Abstract
Restoration of normal DNA promoter methylation and expression states of cancer-related genes may be an option for the prevention as well as the treatment of several types of cancer. Constitutional promoter methylation of BRCA1 DNA repair associated (BRCA1) gene is linked with a high risk of developing breast and ovarian cancer. Furthermore, hypomethylation of the proto-oncogene γ synuclein (SNCG) is associated with the metastasis of breast and ovarian cancer and reduced disease-free survival (DFS). In the present study, we evaluated the potential of curcumin to re-express hypermethylated BRCA1 and to suppress hypomethylated SNCG in triple-negative breast cancer (TNBC) cell line HCC-38, the estrogen receptor-negative/progesterone receptor-negative (ER−/PR−) cell line UACC-3199, and the ER+/PR+ cell line T47D. The cells were treated with 5 and 10 µM curcumin for 6 days and with 5-aza-2′-deoxycytidine (5′-aza-CdR) for 48 h. Methylation-specific PCR and bisulfite pyrosequencing assays were used to assess DNA promoter methylation while gene expression levels were analyzed using quantitative real-time PCR and immunoblotting. We found that curcumin treatment restored BRCA1 gene expression by reducing the DNA promoter methylation level in HCC-38 and UACC-3199 cells and that it suppressed the expression of SNCG by inducing DNA promoter methylation in T47D cells. Notably, 5′-aza-CdR restored BRCA1 gene expression only in UACC-3199, and not in HCC-38 cells. Curcumin-induced hypomethylation of the BRCA1 promoter appears to be realized through the upregulation of the ten-eleven translocation 1 (TET1) gene, whereas curcumin-induced hypermethylation of SNCG may be realized through the upregulation of the DNA methyltransferase 3 (DNMT3) and the downregulation of TET1. Notably, miR-29b was found to be reversely expressed compared to TET1 in curcumin- and 5′-aza-CdR-treated cells, suggesting its involvement in the regulation of TET1. Overall, our results indicate that curcumin has an intrinsic dual function on DNA promoter methylation. We believe that curcumin may be considered a promising therapeutic option for treating TNBC patients in addition to preventing breast and ovarian cancer, particularly in cancer-free females harboring methylated BRCA1.
Keywords: curcumin, BRCA1, SNCG, methylation, demethylation, miR-29b, TET1
Introduction
Both genetic and epigenetic alterations play critical roles in the initiation and progression of carcinogenesis. Compared to genetic defects, epigenetic modifications are more dynamic and therefore more influenced by the environment (e.g., lifestyle, dietary factors) (1,2). DNA methylation is among the most studied epigenetic mechanisms. It involves the addition of a methyl (CH3) group to the cytosine in the CpG dinucleotide, resulting in the formation of 5-methylcytosine. This process is catalyzed by the DNA methyltransferase (DNMT) family of enzymes: DNMT1, DNMT3a and DNMT3b. DNMT1 is a maintenance enzyme that guards existing methylated sites through its preference for hemimethylated DNA (3). DNMT3a and DNMT3b are de novo methyltransferases responsible for establishing DNA methylation patterns during embryogenesis. Any defects in DNMTs will induce imbalances in DNA modification, resulting in genomic instability and gene dysregulation (4,5).
However, DNA demethylation involves the hydroxylation of 5-methylcytosine to 5-hydroxymethylcytosine (6,7). It is mediated by the ten-eleven translocation (TET) family of proteins: TET1, TET2 and TET3 (8). TET1 is a maintenance DNA demethylase enzyme that protects against aberrant demethylation (9). It acts both as a tumor suppressor preventing cell proliferation and tumor metastasis and as an oncogene contributing to aberrant hypomethylation. The delicate balance between DNA methylation and demethylation is known to be regulated by a specific class of microRNAs, termed epi-miRNAs, which target both families of epigenetic enzymes DNMTs and TETs (10).
MicroRNAs (miRs) are short non-coding RNAs that are a novel class of cancer-relevant molecules. The miR-29 family, which consists of miR-29a, miR-29b, and miR-29c, is abnormally expressed in multiple cancers (10). miR-29b is the most highly expressed family member. It is classified as an epi-miRNA, regulating the balance between DNA methylation and demethylation as a regulator for TET1 and DNMTs (10,11). In breast cancer, miR-29b has been reported to be both a suppressor and a promoter of proliferation and metastasis through its regulation of the TET1 gene (12,13).
The BRCA1 gene is a critical DNA repair-related gene that plays an essential role in the mechanisms of DNA repair, cell cycle checkpoints, and transcription. Cells lacking BRCA1 protein are susceptible to mutations and genomic instability, which can lead to early carcinogenesis. The pathogenic germline mutations of the BRCA1 gene are highly associated with familial breast cancers. However, loss-of-function in BRCA1 resulting from aberrant promoter methylation is associated with sporadic breast cancer. BRCA1 promoter methylation has been detected in DNA extracted from white blood cells (WBCs). Several studies have shown that constitutional BRCA1 promoter methylation is linked to a high risk of developing early-onset breast and ovarian cancers (14–19). The promoter region of the BRCA1 gene contains 30 CpG sites covering the area from −567 to +44 relative to the transcription start site (20). This area includes the binding sites of several transcription factors, including SP1, E2F and CTCF. The binding of these factors to the BRCA1 promoter keeps the promoter in a methylation-free state (21,22). The CTCF and E2F factors are enriched at the unmethylated BRCA1 promoter, such as in MCF-7, but not at the methylated promoter in UACC-3199 and HCC-38 cells (22).
γ synuclein is a member of the synuclein family of proteins. It is encoded by the gene SNCG, which is also known as breast cancer-specific gene 1 (BCSG1) (23). This gene is a proto-oncogene that is highly expressed in stages III and IV of breast ductal carcinomas but not in normal breast tissues. The expression of SNCG in the primary breast tumor is associated with metastasis and reduced disease-free survival (DFS) (24). Exon 1 of SNCG contains 15 CpG sites covering the region from −169 to +81 relative to the translation start codon. The demethylation of these CpG sites is responsible for the aberrant expression of SNCG in breast carcinomas (25). The inhibition of SNCG reverses the malignant phenotype of the highly SNCG-hypomethylated cell line T47D (26).
One of the main differences between genetic and epigenetic alterations is the reversibility of the latter process. Accordingly, restoration of the function of defective tumor-suppressor genes and suppression of constitutively activate oncogenes is an attractive clinical option for the prevention and treatment of cancer. Although synthetic demethylating agents, such as 5-azacytidine and 5-aza-2′-deoxycytidine, are effective DNA methylation inhibitors, they have unselective demethylation effects that can lead to the activation of silenced pro-oncogenes. Notably, no DNA methylation inducers have hitherto been identified.
Curcumin is the active component of the herb Curcuma longa. It is believed to have chemopreventive and chemotherapeutic properties. Several studies have shown that this herb can exert anticancer effects, on breast cancer in particular, by targeting various signaling pathways (27). Furthermore, it has been reported that curcumin can function as a DNA methylation inhibitor in breast cancer cells (28–32). Nevertheless, curcumin's potential as a DNA methylation inducer remains to be fully explored. In the present study, we attempted to evaluate the potential of curcumin molecules to restore normal DNA methylation and gene expression patterns to the BRCA1 and SNCG genes in breast cancer cells.
Materials and methods
Cell culture and treatment
The HCC-38, UACC-3199, and T47D breast cancer cell lines were purchased from the American Type Culture Collection (ATCC). The cells were tested for mycoplasma. The cells were cultured in RPMI-1640 media supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. The supplements were obtained from Gibco/Life Technologies (Thermo Fisher Scientific, Inc.). The cells were treated with 5 and 10 µM curcumin (Sigma-Aldrich; Merck KGaA) when they reached 40–60% confluence and incubated in a humidified atmosphere at 37°C and 5% CO2 for 6 days. The HCC-38 and UACC-3199 cells were treated with 5 µM 5-aza-2′-deoxycytidine (5′-aza-CDR) (Sigma-Aldrich; Merck KGaA) when they reached 70% confluence and were incubated at 37°C for 48 h. The cells were then collected for DNA, RNA and protein extraction.
Cell proliferation
The cells were treated with 5 and 10 µM of curcumin for 3 days, after which they were re-seeded at a density of 5,000 cells/E-16 plate and re-incubated with the same doses of curcumin for a further 3 days. The proliferation rate was measured using the RTCA-DP xCELLigence system (Roche-Germany). The cell index represents the cell status based on the measured electrical impendence change divided by a background value.
Methylation-specific PCR
Approximately 2 µg of DNA was treated with sodium bisulfate and purified using the EpiTect Bisulfite kit (Qiagen, Inc.) in accordance with the manufacturer's recommendations. The DNA was then amplified using published PCR primers for BRCA1 and SNCG (33,34) that distinguish between methylated and unmethylated DNA. PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide. Totally methylated bisulfite-treated DNA was used as a positive control. All PCR reactions were repeated at least twice.
Bisulfite pyrosequencing
Bisulfite pyrosequencing was used for DNA methylation quantification. Five different assays (35) were used to assess the methylation status of 23 CpG sites across the BRCA1 promoter. The PCR and pyrosequencing reactions were performed using PyroMark products and reagents (Qiagen, Inc.), as previously described (36). Methylation quantification was performed using PyroMark Q24 software (Qiagen, Inc.).
Real-time PCR
Superscript III (Invitrogen; Thermo Fisher Scientific, Inc.) reverse transcriptase and random hexamers were used for cDNA synthesis. Quantitative real-time PCR was then performed using primers specific to BRCA1, SNCG, and TET1 transcripts, using GAPDH as an internal control. Primers are listed in Table I. PCR was performed with SYBR Green using the CFX96 Real-Time system (Bio-Rad Laboratories, Inc.). For miR-29b, qPCR was performed using the stem-loop reverse transcription primer, and the TaqMan microRNA reverse transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 snRNA was used for normalization. The relative gene expression was calculated based on the threshold cycle (Ct) value using the 2−ΔΔCq method (37). The fold change of mRNA expression was performed relative to DMSO-treated cells.
Table I.
Real-time and MSP PCR primers.
| Primers | Sequence | Annealing temperature (°C) |
|---|---|---|
| BRCA1 | F 5′-TGTAGGCTCCTTTTGGTTATATCATTC-3′ | 59 |
| R 5′-CATGCTGAAACTTCTCAACCAGAA-3′ | ||
| SNCG | F 5′-GGAGGACTTGAGGCCATCTG-3′ | 60 |
| R 5′-CTCCTCTGCCACTTCTCTTTTC-3′ | ||
| TET1 | F 5′-CCCGGGCTCCAAAGTTGTG-3′ | 59 |
| R 5′-GCAGGAAACAGAGTCATTGGTCCT-3′ | ||
| GAPDH | F 5′-TTCAACGGCACAGTCAAGG-3′ | 60 |
| R 5′-CTCAGCACCAGCATCACC-3′ | ||
| M. BRCA1 | F 5′-GGTTAATTTAGAGTTTCGAGAGACG-3′ | 65 |
| R 5′-TCAACGAACTCACGCCGCGCAATCG-3′ | ||
| U. BRCA1 | F 5′-GGTTAATTTAGAGTTTTGAGAGATG-3′ | 65 |
| R 5′-TCAACAAACTCACACCACACAATCA-3′ | ||
| M. SNCG | F 5′-TCGTATTAATATTTTATCGGCGT-3′ | 60 |
| R 5′-CCGCACCCACCACGCCCTCCTTAACGA-3′ | ||
| U. SNCG | F 5′-TTGGTGTTAATAGGAGGTATTGGGGATAGTTGTTGTG-3′ | 59 |
| R 5′-CACACCCACCACACCCTCCTTAACAAT-3′ |
MSP, methylation-specific PCR; BRCA1, BRCA1 DNA repair associated; SNCG, γ synuclein; TET1, ten-eleven translocation 1; F, forward; R, reverse; M, methylated; U, unmethylated.
Western blot analysis
Protein was extracted from the cells using RIPA lysis buffer (R0278; Sigma-Aldrich; Merck KGaA). Bradford method was used for protein quantification. Protein (50 µg) was subjected to 10 and 12% SDS-PAGE and then transferred to PVDF membranes. After blocking at room temperature with 5% non-fat milk for 1 h, the membranes were incubated at 4°C overnight with primary antibodies (dilution 1:1,000), BRCA1 (ab9141), SNCG (ab55424), TET1 (Ab156993), DNMT3a (Ab13888) and DNMT3b (Ab2851) (purchased from Abcam); DNMT1 (D63A6) and GAPDH (purchased from Cell Signaling Technology, Inc.). The membranes were visualized using ECL Detection reagents (Pierce; Thermo Fisher Scientific, Inc.). Images were visualized using a LAS-4000 Imager (Fujifilm). Band quantification was carried out using the GelQuant.NET program (version 1.8.2; Biochem Lab Solutions).
Statistical analysis
For gene expression levels, statistical analysis was performed using Single Factor ANOVA and Tukey's multiple comparisons test (GraphPad Prism version 8.3; GraphPad Software, Inc.) to determine the statistical significance between multiple-dose curcumin-treated and untreated cells. Student's t-test was used to determine the statistical significance between single dose curcumin-treated and untreated cells. All observed differences were considered significant when associated with P-values <0.05.
Results
Curcumin suppresses the proliferation of HCC-38, UACC-3199, and T47D cell lines
In the present study, we used three breast cancer cell lines, HCC-38 and UACC-3199, which are highly BRCA1-hypermethylated cell lines (22), and T47D, which is a highly SNCG-hypomethylated cell line. HCC-38 is a triple-negative breast cancer (TNBC) cell line, UACC-3199 is estrogen receptor-negative/progesterone receptor-negative (ER−/PR−), and T47D is ER+/PR+. Based on previous studies (29,30,38,39), we treated the cells with 5 and 10 µM of curcumin. To investigate the effect of these curcumin concentrations on cell proliferation, we monitored cell proliferation in the presence of curcumin over a total period of 6 days. First, we treated the cells with curcumin for 72 h. Real-time cell proliferation was then monitored over a further 72 h in the presence of curcumin using the xCELLigence system E-Plate. Although curcumin reduced the proliferation of HCC-38 and UACC-3199 cells in a dose-independent manner, the proliferation of T47D cells was dose-dependently inhibited compared to the control (Fig. 1). These results demonstrated that curcumin influenced the proliferation of all three cell lines studied.
Figure 1.
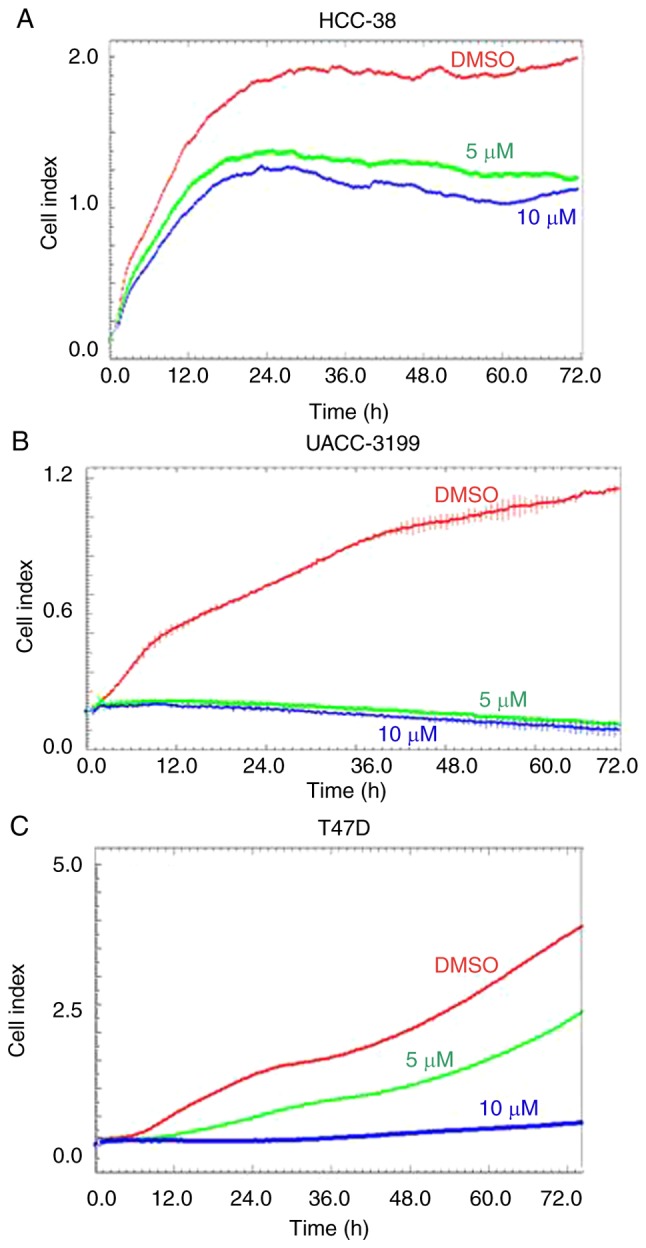
Effect of curcumin on the proliferation of HCC-38, UACC-3199, and T47D cell lines. Representative graphs comparing the rate of proliferation over a total period of 6 days for the (A) HCC-38, (B) UACC-3199 and (C) T47D cell lines when incubated with DMSO (red line), 5 µM curcumin (green line) and 10 µM curcumin (blue line). DMSO, dimethyl sulfoxide.
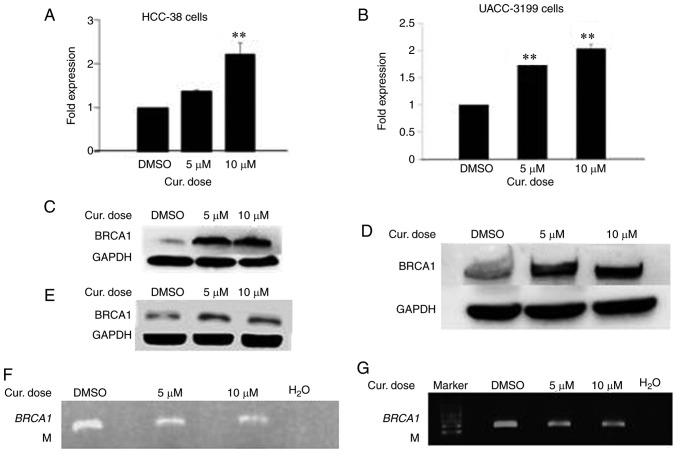
Curcumin increases the mRNA and protein levels of BRCA1 in HCC-38 and UACC-3199 cells by reducing promoter methylation
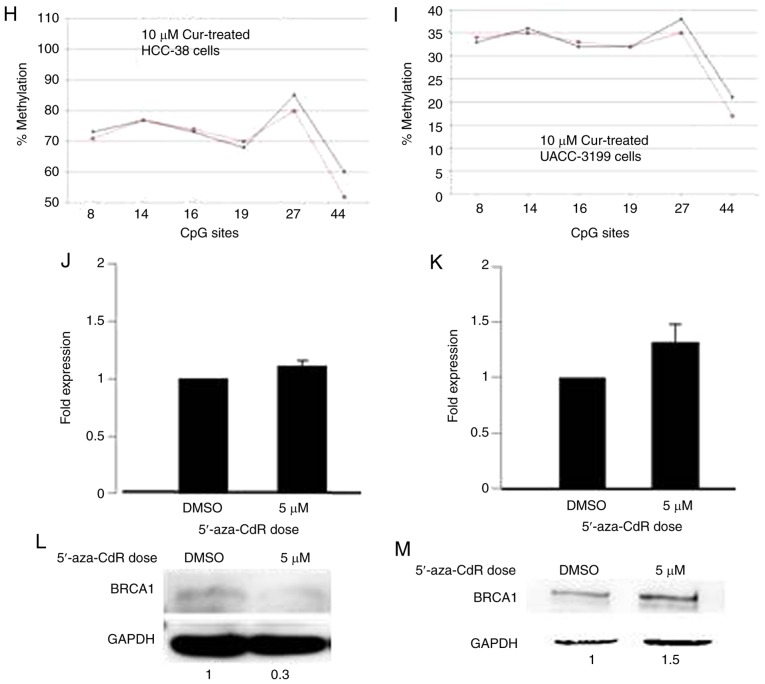
The reactivation of silenced tumor-suppressor genes is an attractive clinical option for the prevention and treatment of cancer. Epigenetic silencing of BRCA1 by promoter hypermethylation results in the low expression of BRCA1 mRNA and BRCA1 protein in sporadic breast cancer. To ascertain whether curcumin can re-express BRCA1 in HCC-38 and UACC-3199 cell lines, the cells were treated with 5 and 10 µM of curcumin for 6 days. Notably, curcumin increased the level of BRCA1 mRNA up to 2-fold in both cell lines in a dose-dependent manner (Fig. 2A and B). Additionally, a consequent high increase was observed in the level of BRCA1 protein (Fig. 2C and D). Then, we evaluated whether curcumin-induced BRCA1 re-expression is associated with the hypomethylation of its promoter. To this end, we assessed the status of BRCA1 promoter methylation in curcumin-treated HCC-38 and UACC-3199 cells using the methylation-specific PCR (MSP) assay. As shown in Fig. 2F and G, the intensity of the methylated band was reduced in the curcumin-treated cells, compared to the control. As further verification of the hypomethylation of the BRCA1 promoter, the promoter region spanning six CpG sites (+8, +14, +16, +19, +27 and +44, in which the reverse MSP primer is located) (Fig. 3A) was analyzed using pyrosequencing. As shown in Fig. 2H and I, the levels of methylation at +27 and +44 CpG sites were reduced in the curcumin-treated cells, compared to the control. These results suggest that the re-expression of BRCA1 in curcumin-treated cells may be associated with the partial hypomethylation of the BRCA1 promoter. Next, we investigated whether the re-expression of the BRCA1 protein is transient or persistent. To this end, curcumin-treated HCC-38 cells were grown in curcumin-free media for a further 10 days, with the media changed every 5 days. Interestingly, BRCA1 protein continued to be highly expressed in the curcumin-free medium, compared to the control, indicating a persistent effect of curcumin (Fig. 2E). Next, to compare the demethylating effect of curcumin to that of the demethylating agent 5′-aza-CdR, we treated the two cell lines with 5 µM 5′-aza-CdR for 48 h. Notably, 5′-aza-CdR was able to re-express BRCA1 mRNA and BRCA1 protein only in UACC-3199 (Fig. 2K and M) and not in HCC-38 cells (Fig. 2J). Intriguingly, the expression of BRCA1 protein was reduced in 5′-aza-CdR-treated HCC-38 cells (Fig. 2L).
Figure 2.
Curcumin induces the expression of BRCA1 in HCC-38 and UACC-3199 cells. The cells were treated with 5 and 10 µM curcumin for 6 days, and the effects of curcumin on BRCA1 mRNA and protein expression in (A and C) HCC-38 and (B and D) UACC-3199 cells, respectively, are shown. (E) Effect of curcumin-free media on the expression of BRCA1 protein in HCC-38 cells. (F and G) MSP analysis of BRCA1 promoter methylation. M, only the methylated bands are shown. (H and I) Methylation plots for the BRCA1 promoter as determined by bisulfite pyrosequencing assay. Black lines represent values for control DMSO-treated cells, and red lines represent 10 µM curcumin-treated cells. Numbers represent CpG sites relative to the transcription start site. (J-M) Cells were treated with 5 µM 5′-aza-CdR for 48 h. Effect of 5′-aza-CdR on BRCA1 mRNA and protein expression in (J and L) HCC-38 and (K and M) UACC-3199 cells, respectively. **P<0.01, vs. the control. Error bars represent the mean ± SD. BRCA1, BRCA1 DNA repair associated; DMSO, dimethyl sulfoxide; 5′-aza-CdR, 5-aza-2′-deoxycytidine; Cur, curcumin.
Figure 3.
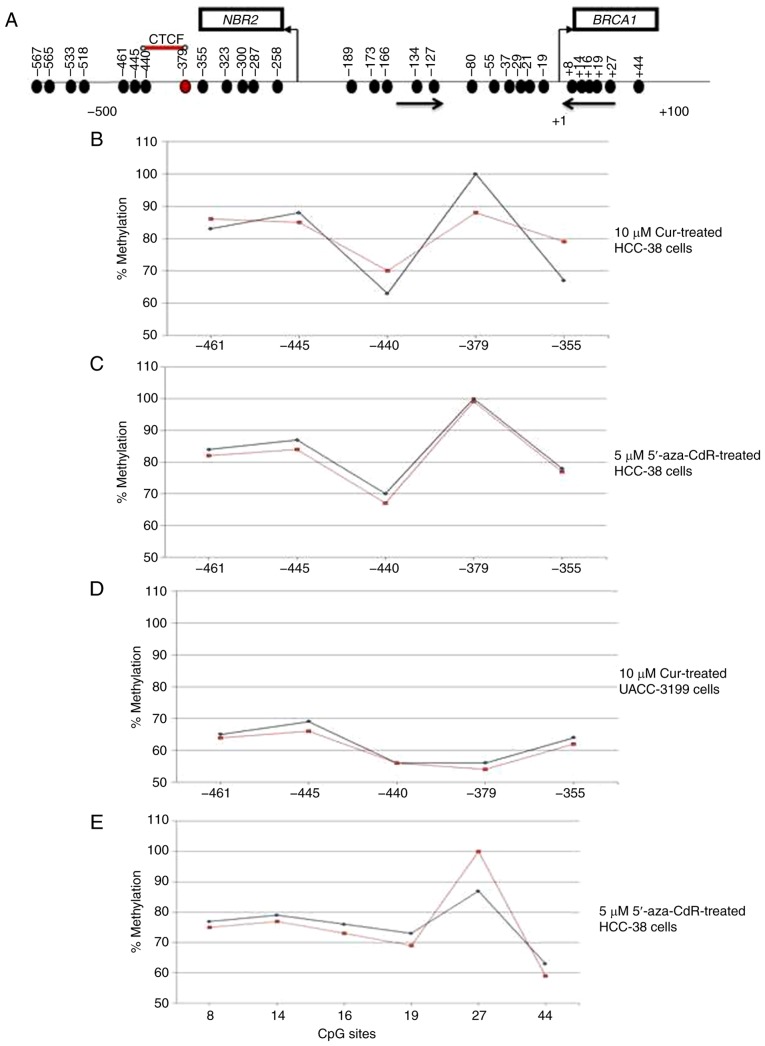
Pyrosequencing analysis of BRCA1 promoter methylation in HCC-38 and UACC-3199 cells. (A) Schematic representation of the BRCA1 CpG island. Vertical arrows indicate the transcription start sites and directions. Horizontal arrows represent forward and reversed MSP primers. Black circles indicate methylated CpG sites. A red circle marks the −379 CpG site. The red line indicates the binding site of the CTCF transcription factor. The numbers refer to the positions of the CpG sites relative to the BRCA1 transcription start site. (B-E) Levels of methylation of CpG sites along the BRCA1 promoter region measured by bisulfite pyrosequencing assay from (B) HCC-38 cells treated with 10 µM curcumin for 6 days, (C and E) HCC-38 cells treated with 5 µM 5′-aza-CdR for 48 h, and (D) UACC-3199 cells treated with 10 µM curcumin for 6 days. Black lines represent values for control DMSO-treated cells, and red lines represent treated cells. BRCA1, BRCA1 DNA repair associated; DMSO, dimethyl sulfoxide; 5′-aza-CdR, 5-aza-2′-deoxycytidine; Cur, curcumin.
Curcumin and not 5′-aza-CDR induces demethylation of the −379 CpG site in the BRCA1 promoter in the HCC-38 cells
To investigate the difference between the effect of curcumin and 5′-aza-CdR on the re-expression of BRCA1 in the HCC-38 cells, we studied the methylation status of 23 CpG sites located in the BRCA1 promoter region (Fig. 3A) by sodium bisulfite pyrosequencing in curcumin-treated cells as compared to 5′-aza-CDR. We found that the CpG site located at −379 was 100% methylated. In comparison to the control, the methylation level of this CpG site was reduced by 12% in curcumin-treated cells, while in 5′-aza-CdR-treated cells, the methylation level was only reduced by 1% (Fig. 3B and C). These results suggest that the methylation of the −379 CpG site is instrumental in controlling the expression of BRCA1 in the HCC-38 cell line. Notably, the −379 CpG site was partially methylated (56%) in the UACC-3199 cells, and the methylation level was only reduced by 2% in the curcumin-treated cells, compared to the control (Fig. 3D). Interestingly, we found also that the level of methylation at the +27 CpG site was increased by 13% in 5′-aza-CdR-treated HCC-38 cells, compared to the control (Fig. 3E).
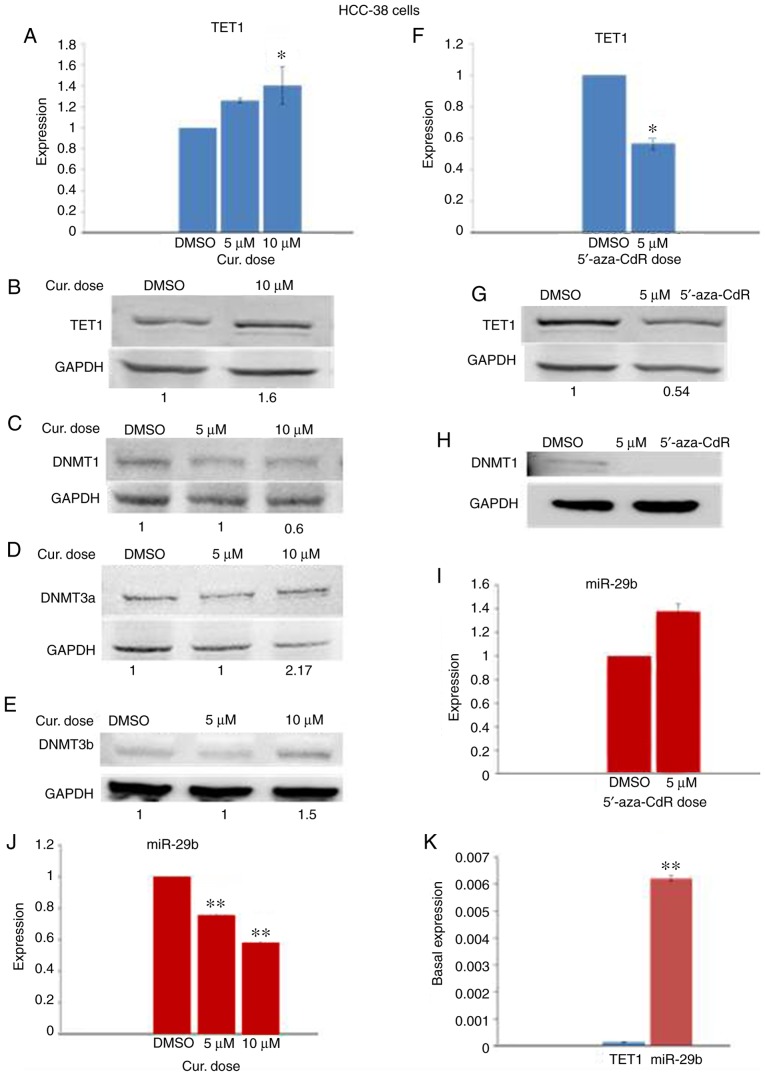
Curcumin downregulates the expression of DNMT1 and upregulates TET1 and DNMT3 in HCC-38 cells
DNMTs and TETs are epigenetic enzyme families responsible for the regulation of DNA methylation and demethylation, respectively. It has been reported that TET1 acts both as a tumor suppressor, reducing breast tumor development through demethylating essential genes (13), and as an oncogene, leading to hypomethylation and activation of oncogenic pathways (8). However, it has been demonstrated that curcumin re-activates methylated tumor-suppressor genes by downregulating the protein level of DNMT1 (30). To investigate which enzyme participates in the curcumin-induced hypomethylation of the BRCA1 promoter in HCC-38 cells, the mRNA and protein levels of TET1, DNMT1, DNMT3a and DNMT3b were analyzed by real-time RT-PCR and immunoblotting in curcumin-treated cells and compared to those of the 5′-aza-CdR-treated cells. Notably, while curcumin reduced the level of DNMT1 protein, it increased TET1 mRNA and TET1 protein in addition to the protein levels of DNMT3a and DNMT3b, compared to the control (Fig. 4A-E). However, in 5′-aza-CdR-treated cells, the TET1 mRNA and TET1 protein levels were decreased while DNMT1 protein was depleted (Fig. 4F-H). These results suggest that curcumin-induced hypomethylation of the BRCA1 promoter in HCC-38 cells may be achieved through the upregulation of TET1.
Figure 4.
Effect of curcumin and 5′-aza-CdR on the expression of TET1, DNMTs, and miR-29b in HCC-38 cells. The cells were treated with 5 and 10 µM curcumin for 6 days, and with 5 µM 5′-aza-CdR for 48 h. (A) Effect of curcumin on TET1 mRNA expression. (B-E) Western blots for the expression of TET1, DNMT1, DNMT3a, and DNMT3b, respectively, in curcumin-treated cells. (F) Effect of 5′-aza-CdR on TET1 mRNA expression. (G and H) Western blots for the expression of TET1 and DNMT1, respectively, in 5′-aza-CdR-treated cells. (I and J) Effect of curcumin and 5′-aza-CdR on miR-29b expression, respectively. (K) Basal expression of TET1 and miR-29b in control HCC-38 cells. *P<0.05 and **P<0.01, vs. the control. Error bars represent the mean ± SD. TET1, ten-eleven translocation 1; DNMTs, DNA methyltransferases; DMSO, dimethyl sulfoxide; 5′-aza-CdR, 5-aza-2′-deoxycytidine; Cur, curcumin.
Curcumin downregulates miR-29b in HCC-38 cells
It has been reported that TET1 is a target of miR-29b (13). To investigate whether the expression of TET1 in HCC-38 cells could be regulated by miR-29b, we analyzed the expression levels of miR-29b by real-time RT-PCR in curcumin- and 5′-aza-CdR-treated cells. Compared to the control, miR-29b was elevated in the 5′-aza-CdR-treated HCC-38 cells (Fig. 4I), and significantly reduced in the curcumin-treated HCC-38 cells (Fig. 4J). These results revealed a reverse expression pattern of miR-29b and TET1 in cells that had been treated with curcumin or 5′-aza-CdR, suggesting that TET1 may be controlled by miR-29b. To support this result, we evaluated the basal expression levels of TET1 and miR-29b in control HCC-38 cells. Interestingly, we found that the basal expression of miR-29b was significantly higher than that of TET1 (Fig. 4K), revealing the possible involvement of miR-29b in the regulation of TET1 in the HCC-38 cell line.
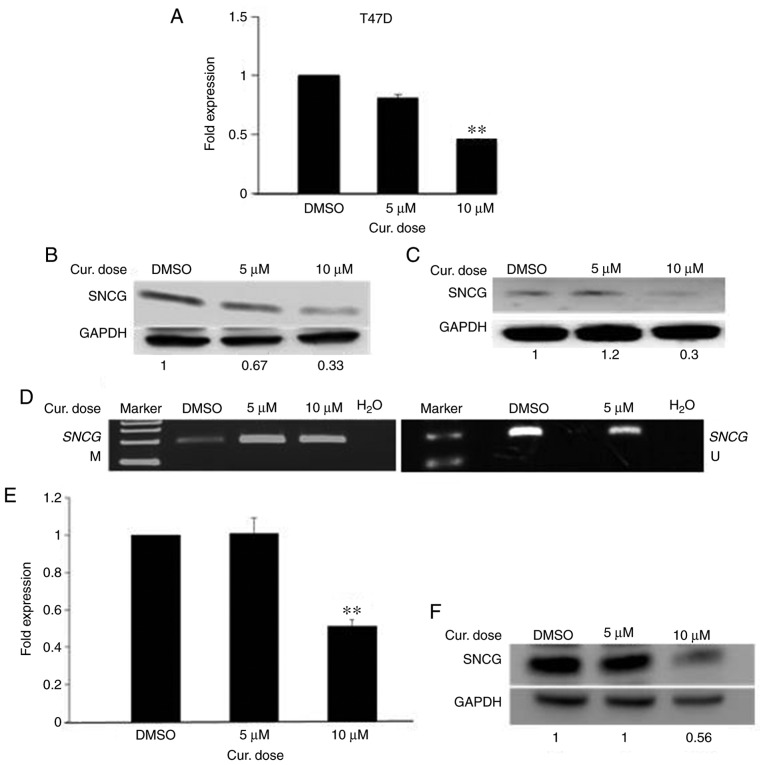
Curcumin decreases the mRNA and protein levels of SNCG in T47D and HCC-38 cells by inducing promoter methylation
The deactivation of an active oncogene is the other side of the coin for cancer treatments. Epigenetic activation of SNCG by promoter hypomethylation results in the high expression of SNCG mRNA and SNCG protein in breast and ovarian cancers (40). To ascertain whether curcumin can deactivate SNCG in the highly SNCG-hypomethylated cell line T47D, the cells were treated with 5 and 10 µM curcumin for 6 days. As shown in Fig. 5A, curcumin decreased the level of SNCG mRNA down to 2-fold in a dose-dependent manner. Additionally, a reduction was observed in the level of SNCG protein (Fig. 5B). Then, we evaluated whether the curcumin-induced reduction of SNCG is associated with the hypermethylation of its promoter. To this end, we assessed the methylation status of the SNCG promoter in curcumin-treated T47D cells using the MSP assay. As Fig. 5D illustrates, a high increase was noted in the intensity of the methylated band with a decrease in that of the unmethylated band, compared to the control. These results suggest that the decreased expression of SNCG in curcumin-treated T47D cells is associated with the hypermethylation of the SNCG promoter. Next, we assessed whether the reduction of SNCG protein in curcumin-treated cells is transient or persistent. To this end, the growth of curcumin-treated T47D cells in curcumin-free media was continued for a further 10 days, with the media changed every 5 days. Importantly, as Fig. 5C illustrates, low expression of SNCG protein continued in the curcumin-free medium, compared to the control, indicating that curcumin had a sustained effect. Next, to ascertain whether curcumin can deactivate SNCG in the HCC-38 cell line, we measured the expression of SNCG in the curcumin-treated HCC-38 cells. In addition to re-expressing BRCA1 in HCC-38, curcumin also decreased SNCG mRNA and SNCG protein down to 2-fold, with the 10 µM dose (Fig. 5E and F). These results demonstrate that curcumin has opposing roles in DNA methylation in the same cell line.
Figure 5.
Curcumin suppresses the expression of SNCG in T47D and HCC-38 cell lines. The cells were treated with 5 and 10 µM curcumin for 6 days. (A and B) Effect of curcumin on SNCG mRNA and SNCG protein expression in T47D cells, respectively. (C) Effect of curcumin-free media on the expression of SNCG protein in T47D cells. (D) MSP analysis of SNCG promoter methylation in T47D cells. M, methylated bands; U, unmethylated bands. (E and F) Effect of curcumin on SNCG mRNA and SNCG protein expression in HCC-38 cells, respectively. **P<0.01, vs. the control. Error bars represent the mean ± SD. SNCG, γ synuclein; Cur, curcumin; DMSO, dimethyl sulfoxide.
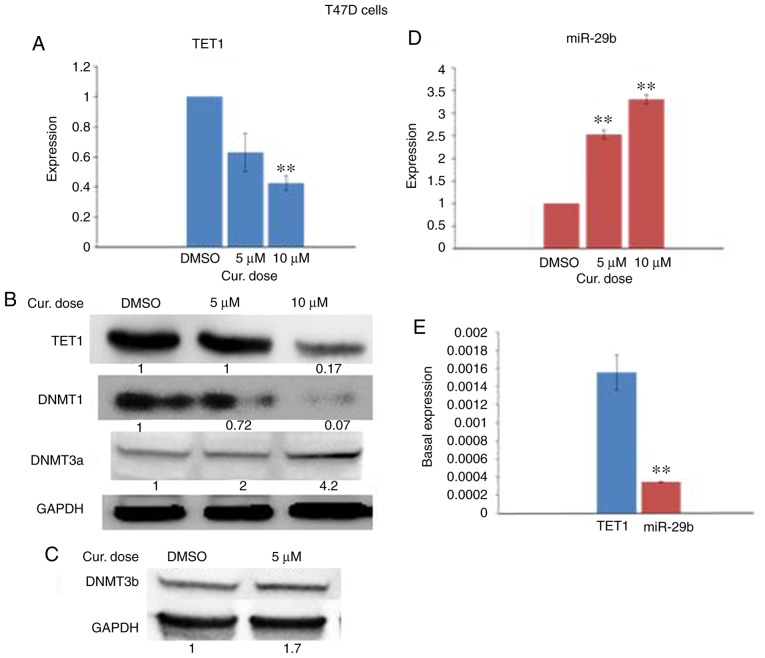
Curcumin downregulates the expression of DNMT1 and TET1 and upregulates DNMT3 in T47D cells
Next, we sought to investigate which enzyme is responsible for the curcumin-induced hypermethylation of the SNCG promoter. To this end, we analyzed the expression levels of TET1, DNMT1, DNMT3a, and DNMT3b by real-time RT-PCR and immunoblotting in curcumin-treated T47D cells. In comparison to the controls, curcumin reduced the expression of TET1 and DNMT1 (Fig. 6A and B) and elevated that of DNMT3a and DNMT3b (Fig. 6B and C). These results suggest that both TET1 and DNMT3 may be involved in the curcumin-induced hypermethylation of the SNCG promoter in T47D cells.
Figure 6.
Effect of curcumin on the expression of TET1, DNMTs and miR-29b in T47D cells. The cells were treated with 5 and 10 µM curcumin for 6 days. (A) Effect of curcumin on TET1 mRNA expression. (B and C) Western blots for TET1, DNMT1, DNMT3a, and DNMT3b, respectively. (D) Effect of curcumin on miR-29b expression. (E) Basal expression of TET1 and miR-29b in control T47D cells. **P<0.01, vs. the control. Error bars represent the mean ± SD. TET1, ten-eleven translocation 1; DNMTs, DNA methyltransferases; DMSO, dimethyl sulfoxide; Cur, curcumin.
Curcumin upregulates miR-29b in T47D cells
To determine whether miR-29b could regulate the expression of TET1 in curcumin-treated T47D cells, we analyzed the expression level of miR-29b by real-time RT-PCR in curcumin-treated cells. In contrast to the curcumin-treated HCC-38 cells, the expression of miR-29b was significantly elevated in the T47D cells (Fig. 6D). However, these results revealed a reverse expression pattern of miR-29b and TET1 in the curcumin-treated T47D cells, suggesting that TET1 may be a target of miR-29b in these cells. To support this result, we evaluated the basal expression levels of TET1 and miR-29b in control T47D cells. In contrast to the HCC-38 cells, we found that the basal expression of miR-29b was significantly lower than that of TET1 (Fig. 6E), also revealing the possible involvement of miR-29b in the regulation of TET1 in T47D cells.
Discussion
In the present study, it was demonstrated that curcumin exhibits contradictory functions in regards to DNA methylation, demethylation and re-expression of the tumor-suppressor gene BRCA1 DNA repair associated (BRCA1), as well as the methylation and suppression of the expression of oncogene γ synuclein (SNCG) in breast cancer cells. We also found that curcumin is a potent inhibitor of cell proliferation. This result is consistent with a previous study that demonstrated that curcumin is a potent growth inhibitor of various breast cancer cell lines (41). Previous studies have found that BRCA1 deficiency and the aberrant expression of SNCG enhance the proliferation of cancer cells (42–44) and that the restoration of normal expression patterns to these two genes reduces cell proliferation. In the present study, the re-expression of BRCA1 in HCC-38 and UACC-3199 cells and the suppression of SNCG in T47D may have been one mechanism by which curcumin influenced the inhibition of cell proliferation in the three cell lines.
Both curcumin and 5′-aza-CdR demethylated and re-activated BRCA1 in the UACC-3199 cell line. Remarkably, in HCC-38 cells, only curcumin re-activated BRCA1, while 5′-aza-CdR did not. We may attribute this to curcumin's ability to reduce the methylation status of the −379 CpG site in the BRCA1 promoter region. This site flanks the binding site of the CTCF transcription factor in the BRCA1 promoter. It has been reported that CTCF binds to the unmethylated BRCA1 promoter simply to function as an insulator, maintaining the BRCA1 promoter region in a methylation-free state (21,22). The binding of this transcription factor was found to be affected by the methylation status of the flanking CpG sites (−440 and −379) (22). Hence, it is plausible that the partial demethylation of the −379 CpG site in the curcumin-treated HCC-38 cells increases the promoter's accessibility to CTCF leading to the re-expression of BRCA1. Notably, the −379 CpG site was not affected by 5′-aza-CdR in either cell line, HCC-38 or UACC-3199. Furthermore, it has been reported that only E2F1, and not CTCF, was enriched in 5′-aza-CdR-treated UACC-3199 cells (22). However, further studies are needed to verify this claim. Remarkably, it has been reported that zebularine (another demethylating agent) and the non-nucleoside demethylation drugs EGCG and procaine were also unable to restore the BRCA1 gene expression in HCC-38 cells (22). Intriguingly, treating the HCC-38 cells with 5′-aza-CdR downregulated the expression of BRCA1. Our findings are consistent with those of a previous study, which demonstrated that the expression of BRCA1 mRNA was reduced by 30–40% in 5′-aza-CdR-treated HCC-38 cells, compared to the control (22).
Previous studies have shown that curcumin demethylates hypermethylated genes by inhibiting DNA methyltransferases (DNMTs) (29,30). In the present study, we found that, while DNMT1 expression was downregulated in the curcumin-treated HCC-38 cells, DNMT3a and DNMT3b were upregulated. This result suggests that the re-expression of BRCA1 in HCC-38 may not result from the modulation of DNMTs. This suggestion is supported by the fact that 5′-aza-CdR failed to re-express BRCA1 in HCC-38 despite the depletion of DNMTs, indicating that another mechanism is involved in the re-expression of BRCA1. Indeed, we found that TET1 was upregulated in the curcumin-treated cells, which may indicate its involvement in the re-expression of BRCA1. This is supported by the fact that in gastric cancer cells, TET1 binds to the hypermethylated PTEN and re-activates its transcription through the demethylation of its promoter (45). Furthermore, it has been suggested that the decreased expression of TET1 may induce aberrant DNA methylation (46). Indeed, we observed an increase in the level of methylation at the +27 CpG site. This finding may explain the further reduction of the expression of BRCA1 protein in the 5′-aza-CdR-treated cells, as TET1 was downregulated in these cells.
The fact that curcumin exerts an effect on both epigenetic enzyme families DNMTs and TETs suggest that curcumin plays contradictory roles in the control of DNA methylation status. Indeed, we found that curcumin could induce methylation to the hypomethylated SNCG promoter in the breast cancer cell lines T47D and HCC-38 with a corresponding decrease in its mRNA and protein expression levels. To the best of our knowledge, our study is the first to demonstrate the methylation-inducing properties of curcumin in breast cancer cell lines. However, with regard to multiple myeloma cells, it has recently been reported that curcumin-induced promoter methylation to the mTOR gene was associated with a corresponding downregulation of its expression. The authors suggested that curcumin-induced hypermethylation of mTOR may be associated with the upregulation of DNMT3a and DNMT3b (47). Here, we found that curcumin downregulated TET1 in T47D and upregulated DNMT3a and DNMT3b in T47D and HCC-38 cells. This suggests that the curcumin-induced hypermethylation of SNCG may be associated with the upregulation of DNMT3. Interestingly, it has been reported that, in SNCG-positive lung cancer cells H292 endogenous overexpression of DNMT3b, but not of DNMT3a or DNMT1, suppressed SNCG expression by inducing DNA methylation of the SNCG CpG island (48). However, the possibility that the curcumin-induced hypermethylation of SNCG may be achieved through the downregulation of TET1 cannot be excluded. This is supported by the finding that the activation of the oncogenic pathway in breast and ovarian cancers is TET1 overexpression-dependent and that the deletion of TET1 attenuated the effect of the oncogenic pathway (8). Thus, it is plausible that the curcumin-induced hypermethylation of SNCG in breast cancer cells may be achieved through the downregulation of TET1. However, further studies are needed to clarify the exact mechanisms of this process.
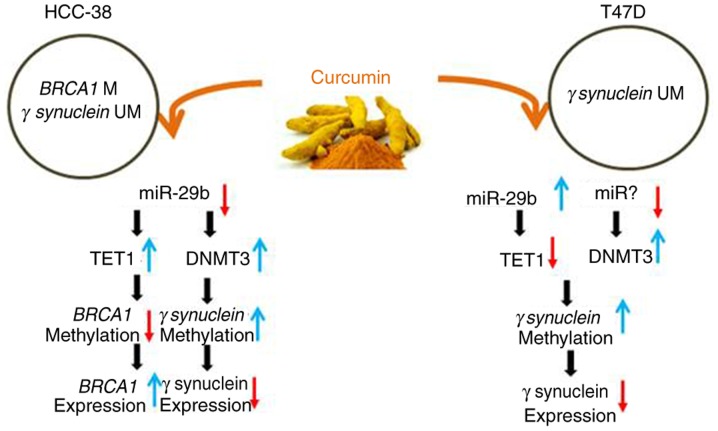
miR-29b is an epi-miRNA, being a regulator for DNMTs and TETs by direct inhibition of these enzymes (49). Thus, it has been suggested that miR-29b may act as a stabilizer of DNA methylation, balancing between methylation and demethylation (10). Notably, it has been shown that curcumin re-expressed PTEN in hepatic stellate cells by downregulating DNMT3b through the upregulation of miR-29b (31). However, miR-29b has also been shown to affect breast cancer proliferation and metastasis by targeting TET1 (12,13). Here, we found that curcumin appears to re-express BRCA1 and suppress SNCG in HCC-38 cells by upregulating TET1 and DNMT3, respectively, which may be realized through the downregulation of miR-29b. However, in T47D cells, curcumin appears to suppress SNCG by downregulating TET1, which may be realized through the upregulation of miR-29b and the upregulation of DNMT3, which may be achieved through another miRNA (Fig. 7 summarizes the results).
Figure 7.
Diagram summarizing the effects of curcumin on the expression of BRCA1 and SNCG in HCC-38 and T47D cells. BRCA1, BRCA1 DNA repair associated; SNCG, γ synuclein; TET1, ten-eleven translocation 1; DNMT, DNA methyltransferase.
Overall, our data suggest that curcumin may act as a stabilizer of DNA methylation balancing between methylation and demethylation by regulating TET1 and DNMT3, which may be achieved through the modulation of miR-29b. However, further studies are needed to confirm the direct role of miR-29b in this epigenetic regulation.
In conclusion, in the present study, we demonstrated that curcumin performs a dual function in DNA methylation. As curcumin is an activator for the hypermethylated BRCA1 promoter, we, therefore, believe that it holds the potential to be an effective therapeutic option for triple-negative breast cancer as well as for the prevention of breast and ovarian cancer, particularly for BRCA1-promoter methylation carriers.
Acknowledgements
We would like to acknowledge Dr B. Karakas and Dr A. Qattan (Molecular Oncology Department, King Faisal Specialist Hospital and Research Centre) for revising the article, and for helping with the statistical analysis, respectively.
Glossary
Abbreviations
- SNCG
γ synuclein
- BRCA1
BRCA1 DNA repair associated
- TETs
ten-eleven translocations
- DNMTs
DNA methyltransferases
- 5′-aza-CdR
5-aza-2′-deoxycytidine
Funding
The King Faisal Specialist Hospital and Research Center supported this research under the RAC #2150 020.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
NAY, ZS, BAS and MAS performed the experiments. NAY and NAM performed the data analysis. NAM conceived and designed the study and drafted the manuscript with the help from NAY. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Doll R, Peto R. The causes of cancer: Quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst. 1981;66:1191–1308. doi: 10.1093/jnci/66.6.1192. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 4.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scourzic L, Mouly E, Bernard OA. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015;7:9. doi: 10.1186/s13073-015-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Good CR, Panjarian S, Kelly AD, Madzo J, Patel B, Jelinek J, Issa JJ. TET1-mediated hypomethylation activates oncogenic signaling in triple-negative breast cancer. Cancer Res. 2018;78:4126–4137. doi: 10.1158/0008-5472.CAN-17-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 10.Yan B, Guo Q, Fu FJ, Wang Z, Yin Z, Wei YB, Yang JR. The role of miR-29b in cancer: Regulation, function, and signaling. Onco Targets Ther. 2015;8:539–548. doi: 10.2147/OTT.S75899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Cao Y, Zhai Y, Ma X, An X, Zhang S, Li Z. MicroRNA-29b regulates DNA methylation by targeting Dnmt3a/3b and Tet1/2/3 in porcine early embryo development. Dev Growth Differ. 2018;60:197–204. doi: 10.1111/dgd.12537. [DOI] [PubMed] [Google Scholar]
- 12.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, An X, Yu H, Zhang S, Tang B, Zhang X, Li Z. MiR-29b/TET1/ZEB2 signaling axis regulates metastatic properties and epithelial-mesenchymal transition in breast cancer cells. Oncotarget. 2017;8:102119–102133. doi: 10.18632/oncotarget.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong EM, Southey MC, Fox SB, Brown MA, Dowty JG, Jenkins MA, Giles GG, Hopper JL, Dobrovic A. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early-onset breast cancer. Cancer Prev Res (Phila) 2011;4:23–33. doi: 10.1158/1940-6207.CAPR-10-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S, Jaworska-Bieniek K, Narod SA, Lubinski J, Wojdacz TK, Jakubowska A. Methylation of the BRCA1 promoter in peripheral blood DNA is associated with triple-negative and medullary breast cancer. Breast Cancer Res Treat. 2014;148:615–622. doi: 10.1007/s10549-014-3179-0. [DOI] [PubMed] [Google Scholar]
- 16.Iwamoto T, Yamamoto N, Taguchi T, Tamaki Y, Noguchi S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res Treat. 2011;129:69–77. doi: 10.1007/s10549-010-1188-1. [DOI] [PubMed] [Google Scholar]
- 17.Al-Moghrabi N, Nofel A, Al-Yousef N, Madkhali S, Bin Amer SM, Alaiya A, Shinwari Z, Al-Tweigeri T, Karakas B, Tulbah A, Aboussekhra A. The molecular significance of methylated BRCA1 promoter in white blood cells of cancer-free females. BMC Cancer. 2014;14:830. doi: 10.1186/1471-2407-14-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobrovic A, Mikeska T, Alsop K, Candiloro I, George J, Mitchell G, Bowtell D. Constitutional BRCA1 methylation is a major predisposition factor for high-grade serous ovarian cancer. Cancer Res. 2014:74. [Google Scholar]
- 19.Al-Moghrabi NM. BRCA1 promoter methylation in peripheral blood cells and predisposition to breast cancer. J Taibah Univ Med Sci. 2017;12:189–193. doi: 10.1016/j.jtumed.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei M, Grushko TA, Dignam J, Hagos F, Nanda R, Sveen L, Xu J, Fackenthal J, Tretiakova M, Das S, Olopade OI. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res. 2005;65:10692–10699. doi: 10.1158/0008-5472.CAN-05-1277. [DOI] [PubMed] [Google Scholar]
- 21.Butcher DT, Mancini-DiNardo DN, Archer TK, Rodenhiser DI. DNA binding sites for putative methylation boundaries in the unmethylated region of the BRCA1 promoter. Int J Cancer. 2004;111:669–678. doi: 10.1002/ijc.20324. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Huo D, Chen Y, Nwachukwu C, Collins C, Rowell J, Slamon DJ, Olopade OI. CpG island methylation affects accessibility of the proximal BRCA1 promoter to transcription factors. Breast Cancer Res Treat. 2010;120:593–601. doi: 10.1007/s10549-009-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, Liu YE, Jia T, Wang M, Liu J, Xiao G, Joseph BK, Rosen C, Shi YE. Identification of a breast cancer-specific gene, BCSG1, by direct differential cDNA sequencing. Cancer Res. 1997;57:759–764. [PubMed] [Google Scholar]
- 24.Wu K, Quan Z, Weng Z, Li F, Zhang Y, Yao X, Chen Y, Budman D, Goldberg ID, Shi YE. Expression of neuronal protein synuclein gamma gene as a novel marker for breast cancer prognosis. Breast Cancer Res Treat. 2007;101:259–267. doi: 10.1007/s10549-006-9296-7. [DOI] [PubMed] [Google Scholar]
- 25.Lu A, Gupta A, Li C, Ahlborn TE, Ma Y, Shi EY, Liu J. Molecular mechanisms for aberrant expression of the human breast cancer specific gene 1 in breast cancer cells: Control of transcription by DNA methylation and intronic sequences. Oncogene. 2001;20:5173–5185. doi: 10.1038/sj.onc.1204668. [DOI] [PubMed] [Google Scholar]
- 26.Lu A, Zhang F, Gupta A, Liu J. Blockade of AP1 transactivation abrogates the abnormal expression of breast cancer-specific gene 1 in breast cancer cells. J Biol Chem. 2002;277:31364–31372. doi: 10.1074/jbc.M201060200. [DOI] [PubMed] [Google Scholar]
- 27.Song X, Zhang M, Dai E, Luo Y. Molecular targets of curcumin in breast cancer (Review) Mol Med Rep. 2019;19:23–29. doi: 10.3892/mmr.2018.9665. [DOI] [PubMed] [Google Scholar]
- 28.Jiang A, Wang X, Shan X, Li Y, Wang P, Jiang P, Feng Q. Curcumin reactivates silenced tumor suppressor Gene RARβ by reducing DNA methylation. Phytother Res. 2015;29:1237–1245. doi: 10.1002/ptr.5373. [DOI] [PubMed] [Google Scholar]
- 29.Kumar U, Sharma U, Rathi G. Reversal of hypermethylation and reactivation of glutathione S-transferase pi 1 gene by curcumin in breast cancer cell line. Tumour Biol. 2017;39:1010428317692258. doi: 10.1177/1010428317692258. [DOI] [PubMed] [Google Scholar]
- 30.Du L, Xie Z, Wu LC, Chiu M, Lin J, Chan KK, Liu S, Liu Z. Reactivation of RASSF1A in breast cancer cells by curcumin. Nutr Cancer. 2012;64:1228–1235. doi: 10.1080/01635581.2012.717682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J, Wu C, Lin Z, Guo Y, Shi L, Dong P, Lu Z, Gao S, Liao Y, Chen B, Yu F. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation-a novel mechanism suppressing liver fibrosis. FEBS J. 2014;281:88–103. doi: 10.1111/febs.12574. [DOI] [PubMed] [Google Scholar]
- 32.Link A, Balaguer F, Shen Y, Lozano JJ, Leung HC, Boland CR, Goel A. Curcumin modulates DNA methylation in colorectal cancer cells. PLoS One. 2013;8:e57709. doi: 10.1371/journal.pone.0057709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birgisdottir V, Stefansson OA, Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG, Eyfjord JE. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res. 2006;8:R38. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Liu W, Wu Y, Zhou Y, Xue R, Luo C, Wang L, Zhao W, Jiang JD, Liu J. Loss of epigenetic control of synuclein-gamma gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 2005;65:7635–7643. doi: 10.1158/0008-5472.CAN-05-1089. [DOI] [PubMed] [Google Scholar]
- 35.Al-Moghrabi N, Al-Showimi M, Al-Yousef N, Al-Shahrani B, Karakas B, Alghofaili L, Almubarak H, Madkhali S, Al Humaidan H. Methylation of BRCA1 and MGMT genes in white blood cells are transmitted from mothers to daughters. Clin Epigenetics. 2018;10:99. doi: 10.1186/s13148-018-0529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Hendrayani SF, Al-Khalaf HH, Aboussekhra A. Curcumin triggers p16-dependent senescence in active breast cancer-associated fibroblasts and suppresses their paracrine procarcinogenic effects. Neoplasia. 2013;15:631–640. doi: 10.1593/neo.13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaughan RA, Garcia-Smith R, Dorsey J, Griffith JK, Bisoffi M, Trujillo KA. Tumor necrosis factor alpha induces Warburg-like metabolism and is reversed by anti-inflammatory curcumin in breast epithelial cells. Int J Cancer. 2013;133:2504–2510. doi: 10.1002/ijc.28264. [DOI] [PubMed] [Google Scholar]
- 40.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 41.Hu S, Xu Y, Meng L, Huang L, Sun H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp Ther Med. 2018;16:1266–1272. doi: 10.3892/etm.2018.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feilotter HE, Michel C, Uy P, Bathurst L, Davey S. BRCA1 haploinsufficiency leads to altered expression of genes involved in cellular proliferation and development. PLoS One. 2014;9:e100068. doi: 10.1371/journal.pone.0100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Z, Niu J, Sun E, Rong X, Zhang X, Ju Y. Gamma-synuclein binds to AKT and promotes cancer cell survival and proliferation. Tumour Biol. 2016;37:14999–15005. doi: 10.1007/s13277-016-5371-9. [DOI] [PubMed] [Google Scholar]
- 44.He JS, Xie N, Yang JB, Guan H, Chen WC, Zou C, Ouyang YW, Mao YS, Luo XY, Pan Y, Fu L. BCSG1 siRNA delivered by lentiviral vector suppressed proliferation and migration of MDA-MB-231 cells. Int J Mol Med. 2018;41:1659–1664. doi: 10.3892/ijmm.2017.3355. [DOI] [PubMed] [Google Scholar]
- 45.Pei YF, Tao R, Li JF, Su LP, Yu BQ, Wu XY, Yan M, Gu QL, Zhu ZG, Liu BY. TET1 inhibits gastric cancer growth and metastasis by PTEN demethylation and re-expression. Oncotarget. 2016;7:31322–31335. doi: 10.18632/oncotarget.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kai M, Niinuma T, Kitajima H, Yamamoto E, Harada T, Aoki H, Maruyama R, Toyota M, Sasaki Y, Sugai T, et al. TET1 Depletion induces aberrant CpG methylation in colorectal cancer cells. PLoS One. 2016;11:e0168281. doi: 10.1371/journal.pone.0168281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Ying Y, Zhu H, Zhu T, Qu C, Jiang J, Fang B. Curcumin-induced promoter hypermethylation of the mammalian target of rapamycin gene in multiple myeloma cells. Oncol Lett. 2019;17:1108–1114. doi: 10.3892/ol.2018.9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Zhou Y, Boggs SE, Belinsky SA, Liu J. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26:5900–5910. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- 49.Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int J Mol Sci. 2013;14:14647–14658. doi: 10.3390/ijms140714647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.