Abstract
目的
探讨miR-449a/b在胃癌发生中的可能作用机制。
方法
采用qRT-PCR方法检测了miR-449a/b和E2F1基因在胃癌细胞BGC-823和胃粘膜细胞GES-1表达情况;采用瞬时转染技术将miR-449a/b模拟物(mimic)以及mimic阴性对照分别转入胃癌细胞BGC-823中,qRT-PCR验证转染成功后,通过CCK-8法检测胃癌细胞的增殖能力,流式细胞术检测胃癌细胞凋亡的变化,细胞划痕实验检测胃癌细胞的迁移能力,Western blot方法检测miR-449a/b上调后其靶基因蛋白的表达水平;并通过转染E2F1过表达质粒和空质粒对照入胃癌细胞BGC-823,Western blot确定转染效率后,分别用qRT-PCR和数字PCR检测miR-449a/b的表达水平。
结果
相比于正常胃粘膜细胞株GES-1,miR-449a/b在胃癌细胞株BGC-823中表达均下调(P < 0.01);过表达miR-449a/b可抑制胃癌细胞BGC-823增殖和迁移,并促进其凋亡(P < 0.05);过表达的E2F1可上调miR-449a/b的表达(P < 0.001),而上调miR-449a/b的表达,其直接靶基因CDK4、CDK6表达下调,并可通过CDKs-pRb-E2F1信号通路,间接下调E2F1蛋白的表达(P < 0.01)。
结论
miR-449a/b的低表达和E2F1的高表达均参与了胃癌的发生、发展,并且miR-449a/b能负反馈调节E2F1抑制胃癌细胞增殖。
Keywords: 胃癌, miR-449a/b, E2F1, 细胞增殖
Abstract
Objective
To investigate the possible role of miR-449a/b in the occurrence of gastric cancer.
Methods
The expression of miR-449a/b and E2F1 mRNA in gastric cancer cells BGC-823 and gastric mucosal cells GES-1 were detected with qRT-PCR. miR-449a/b mimics or a negative control was transiently transfected into BGC-823 cells, and the changes in cell proliferation, apoptosis, and migration ability were assessed using CCK-8 assay, flow cytometry, and scratch wound healing assay, respectively. Western blotting was used to observe the effects of miR-449a/b upregulation on its target gene expression. The effects of transfection with an E2F1-over-expressing plasmid or an empty plasmid were analyzed on the expression level of miR-449a/b in BGC-823 cells using qRT-PCR and digital PCR.
Results
The gastric cancer cell line BGC-823 showed significantly a lowered expression of miR-449a/b compared with the normal gastric mucosal cell line GES-1 (P < 0.01). Overexpression of miR-449a/b obviously inhibited the proliferation and migration and promoted apoptosis of BGC-823 cells (P < 0.05). Overexpression E2F1 in the cells resulted in significantly up-regulated expression of miR-449a/b (P < 0.001). Upregulation of miR-449a/b caused significant down-regulation of its direct target genes CDK4 and CDK6 and also of the expression of E2F1 protein via the CDKs-pRb-E2F1 signaling pathway.
Conclusion
The low expression of miR-449a/b and the high expression of E2F1 are both involved in the occurrence and progression of gastric cancer, and miR-449a/b negatively regulates E2F1 to inhibit the proliferation of gastric cancer cells.
Keywords: gastric cancer, miR-449a/b, E2F1, cell proliferation
胃癌发生于胃粘膜,是最常见的消化道肿瘤,尽管在西方国家胃癌的发病率正在逐渐下降,但在大多数发展中国家(包括中国),胃癌的死亡率和发病率仍然很高[1-2]。由于缺乏早期胃癌诊断的有效生物标志物,大约三分之二的患者被确诊时已发展到晚期或伴有广泛淋巴转移[3],已经严重威胁了人类的健康。目前,胃癌的治疗主要有手术治疗、化疗和靶向治疗等,但由于胃癌的发生是一个多步骤、多因素作用的过程,涉及到许多遗传和环境因素,致使其治疗效果不佳。近年来,有关表观遗传学的研究是肿瘤研究的热点,微小RNA(miRNA)作为表观遗传学的范畴之一,其主要通过结合靶基因mRNA 3'-非翻译区部分功能互补的序列引起mRNA的不稳定或抑制翻译发挥转录后调节功能[4]。最近研究表明,miRNA在胃癌中扮演着致癌和抑癌的双重作用,是肿瘤发生的关键因素,且miRNA的表达失调与胃癌的发生率、早期诊断和预后有关[5-6]。因此,对miRNA的生物学方面研究可能有助于我们更好地了解胃癌的发病机制,促进胃癌的分子生物学诊断与靶向治疗。
miRNA-449的表达首次在胚胎鼠脑组织中被发现[7],后续研究表明其可能通过直接或间接调控一些与细胞周期相关的靶基因影响细胞的增殖、分化、凋亡,以及导致细胞周期紊乱,与癌症的发生和发展密切相关[8]。而以CDKs-pRb-E2F1为信号转导通路的细胞周期调控系统是肿瘤发生和发展的重要通路,作为此通路的下游调节因子E2F1是细胞周期转录因子E2F家族的重要成员。E2F1转录因子是公认的细胞周期调节因子,同时也是DNA损伤诱导的凋亡和检查点反应的有效中介因子。理解E2F1活动的多样和看似二分法的功能一直是研究的重点。虽然E2F途径在癌症中经常被解除,但E2F1自身作为增殖或细胞死亡的启动子对肿瘤发生的作用,目前还远未被了解[9-10]。E2F1转录因子可以控制多个miRNA簇的表达,其自身也被许多miRNA靶向,作为miR-449a/b的间接靶基因,两者关系较为复杂,Yan等[11]通过动力学实验发现miR-449通过影响细胞周期阻滞和凋亡以避免过多的E2F1诱导的细胞增殖,从而在细胞周期进程的调节中发挥重要作用。
目前,miR-449a/b在胃癌中的研究较少,Bou等[12]研究发现miR-449a/b通过激活p53信号途径抑制细胞增殖,但miR-449a/b与E2F1之间的交互调控关系仍不明确。本项目组前期研究结果提示miR-449a/b、E2F1在胃癌组织中相对于癌旁胃组织有异常表达,且miR-449a/b的表达变化与淋巴结转移相关[13],提示两者都参与了胃癌的发生、发展过程。为进一步探讨miR-449a/b在胃癌发生中的作用及其分子机制,本研究以前期工作为基础,运用分子生物学和生物信息学技术,通过细胞实验、miRNA过表达技术和RNA干扰技术,一方面分析miR-449a/b过表达后对胃癌细胞生物学行为的影响;另一方面生物信息学预测miR-449a/b的靶基因,通过qPCR及Western blot等技术检测分析miR-449a/b模拟物(mimic)转染后与细胞周期有关基因的蛋白表达水平,并用重组E2F1表达质粒转染培养的胃癌细胞,增强E2F1的表达,检测转染后miR-449a/b的表达水平,明确miR-449a/b与E2F1之间的交互调控关系。
1. 材料和方法
1.1. 主要试剂
DMEM培养基(HyClone);0.25%胰酶(Gibco);Trizol试剂(Invitrogen, Ambin);SYBR Green qPCR Master Mix(Fermentas);胎牛血清(FBS,Clack);转染试剂riboFECTTM CP Reagent、miR-449a/b模拟物(mimic)和阴性对照(NC)(锐博公司);兔单克隆抗体E2F1(1:1000,CST);兔单克隆抗体CDK4(1:1000,CST);兔单克隆抗体CDK6(1:1000,CST);兔单克隆抗体CDC25A(1:1000,abclonal);兔单克隆抗体pRB(1:1000,abclonal);Actin抗体(1:1000,碧云天);辣根过氧化物酶标记山羊抗小鼠IgG(H+L)、辣根过氧化物酶标记山羊抗兔IgG(H + L)(1:1000,biosharp);ECL发光液(Thermo);E2F1过表达质粒(维真);预染蛋白marker(Fermentas);Annexin-X-FITC凋亡检测试剂盒、CCK-8试剂盒、高纯度质粒小提试剂盒(碧云天)。
1.2. 细胞培养
胃癌细胞株BGC-823、正常胃粘膜细胞GES-1来源于皖南医学院中心实验室保种的细胞株,细胞培养在含10%胎牛血清和100 U/mL青链双抗的完全培养基(DMEM)中,置于5% CO2,37 ℃培养箱中,隔天更换新鲜全血培养基。待细胞贴壁呈倍数生长至80%时,用0.25%胰酶消化传代,以3×105/孔的速度接种于6孔板中。
1.3. 细胞转染
待6孔板细胞生长至30%~50%时,更换含血清无双抗培养基,按照说明书指示使用转染试剂(riboFECTTM CP Reagent)将miR-449a/b mimic(50 nmol/L)和阴性对照(50 nmol/L)转染至BGC-823细胞,48 h后提取RNA,采用qRT-PCR方法检测,确定转染效率。待6孔板细胞生长至80%左右时,更换新鲜完全培养基,按照说明书提示使用转染试剂polyjet将E2F1过表达质粒转染至BGC-823细胞,6 h后更换含血清无双抗培养基,48 h后提取总蛋白,Western blot方法检测E2F1蛋白表达,确定转染效率。
1.4. RNA提取和qRT-PCR分析
按照说明书要求使用Trizol试剂提取细胞中总RNA。用超微量核酸蛋白测定仪(美国Denovix)检测RNA样品的浓度和纯度,琼脂糖凝胶电泳检测RNA完整性。采用反转录试剂盒反转录成cDNA,以U6为内参基因,SYBR®GreenqPCR扩增相关基因,定量表达由2-ΔΔct方法统计。
1.5. Western blot
冷裂解法(Western及IP细胞裂解液,碧云天)从细胞中分离蛋白,BCA试剂盒测定蛋白浓度后,加SDSPAGE蛋白上扬缓冲液(5×)混匀后,100 ℃,5 min使其变性。取变性后蛋白30 μg上样,聚丙烯酰胺凝胶电泳(SDS-PAGE)分离蛋白质,并使用湿转法转移至NC膜(Millipore)。转膜后,用含5%脱脂奶粉的TBST封闭2 h,特异性一抗结合(4 ℃摇床过夜),以Actin为内参基因。次日,用辣根过氧化物酶标记的二抗室温孵育1.5 h,蛋白条带由荧光、化学发光凝胶成像系统(ProteinSimple)检测。
1.6. 细胞增殖实验
取对数期生长细胞,铺于6孔细胞培养板中,24 h后进行细胞转染。转染6 h后,胰酶消化,细胞计数,在96孔板中接种细胞悬液(100 μL/孔),2000/孔,分组为miRNA-449a mimic转染组、miRNA-449b mimic转染组以及转染对照组。分别在转染后12、24、36、48、72 h,向每孔中直接加入10 μL CCK-8试剂,继续放置于5% CO2,37 ℃培养箱中培养1 h,使用酶标仪(TECAN)检测各孔的吸光度A450 nm,并绘制细胞增殖曲线。
1.7. 细胞侵袭实验(划痕实验)
在6孔细胞板中培养胃癌细胞,当融合度接近70%左右时按上述瞬时转染方法转染miR-449a/b mimic和miR-NC。转染24 h后,使用1 mL枪头在每孔贴壁细胞中行垂直划痕,弃去原培养基,加入37 ℃预热的PBS缓冲液漂洗细胞后,加入含2%胎牛血清的培养基。使用倒置显微镜观察划痕后的0、24、48 h的细胞迁移情况,并随机记录下3个不同的视野,每组实验设3个平行重复。
1.8. 细胞凋亡实验
取对数期生长细胞BGC-823,铺于6孔细胞培养板中,待细胞生长至30%~50%时按上述方法分别转染miR-449a/bmimic和阴性对照,转染后48 h,按凋亡检测试剂盒(Annexin-X-FITC,碧云天)说明书提示操作,随即进行流式细胞仪检测,Annexin V-FITC为绿色荧光,PI为红色荧光。同时以不加Annexin V-FITC及PI的一管作为阴性对照,记录胃癌细胞BGC-823凋亡率(可设3个对照:空白、单加Annexin V-FITC、单加PI)。
1.9. 统计学方法
每组实验至少重复3次,采用SPSS 18.0统计学软件分析。结果以均数±标准差表示,两组间的比较采用t检验,以P < 0.05为差异有统计学意义。
2. 结果
2.1. miR-449a/b在胃癌细胞BGC-823中表达下调
通过RT-qPCR技术对培养的正常胃粘膜细胞株GES-1和胃癌细胞株BGC-823中miR-449a/b、E2F1的表达进行检测,分别以U6、β-actin为内参,结果显示:相比于正常胃粘膜细胞株GES-1,miR-449a/b在胃癌细胞株BGC-823中表达均下调,其中miR-449a表达量下调约1/3,差异有统计学意义(P < 0.001),miR-449b表达量下调约1/2,差异有统计学意义(P < 0.01),基因E2F1的表达上调近4倍,差异有统计学意义(P < 0.001,图 1)。
1.

miR-449a/b、E2F1在胃癌细胞中的相对表达量
Relative expression levels of miR-449a/b and E2F1 in gastric cancer cell lines compared with normal gastric mucosal cells (**P < 0.01, ***P < 0.001).
2.2. 过表达miR-449a/b抑制胃癌细胞BGC-823增殖
在转染miR-449a/bmimics及miR-NC并稳定培养后,应用RT-qPCR技术对转染后胃癌细胞中miR-449a/b的表达量进行检测,结果显示miR-449a/b均明显表达上调(P < 0.001,图 2)。使用CCK-8试剂盒检测miR- 449a/bmimics转染后12、24、36、48、72 h的BGC-823细胞增殖能力,结果显示:实验组在转染miR-449a/b mimics后24 h,细胞生长开始受到抑制,在至转染后72 h时抑制细胞生长作用逐渐显著(P < 0.01,图 3)。
2.
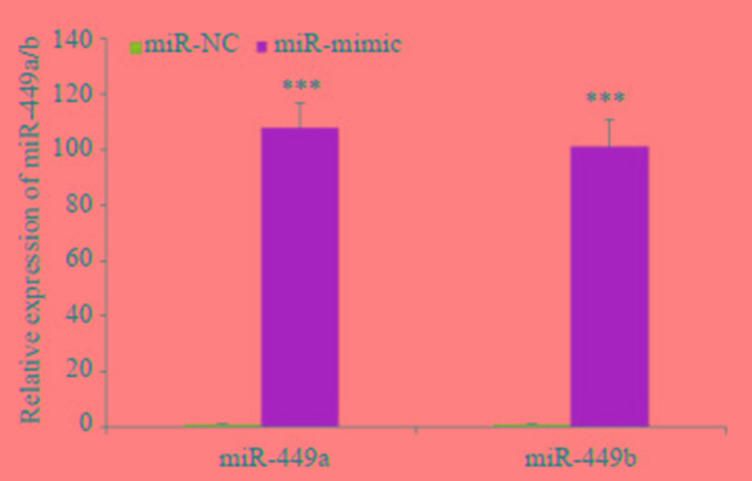
miR-449a/b mimic转染后miR-449a/b在胃癌细胞中的相对表达量
Relative expression of miR-449a/b in gastric cancer cells after transfection with the miR-449a/b mimic (***P < 0.001).
3.

CCK-8法检测过表达miR-449a/b胃癌细胞增殖能力
Detection of the proliferation of gastric cancer cells over-expressing miR-449a/b by CCK-8 assay (**P < 0.01) vs mimics after 72 h.
2.3. 过表达miR-449a/b促进胃癌细胞BGC-823凋亡
转染miR-449a/b mimics 48 h后,流式细胞术检测胃癌细胞BGC-823凋亡率,结果显示:相比于对照组,miR-449a/b mimics转染组细胞凋亡率增加(图 4),差异有统计学意义(P < 0.05)。
4.
流式细胞术检测过表达miRNA-449a/b胃癌细胞凋亡率
Apoptosis of gastric cancer cells over-expressing miR-449a/b detected by flow cytometry. A: miR-449a mimics; B: miR-449b mimics; C: miR-NC; D: Apoptosis rate of gastric cancer cells increases after transfection with miR-449a/b mimics (*P < 0.05) vs miR-NC.
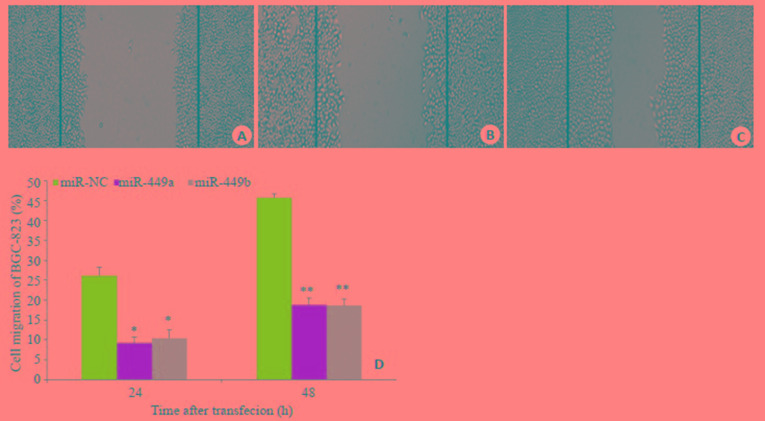
2.4. 过表达miR-449a/b抑制胃癌细胞BGC-823迁移
转染miR-449a/b mimics使miR-449a/b在胃癌细胞BGC-823中过表达后,通过细胞划痕实验检测胃癌细胞迁移能力,结果显示,相比于对照组,miR-449a/b过表达24 h后胃癌细胞迁移能力下降,差异有统计学意义(P < 0.05),48 h后胃癌细胞迁移能力明显下降(P < 0.01,图 5)。
5.
细胞划痕法检测过表达miRNA-449a/b胃癌细胞迁移能力
Migration ability of gastric cancer cells overexpressing miR-449a/b detected by cell scratch assay. A: miR-449a mimics; B: miR-449b mimics; C: miR-NC; D: Migration of gastric cancer cells is inhibited after transfection with miR-449a/b mimics (*P < 0.05, **P < 0.01) vs miR-NC.
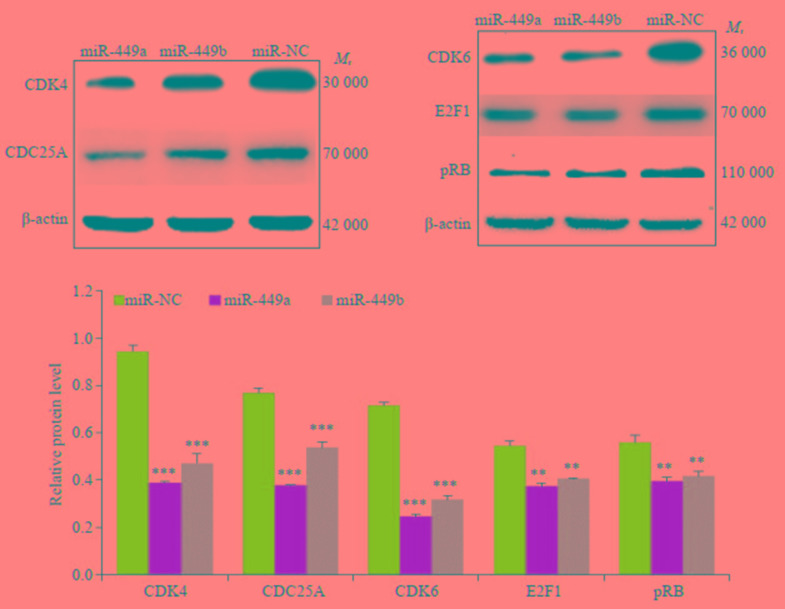
2.5. 过表达miR-449a/b后其靶基因的蛋白表达水平变化
转染miR-449a/b mimics使miR-449a/b在胃癌细胞BGC-823中过表达后,Western blot方法检测miR-449a/b靶基因及CDK-pRb-E2F1通路相关基因的蛋白表达,结果发现,与对照组相比,胃癌细胞BGC-823中CDK4、CDK6、CDC25A、E2F1、pRB的蛋白表达均增高,差异有统计学意义(P < 0.01,图 6)。
6.
miR-449a/b mimic转染后CDK-Rb-E2F1信号通路中相关蛋白的表达
Expression of the related proteins of CDK-Rb-E2F1 signaling pathway after transfection of the cells with miR-449a /b mimics (**P < 0.01, ***P < 0.001) vs miR-NC.
2.6. E2F1过表达质粒转染后miR-449a/b的表达变化
将E2F1过表达质粒和空质粒对照分别转染至胃癌细胞BGC-823,48 h后RT-qPCR及Western blot方法确定其转染效率,qPCR检测miR-449a/b的表达水平,结果显示,miR-449a/b表达均上调,与对照组相比,差异有统计学意义(P < 0.001,图 7A)。miR-449a/b表达均上调,与qPCR结果一致(P < 0.001,图 7B)。
7.
qPCR/ddPCR检测E2F1过表达质粒转染后miR-449a/b的表达
Expression of miR-449a/b after transfection with the E2F1-over-expressing plasmid detected by qPCR/ddPCR. A: Expression of miR-449a/b after transfection with E2F1-over-expressing plasmid detected by qPCR (***P < 0.001); B: Expression of miR-449a/b after transfection of E2F1-over-expressing plasmid detected by ddPCR (***P < 0.001); C: Western blotting for determining the transfection efficiency of the E2F1-over-expressing plasmid; D, E: miR-449a/b microdroplets (C05, C06, D05 and D06 were the transfection group, and C9, C10, D9 and D10 were the control group); F, G: ddPCR was used to quantitatively detect the expression concentration of miR-449a/b with U6 as the internal reference gene. E2F1 represents the transfection group, and NC represents the control group.
3. 讨论
胃癌的发生是一个多步骤的过程,涉及蛋白编码原癌基因和抑癌基因的表达及表观遗传学改变[14]。随着非编码RNA研究的进展,作为表观遗传学研究范畴之一的miRNA可通过调节靶基因的表达与翻译抑制,几乎参与胃癌发生的每一个过程[15-16]。如Zhu等[17]发现,miR-106a靶向基质金属蛋白酶抑制因子-2(TIMP-2)调控胃癌的侵袭和转移,抑制miR-106a的表达可显著降低胃癌细胞的增殖、迁移和侵袭能力。Zhu等[18]研究表明miR-383在胃癌组织中表达显著降低,并与这些癌症患者的晚期进展相关,且过表达miR-383靶向细胞周期蛋白E2(Cyclin E2)抑制胃癌细胞增殖,促进胃癌细胞凋亡。Li等[19]通过AUC分析显示血清miR-381对胃癌的诊断敏感性和特异性优于其他肿瘤标志物,且Kaplan-Meier生存分析显示miR-381的下调与淋巴结转移及胃癌的发展有关,其可能是胃癌早期诊断的潜在标志物。越来越多的研究[20-22]显示miRNA与胃癌的关系密切,并在胃癌的发生和肿瘤进展中发挥重要作用。
本研究首先比较miR-449a/b在胃癌细胞BGC-823与正常胃粘膜细胞GES-1中表达,结果发现,胃癌细胞明显低于正常胃粘膜细胞,这一结果与前期研究miR-449a/b在胃癌组织中的表达相一致,提示miR-449a/b参与了胃癌的发生;其次,为探讨miR-449a/b对胃癌细胞生物学行为影响,本研究运用转染技术使miR-449a/b在胃癌细胞中过表达,通过CCK-8法、流式细胞术、细胞划痕法分别对其增殖、凋亡和迁移的影响进行检测,结果发现,miR-449a/b不仅可抑制胃癌细胞的增殖与迁移,还能促进其凋亡,我们前期在胃癌组织中研究也发现miR-449a/b的表达变化与淋巴结转移相关,提示其可能作为抑癌基因靶向调控某些基因抑制胃癌细胞的生长与迁移能力。
进一步探讨miR-449a/b在胃癌中发挥作用的可能机制,我们发现在胃癌细胞中过表达的miR-449a/b可直接下调与细胞周期调控相关的靶基因(CDK4、CDK6等),作用于CDKs-pRb-E2F1信号通路间接下调细胞转录因子E2F1,这与之前Bou等[12]研究推测的结果相一致。有趣的是,miR-449a/b与基因E2F1的关系甚为密切。有研究发现[23],E2F1能激活miR-449a/b,而在几个被检测的癌细胞系中,如乳腺癌MCF-7、肺癌H1299和非癌性乳腺上皮细胞MCF-10A,miR-449a模拟物的引入几乎完全抑制pRb磷酸化,间接抑制E2F1的表达,呈现一个pRb-E2F1负反馈调控通路。Lizé等[24]研究也表明E2F1能直接上调miR-449a/b以及它们的宿主基因CDC20B的表达,从而抑制细胞增殖,促进细胞凋亡。本研究通过在胃癌细胞中转染E2F1过表达质粒,qPCR和数字PCR两种方法检测miR-449a/b的表达,结果一致表明过表达的E2F1可上调miR-449a/b的表达。因此,我们推测,miR-449a/b的转录失活可能是要维持E2F1的高活性,从而促进在特定的环境中发生肿瘤的转变。
由于细胞周期元素分泌的不平衡导致细胞周期紊乱是肿瘤细胞的特征之一。miR-449a/b,其功能类似于抑癌基因,与多种细胞周期相关控制基因关系密切[25-26]。而E2F1作为周期调控的重要转录因子,不仅与miR-449形成反馈回路,与很多miRNA均形成复杂的网络调控系统。有研究发现[27]miRNA可与转录因子(TFs)联合调控胃癌基因表达,并与胃癌的进展相关。在此研究中,胃癌组织中这些差异表达的mRNA和miRNA主要与细胞周期进展有关,尤其是E2F家族,说明miRNA改变的E2Fs蛋白表达是胃癌发生发展的重要因素。在胃癌中,先前的研究表明,E2F1异常表达并调控miRNA作用于TGF-β途径依赖的胃癌细胞周期阻滞和凋亡[28]。此外,在恶性B淋巴细胞中,miR-155通过靶向TGF-β1/SMAD5信号通路抑制Rb磷酸化调控细胞周期[29];在卵巢子宫内膜异位症研究中,整合分析miRNA和mRNA配对表达谱发现,miRNA-TF-gene调控网络由22个miRNA、12个TFs和430个相关基因组成,其中,miR-449和miR-34b/c簇、FOX家族、GATA家族、E2F家族等一些关键调控因子被认为在子宫内膜异位症的发病机制中发挥重要的调控作用[30]。这些研究表明miRNA与mRNA之间的交互调节作用在疾病,尤其是肿瘤的发生、发展中扮演着越来越重要的角色。
本研究揭示了miRNA-449a/b的肿瘤抑制功能是通过调节pRb/E2F1活性起作用,并表明由于表观遗传学异常导致E2F1的调节失控,从而引起人类癌症中细胞的无限制增殖。因此,我们推测胃癌与miRNA的相关性是否会成为未来胃癌研究的一个新方向,这将有利于胃癌的早期诊断与靶向治疗,以提高胃癌患者的生存率。
Biography
林爱琴,实验师,E-mail: linaiqin945@163.com
Funding Statement
安徽省高校自然科学研究重点项目(KJ2017A252、KJ2019A0426);安徽省重点实验室自主研究课题(LAB201605);皖南医学院校中青年课题(WK201904)
Contributor Information
林 爱琴 (Aiqin LIN), Email: linaiqin945@163.com.
李 铁臣 (Tiechen LI), Email: tiechenli@126.com.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [Jemal A, Bray F, Center MM, et al.Global cancer statistics[J].CA Cancer J Clin, 2011, 61(2):69-90.] [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CACancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [Torre LA, Bray F, Siegel RL, et al.Global cancer statistics, 2012[J].CACancer J Clin, 2015, 65(2):87-108.] [DOI] [PubMed] [Google Scholar]
- 3.Coburn NG. Lymph nodes and gastric cancer. http://d.old.wanfangdata.com.cn/Periodical/zgazyj201605006. J SurgOncol. 2009;99(4):199–206. doi: 10.1002/jso.21224. [Coburn NG.Lymph nodes and gastric cancer[J].J SurgOncol, 2009, 99(4):199-206.] [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs:genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5. [Bartel DP.MicroRNAs:genomics, biogenesis, mechanism, and function[J].Cell, 2004, 116(2):281-97.] [DOI] [PubMed] [Google Scholar]
- 5.Ma YY, Tao HQ. Microribonucleic acids and gastric cancer. Cancer Sci. 2012;103(4):620–5. doi: 10.1111/j.1349-7006.2011.02185.x. [Ma YY, Tao HQ.Microribonucleic acids and gastric cancer[J].Cancer Sci, 2012, 103(4):620-5.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143(1):35–47. doi: 10.1053/j.gastro.2012.05.003. [Song JH, Meltzer SJ.MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers[J].Gastroenterology, 2012, 143(1):35-47.] [DOI] [PubMed] [Google Scholar]
- 7.Wheeler G, Ntounia-Fousara S, Granda B, et al. Identification of new central nervous system specific mouse microRNAs. FEBS Lett. 2006;580(9):2195–200. doi: 10.1016/j.febslet.2006.03.019. [Wheeler G, Ntounia-Fousara S, Granda B, et al.Identification of new central nervous system specific mouse microRNAs[J].FEBS Lett, 2006, 580(9):2195-200.] [DOI] [PubMed] [Google Scholar]
- 8.Lizé M, Klimke A, Dobbelstein M. MicroRNA-449 in cell fate determination. Cell Cycle. 2011;10(17):2874–82. doi: 10.4161/cc.10.17.17181. [Lizé M, Klimke A, Dobbelstein M.MicroRNA-449 in cell fate determination[J].Cell Cycle, 2011, 10(17):2874-82.] [DOI] [PubMed] [Google Scholar]
- 9.Zhang XP, Liu F, Wang W. Coordination between cell cycle progression and cell fate, decision by the p53 and E2F1 pathways in response to DNA damage. J Biol Chem. 2010;285(41):31571–80. doi: 10.1074/jbc.M110.134650. [Zhang XP, Liu F, Wang W.Coordination between cell cycle progression and cell fate, decision by the p53 and E2F1 pathways in response to DNA damage[J].J Biol Chem, 2010, 285(41):31571-80.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poppy Roworth A, Ghari F, La Thangue NB. To live or let diecomplexity within the E2F1 pathway. Mol Cell Oncol. 2015;2(1):e970480. doi: 10.4161/23723548.2014.970480. [Poppy Roworth A, Ghari F, La Thangue NB.To live or let diecomplexity within the E2F1 pathway[J].Mol Cell Oncol, 2015, 2(1):e970480.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan F, Liu HH, Hao JJ, et al. Dynamical behaviors of Rb-E2F pathway including negative feedback loops involving miR-449. PLoS One. 2012;7(9):e43908. doi: 10.1371/journal.pone.0043908. [Yan F, Liu HH, Hao JJ, et al.Dynamical behaviors of Rb-E2F pathway including negative feedback loops involving miR-449[J].PLoS One, 2012, 7(9):e43908] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bou KheirT, Futoma-Kazmierczak E, Jacobsen A, et al. MiR-449 inhibits cell proliferation and is down-regulated in gastric cancer. http://d.old.wanfangdata.com.cn/OAPaper/oai_doaj-articles_43c5664447c567ac128b15a1ce0b5309. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [Bou KheirT, Futoma-Kazmierczak E, Jacobsen A, et al.MiR-449 inhibits cell proliferation and is down-regulated in gastric cancer[J].Mol Cancer, 2011, 10:29.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.林 爱琴, 汪 根莲, 段 仁杰, et al. miRNA-449b在胃癌中的表达及与淋巴结转移的关系. 皖南医学院学报. 2015;34(2):113–6. doi: 10.3969/j.issn.1002-0217.2015.02.004. [林爱琴, 汪根莲, 段仁杰, 等.miRNA-449b在胃癌中的表达及与淋巴结转移的关系[J].皖南医学院学报, 2015, 34(2):113-6.] [DOI] [Google Scholar]
- 14.Rudnicka K, Backert S, Chmiela M. Genetic polymorphisms in inflammatory and other regulators in gastric cancer:risks and clinical consequences. Curr Top MicrobiolImmunol. 2019;421:53–76. doi: 10.1007/978-3-030-15138-6_3. [Rudnicka K, Backert S, Chmiela M.Genetic polymorphisms in inflammatory and other regulators in gastric cancer:risks and clinical consequences[J].Curr Top MicrobiolImmunol, 2019, 421:53-76.] [DOI] [PubMed] [Google Scholar]
- 15.Gao M, Yin H, Fei ZW. Clinical application of microRNA in gastric cancer inEastern asian area. World J Gastroenterol. 2013;19(13):2019–27. doi: 10.3748/wjg.v19.i13.2019. [Gao M, Yin H, and Fei ZW, Clinical application of microRNA in gastric cancer inEastern asian area[J].World J Gastroenterol, 2013, 19(13):2019-27.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer:miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. doi: 10.1017/erm.2013.16. [Wu HH, Lin WC, Tsai KW.Advances in molecular biomarkers for gastric cancer:miRNAs as emerging novel cancer markers[J].Expert Rev Mol Med, 2014, 16:e1.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu M, Zhang N, He SX, et al. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastriccance. FEBS Lett. 2014;588(4):600–7. doi: 10.1016/j.febslet.2013.12.028. [Zhu M, Zhang N, He SX, et al.MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastriccance[J]r.FEBS Lett, 2014, 588(4):600-7.] [DOI] [PubMed] [Google Scholar]
- 18.Zhu C, Huang Q, Zhu H. miR-383 Inhibited the cell cycle progression of gastric cancer cells via targeting cyclin E2. DNA Cell Biol. 2019;38(8):849–56. doi: 10.1089/dna.2019.4624. [Zhu C, Huang Q, Zhu H.miR-383 Inhibited the cell cycle progression of gastric cancer cells via targeting cyclin E2[J].DNA Cell Biol, 2019, 38(8):849-56.] [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Sun H, Guan J, et al. Serum microRNA-381:a potential marker for early diagnosis of gastric cancer. Yonsei Med J. 2019;60(8):720–6. doi: 10.3349/ymj.2019.60.8.720. [Li Y, Sun H, Guan J, et al.Serum microRNA-381:a potential marker for early diagnosis of gastric cancer[J].Yonsei Med J, 2019, 60(8):720-6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhat SA, Majid S, Rehman MU. Scenario and future prospects of microRNAs in gastric cancer:A review. Iran J Basic Med Sci. 2019;22(4):345–52. doi: 10.22038/ijbms.2019.32399.7765. [Bhat SA, Majid S, Rehman MU.Scenario and future prospects of microRNAs in gastric cancer:A review[J].Iran J Basic Med Sci, 2019, 22(4):345-52.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20(30):10432–9. doi: 10.3748/wjg.v20.i30.10432. [Shin VY, Chu KM.MiRNA as potential biomarkers and therapeutic targets for gastric cancer[J].World J Gastroenterol, 2014, 20(30):10432-9.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.王 健, 陈 文静, 林 慧娟, et al. 胃癌中miRNA-340的体内外增殖能力检测及生物信息分析. http://www.j-smu.com/oa/pdfdow.aspx?Sid=201907784. 南方医科大学学报. 2019;39(7):784–90. doi: 10.12122/j.issn.1673-4254.2019.07.06. [王健, 陈文静, 林慧娟, 等.胃癌中miRNA-340的体内外增殖能力检测及生物信息分析[J].南方医科大学学报, 2019, 39(7):784-90.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Feng M, Jiang X, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. http://d.old.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_2764491. GenesDev. 2009;23(20):2388–93. doi: 10.1101/gad.1819009. [Yang X, Feng M, Jiang X, et al.miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A[J].GenesDev, 2009, 23(20):2388-93.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lizé M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA-449a/b suppresses cell proliferation and promotes apoptosis. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=c3869e809938aac7bd336cbdf9de63d1. Cell Death Differ. 2010;17(3):452–8. doi: 10.1038/cdd.2009.188. [Lizé M, Pilarski S, Dobbelstein M.E2F1-inducible microRNA-449a/b suppresses cell proliferation and promotes apoptosis[J].Cell Death Differ, 2010, 17(3):452-8.] [DOI] [PubMed] [Google Scholar]
- 25.Wu WJ, Sun DY, Xie ZY, et al. miR-449a and CDK6 in gastric carcinoma. Oncol Lett. 2014;8(4):1533–8. doi: 10.3892/ol.2014.2370. [Wu WJ, Sun DY, Xie ZY, et al.miR-449a and CDK6 in gastric carcinoma[J].Oncol Lett, 2014, 8(4):1533-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang J, Yang X, He X, et al. MicroRNA-449b-5p suppresses the growth and invasion of breast cancer cells via inhibiting CREPTmediated Wnt/β-catenin signaling. Chem Biol Interact. 2019;302:74–82. doi: 10.1016/j.cbi.2019.02.004. [Jiang J, Yang X, He X, et al.MicroRNA-449b-5p suppresses the growth and invasion of breast cancer cells via inhibiting CREPTmediated Wnt/β-catenin signaling[J].Chem Biol Interact, 2019, 302:74-82.] [DOI] [PubMed] [Google Scholar]
- 27.Zhang XT, Ni ZH, Duan ZP, et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA Co-regulatory network analysis. PLoS One. 2015;10(2):e0116979. doi: 10.1371/journal.pone.0116979. [Zhang XT, Ni ZH, Duan ZP, et al.Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA Co-regulatory network analysis[J].PLoS One, 2015, 10(2):e0116979.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrocca F, Visone, Onelli MR, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apo-ptosis in gastric cancer. Cancer Cell. 2008;13(3):272–86. doi: 10.1016/j.ccr.2008.02.013. [Petrocca F, Visone, Onelli MR, et al.E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apo-ptosis in gastric cancer[J].Cancer Cell, 2008, 13(3):272-86.] [DOI] [PubMed] [Google Scholar]
- 29.Jiang D, Aguiar RC. MicroRNA-155 controls RB phosphorylation in normal and malignant B lymphocytes via the noncanonical TGF-β1/SMAD5 signaling module. Blood. 2014;123(1):86–93. doi: 10.1182/blood-2013-07-515254. [Jiang D, Aguiar RC.MicroRNA-155 controls RB phosphorylation in normal and malignant B lymphocytes via the noncanonical TGF-β1/SMAD5 signaling module[J].Blood, 2014, 123(1):86-93.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao LY, Gu CL, Ye M, et al. Integration analysis of microRNA and mRNApaired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reprod Biol Endocrinol. 2018;16(1):4. doi: 10.1186/s12958-017-0319-5. [Zhao LY, Gu CL, Ye M, et al.Integration analysis of microRNA and mRNApaired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis[J].Reprod Biol Endocrinol, 2018, 16(1):4.] [DOI] [PMC free article] [PubMed] [Google Scholar]