Summary
Although developmental mechanisms driving an increase in brain size during vertebrate evolution are actively studied, we know less about evolutionary strategies allowing accelerated brain growth. In zebrafish and other vertebrates studied to date, apical radial glia (RG) constitute the primary neurogenic progenitor population throughout life [1]; thus, RG activity is a determining factor of growth speed. Here, we ask whether enhanced RG activity is the mechanism selected to drive explosive growth, in adaptation to an ephemeral habitat. In post-hatching larvae of the turquoise killifish, which display drastic developmental acceleration, we show that the dorsal telencephalon (pallium) grows three times faster than in zebrafish. Rather than resulting from enhanced RG activity, we demonstrate that pallial growth is the product of a second type of progenitors (that we term NGPs for non-glial progenitors) that actively sustains neurogenesis and germinal zone self-renewal. Intriguingly, NGPs appear to retain, at larval stages, features of early embryonic progenitors. In parallel, RGs enter premature quiescence and express markers of astroglial function. Altogether, we propose that mosaic heterochrony within the neural progenitor population might permit rapid pallial growth by safeguarding both continued neurogenesis and astroglial function.
Keywords: neurogenesis, juvenile, killifish, zebrafish, radial glia, neural progenitor, quiescence, heterochrony, brain size, Notch
Graphical Abstract
Highlights
-
•
Two types of apical progenitors exist in the pallium of the fast-growing killifish
-
•
Killifish pallial RGs enter precociously into an adult-like quiescent state
-
•
NGPs, both self-renewing and neurogenic, resemble early neuroepithelial progenitors
-
•
Mosaic heterochrony among progenitors sustains rapid killifish pallial growth
Coolen et al. unravel in a fast-growing killifish a striking mosaic heterochrony within the pallial germinal zone, where progenitors at different maturation stages cohabit: radial glia, which enter precociously in an adult-like Notch-dependent quiescence and non-glial progenitors, which resemble early embryonic progenitors and ensure neurogenesis.
Results
The Killifish as A Novel Model to Study Progenitor Dynamics Driving Accelerated Brain Growth
The general scheme of neurogenesis underlying the growth of the dorsal part of the vertebrate telencephalon, or pallium, is well conserved between species studied to date. Early during embryonic development, neuroepithelial cells convert to apical radial glia (RG) cells, the primary progenitors that will generate the large bulk of neurons and glial cells [1]. Nevertheless, the pallium varies substantially in size and organization between vertebrate species, an evolutionary plasticity most likely linked to the crucial adaptive functions of high integrative centers hosted by this brain region. Many studies have focused on developmental mechanisms contributing to the expansion of the pallium, at the origin of the mammalian cortex diversification [2, 3]. They notably show that the emergence of basally located intermediate progenitors, which amplify neuronal production downstream of RGs, contributed to pallium enlargement [3]. In contrast, however, we know little about developmental mechanisms that would allow speeding up pallial development. Teleost fish are advantageous for comparisons of pallial growth mechanisms, because they exhibit an overall similar pallial morphology between species. We and others have extensively characterized neural progenitor lineages underlying pallial construction in zebrafish [4, 5, 6]. In search of a comparative model with rapid pallial growth, we turned to the African turquoise killifish Nothobranchius furzeri (N. furzeri). N. furzeri evolved an extremely fast post-hatching larval development as an adaptation to its ephemeral savannah pool habitat, to engender a new generation before ponds dry out [7, 8]. N. furzeri body size is similar to zebrafish at sexual maturity (∼25 mm), but it reaches this stage in 5 weeks, whereas it takes 3 months to zebrafish [7].
We first located dividing neural progenitors in the killifish pallium, focusing on the first 2 weeks after hatching. We used the marker Sox2, a highly conserved neural progenitor master transcription factor, in combination with the proliferation marker proliferating cell nuclear antigen (PCNA) (Figure 1A). We observed that, like in zebrafish, Sox2+ and PCNA+ cells are confined to the apical surface, overlying differentiated neuronal cells located just underneath in the parenchyme (Figures S1A–S1C'). Thus, accelerated pallial growth in killifish does not correlate with the emergence of basal progenitors as seen in mammals. Consistent with this, eomesa (also known as tbr2), a marker of intermediate progenitors in mammals, is expressed mostly in differentiated neurons and a few Sox2− cells at the ventricle (Figures S1D–S1F). In contrast, we observed a rapid expansion of the apical compartment in killifish. With linear increase, the pallial surface thus increases 5 times over these 2 weeks (Figure S1G), therefore 3 times faster than zebrafish, whereas cellular density remains rather constant (Figure S1H). Unique adaptations must therefore mediate accelerated neurogenesis and pallial growth in the fast-growing killifish.
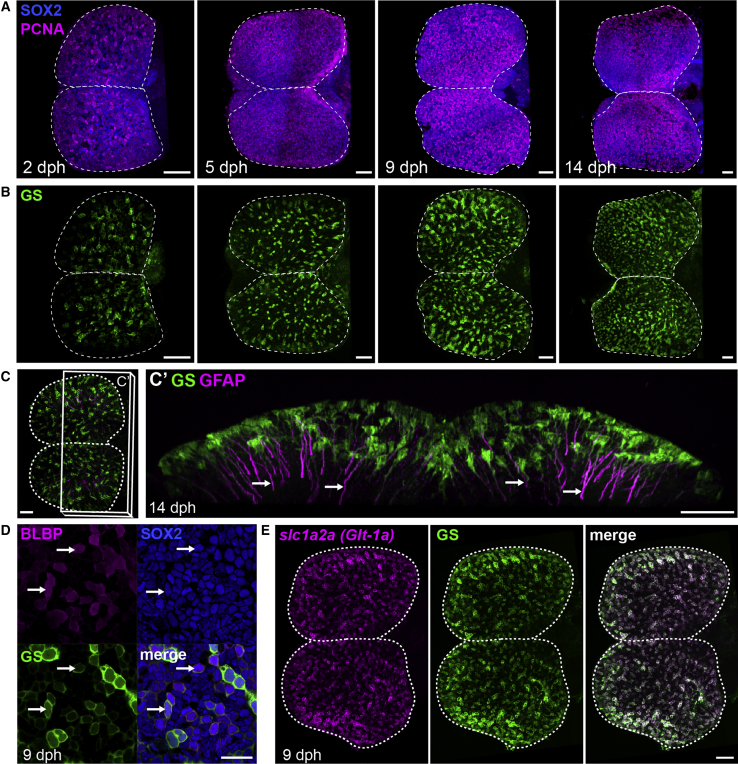
Figure 1.
Apical Progenitors at the Pallial Surface of the Killifish Larval Pallium
(A) Dorsal 3D views of killifish pallium (anterior left) at 2, 5, 9, and 14 days post-hatching (dph) with a whole-mount immunostaining for Sox2 (blue) and PCNA (magenta) highlighting neural progenitors. A dotted line contours the two pallial hemispheres.
(B) Dorsal 3D views of the same brains as in (A), showing immunostaining signal for GS (green) to identify RGs.
(C) Immunostaining for GS (green) and GFAP (magenta) at 14 dph indicating GFAP-enriched processes (arrows). Shown in (C) is a dorsal 3D view, and in (C’) is a frontal view of the 3D reconstruction with a transverse hemisection along the plane shown in (C).
(D) Triple immunostaining for BLBP, Sox2, and GS at 9 dph. Images show high magnifications of the pallial surface on a single optical z-plane. Arrows point to RG cells (GS+BLBP+Sox2+). Scale bar, 20 μm.
(E) ISH for slc1a2a (encoding an ortholog of glutamate transporter GLT-1, magenta) combined with GS immunostaining at 9 dph. Images are dorsal views of a 3D reconstruction. Scale bar, 50 μm.
See also Figure S1.
Non-glial Apical Progenitors Make the Majority of the Killifish Pallial Germinal Zone
To characterize further killifish pallial progenitors, we used the RG marker glutamine synthetase (GS). We detected the presence of RG among Sox2+ cells at all stages studied (Figure 1B). GS+ cells display a typical vertebrate RG morphology in which cell bodies line the ventricle and long basal processes are enriched in the intermediate filament protein glial fibrillary acid protein (GFAP) (Figures 1C and 1C’). As in other vertebrate species, we show that killifish pallial RGs express, in addition, a large set of astroglial-specific genes, also shared by adult neural stem cells and differentiated astrocytes in mammals [9, 10]. These include genes encoding glutamate transporters (Slc1a2a [also known as Glt-1a] and Slc1a3a [also known as Glasta]), brain lipid binding protein (BLBP), calcium-binding protein (S100b), and Aldh1l1 (Figures 1D, 1E, and S1I–S1K).
Surprisingly, however, RGs do not account for the majority of Sox2+ cells and instead distribute sparsely in small clusters (Figures 2A–2A″). RG cell clusters are intermingled with a major pool of GS−Sox2+ progenitors, which we thereafter refer to as non-glial progenitors (NGPs). This is in remarkable contrast with zebrafish, in which RG cells are the major constituents of the pallial ventricular surface and less than 10% of Sox2+ cells appear devoid of glial marker expression (Figures S2A–S2C). Both RG and NGPs express, in addition to Sox2, the primary neural progenitor markers Musashi1 (Msi1) and Nestin (Figures S2D and S2E). Immunostaining against the apical tight-junction protein zona occludens-1 (ZO-1) demonstrated that both RG and NGPs possess an apical membrane domain, together making up the epithelial surface of the pallium (Figure S2F). Staining for phospho-histone 3 (pH3) indicates that NGPs enter mitosis at the apical surface (Figure 2A). NGPs thus qualify as bona fide apical neural progenitors.
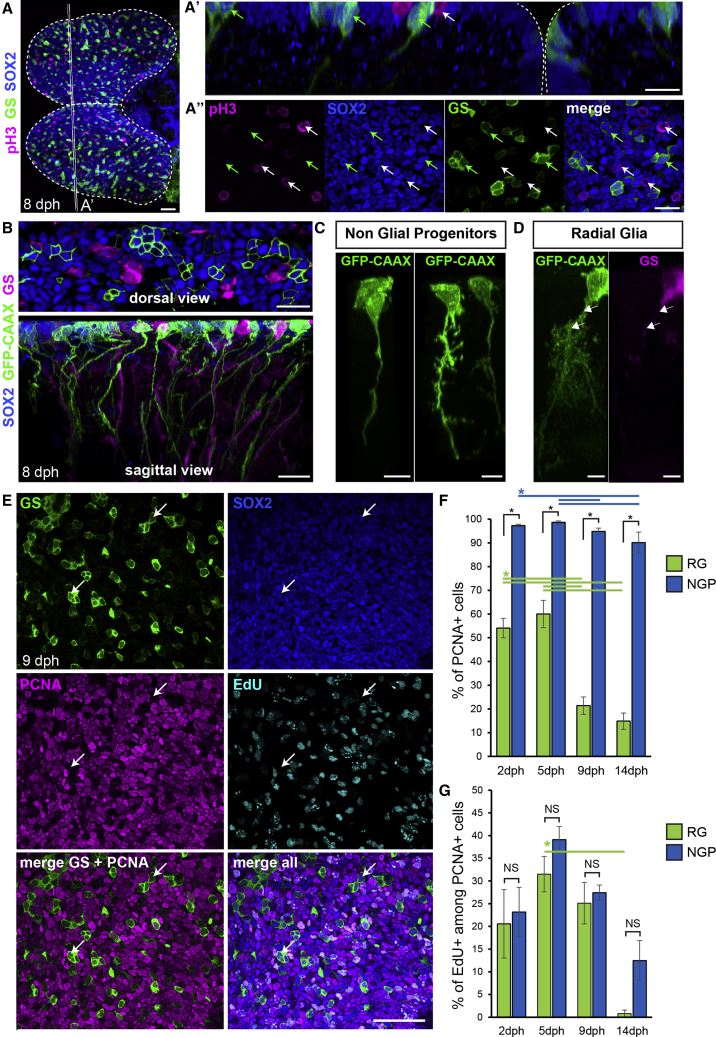
Figure 2.
A Population of Apical Non-glial Progenitors Accounts for the Majority of Proliferating Progenitors in the Killifish Pallium
(A) Immunostaining for GS (green), Sox2 (blue), and pH3 (magenta) at 8 dph. Shown is a dorsal 3D view. Shown in (A’) is high magnification of a 5 μm transverse section through the 3D reconstruction. Shown in (A″) is high magnification of a single optical z-plane. White arrows point to non-glial NGP progenitors entering mitosis at the apical surface and green arrows point to sparsely distributed RGs.
(B) Shown at the top is the optical z-plane showing NGP electroporated with a GFP-CAAX-encoding plasmid (green), together with an immunostaining for GS (magenta) and Sox2 (blue). On the bottom is a sagittal view through a 3D reconstruction, highlighting the presence of a basal process on electroporated NGPs (green) and GS+ RGs (magenta).
(C) Examples of NGP cell morphology.
(D) Typical RG morphology, showing the ramified pattern of the basal process. Individual cells were manually detoured on a 3D reconstruction to visualize their morphologies.
(E) Optical z-plane showing immunostaining for GS (green), Sox2 (blue) and PCNA (magenta), and EdU detection (cyan), on the high magnification of the pallial surface at 9 dph. White arrows point to non-cycling RG cells.
(F) Proportion of PCNA+ cells among RGs (green bars) and NGPs (blue bars) at 2, 5, 9, and 14 dph.
(G) Proportion of EdU+ cells among PCNA+ cells among RGs (green bars) and NGPs (blue bars).
∗Corrected p value < 0.05; two-way ANOVA, followed by pairwise comparisons using Holm’s procedure. Data were rank transformed prior to analysis. Data are represented as mean ± SEM; n = 6, 5, 5, and 3 for 2, 5, 9, and 14 dph, respectively. Scale bars, 50 μm in (A) and (E); 20 μm in (A’), (A’’), and (B); and 10 μm in (C) and (D). See also Figure S2.
To unravel the morphology of NGPs, we electroporated a plasmid encoding a membrane-bound green fluorescent protein (GFP) (GFP-CAAX) into the pallial ventricular zone. Viewing Sox2+ NGPs from the dorsal (ventricular) side confirmed that they possess an apical membrane domain (Figure 2B, top). Unexpectedly, we also found that NGPs, like GS+ RGs, possess a long basal process (Figure 2B, bottom). However, we noticed that NGP processes appear rather straight with only a few short branches (Figures 2B and 2C). In contrast, RG processes show a much more ramified pattern (Figure 2D), reminiscent of protoplasmic astrocytes, as previously noticed for zebrafish adult RGs [11]. Thus, two types of radial apical progenitors compose the ventricular surface of the killifish pallium: prototypical RGs, expressing a full set of astroglial markers and NGPs, negative for astroglial markers and with a more immature morphology.
NGPs Account for the Majority of Proliferating Progenitors in the Killifish Pallium
Pallial growth in zebrafish is driven by the continued neurogenic capacity of RGs [5, 6], which present a sustained cycling activity during larval and juvenile stages (Figures S2G and S2H). Accelerated pallial growth in killifish might therefore result from intensified RG cycling activity. Alternatively, it might also involve the recruitment of additional neurogenic lineages. To test the first hypothesis, we analyzed the proliferation rates of RGs and NGPs in the killifish pallium along the first 2 weeks after hatching by using the PCNA marker (Figure 2E). Larvae were subjected, in addition, to a 4 h 5‐ethynyl‐2′‐deoxyuridine (EdU) pulse prior to fixation to label cells in synthesis (S) phase. Our quantifications reveal that NGPs and RGs differ significantly in their temporal proliferation profile (Figure 2F). The majority of NGPs are labeled by PCNA for the whole period; only a minor decrease of proliferation is observed at late time points. RG cells are already less proliferative than NGPs at 2 days post-hatching (dph) (54.1% ± 4.1% versus 97.3% ± 0.6% of PCNA+ RGs versus NGPs, respectively; p < 0.001). In addition, RG proliferation rate drops dramatically after 9 dph, reaching very low levels at 14 dph (14.9% ± 3.4%). Among PCNA+ cells, ratios of EdU-labeled cells tend to decrease over time in both NGPs and RGs without revealing any statistical differences between the two types of progenitors (Figure 2G). This suggests a parallel trend toward increasing length of growth phases of the cell cycle as pallial development progresses. We additionally analyzed the proliferative activity of pallial progenitors at the adult stage (10 weeks after hatching) (Figures S2I and S2J). At this stage, RGs are rarely found PCNA+ (0.7% ± 0.2%), whereas NGPs are still proliferating, albeit at a lower rate than during larval growth (24.2% ± 2.3%). Altogether, these data indicate that NGPs cycle actively and continuously even up to adulthood, whereas RGs markedly reduce their proliferative activity already during the early post-hatching period.
RG Enter Premature Notch-Dependent Quiescence in Killifish
Reduction of RG proliferation in killifish juveniles is reminiscent of the adult stage in the zebrafish pallium, when RGs transition to a quiescent state, only occasionally activating to cycle [12]. Alternatively, it could correspond to a definitive cell cycle exit for terminal glial differentiation. To decipher between these hypotheses, we first analyzed the expression of highly conserved molecular players of the RG quiescence-to-activation cycle. Zebrafish adult RG cells are maintained in quiescence by the specific activation of one Notch receptor ortholog, Notch3 [12, 13, 14]. A comparable function of Notch3-mediated signaling has been demonstrated in adult mouse neural stem cells [15, 16]. Upon exit of quiescence, in both mice and zebrafish, activated cells induce the expression of the pro-neural gene ascl1 [14, 17]. We therefore analyzed the expression pattern of killifish orthologs of notch3, ascl1 (ascl1a and ascl1b), and of the canonical Notch target her4.2 (ortholog of Hes5 in mammals). All these genes are expressed at the pallial ventricle in a salt-and-pepper fashion (Figures S3A–S3C). Using fluorescence in situ hybridization (FISH), we compared their transcript distribution to the localization of GS+ RGs. We observed that notch3 expression is confined to RGs (Figure 3A). her4.2 expression is highly enriched in RGs, although a lower signal is detected in some NGPs. Finally, only a few scattered RGs express ascl1 orthologs. These expression profiles are thus compatible with killifish RGs being subjected to Notch3-dependent quiescence.
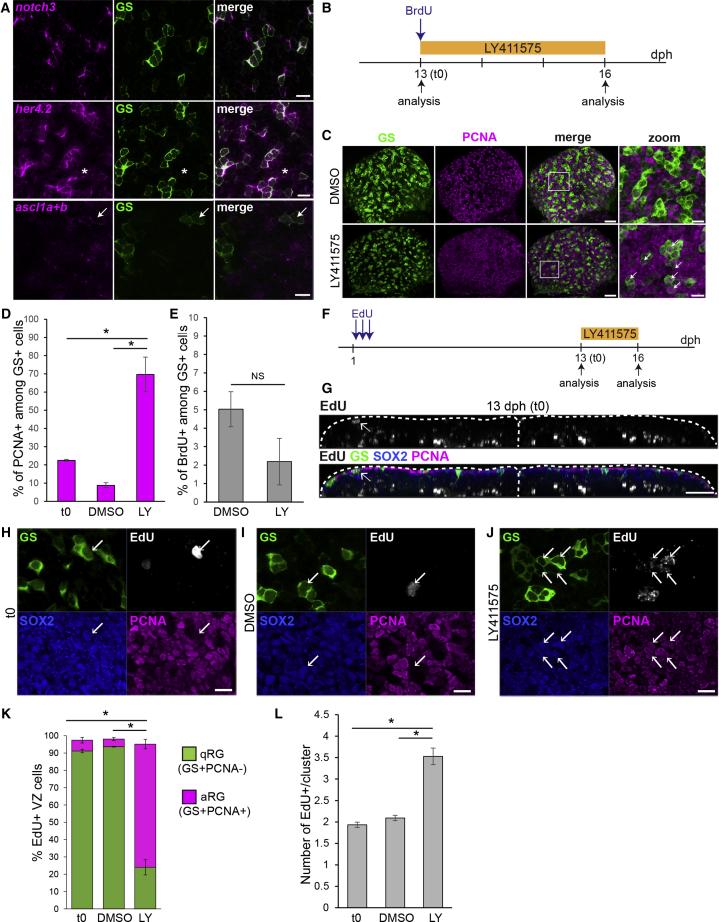
Figure 3.
Killifish RG Enter in a Notch3-Dependent Quiescent State
(A) ISH for notch3, her4.2, and ascl1 orthologs (ascl1a + ascl1b) (magenta) combined with GS immunostaining (green) at 8 dph. Images are high magnifications of the pallial surface on a single optical z-plane. Asterisks indicate NGP cells expressing low amounts of her4.2. White arrows point to RG cells expressing ascl1a and/or ascl1b. Scale bar, 10 μm.
(B) Experimental design for the data shown in (C)–(E).
(C) Double immunostaining for GS (green) and PCNA (magenta) in DMSO control (top) and LY-treated (bottom) brains, illustrating the increase in PCNA+ RG cells (white arrows). Images on the right are high magnifications of the pallial surface. Scale bars, 30 μm (left) and 10 μm (right).
(D) Proportion of PCNA+ cells among GS+ RGs before and after DMSO or LY treatment.
(E) Proportion of BrdU-labeled cells among RG after DMSO or LY treatment
(F) Experimental design for the data shown in (G)–(L).
(G) Immunostaining for GS (green), Sox2 (blue) and PCNA (magenta), and EdU detection (gray) at the end of the chase (13 dph, t0). A 2 μm transverse section through a 3D reconstruction is shown. Arrows point to EdU+ RG cells at the ventricle.
(H–J) Examples of EdU+ RG clusters observed at t0 (H), in control conditions (I), and after LY treatment (J). Optical z-planes showing immunostaining for GS (green), Sox2 (blue) and PCNA (magenta), and EdU detection (cyan) on the high magnification of the pallial surface are shown. Arrows point to EdU+ RG. Scale bar, 10 μm.
(K) Proportion of EdU-labeled cells that are quiescent RGs (qRGs) (green, Sox2+GS+PCNA−) or activated RGs (aRGs) (purple, Sox2+GS+PCNA+). Only EdU+ cells located close to the ventricular zone (VZ) were considered for this analysis.
(L) Average number of EdU+ cells inside each cluster of EdU+ cells.
∗Corrected p value < 0.05; one-way ANOVA, followed by pairwise comparisons using Holm’s procedure. Data were rank transformed prior to analysis. Data are represented as mean ± SEM; n = 3 for each treatment condition. See also Figure S3.
In zebrafish, application of the gamma-secretase inhibitor LY411575 (LY) in the fish water efficiently blocks Notch activity and leads to a massive cell cycle re-entry of quiescent RG cells [13]. We thus tested whether LY would trigger a similar effect on killifish juvenile RG cells. We first verified that LY effectively inhibits the expression of the Notch target her4.2 (Figure S3D). Notably, we observed that her4.2 downregulation is paralleled by an increased expression of ascl1 orthologs, already suggesting an activation of RG cells (Figure S3E). To further probe this, we treated 13-dph larvae for 3 days with LY (or dimethylsulfoxide [DMSO] as a control) and quantified the proportion of PCNA+ RG cells (Figure 3B). In control brains, the proportion of PCNA+ cells stays very low, in line with the low cycling activity of RG cells at this stage (Figures 3C and 3D). Under Notch blockade, in contrast, the proportion of PCNA+ RGs drastically increases (DMSO: 8.8% ± 2.5%; LY: 69.7% ± 9.6% SEM; p = 0.0027). In this experiment, we also applied a pulse of 5-bromo-2'-deoxyuridine (BrdU) prior to treatment (Figure 3B). In both control and LY treatment conditions, most RG cells were devoid of BrdU staining after the chase (Figure 3E). This demonstrates that RGs that activate upon LY treatment do not derive from previously cycling cells. Rather, they re-entered the cell cycle upon Notch blockade.
Additionally, we observed an increase in the number of RGs at the ventricular surface upon Notch blockade (Figure S3F), suggesting that activated RG cells might undergo self-amplifying symmetric divisions under these conditions. To test this hypothesis and further probe the fate of re-activated RGs, we designed an experiment to specifically label and track RG cells. We applied repeated EdU pulses at early stages after hatching, from 1 dph to 2 dph, followed by a long chase period until 13 dph (Figure 3F). After this long chase, most EdU+ cells correspond to early-born differentiated neurons in the parenchyme (Figure 3G). But we also observed some EdU+ cells at the ventricular surface distributed as either single cells or small clusters of cells (Figures 3G–3H, white arrow). Most of these label-retaining cells are GS+PCNA− RGs, confirming that RGs slow down their proliferation early on during the post-hatching phase (Figure 3K). We then took advantage of this RG-specific label retention to follow RG fate upon Notch blockade. We first observed that the average number of cells per cluster significantly increases in contrast to control conditions (Figure 3L). However, EdU+ cell clusters are still almost exclusively composed of RGs, although these are now found proliferative (Figure 3K). This experiment thus confirms that long-lasting quiescent RGs, set aside at early stages, can re-enter the cell cycle upon Notch blockade. Further, it shows that, in these conditions, RGs undergo mostly self-amplifying symmetric gliogenic divisions. We conclude that killifish RG cells enter a Notch-dependent quiescent state, similarly to adult zebrafish RGs. However, they do so surprisingly precociously, in a context in which the pallium is still rapidly growing, and their reactivation is gliogenic.
NGPs Account for the Bulk of Neurogenesis and Pallial Growth in Killifish
Given that RGs enter into quiescence early, neuronal production in the killifish pallium likely relies mostly on their actively dividing neighbors, NGPs. We therefore examined NGP fate potential and self-renewing capacity, both in control conditions and upon Notch blockade. We specifically traced their progeny by applying a pulse of EdU at 13 dph (Figure 4A). At this stage, because of their high proliferation rate in relation to RGs, NGPs indeed constitute the majority of EdU-incorporating cells (Figures S4A and S4B). After the EdU pulse, fish were incubated for 3 days in DMSO or LY, and after a second pulse with BrdU, returned to fish water for an additional period of 3 days. In control conditions, we observed, after 3 days of chase, Sox2− EdU-labeled cells located basally in the parenchyme (Figures 4B and 4C). We identify these cells as neurons. Notably, a substantial fraction of EdU-labeled cells is maintained as Sox2+ NGPs after 3 and even 6 days of chase (68.2% ± 11.9% and 31.9% ± 12.6%; Figure 4C) and still incorporates BrdU after 3 days (Figure S4C). In contrast, the number of RGs labeled by EdU is very low and constant over the whole period (Figure 4D). This tracing analysis overall shows that NGPs are both neurogenic and—at least over this experiment time frame—self-renewing. NGPs, however, do not generate RGs, suggesting that they form an independent lineage.
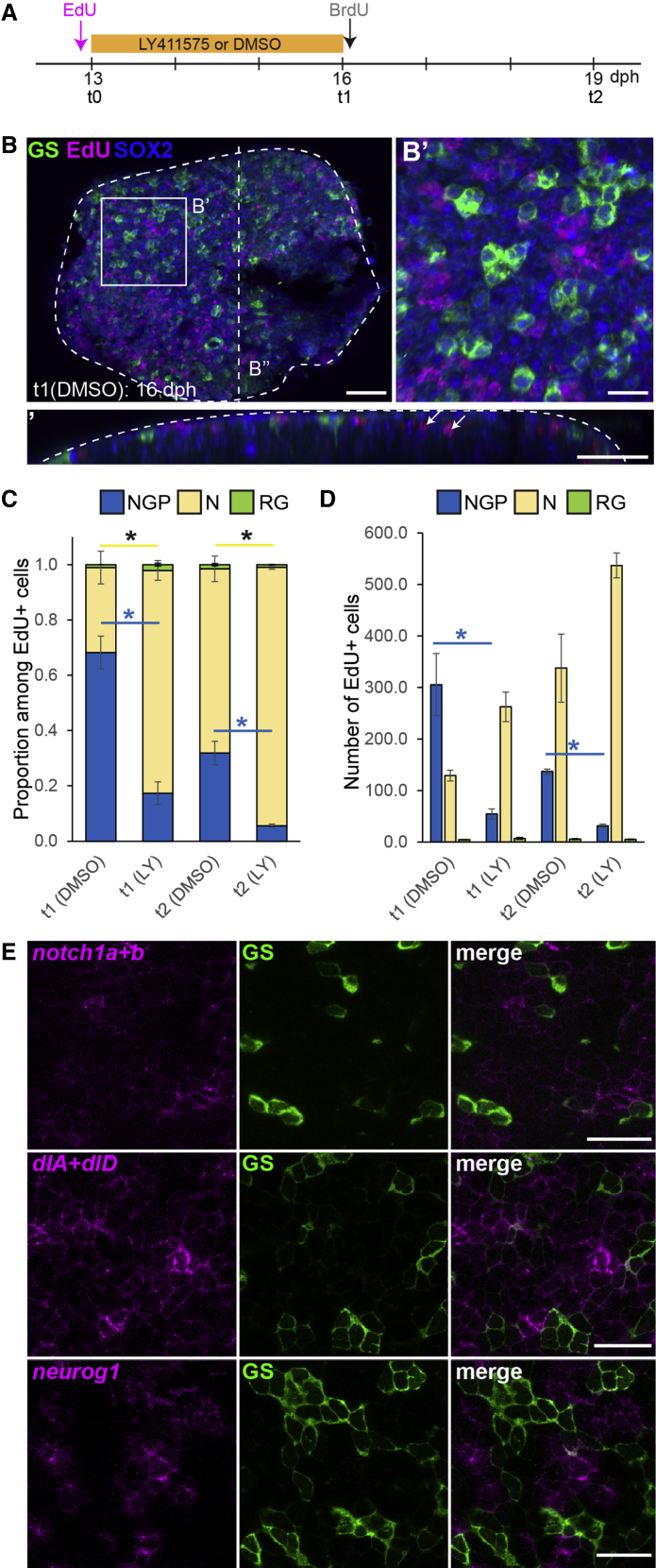
Figure 4.
NGPs Behave Like Early Embryonic Progenitors
(A) Experimental design.
(B) Immunostaining for GS (green) and Sox2 (blue) combined with EdU detection (magenta) on a DMSO control 16 dph killifish pallium. Shown in (B’) is a high magnification of the dorsal view of the 3D reconstruction shown in (B). In (B’’) is 5 μm transverse section through the 3D reconstruction shown in (B). White arrows in (B’’) point to EdU+ cells in the parenchyme that we identified as neurons. Scale bars, 50 μm in (B) and (B’’) and 20 μm in (B’).
(C) Proportion of EdU-labeled cells with NGP (blue, Sox2+GS−), neuronal (yellow, Sox2−GS−), or RG (green, Sox2+GS+) identity. ∗Corrected p value < 0.05. Data were analyzed for each cell type with a two-way ANOVA, followed by pairwise comparisons using Holm’s procedure. Proportions were arcsine transformed prior to analysis. Data are represented as mean ± SEM; n = 2 (t1) and n = 3 (t2) for each treatment condition for each treatment condition at t1 and t2, respectively.
(D) Number of EdU-labeled cells per counted area with NGP (blue, Sox2+GS−), neuronal (yellow, Sox2− and GS−) or RG (green, Sox2+GS+) identity. ∗Corrected p value < 0.05. Data were analyzed for each cell type with a two-way ANOVA, followed by pairwise comparisons using Holm’s procedure. Data was rank transformed prior to analysis. Data are represented as mean ± SEM; n = 2 (t1) and n = 3(t2) for each treatment condition.
(E) ISH for Notch1 orthologs (notch1a + notch1b), Dll1 orthologs (dlA + dlD), and neurog1 (magenta) combined with GS immunostaining (green) at 8 dph. Images are high magnifications of the pallial surface on a single optical z-plane. Scale bar, 20 μm.
See also Figure S4.
NGPs Use the Pro-neural Gene neurog1 and Rely on Notch1 Signaling for Their Maintenance, Similarly to Embryonic Neural Progenitors
In this experiment, we also observed that Notch blockade disturbs the relative proportions of EdU-traced neurons versus NGPs (Figure 4C). This is mainly due to a significant decrease in the number of EdU+ cells maintained as NGPs, paralleled by an increase in the production of EdU+ neurons (Figure 4D). Notch activity is therefore necessary to balance NGP self-renewal and differentiation. This situation is reminiscent of the prominent role of Notch signaling in vertebrate embryonic neural progenitors, a function mainly relayed by Notch1 receptors [13, 18, 19, 20]. The balance relies on antiphase oscillations in neighboring progenitors of Hes-related genes on the one hand versus Notch-ligands and pro-neural genes on the other hand [21]. Consistently, we also show that NGPs express notch1 orthologs (notch1a and notch1b) (Figure 4E). Additionally, we observed among NGPs a salt-and-pepper pattern of expression of the Hes-related gene her4.2 (Figure 3A), genes encoding Dll1-related Notch ligands (dla and dld), and the pro-neural gene neurog1 (Figures 4E and S4D). Of note, the expression of neurog1 in NGPs contrasts with RGs, which use ascl1 orthologs as pro-neural factors when activated (Figure S3B), but also with zebrafish, in which neurog1 expression is restricted to early embryonic stages. Although neurog1 expression in the early embryonic zebrafish pallium has been previously reported [22, 23], we did not detect any at post-embryonic stages (Figure S4E). Our results therefore indicate that neurogenic NGPs rely on Notch1-mediated signaling for their self-renewal and show characteristics of early-embryonic neurogenic progenitors that would be retained at larval stages.
Discussion
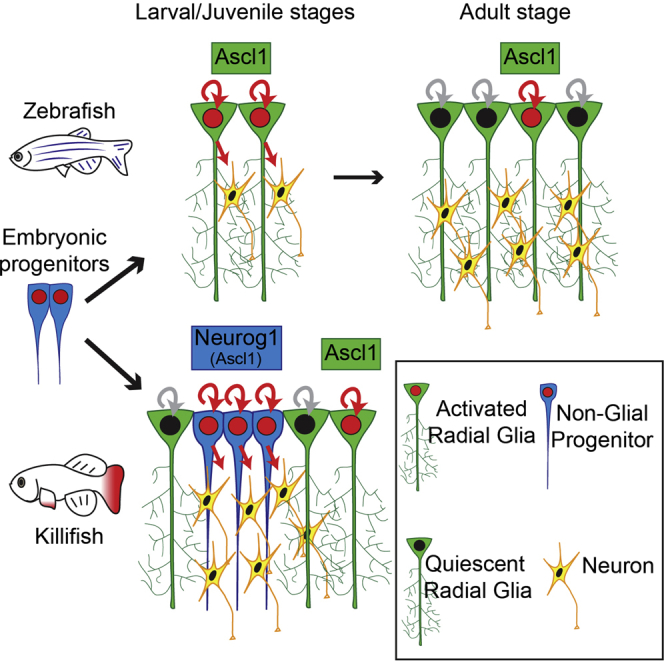
Altogether, our study unravels a striking mosaic heterochrony within the killifish pallial germinal zone, where progenitors at different stages of maturation cohabit. On the one hand, RG cells enter precociously into an adult-like Notch3-dependent quiescence; on the other hand, NGPs—characterized by a more immature morphology, by the expression of primary progenitor markers (Sox2, Nestin, and Msi1), and by their reliance on the notch1 and neurog1 interplay—resemble early neuroepithelial embryonic progenitors. This contrasts with zebrafish, in which the pallial surface is composed mainly of RGs acting as neural progenitors from embryonic to adult stages. The zebrafish situation corresponds likely to the ancestral condition in teleosts. Indeed, we observed GS+ RG cells covering the whole pallial surface in medaka fish (Figure S4F), which are more closely related to killifish (75 my divergence [24]). We propose, therefore, that the presence of an NGP lineage is a killifish adaptation accommodating the explosive growth imposed by its ephemeral habitat.
We show that both NGPs and RGs have an apical contact and a basal process; this raises the question of how these two types of progenitors can achieve such different temporal behaviors while residing in the same local niche. This must rely on a differential sensitivity or accessibility to temporal cues, but the underlying mechanisms remain to be assessed. Our results also indicate that in killifish, in contrast to other vertebrates, only a fraction of pallial progenitors switch on the astroglial program. What licenses this mosaic induction remains to be further investigated. But given that the dual composition of the pallial germinal zone is already present at the pre-hatching diapause III stage (not shown), e.g., prior to the rapid growth phase, it is not a mere consequence of an intensification of growth-promoting signals. Rather, an early change in the encoded neural progenitor program must have occurred in the killifish lineage.
The killifish is a newly co-opted vertebrate model for aging studies, because of its exceptionally short lifespan [25, 26]. Recent studies are beginning to explore causal factors of its accelerated aging, including in the brain [27, 28, 29, 30, 31]. The fast aging in N. furzeri is hypothesized by some authors to be a trade-off to rapid larval and juvenile growth, but this lead remains mechanistically unexplored [32]. Killifish are not paedomorphic and are considered to simply globally speed up body development [33]. However, we demonstrate here that developmental timing at the individual organ level is actually affected. This might have profound implications for pallial structure and physiology. Indeed, in both vertebrate and invertebrate models, neural progenitors have been shown to sequentially produce distinct neuronal subtypes during development [34]. The maintenance of embryonic-like NGPs in killifish could therefore have major consequences on the neuronal cell types composing the pallium. Importantly, our study also implies that the reduction in neurogenic activity observed in killifish upon aging [29] is because of a reduced proliferation of NGPs rather than RGs, because these are the main proliferating progenitors we detected in adult samples. In addition, the sparse distribution of RGs at the pallial surface implies a reduced astroglia to neurons ratio. Considering the prominent role of astroglial cells in neuronal function, this change could deeply affect brain physiology [35] but also contribute to brain aging phenotypes [36].
Interspecies alterations in neural progenitor lineages have been observed in mammals, albeit to control brain size. In mammals, amplification and diversification of basal progenitors indeed contribute to increase pallial size. Some basal progenitors have also been observed in a shark, highlighting a recurrent point of flexibility in pallial lineage patterns [37]. Our work shows that a different strategy has been deployed in killifish to boost neuronal production. However, in both cases, neuronal production is augmented without altering RG division rate. This suggests an inherent constraint imposed to vertebrate brain development, demanding the maintenance of an unwavering RG population. This could be linked to astroglial functions fulfilled by RGs, in particular in teleost fish, in which no parenchymal astrocytes are present. We show notably that killifish RGs express FABP7 (also known as BLBP), which is critical in astrocytes to facilitate synaptogenesis [38]. In addition, RGs express genes encoding components of the synaptic glutamate recycling system (glutamine synthetase [GS] and glutamate transporters [Glast and Glt-1]). In conclusion, our work reveals substantial modifications of anterior brain development associated with increased developmental speed and highlights nodal points of evolutionary plasticity and constraints in vertebrate neural progenitor lineages.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sheep Anti-Digoxigenin Fab fragments Antibody, AP Conjugated | Sigma-Aldrich | Cat#11093274910; RRID: AB_514497 |
| Mouse monoclonal (IgG2a) anti-GS 1/1000Clone GS-6 | Millipore | Cat#MAB302; RRID: AB_2110656 |
| Mouse monoclonal (IgG1) anti-SOX2 antibody [20G5] | Abcam | Cat#ab171380; RRID: AB_2732072 |
| Mouse Monoclonal (IgG1) anti-ZO-1 Antibody (ZO1-1A12) | Thermo Fisher Scientific | Cat#33-9100; RRID: AB_2533147 |
| Mouse Monoclonal (IgG1) Zrf-1 antibody (target antigen: zebrafish GFAP) | ZIRC | Cat#zrf-1; RRID: AB_10013806 |
| Rabbit polyclonal anti-BLBP antibody | Millipore | Cat#ABN14; RRID: AB_10000325 |
| Rabbit Polyclonal anti-PCNA antibody | GeneTex | Cat#GTX124496; RRID: AB_11161916 |
| Chicken anti-GFP Antibody | Aves lab | Cat#GFP-1020; RRID: AB_10000240 |
| Mouse monoclonal (IgG2b) anti-HuC/D, Clone 16A11 | Thermo Fisher Scientific | Cat#A-21271; RRID: AB_221448 |
| Rat monoclonal anti-BrdU antibody [BU1/75 (ICR1)] | Abcam | Cat#ab6326; RRID: AB_305426 |
| Rabbit polyclonal anti-MCM2 antibody | Abcam | Cat#ab4461; RRID: AB_304470 |
| Rabbit polyclonal anti-Msi1 antibody | [39] | N/A |
| See Table S2 for a list of secondary antibodies used in this study | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LY-411575 | Sigma-Aldrich | Cat#SML0506; CAS: 209984-57-6 |
| 5-Bromo-2-deoxyuridine (BrdU) | Sigma-Aldrich | Cat#B5002; CAS: 59-14-3 |
| Antigen Retrieval (HistoVT One) | Nacalai Tesque | Cat#06380-05 |
| Critical Commercial Assays | ||
| Invitrogen™ Molecular Probes™ Click-iT™ EdU Alexa Fluor™ 647 Imaging Kit | Thermo Fisher Scientific | Cat#10063724 |
| Experimental Models: Organisms/Strains | ||
| Killifish: Nothobranchius furzeri MCZS-08/122 | [40] | N/A |
| Zebrafish: Tg(gfap:GFP)mi2001 | [41] | ZFIN: ZDB-ALT-060623-4 |
| Zebrafish: AB WT strain | N/A | ZFIN: ZDB-GENO-960809-7 |
| Oligonucleotides | ||
| See Table S1 for a list of oligonucleotides used in this study | This paper | N/A |
| Recombinant DNA | ||
| ubi:GFP-CAAX | This paper | N/A |
| pENTR5′_ubi | [42] | N/A |
| Tol2kit entry plasmids | [43] | N/A |
| pBS-Zash1a (Zebrafish ascl1a) | [44] | N/A |
| pBS-Zash1b (Zebrafish ascl1b) | [44] | N/A |
| pBS-ngn1 (Zebrafish neurog1) | [22] | N/A |
| pSCA-Nf_aldh1l1 | This paper | N/A |
| pSCA-Nf_ascl1a | This paper | N/A |
| pSCA-Nf_ascl1b | This paper | N/A |
| pSCA-Nf_dlA | This paper | N/A |
| pSCA-Nf_dlD | This paper | N/A |
| pSCA-Nf_eomesa | This paper | N/A |
| pSCA-Nf_her4.2 | This paper | N/A |
| pSCA-Nf_nestin | This paper | N/A |
| pSCA-Nf_neurog1 | This paper | N/A |
| pSCA-Nf_notch1a | This paper | N/A |
| pSCA-Nf_notch1b | This paper | N/A |
| pSCA-Nf_notch3 | This paper | N/A |
| pSCA-Nf_slc1a2a | This paper | N/A |
| pSCA-Nf_slc1a3a | This paper | N/A |
| Software and Algorithms | ||
| ZEN | Carl Zeiss |
http://www.zeiss.com/microscopy/en_us/products/microscope-software/zen.html#introduction; RRID: SCR_013672 |
| Imaris | Bitplane | http://www.bitplane.com/imaris/imaris; RRID: SCR_007370 |
| Fiji | Fiji | http://fiji.sc; RRID: SCR_002285 |
| In Vivo Stat (version 3.7.0.0) | [45] | http://invivostat.co.uk |
Lead Contact and Materials Availability
Further information and requests for resources and reagents (all probes and constructs) should be directed to and will be fulfilled by the Lead Contact, Laure Bally-Cuif (laure.bally-cuif@pasteur.fr).
Experimental Model and Subject Details
Killifish
The Nothobranchius furzeri strain MZCS‐08/122 was used for experiments [39]. Adult fish were fed with frozen red bloodworms (Chironomidae) once a day. Fish were kept in 8 L breeding tanks housing one male and two females. Breeding tanks were equipped with sand boxes from which eggs were collected once a week. Eggs were bleached using a solution of Sodium hypochlorite (78 μL of NaOCl (Sigma, 425044) diluted in 100 mL of 0.3x Danieau’s Medium). Embryos were placed on Petri dishes filled with moist coconut fiber and stored in the dark at room temperature. In these conditions, embryos arrest in diapause II. Plates were checked at least twice a week to remove dead embryos and avoid fungal contamination. To stimulate diapause II exit, plates were placed in an incubator at 26°C with a 12 h/12 h light/dark cycle until the embryos reached the diapause III stage, characterized by the “golden eyes” phenotype [40]. For hatching, a peat moss extract solution was prepared: boiling water was poured on coconut fiber, and the solution was filtered on filter paper and autoclaved. Eggs were placed in a small plastic tray and immersed in cold peat moss extract. To ensure sufficient oxygenation, oxygen tablets were broken into small pieces and added to the hatching tray. Hatched larvae were transferred to a larger box with system water and immediately fed with freshly hatched brine shrimp (Artemia) twice a day. Larvae were raised at 26°C, with a daily 50% water change, and fed twice a day with artemia. Water levels were progressively increased as fish were growing.
Zebrafish
Wild-type zebrafish from the AB line were used for the experiments. Zebrafish were maintained using standard fish-keeping protocols and staged according to [41].
Medaka
Fixed 10 dpf medaka larvae were kindly provided by Matthieu Simion (J.S. Joly laboratory, NeuroPSI, Gif-sur-Yvette).
All animal experiments were carried out in accordance with the official regulatory standards of the Department of Paris (agreement number B75-15-22 to L.B.-C. and A-75-05-25 to A.M.) and conformed to French and European ethical and animal welfare directives (project authorization from the Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation to L.B.-C.).
Method Details
Tissue fixation
Fish larvae were euthanized with tricaine on ice and fixed overnight at 4°C in a 4% solution of paraformaldehyde (PFA) in PBS. After two PBS washes, brains were dissected out in PBS and bleached with a peroxide-containing bleaching solution (5% Formamide, 0.5X SSC, 3% H202) until dark pigments are no longer visible. Brains were subsequently dehydrated in serial dilutions of methanol/PBS. Brains were stored in 100% methanol at −20°C until use.
BrdU and EdU pulse labeling
BrdU and EdU were applied in the fish swimming water for 4 h, except for the experiment presented in Figures 3F–3L were the EdU was refreshed 3 times over the course of 24 h. The final concentrations used were 1 mM for BrdU and 400 μM for EdU. After the pulse, fish were transferred to a tank with fresh fish water during chase periods.
LY411575 treatment
Stock solutions of LY411575 (LY) at 10 mM were prepared by solving 5 mg of LY in 1.05 mL DMSO and stored at −80°C until use. To block Notch signaling, LY was applied in the fish swimming water at a final concentration of 10 μM. The solution was refreshed every 24 h. Control fish were treated with the same final concentration (0.1%) of DMSO carrier.
Electroporation
Fish larvae were anesthetized with tricaine in system water. With a thin needle, a solution containing the DNA plasmid construct (concentration 2 μg/μL) was injected into the forebrain ventricle. The DNA construct was a vector encoding a membrane targeted GFP (GFP-CAAX) under the control of the zebrafish ubiquitin promoter (Mosimann et al., 2011). This construct was generated by performing a gateway LR reaction (LR II clonase, Invitrogen) using the pENTR5′_ubi [42] pME-GFP-CAAX, p3E-polyA and pDest-Tol2pA2 entry vectors (Tol2kit [43]). After intraventricular injection of DNA, three short electropulses (width 50 ms, space 500 ms) at 50 mV were immediately applied. Fish were transferred to system water for recovery and sacrificed 24 h later for immunohistochemistry.
In silico searches and primer design for cloning
For each candidate gene, the protein sequence from the zebrafish ortholog was retrieved from the NCBI and blasted against transcriptomic database of Nothobranchius furzeri (http://nfintb.leibniz-fli.de/nfintb/blast.php). Potential orthologs were selected based on E-value and reverse blast hits results. Orthology relationships were confirmed by phylogenetic analyses using available vertebrate protein sequences and a Maximum-Likehood tree reconstruction method (not shown) (http://www.phylogeny.fr/). Primers were designed using the primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi).
RNA extraction
15dph killifish brains were dissected in cold PBS and placed in an Eppendorf tube. Brain tissue was homogenized with a pestle in Trizol solution (Thermofisher 12044977, 1 mL Trizol for 50–100 mg tissue). After a 5-minute incubation at RT, chloroform was added (200 μL per mL of Trizol). The Eppendorf tube was shaken for 15 s and centrifuged at 12000 g for 15 min. The upper phase was transferred to a fresh tube containing 500 μL Isopropanol and incubated at RT for 10 min. The tube was centrifuged at 12000 g at 4°C for 10 min. After removal of supernatant, the pellet was washed with 70% EtOH and dissolved in 44 μL RNase-free water (Ambion). Genomic DNA was removed using a RQ1 RNase-free DNase treatment (Promega): 5 μL of 10x DNase buffer and 1 μL of RNase-free DNase was added to the 44 μL of RNA and the mix was incubated for 30 min at 37°C. To stop the reaction, 1 μL of 0.25 mM EDTA was added and the tube was incubated at 65°C for 10 min. RNA concentration was measured using a nanodrop and stored at −80°C.
Reverse Transcription
DNase-treated RNA was reverse transcribed with Superscript II reverse transcriptase (Invitrogen) using random hexamer primers and according to the manufacturer’s instruction. Briefly 1 μg of DNase-treated RNA was diluted in 9 μL of RNase-free water. 1 μL of hexamer random primers (100 μM stock) was added and the mix was incubated at 70°C for 5 min. Following the addition on ice of 4 μL of 5x buffer, 2 μL of DTT (0.1 M), 2 μL of dNTPs (10 mM each) and 1 μL of RNase inhibitor (Promega), the mix was incubated at 25°C for 5 min. 1 μL Superscript II enzyme was added, and reverse transcription was performed by incubation at 25°C for 10 min, at 42°C for 1 h, and at 70°C for 10 min. The cDNA was stored at −20°C and used as a template for PCR reactions.
Cloning
Nothobranchius furzeri transcripts were amplified by PCR using primers listed in table S1 and either GoTaq (Promega) or Platinum Taq polymerase (Invitrogen). Fragments were cloned using Strataclone PCR cloning kit (Agilent technologies) according to the manufacturer’s instructions. Clones sequences were verified by sequencing with universal M13 primers (GATC biotech).
RNA probes synthesis
For RNA probes synthesis, 10 μg of plasmid DNA was linearized using the appropriate enzyme, purified using the NucleoSpin Gel and PCR Clean-up kit (Machery-Nagel) and eluted in 20 μL of RNase-free water (Ambion). To generate DIG-labeled probes, 1 μg linearized plasmid was used as a template for transcription reaction using T3 or T7 RNA polymerase (Promega) and DIG-labeling mix (Roche) according to the manufacturer’s instructions. Transcription reaction was carried out for 3 h at 37°C and a final incubation with Turbo DNase I was performed to remove template DNA. Unincorporated nucleotides were removed with the ProbeQuant G-50 Micro Columns Kit (GE Healthcare). The RNA probe was stored at −80°C.
Whole-mount in situ hybridization
In situ hybridization were carried out according to standard protocols as previously described [6]. Zebrafish probes used in this study are neurog1 [22], ascl1a and ascl1b [44]. Killifish probes were generated in this study as described above (Table S1). Briefly brains were rehydrated, washed with PBST and incubated in hybridization buffer at 65°C for 3 h. Brains were then incubated overnight at 65°C with DIG-labeled RNA probes diluted at 1/100 in hybridization buffer. Brains were washed in serial dilutions of hybridization buffer/2XSSC at 65°C to remove excess probe. An immunohistochemistry with anti-DIG antibody coupled to alkaline phosphatase (Roche, 1/5000 dilution) was then performed. In situ signals were revealed either with NBT/BCIP (Roche) or Fast Red (Sigma) for fluorescent visualization.
Whole-mount IHC on brains
Brains stored in 100% MeOH were rehydrated and washed 3 times with PBST (0.1% Tween-20 in PBS). When immunostaining included a nuclear marker, an incubation in HistoVT One (Nacalai Tesque) buffer for 1 h at 65°C was performed. If the immunostaining included BrdU staining, an incubation in HCl 2N at RT for 25 min was performed. Brains were washed three times for 5 min each with PBST. The brains were incubated into Blocking Solution (5% Normal Goat Serum, 0.1% Triton X-100 in PBS) for at least 1 h at RT. Primary antibodies were diluted in blocking Solution and incubated for 24 h at 4°C. Brains were subsequently washed six times for 15 min with PBST and incubated overnight at 4°C with secondary antibodies diluted 1:1000 in Blocking Solution (see Table S2). The brains were then washed six times for 15 min in PBST. For EdU detection the Invitrogen Molecular Probes Click-iT EdU Alexa Fluor 647 Imaging Kit was used, following the manufacturer’s instructions. Brains were mounted in Vectashield solution for confocal imaging.
Image acquisition
Fluorescent images of whole-mount telencephali were acquired on confocal microscopes (LSM700 and LSM710, Zeiss), using either a 20X air objective (Plan Apochromat 20x/0.8 M27) or a 40X oil objective (Plan-Apochromat 40x/1.3 Oil M27). For large samples, a tile scan was performed and stitching was done with the ZEN software after imaging. 3D renderings were generated using the Imaris software (versions 8 and 9, Bitplane). The 3D image was cropped to feature only the pallium as our region of interest and contrast was adjusted for each channel. For Sox2 immunostaining signals, background was substracted using the “remove outliers” function of the Fiji image analysis software. For in situ hybridization experiments revealed in NBT/BCIP images were acquired using a macroscope (Zeiss).
Quantification and Statistical Analysis
3D Images were segmented manually using semi-automatic detection with the Imaris spots function followed by manual curation. For quantification of proliferation in zebrafish RG at post-embryonic stages, the whole pallium was segmented (Figure S2). For quantification of proliferating cells along killifish post-hatching development and at adult stage (Figure 2), one hemisphere per brain was segmented. For the other experiments counting was performed on a region of interest of the ventricular zone (69 × 122 μm and 170 × 240 μm for Figure 3 and 4, respectively). Data are presented as mean ± standard error of the mean (SEM). Statistical analyses were carried out using InVivoStat [45, 46] and graphical plots were generated using Microsoft Excel. The normality of responses was assessed using normality probability plots and the homogeneity of the variance was inspected on a predicted versus residual plot. When the responses deviated noticeably from either criterion, they were arcsine or rank-transformed. Data displaying an approximately Gaussian distribution of residuals and homoscedastic responses with or without transformation were analyzed using parametric tests. Overall factor effects were determined by analysis of variance (ANOVA) and pairwise comparisons were carried out with least significant difference tests (LSD). p values were adjusted for multiple comparisons according to the Holm’s procedure. All the statistical tests performed were two-tailed and their significance level was set at 5% (α = 0.05).
Data and Code Availability
This study did not generate any unique datasets or code.
Acknowledgments
We thank members of the L.B.-C. lab for their critical input, S. Bedu for providing expert zebrafish care, and Kasia Banasiak for help in zebrafish data collection. We thank Sylvie Rétaux (NeuroPSI) for her careful and critical reading of the manuscript. We thank our collaborators Beate Hoppe, Mario Baumgart, and Alessandro Cellerino from the Fritz Lipmann Institute in Jena for providing killifish samples and sharing their expertise in the killifish model. We are grateful to Sylvie Schneider-Maunoury (IBPS) for her support in the establishment of the killifish facility. We thank Matthieu Simion (NeuroPSI) for providing Medaka larval brains. M.C. is supported by INSERM. Work in the L.B.-C. lab was funded by the ANR (ANR-2012-BSV4-0004-01), Labex Revive, Centre National de la Recherche Scientifique, Ecole des Neurosciences de Paris (ENP), Institut Pasteur and the European Research Council (AdG 322936).
Author Contributions
Conceptualization, M.C. and L.B.-C.; Investigation, M.C. and M.L.; Resources, A.M., B.H., and M.B.; Supervision, M.C. and L.B.-C.; Writing – Original draft, M.C.; Writing – Review and Editing, M.C. and L.B.-C.; Funding Acquisition, L.B.-C.
Declaration of Interests
The authors declare no competing interests.
Published: January 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.12.046.
Contributor Information
Marion Coolen, Email: marion.coolen@inserm.fr.
Laure Bally-Cuif, Email: laure.bally-cuif@pasteur.fr.
Supplemental Information
References
- 1.Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lui J.H., Hansen D.V., Kriegstein A.R. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Juan Romero C., Borrell V. Coevolution of radial glial cells and the cerebral cortex. Glia. 2015;63:1303–1319. doi: 10.1002/glia.22827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Z., Yang N., Yeo S.-Y., Chitnis A., Guo S. Intralineage directional Notch signaling regulates self-renewal and differentiation of asymmetrically dividing radial glia. Neuron. 2012;74:65–78. doi: 10.1016/j.neuron.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dirian L., Galant S., Coolen M., Chen W., Bedu S., Houart C., Bally-Cuif L., Foucher I. Spatial regionalization and heterochrony in the formation of adult pallial neural stem cells. Dev. Cell. 2014;30:123–136. doi: 10.1016/j.devcel.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Furlan G., Cuccioli V., Vuillemin N., Dirian L., Muntasell A.J., Coolen M. Life-Long neurogenic activity of individual neural stem cells and continuous growth establish an outside-in architecture in the teleost pallium. Curr. Biol. 2017;27:3288–3301.e3. doi: 10.1016/j.cub.2017.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blažek R., Polačik M., Reichard M. Rapid growth, early maturation and short generation time in African annual fishes. Evodevo. 2013;4:24. doi: 10.1186/2041-9139-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrtílek M., Žák J., Pšenička M., Reichard M. Extremely rapid maturation of a wild African annual fish. Curr. Biol. 2018;28:R822–R824. doi: 10.1016/j.cub.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 9.Molofsky A.V., Deneen B. Astrocyte development: a guide for the perplexed. Glia. 2015;63:1320–1329. doi: 10.1002/glia.22836. [DOI] [PubMed] [Google Scholar]
- 10.Than-Trong E., Bally-Cuif L. Radial glia and neural progenitors in the adult zebrafish central nervous system. Glia. 2015;63:1406–1428. doi: 10.1002/glia.22856. [DOI] [PubMed] [Google Scholar]
- 11.Poskanzer K.E., Molofsky A.V. Dynamism of an astrocyte in vivo: perspectives on identity and function. Annu. Rev. Physiol. 2018;80:143–157. doi: 10.1146/annurev-physiol-021317-121125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapouton P., Skupien P., Hesl B., Coolen M., Moore J.C., Madelaine R. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alunni A., Krecsmarik M., Bosco A., Galant S., Pan L., Moens C.B., Bally-Cuif L. Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Development. 2013;140:3335–3347. doi: 10.1242/dev.095018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Than-Trong E., Ortica-Gatti S., Mella S., Nepal C., Alunni A., Bally-Cuif L. Neural stem cell quiescence and stemness are molecularly distinct outputs of the Notch3 signalling cascade in the vertebrate adult brain. Dev. Camb. Engl. 2018;145:dev161034. doi: 10.1242/dev.161034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehret F., Vogler S., Pojar S., Elliott D.A., Bradke F., Steiner B., Kempermann G. Mouse model of CADASIL reveals novel insights into Notch3 function in adult hippocampal neurogenesis. Neurobiol. Dis. 2015;75:131–141. doi: 10.1016/j.nbd.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Kawai H., Kawaguchi D., Kuebrich B.D., Kitamoto T., Yamaguchi M., Gotoh Y. Area-specific regulation of quiescent neural stem cells by Notch3 in the adult mouse subependymal zone. J. Neurosci. 2017;37:11867–11880. doi: 10.1523/JNEUROSCI.0001-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen J., Urbán N., Achimastou A., Ito A., Simic M., Ullom K., Martynoga B., Lebel M., Göritz C., Frisén J. A transcriptional mechanism integrating inputs from extracellular signals to activate hippocampal stem cells. Neuron. 2014;83:1085–1097. doi: 10.1016/j.neuron.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers C.B., Peng Y., Nguyen H., Gaiano N., Fishell G., Nye J.S. Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development. 2001;128:689–702. doi: 10.1242/dev.128.5.689. [DOI] [PubMed] [Google Scholar]
- 19.Hatakeyama J., Kageyama R. Notch1 expression is spatiotemporally correlated with neurogenesis and negatively regulated by Notch1-independent Hes genes in the developing nervous system. Cereb. Cortex. 2006;16:i132–i137. doi: 10.1093/cercor/bhj166. [DOI] [PubMed] [Google Scholar]
- 20.Basak O., Giachino C., Fiorini E., Macdonald H.R., Taylor V. Neurogenic subventricular zone stem/progenitor cells are Notch1-dependent in their active but not quiescent state. J. Neurosci. 2012;32:5654–5666. doi: 10.1523/JNEUROSCI.0455-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kageyama R., Ohtsuka T., Shimojo H., Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- 22.Blader P., Fischer N., Gradwohl G., Guillemot F., Strähle U. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development. 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- 23.Korzh V., Sleptsova I., Liao J., He J., Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev. Dyn. 1998;213:92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 24.Betancur-R R., Broughton R.E., Wiley E.O., Carpenter K., López J.A., Li C., Holcroft N.I., Arcila D., Sanciangco M., Cureton Ii J.C. The tree of life and a new classification of bony fishes. PLoS Curr. 2013 doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cellerino A., Valenzano D.R., Reichard M. From the bush to the bench: the annual Nothobranchius fishes as a new model system in biology. Biol. Rev. Camb. Philos. Soc. 2016;91:511–533. doi: 10.1111/brv.12183. [DOI] [PubMed] [Google Scholar]
- 26.Harel I., Brunet A. The African turquoise killifish: a model for exploring vertebrate aging and diseases in the fast lane. Cold Spring Harb. Symp. Quant. Biol. 2015;80:275–279. doi: 10.1101/sqb.2015.80.027524. [DOI] [PubMed] [Google Scholar]
- 27.Harel I., Benayoun B.A., Machado B., Singh P.P., Hu C.-K., Pech M.F., Valenzano D.R., Zhang E., Sharp S.C., Artandi S.E., Brunet A. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell. 2015;160:1013–1026. doi: 10.1016/j.cell.2015.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui R., Medeiros T., Willemsen D., Iasi L.N.M., Collier G.E., Graef M., Reichard M., Valenzano D.R. Relaxed selection limits lifespan by increasing mutation load. Cell. 2019;178:385–399. doi: 10.1016/j.cell.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Tozzini E.T., Baumgart M., Battistoni G., Cellerino A. Adult neurogenesis in the short-lived teleost Nothobranchius furzeri: localization of neurogenic niches, molecular characterization and effects of aging. Aging Cell. 2012;11:241–251. doi: 10.1111/j.1474-9726.2011.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripa R., Dolfi L., Terrigno M., Pandolfini L., Savino A., Arcucci V., Groth M., Terzibasi Tozzini E., Baumgart M., Cellerino A. MicroRNA miR-29 controls a compensatory response to limit neuronal iron accumulation during adult life and aging. BMC Biol. 2017;15:9. doi: 10.1186/s12915-017-0354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsui H., Kenmochi N., Namikawa K. Age- and α-synuclein-dependent degeneration of dopamine and noradrenaline neurons in the annual killifish Nothobranchius furzeri. Cell Rep. 2019;26:1727–1733. doi: 10.1016/j.celrep.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Reichard M., Polačik M. Nothobranchius furzeri, an ‘instant’ fish from an ephemeral habitat. eLife. 2019;8:e41548. doi: 10.7554/eLife.41548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichard M., Cellerino A., Valenzano D.R. Turquoise killifish. Curr. Biol. 2015;25:R741–R742. doi: 10.1016/j.cub.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Rossi A.M., Fernandes V.M., Desplan C. Timing temporal transitions during brain development. Curr. Opin. Neurobiol. 2017;42:84–92. doi: 10.1016/j.conb.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barres B.A. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 36.Palmer A.L., Ousman S.S. Astrocytes and aging. Front. Aging Neurosci. 2018;10:337. doi: 10.3389/fnagi.2018.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Docampo-Seara A., Lagadec R., Mazan S., Rodríguez M.A., Quintana-Urzainqui I., Candal E. Study of pallial neurogenesis in shark embryos and the evolutionary origin of the subventricular zone. Brain Struct. Funct. 2018;223:3593–3612. doi: 10.1007/s00429-018-1705-2. [DOI] [PubMed] [Google Scholar]
- 38.Ebrahimi M., Yamamoto Y., Sharifi K., Kida H., Kagawa Y., Yasumoto Y., Islam A., Miyazaki H., Shimamoto C., Maekawa M. Astrocyte-expressed FABP7 regulates dendritic morphology and excitatory synaptic function of cortical neurons. Glia. 2016;64:48–62. doi: 10.1002/glia.22902. [DOI] [PubMed] [Google Scholar]
- 39.Dorn A., Ng’oma E., Janko K., Reichwald K., Polačik M., Platzer M., Cellerino A., Reichard M. Phylogeny, genetic variability and colour polymorphism of an emerging animal model: the short-lived annual Nothobranchius fishes from southern Mozambique. Mol. Phylogenet. Evol. 2011;61:739–749. doi: 10.1016/j.ympev.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Polačik M., Blažek R., Reichard M. Laboratory breeding of the short-lived annual killifish Nothobranchius furzeri. Nat. Protoc. 2016;11:1396–1413. doi: 10.1038/nprot.2016.080. [DOI] [PubMed] [Google Scholar]
- 41.Parichy D.M., Elizondo M.R., Mills M.G., Gordon T.N., Engeszer R.E. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dyn. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosimann C., Kaufman C.K., Li P., Pugach E.K., Tamplin O.J., Zon L.I. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.-B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 44.Allende M.L., Weinberg E.S. The expression pattern of two zebrafish achaete-scute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev. Biol. 1994;166:509–530. doi: 10.1006/dbio.1994.1334. [DOI] [PubMed] [Google Scholar]
- 45.Bate S.T., Clark R.A. The design and statistical analysis of animal experiments by Simon T. Bate. Camb. Core. 2014 /core/books/design-and-statistical-analysis-of-animal-experiments/BDD758F3C49CF5BEB160A9C54ED48706. [Google Scholar]
- 46.Clark R.A., Shoaib M., Hewitt K.N., Stanford S.C., Bate S.T. A comparison of InVivoStat with other statistical software packages for analysis of data generated from animal experiments. J. Psychopharmacol. (Oxford) 2012;26:1136–1142. doi: 10.1177/0269881111420313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.