Abstract
Vulnerable populations of wild yak (Bos mutus), the wild ancestral species of domestic yak, survive in extremely cold, harsh and oxygen-poor regions of the Qinghai-Tibetan Plateau (QTP) and adjacent high-altitude regions. In this study, we sequenced and assembled its genome de novo. In total, six different insert-size libraries were sequenced, and 662 Gb of clean data were generated. The assembled wild yak genome is 2.83 Gb in length, with an N50 contig size of 63.2 kb and a scaffold size of 16.3 Mb. BUSCO assessment indicated that 93.8% of the highly conserved mammal genes were completely present in the genome assembly. Annotation of the wild yak genome assembly identified 1.41 Gb (49.65%) of repetitive sequences and a total of 22,910 protein-coding genes, including 20,660 (90.18%) annotated with functional terms. This first construction of the wild yak genome provides a variable genetic resource that will facilitate further study of the genetic diversity of bovine species and accelerate yak breeding efforts.
Subject terms: DNA sequencing, Genome, Zoology
| Measurement(s) | DNA • genome • sequence_assembly • sequence feature annotation |
| Technology Type(s) | DNA sequencing assay • sequence assembly process • sequence annotation |
| Sample Characteristic - Organism | Bos mutus |
| Sample Characteristic - Environment | Qinghai-Tibetan Plateau |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.11778669
Background & Summary
The yak, which can survive extremely cold, harsh and oxygen-poor conditions, is endemic to the Qinghai-Tibetan Plateau (QTP) and adjacent high-altitude regions1. Yak were domesticated by nomadic people from wild yak at least 7300 years ago2. Nowadays, more than 22 million domestic yak (Bos grunniens) provide necessities, such as food, transport, shelter and fuel, for Tibetans and other humans in high-altitude areas1. In addition, there are still 15–20 thousand wild yak (Bos mutus) surviving in northwestern parts of the QTP3,4. Due to long-term over-breeding and inbreeding caused by traditional yak breeding practices, the reproductive capacity, growth rate, adult size and milk production of domestic yak have declined and mortality has increased, especially among the newborn and young1. However, Datong yak, the only artificially cultivated yak breed that is a cross between wild yak and domestic yak, shows excellent growth characteristics and production performance. Datong yak are generally 30 and 50% heavier than domestic yak at birth and six months of age, respectively, produce more than 15% milk, and have 25 and 31% higher carcass weights at ages of 6 and 18 months, respectively5,6. The high growth, development and production rates of the Datong yak show the feasibility of improving traits of domestic yak with wild yak resources, and potential importance of exploiting wild yak genetic resources for yak breeding in the future. However, the wild yak genome has not been previously sequenced, which has impeded both research and breeding efforts.
Thus, to elucidate genomic features of this vulnerable species, we have constructed a draft genome for wild yak. We extracted genomic DNA from blood tissues, constructed 3 Paired-End (PE) and 3 Mate-Pair (MP) libraries, which were sequenced using the Illumina HiSeq. 2000 platform. After quality filtering and trimming of raw data, Genome Characteristics Estimation (GCE, v1.0.0)7 software was employed to evaluate the genome size using PE reads, and Platanus v1.2.48 to assemble the genome using all clean data. In addition, GapCloser v1.129 was used to perform another round of gap closure based on the assembly results. The final genome assembly size was 2.83 Gb, containing 808,541 contigs (N50 = 63.2 kb) and 734,073 scaffolds (N50 = 16.3 Mb), representing 91.5% of the estimated genome. Structural annotation of the genome yielded 22,910 genes, 90.18% of which could be functionally annotated with at least one of the five protein databases (TrEMBL, SwissProt, InterPro, GO and KEGG). The wild yak genome assembled in this study provides a valuable genetic resource for future efforts to protect the vulnerable wild yak and further comparative analysis of genome biology among bovine species to promote breeding research.
Methods
Sample collection, library construction and sequencing
Genomic DNA was extracted from a blood sample collected from a female wild yak originally captured from the wild and reared at the Datong Yak Farm of Qinghai Province (37°15′35.6″N, 101°22′24.0″E, altitude around 3200 m) using a standard phenol/chloroform method. The quality and integrity of the extracted DNA were checked by measuring its A260/A280 ratio and agarose gel electrophoresis. For paired-end libraries with insert sizes of 280, 500 and 800 bp, 6 μg portions of genomic DNA were used to generate the corresponding libraries using Illumina TruSeq DNA Nano Preparation Kit (Illumina, San Diego, CA, USA). For mate-pair libraries with insert sizes of 2, 5 and 10 kb, 60 μg portions of DNA were used for circularization and further library construction using Nextera Mate Pair Library Preparation Kit (Illumina, San Diego, CA, USA). Both the sample collection and experimental library construction protocol were approved by the Ethical Committees of Lanzhou University. All libraries were sequenced on an Illumina HiSeq. 2000 platform with 150 bp read length, following the manufacturer’s instructions. Finally, 760.85 Gb of raw data were generated in total (Table 1).
Table 1.
Summary statistics of wild yak sequenced reads.
| Library Insert Size (bp) | Raw reads | Qualified reads | ||||
|---|---|---|---|---|---|---|
| Total Data (Gbp) | Reads Length (bp) | Sequence coverage (×) | Total Data (Gbp) | Reads Length (bp) | Sequence coverage (×) | |
| 280 | 106.81 | 150.00 | 35.60 | 103.96 | 145.26 | 34.65 |
| 500 | 89.10 | 150.00 | 29.70 | 86.03 | 144.27 | 28.68 |
| 800 | 109.24 | 150.00 | 36.41 | 102.80 | 140.32 | 34.27 |
| 2,000 | 117.87 | 150.00 | 39.29 | 108.44 | 150.00 | 36.15 |
| 5,000 | 166.14 | 150.00 | 55.38 | 147.18 | 150.00 | 49.05 |
| 10,000 | 171.69 | 150.00 | 57.23 | 113.89 | 150.00 | 37.96 |
| Total | 760.85 | — | 253.62 | 662.30 | — | 220.77 |
Preprocessing and genome size estimation
All the sequencing reads were preprocessed for quality control and filtered with stringent criteria using Lighter v1.1.110 software. Firstly, raw data were filtered by removing reads with >10% unknown bases. Then, paired reads with low-quality bases (quality scores ≤7) covering more than 65% of the read length were filtered out. Reads with PCR duplicates or adapter contamination were also removed. Finally, both read 1 and read 2 files were filtered out if they had >10 bp overlap, allowing 10% mismatch. In total, 662.3 Gb of clean reads were obtained after filtering (Table 1).
Prior to genome assembly, all the preprocessed sequences from the short-insert library were subjected to genome size estimation using Genome Characteristics Estimation (GCE) with a k value of 21. The genome size of wild yak was estimated to be around 3.09 Gb, using the following formula: genome size = k-mer number/k-mer depth, where the k-mer number refers to the total number of k-mers, and k-mer depth is the depth of the main peak in the k-mer frequency distribution (Fig. 1).
Fig. 1.
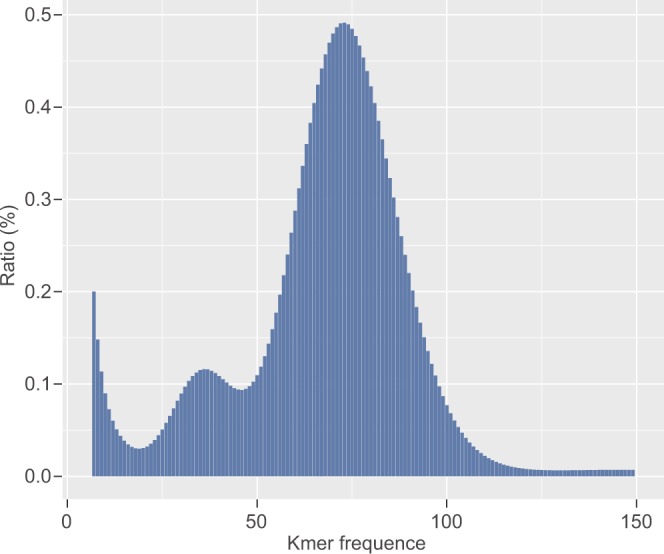
21-mer distribution in the wild yak genome.
Genome assembly
For de novo genome assembly, Platanus software was used for constructing contigs and scaffolds with default parameters, and GapCloser was employed to fill the remaining gaps in the scaffolds with all sequencing reads. These steps finally yielded a wild yak draft genome with a total length of 2.83 Gb, accounting for 91.5% of the estimated genome size (contig and scaffold N50 sizes: 63.2 kb and 16.3 Mb, respectively) (Table 2).
Table 2.
Summary statistics of the wild yak genome assembly.
| Contig | Scaffold | |||
|---|---|---|---|---|
| Size (bp) | Number | Size (bp) | Number | |
| N90 | 8,406 | 52,665 | 418,892 | 355 |
| N80 | 22,366 | 33,612 | 3,980,782 | 155 |
| N70 | 35,157 | 23,897 | 6,690,007 | 102 |
| N60 | 48,527 | 17,241 | 12,581,129 | 71 |
| N50 | 63,194 | 12,274 | 16,301,239 | 50 |
| Longest | 811,040 | 75,900,441 | ||
| Total Size | 2,751,522,574 | 2,831,279,091 | ||
| Total Number (>100 bp) | 808,541 | 734,073 | ||
| Total Number (>2 kb) | 83,689 | 19,451 | ||
| GC content | 1,154,582,425 (41.96%) | |||
To evaluate the completeness of our assembly, we carried out BUSCO11 analyses and the results indicated that 3,974 of the 4,104 conserved single-copy genes in mammals were present in our assembly, of which 3,799 were single, 55 were duplicated, and 120 fragmented matches (Table 3). To validate the single-base accuracy of the genome assembly, we aligned the high-quality reads of short-insert libraries to the assembly using Burrows-Wheeler Aligner (BWA, v0.7.15-r1140)12 software, and the alignment outputs were converted to Binary Alignment Map (BAM) format via SAMtools v1.313. The genome coverage was then calculated by a custom Perl script, which indicated that more than 93.9% of the assembly had >20-fold coverage.
Table 3.
Summary of BUSCO analysis results: matches to 4104 single-copy orthologs in mammalia_obd9.
| BUSCO mode | Species | Complete one-to-one match to ortholog | Complete match of multi gene copies to ortholog | Fragmented match to ortholog | Total number of matches to ortholog | No match to ortholog |
|---|---|---|---|---|---|---|
| Genome | Wild yak | 3792 (92.4%) | 59 | 123 | 3973 (96.8%) | 130 |
| Domestic yak (version 1.1) | 3809 (92.8%) | 32 | 138 | 3979 (97.0%) | 125 | |
| Bos taurus (UMD3.1) | 3794 (92.4%) | 53 | 124 | 3971 (96.8%) | 133 | |
| Wisent (version 1.0) | 3763 (91.7%) | 31 | 180 | 3979 (96.8%) | 130 | |
| OGS | Wild yak | 3821 (93.1%) | 70 | 119 | 4010 (97.7%) | 94 |
| Domestic yak (version 1.1) | 3987 (97.2%) | 27 | 59 | 4073 (99.2%) | 31 | |
| Bos taurus (UMD3.1) | 4009 (97.7%) | 24 | 50 | 4083 (99.5%) | 21 | |
| Wisent (version 1.0) | 3840 (93.6%) | 63 | 165 | 4068 (99.1%) | 36 |
Repeat annotation
Repetitive regions of the wild yak genome were identified using a combination of de novo and homology-based approaches, as applied in a previous analysis of the Ovis ammon polii genome14. For the de novo prediction, RepeatModeler v1.0.11 was employed first to construct a de novo repeat library, then RepeatMasker v4.0.715 was used to identify repeats using both the RepBase16 library of known transposable elements (TEs) and a self-trained repeat database. Next, we applied RepeatProteinMask (a package in RepeatMasker) to identify repeats at the protein level using the TE protein database. In addition, tandem repeats were further annotated using Tandem Repeat Finder (TRF, v4.0.9)17. Finally, the non-redundant repeats were checked according to their coordinates in the genome. Overall, we identified 1.41 Gbp of non-redundant repetitive sequences, representing 49.65% of the wild yak genome assembly; of which long interspersed elements (LINE) were the most abundant, accounting for 35.98% of the whole genome (Fig. 2; Table 4).
Fig. 2.
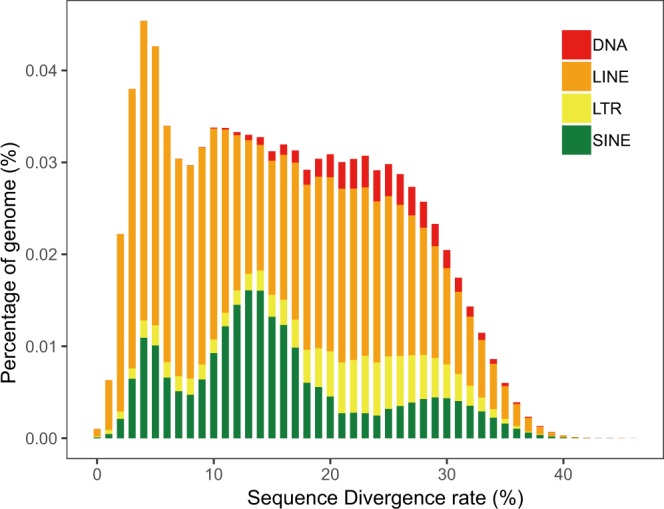
Sequences divergence rate of repeats annotated by RepeatMasker in wild yak. The x-axis represents the sequence divergence rate of repeats. The y-axis represents the percentage of repeat sequences in the genome.
Table 4.
Summary statistics of interspersed repeats in the assembled wild yak genome.
| Type | Repbase TEs | TE proteins | De novo | Combined TEs | ||||
|---|---|---|---|---|---|---|---|---|
| Length (bp) | % in genome | Length (bp) | % in genome | Length (bp) | % in genome | Length (bp) | % in genome | |
| DNA | 58,420,396 | 2.06 | 2,646,996 | 0.09 | 33,460,579 | 1.18 | 68,427,926 | 2.42 |
| LINE | 758,982,393 | 26.81 | 551,346,462 | 19.47 | 680,109,554 | 24.02 | 1,018,625,838 | 35.98 |
| LTR | 130,528,749 | 4.61 | 6,744,768 | 0.24 | 84,503,020 | 2.98 | 141,521,674 | 5 |
| SINE | 279,268,337 | 9.86 | 0 | 0 | 63,907,283 | 2.26 | 324,361,957 | 11.46 |
| Other | 36,651,120 | 1.29 | 123 | 0 | 63,897,730 | 2.26 | 64,143,395 | 2.27 |
| Unknown | 496,273 | 0.02 | 0 | 0 | 227,519,165 | 8.04 | 228,015,202 | 8.05 |
| Total | 1,264,055,747 | 44.65 | 560,660,398 | 19.8 | 1,128,847,407 | 39.87 | 1,396,485,261 | 49.32 |
Gene prediction and annotation
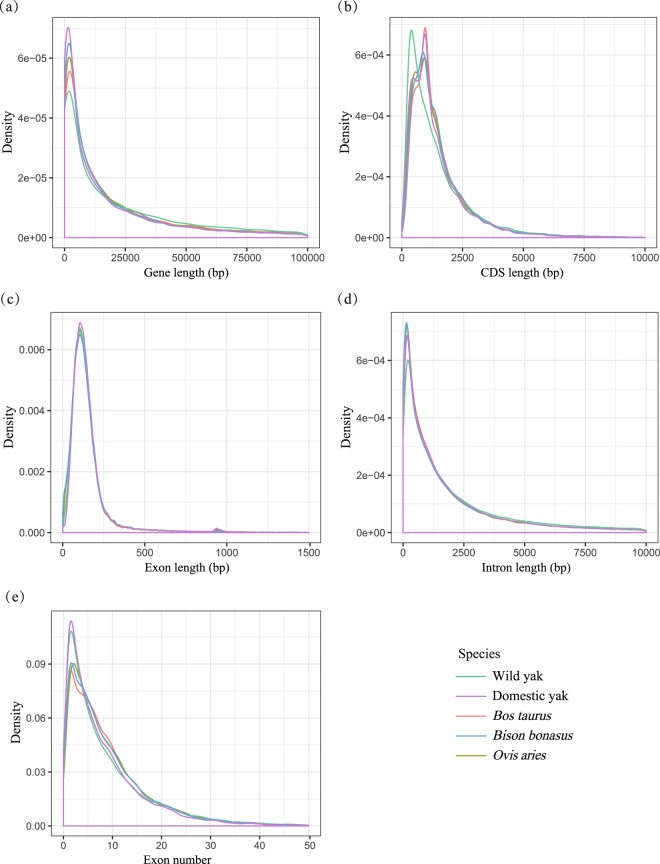
We employed a combination of homology-based and de novo prediction methods to identify protein-coding genes. For homology-based prediction, protein sequences of seven species (Bos taurus, Equus caballus, Homo sapiens, Ovis aries, Sus scrofa, Bison bonasus, Bos grunniens) downloaded from Ensembl18 and GigaDB19,20 were aligned to the wild yak genome using TBLASTN21. Then GeneWise v2.4.122 software was applied to search for accurately spliced alignments based on the filtered homologous genome sequences. For de novo prediction, we used Augustus23, Geneid24, GeneMark, GlimmerHMM25 and SNAP26 to predict genes with parameters trained on wild yak and human repeat-masked genomes. EVidenceModeler software (EVM, v1.1.1)27 was employed to generate a consensus gene set by integrating the genes predicted by the homology and de novo approaches. Low-quality genes of short length (proteins with fewer than 30 amino acids) and/or exhibiting premature termination were removed to produce the final gene set, which is composed of 22,910 genes (Fig. 3; Table 5).
Fig. 3.
Comparison of structural characteristics of the wild yak genes with those of other mammals. (a) mRNA length, (b) CDS length, (c) Exon length, (d) Intron length, (e) Exon number per gene of wild yak, domestic yak, Bos taurus (UMD3.1), Ovis aries (Oar v3.1) and Bison bonasus (version 1.0). The x-axis represents length or number and the y-axis represents the density of genes.
Table 5.
Summary statistics of predicted protein-coding genes in the wild yak genome.
| Gene set | Total Genes Predicted | Average Gene Length (bp) | Average CDS Length (bp) | Average Exons per Gene | Average Exon Length (bp) | Average Intron Length (bp) |
|---|---|---|---|---|---|---|
| Augustus | 21,211 | 50,645.17 | 1,531.32 | 9.04 | 169.39 | 6,096.22 |
| Geneid | 43,221 | 49,433.27 | 1,165.53 | 8.92 | 130.65 | 6,093.69 |
| Genemark | 159,214 | 1,442.59 | 356.34 | 2.52 | 141.29 | 713.63 |
| GlimmerHMM | 4,944 | 8,587.79 | 849.35 | 5.14 | 165.21 | 1,868.66 |
| SNAP | 120,892 | 11,877.32 | 799.25 | 5.36 | 149.13 | 2,541.06 |
| Bos taurus (UMD3.1) | 22,964 | 21,477.93 | 1,260.50 | 7.06 | 178.47 | 3,377.48 |
| Equus caballus (EquCab2) | 20,940 | 20,200.57 | 1,236.63 | 6.95 | 177.96 | 3,239.26 |
| Capra hircus (ARS1) | 23,252 | 24,941.52 | 1,303.75 | 7.25 | 179.78 | 3,834.31 |
| Homo sapiens (GRCH38) | 20,889 | 21,860.19 | 1,294.84 | 7.10 | 182.48 | 3,406.80 |
| Ovis aries (Oar v3.1) | 23,966 | 20,450.21 | 1,205.62 | 6.72 | 179.28 | 3,401.54 |
|
Bison bonasus (version 1.0) |
23,422 | 22,271.56 | 1,279.32 | 7.20 | 177.62 | 3,422.69 |
| Bos grunniens (version 1.1) | 24,478 | 19,939.52 | 1,192.05 | 6.66 | 179.01 | 3,355.53 |
| Final set | 22,910 | 47,211.29 | 1,547.06 | 9.28 | 166.73 | 5,515.67 |
Putative biological functions of these predicted high-quality genes were assigned by searching against five publicly available databases: TrEMBL, Swiss-Prot28, InterPro29, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)30. Approximately 90.18% of these genes were functionally annotated with at least one of these databases, with 90.05, 88.15, 83.51, 64.20 and 53.01% scoring positive hits in TrEMBL, SwissProt, InterPro, GO and KEGG, respectively (Table 6).
Table 6.
Number of predicted genes in wild yak functionally annotated using indicated databases.
| Database | Number | Percent(%) |
|---|---|---|
| Kegg | 12,145 | 53.01 |
| InterProScan | 19,132 | 83.51 |
| GO | 14,709 | 64.2 |
| Swissprot | 20,195 | 88.15 |
| Trembl | 20,631 | 90.05 |
| Annotated | 20,660 | 90.18 |
| Total Gene | 22,910 | — |
Data Records
The whole genome sequencing data were submitted to the NCBI Sequence Read Archive (SRA) database with accession number SRP194583 and Bioproject accession PRJNA53139831. The assembled draft genome of wild yak has been deposited at GenBank under the accession number of VBQZ0000000032. The annotation results of repeated sequences, gene structure and functional prediction were deposited in the Figshare database33.
Technical Validation
Quality assessment of the genome assembly
The assembly presented here is the first wild yak genome version. The contig N50 and scaffold N50 sizes were 63.2 kb and 16.3 Mb respectively, with the longest scaffold 75,900,441 bp. There are 258 scaffolds more than 1 Mb long, with a total length of 2,486,540,864 bp, representing 87.83% of the wild yak genome. By aligning the reads of short insert libraries to the wild yak assembly, we found more than 93.9% of the genome had >20-fold coverage, indicating high accuracy at the nucleotide level. BUSCO analysis carried out to assess the completeness of our assembly resulted in a BUSCO score of 96.8% (complete = 93.8%, single = 92.4%, duplicated = 1.4%, fragmented = 3.0%, missed = 3.2%, genes = 4,104). These results are comparable with those for the published European bison (wisent)34 and domestic yak4 genomes, suggesting our assembly has high quality and is quite complete.
Gene prediction and annotation validation
Gene models in the wild yak assembly were predicted using a combination of homology-based and ab initio gene approaches. Then EVM software was employed to integrate the gene prediction results to produce a consensus gene set. To enhance the quality of the gene prediction, we removed low-quality genes of short length (proteins with fewer than 30 amino acids) and/or exhibiting premature termination. The final gene set consisted of 22,910 genes, and the distributions of gene length, CDS length, exon length, intron length and exon number were similar to those of other mammals (Fig. 3). BUSCO analysis was also performed to assess the completeness of these predicted genes, resulting in a BUSCO value of 97.7% (complete = 94.8%, single = 93.1%, duplicated = 1.7%, fragmented = 2.9%, missed = 2.3%, genes = 4,104) (Table 6). In addition, functional annotation of these predicted genes indicated that 90.18% of them could be assigned to at least one functional term (Table 5). These results clearly indicate that the annotated gene set of the wild yak genome is quite complete.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant nos. 31661143020, 41620104007, 31801089), the National Program for Support of Top-notch Young Professionals, and the Fok Ying Tung Education Foundation (151105).
Author contributions
Q. Qiu and K. Wang designed and supervised the project; B. Yan and Q. Ren prepared the samples; Y. Liu, J. Luo, J. Dou and B. Tang analyzed the data; Y. Liu wrote the manuscript with other authors’ help and Q. Qiu and K. Wang revised the manuscript. All authors read and approved the final manuscript.
Code availability
The software versions, settings and parameters used are described below.
(1) GCE, version 1.0.0, parameters used: kmer_freq_hash -k 21 -l reads.list -t 24 -i 5000000 -o 0 -p wild_yak & > kmer_freq.log; gce -f wild_yak.freq.stat -c 46 -g 155866493014 -m 1 -D 8 -b 1 > wild_yak.Table 2> wild_yak.log.
(2) Lighter, version 1.1.1, parameters used: -k 17 3000000000 -trim -t 20.
(3) Platanus, version 1.2.4, parameters used: platanus assemble -o wild_yak -f <insert size 280 bp pair-end reads> <insert size 500 bp pair-end reads> <insert size 800 bp pair-end reads> -t 30 -m 500 -tmp temp; platanus scaffold -o wild_yak -c wild_yak_contig.fa -b wild_yak_contigBubble.fa -IP1 < insert size 280 bp pair-end reads> -a1 280 -IP2 <insert size 500 bp pair-end reads> -a2 500 -IP3 < insert size 800 bp pair-end reads > -a3 800 -OP4 <insert size 2 k pair-end reads> -a4 2000 -OP5 <insert size 5 k pair-end reads> -a5 5000 -OP6 <insert size 10 k pair-end reads> -a6 10000; platanus gap_close -o wild_yak -c wild_yak_scaffold.fa -IP1 <insert size 280 bp pair-end reads> -IP2 <insert size 500 bp pair-end reads> -IP3 <insert size 800 bp pair-end reads> -OP4 < insert size 2 k pair-end reads > -OP5 <insert size 5 k pair-end reads> -OP6 <insert size 10 k pair-end reads>.
(4) Gap Closer, version 1.12, parameters used: -l 150 -t 30, in configFile: max_rd_len = 100; Paired-end libs: reverse_seq = 0, asm_flags = 3; Mate-pair libs: reverse_seq = 11, asm_flags = 2.
(5) BUSCO, version 3: mammal default parameters, mammalia_odb9.
(6) BWA, version 0.7.15-r1140: default parameters.
(7) SAMtools, version 1.3; default parameters.
(8) RepeatMasker, version 4.0.7 (with RepBase library release-20170127).
(9) RepeatModeler, RepeatModeler-open-1.0.11.
(10) TRF, version 4.09, parameters used: trf wild_yak.gapclose.fa 2 7 7 80 10 50 500 -d -h.
(11) TBLASTN, version 2.5.0, parameter used: –e 1E-5.
(12) GeneWise, version 2.4.1, parameters used: -tfor/-trev (-rfor for genes on forward strand and -trev for reverse strand) -gff.
(13) Augustus, version 3.2.3, parameter used: -species = human.
(14) Geneid, version 1.0, parameters used: -3 -P.
(15) Genemark, version 3.9, parameter used: -f gff3.
(16) Snap, version 2006-07-28, parameter used: –gff.
(17) GlimmerHMM, version 3.0.4, default parameters.
(18) EVM, version 1.1.1, default parameters.
(19) InterProScan, version 5.25-64.0, parameters: -f tsv -iprlookup -goterms -pa -t p.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kun Wang, Email: wangkun@nwpu.edu.cn.
Qiang Qiu, Email: qiuqiang@lzu.edu.cn.
References
- 1.Wiener, G., Han, J. & Long, R. The Yak 2nd edn. (Regional Office for Asia and the Pacific, Food and Agriculture Organization of the United Nations, Bangkok, 2003).
- 2.Qiu Q, et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 2015;6:10283. doi: 10.1038/ncomms10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaller GB, Liu W. Distribution, status, and conservation of wild yak Bos grunniens. Biol. Conserv. 1996;76:1–8. doi: 10.1016/0006-3207(96)85972-6. [DOI] [Google Scholar]
- 4.Qiu Q, et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012;44:946–949. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- 5.Jialin B, Mingqiang W, Zhonglin L, Chesworth JM. Meat production from crossbred and domestic yaks in China. Anim. Sci. 1998;66:465–469. doi: 10.1017/S1357729800009620. [DOI] [Google Scholar]
- 6.Jialin B, Mingqiang W, Zhonglin L, Chesworth JM. The milking performance of dual-purpose crossbred yaks. Anim. Sci. 1998;66:471–473. doi: 10.1017/S1357729800009632. [DOI] [Google Scholar]
- 7.Liu, B. et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant. Biol. (2013).
- 8.Kajitani R, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song L, Florea L, Langmead B. Lighter: fast and memory-efficient sequencing error correction without counting. Genome Biol. 2014;15:1. doi: 10.1186/s13059-014-0509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, et al. Draft genome of the Marco Polo Sheep (Ovis ammon polii) Gigascience. 2017;6:1–7. doi: 10.1093/gigascience/gix106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. in Bioinformatics. 2009;Chapter 4:4.10.11–14.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 16.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 17.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu Q, 2013. Genomic data from the domestic yak (Bos grunniens) GigaScience Database. [DOI]
- 20.Wang K, 2017. Draft genome of European bison (wisent), Bison bonasus. GigaScience Database. [DOI]
- 21.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco, E., Parra, G. & Guigo, R. Using geneid to Identify Genes. Curr. Protoc. in Bioinform. 4.3.1–4.3.28 (2007). [DOI] [PubMed]
- 25.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 26.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.2019. NCBI Sequence Read Archive. SRP194583
- 32.Liu Y. 2019. Bos mutus breed Datong Yak isolate WY2019, whole genome shotgun sequencing project. GenBank. VBQZ00000000
- 33.Liu Y. 2019. The sequence and de novo assembly of the wild yak genome. figshare. [DOI] [PMC free article] [PubMed]
- 34.Wang K, et al. The genome sequence of the wisent (Bison bonasus) Gigascience. 2017;6:1–5. doi: 10.1093/gigascience/gix016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Qiu Q, 2013. Genomic data from the domestic yak (Bos grunniens) GigaScience Database. [DOI]
- Wang K, 2017. Draft genome of European bison (wisent), Bison bonasus. GigaScience Database. [DOI]
- 2019. NCBI Sequence Read Archive. SRP194583
- Liu Y. 2019. Bos mutus breed Datong Yak isolate WY2019, whole genome shotgun sequencing project. GenBank. VBQZ00000000
- Liu Y. 2019. The sequence and de novo assembly of the wild yak genome. figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The software versions, settings and parameters used are described below.
(1) GCE, version 1.0.0, parameters used: kmer_freq_hash -k 21 -l reads.list -t 24 -i 5000000 -o 0 -p wild_yak & > kmer_freq.log; gce -f wild_yak.freq.stat -c 46 -g 155866493014 -m 1 -D 8 -b 1 > wild_yak.Table 2> wild_yak.log.
(2) Lighter, version 1.1.1, parameters used: -k 17 3000000000 -trim -t 20.
(3) Platanus, version 1.2.4, parameters used: platanus assemble -o wild_yak -f <insert size 280 bp pair-end reads> <insert size 500 bp pair-end reads> <insert size 800 bp pair-end reads> -t 30 -m 500 -tmp temp; platanus scaffold -o wild_yak -c wild_yak_contig.fa -b wild_yak_contigBubble.fa -IP1 < insert size 280 bp pair-end reads> -a1 280 -IP2 <insert size 500 bp pair-end reads> -a2 500 -IP3 < insert size 800 bp pair-end reads > -a3 800 -OP4 <insert size 2 k pair-end reads> -a4 2000 -OP5 <insert size 5 k pair-end reads> -a5 5000 -OP6 <insert size 10 k pair-end reads> -a6 10000; platanus gap_close -o wild_yak -c wild_yak_scaffold.fa -IP1 <insert size 280 bp pair-end reads> -IP2 <insert size 500 bp pair-end reads> -IP3 <insert size 800 bp pair-end reads> -OP4 < insert size 2 k pair-end reads > -OP5 <insert size 5 k pair-end reads> -OP6 <insert size 10 k pair-end reads>.
(4) Gap Closer, version 1.12, parameters used: -l 150 -t 30, in configFile: max_rd_len = 100; Paired-end libs: reverse_seq = 0, asm_flags = 3; Mate-pair libs: reverse_seq = 11, asm_flags = 2.
(5) BUSCO, version 3: mammal default parameters, mammalia_odb9.
(6) BWA, version 0.7.15-r1140: default parameters.
(7) SAMtools, version 1.3; default parameters.
(8) RepeatMasker, version 4.0.7 (with RepBase library release-20170127).
(9) RepeatModeler, RepeatModeler-open-1.0.11.
(10) TRF, version 4.09, parameters used: trf wild_yak.gapclose.fa 2 7 7 80 10 50 500 -d -h.
(11) TBLASTN, version 2.5.0, parameter used: –e 1E-5.
(12) GeneWise, version 2.4.1, parameters used: -tfor/-trev (-rfor for genes on forward strand and -trev for reverse strand) -gff.
(13) Augustus, version 3.2.3, parameter used: -species = human.
(14) Geneid, version 1.0, parameters used: -3 -P.
(15) Genemark, version 3.9, parameter used: -f gff3.
(16) Snap, version 2006-07-28, parameter used: –gff.
(17) GlimmerHMM, version 3.0.4, default parameters.
(18) EVM, version 1.1.1, default parameters.
(19) InterProScan, version 5.25-64.0, parameters: -f tsv -iprlookup -goterms -pa -t p.