Short abstract
Background
Recent studies have found that microRNAs (miRNAs) play a critical role in development and progression of intervertebral disc degeneration. In the present study, we examined the role of miR-185 in nucleus pulposus cell behavior in vitro and the histological changes of intervertebral disc tissue in intervertebral disc degeneration rat models in vivo.
Methods
Intervertebral disc degeneration models were developed in Sprague-Dawley rats. Intervertebral disc tissue was collected for histological evaluation after miR-185 agomir/agomir transduction. Next, nucleus pulposus tissues were collected from lumbar intervertebral discs to isolate nucleus pulposus cells, which were treated with miR-185 mimic/inhibitor and an inhibitor of the Wnt signaling pathway to assess cell viability and apoptosis.
Results
We observed a high expression of Galectin-3 in nucleus pulposus cells of rats with intervertebral disc degeneration. Bioinformatics prediction and dual-luciferase reporter assay confirmed that miR-185 specifically binds to and negatively regulates Galectin-3. Furthermore, we found that miR-185 inhibition resulted in increased expression of Galectin-3, pro-autophagy factors (LC3 and Beclin-1), and pro-apoptosis factors (caspase-3 and Bax), along with the activation of the Wnt/β-catenin signaling pathway. Moreover, the gain- and loss-of-function studies suggested that miR-185 overexpression promoted cell viability and inhibited nucleus pulposus cell apoptosis and autophagy via inactivation of the Wnt/β-catenin signaling pathway. Moreover, miR-185 agomir alleviated the histological changes observed in intervertebral disc tissues in intervertebral disc degeneration rats, which helped us validate the results observed in vitro.
Conclusions
Overexpression of miR-185 promotes nucleus pulposus cell viability and reduces the histological changes observed in intervertebral disc tissues in rats with intervertebral disc degeneration via inactivation of the Wnt/β-catenin signaling pathway and Galectin-3 inhibition. Our findings also highlight the potential of miR-185 as a promising novel therapeutic target to prevent and control intervertebral disc degeneration.
Keywords: MicroRNA-185, Galectin-3, Wnt/β-catenin signaling pathway, intervertebral disc degeneration, nucleus pulposus cells, autophagy, apoptosis
Introduction
Lower back pain is the leading factor resulting in disability across the globe, intervertebral disc degeneration (IDD) being the main cause for it.1,2 The risk factors for IDD include aging and excessive manual labor, and some genetic factors including defects in the Vitamin D receptor and interleukin-1.3,4 One of the main features of IDD is a decreased number of nucleus pulposus cells.5 Disc degeneration makes the nucleus pulposus cells less gelatinous and presents more fibrous cracks and fissures in the annulus fibrosus.6 Moreover, fusion surgery has emerged as one of the most common ways to alleviate back pain due to IDD; however, it can have an adverse impact on the surrounding discs.7 Therefore, a great number of patients hope for better treatments. Interestingly, a previous study found that some microRNAs (miRNAs) are involved in the pathogenesis of IDD.8
miR-185 is found at the genomic location 22q11.21.9 A low miR-185 expression has been observed in samples of pediatric renal tumors, ovarian cancers, and prostate cancers, indicating that miR-185 may be associated with tumor initiation and progression.10 Also, overexpression of miR-185 can inhibit cell autophagy.11 Galectin-3, a β-galactosidase-binding lectin, plays an important role in the regulation of cell proliferation and apoptosis.12 Previous studies have found that the recovery of function in spinal cord injury can be improved by pharmacological suppression of the Galectin-3 activity.13 Moreover, an earlier report indicated that Galectin-3 is a key regulator in the Wnt/β-catenin signaling pathway.14 Previous studies discovered that the organization of the annulus fibrosus and endplates can be supported by the Wnt/β-catenin signaling pathway and that appropriate control of this signaling pathway is essential for the maintenance of intervertebral disc structures.15 Although the effects of the factors mentioned above on IDD have been explored, there are few studies that combine these results to address the problem of IDD. Considering all of the existent evidence, in this study, we hypothesized that miR-185 might regulate the autophagy and apoptosis of nucleus pulposus cells in IDD through modulation of the Wnt/β-catenin signaling pathway via Galectin-3.
Materials and methods
Ethics statement
Animal use and experiments were approved by the Experimental Animal Ethics Committee of the First Hospital of Jilin University. All of the animal experimental operating procedures were performed following the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Model establishment
A total of 80 male Sprague-Dawley rats (Guangzhou Geneseed Biotech Co., Ltd., Guangzhou, Guangdong, China), aged 2–3 months and weighing 200–250 g, were used in this study. After a week of normal feeding, the rats were randomly divided into the IDD (n = 60) and the sham groups (n = 20) and injected with 5% pentobarbital sodium (5 mg/100 g of body weight). The rats in the IDD group were fixed in a prone position and disinfected. The lower thoracic region, lumbar vertebra, and upper sacrum were exposed using a posterior midline approach. Next, the lumbar erector spinae, whole lumbar spinous process, supraspinous ligament, interspinal ligaments, and half of the bilateral lumbar zygapophyseal joints in rats were resected and the wounds were irrigated with a normal saline solution containing 100 IU/mL penicillin. The deep fascia and skin were sutured without suturing the erector spinae to establish the rat model of imbalance of dynamic and static forces. The rats in the sham group used as control were only subjected to an incision of the skin and immediate suturing and then given gentamicin to prevent infection three days after treatment with the freedom to move. All of the rats were anesthetized by an intraperitoneal injection of pentobarbital sodium, followed by magnetic resonance imaging examination. The severely degenerated and normal intervertebral discs were excluded and the intervertebral discs in grades II–III degeneration evaluated by Thompson were recorded and improved. The rats were euthanized using an overdose of pentobarbital sodium by intraperitoneal injection after two, four, and eight weeks of operation. The whole L4–5 and L5–6 segments were collected, with the paraspinal muscle cleared and the posterior spines of these two segments removed. Then, the remaining specimens were fixed for 48 h, decalcified with 20% ethylene diamine tetraacetic acid for four weeks, routinely treated, and stained with hematoxylin–eosin (HE) staining and Masson staining. The successful IDD rat models were injected with miR-185 agomir/agomir-NC (n = 20) (Shanghai GenePharma Co., Ltd., Shanghai, China) on the days 1, 7, and 14 via tail vein, while the remaining 20 rats with IDD were left untreated. At week 8 after the operation, the intervertebral disc tissues were collected for histological evaluation.16,17
Isolation and culture of nucleus pulposus cells
Whole lumbar spines of rats were collected, the ligaments adjacent to the intervertebral disc and the attached muscles were cleared, and the nucleus pulposus tissues from the lumbar intervertebral discs were carefully collected using scalpel blades and curettes. Next, the collected nucleus pulposus tissues were cut into 1 mm3 tissue blocks. The blocks were detached using 0.1% type II collagenase at 37°C for 4 h and then added with Dulbecco’s modified Eagle medium (DMEM)-F12 containing 20% fetal bovine serum (FBS) to resuspend the lower cells and the incompletely detached nucleus pulposus tissues. Next, the cells were cultured with 5% CO2 at 37°C in DMEM-F12 containing 10% FBS, with the medium renewed every 48 h. Upon reaching 90% confluence after 15 days of culture, the cells were detached with trypsin and subcultured at 1:4. The cells at passage 2 were used in this study.
HE staining
The tissue sections were subjected to xylene dewaxing and gradient ethanol hydration, followed by hematoxylin staining (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) for 2 min, and color separation with 1% hydrochloric acid ethanol for 10 s. Next, the sections were stained with eosin for 1 min, dehydrated, cleared with xylene, and mounted. Lastly, the morphological changes of intervertebral disc tissues were observed using an optical microscope (XP-330, Shanghai Bingyu Optical Instrument Co., Ltd., Shanghai, China).
Masson staining
After dewaxing and hydration, the tissue sections were stained by Regaud hematoxylin for 5–10 min, then stained by Masson Ponceau acid fuchsin for 5–10 min, soaked in 2% glacial acetic acid, and differentiated with 1% phosphomolybdic acid for 3–5 min. Next, the sections were stained directly in aniline blue or light green for 5 min, soaked in 2% glacial acetic acid, dehydrated, cleared, and fixed. Lastly, the sections were observed under the microscope.
Immunohistochemistry
The sections were baked at 60°C overnight, dewaxed, and dehydrated, followed by antigen retrieval in 0.01 M citrate buffer for 10 min. Next, the sections were immersed in a 0.3% H2O2-methanol solution for 20 min, blocked with 10% goat serum for 10 min, and incubated with the rabbit anti-rat antibody to Galectin-3 (1:500, ab53082, Abcam Inc., Cambridge, MA, USA) at 4°C overnight. Phosphate-buffered saline was used instead of the primary antibody as a negative control (NC). A day later, the sections were incubated with the goat anti-rabbit secondary antibody to immunoglobulin G (IgG) (1:3000, ab205718, Abcam) for 30 min. The sections were developed with diaminobenzidine for 5 min, counter-stained with hematoxylin, differentiated by 1% hydrochloric alcohol, returned to blue, and mounted. Lastly, the sections were photographed using an optical microscope. Cells with a pale brown-yellow cytoplasm or membrane were regarded as positive cells.
Dual-luciferase reporter gene assay
The target fragments were introduced into a pmiR-reporter. The complementary sequence mutation site of the seed sequence was designed based on the Galectin-3 wild-type (WT) gene and then inserted into a pmiR-reporter plasmid by restriction endonuclease digestion and T4 DNA ligase treatment. The correctly sequenced luciferase plasmids were co-treated with miR-185 into HEK-293T cells (CRL-1415, Shanghai Xin Yu Biotech Co., Ltd., Shanghai, China). Then, a luciferase detection kit (RG005, Beyotime Institute of Biotechnology, Shanghai, China) was used to determine the luciferase activity.
Cell grouping and transfection
Nucleus pulposus cells of rats with IDD were grouped into blank, NC (nucleus pulposus cells delivered with blank plasmid), miR-185 mimic (100 nM), miR-185 inhibitor (100 nM), Dickkopf-1 (DKK-1) (inhibitor of the Wnt/β-catenin signaling pathway), and miR-185 mimic + DKK-1. All of the plasmids were purchased from Merck (Merck Serono, Geneva, Switzerland).
Reverse transcription-quantitative polymerase chain reaction
Total RNA was extracted and reversely transcribed into cDNA. The primers were designed and synthesized by TaKaRa (TaKaRa Bio Inc, Shiga, Japan; Table 1). The quantitative PCR was conducted using an ABI7500 Real-Time PCR instrument (7500, Applied Biosystems Inc., CA, USA). U6 was used as the internal reference for miR-185, and β-actin was used as the internal reference for the other genes. The fold changes were calculated by the 2−ΔΔCt method.
Table 1.
Primer sequences for RT-qPCR.
| Genes | Primer sequences (5′-3′) |
|---|---|
| miR-185 | F: 5′-CAATGGAGAGAAAGGCAGTTCC-3′ |
| R: 5′-AATCCATGAGAGATCCCTACCG-3′ | |
| Galectin-3 | F: 5′-ATGGCAGACGGCTTCTCACT-3′ |
| R: 5′-CGCGAAGGGTGCGGTACTAG-3′ | |
| β-catenin | F: 5′-CCACGACTAGTTCAGCTGCTTGTAC-3′ |
| R:5′-ACTGCACAAACAGTGGAATGGTATT-3′ | |
| Wnt3a | F: 5′-GAGCTCTGCCATTCACCCTA-3′ |
| R: 5′-GGCTGCAGAGCACTAAGAGG-3′ | |
| Caspase-3 | F: 5′-CAGAGCTGGACTGCGGTATTGA-3′ |
| R: 5′-AGCATGGCGCAAAGTGACTG-3′ | |
| Bax | F: 5′-CGAGCTGATCAGAACCATCA-3′ |
| R: 5′-GGTCCCGAAGTAGGAGAGGA-3′ | |
| Bcl-2 | F: 5′-GGAGGATTGTGGCCTTCTTTG-3′ |
| R: 5′-CATCCCAGCCTCCGTTATCC-3′ | |
| LC3 | F: 5′-CTTCGCCGACCGCTGTTA-3′ |
| R: 5′-ATCCGTCTTCATCCTTCTTCCTG-3′ | |
| Beclin-1 | F: 5′-ACAGCTCCATTACTTACCACAGCCC-3′ |
| R: 5′-AATCTTCGAGAGACACCATCCTGGC-3′ | |
| β-actin | F: 5′-CCGTCAAAAGATGACCCAGATC-3′ |
| R: 5′-CACAGCCTGGATGGCTACGT-3′ | |
| U6 | F: 5′-ATGACGTCTGCCTTGGAGAAC-3′ |
| R: 5′-TCAGTGTGCTACGGAGTTCAG-3′ |
RT-qPCR: reverse transcription quantitative polymerase chain reaction; miR: microRNA; Bcl-2: B-cell lymphoma-2; Bax: Bcl-2 associated protein X; LC3: light chain 3; F: forward; R: reverse.
Western blot analysis
The nucleus pulposus cells of rats with IDD were lysed using a protein lysate for total protein isolation. The protein samples were mixed with the loading buffer, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto a nitrocellulose membrane. The membrane was then incubated at 4°C overnight with the primary antibodies: Galectin-3 (1:500, ab53082), β-catenin (1:5000, ab32572), wnt3a (1:1 0000, ab172612), caspase-3 (1:500, ab13847), Bcl-2 associated protein X (Bax) (1: 1000, ab32503), B-cell lymphoma-2 (Bcl-2) (1:500, ab59348), light chain 3B (LC3B) (0.5 µg/mL, ab48394), and Beclin-1 (0.5 µg/mL, ab62557). Next, the membrane was washed with tris-buffered saline tween and incubated with the horseradish peroxidase-conjugated goat anti-rabbit secondary antibody to IgG (1:3000, ab205718) for 1 h. The membrane was developed using an enhanced chemiluminescence reagent (Pierce, Waltham, MA, USA) and then observed.
TOP-Flash/FOP-Flash assay
Nucleus pulposus cells were inoculated in a 96-well plate. The treated cells, TOP-Flash/FOP-Flash and PRL-TK plasmids, were added into each well after 24 h. A total of 100 μL of low glucose-DMEM (L-DMEM) was added with plasmid and allowed to stand for 5 min. Then, 0.5 μL of lipofectamine 2000 (11668019, Thermo Fisher Scientific, Waltham, MA, USA) was mixed with 100 μL of L-DMEM. The previous medium was removed and then the cells were washed with L-DMEM, cultured with the mixed transfection liquid for 6 h, and cultured in complete medium for 24–28 h, with the medium removed after incubation. The luminescent signal reflecting the activation of the transcription factors related to the Wnt/β-catenin signaling pathway was compared based on the ratio of the firefly relative light units (RLUs) to the firefly RLU value measured by the dual-luciferase reporter assay system (E1910, Promega, Madison, WI, USA).
TEM observation and acridine orange (AO)/monodansylcadaverine (MDC) staining
After removal of the culture media, the cells in each group were collected and fixed for 10 min with pre-chilled glutaric dialdehyde at 4°C. The cells were scraped with a scraping panel and then added into a centrifuge tube at 4°C overnight. After gradient centrifugation, the cells were dehydrated, embedded with DurcopanACM resin, and cut into 80 nm slices. The slices were then stained for 30 min with uranium acetate, washed 3 times, stained for 30 min with lead citrate, and dried at room temperature. The morphology and organelles of the nucleus pulposus cells were observed using transmission electron microscope (TEM; HT7700 Exalens, Hitachi, Japan), and the number of podocyte autophagosomes was counted in every 10 visual fields with equal magnification.
A small amount of the Lyso-Tracker Red, AO, and MDC was added into the cell culture fluid proportionally to prepare the dyeing solution with a final concentration of 75 nM, 500 M, and 0.5 μg/mL, respectively. The above dyeing solutions were pre-warmed at 37°C and then cocultured with the cells for 15 and 60 min at 37°C. Lastly, after removal of the dyeing solution, the cells were added with fresh culture fluid and observed under a fluorescence microscope.
MTT assay
The cells were inoculated in a 96-well plate (2 × 103 cells/well), cultured with 120 mL of medium, and detected at different time points (0, 12, 24, and 36 h) under the pressure of 1.0 Mpa. After that, the cells were cultured with 20 μL of MTT medium (0.5 mg/mL) for 4 h at 37°C. After the medium in the wells was removed, the cells were added with 150 mL of dimethyl sulfoxide and oscillated for 10 min. Then, the optical density value was measured at 570 nm using a Quant Universal Microplate Spectrophotometer (Biotek Instruments, Winooski, VT, USA) and recorded.
Flow cytometry
The nucleus pulposus cells were inoculated in a six-well plate (2 × 105 cells/well) and divided into blank, NC, and cell transfection groups. After treatment for 72 h, the cells were detached with trypsin, centrifuged, and the supernatant was discarded. Then, the cell precipitates were re-suspended in 500 μL of the binding buffer using an Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit I (556547, Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Following the addition of 5 μL of FITC and 5 μL of propidium iodide, the samples were mixed and incubated for 15 min. The cell apoptosis was analyzed using a flow cytometer.
Statistical analysis
All data were analyzed using the SPSS 21.0 software (IBM, Armonk, NY, USA). Measurement data were expressed as mean ± standard deviation. Comparisons between two groups were analyzed using an independent-sample t-test. Data correction was conducted by Welch. The normality test for data among multiple groups was performed using the Shapiro–Wilk method. The measurement data in normal distribution were analyzed by one-way analysis of variance (ANOVA), and the data with skewed distribution were compared by nonparametric Kruskal–Wallis. The viability of the cells at different time points was analyzed by repeated-measures ANOVA. A p-value < 0.05 was considered statistically significant.
Results
Successful establishment of the IDD rat model
HE staining and Masson staining revealed that the rats in the sham group (Figure 1(a)) exhibited a clear tissue structure of the intervertebral discs, a clear boundary between the annulus fibrosus and the nucleus pulposus, orderly arranged collagenous fibers without obvious fractures, and a great number of visible cells in the nucleus pulposus with clear structure. The results showed that two weeks after the operation (Figure 1(a)), the rats in the IDD group presented relatively clear tissue structure of intervertebral disc with a clear boundary between annulus fibrosus and the nucleus pulposus, relatively orderly arranged collagenous fibers without obvious fractures, and many visible cells in the nucleus pulposus with clear structure. Four weeks after the operation (Figure 1(a)), the tissue structures of the intervertebral discs of rats were unclear, and there was no clear boundary between the annulus fibrosus and the nucleus pulposus, with no obvious fracture of the collagenous fibers, a reduced volume of nucleus pulposus, decreased number of cells in the nucleus pulposus, and disordered tissue structure. Eight weeks after the operation (Figure 1(a)), the tissue structures of the intervertebral discs of rats were much more disordered and the collagenous fibers in the annulus fibrosus were differentiated and fractured with a reduced volume of the nucleus pulposus and the number of cells in the nucleus pulposus. These findings indicated that with time, the collagen fibers of the annulus fibrosus started cracking and even fracturing, the tissue structures of the nucleus pulposus were gradually in disorder and the number of cells in the nucleus pulposus decreased.
Figure 1.
Pathomorphological changes in the nucleus pulposus, intervertebral discs, and annulus fibrosus. (a) Intervertebral disc tissues in the sham group and the IDD group 2 W, 4 W, and 8 W after operation were stained by HE (sagittal) (×100). (b) The annulus fibrosus and nucleus pulposus in the sham group and the IDD group 2 W, 4 W, and 8 W after operation were stained by Masson (×100). HE: hematoxylin–eosin.
The Masson staining results were also observed under the Masson staining light microscope. Collagenous fibers presented in blue by Masson staining and were used to reflect the morphology of the collagen fibers in the intervertebral discs. The results indicated that in the sham and IDD groups two weeks after the operation, the collagenous fibers in the intervertebral discs were orderly arranged, the shape of the nucleus pulposus was normal, the boundary between the annulus fibrosus and the nucleus pulposus was clear, and many notochord cells were visible. Four weeks after the operation, the collagen fibers were disorderly arranged and presented cracks, most cells in the nucleus pulposus were chondroid cells, and the number of notochord cells decreased. Eight weeks after the operation, the collagenous fibers were much more disordered with obvious cracks and even fractures, the cells in nucleus pulposus were very few, and the nucleus pulposus tissues appeared with fibrosis (Figure 1(b)).
The results above revealed that with the modeling time extending, the collagenous fibers in the annulus fibrosus became more disordered, presented cracks, and even fractures appeared. The cells in the nucleus pulposus gradually decreased with disappearing vacuoles.
The Galectin-3 expression is increased in nucleus pulposus cells of rats with IDD
Immunohistochemistry showed positive expression of the Galectin-3 protein mainly in the cytoplasm, observed as yellow or brown-yellow granules. It was also observed that only a few nucleus pulposus cells presented weak Galectin-3 expression in normal nucleus pulposus tissues, while the nucleus pulposus tissues of rats with IDD showed increased expression of Galectin-3 and a larger number of positive cells. The rate of Galectin-3-positive cells was 22.45% ± 3.78% in the sham group and 61.56% ± 6.02% in the IDD group (p < 0.05) (Figure 2). Therefore, the rats with IDD showed a higher expression of Galectin-3.
Figure 2.
Positive expression of Galectin-3 increases in nucleus pulposus cells of rats with IDD. (a) Immunohistochemistry was used to examine the Galectin-3 expression of rats in the sham, IDD, and NC groups (× 100). (b) The quantitation for a positive rate of Galectin-3 expression in the sham, IDD, and NC groups. *p < 0.05 versus the sham group. All data were presented as mean ± standard deviation, and comparisons among multiple groups were analyzed by one-way ANOVA, n = 20.
NC: negative control.
miR-185 targets Galectin-3
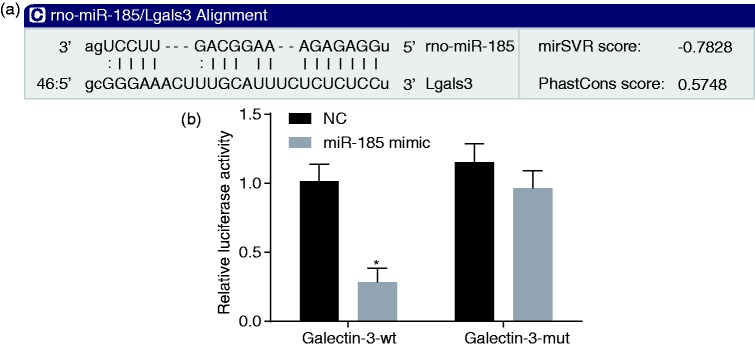
The online analysis software indicated that Galectin-3 was a target of miR-185 (Figure 3(a)) (Lgals3 referred to Galectin-3 in rats), which was confirmed by our dual-luciferase reporter assay (Figure 3(b)) which showed a reduced luciferase activity of Galectin-3-WT by a miR-185 mimic.
Figure 3.
Galectin-3 is targeted by miR-185. (a) The binding sites between Galectin-3 and miR-185 were predicted by the online software. (b) Dual-luciferase reporter gene assay was applied to detect luciferase activity. *p < 0.05 versus the NC group. All data were presented as mean ± standard deviation of three independent biological replicates and comparisons between two groups were analyzed by unpaired t-test.
NC: negative control.
Overexpression of miR-185 increases Bcl-2 expression and decreases the expression of Galectin-3, wnt3a, β-catenin, caspase-3, LC3, Bax, and Beclin-1
The results from the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (Figure 4(a)) and Western blot analysis (Figure 4(b) and (c)) showed that compared with the blank and NC groups, the miR-185 mimic and miR-185 mimic + DKK-1 groups exhibited an increased miR-185 and Bcl-2 expression, and decreased Galectin-3, wnt3a, β-catenin, caspase-3, LC3, Bax, and Beclin-1 expression. However, the miR-185 inhibitor group showed the opposite trend (p < 0.05). Moreover, the DKK-1 group exhibited decreased wnt3a, β-catenin, caspase-3, LC3, Bax, and Beclin-1 expression, and increased Bcl-2 expression without a significantly differed expression of miR-185 and Galectin-3. Compared with the miR-185 mimic and DKK-1 groups, the miR-185 mimic + DKK-1 group showed increased miR-185 and Bcl-2 expression, and decreased Galectin-3, wnt3a, β-catenin, caspase-3, LC3, Bax, and Beclin-1 expression.
Figure 4.
Inhibition of miR-185 increases the mRNA levels and the protein expression of Galectin-3, wnt3a, β-catenin, caspase-3, LC3, Bax, and Beclin-1 and decreases the mRNA levels and protein expression of Bcl-2. (a) The miR-185 expression and mRNA expression of Galectin-3, wnt3a, β-catenin, caspase-3, LC3, Bax, and Beclin-1 were determined by RT-qPCR. (b and c) Western blot analysis of the protein bands (b) and the protein levels (c) of Galectin-3, wnt3a, β-catenin, caspase-3, LC3, Bax, and Beclin-1 normalized to GAPDH. *p < 0.05 versus the blank and NC groups; #p < 0.05 versus the miR-185 mimic and DKK-1 groups. All data were presented as mean ± standard deviation of three independent biological replicates, and comparisons among multiple groups were analyzed by one-way ANOVA.
NC: negative control; DKK-1: Dickkopf-1.
Overexpression of miR-185 promotes cell viability and inhibits apoptosis through suppression of the Wnt/β-catenin signaling pathway
The results obtained from the MTT assay (Figure 5(a)) showed that after 48 h of treatment, compared with the blank and NC groups, the cell viability slightly increased in the miR-185 mimic and miR-185 mimic + DKK-1 groups, but slightly decreased in the miR-185 inhibitor group. Compared with the miR-185 mimic + DKK-1 group, the cell viability slightly decreased in the miR-185 mimic and DKK-1 groups and significantly decreased in the DKK-1 group. After 72 h, compared with the blank and NC groups, the cell viability increased in the miR-185 mimic and miR-185 mimic + DKK-1 groups and decreased in the miR-185 inhibitor group. Compared with the miR-185 mimic + DKK-1 group, the cell viability in the miR-185 mimic group decreased, being even lower in the DKK-1 group. Moreover, flow cytometry was used to detect cell apoptosis. The results (Figure 5(b) and (c)) showed that compared with the blank and NC groups, the cell apoptosis was significantly inhibited in the miR-185 mimic + DKK-1 group, slightly inhibited in the miR-185 mimic group, and promoted in the miR-185 inhibitor group. Compared with the miR-185 mimic + DKK-1 group, the apoptosis was enhanced in the miR-185 mimic and DKK-1 groups (p < 0.05). These results above showed that increased expression of miR-185 inhibited cell apoptosis, which could also be triggered by inhibition of the Wnt/β-catenin signaling pathway.
Figure 5.
Overexpression of miR-185 promotes cell viability and inhibits apoptosis through suppression of the Wnt/β-catenin signaling pathway. (a) Cell viability after transfection examined by MTT assay. (b and c) Cell apoptosis of each group after transfection detected by flow cytometry. *p < 0.05 versus the blank and NC groups; #p < 0.05 versus the miR-185 mimic + DKK-1 group. All data were presented as mean ± standard deviation of three independent biological replicates, data of cell viability in different time were analyzed by ANOVA of repeated measurement and cell apoptosis rate was analyzed by one-way ANOVA. NC: negative control; DKK-1: Dickkopf-1; FITC: fluorescein isothiocyanate; OD: optical density.
Overexpression of miR-185 inhibits cell autophagy by suppression of the Wnt/β-catenin signaling pathway via Galectin-3
Cell autophagy was characterized by autophagosome and autophagic vacuole appeared in cytoplasm. Autophagosomes and autophagic vacuoles were detected in the cytoplasm by TEM and observed as bilayered vesicles of cytoplasmic debris containing damaged organelles or polyproteins. Compared with the blank and NC groups, the number of vesicles was significantly reduced in the miR-185 mimic and miR-185 mimic + DKK-1 groups. However, it was elevated in the miR-185 inhibitor group. These results showed that miR-185 suppressed cell autophagy by inhibiting Galectin-3 (Figure 6(a)).
Figure 6.
Overexpression of miR-185 inhibits cell autophagy by suppression of the Wnt/β-catenin signaling pathway via Galectin-3. (a) Cell autophagy was observed under TEM (scale bar = 2 μm) and after transfection with Lyso-Tracker Red, MDC, and AO staining (×400). (b) Quantitative analysis for the Lyso-Tracker Red fluorescence intensity. *p < 0.05 versus the blank and NC groups. All data were presented as mean ± standard deviation of three independent biological replicates, and comparisons among multiple groups were analyzed by one-way ANOVA.
NC: negative control; DKK-1: Dickkopf-1; MDC: monodansylcadaverine; MFI: Median Fluorescence Intensity; AO: acridine orange.
The cells were treated with Lyso-Tracker Red, MDC, and AO staining. It was observed under the fluorescence microscope that the volume of the acid chamber was larger when analyzed with the Lyso-Tracker Red staining and the MDC staining revealed the presence of autophagosomes. The acidic vesicular organelles (AVOs) with red fluorescence detected by AO staining further demonstrated the existence of autophagosomes (Figure 6(a) and (b)). The data revealed that compared with the blank and NC groups, the AVOs were suppressed in the miR-185 mimic, DKK-1, and miR-185 mimic + DKK-1 groups, while promoted in the miR-185 inhibitor group. The results above showed that overexpression of miR-185 could inhibit cell autophagy by suppression of the Wnt/β-catenin signaling pathway.
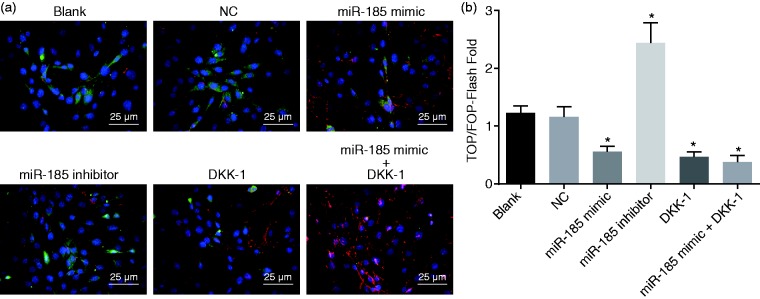
Overexpression of miR-185 inhibits the Wnt/β-catenin signaling pathway
TOP-Flash/FOP-Flash showed that the TOP-Flash plasmid contained the firefly luciferase reporter gene. Three repeated T-cell factor (TCF) binding sequences existed upstream of the luciferase promoter, which could regulate the expression of the luciferase downstream gene based on the activity of β-catenin. Except for the mutant TCF binding sequence, all other sequences were consistent with TOP-Flash and not influenced by the β-catenin activity. Therefore, TOP/TOP-Flash was usually used as the detection index for Wnt/β-catenin signaling in cells, and the key for the activation of the Wnt/β-catenin signaling pathway was that β-catenin entered into the nucleus and bound to the transcription factor TCF/lymphoid enhancer-binding factor to co-regulate the gene expression. The results of TOP-Flash revealed that the activity of TOP-Flash increased in the miR-185 inhibitor group, but decreased after overexpression of miR-185 (Figure 7), suggesting that the upregulation of miR-185 exerted an inhibitory effect on the Wnt/β-catenin signaling pathway.
Figure 7.
Overexpression of miR-185 inhibits the Wnt/β-catenin signaling pathway. (a) TOP/TOP-Flash plasmids were observed (×400). (b) Quantitative analysis of the activity of the TOP/TOP-Flash plasmids after transfection. *p < 0.05 versus the blank and NC groups. All data were presented as mean ± standard deviations of three independent biological replicates, and comparisons among multiple groups were analyzed by one-way ANOVA. NC: negative control; DKK-1: Dickkopf-1.
Overexpression of miR-185 alleviates IDD in rats in vivo
Lastly, we decided to verify our in vitro findings through in vivo experiments. The functions of miR-185 were assessed on histological changes of intervertebral disc tissues in rats with IDD. Both agomir-NC and miR-185 agomir were injected into rats with IDD, followed by determination of the expression of miR-185 and Galectin-3 in nucleus pulposus tissues using RT-qPCR (Figure 8(a)). Compared with the sham group, the miR-185 expression was diminished and the Galectin-3 expression was elevated in the model group. In comparison to the model + agomir-NC group, the results were the opposite in the model + miR-185 agomir group. Then, HE (Figure 8(b)) and Masson staining (Figure 8(c)) were performed to observe the histological changes of degenerative discs in rats eight weeks after modeling. In the agomir-NC group, the intervertebral disc tissues were disordered and integrated, dysplasia occurred in some chondrocytes, a large number of degenerative cell clusters were formed, and the intervertebral space narrowed. The intervertebral disc tissues in the miR-185 agomir group were neatly arranged with few deformities in the discs and a small number of abnormal chondrocytes and degenerative cells. Taken together, the in vivo results provided further validation that the upregulation of miR-185 alleviates IDD at the histological level.
Figure 8.
Overexpression of miR-185 alleviates IDD on the histological level. (a) The expression of miR-185 and Galectin-3 in the nucleus pulposus tissues of rats determined by RT-qPCR. (b) The pathological condition of intervertebral disc tissues detected by HE staining (×100). (c) The pathological condition of nucleus pulposus tissues detected by Masson staining (×200). NC: negative control.
Discussion
IDD is a fairly common phenomenon attributed to aging and excessive physical load and remains a major cause of biochemical disc degradation and structural compromise.18 It is urgent to develop new treatments for IDD due to the inefficiency of the therapies currently available.19 A previous study reported that miRNAs, a newly identified class of gene regulators, are associated with a wide range of pathophysiological, neurodegenerative, and reproductive disorders.20 Therefore, we hypothesized that the overexpression of miR-185 could promote nucleus pulposus cell viability and inhibit nucleus pulposus cell autophagy and apoptosis by suppressing Galectin-3 and blocking the Wnt/β-catenin signaling pathway in IDD, thus impeding the development of IDD.
The expression of Galectin-3 in nucleus pulposus cells obtained from rats with IDD was determined, showing that Galectin-3 was highly expressed. Previous evidence has suggested that overexpression of intracellular Galectin-3 is present in the period of neoplastic progression and metastasis in many human tumors including liver, brain, and colon.21–23 Another study also reported the upregulation of Galectin-3 in pancreatic cancers and cancer cells.24 Unlike the results observed for Galectin-3 in IDD, we found that miR-185 was deregulated in nucleus pulposus cells in IDD, which is highly suggestive of an association with IDD pathology. With time following injury, miR-185 was found to be downregulated in patients with spinal cord injuries due to thoracolumbar spine compression fractures.25 Furthermore, a decreased expression of miR-185 has been found in breast cancer tissues when compared to the healthy control groups.26 Moreover, we also proved that Galectin-3 was a target of miR-185. The expression of some genes is regulated by miRNAs to control cellular and biological functions; therefore, it is important to determine the functions of the targeted genes.27
The Wnt/β-catenin signaling pathway is an important tool for therapeutic development for osteoporosis and fracture healing.28 We found that the upregulation of miR-185 can inhibit the Wnt/β-catenin signaling pathway through its indirect effect on the wnt3a and β-catenin genes. A previous study reported that the overexpression of miR-185 decreases the expression of β-catenin, c-Myc, and SOX9, indicating that the Wnt/β-catenin signaling pathway is involved in the regulatory role of miR-185 in non-small cell lung cancer cell proliferation, invasion, and migration.29
Ectopic miR-185 expression promoted the viability and inhibited the apoptosis of nucleus pulposus cells in IDD in our study according to the decreased expression of caspase-3 and Bax, and the increased expression of Bcl-2. A previous study reported that the upregulation of miR-185 can significantly decrease the expression of caspase-3, which is a pro-apoptotic marker,30 suggesting that miR-185 may regulate cell apoptosis. Additionally, the upregulation of miR-185 targeting SOCS3 can stimulate cell proliferation and suppress cell apoptosis in diabetes by enhancing insulin secretion in pancreatic β-cells.31 Moreover, we also explored the relationship between miR-185 and cell autophagy, showing that miR-185 might inhibit nucleus pulposus cell autophagy in IDD based on LC3 and Beclin-1 expression. In a study performed by Wen et al., the expression of Beclin-1 increases in the SH-SY5Y dopaminergic neuroblastoma cells, and the overexpression of miR-185 inhibits the expression of Beclin-1.11 Another study showed that miR-185 results in increased LC3 expression in nasopharyngeal carcinoma cells.32
In conclusion, miR-185 negatively regulates Galectin-3 and disrupts the Wnt/β-catenin signaling pathway, leading to suppressed autophagy and apoptosis of nucleus pulposus cells in IDD (Figure 9). Therefore, our findings provide an insight into new potential therapeutic targets and strategies for the prevention and treatment of IDD. The question of whether different concentrations of miR-185 mimic yields different outcomes remains to be explored for clinical applications in the future. Also, the specific mechanism explaining this regulatory process remains unclear and further work is required to consolidate this model.
Figure 9.
The regulatory mechanism of miR-185 in IDD. Overexpression of miR-185 inhibits nucleus pulposus cell autophagy and apoptosis through suppression of the Wnt/β-catenin signaling pathway via downregulation of Galectin-3 in rats with IDD. IDD: intervertebral disc degeneration; miR: microRNA.
Acknowledgments
The authors would like to express their gratitude for the helpful technical assistances provided by their colleagues.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD
Junting Zang https://orcid.org/0000-0003-3091-8229
References
- 1.Daly C, Ghosh P, Jenkin G, Oehme D, Goldschlager T. A review of animal models of intervertebral disc degeneration: pathophysiology, regeneration, and translation to the clinic. Biomed Res Int 2016; 2016: 5952165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaudreuil N, Kadow T, Yurube T, Hartman R, Ngo K, Dong Q, Pohl P, Coelho JP, Kang J, Vo N, Sowa G. NSAID use in intervertebral disc degeneration: what are the effects on matrix homeostasis in vivo? Spine J 2017; 17: 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol 2014; 10: 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Y, Egan B, Wang J. Genetic factors in intervertebral disc degeneration. Genes Dis 2016; 3: 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X, He Y. Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci Rep 2017; 7: 13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Z, Liu ZH, Zhao XH, Sun L, Chen YF, Zhang WL, Gao Y, Zhang YZ, Wan ZY, Samartzis D, Wang HQ, Luo ZJ. Impact of direct cell co-cultures on human adipose-derived stromal cells and nucleus pulposus cells. J Orthop Res 2013; 11: 1804–1813. [DOI] [PubMed] [Google Scholar]
- 7.Choi H, Johnson ZI, Risbud MV. Understanding nucleus pulposus cell phenotype: a prerequisite for stem cell based therapies to treat intervertebral disc degeneration. Curr Stem Cell Res Ther 2015; 10: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Zhang Y, Feng X, Li S, Gao Y, Wang K, Song Y, Yang S, Tu J, Shao Z, Yang C. Inhibition of microRNA-34a prevents IL-1beta-induced extracellular matrix degradation in nucleus pulposus by increasing GDF5 expression. Exp Biol Med (Maywood) 2016; 17: 1924–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, Chen Y, Rao MK. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene 2010; 35: 4971–4979. [DOI] [PubMed] [Google Scholar]
- 10.Ma X, Shen D, Li H, Zhang Y, Lv X, Huang Q, Gao Y, Li X, Gu L, Xiu S, Bao X, Duan J, Zhang X. MicroRNA-185 inhibits cell proliferation and induces cell apoptosis by targeting VEGFA directly in von Hippel-Lindau-inactivated clear cell renal cell carcinoma. Urol Oncol 2015; 4: 169.e1–169.e11. [DOI] [PubMed] [Google Scholar]
- 11.Wen Z, Zhang J, Tang P, Tu N, Wang K, Wu G. Overexpression of miR185 inhibits autophagy and apoptosis of dopaminergic neurons by regulating the AMPK/mTOR signaling pathway in Parkinson’s disease. Mol Med Rep 2018; 1: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hisamatsu K, Niwa M, Kobayashi K, Miyazaki T, Hirata A, Hatano Y, Tomita H, Hara A. Galectin-3 expression in hippocampal CA2 following transient forebrain ischemia and its inhibition by hypothermia or antiapoptotic agents. Neuroreport 2016; 5: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pajoohesh-Ganji A, Knoblach SM, Faden AI, Byrnes KR. Characterization of inflammatory gene expression and galectin-3 function after spinal cord injury in mice. Brain Res 2012; 1475: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimura T, Takenaka Y, Fukumori T, Tsutsumi S, Okada K, Hogan V, Kikuchi A, Kuwano H, Raz A. Implication of galectin-3 in Wnt signaling. Cancer Res 2005; 9: 3535–3537. [DOI] [PubMed] [Google Scholar]
- 15.Kondo N, Yuasa T, Shimono K, Tung W, Okabe T, Yasuhara R, Pacifici M, Zhang Y, Iwamoto M, Enomoto-Iwamoto M. Intervertebral disc development is regulated by Wnt/beta-catenin signaling. Spine (Phila Pa 1976) 2011; 8: E513–E518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji ML, Jiang H, Zhang XJ, Shi PL, Li C, Wu H, Wu XT, Wang YT, Wang C, Lu J. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat Commun 2018; 9: 5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Shi X, Peng X, Li Y, Ma H, Li D, Cao X. MicroRNA-181a exerts anti-inflammatory effects via inhibition of the ERK pathway in mice with intervertebral disc degeneration. J Cell Physiol 2020; 235: 2676–2686. [DOI] [PubMed] [Google Scholar]
- 18.Cheung KM, Samartzis D, Karppinen J, Mok FP, Ho DW, Fong DY, Luk KD. Intervertebral disc degeneration: new insights based on “skipped” level disc pathology. Arthritis Rheum 2010; 8: 2392–2400. [DOI] [PubMed] [Google Scholar]
- 19.Vasiliadis ES, Pneumaticos SG, Evangelopoulos DS, Papavassiliou AG. Biologic treatment of mild and moderate intervertebral disc degeneration. Mol Med 2014; 20: 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B, Yu Q, Li H, Guo X, He X. Characterization of microRNA expression profiles in patients with intervertebral disc degeneration. Int J Mol Med 2014; 1: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MK, Sung CO, Do IG, Jeon HK, Song TJ, Park HS, Lee YY, Kim BG, Lee JW, Bae DS. Overexpression of Galectin-3 and its clinical significance in ovarian carcinoma. Int J Clin Oncol 2011; 4: 352–358. [DOI] [PubMed] [Google Scholar]
- 22.Wu KL, Huang EY, Jhu EW, Huang YH, Su WH, Chuang PC, Yang KD. Overexpression of galectin-3 enhances migration of colon cancer cells related to activation of the K-Ras-Raf-Erk1/2 pathway. J Gastroenterol 2013; 3: 350–359. [DOI] [PubMed] [Google Scholar]
- 23.Cheung KJ, Libbrecht L, Tilleman K, Deforce D, Colle I, Van Vlierberghe H. Galectin-3-binding protein: a serological and histological assessment in accordance with hepatitis C-related liver fibrosis. Eur J Gastroenterol Hepatol 2010; 9: 1066–1073. [DOI] [PubMed] [Google Scholar]
- 24.Song S, Ji B, Ramachandran V, Wang H, Hafley M, Logsdon C, Bresalier RS. Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PLoS One 2012; 7: e42699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao P, Wang S, Zhou Y, Zheng H, Zhao G. MicroRNA-185 regulates spinal cord injuries induced by thoracolumbar spine compression fractures by targeting transforming growth factor-beta1. Exp Ther Med 2017; 13: 1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R, Tian S, Wang HB, Chu DP, Cao JL, Xia HF, Ma X. MiR-185 is involved in human breast carcinogenesis by targeting Vegfa. FEBS Lett 2014; 23: 4438–4447. [DOI] [PubMed] [Google Scholar]
- 27.Fu P, Du F, Yao M, Lv K, Liu Y. MicroRNA-185 inhibits proliferation by targeting c-Met in human breast cancer cells. Exp Ther Med 2014; 8: 1879–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regard JB, Zhong Z, Williams BO, Yang Y. Wnt signaling in bone development and disease: making stronger bone with Wnts. Cold Spring Harb Perspect Biol 2012; 4: a007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei Z, Shi H, Li W, Yu D, Shen F, Yu X, Lu D, Sun C, Liao K. miR185 inhibits nonsmall cell lung cancer cell proliferation and invasion through targeting of SOX9 and regulation of Wnt signaling. Mol Med Rep 2018; 17: 1742–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JO, Kwon EJ, Song DW, Lee JS, Kim DH. miR-185 inhibits endoplasmic reticulum stress-induced apoptosis by targeting Na+/H+ exchanger-1 in the heart. BMB Rep 2016; 49: 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bao L, Fu X, Si M, Wang Y, Ma R, Ren X, Lv H. MicroRNA-185 targets SOCS3 to inhibit beta-cell dysfunction in diabetes. PLoS One 2015; 10: e0116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng JZ, Chen JJ, Wang ZG, Yu D. MicroRNA-185 inhibits cell proliferation while promoting apoptosis and autophagy through negative regulation of TGF-beta1/mTOR axis and HOXC6 in nasopharyngeal carcinoma. Cancer Biomark 2018; 23: 107–123. [DOI] [PubMed] [Google Scholar]