Abstract
Background:
NMAAP1 plays a role in regulating macrophage differentiation to the M1 type and exerting antitumoral functions. It is not clear what role and mechanism NMAAP1 does play in the reversal of macrophages from M1 to M2.
Methods:
We detected the typing of macrophages with high or low expression of NMAAP1 by QPCR and ELISA, and detected the colocalization of NMAAP1 and endogenous IP3R by laser confocal microscopy, and detected the protein expression in cells by Western-blotting.
Results:
Our study found that knockdown NMAAP1 in RAW264.7 cells induced macrophage polarization to the M2 type and up-regulation of NMAAP1 in RAW264.7 cells maintain M1 Phenotype even in the presence of IL-4, a stronger inducer of the M2 type. Additionally, Co-immunoprecipitation revealed a protein-protein interaction between NMAAP1 and IP3R and then activates key molecules in the PKC-dependent Raf/MEK/ERK and Ca2+/CaM/CaMKII signaling pathways. Activation of PKC (Thr638/641), ERK1/2 (Thr202/Tyr204) and CaMKII (Thr286) is involved in the regulation of cell differentiation.
Conclusion:
NMAAP1 interacts with IP3R, which in turn activates the PKC-dependent Raf/MEK/ERK and Ca2+/CaM/CaMKII signaling pathways. These results provide a new explanation of the mechanism underlying M1 differentiation.
Keywords: NMAAP1, macrophage, M1, polarization, IP3R, Signaling Pathway
1. INTRODUCTION
As an important part of the innate immune system, macrophages play an integral role in maintaining homeostasis, defending against microbes, and directly killing tumoral cells and presenting tumor-associated antigens [1].
Macrophages also display significant heterogeneity and plasticity. Interferon γ (IFN-γ), Lipopolysaccharides (LPS), and Bacillus Calmette-Guerin (BCG) can induce the M1 phenotype in macrophages through classical pathways. This type of cell secretes a large number of pro-inflammatory cytokines (such as TNF-α, IL-6, IL-12), as well as Nitric Oxide (NO), Reactive Oxygen Species (ROS) [2], and chemokines CXCL9 and CXCL10 [3]; along with high expression of Major Histocompatibility Complex class II (MHC-II) molecules and costimulatory molecules CD80 and CD86. IL-4/IL-13, Transforming Growth Factor β (TGF-β) or glucocorticoids activate alternative pathways that differentiate macrophages into the M2 phenotype, which exhibit elevated secretion of IL-10 and high expression of scavenger, mannose and galactose-type receptors, as well as chemokines CCL17, CCL22 and CCL24 [4]. M2 macrophages can inhibit cell proliferation and activity, promote angiogenesis and wound healing, and regulate inflammatory responses and adaptive Th2 immune responses, which may promote tumor growth [5, 6].
The Novel Macrophage Activation-Associated Protein 1 (NMAAP1) is a molecule specifically expressed in BCG-Activated Macrophages (BAM), which shares similar sequences with DANGER, a newly discovered triphosphate receptor-binding protein, which can bind Inositol 1,4,5-trisphosphate receptor (IP3R) to regulate cell differentiation [7]. NMAAP1 plays a role in regulating macrophage differentiation to the M1 type and exerting antitumoral functions [8]. However, tumoral cells secrete TGF-β, IL-4, IL-10 and IL-17, which prompt M1 macrophages to transform into the M2 type. As a result, TNF-α, IL-1, IFN-γ release and NO synthesis are decreased, weakening their antitumoral and antigen-presenting functions, which may promote tumoral proliferation and metastasis [9]. It is not clear what role NMAAP1 does play in the reversal of macrophages from M1 to M2.
The DANGER protein superfamily is a closely related protein group that regulates the differentiation of nerve cells and the development of tissues and organs. As a member of this family, NMAAP1 may regulate intracellular calcium concentration through regulation of inositol 1,4,5-Trisphosphate Receptors (IP3R) [10], though its role in macrophage differentiation and modulation remains unclear. Binding between IP3Rs and IP3 in the Peritoneal Macrophage (PM) will stimulate Phospholipase C (PLC) and promote Ca2+ release from the Endoplasmic Reticulum (ER) [11, 12]. While Ca2+ can regulate some signal transduction through Ca2+/CaM [13, 14] and Ca2+/Raf/MEK/ERK in suppression of inflammatory reactions [15].
Maintaining a dynamic balance of intracellular calcium ions (Ca2+) is essential, as it is an important multi-function second messenger which regulates a variety of cellular physiological activities, including cell differentiation, proliferation, growth [16], apoptosis [17], cytokine secretion, gene expression, transcription, biofilm fusion, embryo development, and morphogenesis [18].
In this study, we evaluated the effects and mechanisms associated with NMAAP1 activity on macrophage differentiation and transformation. We found NMAAP1 could maintain the M1 type and prevent macrophage shift from M1 to M2 by interacting with IP3R and modulating calcium-related signaling pathways.
2. MATERIALS AND METHODS
2.1. Overexpression of NMAAP1 in RAW264.7 Cells
2.2. RNA Interference by siRNA
Transient transfection of siRNA was performed using LipofectamineTM RNAiMax (Invitrogen) according to the manufacturer’s protocol. 1×105 ON/ RAW264.7 cells were seeded in DMEM containing 10% FBS. After moving the culture to a fresh medium the next day, the transfection solution contained 6 μl of transfection reagent. 2.5 μl of 50 nM SiRNA were added to the cells for 24 h and then harvested for further investigation.
2.3. Reverse Transcription and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was directly isolated from adherent macrophages using Trizol reagent (Takara, Japan), according to the manufacturer′s protocol. RNA quantity and quality were determined using an absorbance microplate reader TQuant (BioTEK, USA). cDNA for reverse transcription was prepared from 1 μg of total RNA using oligo(dT) primers and reverse transcriptase M-MLV in a total volume of 20 μl, according to the manufacturer′s instructions (Takara, Japan). The resulting cDNA was used for real-time PCR for NMAAP1, Inducible Nitric Oxide Synthase (iNOS) and cytokines (IL-1β,TNF-α, arginase 1(ARG-1), IL-10, and TGF-β) using an ABI PRISM 7300 sequence detection system (Applied Biosystems, USA). Quantitative PCR was carried out using the following conditions: initial denaturation at 95 °C for 5 min, followed by 40 cycles of 5 s for denaturation at 95 °C, then 30 s at 60 °C. Expression of each gene was normalized to the expression of GAPDH. The forward and reverse specific primer sequences were used in Table 1.
Table 1. The primers of genes.
| Name of Gene | Primers |
|---|---|
| NMAAP1 | Forward 5′-CCTTCCCCTGCCCAATA-3′ Reverse 5′-TCCACGAAACCTTCCACA-3′ |
| TNF-α | Forward 5′-ACTGAACTTCGGGGTGATCG-3′ Reverse 5′-CCACTTGGTGGTTTGCTACG-3′ |
| IL-1β | Forward 5′-TGGACCTTCCAGGATGAGGACA-3′ Reverse 5′-GTTCATCTCGGAGCCTGTAGTG-3′ |
| iNOS | Forward 5′-ATGGCAACATCAGGTCGG-3′ Reverse 5′-GCACAACTGGGTGAACTCC-3′ |
| MCP-1 | Forward 5′-TTAAAAACCTGGATCGGAACC-3′ Reverse 5′-GCATTAGCTTCAGATTTACGG-3′ |
| IL-10 | Forward 5′-GTTGCCAAGCCTTATCGG-3′ Reverse 5′-GCTCTTATTTTCACAGGGGAG-3′ |
| Arg1 | Forward 5′-CAGTCTGGCAGTTGGAAGC-3′ Reverse 5′-GGTTGTCAGGGGAGTGTTG-3′ |
| TGF-β | Forward 5′-GAGGCGGTGCTCGCTTTGTA-3′ Reverse 5′-CGTTGTTGCGGTCCACCATTA-3′ |
| GAPDH | Forward 5′-GACTTCAACAGCAACTCCCACTC-3′ Reverse 5′-TAGCCGTATTCATTGTCATACCAG-3′ |
p-CMV-N-Flag/NMAAP1 eukaryotic expression vector were constructed through regular methods, and RAW264.7 cells with overexpression of NMAAP1 (ON/RAW264.7) were maintained at 37 °C and 5% CO2 in a complete medium containing DMEM (Gibco, Grand Island, New York, USA) supplemented with 10% fetal calf serum (Gibco), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco). Then, the ON/ RAW264.7 cells were treated with IL-4 (25 ng/ml) for 48 h.
2.4. ELISA
The supernatants were collected for analysis. The concentrations of TNF-α, IL-1β, IL-12p40, IL-10 and TGF-β in the medium were determined by ELISA analysis, according to the manufacturer’s instructions (eBioscience, USA). TNF-α, IL-1β and IL-12p40 were used as biomarkers for M1 macrophages, and TGF-β and IL-10 were used as biomarkers for M2 macrophages.
2.5. Immunofluorescence
ON/RAW264.7 cells were cultured on slides at a seeding density of 50-60%. After the cells were attached, the slides were rinsed twice with pre-cooled PBS. After fixing with 4% paraformaldehyde for 30 min at room temperature and washing the cells twice with PBS, the cells were poled using 0.1% Triton X-100 at room temperature for 10 min. After blocking the cells with normal goat serum at room temperature for 1-2 h, the rabbit anti-mouse IP3R antibodies and goat anti-rabbit-PE-Flag mouse monoclonal antibodies were added and cultured for 2 h at 37 °C. After washing, goat anti-rabbit-FITC were added and cultured for another 2 h. Then slide was then washed with PBST 3 times for 10 minutes on each occasion. The slides were dyed with DAPI for 3-5 min, then washed with PBS 3-5 times and observed under a fluorescence microscope.
2.6. Co-Immunoprecipitation (CO-IP)
The cells were lysed with pre-cooled protein lysate and placed on ice for 30 min, vortex-mixed once every 10 min. The protein supernatant was collected by centrifugation at 18630 g (Dragonlab D3024R) in a centrifuge at 4 °C for 20 minutes. After determining the protein concentration, 30 μl of Protein A/G Agarose and 2 μg of the same common IgG antibody was added to the supernatant and then placed on the mixer for 4 h at 4 °C. After centrifugation at 590 g (Dragonlab D3024R) for 5 minutes at 4 °C, the supernatant was transferred to a new EP tube for subsequent immunoprecipitation. 2 μg of primary antibody and protein lysate were added for IP and, the mix was then shaken slowly at 4 °C overnight. Next, 30 μl of resuspended Protein A/G Agarose were added to the sample and shaken slowly at 4 °C for 2 hours. After centrifuged at 590 g (Dragonlab D3024R) for 5 min at 4 °C, the supernatant was carefully discarded, retaining the Protein A/G Agarose precipitate, and rinsing with protein lysate 5 times. The supernatant was discarded, and the pellet was resuspended by adding 30 μl of protein lysate. After adding 5x protein loading buffer, the sample was mixed and suspended in a boiling water bath for 10 min. The samples were then subjected to sodium salt (SDS)-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western blotting.
2.7. Western-Blotting
For the preparation of the protein samples, 0.5 ml of protein lysate was added to 1×107 cells, along with a protease inhibitor at 1:100. The sample was then incubated on ice for 30 min and mixed once every 10 min to ensure cell lysis is complete. Next centrifugation was performed at 18630 g (Dragonlab D3024R) and 4 °C for 20 minutes. The supernatant was then moved into an EP tube. After determination of protein concentration, the sample was added into SDS-PAGE, run in 80 V constant pressure electrophoresis and transferred to the Polyvinylidene Fluoride (PVDF) membrane. Then, the PVDF membrane was placed in a TBST buffer containing 5% skim milk, and placed on a shaker for 1 hour. The blocked PVDF membrane was placed in a hybridization bag containing the primary antibody dilution and incubated overnight on a 4 °C shaker. After incubation the PVDF membrane with the primary antibody was removed and rinsed 3 times with TBST buffer for 10 min on each occasion. The PVDF membrane was placed in a hybrid bag containing the secondary antibody dilution and incubated for 1.5 h on a shaker at room temperature. After washing, the PVDF membrane was placed on a chemiluminescence developer using Thermo's Electrochemiluminescence (ECL) Plus. The chromogenic solution was prepared in a dark-protected EP tube and uniformly added to the PVDF membrane, which was then placed in the gel for 1-2 min, in a gel imager. A photograph was taken for grayscale analysis of protein bands using Image J software.
2.8. Statistical Analysis
Data were expressed as mean±SEM and analyzed by one-way analysis of variance. P values <0.05 (95% confidence level) were considered statistically significant.
3. RESULTS
3.1. NMAAP1 Prevents Macrophage Shift from M1 to M2
Firstly, we found that downregulated expression of NMAAP1 shifted macrophages from M1 to M2. The expression of IL-1β,iNOS and Monocyte Chemotactic Protein (MCP1) decreased, while Arg1, IL-10 and TGF-β were up-regulated. The secretion of TNF-α and IL-1β decreased and IL-10 increased, respectively (Figure 1).
Figure 1.
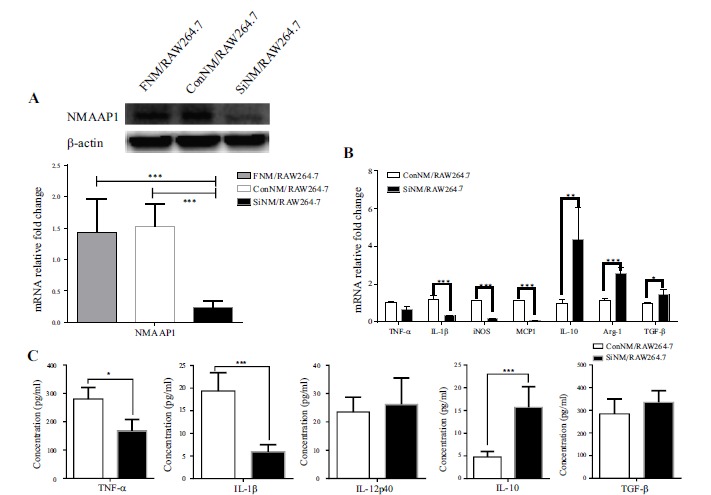
Down-regulation of NMAAP1 promotes macrophage shift from M1 to M2. (A) Gene and protein expression of NMAAP1 in ConNM/RAW264.7 and SiNM/RAW264.7 cells was detected by qRT-PCR and Western-blotting. (B) Genes expression in ConNM/RAW264.7 and SiNM/RAW264.7 cells was detected by qRT-PCR. (C) ELISA was used to detect the secretion of TNF-α, IL-1, IL-12p40, IL-10 and TGF-β by ConNM/RAW264.7 and SiNM/RAW264.7 cells. (Note: ConNM/RAW264.7: siRNA control sequence transfected cells; SiNM/RAW264.7: NMAAP1 siRNA sequence transfected cells). * P < 0.05, ** P < 0.01, *** P < 0.001.
Secondly, we used IL-4 (25 ng/ml) to act on macrophages with high expression of NMAAP1. After 48 h, qRT-PCR revealed increased expression of Arg1. Although the expression of iNOS and TNF-α decreased, it was still significantly higher than that of the control group, with an iNOS/Arg1 ratio >1, indicating macrophages were not transformed into the M2 type and were in a state of biased M1 (Figure 2). These results suggest NMAAP1 prevented IL-4-induced macrophage transformation from M1 to M2.
Figure 2.
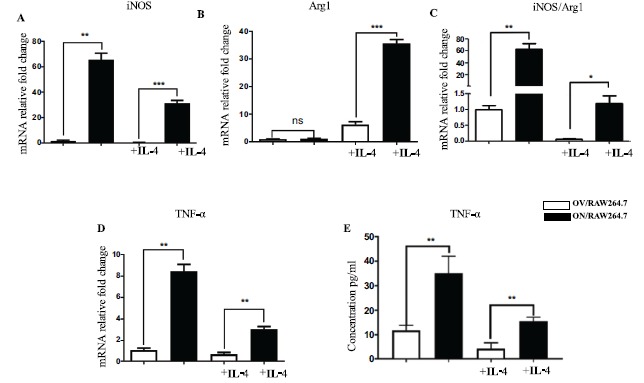
NMAAP1 prevents macrophage shift from M1 to M2. Expression of iNOS(A), Arg1(B)in OV/RAW264.7 and ON/RAW264.7 cells were measured through qRT-PCR. The ratio of iNOS/Arg1 showed in (C). TNF-αin OV/RAW264.7 and ON/RAW264.7 cells was measured through qRT-PCR (D). The concentration of TNF-α detected by ELISA was showed in (E). (Note: OV/RAW264.7: empty vector transfected cells; ON/RAW264.7: NMAAP1 overexpression vector transfected cells). * P < 0.05, ** P < 0.01, *** P < 0.001
3.2. Co-Localization of NMAAP1 and IP3R
Studies have confirmed NMAAP1 interacts with other synergistic molecules to regulate cell growth and differentiation. To demonstrate the interaction of NMAAP1 with IP3R, we analyzed the distribution of expression of these two proteins in ON/RAW264.7 cells. As shown in Figure 3A, co-localization of NMAAP1 and endogenous IP3R was detected by laser confocal microscopy. Both NMAAP1 and IP3R were expressed in the nucleus and cytoplasm, with NMAAP1 being more predominant in the nucleus, and IP3R in the cytoplasm.
Figure 3.
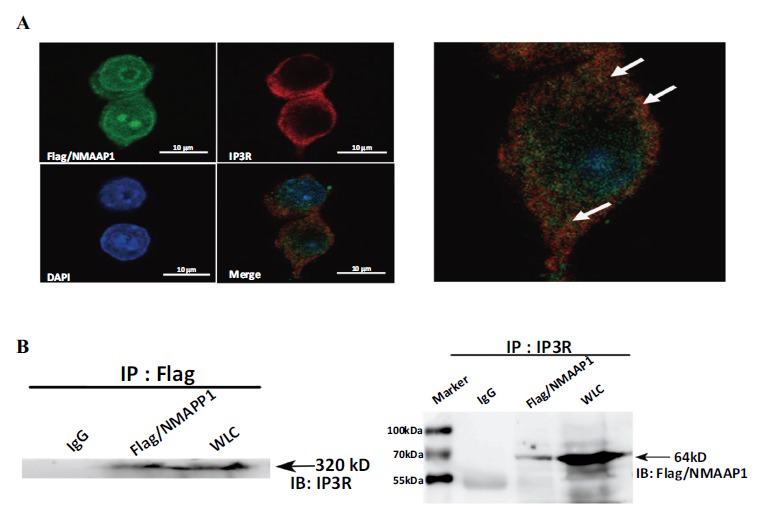
NMAAP1 co-localizes and interacts with IP3R in Flag/NMAAP1-transfected RAW264.7 cells. (A) Immunofluorescence staining for Flag/NMAAP1 (green, upper left), IP3R (red, upper right), DAPI (blue, lower left), and three recombinant overlay images of Flag/NMAAP1 transfected RAW264.7 macrophages (Bottom right). (B) Co-immunoprecipitation experiments were performed using RAW264.7 cells transfected with Flag/NMAAP1. The experimental samples were as follows: IgG: immunoprecipitated product of the isotype control group; Flag/NMAAP1: immunoprecipitated product of the target group; WLC: Total protein extracted from FN/RAW264.7 cells. (The color version of the figure is available in the electronic copy of the article).
3.3. NMAAP1 Interacts with IP3R
To assess protein-protein interactions, we performed Co-Immunoprecipitation analysis (CO-IP) on ON/RAW264.7 cells. As shown in Figure 3, anti-Flag monoclonal antibodies were used on ON/RAW264.7 cells. The source IP3R protein was precipitated, and IP3R expression was detected in the NMAAP1 interacting protein immunoprecipitate complex: The anti-IP3R polyclonal antibody precipitated the NMAAP1 fusion protein on ON/RAW264.7 cells, and expression of the NMAAP1 fusion protein was detected in the endogenous IP3R-interacting protein immunoprecipitate complex (Figure 3). In the control group (IgG group), no protein binding was detected in the immunoprecipitated samples with the corresponding isotype control antibodies. These results indicate NMAAP1 interacts with IP3R in macrophages.
3.4. NMAAP1 Induces Activation of Calcium-Related Signaling Pathways
To demonstrate the regulation of cellular signaling pathways by increased intracellular calcium, we used Western blot to detect activation of key molecules in calcium signaling pathways. As shown in Figure 4, elevated intracellular calcium activates key molecules in the PKC-dependent Raf/MEK/ERK and Ca2+/CaM/CaMKII signaling pathways, such as PKC (Thr638/641), ERK1/2 (Thr202) /Tyr204), and CaMKII (Thr286). This confirms elevated calcium ions in these cells can induce activation of calcium-related signaling pathways.
Figure 4.
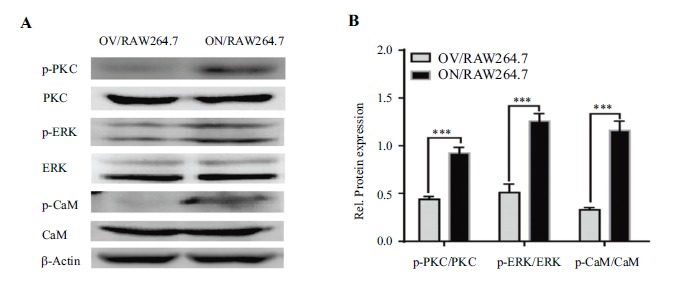
NMAAP1 enhances Ca2+ signaling pathways in ON/RAW264.7 cells. (A) Western Blot was used to detect the activation of p-PKC, total PKC, p-ERK1/2, total ERK1/2, p-CaMK, total CaMK in OV/RAW264.7 and ON/RAW264.7 cells. (B) is the analysis result of Figure 4A. *** P <0.001.
4. DISCUSSION
NMAAP1 is a membrane-associated protein which contains the Mab-21 domain and is a member of the DANGER superfamily. Our results showed NMAAP1 maintains the M1 phenotype in macrophages, and this effect is mediated through binding with IP3R and activate calcium-related signaling pathways in RAW264.7 cells.
Structural analogy analysis has determined most molecules in the DANGER family contain a conserved Mab-21 region, which assures an evolutionary similarity of up to 70% in this group. Most members of this family participate in the regulation of tissue development and cell differentiation [19-21]. NMAAP1 has this Mab-21 domain, and induces macrophage polarization to the M1 phenotype and promotes macrophage to secret some cytokines to kill tumor cells such as TNF-α, even in the presence of IL-4, a stronger inducer of the M2 type [2, 21, 22].
Studies have shown that DANGER molecules expressed in neurons can be physiologically associated with IP3R molecules and regulate IP3R-mediated intracellular calcium release [10, 23, 24]. IP3R is the most common calcium channel in cells, mainly located on the endoplasmic reticulum and mitochondria to control the release of calcium ions, thereby maintaining the dynamic balance of intracellular calcium ions. We found NMAAP1 was expressed in the cytoplasm, cell membrane and nucleus, mainly the nuclear membrane; while IP3R was chiefly expressed on the rough endoplasmic reticulum, cytoplasm and outer nuclear membrane. Co-immunoprecipitation revealed a protein-protein interaction between NMAAP1 and IP3R. These results indicate that NMAAP1 may regulate the calcium channel of IP3R, which is consistent with the function of DANGER in nerve cells [25, 26].
CaM is the most important calcium signal receptor and transducer in cells. Ca2+/CaM can bind to a subunit of CaMKII, causing autophosphorylation of its Thr286 residue, thereby participating in the regulation of gene expression. Studies on the mechanism of M1-type differentiation of IFNγ-induced macrophages indicate activation of PKC and elevation of intracellular calcium concentration are two key factors in this process [27]. Our study found RAW264.7 cells stably expressing NMAAP1 interact with IP3R to activate key molecules in the PKC-dependent Raf/MEK/ERK and Ca2+/CaM/CaMKII signaling pathways. Activation of PKC (Thr638/641), ERK1/2 (Thr202/Tyr204), CaMKII (Thr286) and p65 (Thr276) is involved in the regulation of cell differentiation [28, 29].
CONCLUSION
NMAAP1 interacts with IP3R, which in turn activates the PKC-dependent Raf/MEK/ERK and Ca2+/CaM/CaMKII signaling pathways. These results provide a new explanation of the mechanism underlying M1 differentiation. However, NMAAP1 may also regulate this process through other molecular pathways.
ACKNOWLEDGEMENTS
Declared none.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This work is financially supported by National Natural Science Foundation of China. (No. 81871245) and the Fundamental Research Funds for the Central Universities.
CONFLICT OF INTEREST
All the authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Shapouri-Moghaddam A., Mohammadian S., Vazini H., Taghadosi M., Esmaeili S.A., Mardani F., Seifi B., Mohammadi A., Afshari J.T., Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018;233(9):6425–6440. doi: 10.1002/jcp.26429. [http://dx.doi.org/10.1002/jcp.26429]. [PMID: 29319160]. [DOI] [PubMed] [Google Scholar]
- 2.Ruytinx P., Proost P., Van Damme J., Struyf S., Struyf S. Chemokine-induced macrophage polarization in inflammatory conditions. Front. Immunol. 2018;9:1930. doi: 10.3389/fimmu.2018.01930. [http://dx.doi.org/10.3389/fimmu.2018.01930]. [PMID: 30245686]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang N., Liang H., Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [http://dx.doi.org/10.3389/fimmu.2014.00614]. [PMID: 25506346]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/s1471-4906(02)02302-5. [http://dx.doi.org/10.1016/S1471-4906(02)02302-5]. [PMID: 12401408]. [DOI] [PubMed] [Google Scholar]
- 5.Benoit M., Desnues B., Mege J.L. Macrophage polarization in bacterial infections. J. Immunol. 2008;181(6):3733–3739. doi: 10.4049/jimmunol.181.6.3733. [http://dx.doi.org/10.4049/jimmunol.181.6.3733]. [PMID: 18768823]. [DOI] [PubMed] [Google Scholar]
- 6.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14(7):399–416. doi: 10.1038/nrclinonc.2016.217. [http://dx.doi.org/10.1038/nrclinonc.2016.217]. [PMID: 28117416]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho S.H., So G.M., Chow K.L. Postembryonic expression of Caenorhabditis elegans mab-21 and its requirement in sensory ray differentiation. Dev. Dyn. 2001;221(4):422–430. doi: 10.1002/dvdy.1161. [http://dx.doi.org/10.1002/dvdy.1161]. [PMID: 11500979]. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q., Tian Y., Zhao X., Jing H., Xie Q., Li P., Li D., Yan D., Zhu X. NMAAP1 expressed in BCG-activated macrophage promotes M1 macrophage polarization. Mol. Cells. 2015;38(10):886–894. doi: 10.14348/molcells.2015.0125. [http://dx.doi.org/10.14348/molcells.2015.0125]. [PMID: 26429502]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baxevanis C.N., Perez S.A. Cancer dormancy: A regulatory role for endogenous immunity in establishing and maintaining the tumor dormant state. Vaccines (Basel) 2015;3(3):597–619. doi: 10.3390/vaccines3030597. [http://dx.doi.org/10.3390/vaccines3030597]. [PMID: 26350597]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang B.N., Ahmad A.S., Saleem S., Patterson R.L., Hester L., Doré S., Snyder S.H. Death-associated protein kinase-mediated cell death modulated by interaction with DANGER. J. Neurosci. 2010;30(1):93–98. doi: 10.1523/JNEUROSCI.3974-09.2010. [http://dx.doi.org/10.1523/JNEUROSCI.3974-09.2010]. [PMID: 20053891]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roest G., La Rovere R.M., Bultynck G. IP 3 receptor properties and function at membrane contact sites. Adv. Exp. Med. Biol. 2017;981:149–178. doi: 10.1007/978-3-319-55858-5_7. [https://doi.org/10.1007/978-3-319-55858-5_7]. [PMID: 29594861]. [DOI] [PubMed] [Google Scholar]
- 12.Thillaiappan N.B., Chakraborty P., Hasan G., Taylor C.W. IP3 receptors and Ca2+ entry. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866(7):1092–1100. doi: 10.1016/j.bbamcr.2018.11.007. [https://doi.org/10.1016/j.bbamcr.2018.11.007]. [PMID: 30448464]. [DOI] [PubMed] [Google Scholar]
- 13.Nakanishi A., Hatano N., Fujiwara Y., Sha’ri A., Takabatake S., Akano H., Kanayama N., Magari M., Nozaki N., Tokumitsu H. AMP-activated protein kinase-mediated feedback phosphorylation controls the Ca2+/calmodulin (CaM) dependence of Ca2+/CaM-dependent protein kinase kinase β. J. Biol. Chem. 2017;292(48):19804–19813. doi: 10.1074/jbc.M117.805085. [http://dx.doi.org/10.1074/jbc.M117.805085]. [PMID: 28974582]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wayman G.A., Lee Y.S., Tokumitsu H., Silva A.J., Soderling T.R. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59(6):914–931. doi: 10.1016/j.neuron.2008.08.021. [http://dx.doi.org/10.1016/j.neuron.2008.08.021]. [PMID: 18817731]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayush O., Jin Z.W., Kim H.K., Shin Y.R. Im, S.Y.; Lee, H.K. Glutamine up-regulates MAPK phosphatase-1 induction via activation of Ca2+→ ERK cascade pathway. Biochem. Biophys. Rep. 2016;7:10–19. doi: 10.1016/j.bbrep.2016.05.011. [http://dx.doi.org/10.1016/j.bbrep.2016.05.011]. [PMID: 28955885]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1(1):11–21. doi: 10.1038/35036035. [http://dx.doi.org/10.1038/35036035]. [PMID: 11413485]. [DOI] [PubMed] [Google Scholar]
- 17.Zhivotovsky B., Orrenius S. Calcium and cell death mechanisms: a perspective from the cell death community. Cell Calcium. 2011;50(3):211–221. doi: 10.1016/j.ceca.2011.03.003. [http://dx.doi.org/10.1016/j.ceca.2011.03.003]. [PMID: 21459443]. [DOI] [PubMed] [Google Scholar]
- 18.La Rovere R.M., Roest G., Bultynck G., Parys J.B. Intracellular Ca(2+) signaling and Ca(2+) microdomains in the control of cell survival, apoptosis and autophagy. Cell Calcium. 2016;60(2):74–87. doi: 10.1016/j.ceca.2016.04.005. [http://dx.doi.org/10.1016/j.ceca.2016.04.005]. [PMID: 27157108]. [DOI] [PubMed] [Google Scholar]
- 19.Nikolaidis N., Chalkia D., Watkins D.N., Barrow R.K., Snyder S.H., van Rossum D.B., Patterson R.L. Ancient origin of the new developmental superfamily DANGER. PLoS One. 2007;2(2):e204–e204. doi: 10.1371/journal.pone.0000204. [http://dx.doi.org/10.1371/journal.pone.0000204]. [PMID: 17301879]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray P.J. Macrophage polarization. Annu. Rev. Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [http://dx.doi.org/10.1146/annurev-physiol-022516-034339]. [PMID: 27813830]. [DOI] [PubMed] [Google Scholar]
- 21.Zhou D., Huang C., Lin Z., Zhan S., Kong L., Fang C., Li J. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell. Signal. 2014;26(2):192–197. doi: 10.1016/j.cellsig.2013.11.004. [http://dx.doi.org/10.1016/j.cellsig.2013.11.004]. [PMID: 24219909]. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Lu Y., Han F., Chang Y., Li X., Han Z., Xue M., Cheng Y., Sun B., Chen L. Saxagliptin regulates M1/M2 macrophage polarization via CaMKKβ/AMPK pathway to attenuate NAFLD. Biochem. Biophys. Res. Commun. 2018;503(3):1618–1624. doi: 10.1016/j.bbrc.2018.07.090. [http://dx.doi.org/10.1016/j.bbrc.2018.07.090]. [PMID: 30060948]. [DOI] [PubMed] [Google Scholar]
- 23.van Rossum D.B., Patterson R.L., Cheung K.H., Barrow R.K., Syrovatkina V., Gessell G.S., Burkholder S.G., Watkins D.N., Foskett J.K., Snyder S.H. DANGER, a novel regulatory protein of inositol 1,4,5-trisphosphate-receptor activity. J. Biol. Chem. 2006;281(48):37111–37116. doi: 10.1074/jbc.M608760200. [http://dx.doi.org/10.1074/jbc.M608760200]. [PMID: 16990268]. [DOI] [PubMed] [Google Scholar]
- 24.Bandyopadhyay B.C., Ong H.L., Lockwich T.P., Liu X., Paria B.C., Singh B.B., Ambudkar I.S. TRPC3 controls agonist-stimulated intracellular Ca2+ release by mediating the interaction between inositol 1,4,5-trisphosphate receptor and RACK1. J. Biol. Chem. 2008;283(47):32821–32830. doi: 10.1074/jbc.M805382200. [http://dx.doi.org/10.1074/jbc.M805382200]. [PMID: 18755685]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasri N.N., Bultynck G., Smyth J., Szlufcik K., Parys J.B., Callewaert G., Missiaen L., Fissore R.A., Mikoshiba K., de Smedt H. The N-terminal Ca2+-independent calmodulin-binding site on the inositol 1,4,5-trisphosphate receptor is responsible for calmodulin inhibition, even though this inhibition requires Ca2+. Mol. Pharmacol. 2004;66(2):276–284. doi: 10.1124/mol.66.2.276. [http://dx.doi.org/10.1124/mol.66.2.276]. [PMID: 15266018]. [DOI] [PubMed] [Google Scholar]
- 26.Taylor C.W., Prole D.L. Ca(2+) signalling by IP(3) receptors. Subcell. Biochem. 2012;59:1–34. doi: 10.1007/978-94-007-3015-1_1. [http://dx.doi.org/10.1007/978-94-007-3015-1_1]. [PMID: 22374086]. [DOI] [PubMed] [Google Scholar]
- 27.Celada A., Schreiber R.D. Role of protein kinase C and intracellular calcium mobilization in the induction of macrophage tumoricidal activity by interferon-gamma. J. Immunol. 1986;137(7):2373–2379. [PMID: 3093574]. [PubMed] [Google Scholar]
- 28.Leon C.M., Barbosa C.M., Justo G.Z., Borelli P., Resende J.D., Jr, de Oliveira J.S., Ferreira A.T., Paredes-Gamero E.J. Requirement for PLCγ2 in IL-3 and GM-CSF-stimulated MEK/ERK phosphorylation in murine and human hematopoietic stem/progenitor cells. J. Cell. Physiol. 2011;226(7):1780–1792. doi: 10.1002/jcp.22507. [http://dx.doi.org/10.1002/jcp.22507]. [PMID: 21506110]. [DOI] [PubMed] [Google Scholar]
- 29.Mohanraj M., Sekar P., Liou H.H., Chang S.F., Lin W.W. The mycobacterial adjuvant analogue TDB Attenuates neuro-inflammation via mincle-independent PLC-γ1/PKC/ERK signaling and microglial polarization. Mol. Neurobiol. 2019;56(2):1167–1187. doi: 10.1007/s12035-018-1135-4. [http://dx.doi.org/10.1007/s12035-018-1135-4]. [PMID: 29876879]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


