Abstract
A long-standing mystery shrouds the mechanism by which catalytically repressed receptor tyrosine kinase domains accomplish transphosphorylation of activation loop (A-loop) tyrosines. Here we show that this reaction proceeds via an asymmetric complex that is thermodynamically disadvantaged because of an electrostatic repulsion between enzyme and substrate kinases. Under physiological conditions, the energetic gain resulting from ligand-induced dimerization of extracellular domains overcomes this opposing clash, stabilizing the A-loop-transphosphorylating dimer. A unique pathogenic fibroblast growth factor receptor gain-of-function mutation promotes formation of the complex responsible for phosphorylation of A-loop tyrosines by eliminating this repulsive force. We show that asymmetric complex formation induces a more phosphorylatable A-loop conformation in the substrate kinase, which in turn promotes the active state of the enzyme kinase. This explains how quantitative differences in the stability of ligand-induced extracellular dimerization promotes formation of the intracellular A-loop-transphosphorylating asymmetric complex to varying extents, thereby modulating intracellular kinase activity and signaling intensity.
Receptor tyrosine kinase (RTK) signaling regulates a myriad of biological processes in metazoan organisms by universally mediating intercellular communication1–3. A prototypical RTK consists of an extracellular ligand-binding region, a single-pass transmembrane helix and an intracellular region harboring a conserved tyrosine kinase domain. In the resting (unliganded) state, RTKs exist either as monomers or as preformed dimers in which their kinase domains are catalytically repressed through a variety of autoinhibitory mechanisms4–7. Binding of extracellular stimuli either induces dimerization of monomeric RTKs or causes reorientation of monomers within preformed RTK dimers, in each case resulting in the derepression of autoinhibited intracellular kinase domains8. In the case of the epidermal growth factor receptor (EGFR) family, extracellular dimerization promotes formation of an asymmetric intracellular kinase dimer in which one EGFR kinase (the activator) allosterically drives the other kinase (the receiver) into an active state without the need for phosphorylation of A-loop tyrosines9–11. By contrast, in all other RTK family members, extracellular dimerization leads to kinase activation by enabling kinase transphosphorylation on A-loop tyrosine residues2,12–15. Kinase activation that is dependent on phosphorylation of A-loop tyrosines is a prerequisite for all subsequent tyrosine transphosphorylation events within RTKs and their downstream substrates16.
The molecular mechanism whereby unphosphorylated (and hence catalytically repressed) RTKs accomplish the initial A-loop-tyrosine transphosphorylation reaction is an enigma. A twofold symmetric dimer poised for A-loop transphosphorylation has been observed in the crystal structure of inhibitor-bound IGF1R kinase17, although there is no evidence that A-loop transphosphorylation can occur simultaneously in both kinases. Here we present the remarkable finding that a particular pathogenic FGFR2 substitution (R678G)18 differs from all other known substitutions in that it does not act by shifting the kinase equilibrium to the active state. Rather, it imparts a gain-of-function by promoting phosphorylation of A-loop tyrosines, which then leads to kinase activation. Indeed, our X-ray crystallographic, solution NMR and cell-based experiments show that FGFR2R678G facilitates formation of an induced-fit asymmetric kinase dimer that mediates A-loop transphosphorylation. We present evidence that this mode of A-loop-tyrosine transphosphorylation is shared among multiple members of the RTK superfamily.
Results
FGFR2R678G accelerates phosphorylation of A-loop tyrosines
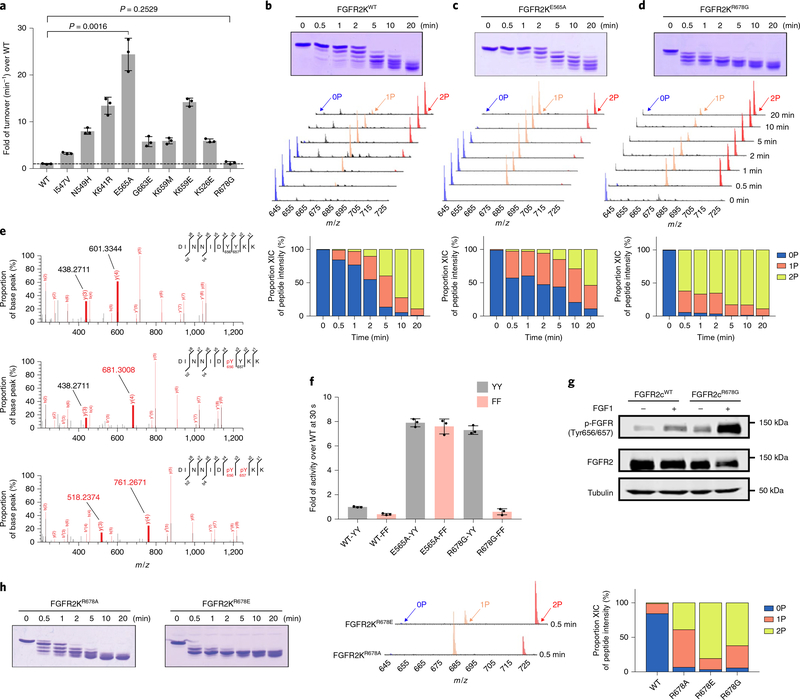
Four pathogenic gain-of-function subsitutions (I547V, K526E, G663E and R678G) in the FGFR2 kinase (FGFR2K) domain map to the kinase hinge, αC helix, A-loop and P + 1 pocket of the kinase, respectively19,20. We assessed the intrinsic kinase activities of the corresponding FGFR2Ks by measuring their catalytic turnover rates under initial rate conditions using a minimal enzyme concentration and an excess of substrate peptide to minimize bimolecular collisions that would otherwise lead to A-loop-tyrosine transphosphorylation and kinase activation. For comparison, wild-type FGFR2K (FGFR2KWT) and five FGFR2Ks, each harboring a distinct pathogenic activating substitution at either the molecular brake near the kinase hinge (N549H, K641R, E565A) or the A-loop (K659M, K659E), were included in this experiment. FGFR2KN549H, FGFR2KK641R, FGFR2KE565A, FGFR2KK659M and FGFR2K659E had greater turnover rates, ranging from 5.9- to 24.4-fold enhancement relative to FGFR2KWT (Fig. 1a). These elevated intrinsic kinase activities are consistent with our previous structural data, which show that these substitutions facilitate the active-state conformation21,22. The catalytic rates and intrinsic activities of the FGFR2KI547V, FGFR2KK526E and FGFR2KG663E mutants were also greater than FGFR2KWT (in the range 3.2- to 5.9-fold enhancement); thus they also more readily adopt an active-state conformation. Surprisingly, however, there was no measurable increase in the catalytic rate and intrinsic activity of the FGFR2KR678G mutant (Fig. 1a), implying that, unlike other gain-of-function mutations, R678G does not act by enhancing the propensity of the kinase to adopt an active-state conformation.
Fig. 1 |. The FGFR2KR678G Crouzon syndrome substitution accelerates phosphorylation of A-loop tyrosines without elevating intrinsic kinase activity.
a, Turnover rates (min−1) of unphosphorylated FGFR2KWT and nine variants harboring distinct pathogenic mutations. Note that the turnover rate of the R678G mutant is indistinguishable from the wild type. Data are mean ± s.d. (n = 3). Statistical analysis was performed via a two-tailed unpaired Student’s t test. b–d, Top, kinetics of overall tyrosine transphosphorylation in FGFR2KWT, FGFR2KE565A and FGFR2KR678G assayed by native gel electrophoresis. Middle, LC–MS spectra showing transphosphorylation on A-loop tandem tyrosines (Y656 and Y657) in samples corresponding to those analyzed above. Bottom, quantitation of LC–MS relative ion intensities. Kinase assays were done independently twice with similar results. e, Phosphorylation on Y656 precedes that of Y657. MS/MS spectra of 0P (top), 1P (middle) and 2P (bottom) FGFR2KWT A-loop tryptic peptides. Note the increase by 80 Da in the mass of the y4 ion (but not the y3 ion) in the mono-phosphorylated peptide as compared to the non-phosphorylated peptide, demonstrating phosphorylation on Y656 (but not Y657). f, MALDI–TOF mass spectrometry analysis of the effects of substituting A-loop tandem tyrosines (YY) with phenylalanines (FF) on the substrate phosphorylation activities of FGFR2KWT, FGFR2KE565A and FGFR2KR678G, respectively, in each case shown relative to unphosphorylated FGFR2KWT as measured at 0.5 min. Data are mean ± s.d. (n = 3). g, Immunoblot analyses of whole extracts of untreated or FGF1-treated L6 myoblasts stably expressing wild-type FGFR2c or its R-to-G variant probed with an anti-p-FGFR (Y656/Y657), an anti-FGFR2 or an anti-β-tubulin antibody. Experiments were performed in biological triplicates with similar results. Full-length gels are shown in Supplementary Fig. 15a. h, Left, kinetics of overall tyrosine transphosphorylation in FGFR2KR678A and FGFR2KR678E assayed by native gel electrophoresis. Middle and right, LC–MS analysis of transphosphorylation on A-loop tandem tyrosines of samples at 0.5 min (middle) and corresponding quantitation (right). Experiments were done independently twice with similar results.
We suspected that the R678G mutation conferred its gain-of-function by facilitating transphosphorylation on A-loop tyrosines. To test this, we used time-resolved LC–MS to compare the A-loop-tyrosine phosphorylation capacities of FGFR2KWT, FGFR2KE565A and FGFR2KR678G in vitro in the absence of any substrate peptide. For this experiment, we intentionally used a high enzyme concentration (that is, 67.5 μM) to encourage productive bimolecular collisions between kinase molecules and subsequent A-loop transphosphorylation. The FGFR2KE565A molecular brake mutant21 was used as a representative of those gain-of-function mutants that act by stabilizing the active kinase conformation. The A-loop tyrosine phosphorylation rate in FGFR2KR678G was approximately tenfold faster than in both FGFR2KWT and the FGFR2KE565A mutant (Fig. 1b–d). After 0.5 min, essentially all non-phosphorylated (0P) A-loop in FGFR2KR678G was converted to mono-phosphorylated (1P) and bis-phosphorylated (2P) forms (Fig. 1d), whereas it took >5 min for either FGFR2KWT or the FGFR2KE565A mutant to reach this state (Fig. 1b,c). Notably, MS/MS analysis showed that the 1P form of the kinase was phosphorylated exclusively on Y656 (Fig. 1e), implying that phosphorylation on Y656 precedes phosphorylation on Y657. It follows that—in contrast to other pathogenic substitutions—R678G does not act by increasing the intrinsic activity of the kinase; rather, it accelerates phosphorylation of A-loop tyrosines, which then leads to kinase activation. Indeed, substitution of the tandem A-loop tyrosines of FGFR2KR678G with phenylalanines (Y656F/Y657F) almost completely eliminated the elevated activity of FGFR2KR678G, while a corresponding YY-to FF substitution had little impact on the elevated activity of FGFR2KE565A (Fig. 1f). The insensitivity of FGFR2KE565A to A-loop tyrosine substitutions is to be expected because the E565A substitution directly drives the kinase into an active-state conformation21, thus bypassing the need for A-loop-tyrosine phosphorylation. By contrast, the R678G mutation does not confer a gain-of-function by encouraging the kinase to adopt an active-state conformation; rather, it indirectly stabilizes the active-state conformation by facilitating A-loop-tyrosine phosphorylation, and thus remains dependent on A-loop phosphorylation. We also considered the possibility that FGFR2KR678G might possess a higher intrinsic activity than FGFR2KWT after phosphorylation of A-loop tyrosines. However, measurement of catalytic turnover rates showed that the specific activity of A-loop phosphorylated FGFR2KR678G does not exceed that of A-loop-phosphorylated FGFR2KWT (Supplementary Fig. 1).
Consistent with the conservation of R678 within the FGFR subfamily, substitution of corresponding arginines in the isolated kinase domains of FGFR1 (R675), FGFR3 (R669) and FGFR4 (R664) with glycine accelerated their A-loop-tyrosine phosphorylation activity in vitro (Supplementary Fig. 2a–c). Moreover, all four full-length FGFRs carrying an R-to-G substitution elicited greater degrees of ligand-induced A-loop transphosphorylation as compared to their wild-type counterparts when ectopically expressed in L6 myoblasts (Fig. 1g and Supplementary Fig. 2d–f). We conclude that the mechanism by which the FGFR2 R678G substitution accelerates A-loop phosphorylation is conserved throughout the FGFR family.
As the R-to-G substitution removes a positive charge, we speculated that the presence of an arginine residue at the 678th position of FGFR kinases inhibits A-loop-tyrosine transphosphorylation activity. We therefore substituted R678 in FGFR2K with either an alanine or a glutamic acid (that is, an opposite charge), and compared the phosphorylation rates of A-loop tyrosines of the resulting altered kinases (that is, FGFR2KR678A and FGFR2KR678E) with those of FGFR2KWT and FGFR2KR678G. As in the case of FGFR2KR678G, both FGFR2KR678A and FGFR2KR678E had dramatically accelerated rates of transphosphorylation of A-loop tyrosines relative to FGFR2KWT, with FGFR2KR678E showing the greatest increase (Fig. 1h). We conclude that the presence of a positively charged residue at this locus in the FGFR family inhibits transphosphorylation of A-loop tyrosines.
Mechanism of transphosphorylation of A-loop tyrosines
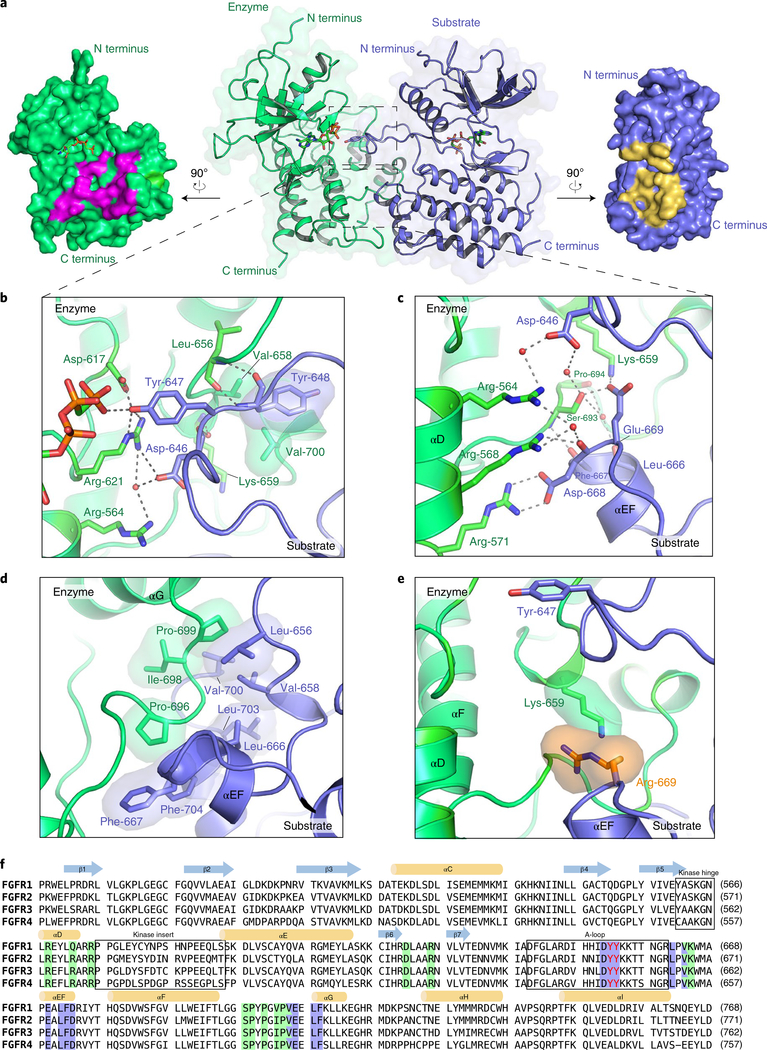
We set out to solve the crystal structure of FGFR2KR678E because of its pronounced A-loop-tyrosine phosphorylation activity. Although this protein proved refractory to crystallization, we successfully crystallized the FGFR3K isoform containing the analogous R669E mutation (FGFR3KR669E) and solved its structure at a resolution of 2.2 Å (Supplementary Table 1). The structure reveals an asymmetric complex of two FGFR3KR669E molecules trapped in the act of transphosphorylation, with one molecule serving as enzyme and the other as substrate, the latter offering one of its A-loop tyrosines (Y647) for phosphorylation (corresponding to Y656 of FGFR2) (Fig. 2). Both kinase molecules contain a bound AMP-PCP molecule (a nonhydrolyzable ATP analog) in the cleft between kinase N and C lobes (Fig. 2a); however, they do not appear to play any role in facilitating the kinase A-loop conformation. At the dimer interface, the C lobe of the substrate kinase engages both the catalytic pocket of the enzyme (Fig. 2b), as well as a second site distal to it (Fig. 2c,d).
Fig. 2 |. FGFR3R669E promotes formation of an A-loop-transphosphorylating asymmetric complex.
a, Middle, overall view of the crystal structure of the FGFR3KR669E asymmetric complex shown as a cartoon superimposed on a semitransparent surface. Enzyme- and substrate-acting kinases are in green and blue, respectively. Bound AMP-PCP molecules are shown as sticks. Left and right, surface regions mediating asymmetric complex formation are highlighted in magenta (enzyme) and yellow (substrate), respectively. b, Close-up view of contacts at the enzyme’s catalytic site. c,d, Expanded views of the dimer interface distal to the active site, with hydrogen bonds and hydrophobic interactions shown in c and d, respectively. e, Reversion of the engineered glutamic acid at position 669 to an arginine residue introduces an electrostatic clash with K659 of the enzyme kinase. f, Sequence alignment of the kinase domains of FGFR1–FGFR4. Residues of enzyme and substrate kinases that mediate the FGFR3KR669E A-loop-transphosphorylating asymmetric complex interface are highlighted in blue and green, respectively. Tandem tyrosine phosphorylation sites in the A-loop are in red.
At the active site of the enzyme, residues 646Asp–Tyr–Tyr648 of the A-loop of the substrate kinase form a short antiparallel β strand with residues 655Arg–Leu656 at the C-terminal end of the A-loop of the enzyme kinase (Fig. 2b). Residues 646Asp–Tyr–Tyr648 within the substrate kinase have a high temperature factor, implying that they interact weakly with the enzyme kinase (Supplementary Fig. 3a,e). Y647 (P0) inserts into the active site of the enzyme kinase, where its hydroxyl group makes hydrogen bonds with both D617 (the catalytic base) and R621 in the catalytic loop of the enzyme. These hydrogen bonds presumably act in concert to abstract a proton from Y647, priming it for a nucleophilic attack on the γ-phosphate of ATP located 2.8 Å away from it. This structural observation is consistent with our MS/MS data on FGFR2K showing that phosphorylation of Y656 precedes that of Y657 (Fig. 1e). Coordination of Y647 in the active site is also buttressed by interactions of D646 (P − 1) and Y648 (P + 1) with the enzyme kinase near the active site (Fig. 2b). Specifically, D646 (P − 1) makes water-mediated hydrogen bonds with R621 and R564 (in helix αD) of the enzyme, while Y648 (P + 1) loosely engages V658 and V700 at the periphery of the enzyme P + 1 pocket. The remainder of the enzyme and substrate A-loops do not participate in asymmetric complex formation and are consequently either highly flexible or altogether disordered. Consequently, the A-loops have much higher temperature factors relative to the rest of the protein, inflating the overall temperature factor. Indeed, the middle section of the enzyme A-loop (residues L645 to N653) has scattered electron density and was largely modeled on the basis of known crystal structures of activated FGFRKs21 (Supplementary Fig. 3d).
Distal to the enzyme active site, αEF and αG helices from the P + 1 pocket of the substrate kinase engulf the glycine/proline-rich loop between helices αF and αG of the enzyme kinase (Fig. 2c,d). Notably, substrate residues involved in the distal interface have much lower temperature factors (Supplementary Fig. 3b,c) relative to those engaged at the active site of the enzyme (Supplementary Fig. 3a). This implies that contacts at the distal site are the principal stabilizing forces of the asymmetric complex. A total of nine direct and water-mediated hydrogen bonds are formed between enzyme and substrate (Fig. 2c). Among these, two are mediated by the mutationally introduced E669 at the center of the interface; these evidently encourage formation of the A-loop-transphosphorylating asymmetric complex. D668 of the substrate kinase plays a prominent role in supporting this distal interface by forming a salt bridge with R571 of the enzyme kinase, while also making a backbone-mediated hydrogen bond with R568 of the enzyme kinase (Fig. 2c). Another critical hydrogen bond is formed by the backbone atoms of F667 (from the substrate kinase) and S693 (from the enzyme kinase). The few notable hydrophobic and van der Waals contacts involve L656, V658, L666, F667, V700, L703 and F704 from the substrate kinase and P696, I698 and P699 from the enzyme kinase (Fig. 2d). The kinase insert region—defined structurally as the loop between the αD and αE helices—does not contribute to the asymmetric complex interface (Supplementary Fig. 4).
The A-loop-transphosphorylating dimer is suppressed
The amino acid composition of the FGFR3KR669E asymmetric complex interface is strictly conserved among human FGFRs (Fig. 2f). Modeling showed that reversion of the engineered E669 in FGFR3 to an arginine in the asymmetric complex would create an electrostatic clash with an FGFR-invariant lysine (K659 in FGFR3) in the enzyme kinase (Fig. 2e), thereby suppressing formation of the A-loop-transphosphorylating asymmetric complex. It follows that the naturally occurring pathogenic R678G FGFR2 substitution (or our engineered R678A and R678E substitutions) promotes formation of the A-loop-tyrosine-phosphorylating dimer by eliminating the native electrostatic repulsion. In comparison to R678G and R678A substitutions, the engineered R678E substitution both abolishes the electrostatic repulsion and adds two hydrogen bonds to the interface. This explains why the R-to-E substitution in FGFR2 imparts faster phosphorylation of A-loop tyrosines as compared to the R-to-A and R-to-G substitutions at the same locus.
An asymmetric complex exists in solution
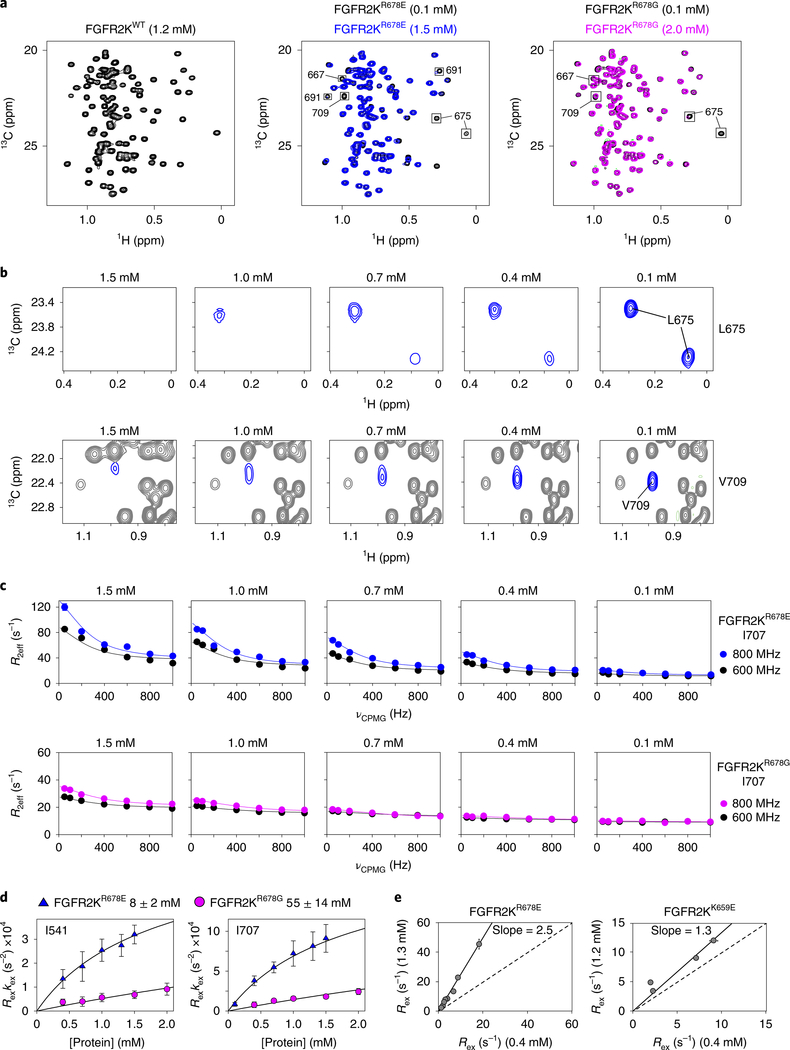
We applied NMR spectroscopy to detect the existence of an A-loop-transphosphorylating asymmetric complex in solution. Specifically, we acquired 1H/15N transverse relaxation-optimized (TROSY) and 13C heteronuclear multiple quantum coherence (HMQC) spectra23,24 on FGFR2KWT, FGFR2KR678E and FGFR2KR678G over a range of kinase concentrations. Regardless of concentration, spectra of FGFR2KWT showed well-resolved peaks of uniform intensity and peak height (Fig. 3a and Supplementary Fig. 5a). By contrast, FGFR2KR678E and FGFR2KR678G spectra contained less intense peaks at the highest concentration tested; moreover, several peaks corresponding to V667, K668, L675, I707, V709, E710, E711, L712 and F713 were completely absent in the FGFR2KR678E sample (Fig. 3a and Supplementary Fig. 5b). Successive dilutions of FGFR2KR678E and FGFR2KR678G led to the reappearance of missing peaks in FGFR2KR678E and an overall improvement in spectral quality (Fig. 3a,b and Supplementary Fig. 5b,c). Because chemical exchange between kinase monomers and dimers would be expected to cause peak broadening and intensity reduction, these data imply that FGFR2KR678E and FGFR2KR678G have a propensity to reversibly dimerize, while FGFR2KWT does not. Importantly, several of the missing and attenuated peaks correspond precisely to residues at the asymmetric complex interface in the crystal structure, including V667 (V658 in FGFR3K), L675 (L666 in FGFR3K) and V709 (V700 in FGFR3K) (Fig. 2). We conclude that FGFR2KR678E and FGFR2KR678G dimerize in solution via an interface identical to that observed in the crystal structure of the asymmetric FGFR3KR669E complex. Consistent with our NMR data, molecular dynamics simulation analyses showed that the crystallographically observed complex can stably persist on a timescale of at least 100 ns in silico. Notably, salt-bridge and hydrogen-bond interactions at the distal portion of the dimer interface were stable throughout three independent and unrestrained simulations (Supplementary Fig. 6a,b). Moreover, in agreement with the observed high temperature factors of A-loop residues in the crystal structure, A-loops underwent large r.m.s.d. fluctuations during simulations.
Fig. 3 |. The crystallographically deduced A-loop-transphosphorylation asymmetric complex forms in solution.
a, Overlays of leucine/valine regions of 1H/13C methyl HMQC spectra for FGFR2KR678E (middle) and FGFR2KR678G (right) mutants acquired at either high (1.5 mM for R678E; 2.0 mM for FGFR2KR678G) or low (0.1 mM) concentrations. Peaks sustaining >20% intensity loss are boxed. Left, corresponding spectrum of FGFR2KWT at 1.2 mM is shown for comparison. HMQC experiments were performed independently twice with similar results. b, Dilution-dependent reappearance of peaks corresponding to L675 (top) and V709 (bottom) for FGFR2KR678E. c, CPMG dispersion curves for I707 in FGFR2KR678E (top) and FGFR2KR678G (bottom) at the protein concentrations shown. Curves plotted in blue and black represent data collected at 800 MHz and 600 MHz, respectively. Note that the 0.1 mM FGFR2KR678E dataset in blue was collected at 900 MHz. d, Plots of kex × Rex derived from CPMG relaxation dispersion experiments for FGFR2KR678E and FGFR2KR678G as a function of protein concentration. Plots were globally fitted using multiple residues to estimate dimerization Kd values (boxed above). Error bars for kex × Rex and Kd values reflect errors from non-linear least squares fits. e, Correlation plots of Rex values for FGFR2KR678E (left) and FGFR2KK659E (right) determined at 1.3 mM and 0.4 mM (FGFR2KR678E) and 1.2 mM and 0.4 mM (FGFR2KK659E), respectively. A slope of 1.0 is indicated by the dashed line. For d and e, n = 1 using independent samples; two technical replicates were acquired for select CPMG frequencies. The center value is the optimal fit to the data using equation (2). For e, the solid line is a linear correlation with the best fit slope to the data reported and a y-intercept of 0.
To estimate the binding affinity between enzyme and substrate kinases within the asymmetric kinase dimer, we acquired Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion data25,26 on FGFR2KR678E and FGFR2KR678G over a range of concentrations and at two magnetic field strengths. FGFR2KWT and the gain-of-function FGFR2KK659E mutant were used as controls; we previously showed that FGFR2KK659E has a strong propensity to adopt an active-state conformation because the K659E substitution introduces intramolecular hydrogen bonds with R635 in the catalytic loop21,22,27,28. We formulated an equation relating CPMG-derived Rex and kex values to the protein concentration, which enabled us to derive the dissociation constant (Kd) of the dimer (equation (13)). FGFR2KWT showed no change in CPMG values, consistent with the absence of any tendency to dimerize. On the other hand, we did observe concentration dependent changes in the Rex and kex values for FGFR2KR678E and FGFR2KR678G, and were therefore able to calculate respective Kd values in the millimolar range (that is, 8 ± 2 mM and 55 ± 14 mM) (Fig. 3c,d, Supplementary Fig. 7 and Supplementary Table 2). By contrast, the intrinsically active FGFR2KK659E mutant showed only a modest protein-concentration-dependent change in CPMG-derived Rex values as compared to the strong concentration dependence displayed by FGFR2KR678E (Fig. 3e). These data underscore the unique capacity of the R678G/E substitution to encourage formation of the asymmetric complex.
Validation of the A-loop-transphosphorylating dimer
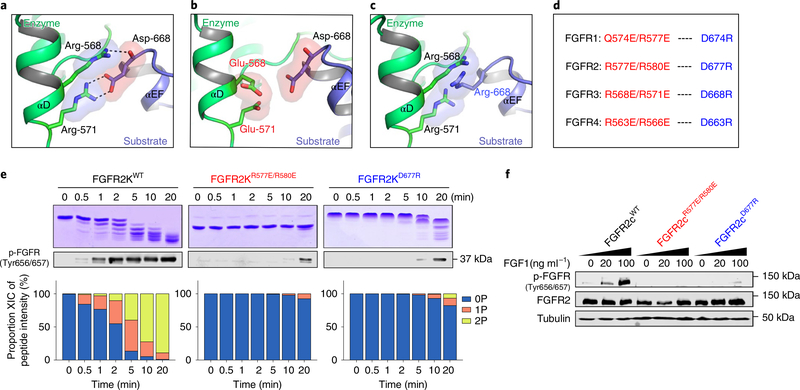
To test the functional validity of our structurally deduced A-loop-tyrosine-transphosphorylating asymmetric complex, we disrupted conserved salt-bridge and hydrogen-bonding interactions between the two arginines from the αD helix of the enzyme kinase and the aspartic acid residue in the αEF helix of the incoming substrate kinase in each of four recombinant FGFRKs (FGFR1K–FGFR4K) and their respective transmembrane forms (Fig. 4a). To maximally inhibit asymmetric complex formation, we replaced each of these three selected residues with oppositely charged amino acids so as to eliminate critical salt-bridge and hydrogen-bonding interactions and create an electrostatic clash between enzyme and substrate kinases (Fig. 4b–d). Consistent with structural predictions, as compared to their wild-type counterparts, all salt-bridge mutants were severely compromised in their ability to transphosphorylate A-loop tyrosines in vitro (Fig. 4e and Supplementary Fig. 8a–c). More importantly, these mutations completely obliterated the ligand-dependent A-loop-tyrosine phosphorylation activity of all four full-length FGFRs ectopically expressed on the surface of L6 myoblasts (Fig. 4f and Supplementary Fig. 8d–f). We conclude that the crystallographically deduced A-loop-transphosphorylating asymmetric complex is not reflective of a pathological phenomenon, but rather represents a bona fide mechanism whereby all four FGFR family members conduct A-loop-tyrosine phosphorylation in the context of ligand-induced dimers in living cells.
Fig. 4 |. Functional validation of the crystallographically deduced A-loop-transphosphorylating mechanism in vitro and in vivo.
a, Expanded view of the FGFR3KR669E asymmetric complex interface highlighting the key contribution of (i) the salt bridge between R571 (enzyme) and D668 (substrate), and (ii) the hydrogen bond between R568 (enzyme) and D668 (substrate) backbone (in each case shown as dashed lines). b,c, Introduction of a R568E/R571E double substitution in the enzyme kinase (b) or a D668R single substitution in the substrate kinase (c) are predicted to inhibit A-loop-transphosphorylating asymmetric complex formation by eliminating both salt-bridge and hydrogen-bonding interactions and by introducing electrostatic clashes. d, Equivalent residues in FGFR1–FGFR4 that mediate salt bridges and hydrogen bonds at the asymmetric complex interface and their corresponding substitution to residues with opposite charge, engineered to abolish dimerization. e, Kinetic analyses by native gel electrophoresis (top), immunoblotting (middle) and time-resolved LC–MS of A-loop-tyrosine phosphorylation (bottom) in wild-type FGFR2Ks and its variants harboring mutations predicted to disrupt the asymmetric complex. Kinase assays were done independently twice with similar results. f, Immunoblot analyses of whole lysates of buffer-treated or FGF1-stimulated L6 myoblasts overexpressing either full-length wild-type FGFR2 or corresponding variants harboring dimer-breaking substitutions. Blots were probed with anti-p-FGFR (Y656/Y657), anti-FGFR2 and anti-β-tubulin antibodies. Experiments were performed in biological triplicates with similar results. Full-length gels are shown in Supplementary Fig. 15b,c.
Asymmetry of the A-loop transphosphorylation complex
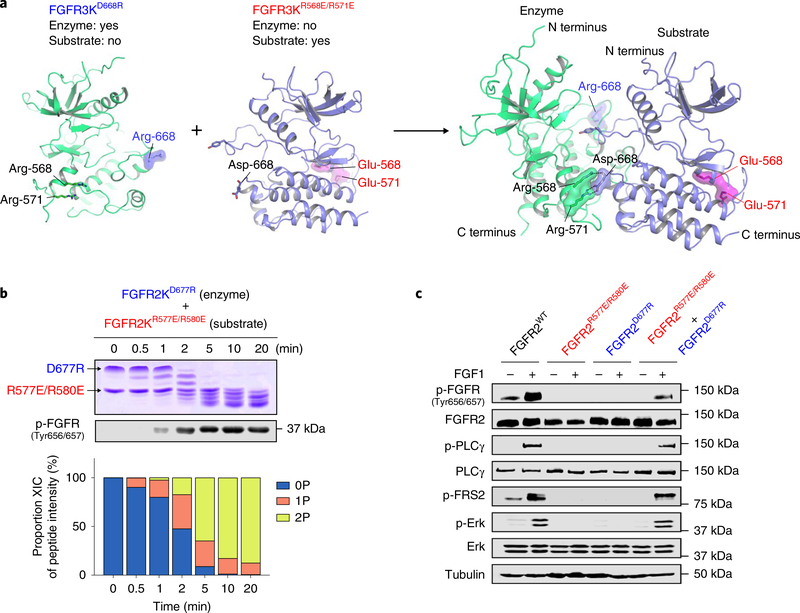
Given the asymmetry of the A-loop-transphosphorylating dimer, the two arginines from the αD helix, although essential for the function of the enzyme kinase, are dispensable for the ability of the substrate kinase to interact with the enzyme kinase. Conversely, the conserved aspartic acid in the αEF helix is essential for the ability of the substrate kinase to engage the enzyme kinase, but is dispensable for the function of the enzyme kinase (Fig. 5a). With these considerations in mind, we functionally tested the asymmetry of the A-loop-transphosphorylation complex by comparing the kinetics of A-loop transphosphorylation in a 1:1 mixture of FGFR2KR577E/R580E and FGFR2KD677R with the corresponding kinetics of FGFR2KR577E/R580E and FGFR2KD677R alone. We reasoned that in such mixtures, FGFR2KD677R and FGFR2KR577E/R580E should complement each other and form a productive A-loop-transphosphorylating asymmetric heterodimer in which FGFR2KD677R acts as the enzyme and FGFR2KR577E/R580E presents its A-loop tyrosine for phosphorylation. Indeed, we detected robust A-loop phosphorylation in the FGFR2KD677R:FGFR2KR577E/R580E mixture within 2 min (Fig. 5b), whereas neither FGFR2KR577E/R580E nor FGFR2KD677R alone showed any measurable A-loop-transphosphorylation activity (compare with Fig. 4e). A similar complementation took place in 1:1 mixtures containing corresponding salt-bridge mutants of FGFR1, FGFR3 and FGFR4 (Supplementary Fig. 9).
Fig. 5 |. In vitro and in vivo complementation assays reinforce the existence of an asymmetric A-loop-tyrosine transphosphorylation complex.
a, Cartoon representation of heterodimerization of FGFR3D668R and FGFR3R568E/R571E in which FGFR3D668R assumes the role of enzyme, while FGFR3R568E/R571E acts as substrate. Locations of mutated residues (E568 and E571, red; R668, blue) are highlighted. b, Kinetic analysis of phosphorylation of A-loop tyrosines in reactions containing equimolar amounts of FGFR2KD677R and FGFR2KR577E/R580E by native gel electrophoresis (top), immunoblotting with an anti-p-FGFR antibody (middle) and time-resolved LC–MS (bottom). Kinase assays were done independently twice with similar results. c, Lysates from buffer-treated or FGF1-treated L6 myoblasts stably expressing either wild-type FGFR2c, FGFR2cR577E/R580E or FGFR2cD677R alone, or co-expressing FGFR2cR577E/R580E and FGFR2cD677R, in each case analyzed by immunoblotting using antibodies specific for selected target proteins and their phosphorylated forms. An anti-β-tubulin antibody was used as a loading control. Experiments were performed in biological triplicates with similar results. Full-length gels are shown in Supplementary Fig. 15d,e.
We further interrogated the asymmetry of the A-loop-transphosphorylating complex by co-expressing full-length FGFR2KR577E/R580E and FGFR2KD677R in L6 cells. We reasoned that treatment of such cells with FGF1 should induce heterodimerization of FGFR2KR577E/R580E and FGFR2KD677R. We found that FGF1 stimulation of cells co-expressing these mutants led to clear phosphorylation of FGFR on A-loop tyrosines. This was mirrored by robust phosphorylation of two direct downstream FGFR substrates, namely PLCγ1 (on Y783) and FRS2α (on Y436), with subsequent activation of the Ras–MAP kinase cascade as measured by phosphorylation of MAPKs on T202/Y204 (Fig. 5c). These data provide compelling validation of our crystallographically deduced asymmetric mode of A-loop-tyrosine transphosphorylation in a physiological context.
Allosteric changes in enzyme and substrate kinases
To explore the existence of long-range allostery within the enzyme and substrate kinases, we engineered a double mutant (FGFR2KR678E/R577E) that primarily functions as a substrate for phosphorylation. As shown above, FGFR2KR678E is more proficient than its FGFR2KWT counterpart in serving as a substrate. The introduction of a R577E substitution further biases this double mutant to predominantly function as a substrate by impairing its ability to act as an enzyme. Thus, when mixed with FGFR2KWT, FGFR2KR678E/R577E should preferentially form a heterodimeric A-loop-transphosphorylating complex with FGFR2KWT as the enzyme.
With these considerations in mind, we interrogated enzyme-induced allostery in the substrate kinase via HMQC spectral analysis and methyl multiple quantum CPMG relaxation dispersion experiments29 on isotopically 13C-ILV methyl-labeled FGFR2KR678E/R577E in the presence of a twofold molar excess of unlabeled FGFR2KWT. We detected significant peak intensity reductions in HMQC spectra or enhancements in Rex values for residues in FGFR2KR678E/R577E distal to the dimer interface, including around the DFG motif (I541, L647), A-loop (I651, I654) and molecular brake (I548) (Supplementary Fig. 10). These data imply that binding of the enzyme kinase (that is, FGFR2KWT) induces conformational changes on a microsecond-to-millisecond timescale within the substrate kinase.
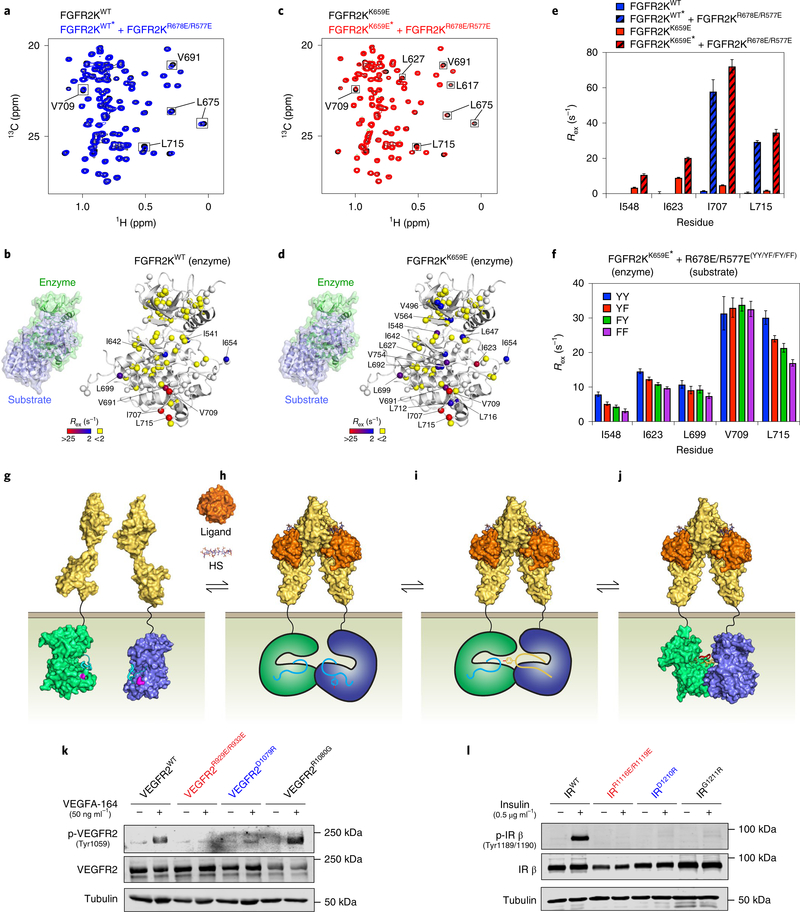
To probe for substrate-induced allostery in the enzyme kinase, we did the converse experiment in which an excess of unlabeled FGFR2KR577E/R678E was added to isotopically labeled FGFR2KWT. In comparison to the HMQC spectrum of FGFR2KWT alone, the corresponding spectrum of FGFR2KWT in the presence of FGFR2KR577E/R678E showed reduced peak intensities. Moreover, CPMG relaxation dispersion experiments gave increased Rex values exclusively for residues at the dimer interface, including I707, V709, and L715 (Fig. 6a,b and Supplementary Fig. 11a,b). Because FGFR2KWT is conformationally rigid, we attribute the absence of significant substrate-induced allosteric perturbations in the enzyme kinase to the repressed nature of the autoinhibitory state. To enhance sensitivity, we used the less autoinhibited and conformationally more dynamic FGFR2KK659E mutant as the enzyme kinase. We found that addition of a twofold molar excess of the FGFR2KR577E/R678E substrate kinase to an isotopically enriched FGFR2KK659E enzyme kinase led to significant reductions in HMQC peak intensities and enhancements in relaxation dispersions for residues well beyond those at the asymmetric complex interface (Fig. 6c and Supplementary Fig. 11c–e). Specifically, the FGFR2KK659E mutant incurred large increases in Rex values for residues in the catalytic loop (I623, L627), DFG motif (I642, L647) and molecular brake (I548) (Fig. 6d,e); the latter two regions are known to regulate the equilibrium between inhibited and active FGFR kinase states22. These structural changes distal to the dimer interface suggest that substrate binding promotes an active conformation in the enzyme kinase, facilitating transphosphorylation of the A-loop tyrosine of the substrate.
Fig. 6 |. Asymmetric complex formation induces reciprocal allosteric changes in enzyme and substrate kinases.
a,c, Overlays of 1H/13C HMQC (leucine/valine region) spectra of 0.4 mM isotopically labeled FGFR2KWT (a; blue) or FGFR2KK659E (c; red) either alone or together with 0.8 mM unlabeled substrate kinase (that is, FGFR2KR577E/R678E). Peaks sustaining >20% loss of intensity are boxed. Experiments were performed independently twice with similar results. b,d, Rex values (with range depicted by a boxed colored bar) derived from CPMG relaxation dispersion experiments for FGFR2KWT (b) or FGFR2KK659E (d) mixed with unlabeled FGFR2KR577E/R678E mapped onto the enzyme-acting kinase in the asymmetric complex crystal structure. e, Changes in Rex values of selected residues in FGFR2KWT or FGFR2KK659E enzyme kinase induced upon addition of substrate (that is, FGFR2KR577E/R678E). f, Reductions in CPMG-derived Rex values in FGFR2KK659E enzyme kinase when A-loop tyrosines (annotated YY) of FGFR2KR577E/R678E substrate kinase are substituted to YF, FY and FF. e,f, n = 1 using independent samples for each set of CPMG measurements acquired at two magnetic field strengths; error bars reflect the fitted error to equation (2). a,c,e,f, Isotopically enriched kinases contained in mixtures are indicated by asterisks. g–j, Induced-fit model for A-loop-tyrosine transphosphorylation. g, Asymmetric complex formation of FGFR kinases (enzyme and substrate in green and blue, respectively) is thermodynamically inhibited by a charge repulsion between K659 in the enzyme-acting kinase and R669 in the incoming substrate-acting kinase (both residues highlighted in pink). h, Energetic gains in extracellular FGF-induced FGFR dimerization offset these repulsive forces, facilitating formation of a C lobe–C lobe-mediated asymmetric kinase dimer. HS, heparan sulfate. i, Asymmetric complex formation imparts upon the substrate A-loop a more phosphorylatable conformation (indicated as a change in color to yellow). j, This encourages the A-loop of the enzyme to adopt the active state (depicted by a change in color to red), resulting in the formation of an A-loop-tyrosine transphosphorylation complex as revealed by the crystal structure. k,l, Immunoblot analyses of L6 myoblast cell lines overexpressing either full-length mouse wild-type VEGFR2 (k) or human wild-type insulin receptor (IR) (l), together with variants harboring either a R1080G substitution (k) or a G1211R substitution (l) (in each case corresponding to FGFR2 R678) plus dimer-disrupting substitutions R929E/R932E and D1079R (k) or R1116E/R1119E and D1210R substitutions (l) (in each case corresponding to FGFR2 R577E/R580E and D677R). Cells were stimulated with either VEGF (k) or insulin (l) at the concentrations shown. Whole-cell lysates were analyzed by immunoblotting using antibodies specific for p-VEGFR2, VEGFR2 (k) or antibodies specific for phosphorylated human insulin receptor (p-hIR) or human insulin receptor (hIR) (l). k,l, An antibody to β-tubulin was used as a loading control. Experiments were performed in biological triplicates with similar results. Full-length gels are shown in Supplementary Fig. 15f,g.
On the basis of the crystal structure, substrate-induced allosteric changes in the enzyme kinase could arise from enzyme–substrate contacts either at the active site of the enzyme and/or distal to it. To assess the relative contribution of contacts proximal to the active site of the enzyme to substrate-induced enzyme allostery, we replaced the tandem A-loop tyrosines (that is, Y656/Y657) of FGFR2KR678E/R577E either individually or in combination with phenylalanine: R678E/R577EYF, R678E/R577EFY and R678E/R577EFF (Fig. 6f and Supplementary Fig. 12). Addition of a twofold excess of each of these three unlabeled substrate kinases to an isotopically enriched FGFR2KK659E enzyme kinase led to reductions in Rex values for I548, I623, L699 and L715 in the order R577E/R678EYF > R577E/R678EFY > R577E/R678EFF (Fig. 6f). These data imply that contacts at the active site of the enzyme as well as contacts at the distal site act together to facilitate the active-state conformation of the enzyme kinase. On the basis of these data, we propose an induced-fit model in which asymmetric complex formation imparts upon the substrate kinase a more phosphorylatable A-loop conformation. This in turn supports the active conformation of the enzyme kinase, thus enabling the kinase to transphosphorylate the A-loop tyrosine of the substrate (Fig. 6g–j).
Generality of transphosphorylating asymmetric complex
Of a total of 58 human RTK superfamily members, 24 have an arginine or lysine residue at the locus corresponding to FGFR-invariant R678 and K668 (FGFR2 numbering) (Supplementary Fig. 13a). Besides FGFR1–FGFR4, these include all three members of the VEGFR family (VEGFR1–VEGFR3), TRK family (TRKA, TRKB and TRKC), two members of the TAM (TYRO3-, AXL- and MER-TK) receptor family (that is, AXL and MER), eight ephrin type A (EphA1–EphA8) and four ephrin type B (EphB1–EphB4) receptors (Supplementary Fig. 13a). This implies that A-loop-tyrosine transphosphorylation in these RTKs is also suppressed by antagonizing electrostatic forces. Notably, VEGFR1–VEGFR3, AXL and MER also conserve the two residues that mediate the FGFR-invariant R571:D668 salt bridge (in FGFR2), a key contributor of binding energy for asymmetric complex formation (Supplementary Fig. 13b). It therefore seemed highly likely that in common with FGFRs, VEGFR1–VEGFR3, AXL and MER also form an asymmetric complex to conduct A-loop-tyrosine phosphorylation. To test this conjecture, we selected VEGFR2 as representative of this group of RTKs and established L6 cell lines expressing either full-length wild-type VEGFR2 or variants thereof harboring either a R1080G substitution (corresponding to the R678G gain-of-function substitution in FGFR2) or the R929E/R932E and D1079R substitutions (corresponding to dimer-disrupting R577E/R580E and D677R substitutions in FGFR2). We found that the R1080G substitution enhanced VEGF-induced A-loop transphosphorylation of VEGFR2, whereas the R929E/R932E and D1079R substitutions completely abolished it (Fig. 6k). We conclude that formation of asymmetric A-loop-transphosphorylating dimers is a shared feature of multiple members of the RTK superfamily.
The dimer-suppressing FGFR-invariant R678 (in FGFR2) is not conserved in ten RTKs, namely insulin receptor, IGF1R, PDGFRα, TYRO3, RET, ROS, ALK, LTK, PTK7 and CCK4. Notably, insulin receptor, IGF1R, ROS, ALK, LTK, PTK7 and CCK4 all possess a glycine at this locus, which corresponds to the R678G FGFR2 pathogenic substitution. In PDGFRα and TYRO3, this locus is occupied by an asparagine, while in RET it is replaced by a histidine. Intriguingly, however, these ten RTKs still conserve the salt-bridge-forming residues that mediate the asymmetric complex in FGFR1–FGFR4, VEGFR1–VEGFR3, AXL and MER. On the basis of these observations, we hypothesized that A-loop-tyrosine phosphorylation in these RTKs also proceeds via an asymmetric kinase complex. We therefore selected insulin receptor as an example of this set of RTKs and engineered L6 cell lines expressing either full-length wild-type insulin receptor or variants thereof harboring either the R1116E/R1119E or D1210R substitutions that correspond to the dimer-disrupting R577E/R580E and D677R substitutions in FGFR2. As an additional test, we established an L6 cell line expressing an insulin receptor variant harboring a G1211R substitution to introduce an electrostatic repulsion between enzyme and substrate kinases as occurs in FGFR1–FGFR4, VEGFR1–VEGFR3, AXL and MER. In contrast to wild-type insulin receptor, both R1116E/R1119E and D1210R insulin receptor mutants completely failed to undergo A-loop-tyrosine phosphorylation in response to insulin stimulation (Fig. 6l). The G1211R substitution also incurred a major loss in its ability to undergo insulin-induced A-loop-tyrosine phosphorylation. These results strongly suggest that asymmetric complex formation is a general mechanism for A-loop-tyrosine phosphorylation in multiple RTKs.
Discussion
In comparison to other biological complexes (such as ligand–receptor complexes, whose interfaces typically bury a surface area ranging from 2,000 to 5,000 Å2), the dimer interface is overwhelmingly hydrophilic and buries a modest total surface area of only 1,112 Å2. Furthermore, contacts between enzyme and substrate kinases at the active site of the enzyme are rather transient, thus contributing minimally to asymmetric complex stability. Indeed, stable dimers of FGFR kinases or their R-to-E derivatives were undetectable using conventional techniques such as size-exclusion chromatography, multiangle light scattering or surface plasmon resonance spectroscopy in solution. The thermodynamically weak nature of the asymmetric complex that drives A-loop-tyrosine phosphorylation makes perfect physiological sense: it safeguards against undesired ligand-independent A-loop-tyrosine transphosphorylation that has pathological consequences (as exemplified by the R678G substitution responsible for Crouzon syndrome) and ensures that A-loop-tyrosine transphosphorylation—and hence RTK signaling—is fastidiously controlled by ligand-induced extracellular dimerization. Specifically, intracellular kinase domains will only assemble into A-loop-transphosphorylating asymmetric complex when they are forced into proximity upon ligand-induced dimerization of the receptor extracellular domains. A salient feature of this process is a delicate interplay between energetic gains in extracellular ligand–receptor dimerization on the one hand and a weak propensity of the intracellular kinase to form asymmetric complex on the other. It follows that differences in the abilities of various FGF ligands to bind and dimerize the extracellular domains of cognate FGFRs lead to differential stabilization of A-loop-transphosphorylating asymmetric complex, resulting in corresponding differences in A-loop-tyrosine phosphorylation and hence kinase activation and signaling. The pathogenic FGFR2 R678G substitution subverts this delicate balance by lowering the energetic barrier that impedes formation of asymmetric complexes.
The asymmetric A-loop-transphosphorylation model we present is not confined to the FGFR family alone; we present compelling evidence that it is applicable to multiple other RTK family members that rely on A-loop-tyrosine transphosphorylation for activation. Notably, the reduced A-loop-transphopsphorylating activity of the G1211R insulin receptor variant implies that formation of an A-loop-transphosphorylating asymmetric complex is suppressed to different extents among different RTK members. Thus, the 678 locus (FGFR2 nomenclature) in RTKs serves as a critical nexus for the regulation of RTK signaling by controlling the rate of A-loop transphosphorylation.
Methods
Bacterial and mammalian expression constructs
cDNA fragments encoding minimal kinase domains of human FGFR1–FGFR4 were amplified by PCR and subcloned into appropriate restriction sites in the pETDuet-1 bacterial expression vector (69909–3, Novagen) in-frame with an N-terminal 6×His tag as an aid in protein purification. cDNA fragments encoding full-length human FGFR1–FGFR4, VEGFR2 and insulin receptor were amplified by PCR and subcloned into lentiviral transfer plasmids pEF1α-IRES-Neo or pEF1α-IRES-Hygro using a ligation-independent In-Fusion HD cloning kit (639648, Clontech Laboratories). The resulting constructs were then served as templates to introduce single- or multiple-site mutations using a QuikChange mutagenesis kit (Stratagene), Q5 Site-Directed Mutagenesis kit (E0554S, New England Biolabs) or an In-Fusion HD cloning kit. To prevent disulfide-linked dimerization of FGFR kinases, the conserved surface-exposed cysteine in the nucleotide-binding loop (also termed the glycine-rich loop) of each FGFR kinase was substituted to alanine (FGFR1K, C488A; FGFR2K, C491A; FGFR3K, C482A; FGFR4K, C477A]. In the case of FGFR1K and FGFR3K, an additional surface-exposed cysteine in the kinase insert region of these kinases were replaced with serine (FGFR1K, C584S; FGFR3K, C582S). All PCR primers were designed using NEBaseChanger software v.1.2.6 (New England Biolabs) or the In-Fusion cloning primer design tool (Clontech Laboratories). The authenticity of each expression construct was confirmed by restriction enzyme digestion and DNA sequencing. Construct information are provided in Supplementary Table 3.
Expression and purification of FGFR kinases
Competent BL21 (DE3) Escherichia coli cells were transformed with kinase expression constructs, cultured at 37 °C to an optical density at 600 nm of between 0.6 and 0.8, and protein expression was induced with 1 mM IPTG overnight at 20 °C. Cells were collected by centrifugation at 5,000g and 4 °C (Beckman Coulter, J6-M1) and lyzed in 25 mM HEPES, pH 7.5 buffer containing 150 mM NaCl and 10% glycerol using an Emulsiflex-C3 homogenizer (Avestin), followed by centrifugation at 40,000g for 60 min at 4 °C (Beckman Coulter, Avanti J-25). Supernatants containing the N-terminal His-tagged kinase proteins were filtered through a 0.45-μm membrane (295–3345, Nalgene), diluted with 25 mM HEPES, pH 7.5 buffer containing 150 mM NaCl and applied to a 5-ml prepacked HisTrap excel Ni2+ metal affinity chromatography column (17371206, GE Healthcare Life Sciences). Bound kinase proteins were eluted with a linear gradient of 18 column volumes of imidazole (0–0.5 M). Fractions containing kinase proteins as determined by SDS-PAGE were loaded onto a 20-ml Source 15Q anion exchange chromatography column (17094701, GE Healthcare Life Sciences) equilibrated in 25 mM Tris-HCl buffer, pH 8.0. Kinase proteins were eluted using a linear gradient of 13 column volumes of NaCl (0–0.5 M) in the same buffer. Fractions containing kinase proteins were pooled, concentrated to about 5 mg ml−1 and treated overnight at 18 °C with FastAP Thermosensitive Alkaline Phosphatase (EF0651, Thermo Scientific Fisher) to remove all traces of phosphorylation. A further round of Source Q column chromatography yielded highly homogenous kinase preparations as judged by native gel electrophoresis (Supplementary Fig. 14). All column chromatography purifications were performed on an AKTA Pure 25 system (GE Healthcare) at 4 °C. Purified kinase proteins were flash frozen in small aliquots in liquid nitrogen and stored at −80 °C until use. Protein concentrations were determined using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies) at 280 nm.
Crystallization and X-ray crystal structure determination
FGFR3R669E was crystallized by mixing 2 μl of protein (20 mg ml−1) with 2 μl of crystallization buffer consisting of 0.1 M HEPES, pH 7.5 and 1.8 M (NH4)2SO4 using the hanging drop vapor diffusion method at 18 °C. Crystals grew over a period of 3–4 weeks and were cryoprotected by stepwise transfer into mother liquor supplemented with an increasing amount of glycerol up to 25%. These were then mounted on CryoLoops (Hampton Research) and flash frozen in liquid nitrogen. Diffraction data were collected to 2.2 Å on a single protein crystal at beamline X4A at the Brookhaven National Laboratory at 100 K with an ADSC Quantum4 CCD detector, a wavelength of 0.97910 Å, and a crystal-to-detector distance of 200 mm. FGFR3R669E crystals belonged to the monoclinic space group P21 and contained two molecules in the asymmetric unit. X-ray diffraction data were indexed, integrated and scaled using XDS and SCALA modules from the CCP4 software suite30. A clear molecular replacement solution was found for both copies of FGFR3R669E using the Phaser module of PHENIX31 and the crystal structure of mutationally activated FGFR3KK650E (Protein Data Bank accession 4K33)28 as search model. To avoid any bias, A-loops were omitted from the search model. Iterative rounds of model building and refinement were carried out using Coot32 and the Phenix.Refine module of PHENIX31. The structure was refined to a resolution of 2.2 Å with working and free R factors of 19.28% and 22.99%, respectively. X-ray diffraction data collection and structure refinement statistics are summarized in Supplementary Table 1. On the basis of MolProbity analysis performed within PHENIX31, the final model has an overall score of 1.7, an all-atom clash score of 2.1, no Cβ deviations, with the Ramachandran plot showing 99.83% of residues in favored and allowed regions, and 2.8% of residues flagged as rotamer outliers.
Catalytic turnover rate measurements via radiolabeled kinase assay
Intrinsic activities of FGFR2KWT and its various mutants were determined by measuring their ability to phosphorylate an optimal octapeptide substrate of FGFR, that is, AEEEYFFL33, fused to the C terminus of glutathione S-transferase. The fusion protein substrate was expressed in DH5α E. coli cells and purified via single-step glutathione affinity chromatography. To generate a phosphorylation signal under initial rate kinetics (that is, to minimize A-loop-tyrosine phosphorylation), a low concentration of kinase (10–50 nM) was incubated in 50 μl of reaction buffer (50 mM EPPS-NaOH, pH 8.0, 5% glycerol and 0.05% β-mercaptoethanol) containing 30 μM peptide substrate, 0.2 mM γ−32P-ATP (specific activity of 0.6 mCi mMol−1) and 12 mM MgCl2 for 10 min at 30 °C. Thirty-five microliters of each reaction mixture was spotted onto phosphocellulose filter paper strips (2 × 1 cm) and the substrate immobilized by rinsing the filters in 5% trichloracetic acid at 65 °C three times for 10 min each. Incorporation of 32P into the peptide substrate was quantified by liquid scintillation counting.
A-loop phosphorylated forms of kinases were generated by incubating kinase (25 μM) with 2 mM ATP and 12 mM MgCl2 for 2 h at 30 °C. Reactions were quenched by addition of 12 mM EDTA followed by 100-fold dilution into kinase assay buffer. The respective unphosphorylated forms were prepared identically except that MgCl2 was omitted from the reaction. The specific activity of unphosphorylated (that is, −MgCl2) and A-loop-phosphorylated (that is, +MgCl2) kinases was determined via the same peptide substrate phosphorylation assay as described above, except that 50 μM substrate was used.
Quantitative analysis of A-loop tyrosine transphosphorylation by LC–MS
Transphosphorylation on A-loop tyrosines was initiated by mixing wild-type or mutated FGFR kinases with reaction buffer containing ATP and MgCl2 to final concentrations of 67.5 μM (kinase), 25 mM (ATP) and 50 mM MgCl2. Reactions were quenched at different times by adding EDTA (final concentration 50 mM) to the reaction mixture. The progress of FGFR kinase transphosphorylation was monitored by native PAGE (17062401, GE Healthcare) and immunoblot using anti-p-FGFR antibody specific for phosphorylated A-loop tyrosine. To accurately quantitate phosphorylation on A-loop tyrosines, kinase reaction products were resolved by SDS-PAGE (14%), the proteins were stained with Bio-safe Coomassie G250 (Bio-Rad) and bands were excised. Following complete destaining with 50% methanol, 25 mM NH4HCO3 and 30% acetonitrile, each gel section was diced into small pieces, dehydrated with acetonitrile and dried by vacuum centrifugation. Gel pieces were rehydrated with 12.5 ng μl−1 protease trypsin solution (Trypsin Gold (mass spectrometry grade), Promega) in 50 mM NH4HCO3 and incubated at 37 °C for 4 h. The resulting peptides were extracted twice with 5% formic acid and 50% acetonitrile followed by a final extraction with acetonitrile. Samples were concentrated by vacuum centrifugation and peptides were desalted using a Stage Tip manually packed with Empora C18 High Performance Extraction Disks. LC–MS/MS analysis was performed using a Thermo Scientific Q Exactive High Field mass spectrometer coupled to a Thermo Scientific EASY-nLC 1000 (Thermo Fisher Scientific) equipped with a self-packed 75 μm × 20 cm reverse phase column (New Objective PicoTip Emitter) packed with Reprosil C18 (3 μm; Dr. Maisch) for peptide separation. The analytical column was placed in a column heater (Sonation) set to 45 °C. Peptide mixtures were loaded onto the analytical column with buffer A (0.1% formic acid) at a maximum back-pressure of 300 bar; they were then eluted with a 3–40% acetonitrile gradient in 0.1% formic acid over 60 min at a flow rate of 250 nl min−1. The mass spectrometer was operated in data-dependent (DDA) mode with survey scans acquired at a resolution of 120,000 over a scan range of 300–1,750 m/z. Up to ten of the most abundant precursors from the survey scan were selected with an isolation window of 1.6 Th and fragmented by higher-energy collisional dissociation with a normalized collision energy of 27. The maximum ion injection time for the survey and MS/MS scans was 60 ms and the ion target value for both scan modes was set to 3e34.
All mass spectra were converted to mgf peak list format using Proteome Discoverer v.1.4 (Thermo Fisher Scientific) and generated mgf files were searched against a human Uniprot protein database using Mascot (Matrix Science; v.2.5.0; http://www.matrixscience.com). Decoy proteins were added to the search to allow for the calculation of false-discovery rates (FDR). The search parameters were as follows: (i) two missed cleavage tryptic sites are allowed; (ii) precursor ion mass tolerance = 5 ppm; (iii) fragment ion mass tolerance = 0.1 Da; and (iv) variable protein modifications are allowed for phosphoserine, phosphothreonine and phosphotyrosine, for methionine oxidation, deamidation of asparagine and glutamines, and protein N-terminal acetylation. MudPit scoring was typically applied using a significance threshold score of P < 0.01. A decoy database search was always activated and, in general, with P < 0.01, the FDR averaged around 1% for peptide identifications. The Mascot search result was finally imported into Scaffold (Proteome Software, v.4.7.3) to further analyze MS/MS on the basis of protein and peptide identifications. X! Tandem (The GPM, https://thegpm.org; version CYCLONE (2010.12.01.1)) was performed and its results were merged with those from Mascot. The two search engine results were combined and displayed at 1% FDR with a minimum peptide requirement of 2. Peptide spectral matches to tyrosine-phosphorylated FGFR peptides were listed and further analyzed with Thermo Fisher Scientific Xcalibur (v.4.1.31.9) software. Layouts containing predicted experimental masses with a mass accuracy set to 5 ppm were constructed. Finally, phosphotyrosine peptide intensities were manually extracted and tabulated. The intensities of peptides as (i) unmodified, (ii) modified with one phosphate and (iii) modified with two phosphates were recorded and the percentage of each modification (calculated as a fraction of the total ion intensities for all three peptide states, including tryptic and one missed cleavage tryptic peptide for each state) was calculated for each time point. The plateau plot illustration in Fig. 1e shows spectra for the missed cleavage peptide of sequence DINNIDYYKK including the unmodified (0P) and those with one (1P) and two phosphates (2P).
Quantitative analysis of substrate peptide phosphorylation by MALDI–TOF
An N-terminally His-tagged substrate peptide consisting of residues L761 to T821 of FGFR2 was expressed and purified using sequential Ni2+ metal affinity chromatography and size-exclusion chromatography. This substrate peptide corresponds to the C-terminal tail of FGFR2 and contains five authentic tyrosine phosphorylation sites (Y769, Y779, Y783, Y805 and Y812). Wild-type and mutant FGFR2 kinases and their respective YY-to-FF derivatives were mixed with kinase reaction buffer containing ATP, MgCl2 and the substrate peptide to final concentrations of 13.5 μM (kinase), 262 μM (substrate), 10 mM (ATP) and 20 mM (MgCl2). The reactions were quenched at different time points by adding EDTA to the reaction mix to a final concentration of 50 mM. The progress of substrate phosphorylation was monitored by native PAGE and phosphate incorporation into the substrate peptide was quantified by time-resolved MALDI–TOF mass spectrometry (Autoflex MALDI–TOF mass spectrometry operated in linear ion mode; Bruker Daltonics) by comparative analysis of signals from phosphorylated and cognate non-phosphorylated peptides as previously described35.
Molecular dynamics simulations
Molecular dynamics simulations were done using the AMBER 16.06 package with the ff14SB36 force field for the protein and the TIP3P model for water. Force-field parameters for ATP37 and magnesium38 were used to simulate ATP-bound complexes. Three models for the enzyme kinase were prepared with Modeller39 using the crystal structure of the FGFR3R669E asymmetric complex and filtered on the basis of any steric clashes toward the enzyme kinase. The kinase insert region was modeled using the complete kinase insert region of FGFR1-inhibited structure 3KY2 (ref. 40). The three models were neutralized using Na+ ions and solvated in a cubic TIP3P water box with a 15 Å buffer between protein and boundary. Each model was equilibrated using repeated minimization and restrained dynamics. Solvated complexes were initially minimized using a conjugate gradient minimization for 2,000 steps followed by a 200 ps constant volume simulation at 300 K with a tight restraint of 500 kcal mol−1 on crystal waters, ATP and heavy atoms of the proteins. This was followed by a second round of conjugate gradient minimization for 2,000 steps and a 200 ps restrained constant volume simulation with a reduced restraint of 50 kcal mol−1. Iterative 200 ps constant volume simulations were performed with reduced restraint from 50, to 10, to 2 kcal mol−1 at 300 K. Two constant pressure simulations were then run for 200 ps with restraints of 2.0 and 0.5 kcal mol−1 at 300 K. Finally, 150 ns unrestrained molecular dynamics simulations were carried out for each model, in which the first 50 ns were considered as equilibrium simulations and the last 100 ns simulations were considered as production runs. Snapshots from unrestrained production runs were collected every 10 ps for analysis. For all molecular dynamics simulations, time steps were set at 2 fs with SHAKE constraints, with the particle mesh Ewald41 implementation for electrostatics and a 12 Å cutoff for non-bonded interactions.
Cell culture, lentivirus production and generation of stable cell lines
A lentiviral expression system was used to stably express full-length wild-type human FGFR1–FGFR4, mouse VEGFR2 or human insulin receptor and their various mutated versions in (rat) L6 cells; the latter is a myoblast cell line (CRL-1458, ATCC) with negligible expression of either FGFR or VEGFR2 and a low level of expression of insulin receptor. For virus production, HEK293T cells were seeded in 100-mm culture dishes and co-transfected with 5 μg of lentiviral transfer plasmid encoding wild-type receptors and mutants thereof, 1.6 μg of pMD2.G envelope plasmid and 2.5 μg of psPAX2 packaging plasmid using the calcium phosphate co-precipitation method. Fresh medium was added to the cells for a 3-d period after transfection. Cell culture supernatants containing recombinant lentivirus particles were collected, centrifuged and filtered. L6 cells were infected by addition of 2 ml of viral stock and 5 μg ml−1 polybrene (134220, Santa Cruz Biotechnology) in six-well cell culture dishes overnight. Following infection, cells were subjected to selection by addition of 0.5 mg ml−1 G418 (6483, KSE Scientific) or 100 μg ml−1 hygromycin B (ant-hg-1, InvivoGen) for 7–10 d. Stable expression of recombinant receptor proteins in all cell lines was verified by immunoblotting. Stable cell lines were maintained in DMEM (10–017-CV, Corning) supplemented with 10% FBS (FBS-01, LDP), 100 U ml−1 penicillin plus 100 μg ml−1 streptomycin (15140–122, Gibco) and 0.5 mg ml−1 G418 or 100 μg ml−1 hygromycin B.
Cell stimulation and phosphorylation analysis
L6 myoblasts stably expressing FGFRs, VEGFR2 or insulin receptor were grown in 100-mm culture plates until 80–90% confluence and serum starved in DMEM/F12 medium 1:1 (SH30023.02, HyClone) overnight. Cells were stimulated with FGF1, VEGFA-164 (493-MV-005, R&D) or insulin (I2643, Sigma) at the concentrations stated in the text for 10 min so as to induce receptor transphosphorylation. Cells were then lyzed in RIPA buffer (89900, Thermo Fisher Scientific) containing protease (88665, Thermo Fisher Scientific) and phosphatase (88667, Thermo Fisher Scientific) inhibitors. Samples were subjected to 8% SDS-PAGE and transferred onto nitrocellulose membranes (1620115, Bio-Rad). The membrane was blocked in Tris-buffered saline, pH 7.6 containing 0.05% Tween-20 and 5% BSA (BP1600–100, Fisher BioReagents) for 1 h. Phosphorylation on FGFR A-loop tyrosines was detected using a specific anti-p-Y653/654 antibody (3471, Cell Signaling Technologies). A rabbit anti-FGFR1 antibody was raised by immunizing rabbits with C-terminal tail peptide of FGFR1 fused to the C terminus of glutathione S-transferase (Cocalico Biologicals). Anti-FGFR2 (11835), anti-FGFR3 (4574), anti-FGFR4 (8562), anti-p-PLCγ (Y783; 2821), anti-PLCγ (2822), anti-p-FRS2 (Y436; 3861), anti-p-ERK1/2 (T202/Y204; 4370) and anti-p-VEGFR2 (Y1059; 3817), anti-phosphorylated insulin receptor β (Y1146; 3021) and anti-insulin receptor β (3025) antibodies were purchased from Cell Signaling Technologies; anti-ERK2 (sc-153) was obtained from Santa Cruz and an anti-β-tubulin antibody (PA1–41331) was purchased from Thermo Fisher Scientific. An anti-VEGFR2 antibody was generously provided by N. Rahimi (Boston University). Incubation with all primary antibodies was followed by incubation with an anti-rabbit IRDye secondary antibody (926–32211, LI-COR) for 60 min. Membranes were imaged using an Odyssey Fc Dual-mode Imaging System (LI-COR).
NMR dilution experiments
Protein-concentration-dependent changes in peak height/intensity were examined in 4-mm Shigemi tubes at 25 °C using a 600 MHz Bruker Avance III spectrometer equipped with a 5-mm TCI cryogenic probe. 1H/15N TROSY and/or 1H/13C HMQC spectra were acquired on the following samples at two concentrations: FGFR2KR678E (1.5 mM and 0.1 mM), FGFR2KR678G (2.0 mM and 0.1 mM) and mixed dimers of FGFR2KWT/FGFR2KR678E/R577E and FGFR2KK659E/FGFR2KR678E/R577E (0.4 mM isotopically enriched protein, 0.8 mM natural abundance protein). Spectral widths in the direct and indirect dimensions for each experiment were 12,019.2 Hz and 2,190.1 Hz for 1H/15N TROSY, and 10,000 Hz and 3,017.5 Hz for 1H/13C HMQC, respectively. An acquisition time of 59.8 ms was used in the direct dimension and an evolution time of 33.8 ms (15N) or 32.8 ms (13C) was used in indirect dimensions. A recycle delay of 1 s was used for all experiments with 4–64 scans implemented depending on the protein concentration. Intensity ratios were calculated by dividing the peak heights for each residue at the higher concentration by the corresponding value at the lower concentration. Errors were propagated using the s.d. of the noise within each spectrum acquired at the two concentrations.
CPMG relaxation dispersion experiments
Methyl CPMG experiments29 were acquired on Bruker 600, 800, and 900 MHz (600 MHz at New York University; 800 and 900 MHz at the New York Structural Biology Center) instruments equipped with 5-mm TCI cryogenic probes. All data were acquired at 25 °C using a constant time period of 40 ms. Frequencies of 180° pulses (νCPMG) used during this constant time were 0, 50, 100, 200, 400, 600, 800 and 1,000 Hz. In each case, the 50 and 1,000 Hz data were acquired twice. Direct and indirect spectral widths for methyl CPMG experiments at 600, 800, and 900 MHz were 10,000 and 3,017.5 Hz, 12,019.2 and 4,025.8 Hz, and 12,019.2 and 4,524.9 Hz, respectively, with an acquisition time of 59.8 ms in the direct dimension and ~32.8 ms in the indirect dimension. Recycle delays used were 2.5, 2.2, and 2.25 sec at 600, 800, and 900 MHz, respectively, and the number of scans used was either 4 or 16. All data were processed using NMRPipe42 and analyzed using Sparky (https://www.cgl.ucsf.edu/home/sparky/). R2eff values were calculated using the following equation:
| (1) |
where Iν is the peak intensity at a given CPMG frequency, I0 is the peak intensity with no constant time delay and T is the constant time period.
Rex and kex values were fitted according to the fast-exchange approximation of the Carver–Richards equation43,44 on a residue-by-residue basis:
| (2) |
where is the intrinsic relaxation rate of the system, pA and pB are the populations of A and B, respectively, Ikex is the sum of the forward and reverse rate constants corresponding to the interconversion of populations A and B, and Δω is the difference in chemical shift between populations. Note that the above analysis assumes no contribution from 1H chemical shift dispersion (that is, Δω for 1H was equal to zero). CPMG-derived Rex and kex values used for Kd determination were obtained by simultaneous fitting equation (2) using data from two magnetic field strengths. CPMG-derived Rex values for FGFR2KK659E at 1.2 mM and 0.4 mM were obtained from fits to data at 600 MHz only.
Determination of binding affinity using fitted parameters from CPMG relaxation dispersion experiments
The following derivation relates the dissociation constant (Kd) and the total kinase concentration (PT) to the fitted Rex and kex values obtained from CPMG relaxation dispersion experiments. [D] and [M] are the concentrations of dimer and monomer, respectively.
| (3) |
| (4) |
Solve for [M]
| (5) |
Substitute equation (5) into equation (3)
| (6) |
Solve for [D] and use the following quadratic solution
| (7) |
For CPMG relaxation dispersion experiments, we found that in most cases, only the enzyme or substrate kinase displayed relaxation dispersion. Furthermore, the relaxation dispersion curves fit well to a two-state fast-exchange equation (equation 2). For this reason, we define the two states, A and B, to correspond to the monomer plus the enzyme kinase (or substrate kinase) and the substrate kinase (or enzyme kinase). As the substrate kinase and enzyme kinase in the dimer are equal to the dimer concentration, we can write the fraction of population A (pA) and B (pB) as follows
| (8) |
| (9) |
The relaxation owing to chemical exchange (Rex) is given by the following expression
| (10) |
Substitute the value of pA from equation (8) into equation (10)
| (11) |
Substitute the expression for pB in equation (9) into equation (11)
| (12) |
Substitute the expression for [D] in equation (7) into equation (12)
| (13) |
The fitted Rex and kex values from CPMG curves at each PT value were used to obtain the value of Kd from equation (13) (Supplementary Table 2). To generate the Kd values listed in Fig. 3, a global fit to equation (13) was used for residues I541 and I707 that displayed relaxation dispersions in only the substrate or enzyme kinase, respectively. This was needed to ensure that the assumption made in equation (9) was accurate (that is, pA = [M] + [D]). The global fits to derive the Kd values assumed that Δω was the same for I541 and I707 among FGFR2KR678E and FGFR2KR678G, and that the CPMG relaxation dispersion values for I541 and I707 each reported on the same Kd value for the respective mutant. Errors from the fits of Rex × kex were used as weights for Kd fitting obtained with the NonlinearModelFit function in Mathematica v.10.3.1.0.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Atomic coordinates and structure factors of the FGFR3R669E asymmetric complex have been deposited in the Protein Data Bank under accession 6PNX. Raw mass spectrometry files and Mascot generic format files have been deposited in the MassIVE database under accession MSV000084018. All other data generated or analyzed during this study are included in this published article and its associated Supplementary Information.
Supplementary Material
Acknowledgements
The authors are indebted to N. Cowan for critically reading and editing the manuscript. This work was supported by National Institute of Dental and Craniofacial Research (NIDCR) grant R01 DE13686 (to M.M.), National Institute of General Medical Sciences (NIGMS) grant R01 GM117118 (to N.J.T.), NIGMS grant R35 GM127040 (to Y.Z.), National Institute of Neurological Disorders and Stroke (NINDS) grant P30 NS050276 and Shared Instrumentation Grant RR027990 (to T.A.N.), China Scholarship Council (CSC) and China Association for Science and Technology (CAST) (to L.C.), National Cancer Institute (NCI) predoctoral grant F99CA212474 (to W.M.M.) and the Natural Science Foundation of China (NSFC) grant 81930108 (to G.L.). An NMR cryoprobe at New York University was supported by an NIH S10 grant (OD016343). Data collection at the New York Structural Biology Center was made possible by a grant from NYSTAR. Computing resources were provided by New York University-ITS. We dedicate this work to the memory of J.M., who died suddenly before submission.
Footnotes
Competing interests
The authors have no conflicting interest to report
online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41589019-0455-7.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information is available for this paper at https://doi.org/10.1038/s41589-019-0455-7.
References
- 1.Hunter T Signaling–2000 and beyond. Cell 100, 113–127 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA & Schlessinger J Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blume-Jensen P & Hunter T Oncogenic kinase signalling. Nature 411, 355–365 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Hubbard SR Autoinhibitory mechanisms in receptor tyrosine kinases. Front Biosci. 7, d330–d340 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Hubbard SR Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol 5, 464–471 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Schlessinger J Signal transduction. Autoinhibition control. Science 300, 750–752 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Wybenga-Groot LE et al. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell 106, 745–757 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Hubbard SR & Miller WT Receptor tyrosine kinases: mechanisms of activation and signaling. Curr. Opin. Cell Biol 19, 117–123 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Gureasko J, Shen K, Cole PA & Kuriyan J An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Kovacs E, Zorn JA, Huang Y, Barros T & Kuriyan J A structural perspective on the regulation of the epidermal growth factor receptor. Annu. Rev. Biochem 84, 739–764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jura N et al. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell 42, 9–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolen B, Taylor S & Ghosh G Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Pellicena P & Kuriyan J Protein–protein interactions in the allosteric regulation of protein kinases. Curr. Opin. Struct. Biol 16, 702–709 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Boggon TJ & Eck MJ Structure and regulation of Src family kinases. Oncogene 23, 7918–7927 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Binns KL, Taylor PP, Sicheri F, Pawson T & Holland SJ Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of Eph receptors. Mol. Cell. Biol 20, 4791–4805 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furdui CM, Lew ED, Schlessinger J & Anderson KS Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21, 711–717 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Wu J et al. Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor. EMBO J. 27, 1985–1994 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kan SH et al. Genomic screening of fibroblast growth-factor receptor 2 reveals a wide spectrum of mutations in patients with syndromic craniosynostosis. Am. J. Hum. Genet 70, 472–486 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonell LM, Kernohan KD, Boycott KM & Sawyer SL Receptor tyrosine kinase mutations in developmental syndromes and cancer: two sides of the same coin. Hum. Mol. Genet 24, R60–R66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkie AO Bad bones, absent smell, selfish testes: the pleiotropic consequences of human FGF receptor mutations. Cytokine Growth Factor Rev. 16, 187–203 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Chen H et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell 27, 717–730 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H et al. Elucidation of a four-site allosteric network in fibroblast growth factor receptor tyrosine kinases. Elife 6, e21137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosen MK et al. Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol 263, 627–636 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Pervushin K, Riek R, Wider G & Wuthrich K Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA 94, 12366–12371 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mittermaier A & Meneses E Analyzing protein–ligand interactions by dynamic NMR spectroscopy. Methods Mol. Biol 1008, 243–266 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Mittermaier AK & Kay LE Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci 34, 601–611 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Chen H et al. Cracking the molecular origin of intrinsic tyrosine kinase activity through analysis of pathogenic gain-of-function mutations. Cell Rep. 4, 376–384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Z et al. Structural mimicry of A-loop tyrosine phosphorylation by a pathogenic FGF receptor 3 mutation. Structure 21, 1889–1896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korzhnev DM, Kloiber K, Kanelis V, Tugarinov V & Kay LE Probing slow dynamics in high molecular weight proteins by methyl-TROSY NMR spectroscopy: application to a 723-residue enzyme. J. Am. Chem. Soc 126, 3964–3973 (2004). [DOI] [PubMed] [Google Scholar]
References
- 30.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Songyang Z et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature 373, 536–539 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Rappsilber J, Mann M & Ishihama Y Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Byron SA et al. The N550K/H mutations in FGFR2 confer differential resistance to PD173074, dovitinib, and ponatinib ATP-competitive inhibitors. Neoplasia 15, 975–988 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier JA et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meagher KL, Redman LT & Carlson HA Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem 24, 1016–1025 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Allner O, Nilsson L & Villa A Magnesium ion–water coordination and exchange in biomolecular simulations. J. Chem. Theory Comput 8, 1493–1502 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Webb B & Sali A Protein structure modeling with MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae JH et al. Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells. Proc. Natl Acad. Sci. USA 107, 2866–2871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S & Walker RC Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. explicit solvent particle mesh ewald. J. Chem. Theory Comput 9, 3878–3888 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Delaglio F et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Carver JP & Richards RE General 2-site solution for chemical exchange produced dependence of T2 upon Carr–Purcell pulse separation. J. Magn. Reson 6, 89–105 (1972). [Google Scholar]
- 44.Luz Z & Meiboom S Nuclear magnetic resonance study of the protolysis of trimethylammonium ion in aqueous solution-order of the reaction with respect to solvent. J. Chem. Phys 39, 366–370 (1963). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors of the FGFR3R669E asymmetric complex have been deposited in the Protein Data Bank under accession 6PNX. Raw mass spectrometry files and Mascot generic format files have been deposited in the MassIVE database under accession MSV000084018. All other data generated or analyzed during this study are included in this published article and its associated Supplementary Information.