Abstract
Shiga toxin-producing Escherichia coli (STEC) is an important foodborne pathogen. The increasing incidence of non-O157 STEC has posed a great risk to public health. Besides the Shiga toxin (Stx), the adherence factor, intimin, coded by eae gene plays a critical role in STEC pathogenesis. In this study, we investigated the prevalence and polymorphisms of eae gene in non-O157 STEC strains isolated from different sources in China. Among 735 non-O157 STEC strains, eae was present in 70 (9.5%) strains. Eighteen different eae genotypes were identified in 62 eae-positive STEC strains with the nucleotide identities ranging from 86.01% to 99.97%. Among which, seven genotypes were newly identified in this study. The eighteen eae genotypes can be categorized into five eae subtypes, namely β1, γ1, ε1, ζ3 and θ. Associations between eae subtypes/genotypes and serotypes as well as origins of strains were observed in this study. Strains belonging to serotypes O26:H11, O103:H2, O111:H8 are associated with particular eae subtypes, i.e., β1, ε1, θ, respectively. Most strains from diarrheal patients (7/9, 77.8%) carried eae-β1 subtype, while most isolates from cattle (23/26, 88.5%) carried eae-ζ3 subtype. This study demonstrated a genetic diversity of eae gene in non-O157 STEC strains from different sources in China.
Subject terms: Bacteria, Bacteria, Public health, Public health
Introduction
Shiga toxin-producing Escherichia coli (STEC) is a group of food-borne pathogens that can cause non-bloody diarrhea, hemorrhagic colitis (HC), and the fatal hemolytic uremic syndrome (HUS) in humans1. It has been estimated that there are more than 470 STEC serotypes, among which O157:H7 serotype is usually associated with more severe clinical outcomes2,3. However, non-O157 STEC strains such as O26, O45, O103, O111, O121, and O145 (referred to as the ‘top six’ non-O157 STEC) have been increasingly recognized to cause food poisoning, bloody diarrhea, HUS, and other gastrointestinal illnesses in recent years4,5. Domestic and wild animals, including cattle, sheep and goats are considered to be the most important reservoirs of STEC6. Human infections mainly occur through ingestion of contaminated food or water, exposure to the environment or direct contact with animals7.
Shiga toxin (Stx) is considered to be the primary virulence factor of STEC that is responsible for immunopathologies such as HC and HUS8. However, Stx alone is insufficient to cause severe illness without the adherence of bacteria to gut epithelial cells9. Multiplex genes that enable STEC strains to attach, colonize, produce and secrete toxin proteins have been identified9. In a subset of STEC strains, intimin plays a critical role in intestinal colonization, which is encoded by the eae gene that resides on the locus of enterocyte effacement (LEE) pathogenicity island. The LEE island encodes a type III secretion system that is responsible for the attaching and effacing (A/E) lesions on intestinal epithelia10. A/E lesions is characterized by the local effacement of microvilli, the tight attachment of bacteria to the eukaryotic surface and the subsequent reorganization of filamentous actin to pedestal-like structures11. Intimin is also an important virulence factor of other bacteria, such as enteropathogenic E. coli (EPEC), E. albertii, and Citrobacter rodentium12.
The full length of eae gene is about 2800 nucleotides. The N-terminal of intimin from different sources is highly conserved, while the C-terminal where cellular binding activity is highly variable13. Based on the difference of the C-terminal, at least 30 intimin subtypes have been defined, namely, α1, α2, α8, β1, β2, β3, γ1, γ2, ε1, ε2, ε3, ε4, ξ, z, z3, η, η2, θ, τ, ι1, ι2, κ, λ, μ, ν, υ, ο, π, ρ, and σ14. Intimin subtypes are correlated with host specificity and tissue tropism15. Intimin subtype β1 appears to be the most frequent among atypical EPEC strains from diarrheal patients and animals in China16. Several studies have shown the association between serotypes and specific intimin subtypes17. For example, O157:H7 and O145:H28 serotypes are associated with the eae-γ1 subtype, whereas O26:H11 often carries eae-β1, O103:H2 and O121:H19 harbor eae-ε, and O111:H8 harbors eae-θ subtype18. These serotypes were most frequently reported in global dysentery and HUS cases caused by STEC5. Thus, eae subtyping would be a valuable tool for risk assessment and prediction of disease outcome. However, there are limited studies of intimin characteristics among other non-O157 serotypes. In this study, we investigated the prevalence of eae gene and analyzed eae subtypes and polymorphisms in non-O157 STEC strains isolated from diarrheal patients, healthy carriers, animals, and raw meats in China.
Results
Prevalence of eae in the non-O157 STEC collection
Among 735 non-O157 STEC strains, eae was present in 70 (9.5%) strains, which were isolated from cattle (n = 41), yak (n = 2), raw beef (n = 3), raw mutton (n = 1), and diarrheal patients (n = 9). Fourteen eae-positive strains were identified from unknown sources. All strains recovered from goat, pig, plateau pika, marmot, Tibetan antelope, water, and healthy carriers were eae-negative (Table 1 and Table S1).
Table 1.
The origin and location of 735 non-O157 STEC isolates used in this study.
| Source | Location | Year | No. of isolates | No. of eae-positive (%) |
|---|---|---|---|---|
| Cattle | Shandong, Sichuan, Heilongjiang | 2009, 2012, 2015, 2017 | 154 | 41 (26.6) |
| Goat | Henan, Sichuan, Shandong | 2009, 2017 | 156 | 0 (0) |
| Pig | Chongqing, Beijing, Guizhou, Shandong, Heilongjiang | 2011, 2012, 2013, 2015 | 135 | 0 (0) |
| Yak | Qinghai | 2012 | 128 | 2 (1.6) |
| Plateau Pika | Qinghai | 2012, 2012, 2015 | 22 | 0 (0) |
| Marmot | Qinghai | 2012, 2013 | 8 | 0 (0) |
| Tibetan antelope | Qinghai | 2014 | 5 | 0 (0) |
| Raw meats | Beijing, Sichuan | 2013, 2014 | 60 | 4 (6.7) |
| Diarrheal patient | Henan, Shenzhen, Shanghai, Sichuan, Beijing | 2010, 2012, 2013, 2014, 2016, 2018 | 31 | 9 (29.0) |
| Healthy carrier | Qinghai, Shenzhen | 2013, 2014 | 4 | 0 (0) |
| Water | Shandong | 2017 | 1 | 0 (0) |
| Unknown | Heilongjiang, Guangxi, others | 2014 | 31 | 14 (45.2) |
| Total | 735 | 70 (9.5) |
The diversity and subtypes of eae in non-O157 STEC strains
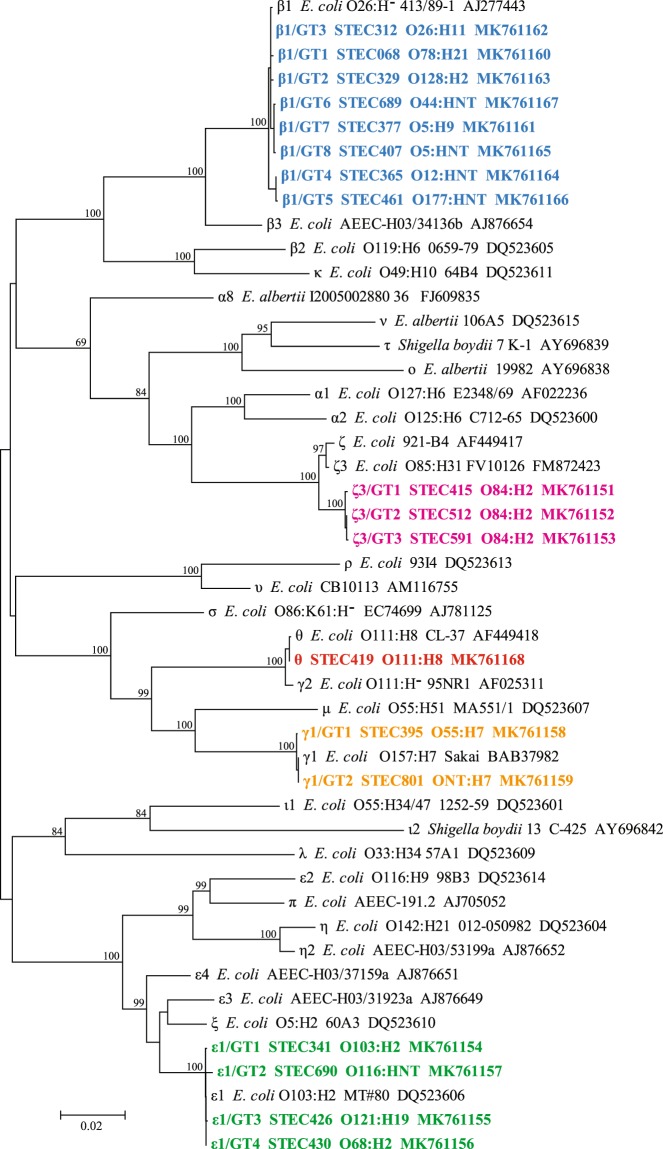
The complete eae sequences were obtained from 62 out of 70 strains, eight strains which failed to yield the complete eae sequences were excluded in this study. Among the 62 eae sequences, 18 unique eae sequences were identified (Table 2), the nucleotide identities among the 18 unique eae sequences ranged from 86.01% to 99.97% based on pairwise comparisons. Five eae subtypes, namely, β1, γ1, ε1, ζ3 and θ, were assigned based on phylogenic analysis. eae sequence polymorphism, designated as genotypes (GTs) were identified in each eae subtype to represent the diversity within a subtype. Except θ, the other four subtypes contained 2 to 8 genotypes respectively (Fig. 1). The BLASTn search against GenBank database (nr/nt) showed that 7 eae genotypes (β1/GT5, β1/GT6, β1/GT8, ε1/GT1, ζ1/GT2, ζ1/GT2 and ζ1/GT3) in this study are unique comparing with all available eae sequences in the database (accessed 25/7/2019).
Table 2.
eae subtypes of 62 eae-positive non-O157 STEC strains.
| eae subtype /genotype | Origin | Serotype | stx subtype | Sequence type |
|---|---|---|---|---|
| β1 /GT1 (2) | Yak | O78:H21 (1), O78:HNT (1) | stx2a | ST3884 (1), ST40 (1) |
| β1 /GT2 (1) | Unknown | O128:H2 | stx2f | N3 |
| β1 /GT3 (8) | Diarrheal patient (5), Unknown (3) | O26:H11 | stx1a (6), stx2a (2) | ST21 |
| β1 /GT4 (1) | Raw beef | O12:HNT | stx2c | ST659 |
| β1 /GT5 (8) | Cattle | O177:HNT | stx2c | ST659 (1), ST7220 (7) |
| β1 /GT6 (1) | Cattle | O44:HNT | stx1a | N1 |
| β1 /GT7 (2) | Diarrheal patient (1), Raw mutton (1) | O5:H9 (1), O5:HNT (1) | stx1a | ST342 |
| β1 /GT8 (1) | Diarrheal patient | O5:HNT | stx1a | ST342 |
| ε1 /GT1 (1) | Unknown | O103:H2 | stx1a | N4 |
| ε1 /GT2 (3) | Cattle | O5:HNT (1), O116:HNT (1), ONT:HNT (1) | stx2c (2), stx1a+ stx2c (1) | ST119 (2), N2 |
| ε1 /GT3 (1) | Unknown | O121:H19 | stx2a | ST655 |
| ε1 /GT4 (1) | Unknown | O68:H2 | stx1a | N5 |
| γ1 /GT1 (2) | Unknown | O55:H7 | stx1a | ST335 |
| γ1 /GT2 (1) | Diarrheal patient | ONT:H7 | stx1a+ stx2a | ST11 |
| θ (5) | Raw beef (2), Unknown (3) | O103:H25 (2), O111:H8 (2), O111:HNT (1) | stx1a | ST16 (3), ST343 (2) |
| ζ3 /GT1 (1) | Diarrheal patient | O84:H2 | stx1a | ST306 |
| ζ3 /GT2 (22) | Cattle (22) | O84:H2 | stx1a | ST306 |
| ζ3 /GT3 (1) | Cattle (1) | O84:H2 | stx1a | ST306 |
Figure 1.
Phylogenetic relationships of 18 different eae sequences obtained in this study and 30 eae subtypes reference sequences based on Neighbor-Joining method. The corresponding eae subtype, strain name, and GenBank accession number are listed on the right. The eae subtypes/genotypes identified in this study are indicated in bold and different colors. Scale bar indicates genetic distance.
Three major eae genotypes (ζ3/GT2, β1/GT3 and β1/GT5) contained 22, 8 and 8 strains respectively, and 10 genotypes contained only one strain, while the rest contained two to five strains (Table 2).
eae genotypes in correlation with serotypes
Fifteen different O serogroups and 9 different H types were identified among the 62 STEC strains, which belonged to 19 serotypes: ONT:H7, O103:H25, O103:H2, O111:HNT, O111:H8, O116:HNT, O12:HNT, O121:H19, O128:H2, O117:HNT, O26:H11, O44:HNT, O5:HNT, O5:H9, O55:H7, O68:H2, O78:HNT, O78:H21 and O84:H2. Two and 17 strains were O and H untypable, respectively. The most predominant serotype was O84:H2 (24/62, 38.7%), followed by O26:H11 (8/62, 12.9%), and O177: HNT (7/62, 11.3%).
A link was observed between serotypes and eae genotypes. Each eae genotype contained one to three different serotypes, with the exception of serotypes O5:HNT and O84:H2. Strains of O5:HNT serotype were assigned to β1/GT7, β1/GT8 or ε1/GT10 eae genotype, while O84:H2 strains carried eae ζ3/GT1, ζ3/GT2, or ζ3/GT3 genotypes (Table 2).
stx type/subtypes in eae positive non-O157 STEC strains
Overall, 43 strains harbored stx1 only, 17 strains contained stx2 only and 2 strains possessed both stx1 and stx2. Only one stx1 subtype, stx1a, was identified, while three stx2 subtypes (stx2a, stx2c and stx2f) were detected among the 62 STEC strains.
Among 24 eae-β1 harboring strains, 10 strains carried stx1a, nine carried stx2c and the other possessed stx2a, or stx2f subtypes. The strains with eae-ε1 subtype carried diverse stx subtypes: two strains contained stx1a, two strains carried stx2c, one carried stx2a, and one harbored both stx1a and stx2c. Two of eae-γ1 strains carried stx1a and one harbored both stx1a and stx2a. All the 24 eae-ζ3 and 5 eae-θ containing strains possessed stx1a (Table 2).
STEC origin correlated with the eae genotypes
STEC strains carrying eae-β1 subtype were detected from all sources investigated in this study. A concordance was observed between STEC origin and eae genotypes. Each eae genotype contained strains from a specific host source with the exception of three genotypes. The β1/GT7 genotype was detected from strains isolated from human and mutton. The β1/GT3 or θ genotype contain strains from two different sources (Table 2).
Human-derived strains belonged to five eae genotypes, among which, three (β1/GT8, γ1/GT2, and ζ1/GT3) are unique. Furthermore, all cattle-derived strains belonged to five unique eae genotypes, i.e. β1/GT5, β1 /GT6, ε1/GT2, ζ3/GT2, and ζ3/GT3 (Table 2).
MLST analysis of eae-positive non-O157 STEC
The 62 strains were typed into 18 sequence types (STs) with 5 novel STs named as N1-N5. One new ST (strain STEC430) was resulted from a novel allele in icd. The other four new STs (strain STEC329, STEC341, STEC689, STEC790) were due to the new combinations of previously known alleles. An eae genotype was corresponding to one or more STs and vice versa (Table 2).
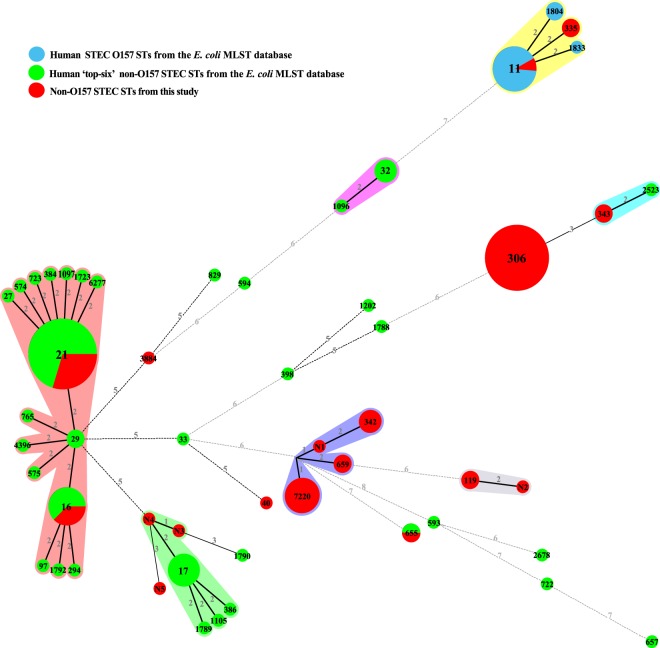
A minimum spanning tree was constructed with the STs from this study and those of O157 and ‘top six’ non-O157 STEC from MLST database (Fig. 2). Most STs differed from each other by 2 or more alleles, while two pairs of STs (N1 and ST7220, N3 and N4) differed from each other by only 1 allele. STs of human O157 STEC are different from those of ‘top six’ non-O157 STEC. Only three STs (ST16, ST21 and ST655) were shared by strains from this study and human ‘top six’ non-O157 STEC assigned as O111, O26 and O121 serogroups, respectively. However, STs of O103 serogroup are diverse, which were different from those of MLST database. An ONT:H7 strain obtained in this study was typed as ST11, which is often recognized as ST of STEC O157:H7 (Table 2 and Table S1).
Figure 2.
Minimum spanning tree of 62 STs from this study (red), 13 STs from human O157 STEC (blue), and 65 STs from human ‘top six’ non-O157 STEC (green). Each circle represents a ST, with the pie divided proportionally to the number of isolates in that ST from different sources. The number in a circle indicates the ST number. The numbers on connecting lines represent the number of allelic differences between two STs.
Discussion
Intimin encoded by eae is an important virulence factor in many STEC strains, which plays a critical role in intestinal colonization. Previous studies revealed that most clinical STEC strains possessed eae19,20. Moreover, the presence of eae was significantly associated with a higher risk for HUS development21. STEC O157 isolates were strongly associated with the simultaneous presence of both stx2 and eae, forming the basis of why STEC O157 predominates in patients with HUS, when compared with non-O157 strains22. However, a subsequent study reported that eae was detected in the majority (52.5%) of non-O157 STEC strains in England23. A recent investigation of STEC infections in the south east of England revealed that 76% of non-O157 HUS-associated STEC isolates possessed eae gene24. These data indicated that the presence of eae is strongly related with disease severity irrespective of seropathotypes. In this study, we observed a lower prevalence of eae (29.0%) among non-O157 STEC strains from diarrheal patients, which might partially be due to limitation of the current study where the source of 14 eae-positive isolates is unavailable. On the other hand, the low prevalence of eae in diarrheal patients could account for the less severe disease in this study. Among the nine eae-positive isolates from diarrheal patients, only one was both stx1a and stx2a positive, while the rest all harbored stx1a only (Table 2). Stx2 positive strain especially Stx2a induces more severe clinical symptoms and higher mortality than other Stx subtypes25. Previous study reported that 20% of meat STEC isolates carried eae19, 36% of the cattle isolates possessed eae26. In this study, we found that 6.7% of raw meat isolates possessed eae, the prevalence of eae in cattle isolates is 26.6%. The variation of eae prevalence among different studies possibly due to several factors including sample sources, isolation/detections assays, or geographic distribution.
The eae gene was classified into different subtypes based on the variety of the 280 amino acids C-terminal region13. The highly divergent C-terminal region of eae constitutes the molecule that binding to receptors on the epithelial cell27. Various eae subtypes may confer distinct colonization patterns within the human intestine, thus leading to distinct pathogenic capacity. Among the known eae subtypes described so far, four subtypes (β, ε, γ1, θ) have been reported to be associated with more virulent STEC and thus posing greater health risk28. In a previous study, the eae subtypes of STEC strains recovered from children with HUS in Uruguay, include γ1, γ2 and β129, highlighting the clinical significance of these eae subtypes. In this study, we found that β1 and ζ3 were the most prevalent eae subtypes, which were detected in strains from different sources. Notably, eae-β1 was the most predominant subtype among strains from diarrheal patients30.
Studies have indicated association between serotypes and eae subtypes in STEC strains, particularly predominant serotypes. Strains belonging to serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 are associated with particular eae subtypes, i.e., γ1, β1, ε, θ, and γ1, respectively31,32. Consistent with previous studies, we found that all eight O26:H11 strains (five from diarrheal patients and three from unknown origin) in this study carried the eae-β1 subtype, all three O111 strains harbored the eae-θ subtype, and one O103 strain possessed the eae-ε1 subtype. However, the origin of three O111 strains and one O103 strain were unavailable. Notably, the three O111 strains share identical eae sequences with the outbreak strain O111:H8 in the United States33.The eae sequence of strain STEC801 (ONT:H7) isolated from a diarrheal patient was identical to that of strain Sakai, which caused a notorious outbreak in Japan in 199634. Comparison with STs observed in human non-O157 STEC infections gives an indication of the potential risk for different eae subtype STEC. Strains of the three STs (ST16, ST21 and ST655) in this study had the same O serogroups with ‘top six’ non-O157 STEC (O111, O26 and O121). The three eae subtypes were θ, β1 and ε1, respectively. These results indicated that determination of eae subtypes could be used as a valuable tool in combinations with serotypes, and other virulence factors in risk assessment and prediction of severe disease outcomes.
Shiga toxin subtypes have been found to differ in toxin potency. Strains that carry Stx2a (with or without Stx2c) are often associated with severe symptoms such as HC and HUS35,36. The stx2a gene is most often present in eae positive STEC strains and has consistently been associated with HUS37. In this study, 6 out of 62 isolates carried stx2a subtypes, which were isolated from diarrheal patients, yak and other unknown sources. Notably, the 6 stx2a isolates harbored eae-β1, eae-ε1 and eae-γ1 subtypes, all of which were associated with high virulence, thus they were likely to have high pathogenic potential to humans.
Besides intimin gene eae, the plasmid-carried enterohemolysin gene (ehxA) also plays an important role in STEC pathogenicity and frequently associated with diarrheal disease and HUS38. In a previous study, we described the presence and genetic diversity of the ehxA gene in 434 non-O157 STEC isolates. The ehxA gene was positive in 138 (31.8%) isolates, and 15 (10.9%) ehxA-positive isolates harbored eae, which were grouped into ehxA group I (n = 2) and group II (n = 13). All strains from diarrheal patients belonged to ehxA group II and most of those strains harbored eae. Thus, ehxA group II and eae-positive strains were clinically related39.
In this study, we observed the association between eae genotypes and host sources. Most strains from diarrheal patients (7/9, 77.8%) carried eae-β1 subtype, and most isolates from cattle (23/26, 88.5%) carried eae-ζ3 subtype. However, the prevalence of eae subtypes in a specific source varied significantly among studies. For instance, Tostes et al.28 reported the predominance of eae subtypes λ/ γ1 and β in cattle isolates, and λ/ γ1 in human isolates. Similarly, Blanco et al.39 reported eae-ζ as most frequent subtype in E. coli isolates from sheep in Spain. Whereas, Aktan et al.40 identified eae-β and eae-γ as the most frequent subtypes among E. coli from sheep in England and Wales. The distribution of eae subtypes in the same source may vary among different regions.
In conclusion, the current study reports the prevalence and subtype of eae gene among non-O157 STEC strains from a wide variety of sources in China. Among 735 non-O157 STEC strains, eae was present in 70 (9.5%) strains isolated from diarrheal patients, animals, raw meats and other unknown sources. Five eae subtypes and 18 different eae genotypes were identified, suggesting the high diversity of eae among different sources. To our knowledge, this is the first study investigating the prevalence of eae subtypes among non-O157 STEC strains from a wide range of sources. Our study suggests associations between eae subtypes and serotypes as well as host sources. Furthermore, it can be inferred from this study that the determination of eae subtype could be considered together with seropathotypes and other virulence factors in risk assessment of STEC infections.
Materials and Methods
Ethics statement
The current study and all experimental protocols were approved by the ethic committee of the National Institute for Communicable Diseases Control and Prevention, China CDC, with the number ICDC-2017006. This work was part of STEC surveillance program conducted in China, the informed permission has been obtained from patients and the owners of animals to use fecal samples and conduct relevant studies. All methods used in this study were performed in accordance with the relevant guidelines and regulations.
Bacterial strains and detection of eae gene
A total of 735 non-O157 STEC strains collected during April 2009 to December 2018 were used in this study. Most strains were isolated from different samples through the STEC surveillance conducted in China. Others were obtained from local Centers for Disease Control and Prevention (CDC) in eleven geographical regions in China. Some strains were reported in our previous studies41–43. Of the 735 STEC strains, 608, 60, 35, and 1 were isolated from animals, raw meats, humans and environmental water, respectively. The sources of 31 strains were unavailable (Table 1 and Table S1). All strains were confirmed to be STEC by using previously described methods41.
The presence of eae gene among all 735 non-O157 STEC strains were screened using PCR method with primer eae-F (5′-TCAATGCAGTTCCGTTATCAGTT-3′) and eae-R (5′-GTAAAGTCCGTTACCCCAACCTG-3′)44.
Serotyping and stx subtyping
The O serogroup of each isolate was initially screened by an O-genotyping PCR method designed by Iguchi et al.45, and further confirmed by using E. coli antisera, O1–O188 (Statens Serum Institut, Hillerød, Denmark). The entire coding sequence of fliC was amplified by PCR with the primers fliC-F (5′-ATGGCACAAGTCATTAATACCCAAC-3′) and fliC-R (5′-CTAACCCTGCAGCAGAGACA-3′) as reported by Fields et al. and then sequenced with Sanger sequencing46. The sequences were analyzed with the SerotypeFinder database (https://cge.cbs.dtu.dk/services/SerotypeFinder/) to determine H types47.
The stx1 subtypes (stx1a, stx1c, stx1d) and stx2 subtypes (stx2a to stx2h) were determined by a PCR-based subtyping protocol in combination with a phylogeny scheme as described previously48,49.
Sequencing of the complete eae gene
The complete eae gene was obtained by PCR using previously described method16. Two additional primer designed in this study were used for sequencing: eaeR3-A (5′-TCCATGTGTATTTTCCATTGCC-3′) and eaeR3-B (5′-TATATTTCCCATCACCTCCAC-3′). All PCR products were visualized by agarose gel electrophoresis and purified by using a QIAquick PCR purification kit (Qiagen, Germany), and then sequenced using BigDye™ Terminator V3.1 Cycle Sequencing kit (Applied Biosystems, USA).
eae subtyping and polymorphism analysis
Each of the sequenced ~3.2 kb LEE region that contained the complete eae gene was checked and assembled by SeqMan II (DNASTAR Inc., USA). The eae subtypes reference sequences were downloaded from GenBank. The MEGA 7 software (www.megasoftware.net)50 was used to align the complete eae sequences obtained in this study and the reference eae sequences. A Neighbor-Joining tree was constructed with maximum composite likelihood model. Bootstrap analyses were performed (1,000 replicates) to estimate the stability and genetic distances were calculated by the maximum composite likelihood method. A novel subtype was defined by a cutoff value of 95% nucleotide sequence identity as described previously51.
Multilocus sequence typing (MLST)
Seven housekeeping genes (adk, fumC, gyrB, icdF, mdh, purA, and recA) were amplified by PCR and sequenced according to the scheme provided by the E. coli MLST website (http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search). Each locus was assigned an allele number by comparing sequences against the E. coli MLST database. The allelic profile of seven housekeeping genes was used to generate a specific sequence type (ST) for each isolate. STs of human STEC O157 and the ‘top six’ non-O157 serogroups were downloaded from the E. coli MLST database for comparison. A minimum spanning tree was generated with BioNumerics software version 4.0 (Applied Maths, Belgium).
Nucleotide sequence accession numbers
The 18 diverse eae sequences obtained in this study were submitted to GenBank (Acc. MK761151–MK761168).
Supplementary information
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2018YFC1603800), the National Natural Science Foundation of China (81701977, 81772152), and the National Science and Technology Major Project (2018ZX10201001-006 and 2018ZX10301407-002). J.Z. was funded by the New Zealand Food Safety Science & Research Centre.
Author contributions
X.B. and Y.X. designed the research. H.S., R.F. and S. Fu. prepared the samples. X.Y. conducted the experiments. X.Y., J.Z. and A.M. analyzed the data. X.Y., X.B. and Y.X. drafted the manuscript. J.Z. and A.M. polished the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60225-w.
References
- 1.Gonzalez-Escalona N, Kase JA. Virulence gene profiles and phylogeny of Shiga toxin-positive Escherichia coli strains isolated from FDA regulated foods during 2010-2017. Plos One. 2019;14:e0214620. doi: 10.1371/journal.pone.0214620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mora A, et al. Seropathotypes, Phylogroups, Stx subtypes, and intimin types of wildlife-carried, Shiga toxin-producing Escherichia coli strains with the same characteristics as human-pathogenic isolates. Appl. Env. Microbiol. 2012;78:2578–2585. doi: 10.1128/aem.07520-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karmali MA. Factors in the emergence of serious human infections associated with highly pathogenic strains of Shiga toxin-producing Escherichia coli. Int. J. Med. Microbiol. 2018;308:1067–1072. doi: 10.1016/j.ijmm.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Tseng M, et al. Increasing incidence of non-O157 Shiga toxin-producing Escherichia coli (STEC) in Michigan and association with clinical illness. Epidemiol. Infect. 2016;144:1394–1405. doi: 10.1017/s0950268815002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valilis E, Ramsey A, Sidiq S, DuPont HL. Non-O157 Shiga toxin-producing Escherichia coli-A poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. 2018;76:82–87. doi: 10.1016/j.ijid.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 2006;43:1587–1595. doi: 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 7.Schlager S, et al. Petting zoos as sources of Shiga toxin-producing Escherichia coli (STEC) infections. Int. J. Med. Microbiol. 2018;308:927–932. doi: 10.1016/j.ijmm.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Lee Moo-Seung, Tesh Vernon. Roles of Shiga Toxins in Immunopathology. Toxins. 2019;11(4):212. doi: 10.3390/toxins11040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farfan MJ, Torres AG. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 2012;80:903–913. doi: 10.1128/iai.05907-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens, M. P. & Frankel, G. M. The Locus of Enterocyte Effacement and Associated Virulence Factors of Enterohemorrhagic Escherichia coli. Microbiol. Spectr. 2, EHEC-0007-2013, 10.1128/microbiolspec.EHEC-0007-2013 (2014). [DOI] [PubMed]
- 11.Donnenberg MS, Kaper JB, Finlay BB. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/s0966-842x(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 12.Lacher DW, Steinsland H, Whittam TS. Allelic subtyping of the intimin locus (eae) of pathogenic Escherichia coli by fluorescent RFLP. FEMS Microbiol. Lett. 2006;261:80–87. doi: 10.1111/j.1574-6968.2006.00328.x. [DOI] [PubMed] [Google Scholar]
- 13.Hernandes RT, Elias WP, Vieira MA, Gomes TA. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 2009;297:137–149. doi: 10.1111/j.1574-6968.2009.01664.x. [DOI] [PubMed] [Google Scholar]
- 14.Ooka T, et al. Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 2012;18:488–492. doi: 10.3201/eid1803.111401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang WL, et al. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 2002;40:4486–4492. doi: 10.1128/jcm.40.12.4486-4492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, et al. Genetic Diversity of Intimin Gene of Atypical Enteropathogenic Escherichia coli Isolated from Human, Animals and Raw Meats in China. Plos One. 2016;11:e0152571. doi: 10.1371/journal.pone.0152571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McWilliams BD, Torres AG. EHEC Adhesins. Microbiol. Spectr. 2014;2:EHEC00032013. doi: 10.1128/microbiolspec.EHEC-0003-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bibbal D, et al. Intimin gene (eae) subtype-based real-time PCR strategy for specific detection of Shiga toxin-producing Escherichia coli serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 in cattle feces. Appl. Env. Microbiol. 2014;80:1177–1184. doi: 10.1128/aem.03161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang Minh S, Kimura E, Hoang Minh D, Honjoh K, Miyamoto T. Virulence characteristics of Shiga toxin-producing Escherichia coli from raw meats and clinical samples. Microbiol. Immunol. 2015;59:114–122. doi: 10.1111/1348-0421.12235. [DOI] [PubMed] [Google Scholar]
- 20.Fierz, L., Cernela, N., Hauser, E., Nuesch-Inderbinen, M. & Stephan, R. Characteristics of Shiga toxin-Producing Escherichia coli Strains Isolated during 2010–2014 from Human Infections in Switzerland. Front. Microbiol.8, 1471, 10.3389/fmicb.2017.01471 (2017). [DOI] [PMC free article] [PubMed]
- 21.De Rauw, K., Buyl, R., Jacquinet, S. & Pierard, D. Risk determinants for the development of typical haemolytic uremic syndrome in Belgium and proposition of a new virulence typing algorithm for Shiga toxin-producing Escherichia coli. Epidemiol Infect, 1-5, 10.1017/S0950268818002546 (2018). [DOI] [PubMed]
- 22.Werber D, et al. Strong association between Shiga toxin-producing Escherichia coli O157 and virulence genes stx2 and eae as possible explanation for predominance of serogroup O157 in patients with haemolytic uraemic syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 2003;22:726–730. doi: 10.1007/s10096-003-1025-0. [DOI] [PubMed] [Google Scholar]
- 23.Byrne L, et al. Epidemiology and microbiology of Shiga toxin-producing Escherichia coli other than serogroup O157 in England, 2009-2013. J. Med. Microbiol. 2014;63:1181–1188. doi: 10.1099/jmm.0.075895-0. [DOI] [PubMed] [Google Scholar]
- 24.Carroll KJ, Harvey-Vince L, Jenkins C, Mohan K, Balasegaram S. The epidemiology of Shiga toxin-producing Escherichia coli infections in the South East of England: November 2013-March 2017 and significance for clinical and public health. J. Med. Microbiol. 2019;68:930–939. doi: 10.1099/jmm.0.000970. [DOI] [PubMed] [Google Scholar]
- 25.Shringi S, et al. Differential virulence of clinical and bovine-biased enterohemorrhagic Escherichia coli O157:H7 genotypes in piglet and Dutch belted rabbit models. Infect. Immun. 2012;80:369–380. doi: 10.1128/IAI.05470-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jajarmi M, Imani Fooladi AA, Badouei MA, Ahmadi A. Virulence genes, Shiga toxin subtypes, major O-serogroups, and phylogenetic background of Shiga toxin-producing Escherichia coli strains isolated from cattle in Iran. Microb. Pathog. 2017;109:274–279. doi: 10.1016/j.micpath.2017.05.041. [DOI] [PubMed] [Google Scholar]
- 27.Frankel G, Candy DC, Everest P, Dougan G. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect. Immun. 1994;62:1835–1842. doi: 10.1128/IAI.62.5.1835-1842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tostes R, et al. Subtyping Escherichia coli Virulence Genes Isolated from Feces of Beef Cattle and Clinical Cases in Alberta. Foodborne Pathog. Dis. 2017;14:35–42. doi: 10.1089/fpd.2016.2199. [DOI] [PubMed] [Google Scholar]
- 29.Perez L, et al. Hemolytic uremic syndrome with mild renal involvement due to Shiga toxin-producing Escherichia coli (STEC) O145 strain. Rev. Argent. Microbiol. 2014;46:103–106. doi: 10.1016/s0325-7541(14)70056-2. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins C, et al. Subtyping of virulence genes in verocytotoxin-producing Escherichia coli (VTEC) other than serogroup O157 associated with disease in the United Kingdom. J. Med. Microbiol. 2003;52:941–947. doi: 10.1099/jmm.0.05160-0. [DOI] [PubMed] [Google Scholar]
- 31.Oswald E, et al. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 2000;68:64–71. doi: 10.1128/iai.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarr CL, Whittam TS. Molecular evolution of the intimin gene in O111 clones of pathogenic. Escherichia coli. J. Bacteriol. 2002;184:479–487. doi: 10.1128/jb.184.2.479-487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Escherichia coli O111:H8 outbreak among teenage campers-Texas, 1999. MMWR Morb. Mortal. Wkly. Rep.49, 321–324 (2000). [PubMed]
- 34.Watanabe H, Wada A, Inagaki Y, Itoh K, Tamura K. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet. 1996;348:831–832. doi: 10.1016/s0140-6736(05)65257-9. [DOI] [PubMed] [Google Scholar]
- 35.Fuller CA, Pellino CA, Flagler MJ, Strasser JE, Weiss AA. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011;79:1329–1337. doi: 10.1128/iai.01182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orth D, et al. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn. Microbiol. Infect. Dis. 2007;59:235–242. doi: 10.1016/j.diagmicrobio.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Fao/Who Stec Expert, G Hazard Identification and Characterization: Criteria for Categorizing Shiga Toxin-Producing Escherichia coli on a Risk Basis(dagger) J. Food Prot. 2019;82:7–21. doi: 10.4315/0362-028X.JFP-18-291. [DOI] [PubMed] [Google Scholar]
- 38.Lorenz SC, et al. Prevalence of hemolysin genes and comparison of ehxA subtype patterns in Shiga toxin-producing Escherichia coli (STEC) and non-STEC strains from clinical, food, and animal sources. Appl. Env. Microbiol. 2013;79:6301–6311. doi: 10.1128/AEM.02200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanco M, et al. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 2003;41:1351–1356. doi: 10.1128/jcm.41.4.1351-1356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aktan I, et al. Characterisation of attaching-effacing Escherichia coli isolated from animals at slaughter in England and Wales. Vet. Microbiol. 2004;102:43–53. doi: 10.1016/j.vetmic.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Bai X, et al. Molecular and Phylogenetic Characterization of Non-O157 Shiga Toxin-Producing Escherichia coli Strains in China. Front. Cell Infect. Microbiol. 2016;6:143. doi: 10.3389/fcimb.2016.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu S, et al. Genetic diversity of the enterohaemolysin gene (ehxA) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci. Rep. 2018;8:4233. doi: 10.1038/s41598-018-22699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan R, et al. Tellurite resistance profiles and performance of different chromogenic agars for detection of non-O157 Shiga toxin-producing Escherichia coli. Int. J. Food Microbiol. 2018;266:295–300. doi: 10.1016/j.ijfoodmicro.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 44.Bai X, et al. Shiga toxin-producing Escherichia coli in yaks (Bos grunniens) from the Qinghai-Tibetan Plateau, China. Plos One. 2013;8:e65537. doi: 10.1371/journal.pone.0065537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iguchi A, et al. Escherichia coli O-Genotyping PCR: a Comprehensive and Practical Platform for Molecular O Serogrouping. J. Clin. Microbiol. 2015;53:2427–2432. doi: 10.1128/jcm.00321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fields PI, et al. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J. Clin. Microbiol. 1997;35:1066–1070. doi: 10.1128/JCM.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015;53:2410–2426. doi: 10.1128/jcm.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai X, et al. Identification and pathogenomic analysis of an Escherichia coli strain producing a novel Shiga toxin 2 subtype. Sci. Rep. 2018;8:6756. doi: 10.1038/s41598-018-25233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheutz F, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012;50:2951–2963. doi: 10.1128/jcm.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanco M, et al. Identification of two new intimin types in atypical enteropathogenic Escherichia coli. Int. Microbiol. 2006;9:103–110. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.