosol, polyubiquitination, and finally degradation by the 26S proteasome (138). The ERAD sensors that recognize terminal misfolding in the ER and the retro-translocation/degradation machinery must handle different protein classes, including glycosylated and non-glycosylated clients, which can be luminal or integral transmembrane proteins and expose various folding defects. In mammalian cells and yeast, three different ERAD pathways have been distinguished depending on the location of the misfolded domain and the involved membrane-embedded E3 ubiquitin ligase complex. ERAD-L substrates can be soluble or membrane bound and display lesions in the luminal domain. ERAD-M substrates expose a defect in the transmembrane domain and ERAD-C substrates are categorized by a defect in the cytosolic region of a membrane-anchored protein (114; 138). In S. cerevisiae the two major ubiquitin ligase complexes involved in ERAD include 3-HYDROXY-3-METHYLGLUTARYL-COA REDUCTASE (HMGR) DEGRADATION 1 (HRD1 in yeast; mammals have HRD1 and an additional homolog called GP78) and DEGRADATION OF α FACTOR (DOA10 in yeast; TEB4 in mammals). ERAD-C substrates are eliminated by the DOA10 ubiquitin ligase in cooperation with the ubiquitin-conjugating enzymes UBC6 and UBC7 (45). ERAD-L and ERAD-M substrates are, in most cases, degraded by the HRD1 complex. HRD1 is the core structural component of the membrane-embedded HRD1 ligase complex. In addition to HRD1 and luminal factors such as KAR2 (the yeast BiP homolog) and the lectin YOS9, the yeast HRD1 complex consists of the membrane-bound adaptor HRD3 (SEL1L in mammals), USA1 (HERP in mammals) and DER1 (DER1 to DER3 in mammals). In addition to ubiquitination, HRD1 functions in the retro-translocation of ERAD clients from the ER across the membrane bilayer into the cytosol. Using cryoelectron microscopy it was recently shown that a complex of HRD1 and HRD3 forms a membrane channel that resembles the structure of the Sec61 complex for the transport of nascent proteins into the ER (119). HRD3/SEL1L has a large luminal domain composed of multiple tetratricopeptide repeats for substrate protein binding and their recruitment to HRD1. USA1 facilitates the assembly and oligomerization of the complex in yeast (49). The multi-pass transmembrane protein DER1 is involved in the recognition and turnover of certain luminal substrates and retro-translocation (91). The requirement for the CDC48 ATPase complex (P97 in mammals) on the cytosolic side to extract polyubiquitinated proteins from the ER membrane and transfer them to the proteasome is common to both the HRD1 and DOA10 complexes (114).
The HRD1 complex in plants
Compared to metazoans and yeast, the composition and molecular function of the plant HRD1 complex are not well studied. The interactions of ERAD components with each other and with ERAD substrates have been reported by Co-IP experiments (16; 17; 55; 56; 81), but the composition of the HRD1 complex and its dynamics are unknown. The core ERAD component HRD1 is ubiquitously expressed in plant tissues/organs. Moreover, ERAD is active in different plant parts, as observed by studies on bri1-5/bri1-9 and the suppression of a floral abscission defect caused by a misfolded form of the receptor-like protein kinase HAESA-LIKE 2 (5; 47; 48; 55; 57; 61; 81; 128). Because protein folding is inherently error-prone even under non-stressed conditions, a basal level of ERAD is most likely maintained in all tissues as a safeguard to prevent the accumulation of misfolded proteins and restore protein homeostasis. Arabidopsis has two HRD1 homologs (HRD1A and HRD1B) with a redundant role for the degradation of the ERAD substrate BRI1-9 (127). By contrast, Arabidopsis has only one functional HRD3/SEL1L homolog implicated in the elimination of misfolded proteins (56; 78; 127). In the powerful bri1-9 suppressor screen that revealed many key ERAD components, Li and colleagues identified a protein called EBS7 that blocks the degradation of BRI1-9. Although, EBS7 could not clearly complement ERAD in the yeast Δusa1 mutant and a HERP transgene did not affect the growth of ebs7 bri1-9 plants, it is likely that EBS7 has a function similar to USA1/HERP (81). Specifically, EBS7 interacts with HRD1A and regulates its stability. DER1 mutants have not been reported in the bri1-9 genetic screen, presumably because Arabidopsis has more than one functional DER1 homolog. The Arabidopsis DER1 proteins are uncharacterized, but an earlier study showed that maize DER1s complement the growth defect of a yeast Δder1 strain (66). Affinity purification of the native complex coupled with quantitative mass spectrometry as well as in vivo real-time non-invasive fluorescence-based protein assays will help to better understand the HRD1 complex and its interaction network (53; 118).
In addition to their central role in ERAD, E3 ubiquitin ligases regulate endogenous proteins that lack any folding defect, such as 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), the rate-limiting enzyme in sterol biosynthesis. A genetic screen to identify regulatory components of the mevalonate pathway revealed DOA10A as the regulator of HMGR in Arabidopsis (30). Interestingly, in legumes, HMGR is regulated by MAKIBISHI 1 (a homolog of mammalian RMA1), another ER-resident E3 ubiquitin ligase (112), suggesting that a divergent set of E3 ubiquitin ligases has evolved to control the fate of metabolic enzymes. Arabidopsis DOA10A (alternative name ECERIFERUM9 – CER9) is also implicated in the regulation of plant thermotolerance, plays a role in cuticle lipid biosynthesis, the maintenance of plant water status and abscisic acid signaling (18; 75; 85).
Compared to the glycoprotein substrates, little is known about the complex and components involved in ERAD of non-glycosylated luminal and membrane-bound proteins. A mutant variant of the polytopic membrane protein barley mildew resistance locus O (MLO-1) is degraded in an HRD1- and HRD3-dependent manner in S. cerevisiae (95). MLO-1 is a non-glycosylated protein with a lesion in a cytosolic loop domain (ERAD-C substrate). The requirement of HRD3/SEL1L for MLO-1 degradation was also shown in Arabidopsis (78). Furthermore, transient expression in protoplasts demonstrated that MLO-1 degradation can be blocked upon the co-expression of a dominant negative variant of CDC48 suggesting the involvement of ubiquitination and extraction from the membrane (95).
One of the best characterized glycoprotein ERAD substrates in S. cerevisiae is a misfolded variant of the vacuolar enzyme carboxypeptidase Y (CPY*) that is degraded in a glycan-dependent manner (59). The introduction of the homologous mutation into Arabidopsis CPY generated a misfolded protein that is subjected to ERAD in yeast and Arabidopsis (144). Although, AtCPY* carries a single N-glycan, the glycosylated and non-glycosylated variants display similar degradation kinetics in Arabidopsis protoplasts. Moreover, AtCPY* degradation is blocked in the yeast Δhrd1 mutant but not in Δos9, suggesting that AtCPY* is degraded in a glycan-independent manner. Contrastingly, the elimination of all three N-glycans from a misfolded variant of the extracellular domain from the receptor-like kinase STRUBBELIG (called SUBEX-C57Y) blocked its degradation in Arabidopsis, suggesting a glycan-dependent pathway for this ERAD substrate (56). The expression of different types of misfolded substrates with defined features will help to better understand the mechanisms and requirements for ERAD in the future.
Physiological function of the HRD1 ERAD components
The commonly used ERAD inhibitor kifunensine blocks the action of the five class I α-mannosidases (MNS1-MNS5) in Arabidopsis and causes the formation of short and swollen roots (77). However, the observed kifunensine-dependent phenotype is not caused by the inhibition of MNS4 and MNS5, which have redundant function and are involved in ERAD, but by the complete blockage of the N-glycan-processing enzymes MNS1, MNS2 and MNS3 that are dispensable for ERAD (57; 77).
Consistent with this finding, the genetic disruption of key ERAD components such as HRD1 or HRD3/SEL1L does not lead to developmental phenotypes. However, different phenotypes have been described under non-physiological conditions. In addition to increased salt sensitivity as observed in hrd3/sel1l, os9, hrd1a hrd1b and mns4 mns5 (55; 57; 78; 128), hrd3/sel1l seedlings are less tolerant towards ER stress-inducing agents such as tunicamycin (17). Arabidopsis hrd3/sel1l and hrd1a hrd1b plants are more sensitive to selenate (135), and hrd1a hrd1b displays increased heat tolerance (75).
Disruption of the Arabidopsis E2 ubiquitin-conjugating enzyme UBC32 (a homolog of yeast UBC6) also partially suppressed the bri1-5 and bri1-9 growth phenotypes. Strikingly, UBC32-deficient plants displayed an opposite salt stress response that did not fit the overall picture of the ERAD function in plants (22). However, recently it was shown that UBC32 negatively regulates OS9 protein levels by targeting OS9 to proteasomal degradation (16). Similar to transgenic OS9 overexpression, UBC32 deficiency increases OS9 levels and causes a hyperactive ERAD, which results in reduced salt sensitivity. Mechanistically, the interaction between UBC32 and OS9 is less clear, because OS9 is a luminal ERAD factor associated with the HRD1 complex, and UBC32 is a proposed factor of the DOA10 complex. UBC32 is membrane-anchored, but only few amino acids are located in the ER lumen. Interestingly, either UBC32 or OS9 deficiency can suppress the bri1-5/bri1-9 phenotypes (22; 55; 128), which is difficult to explain in the current model for UBC32 and OS9 function. Regardless of this uncertainty, the role of UBC32 in mediating OS9 levels, as well as the HRD1-dependent regulation of UBC32 protein levels, are examples demonstrating that the abundance of the individual ERAD components is tightly regulated in plants, as observed in mammalian cells (41).
A defective ERAD pathway in os9, hrd3/sel1l, mns4 mns5 and ebs7 activates the UPR (55; 57; 78; 81) because misfolded proteins are not efficiently cleared and their accumulation causes ER stress. Consistent with this observation, ERAD components such as OS9 (55; 88), HRD3/SEL1L, HRD1 (63) and EBS7 (81) are upregulated in response to UPR-inducing agents such as tunicamycin and dithiothreitol, suggesting that enhanced activity of the complex is beneficial under ER stress conditions. Moreover, a recent study demonstrated that transgenic overexpression of the UPR- and salt stress-associated transcription factors bZIP17, bZIP28 and bZIP60 restored the tolerance of the Arabidopsis hrd3/sel1l mutant to abiotic stress. Likely, due to an increased HRD1 expression, the UPR induction compensated for loss of the HRD3/SEL1L ERAD component highlighting once more the close interrelation between UPR and ERAD (76).
Glycan-dependent ERAD: the recognition process
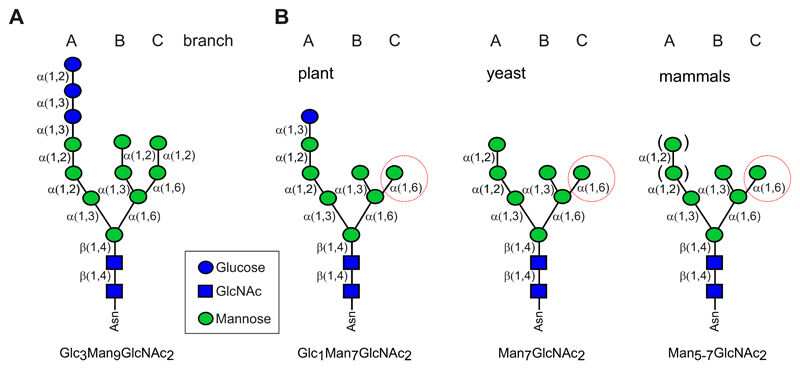
A hallmark of the glycan-dependent ERAD pathway is the recognition of a bipartite signal composed of a non-native protein conformation and an N-glycan displaying a specific composition (20; 56; 59; 111; 130). The glycan signal for degradation was initially identified in yeast and is characterized by the presence of a free α1,6-linked mannose residue on the C-branch of the oligosaccharide (20). This particular glycan determinant for degradation appears to be conserved in mammalian cells (100) as well as in plants (48; 57) (Figure 3). The class I α-mannosidases HTM1 in yeast and its Arabidopsis counterparts MNS4/MNS5 are required to generate the free α1,6-mannose. In the current ERAD model in plants, this glycan signal is recognized by OS9 (YOS9 in yeast; OS-9 and XTP3-B in mammals), a luminal ER-resident protein with a mannose 6-phosphate receptor homology domain (55). HRD3/SEL1L recognizes the unstructured polypeptide domain and together with OS9 brings non-native glycoproteins to the HRD1 complex (27).
Figure 3. N-glycan structures.
(A) The transferred N-glycan is composed of three glucose, nine mannose, and two GlcNAc residues. The three branches (A, B, and C) and the linkage between the individual sugar residues are indicated. (B) Schematic representation of the proposed prevailing N-glycan structure that is present on plant, yeast or mammalian glycoprotein ERAD substrates. The exposed free α1,6-mannose residue (red circle) serves as the common glycan determinant for degradation of misfolded glycoproteins.
A fundamental question in ERQC is the discrimination of immature folding intermediates that transiently display non-native characteristics from terminally misfolded proteins. Folding-defective glycoproteins are retrieved from the CNX/CRT cycle and directed to ERAD by a process that is currently not well understood. The evidence from mammalian cells and yeast strongly indicates that mannose trimming reactions mediated by members of the class I α-mannosidases family (EDEM1-3 in mammals, HTM1 in yeast, MNS4 and MNS5 in plants) are crucial for process and determine the exit from CNX/CRT (72; 111). The overexpression of EDEM1 accelerated the release of a misfolded glycoprotein from calnexin (94). Importantly, in this study the presence of increased EDEM1 levels was accompanied by reduced amounts of mono-glucosylated N-glycans on the misfolded glycoprotein, indicating that non-glucosylated glycoproteins are released from CNX and re-glucosylation by UGGT is abolished due to EDEM1 function. More recent studies in yeast revealed that a complex consisting of HTM1 and PDI discriminates between protein conformations and preferentially processes N-glycans on partially structured proteins to mark them for degradation (39; 82; 109). The HTM1-PDI complex, mammalian EDEMs and MNS4/MNS5 (with or without additional partners) could, therefore, represent a novel class of folding-status-sensing enzymes that extract terminally misfolded proteins from the CNX/CRT cycle and initiate ERAD.
Although the role of CNX/CRT and de/re-glucosylation cycle of glycoproteins appears comparable to that in mammalian cells, the structural analysis of N-glycans from glycan-dependent ERAD substrates indicates a clear difference compared to mammalian cells and yeast. Glc1Man9GlcNAc2 has been detected on misfolded glycoproteins, but no mono-glucosylated N-glycans with trimmed mannose residues (Glc1Man7GlcNAc2) have been reported (36; 50; 130). In plants, this particular mono-glucosylated N-glycan was detected on the misfolded ERAD substrates BRI1-5 (57) and SUBEX-C57Y (56), suggesting a different mechanism to terminate futile CNX/CRT cycles and the targeting of glycan-dependent ERAD substrates for subsequent clearance (Figure 3).
How are misfolded glycoproteins released from CNX/CRT? A specific trigger may liberate the ERAD substrate carrying the mono-glucosylated N-glycans derived from the CNX/CRT cycle and lead to the degradation of the glycoprotein substrate. For example, trimming of terminal mannose residues from the B- and/or C-branches may reduce the affinity of the CNX/CRT to the oligosaccharide and shift the binding equilibrium away from CNX/CRT (Figure 4) (94). In Schizosaccharomyces pombe, it has been shown that a reduced mannose content on the B- and C-branches decreases the de-glucosylation activity of GCSII by affecting mannose-binding of the GCSII beta-subunit (125). In vitro activity assays support these data (133), and the determination of the GCSII beta-subunit structure, revealed a shallow binding pocket that accommodates only a single mannose residue (103). In this model, the plant α-mannosidases MNS4 and MNS5 that act on the C-branch (57) are important players that regulate alone or in combination with an unknown factor the release from CNX/CRT and initiate ERAD. The conformational changes that promote the dissociation from CNX/CRT are unclear and in vitro binding experiments with an immobilized CNX domain do not indicate that carbohydrate binding of CNX is influenced by the presence or absence of the two outermost mannose residues on the B- and C-branches (136).
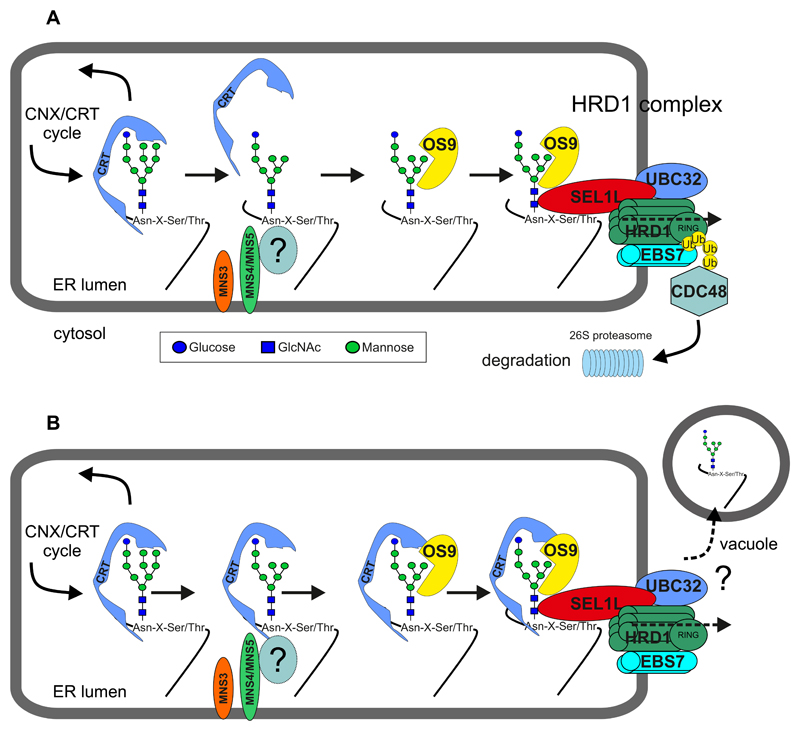
Figure 4. Proposed models for glycan-dependent ERAD via the HRD1 complex.
Terminally misfolded glycoproteins are extracted from the CNX/CRT cycle and sent for degradation by a poorly understood mechanism, which involves the detection of a non-native protein conformation and mannose trimming. The two ER-resident α-mannosidases MNS4 and MNS5 remove the terminal mannose residue from the C-branch and generate the free α1,6-mannose that is recognized by the lectin OS9 in combination with SEL1L. Additional luminal factors like BiP (not shown) may help to direct the misfolded protein to the HRD1 complex. The mannose on the B-branch is removed by MNS3. In contrast to mammals and yeast, this trimming reaction is dispensable for plant ERAD (57). A yet unknown factor (“?”) may act as a folding sensor and assist MNS4/MNS5 in the termination of futile folding attempts. (A) MNS4/MNS5 activity diverts misfolded proteins out of the CNX/CRT cycle leading to dissociation of the lectin. The disposal of misfolded glycoproteins requires the HRD1 complex. HRD1 is implicated in ERAD substrate retro-translocation to the cytosol (presumably by forming a channel) and polyubiquitination (Ub) mediated by the RING domain of HRD1. Extraction of the polyubiquitinated protein and subsequent degradation by the 26S proteasome involves the cytosolic ATPase CDC48 and accessory proteins (not shown). UBC32 associates with the HRD1 complex or is part of the plant DOA10 complex (not indicated here) and involved in the regulation of the ERAD component OS9. EBS7 is involved in the maintenance of HRD1 stability. (B) In the “persistent binding model”, de-mannosylation of the B- and C-branches attenuates GCSII-mediated de-glucosylation leading to enhanced CNX/CRT-binding. The CNX/CRT-bound glycoprotein is escorted to the HRD1 complex. The CNX/CRT-binding is either lost during translocation and degradation following the pathway as shown in (A) or the misfolded glycoprotein is directed to an alternative degradation route involving transport to the vacuole.
In an alternative model, there is persistent binding of CNX/CRT to the N-glycans on ERAD substrates, and the whole CNX/CRT-ERAD substrate complex is subsequently targeted in a MNS4/MNS5-, OS9 and HRD3/SEL1L-dependent way to the HRD1 complex for degradation. The persistent binding of CNX/CRT could ensure that the misfolded protein remains soluble in the ER and competent for ERAD (34) and/or may be required to concentrate the aberrant glycoprotein in a distinct subregion of the ER dedicated to ERAD as has been proposed for mammalian cells (8). In the prevailing models for the CNX/CRT-glycoprotein interaction in mammalian cells, the de-glucosylation on the A-branch of the N-glycan terminates the CNX/CRT association and dictates the fate (secretion of folded proteins and degradation of misfolded ones) of the glycoproteins (14; 23). Human ER-α-mannosidase I is more active on the A branch than the Arabidopsis couterpart MNS3 (4; 36; 77). In addition, a mammalian endomannosidase can irreversibly remove the Glcα1,3-Man disaccharide from the A branch of mono-glucosylated N-glycans to impede reglucosylation (150). Plants are devoid of endomannosidases or ER-resident enzymes that perform trimming of the A branch (77) and act as a “timer” to prevent the reglucosylation and subsequent re-association with CNX/CRT (43). Therefore, the presence of trimmed mono-glucosylated oligomannosidic N-glycans on plant glycoprotein ERAD substrates could be indicative of a plant-specific mechanism. However, a recent study showed that trapping of ERAD substrates in the mono-glucosylated state did not interfere with the substrate degradation in mammalian cells (132), suggesting that the plant-type of glycan-dependent ERAD may also function in mammals.
Glycan-dependent ERAD: the retro-translocation and subsequent disposal process
There has been controversy regarding the nature of the retro-translocation channel (7; 91; 115). Recently, a channel structure formed by the yeast HRD1 in complex with HRD3 was revealed, which may represent the long-sought retro-translocation channel of the ERAD substrates (119). The HRD1 structure is conserved in plants, suggesting a similar role in misfolded protein dislocation. The monitoring of a GFP variant fused to the P-region of CRT in Nicotiana benthamiana leaf epidermal cells over time revealed the accumulation of the ER-targeted protein in the cytoplasm (12), suggesting the presence of an active retro-translocation process in plants. Furthermore, biochemical studies following the degradation route of castor bean toxins ricin and Ricinus communis agglutinin revealed the accumulation and proteasomal degradation of these glycoproteins in the cytosol (29; 87). In sharp contrast to other glycoprotein ERAD substrates, their degradation was not blocked by specific class I α-mannosidase inhibitors, indicating targeting to a glycan-independent ERAD pathway involving retro-translocation. BRI1-9 is ubiquitinated and stabilized by the proteasome inhibitor MG132 (46; 81), suggesting that it is degraded by the classic proteasome-mediated pathway after retro-translocation from the ER into the cytosol. BRI1-5, in contrast, is degraded in a proteasome-independent manner, as observed for SUBEX-C57Y (46; 56). Moreover, no ubiquitination has been described for these two misfolded glycoproteins, indicating a different degradation pathway like transport to the lytic vacuole. Remarkably, BiP and other ER-resident proteins have been found in the vacuole (2; 110; 131). This trafficking route, which may bypass processing in the Golgi, may represent the normal pathway for the turn-over of ER-resident proteins. When misfolded proteins accumulate, the pathway might be used to deliver ERAD substrates for subsequent degradation in the vacuole. Alternatively, ERQC-autophagy-like processes may be involved. More detailed cell biological and biochemical studies are needed to understand these final steps of the ERQC pathway and the involved regulators.
Summary and Outlook
Due to the identification of key players of ERQC processes such as ERAD and identification of first defined ERAD substrates in plants (Table 1), it is now possible to move towards elucidation of the molecular mechanisms underlying these processes. Although, some basic concepts are emerging, our overall picture of CNX/CRT and UGGT function is still incomplete. We don’t know the structural basis of CNX/CRT-UGGT client recognition. We also don’t know which type of glycoproteins is subjected to repeated cycles of CNX/CRT interactions and whether a certain number of cycles or a particular time spent in the cycle is involved in permanent partitioning of the substrate to ERAD. In addition to the yet unclear CNX/CRT cycle termination event, the initial step of ERAD substrate selection is still elusive. In other words, the decisive step leading to the generation of the defined glycan signal for subsequent degradation is unknown. MNS4 and MNS5 are important players, but whether they can themselves sense misfolding or act in concert with another folding sensor remains to be demonstrated. The composition of the native HRD1 complex as well as other unknown E3 ubiquitin ligase complexes involved in ERAD must be identified and studied in plants. Furthermore, the final steps of ERAD including retro-translocation and proteasomal degradation are for most ERAD substrates unclear. Likewise, the molecular function of most chaperones involved in non-glycoprotein folding and maturation such as SHEPHERD has not been explored. Understanding the underlying molecular players of ERQC and the dynamic processes leading to protein homeostasis will be essential for future attempts to manipulate the stress responses of plants and other cellular pathways related to protein biogenesis in the ER.
Acknowledgements
Work on ERQC in the Strasser laboratory is supported by a grant from the Austrian Science Fund (FWF): P28218-B22.
References
- 1.Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 2013;1833:2430–7. doi: 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Andème Ondzighi C, Christopher D, Cho E, Chang S, Staehelin L. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell. 2008;20:2205–20. doi: 10.1105/tpc.108.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 4.Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer G. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell. 2008;19:216–25. doi: 10.1091/mbc.E07-05-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer J, Taylor I, Walker JC. Disrupting ER-associated protein degradation suppresses the abscission defect of a weak hae hsl2 mutant in Arabidopsis. J Exp Bot. 2016;67:5473–84. doi: 10.1093/jxb/erw313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353 doi: 10.1126/science.aac4354. aac4354. [DOI] [PubMed] [Google Scholar]
- 7.Baldridge RD, Rapoport TA. Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell. 2016;166:394–407. doi: 10.1016/j.cell.2016.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benyair R, Ogen-Shtern N, Mazkereth N, Shai B, Ehrlich M, Lederkremer GZ. Mammalian ER mannosidase I resides in quality control vesicles, where it encounters its glycoprotein substrates. Mol Biol Cell. 2015;26:172–84. doi: 10.1091/mbc.E14-06-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bies C, Blum R, Dudek J, Nastainczyk W, Oberhauser S, et al. Characterization of pancreatic ERj3p, a homolog of yeast DnaJ-like protein Scj1p. Biol Chem. 2004;385:389–95. doi: 10.1515/BC.2004.043. [DOI] [PubMed] [Google Scholar]
- 10.Blanco-Herrera F, Moreno AA, Tapia R, Reyes F, Araya M, et al. The UDP-glucose: glycoprotein glucosyltransferase (UGGT), a key enzyme in ER quality control, plays a significant role in plant growth as well as biotic and abiotic stress in Arabidopsis thaliana. BMC Plant Biol. 2015;15:127. doi: 10.1186/s12870-015-0525-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boisson M, Gomord V, Audran C, Berger N, Dubreucq B, et al. Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 2001;20:1010–9. doi: 10.1093/emboj/20.5.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandizzi F, Hanton S, DaSilva L, Boevink P, Evans D, et al. ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 2003;34:269–81. doi: 10.1046/j.1365-313x.2003.01728.x. [DOI] [PubMed] [Google Scholar]
- 13.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–51. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Caramelo J, Parodi A. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–5. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaiwanon J, Garcia VJ, Cartwright H, Sun Y, Wang ZY. Immunophilin-like FKBP42/TWISTED DWARF1 Interacts with the Receptor Kinase BRI1 to Regulate Brassinosteroid Signaling in Arabidopsis. Mol Plant. 2016;9:593–600. doi: 10.1016/j.molp.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, Liu R, Wang Q, Xie Q. ERAD Tuning of the HRD1 Complex Component AtOS9 Is Modulated by an ER-Bound E2, UBC32. Mol Plant. 2017;10:891–4. doi: 10.1016/j.molp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Chen Q, Zhong Y, Wu Y, Liu L, Wang P, et al. HRD1-mediated ERAD tuning of ER-bound E2 is conserved between plants and mammals. Nat Plants. 2016;2:16094. doi: 10.1038/nplants.2016.94. [DOI] [PubMed] [Google Scholar]
- 18.Chong LP, Wang Y, Gad N, Anderson N, Shah B, Zhao R. A highly charged region in the middle domain of plant endoplasmic reticulum (ER)-localized heat-shock protein 90 is required for resistance to tunicamycin or high calcium-induced ER stresses. J Exp Bot. 2015;66:113–24. doi: 10.1093/jxb/eru403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen A, Svensson K, Thelin L, Zhang W, Tintor N, et al. Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS One. 2010;5:e11342. doi: 10.1371/journal.pone.0011342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clerc S, Hirsch C, Oggier D, Deprez P, Jakob C, et al. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–72. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crofts AJ, Leborgne-Castel N, Pesca M, Vitale A, Denecke J. BiP and calreticulin form an abundant complex that is independent of endoplasmic reticulum stress. Plant Cell. 1998;10:813–24. doi: 10.1105/tpc.10.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui F, Liu L, Zhao Q, Zhang Z, Li Q, et al. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell. 2012;24:233–44. doi: 10.1105/tpc.111.093062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Alessio C, Dahms NM. Glucosidase II and MRH-domain containing proteins in the secretory pathway. Curr Protein Pept Sci. 2015;16:31–48. doi: 10.2174/1389203716666150213160438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Oliveira MVV, Xu G, Li B, de Souza Vespoli L, Meng X, et al. Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nature Plants. 2016:15218. doi: 10.1038/nplants.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dejgaard K, Theberge JF, Heath-Engel H, Chevet E, Tremblay ML, Thomas DY. Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J Proteome Res. 2010;9:1763–71. doi: 10.1021/pr900900x. [DOI] [PubMed] [Google Scholar]
- 26.Denecke J, Goldman M, Demolder J, Seurinck J, Botterman J. The tobacco luminal binding protein is encoded by a multigene family. Plant Cell. 1991;3:1025–35. doi: 10.1105/tpc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–59. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 28.Deprez P, Gautschi M, Helenius A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell. 2005;19:183–95. doi: 10.1016/j.molcel.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Di Cola A, Frigerio L, Lord J, Ceriotti A, Roberts L. Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc Natl Acad Sci U S A. 2001;98:14726–31. doi: 10.1073/pnas.251386098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doblas VG, Amorim-Silva V, Posé D, Rosado A, Esteban A, et al. The SUD1 gene encodes a putative E3 ubiquitin ligase and is a positive regulator of 3-hydroxy-3-methylglutaryl coenzyme a reductase activity in Arabidopsis. Plant Cell. 2013;25:728–43. doi: 10.1105/tpc.112.108696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunkley T, Hester S, Shadforth I, Runions J, Weimar T, et al. Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci U S A. 2006;103:6518–23. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farid A, Malinovsky FG, Veit C, Schoberer J, Zipfel C, Strasser R. Specialized roles of the conserved subunit OST3/6 of the oligosaccharyltransferase complex in innate immunity and tolerance to abiotic stresses. Plant Physiol. 2013;162:24–38. doi: 10.1104/pp.113.215509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farid A, Pabst M, Schoberer J, Altmann F, Glössl J, Strasser R. Arabidopsis thaliana alpha1,2-glucosyltransferase (ALG10) is required for efficient N-glycosylation and leaf growth. Plant J. 2011;68:314–25. doi: 10.1111/j.1365-313X.2011.04688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferris SP, Jaber NS, Molinari M, Arvan P, Kaufman RJ. UDP-glucose: glycoprotein glucosyltransferase (UGGT1) promotes substrate solubility in the endoplasmic reticulum. Mol Biol Cell. 2013;24:2597–608. doi: 10.1091/mbc.E13-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foresti O, Frigerio L, Holkeri H, de Virgilio M, Vavassori S, Vitale A. A phaseolin domain involved directly in trimer assembly is a determinant for binding by the chaperone BiP. Plant Cell. 2003;15:2464–75. doi: 10.1105/tpc.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frenkel Z, Gregory W, Kornfeld S, Lederkremer G. Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6-5GlcNAc2. J Biol Chem. 2003;278:34119–24. doi: 10.1074/jbc.M305929200. [DOI] [PubMed] [Google Scholar]
- 37.Fujimori T, Suno R, Iemura SI, Natsume T, Wada I, Hosokawa N. Endoplasmic reticulum proteins SDF2 and SDF2L1 act as components of the BiP chaperone cycle to prevent protein aggregation. Genes Cells. 2017;22:684–98. doi: 10.1111/gtc.12506. [DOI] [PubMed] [Google Scholar]
- 38.Gao M, Wang X, Wang D, Xu F, Ding X, et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe. 2009;6:34–44. doi: 10.1016/j.chom.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 39.Gauss R, Kanehara K, Carvalho P, Ng DT, Aebi M. A complex of pdi1p and the mannosidase htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol Cell. 2011;42:782–93. doi: 10.1016/j.molcel.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 40.Gillmor C, Poindexter P, Lorieau J, Palcic M, Somerville C. Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol. 2002;156:1003–13. doi: 10.1083/jcb.200111093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagiwara M, Ling J, Koenig PA, Ploegh HL. Posttranscriptional Regulation of Glycoprotein Quality Control in the Endoplasmic Reticulum Is Controlled by the E2 Ub-Conjugating Enzyme UBC6e. Mol Cell. 2016;63:753–67. doi: 10.1016/j.molcel.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson SR, Culyba EK, Hsu TL, Wong CH, Kelly JW, Powers ET. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci U S A. 2009;106:3131–6. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–65. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–49. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 45.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–60. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 46.Hong Z, Jin H, Fitchette A, Xia Y, Monk A, et al. Mutations of an alpha1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell. 2009;21:3792–802. doi: 10.1105/tpc.109.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–29. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong Z, Kajiura H, Su W, Jin H, Kimura A, et al. Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:11437–42. doi: 10.1073/pnas.1119173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horn SC, Hanna J, Hirsch C, Volkwein C, Schütz A, et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36:782–93. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 50.Hosokawa N, Tremblay LO, Sleno B, Kamiya Y, Wada I, et al. EDEM1 accelerates the trimming of alpha1,2-linked mannose on the C branch of N-glycans. Glycobiology. 2010;20:567–75. doi: 10.1093/glycob/cwq001. [DOI] [PubMed] [Google Scholar]
- 51.Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, et al. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell. 2014;54:166–79. doi: 10.1016/j.molcel.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howell SH. Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol. 2013;64:477–99. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- 53.Hwang J, Walczak CP, Shaler TA, Olzmann JA, Zhang L, et al. Characterization of protein complexes of the endoplasmic reticulum-associated degradation E3 ubiquitin ligase Hrd1. J Biol Chem. 2017;292:9104–16. doi: 10.1074/jbc.M117.785055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Häweker H, Rips S, Koiwa H, Salomon S, Saijo Y, et al. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem. 2010;285:4629–36. doi: 10.1074/jbc.M109.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hüttner S, Veit C, Schoberer J, Grass J, Strasser R. Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins. Plant Mol Biol. 2012;79:21–33. doi: 10.1007/s11103-012-9891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hüttner S, Veit C, Vavra U, Schoberer J, Dicker M, et al. A context-independent N-glycan signal targets the misfolded extracellular domain of Arabidopsis STRUBBELIG to endoplasmic-reticulum-associated degradation. Biochem J. 2014;464:401–11. doi: 10.1042/BJ20141057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hüttner S, Veit C, Vavra U, Schoberer J, Liebminger E, et al. Arabidopsis Class I α-Mannosidases MNS4 and MNS5 Are Involved in Endoplasmic Reticulum-Associated Degradation of Misfolded Glycoproteins. Plant Cell. 2014;26:1712–28. doi: 10.1105/tpc.114.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishiguro S, Watanabe Y, Ito N, Nonaka H, Takeda N, et al. SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 2002;21:898–908. doi: 10.1093/emboj/21.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jakob C, Burda P, Roth J, Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142:1223–33. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin H, Hong Z, Su W, Li J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2009;106:13612–7. doi: 10.1073/pnas.0906144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin H, Yan Z, Nam K, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–30. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin Y, Zhuang M, Hendershot LM. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry. 2009;48:41–9. doi: 10.1021/bi8015923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamauchi S, Nakatani H, Nakano C, Urade R. Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 2005;272:3461–76. doi: 10.1111/j.1742-4658.2005.04770.x. [DOI] [PubMed] [Google Scholar]
- 64.Kelleher D, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 65.Kim JH, Nguyen NH, Nguyen NT, Hong SW, Lee H. Loss of all three calreticulins, CRT1, CRT2 and CRT3, causes enhanced sensitivity to water stress in Arabidopsis. Plant Cell Rep. 2013;32:1843–53. doi: 10.1007/s00299-013-1497-z. [DOI] [PubMed] [Google Scholar]
- 66.Kirst ME, Meyer DJ, Gibbon BC, Jung R, Boston RS. Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol. 2005;138:218–31. doi: 10.1104/pp.105.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein EM, Mascheroni L, Pompa A, Ragni L, Weimar T, et al. Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J. 2006;48:657–73. doi: 10.1111/j.1365-313X.2006.02904.x. [DOI] [PubMed] [Google Scholar]
- 68.Koiwa H, Li F, McCully M, Mendoza I, Koizumi N, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15:2273–84. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kozlov G, Pocanschi CL, Rosenauer A, Bastos-Aristizabal S, Gorelik A, et al. Structural basis of carbohydrate recognition by calreticulin. J Biol Chem. 2010;285:38612–20. doi: 10.1074/jbc.M110.168294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kreibich G, Ulrich BL, Sabatini DD. Proteins of rough microsomal membranes related to ribosome binding. I. Identification of ribophorins I and II, membrane proteins characteristics of rough microsomes. J Cell Biol. 1978;77:464–87. doi: 10.1083/jcb.77.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumari S, Roy S, Singh P, Singla-Pareek SL, Pareek A. Cyclophilins: proteins in search of function. Plant Signal Behav. 2013;8:e22734. doi: 10.4161/psb.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lamriben L, Graham JB, Adams BM, Hebert DN. N-Glycan-based ER Molecular Chaperone and Protein Quality Control System: The Calnexin Binding Cycle. Traffic. 2016;17:308–26. doi: 10.1111/tra.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lerouxel O, Mouille G, Andème-Onzighi C, Bruyant M, Séveno M, et al. Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 2005;42:455–68. doi: 10.1111/j.1365-313X.2005.02392.x. [DOI] [PubMed] [Google Scholar]
- 74.Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci U S A. 2009;106:15973–8. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li LM, Lü SY, Li RJ. The Arabidopsis endoplasmic reticulum associated degradation pathways are involved in the regulation of heat stress response. Biochem Biophys Res Commun. 2017;487:362–7. doi: 10.1016/j.bbrc.2017.04.066. [DOI] [PubMed] [Google Scholar]
- 76.Li Q, Wei H, Liu L, Yang X, Zhang X, Xie Q. Unfolded protein response activation compensates endoplasmic reticulum-associated degradation deficiency in Arabidopsis. J Integr Plant Biol. 2017;59:506–21. doi: 10.1111/jipb.12544. [DOI] [PubMed] [Google Scholar]
- 77.Liebminger E, Hüttner S, Vavra U, Fischl R, Schoberer J, et al. Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell. 2009;21:3850–67. doi: 10.1105/tpc.109.072363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu L, Cui F, Li Q, Yin B, Zhang H, et al. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 2011;21:957–69. doi: 10.1038/cr.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012;24:4635–51. doi: 10.1105/tpc.112.101535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y, Li J. A conserved basic residue cluster is essential for the protein quality control function of the Arabidopsis calreticulin 3. Plant Signal Behav. 2013;8:e23864. doi: 10.4161/psb.23864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, Zhang C, Wang D, Su W, Liu L, et al. EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc Natl Acad Sci U S A. 2015;112:12205–10. doi: 10.1073/pnas.1511724112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu YC, Fujimori DG, Weissman JS. Htm1p-Pdi1p is a folding-sensitive mannosidase that marks N-glycoproteins for ER-associated protein degradation. Proc Natl Acad Sci U S A. 2016;113:E4015–24. doi: 10.1073/pnas.1608795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loibl M, Wunderle L, Hutzler J, Schulz BL, Aebi M, Strahl S. Protein O-mannosyltransferases associate with the translocon to modify translocating polypeptide chains. J Biol Chem. 2014;289:8599–611. doi: 10.1074/jbc.M113.543116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luan S, Kudla J, Gruissem W, Schreiber SL. Molecular characterization of a FKBP-type immunophilin from higher plants. Proc Natl Acad Sci U S A. 1996;93:6964–9. doi: 10.1073/pnas.93.14.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lü S, Zhao H, Des Marais DL, Parsons EP, Wen X, et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012;159:930–44. doi: 10.1104/pp.112.198697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma ZX, Leng YJ, Chen GX, Zhou PM, Ye D, Chen LQ. The THERMOSENSITIVE MALE STERILE 1 Interacts with the BiPs via DnaJ Domain and Stimulates Their ATPase Enzyme Activities in Arabidopsis. PLoS One. 2015;10:e0132500. doi: 10.1371/journal.pone.0132500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marshall RS, Jolliffe NA, Ceriotti A, Snowden CJ, Lord JM, et al. The role of CDC48 in the retro-translocation of non-ubiquitinated toxin substrates in plant cells. J Biol Chem. 2008;283:15869–77. doi: 10.1074/jbc.M709316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martínez I, Chrispeels M. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15:561–76. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maruyama D, Endo T, Nishikawa S. BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:1684–9. doi: 10.1073/pnas.0905795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maruyama D, Sugiyama T, Endo T, Nishikawa S. Multiple BiP genes of Arabidopsis thaliana are required for male gametogenesis and pollen competitiveness. Plant Cell Physiol. 2014;55:801–10. doi: 10.1093/pcp/pcu018. [DOI] [PubMed] [Google Scholar]
- 91.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16:77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 92.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–69. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mohorko E, Glockshuber R, Aebi M. Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J Inherit Metab Dis. 2011;34:869–78. doi: 10.1007/s10545-011-9337-1. [DOI] [PubMed] [Google Scholar]
- 94.Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- 95.Müller J, Piffanelli P, Devoto A, Miklis M, Elliott C, et al. Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell. 2005;17:149–63. doi: 10.1105/tpc.104.026625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Müller LM, Lindner H, Pires ND, Gagliardini V, Grossniklaus U. A subunit of the oligosaccharyltransferase complex is required for interspecific gametophyte recognition in Arabidopsis. Nat Commun. 2016;7 doi: 10.1038/ncomms10826. 10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nekrasov V, Li J, Batoux M, Roux M, Chu Z, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–38. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Neubert P, Strahl S. Protein O-mannosylation in the early secretory pathway. Curr Opin Cell Biol. 2016;41:100–8. doi: 10.1016/j.ceb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 99.Niemann MC, Bartrina I, Ashikov A, Weber H, Novák O, et al. Arabidopsis ROCK1 transports UDP-GlcNAc/UDP-GalNAc and regulates ER protein quality control and cytokinin activity. Proc Natl Acad Sci U S A. 2015;112:291–6. doi: 10.1073/pnas.1419050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ninagawa S, Okada T, Sumitomo Y, Kamiya Y, Kato K, et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J Cell Biol. 2014;206:347–56. doi: 10.1083/jcb.201404075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Noh S, Kwon C, Oh D, Moon J, Chung W. Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene. 2003;311:81–91. doi: 10.1016/s0378-1119(03)00559-6. [DOI] [PubMed] [Google Scholar]
- 102.Ohta M, Takaiwa F. Emerging features of ER resident J-proteins in plants. Plant Signal Behav. 2014;9:e28194. doi: 10.4161/psb.28194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olson LJ, Orsi R, Alculumbre SG, Peterson FC, Stigliano ID, et al. Structure of the lectin mannose 6-phosphate receptor homology (MRH) domain of glucosidase II, an enzyme that regulates glycoprotein folding quality control in the endoplasmic reticulum. J Biol Chem. 2013;288:16460–75. doi: 10.1074/jbc.M113.450239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park CJ, Bart R, Chern M, Canlas PE, Bai W, Ronald PC. Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One. 2010;5:e9262. doi: 10.1371/journal.pone.0009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park CJ, Sharma R, Lefebvre B, Canlas PE, Ronald PC. The endoplasmic reticulum-quality control component SDF2 is essential for XA21-mediated immunity in rice. Plant Sci. 2013;210:53–60. doi: 10.1016/j.plantsci.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 106.Pearse BR, Gabriel L, Wang N, Hebert DN. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 2008;181:309–20. doi: 10.1083/jcb.200712068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Persson S, Rosenquist M, Svensson K, Galvão R, Boss WF, Sommarin M. Phylogenetic analyses and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 2003;133:1385–96. doi: 10.1104/pp.103.024943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, et al. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat Commun. 2014;5 doi: 10.1038/ncomms4072. 3072. [DOI] [PubMed] [Google Scholar]
- 109.Pfeiffer A, Stephanowitz H, Krause E, Volkwein C, Hirsch C, et al. A Complex of Htm1 and the Oxidoreductase Pdi1 Accelerates Degradation of Misfolded Glycoproteins. J Biol Chem. 2016;291:12195–207. doi: 10.1074/jbc.M115.703256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pimpl P, Taylor J, Snowden C, Hillmer S, Robinson D, Denecke J. Golgi-mediated vacuolar sorting of the endoplasmic reticulum chaperone BiP may play an active role in quality control within the secretory pathway. Plant Cell. 2006;18:198–211. doi: 10.1105/tpc.105.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pisoni GB, Molinari M. Five Questions (with their Answers) on ER-Associated Degradation. Traffic. 2016;17:341–50. doi: 10.1111/tra.12373. [DOI] [PubMed] [Google Scholar]
- 112.Pollier J, Moses T, González-Guzmán M, De Geyter N, Lippens S, et al. The protein quality control system manages plant defence compound synthesis. Nature. 2013;504:148–52. doi: 10.1038/nature12685. [DOI] [PubMed] [Google Scholar]
- 113.Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 2005;24:1730–8. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–79. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Römisch K. A Case for Sec61 Channel Involvement in ERAD. Trends Biochem Sci. 2017;42:171–9. doi: 10.1016/j.tibs.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 116.Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–49. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schmitz A, Herzog V. Endoplasmic reticulum-associated degradation: exceptions to the rule. Eur J Cell Biol. 2004;83:501–9. doi: 10.1078/0171-9335-00412. [DOI] [PubMed] [Google Scholar]
- 118.Schoberer J, Botchway SW. Investigating protein-protein interactions in the plant endomembrane system using multiphoton-induced FRET-FLIM. Methods Mol Biol. 2014;1209:81–95. doi: 10.1007/978-1-4939-1420-3_6. [DOI] [PubMed] [Google Scholar]
- 119.Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature. 2017 doi: 10.1038/nature23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schott A, Ravaud S, Keller S, Radzimanowski J, Viotti C, et al. Arabidopsis stromal-derived Factor2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum. J Biol Chem. 2010;285:18113–21. doi: 10.1074/jbc.M110.117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schulz BL, Stirnimann CU, Grimshaw JP, Brozzo MS, Fritsch F, et al. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci U S A. 2009;106:11061–6. doi: 10.1073/pnas.0812515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci U S A. 2008;105:8256–61. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Soussilane P, Soussillane P, D'Alessio C, Paccalet T, Fitchette A, et al. N-glycan trimming by glucosidase II is essential for Arabidopsis development. Glycoconj J. 2009;26:597–607. doi: 10.1007/s10719-008-9201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Srivastava R, Deng Y, Shah S, Rao AG, Howell SH. BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell. 2013;25:1416–29. doi: 10.1105/tpc.113.110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stigliano ID, Alculumbre SG, Labriola CA, Parodi AJ, D'Alessio C. Glucosidase II and N-glycan mannose content regulate the half-lives of monoglucosylated species in vivo. Mol Biol Cell. 2011;22:1810–23. doi: 10.1091/mbc.E11-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Strasser R. Plant protein glycosylation. Glycobiology. 2016;26:926–39. doi: 10.1093/glycob/cww023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Su W, Liu Y, Xia Y, Hong Z, Li J. Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:870–5. doi: 10.1073/pnas.1013251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Su W, Liu Y, Xia Y, Hong Z, Li J. The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol Plant. 2012;5:929–40. doi: 10.1093/mp/sss042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun T, Zhang Q, Gao M, Zhang Y. Regulation of SOBIR1 accumulation and activation of defense responses in bir1-1 by specific components of ER quality control. Plant J. 2014;77:748–56. doi: 10.1111/tpj.12425. [DOI] [PubMed] [Google Scholar]
- 130.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–75. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 131.Tamura K, Yamada K, Shimada T, Hara-Nishimura I. Endoplasmic reticulum-resident proteins are constitutively transported to vacuoles for degradation. Plant J. 2004;39:393–402. doi: 10.1111/j.1365-313X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- 132.Tannous A, Patel N, Tamura T, Hebert DN. Reglucosylation by UDP-glucose: glycoprotein glucosyltransferase 1 delays glycoprotein secretion but not degradation. Mol Biol Cell. 2015;26:390–405. doi: 10.1091/mbc.E14-08-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Totani K, Ihara Y, Matsuo I, Ito Y. Substrate specificity analysis of endoplasmic reticulum glucosidase II using synthetic high mannose-type glycans. J Biol Chem. 2006;281:31502–8. doi: 10.1074/jbc.M605457200. [DOI] [PubMed] [Google Scholar]
- 134.Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–72. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 135.Van Hoewyk D. Defects in endoplasmic reticulum-associated degradation (ERAD) increase selenate sensitivity in Arabidopsis. Plant Signal Behav. 2016 doi: 10.1080/15592324.2016.1171451. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vassilakos A, Michalak M, Lehrman MA, Williams DB. Oligosaccharide binding characteristics of the molecular chaperones calnexin and calreticulin. Biochemistry. 1998;37:3480–90. doi: 10.1021/bi972465g. [DOI] [PubMed] [Google Scholar]
- 137.Vasudevan D, Haltiwanger RS. Novel roles for O-linked glycans in protein folding. Glycoconj J. 2014;31:417–26. doi: 10.1007/s10719-014-9556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vembar S, Brodsky J. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–57. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vu KV, Nguyen NT, Jeong CY, Lee YH, Lee H, Hong SW. Systematic deletion of the ER lectin chaperone genes reveals their roles in vegetative growth and male gametophyte development in Arabidopsis. Plant J. 2017;89:972–83. doi: 10.1111/tpj.13435. [DOI] [PubMed] [Google Scholar]
- 140.Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–40. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 141.Wu G, Otegui MS, Spalding EP. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell. 2010;22:3295–304. doi: 10.1105/tpc.110.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu C, Ng DT. O-mannosylation: The other glycan player of ER quality control. Semin Cell Dev Biol. 2015;41:129–34. doi: 10.1016/j.semcdb.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 143.Xu C, Wang S, Thibault G, Ng DT. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science. 2013;340:978–81. doi: 10.1126/science.1234055. [DOI] [PubMed] [Google Scholar]
- 144.Yamamoto M, Kawanabe M, Hayashi Y, Endo T, Nishikawa S. A vacuolar carboxypeptidase mutant of Arabidopsis thaliana is degraded by the ERAD pathway independently of its N-glycan. Biochem Biophys Res Commun. 2010;393:384–9. doi: 10.1016/j.bbrc.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 145.Yamamoto M, Maruyama D, Endo T, Nishikawa S. Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol. 2008;49:1547–62. doi: 10.1093/pcp/pcn119. [DOI] [PubMed] [Google Scholar]
- 146.Yang X, Srivastava R, Howell SH, Bassham DC. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 2016;85:83–95. doi: 10.1111/tpj.13091. [DOI] [PubMed] [Google Scholar]
- 147.Yuen CY, Wong K, Christopher DA. Phylogenetic characterization and promoter expression analysis of a novel hybrid protein disulfide isomerase/cargo receptor subfamily unique to plants and chromalveolates. Mol Genet Genomics. 2016;291:455–69. doi: 10.1007/s00438-015-1106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yuen CYL, Wang P, Kang BH, Matsumoto K, Christopher DA. A Non-Classical Member of the Protein Disulfide Isomerase Family, PDI7 of Arabidopsis thaliana, Localizes to the cis-Golgi and Endoplasmic Reticulum Membranes. Plant Cell Physiol. 2017;58:1103–17. doi: 10.1093/pcp/pcx057. [DOI] [PubMed] [Google Scholar]
- 149.Zhao B, Lv M, Feng Z, Campbell T, Liscum E, Li J. TWISTED DWARF 1 Associates with BRASSINOSTEROID-INSENSITIVE 1 to Regulate Early Events of the Brassinosteroid Signaling Pathway. Mol Plant. 2016;9:582–92. doi: 10.1016/j.molp.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 150.Zuber C, Spiro MJ, Guhl B, Spiro RG, Roth J. Golgi apparatus immunolocalization of endomannosidase suggests post-endoplasmic reticulum glucose trimming: implications for quality control. Mol Biol Cell. 2000;11:4227–40. doi: 10.1091/mbc.11.12.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]