Abstract
Nature uses 64 codons to encode the synthesis of proteins from the genome, and chooses 1 sense codon—out of up to 6 synonyms—to encode each amino acid. Synonymous codon choice has diverse and important roles, and many synonymous substitutions are detrimental. Here we demonstrate that the number of codons used to encode the canonical amino acids can be reduced, through the genome-wide substitution of target codons by defined synonyms. We create a variant of Escherichia coli with a four-megabase synthetic genome through a high-fidelity convergent total synthesis. Our synthetic genome implements a defined recoding and refactoring scheme—with simple corrections at just seven positions—to replace every known occurrence of two sense codons and a stop codon in the genome. Thus, we recode 18,214 codons to create an organism with a 61-codon genome; this organism uses 59 codons to encode the 20 amino acids, and enables the deletion of a previously essential transfer RNA.
Nature uses 64 triplet codons to encode the synthesis of proteins that are composed of the canonical 20 amino acids; 18 of these amino acids are encoded by more than 1 synonymous codon1. Synonymous codon choice can influence mRNA folding2, gene expression3–6, co-translational folding and protein levels2,7,8, and has emerging roles9,10. In addition, synonymous codons may have different roles at different positions in the genome11.
Reducing the number of sense codons used to encode the canonical amino acids—through genome-wide replacement of a target codon with synonymous codons (which we term synonymous codon compression)—would address whether all synonymous codons are necessary, and may also provide a foundation for the in vivo biosynthesis of genetically encoded non-canonical biopolymers12.
Up to 321 amber stop codons have been removed from the E. coli genome, using site-directed mutagenesis approaches that commonly introduce large numbers of off-target mutations13–15. Sense codons are commonly more abundant than stop codons by several orders of magnitude, and—in principle—high-fidelity genome synthesis would be the preferred route for tackling their removal. Efforts to alter synonymous codons in individual genes16, genomic regions and essential operons9,16–21 have provided insight into synonymous codon choice, and a subset of these studies have attempted to alter synonymous codons in ways that are consistent with synonymous codon compression17–19,21. However, these previous studies have mutated only a small fraction of targeted sense codons in the genome of a single strain.
There are an extremely large number of theoretical genomes that are formally compatible with synonymous codon compression (np, in which n is the number of synonyms for a target codon (n = 2–6) and P is the number of target-codon positions (P = 103 to 105)), and it is not possible to experimentally test the viability of np genomes. Defined synonymous codons have previously been used17 to replace the target codons in a 20-kb region of the E. coli genome that is rich in both essential genes and target codons; these studies identified simple defined ‘recoding schemes’ that permit synonymous codon compression in this region17. However, it remained unclear whether these schemes could be applied for genome-wide recoding.
DNA synthesis and assembly methods enabled the creation of a Mycoplasma mycoides with a 1.08-Mb synthetic genome22,23, and the creation of 9 strains of Saccharomyces cerevisiae in which 1 or 2 of the 16 chromosomes is replaced by synthetic DNA24–31 (up to 0.99 Mb, 8% of the yeast genome). Replicon excision for enhanced genome engineering through programmed recombination (REXER)—an approach for replacing more than 100 kb of the E. coli genome with synthetic DNA in a single step—has recently been reported17, and it has been demonstrated that REXER can be iterated via genome stepwise interchange synthesis (GENESIS)17. Here we implement a convergent total synthesis to replace the 4-Mb E. coli MDS42 (ref. 32) genome with a synthetic genome. The synthetic genome is refactored33 and recoded for the genome-wide removal of two sense codons and a stop codon, which creates a synthetic E. coli that uses 61 codons for protein synthesis.
Design of a recoded genome
We designed a genome in which the serine codons TCG and TCA, and the stop codon TAG, in open reading frames (ORFs) of MDS42 E. coli (Supplementary Data 1) are systematically replaced by their synonyms AGC, AGT and TAA, respectively (Fig. 1a, Supplementary Data 2, 3). It has previously been shown that this defined recoding scheme is allowed in a 20-kb region of the genome17.
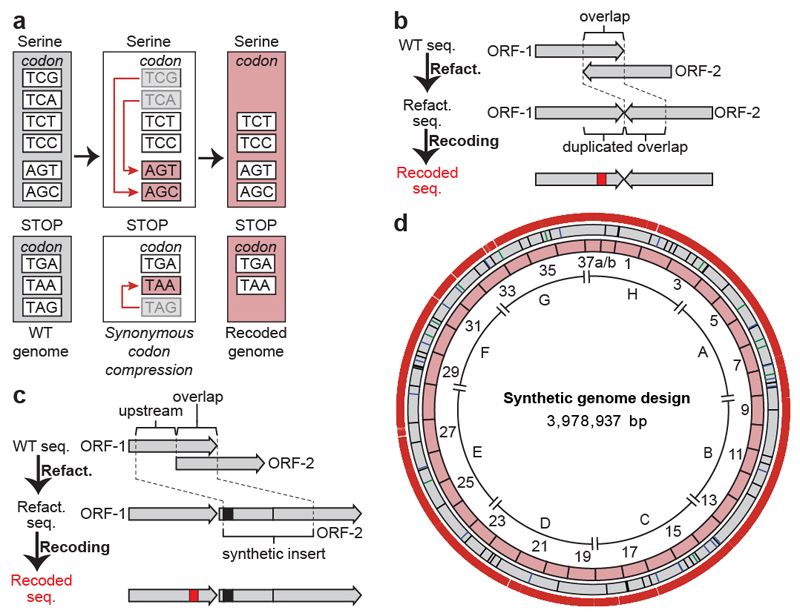
Fig. 1. Design of the synthetic genome, implementing a defined recoding scheme for synonymous codon compression.
a, The defined recoding scheme for synonymous codon compression. Synonymous serine codons and three stop codons used in the genome of wild-type E. coli are shown (grey boxes). Systematically implementing a defined recoding scheme for synonymous codon compression (red arrows) recodes target codons to defined synonyms, and replaces the amber stop codon TAG with the ochre stop codon TAA. This creates an organism with a recoded genome that uses a reduced number of serine and termination codons (pink boxes).
b, Refactoring of 3′, 3′ overlaps enables their independent recoding. The overlap between two ORFs (ORF1 and ORF2) is duplicated, which enables independent recoding (red box) of these ORFs.
c, Refactoring 5′, 3′ overlaps. The overlap plus 20 bp upstream is duplicated to generate a synthetic insert. When the overlap is longer than 1 bp at the end of the upstream ORF, an in-frame TAA (black box) is introduced in the beginning of the synthetic insert; this in-frame stop codon ensures the termination of translation from the original ribosome-binding site. Thus, all full-length translation of the downstream ORF is initiated from the reconstructed ribosome-binding site in the synthetic insert. This refactoring enables the independent recoding (red box) of ORFs.
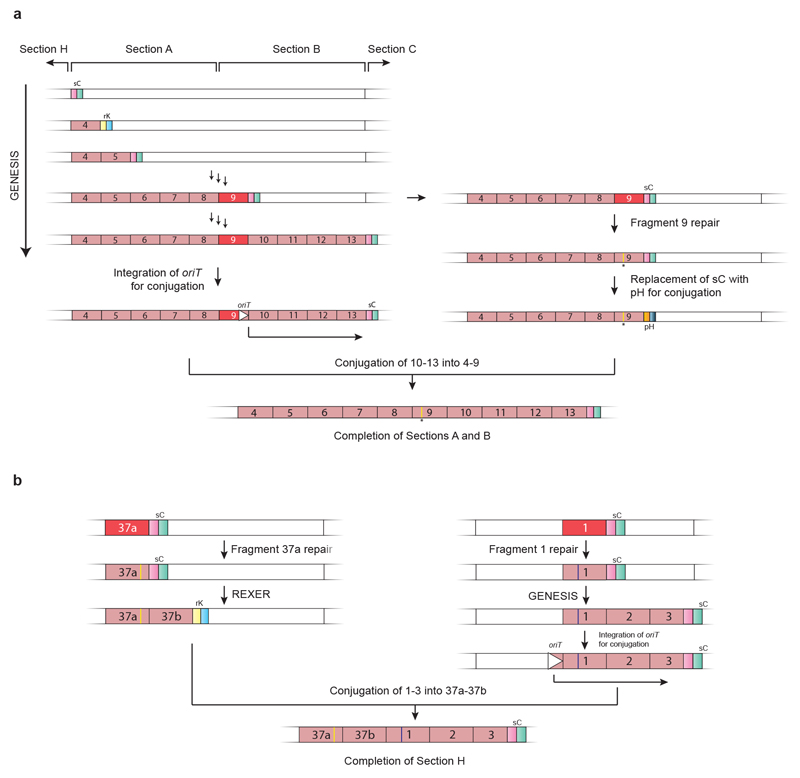
d, Map of the synthetic genome design with all TCG, TCA and TAG codons removed. Outer ring shows positions (18,218 red bars) of all TCG to AGC, TCA to AGT and TAG to TAA recodings. Grey ring shows positions of designed silent mutations in overlaps (12 green bars), refactoring of 3′, 3′ overlaps (schematic shown in b, 21 blue bars) and refactoring of 5′, 3′ overlapping regions (schematic shown in c, 58 black bars). Pink ring shows 37 fragments of approximately 100 kb in size each. Fragment 37 is shown as 37a and 37b to reflect the final assembly. The sections A to H are indicated.
Many target codons are found in areas of overlap between ORFs. We classified these overlaps as 3′, 3′ (between ORFs in opposite orientations) or 5′, 3′ (between ORFs in the same orientation). When the recoding of a 3′, 3′ overlap could be achieved without changing the encoded protein sequences, the structure of the overlap was maintained and the sequences were directly recoded. Otherwise, we duplicated the overlap and individually recoded each ORF (Fig. 1b, Supplementary Data 4). For 5′, 3′ overlaps, we separated the ORFs by duplicating both the overlap between the ORFs and the 20-bp sequence upstream of the overlap, which enabled independent recoding of each ORF (Fig. 1c, Supplementary Data 4). Using the defined rules for synonymous codon compression and refactoring, we designed a genome in which all 18,218 target codons are recoded to their target synonyms (Fig. 1d, Supplementary Data 3).
Synthesis of recoded sections
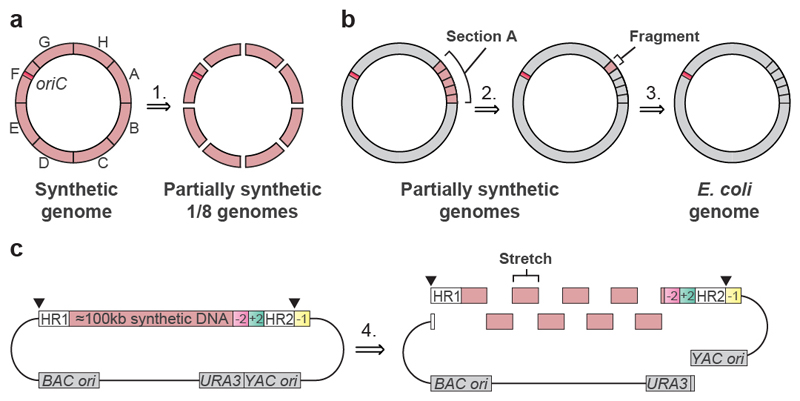
We performed a retrosynthesis—analogous to that commonly used for designing synthetic routes in chemistry 34—on the designed genome (Fig. 2). We disconnected the genome into eight sections, each of approximately 0.5 Mb in length, which were labelled A to H (Figs. 1d, 2a, Supplementary Data 2); we then disconnected each section into 4 or 5 fragments (Fig. 2b). This yielded 37 fragments (Fig. 1d, Supplementary Data 2) that were between 91 kb and 136 kb in length. We placed the boundaries between fragments or sections in intergenic regions that are between non-essential genes. The fragments were further disconnected into 9–14 stretches that were approximately 10 kb in length (Fig. 2c, Supplementary Data 5).
Fig. 2. Retrosynthesis of the synthetic genome.
a, Disconnecting the genome into eight sections. The synthetic genome was disconnected into sections A to H, with each section corresponding to approximately 0.5 Mb (step 1). The position of the replication origin oriC (orange square) is indicated. Sections were assembled into a completely recoded genome (in the forward sense, opposite to the direction of the retrosynthesis arrow) by directed conjugation (Fig. 3, Extended Data Fig. 7).
b, Disconnecting genome sections into 100-kb fragments. Sections are further disconnected into 4 or 5 fragments of around 100 kb in length each. Section A is depicted, and other sections were treated similarly. Nearly all sections were constructed entirely through consecutive REXER steps, by GENESIS (Extended Data Fig. 1). Each step replaced around 100 kb of wild-type genomic sequence with 100 kb of synthetic fragment (steps 2 and 3). c, Disconnecting each 100-kb synthetic fragment into 10-kb synthetic stretches. Each 100-kb synthetic fragment is further disconnected into 9 to 14 short synthetic stretches of around 10 kb in length (step 4). The BACs that carry 100-kb synthetic fragments (pink) were assembled by homologous recombination in yeast. Each BAC contains Cas9 cleavage sites (black triangles) that enable excision of the synthetic DNA in vivo, homology regions 1 and 2 (HR1 and HR2) for targeting recombination, and the appropriate double-selection cassette. The −2 (sucrose sensitivity, encoded by sacB),+2 (chloramphenicol resistance, encoded by cat) double selection cassette is indicated. However different double selection cassettes are used for selection in different steps of REXER. A negative-selection marker (rpsL; indicated as −1) is used to enable loss of the backbone after REXER. BAC and yeast artificial chromosome (YAC) origins and a URA3 marker, all for maintenance in E. coli and S. cerevisiae, are indicated.
We assembled bacterial artificial chromosomes (BACs) for REXER (Fig. 2c, Supplementary Data 6–9) that contained each fragment, using homologous recombination in S. cerevisiae17,35. For 36 of the fragments, BAC assembly proceeded smoothly (Supplementary Data 10). Fragment 37 was challenging to assemble and we therefore split it into two 50-kb fragments (labelled 37a and 37b), which were straightforward to assemble (Supplementary Data 10).
We initiated genome replacement in seven distinct strains using REXER (Extended Data Fig. 1a). The start point for REXER in each strain corresponds to the beginning of sections A, C, D, E, F, G or H (Figs. 1d, 2a); section B was subsequently built on section A. In each strain, the positive and negative selection markers that are introduced in the first REXER provide a template for the next round of REXER, which enables GENESIS17 (Fig. 2b, Extended Data Fig. 1b). We found that REXER could be initiated by the electroporation of linear double-stranded spacers generated by PCR (Supplementary Data 11–13) rather than plasmid-encoded spacers17, which accelerated GENESIS. For sections A, C, D, E, F and G, we proceeded with GENESIS in a clockwise direction for 4 or 5 steps of REXER and replaced approximately 0.5 Mb of genomic DNA with synthetic DNA. We sequenced the genomes of cells after each step of REXER and identified clones that were fully recoded over the targeted genomic region (Supplementary Data 11). Section A was completed first, and we therefore proceeded with GENESIS through section B in a strain that contained recoded section A.
We carried out numerous single-step REXERs with individual fragments (Supplementary Data 11), in parallel with GENESIS, to accelerate the identification of genomic regions that may be challenging to recode. For 35 steps, including all of sections A, C, D, E, F and G, we completely recoded the targeted genomic sequence by GENESIS. However, we observed incomplete replacement of the corresponding genomic region by synthetic DNA for fragment 9 (in section B), and for fragments 37a and 1 (in section H) (Supplementary Data 11).
Identifying and repairing design flaws
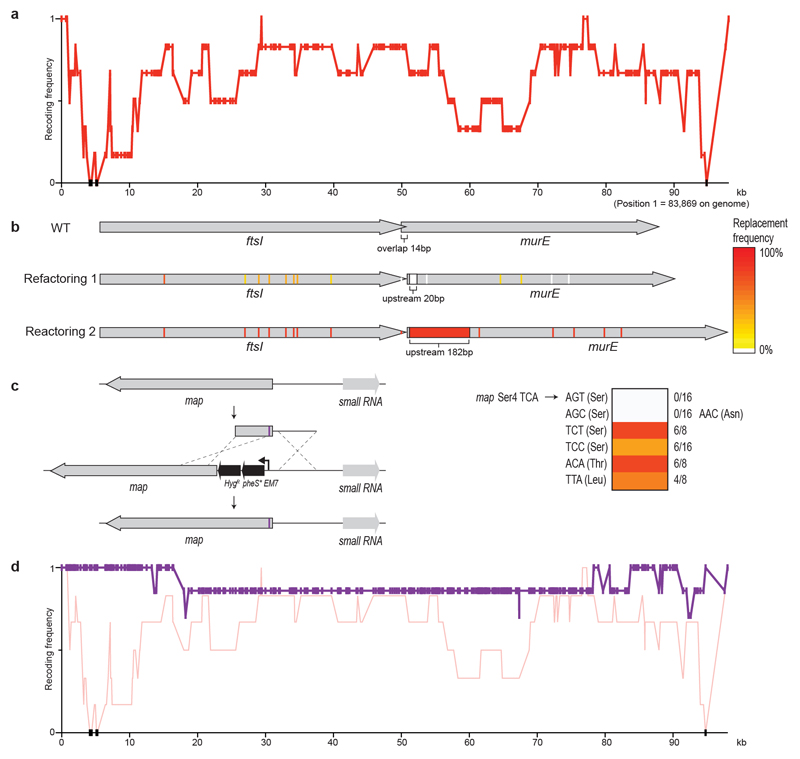
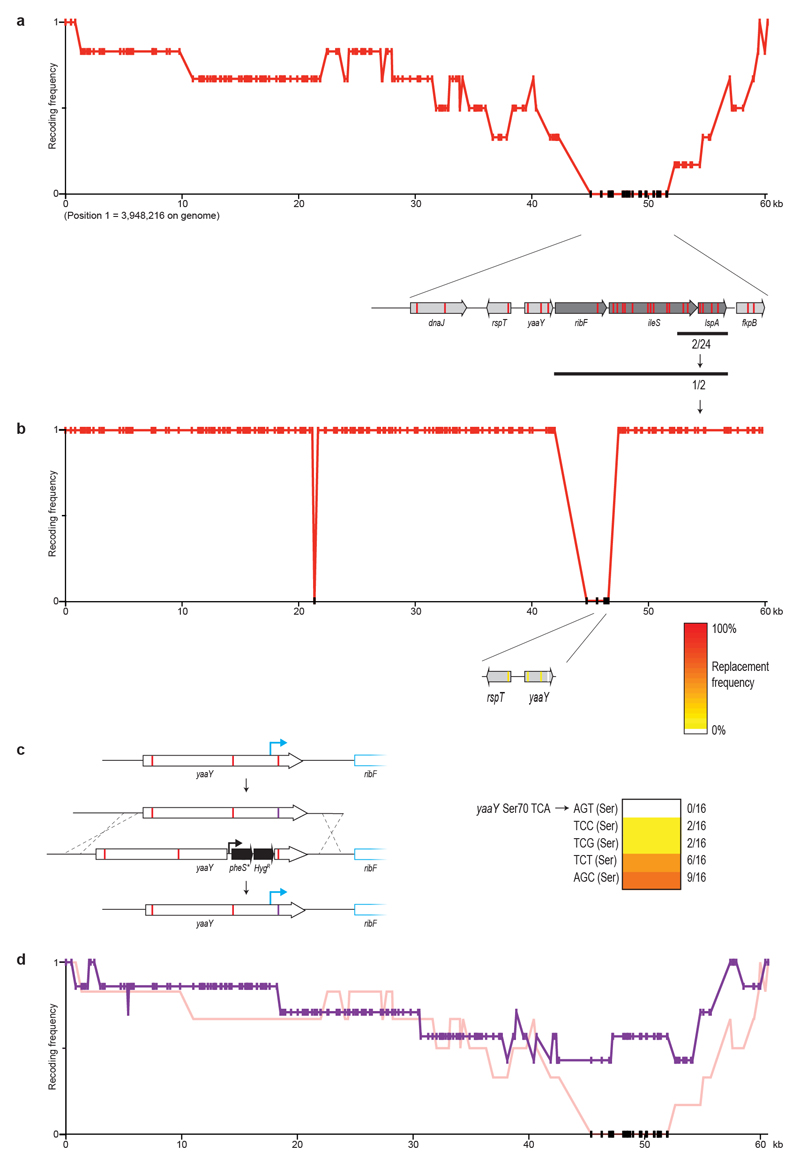
Sequencing several clones following REXER enabled us to score the frequency with which each target codon is recoded, and thereby to compile a recoding landscape for the genomic region 17. From the recoding landscape with fragment 1, we identified the fourth codon (TCA, Ser4) in map, which is an essential gene that encodes methionine amino peptidase, as recalcitrant to recoding by our defined scheme (Extended Data Fig. 2). We also identified a second region—which encompasses a 14-bp overlap of the essential genes ftsI and murE, and several serine codons in ftsI and murE—that was not replaced by our recoded and refactored sequence. As this region has previously been recoded with the same recoding scheme, duplicating the overlap plus 182 bp rather than the 20 bp used in our synthetic genome design 17 (Fig. 1c), the defect in the synthetic DNA for this region is in its refactoring. REXER using a new fragment-1 BAC—which contained both the extended refactoring (Extended Data Fig. 2) and a TCA-to-TCT mutation at Ser4 in map (Extended Data Fig. 2, Supplementary Data 14)—enabled complete recoding of the targeted 100-kb region of the genome (Extended Data Fig. 2).
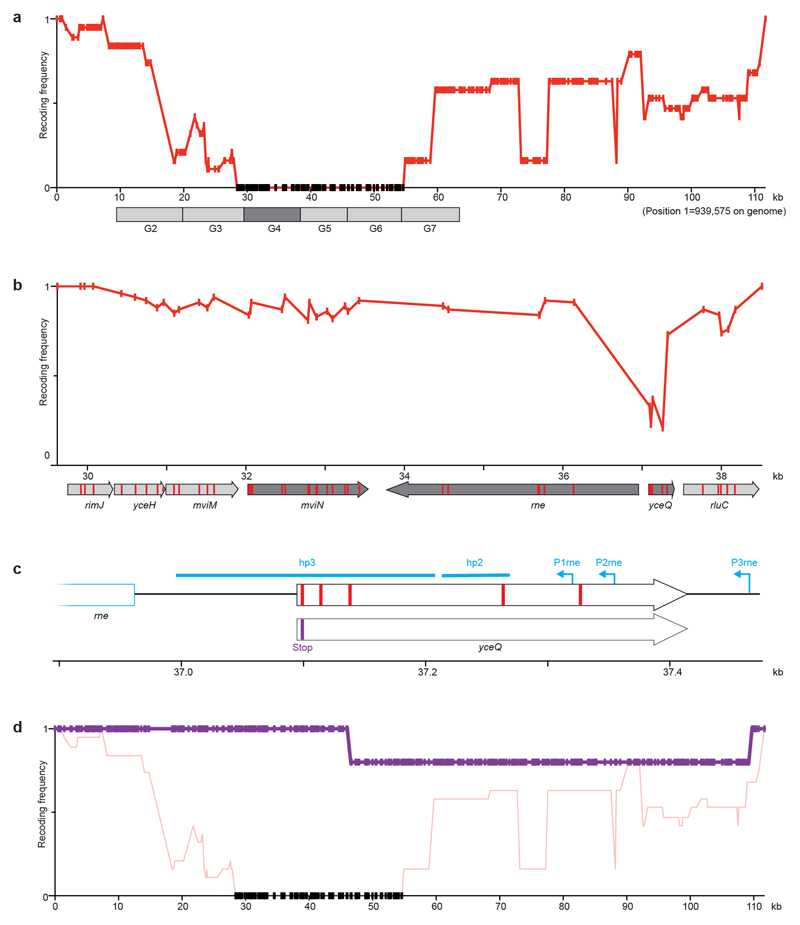
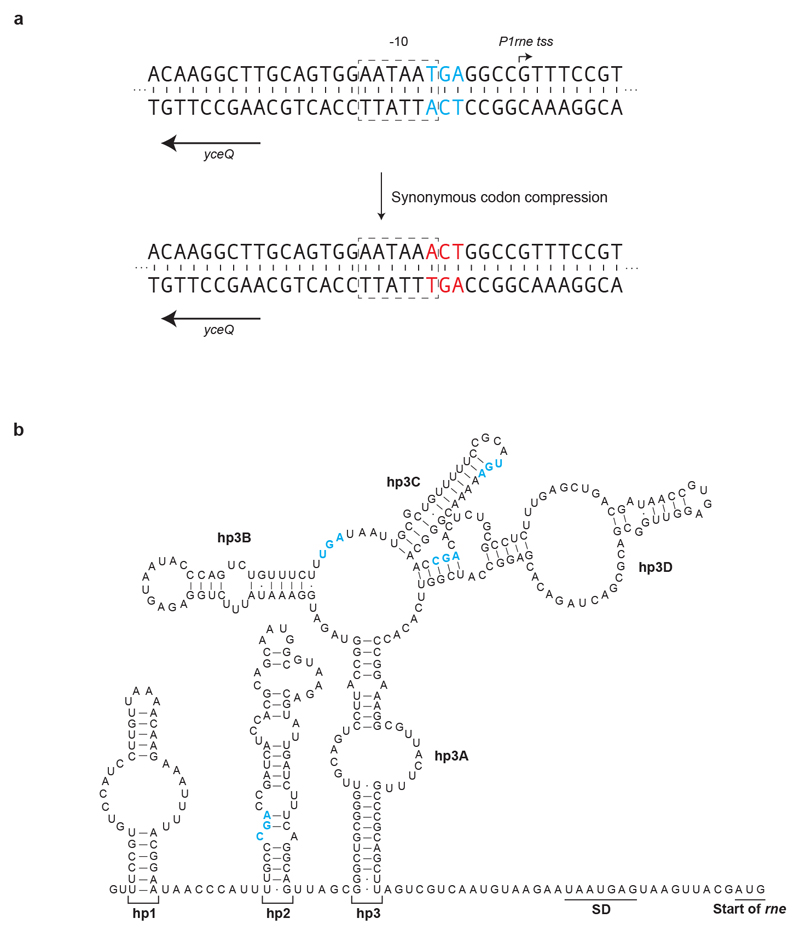
From the post-REXER recoding landscape of fragment 9 and additional experiments, we identified five target codons within yceQ as being problematic to recode (Extended Data Fig. 3). Similarly, we identified a single codon at the 3′ end of yaaY in fragment 37a, which was never recoded (Extended Data Fig. 4). yceQ and yaaY both encode ‘predicted proteins’, multiple insertions in yceQ are viable36 and there are no reports in the Universal Protein Resource (UniProt) of mRNA production and/or protein synthesis from these predicted genes37. Notably, the codons that are recalcitrant to recoding within yceQ and yaaY all lie within the 5′ untranslated regions of adjacent essential genes, and altering these sequences probably has negative effects on the regulation of these essential genes. Indeed, the target codons in yceQ map to RNA secondary structures and promoter elements within the 5′ untranslated region of rne (which encodes the essential RNase, RNase E)38–41 (Extended Data Fig. 5), and these sequences are essential for controlling RNase E homeostasis41.
We fixed fragment 9 by introducing a stop codon into the 5′ sequence of yceQ, thus minimizing translation but retaining native sequences for regulating rne transcription (Extended Data Fig. 3, Supplementary Data 14). REXER using this new BAC led to a complete recoding of the corresponding genomic region (Extended Data Fig. 3, Supplementary Data 11). REXER using a new BAC that contained fragment 37a with a TCA-to-AGC substitution at the problematic codon in yaaY led to a complete recoding of the corresponding region of the genome (Extended Data Fig. 4, Supplementary Data 14).
Having pinpointed and fixed all the initially problematic sequences, we completed the assembly of a strain in which sections A and B are fully recoded (Extended Data Fig. 6), and the assembly of a strain in which section H is entirely recoded (Extended Data Fig. 6, Supplementary Data 11). This completed the assembly of all the sections in seven distinct strains.
Assembly of a recoded genome
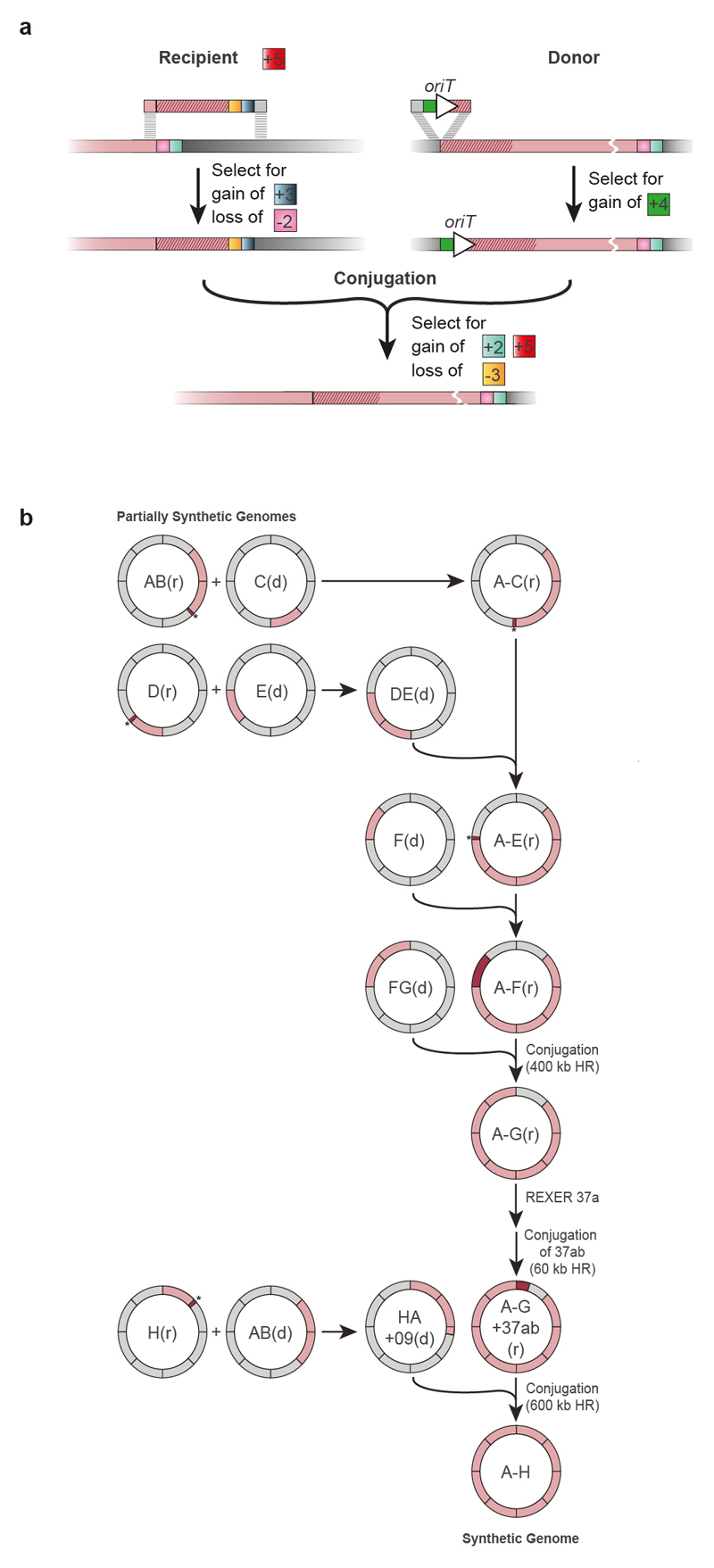
We developed a conjugation-based strategy42–44 to assemble the recoded sections into a single genome (Fig. 3). Our strategy assembles the recoded genome in a clockwise manner, by conjugating recoded ‘donor’ sections that contain the origin of transfer (oriT), into adjacent recoded ‘recipient’ sections that have been extended to provide homology to the donor (Extended Data Fig. 7, Supplementary Data 15, 16). Following conjugation between the donor and the recipient cells, we selected for recipient cells; we then selected for those recipients that had gained the positive marker at the end of the recoded sequence from the donor and lost the negative marker at the end of the extension in the recipient (Extended Data Fig. 7).
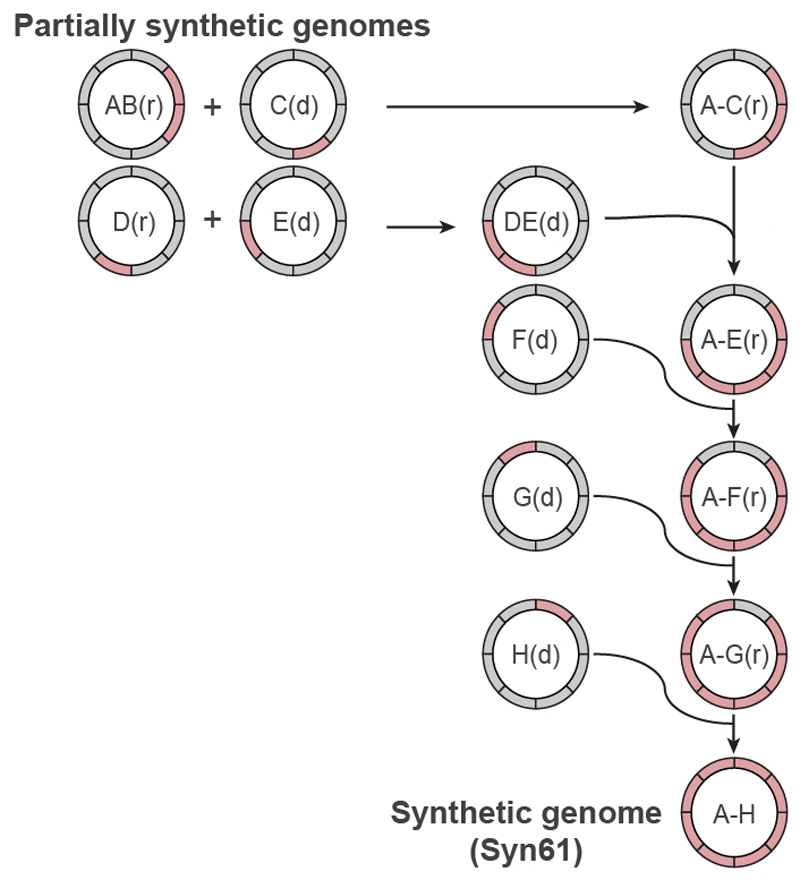
Fig. 3. Assembly of recoded genome sections to create Syn61.
Synthetic genomic sections (pink) from multiple individual partially recoded genomes were assembled into a single fully recoded genome in the indicated sequence of conjugations. The donor (d) and recipient (r) strains contain unique recoded genomic sections, denoted in pink. The recoded genomic content from the donor was conjugated in a clockwise manner to replace the corresponding wild-type genomic section (grey) in the recipient. Conjugation proceeded until the final fully recoded A to H strain (that is, Syn61) was assembled. Extended Data Figure 7 shows the process in more detail, including all homology regions.
The resulting cells, which contain the recoded sections of both the donor and the recipient, can then be used as a recipient for the next recoded donor, and iteration of the process enables the recoded genome to be assembled through the addition of recoded sections to an increasingly recoded recipient (Fig. 3, Extended Data Fig. 7). Donor cells contained a version of the F′ plasmid that facilitates transfer of the donor genome to the recipient cells, but which is not competent to transfer itself to recipient cells (Supplementary Data 17). As a result, this F′ plasmid does not have to be lost from the recipient cells after every conjugation; this accelerated our workflow.
Conjugative assembly (Fig. 3, Extended Data Fig. 7) enabled the synthesis of a synthetic E. coli that we named ‘Syn61’, in which all 1.8x 104 target codons in the genome are recoded (Supplementary Data 18). The synthesis introduced only 8 non-programmed mutations, and none of these non-programmed mutations affects recoding (Supplementary Data 19); 4 of these mutations arose during the preparation of the 100-kb BACs, and 4 arose during the recoding process.
Properties of Syn61
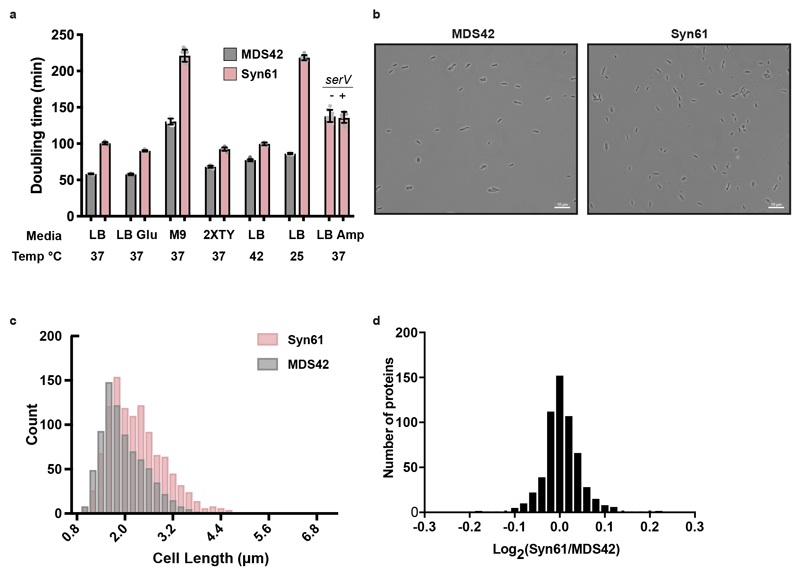
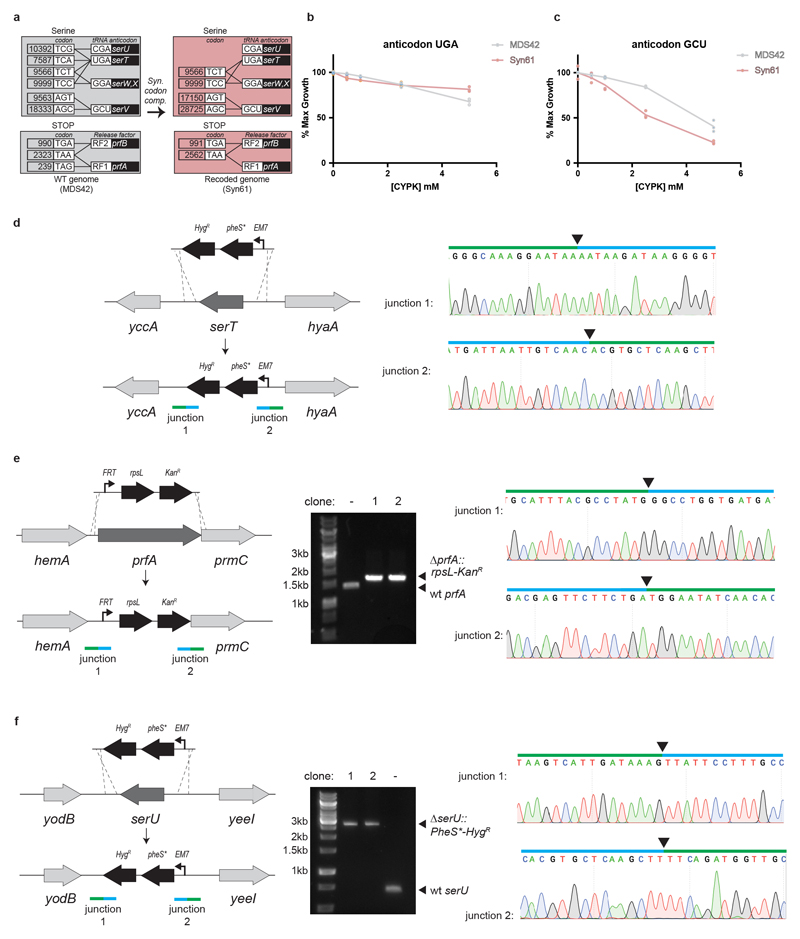
Syn61 doubled only 1.6x slower than MDS42 in lysogeny broth (LB) plus glucose at 37 °C, and this ratio increased at 25 °C and decreased at 42 °C (Extended Data Fig. 8a). Syn61 contains 65% more AGT and AGC codons than are present in MDS42; however, providing additional copies of serV—the transfer RNA (tRNA) that decodes these codons (Fig. 4a)—did not increase growth (Extended Data Fig. 8a). This suggests serV is not limiting. Imaging Syn61 cells suggests that they are slightly longer than MDS42 (Extended Data Fig. 8b, c). We observed minimal differences in the proteomes quantified in both Syn61 and MDS42 (Extended Data Fig. 8d, Supplementary Data 20). Co-translational incorporation of a non-canonical amino acid, using an orthogonal aminoacyl-tRNA synthetase/tRNACGA pair45–47 targeted to TCG codons, was extremely toxic in MDS42 but non-toxic in Syn61; this validates the removal of TCG codons in Syn61 (Fig. 4b). This approach also provided additional insights (Extended Data Fig. 9a–c). serT encodes the tRNASerUGA, which is the only tRNA predicted to decode TCA codons in E. coli and is therefore essential 48. Because Syn61 does not contain TCA codons, serT is dispensable in this strain (Fig. 4c, Extended Data Fig. 9d, Supplementary Data 21), as expected. serU and prfA could also be deleted in Syn61 (Extended Data Fig. 9e, f, Supplementary Data 21). These data provide functional confirmation that we have removed the target codons from the genome, show that the cognate tRNAs and release factor can be removed in Syn61, and demonstrate the unique properties of Syn61 that arise from recoding.
Fig. 4. Functional consequences of synonymous codon compression in Syn61.
a, Synonymous codon compression and deletion of prfA, serU and serT. The grey boxes show the serine codons and stop codons, together with the tRNAs and release factors that decode them in wild-type E. coli (wild-type genome). tRNA anticodons and release factors are connected to the codons that they are predicted to read by black lines. The tRNA and release factor genes are shown in the black boxes. Synonymous codon compression leads to a recoded genome (pink boxes), in which tRNAs with CGA anticodons should have no cognate codons and serT should be dispensable. All factors that read the target codons should be dispensable in Syn61.
b, Co-translational incorporation of the non-canonical amino acid Nε-(((2-methylcycloprop-2-en-1-yl) methoxy) carbonyl)-l-lysine (CYPK), using the orthogonal Methanosarcina mazei pyrrolysyl-tRNA synthetase (PylRS)/tRNAPylCGA pair, was toxic in MDS42, but not in Syn61. When provided with CYPK, this pair will incorporate the noncanonical amino acid in response to TCG codons in a dose-dependent manner. In MDS42 (grey), this incorporation leads to mis-synthesis of the proteome, and toxicity. In Syn61 (pink) (which does not contain TCG codons), this is non-toxic. The lines follow the mean of three biological replicates (each shown as a dot) at each CYPK concentration (0 mM, 0.5 mM, 1 mM, 2.5 mM and 5 mM). Percentage of maximum growth was determined by the final optical density at 600 nm (OD600) with the indicated concentration of CYPK divided by the final OD600 in the absence of CYPK. Final OD600 values were determined after 600 min.
c, Synonymous codon compression enables deletion of serT in Syn61. PCR flanking the serT locus before (−) and after (clones 1 and 2) replacement with a PheS*-HygR double selection cassette; HygR denotes hygromycin resistance (aph(4)-Ia), PheS* denotes a mutant of PheS that encodes a Thr251Ala, Ala294Gly mutant of phenylalanyl-tRNA synthetase. The experiment was performed once. See Extended Data Fig. 9. Full gels are in Supplementary Fig. 1.
Discussion
We have created E. coli in which the entire 4-Mb genome is replaced with synthetic DNA; to our knowledge, the scale of genomic replacement in Syn61 is approximately 4x larger than previously reported for genome or chromosome replacement in any organism (Extended Data Fig. 10a).
We have demonstrated the genome-wide removal of all 1.8x 104 target codons, and thereby removed orders-of-magnitude more codons than previous efforts (Extended Data Fig. 10b). Our synthetic genome contains only 2x 10−4 non-programmed mutations per target codon (Extended Data Fig. 10c), which is orders-of-magnitude lower than the non-programmed mutation frequency in previous recoding efforts14 (Extended Data Fig. 10c).
The creation of an organism that uses a reduced number of sense codons (59) to encode the 20 canonical amino acids demonstrates that life can operate with a reduced number of synonymous sense codons. Our final synthetic genome was recoded using defined refactoring and recoding schemes, and a recoding rule that was previously determined on just 83 (0.43%) of the target codons in the genome17. There are a vast number of theoretical recoding schemes and previous work has established that not all recoding schemes are viable16–20; it is therefore notable that it is possible to identify a single defined recoding scheme that—with a small number of simple corrections—allows genome-wide synonymous codon compression.
The strategies that we have developed for disconnecting a designed genome into sections, fragments and stretches, and realizing the design through the convergent, seamless and robust integration of REXER, GENESIS and directed conjugation, provides a blueprint for future genome syntheses. In future work, we will further characterize the consequences of synonymous codon compression in Syn61 and investigate additional recoding schemes. In addition, we will investigate the extent to which our approach enables sense-codon reassignment for non-canonical biopolymer synthesis12.
Methods
Recoded genome design
We based our synthetic genome design on the sequence of the E. coli MDS42 genome (accession number AP012306.1, released 07-Oct-2016), which has 3547 annotated CDS (Supplementary Data 1). We manually curated the starting genome annotation to remove three CDS and add another twelve. The three predicted CDS removed were htgA, ybbV, and yzfA; there is no evidence that these sequences encode proteins37, and these sequences completely or largely overlap with better characterised genes, which would make it difficult to recode them without disrupting their overlapping genes or creating large repetitive regions. Conversely, the pseudogenes ydeU, ygaY, pbl, yghX, yghY, agaW, yhiK, yhjQ, rph, ysdC, glvG, and cybC were promoted to CDS. To enable negative selection with rpsL, we mutated the genomic copy of rpsL to rpsLK43R. Finally, deep sequencing of our in-house MDS42 revealed a 51 bp insertion between mrcB and hemL which had not been reported in AP012306.1. We manually introduced and annotated this insertion in our starting genome sequence.
We produced a custom Python script that i) identifies and recodes all target codons, and ii) identifies and resolves overlapping gene sequences that contain target codons (available at https://github.com/TiongSun/genome_recoding). From our curated MDS42 starting sequence, we used the script to generate a new synthetic genome in which all TCG, TCA and TAG codons were replaced with AGC, AGT and TAA respectively. The script reported 91 CDS with overlaps containing target codons. In 33 instances, genes were overlapping tail-to-tail (3', 3') (Supplementary Data 4); 12 of these could be recoded by introducing a silent mutation in the overlapping gene, while the remaining 21 were duplicated to separate the genes (Fig. 1b). 58 instances of genes overlapping head-to-tail (5', 3') were resolved by duplicating the overlap plus 20 bp of upstream sequence to allow endogenous expression of the downstream gene (Fig. 1c). For overlaps longer than 1 bp, an in-frame TAA was introduced to terminate expression from the original RBS for the downstream gene. prfB (release-factor RF-2) was not annotated as a CDS in our starting MDS42 genome due to its regulatory internal stop codon, and we therefore recoded all the target codons in the gene manually, thereby maintaining the internal stop codon. The resulting genome design contained 3556 CDS with 1,156,625 codons of which 18,218 were recoded (Supplementary Data 2, Supplementary Data 3).
Retrosynthesis of recoded stretches
We divided the designed genome into 37 fragments of between 91 and 136 kb. We chose the boundary sequences that delimit these fragments so that: i) they consist of a 5’-NGG-3’ PAM to allow REXER4 to be used for integration if necessary, ii) the PAM does not sit within 50 bp of a target codon, iii) the PAM is in-between non-essential genes and iv) the PAM does not disturb any annotated features such as promoters. We called the regions ~50-100 bp upstream and downstream of these boundaries ‘landing sites’, and these are annotated as Lxx, where xx is the number of the upstream fragment, e.g. L01 is the landing site between fragment 1 and 2 (Supplementary Data 2). In our design, a landing site sequence is contained in the 3’ end of a fragment and the 5’ end of the next – as a result all 37 fragments contain overlapping homologies of 54-155 bp with their neighbouring fragment.
Each fragment was further broken down to 7-14 stretches of 4-15 kb. We designed the stretches so that they contain overlaps of 80-200 bp with each other, and the overlap regions were defined at intergenic regions free of any recoding targets. A total of 409 stretches were synthesised (GENEWIZ, USA) and supplied in pSC101 or pST vectors flanked by BsaI, AvrII, SpeI, or XbaI restriction sites. The synthetic stretches naturally did not contain at least one of these restriction sites.
Construction of selection cassettes and plasmids for REXER/GENESIS
The cloning procedures described in this section were performed in E. coli DH10b, which is resistant to streptomycin by virtue of an rpsLK43R mutation. The plasmid pKW20_CDFtet_pAraRedCas9_tracrRNA used throughout this study encodes Cas9 and the lambda-red recombination components alpha/beta/gamma under the control of an arabinose-inducible promoter, as well as a tracrRNA under its native promoter, as previously described17.
The protospacers for REXER are encoded in the plasmid pKW1_MB1Amp_Spacer (Supplementary Data 13), which contains a pMB1 origin of replication, an ampicillin resistance marker and the protospacer array under the control of its endogenous promoter as previously described17. From this plasmid we constructed the derivative pKW3_MB1Amp _TracrK_Spacer (Supplementary Data 14), which additionally contains a tracrRNA upstream of the protospacer array. For this we introduced a PCR product containing tracrRNA with its modified endogenous promoter into the BamHI site of pKW1_MB1Amp_Spacer via Gibson assembly using the NEBuilder HiFi Master Mix. From this plasmid a derivative that additionally encodes Cas9 was constructed, also by Gibson assembly, and named pKW5_MB1Amp_TracrK_Cas9_Spacer.
For each REXER step, a derivative of one of these three plasmids was constructed to harbour a protospacer/direct repeat array containing 2 (REXER2) or 4 (REXER4) protospacers, corresponding to the target sequences for cutting the BAC and genome. The different protospacer arrays were constructed from overlapping oligos through multiple rounds of PCR – the products were inserted by Gibson assembly between restriction sites AccI and EcoRI in the backbone of pKW1_MB1Amp_Spacer, pKW3_MB1Amp_TracrK_Spacer or pKW5_MB1Amp_TracrK_Cas9_Spacer. The protospacer arrays resulting from each assembly were verified to be mutation-free by Sanger sequencing. Supplementary Data 9 contains a table indicating which backbone was used for each REXER step together with the protospacer sequences they contain.
The positive-negative selection cassettes used in REXER and GENESIS are -1/+1 (rpsL-KanR), -2/+2 (sacB-CmR) and -3/+3 (pheST251A_A294G-HygR). -1/+1 and -2/+2 are as previously described17. In -3/+3, pheST251A_A294G is dominant lethal in the presence of 4-chlorophenylalanine, and HygR confers resistance to hygromycin. Both proteins are expressed polycistronically under control of the EM7 promoter. The -3/+3 cassette was synthesised de novo. The -3/+3 cassette is also referred to as pheS*-HygR.
Constructing strains containing double selection cassettes at genomic landing sites
According to our design, each region of the genome that is targeted for replacement by a synthetic fragment is flanked by an upstream landing site and a downstream landing site; these genomic landing site sequences are the same as the landing site sequences described above. Initiation of REXER/GENESIS requires the insertion of a double selection cassette in the upstream genomic landing site. We inserted double selection cassettes at the landing sites through lambda-red mediated recombination. Briefly, either the sacB-CmR or the rpsL-KanR cassettes were PCR amplified with primers containing homology regions to the genomic landing sites of interest. For recombination experiments, we prepared electrocompetent cells as described previously17 and electroporated 3 μg of the purified PCR product into 100 μL of MDS42rpsLK43R cells harbouring the pKW20_CDFtet_pAraRedCas9_tracrRNA plasmid expressing the lambda-red alpha/beta/gamma genes. The recombination machinery was induced, under control of the arabinose promoter (pAra), with L-arabinose added at 0.5% for 1 hour starting at OD600 = 0.2. Pre-induced cells were electroporated and then recovered for 1 hour at 37 °C in 4 mL of super optimal broth (SOB) medium. Cells were then diluted into 100 mL of LB medium with 10 μg/mL tetracycline and grown for 4 hours at 37 °C, 200 rpm. The cells were subsequently spun down, resuspended in 4 mL of H2O, serially diluted, plated and incubated overnight at 37 °C on LB agar plates containing 10 μg/mL tetracycline, 18 μg/mL chloramphenicol (for sacB-CmR) or 50 μg/mL kanamycin (for rpsL-KanR).
BAC assembly and delivery
We constructed Bacterial Artificial Chromosomes (BACs) shuttle vectors that contained 97-136 kb of synthetic DNA. On the 5' side, the synthetic DNA was flanked by a region of homology to the genome (HR1), and a Cas9 cut site. On the 3' side the synthetic DNA was flanked by a double selection cassette, a region of homology to the genome (HR2), and a second Cas9 cut site. The BAC also contained a negative selection marker, a BAC origin, a URA marker and YAC origin (CEN6 centromere fused to an autonomously replicating sequence (CEN/ARS)) (Fig. 2c, Supplementary Data 6-8 provides maps with these features annotated).
BACs were assembled by homologous recombination in S. cerevisiae. Each assembly combined i) 7-14 stretches of synthetic DNA, each 6-13 kb in length, with ii) a selection construct (see below) and iii) a BAC shuttle vector backbone (Supplementary Data 6-8)17.
Synthetic DNA stretches were excised by digestion with BsaI, AvrII, SpeI, or XbaI restriction sites from their source vectors provided by GENEWIZ. In the case of AvrII, SpeI, and XbaI, restriction digests were followed by Mung Bean nuclease treatment to remove sticky ends.
Selection constructs contained a region of homology to the 3' most stretch of the fragment, a double selection cassette (sacB-CmR or rpsL-KanR) a region of homology (HR2) to the targeted genomic locus, a negative selection marker (rpsL, sacB or pheS*- HygR) and YAC. For specific double selection cassettes, negative selection markers, and homology region sequences see Supplementary Data 9. We assembled episomal versions of the selection constructs in a pSC101 backbone from 3 PCR fragments with NEBuilder HiFi DNA Assembly Master Mix. The episomal versions were designed so that restriction digestion with BsaI yielded a DNA fragment for BAC assembly.
The BAC backbone containing a BAC origin and a URA3 marker was amplified by PCR using a previously described BAC17 as a template, and the PCR product used for BAC assembly. The primers used for these PCR assemblies are listed in Supplementary Data 9.
To assemble the stretches, selection construct, and BAC backbone, 30-50 fmol of each piece of DNA was transformed into S. cerevisiae spheroplasts; these were prepared as previously described35. Following assembly we identified yeast clones potentially harbouring correctly assembled BACs by colony PCR at the junctions of overlapping fragments and vector-insert junctions. Clones that appeared correct by colony PCR were sequence verified by NGS after transformation into E. coli, as described below.
The assembled BACs were extracted from yeast with the Gentra Puregene Yeast/Bact. Kit (Qiagen) following the manufacturer’s instructions. MDS42rpsLK43R cells were transformed with the assembled BAC by electroporation. Due to the large size of the BACs we sometimes observed inefficient electroporation into target cells. Consequently, we introduced an oriT-Apramycin cassette provided as a PCR product with 50 bp homology regions by lambda-red-mediated recombination (as described above) into some BACs post assembly (Supplementary Data 6-8). This facilitated transfer of BACs, from E. coli that had been successfully transformed, to other strains by conjugation.
Synthesis of recoded sections
We used various genomic and plasmid selection markers for sequential REXER experiments (GENESIS) (Supplementary Data 11). We used an rpsL-KanR (-1/+1) or sacB-CmR (-2/+2) cassette at genomic landing sites for selection. We used rpsL-KanR-sacB (-1/+1,-2), rpsL-KanR-pheS*- HygR (-1/+1,-3/+3) or sacB-CmR-rpsL (-2/+2,-1) cassettes as episomal selection markers.
For each REXER, MDS42rpsLK43R cells containing pKW20_ CDFtet_pAraRedCas9_tracrRNA and a double selection cassette at the relevant upstream genomic landing site were transformed with the relevant BAC. We plated cells on LB agar supplemented with 2% glucose, 5 μg/ml tetracycline and antibiotic selecting for the BAC (i.e. 18 μg/ml chloramphenicol or 50 μg/ml kanamycin). We inoculated individual colonies into LB medium with 5 μg/ml tetracycline and the BAC specific antibiotic and grew cells overnight at 37 °C, 200 rpm. The overnight culture was diluted in LB medium with 5 μg/ml tetracycline, and the BAC specific antibiotic, to OD600 = 0.05 and grown at 37 °C with shaking for about 2 h, until OD600 ≈ 0.2. To induce lambda-red expression we added arabinose powder to the culture to a final concentration of 0.5% and the incubated the culture for one additional hour at 37 °C with shaking. We harvested the cells at OD600 ≈ 0.6, and made the cells electro-competent as described previously17.
For each REXER experiment a linear dsDNA protospacer array was PCR amplified from pKW1_MB1Amp_Spacers using universal primers (Supplementary Data 12). Approximately 5-10 μg of the resulting DpnI digested and purified PCR product was transformed into 100 μL electro-competent and induced cells. Cells were recovered in 4 ml SOB medium for 1 h at 37 °C and then diluted to 100 mL LB supplemented with 5 μg/mL tetracycline and antibiotic selecting for the BAC and incubated for another 4 h at 37 °C with shaking. Alternatively, electrocompetent and induced cells were transformed with 5 μg of circular protospacer array (pKW1_MB1Amp_Spacers or pKW3_MB1Amp_Spacers plasmid) and after 1 h recovery in SOB medium at 37°C transferred into 100 mL LB supplemented with 100 μg/mL ampicillin for another 4 h at 37 °C with shaking (Supplementary Data 12, 13). If REXER2 was not sufficient we performed REXER4 using pKW5_MB1Amp_Spacers plasmid as previously described17.
We spun down the culture and resuspended it in 4 ml Milli-Q filtered water and spread in serial dilutions on selection plates of LB agar with 5 μg/ml tetracycline, an agent selecting against the negative selection marker and an antibiotic selecting for the positive marker originating from the BAC. The plates were incubated at 37 °C overnight. Multiple colonies were picked, resuspended in Milli-Q filtered water, and arrayed on several LB agar plates supplemented with 50 μg/ml kanamycin, 18 μg/ml chloramphenicol, 200 μg/ml streptomycin, 7.5% sucrose or 2.5 mM 4-chloro-phenylalanine. Colony PCR was also performed from resuspended colonies using both a primer pair flanking the genomic locus of the landing site and the position of the newly integrated selection cassette from the BAC. REXER-mediated recombination results in an approximately 500 bp band at the upstream genomic locus with a 2.5 kb (rK-landing site) or 3.5 kb (sC-landing site) band for the control MDS42rk/ MDS42sC strain indicating successful removal of the landing site from the genome. Primer pairs flanking the 3’ end of the replaced DNA generate an approximately 2.5 kb (rK selection cassette on pBAC) or 3.5 kb (sC selection cassette on pBAC) band and a 500 bp band for the control MDS42rk/ MDS42sC strain indicating successful integration of the selection markers.
If a plasmid based circular protospacer array was used in the previous REXER experiment the plasmid had to be lost before the next experiment. Thus, a successful clone from the first REXER experiment was grown in LB supplemented with 2% glucose, 5 μg/mL tetracycline and antibiotic selecting for the positive marker in the genome to a dense culture at 37 °C with shaking. 2 μL of the culture were then streaked out on an LB agar plate with the same supplements and incubated at 37°C overnight. Several colonies were arrayed in replica on LB agar plate and LB agar plate supplemented with 100 μg/mL ampicillin to screen for the loss of the plasmid.
BAC editing
When encountering loss-of-function mutations in a selection cassette on BACs in E. coli, the faulty cassette was replaced with a suitable double selection cassette provided (Supplementary Data 9) as a PCR-product flanked by 50 bp homology regions and integrated by lambda-red-mediated recombination.
Changes in the synthetic, recoded sequence of a BAC, either to correct spontaneous mutations or change recoded codons, were introduced by a two-step replacement approach; For BACs containing the selection cassettes -2/+2 and -1 in the end of the recoded sequence, the -3/+3 cassette was provided as a PCR-product flanked by 50bp-homology regions targeting the desired locus and integrated by lambda-red-mediated recombination followed by selection for +3. Due to the homology between the recoded DNA and the genome, some of the resulting clones would contain -3/+3 on the BAC and some on the genome. To identify clones with the cassette on the BAC, clones were plated in replica on agar plates selecting (1) for +3, (2) against -3, and (3) for +2 and against -3; Only clones surviving on plate (1) and (2) but not on (3) have the -3/+3 cassette integrated on the BAC. The location of the cassette was verified by purifying the BAC using QIAprep Spin Miniprep Kit followed by genotyping. In a second step, the -3/+3 cassette was replaced by providing a PCR-product of the desired sequence flanked by 50 bp-homology regions and integrated by lambda-red-mediated recombination followed by selection for +2 and against -3. The BAC was genotyped as above and sequence-verified by NGS.
Preparing a non-transferable F’ plasmid and conjugative transfer of episomes
We created the version of the F’ plasmid used for conjugation of genomic DNA, as well as transfer of BACs between strains, to enable transfer of sequences bearing oriT without transfer of the F’ plasmid itself (Supplementary Data 17). We achieved this by deleting the nick-site in the origin of transfer (oriT) within the F’ plasmid itself, a related approach was previously reported49. The F’ plasmid derivative, pRK24 (addgene #51950), was modified by integrating desired markers as PCR-products flanked by 50 bp-homology regions and integration was performed by lambda-red-mediated recombination using a variant of pKW20 carrying KanR instead of TetR. First, the β-lactamase gene, conferring ampicillin resistance in pRK24, was replaced with the artificial T5-luxABCDE operon50, which generates bioluminescence that allows visual identification of infected bacterial cells. Next, TetR was replaced with T3-aac3 that produces aminoglycoside 3-N-acetyltransferase IV for selection with 50 μg/mL apramycin. Finally, a 24 bp deletion of the nick-site in oriT was made by integrating EM7-bsd that expresses blasticidin-S deaminase, and can be selected for with 50 μg/mL blasticidin in low-salt TYE/LB. The resulting F’-plasmid called pJF146 (Supplementary Data 17), was extracted using QIAprep Spin Miniprep Kit (QIAgen) and transformed by electroporation into donor strains for subsequent conjugation.
Transfer of episomal DNA containing oriT was performed by conjugation42,43. A donor strain was double transformed with pJF146 and an assembled BAC with oriT (see above). A recipient strain was transformed with pKW20. 5 ml of donor and recipient culture were grown to saturation overnight in selective LB media and subsequently washed 3 times with LB media without antibiotics. The resuspended donor and recipient strains were combined in a 4:1 ratio, spotted on TYE agar plates and incubated for 1h at 37°C. The cells were washed off the plate and spread in serial dilutions on LB agar plates with 2% glucose, 5 μg/ml tetracycline selecting for the recipient strain and antibiotic selecting for the BAC. Successful transfer of the BAC was confirmed by colony PCR of the BAC-vector insert junctions.
Assembling a synthetic genome from recoded sections
Transfer of genomic DNA was combined with subsequent recBCD-mediated recombination to assemble partially synthetic E. coli genomes into a synthetic genome. In preparation of the donor and recipient strains a rpsL-HygR-oriT or GmR-oriT cassette was supplied as PCR product and integrated into the donor strain genome via lambda-red-mediated recombination (Supplementary Data 15, 16). Separately, a pheS*-HygR cassette was integrated approximately 3 kb downstream of the synthetic DNA in the donor strains. This provided a template genomic DNA for PCR amplification of a 3 kb synthetic DNA segment with 3’ pheS*- HygR selection cassette. This PCR product was provided to the recipient strains to replace the WT DNA in a lambda-red-mediated recombination. Thereby, the selection marker at the 3’ end of the synthetic segment was replaced and a 3 kb homology region to the donor synthetic DNA was generated. This strategy served to systematically generate recipient strains with 3 kb of homology with their respective donors, always with a pheS-HygR at the 3’ end. Additionally, the donor strains were transformed with pJF146 and sensitivity to tetracycline was confirmed. In contrast, pKW20 was maintained in the donor strains to confer tetracycline resistance.
For conjugation, donor and recipient strain were grown to saturation overnight in LB medium with 2% glucose, 5 μg/ml tetracycline and 50 μg/ml kanamycin or 20 μg/ml chloramphenicol (donor) and 50 μg/ml apramycin and 200 μg/mL hygromycin B (recipient). The overnight cultures were diluted 1:10 in the same selective LB medium and grown to OD600 = 0.5. 50 ml of both donor and recipient culture were washed 3 times with LB medium with 2% glucose and then each resuspended in 400 μl LB medium with 2% glucose. 320 μl of donor was mixed with 80 μl of recipient, spotted on TYE agar plates and incubated at 37°C. The incubation time depended on the length of transferred synthetic DNA and doubling time of the recipient strain and varied from 1h to 3h. Cells were washed off the plate and transferred into 100 ml LB medium with 2% glucose and 5 μg/ml tetracycline and incubated at 37°C for 2h with shaking. Subsequently 50 μg/ml kanamycin or 20 μg/ml chloramphenicol (selecting for the transferred positive selection marker of the donor) was added, followed by another 2 h incubation at 37°C. The culture was spun down and resuspended in 4 ml Milli-Q filtered water and spread in serial dilutions on selection plates of LB agar with 2% glucose, 5 μg/ml tetracycline, 2.5 mM 4-chloro-phenylalanine and 50 μg/ml kanamycin or 20 μg/ml chloramphenicol. Successful DNA transfer and recombination was determined by colony PCR for the loss of the pheS*- HygR cassette, integration of the donor’s selection cassette and absence of the Gm-oriT cassette.
We performed a convergent synthesis of a genome recoded through sections A-E (Extended Data Fig. 7). We then used the A-E strain as a recipient for F, generating a recoded strain, A-F. A-F was then used as a recipient for F-G, generating A-G; this conjugation used a much longer shared recoded sequence (0.4 Mb) between the donor and recipient strains to increase conjugation efficiency.
To create a completely recoded genome we first created a recipient strain by introducing 37a and 37b into A-G to create A-G-37ab (providing a 115 kb homology region with the final donor). We created the final donor strain by conjugation between strain H and strain AB, which yielded strain H-A-09, in which H, A and fragment 9 from section B are recoded. The additional sequence from A and B was added to H to ensure that we did not erase the recoding in A in the final conjugation. The final conjugation between the H-A-09 donor strain and A-G-37ab recipient strain led to the synthesis of E. coli, which we name E.coli Syn61, in which all 1.8 x104 target codons in the genome are recoded.
Preparation of whole-genome and BAC libraries for next-generation sequencing
E. coli genomic DNA was purified using the DNEasy Blood and Tissue Kit (QIAgen) as per manufacturer’s instructions. BACs were extracted from cells with the QIAprep Spin Miniprep Kit (QIAgen) as per manufacturer’s instructions. We found that this kit was suitable for purification of BACs in excess of 130 kb. We avoided vigorous shaking of the samples throughout purification so as to reduce DNA shearing.
Paired-end Illumina sequencing libraries were prepared using the Illumina Nextera XT Kit as per manufacturer’s instructions. Sequencing data was obtained in the Illumina MiSeq, running 2 x 300 or 2 x 75 cycles with the MiSeq Reagent kit v3.
Sequencing data analysis
The standard workflow for sequence analysis in this work is compiled in the iSeq package, available at https://github.com/TiongSun/iSeq. In short, sequencing reads were aligned to a reference recoded or wild-type genome using bowtie2 with soft-clipping activated51. Aligned reads were sorted and indexed with samtools52. A customised Python script combines functionalities of samtools and igvtools to yield a variant calling summary. This script was used to assess mutations, indels and structural variations, in combination with visual analysis in the Integrative Genomics Viewer53.
We produced a custom Python script to generate recoding landscapes across a target genomic region (available at https://github.com/TiongSun/recoding_landscapes). Briefly, the script takes a BAM alignment file, a reference in fasta and a GeneBank annotation file as inputs. It identifies the target codons for recoding, and compiles the reads that align to these target codons in the alignment file. It then outputs the frequency of recoding at each target codon, and plots these frequencies across the length of the genomic region of interest.
Growth rate measurement and analysis
Bacterial clones were grown overnight at 37 °C in LB with 2 % glucose and 100 μg/mL streptomycin. Overnight cultures were diluted 1:50 and monitored for growth while varying temperature (25 °C, 37 °C, or 42 °C) and media conditions (LB, LB with 2 % glucose, M9 minimal media, 2XTY). Measurements of OD600 were taken every 5 min for 18 h on a Biomek automated workstation platform with high speed linear shaking.
To determine doubling times, the growth curves were log2-transformed. At a linear phase of the curve during exponential growth, the first derivative was determined (d(log2(x))/dt) and ten consecutive time-points with the maximal log2-derivatives were used to calculate the doubling time for each replicate. A total of 10 independently grown biological replicates were measured for the recoded Syn61 strain and wt MDS42rpsLK43R. The mean doubling time and standard deviation from the mean were calculated for all n=10 replicates.
Microscopy and cell size measurement
Cells were grown with shaking in LB supplemented with 100 μg/mL streptomycin to approximately OD600=0.2. A thin layer of bacteria was sandwiched between an agarose pad and a coverslip. A standard microscope slide was prepared with a 1% agarose pad (Sigma-Aldrich A4018-5G). A sample of 2 μl to 4 μl of bacterial culture was dropped onto the top of the pad. This was covered by a #1 coverslip supported on either side by a glass spacer matched to the ~1 mm height of the pad. Samples were imaged on an upright Zeiss Axiophot phase contrast microscope using a 63X 1.25NA Plan Neofluar phase objective (Zeiss UK, Cambridge, UK). Images were taken using an IDS ueye monochrome camera under control of ueye cockpit software (IDS Imaging Development Systems GmbH, Obersulm, Germany). 10 fields were taken of each sample. Images were loaded in to Nikon NIS Elements software for further quantitation (Nikon Instruments Surrey UK). The General analysis tool was used to apply an intensity threshold to segment the bacteria. A one micron lower size limit was imposed to remove background particulates and dust. Length measurements were subsequently made on the segmented bacteria using the General Analysis quantification tools.
Mass Spectrometry
Three biological replicates were performed for each strain. Proteins from each Escherichia coli lysates were solubilized in a buffer containing 6 M urea in 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, proteins were diluted to 1 M urea with 50 mM ammonium bicarbonate, digested with Lys-C (Promega, UK) at a protein to enzyme ratio of 1:50 for 2 hours at 37 °C, followed by digestion with Trypsin (Promega, UK) at a protein to enzyme ratio of 1:100 for 12 hours 37 °C. The resulting peptide mixtures were acidified by the addition formic acid to a final concentration of 2% v/v. The digests were analysed in duplicate (1 ug initial protein/injection) by nano-scale capillary LC-MS/MS using a Ultimate U3000 HPLC (ThermoScientific Dionex, San Jose, USA) to deliver a flow of approximately 300 nL/min. A C18 Acclaim PepMap100 5 μm, 100 μm x 20 mm nanoViper (ThermoScientific Dionex, San Jose, USA), trapped the peptides prior to separation on a C18 Acclaim PepMap100 3 μm, 75 μm x 250 mm nanoViper (ThermoScientific Dionex, San Jose, USA). Peptides were eluted with a 100 minute gradient of acetonitrile (2% to 60%). The analytical column outlet was directly interfaced via a nano-flow electrospray ionisation source, with a hybrid dual pressure linear ion trap mass spectrometer (Orbitrap Velos, ThermoScientific, San Jose, USA). Data dependent analysis was carried out, using a resolution of 30,000 for the full MS spectrum, followed by ten MS/MS spectra in the linear ion trap. MS spectra were collected over a m/z range of 300–2000. MS/MS scans were collected using a threshold energy of 35 for collision induced dissociation. All raw files were processed with MaxQuant 1.5.5.154 using standard settings and searched against an Escherichia coli strain K-12 with the Andromeda search engine55 integrated into the MaxQuant software suite. Enzyme search specificity was Trypsin/P for both endoproteinases. Up to two missed cleavages for each peptide were allowed. Carbamidomethylation of cysteines was set as fixed modification with oxidized methionine and protein N-acetylation considered as variable modifications. The search was performed with an initial mass tolerance of 6 ppm for the precursor ion and 0.5 Da for CID MS/MS spectra. The false discovery rate was fixed at 1% at the peptide and protein level. Statistical analysis was carried out using the Perseus (1.5.5.3) module of MaxQuant. Prior to statistical analysis, peptides mapped to known contaminants, reverse hits and protein groups only identified by site were removed. Only protein groups identified with at least two peptides, one of which was unique and two quantitation events were considered for data analysis. For proteins quantified at least once in each strain, the average abundance of each protein across replicates of Syn61 was divided by the abundance in MDS42 replicates, and then log2-transformed. A P-value for the difference in abundance between strains was calculated by two-sample T-test (Perseus).
Toxicity of CYPK incorporation using orthogonal aminoacyl-tRNA synthetases tRNAXXXs
We used a variant of stochastic orthogonal recoding of translation (SORT) to investigate the toxicity of a non--canonical amino acid using tRNAs with different anticodons45–47. Electrocompetent MDS42 and Syn61 cells were transformed with plasmid pKW1_MmPylS_PylTXXX for expression of PylRS and tRNAPylXXX, where XXX is the indicated anticodon. Three variants of this plasmid were used, with the anticodon of tRNAPyl mutated to CGA (pKW1_MmPylS_PylTCGA), UGA (pKW1_MmPylS_PylTUGA) or GCU (pKW1_MmPylS_PylTGCU). Cells were grown over night in LB medium with 75 μg/ml spectinomycin. Overnight cultures were diluted 1:100 into LB supplemented with Nε-(((2-methylcycloprop-2-en-1-yl) methoxy) carbonyl)-L-lysine (CYPK) at 0 mM, 0.5 mM, 1 mM, 2.5 mM and 5 mM and growth was measured as described above. “% Max Growth” was determined as the final OD600 in the presence of the indicated concentration of CYPK divided by the final OD600 in the absence of CYPK. Final OD600s were determined after 600 min.
Deletion of prfA, serU and serT by homologous recombination
Recoded versions of the pheS*-HygR and rpsL-KanR cassettes, according to the recoding scheme described in Fig. 1a, were synthesised de novo, so that expression of the selection proteins would not rely on decoding by serU or serT. For deleting prfA, the recoded rpsL-KanR was amplified with oligos containing ~50 bp homology to the prfA flanking genomic sequences. The same was done for serU and serT with recoded selection cassette pheS*-HygR. Oligonucleotide sequences are provided in Supplementary Data 21. Syn61 cells harbouring the plasmid pKW20_CDFtet_pAraRedCas9_tracrRNA were made competent as described above, using 2xTY instead of LB. Cells were electroporated with ~8 μg of PCR product, and recovered for 1 hour in 4 mL SOB, then transferred to 100 mL 2xTY supplemented with 5 μg/ml tetracycline. After 4 hours cells were spun down, resuspended in 500 μL H2O and plated in serial dilutions in 2xTY agar plates supplemented with 5 μg/ml tetracycline and 200 μg/ml hygromycin B (for pheS*-HygR) or 50 μg/ml kanamycin (for rpsL-KanR). Deletions were verified in each case by colony PCR with primers flanking the locus of interest.
Extended Data
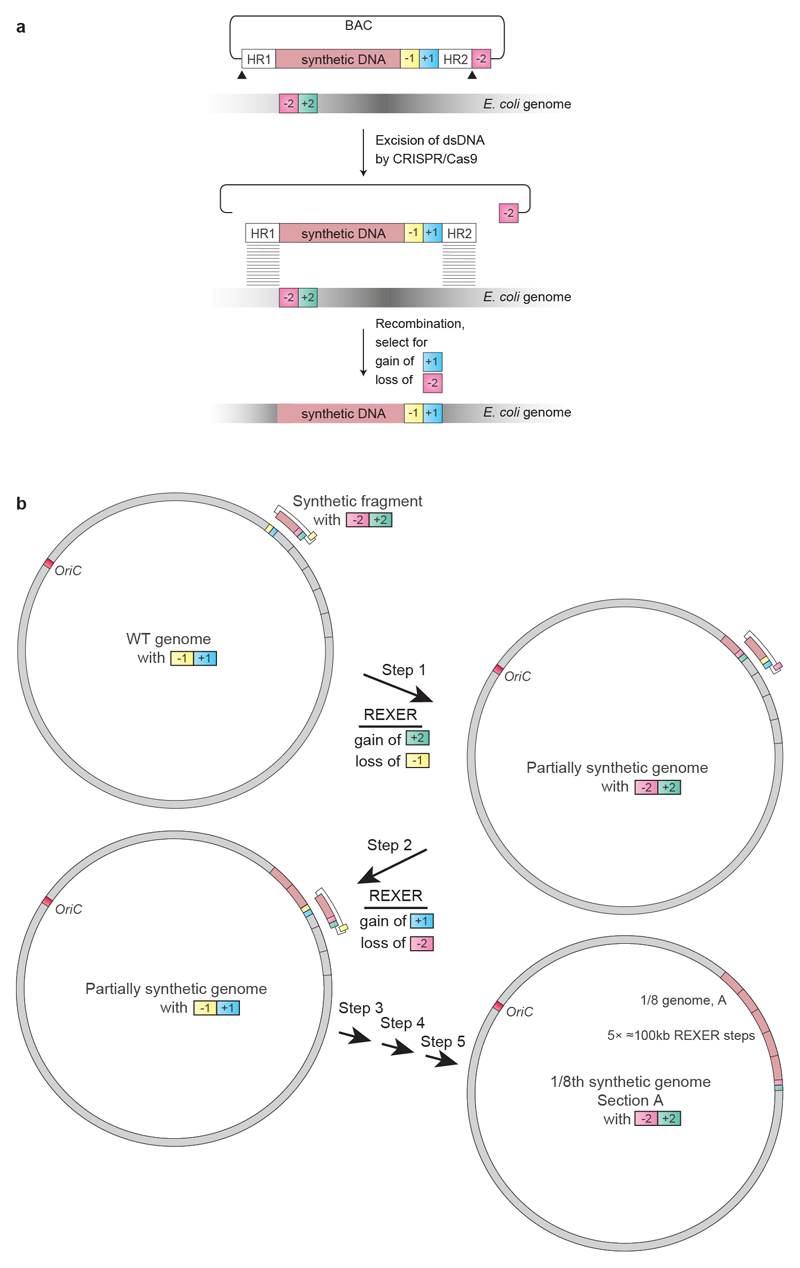
Extended Data Fig. 1. Using 100-kb fragments of synthetic DNA to replace the corresponding regions in the genome through REXER, and using GENESIS for the stepwise replacement of genomic DNA by synthetic DNA to generate recoded sections.
a, REXER uses CRISPR–Cas9- and lambda-red-mediated recombination to replace genomic DNA with synthetic DNA provided from an episome (BAC). This enables large regions of the genome (>100 kb) to be replaced by synthetic DNA17. The black triangles denote the location of CRISPR protospacers, which are cleaved by Cas9 to liberate the synthetic DNA (pink) cassette from the BAC flanked by homology regions. Homology regions 1 and 2 program the location of recombination into the E. coli genome. The double-selection cassette (−1, +1) ensures the integration of the synthetic DNA, and the double-selection cassette (−2, +2) on the genome ensures the removal of the corresponding wild-type DNA. In the example shown in the figure, +1 is kanR, −1 is rpsL, +2 is cat and −2 is sacB.
b, Iterative cycles of REXER, with alternating choices of positive- and negative-selection cassettes, enables GENESIS17. This enables large sections of the synthetic genome to be assembled through the iterative addition of fragments, which replace the corresponding genomic sequences, in a clockwise manner. The first REXER of a 100-kb synthetic fragment of DNA leaves a −1, +1 double-selection cassette on the genome, which acts as a landing site for the downstream integration of a second fragment of synthetic DNA that contains a −2, +2 double-selection cassette. In the example shown, +1 is kanR, −1 is rpsL, +2 is cat and −2 is sacB, but the same logic can be used with different permutations of positive and negative selection markers on the genome and the BAC.
Extended Data Fig. 2. Recoding ftsI-murE and map in fragment 1.
a, Recoding landscape of fragment 1. We sequenced six clones after REXER. Each dot represents the frequency of recoding within the sequenced clones (y axis) for a target codon at the indicated position in the genome (x axis). Black dots indicate positions at which we did not observe recoding. Four codons and a refactoring of ftsI-murE, and one codon in map, were rejected.
b, Refactoring the 14-bp overlap of ftsI and murE. The codons and overlaps are colour-coded by their post-REXER replacement frequency in the clones sequenced. Using our initial refactoring scheme (refactoring 1) (in which the overlap plus 20 bp of upstream sequence was duplicated), we did not observe replacement of the overlap by synthetic DNA (in the six clones sequenced after REXER). Refactoring scheme 2 (refactoring 2) (which duplicates the overlap plus 182 bp of upstream sequence) resulted in complete recoding of this region in 12 of the 16 post-REXER clones that we sequenced.
c, Testing alternative codons at Ser4 in map. A double-selection cassette, pheS*-HygR, on a constitutive EM7 promoter was introduced upstream of map, followed by a ribosome-binding site. We replaced the cassette using linear double-stranded DNA that introduces alternative codons (purple bar) at position four, via lambda-red recombination and negative selection for loss of pheS*. DNA with AGC and AGT did not integrate (0/16 clones); we recovered one clone for AGC but sequencing revealed that it contained a mutant AAC (Asn) codon. TCT (6/8), TCC (6/16), ACA (6/8) and TTA (4/8) were allowed.
d, Recoding landscape (purple) over the genomic region shown in a, following REXER with a BAC that contained refactoring scheme 2 for the ftsI-murE overlap and TCT at position 4 in map. In total, 2/7 post-REXER clones were completely refactored and recoded, and each target codon was replaced in at least 5/7 clones. The data from a are shown in red for comparison.
Extended Data Fig. 3. Recoding rne and yceQ in fragment 9.
a, Recoding landscape of fragment 9. Our designed synthetic sequence of fragment 9 was integrated into the genome by REXER, and 19 clones were completely sequenced by next-generation sequencing. The recoding landscape graph shows the frequency at which each target codon was recoded across the 19 clones. Although most codon replacements were accepted, recoding of a 26-kb region was consistently rejected; codon positions with a recoding frequency of zero in all the sequenced clones are indicated by black dots. To pinpoint the problematic sequence, 10-kb stretches of the genome (labelled G2 to G7) were deleted in the presence of the episomal copy of synthetic fragment 9. The synthetic sequence was sufficient to support deletion of all stretches except G4 (dark grey box), which suggests that an underlying problem is within this stretch. None of the nineteen clones was completely recoded.
b, Recoding landscape of stretch G4. After REXER across the 10-kb G4 stretch, and sequencing of 10 clones, the recoding landscape shown was generated. This revealed a clear recoding minimum at yceQ—a ‘gene’ that encodes a predicted protein for which there is little evidence of transcription, protein synthesis or homologues37. All target codons in yceQ were recoded at least once in individual clones, but never simultaneously; thus, the minimum of the recoding landscape does not reach zero, and 0/10 clones were completely recoded. This is consistent with epistasis between the targeted positions. In the map below the recoding landscape, sequences annotated as essential are shown in dark grey and target codons are shown in red. The sequence position (x axis) is with reference to a.
c, Altered design of the region surrounding rne in fragment 9. Top, original design of yceQ recoding and rne (which encodes RNase E) regulatory sequences. Target codons are shown in red. P1rne, P2rne and P3rne are the promoters (blue arrows) for the essential gene rne; these are found in and around the hypothetical gene yceQ. The −10 sequence of the major promoter P1rne is mutated by our initial design. The sequences that contains hairpin 1 (hp1) and hairpin 2 (hp2), which bind to RNase E to mediate transcript degradation, are shown as blue bars; these sequences encompass the remaining target codons and are also mutated by our initial design. Bottom, the second codon in yceQ was replaced with a stop codon (purple) and the remaining target codons retained their original sequence. The sequence position (x axis) is with reference to a.
d, The modified fragment 9 (from c) was integrated on the genome, which resulted in complete recoding in 4/5 clones that we sequenced. The axes of the graph are the same as in a. The recoding landscape for the modified fragment 9, derived from sequencing five clones, is shown in purple. The data from a are reproduced for comparison.
Extended Data Fig. 4. Recoding yaaY in fragment 37a.
a, Recoding landscape of fragment 37a. Our designed synthetic sequence of fragment 37a was integrated into the genome by REXER, and six clones were completely sequenced by next-generation sequencing. Although most codon replacements were accepted, recoding of a 6.5-kb region was consistently rejected. Target-codon positions that were never recoded in the six clones sequenced are indicated by black dots.
b, Identification of the problematic target codon. Within the identified 6.5-kb problematic region, we first focused on codons in essential genes (dark grey arrows) rather than non-essential genes (light grey arrows). Sanger sequencing (black bar) of 24 clones showed that 2 clones were recoded in all 6 target codons within a sub-section of the essential genes. Further Sanger sequencing of the remaining target codons in essential genes in these two clones revealed that 1 clone was recoded at all 17 target codons. This clone was completely sequenced by next-generation sequencing and used to generate a recoding landscape, in which each target codon is either recoded (red) or not recoded (black). In combination with the recoding landscape in a, this enabled us to identify a problematic region 1.8-kb upstream of ribF. Here we focused on the four target codons in the genes rpsT and yaaY as the nearest codons to the essential ribF gene. Sanger sequencing of 33 clones across this sequence revealed only 1 codon that was never recoded—the codon for Ser70 in the hypothetical gene yaaY (sequencing results are shown as colour-coded on the gene map of rspT and yaaY). We therefore investigated alternative codon replacements in yaaY.
c, Alternative codon replacement in the hypothetical gene yaaY. At position Ser70 in this gene, replacement of TCA with AGT was not successful. To investigate alternative codon replacement schemes, a double-selection marker (pheS*-HygR) on a constitutive EM7 promoter, followed by a ribosome-binding site, was introduced into yaaY, 12 bp upstream of the codon for Ser70. The negative-selection marker was then used to select for clones that had replaced the cassette using linear doublestranded DNA that introduces alternative codons (purple bar) at position 70, via lambda-red recombination. Although linear double-stranded DNA with AGT did not integrate (0/16 clones), integration of double-stranded DNA with TCC (2/16), TCG (2/16), TCT (6/16) and AGC (9/16) proved viable.
d, Recoding landscape following REXER with a BAC that contains a corrected version of fragment 37a, bearing AGC at position Ser70 in the hypothetical gene yaaY (purple). When integrated by REXER, we identified 1/7 completely recoded clones. AGC at position Ser70 in yaaY was introduced in 4/7 clones.
Extended Data Fig. 5. Substitutions in the hypothetical gene yceQ overlap with regulatory elements in rne.
a, In our original design, a programmed substitution of a TCA (blue) to AGT (red) in the hypothetical gene yceQ leads to mutation of the −10 region of the P1rne promoter (boxed). The transcriptional start site (tss) of this promoter for rne transcription is indicated by an arrow; this is the major promoter for rne transcription.
b, Target-codon substitutions overlap with and may potentially disrupt the key regulatory hairpins (hp2 and hp3) in the long 5′ untranslated region of the rne transcript. hp2 and hp3 mediate a regulatory feedback loop, in which RNase E is recruited to the mRNA to promote degradation of its own transcript. A schematic of the wild-type secondary structure of the rne 5′ untranslated region is shown40. The target codons for synonymous replacement are highlighted in blue.
Extended Data Fig. 6. Completing sections A, B and H.
a, GENESIS was initiated with fragment 4 and proceeded smoothly until fragment 9, in which we were unable to recode yceQ. Identifying and fixing the problems with our initial design of fragment 9 was carried out as described in Extended Data Fig. 3, by introducing a stop codon (yellow line) at the start of the predicted yceQ ORF. Following a swap of the sacB-cat (sC) double-selection cassette at the end of fragment 9 for a pheS*-HygR (pH) double selection cassette, this strain was ready to act as the recipient for conjugation to assemble a strain in which fragments 4–13 (section A plus section B) are fully recoded. In parallel, we continued to recode the strain that contains the recoded fragment 4 to incomplete fragment 9 by GENESIS; this generated a second strain for assembly in which fragments 4–8 and 10–13 were completely recoded, and fragment 9 was partially recoded. We then integrated oriT (white triangle) 3 kb upstream of the start of fragment 10 in the second strain to generate a donor for conjugation, to assemble a strain in which fragments 4–13 (section A plus section B) are fully recoded. Conjugation of the donor and recipient strains resulted in a strain in which sections A and B are fully recoded. rK, rpsL-kanR double-selection cassette.
b, Individual REXER of fragments 37a and 1 led to incomplete recoding. We carried out troubleshooting of both fragments independently (Extended Data Figs. 2, 4). The repairs are indicated with yellow and purple lines in fragment 37a and fragment 1, respectively. Each strain then served as a starting point for two independent sets of GENESIS; one generated 37a–37b (on the left) and ended in an rpsL-kanR double-selection cassette, and one generated 1–3 (on the right) and ended in a sacB-cat double-selection cassette. We integrated an oriT (white triangle) 3 kb upstream of the start of fragment 1, and this strain served as a donor for the directed conjugation of 1–3 into 37a–37b. The correct product was selected for by the gain of cat and the loss of rpsL. This resulted in the completion of section H in a single strain.
Extended Data Fig. 7. Assembly of an organism with a fully synthetic genome through conjugation of recoded genome sections.
a, Schematic assembly of partially synthetic donor and recipient genomes into a more-synthetic genome, through conjugation. In the recipient cell, the recoded genome section (pink) is extended with recoded DNA (dark pink)—commonly, 3–4 kb—by a lambda-red-mediated recombination and positive and negative selection; this step takes advantage of the genomic markers at the end of the recoded sequence that are introduced by GENESIS, and provides a homology region with the end of the recoded fragment in the donor strain. The donor strain is prepared by integration of an oriT at the end of the recoded DNA. The indicated positive and negative selection ensures the survival of recipient strains, and selects for recipients that have successfully integrated the synthetic DNA from the donor. An F′ plasmid that contains a mutation in the oriT sequence that makes it non-transferrable was used to facilitate conjugation of the donor genome to the recipient. +2, cat; −2, sacB; +3, HygR; −3, pheS*; +4, aacC1 (a gene conferring gentamycin resistance); +5, tetA (a gene conferring tetracycline resistance). The homologous regions in the donor and recipient are both shown in dark pink.
b, Synthetic genomic sections (pink) from multiple individual partially recoded genomes were assembled into a single fully recoded genome using conjugative assembly. The donor (d) and recipient (r) strains contain unique recoded genomic sections labelled in pink; recoded overlapping homology regions (3 kb to 400 kb in size) were used to seamlessly recombine the strains, and are shown in dark pink. Small homology regions ranging from 3 to 5 kb in size are denoted with an asterisk. Conjugations for which we used greater than 5-kb homology (HR) are indicated. For assembly, the recoded genomic content from the donor was conjugated in a clockwise manner to replace the corresponding wild-type genomic section (grey) in the recipient. The origin of strain AB and strain H is described in detail in Extended Data Fig. 6; all other individual synthetic genomes were generated by GENESIS (Extended Data Fig. 1). Conjugation followed by recombination proceeded until the final fully recoded A–H strain was assembled and sequence-verified by next-generation sequencing.
Extended Data Fig. 8. Characterization of an organism with a fully synthetic genome.
a, Doubling times for Syn61 and MDS42. Our fully synthetic recoded E. coli Syn61 has a doubling time that is 1.6x longer than that of MDS4232, when grown in standard medium conditions (90.1 min versus 57.6 min in lysogeny broth (LB) + 2% glucose). The ratio of growth rates between Syn61 and MDS42 in LB (decreased carbon catabolite repression) at 37 °C is 1.7, in M9 minimal medium is 1.7, in richer medium (2XTY) is 1.4, in LB at 25 °C is 2.5 and in LB at 42 °C is 1.3. The doubling times in different medium conditions are: LB at 37 °C, 58.3 min and 100.6 min; LB + 2% glucose, 57.6 min and 90.1 min; M9 minimal medium, 130.5 min and 221.1 min; 2XTY, 68.2 min and 92.6 min; LB at 25 °C, 86.3 min and 218.4 min; LB at 42 °C, 77.4 min and 99.7 min, for MDS42 and Syn61, respectively. Syn61 containing a plasmid without (−) or with (+) serV exhibited a growth-rate ratio of 0.99 (138.3 min versus 136.2 min). Doubling times represent the average of ten independently grown biological replicates of each strain, and are shown as mean ± s.d. (see Supplementary Methods). The data for individual experiments are represented by dots.
b, Representative microscopy images of E. coli strain MDS42 and Syn61. Samples were imaged on an upright Zeiss Axiophot phase-contrast microscope using a 63X 1.25 NA Plan Neofluar phase objective (see Supplementary Methods). The experiment was performed twice with similar results.
c, Histogram of cell lengths quantified from microscopy images of strains MDS42 and Syn61. The mean cell length (±s.d.) for MDS42 was 1.97 ± 0.57 μm and for Syn61 was 2.3 ± 0.74 μm. Images of n = 500 cells were taken during exponential growth phase for both strains. Cell-length measurements were made using Nikon NIS Elements software (see Supplementary Methods). A 1-μm lower size limit was imposed to remove background particulates and dust from quantification; this also precludes quantification of extracellular vesicles.
d, Label-free quantification of the MDS42 and Syn61 proteomes. Each strain was grown in three biological replicates. Each biological replicate was analysed by tandem mass spectrometry in technical duplicate. Technical duplicates of biological replicates were merged. A total of 1,084 proteins was quantified across the samples. No protein quantified in both MDS42 and Syn61 differed in abundance—as judged by label-free quantification values—by more than 1.16-fold.
Extended Data Fig. 9. Consequences of synonymous codon compression in Syn61.
a, Synonymous codon compression and deletion of prfA, serU and serT in E. coli. The grey boxes shows the E. coli serine codons and stop codons, together with the tRNAs and release factors that decode them in wild-type E. coli (WT genome). tRNA anticodons and release factors are connected to the codons that they are predicted to read by black lines. The tRNA and release factor genes are shown in the black boxes. Synonymous codon compression (syn. codon. comp.) leads to Syn61 cells with a recoded genome (pink boxes), in which TCG and TCA codons are removed. The abundance of each codon is listed in its box.
b, As in Fig. 4b, but with the M. mazei PylRS/tRNAPylUGA pair (anticodon UGA). There are fewer cognate codons to this anticodon in Syn61 than in MDS42; CYPK addition might therefore be expected to be less toxic in Syn61, as observed.
c, As in Fig. 4b, but with the M. mazei PylRS/tRNAPylGCU pair (anticodon GCU). There are a greater number of cognate codons to this anticodon in Syn61 than in MDS42; CYPK addition might therefore be expected to be more toxic in Syn61, as observed.
d, serT (dark grey) is deleted by insertion of a PheS*-HygR double-selection cassette (black) via lambda-red-mediated recombination. Recombination yields new junctions 1 and 2, indicated by green and blue bars. For each recombination, both junctions were sequence-verified by Sanger sequencing. Above the Sanger chromatograms, the arrows indicate the precise location of the junction, the blue bar indicates the sequence that corresponds to the selection cassette and the green bar corresponds to the genomic sequence that flanks the selection cassette. The primers used to generate selection cassettes with suitable homologies to serU, serT and prfA for recombination are provided in Supplementary Data 21. The experiment was performed once.
e, prfA (dark grey) is deleted by the insertion of an rpsL-kanR double-selection cassette (in black) via lambda-red-mediated homologous recombination. The agarose gels are annotated as described in Fig. 4c, and the rest of the data are annotated as described in d. The experiment was performed once.
f, serU (dark grey) is deleted by insertion of a PheS*-HygR double-selection cassette (in black) via lambda-red-mediated recombination. The agarose gels are annotated as described in Fig. 4c, and the rest of the data are annotated as described in d. The experiment was performed once. The full gels are available in Supplementary Fig. 1.
Extended Data Fig. 10. The scale of genome synthesis, and scale and fidelity of recoding.
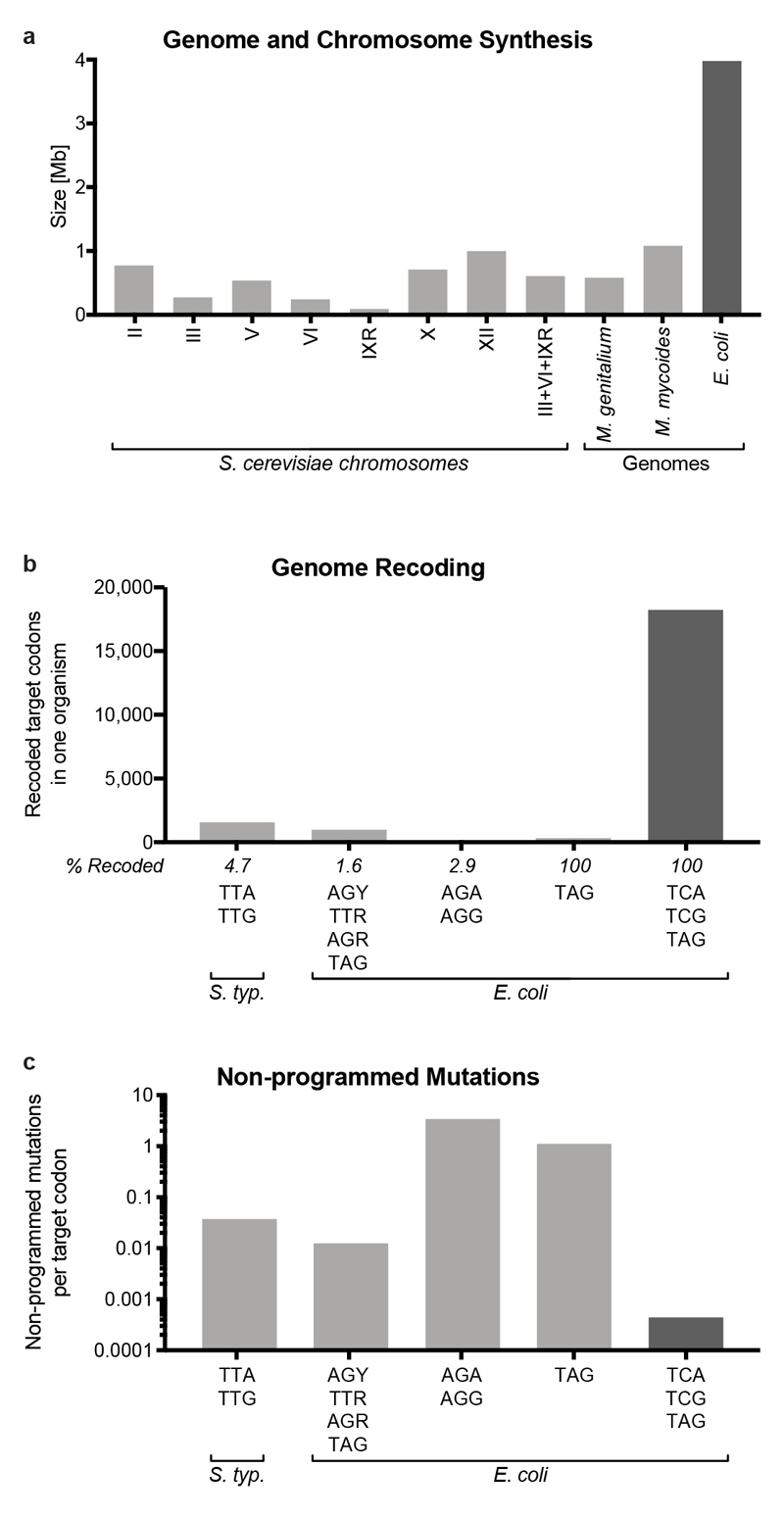
a, Genome and chromosome synthesis. The size (in Mb) of synthetic genomes that have been produced for M. genitalium and M. mycoides22,23, and several S. cerevisiae chromosomes24–31 (light grey). The size of the synthetic E. coli genome presented here is shown in dark grey.
b, Genome recoding efforts. Attempts to recode target codons TTA and TTG in Salmonella enterica serovar Typhimurium LT218; AGC, AGT, TTG, TTA, AGA, AGG and TAG in E. coli19; AGA and AGG in E. coli16, as well as recoding of all TAG in E. coli14 (light grey), compared to the removal of all TCA, TCG and TAG in E. coli presented here (dark grey). The total number of codons recoded in a single strain is shown on the graph, and the maximum percentage of target codons recoded in a single strain in each effort is indicated.
c, Number of reported non-programmed mutations and indels as a function of the number of target codons recoded for the experiments shown in b.
Supplementary Material
Acknowledgements
This work was supported by the Medical Research Council (MRC), UK (MC_U105181009 and MC_UP_A024_1008), the Medical Research Foundation (MRF-109-0003-RG-CHIN/C0741) and an ERC Advanced Grant SGCR, all to JWC, and by the Lundbeck Foundation (R232-2016-3474) to JF. J.W.C. thanks H. Pelham for supporting this project. We are grateful to Mark Skehel and the MRC-LMB mass spectrometry service for LFQ-based proteomics, Nick Barry for microscopy, and Alastair Crisp for helping with Python scripts. We also thank Christopher J. K. Wan, Samuel H. Kim, Lavinia Dunsmore, Nicolas Huguenin-Dezot, and Stephen D. Fried for their support in experimental work.
Footnotes
Author contributions:
K.W. and T.C. designed the target genome sequence. T.C. generated scripts for data analysis. All authors, except T.S.E., contributed to assembly of sections. J.F., L.F.H.F., K.W. and A.G.L. led the fixing of deleterious synthetic sequences. J.F., D.d.l.T., L.F.H.F., W.E.R. and Y.C. led the assembly of sections into Syn61 and characterized the strain with the assistance of T.S.E. J.W.C. supervised the project and wrote the paper with the other authors.
Competing interests
The authors declare no competing interests.
Author Information
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
The sequences and genome design details used in this study are available in the Supplementary Data. Supplementary Data 1 provides the GenBank file of the E. coli MDS42 genome (NCBI accession number AP012306.1); Supplementary Data 2 provides the GenBank file of the designed synthetic E. coli genome with codon replacements and refactorings; Supplementary Data 3 provides the table of target codons; Supplementary Data 4 provides the table of overlaps and refactoring; Supplementary Data 5 provides the table of 10-kb stretches; Supplementary Data 6 provides the GenBank file of the BAC sacB-cat-rpsL; Supplementary Data 7 provides the GenBank file of BAC-rpsL-kanR-sacB; Supplementary Data 8 provides the GenBank file of the BAC rpsL-kanR-pheS*-HygR; Supplementary Data 9 provides the table of BAC construction; Supplementary Data 10 provides the table of BAC assembly; Supplementary Data 11 provides the table of REXER experiments; Supplementary Data 12 provides the GenBank file of spacer plasmids without trans-activating CRISPR RNA (tracrRNA) and annotation for linear spacers; Supplementary Data 13 provides the GenBank file of spacer plasmids with tracrRNA and annotation for linear spacers; Supplementary Data 14 provides the table of oligonucleotides used for recoding fixing experiments; Supplementary Data 15 provides the GenBank file of the gentamycin-resistance oriT cassette; Supplementary Data 16 provides the oligonucleotide primers used for conjugation; Supplementary Data 17 provides the GenBank file of the pJF146 F′ plasmid that does not self-transfer; Supplementary Data 18 provides the GenBank file of the fully recoded genome of Syn61, verified by next-generation sequencing; Supplementary Data 19 provides the table of design optimizations and non-programmed mutations; Supplementary Data 20 provides a list of the proteins identified by tandem mass spectrometry; and Supplementary Data 21 provides a list of the primers used for deletion experiments. All other datasets generated and/or analysed in this study are available from the corresponding author upon reasonable request. All materials (Supplementary Data 9, 12, 13, 17, 18) from this study are available from the corresponding author upon reasonable request.
Code availability
Code used for genome design is available at https://github.com/TiongSun/genome_recoding; for sequencing at https://github.com/TiongSun/iSeq; and for generating recoding landscapes at https://github.com/TiongSun/recoding_landscape.
References
- 1.Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 2.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho BK, et al. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 6.Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- 7.Kimchi-Sarfaty C, et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 8.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 9.Mittal P, Brindle J, Stephen J, Plotkin JB, Kudla G. Codon usage influences fitness through RNA toxicity. Proc Natl Acad Sci U S A. 2018;115:8639–8644. doi: 10.1073/pnas.1810022115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cambray G, Guimaraes JC, Arkin AP. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli. Nat Biotechnol. 2018;36:1005–1015. doi: 10.1038/nbt.4238. [DOI] [PubMed] [Google Scholar]
- 11.Quax TE, Claassens NJ, Soll D, van der Oost J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol Cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin JW. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 13.Mukai T, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38:8188–8195. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukai T, et al. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci Rep. 2015;5 doi: 10.1038/srep09699. 9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Napolitano MG, et al. Emergent rules for codon choice elucidated by editing rare arginine codons in Escherichia coli. Proc Natl Acad Sci U S A. 2016;113:E5588–5597. doi: 10.1073/pnas.1605856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau YH, et al. Large-scale recoding of a bacterial genome by iterative recombineering of synthetic DNA. Nucleic Acids Res. 2017;45:6971–6980. doi: 10.1093/nar/gkx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 20.Mukai T, et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res. 2015;43:8111–8122. doi: 10.1093/nar/gkv787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchison CA, 3rd, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351 doi: 10.1126/science.aad6253. aad6253. [DOI] [PubMed] [Google Scholar]
- 22.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 23.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355 doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie ZX, et al. "Perfect" designer chromosome V and behavior of a ring derivative. Science. 2017;355 doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell LA, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355 doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 28.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science. 2017;355 doi: 10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science. 2017;355 doi: 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- 31.Richardson SM, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 32.Posfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 33.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100025. 2005 0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corey EJ, Cheng X-m. The logic of chemical synthesis. John Wiley; 1989. [Google Scholar]
- 35.Kouprina N, Noskov VN, Koriabine M, Leem SH, Larionov V. Exploring transformation-associated recombination cloning for selective isolation of genomic regions. Methods Mol Biol. 2004;255:69–89. doi: 10.1385/1-59259-752-1:069. [DOI] [PubMed] [Google Scholar]
- 36.Goodall ECA, et al. The Essential Genome of Escherichia coli K-12. MBio. 2018;9 doi: 10.1128/mBio.02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pundir S, Martin MJ, O'Donovan C. UniProt Protein Knowledgebase. Methods Mol Biol. 2017;1558:41–55. doi: 10.1007/978-1-4939-6783-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claverie-Martin F, Diaz-Torres MR, Yancey SD, Kushner SR. Analysis of the altered mRNA stability (ams) gene from Escherichia coli. Nucleotide sequence, transcriptional analysis, and homology of its product to MRP3, a mitochondrial ribosomal protein from Neurospora crassa. J Biol Chem. 1991;266:2843–2851. [PubMed] [Google Scholar]
- 39.Jain C, Belasco JG. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- 40.Diwa A, Bricker AL, Jain C, Belasco JG. An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes Dev. 2000;14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- 41.Schuck A, Diwa A, Belasco JG. RNase E autoregulates its synthesis in Escherichia coli by binding directly to a stem-loop in the rne 5' untranslated region. Mol Microbiol. 2009;72:470–478. doi: 10.1111/j.1365-2958.2009.06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma NJ, Moonan DW, Isaacs FJ. Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering. Nat Protoc. 2014;9:2285–2300. doi: 10.1038/nprot.2014.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 45.Elliott TS, Bianco A, Townsley FM, Fried SD, Chin JW. Tagging and Enriching Proteins Enables Cell-Specific Proteomics. Cell chemical biology. 2016;23:805–815. doi: 10.1016/j.chembiol.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elliott TS, et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nature biotechnology. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krogager TP, et al. Labeling and identifying cell-specific proteomes in the mouse brain. Nat Biotechnol. 2018;36:156–159. doi: 10.1038/nbt.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neidhardt FC. Escherichia coli and Salmonella typhimurium : cellular and molecular biology. American Society for Microbiology; 1987. [Google Scholar]
- 49.Strand TA, Lale R, Degnes KF, Lando M, Valla S. A new and improved host-independent plasmid system for RK2-based conjugal transfer. PLoS One. 2014;9:e90372. doi: 10.1371/journal.pone.0090372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryksin AV, Matsumura I. Rational design of a plasmid origin that replicates efficiently in both gram-positive and gram-negative bacteria. PLoS One. 2010;5:e13244. doi: 10.1371/journal.pone.0013244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 55.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.