Abstract
Increased atmospheric CO2 is driving ocean acidification (OA), and potential changes in marine ecosystems. Research shows that both planktonic and benthic communities are affected, but how these changes are linked remains unresolved. Here we show experimentally that decreasing seawater pH (from pH 8.1 to 7.8 and 7.4) leads to reduced biofilm formation and lower primary producer biomass within biofilms. These changes occurred concurrently with a re-arrangement of the biofilm microbial communities. Changes suggest a potential shift from autotrophic to heterotrophic dominated biofilms in response to reduced pH. In a complimentary experiment, biofilms reared under reduced pH resulted in altered larval settlement for a model species (Galeolaria hystrix). These findings show that there is a potential cascade of impacts arising from OA effects on biofilms that may drive important community shifts through altered settlement patterns of benthic species.
Subject terms: Microbial ecology, Climate-change impacts, Marine biology
Introduction
Since the start of the industrial revolution, an additional 555 Pg of carbon has been released into the atmosphere, of which 155 Pg C (≈30%) has entered the ocean. This process is predicted to decrease seawater pH by 0.2 to 0.4 units on average by the year 21001. It is estimated that average surface ocean pH has already decreased from near 8.25 to 8.1 over the past 250 years and is forecasted to decrease to near or below 7.85 by the end of the century1. Acidification of the oceans is expected, and has been shown experimentally, to affect marine ecosystems in a myriad of ways with stimulative, inhibitive, or neutral effects depending on organism or location2–5.
Complex lifecycles in most marine invertebrates involve a long-lived benthic adult stage, and a shorter-lived planktonic larval stage that are thought to be most susceptible to climate change6–8. At the completion of the free-living larval stage, the competent larvae of benthic species must attach to a substrate (settle) and metamorphose. The transition is likely to affect settlement success of calcifying species under ocean acidification regimes; the first is a direct effect where lowered pH and lower saturation states of calcium carbonate (calcite and aragonite) ions which reduce calcification during the transition from planktonic to benthic life stages9,10; the second is an indirect effect through the disturbance of settlement cues between biofilms and larvae11. While evidence supports impacts of OA on calcification, recent work shows that these effects are likely driven by changes in saturation state and not directly by pH12,13. Less is known on the responses across life-history stages to OA scenarios that might be related to a loss of interaction (settlement cues) between larvae and microbial communities. While reduced pH has been linked to decreases in the settlement success of vermetids14, corals14–18, sea stars11, and sea urchins19, our understanding regarding the OA effects on the settlement processes of marine invertebrates remains limited. In this respect, a recent review by Espinel et al.20 found less than 50 published studies, with the majority indicated neutral or negative changes in settlement under reduced pH. Within these studies, only a small number examined the outcomes of pH induced changes of substrates (biofilms and CCAs) on settlement, and these indicated a reduction in settlement rate in taxa such as coral and sea stars20.
Settlement success has in part been attributed to biofilm recognition or quorum sensing21,22. These cues have been strongly linked to biofilm bacteria of the genus Pseudoalteromonas22,23 and the production of brominated compounds, with their effect potentially modified by factors including carotenoids24. It is likely, however, that a large number of microorganisms and metabolites are involved in promoting settlement of marine species, yet to be described. Altered settlement rates due to OA driven changes in biofilm communities and will be important as they could shape the future distributions, abundances and ecology of marine benthic communities. In addition, changes in settlement may impact the sustainability of species that are cultured or harvested, where recruitment and the supply of juveniles for recruitment and on-growing may be reduced.
The present research seeks to better understand the effects of reduced pH on the development of microbial biofilm communities, and the potential effects of changes in biofilms on the settlement of marine larvae. This is examined in two independent, but complimentary experiments. Firstly, we developed biofilms in flow-through aquaria for up to 69 days at ambient (pH 8.1) or reduced seawater pH (7.8 and 7.4) to determine their general characteristics (biomass, chlorophyll and carotenoid levels) and associated microbial community (diversity, composition). Secondly, we later assayed biofilms for settlement cues using competent larvae of a common polychaete tubeworm (Galeolaria hystrix). This species is an abundant, intertidal suspension feeder that typically settle as individuals in the shallow waters of low tidal zones at Portobello Beach, New Zealand25. The complex, heavily biofilmed and highly variable environment in which G. hystrix thrives made this serpulid polychaete a preferable model species to illustrate the cascading effects of ocean acidification on micro- to macro-community formation.
Results
Environmental seawater
Over the 42-day deployment of the SeaFET in Otago Harbour, the seawater pH ranged from 8.04 to 8.19 pH units (Fig. 1a). Within this period a strong diel pattern was detected, with single fluctuations ranging up to 0.12 pH units (max = 8.19, min = 8.07). The daily variation in pH is most tightly coupled with daily light cycles (Fig. 1b), with seawater pH increasing from an overnight minimum and reaching a peak mid-afternoon, 2–3 hours after maximum mid-day irradiance. In contrast, tidal water exchange (which is semidiurnal at our study site) has a secondary and smaller influence on pH (Fig. 1c), with an incoming tide and tidal mixing associated with periods of variable and increasing pH.
Figure 1.
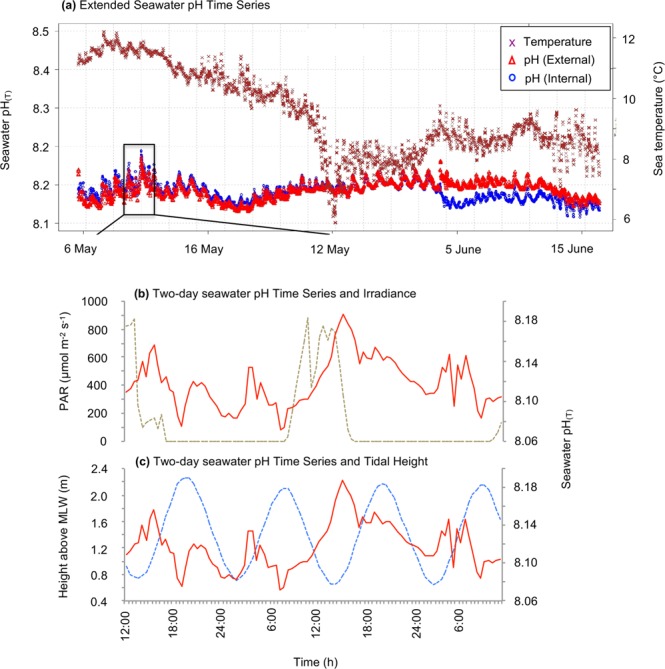
(a) Seawater pH(T) and sea temperature (°C) from 42-day deployment of the SeaFET at Portobello Marine Lab (PML) Wharf, Otago Harbour at 1 m depth. (b) Seawater pH and PAR (photosynthetically active radiation, 700–400 nm) light time series at PML from 9 to 11 May 2015. (c) Seawater pH and tidal height time series at PML from 9 to 11 May 2015. The pH (Internal) and pH (External) are the internal and external reference electrode measurements, respectively, which are equivalent when the instrument is correctly calibrated for accurate measurement.
Experiment 1: Biofilm development under pH treatment
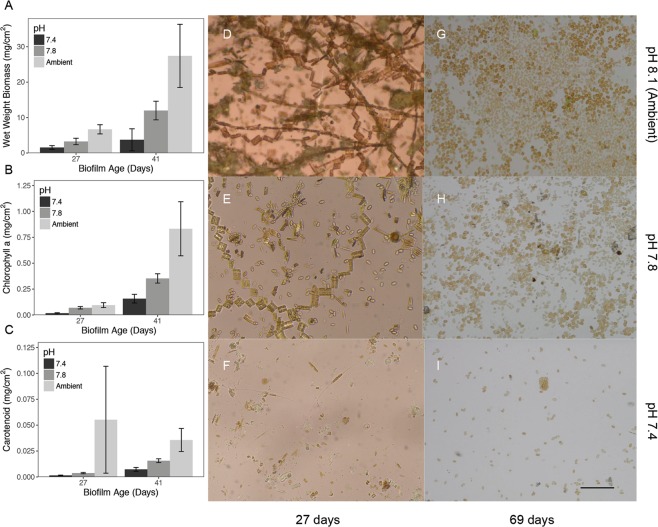
Biofilm wet weight biomass was significantly (ANOVA, F2,9 = 5.419 p < 0.05) different among pH treatments, with a more pronounced difference in older biofilms (Fig. 2). Biomass increased 12-fold under ambient pH conditions from young to older biofilms, while the increase over time was less pronounced under both reduced pH treatments. The biomass of old biofilms measured in the slides was 7-fold lower when these were developed at the lowest pH than at ambient conditions. This trend was consistent with the (5-fold) lower chlorophyll-a concentration for both young (ANOVA, F2,9 = 6.592, p < 0.05) and older biofilms (ANOVA, F2,9 = 12.725, p < 0.05), under the pH 7.4 treatment (Fig. 2). A similar trend was observed for carotenoid concentrations (Fig. 2) in young (ANOVA, F2,9 = 5.949, p < 0.05) and old biofilms (ANOVA, F2,9 = 14.001, p < 0.05).
Figure 2.
Effects of ocean acidification on biofilm development. (A–C) Changes in biomass and pigments associated to primary producers in response to pH treatment for young and old biofilms (n = 5 for each bar). (D–I) Representative images for biofilms at different pH for young and old biofilms. Ambient pHNIST was 8.1. Scale bar 50 μm.
Microbial community analysis
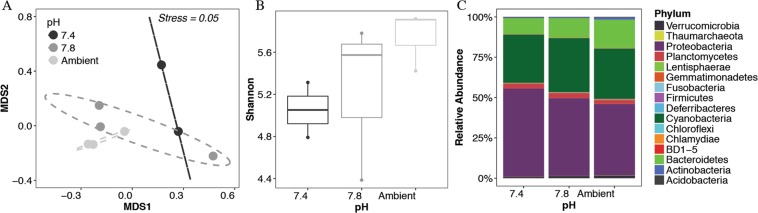
Reduced pH also led to clear and simultaneous shifts in microbial communities (Fig. 3) for both young and old biofilms (Figs. 3 and 4, Supplementary Table 3). In young biofilms (<30 days old), microbial communities observed under both pH scenarios were statistically distinct from those raised in ambient pH (Fig. 3A and Supplementary Fig. 4) (ANOSIM: 16S, R = 0.54, p = 0.04; invertebrates, R = 0.49, p < 0.01), with the lowest observed variance in community changes at the lowest pH. Despite changes in community structure and a trend of reduced alpha diversity at lower pH for microbial communities (Fig. 3B), no significant differences (ANOVA p > 0.05) in diversity (both richness and Shannon diversity) were observed among microbial communities. A re-arrangement was also observed at a broad taxonomic level in the microbial community with fold decreases in Actinobacteria (−2.8x), Bacteroidetes (−1.7x), Firmicutes (−3.3x), Fusobacteria (−1.5x), Gemmatimonadetes (−2.2x), Nitrospiraea (−8x), Proteobacteria (−1.3x) anderrucomicrobia (−2x). In contrast, fold increases were observed for BD1–5 (3.5x) and Planctomycetes (1.4x). Not all microbial phyla responded in gradual fashion, however, with pH 7.8 being preferred (e.g. Candidate division BRC1) or avoided (e.g. Chlamydiae) by certain taxa. These groups were detectable across various pH levels, although certain groups went from undetected to regularly detected under OA conditions (Tenericutes, Candidate division SR1, NPL-UPA2, Tenericutes), and others were no longer detected at reduced pH (Deinococcus-Thermus).
Figure 3.
Microbial community response of 27-day old biofilms to ocean acidification. (A) Differences in communities, (B) Shannon diversity and (C) composition. Ambient pH was 8.1. Supplemental information and full data can be found in Figs. S1–4.
Figure 4.
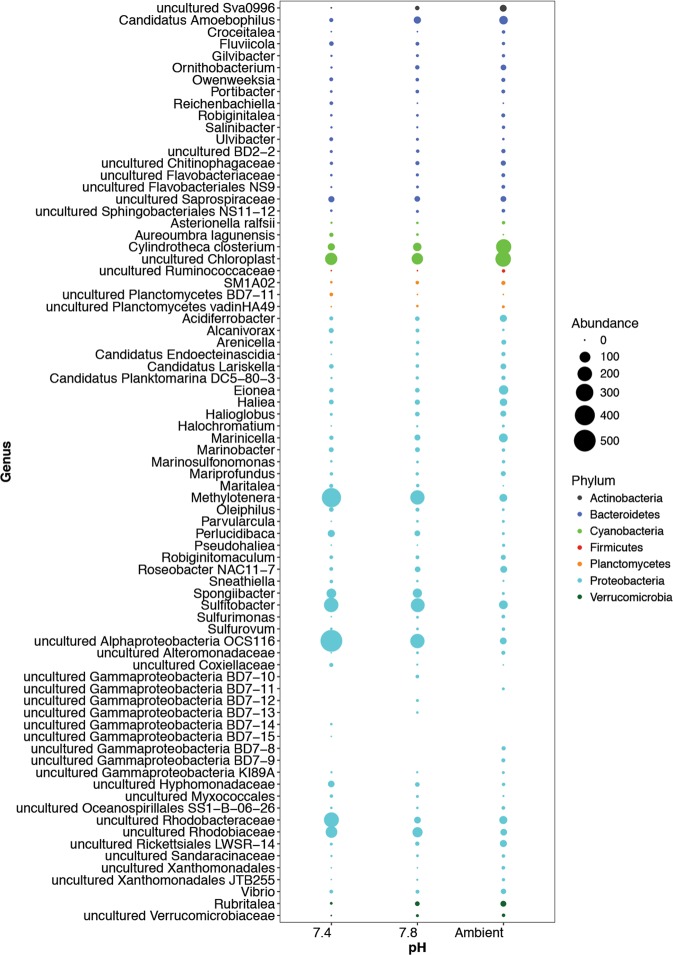
Summary of significantly affected microbial genera based on OTUs correlated to pH. Data represents 198 OTUs with a significant (p < 0.05) Spearmans correlation (Rho <−0.5 and >0.5).
While patterns at high taxonomic levels (phylum) suggest strong effects for certain microbial groups, we aimed to identify specific organisms (operational taxonomic units; OTUs) significantly associated with changing pH. A Spearmans correlation against pH identified 198 OTUs with strong (p < 0.05, Rho < −0.5 and >0.5) responses to pH (Fig. 4, Supplementary Figs. 5–7). Within responsive OTUs, certain taxa were negatively influenced by pH (Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia) while others were positively affected (Chlamydiae, Lentisphaerae, and Proteobacteria). Interestingly the number of responsive organisms was highly variable across phyla with most organisms belonging to the Proteobacteria. Within this phyla, both positively (e.g. Methylotenera) and negatively (e.g. Haliea) affected organisms were detected. While OTUs closely associated to the gammaproteobacterial, Pseudoalteromonas were not detected, organisms within the gammaproteobacteria were amongst the most commonly associated with changes in pH, with 21 representatives from the Order Alteromonadales (were Pseudoalteromonas belongs), including 17 organisms negatively responsive to a decrease in pH. These low pH sensitive organisms (Spearmans correlation to pH > 0.72) included members of the genera Haliea, Eionea, Halioglobus, Candidatus Endobugula, Pseudohaliea, OM60(NOR5) clade, and several uncultured groups. Overall, this represents a gradual shift in the biofilm from a primary producer dominated community (e.g. Cyanobacteria and diatoms [Cylindrotheca]) to a heterotrophic (e.g. Methylotenera and Roseobacter OCS116 clade) dominated one, and is consistent with the observed decrease in biomass and pigments of primary producers (see Fig. 2).
Experiment 2: Settlement assays on biofilms developed under pH treatment
Average Galeolaria hystrix settlement success ranged from 0.8% on clean control slides, up to 29% observed on 60-day old biofilms raised in pH 7.8 (Fig. 5). On 23-day old biofilm, a two-way ANOVA indicated a significant difference in settlement rate among pH treatments (F(3, 31) = 8.838, p < 0.001). A Tukey’s Post hoc test indicated settlement was significantly greater on the ambient and pH 7.8 biofilms than on the control slides, but the difference was not significant among the pH treated biofilms, or between pH 7.4 and the control slides. This difference was consistent at both 24 and 48 h during the settlement assay.
Figure 5.
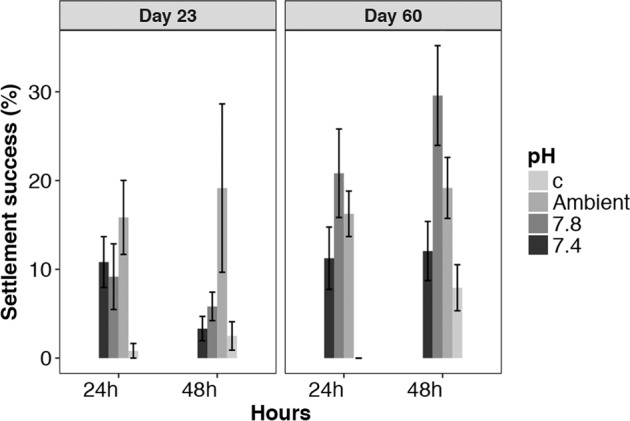
Settlement success of the model invertebrate (tubeworm) Galeolaria hystrix on biofilms grown at different pH. (a) The proportion of competent larvae settled 24 h and 48 h after being offered 23-day old biofilms developed under pHNIST 7.4, 7.8 and 8.1. (b) The proportion of competent larvae settled 24 h and 48 h after being offered 60-day old biofilms developed under pHNIST 7.4, 7.8 and 8.1. For each settlement experiment a control (sterile glass slide) was used to determine non-selective settlement.
On 60-day old biofilm (Fig. 5), a two-way ANOVA indicated settlement was significantly greater (F(1, 63) = 7.479, p = 0.008) at 48 h than at 24 h, and was significantly different (F(3, 63) = 19.614, p < 0.001) among pH treatments/control. A Tukey’s Post hoc test indicated settlement was significantly lower on the control slides. Within the pH treatments, settlement was not significantly different between the ambient and pH 7.8, nor between the ambient and pH 7.4. Settlement was, however, significantly greater on pH 7.8 compared with pH 7.4, with this pattern of settlement among pH treatments consistent at both 24 h and 48 h (Time × pH, F(3, 63) = 1.624, p = 0.194).
Discussion and Conclusions
Ocean acidification (OA) will alter marine biological processes, and while a wide range of individual responses are reported4, we aimed to address currently unresolved questions related to impacts across interactive trophic or taxonomic levels. In this study, using two independent but complimentary experiments, we showed that even a short term reduction in seawater pH (<69 days) can influence biofilm development and microbial community composition, and that modified biofilms may alter settlement rates in invertebrate larvae such as Galeolaria hystrix.
Environmental pH variability and experimental treatments
In order to conduct ecologically meaningful in vitro ocean acidification experiments, relevant pH targets must consider site/ecosystem-specific carbonate systems and natural pH fluctuations of in situ habitats. Our in situ measurements show that seawater pH at our study, and where our biological experiments were conducted, has a relatively high degree of variability, over relatively short time scales (i.e. hours). This variability is associated with diurnal light cycles driving the photosynthetic uptake of CO2 (and a resulting reduction in pH) during daylight, while semi-diurnal tidal cycles mix water at the site with relatively high pH coastal seawater. Our observations of short term (hourly) and longer-term variability are consistent with measurements of pH for nearshore and estuarine environments26 where pH varies on the order of 0.2 to 0.3 pH units associated with biological activity and water exchange.
Over the measurement period, the average seawater pHT (pH total scale) at the in situ collection site at Portobello Marine Laboratory was pHT 8.13 ± 0.01 and was consistent with the 2010 global open-ocean estimate of pHT 8.07 (NOAA, 2010). Therefore, the near-future, 2100 IPCC1 projection of pHNIST 7.827 was an appropriate near-future pH target. Although current research supports a pH of approximately pH < 7.5 if atmospheric CO2 reaches levels of 1900 ppm by 23001,27, observed diel fluctuations (up to pHT 0.116, Fig. 1) justified a slightly reduced target of pHNIST 7.4 (pH National Institute of Standards and Technology scale) as a more ecologically relevant, extreme 2300 pH target. These pH targets are consistent with a number of in vitro studies16,18,28–31 and overlap with a number of in situ studies at CO2 vent sites32–37 allowing for comparison among observations.
Experiment 1: Microbial community responses to reduced pH
In the first experiment, we observed shifts in biofilm community and structure across reduced pH suggesting a loss of cells, however it does not indicate a loss of diversity or potential functions encoded within biofilm organisms. Instead we saw a re-arrangement of the microbial community already detectable at a broad taxonomic level. The observed negative effect of OA of Nitrospiraea and Actinobacteria in this study is consistent with the decrease of Nitrospiraea sequences observed in response to natural pCO2 increases in coastal sediments along natural CO2 gradients at a volcanic vent in Papua New Guinea38. However, contrasting results were reported for Bacteriodetes and Proteobacteria in response to pH; in our study we found that they decreased in response to OA whereas Raulf et al.38 found a linear increase in relative sequence abundance with decreasing pH along their pH gradient. Consistent to our results, a decrease of Bacteroidetes sequence in response to OA was also found in microbial biofilms of crustose coralline algae16. These results could indicate differences in the response to OA of microbial communities living on sediments versus on other solid surfaces, or a differences related to the experimental conditions (i.e., mesocoms experiment vs natural pCO2 gradient). Nevertheless, the confirmation of Nitrospiraea and Actinobacteria changes suggests a key role of these organisms in the light of the responses of biofilm microbial communities to OA.
It is important to acknowledge that while certain taxa decreased, others had positive responses to changes in pH, including several groups from within the proteobacteria (e.g. Methylotenera, Sulfitobacter and unclassified alphaproteobacteria from the OCS116 cluster). Further studies are needed to confirm which key members of microbial biofilm communities are likely to have consistent responses to climate change pressures such as OA, and this would include examining multiple stressors simultaneously (e.g. pH, temperature, etc.). For example, it has been shown that in planktonic microbial communities the combination of OA and temperature39, or OA and eutrophication40 can select for specific members of the microbial communities which might differ from OA alone.
Experiment 2: Biofilm modification and larval settlement
In the second experiment, settlement of Galeolaria was significantly different on glass slides that had been developed under the three pH treatments. The most likely cause of this pattern is a change in settlement cues associated with biofilms that had modified physical or biological/chemical characteristics. A loss of microorganisms known to trigger settlement21,23, as well as the reduction in other factors (e.g. carotenoids) known to enhance settlement24 suggest that OA scenarios, especially conditions such as acidification to pH 7.4, could lead to regime shifts that can alter key ecosystems processes such as recruitment and their services41.
While it is clear that microbes contribute to chemotaxis and settlement success21,23, and that acidification can influence invertebrate settlement15–18, a definitive link between these two processes remains unresolved. Microbial biofilms are likely critical for settlement induction in marine species42–44, and changes in biofilm composition, such as those related to age21,45,46 or pH16, can affect settlement rates in marine invertebrates. For example, Webster et al.16 reported decreases in dominant microbial populations (Alphaproteobacteria and Bacteroidetes) and emergence of Proteobacteria as a result of lower seawater pH. This coincided with reduced coral larvae settlement, although an explicit mechanism (e.g. altered settlement on CCA is due to changes in the chemical inducer of the algae or altered associated microbes) was not identified.
Conclusions
There is an important caveat when interpreting our findings, namely that biofilm microbial assessments and settlement experiments were carried out on separately reared biofilms. Nevertheless, our results are consistent with prior work and support the hypothesis that alterations in microbial biofilms are directly linked to both changes in settlement and abundance of invertebrate communities. This may represent a model scenario to OA, but further studies are needed to directly provide a mechanism to explain how settlement cues may be influenced. Observed shifts in the settlement of Galeolaria hystrix suggest that changes in community structure for invertebrates were at least in part linked to changes in microbial biofilm composition. This has broader implications as altered settlement and reduction in biofilms could drive important changes in benthic communities’ composition and distribution.
Presently, there is limited information on the potential effects of OA on the settlement process20,47. Mostly, it has shown that OA has a negative impact on settlement in marine invertebrates, however these conclusions are based on relatively few taxa, and mostly within the Cnidaria and Echinoderm phyla. In addition, settlement is a complex process that involves larval supply and behaviour, substrate characteristics and early post-settlement growth8,48, and the relative importance of OA-induced changes in each step required further research20. Similarly, settlement characteristics are likely species-species specific, with, for example in the degree of substrate selectivity. The present study provides investigations of the effects of OA on settlement substrates and settlement efficiency, and may provide the basis for future studies across a broader range of taxa.
Methods
Detailed methods are provided in the supplementary materials including experimental set up, sea water chemistry, microbial community analyses, larval rearing and statistical analyses.
Environmental pH measurements
In situ seawater pHT was measured using a SeaFET ocean pH sensor (Sea Bird Scientific, USA) deployed continuously for 42 days from 4 May to 15 June 2015 at 1.0 m depth, coinciding with our period of Experiment 1. The SeaFET instrument utilizes both an internal and an external reference electrode for pH measurements (i.e. pH (Internal) and pH (external), respectively) and if the instrument is correctly calibrated and there will be close agreement between both pH measurement (SeaFET user manual, Sea Bird Scientific Document SeaFET170601, March 2019). The SeaFET was set to record an average pH every 30 minutes from 20 rapid recordings containing a burst of 30 measurements. Post-deployment processing is required to calculate seawater pH, and this was made using SeaFETCom software version 1.2. These calculations use salinity (recorded independently at an average of 35 PSU) and the ambient sea temperature (°C) simultaneously recorded by the SeaFET. Calculated pH over time was graphed using R-code developed by Andrew Marriner, Ocean Atmosphere Technician at National Institute of Water and Atmospheric Research.
Experiment 1: Biofilm development under pH treatment
Biofilms were developed in flow-through seawater aquarium supplied with the desired pH seawater. Each pH level had an independent 70 L header tank in which the supplied seawater pH was adjusted, before flowing into four replicate 5 L aquaria which contained the biofilm slides (see Supplementary Fig. S8). The three pH treatments were ambient pHNIST 8.1 (pCO2 = 384 ppm), pHNIST 7.8 (near future, 2100, pCO2 = 1108 ppm) and pHNIST 7.4 (extreme, 2300, pCO2 = 2465 ppm). Biofilms were developed on glass microscope slides suspended below the water’s surface at a 45° angle perpendicular to flow-through direction. Samples for microbial community analysis were collected from an initial set of incubated slides incubated for up to 69 days in Experiment 1, while a second development of slides up to 60-days period was used for development of biofilms for settlement assays in Experiment 2. Biofilm wet weight biomass and microbial community analysis were collected from Experiment 1, and chlorophyll-a and carotenoid analysis were collected from Experiment 2. Biofilm and pigment analysis were made for 5 replicate slides from each treatment.
Experiment 1: Microbial biofilm characteristics and community analysis
Biofilm material was carefully removed from both sides of the lower 15 cm2 section of each slide (total area = 30 cm2) using a sterilised metal laboratory spatula. Samples were stored at −4 °C until transfer to the lab and stored until processing at −20 °C. Further processing was performed as previously described49. Total DNA was extracted from each individual biofilm sample using a MoBio PowerSoil DNA Isolation Kit (MO BIO Laboratories Inc., Solana Beach, CA, USA) following manufacturer’s protocol with the following modification: Bead-beating (2 × 15 s) cycles was performed using a 2010 GenoGrinder (SPEX SamplePrep, Metuchen, NJ, USA). After extraction, a Nanodrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to assess DNA quantity and quality.
The 16S rRNA gene amplicon sequencing was performed using primers 515 F/806 R (V4 region of the 16S gene) and the Earth Microbiome Project protocol (Version 4_13)50. All samples were sequenced on a single Illumina MiSeq run. Sequences were first processed in Qiime (version 1.9.1) using default parameters51 including minimum read length of 75 bp, min number of consecutive high quality base calls to include a read as a fraction of the input read length of 0.75, Phred quality score of 3, no ambiguous bases allowed, and no mismatches allowed in primer sequence52. All sequences kept for analysis were 151 bp. Sequence clustering (97% sequence similarity) into Operational Taxonomic Units (OTUs) was done using the SILVA (version 119) reference library53 and UCLUST54 following the open-reference Operational Taxonomic Unit (OTU) picking protocol. Taxonomy assignments were done using BLAST against the SILVA database (max-e value = 0.001)55. Subsampling and rarefactions (10 times) were performed to equal read depths of 8,000 per sample, and samples below that threshold were removed. After rarefaction, all 10 OTU tables were merged and exported for further processing in R56. The rarified OTU table (in biom format) was processed using the phyloseq package57. To account for the multiple rarefication (10 total) abundances, a mean was calculated (dividing by 10) and results were rounded to whole integers using the transform sample_counts() command. Taxa (OTUs) with less than 1 count were deleted using the prune_taxa() command. Alpha diversity (Shannon and richness) were calculated using the estimate_richness() command.
The NMDS plot was created using a Bray-Curtis distance matrix through “phyloseq” and “vegan”58 packages. Significant treatment and age effects were determined using an Anosim test. To determine samples forming statistically significant groups, a cluster analysis was performed using the pvclust package (method = Ward; distance matrix = Bray-Curtis; bootstrap value, n = 1000)59. Significant groups (representing 95% confidence) were marked with boxes (red). All data analyzed in this paper along with analysis code can be found at: https://github.com/semorales/Nelson_OA_2019.
Experiment 2: Biofilm modification and larval settlement
We examined the effect of biofilms developed under experimental pH conditions on the settlement success of the polychaete tubeworm Galeolaria hystrix. Settlement assays were conducted using G. hystrix larvae reared to competency using methods described by Nelson et al.25, and placed with 23-day old and 60-day old biofilms developed on glass slides in the flow-through system during experimental period 2. Settlement was measured and scored at 24 h and 48 h after the introduction of competent larvae to the substrates. Clean glass slides with no biofilm developed on the surface was used as a negative control for non-specific background settlement.
Supplementary information
Acknowledgements
We would like to acknowledge the technical staff at the Portobello Marine Laboratory, and Kim Currie for Carbonate Chemistry of water samples. KN was supported by the Duffus Lubecki Postgraduate Scholarship for Applied Science at Otago University and a discretionary scholarship from Ngai Tahu. FB was supported by a Rutherford Discovery Fellowship (Royal Society of NZ). ML was supported by the MBIE (Ministry for Business, Innovation and Employment) through the CARIM Programme (Coastal Ocean Acidification: Rate, Impacts, Management).
Author contributions
K.N. conceived of the study, designed the study, collected field data, carried out laboratory work and analyses, participated in data analysis, helped draft manuscript; F.B. lead data analysis and drafting of manuscript, S.M. lead graphical and statistical analyses, participated in data analysis, helped draft manuscript, coordinated completion of manuscript, M.L. supervised the study, helped design the study, participated in data analysis, helped draft manuscript, helped with laboratory work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Katie S. Nelson and Federico Baltar.
Contributor Information
Miles D. Lamare, Email: miles.lamare@otago.ac.nz
Sergio E. Morales, Email: sergio.morales@otago.ac.nz
Supplementary information
is available for this paper at 10.1038/s41598-020-60023-4.
References
- 1.IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. (Cambridge University Press, 2013).
- 2.Mandalakis M, et al. Microbial strains isolated from CO2-venting Kolumbo submarine volcano show enhanced co-tolerance to acidity and antibiotics. Mar. Env. Res. 2019;144:102–110. doi: 10.1016/j.marenvres.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Mostofa KMG, et al. Reviews and Syntheses: Ocean acidification and its potential impacts on marine ecosystems. Biogeosciences. 2016;13:1767–1786. doi: 10.5194/bg-13-1767-2016. [DOI] [Google Scholar]
- 4.Kroeker KJ, et al. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob. Chang. Biol. 2013;19:1884–1896. doi: 10.1111/gcb.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kroeker KJ, Kordas RL, Crim RN, Singh GG. Meta-analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecol. Lett. 2010;13:1419–1434. doi: 10.1111/j.1461-0248.2010.01518.x. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann GE, et al. The Effect of Ocean Acidification on Calcifying Organisms in Marine Ecosystems: An Organism-to-Ecosystem Perspective. Annu. Rev. Ecology, Evolution, Syst. 2010;41:127–147. doi: 10.1146/annurev.ecolsys.110308.120227. [DOI] [Google Scholar]
- 7.Przeslawski R, Byrne M, Mellin C. A review and meta-analysis of the effects of multiple abiotic stressors on marine embryos and larvae. Glob. Chang. Biol. 2015;21:2122–2140. doi: 10.1111/gcb.12833. [DOI] [PubMed] [Google Scholar]
- 8.Byrne M. Impact of Ocean Warming and Ocean Acidification on Marine Invertebrate Life History Stages: Vulnerabilities and Potential for Persistence in a Changing Ocean. Oceanography Mar. Biology: An. Annu. Rev. 2011;49:1–42. [Google Scholar]
- 9.Wolfe K, Dworjanyn SA, Byrne M. Effects of ocean warming and acidification on survival, growth and skeletal development in the early benthic juvenile sea urchin (Heliocidaris erythrogramma) Glob. Chang. Biol. 2013;19:2698–2707. doi: 10.1111/gcb.12249. [DOI] [PubMed] [Google Scholar]
- 10.Byrne M, Lamare M, Winter D, Dworjanyn SA, Uthicke S. The stunting effect of a high CO2 ocean on calcification and development in sea urchin larvae, a synthesis from the tropics to the poles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120439. doi: 10.1098/rstb.2012.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uthicke S, et al. Impacts of ocean acidification on early life-history stages and settlement of the coral-eating sea star Acanthaster planci. PLoS One. 2013;8:e82938. doi: 10.1371/journal.pone.0082938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldbusser GG, et al. Saturation-state sensitivity of marine bivalve larvae to ocean acidification. Nat. Clim. Change. 2014;5:273–280. doi: 10.1038/nclimate2479. [DOI] [Google Scholar]
- 13.Stumpp M, et al. Acidified seawater impacts sea urchin larvae pH regulatory systems relevant for calcification. Proc. Natl Acad. Sci. USA. 2012;109:18192–18197. doi: 10.1073/pnas.1209174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milazzo M, et al. Ocean acidification impairs vermetid reef recruitment. Sci. Rep. 2014;4:4189. doi: 10.1038/srep04189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doropoulos C, Diaz-Pulido G. High CO2 reduces the settlement of a spawning coral on three common species of crustose coralline algae. Mar. Ecol. Prog. Ser. 2013;475:93–99. doi: 10.3354/meps10096. [DOI] [Google Scholar]
- 16.Webster NS, Uthicke S, Botte ES, Flores F, Negri AP. Ocean acidification reduces induction of coral settlement by crustose coralline algae. Glob Chang. Biol. 2013;19:303–315. doi: 10.1111/gcb.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albright R, Mason B, Miller M, Langdon C. Ocean acidification compromises recruitment success of the threatened Caribbean coral Acropora palmata. Proc. Natl Acad. Sci. USA. 2010;107:20400–20404. doi: 10.1073/pnas.1007273107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura M, Ohki S, Suzuki A, Sakai K. Coral larvae under ocean acidification: survival, metabolism, and metamorphosis. PLoS One. 2011;6:e14521. doi: 10.1371/journal.pone.0014521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupont S, Dorey N, Stumpp M, Melzner F, Thorndyke M. Long-term and trans-life-cycle effects of exposure to ocean acidification in the green sea urchin Strongylocentrotus droebachiensis. Mar. Biol. 2013;160:1835–1843. doi: 10.1007/s00227-012-1921-x. [DOI] [Google Scholar]
- 20.Espinel-Velasco N, et al. Effects of ocean acidification on the settlement and metamorphosis of marine invertebrate and fish larvae: a review. Mar. Ecol. Prog. Ser. 2018;606:237–257. doi: 10.3354/meps12754. [DOI] [Google Scholar]
- 21.Whalan S, Webster NS. Sponge larval settlement cues: the role of microbial biofilms in a warming ocean. Sci. Rep. 2014;4:4072. doi: 10.1038/srep04072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadfield MG. Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Ann. Rev. Mar. Sci. 2011;3:453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 23.Sneed Jennifer M., Sharp Koty H., Ritchie Kimberly B., Paul Valerie J. The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proceedings of the Royal Society B: Biological Sciences. 2014;281(1786):20133086. doi: 10.1098/rspb.2013.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura M, Koyama T, Nakano Y, Uemura D. Characterization of a natural inducer of coral larval metamorphosis. J. Exp. Mar. Biol. Ecol. 2007;340:96–102. doi: 10.1016/j.jembe.2006.08.012. [DOI] [Google Scholar]
- 25.Nelson KS, Liddy M, Lamare MD. Embryology, larval development, settlement and metamorphosis in the New Zealand Serpulid Polychaete Galeolaria hystrix. Invertebrate Reprod. Dev. 2017;61:207–217. doi: 10.1080/07924259.2017.1318183. [DOI] [Google Scholar]
- 26.Hofmann GE, et al. High-frequency dynamics of ocean pH: a multi-ecosystem comparison. PLoS One. 2011;6:e28983. doi: 10.1371/journal.pone.0028983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caldeira K, Wickett ME. Oceanography: anthropogenic carbon and ocean pH. Nat. 2003;425:365. doi: 10.1038/425365a. [DOI] [PubMed] [Google Scholar]
- 28.McDonald MR, et al. Effects of ocean acidification over the life history of the barnacle Amphibalanus amphitrite. Mar. Ecol. Prog. Ser. 2009;385:179–187. doi: 10.3354/meps08099. [DOI] [Google Scholar]
- 29.García E, et al. Robustness of Paracentrotus lividus larval and post-larval development to pH levels projected for the turn of the century. Mar. Biol. 2015;162:2047–2055. doi: 10.1007/s00227-015-2731-8. [DOI] [Google Scholar]
- 30.Jansson A, Lischka S, Boxhammer T, Schulz KG, Norkko J. Larval development and settling of Macoma balthica in a large-scale mesocosm experiment at different fCO2 levels. Biogeosciences Discuss. 2015;12:20411–20435. doi: 10.5194/bgd-12-20411-2015. [DOI] [Google Scholar]
- 31.Pecquet A, Dorey N, Chan KYK. Ocean acidification increases larval swimming speed and has limited effects on spawning and settlement of a robust fouling bryozoan, Bugula neritina. Mar. Pollut. Bull. 2017;124:903–910. doi: 10.1016/j.marpolbul.2017.02.057. [DOI] [PubMed] [Google Scholar]
- 32.Hall-Spencer JM, et al. Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nat. 2008;454:96–99. doi: 10.1038/nature07051. [DOI] [PubMed] [Google Scholar]
- 33.Cigliano M, Gambi MC, Rodolfo-Metalpa R, Patti FP, Hall-Spencer JM. Effects of ocean acidification on invertebrate settlement at volcanic CO2 vents. Mar. Biol. 2010;157:2489–2502. doi: 10.1007/s00227-010-1513-6. [DOI] [Google Scholar]
- 34.Lidbury I, Johnson V, Hall-Spencer JM, Munn CB, Cunliffe M. Community-level response of coastal microbial biofilms to ocean acidification in a natural carbon dioxide vent ecosystem. Mar. Pollut. Bull. 2012;64:1063–1066. doi: 10.1016/j.marpolbul.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Ricevuto E, Kroeker KJ, Ferrigno F, Micheli F, Gambi MC. Spatio-temporal variability of polychaete colonization at volcanic CO2 vents indicates high tolerance to ocean acidification. Mar. Biol. 2014;161:2909–2919. doi: 10.1007/s00227-014-2555-y. [DOI] [Google Scholar]
- 36.Fabricius KE, Kluibenschedl A, Harrington L, Noonan S, De’ath G. In situ changes of tropical crustose coralline algae along carbon dioxide gradients. Sci. Rep. 2015;5:9537. doi: 10.1038/srep09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamare Miles D., Liddy Michelle, Uthicke Sven. In situ developmental responses of tropical sea urchin larvae to ocean acidification conditions at naturally elevated p CO 2 vent sites. Proceedings of the Royal Society B: Biological Sciences. 2016;283(1843):20161506. doi: 10.1098/rspb.2016.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raulf FF, et al. Changes in microbial communities in coastal sediments along natural CO2 gradients at a volcanic vent in Papua New Guinea. Env. Microbiol. 2015;17:3678–3691. doi: 10.1111/1462-2920.12729. [DOI] [PubMed] [Google Scholar]
- 39.Lindh MV, et al. Consequences of increased temperature and acidification on bacterioplankton community composition during a mesocosm spring bloom in the Baltic Sea. Env. Microbiol. Rep. 2013;5:252–262. doi: 10.1111/1758-2229.12009. [DOI] [PubMed] [Google Scholar]
- 40.Baltar, F. et al. Response of rare, common and abundant bacterioplankton to anthropogenic perturbations in a Mediterranean coastal site. FEMS Microbiol Ecol91, 10.1093/femsec/fiv058 (2015). [DOI] [PubMed]
- 41.Kroeker KJ, Micheli F, Gambi MC. Ocean acidification causes ecosystem shifts via altered competitive interactions. Nat. Clim. Change. 2012;3:156–159. doi: 10.1038/nclimate1680. [DOI] [Google Scholar]
- 42.Johnson CR, Sutton DC. Bacteria on the Surface of Crustose Coralline Algae Induce Metamorphosis of the Crown-of-Thorns Starfish Acanthaster Planci. Mar. Biol. 1994;120:305–310. doi: 10.1007/Bf00349692. [DOI] [Google Scholar]
- 43.Hadfield, M. & Paul, V. in Marine Chemical Ecology Marine Science 431–461 (2001).
- 44.Huggett MJ, Williamson JE, de Nys R, Kjelleberg S, Steinberg PD. Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia. 2006;149:604–619. doi: 10.1007/s00442-006-0470-8. [DOI] [PubMed] [Google Scholar]
- 45.Pearce CM, Scheibling RE. Induction of Metamorphosis of Larvae of the Green Sea Urchin, Strongylocentrotus droebachiensis, by Coralline Red Algae. Biol. Bull. 1990;179:304–311. doi: 10.2307/1542322. [DOI] [PubMed] [Google Scholar]
- 46.Toupoint N, et al. Effect of biofilm age on settlement of Mytilus edulis. Biofouling. 2012;28:985–1001. doi: 10.1080/08927014.2012.725202. [DOI] [PubMed] [Google Scholar]
- 47.Ross PM, Parker L, O’Connor WA, Bailey EA. The Impact of Ocean Acidification on Reproduction, Early Development and Settlement of Marine Organisms. Water. 2011;3:1005–1030. doi: 10.3390/w3041005. [DOI] [Google Scholar]
- 48.Kurihara H. Effects of CO2-driven ocean acidification on the early developmental stages of invertebrates. Mar. Ecol. Prog. Ser. 2008;373:275–284. doi: 10.3354/meps07802. [DOI] [Google Scholar]
- 49.Samad MS, et al. Response to nitrogen addition reveals metabolic and ecological strategies of soil bacteria. Mol. Ecol. 2017;26:5500–5514. doi: 10.1111/mec.14275. [DOI] [PubMed] [Google Scholar]
- 50.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinforma. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 55.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 56.R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2018).
- 57.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oksanen, J., et al Vegan: Community Ecology Package, R package, v. 2.4–6 (2018).
- 59.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinforma. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.