Abstract
Background
ABO gene polymorphisms have been reported to be associated with the risk of multiple cancers and cardiocerebrovascular diseases. However, the results remained controversial. In this study, we conducted a systematic review and meta-analysis to clarify the association between two SNPs (rs505922 and rs657152) in ABO gene and cancers/cardiocerebrovascular diseases.
Method
All eligible case-control studies come from PubMed, Embase and Web of Science up to Jan. 1, 2019. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the corresponding associations. Sensitivity analysis, publication bias assessment, and heterogeneity test were performed using STATA 12.0.
Results
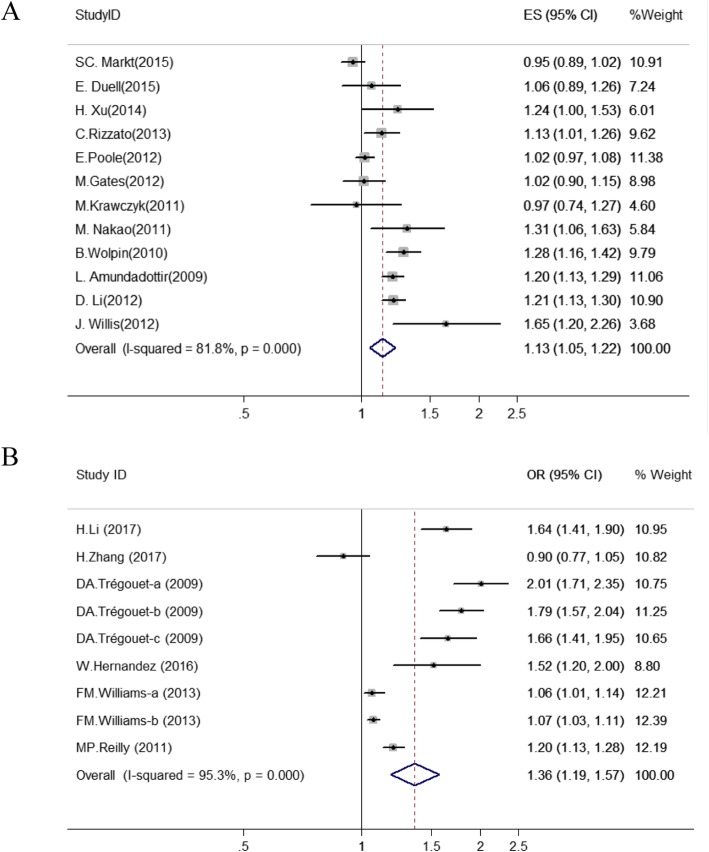
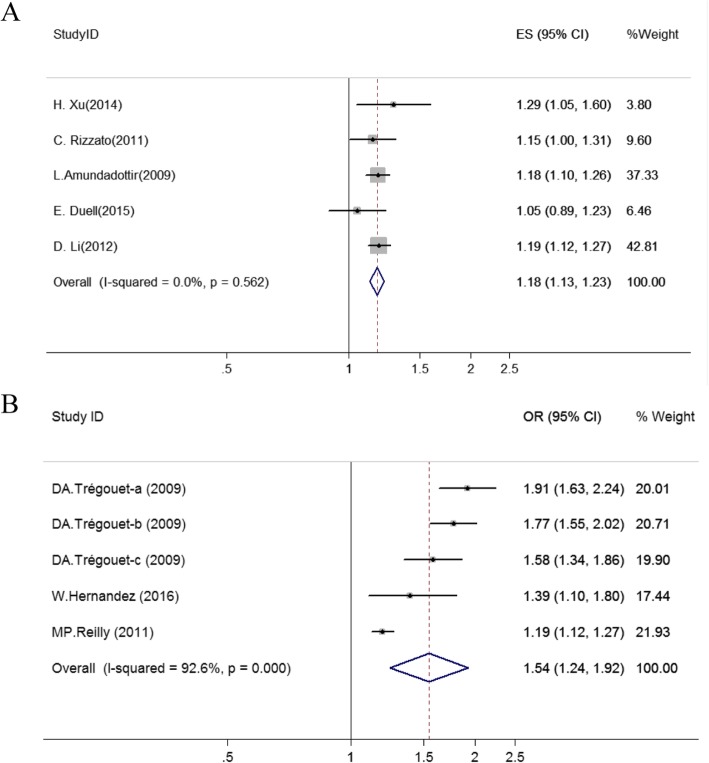
A total of nineteen articles involving twenty-two case-control populations were included according to inclusion and exclusion criteria. Twelve populations (20,820 cases and 27,837 controls) were used to evaluate the relationship between rs505922 and overall cancers and nine populations (22,275 cases and 71,549 controls) were included to assess the association between rs505922 and cardiocerebrovascular diseases. The results showed a significant association between the rs505922 polymorphism and cancers (CvsT: OR = 1.13, 95%CI = 1.05–1.22, P = 0.001), and cardiocerebrovascular diseases (OR = 1.36, 95%CI = 1.19–1.57, P < 0.001). Five populations (8660 cases and 10,618 controls) were included to evaluate association between rs657152 and cancers and five populations (8105 cases and 6712 controls) were included to estimate the relationship between rs657152 and cardiocerebrovascular diseases. The result of meta-analysis reveals that rs657152 was significantly associated with cancers (OR = 1.18, 95%CI = 1.13–1.23, P < 0.001) and cardiocerebrovascular diseases (OR = 1.54, 95%CI = 1.24–1.92, P < 0.001).
Conclusion
Our study suggested that ABO polymorphisms might serve as a risk factor of pancreatic cancers and cardiocerebrovascular diseases.
Keywords: ABO gene polymorphism, rs505922, rs657152, Cardiocerebrovascular diseases, Cancer, Meta-analysis
Background
The incidence of cancer and cardiovascular disease increases with age and both of them are related to inflammation and thrombosis et al. [1, 2]. Epidemiological studies have demonstrated that ABO blood groups were associated with several chronic inflammation related diseases, including cancers and cardiocerebrovascluar diseases [3–5]. Although a possible common pathogenic mechanism involving the von Willebrand factor between ABO blood group system and cancer/cardiovascular disease has been discussed [6], the relationship between histoblood group antigens and inflammation related diseases remains unknown and the regulatory mechanisms underlying ABO expression was still unclear.
The ABO blood group system is composed of complex carbohydrate structures that are biosynthesized by A- and B- transferases encoded by ABO gene. ABO gene is located at the 9q34 region of the chromosome and encodes enzyme glycosyltransferase with two main allele (A and B), a specific glycosyltransferase catalyzes the covalent attachment of N-acetylgalactosamine or D-galactose to a common precursor side chain (H determinant), ultimately to an A or B antigen [7]. Several SNPs in the ABO gene have been suggested to be associated with increased risks to the development of cancer and cardiocerebrovascluar disease by genome-wide association studies (GWAS) and candidate gene studies [8–11]. Particularly, the most widely investigated SNPs of the ABO gene were rs505922 and rs657152 [12–15]. Meta-analysis that evaluated the relationship between rs505922 SNP and overall cancer have already been reported by Duan et al. [16] four years ago. However, two new studies of cancer risk [11, 17] and several studies of cardiocerebrovascluar diseases risk [14, 15, 18–21] have been reported in recent years and the relationship between SNPs in ABO gene and cancer/cardiocerebrovascular diseases risk was still unclear. Here, we have performed a meta-analysis including the newly published studies and evaluated the associations between polymorphisms of ABO gene and cancers/cardiocerebrovascular diseases.
Methods
Search strategy
Two investigators (Yanxia Li and Luyang Liu) performed a systematic literature search in three databases: PubMed, Web of Science and Embase to identify relevant articles published from the initial to Jan 1, 2019. The following search terms were used either separately or in combination: “SNP” “Polymorphism” and “Polymorphism, Single Nucleotide” “rs505922, rs657152” and “neoplasms” “carcinoma” “tumor” “cancer” and “vascular diasease” “cardiovascular disease” “cardiac-cerebral disease” and “ABO Blood-Group System” “Lewis Blood-Group System” “ABO”. Once suitable studies were singled out from the search results, other potentially relevant articles were identified by cross-references within eligible studies. The references of each identified articles were also searched manually to identify eligible studies.
Selection criteria
The inclusion criteria for these studies were as follows: 1) Studies evaluating the association between rs505922/rs657152 variants and cancers or cardiocerebrovascular diseases; 2) The study design was a case-control study in humans (both hospital based case-control study and cohort based nested case-control study were included); 3) Researches containing universal allele and genotype data; 4) Studies written in English. The exclusion criteria were as follows: 1) Studies did not describe the association of ABO gene polymorphism with cancers or cardiocerebrovascular diseases; 2) Systematic reviews or articles focusing on animals; 3) Studies that did not provide usable data for meta-analysis.
Data extraction and quality assessment
Two investigators (Yanxia Li and Luyang Liu) independently extract data and verify the accuracy of the data. The following information was extracted from each article: first authors, publication years, ethnicity, cancer types, and study design (hospital- or population-based), sample size of subject, number of cases and controls for each genotype, OR and 95% CI in allele model. When there was no data of allele model in the article, we calculated the data of allele model by using the formula. To assess the quality of the study, we used the Newcastle-Ottawa Scale (NOS) with a nine-star system [22]; this scale assesses the quality of cohort and case-control studies. The highest score of NOS is nine stars: four stars for the selection process, three stars for exposure/outcome, and two stars for comparability. A score of seven or above was considered to be high-quality study.
Statistical analysis
We used odds ratios (ORs) and 95% confidence intervals (CIs) to assess the relationship between each SNP and the risk of cancers or cardiocerebrovascular diseases. The association was examined using allele model. The significance of the pooled OR was determined by the Z-test. Subgroup analyses were conducted according to cancer types, ethnic groups and sources of controls. Heterogeneity between articles was identified with Q-test and I2 index, I2 values of > 50% indicated heterogeneity among studies. If there was heterogeneity (I2 > 50%) between studies, we used a random effect model (DerSimonian-Laird method), otherwise we used a fixed effect model (Mantel-Haenszel method). Sensitivity analysis was performed to assess the effects of individual studies on pooled results and the stability of the results. We used Egger’s and Begg’s test to evaluate the publication bias, with a P > 0.05 considered to be evidence for no potential publication bias. Trim and fill method was also applied in detecting publication bias. All the tests were two-sided and P < 0.05 was considered to be statistically significant. Stata version 12.0 SE was applied to carry out the statistical analysis.
Results
Study characteristics
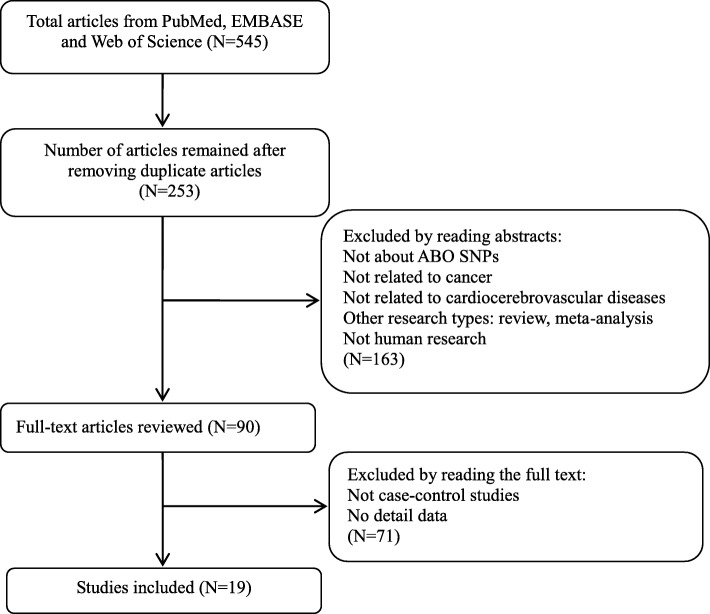
After retrieving the database, a total of 545 records [PubMed (n = 146), Web of Science (n = 149), EMBASE (n = 250)] were obtained. According to the inclusion/exclusion criteria, nineteen articles were included and the detailed flowchart of study selection process was presented in Fig. 1. Twelve studies reported the association between rs505922 and cancer risk [11–13, 17, 23–30] and five studies reported the association between rs657152 and cancer risk [12, 13, 17, 30, 31]. Six studies included nine populations reported the association of cardiocerebrovascular diseases with rs505922 [14, 15, 18–21], and three studies with rs657152 [14, 15, 20]. Detailed characteristics and genotype distribution of included articles for two SNPs were shown in Table 1 and Table 2. In addition, seven additional SNPs which had strong linkage disequilibrium (LD) with rs525922 were reported to be associated with cardiocerebrovascular disease (Table S1) [8, 14, 15, 21, 32–35]. However, the association between these SNPs with cancer risk has not been reported. According to the source of control groups, nine studies were population-based (PB), six studies were hospital-based (HB), and three studies were population and hospital-based(PB/HB) control. For SNP rs505922, ten studies were from Caucasian population [11, 13, 15, 19, 20, 23–26, 29], four studies were from Asian [12, 18, 21, 27] and four studies were from Mixed and African [14, 17, 28, 30]. For the rs657152 polymorphism, there were four studies originating from Caucasian [13, 15, 20, 31], and four studies were from Asian and African or Mixed population [12, 14, 17, 30]. Each study was scored based on the Newcastle-Ottawa Scale (NOS) and detailed study qualities were presented in Table S2 and Table S3.
Fig. 1.
Flow chart of the study selection process
Table 1.
Characteristics of the studies included in the meta-analysis of the association between SNPs and cancers
| SNP loci | Year | First author | Ethnicity | Cancer/disease | Controla | Sample size (case/control) | Case | Control | OR(95%CI) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hom† wild |
Het† | Hom variant |
Hom† wild |
Het† | Hom variant |
||||||||
| rs505922 | 2015 | SC. Markt | Caucasian | Prostate cancer | – | 2774/4443 | 1172 | 1226 | 376 | 1772 | 2070 | 601 | 0.95(0.89–1.02) |
| 2015 | E. Duell | Caucasian | Gastric cancer | PB | 365/1284 | 137 | 172 | 49 | 533 | 541 | 188 | 1.06(0.89–1.26) | |
| 2014 | H. Xu | Asian | Pancreatic cancer | PB | 256/548 | 69 | 124 | 63 | 186 | 246 | 115 | 1.24(1.00–1.53) | |
| 2013 | C.Rizzato | Caucasian | Pancreatic cancer | HB/PB | 1028/2257 | 342 | 518 | 163 | 860 | 1055 | 336 | 1.13(1.01–1.26) | |
| 2012 | E.Poole | Caucasian | Ovarian cancer | HB/PB | 5233/6838 | 2222 | 2407 | 603 | 2987 | 3044 | 806 | 1.02(0.97–1.08) | |
| 2012 | D. Li | Mixed | Pancreatic cancer | HB | 3851/3934 | – | – | – | – | – | – | 1.21(1.13–1.30) | |
| 2012 | J. Willis | Caucasian | Pancreatic cancer | HB | 385/149 | – | – | – | – | – | – | 1.65(1.20–2.26) | |
| 2012 | M.Gates | Caucasian | Breast cancer | PB | 1138/1090 | 489 | 505 | 144 | 471 | 487 | 132 | 1.02(0.90–1.15) | |
| 2011 | M.Krawczyk | Caucasian | Cholangiocarcinoma | PB | 180/350 | 84 | 68 | 28 | 154 | 146 | 50 | 0.97(0.74–1.27) | |
| 2011 | M. Nakao | Asian | Pancreatic cancer | HB | 185/1465 | 38 | 101 | 46 | 428 | 745 | 292 | 1.31(1.06–1.63) | |
| 2010 | B.Wolpin | Mixed | Pancreatic cancer | PB | 1534/1583 | 511 | 752 | 271 | 657 | 705 | 221 | 1.28(1.16–1.42) | |
| 2009 | L.Amundadottir | Mixed | Pancreatic cancer | HB/PB | 3891/3932 | 1436 | 1856 | 599 | 1667 | 1785 | 480 | 1.20(1.13–1.29) | |
| rs657152 | 2015 | E. Duell | Caucasian | Gastric cancer | PB | 365/1284 | – | – | – | – | – | – | 1.05(0.89–1.23) |
| 2014 | H. Xu | Asian | Pancreatic cancer | PB | 256/548 | 71 | 124 | 61 | 199 | 240 | 108 | 1.29(1.05–1.60) | |
| 2012 | D. Li | Caucasian | Pancreatic cancer | HB | 3851/3934 | – | – | – | – | – | – | 1.19(1.12–1.27) | |
| 2011 | C. Rizzato | Caucasian | Pancreatic cancer | PB | 686/1255 | 199 | 357 | 130 | 437 | 591 | 227 | 1.15(1.00–1.31) | |
| 2009 | L.Amundadottir | Mixed | Pancreatic cancer | HB/PB | 3502/3597 | 1191 | 1702 | 609 | 1395 | 1691 | 511 | 1.18(1.13–1.23) | |
aPB population based control, HB hospital based control; †Hom homozygous, Het heterozygote
Table 2.
Characteristics of the studies included in the meta-analysis of the association between SNPs and cardiocerebrovascular diseases
| SNP loci | Year | First author | Ethnicity | Cancer/disease | Controla | Sample size (case-control) | Case | Control | OR(95%CI) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hom† wild |
Het† | Hom variant |
Hom† wild |
Het† | Hom variant |
||||||||
| rs505922 | 2017 | H. Li | Asian | Ischemic stroke | PB | 991/1002 | 511 | 406 | 74 | 657 | 306 | 39 | 1.64(1.41–1.90) |
| 2017 | H. Zhang | Asian | Large artery atherosclerotic stroke | PB | 644/642 | 146 | 325 | 173 | 129 | 322 | 191 | 0.90(0.77–1.05) | |
| 2016 | W. Hernandez | African | Venous Thrombosis | HB | 146 /432 | – | – | – | – | – | – | 1.52(1.20–2.00) | |
| 2013 | FM.Williams-a‡ | Caucasian | Ischemic stroke | HB | 4092/8383 | – | – | – | – | – | – | 1.06(1.01–1.14) | |
| 2013 | FM.Williams-b‡ | Caucasian | Ischemic stroke | HB | 8443 /54810 | – | – | – | – | – | – | 1.07(1.03–1.11) | |
| 2011 | MP. Reilly | Caucasian | Myocardial infarction | HB | 5783/3644 | – | – | – | – | – | – | 1.20(1.13–1.28) | |
| 2009 | DA.Trégouet-a§ | Caucasian | Venous thromboembolism | PB | 419/1228 | 97 | 209 | 113 | 519 | 559 | 150 | 2.01(1.71–2.35) | |
| 2009 | DA.Trégouet-b§ | Caucasian | Venous thromboembolism | PB | 1150/801 | 299 | 575 | 276 | 339 | 364 | 98 | 1.79(1.57–2.04) | |
| 2009 | DA.Trégouet-c§ | Caucasian | Venous thromboembolism | PB | 607/607 | 177 | 302 | 128 | 265 | 272 | 70 | 1.66(1.41–1.95) | |
| rs657152 | 2016 | W. Hernandez | African | Venous Thrombosis | HB | 146 /432 | – | – | – | – | – | – | 1.39(1.10–1.80) |
| 2011 | MP. Reilly | Caucasian | Myocardial infarction | HB | 5783/3644 | – | – | – | – | – | – | 1.19(1.12–1.27) | |
| 2009 | DA.Trégouet-a§ | Caucasian | Venous thromboembolism | PB | 419/1228 | 89 | 208 | 122 | 472 | 579 | 177 | 1.91(1.63–2.24) | |
| 2009 | DA.Trégouet-b§ | Caucasian | Venous thromboembolism | PB | 1150/801 | 276 | 575 | 299 | 318 | 373 | 110 | 1.77(1.55–2.02) | |
| 2009 | DA.Trégouet-c§ | Caucasian | Venous thromboembolism | PB | 607/607 | 171 | 302 | 134 | 249 | 280 | 78 | 1.58(1.34–1.86) | |
aPB population based control, HB hospital based control; †Hom homozygous, Het heterozygote; a‡ represent the MOnica Risk, Genetics, Archiving and Monograph(MORGAM) and the Wellcome Trust Case Control Consortium 2(WTCCC2) population, b‡ represent MetaStroke population; a§ represent GWAS population from 4 different French centers, b§represent MARseille THrombosis Association study (MARTHA) population, c§represent FARIVE that is a multicenter case-control study
Meta-analysis of rs505922 polymorphism
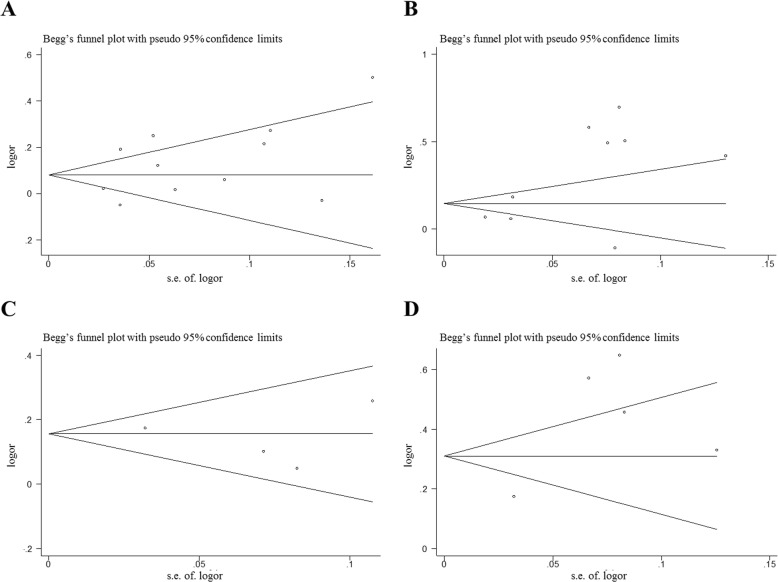
Meta-analysis was conducted to estimate the associations between rs505922 and cancer risk (Fig. 2a; Table S4) in 20,820 cancer cases and 27,837 controls. The rs505922 polymorphism was significantly associated with an increased cancers risk in the allele model (OR = 1.13, 95%CI = 1.05–1.22, P = 0.001). Subgroup analysis was conducted based on ethnicity, type of cancer, and source of control. The association between rs505922 and cancer risk was identified in Asian population subgroup (OR = 1.27, 95%CI = 1.10–1.48, P = 0.002), Mixed population subgroup (OR = 1.22, 95%CI = 1.17–1.27, P < 0.001), Pancreatic cancers subgroup (OR = 1.23, 95%CI = 1.16–1.31, P < 0.001), and Hospital based control groups (OR = 1.30, 95%CI = 1.12–1.51, P = 0.003) (Table S5; Fig. S1). However, no significant association was observed in Caucasian population(OR = 1.05, 95%CI = 0.97–1.13, P = 0.232). Sensitivity analyses were conducted by omitting each individual article to measure its specific effect on the pooled ORs (Fig. S2a). Sensitivity analysis plot indicated that no single study significantly affected the combined OR of SNP loci. Because of the heterogeneity of the research, we use the random effect model (allele model: I 2 = 81.8%) (Table S4). No significant publication bias was observed in any studies of SNPs (Fig. 4a; Table S4). After the applying the trim and fill method, there is no change in the OR value after the combination, also indicating that the original result is stable (Fig. S3).
Fig. 2.
Forest plot of the relationship between rs505922 polymorphisms and cancer (a) and cardiocerebrovascular disease (b) risk (allele model and random-effect model). The circle and horizontal lines correspond to OR and 95% CI and the area of the squares reflects the weight of individual studies included in the meta-analysis. The diamond represents the pooled ORs and 95% CI. The dotted red line represents the total OR value
Fig. 4.
Begg’s funnel plot of publication bias test for rs505922 and rs657152 (allele model and random-effect model); Each point represents a separate study for the indicated association between rs505922 and cancer (a)/cardiocerebrovascular disease (b) risk, and rs657152 and cancer (c)/cardiocerebrovascular disease (d) risk, respectively. Each point stands for an individual article in overall population under allele model. s.e., standardized effect
We also performed a meta-analysis to evaluate the association between rs505922 SNP and cardiocerebrovascular diseases (Fig. 2b; Table S4). The rs505922 SNP was significantly associated with an increased cardiocerebrovascular diseases risk in the allele model (C/T: OR = 1.36, 95%CI = 1.19–1.57, P < 0.001). Subgroup analysis indicated that rs505922 was associated with a significantly higher risk of cardiocerebrovascular diseases in Caucasian population subgroup (OR = 1.39, 95%CI = 1.19–1.64, P < 0.001, allele model), African population subgroup(OR = 1.52, 95%CI = 1.18–1.96, P = 0.001), Hospital based control(OR = 1.14, 95%CI = 1.05–1.23, P = 0.003) and Population based control(OR = 1.54, 95%CI = 1.18–2.02, P = 0.002) (Table S5; Fig. S4). However, no significant association was observed in Asian subgroup (OR = 1.21, 95%CI = 0.67–2.19, P = 0.524) (Table S5; Fig. S4). Sensitivity analysis showed the results of this study were stable (Fig. S2b). There was no significant publication bias among the enrolled studies in Begg’s and Egger’s test (Fig. 4b; Table S4). After applying the trim and fill method, no new literature has been added, indicating that the result is stable (Fig. S5).
Meta-analysis of rs657152 polymorphisms
Five studies reported the association between rs657152 and cancers risk. Our results showed that rs657152 was significantly associated with increased cancers risk in allele model (OR = 1.18, 95%CI = 1.13–1.23, P < 0.001) (Fig. 3a; Table S4). Sensitivity analysis revealed no significant influence on the pooled OR by any individual study (Fig. S6a). We conducted Begg’s and Egger’s tests to assess the publication bias for these studies and no evidence of publication bias was detected among the enrolled studies (Fig. 4c; Table S4). According to the results of trim and fill method, the result was stable (Fig. S7).
Fig. 3.
Forest plot of the relationship between rs657152 polymorphisms and cancer (a) and cardiocerebrovascular disease (b) risk (allele model and random-effect model). The circle and horizontal lines correspond to OR and 95% CI and the area of the squares reflects the weight of individual studies included in the meta-analysis. The diamond represents the pooled ORs and 95% CI. The dotted red line represents the total OR value
We also evaluated the relationship between rs657152 and cardiocerebrovascular diseases. A significant association between rs657152 and cardiocerebrovascular diseases under allele model were observed (OR = 1.54, 95%CI = 1.24–1.92, P < 0.001) (Fig. 3b; Table S4). Sensitivity analysis suggested that there was no significant change in the overall outcomes after removing any of the studies (Fig. S6b). The Begg’s and Egger’s tests also showed that no meaningful publication bias were found (Fig. 4d; Table S4). The result of the trim and fill method proves that the result was stable (Fig. S8).
Discussion
In this study, we conducted a meta-analysis to clarify the relationship between ABO SNPs (rs505922 and rs657152) and cancer/cardiocerebrovascular diseases risk. Our results showed that these two SNPs were associated with pancreatic cancer risk and also increased the risk of cardiocerebrovascular diseases.
Regarding to rs505922 SNP, our results showed that the variant type of rs505922 could increase the risk of overall cancer, suggesting a potential predictive ability of this SNP for cancer risk. When we conducted a subgroup analysis of rs505922 based on cancer sites and ethnicity, we found that there was no significant association between rs505922 and Non-pancreatic cancer subgroup or Caucasian subgroup. This result may be due to heterogeneity of cancer types or insufficient statistical power. Therefore, further studies with large samples size are warrant to evaluate the association between the rs505922 polymorphisms in Non-pancreatic cancers. On the other side, our data also showed that rs505922 was associated with cardiocerebrovascular diseases. In subgroup analysis, we found that there was no association between rs505922 and cardiocerebrovascular diseases in Asian subgroup. Only two studies with small population were included in this analysis, further studies are needed in Asian population. The SNP is located in the first intron region of ABO gene, the protective T allele of rs505922 is in complete linkage disequilibrium (r2 = 1.0) with the O allele, is marks of O allele. However, the regulatory mechanism underlying the expression of histoblood group antigens was unclear.
For rs657152, our study demonstrated that this SNP was associated with cancer/ cardiocerebrovascular diseases risk. However, only eight studies [12–15, 17, 20, 30, 31] were reported and most of studies were conducted in Caucasian population. Therefore, more studies with different ethnic background and larger sample size are needed in the future. Rs657152 is located in the intron area of ABO, the possible function has not been revealed yet. Rs657152 has been found to be associated with several biological molecule, including LDL cholesterol [36], liver derived alkaline phosphatas [37], and IL-6 [38]. This implied that rs657152 may affect the occurrence and development of disease by influencing these biological molecule. Further subsequent functional studies are warrant.
The underlying mechanism for the relationship between ABO blood group and cancer risk is still poorly understood [39]. It is reported that blood type may affect the progression and expansion of malignant tumors by altering the systemic inflammatory response [40]. Recent studies reported an association between polymorphisms at the ABO gene locus and circulating levels of tumour necrosis factor-alpha [41], soluble intercellular adhesion molecule (ICAM)-1 [42, 43], E-selectin [44, 45], and P-selectin [43]. These adhesion molecules were important mediators of chronic inflammation and immune cell recruitment [46]. They may provide a biological basis for the postulated influence of ABO on cancer survival, by linking ABO blood group and tumour initiation and spread [39]. In addition, some researches have shown that the structure of certain tumor antigens was similar to the structure of antigens of ABO blood group system. Smith and Prieto [47] suggested the Forssmann antigen which predominant in stomach and colon tumors, was almost structurally identical to the A antigen determinant. Blood group A carrier may have diminished tumor immune response due to reduced ability to recognize and attack tumor cells [48].
There was some evidence linking ABO blood group and cardiocerebrovascular diseases. Jenkins, P.V. et al. reported an association between ABO blood types and von Willebrand factor(vWF) and factor VIII(FVIII), both of which play crucial roles in the coagulation pathway [49]. Higher levels of vWF and FVIII has been observed in non-O blood type than O blood type [50]. Therefore, type O blood may be a risk factor for bleeding [51]. In addition, the non-O blood group has been shown to be correlated with higher total cholesterol and LDL-C levels [52], and the latest study proposed that approximately 10% of the effect of ABO blood group on coronary artery disease (CAD) susceptibility was mediated by plasma cholesterol levels [53].
Limitations in this study should be mentioned. First, the studies included in our meta-analysis were limited to published reports and English language studies. Unpublished reports or those published in non-English language studies were not included in the analysis. It would limit our sample size and publication bias might be exist. Second, both of the hospital based and population based case-control studies were included in our study. Therefore, selection bias would be exist compared to the meta-analysis only included population based case-control studies. Third, the limited number of published studies may influence the reliability of our results. Finally, the lack of original data limited further evaluations of the potential gene-gene and gene-environment interactions.
Conclusion
In summary, the results of our meta-analysis revealed that rs505922 and rs657152 were correlated with an increased pancreatic cancers risk. Due to most of the studies were conducted in pancreatic cancer type and Caucasian populations, further studies in multiple cancer types and multiple ethnic populations are needed. In addition, our meta-analysis also revealed that rs505922 and rs657152 associated with cardiocerebrovascular diseases. However, owing to limited number of studies, further studies with larger samples size are warrant. This study can provide clues for further exploration of novel biomarkers with cancer/cardiocerebrovascular early-warning function.
Supplementary information
Additional file 1. Figure S1 Subgroup analysis of rs505922 and cancer risk. Figure S2 Sensitivity analysis diagram of rs505922 and cancer (a) and cardiocerebrovascular disease (b) risk. Figure S3 Trim and fill method results (a) and filled funnel plot (b) of rs505922 and cancer risk. Figure S4 Subgroup analysis of rs505922 and cardiocerebrovascular disease risk. Figure S5 Trim and fill method results (a) and filled funnel plot (b) of rs505922 and cardiocerebrovascular disease risk. Figure S6 Sensitivity analysis of rs657152 and cancer (a) and cardiocerebrovascular disease (b) risk. Figure S7 Trim and fill method results (a) and filled funnel plot (b) of rs657152 and cancer risk. Figure S8 Trim and fill method results (a) and filled funnel plot (b) of rs657152 and cardiocerebrovascular disease risk. Table S1. Previous reported SNPs had strong linkage disequilibrium with rs505922. Table S2 Quality assessment of case-control studies according to NOS for rs505922. Table S3 Quality assessment of case-control studies according to NOS for rs657152. Table S4 Results for meta-analysis of ABO gene polymorphisms with cancer and cardiocerebrovascular disease risk. Table S5 Subgroup analysis of rs505922 polymorphism
Acknowledgements
Not applicable
Abbreviations
- HB
Hospital based
- NOS
Newcastle-Ottawa Scale
- PB
Population based
Authors’ contributions
Study conception and design: LL and HZ. Acquisition, analysis, or interpretation of data: YXL and LYL. Statistical analysis: YXL and YBH. Drafting of the manuscript: YXL and LYL. Administrative, technical, or material support: HZ. Study supervision: LL and HZ. All authors have read and approved the final version of manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81973113, 81320108022, 81470153, and 81502877), Tianjin Science and Technology Committee Foundation (18YFZCSY00520 and 16JCYBJC26600), the National Key Research and Development Program of China (2016YFC1302703).
The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanxia Li and Luyang Liu contributed equally to this work.
Contributor Information
Hong Zheng, Email: zhengh64@aliyun.com.
Lian Li, Email: lilian@tmu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-0975-8.
References
- 1.Falanga A, Russo L, Milesi V, et al. Mechanisms and risk factors of thrombosis in cancer. Crit Rev Oncol Hematol. 2017;118:79–83. doi: 10.1016/j.critrevonc.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Meng X, Chen Y, et al. The biology of aging and Cancer: frailty, inflammation, and immunity. Cancer J. 2017;23(4):201–205. doi: 10.1097/PPO.0000000000000270. [DOI] [PubMed] [Google Scholar]
- 3.Zhang BL, He N, Huang YB, et al. ABO blood groups and risk of cancer: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2014;15(11):4643–4650. doi: 10.7314/APJCP.2014.15.11.4643. [DOI] [PubMed] [Google Scholar]
- 4.Zu B, You G, Fu Q, et al. Association between ABO blood group and risk of congenital heart disease: a 6-year large cohort study. Sci Rep. 2017;7:42804. doi: 10.1038/srep42804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasan SK, Rostgaard K, Majeed A, et al. ABO blood group and risk of thromboembolic and arterial disease: a study of 1.5 million blood donors. Circ. 2016;133(15):1449–1457. doi: 10.1161/CIRCULATIONAHA.115.017563. [DOI] [PubMed] [Google Scholar]
- 6.Franchini M, Lippi G. The intriguing relationship between the ABO blood group, cardiovascular disease, and cancer. BMC Med. 2015;13:7. doi: 10.1186/s12916-014-0250-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto F, Clausen H, White T, et al. Molecular genetic basis of the histo-blood group ABO system. Nat. 1990;345(6272):229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 8.Ling X, Zheng Y, Tao J, et al. Association study of polymorphisms in the ABO gene with ischemic stroke in the Chinese population. BMC Neurol. 2016;16:146. doi: 10.1186/s12883-016-0671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heit JA, Armasu SM, Asmann YW, et al. A genome-wide association study of venous thromboembolism identifies risk variants in chromosomes 1q24.2 and 9q. J Thromb Haemost. 2012;10(8):1521–1531. doi: 10.1111/j.1538-7836.2012.04810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao M, Tang H, Zheng X, et al. Association analysis of GWAS and candidate gene loci in a Chinese population with coronary heart disease. Int J Clin Exp Med. 2015;8(5):7497–7506. [PMC free article] [PubMed] [Google Scholar]
- 11.Markt SC, Shui IM, Unger RH, et al. ABO blood group alleles and prostate cancer risk: results from the breast and prostate cancer cohort consortium (BPC3) Prostate. 2015;75(15):1677–1681. doi: 10.1002/pros.23035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H-L, Cheng J-R, Zhang W, et al. Re-evaluation of ABO gene polymorphisms detected in a genome-wide association study and risk of pancreatic ductal adenocarcinoma in a Chinese population. Chin J Cancer. 2014;33(2):68–73. doi: 10.5732/cjc.013.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duell EJ, Bonet C, Munoz X, et al. Variation at ABO histo-blood group and FUT loci and diffuse and intestinal gastric cancer risk in a European population. Int J Cancer. 2015;136(4):880–893. doi: 10.1002/ijc.29034. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez W, Gamazon ER, Smithberger E, et al. Novel genetic predictors of venous thromboembolism risk in African Americans. Blood. 2016;127(15):1923–1929. doi: 10.1182/blood-2015-09-668525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reilly MP, Li M, He J, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377(9763):383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan YF, Zhu F, Li XD, et al. Association between ABO gene polymorphism (rs505922) and cancer risk: a meta-analysis. Tumour Biol. 2015;36(7):5081–5087. doi: 10.1007/s13277-015-3159-y. [DOI] [PubMed] [Google Scholar]
- 17.Li D, Ej D, Yu K, et al. pathway analysis of genome-wide association study data highlights pancreatic development genes as susceptibility factors for pancreatic cancer. Carcinog. 2012;33(7):1384–1390. doi: 10.1093/carcin/bgs151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Zhang Z, Zhang J, et al. Fine-mapping of ABO gene identifies two novel SNPs associated with large artery atherosclerotic stroke in a Chinese han population. Mol Neurobiol. 2017;54(3):2107–2113. doi: 10.1007/s12035-016-9794-5. [DOI] [PubMed] [Google Scholar]
- 19.Williams FM, Carter AM, Hysi PG, et al. Ischemic stroke is associated with the ABO locus: the EuroCLOT study. Ann Neurol. 2013;73(1):16–31. doi: 10.1002/ana.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tregouet DA, Heath S, Saut N, et al. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood. 2009;113:5298–5303. doi: 10.1182/blood-2008-11-190389. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Cai Y, Xu AD. Association study of polymorphisms in the ABO gene and their gene-gene interactions with ischemic stroke in Chinese population. J Clin Lab Anal. 2018;32(4):e22329. doi: 10.1002/jcla.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 23.Krawczyk M, Mihalache F, Hoblinger A, et al. Pancreatic cancer risk variant ABO rs505922 in patients with cholangiocarcinoma. World J Gastroenterol. 2011;17(41):4640–4642. doi: 10.3748/wjg.v17.i41.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gates MA, Xu M, Chen WY, et al. ABO blood group and breast cancer incidence and survival. Int J Cancer. 2012;130(9):2129–2137. doi: 10.1002/ijc.26220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizzato C, Campa D, Pezzilli R, et al. ABO blood groups and pancreatic cancer risk and survival: results from the PANcreatic disease ReseArch (PANDoRA) consortium. Oncol Rep. 2013;29(4):1637–1644. doi: 10.3892/or.2013.2285. [DOI] [PubMed] [Google Scholar]
- 26.Poole EM, Gates MA, High BA, et al. ABO blood group and risk of epithelial ovarian cancer within the ovarian Cancer association consortium. Cancer Causes Control. 2012;23(11):1805–1810. doi: 10.1007/s10552-012-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakao M, Matsuo K, Hosono S, et al. ABO blood group alleles and the risk of pancreatic cancer in a Japanese population. Cancer Sci. 2011;102(5):1076–1080. doi: 10.1111/j.1349-7006.2011.01907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolpin BM, Kraft P, Gross M, et al. Pancreatic cancer risk and ABO blood group alleles: results from the pancreatic cancer cohort consortium. Cancer Res. 2010;70(3):1015–1023. doi: 10.1158/0008-5472.CAN-09-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willis JA, Olson SH, Orlow I, et al. A replication study and genome-wide scan of single-nucleotide polymorphisms associated with pancreatic cancer risk and overall survival. Clin Cancer Res. 2012;18(14):3942–3951. doi: 10.1158/1078-0432.CCR-11-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzato C, Campa D, Giese N, et al. Pancreatic cancer susceptibility loci and their role in survival. PLoS One. 2011;6(11):e27921. doi: 10.1371/journal.pone.0027921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinds DA, Buil A, Ziemek D, et al. Genome-wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum Mol Genet. 2016;25(9):1867–1874. doi: 10.1093/hmg/ddw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germain M, Chasman DI, de Haan H, et al. Meta-analysis of 65,734 individuals identifies TSPAN15 and SLC44A2 as two susceptibility loci for venous thromboembolism. Am J Hum Genet. 2015;96(4):532–542. doi: 10.1016/j.ajhg.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heit JA, Cunningham JM, Petterson TM, et al. Genetic variation within the anticoagulant, procoagulant, fibrinolytic and innate immunity pathways as risk factors for venous thromboembolism. J Thromb Haemost. 2011;9(6):1133–1142. doi: 10.1111/j.1538-7836.2011.04272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruzelius M, Strawbridge RJ, Tregouet DA, et al. Influence of coronary artery disease-associated genetic variants on risk of venous thromboembolism. Thromb Res. 2014;134(2):426–432. doi: 10.1016/j.thromres.2014.03.054. [DOI] [PubMed] [Google Scholar]
- 36.Teupser D, Baber R, Ceglarek U, et al. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ Cardiovasc Genet. 2010;3(4):331–339. doi: 10.1161/CIRCGENETICS.109.907873. [DOI] [PubMed] [Google Scholar]
- 37.Yuan X, Waterworth D, Perry JRB, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83(4):520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naitza S, Porcu E, Steri M, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8(1):e1002480. doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franchini M, Favaloro EJ, Targher G, et al. ABO blood group, hypercoagulability, and cardiovascular and cancer risk. Crit Rev Clin Lab Sci. 2012;49(4):137–149. doi: 10.3109/10408363.2012.708647. [DOI] [PubMed] [Google Scholar]
- 40.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melzer D, Perry JR, Hernandez D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4(5):e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pare G, Chasman DI, Kellogg M, et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4(7):e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbalic M, Dupuis J, Dehghan A, et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet. 2010;19(9):1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paterson AD, Lopes-Virella MF, Waggott D, et al. Genome-wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscler Thromb Vasc Biol. 2009;29(11):1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi L, Cornelis MC, Kraft P, et al. Genetic variants in ABO blood group region, plasma soluble E-selectin levels and risk of type 2 diabetes. Hum Mol Genet. 2010;19(9):1856–1862. doi: 10.1093/hmg/ddq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franchini M, Liumbruno GM, Lippi G. The prognostic value of ABO blood group in cancer patients. Blood Transfus. 2016;14(5):434–440. doi: 10.2450/2015.0164-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith DF, Prieto PA. Forssmann antigen. In: Roitt IM, Delves PH, editors. Encyclopedia of Immunology. Academic Press. 1992. pp. 591–592. [Google Scholar]
- 48.AE SRY, Kutikhin AG. ABO and Rh blood groups in relation to ovarian, endometrial and cervical cancer risk among the population of South-East Siberia. Asian Pac J Cancer Prev. 2012;13(10):5091–5096. doi: 10.7314/APJCP.2012.13.10.5091. [DOI] [PubMed] [Google Scholar]
- 49.Jenkins PV, O'Donnell JS. ABO blood group determines plasma von Willebrand factor levels: a biologic function after all? Transfus. 2006;46(10):1836–1844. doi: 10.1111/j.1537-2995.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- 50.Gill JC, Endres-Brooks J, Bauer PJ, et al. The effect of ABO blood group on the diagnosis of von Willebrand disease. Blood. 1987;69(6):1691–1695. doi: 10.1182/blood.V69.6.1691.1691. [DOI] [PubMed] [Google Scholar]
- 51.Dentali F, Sironi AP, Ageno W, et al. Relationship between ABO blood group and hemorrhage: a systematic literature review and meta-analysis. Semin Thromb Hemost. 2013;39(1):72–82. doi: 10.1055/s-0032-1329550. [DOI] [PubMed] [Google Scholar]
- 52.Silbernagel G, Chapman MJ, Genser B, et al. High intestinal cholesterol absorption is associated with cardiovascular disease and risk alleles in ABCG8 and ABO: evidence from the LURIC and YFS cohorts and from a meta-analysis. J Am Coll Cardiol. 2013;62(4):291–299. doi: 10.1016/j.jacc.2013.01.100. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, Chen C, Ke X, et al. Analysis of circulating cholesterol levels as a mediator of an association between ABO blood group and coronary heart disease. Circ Cardiovasc Genet. 2014;7(1):43–48. doi: 10.1161/CIRCGENETICS.113.000299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figure S1 Subgroup analysis of rs505922 and cancer risk. Figure S2 Sensitivity analysis diagram of rs505922 and cancer (a) and cardiocerebrovascular disease (b) risk. Figure S3 Trim and fill method results (a) and filled funnel plot (b) of rs505922 and cancer risk. Figure S4 Subgroup analysis of rs505922 and cardiocerebrovascular disease risk. Figure S5 Trim and fill method results (a) and filled funnel plot (b) of rs505922 and cardiocerebrovascular disease risk. Figure S6 Sensitivity analysis of rs657152 and cancer (a) and cardiocerebrovascular disease (b) risk. Figure S7 Trim and fill method results (a) and filled funnel plot (b) of rs657152 and cancer risk. Figure S8 Trim and fill method results (a) and filled funnel plot (b) of rs657152 and cardiocerebrovascular disease risk. Table S1. Previous reported SNPs had strong linkage disequilibrium with rs505922. Table S2 Quality assessment of case-control studies according to NOS for rs505922. Table S3 Quality assessment of case-control studies according to NOS for rs657152. Table S4 Results for meta-analysis of ABO gene polymorphisms with cancer and cardiocerebrovascular disease risk. Table S5 Subgroup analysis of rs505922 polymorphism
Data Availability Statement
Not applicable.