Abstract
Inflammatory bowel diseases (IBD) are chronic and relapsing inflammatory conditions of the gut that include Crohn's disease and ulcerative colitis. The pathogenesis of IBD is not completely unraveled, IBD are multi-factorial diseases with reported alterations in the gut microbiota, activation of different immune cell types, changes in the vascular endothelium, and alterations in the tight junctions’ structure of the colonic epithelial cells. Proteomics represents a useful tool to enhance our biological understanding and to discover biomarkers in blood and intestinal specimens. It is expected to provide reproducible and quantitative data that can support clinical assessments and help clinicians in the diagnosis and treatment of IBD. Sometimes a differential diagnosis of Crohn's disease and ulcerative colitis and the prediction of treatment response can be deducted by finding meaningful biomarkers. Although some non-invasive biomarkers have been described, none can be considered as the “gold standard” for IBD diagnosis, disease activity and therapy outcome. For these reason new studies have proposed an “IBD signature”, which consists in a panel of biomarkers used to assess IBD. The above described approach characterizes “omics” and in this review we will focus on proteomics.
Keywords: Proteomics, Inflammatory bowel disease, Cronh’s disease, Ulcerative colitis, Proteins, Biomarkers discovery
Core tip: Patients' heterogeneity is a hallmark for inflammatory bowel diseases (IBD). Some patients present limited bowel involvement and a mild course of the disease, others develop very extensive, aggressive disease and variable response to therapy. In IBD, there is a great need of patient stratification and of new biomarkers as part of a personalized medicine approach to patient care. Biological therapies are more and more widely used for IBD patients, because of their efficacy in patient’s refractory to other drugs; still, biological treatments fail in 20%-40% of patients and, to date, no reliable clinical or molecular predictor of response to biological therapeutic strategy has been described. This review aims to collect the "omics" approach for research of serological biomarkers of diagnosis, response to specific biological therapies in the IBD field.
INFLAMMATORY BOWEL DISEASE
Ulcerative colitis (UC) and Crohn’s disease (CD) are the two main inflammatory bowel diseases (IBD)[1-4]. Despite some shared characteristics, they can be distinguished by differences in genetic predisposition, risk factors, and clinical, endoscopic and histological features. CD is characterized by diffuse chronic inflammation throughout the gastrointestinal tract, in a non-continuous manner[5]; UC presents with inflammation limited to the colon, spreading continuously from the rectum[6]. The pathogenesis of IBD is at present not completely unraveled; however, genetically susceptible individuals seem to have a dysregulated mucosal immune response to the commensal gut flora, which results in bowel inflammation[7]. IBD are multi-factorial diseases[8] with reported alterations in the gut microbiota[9-12], activation of different immune cell types[13-15], changes in the vascular endothelium[16,17], and alterations in the tight junctions structure of colon epithelial cells[18-20].
Nowadays, the diagnostic and prognostic tools for IBD and the outcome of therapy are largely based on evaluation of clinical symptoms in combination with endoscopy, histology, radiology and non-specific biomarkers from serum or stools[21].
BIOMARKERS IN INFLAMMATORY BOWEL DISEASE
Inflammation in IBD is characterized by the increased levels of some molecules extensively validated but not all included in the laboratory routine. Some of them are related to the inflammatory acute-phase response, coagulation and fibrinolysis (fibrinogen, plasminogen, complement components), proteinase inhibitors (α1-antitrypsin and α1-anti-chymotrypsin), transport proteins (haptoglobin and ceruplasmin) and other serum proteins[22] and cytokines[23]. Elevated platelet and white blood cell counts may also indicate inflammation but they cannot be considered strictly related to bowel inflammation[23]. C-reactive protein (CRP), anti-Saccaromyces cerevisiae (ASCA) and anti-neutrophil cytoplasmic antibody are the most widely used indicators. CRP has a short reaction time (6-10 h) and it is useful for the identification of inflammatory disease activity especially in CD, but not in UC[24]. CRP has low specificity enabling to differentiate between CD, UC and infectious colitis[21], and also the 25% of IBD patients with demonstrable disease activity have CRP levels above the normal threshold[22]. ASCA is an antibody used for the identification of CD patients who are often positive (39%-79% of CD patients, 5%-15% UC patients)[25,26], however a large part of healthy controls is also positive (14%-18%) to this antibody, limiting the diagnostic value of its detection[27]. anti-neutrophil cytoplasmic antibodies are antibodies found in immune-mediated pathologies, such as rheumatoid arthritis and Wegener’s granulomatosis[28], and have shown a different staining pattern in UC and CD patients[29-31], but as for ASCA 32% of healthy population is also positive to them[32].
Another explored field in the search for IBD biomarkers is the analysis of stool proteins, which can be dysregulated or abnormally present in patients. Stool markers have the advantage of increased specificity for bowel inflammation and reflect any mucosal barrier disruption. Fecal markers can be useful to diagnose CD, where inflammation is patchy and is possibly missed at endoscopy[33]. Fecal calprotectin (FC) accounts for up to 5% of the neutrophil granulocytes’ protein content with chemotactic and antimicrobial activities. It is stable in stool for more than a week and can resists to bacterial degradation[34]. FC is not a specific marker for IBD, but it correlates with increased disease activity at least in adults[35], but not in pediatric patients where was found with high sensitivity (98%), but only modest specificity (68%)[36]. Disease location should also be taken into account when interpreting FC levels. Patients with ileal CD may have ulcers even in the absence of markedly elevated FC levels. Consequently, the cut-off values for ileal CD may differ from those with ileocolic disease[37,38]. A study conducted by De Vos et al[39] has demonstrated that Calprotectin decreased 2 wk after Infliximab administration predicts remission in anti-TNF-naïve patients with UC. The increase of FC can also be a suitable marker for the identification of relapse, given the fact that the levels are increased as early as 6 mo before clinical and endoscopic relapse[40]. Lactoferrin is an iron-binding protein expressed by neutrophils during inflammation and represents a defense against infection as part of the innate immune system[41,42]. As a biomarker, Lactoferrin can distinguish IBD from Irritable Bowel Syndrome, but not between CD and UC[27].
Although many non-invasive biomarkers have been described, none can be considered as the “gold standard” for IBD diagnosis, disease activity and therapy outcome. A single ideal biomarker is very unlikely to be found. As for other pathologies as pancreatic cancer[43-46], non-small cell lung cancer[47] and colorectal cancer[48] new studies have proposed the idea of a “Biomarker Signature”, which consists in a panel of biomarkers used to assess various pathological conditions and response to therapy[49], and which is applicable also to IBD diagnosis and prognosis. Table 1 summarizes the biomarkers commonly used for IBD.
Table 1.
Biomarkers in inflammatory bowel disease
| Marker | Setting | Diagnostic accuracy | Ref. |
| C-Reactive Protein (CRP) | Serum | Higher in CD vs UC | Henriksen et al[24], 2008 |
| 25% IBD patients have levels above normal | Vermeire et al[22], 2004 | ||
| Anti-Saccharomyces cerevisiae Antibodies (ASCA) | Serum | 39%-79% CD positive | Peyrin-Biroulet et al[25], 2015; |
| 5%-15% UC positive | Reumaux et al[26], 2004 | ||
| 14%-18% HC positive | Bennike et al[27], 2014 | ||
| Anti-neutrophil cytoplasmic antibodies (ANCA) | Serum | Different pattern in CD and UC | Peeters et al[31], 2001; |
| Peyrin-Biroulet et al[30], 2007; | |||
| Reumaux et al[29], 2003 | |||
| 32% HC positive | Bernstein et al[32], 2011 | ||
| Calprotectin | Colorectal mucus | Higher in IBD vs HC | Loktionov et al[79], 2016 |
| Higher in UC vs CD | |||
| Calgranulin C (S100A12) | Higher in UC vs CD | ||
| Eosinophil-derived neurotoxin (EDN) | Higher in IBD vs HC | ||
| Higher in UC vs CD | |||
| Fecal calprotectin (FC) | Stool | It correlates with disease activity in adults | Gisbert et al[35], 2009 |
| Lactoferrin | Stool | It distinguishes IBD from IBS | Bennike et al[27], 2014 |
CD: Crohn’s disease; UC: Ulcerative colitis; HC: Healthy controls; IBS: Irritable bowel syndrome; IBD: Inflammatory bowel disease.
PROTEOMIC APPROACH TO INFLAMMATORY BOWEL DISEASE RESEARCH
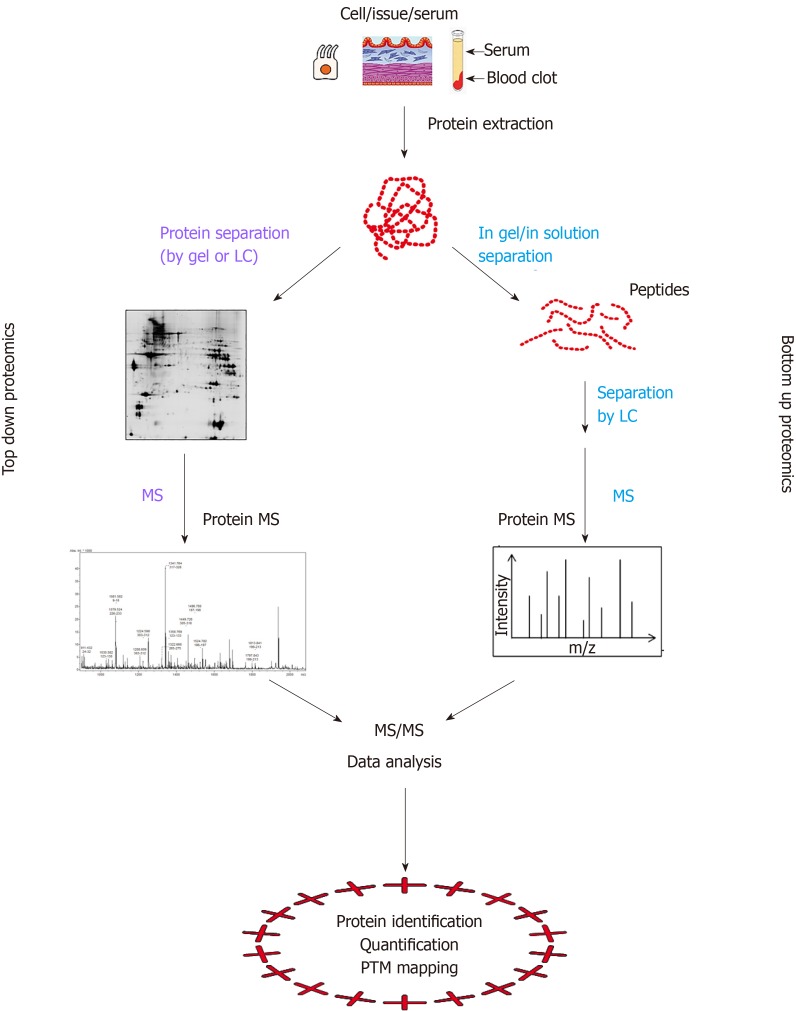
Proteomics comprehensively studies the protein composition and abundance in a given cell population and its changes under biological perturbations[50,51]. The proteome may be considered the signature of a disease, in fact it is the result of the interactions between the genetic background and environmental factors[49]. The novel proteomic technologies now facilitate the analysis of transcriptome variations also in the IBD context and have already provided with new candidate biomarkers[52]. They help to investigate the inflammatory response, epithelial barrier function and gut microbiome from different biological samples, i.e., serum/blood, colon samples and feces. The proteomic strategies can be bottom-up and top-down (Figure 1). In the bottom-up approach, purified proteins or complex protein mixtures are subjected to proteolytic cleavage and the peptide products are analyzed by mass spectrometry (MS). Conversely, the top-down approach is based either on the analysis of intact proteins followed by the direct measurement of fragment ions by MS or on the isolation of the protein by gel-based separative methods, protein gel elution and MS analysis.
Figure 1.
Schematic illustration of the difference between protein-based top-down and peptide-based bottom-up proteomics.
Proteomics in the study of IBD pathogenesis
By LC-MS analysis of colon mucosal biopsies from 10 patients with UC, Bennike et al[53] identified 5711 quantifiable proteins classified by biological function, sub-cellular location and molecular function. Forty-six proteins demonstrated statistically significant changes in mean abundance between UC biopsies and control biopsies; among those proteins, the one with the largest mean fold abundance change was lactotransferrin, which was 219 times more abundant in the UC group. The relative abundance of lactotransferrin also correlated to the severity of tissue inflammation in the patients with UC, as determined by the colon inflammation grade score based on histology. Good correlation was found between the colon inflammation grade score and the relative abundance of lactotransferrin in the tissue (0.82)[53]. Eleven of the 46 proteins identified in the UC biopsies are present in neutrophils and are associated with the formation of neutrophil extra-cellular traps which are released from neutrophils in response to inflammatory stimuli[54,55], and are a sign of chronic inflammation even in the absence of visible inflammation[54,56,57].
Proteomics has also investigated IBD-related immune-cell responses. Riaz et al[58] compared Th1 and Th17 clones isolated from the intestinal mucosa of CD patients by means of label-free quantitative mass-spectrometry analysis, which led to the identification of a total number of 7401 unique protein groups and demonstrated that 334 proteins were differentially expressed. The largest differences between the two phenotypes were observed in such proteins with cytotoxic function as Granzyme B and perforin, which are lower in Th17 cells than in Th1 cells. Other differentially expressed proteins with higher expression in the Th1 clones included several transcription factors with both known and unknown functions in CD4+ T-cells. The most striking differences at quantitative analysis are about CD4+ T cells with Th1 phenotype having a much higher degree of cytotoxic features as compared with Th1/Th17 phenotype[58].
As discussed above, the disruption of the intestinal barrier is a typical event in IBD pathogenesis. The intestinal epithelium is the largest surface exposed and coming into contact with the external environment. The intestinal epithelial cells (IECs) are the main component of the physical barrier between the luminal micro-environment and the host and act as the host’s first line-of-defense against potential harmful stimulants. They also represent the innate immunity within the gut mucosa[59]. Normally, the intestinal epithelium is covered by a single layer of IECs, which are characterized by a fast renewal rate, and act as a protective barrier against luminal antigens, but this barrier can be damaged, thus promoting a state of chronic inflammation due to mucosal immune cell infiltration, as is typically observed in IBD patients[59]. The molecular changes in the epithelial layer, extra-cellular matrix and junction proteins in inflamed and non-inflamed intestinal tissue have been only partially addressed to date. In 2012 Poulsen et al[60] analyzed the proteomic profiles of whole colonic biopsies from UC patients using 2D-gel electrophoresis and MALDI-TOF MS for the identification of differently expressed protein spots. Forty-three proteins were identified differentially expressed between UC inflamed and non-inflamed tissue, including proteins involved in the energy metabolism and in oxidative stress[60]. Proteomic studies on isolated IECs obtained from surgical specimens of full-thickness colonic tissues from UC-, CD-affected patients and non-inflamed controls were analyzed by gel-based stable-isotope label technologies (2D-DIGE and ICPL LC-MS/MS) and immunoblot assay to evaluate any proteome changes. Moreover, the results were verified on a group of patients not participating in the discovery phase[61]. The differential proteomic approaches have revealed changes in several molecules involved in extracellular matrix, mechano-transduction, metabolic rewiring and autophagy that characterize quiescent UC and quiescent CD epithelial cells and they may help understanding the complex mechanisms associated to IBD. UC patients are characterized by cytoskeletal rearrangement and increased level of specific enzymes that contribute to cell homeostasis, enabling cells to cope with energy requirements and macro-autophagy. CD patients are characterized by metabolic rewiring to sustain the cell metabolism, whereas autophagy and cell renewal are blunted[61-63]. Table 2 provides a summary of the proteins and pathways identified by the proteomic approach as involved in IBD pathogenesis.
Table 2.
Proteomics in inflammatory bowel disease pathogenesis
| Protein | Setting | Diagnostic accuracy | Ref. |
| Lactotransferrin | UC vs HC biopsies | It correlates to the colon inflammation grade score | Bennike et al[53], 2015 |
| Neutrophil extracellular traps (NETs) | Sign of chronic inflammation | ||
| Granzyme B and Perforin | CD Th1 and Th17 clones from intestinal mucosa | Higher in Th1 vs Th17 | Riaz et al[58], 2016 |
| RORC and FOXP3 | |||
| Glycerol-3-phosphatedehydrogenase | UC biopsies inflamed vs non-inflamed | Higher in inflamed vs non-inflamed tissue | Poulsen et al[60], 2012 |
| Alphaenolase | Lower in inflamed vs non- inflamed tissue | ||
| Keratins 10, 14, 19 | UC intestinal epithelial cells | Higher in QUC vs HC | Moriggi et al[61], 2017 |
| Keratin 8 | Lower in QUC vs HC | ||
| Tricarboxylic acid cycle enzymes | |||
| Oxidative phosphorylation enzymes | |||
| Vinculin and α-tubulin | |||
| Keratin 8, 18 | CD intestinal epithelial cells | Lower in QCD vs HC | |
| Heat shock cognate-70 (HSC70) | |||
| Vinculin and α-tubulin | Higher in QCD vs HC | ||
| Fibrinopeptide A (FPA) | CD serum | Higher in CD vs HC | Nanni et al[62], 2009 |
| Complement 3 protein (C3) | |||
| Apolipoprotein A-IV | |||
| Apolipoprotein E | Lower in CD vs HC | ||
| L-lactate dehydrogenase | IBD and HC intestinal epithelial cells | Higher in IBD vs HC; Higher in CD vs UC | Shkoda et al[63], 2007 |
| Carbonyl reductase | |||
| Keratin 19 | |||
| Rho-GDI dissociation inhibitor α | |||
| Annexin 2 | UC intestinal epithelial cells | Higher in UC vs HC | |
| Programmed cell death protein 8 (PDCD8) |
IBD: Inflammatory bowel disease; CD: Crohn’s disease; UC: Ulcerative colitis; QCD: Quiescent Crohn’s disease; QUC: Quiescent ulcerative colitis; HC: Healthy controls; CRC: Colorectal carcinoma.
Proteomics for the identification of novel biomarkers
Another approach is the identification of biomarkers useful for the diagnosis, treatment selection and response monitoring. A recent study focused on diagnosis has identified a serological panel which demonstrates transmural intestinal injury and is able to indicate complications in CD patients with 70% sensitivity and 72.5% specificity[64]. The increase of circulating epithelial component proteins may be a sign of transmural intestinal injury and stricturing or fistulizing intestinal complications. The serum biomarkers for the stratification of IBD patients are unable to distinguish between CD and UC[65], while the proteomic profiles of colon biopsies can identify a more precise signature of these diseases[61,66,67]. In 2016 Starr et al[68] established two candidate biomarker panels: A 5-protein panel to discriminate IBD from control patients and a 12-protein panel to distinguish CD from UC patients in children with a new IBD diagnosis.
Proteomics has been applied to the identification of treatment-response biomarkers. The anti-TNF drug called Infliximab is one of the most used drugs in IBD, but the factors predicting the response and the molecular mechanisms that are related to the loss of response or non-responsiveness are not completely known. Meuwis et al[69] have analyzed sera from responder and non-responder CD patients at baseline and then comparing sera throughout the induction period (week 4 for non-fistulizing and week 10 for fistulizing patients) and have shown that the platelet aggregation Factor 4 (PF4) was higher in non-responders than responders to Infliximab therapy (both before and after treatment). PF4 is considered as an acute-phase reactant because its level increases with general inflammation, as already observed in the plasma of CD patients[70-72] . Gazouli et al[73] have compared sera before treatment and after IFX induction (week 12) and successfully identified 15 proteins that were differentially accumulated in the sera, most of them modifying the activation of monocytes /macrophages and directly and indirectly regulating the differentiation and activation of CD4+ T-lymphocytes. Also, a recent study by Magnusson et al[74] reported on the proteomic analysis on biopsies obtained from 6 UC patients (3 responders and 3 non-responders) treated in vitro with or without Infliximab and also from 43 UC patients’ sera at different time points: Baseline, week 2 and week 14. Those authors have shown that the response in UC patients is associated with reduced monocyte activation 2 wk after therapy initiation, suggesting that the monocytes of these patients are less responsive to inflammatory stimuli when reaching the intestinal mucosa. In therapy responders Infliximab has had influence on Tenascin C, which might be a down-regulator of the two chemokines CCL2 (mcp-1) and CXCL10 (IP-10)[74], which are produced by inflammatory cells and stromal cells, recruit leucocytes, and are induced in inflamed UC mucosa[75-77]. Table 3 summarized the potential biomarkers identified by proteomics in IBD.
Table 3.
Proteomics in inflammatory bowel disease diagnosis and response to therapy
| Proteins | Setting | Diagnostic accuracy | Ref. |
| Platelet aggregation factor 4 (PF4) | Responder vs non-responder’s CD serum | Higher in non-responders | Mewuis et al[69], 2008 |
| Proteins that regulate CD4+ T-cell activation | Serum before IFX treatment vs serum after IFX induction period | Higher before treatment | Gazouli et al[73], 2013 |
| Proteins that regulate monocytes/macrophages activation | |||
| Tenascin C | Responder vs non-responder’s UC serum | Higher in non-responders | Magnusson et al[74], 2015 |
CD: Crohn’s disease; UC: Ulcerative colitis.
CONCLUSION
In the IBD micro-environment a multitude of components interact. No information about a single gene, a single molecule or microbe can exhaustively explain the events that result from such a complex signaling. Also, the wide range of variability between patients’ disease features and medical histories makes it difficult to understand how every component of IBD acts and influences other components. On the other hand, even if the diagnostic gold standard is endoscopy, the introduction of novel molecular biomarkers in clinical practice has always nurtured hopes for new tools that can lead to improvements in diagnostic accuracy. However, the low diagnostic performance of the available markers strongly limits their use in clinical practice. Still, it is reasonable to hypothesize that combining the modification of several biomarkers may identify a sort of fingerprint for IBD with specific disease features.
Indeed, techniques and methodologies that can deal with a very large volume of data and describe a wide picture, rather than focus on single alteration, are likely to represent the necessary step forward in describing and comprehending IBD[78]. For all these reasons omics can support the discovery of novel molecular interactions through a better definition of relevant biological pathways and interactions, rather than the analysis of the role of the perturbation of a single element. Omics can lead to the identification of representative patterns of disease which may replace simple biomarkers in clinical practice for the diagnosis, monitoring of IBD and for the personalization of therapies and treatments. Exploiting omic techniques and mastering big data analysis will help researchers to embrace the complexity and overcome the limitations of deciphering inflammatory disorders away from any restricted point of view. Table 3 provides a summary of the potential biomarkers identified by proteomics in IBD.
Footnotes
Conflict-of-interest statement: There is no conflict of interest associated with any of the senior author or other coauthors contributed their efforts in this manuscript.
Manuscript source: Invited manuscript
Peer-review started: November 25, 2019
First decision: December 23, 2019
Article in press: February 10, 2020
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Poullis A, Can G S-Editor: Dou Y L-Editor: A E-Editor: Ma YJ
Contributor Information
Laura Francesca Pisani, Gastroenterology and Digestive Endoscopy Unit, IRCCS Policlinico San Donato, San Donato Milanese 20097, Italy.
Manuela Moriggi, Gastroenterology and Digestive Endoscopy Unit, IRCCS Policlinico San Donato, San Donato Milanese 20097, Italy.
Cecilia Gelfi, Department of Biomedical Science for Health, University of the Study of Milan, IRCCS Istituto Ortopedico Galeazzi, Milan 20122, Italy.
Maurizio Vecchi, Gastroenterology and Endoscopy Unit, IRCCS Ca' Granda Foundation, Policlinico Hospital, University of the Study of Milan, Milan 20122, Italy.
Luca Pastorelli, Gastroenterology and Digestive Endoscopy Unit, IRCCS Policlinico San Donato, San Donato Milanese 20097, Italy; Department of Biomedical Science for Health, University of the Study of Milan, Milan 20122, Italy. luca.pastorelli@unimi.it.
References
- 1.Odze R. Diagnostic problems and advances in inflammatory bowel disease. Mod Pathol. 2003;16:347–358. doi: 10.1097/01.MP.0000064746.82024.D1. [DOI] [PubMed] [Google Scholar]
- 2.Bosani M, Ardizzone S, Porro GB. Biologic targeting in the treatment of inflammatory bowel diseases. Biologics. 2009;3:77–97. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Ullman T, Lazarev M. Scope early and often in ulcerative colitis and Crohn's colitis? Gastroenterology. 2009;136:718–9; discussion 719-20. doi: 10.1053/j.gastro.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 4.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 5.Baumgart DC, Sandborn WJ. Crohn's disease. Lancet. 2012;380:1590–1605. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 6.Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380:1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 7.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiocchi C. Genes and 'in-vironment': how will our concepts on the pathophysiology of inflammatory bowel disease develop in the future? Dig Dis. 2012;30 Suppl 3:2–11. doi: 10.1159/000342585. [DOI] [PubMed] [Google Scholar]
- 9.Yu CG, Huang Q. Recent progress on the role of gut microbiota in the pathogenesis of inflammatory bowel disease. J Dig Dis. 2013;14:513–517. doi: 10.1111/1751-2980.12087. [DOI] [PubMed] [Google Scholar]
- 10.Vetrano S, Danese S. Colitis, microbiota, and colon cancer: an infernal triangle. Gastroenterology. 2013;144:461–463. doi: 10.1053/j.gastro.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blumberg R. What are innate and acquired immunity, and why are they important in IBD? Inflamm Bowel Dis. 2008;14 Suppl 2:S93–S94. doi: 10.1002/ibd.20689. [DOI] [PubMed] [Google Scholar]
- 14.Wallace KL, Zheng LB, Kanazawa Y, Shih DQ. Immunopathology of inflammatory bowel disease. World J Gastroenterol. 2014;20:6–21. doi: 10.3748/wjg.v20.i1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cader MZ, Kaser A. Recent advances in inflammatory bowel disease: mucosal immune cells in intestinal inflammation. Gut. 2013;62:1653–1664. doi: 10.1136/gutjnl-2012-303955. [DOI] [PubMed] [Google Scholar]
- 16.D'Alessio S, Tacconi C, Fiocchi C, Danese S. Advances in therapeutic interventions targeting the vascular and lymphatic endothelium in inflammatory bowel disease. Curr Opin Gastroenterol. 2013;29:608–613. doi: 10.1097/MOG.0b013e328365d37c. [DOI] [PubMed] [Google Scholar]
- 17.Rieder F, Kessler SP, West GA, Bhilocha S, de la Motte C, Sadler TM, Gopalan B, Stylianou E, Fiocchi C. Inflammation-induced endothelial-to-mesenchymal transition: a novel mechanism of intestinal fibrosis. Am J Pathol. 2011;179:2660–2673. doi: 10.1016/j.ajpath.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raleigh DR, Boe DM, Yu D, Weber CR, Marchiando AM, Bradford EM, Wang Y, Wu L, Schneeberger EE, Shen L, Turner JR. Occludin S408 phosphorylation regulates tight junction protein interactions and barrier function. J Cell Biol. 2011;193:565–582. doi: 10.1083/jcb.201010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Sadi R, Guo S, Dokladny K, Smith MA, Ye D, Kaza A, Watterson DM, Ma TY. Mechanism of interleukin-1β induced-increase in mouse intestinal permeability in vivo. J Interferon Cytokine Res. 2012;32:474–484. doi: 10.1089/jir.2012.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323–1333. doi: 10.1053/j.gastro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein J, Dignass AU. European Gastroenterology Journal. 2015. Laboratory diagnostics in IBD - What the gastroenterologist should know; pp. 32–47. [Google Scholar]
- 22.Vermeire S, Van Assche G, Rutgeerts P. C-reactive protein as a marker for inflammatory bowel disease. Inflamm Bowel Dis. 2004;10:661–665. doi: 10.1097/00054725-200409000-00026. [DOI] [PubMed] [Google Scholar]
- 23.Cioffi M, Rosa AD, Serao R, Picone I, Vietri MT. Laboratory markers in ulcerative colitis: Current insights and future advances. World J Gastrointest Pathophysiol. 2015;6:13–22. doi: 10.4291/wjgp.v6.i1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henriksen M, Jahnsen J, Lygren I, Stray N, Sauar J, Vatn MH, Moum B IBSEN Study Group. C-reactive protein: a predictive factor and marker of inflammation in inflammatory bowel disease. Results from a prospective population-based study. Gut. 2008;57:1518–1523. doi: 10.1136/gut.2007.146357. [DOI] [PubMed] [Google Scholar]
- 25.Peyrin-Biroulet L, Sandborn W, Sands BE, Reinisch W, Bemelman W, Bryant RV, D'Haens G, Dotan I, Dubinsky M, Feagan B, Fiorino G, Gearry R, Krishnareddy S, Lakatos PL, Loftus EV, Jr, Marteau P, Munkholm P, Murdoch TB, Ordás I, Panaccione R, Riddell RH, Ruel J, Rubin DT, Samaan M, Siegel CA, Silverberg MS, Stoker J, Schreiber S, Travis S, Van Assche G, Danese S, Panes J, Bouguen G, O'Donnell S, Pariente B, Winer S, Hanauer S, Colombel JF. Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE): Determining Therapeutic Goals for Treat-to-Target. Am J Gastroenterol. 2015;110:1324–1338. doi: 10.1038/ajg.2015.233. [DOI] [PubMed] [Google Scholar]
- 26.Reumaux D, Duthilleul P, Roos D. Pathogenesis of diseases associated with antineutrophil cytoplasm autoantibodies. Hum Immunol. 2004;65:1–12. doi: 10.1016/j.humimm.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Bennike T, Birkelund S, Stensballe A, Andersen V. Biomarkers in inflammatory bowel diseases: current status and proteomics identification strategies. World J Gastroenterol. 2014;20:3231–3244. doi: 10.3748/wjg.v20.i12.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iskandar HN, Ciorba MA. Biomarkers in inflammatory bowel disease: current practices and recent advances. Transl Res. 2012;159:313–325. doi: 10.1016/j.trsl.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reumaux D, de Boer M, Meijer AB, Duthilleul P, Roos D. Expression of myeloperoxidase (MPO) by neutrophils is necessary for their activation by anti-neutrophil cytoplasm autoantibodies (ANCA) against MPO. J Leukoc Biol. 2003;73:841–849. doi: 10.1189/jlb.1102567. [DOI] [PubMed] [Google Scholar]
- 30.Peyrin-Biroulet L, Standaert-Vitse A, Branche J, Chamaillard M. IBD serological panels: facts and perspectives. Inflamm Bowel Dis. 2007;13:1561–1566. doi: 10.1002/ibd.20226. [DOI] [PubMed] [Google Scholar]
- 31.Peeters M, Joossens S, Vermeire S, Vlietinck R, Bossuyt X, Rutgeerts P. Diagnostic value of anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease. Am J Gastroenterol. 2001;96:730–734. doi: 10.1111/j.1572-0241.2001.03613.x. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein CN, El-Gabalawy H, Sargent M, Landers C, Rawsthorne P, Elias B, Targan SR. Assessing inflammatory bowel disease-associated antibodies in Caucasian and First Nations cohorts. Can J Gastroenterol. 2011;25:269–273. doi: 10.1155/2011/712350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tibble J, Teahon K, Thjodleifsson B, Roseth A, Sigthorsson G, Bridger S, Foster R, Sherwood R, Fagerhol M, Bjarnason I. A simple method for assessing intestinal inflammation in Crohn's disease. Gut. 2000;47:506–513. doi: 10.1136/gut.47.4.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Røseth AG, Fagerhol MK, Aadland E, Schjønsby H. Assessment of the neutrophil dominating protein calprotectin in feces. A methodologic study. Scand J Gastroenterol. 1992;27:793–798. doi: 10.3109/00365529209011186. [DOI] [PubMed] [Google Scholar]
- 35.Gisbert JP, Bermejo F, Pérez-Calle JL, Taxonera C, Vera I, McNicholl AG, Algaba A, López P, López-Palacios N, Calvo M, González-Lama Y, Carneros JA, Velasco M, Maté J. Fecal calprotectin and lactoferrin for the prediction of inflammatory bowel disease relapse. Inflamm Bowel Dis. 2009;15:1190–1198. doi: 10.1002/ibd.20933. [DOI] [PubMed] [Google Scholar]
- 36.Henderson P, Anderson NH, Wilson DC. The diagnostic accuracy of fecal calprotectin during the investigation of suspected pediatric inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2014;109:637–645. doi: 10.1038/ajg.2013.131. [DOI] [PubMed] [Google Scholar]
- 37.Gecse KB, Brandse JF, van Wilpe S, Löwenberg M, Ponsioen C, van den Brink G, D'Haens G. Impact of disease location on fecal calprotectin levels in Crohn's disease. Scand J Gastroenterol. 2015;50:841–847. doi: 10.3109/00365521.2015.1008035. [DOI] [PubMed] [Google Scholar]
- 38.Manceau H, Chicha-Cattoir V, Puy H, Peoc'h K. Fecal calprotectin in inflammatory bowel diseases: update and perspectives. Clin Chem Lab Med. 2017;55:474–483. doi: 10.1515/cclm-2016-0522. [DOI] [PubMed] [Google Scholar]
- 39.De Vos M, Louis EJ, Jahnsen J, Vandervoort JG, Noman M, Dewit O, Dʼhaens GR, Franchimont D, Baert FJ, Torp RA, Henriksen M, Potvin PM, Van Hootegem PP, Hindryckx PM, Moreels TG, Collard A, Karlsen LN, Kittang E, Lambrecht G, Grimstad T, Koch J, Lygren I, Coche JC, Mana F, Van Gossum A, Belaiche J, Cool MR, Fontaine F, Maisin JM, Muls V, Neuville B, Staessen DA, Van Assche GA, de Lange T, Solberg IC, Vander Cruyssen BJ, Vermeire SA. Consecutive fecal calprotectin measurements to predict relapse in patients with ulcerative colitis receiving infliximab maintenance therapy. Inflamm Bowel Dis. 2013;19:2111–2117. doi: 10.1097/MIB.0b013e31829b2a37. [DOI] [PubMed] [Google Scholar]
- 40.Molander P, Färkkilä M, Ristimäki A, Salminen K, Kemppainen H, Blomster T, Koskela R, Jussila A, Rautiainen H, Nissinen M, Haapamäki J, Arkkila P, Nieminen U, Kuisma J, Punkkinen J, Kolho KL, Mustonen H, Sipponen T. Does fecal calprotectin predict short-term relapse after stopping TNFα-blocking agents in inflammatory bowel disease patients in deep remission? J Crohns Colitis. 2015;9:33–40. doi: 10.1016/j.crohns.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 41.Kane SV, Sandborn WJ, Rufo PA, Zholudev A, Boone J, Lyerly D, Camilleri M, Hanauer SB. Fecal lactoferrin is a sensitive and specific marker in identifying intestinal inflammation. Am J Gastroenterol. 2003;98:1309–1314. doi: 10.1111/j.1572-0241.2003.07458.x. [DOI] [PubMed] [Google Scholar]
- 42.Angriman I, Scarpa M, D'Incà R, Basso D, Ruffolo C, Polese L, Sturniolo GC, D'Amico DF, Plebani M. Enzymes in feces: useful markers of chronic inflammatory bowel disease. Clin Chim Acta. 2007;381:63–68. doi: 10.1016/j.cca.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 43.Nixon AB, Pang H, Starr MD, Friedman PN, Bertagnolli MM, Kindler HL, Goldberg RM, Venook AP, Hurwitz HI Alliance for Clinical Trials In Oncology. Prognostic and predictive blood-based biomarkers in patients with advanced pancreatic cancer: results from CALGB80303 (Alliance) Clin Cancer Res. 2013;19:6957–6966. doi: 10.1158/1078-0432.CCR-13-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ingvarsson J, Wingren C, Carlsson A, Ellmark P, Wahren B, Engström G, Harmenberg U, Krogh M, Peterson C, Borrebaeck CA. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics. 2008;8:2211–2219. doi: 10.1002/pmic.200701167. [DOI] [PubMed] [Google Scholar]
- 45.Vigren E, Hamberg M, Zhaunerchyk V, Kaminska M, Thomas RD, Trippel S, Zhang M, Kashperka I, Ugglas MA, Walsh C, Wester R, Semaniak J, Larsson M, Geppert WD. Dissociative recombination of the acetaldehyde cation, CH(3)CHO(+) Phys Chem Chem Phys. 2010;12:11670–11673. doi: 10.1039/c003857a. [DOI] [PubMed] [Google Scholar]
- 46.Wingren C, Sandström A, Segersvärd R, Carlsson A, Andersson R, Löhr M, Borrebaeck CA. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012;72:2481–2490. doi: 10.1158/0008-5472.CAN-11-2883. [DOI] [PubMed] [Google Scholar]
- 47.Mehan MR, Williams SA, Siegfried JM, Bigbee WL, Weissfeld JL, Wilson DO, Pass HI, Rom WN, Muley T, Meister M, Franklin W, Miller YE, Brody EN, Ostroff RM. Validation of a blood protein signature for non-small cell lung cancer. Clin Proteomics. 2014;11:32. doi: 10.1186/1559-0275-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pommier AJ, Shaw R, Spencer SK, Morgan SR, Hoff PM, Robertson JD, Barry ST, Jürgensmeier JM. Serum protein profiling reveals baseline and pharmacodynamic biomarker signatures associated with clinical outcome in mCRC patients treated with chemotherapy ± cediranib. Br J Cancer. 2014;111:1590–1604. doi: 10.1038/bjc.2014.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viennois E, Zhao Y, Merlin D. Biomarkers of Inflammatory Bowel Disease: From Classical Laboratory Tools to Personalized Medicine. Inflamm Bowel Dis. 2015;21:2467–2474. doi: 10.1097/MIB.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson NL, Anderson NG. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 51.Barnett M, Young W, Cooney J, Roy N. Metabolomics and Proteomics, and What to Do with All These 'Omes': Insights from Nutrigenomic Investigations in New Zealand. J Nutrigenet Nutrigenomics. 2014;7:274–282. doi: 10.1159/000381349. [DOI] [PubMed] [Google Scholar]
- 52.Hong SN, Joung JG, Bae JS, Lee CS, Koo JS, Park SJ, Im JP, Kim YS, Kim JW, Park WY, Kim YH. RNA-seq Reveals Transcriptomic Differences in Inflamed and Noninflamed Intestinal Mucosa of Crohn's Disease Patients Compared with Normal Mucosa of Healthy Controls. Inflamm Bowel Dis. 2017;23:1098–1108. doi: 10.1097/MIB.0000000000001066. [DOI] [PubMed] [Google Scholar]
- 53.Bennike TB, Carlsen TG, Ellingsen T, Bonderup OK, Glerup H, Bøgsted M, Christiansen G, Birkelund S, Stensballe A, Andersen V. Neutrophil Extracellular Traps in Ulcerative Colitis: A Proteome Analysis of Intestinal Biopsies. Inflamm Bowel Dis. 2015;21:2052–2067. doi: 10.1097/MIB.0000000000000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 55.O'Donoghue AJ, Jin Y, Knudsen GM, Perera NC, Jenne DE, Murphy JE, Craik CS, Hermiston TW. Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS One. 2013;8:e75141. doi: 10.1371/journal.pone.0075141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 57.Delgado-Rizo V, Martínez-Guzmán MA, Iñiguez-Gutierrez L, García-Orozco A, Alvarado-Navarro A, Fafutis-Morris M. Neutrophil Extracellular Traps and Its Implications in Inflammation: An Overview. Front Immunol. 2017;8:81. doi: 10.3389/fimmu.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riaz T, Sollid LM, Olsen I, de Souza GA. Quantitative Proteomics of Gut-Derived Th1 and Th1/Th17 Clones Reveal the Presence of CD28+ NKG2D- Th1 Cytotoxic CD4+ T cells. Mol Cell Proteomics. 2016;15:1007–1016. doi: 10.1074/mcp.M115.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pastorelli L, De Salvo C, Mercado JR, Vecchi M, Pizarro TT. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front Immunol. 2013;4:280. doi: 10.3389/fimmu.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poulsen NA, Andersen V, Møller JC, Møller HS, Jessen F, Purup S, Larsen LB. Comparative analysis of inflamed and non-inflamed colon biopsies reveals strong proteomic inflammation profile in patients with ulcerative colitis. BMC Gastroenterol. 2012;12:76. doi: 10.1186/1471-230X-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moriggi M, Pastorelli L, Torretta E, Tontini GE, Capitanio D, Bogetto SF, Vecchi M, Gelfi C. Contribution of Extracellular Matrix and Signal Mechanotransduction to Epithelial Cell Damage in Inflammatory Bowel Disease Patients: A Proteomic Study. Proteomics. 2017;17 doi: 10.1002/pmic.201700164. [DOI] [PubMed] [Google Scholar]
- 62.Nanni P, Mezzanotte L, Roda G, Caponi A, Levander F, James P, Roda A. Differential proteomic analysis of HT29 Cl.16E and intestinal epithelial cells by LC ESI/QTOF mass spectrometry. J Proteomics. 2009;72:865–873. doi: 10.1016/j.jprot.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 63.Shkoda A, Werner T, Daniel H, Gunckel M, Rogler G, Haller D. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114–1125. doi: 10.1021/pr060433m. [DOI] [PubMed] [Google Scholar]
- 64.Yau YY, Leong RWL, Pudipeddi A, Redmond D, Wasinger VC. Serological Epithelial Component Proteins Identify Intestinal Complications in Crohn's Disease. Mol Cell Proteomics. 2017;16:1244–1257. doi: 10.1074/mcp.M116.066506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korolkova OY, Myers JN, Pellom ST, Wang L, M'Koma AE. Characterization of Serum Cytokine Profile in Predominantly Colonic Inflammatory Bowel Disease to Delineate Ulcerative and Crohn's Colitides. Clin Med Insights Gastroenterol. 2015;8:29–44. doi: 10.4137/CGast.S20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M'Koma AE, Seeley EH, Washington MK, Schwartz DA, Muldoon RL, Herline AJ, Wise PE, Caprioli RM. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm Bowel Dis. 2011;17:875–883. doi: 10.1002/ibd.21442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seeley EH, Washington MK, Caprioli RM, M'Koma AE. Proteomic patterns of colonic mucosal tissues delineate Crohn's colitis and ulcerative colitis. Proteomics Clin Appl. 2013;7:541–549. doi: 10.1002/prca.201200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Starr AE, Deeke SA, Ning Z, Chiang CK, Zhang X, Mottawea W, Singleton R, Benchimol EI, Wen M, Mack DR, Stintzi A, Figeys D. Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn's disease from UC. Gut. 2017;66:1573–1583. doi: 10.1136/gutjnl-2015-310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meuwis MA, Fillet M, Lutteri L, Marée R, Geurts P, de Seny D, Malaise M, Chapelle JP, Wehenkel L, Belaiche J, Merville MP, Louis E. Proteomics for prediction and characterization of response to infliximab in Crohn's disease: a pilot study. Clin Biochem. 2008;41:960–967. doi: 10.1016/j.clinbiochem.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 70.Slungaard A. Platelet factor 4 modulation of the thrombomodulin-protein C system. Crit Care Med. 2004;32:S331–S335. doi: 10.1097/01.ccm.0000126359.92825.e9. [DOI] [PubMed] [Google Scholar]
- 71.Bikfalvi A. Platelet factor 4: an inhibitor of angiogenesis. Semin Thromb Hemost. 2004;30:379–385. doi: 10.1055/s-2004-831051. [DOI] [PubMed] [Google Scholar]
- 72.Simi M, Leardi S, Tebano MT, Castelli M, Costantini FM, Speranza V. Raised plasma concentrations of platelet factor 4 (PF4) in Crohn's disease. Gut. 1987;28:336–338. doi: 10.1136/gut.28.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gazouli M, Anagnostopoulos AK, Papadopoulou A, Vaiopoulou A, Papamichael K, Mantzaris G, Theodoropoulos GE, Anagnou NP, Tsangaris GT. Serum protein profile of Crohn's disease treated with infliximab. J Crohns Colitis. 2013;7:e461–e470. doi: 10.1016/j.crohns.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 74.Magnusson MK, Strid H, Isaksson S, Bajor A, Lasson A, Ung KA, Öhman L. Response to infliximab therapy in ulcerative colitis is associated with decreased monocyte activation, reduced CCL2 expression and downregulation of Tenascin C. J Crohns Colitis. 2015;9:56–65. doi: 10.1093/ecco-jcc/jju008. [DOI] [PubMed] [Google Scholar]
- 75.Banks C, Bateman A, Payne R, Johnson P, Sheron N. Chemokine expression in IBD. Mucosal chemokine expression is unselectively increased in both ulcerative colitis and Crohn's disease. J Pathol. 2003;199:28–35. doi: 10.1002/path.1245. [DOI] [PubMed] [Google Scholar]
- 76.Uguccioni M, Gionchetti P, Robbiani DF, Rizzello F, Peruzzo S, Campieri M, Baggiolini M. Increased expression of IP-10, IL-8, MCP-1, and MCP-3 in ulcerative colitis. Am J Pathol. 1999;155:331–336. doi: 10.1016/S0002-9440(10)65128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leal RF, Planell N, Kajekar R, Lozano JJ, Ordás I, Dotti I, Esteller M, Masamunt MC, Parmar H, Ricart E, Panés J, Salas A. Identification of inflammatory mediators in patients with Crohn's disease unresponsive to anti-TNFα therapy. Gut. 2015;64:233–242. doi: 10.1136/gutjnl-2013-306518. [DOI] [PubMed] [Google Scholar]
- 78.Fiocchi C. Inflammatory Bowel Disease: Complexity and Variability Need Integration. Front Med (Lausanne) 2018;5:75. doi: 10.3389/fmed.2018.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Loktionov A, Chhaya V, Bandaletova T, Poullis A. Inflammatory bowel disease detection and monitoring by measuring biomarkers in non-invasively collected colorectal mucus. J Gastroenterol Hepatol. 2017;32:992–1002. doi: 10.1111/jgh.13627. [DOI] [PubMed] [Google Scholar]