Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with a strong genetic component whose knowledge evolves quickly. Next-generation sequencing is the only effective technology to deal with the high genetic heterogeneity of ASD in a clinical setting. However, rigorous criteria to classify rare genetic variants conferring ASD susceptibility are currently lacking. We have performed whole-exome sequencing to identify both nucleotide variants and copy number variants (CNVs) in 253 ASD patients, including 68 patients with intellectual disability (ID) and 90 diagnosed as Asperger syndrome. Using explicit criteria to classify both susceptibility genes and susceptibility variants we prioritized 217 genes belonging to the following categories: syndromic genes, genes with an excess of de novo protein truncating variants and genes targeted by rare CNVs. We obtained a susceptibility variant detection rate of 19.7% (95% CI: [15–25.2%]). The rate for CNVs was 7.1% (95% CI: [4.3–11%]) and 12.6% (95% CI: [8.8–17.4%]) for nucleotide variants. The highest rate (30.1%, 95% CI: [20.2–43.2%]) was obtained in the ASD + ID subgroup. A strong contributor for at risk nucleotide variants was the recently identified set of genes (n = 81) harboring an excess of de novo protein truncating variants. Since there is currently no evidence that the genes targeted here are necessary and sufficient to cause ASD, we recommend to avoid the term “causative of ASD” when delivering the information about a variant to a family and to use instead the term “genetic susceptibility factor contributing to ASD”.
Subject terms: Autism spectrum disorders, Clinical genetics
Introduction
Autism spectrum disorder (ASD), whose prevalence is estimated to be around 1 per 100 children, encompass a wide range of phenotypic manifestations ranging from severe behavioral impairment, often associated with intellectual disability (ID), to mild difficulties in social interaction. The fact that, clinically, autism is not a single disease entity is mirrored by the extreme genetic heterogeneity underlying this condition. Chromosomal microarray analysis is usually used as the first-tier genetic test for individuals with autism with a diagnostic yield of 7–9%1,2. However, such analysis cannot detect single nucleotide variations and small Indels but only loss or gain of genomic DNA material. In light of recent advances in ASD genetics, especially regarding the importance of de novo truncating variants, it is estimated that rare variants within several hundreds of genes may contribute to ASD susceptibility, each of them being present only in a very small proportion of cases3. In this context, next-generation sequencing is the only reasonable and cost-effective approach to search for variants in these genes in a diagnostic perspective.
The aims of the present study were threefold: (i) to establish an accurate list of susceptibility genes from literature data and to provide guidelines to categorize rare susceptibility variants (ii) based on these criteria, to estimate the whole exome sequencing (WES) detection rate of rare susceptibility variants in a sample of ASD patients typical of those attending genetic consultations and (iii) to propose some recommendations for susceptibility factors assessment and interpretation in clinical practice.
Methods
Patient recruitment
During the 2009–2017 period, 679 unrelated subjects received a diagnosis of ASD or Asperger syndrome by clinicians of our local expert center. This study was carried out on a subset of 253 cases, including all patients from whom parental DNA was available (n = 159) and 94 randomly selected patients.
Clinical evaluation was mainly based on ADOS-2. In addition, ADI-R and CARS, were also used for 76 and 145 patients, respectively. All diagnoses were made according to DSM-IV-TR criteria. Accordingly, the diagnosis of Asperger syndrome included the absence of language impairment, as documented during clinical examination. IQ was assessed in subjects with age >5 years with the Wechsler Intelligence Scale for Children (WISC) or the Raven Progressive matrix. PsychoEducational Profil third edition (PEP-3) and Vineland Adaptive Behavior Scales (VABS-2) assessment, when available, were used to refine the classification of cases with heterogeneous cognitive functions or mild cognitive impairment. Subjects with age <5 years were classified in the ASD subgroup, without further specification.
Following national recommendations, diagnostic assessment included a genetic consultation. Every patient was received by a senior clinical geneticist for thorough questioning about development, previous medical records and family history, followed by a complete physical examination.
This genetic study was approved by our legal ethics committee. Parents or legal representative of the children signed an informed consent for genetic analyses.
Molecular genetics
All included patients had previously been negatively screened for Fragile X expansion and 54 had been tested by array Comparative Genomic Hybridization (aCGH) Agilent 180K (Santa Clara, United-States), yielding to the prior identification (subsequently confirmed by exome sequencing) of four CNVs conferring susceptibility for ASD. For each proband, DNA was extracted from fresh blood samples. DNA was also obtained from affected siblings and from parents when available.
Exome sequencing was performed on every proband. Exomes were captured using the Agilent SureSelect All Exons V5-UTR or V6 Kit (Santa Clara, United-States). Sequencing was performed on an Illumina HiSeq4000 (Illumina, San Diego, CA, USA) at the CNRGH (Centre National de Recherche en Génomique Humaine, Evry, France) with paired end mode, 150 base pairs (bp) reads. Nucleotide variants and copy number variants (CNVs) were analyzed through a bioinformatics pipeline to perform variant calling, quality check, annotation and CNVs detection (see Sup Info). When possible, the segregation of retained variants was examined by Sanger sequencing for nucleotide variants and by ddPCR or QMPSF for CNVs. Parenthood was checked by polymorphic microsatellites for patients carrying de novo mutations.
List of genes and interpretation of variants
To establish a panel of genes firmly contributing to ASD susceptibility, we retained three sets of genes:
(i) genes showing a statistically significant excess of de novo protein truncating variants (PTVs), (i.e. nonsense, canonical splice site variants and frameshift indels) in large published cohorts. Since these genes have been identified in several, sometimes redundant recent trio studies1,4–10, we reanalyzed (see Sup Info) all de novo events reported in each of these studies (Sup Table 1). Depending on the level of statistical evidence for the enrichment in de novo PTVs, we ranked these genes into two categories: class A (false discovery rate (FDR) <5%) meaning definitely involved and class B (5% ≤ FDR ≤ 10%), meaning probably involved in ASD susceptibility. This led to a list of 81 genes (Sup Table 2). For these genes, we retained as susceptibility factor all rare PTVs found in patients, whatever the inheritance (i.e. de novo occurrence or not).
(ii) syndromic ASD–related genes, i.e. genes causing monogenic diseases for which ASD can be part of the phenotype (n = 107). This set of genes was carefully curated from the Simon Foundation Autism Research Initiative (Sfari) database and from literature data. For each syndromic gene, the mode of inheritance was specified and rare non-synonymous variants were prioritized. For these genes, pathogenicity of rare variants (MAF < 1% in GnomAD) was evaluated using the guidelines from the American College of Medical Genetics-Association for Molecular Pathology (ACMG-AMP). Only variants considered as likely pathogenic or pathogenic (class 4 or 5) were retained as advised by the guidelines. For our analysis, we considered these variants as probable (class 4) or definite (class 5) ASD-susceptibility variants and then evaluated whether patients carrying them harbored or not the full syndromic presentation typical of each gene.
(iii) genes intersected by a burden of rare CNVs, listed in the Sfari database (n = 85).
Combining these three sets yielded a high confidence list of 217 ASD susceptibility genes (Sup Table 3). Note that some of these genes were simultaneously present in several sets. (Fig. 1).
Fig. 1.
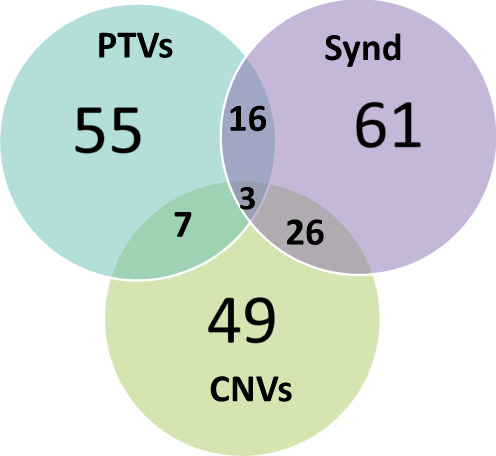
Venn diagram of the gene list stratified in three categories: syndromic genes, genes with an excess of de novo PTVs, genes intersected by a burden of rare CNVs.
Besides this main panel and despite the fact that the interpretation was more problematic, we also examined rare missense variants in genes carrying a burden of de novo and transmitted mutations identified by TADA analyses4,11 as well as in genes recently reported as carrying an excess of de novo missense mutations in ASD and developmental disorders (DD)1,12. This analysis was conducted on 89 genes (61 already included in the main list and 28 additional genes). For this analysis, only de novo missense variants predicted as damaging by three in silico prediction tools were considered.
Finally, for patients with ID or additional sensory impairment, we examined all OMIM referenced genes.
While CNV detection was performed on all genes of the main list, variant interpretation depended on the gene subsets. We retained all previously associated CNVs (i.e. duplications or deletions) within the set of large recurrent CNVs associated with various DDs including ASD13. In contrast, we used a conservative approach to interpret the remaining CNVs, and only retained rare deletions targeting Loss of Function (LoF) intolerant genes as probable ASD susceptibility variants.
Results
Among the 253 ASD probands (81.4% males), 68 were categorized in the ASD+ID subgroup while 90 were classified as having Asperger syndrome. The average age at inclusion was 8.5 ± 5.6 years (range 3–41). Positive family history (among first, second or third-degree relatives) for ASD, DD or ID was reported for 27.7% of probands. Demographic and phenotypic characteristics of the sample are displayed in Table 1.
Table 1.
Demographic and phenotypic characteristics of the sample.
| % (N/253) | |
|---|---|
| Males | 81.4% (206) |
| Family history | 27.7% (70) |
| Consanguinity | 1.6 % (4) |
| Asperger syndrome | 35.6% (90) |
| Intellectual disability | 26.9% (68) |
| Dysmorphic features | 26.5% (67) |
| Premature birth | 3.95% (10) |
| Microcephaly | 3.7% (9) |
| Macrocephaly | 7.9% (20) |
| Epilepsy | 7.1% (18) |
As shown in Table 2, ASD-contributing CNVs were present in 18/253 cases (7.1%, 95% CI: [4.3–11%]) while ASD-contributing nucleotide variants (Table 3) were detected in 32/253 subjects (12.6%, 95% CI: [8.8–17.4%]), yielding to a global detection rate of 50/253 (19.7%, 95% CI: [15–25.2%]). Of note, no patient carried multiple susceptibility variants. A slightly higher detection rate (29.8%, 95% CI: [17.3–44.9%]) was found in females. When the sample was split into three clinical subgroups, the rate of CNVs and nucleotide susceptibility-variants was particularly high in the ASD+ID subgroup (30.1%, 95% CI: [20.2–43.2%]) (Table 4). In the whole sample, two CNVs (NRXN1 E1–3 del and 15q11 BP 1–2 del) were recurrent and four genes were found to carry nucleotide variants more than once: CHD8, SHANK3, NRXN1, and KMT2A. Regarding nucleotide susceptibility variants, the contribution of the set of genes previously shown as enriched in de novo PTVs in large ASD cohorts should be highlighted. Ten genes carrying ASD-susceptibility variants belonged to this category vs seven syndromic genes and nine genes belonging simultaneously to the two categories. Overall, the high frequency of de novo events is noteworthy: we identified 21/28 de novo nucleotide variants and 6/14 de novo CNVs among 42 patients from whom parental DNA was available. Regarding the mode of inheritance of ASD contributing variants, no autosomal recessive inheritance was encountered in this mainly outbred population.
Table 2.
CNVs conferring susceptibility for autism identified among the 253 ASD subjects.
| ID | Chr | Start (hg19) | End (hg19) | Size (bp) | Type | Syndrome/gene | Inheritance | Diag | Age | Sex | Family history |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogenic CNVs | |||||||||||
| 266 | 15 | 22833359 | 23330473 | 497114 | Del | 15q11 del syndrome (BP1-BP2) | Maternal | Asperger | 10 | M | S |
| 175 | 15 | 22833359 | 23330473 | 497114 | Del | 15q11 del syndrome (BP1-BP2) | Maternal | ASD | 6 | M | S |
| 298 | 15 | 22833359 | 23330473 | 497114 | Del | 15q11 del syndrome (BP1-BP2) | NA | Asperger | 13 | F | S |
| 277 | 15 | 22833359 | 23330473 | 497114 | Del | 15q11 del syndrome (BP1-BP2) | NA | Asperger | 16 | M | Fa |
| 207 | 15 | 23686022 | 28567298 | 4881276 | Dup | 15q11.2q13 dup syndrome (BP2-BP3) | DN | ASD | 9 | M | Fa |
| 405 | 15 | 23686022 | 30114707 | 6428685 | Dup | 15q11.2q13 dup syndrome (BP2-BP4)c | DN | ASD | 5 | F | S |
| 225 | 16 | 14968893 | 16267261 | 1298368 | Del | 16p13 del syndrome | Maternal | ASD+ID | 5 | F | S |
| 106 | 16 | 14968893 | 16267261 | 1298368 | Dup | 16p13 dup syndrome | Paternal | ASD+ID | 11 | F | S |
| 40 | 16 | 29674271 | 30198544 | 524273 | Dup | 16p11.2 dup syndrome | DN | Asperger | 11 | M | Fa |
| 392 | 16 | 29674271 | 30198544 | 524273 | Del | 16p11.2 del syndrome | DN | ASD+ID | 7 | M | S |
| 288 | 22 | 51123001 | 51220846 | 97845 | Del | SHANK3 (E9–22)+2 other genes | DN | ASD+ID | 32 | M | S |
| 394 | X | 144899347 | 148678216 | 3778869 | Del | Del Xq27.3-q28 (15 genes including FMR1 and AFF2)c | NA | ASD+ID | 24 | F | Fa |
| 52 | 1 | 146500897 | 147806796 | 1305899 | Dup | 1q21.1 dup syndrome | Not maternal | ASD | 8 | M | S |
| 304 | 2 | 51253508 | 51259674 | 6166 | Del | NRXN1 (E1–3)c | Paternal | ASD+ID | 6 | M | Fa |
| 396 | 2 | 51253508 | 51259674 | 6166 | Del | NRXN1 (E1–3)c | DN | ASD+ID | 7 | M | S |
| 318 | 2 | 51253508 | 51259674 | 6166 | Del | NRXN1 (E1–3) | Paternal | Asperger | 12 | M | S |
| Likely pathogenic CNVs | |||||||||||
| 55 | 18 | 44624417 | 44641759 | 17342 | Del | KATNAL2 (E 16–18) | Paternal | ASD | 6 | M | Faa |
| 365 | 9 | 119373136 | 119495896 | 122760 | Del | ASTN2 (E1–4) | Maternal | Asperger | 19 | F | Fab |
NA not assessed, E exon, BP breakpoint, DN de novo, Del deletion, Dup duplication, F female, M male, S sporadic, Fa familial.
a1stSib (ASD): wild type and 2nd Sib (language delay): variant carrier.
bSib (Asperger): variant carrier.
cAlready identified by aCGH.
Table 3.
Nucleotide variants conferring susceptibility for autism (stratified by category) identified among the 253 ASD subjects. Nucleotide variants in genes harboring a statistically significant excess of de novo PTVs were classified either as susceptibility variants in class A genes (false discovery rate (FDR) <5%) or as probable susceptibility variants in class B genes (10% < FDR < 5%). Nucleotide variants in syndromic ASD–related genes were classified using the guidelines from the American College of Medical Genetics-Association for Molecular Pathology.
| ID | Gene | Variation | Transcript | Inheritance | o/e | GnAD | Diag | Age | Sex | Family history |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) Nucleotide variants in genes harboring a statistically significant excess of de novo PTVs | ||||||||||
| Susceptibility variants | ||||||||||
| 213 | CHD8 | p.Gln1171* c.3511C>T | NM_001170629.1 | Paternal | 3.94% | _ | ASD+ID | 7 | F | S |
| 223 | CHD8 | p.Lys528Glyfs*14 c.1582_1583delAA | NM_001170629.1 | DN | 3.94% | _ | Asperger | 19 | M | S |
| 220 | CHD8 | p.Leu980Argfs*5 c.2937_2939delinsTC | NM_001170629.1 | DN | 3.94% | _ | Asperger | 30 | M | S |
| 98 | NRXN1 | p.Leu158Alafs*29 c.471dupG | NM_001135659.1 | DN | 12.30% | 1/242 430 | Asperger | 12 | M | S |
| 307 | NRXN1 | p.Glu389Serfs*3 c.1165delG | NM_001135659.1 | Maternal | 12.30% | _ | ASD | 3 | M | S |
| 253 | ANK2 | p.Glu3062* c.9184G>T | NM_001148.4 | DN | 6.20% | _ | Asperger | 11 | M | S |
| 325 | GIGYF1 | p.Ter1036Glyext*72 c.3106T>G | NM_022574.4 | DN | 35.64% | _ | ASD | 12 | M | Faa |
| 341 | KDM5B | p.Arg1397* c.4189C>T | NM_006618.4 | NA | 44.95% | 2/247 406 | ASD | 4 | M | S |
| Probable susceptibility variants | ||||||||||
| 122 | RIMS1 | p. ? c.100+1G>A | NM_001168407.1 | Maternal | 18.65% | 1/31 404 | ASD+ID | 14 | F | S |
| 188 | SETD2 | p.Arg1708* c.5122C>T | NM_014159.6 | DN | 13% | _ | Asperger | 8 | F | Fab |
| 301 | DIP2A | p. ? c.1429+2T>G | NM_015151.3 | Maternal | 23.89% | _ | Asperger | 6 | F | S |
| 285 | HECTD4 | p.Ala3108Glyfs*5 c.9322dupG | NM_001109662.3 | Maternal | 7.56% | _ | Asperger | 12 | M | S |
| 168 | SPEN | p.Ser2411* c.7232C>A | NM_015001.2 | DN | 2.31% | _ | Asperger | 6 | M | S |
| (b) Nucleotide variants in syndromic ASD-related genes | ||||||||||
| Susceptibility variants | ||||||||||
| 370 | EBF3 | p.Gln78* c.232C>T | NM_001005463.2 | DN | 3% | _ | ASD | 4 | M | S |
| 79 | TSC2 | p.Val1062Glyfs*7 c.3185_3188del | NM_000548.3 | DN | 2% | _ | ASD+ID | 8 | F | S |
| Probable susceptibility variants | ||||||||||
| 30 | DEAF1 | p.Arg224Gln c.671G>Ac | NM_021008.3 | Maternal | 41% | 1/246 022 | ASD+ID | 14 | M | S |
| 328 | DNMT3A | p.Gln249* c.745C>T | NM_022552.4 | DN | 126% | 1/244 420 | ASD+ID | 10 | M | S |
| 215 | AUTS2 | p.Thr328Argfs*19 c.983_984del | NM_015570.3 | DN | 13% | _ | ASD+ID | 4 | M | S |
| 222 | DYNC1H1 | p.? c.4396–1G>C | NM_001376.4 | DN | 5% | _ | ASD | 27 | F | Faa |
| 121 | NR2F1 | p.Gly395Ala c.1184G>C | NM_005654.5 | DN | 0.00% | _ | ASD+ID | 20 | M | S |
| (c) Nucleotide variants in genes belonging to both categories | ||||||||||
| Susceptibility variants | ||||||||||
| 296 | SHANK3 | p.Pro922Argfs*34 c.2765del | NM_033517.1 | DN | 3.98% | _ | ASD+ID | 8 | F | S |
| 377 | SHANK3 | p.Leu1030Cysfs*48 c.3088delC | NM_033517.1 | DN | 3.98% | _ | ASD+ID | 6 | M | S |
| 353 | SHANK3 | p.Ala1227Glyfs*69 c.3679dupGd | NM_033517.1 | NA | 3.98% | _ | ASD+ID | 31 | F | S |
| 136 | GRIN2B | p.Ser9Phefs*50 c.23_24insC | NM_000834.3 | DN | 0.00% | _ | ASD | 4 | M | S |
| 85 | ASXL3 | p.Ser1246* c.3737C>A | NM_030632.1 | DN | 9.86% | _ | ASD+ID | 11 | M | Fa |
| 78 | SCN2A | p.Phe601Leufs*40 c.1800delC | NM_001040142.1 | DN | 6.33% | _ | ASD+ID | 12 | F | S |
| 382 | CHD2 | p.Val332Glyfs*25 c.995_999delTGAAG | NM_001271.3 | NA | 2.75% | _ | ASD+ID | 7 | M | S |
| 47 | DYRK1A | p.Arg255* c.763C>Te | NM_101395.2 | Not maternal | 8.45% | _ | ASD+ID | 18 | M | S |
| 395 | ANKRD11 | p.Glu1154Glyfs*16 c.3460dup | NM_001256182.1 | DN | 4.72% | _ | Asperger | 11 | M | S |
| Probable susceptibility variants | ||||||||||
| 291 | KMT2A | p. ? c.10835+1G>Af | NM_001197104 | Paternal | 3.12% | _ | Asperger | 6 | M | S |
| 324 | KMT2A | p.Gln3192Pro c.9575A>C | NM_001197104 | DN | 3.12% | _ | ASD | 9 | M | S |
| 157 | MECP2 | p.Glu282Gly c.845A>G | NM _004992.3 | DN | 10.72% | _ | ASD | 13 | F | S |
F female, M male, S sporadic, Fa familial, DN de novo, NA not assessed, o/e observed/expected ratio of PTVs in GnomAD, GnAD allelic frequency in the GnomAD database.
aSib (ASD): wild type.
bSib (Asperger): wild type.
cp.Arg224Trp found as a de novo event in an ASD case27.
dVariation found as de novo event in ASD+ID cases28.
eVariation found as a de novo event in a syndromic case of DYRK1A-haploinsufficiency29.
fVariation found in three related ID patients14..
Table 4.
Detection rate for CNVs and nucleotide variants among the 253 ASD subjects stratified by clinical subgroups.
| ASD+ID n = 68 | ASD n = 95 | Asperger n = 90 | Total n = 253 | |
|---|---|---|---|---|
| CNVs | 7 | 5 | 6 | 18–7.1% [4.3–11%] |
| Nucleotide variants | 14 | 8 | 10 | 32–12.6% [8.8–17.4%] |
| Total | 21–30.1% [20.2–43.2%] | 13–13.7% [7.5–22.3%] | 16–17.7% [10.5–27.3%] | 50–19.7% [15–25.2%] |
Finally, variants in two genes not included in our list, but which cause nevertheless part of the phenotype, were found in two patients presenting ASD, ID, and sensory deficits. A patient with hearing impairment carried compound heterozygous c.2485C > T, p.Gln829*/c.5665T > C, p.Trp1889Arg (NM_194248.2) OTOF (MIM: 601071) pathogenic variants, while a patient with severe visual impairment, born from consanguineous parents, carried a homozygous c.389del, p.Pro130Leu fs*36 (NM_000180.3) GUCY2D (MIM: 204000) pathogenic variant. These mutations explain the sensory deficits of these patients but there is currently no evidence that they are related to the ASD phenotype.
Genotype/phenotype correlations
Consistency with the previously described phenotypes was checked for syndromic genes and was fair to good for most genes (DNMT3A, ASXL3, EBF3, NR2F1, DYRK1A, DEAF1, ANKRD11, SHANK3). (See Sup Info) but in some cases unusual phenotypes were encountered:
Patients carrying MECP2 or KMT2A mutations did not harbor the classical features of Rett or Wiedemann-Steiner syndrome, respectively. A MECP2 mutation (not present in the International Rett database) was identified in a girl who presented with mild ASD. She had no developmental delay (walk at 15 months, no language delay) and was enrolled in an ordinary school at the age of 14. She demonstrated no ID and no dysmorphic features. KMT2A mutations were identified in two patients. One of them carrying the p.Gln3192Pro de novo mutation was the first child of a couple with epilepsy and mild cognitive impairment. He had a global developmental delay with a sitting age at 1 year but a normal walking age at 17 months, a language delay and a motor instability. Clinical evaluation showed a normal growth, an hyperlaxity and a diffuse hypertrichosis but no hairy elbow or morphological features typical of Wiedemann–Steiner Syndrome. The second one was the first child of a couple without medical history. He carried a c.10835 + 1G > A splice mutation already described in 3 related ID patients, not diagnosed as Wiedemann-Steiner. This variant leads to the in frame deletion of exon 28 and likely disrupts the stabilizing interaction between the C terminal KMT2A fragments14. Our patient had no motor delay but a language delay. Electroencephalogram showed left occipital spikes, but he never had seizures. MRI showed an isolated agenesis of the corpus callosum. The diagnosis of Wiedemann-Steiner syndrome was not clinically evoked.
Likewise, although DYNC1H1 missense or truncating variations are usually associated with a severe phenotype including ID, neuronal migration defects and epileptic encephalopathy15, our patient had an unremarkable ASD phenotype without neurological features. CHD2 truncating mutations are mostly associated with pediatric refractory epilepsy16, but our patient had only experienced one tonic-clonic seizure. In the same vein and in line with recent reports underlining the phenotypic heterogeneity of SCN2A17 and GRIN2B18 mutation carriers, none of the patient carrying de novo truncating mutations on these genes had epilepsy, which is a feature generally associated with these mutations.
It should also be noted that besides these well-characterized syndromic genes, another patient carrying the RIMS1 splice variant received a diagnosis of Dravet syndrome but bore no mutation on genes causing this phenotype. Interestingly, the RIM1a protein encoded by the mice ortholog of RIMS1 has been characterized as a presynaptic active zone protein mainly expressed in neurons from the cerebellum19 and controlling epileptogenesis following pharmacologically induced status epilepticus20. This suggests that the RIMS1 truncating variant identified here, which affects a minor transcript expressed in the cerebellum (https://gtexportal.org), could be associated with a peculiar neurological phenotype.
Discussion
In this report, the rate of identified CNVs, largely based on well-known recurrent CNVs, was similar to that obtained in several recent studies1,2. It is notable that four of our patients carry the 15q11.2 recurrent deletion which is known to have a mild effect on developmental disorder. Hence, it is very probable that additional factors are involved in this subset. Regarding nucleotide variants, we used rigorous criteria to classify both ASD susceptibility genes and variants. As a result, we retained only 217 genes as compared for example with the 880 genes present in the Sfari database (october 2018 release), often with minimal evidence of involvement in ASD susceptibility. We nevertheless obtained a detection rate of 12.6% for nucleotide variants. Combining CNVs and nucleotide variants, we reached a higher detection rate (19.7%) than the one obtained in 105 ASD patients previously analyzed by WES and aCGH (15.8%)2. This is partly due to the contribution of the set of genes with an excess of de novo PTVs in ASD patients, which were identified in the recent years thanks to the study of more than 6000 ASD trios1,4–10. Clearly, this huge effort has obvious consequences for clinical practice. The finding of a particularly high rate of de novo PTVs in this set of genes (14/20 PTVs = 70% of the total number of PTVs in patients for whom parental DNA was available) reinforces the conclusion that this set is truly enriched in de novo PTVs in ASD subjects. As seen in Sup Table 2, the gene which is the most frequently hit by de novo PTVs in ASD patients from the literature is CHD821. Interestingly, among the three CHD8 PTVs identified in this study, two were de novo events while the third was inherited from a parent whose sole phenotypic feature was macrocephaly. More generally, the presence of 30% of PTVs inherited from parents with no ASD diagnosis underlines the incomplete penetrance and/or variable expressivity of these variants.
Due to the stringent criteria used to classify variants, it should be stressed that our detection rate is likely to be underestimated. First, concerning CNVs, partial duplications intersecting genes were conservatively considered of uncertain significance (Sup Table 4). However, it is well known that tandem intragenic duplications often disrupt reading frames and can lead to haploinsufficiency. In a recent analysis of all structural variation detected in the gnomAD dataset, the authors showed a significant correlation between constraint metrics reflecting intolerance to Lof mutations and the depletion of partial genic duplications at the gene level22. However, despite these findings, the individual interpretation of partial duplications remains challenging and will require additional investigations. Second, since all missense variants impacting genes exhibiting a statistically significant excess of de novo missense mutations in ASD1,12 were inherited, we decided to consider them as variants of unknown significance (VUS) (Sup Table 5) even when they were absent from gnomAD.
The detection rate was particularly high in the ASD+ID subgroup. It was in the same range as those recently reported in ID or DD patients (25–39%)23–25, or in a cohort of 163 ID/DD patients “with reported ASD or autistic features” (25.8%)26. Of note, all these studies used larger panels of genes than ours. In our study, although ASD+ID patients were more likely hit, susceptibility variants were also found with a high frequency in the Asperger group. Several genes carried susceptibility variants in both ASD+ID and Asperger subjects. For example, in accordance with a previous report outlining that cognitive ability of CHD8 patients ranges from profoundly disabled to age-appropriate21, of the three patients carrying CHD8 PTVs, one had ID associated to ASD while two others received a diagnosis of Asperger syndrome. However, macrocephaly, a shared feature among CHD8 patients, was present in all three patients. Likewise, NRXN1 exon 1–3 deletions were present in patients with each clinical presentation (ASD, ASD+ID, Asperger).
The genotype/phenotype correlation for syndromic genes was generally good, but in some cases was not typical. As already outlined23,25, it is noteworthy that the phenotypic spectrum of mutations in several “syndromic genes” for which a specific phenotype has previously been described is broader than initially thought. Clearly, for a large set of genes, prioritizing by double entries (genes with both an excess of de novo PTVs and syndromic genes) allows the identification of ASD susceptibility factors in more patients than prioritizing upon a single criteria based on strict phenotypic concordance with described syndromic features.
In summary, the strategy we advocate here is an efficient and cost-effective manner to deal with the detection of both rare nucleotide variants and CNVs in a clinical setting. Of course, exome data should be periodically reanalyzed and the list of genes updated. Due to the extreme rarity of variants studied here, it is currently difficult to establish for each of them if they are fully causative of an ASD phenotype. Most likely, only a subset of them are necessary and sufficient to cause ASD. For this reason, we recommend to avoid the term “causative of ASD” when delivering the information about these variants to a family and to use instead the term “genetic susceptibility factor contributing to ASD phenotype”. At the present stage, with the exception of pathogenic variants in syndromic genes causing well-defined monogenic diseases, using these results for prenatal diagnosis is premature.
Supplementary information
Table S1. De novo PTVs among ASD patients from SSC, ASC, ACGC, TASC, MSSNG and AGRE cohorts.
Table S2. List of gene with a significant excess of de novo truncating variants in the literature.
Table S3. List of 217 genes conferring ASD susceptibility prioritized in this study.
Table S4. CNVs of unknown significance found among the 253 ASD subjects.
Table S5. Nucleotide variants of unknown significance found among the 253 ASD subjects.
Acknowledgements
Supported by Fondation Deniker and Inserm grants. We are indebted to families B. and H. for their generous gifts. This study was cosupported by European Union and Région Normandie in the context of Recherche Innovation Normandie (RIN 2018). Europe gets involved in Normandie with the European Regional Development Fund (ERDF). Funding sources did not have any involvement in the study design.
Conflict of interest
The authors have full access to all the data in the study and had final responsibility for the decision to submit the article. The authors declare no conflict of interest regarding the publication of this article.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-020-0760-7).
References
- 1.Yuen C, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tammimies K, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 3.Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat. Rev. Genet. 2014;15:133–141. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- 4.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong S, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krumm N, et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Roak BJ, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stessman HAF, et al. Targeted sequencing identifies 91 neurodevelopmental disorder risk genes with autism and developmental disability biases. Nat. Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 2016;7:13316. doi: 10.1038/ncomms13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders SJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisheker MR, et al. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat. Neurosci. 2017;20:1043–1051. doi: 10.1038/nn.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet. Med. 2013;15:478–481. doi: 10.1038/gim.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogaert DJ, et al. Early-onset primary antibody deficiency resembling common variable immunodeficiency challenges the diagnosis of Wiedeman-Steiner and Roifman syndromes. Sci. Rep. 2017;7:3702. doi: 10.1038/s41598-017-02434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willemsen MH, et al. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J. Med. Genet. 2012;49:179–183. doi: 10.1136/jmedgenet-2011-100542. [DOI] [PubMed] [Google Scholar]
- 16.Lamar K-MJ, Carvill GL. Chromatin remodeling proteins in epilepsy: lessons from CHD2-associated epilepsy. Front Mol. Neurosci. 2018;11:208. doi: 10.3389/fnmol.2018.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff M, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140:1316–1336. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 18.Platzer K, et al. GRIN2B encephalopathy: novel findings on phenotype, variant clustering, functional consequences and treatment aspects. J. Med. Genet. 2017;54:460–470. doi: 10.1136/jmedgenet-2016-104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kintscher M, Wozny C, Johenning FW, Schmitz D, Breustedt J. Role of RIM1α in short- and long-term synaptic plasticity at cerebellar parallel fibres. Nat. Commun. 2013;4:2392. doi: 10.1038/ncomms3392. [DOI] [PubMed] [Google Scholar]
- 20.Pitsch J, et al. The presynaptic active zone protein RIM1α controls epileptogenesis following status epilepticus. J. Neurosci. 2012;32:12384–12395. doi: 10.1523/JNEUROSCI.0223-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins, R. L. et al. An open resource of structural variation for medical and population genetics. bioRxiv 578674 10.1101/578674 (2019).
- 23.Chérot E, et al. Using medical exome sequencing to identify the causes of neurodevelopmental disorders: experience of 2 clinical units and 216 patients. Clin. Genet. 2018;93:567–576. doi: 10.1111/cge.13102. [DOI] [PubMed] [Google Scholar]
- 24.Martínez F, et al. High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J. Med. Genet. 2017;54:87–92. doi: 10.1136/jmedgenet-2016-103964. [DOI] [PubMed] [Google Scholar]
- 25.Redin C, et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J. Med. Genet. 2014;51:724–736. doi: 10.1136/jmedgenet-2014-102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi M, et al. Outcomes of diagnostic exome sequencing in patients with diagnosed or suspected autism spectrum disorders. Pediatr. Neurol. 2017;70:34–43.e2. doi: 10.1016/j.pediatrneurol.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 27.Vulto-van Silfhout AT, et al. Mutations affecting the SAND domain of DEAF1 cause intellectual disability with severe speech impairment and behavioral problems. Am. J. Hum. Genet. 2014;94:649–661. doi: 10.1016/j.ajhg.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earl RK, et al. Clinical phenotype of ASD-associated DYRK1A haploinsufficiency. Mol. Autism. 2017;8:54. doi: 10.1186/s13229-017-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. De novo PTVs among ASD patients from SSC, ASC, ACGC, TASC, MSSNG and AGRE cohorts.
Table S2. List of gene with a significant excess of de novo truncating variants in the literature.
Table S3. List of 217 genes conferring ASD susceptibility prioritized in this study.
Table S4. CNVs of unknown significance found among the 253 ASD subjects.
Table S5. Nucleotide variants of unknown significance found among the 253 ASD subjects.


