Abstract
Introduction
Despite a significant decline in Senegal, malaria remains a burden in various parts of the country. Assessment of multiplicity of Plasmodium falciparum infection and genetic diversity of parasites population could help in monitoring of malaria control.
Objective
To assess genetic diversity and multiplicity of infection in P. falciparum isolates from three areas in Senegal with different malaria transmissions.
Methods
136 blood samples were collected from patients with uncomplicated P. falciparum malaria in Pikine, Kedougou and Thies. Polymorphic loci of msp1 and 2 (Merozoite surface protein-1 and 2) genes were amplified by nested PCR.
Results
For msp1gene, K1 allelic family was predominant with frequency of 71%. Concerning msp2 gene, IC3D7 allelic family was the most represented with frequency of 83%. Multiclonal isolates found were 36% and 31% for msp1et msp2 genes respectively. The MOI found in all areas was 2.56 and was statistically different between areas (P=0.024). Low to intermediate genetic diversity were found with heterozygosity range (He=0,394–0,637) and low genetic differentiation (Fst msp1= 0.011; Fst msp2=0.017) were observed between P. falciparum population within the country.
Conclusion
Low to moderate genetic diversity of P.falciparum strains and MOI disparities were found in Senegal.
Keywords: Senegal, MOI, Genetic diversity, msp1, msp2
Introduction
Malaria control interventions showed a significant progress worldwide1. However, malaria remains a burden in sub-Saharan Africa despite scaling up malaria interventions1. Senegalese national malaria control program (NMCP) intensified malaria control strategies for more than ten years including large distribution of long-lasting insecticide-treated bed nets (LLINs), use of histidine rich protein 2-based rapid diagnostic tests (HRP2-RDT) and artemisinin-based combination therapies (ACTs) for the treatment of uncomplicated Plasmodium(P) falciparum malaria2. Nevertheless in 2015, 492253 cases and 526 deaths due to P. falciparum malaria had been registered in the country3.
In Senegal, the overall population is exposed to malaria with a gradient of transmission increasing from North to South due to malaria epidemiology that occurs as a result of intensified control efforts in these areas. Indeed, malaria is hyperendemic in the South (annual incidence is greater than 100/1000 inhabitants) and hypoendemic in the North (annual incidence rates are now less than 5/1000 inhabitants)3. Malaria transmission pattern affects the multiplicity of infection (MOI) and genetic diversity of parasite populations, the latter's increase when transmission is high and decrease when transmission is low4,5. Thus, assessment of genetic diversity and multiplicity of infection (MOI) provide insight on malaria transmission and genetic differentiation measurements such as heterozygosity (He) and fixation index (Fst) which are important data in monitoring of malaria control and elimination strategies4,6,7,8,9,10.
The polymorphic loci of merozoite surface proteins (msp1 and msp2) are now well established to assess the genetic diversity of P. falciparum population and multiplicity of infection (MOI) which is an indicator of malaria transmission intensity in endemic areas9,11,12,8,13,14,15. Additionally, many studies demonstrated that msp1 and msp2 genes are enough robust polymorphism markers to characterize the parasite population structure12,16 and are the gold standards to evaluate the MOI17 Msp1 gene has three allelic families K1, MAD20 and RO3318 and msp2 gene two allelic families, IC3D7 and FC2719.
The characterization of genetic diversity of P. falciparum through different areas with various endemicities within a country has been extensively looked into in different parts of the world8,9,13,16,20,21. However, a few studies about this topic have been performed in Senegal by Ahouidi et al22 and Konaté et al23. Hence, it is important to have current data on genetic diversity on parasite populations through several areas in the country to better inform malaria control programs. In addition, the monitoring of malaria transmission intensity is a clear priority to malaria elimination. As MOI is an important metric of malaria transmission24,25,26,27,28, it is a robust measure of changing malaria transmission and remains one of the most accurate molecular approaches to evaluate malaria parameters29 in areas moving towards elimination like Senegal. It is important to have insight of malaria transmission intensity using the MOI within the country to know which area need more interventions in order to achieve the malaria elimination target in Senegal. This study aimed to investigate the genetic diversity and the multiplicity of P. falciparum infection of circulating parasites strains from three areas in Senegal having different endemicities by genotyping the highly polymorphic loci of msp1 and msp2 genes. Some genetic measurements such as He and Fst were also analyzed.
Materials and methods
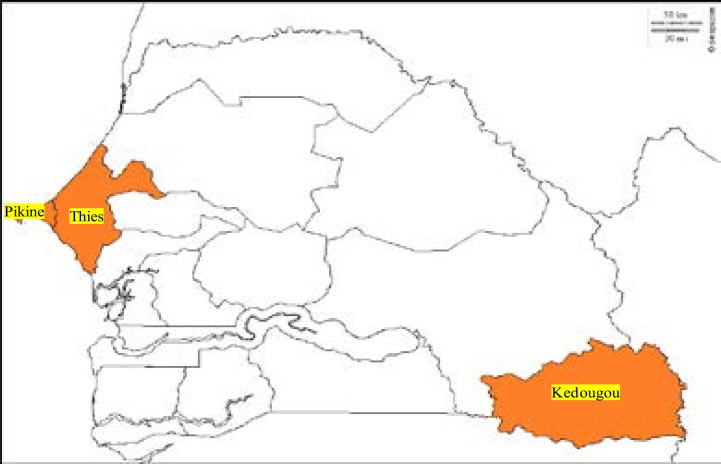
Figure 1. Map
Fig 1.
Map of the three malaria study areas ( Kedougou, Pikine and Thies) in Senegal. This map was generated using online website (http://www.d-maps.com)
Study sites
This study was carried out in three areas of Senegal with different endemicity of malaria: Pikine, Thies and Kedougou. Pikine is a sub-urban area which is 15 km the capital center (Dakar) with a heterogeneous endemicity (related to recurrent floods) with an entomological inoculation rate (EIR) ranging from 0 to 20 infectious bites per years and a malaria incidence greater than 15 malaria cases per 1000 habitants. Thies, in the west of the country at 70 km from Dakar, is a hypo-endemic area with a low EIR varying from 0 to 20 infectious bites per years and an average of 5 to 15 malaria cases per 1,000 habitants. Kedougou, located in the South-East, at 685 Km from Dakar, is hyper-endemic with an incidence higher than 15 malaria cases per 1000 habitants. In Kedougou, EIR is high ranging from 20 to 100 infectious bites/person/year2.
Sample collection
This study was approved by Ethics Committee of the Ministry of Health of Senegal. Febrile patients visiting the NMCP sentinel site in these three areas during malaria transmission season were enrolled in this study after giving informed consent or guardians consent for children depending on the age of the patient. Blood samples were collected on filter-paper from patients who met the following criteria: Living in a 15-km radius of health facilities, having fever (axillary temperature ≥ 37.5 C) or history of fever in the previous 48 h, age ranging from 6 months to 75 years and uncomplicated P. falciparum malaria with parasite density ≥ 1000 asexual forms per microliter. Patients who presented signs or symptoms of severe malaria as defined by World Health Organization (WHO) 30 and pregnant women were not included.
DNA extraction and PCR genotyping
Parasite DNA, from filter paper was extracted using QIAamp DNA Mini kit (Qiagen™, QIAGEN, USA) according to the manufacturer's instructions.
The polymorphic loci of msp1 block 2 (K1, MAD20 and RO33 allelic families) and msp2 central region (IC3D7 and FC27 allelic families) were amplified by nested PCR as described previously31,32. The primer sequences are as shown in Table 1. All PCR reactions were performed out in a total volume of 20 µl containing 6 µl GoTaq Green Master Mix, 0.5 µM of each primer, and 11 µl reagent grade water. In the first round reaction (nest 1), 1 µl of genomic DNA was added as a template. In the second nested reaction (nest 2), 1 µl of the nest 1 PCR product was used as DNA template. Cycling conditions for primary PCR were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min and extension at 72°C for 2 min; a final extension was done at 72°C for 3 min. The cycling conditions for secondary PCR were, initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 61°C for 2 min, extension at 72 °C for 2 min, with a final extension cycle of 72°C for 3 min. Positives (3D7 and Dd2) and negative (reagent grade water) controls were systematically incorporated in each PCR run. The nested PCR product were revealed by electrophoresis on 2% agarose gels stained with ethidium bromide and visualized under UV trans-illumination (VersaDoc®, BIORAD, Hercules, USA). The size of PCR fragments were estimated using 100 bp molecular weight.
Table 1.
Sequences of the primers used to amplify the msp 1 and msp 2 genes of P. falciparum isolates
| Amplification/gene | Primers | Primer sequence |
| Primary PCR | ||
| msp1 | M1-OR | 5′-CTA GAA GCT TTA GAA GAT GCA GTA TTG-3′ |
| M1-OF | 5′-CTT AAA TAG TAT TCT AAT TCA AGT GGA TCA-3′ | |
| msp2 | M2R | 5′-ATG AAG GTA ATT AAA ACA TTG TCT ATT ATA-3′ |
| M2F | 5′-CTT TGT TAC CAT CGG TAC ATT CTT-3′ | |
| Secondary PCR | ||
| K1 | M1K1R | 5′-AAA TGA AGA AGA AAT TAC TAC AAA AGG TGC-3′ |
| M1K1F | 5′-GCT TGC ATC AGC TGG AGG GCT TGC ACC AGA-3′ | |
| MAD20 | M1MAD20R | 5′-AAA TGA AGG AAC AAG TGG AAC AGC TGT TAC-3′ |
| M1MAD20F | 5′-ATC TGA AGG ATT TGT ACG TCT TGA ATT ACC-3′ | |
| RO33 | M1RO33R | 5′-TAAAGG ATG GAG CAA ATA CTC AAG TTG TTG-3′ |
| M1RO33F | 5′-CAT CTG AAG GAT TTG CAG CAC CTG GAG ATC-3′ | |
| IC3D7 | M2ICR | 5′-AATACT AAG AGT GTA GGT GCA TATGCT CCA-3′ |
| M2ICF | 5′-TTT TAT TTG GTG CAT TGC CAG AAC TTG AAC-3′ | |
| FC27 | M2FCR | 5′-AGAAGT ATG GCA GAA AGT AAC CCT TCT ACT-3′ |
| M2FCF | 5′-GAT TGT AAT TCG GGG GAT TCA GTT TGT TCG-3′ | |
Statistical analysis
The online Biostatgv was used for statistical analysis. For all tests, the significance level was p < 0.05. Isolates presenting more than one allele were considered as multiclonal isolates. The chi-square test (X2) was used to compare the frequencies of multiclonal isolates between localities. The multiplicity of infection (MOI) was calculated by dividing the total number of alleles detected for msp1and msp2 genes by the total number of samples11. Student's t test was used to compare MOI between localities. We used the expected heterozygosity (He) and genetic differentiation (FST) to assess population structure of parasites. Heterozygosity was calculated using the following formula He = n/(n-1) (1-∑Pi2), where n = sample size, Pi = allele frequency as described by Nei et al.33 and Fst was calculated as described previously22,34.
Results
The study population characteristics
A total of 13 P. falciparum infected patients were enrolled from three Senegalese localities. Among these patients, 64.7% were male and 35.3% were female with a sex ratio of 1.83. Patients' age ranged from 4 to 75 years with a mean age of patients was 22.3. The parasite density ranged from 1000 to 404000 asexual forms per microliter.
Allelic distribution of msp1and msp2 genes
Overall, the three allelic families (K1, MAD20 and RO33) of msp1gene and two (3D7 and FC27) of msp2 gene were observed in this study.
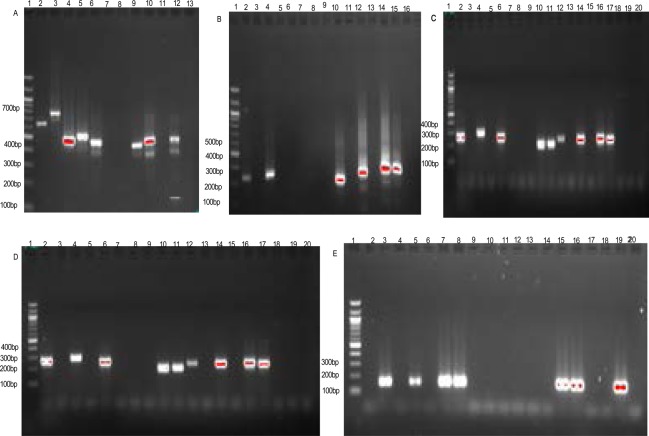
A total of forty-two alleles types were detected for the two genes in all localities: twenty-two for msp1 (Fig 2) and twenty for msp2 (Fig 3).
Fig 2.
Banding pattern of msp1 and msp2 alleles of P. falciparum.
A = IC3D7; Lane 1, 100 bp DNA ladder; Lanes, 2–9, selected samples; Lane 10–12, Positives controls (10,IC3D7; 11,Dd2; 12,HB3); Lane13, negative control.
B = FC27; Lane 1, 100 bp DNA ladder; Lane 10–12, selected samples; Lane,13–15, Positives controls (13, IC3D7; 14, Dd2; 15,HB3); Lane 16, negative control.
C = K1, D = MAD20, E = RO33; Lane 1, 100 bp DNA ladder; Lanes, 2–16, selected samples; Lane 17–19, Positives controls (17,IC3D7; 18,Dd2;19,HB3); Lane 20, negative control.
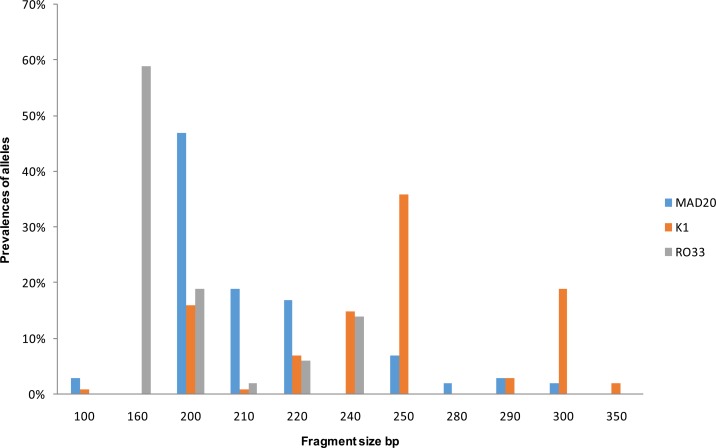
Fig 3.
Prevalence of MAD20, K1, RO33 alleles distribution according to their length or number of base pairs.
In msp1 gene, nine K1 alleles types (range from 100 bp to 350 bp), eight MAD20 alleles types (100 bp-300 bp) and five RO33 alleles types (160 bp-250 bp) were observed while for msp2 gene, eleven IC3D7 (300 bp-800 bp) and nine FC27 (200 bp -600 bp) alleles types were identified. Distribution of the different allelic families of msp1 and msp2 genes and their combinations in different localities are shown in table 2.
Table 2.
Distribution of allelic families of msp1 and msp2 genes of P.falciparum
| Localities | ||||
| Pikine (n=45) | Kédougou (n=50) | Thiès (n=41) | Total (136) | |
| Gene /Allelic families | N(%) | |||
| Msp1 | ||||
| K1 | 22(49) | 14(28) | 25(61) | 61(44) |
| MAD20 | 6(13) | 10(20) | 2(5) | 18(13) |
| RO33 | 1(2) | 3(6) | 4(9) | 8(6) |
| K1+MAD20 | 4(9) | 9(18) | 2(5) | 15(11) |
| K1+ RO33 | 4(9) | 4(8) | 7(17) | 15(11) |
| MAD20+RO33 | 8(18) | 4(8) | 1(3) | 13(10) |
| K1+MAD20+RO33 | 0 | 6(12) | 0 | 6(5) |
| Total K1 | 30(66.66) | 33(66) | 34(82.9) | 97(71) |
| Total MAD20 | 18(40) | 29(59) | 5(12.1) | 52(38) |
| Total RO33 | 13(28.88) | 17(35) | 12(29.2) | 42(31) |
| Multiclonal isolates | 16(36) | 23(46) | 10(25) | 49(36) |
| Msp2 | ||||
| IC3D7 | 20(44) | 26(52) | 25(60) | 71(52) |
| FC27 | 11(24) | 7(14) | 5(13) | 23(17) |
| IC3D7 + FC27 | 14(32) | 17(34) | 11(27) | 42(31) |
| Total IC3D7 | 34(76) | 43(86) | 36(88) | 113(83) |
| Total FC27 | 25(56) | 24(48) | 16(40) | 65(48) |
| Multiclonal isolates | 14(32) | 17(34) | 11(27) | 42(31) |
n: number of isolates
For msp1 gene, K1, Ro33, and Mad20 allelic families were found with frequencies of 71% (97/136), 38% (52/136), 31% (42/136) respectively. In Samples with single allelic family, K1 was the most represented with 44% (61/136). K1+RO33 and K1+MAD20 allelic combination were higher among samples with two different allelic families with equal frequency of 11% (15/136). Samples with all three allelic families (K1+MAD20+RO33) represented 5% and were found only in Kedougou (p = 0.008). Multiclonal isolates found in all study sites were 36% (49/136) with a maximum in Kedougou (46%; 23/50) and minimum in Thies (25%; 10/41), however no statistical difference was found (p = 0.33).
Concerning msp2 gene, IC3D7 and FC27 allelic families were identified with frequencies of 83% (113/136) and 48% (65/136) of the isolates, respectively. The frequency of samples with only the IC3D7 allelic family was more predominant with 52% (71/136) than those harboring only the FC27 allelic family with 17% (23/136). The frequency of multiclonal isolates found in all localities was 31% (42136) with the highest value in Kedougou (34%, 17/50) and the lowest in Thies (27%, 11/41), however the difference was not statistically significant between localities (p=0.865).
Multiplicity of infection, heterozygosity and genetic differentiations
The multiplicity of infection (MOI) and heterozygosity (He) in different localities are shown in Table 3. The number of msp1 and msp2 genotypes per isolates ranged from 1 to 5. A higher value of MOI was found in Pikine 2.97. MOI found in all localities was 2.56 with statistically different between areas (p=0.024).
Table 3.
Multiplicity of infection and heterozygosity of P.falciparum populations
| Localities | |||
| Pikine | Kédougou | Thiès | |
| MOI msp1+msp2 | 2.97 | 2.51 | 2.21 |
| He msp1 | 0.632 | 0.637 | 0.495 |
| He msp2 | 0.509 | 0.457 | 0.394 |
MOI, multiplicity of infection; He, expected heterozygosity
For msp1 gene, heterozygosity (He) and genetic differentiation (Fst) observed in all study areas were 0.588 and 0.011 respectively. We found approximately the same value of He in Kedougou (0.637) and Pikine (0.632).
Regarding msp2 gene, He and Fst found in whole study sites were 0.453 and 0.017 respectively. A low He value was observed in Thies (0.394) while a high value was noticed in Pikine with 0.509.
Discussion
The present study aimed to provide a current overview on P. falciparum population structure in Senegal by analyzing the most polymorphic regions of msp1 and msp2 genes on isolates from areas presenting different malaria transmission patterns. This recent information on the molecular epidemiology of the most virulent plasmodial species in this country could inform malaria program of monitoring, control and to adapt if necessary the interventions to local malaria epidemiology settings.
In this study, K1 and IC3D7 allelic families were most predominant in all study sites. The strong presence of both allelic families has been already reported by studies carried out in West Africa8,15,35, Western Uganda36 and Iran37. Twenty-two alleles for msp1 gene and twenty alleles for msp2 gene were identified. Similar data were found in areas where malaria transmission is low13,38. However, a larger allele's numbers were observed in high malaria transmission areas in Kedougou, Senegal23 and in Burkina Faso14 suggesting that malaria endemicity affects the circulating strains numbers.
P. falciparum multiclonal isolates for msp1 and msp2 allelic families found in all study areas were relatively high with the higher in Kedougou. Similar findings were reported in hyperendemic areas13,14,33,39. The lower multiclonal isolates were found in Thies reflecting the low endemicity level of this study area which is agreement with other studies performed in hypoendemic areas16,40,41. These differences show that multiclonal isolates are more common in high malaria transmission area16.
MOI is an indicator of malaria transmission level because it has been to be higher in high malaria transmission areas and decreased when this latter decline16,42. Our results show moderate to high MOI values and a significant difference on MOIs between these localities (p=0.024). Therefore, our findings found in Thies and Kedougou are in phase with this hypothesis. In this last locality, Niang et al, found the same results in 201743. However, our most important MOI value was obtained in Pikine, an area with particular malaria epidemiological characteristics linked to floods during raining season (artificial water collection reservoirs) which increase opportunities for mosquito breeding growth2. These urban characteristics, reported by many studies performed in African cities44,45,46,47,48,49, could explain the high MOI found in this locality.
Also, low to moderate genetic diversity with heterozygosity were found in our study, ranging from 0.394 to 0.637. The lowest He value was found in Thies (He=0,394) and the highest He values were found in Kedougou (He=0.637) and Pikine (He=0.632). This variability between genetic diversity levels observed within country could be explained by local malaria epidemiology settings. Similar results were reported in Mali where the He values increase with malaria transmission gradient north to South8. The low genetic diversity of P. falciparum population found in Thies is consistent with clonal expansion reported previously in this area50 suggesting a high self-fertilization rate between genetically identical parasites during the sexual stages in mosquito. This low genetic diversity was also reported in countries where malaria decline, due to scale up of interventions9,51. However, moderate heterozygosity was found in Kedougou and Pikine indicating a reduction in clonal expansion and an increase of genetic diversity in these localities. Likewise, it suggest that genetic diversity of P. falciparum is greater in high malaria transmission areas and decreases when transmission regresses4,5,52.
Concerning Fst, we found a very low genetic differentiation (Fst msp1= 0.011; Fst msp2= 0.017) between P. falciparum population within country. Ahouidi et al,22 had already reported this little genetic divergence (Fst msp2 =0.012) in parasites populations in Senegal. These data reveal a gene flow between parasite populations facilitated by extensive human migration events between endemic regions and then causing the vector's displacement. Likewise, a lack of genetic differentiation was reported by others studies, on sites not far as ours studies areas, located approximately from 61 km to 685km8,22. Similar trends have been observed in Mali8 and elsewhere of the world such as in Latin American countries, Malaysia and South-East Asia where malaria transmission declines16,53,54,55,56. Overall, the low to moderate genetic diversity and little genetic differentiation in P. falciparum population found in Senegal shows that the effectiveness of the malaria controls scaling up since 20062. Nonetheless, disparities exist on malaria transmission levels in different areas.
The main limitation of this study was the use of msp genotyping which, as others marker based on DNA fragment size, could reduce the genetic diversity evaluation of the parasites strains. Nevertheless, msp1 and msp2 genes are robust polymorphism markers and can be used successfully to characterize genetic P. falciparum strains populations12,13,14,15.
Conclusion
This study gives current data on genetic diversity of P. falciparum population in Senegal. Low to moderate genetic diversity was found on parasite strains reflecting the decline of malaria transmission as well as interventions effectiveness. A high gene flow of parasites between localities shows rapid genes diffusion on P. falciparum population in this country. Our findings also confirm the disparities on malaria transmission level within Senegal. Thus, to achieve the malaria elimination objectives, Senegalese national malaria control program (NMCP) should readjust the malaria control strategies linked to the malaria epidemiological local patterns of sentinel sites like Pikine and Kedougou due to their endemicities.
Acknowledgements
We would like to express our gratitude to the study population, Younouss Diedhiou, Lamine Ndiaye, Amadou Mactar Mbaye, Nasserdine Papa Nze, Cyrille Diedhiou, Dior Diop and staff for their contribution in this study.
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.WHO, author. World malaria report 2016. World Health Organization; http://www.who.int/malaria/publications/world-malaria-report-2016/report/en/ [Google Scholar]
- 2.PNLP, author. Programme Nationale de Lutte contre le Paludisme (PNLP) Sénégal /Plan stratégique national. 2011–2015. http://www.africanchildforum.org/clr/policy/country/Senegal malaria_2011-2015.pdf.
- 3.PNLP, author. Bulletin épidémiologique annuel du paludisme au Senegal. Dakar: 2015. http://www.pnlp.sn/telechargements/Rapports/Bulletin-Epidemiologique-Annuel-2015-du-Paludisme-au-Senegal.pdf. [Google Scholar]
- 4.Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17(10):1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 5.Su X-Z, Jiang H, Yi M, Mu J, Stephens RM. Large-scale genotyping and genetic mapping in Plasmodium parasites. Korean J Parasitol. 2009;47:83–91. doi: 10.3347/kjp.2009.47.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnott A, Barry AE, Reeder JC. Understanding the population genetics of Plasmodium vivax is essential for malaria control and elimination. Malaria Journal. 2012;11:14. doi: 10.1186/1475-2875-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khaireh BA, Assefa A, Guessod HH, Basco LK, Khaireh MA, Pascual A, Briolant S, Bouh SM, Farah IH, Ali HM, Abdi AAA, Aden MO, Abdillahi Z, Ayeh SN, Darar HY, Koeck JL, Rogier C, Pradines B, Bogreau H. Population genetics analysis during the elimination process of Plasmodium falciparum in Djibouti. Malaria Journal. 2013;12:201. doi: 10.1186/1475-2875-12-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nabet C, Doumbo S, Jeddi F, Konaté S, Manciulli T, Fofana B, L'Ollivier C, Camara A, Moore S, Ranque S, Théra MA, Doumbo OK, Piarroux R. Genetic diversity of Plasmodium falciparum in human malaria cases in Mali. Malaria Journal. 2016;15:353. doi: 10.1186/s12936-016-1397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohd RMAR, Umi RS, Nor AN, Abass AK, Amirrudin M, Prem KM, Jenarun J, Christina R, Mallika I, Rose NM, Noor RA. Genetic Diversity of Plasmodium falciparum Populations in Malaria Declining Areas of Sabah, East Malaysia. Plos One. 2016;11:3. doi: 10.1371/journal.pone.0152415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viriyakosol S, Siripoon N, Petcharapirat C, Petcharapirat P, Jarra W, Thaithong S, Brown KN, Snounou G. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bulletin of the World Health Organization. 1995;73(1):85–95. [PMC free article] [PubMed] [Google Scholar]
- 11.Mayengue PI, Ndounga M, Davy MM, Tandou N, Ntoumi F. In vivo chloroquine resistance and prevalence of the pfcrt codon 76 mutation in Plasmodium falciparum isolates from the republic of Congo. Acta Tropica. 2007;95:219–225. doi: 10.1016/j.actatropica.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Mwingira F, Nkwengulila G, Schoepflin S, Sumari D, Beck HP, Snounou G, Felger I, Olliaro P, Mugittu K. Plasmodium falciparum msp1, msp2 and glurp allele frequency and diversity in sub-Saharan Africa. Malaria Journal. 2011;10:79. doi: 10.1186/1475-2875-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salem MSOA, Ndiaye M, OuldAbdallahi M, Lekweiry KM, Bogreau H, Konaté L, Faye B, Gaye O, Faye O, Boukhary AOMSO. Polymorphism of the merozoite surface protein-1 block 2 region in Plasmodium falciparum isolates from Mauritania. Malaria Journal. 2014;13:26. doi: 10.1186/1475-2875-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soulama I, Nébié I, Ouédraogo A, Gansane A, Diarra A, Tiono AB, Bougouma EC, Konaté AT, Kabré GB, Taylor WRJ, Sirima SB. Plasmodium falciparum genotypes diversity in symptomatic malaria of children living in an urban and a rural setting in Burkina Faso. Malaria Journal. 2009;8:135. doi: 10.1186/1475-2875-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yavo W, Konaté A, Mawili-Mboumba DP, Kassi FK, Tshibola Mbuyi ML, Angora EK, Hervé Menan EI, Bouyou-Akotet MK. Genetic Polymorphism of msp1 and msp2 in Plasmodium falciparum Isolates from Côte d'Ivoire versus Gabon. Journal of Parasitology Research. 2016 doi: 10.1155/2016/3074803. 7 p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atroosh WM, Al-Mekhlafi HM, Mahdy MAK, Saif-Ali R, Al-Mekhlafi AM, Surin J. Genetic diversity of Plasmodium falciparum isolates from Pahang, Malaysia based on msp1 and msp2 genes. Parasites & Vectors. 2011;4:233. doi: 10.1186/1756-3305-4-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ntoumi F, Contamin H, Rogier C, Bonnefoy S, Trape JF, Mercereau-Puijalon O. Age-dependent carriage of multiple Plasmodium falciparum merozoite surface antigen-2 alleles in asymptomatic malaria infections. Am J Trop Med Hyg. 1995:81–88. doi: 10.4269/ajtmh.1995.52.81. [DOI] [PubMed] [Google Scholar]
- 18.Peterson G, Coppel RL, Monoley MB, KEMP DJ. Third form of the precursor to the major merozoite surface antigens of Plasmodium falciparum. Mol Cell Biol. 1988;8:2644–2667. doi: 10.1128/mcb.8.6.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenton B, Clark JT, Wilson CF, Mcbride JS, Walliker D. Polymorphism of the 35 and 48 kilodalton Plasmodium falciparum mérozoïte surface antigen. Mol Biochem Parasitol. 1989;34:79–86. doi: 10.1016/0166-6851(89)90022-4. [DOI] [PubMed] [Google Scholar]
- 20.Apinjoh TO, Tata RB, Anchang-Kimbi JK, Chi HF, Fon EM, Mugri RN, Tangoh DA, Nyingchu RV, Ghogomu SM, Nkuo-Akenji T, Achidi EA. Plasmodium falciparum merozoite surface protein 1 block 2 gene polymorphism in field isolates along the slope of mount Cameroon: a cross — sectional study. BMC Infections Diseases. 2015;15:309. doi: 10.1186/s12879-015-1066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khaminsou N, Kritpetcharat O, Daduang J, Charerntanyarak L, Kritpetcharat P. Genetic analysis of the merozoite surface protein-1 blocks 2 allelic types in Plasmodium falciparum clinical isolates from Lao PDR. Malaria Journal. 2011;10:371. doi: 10.1186/1475-2875-10-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahouidi AD, Bei AK, Neafsey DE, Sarr O, Volkman S, Milner D, Cox-Singh J, Ferreira MU, Ndir O, Premji Z, Mboup S, Duraisingh MT. Population genetic analysis of large sequence polymorphisms in Plasmodium falciparum blood-stage antigens. Infection, Genetics and Evolution. 2010;10:200–206. doi: 10.1016/j.meegid.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konate L, Zwetyenga J, Rogier C, Bischoff E, Fontenille D, Tall A, Spiegelz A, Trape JF, Mercereau-Puijalonl O. Variation of Plasmodium falciparum msp1 block 2 and msp2 allele prevalence and of infection complexity in two neighbouring Senegalese villages with different transmission conditions. Trans R Soc Trop Med Hyg. 1999;93:21–28. doi: 10.1016/s0035-9203(99)90323-1. [DOI] [PubMed] [Google Scholar]
- 24.Beck HP, Felger I, Vounatsou P, Hirt R, Tanner M, Alonso P, Menendez C. Effect of iron supplementation and malaria prophylaxis in infants on Plasmodium falciparum genotypes and multiplicity of infection. Trans R Soc Trop Med Hyg. 1999;93:41–45. doi: 10.1016/s0035-9203(99)90326-7. [DOI] [PubMed] [Google Scholar]
- 25.Kolakovich KA, Ssengoba A, Wojcik K, Tsuboi T, al-Yaman F, Alpers M, Adams JH. Plasmodium vivax: favored gene frequencies of the merozoite surface protein-1 and the multiplicity of infection in a malaria endemic region. Exp Parasitol. 1996;83:11–19. doi: 10.1006/expr.1996.0044. [DOI] [PubMed] [Google Scholar]
- 26.Mbugi EV, Mbugi EV, Mutayoba BM, Balthazary ST, Malisa AL, Nyambo TB, Mshinda H. Multiplicity of infections and level of recrudescence in Plasmodium falciparum malaria in Mlimba, Tanzania. Afr J Biotech. 2006;5:1655–1662. doi: 10.1186/1475-2875-5-94. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnot D. Unstable malaria in Sudan: the influence of the dry season. Clone multiplicity of Plasmodium falciparum infections in individuals exposed to variable levels of disease transmission. Trans R Soc Trop Med Hyg. 1998;92:580–585. doi: 10.1016/s0035-9203(98)90773-8. [DOI] [PubMed] [Google Scholar]
- 28.Schleiermacher D, Rogier C, Spiegel A, Tall A, Trape JF, Mercereau Puijalon O. Increased multiplicity of Plasmodium falciparum infections and skewed distribution of individual msp1 and msp2 alleles during pregnancy in Ndiop, a Senegalese village with seasonal, mesoendemic malaria. Am J Trop Med Hyg. 2001;64:303–309. doi: 10.4269/ajtmh.2001.64.303. [DOI] [PubMed] [Google Scholar]
- 29.Tusting L S, Bousema T, Smith D L, Drakeley C. Measuring changes in Plasmodium falciparum transmission: Precision, accuracy and costs of metrics. Adv Parasitol. 2014;84:151–208. doi: 10.1016/B978-0-12-800099-1.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO, author. Communicable diseases cluster: severe falciparum malaria. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2000;94:0S1–0S9. [PubMed] [Google Scholar]
- 31.Papa Mze N, Ndiaye YD, Diedhiou CK, Rahamatou S, Dieye B, Daniels RF, Hamilton EJ, Diallo M, Bei AK, Wirth DF, Mboup S, Volkman SK, Ahouidi AD, Ndiaye D. RDTs as a source of DNA to study Plasmodium falciparum drug resistance in isolates from Senegal and the Comoros Islands. Malaria Journal. 2015;14:373. doi: 10.1186/s12936-015-0861-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snounou G, Zhu X, Spiripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variant in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 33.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drakeley CJ, Duraisingh MT, Povoa M, Conway DJ, Targett GA, Baker DA. Geographical distribution of a variant epitope of Pfs48/45, a Plasmodium falciparum transmission-blocking vaccine candidate. Mol Biochem Parasitol. 1996;81(2):253–257. doi: 10.1016/0166-6851(96)02718-1. [DOI] [PubMed] [Google Scholar]
- 35.Henry M, Diallo I, Bordes J, Ka S, Pradines B, Diatta B, Mbaye PS, Sané M, Thiam M, Gueye PM, Wade B, Touze JE, Debonne JM, Rogier C, Fusai T. Urban malaria in Dakar, Senegal: chemosusceptibility and genetic diversity of Plasmodium falciparum isolates. Am J Trop Med Hyg. 2006;75:146–151. [PubMed] [Google Scholar]
- 36.Peyerl-Hoffmann G, Jelinek T, Kilian A, Kabagambe G, Metzger WG, Von Sonnenburg F. Genetic diversity of Plasmodium falciparum and its relationship to parasite density in an area with different malaria endemicities in West Ouganda. Trop Med Int Health. 2001;6(8):607–613. doi: 10.1046/j.1365-3156.2001.00761.x. [DOI] [PubMed] [Google Scholar]
- 37.Heidari A, Keshavarz H, B Rokni M, Jelinek T. Genetic diversity in merozoite surface protein msp1 and msp2 genes of Plasmodium falciparum in a major endemic region of Iran. Korean Journal of Parasitology. 2007;45(1):59–63. doi: 10.3347/kjp.2007.45.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auboy A, Migot-Nabias F, Deloron P. Polymorphism in two Merozoite surface protein of Plasmodium falciparum isolates from Gabon. Malaria Journal. 2003;2:12. doi: 10.1186/1475-2875-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wanji S, Kengne-Ouafo AJ, Eyong EEJ, Kimbi HK, Tendongfor N, Ndamukong-Nyanga JL, Nana-Djeunga HC, Bourguinat C, Sofeu-Feugaing DD, Charvet CL. Genetic diversity of plasmodium falciparum merozoite surface protein-1 block 2 in sites of contrasting altitudes and malaria endemicities in the mount Cameroon region. Am J Trop Med Hyg. 2012;86(5):764–774. doi: 10.4269/ajtmh.2012.11-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Druilhe P, Daubersies P, Patarapotikul J, Gentil C, Chene L, Chongsuphajaisiddhi T, Mellouk S, Langsley G. A primary malaria infection is composed of Averyy wide range of genetically diverse but related parasites. Am J Clin Invest. 1998;101:2008–2020. doi: 10.1172/JCI119890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Issifou S, Djikou S, Sanni A, Lekoulo F, Ntoumi F. Pas influence de la saison de transmission ni de l'âge des patients sur la complexité et la diversité génétique des infections dues à Plasmodium falciparum à Cotonou (Bénin) Bull Soc Pathol Exot. 2001;94(2):195–198. [PubMed] [Google Scholar]
- 42.Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F. Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malaria Journal. 2008;7:17. doi: 10.1186/1475-2875-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niang M, Thiam LG, Loucoubar C, Sow A, Sadio BD, Diallo M, Sall AA, Toure-Balde A. Spatio-temporal analysis of the genetic diversity and complexity of Plasmodium falciparum infections in Kedougou, southeastern Senegal. Parasites & Vectors. 2017;10:33. doi: 10.1186/s13071-017-1976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castro MC, Kanamori S, Kannady K, Mkude S, Killeen GF, Fillinger U. The importance of drains for the larval development of lymphatic filariasis and malaria vectors in dares salaam, United Republic of Tanzania. PLoS Neglected Tropical Diseases. 2010;4(5):693. doi: 10.1371/journal.pntd.0000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ceesay SJ, Bojang KA, Nwakanma D, Conway DJ, Koita OA, Doumbia SO, Ndiaye D, Coulibaly TF, Diakité M, Traoré SF, Coulibaly M, Ndiaye JL, Sarr O, Gaye O, Konaté L, Sy N, Faye B, Faye O, Sogoba N, Jawara M, Dao A, Poudiougou B, Diawara S, Okebe J, Sangaré L, Abubakar I, Sissako A, Diarra A, Kéita M, Kandeh B, Long CA, Fairhurst RM, Duraisingh M, Perry R, Muskavitch MA, Valim C, Volkman SK, Wirth DF, Krogstad DJ. Sahel, savana, riverine and urban malaria in West Africa: similar control policies with different outcomes. Acta Tropica. 2012;121(3):166–174. doi: 10.1016/j.actatropica.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaki PP, Govella NJ, Shoo B, Hemed A, Tanner M, Fillinger U, Killeen GF. Achieving high coverage of larval-stage mosquito surveillance: challenges for a community-based mosquito control programme in urban Dares Salaam, Tanzania. Malaria Journal. 2009;8:311. doi: 10.1186/1475-2875-8-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Silva PM, Marshall JM. Factors Contributing to Urban Malaria Transmission in sub-Saharan Africa: A Systematic Review. Journal of Tropical Medicine. 2012 doi: 10.1155/2012/819563. 10p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khaemba BM, Mutani A, Bett MK. Studies of anopheline mosquitoes transmitting malaria in a newly developed highland urban area: a case study of Moi University and its environs. East Afr Med J. 1994;71:159–164. [PubMed] [Google Scholar]
- 49.Wilson DW, Goodman CD, Sleebs BE, Weiss GE, Jong NWM, Angrisano F, Langer C, Baum J, Crabb BS, Gilson PR, McFadden GI, Beeson JG. Macrolides rapidly inhibit red blood cell invasion by the human malaria parasite, Plasmodium falciparum. BMC Biology. 2015;13:52. doi: 10.1186/s12915-015-0162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniels RF, Chang HH, Séne PD, Park DC, Neafsey DE, Schaffner SF, Hamilton EJ, Lukens AK, Tyne DV, Mboup S, Sabeti PC, Ndiaye D, Wirth DF, Hartl DL, Volkman SK. Genetic surveillance detects both clonal and epidemic transmission of malaria following enhanced intervention in Senegal. PLoS One. 2013;8:60780. doi: 10.1371/journal.pone.0060780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escalante AA, Ferreira MU, Vinetz JM, Volkman SK, Cui L, Gamboa D, Krogstad DJ, Barry AE, Carlton JM, Eijk AM, Pradhan K, Mueller I, Greenhouse B, Pacheco MA, Vallejo AF, Herrera S, Felger I. Malaria molecular epidemiology: lessons from the International Centers of Excellence for Malaria Research Network. Am J Trop Med Hyg. 2015;93(3):79–86. doi: 10.4269/ajtmh.15-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conway DJ, Roper C, Oduola AM, Arnot DE, Kremsner PG, Grobusch MP, Curtis CF, Greenwood BM. High recombination rate in natural populations of Plasmodium falciparum. Proc Natl Acad Sci U S A. 1999;96:4506–4511. doi: 10.1073/pnas.96.8.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chenet SM, Schneider KA, Villegas L, Escalante AA. Local population structure of Plasmodium: impact on malaria control and elimination. Malaria Journal. 2012;11:412. doi: 10.1186/1475-2875-11-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larrañaga N, Mejía RE, Hormaza JI, Montoya A, Soto A, Fontecha GA. Genetic structure of Plasmodium falciparum populations across the Honduras-Nicaragua border. Malaria Journal. 2013;12:354. doi: 10.1186/1475-2875-12-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nkhoma SC, Nair S, Al-Saai S, Ashley E, McGready R, Phyo AP, Oisnosten F, Anderson TJ. Population genetic correlates of declining transmission in a human pathogen. Mol Ecol. 2013;22:273–285. doi: 10.1111/mec.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pumpaibool T, Arnathau C, Durand P, Kanchanakhan N, Siripoon N, Suegorn A, Sitthiamorn C, Renaud F, Harnyuttanakorn P. Genetic diversity anpopulation structure of Plasmodium falciparum in Thailand, a low transmission country. Malaria Journal. 2009;8:155. doi: 10.1186/1475-2875-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]