Abstract
Administration of the microtubule inhibitor docetaxel is a common treatment for metastatic castration-resistant prostate cancer (mCRPC) and results in prolonged patient overall survival. Usually, after a short period of time chemotherapy resistance emerges and there is urgent need to find new therapeutic targets to overcome therapy resistance. The lysine-acetyltransferase p300 has been correlated to prostate cancer (PCa) progression. Here, we aimed to clarify a possible function of p300 in chemotherapy resistance and verify p300 as a target in chemoresistant PCa. Immunohistochemistry staining of tissue samples revealed significantly higher p300 protein expression in patients who received docetaxel as a neoadjuvant therapy compared to control patients. Elevated p300 expression was confirmed by analysis of publicly available patient data, where significantly higher p300 mRNA expression was found in tissue of mCRPC tumors of docetaxel-treated patients. Consistently, docetaxel-resistant PCa cells showed increased p300 protein expression compared to docetaxel-sensitive counterparts. Docetaxel treatment of PCa cells for 72 h resulted in elevated p300 expression. shRNA-mediated p300 knockdown did not alter colony formation efficiency in docetaxel-sensitive cells, but significantly reduced clonogenic potential of docetaxel-resistant cells. Downregulation of p300 in docetaxel-resistant cells also impaired cell migration and invasion. Taken together, we showed that p300 is upregulated by docetaxel, and our findings suggest that p300 is a possible co-target in treatment of chemoresistant PCa.
Keywords: prostate cancer, chemotherapy resistance, p300, docetaxel, colony formation ability
Introduction
Although therapy of organ-confined prostate cancer (PCa) by radical prostatectomy or radiotherapy is curative, treatment of advanced PCa is considered merely palliative. Androgen deprivation therapy (ADT) remains the gold standard for patients with prostate-specific antigen (PSA) progression. However, tumor progression is inevitable and leads to the development of castration-resistant prostate cancer (CRPC). Treatment options for non-metastatic and metastatic CRPC (mCRPC) include inhibitors of androgen synthesis and antiandrogens such as enzalutamide and apalutamide as well as the chemotherapeutic compound docetaxel (Taxotere®).
In 2004, the landmark study TAX327 showed a significant survival benefit of 2.4 months for docetaxel treatment compared to mitoxantrone, which was the first study to demonstrate a survival benefit for chemotherapy in CRPC patients (Tannock et al. 2004). Docetaxel treatment resulted in PSA decline, prolonged overall survival (OS), and improved quality of life. Furthermore, the STAMPEDE and CHAARTED trials have utilized docetaxel together with ADT into first-line treatment for hormone sensitive PCa (HSPC) with a survival benefit of 13.4 months compared to ADT alone (James et al. 2016, Kyriakopoulos et al. 2018). Additionally, docetaxel treatment has no negative consequences for subsequent endocrine therapies. Both abiraterone acetate and enzalutamide are used as effective second-line therapies after resistance to docetaxel has evolved (Lavaud et al. 2018). Despite development of novel therapies, treatment options for mCRPC patients are still limited, and there is an urgent need to find new therapeutic targets to overcome chemotherapy resistance.
Transcriptional co-regulators of the AR are involved in therapy resistance with several of them increasingly expressed during ADT (Comuzzi et al. 2004, Heemers et al. 2007, Qin et al. 2014). Two of these well-known coactivators are the histone acetyltransferases p300 and CBP (CREB binding protein) that show elevated expression in advanced PCa and have oncogenic potential (Debes et al. 2003, Comuzzi et al. 2004). While these coactivators show high levels of homology, they play distinctive roles in PCa and other diseases (Kalkhoven 2004, Ramos et al. 2010). Common features of p300 and CBP include regulation of transcription via remodeling chromatin structure by acetylating conserved lysine amino acids of histone proteins. They are also capable of recruiting the basal transcription machinery to gene promoters and acting as adaptor molecules (Chan & La Thangue 2001). In a previous study, it was demonstrated that p300 might be a valid target in PCa cells, as downregulation of p300 induced apoptosis and decreased cell migration in androgen-dependent and CRPC cells (Santer et al. 2011). Based on those previous results, this study aimed to investigate whether p300 is a possible new target in the context of chemotherapy resistance. Therefore, we analyzed (1) p300 expression in patients that received docetaxel, (2) p300 expression in docetaxel-resistant (DR) cells compared to docetaxel-sensitive counterparts, and (3) the effects of short-term docetaxel treatment on p300 expression. To study the functional role of p300, an RNA-interference (RNAi) approach was used and doxycycline-inducible p300 knockdown cell lines were generated. In addition, effects of p300 downregulation on colony formation efficiency, cell migration, and invasion in docetaxel-resistant cells were determined.
Materials and methods
Cell culture and chemicals
Human PCa cell lines PC3 and DU145 were purchased from the American Type Culture Collection (ATCC, LGC Standards, Wesel, Germany). Docetaxel-resistant PC3-DR and DU145-DR were previously established by Puhr et al. (2012). CWR22RV1 and CWR22RV1-DR cells were a kind gift of Prof Dr William Watson (University College Dublin). All cell lines were cultured in RPMI 1640 (PAN Biotech, Aidenbach, Germany) supplemented with 10% (v/v) fetal bovine serum (PAN Biotech, Aidenbach, Germany), 1% (v/v) penicillin/streptomycin, and 1% (v/v) GlutaMAX (both from Lonza, Vienna, Austria). Docetaxel-resistant cell lines were cultured in the presence of 12.5 nmol/L docetaxel (Sigma Aldrich). HEK293FT cells were obtained from Life Technologies and grown according to the manufacturer’s instructions. The authenticity of all cell lines was validated via short tandem repeat (STR) profiling.
Immunohistochemistry (IHC)
For IHC staining, a tissue microarray (TMA) of 14 patients that received neoadjuvant docetaxel therapy before radical prostatectomy and 14 patients with no chemotherapy was used. The use of patient material was approved by the Ethics Committee of the Medical University of Innsbruck (study No AM 3174 including amendment 2). For detailed information about clinical data from patients, see publication of Puhr et al. (2012). IHC staining was performed on a Discovery-XT staining device (Ventana) and the following specific antibody was used: anti-p300 (1:100, D8Z4E, Cell Signaling Technology). Antibody specificity was verified by Western blot and IHC staining of PC3-DR cells with p300 downregulation. For IHC, cells were embedded by coagulation in plasma clots after harvesting, transferred into a biopsy histosette, fixed in formalin, and embedded in paraffin. Importantly, cross staining of CBP was excluded by Western blot analysis.
Transcriptome analysis of patient data
The publicly available transcriptome dataset GSE77930 (Kumar et al. 2016) was downloaded from the GEO database and analyzed with the Qlucore Omics Explorer v3.5. Gene set activity scores for the Hallmark ‘Androgen response’ and ‘Myc targets’ gene sets (Molecular Signatures Database, MSigDB) were calculated in R with the GSVA package (Hanzelmann et al. 2013).
Western blot
Cells were lysed in LDS sample buffer and 50 µg total protein was separated either on 3–8% Tris-Acetate gels (Thermo Fisher Scientific) for analysis of p300 and CBP expression or 4–12% Bis-Tris gels (Expedeon, San Diego, CA, USA) for all other proteins and transferred onto methanol-activated PVDF membranes or 0.2 µm nitrocellulose membranes (GE Healthcare). Blocking of membranes and antibody incubation were performed in 5% BSA in TBS-T. The following antibodies were used: anti-p300 (1:4000, ab10485, Abcam), anti-CBP (1:1000, Cell Signaling Technology), anti-c-Myc (1:1000, Cell Signaling Technology), anti-Histone H3 (1:1000, Cell Signaling Technology), anti-Acetyl-Histone H3 (Lys18, 1:1000, Cell Signaling Technology), anti-Vimentin (1:500, Santa Cruz Biotechnology), anti-Vinculin (1:500, Santa Cruz Biotechnology), anti-Lamin A (1:2000, Abcam), anti-α-tubulin (1:500, Santa Cruz Biotechnology), and anti-GAPDH (1:50000, Merck Millipore). House-keeping controls were selected in a cell line-specific manner on the basis of data showing no change in their expression in that cell line.
RNA isolation and quantitative real-time PCR
Total RNA was isolated using the EXTRACTME TOTAL RNA KIT (LabConsulting, Vienna, Austria) according to the manufacturer’s manual. cDNA synthesis was performed with the iScript Select cDNA Synthesis Kit (Bio-Rad). For real-time PCR a Luna Script RT Super Mix Kit (New England Biolabs, Ipswich, MA, USA) was used. HPRT1 (Fwd: GCTTTCCTTGGTCAGGCAGTA, Rev: GTCTGGCTTATATCCAACACTTCGT, Probe: CAAGGTCGCAAGCTTGCTGGTGAAAAGGA), TATA-Box binding protein (TBP; Fwd: CACGAACCACGGCACTGATT, Rev: TTTTCTTGCTGCCAGTCTGGAC, Probe: TCTTCACTCTTGGCTCCTGTGCACA), and HMBS (TaqMan Gene Expression Assay from Thermo Fisher Scientific; Hs00609297_m1) were used as reference genes. The following Taqman gene expression assays were used: p300 (Hs00914223_m1), CBP (Hs00932878_m1), c-Myc (Hs00153408_m1), and Vimentin (Hs00185584_m1).
Generation of doxycycline-inducible knockdown cell lines
Stable cell lines with inducible p300 knockdown were generated by lentiviral-based transduction of shRNA vectors using the BLOCK-iT HiPerform Lentiviral Pol-II miR RNAi Expression System with emGFP from Invitrogen. Briefly, miR Select oligos (Hmi405238: shp300-1; Hmi405239: shp300-2) were purchased from Life Technologies and ligated into pcDNA 6.2-GW/EmGFP-miR expression vector according to the manufacturer’s protocol. Then the shRNAs were shuttled into the pDONR221 vector to generate entry clones. Entry plasmids together with a pENTR-tetOn (from pHR-TetCMV-eGFP-dest ligated into pENTR 5′/CMVp vector) were then used in MultiSite Gateway reactions with pLenti6.4/R4R2/V5-DEST to generate doxycycline-inducible shp300-1 and shp300-2. To generate cell lines stably expressing the doxycycline activator/repressor cassette, HEK293FT cells were first co-transfected with the packaging vectors pVSV-G and psPAX2 together with pHR-SFFV-rtTAM2-T2A-Puro using X-tremeGENE reagent (Roche) according to the manufacturer’s manual. Supernatants containing virus particles were collected 48 h after transfection, filtered through a 0.45 µm membrane filter, and used for infection of target cells. Cells were selected with 2 µg/mL puromycin and subsequently transfected with packaging vectors together with the pLenti6.4 expression vector as described previously. Selection of infected cells was performed using 2 µg/mL blasticidine. For activation of the system, 100 ng/mL doxycycline was used. Activation status of the system was verified by GFP expression.
Proliferation assay
Cumulative population doubling levels (PDL) were determined by continuously seeding a defined number of cells in T25 flasks. Cell numbers were determined by CASY cell counter (Schärfe System, Reutlingen, Germany) every 3–4 days. Cumulative PDL was calculated with the following formula: PDL = 3.32 × (log10(X) − log10(Y)) + I, where X = number of cells at the end of growth period, Y = number of cells at the beginning of growth period, and I = initial population doubling level. To compare the growth curves, linear fit regression with the test for significance of slopes and intercepts was performed in GraphPad Prism 8.
High-resolution colony formation assay
Cell numbers were determined using CASY cell counter system (Schärfe System). Per 75 cm2 culture flask, 1000 cells were seeded and incubated for 10–14 days in the absence or presence of 100 ng/mL doxycycline. Cells were fixed with 100% ice-cold methanol for 5 min and stained with crystal violet (0.5% dissolved in PBS containing 20% methanol (Sigma)) for 5 min. Colony formation efficiency was determined by the software CATCH-colonies (https://catch-colonies.net). To correct for differences in the seeding density of the different stable shRNA cell lines, the colony formation efficiency was normalized to flasks that were seeded at the same time but not treated with doxycycline.
Wound scratch assay
Cells were seeded until they were nearly confluent in multi-well plates and treated with 100 ng/mL doxycycline or 10 µM CPI-637 (MedChem Express, Monmouth Junction, NJ, USA) for 96 h in total. After the first 24 h, 10 µM of the proliferation inhibitor cytosine β-D-arabinofuranoside was added. After another 24 h, a scratch was made using a 10-µL pipette tip. Images were taken after another 48 h and analyzed using the MRI Wound Healing Tool of ImageJ.
Migration and invasion assay
For migration and invasion assays, Boyden chamber inserts with 8 µm pore size (Fluoroblok System, Becton Dickinson) were used. 3 × 104 cells per well of PC3-DR shCtrl, PC3-DR shp300-1, and PC3-DR shp300-2 were seeded in duplicate in serum-free medium. Bottom chambers were filled with medium containing 10% FCS serving as chemoattractant. For invasion assays, inserts were coated with 30 µl of matrigel (diluted 1:3 in serum-free medium; Corning). Cells were incubated for 48–72 h in the absence or presence of 100 ng/mL doxycycline or 20 µM CPI-637, respectively. Afterwards, cells were stained with 2 µM of calcein AM (Sigma) diluted in HBSS containing 1% FCS. Fluorescent images were taken using a JuLI smart fluorescent cell analyzer (Science Services, Munich, Germany), and extinction/emission at 494/517 was measured using TECAN plate reader (Tecan Group Ltd., Männedorf, Switzerland).
Immunofluorescence
Cells were seeded on glass coverslips in the absence or presence of 100 ng/mL doxycycline for 72 h. Cells were fixed with 4% paraformaldehyde and permeabilized with 1% BSA in PBS containing 0.2% Triton X-100. Washing steps were performed with 1% BSA in PBS. Anti-Vimentin (1:500, Santa Cruz Biotechnology) was used as a primary antibody, together with the fluorescence-labeled secondary antibody goat anti-mouse 555 (ThermoFisher). Coverslips were mounted with Vectashield mounting medium containing DAPI.
Viability assays
PC3-DR cells were seeded in multi-well plates and treated with a range of CPI-637 concentrations. The solvent DMSO served as a control. The viability was measured using RealTime-Glo™ MT Cell Viability Assay (Promega) after 72 h and the IC50 was calculated using a nonlinear fit model with variable slope of log transformed data in GraphPad Prism 8.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software Inc.). Gaussian distribution of patient samples was determined using Kolmogorov–Smirnov test. Comparison of the two groups was performed using Student’s t-test and Mann–Whitney U test, depending on Gaussian distribution. Comparison of multiple groups was performed using one-way ANOVA and correcting for multiple testing using Bonferroni multiple comparison test. For integration of multiple independent replicates from methods that yield relative abundance values (qPCR, Western blot), each replicate was normalized to its average signal intensity. P-values of <0.05 were considered statistically significant and encoded in figure legends as follows: *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001. If not stated otherwise, doxycycline-treated cells were normalized to non-treated control cells, and doxycycline-treated cells are shown in graphs.
Results
p300 expression is increased in docetaxel-treated patients
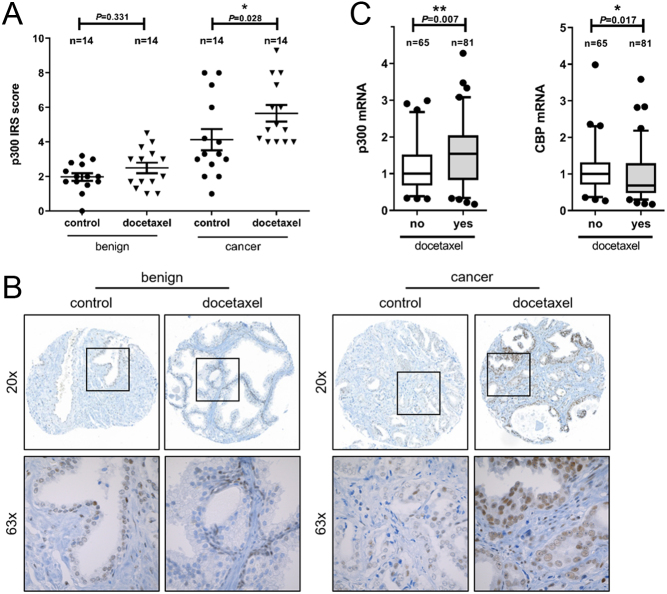
To evaluate whether p300 expression is altered upon docetaxel treatment, we analyzed tissue material of patients who received docetaxel by IHC staining using a specific p300 antibody (Supplementary Fig. 1A, B and C, see section on supplementary materials given at the end of this article). In total, 28 patients were included, 14 of them received neoadjuvant docetaxel therapy before radical prostatectomy (RPE), while the other 14 patients did not receive chemotherapy prior to RPE. For detailed information about selected patients, please see the work of Puhr et al. (2012), where clinical data are fully described. We observed a significantly increased p300 expression in cancerous areas of docetaxel-treated patients compared to control patients (Fig. 1A and B), which was not the case in benign areas. Furthermore, p300 staining was observed mainly in nuclear areas.
Figure 1.
Expression of p300 is increased upon docetaxel treatment. (A) Quantification of p300 immunoreactivity scores (IRS) after IHC staining (Mann–Whitney U test; scatter dot blot with line at mean + s.e.m.). (B) IHC staining of p300 (nuclear localization). Upregulation of p300 expression in cancerous tissue of docetaxel-treated patients compared to control patients. (C) p300 and CBP mRNA expression were determined in samples of docetaxel-treated and docetaxel-untreated mCRPC patients (Mann–Whitney U test; box whisker plot with 5–95 percentile).
Elevated p300 expression was further confirmed in publicly available datasets of mCRPC tissue samples from patients that suffered from relapse after docetaxel treatment (Kumar et al. 2016). Our analysis revealed significantly increased p300 levels (1.5-fold) in patients treated with docetaxel compared to patients that did not receive docetaxel at any time in their treatment course (Fig. 1C). To assess if docetaxel-mediated increase is specific for p300, we included the analysis of the closely related coactivator CBP. Interestingly, CBP expression levels were found slightly decreased in docetaxel-treated patients compared to non-docetaxel-treated patients (Fig. 1C). Furthermore, we analyzed AR expression and activity upon docetaxel treatment to evaluate whether the AR has any impact on docetaxel-mediated upregulation of p300. Of note, AR mRNA expression and androgen response were not significantly changed in patients that relapsed after docetaxel treatment compared to control patients (Supplementary Fig. 2A and B).
Docetaxel-resistant and docetaxel-treated PCa cells show increased p300 expression
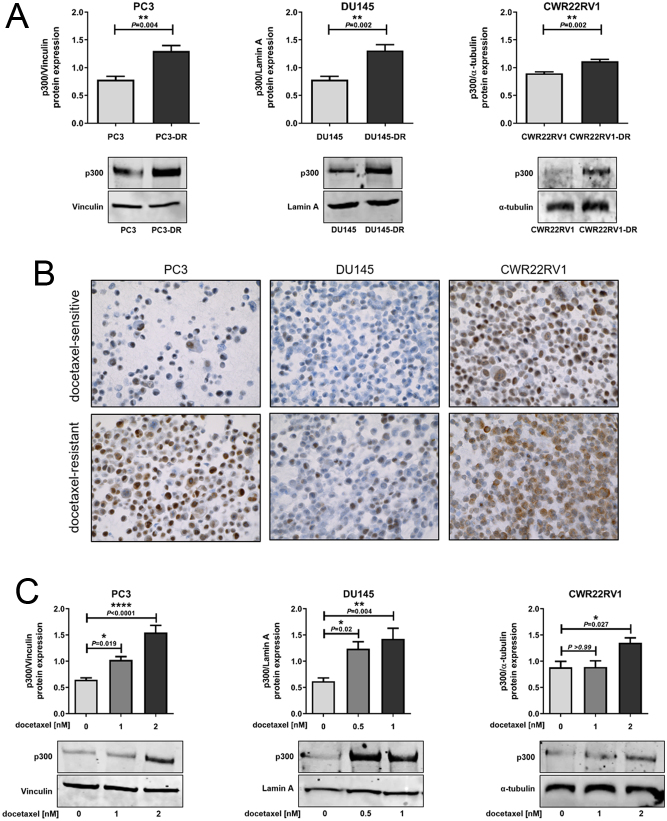
Next, we compared p300 expression in docetaxel-resistant (DR) derivatives of commonly employed PCa cell lines (PC3-DR, DU145-DR, and CWR22RV1-DR) relative to their docetaxel-sensitive counterparts. p300 protein levels were significantly increased (1.5-fold) in all three docetaxel-resistant cell lines tested compared to docetaxel-sensitive counterparts (Fig. 2A), whereas mRNA levels were unchanged (Supplementary Fig. 3A). Increased p300 protein expression in docetaxel-resistant PC3-DR and DU145-DR was confirmed by IHC staining (Fig. 2B). CBP mRNA and protein expression were not significantly regulated in docetaxel-resistant cells compared to sensitive counterparts (except downregulated CBP mRNA expression in PC3-DR cells, Supplementary Fig. 3B and C).
Figure 2.
p300 expression is increased in docetaxel-resistant prostate cancer cells. (A) Comparison of p300 protein expression between docetaxel-sensitive and docetaxel-resistant (DR) PC3 (n = 4), DU145 (n = 5), and CWR22RV1 (n = 4). Data represent mean + s.e.m. (t-test). (B) IHC staining for p300 in docetaxel-resistant PC3-DR, DU145-DR, and CWR22RV1-DR compared to docetaxel-sensitive counterparts. Magnification 40×. (C) PC3 (n = 5), DU145 (n = 5), and CWR22RV1 (n = 4) were treated with the indicated concentrations of docetaxel for 72 h, and p300 protein expression was analyzed by Western blot, and one representative Western blot is shown. Values represent mean + s.e.m. (one-way ANOVA).
To understand the molecular basis of these findings, we analyzed p300 expression upon short-term docetaxel treatment in several PCa cell lines. To this end, we measured p300 mRNA and protein expression in PC3, DU145, and CWR22RV1 cells after treatment with docetaxel for 72 h. Protein expression of p300 significantly increased (2–2.5-fold increase) in all three cell lines upon docetaxel treatment (Fig. 2C), whereas mRNA levels were not significantly changed (Supplementary Fig. 4A). Time-course experiments revealed that p300 protein, but not mRNA expression, increased within 8 h of docetaxel treatment and reached a plateau after 16 h (Supplementary Fig. 4B and C). Interestingly, p300 expression in docetaxel-resistant cells decreased if the cells were cultured without docetaxel (Supplementary Fig. 4D and E). To investigate whether docetaxel influences the protein degradation rate of p300, translation was blocked with cycloheximide (CHX). Of note, p300 had a protein half-life time of around 6 h in untreated PC3 cells, whereas addition of docetaxel (PC3 and PC3-DR) stabilized the protein (Supplementary Fig. 4F).
We also analyzed the expression levels of the well-described p300 downstream target c-Myc, which is a known oncogene in PCa (Koh et al. 2010). Our analysis revealed no significant change of c-Myc mRNA levels (Supplementary Fig. 5A), but a Myc gene expression activity score was significantly increased in docetaxel-treated patients (Supplementary Fig. 5B). Additionally, docetaxel-resistant PC3-DR and DU145-DR showed higher c-Myc protein expression compared to docetaxel-sensitive counterparts (Supplementary Fig. 5C), and c-Myc protein expression was increased (1.5-fold) in DU145 upon treatment with 1 nM docetaxel compared to the control (Supplementary Fig. 5D).
Docetaxel-resistant cells show reduced colony formation ability upon p300 inhibition
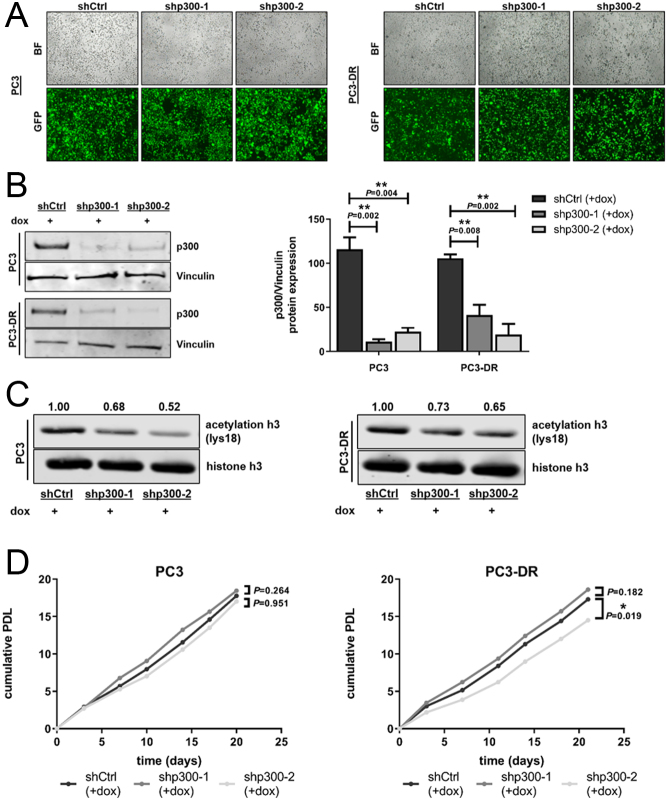
To study a possible mechanistic role of p300 in docetaxel resistance, PC3, CWR22RV1, and their docetaxel-resistant counterparts were stably transduced with doxycycline-inducible short hairpin (sh)p300 vectors. We selected these cell lines to include both AR-negative (PC3 and PC3-DR) as well as AR-positive (CWR22RV1 and CWR22RV1-DR) sublines. Doxycycline treatment activated the inducible system as indicated by homogeneous expression of vector-integrated GFP (Fig. 3A), which led to decreased p300 protein expression (Fig. 3B) and activity by decreased acetylation of histone h3 on lysine 18 (Fig. 3C). Of note, p300 knockdown had no biologically consistent effect on the proliferation rate of docetaxel-sensitive or docetaxel–resistant cells over a period of 20 days (Fig. 3D). The significantly reduced proliferation rate of PC3-DR shp300-2 was not reproducible with the shp300-1 construct and is, thus, likely an artifact or off-target effect. To exclude the possibility that CBP is upregulated by p300 inhibition, thus compensating for the effects of p300 downregulation, we analyzed CBP expression upon p300 knockdown. As expected, specific downregulation of p300 had no impact on CBP expression (Supplementary Fig. 6).
Figure 3.
Establishment and validation of doxycycline-inducible p300 knockdown cell lines using a non-targeting control (shCtrl) sequence and two specific p300-shRNA sequences (shp300-1 and shp300-2). Docetaxel-sensitive PC3 and docetaxel-resistant PC3-DR are shown here, representing all mentioned cell lines. For better visualization in the following experiments, only doxycycline-treated shCtrl, shp300-1, and shp300-2 replicates are shown. Confirmation of (A) uniform expression of shRNA-constructs by fluorescence microscopy (BF = bright field, GFP = green fluorescent protein) and magnification 40× and (B) p300 knockdown by Western blot analysis after activation of shp300-sequences with 100 ng/mL doxycycline for 72 h. Values indicated are mean + s.e.m. (one-way ANOVA, n = 3). (C) Decreased p300 activity was confirmed by analysis of histone h3 acetylation on lysine18 in PC3 and PC3-DR cells. One representative Western blot out of three independent experiments is shown. (D) Cumulative population doubling levels (PDL, n = 1) of PC3 and PC3-DR cells over time with downregulated p300 were calculated by cell number measurement upon each passage (linear fit regression and test for significance of slopes and intercepts).
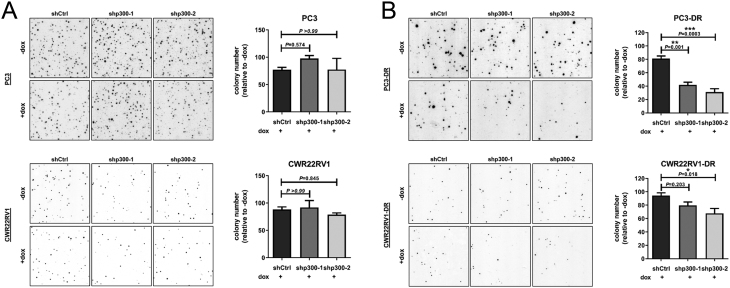
An important characteristic of aggressive tumor cells is the ability to form colonies, thereby assessing a single cell’s ability to undergo unlimited division. In docetaxel-sensitive PC3 and CWR22RV1, knockdown of p300 had no significant effect on the clonogenic potential (Fig. 4A). However, interestingly, p300 inhibition in docetaxel-resistant PC3-DR and CWR22RV1-DR significantly reduced colony formation efficiency (reduction by 40–50% in PC3-DR and 20–30% in CWR22RV1-DR; Fig. 4B).
Figure 4.
p300 inhibition decreases colony formation ability of docetaxel-resistant cells. Representative images and quantification of high-resolution colony formation assays of docetaxel-sensitive PC3 and CWR22RV1 (A) and docetaxel-resistant PC3-DR and CWR22RV1-DR (B). Colony numbers were analyzed by the software CATCH-colonies. Data represent mean + s.e.m. (one-way ANOVA, n = 3).
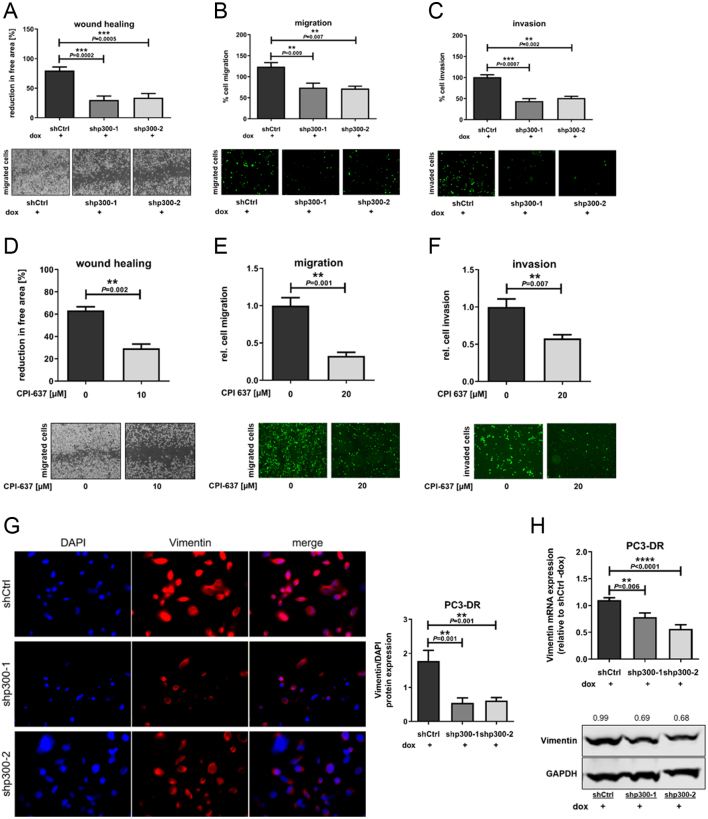
Cell migration and invasion are impaired upon p300 downregulation in docetaxel-resistant cells
It has previously been shown that p300 is involved in migration and invasion in PCa cells (Santer et al. 2011). Therefore, we wanted to test if p300 inhibition might also impair cell migration and invasion in docetaxel-resistant cells. We employed PC3-DR cells for these experiments since the migration ability of CWR22RV1-DR cells is low. We first performed wound scratch assays that revealed a 2.5-fold decrease in the wound healing rate upon p300 downregulation (Fig. 5A). Consistently, significantly decreased cell migration by ~50% was observed in Boyden chambers following p300 downregulation (Fig. 5B). Moreover, invasion assays, which were conducted on matrigel-coated membranes, also showed a significantly reduced invasion ability (reduction by 50%) upon p300 downregulation (Fig. 5C). These findings were confirmed by the use of the dual inhibitor of p300 and CBP CPI-637. Viability assays were performed to calculate the IC50 for PC3-DR cells (Supplementary Fig. 7). It was determined that IC50 for CPI-637 is 17.52 µM. PC3-DR cells treated with CPI-637 for 72 h revealed a significantly decreased migration and invasion ability (reduction by 50–60%, Fig. 5D, E and F). Vimentin is a cytoskeleton component that plays an important role in migration and was therefore analyzed in p300-silenced PC3-DR cells by immunofluorescence, where we observed a significantly decreased vimentin protein expression (by 60–70%, Fig. 5G). Reduced vimentin levels were additionally confirmed by qRT-PCR and Western blot analysis (Fig. 5H).
Figure 5.
p300 downregulation decreases cell migration and invasion. (A) Wound scratch assays were performed on confluent layers of PC3-DR shCtrl, shp300-1, and shp300-2 treated with 100 ng/mL doxycycline. Images were taken 48 h after scratch and analyzed by ImageJ (n = 6). (B) PC3-DR were seeded in Boyden chambers and shp300 sequences were activated with 100 ng/mL doxycycline for 72 h. Cell migration was measured after staining with calcein and visualized by fluorescence microscopy (n = 4). (C) Invasion assays were conducted as in B, except that Boyden chambers were pre-coated with Matrigel (n = 3). Values indicated in A–C denote mean + s.e.m. (one-way ANOVA). (D) Wound scratch assays on PC3-DR treated with 10 µM CPI-637. Data represent mean + s.e.m. (t-Test, n = 3). (E) Migration (n = 4) and (F) invasion assays (n = 5) of PC3-DR treated with 20 µM CPI-637. Values indicated are mean + s.e.m. (t-test). (G) Immunofluorescence staining for vimentin (red) and quantification relative to counterstaining of nuclei (blue). Original magnification 630×. (H) mRNA (qPCR) and protein expression (Western blot) of vimentin. Data in G-H represent mean + s.e.m. (one-way ANOVA, n = 5).
Discussion
Although chemotherapy for CRPC patients confers a clinical benefit for patients, there is no curative treatment available for late stages of PCa. Development of chemotherapy resistance occurs rapidly, and it remains largely unclear which factors are differentially expressed during chemotherapy and might contribute to docetaxel insensitivity. Thus, it is important to understand the molecular mechanisms and to find new therapeutic targets to overcome therapy failure.
As a coactivator of the AR, p300 is involved in many biological processes such as differentiation, proliferation, and cell cycle regulation (Iyer et al. 2004) and has already been associated with tumor progression and poor prognosis (Debes et al. 2003). Furthermore, p300 expression is increased upon androgen deprivation (Heemers et al. 2007) and plays an essential role in ligand-independent transactivation of the AR in androgen-independent PCa cells (Debes et al. 2002).
The main finding of this study is that p300 is upregulated upon docetaxel treatment in primary PCa and mCRPC tissue samples as well as in docetaxel-sensitive and docetaxel-resistant PCa cells. IHC staining was performed in samples of patients who received neoadjuvant docetaxel therapy at the Department of Urology of the Medical University of Innsbruck. Of course, these samples do not reflect the same clinical stage as docetaxel-resistant cells. However, patients who receive chemotherapy for PCa in the Authors’ institution are not subjected to removal of tumor tissue. Therefore, the samples from individuals who received neoadjuvant docetaxel therapy were selected to be able to analyze p300 expression in patient material. Furthermore, metastatic tissue samples obtained by rapid autopsy from a publicly available transcriptome dataset including mCRPC patients that received docetaxel in their treatment course were analyzed. Increased 300 expression was observed in vitro in docetaxel-resistant AR-positive and AR-negative cells (PC3-DR and CWR22RV1-DR) compared to their respective docetaxel-sensitive counterparts. This finding indicates that p300 is upregulated by docetaxel regardless of AR expression and is consistent with previous publications documenting multiple functions of p300 in PCa also independently of the AR (Debes et al. 2003, Santer et al. 2011). This is also in concordance with the finding that AR expression and activity are not changed upon docetaxel treatment; although, in previous studies it has been shown that docetaxel impairs transcriptional activity of the AR (Zhu et al. 2010). Other groups reported an inhibitory effect of docetaxel on AR activity by interfering with AR intracellular trafficking (Martin et al. 2015) and nuclear translocation (Thadani-Mulero et al. 2014).
Contrary to p300, the closely related coactivator CBP was not upregulated upon docetaxel treatment. Thus, we conclude that docetaxel-induced upregulation is specific for p300. Hatano and colleagues reported a role of c-Myc, which is a known downstream target of p300 in docetaxel resistance (Hatano et al. 2013). Concordant with their study, we observed an increased expression of c-Myc in docetaxel-treated patients; however, the upregulation was not statistically significant. Nevertheless, Myc activity was significantly increased in patients who received docetaxel. Ogiwara and colleagues also reported that p300 ablation caused downregulation of Myc expression in CBP-deficient cells and thereby suppressed cancer cell growth (Ogiwara et al. 2016). We assume that p300 upregulation by docetaxel is not mediated by c-Myc, but c-Myc is affected in consequence as a target gene of p300.
We found that p300 mRNA expression was not significantly changed upon docetaxel treatment, which indicates that p300 protein increase is not due to increased transcriptional activity. p300 protein expression also does not increase because of elevated translation, since p300 protein expression in docetaxel-resistant cells was unchanged upon translation inhibition, which suggests that p300 is protected from proteasomal degradation. In a previous study, it was found that androgens downregulate p300 protein but not mRNA expression (Heemers et al. 2007). Those results suggest similarities in regulation of p300 by docetaxel and androgens.
It has been previously shown that the competitive histone acetyltransferase p300/CBP inhibitor C646 reduced colony formation in AML cell lines and primary blasts (Gao et al. 2013). While this highlights the capacities of HAT inhibitors, there are issues with the potency and selectivity of these early inhibitors (Lasko et al. 2017). Meanwhile, more effective bromodomain and extra-terminal (BET) inhibitors that prevent protein-protein interactions between BET proteins and acetylated histones have been developed. The BET inhibitors INCB054329 and INCB057643 have been shown to be effective as single agents in PCa. Likely, novel BET inhibitors will be combined with existing therapies, in particular, for therapy-resistant PCa (Vazquez et al. 2019). In this study, we initially employed a shRNA approach to down-regulate p300 and exclude non-specific effects. We confirmed effects of shRNA-mediated p300 downregulation on migration ability of docetaxel-resistant cells with the bromodomain inhibitor CPI-637 that is specific for p300 and CBP, where we observed similar results as with genetic p300 downregulation.
In contrast to docetaxel-sensitive cells, p300 inhibition in docetaxel-resistant PC3-DR and CWR22RV1-DR significantly reduced colony formation efficiency. Downregulation of p300 had no specific effects on proliferation of docetaxel-resistant cells, indicating that p300 inhibition indeed impaired the colony-initiating capacity and that the reduced colony number is not just a secondary effect. It has already been described that docetaxel-resistant cells show a stem-cell-like phenotype (Puhr et al. 2012, Marin-Aguilera et al. 2014), suggesting that p300 inhibition is effective in conditions in which pathways connected to colony formation and tumor-initiation play a central role. A possible explanation as to why p300 inhibition shows no effects on colony formation of docetaxel-sensitive cells could be the formation of p300-complexes. It has been described that the cysteine protease USP24 stabilizes p300 and thereby increases acetylation of histone h3 (Wang et al. 2018). The CtBP1-p300-FOXO 3a complex was found to repress apoptotic regulators Bax and Bim in osteosarcoma cells (Li et al. 2019). Cell proliferation is regulated by p300 in complex with the transcriptional repressor YY1 and HDAC2 (Tang et al. 2019). Future studies of therapy resistance in PCa should therefore examine and quantitate the complexes relevant to p300. In contrast to PCa, in some models of breast cancer, p300 is considered a tumor inhibitor (Asaduzzaman et al. 2017). However, the histone methyltransferase DOT1L in complex with p300 and c-Myc enhanced cellular stemness (Cho et al. 2015). The reasons for obviously contrasting effects of p300 on stemness in breast cancer are not yet known.
p300 inhibition significantly decreased cell migration and invasion in PC3-DR cells and reduced vimentin expression. These findings indicate that p300 is involved in cellular pathways that regulate migration and invasion in PCa and are in concordance with previous studies, where they reported involvement of p300 in migration in nasopharyngeal carcinoma and breast cancer cells (Fermento et al. 2014, Liao et al. 2017).
Taken together, p300 could be a valid target in docetaxel-resistant PCa as it reduces the metastatic potential of docetaxel-resistant cells by reducing colony formation, migration, and invasion capability. So far, multiple factors that contribute to the development of docetaxel resistance in PCa have been described (Patterson et al. 2006, Deng et al. 2019, Kapur et al. 2019). Therefore, docetaxel resistance in PCa is heterogeneous, thus suggesting that medical intervention may be based on a personalized approach. This issue is particularly relevant because p300 inhibitors enter clinical trials in oncology, thus pointing to appropriate selection of patients who will benefit from specific p300 targeting in combinations with other drugs approved or tested for treatment of PCa.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by the Austrian Science Fund FWF grant W1101-B12 to Z Culig and P31122 to N Sampson.
Author contribution statement
M Gruber and L Ferrone performed experiments. M Gruber analyzed data and wrote the first version of the manuscript. M Puhr organized and supervised immunohistochemical analyses and supervised in vitro experiments with chemoresistance models. T Furlan performed experiments with the bromodomain inhibitor. F R Santer, T Furlan, I Eder, and N Sampson provided experimental suggestions and analyzed data. M Puhr and G Schäfer established TMA. F Handle established the experimental system, performed the bioinformatic analysis of the transcriptome dataset, and supervised in vitro experiments. Z Culig was responsible for conception and design of the study, data analysis, and supervision. All authors contributed to manuscript writing.
Acknowledgements
The authors thank Sarah Peer for immunohistochemical staining; Eberhard Steiner, M.Sc., for patient selection and statistical analysis; Prof Walther Parson for cell line authentication; Prof Dr. William Watson for providing CWR22RV1 and CWR22RV1-DR cells; Dr Martin Hermann for help with immunofluorescence staining; Prof Helmut Klocker and Dr Maximilian Brandt for proofreading the manuscript, and all members of the Division of Experimental Urology at the Medical University of Innsbruck for helpful discussions. F Handle and Z Culig, the joint senior authors.
References
- Asaduzzaman M, Constantinou S, Min H, Gallon J, Lin ML, Singh P, Raguz S, Ali S, Shousha S, Coombes RC, et al. 2017. Tumour suppressor EP300, a modulator of paclitaxel resistance and stemness, is downregulated in metaplastic breast cancer. Breast Cancer Research and Treatment 461–474. ( 10.1007/s10549-017-4202-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. Journal of Cell Science 2363–2373. [DOI] [PubMed] [Google Scholar]
- Cho MH, Park JH, Choi HJ, Park MK, Won HY, Park YJ, Lee CH, Oh SH, Song YS, Kim HS, et al. 2015. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nature Communications 7821 ( 10.1038/ncomms8821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzi B, Nemes C, Schmidt S, Jasarevic Z, Lodde M, Pycha A, Bartsch G, Offner F, Culig Z, Hobisch A. 2004. The androgen receptor co-activator CBP is up-regulated following androgen withdrawal and is highly expressed in advanced prostate cancer. Journal of Pathology 159–166. ( 10.1002/path.1609) [DOI] [PubMed] [Google Scholar]
- Debes JD, Schmidt LJ, Huang H, Tindall DJ. 2002. p300 mediates androgen-independent transactivation of the androgen receptor by interleukin 6. Cancer Research 5632–5636. [PubMed] [Google Scholar]
- Debes JD, Sebo TJ, Lohse CM, Murphy LM, Haugen DA, Tindall DJ. 2003. p300 in prostate cancer proliferation and progression. Cancer Research 7638–7640. [PubMed] [Google Scholar]
- Deng L, Gu X, Zeng T, Xu F, Dong Z, Liu C, Chao H. 2019. Identification and characterization of biomarkers and their functions for docetaxel-resistant prostate cancer cells. Oncology Letters 3236–3248. ( 10.3892/ol.2019.10623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fermento ME, Gandini NA, Salomon DG, Ferronato MJ, Vitale CA, Arevalo J, Lopez Romero A, Nunez M, Jung M, Facchinetti MM, et al. 2014. Inhibition of p300 suppresses growth of breast cancer. Role of p300 subcellular localization. Experimental and Molecular Pathology 411–424. ( 10.1016/j.yexmp.2014.09.019) [DOI] [PubMed] [Google Scholar]
- Gao XN, Lin J, Ning QY, Gao L, Yao YS, Zhou JH, Li YH, Wang LL, Yu L. 2013. A histone acetyltransferase p300 inhibitor C646 induces cell cycle arrest and apoptosis selectively in AML1-ETO-positive AML cells. PLoS ONE e55481 ( 10.1371/journal.pone.0055481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, Guinney J. 2013. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 7 ( 10.1186/1471-2105-14-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano K, Yamaguchi S, Nimura K, Murakami K, Nagahara A, Fujita K, Uemura M, Nakai Y, Tsuchiya M, Nakayama M, et al. 2013. Residual prostate cancer cells after docetaxel therapy increase the tumorigenic potential via constitutive signaling of CXCR4, ERK1/2 and c-Myc. Molecular Cancer Research 1088–1100. ( 10.1158/1541-7786.MCR-13-0029-T) [DOI] [PubMed] [Google Scholar]
- Heemers HV, Sebo TJ, Debes JD, Regan KM, Raclaw KA, Murphy LM, Hobisch A, Culig Z, Tindall DJ. 2007. Androgen deprivation increases p300 expression in prostate cancer cells. Cancer Research 3422–3430. ( 10.1158/0008-5472.CAN-06-2836) [DOI] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C. 2004. p300/CBP and cancer. Oncogene 4225–4231. ( 10.1038/sj.onc.1207118) [DOI] [PubMed] [Google Scholar]
- James ND, Sydes MR, Clarke NW, Mason MD, Dearnaley DP, Spears MR, Ritchie AW, Parker CC, Russell JM, Attard G, et al. 2016. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 1163–1177. ( 10.1016/S0140-6736(15)01037-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E. 2004. CBP and p300: HATs for different occasions. Biochemical Pharmacology 1145–1155. ( 10.1016/j.bcp.2004.03.045) [DOI] [PubMed] [Google Scholar]
- Kapur N, Mir H, Sonpavde GP, Jain S, Bae S, Lillard JW, Singh S. 2019. Prostate cancer cells hyper-activate CXCR6 signaling by cleaving CXCL16 to overcome effect of docetaxel. Cancer Letters 1–13. ( 10.1016/j.canlet.2019.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. 2010. MYC and prostate cancer. Genes and Cancer 617–628. ( 10.1177/1947601910379132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, Etzioni R, Bolouri H, Montgomery B, White T, et al. 2016. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nature Medicine 369–378. ( 10.1038/nm.4053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulos CE, Chen YH, Carducci MA, Liu G, Jarrard DF, Hahn NM, Shevrin DH, Dreicer R, Hussain M, Eisenberger M, et al. 2018. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer: long-term survival analysis of the randomized phase III E3805 CHAARTED trial. Journal of Clinical Oncology 1080–1087. ( 10.1200/JCO.2017.75.3657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, et al. 2017. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 128–132. ( 10.1038/nature24028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavaud P, Gravis G, Foulon S, Joly F, Oudard S, Priou F, Latorzeff I, Mourey L, Soulie M, Delva R, et al. 2018. Anticancer activity and tolerance of treatments received beyond progression in men treated upfront with androgen deprivation therapy with or without docetaxel for metastatic castration-naive prostate cancer in the GETUG-AFU 15 phase 3 trial. European Urology 696–703. ( 10.1016/j.eururo.2017.09.022) [DOI] [PubMed] [Google Scholar]
- Li C, Xiao XQ, Qian YH, Zhou ZY. 2019. The CtBP1-p300-FOXO3a transcriptional complex represses the expression of the apoptotic regulators Bax and Bim in human osteosarcoma cells. Journal of Cellular Physiology 22365–22377. ( 10.1002/jcp.28802) [DOI] [PubMed] [Google Scholar]
- Liao ZW, Zhao L, Cai MY, Xi M, He LR, Yu F, Zhou TC, Liu MZ. 2017. P300 promotes migration, invasion and epithelial-mesenchymal transition in a nasopharyngeal carcinoma cell line. Oncology Letters 763–769. ( 10.3892/ol.2016.5491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Aguilera M, Codony-Servat J, Reig Ò, Lozano JJ, Fernandez PL, Pereira MV, Jimenez N, Donovan M, Puig P, Mengual L, et al. 2014. Epithelial-to-mesenchymal transition mediates docetaxel resistance and high risk of relapse in prostate cancer. Molecular Cancer Therapeutics 1270–1284. ( 10.1158/1535-7163.MCT-13-0775) [DOI] [PubMed] [Google Scholar]
- Martin SK, Banuelos CA, Sadar MD, Kyprianou N. 2015. N-terminal targeting of androgen receptor variant enhances response of castration resistant prostate cancer to taxane chemotherapy. Molecular Oncology 628–639. ( 10.1016/j.molonc.2014.10.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, Kohno T. 2016. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discovery 430–445. ( 10.1158/2159-8290.CD-15-0754) [DOI] [PubMed] [Google Scholar]
- Patterson SG, Wei S, Chen X, Sallman DA, Gilvary DL, Zhong B, Pow-Sang J, Yeatman T, Djeu JY. 2006. Novel role of Stat1 in the development of docetaxel resistance in prostate tumor cells. Oncogene 6113–6122. ( 10.1038/sj.onc.1209632) [DOI] [PubMed] [Google Scholar]
- Puhr M, Hoefer J, Schafer G, Erb HH, Oh SJ, Klocker H, Heidegger I, Neuwirt H, Culig Z. 2012. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. American Journal of Pathology 2188–2201. ( 10.1016/j.ajpath.2012.08.011) [DOI] [PubMed] [Google Scholar]
- Qin J, Lee HJ, Wu SP, Lin SC, Lanz RB, Creighton CJ, Demayo FJ, Tsai SY, Tsai MJ. 2014. Androgen deprivation-induced NCoA2 promotes metastatic and castration-resistant prostate cancer. Journal of Clinical Investigation 5013–5026. ( 10.1172/JCI76412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos YF, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, Van Galen M, Van Dam H, Van Ommen GJ, Den Dunnen JT, Zantema A, et al. 2010. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Research 5396–5408. ( 10.1093/nar/gkq184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer FR, Hoschele PP, Oh SJ, Erb HH, Bouchal J, Cavarretta IT, Parson W, Meyers DJ, Cole PA, Culig Z. 2011. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Molecular Cancer Therapeutics 1644–1655. ( 10.1158/1535-7163.MCT-11-0182) [DOI] [PubMed] [Google Scholar]
- Tang W, Zhou W, Xiang L, Wu X, Zhang P, Wang J, Liu G, Zhang W, Peng Y, Huang X, et al. 2019. The p300/YY1/miR-500a-5p/HDAC2 signalling axis regulates cell proliferation in human colorectal cancer. Nature Communications 663 ( 10.1038/s41467-018-08225-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock IF, De Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Theodore C, James ND, Turesson I, et al. 2004. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. New England Journal of Medicine 1502–1512. ( 10.1056/NEJMoa040720) [DOI] [PubMed] [Google Scholar]
- Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, Corey E, Nanus DM, Plymate SR, Giannakakou P. 2014. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Research 2270–2282. ( 10.1158/0008-5472.CAN-13-2876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez R, Civenni G, Kokanovic A, Shinde D, Cantergiani J, Marchetti M, Zoppi G, Ruggeri B, Liu PCC, Carbone GM, et al. 2019. Efficacy of novel bromodomain and extraterminal inhibitors in combination with chemotherapy for castration-resistant prostate cancer. European Urology Oncology [epub]. ( 10.1016/j.euo.2019.07.013) [DOI] [PubMed] [Google Scholar]
- Wang YC, Wu YS, Hung CY, Wang SA, Young MJ, Hsu TI, Hung JJ. 2018. USP24 induces IL-6 in tumor-associated microenvironment by stabilizing p300 and beta-TrCP and promotes cancer malignancy. Nature Communications 3996 ( 10.1038/s41467-018-06178-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ML, Horbinski CM, Garzotto M, Qian DZ, Beer TM, Kyprianou N. 2010. Tubulin-targeting chemotherapy impairs androgen receptor activity in prostate cancer. Cancer Research 7992–8002. ( 10.1158/0008-5472.CAN-10-0585) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a